Abstract
Among the various pathogenic Escherichia coli strains, enterohemorrhagic E. coli (EHEC) is the most devastating. Although serotype O157:H7 strains are the most prevalent, strains of different serotypes also possess similar pathogenic potential. Here, we present the results of a genomic comparison between EHECs of serotype O157, O26, O111, and O103, as well as 21 other, fully sequenced E. coli/Shigella strains. All EHECs have much larger genomes (5.5–5.9 Mb) than the other strains and contain surprisingly large numbers of prophages and integrative elements (IEs). The gene contents of the 4 EHECs do not follow the phylogenetic relationships of the strains, and they share virulence genes for Shiga toxins and many other factors. We found many lambdoid phages, IEs, and virulence plasmids that carry the same or similar virulence genes but have distinct evolutionary histories, indicating that independent acquisition of these mobile genetic elements has driven the evolution of each EHEC. Particularly interesting is the evolution of the type III secretion system (T3SS). We found that the T3SS of EHECs is composed of genes that were introduced by 3 different types of genetic elements: an IE referred to as the locus of enterocyte effacement, which encodes a central part of the T3SS; SpLE3-like IEs; and lambdoid phages carrying numerous T3SS effector genes and other T3SS-related genes. Our data demonstrate how E. coli strains of different phylogenies can independently evolve into EHECs, providing unique insights into the mechanisms underlying the parallel evolution of complex virulence systems in bacteria.
Keywords: bacteriophage, genome evolution, type III secretion system
The acquisition of virulence determinants through successive horizontal gene transfer is a major force driving the evolution and diversification of pathogenic bacteria, compared with modification of existing DNA (1). A specific genomic background may be required for integration, retention, and expression of foreign DNA (1, 2), and the evolution of pathogenic bacteria often exhibits a strong lineage dependency. Interestingly, strains with the same pathotype have occasionally emerged from multiple lineages, but the genetic mechanisms underlying such parallel evolution are not fully understood. Enterohemorrhagic Escherichia coli (EHEC) strains present a striking example of this phenomenon (3, 4).
Among various pathogenic E. coli strains causing intestinal or extra-intestinal diseases in humans (5), the most devastating are the EHEC strains, which cause diarrhea, hemorrhagic colitis, and life-threatening hemolytic uremic syndrome (6). Typical EHEC strains produce Shiga toxins (Stx1 and Stx2), and possess a pathogenicity island referred to as the “locus of enterocyte effacement” (LEE) and a large plasmid encoding enterohemolysin (6). LEE, which is also found in enteropathogenic E. coli (EPEC) and the mouse pathogen Citrobacter rodentium (7, 8), encodes a set of proteins constituting the type III secretion system (T3SS) machinery and several other T3SS-related proteins, such as the “intimin” adhesin and the effector proteins secreted by the T3SS (9, 10). These proteins enable the bacteria to induce attaching and effacing lesions, which are characterized by effacement of the brush border microvilli and intimate bacterial attachment to intestinal epithelial cells (5).
Among the EHECs of various serotypes, the genome sequences are available for only 2 strains of O157:H7 (11, 12). Strain RIMD 0509952 (referred to as O157 Sakai) contains 1.5 Mb of sequence that is absent in the laboratory E. coli strain K-12 (11). The majority of this unique O157 sequence contains prophages (PPs), integrative elements (IEs; defined here as genetic elements that contain cognate integrases but no other genes related to bacteriophages or conjugal transfer functions), and plasmids. The O157 Sakai contains 18 PPs (Sp1-Sp18), 6 IEs (SpLE1–SpLE6), and 2 plasmids. Most virulence-related genes are encoded within these regions: the LEE corresponds to SpLE4, and lambdoid PPs carry the stx1 and stx2 genes, a number of T3SS effector genes (non-LEE effectors), and other virulence-related genes (13). Importantly, although O157:H7 is the most prevalent EHEC serotype, other serotype strains belonging to non-O157 lineages (non-O157 EHECs) are thought to possess similar pathogenic potential (3). Several studies suggested that non-O157 EHECs share a significant number of virulence genes with O157 (14–17), but the whole virulence gene repertoire has not been determined for any of the non-O157 EHEC strains. To elucidate the mechanism underlying this parallel evolution of EHECs, we sequenced 3 major non-O157 EHECs of O26, O111, and O103 serotypes and performed a robust genomic comparison between these non-O157 EHECs, O157 EHEC, and 15 other fully sequenced E. coli strains, including 9 recently sequenced strains (18–20) and 6 Shigella strains, which are known to be E. coli sublineages [supporting information (SI) Table S1 and all references therein].
Results
General Features of the Non-O157 EHEC Genomes.
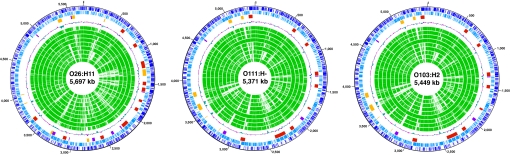
The chromosomes of O26, O111, and O103 were found to be 5,697,240 bp, 5,371,077 bp, and 5,449,314 bp in size, respectively (Table 1, Table S1, and Fig. 1). Each strain harbored various numbers and sizes of plasmids (1–5 plasmids between 4 and 205 kb in size). The chromosome sizes of the non-O157 EHECs were similar to or larger than that of O157 Sakai (5,498 kb) and much larger than that of other sequenced E. coli/Shigella strains (5,231–4,369 kb). O26 possessed the largest chromosome, with a total genome size including plasmids of nearly 6 Mb. The EHECs therefore contained more protein-coding sequences (CDSs) than other strains (see Table S1). The EHECs also possessed significantly larger numbers of transfer RNA (tRNA) genes (98 to 106) than the other strains (81 to 94), although all E. coli/Shigella strains possessed 7 sets of rRNA operons.
Table 1.
General genomic features of the four EHEC strains
| Strain | O157 Sakai | O26 | O111 | O103 |
|---|---|---|---|---|
| Chromosome (kb) | 5,498 | 5,697 | 5,371 | 5,449 |
| CDSs | 5,363 | 5,609 | 5,264 | 5,264 |
| rRNA operons | 7 | 7 | 7 | 7 |
| tRNAs | 105 | 101 | 106 | 98 |
| Prophages | 18 (13) | 21 (13) | 17 (15) | 15 (11) |
| Integrative elements | 6 | 9 | 7 | 6 |
| IS elements | 81 | 104 | 95 | 102 |
| Plasmid (kb) | 93/3 | 85/63/6/4 | 205/98/78/8/7 | 72 |
| CDSs [plasmid total] | 95 | 186 | 468 | 90 |
| IS elements | 17 | 31 | 24 | 14 |
| Total genome size (kb) | 5,594 | 5,856 | 5,766 | 5,525 |
Numbers of lambdoid PPs are indicated in parentheses
Fig. 1.
Circular maps of the O26, O111, and O103 chromosomes. From the outside in: (First circle) nucleotide sequence positions (in Mb); (Second and Third circles) CDSs transcribed clockwise and counterclockwise, respectively; (Fourth circle) locations of PPs and IEs: (red) lambdoid PPs; (purple) other PPs; (yellow) IEs; (Fifth circle) G+C content; (Sixth to Fourteenth circle) CDSs conserved in O157, O26, O111, O103, CFT073, E24377A, Sb227, Sd197, and K-12 MG1655, respectively.
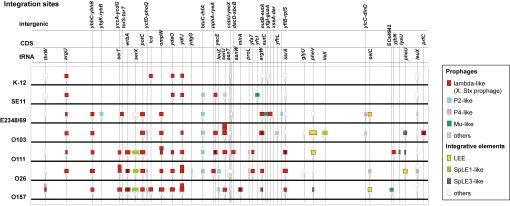
As demonstrated for other E. coli strains (21), the chromosomal backbones (here defined as “chromosome regions other than PPs and IEs”) of non-O157 EHECs were well conserved and exhibited overall genomic synteny, excluding small inversions found in O26 and O103 (Fig. S1A). However, various sizes of strain-specific insertions were present throughout the chromosomes, and most of these were PPs and IEs, as seen in O157 Sakai (see Table 1, Table S2, and Fig. 1). Because the average sizes of CDSs and intergenic regions on the EHEC chromosome backbones do not differ from those of other strains (data not shown), the enlargement of chromosome size in the EHECs is largely attributable to the acquisition of these elements. The elements were integrated into rather limited loci, one-third of which comprised tRNA genes, and 2 or 3 elements were often integrated in tandem into a single site (Fig. 2). Most PPs found in the EHECs were lambdoid phages (see Table S2). Dot-plot analysis revealed that these lambdoid PPs have high levels of intra- and inter-strain DNA sequence similarities, yet also contain remarkable genomic mosaicism (Fig. S2A). Several IEs were also commonly present in the EHECs, as described below.
Fig. 2.
Chromosomal integration sites of PPs and IEs found in the 7 fully sequenced E. coli strains (O157 Sakai, O26, O111, O103, K-12 MG1655, SE11, and E2348/69) are shown schematically. Only the strains in which PPs and IEs have been fully annotated were used in the analysis.
Among the plasmids found in the EHECs, 1 in each strain (pO157, pO26_1, pO111_3, and pO103) was a virulence plasmid generally termed pEHEC (Table S3). O111 contained 4 additional plasmids, including a large multidrug resistance plasmid very similar to plasmid R27 of Salmonella typhi and a PP plasmid nearly identical to bacteriophage P1. O26 also contained 3 additional plasmids, including an R100-like plasmid that carried a kanamycin resistance gene.
Each EHEC contained many insertion sequence (IS) elements (98–135 copies) (Table S4). The IS elements identified in the 4 EHECs were categorized into 38 known or newly identified types, 15 of which were found in all EHECs. IS629 and ISEc8 were the most commonly enriched elements (13–49 copies and 7–11 copies, respectively, in each EHEC).
Genomic Comparison of EHECs and Other Sequenced E. coli/Shigella Strains.
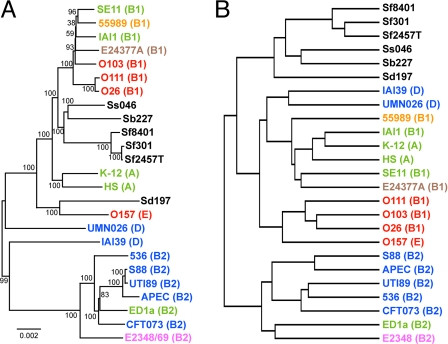
We performed an all-to-all BLASTP analysis of the CDSs in 25 fully sequenced E. coli/Shigella strains. The CDSs were classified into 12,940 groups (defined by ≥90% amino acid sequence identity and ≥60% aligned length coverage of a query sequence). Of these, 1,919 CDS groups were conserved in all strains (Table S5). From these 1,919 groups, we first selected all orthologous CDS groups (926), in which all group members were the same length, and used these 926 CDS groups to analyze the phylogenetic relationship of the 25 strains by the split decomposition method (22). The result of this analysis supported a previous finding that the 3 non-O157 EHECs belong to the E. coli phylogroup B1, whereas O157 belongs to group E (Fig. S1B) (3, 17). However, some conflicting phylogenetic signals were also found, indicating that recombination events occurred between these strains. We therefore further selected 345 CDS groups with very low probability of recombination by using the PHI-test (23), and used them to construct a more precise genome-wide phylogenetic tree. The tree constructed by the neighbor-joining method indicates that O26, O111, and O103 belong to distinct sublineages of group B1, whereas O26 and O111 are closely related (Fig. 3A). The tree constructed by the maximum parsimony method also supports this result (Fig. S1C).
Fig. 3.
Genome-wide phylogenetic analysis and whole gene repertoire comparison of the EHECs and other fully sequenced E. coli/Shigella strains. (A) The neighbor-joining tree constructed by using the concatenated nucleotide sequences of 345 orthologous CDS groups from the 25 sequenced strains. These CDS groups were selected as nonrecombinogenic CDS groups by using the PHI-test (cut off value: P ≥ 0.05), from 926 orthologous CDS groups, in which all members of each group were conserved and of same length in all of the 25 strains. Locus tags of the 345 and 926 CDSs in K-12 MG1655 are listed in the legend of Fig. S1. The reliability of the internal branches was assessed by bootstrapping with 250 pseudoreplicates. The E. coli phylogroup (A, B1, B2, D, or E) of each strain is indicated in brackets. Pathotypes of the strains are indicated by different colors (see Table S1 for the details of the strains). (Scale bar: number of substitutions per site.) (B) The hierarchical clustering tree that was constructed based on a gene repertoire comparison of the 25 strains. The entire gene repertoire of the 25 strains is represented by 12,940 CDS groups that were defined based on the results of an all-to-all BLASTP analysis of CDSs from the 25 strains.
In contrast, the 4 EHECs formed a single cluster in a cluster analysis of the 25 strains based on the conservation patterns of the 12,940 CDS groups (Fig. 3B). This result is not attributable to the genome size effect, because the numbers of CDS families identified in each EHEC (4,705–4,965) are in a range observed for other E. coli strains (5,101–4,033), whereas all Shigella strains contain much lower numbers of CDS families (see Table S1). Furthermore, to avoid biases introduced by different gene prediction criteria, we performed 1-way comparisons by using the TBLASTN homology search (see Fig. 1 and Fig. S3). We found that more O157 CDSs are conserved in the non-O157 EHECs (84–86%) than in the non-EHEC strains (63–77%). Similarly, a higher number of CDSs for each non-O157 EHEC were conserved among the EHECs (including O157) than in the non-EHEC strains. These results indicate that the whole-gene repertoires of the EHECs are more similar to each other than to any of the other strains.
EHEC- or EHEC/EPEC-Specific Genes.
To identify EHEC-specific CDSs (or CDS families), we selected 1,761 CDSs that were present in at least one EHEC or EPEC strain [EHEC and EPEC are known to share many T3SS-related genes (24)] but were absent in all other pathotypes (EHEC/EPEC-specific genes) (see Table S5). Of the 1,761 EHEC/EPEC-specific genes, 87 were present in all EHECs. As expected, 34 of these genes were also present in the EPEC strain and most were T3SS-related. Of the other EHEC/EPEC-specific genes, 228 were present in 2 to 4 EHEC/EPEC strains with various combinations, and the remaining genes were strain-specific (174 to 430 in each strain). Most EHEC/EPEC-specific genes were encoded by mobile elements (see Table S5), and a limited number of EHEC/EPEC-specific genes were present on the chromosome backbone. Importantly, EHEC/EPEC-specific genes with known or predictable functions include not only phage- or plasmid-related genes, but also many virulence-related genes (see Table S5).
EHEC Virulence Factors and Forces Driving Their Acquisition.
The genomic locations of the virulence-related genes shared by the EHEC strains indicate that the major forces driving the acquisition of these genes are mobile genetic elements; we found that the EHEC strains contain many similar mobile elements that carry the same or very similar virulence genes. However, such elements present in each EHEC strain, including LEEs, lambdoid phages, some IEs, and pEHEC plasmids, exhibited significant structural diversities.
Integrative Elements.
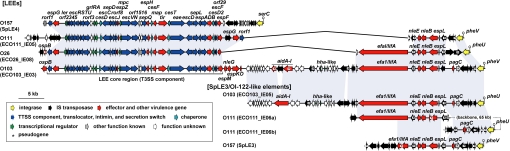
The LEEs of non-O157 EHECs contained cognate integrases and were integrated into the pheU (O26) or pheV (O111 and O103) tRNA loci, whereas the LEE of O157 was integrated into the selC tRNA locus (Fig. 4). As reported previously for several EHEC/EPEC strains (25, 26), the LEE core regions of the 4 EHECs had well-conserved structures, with a minor rearrangement in O111 (an IS-mediated translocation of the espG/rorf1-encoding segment). However, each core region encoded for a different subtype of intimin and other genes exhibited various levels of sequence variation.
Fig. 4.
The genetic organization of the LEEs and SpLE3-like IEs identified in the 4 EHEC genomes is shown. Homologous regions are indicated by purple shading.
By contrast, the LEE accessory regions (LEE-ARs) showed marked structural diversity and an interesting similarity to other IEs of EHECs (see Fig. 4). Although the O157 LEE possessed the simplest LEE-AR, the right LEE-ARs of other EHECs, which are located downstream of the core region in Fig. 4, had complex structures. However, these structures were similar to the SpLE3 of O157 (also known as OI-122), which is integrated into the pheV locus and encodes several virulence-related proteins, including 3 T3SS effectors. O103 had the largest right LEE-AR (approximately 52 kb), encoding 4 additional effectors. Furthermore, O103 contained an additional SpLE3-like element (O103_IE05 at the pheU locus) that was nearly identical to its LEE-AR. O111 also contained a SpLE3-like IE at the pheU locus. This element was nearly identical to a part of the O103 LEE-AR, but had been split into 2 fragments by IS-mediated genomic rearrangements. These findings suggest that the LEEs and SpLE3-like elements have closely related, but complex, evolutionary histories. Although it is known that SpLE3/OI-122 is widely distributed among LEE-positive strains (27, 28), more intensive analyses of LEEs and SpLE3-like IEs may be required to fully understand their evolutionary histories, structural diversities, and implications for the virulence of each strain.
A large IE similar to the SpLE1 of O157 was also found in all non-O157 EHECs (Fig. S2B). Although the genomic structures of these SpLE1-like IEs have diverged somewhat, especially in O103, the urease and tellurium resistance operons were present in all elements (29, 30). Several other virulence factors, such as nonfimbrial adhesins, were also commonly present, although with some variations.
Lambdoid PPs.
As in O157, lambdoid PPs of the non-O157 EHECs contained numerous virulence-related genes, such as those encoding Stxs and various T3SS effectors (see Table S2). Although the Stx1 and Stx2 phages of each EHEC contained almost identical stx1 or stx2 genes (not the variant type) at analogous positions, these PPs were highly divergent with regard to gene organization and chromosomal location (Fig. S2C), as previously suggested (31). This indicates that these PPs have different origins. It is also worth noting that three Stx phages from the non-O157 EHECs encode 1 or more T3SS effectors. In particular, the Stx1 phage of O26 was found to encode as many as 6 effectors.
The PP/IE-encoded T3SS effectors identified in the fully sequenced EHEC/EPECs are summarized in Table 2. The effector repertoires of the EHECs were quite similar, although some variations were observed. Many effectors or their homologs have also been found in the EPEC strain (24), but this strain contains significantly fewer effectors. These PP/IE-encoded effectors were completely absent in the sequenced LEE-negative strains and, thus, can be regarded as substrates of the LEE-related T3SS. Although several effector homologs were also present in the chromosomal backbones (non-PP/IE regions), it has been suggested that they are substrates for, or remnants of, the second E. coli T3SS encoded by the ETT2 gene cluster, which is widely distributed in E. coli but has been degraded to various extents (24).
Table 2.
PP/IE-encoded T3SS effectors of the EHEC/EPEC strains
| EHEC |
EPEC |
||||
|---|---|---|---|---|---|
| O157 | O26 | O111 | O103 | E2348/69 | |
| EspBa | 1 | 1 | 1 | 1 | 1 |
| EspFa | 1 | 1 | 1 | 1 | 1 |
| EspGa | 1 | 1 | 1 | 1 | 2 |
| EspHa | 1 | 1 | 1 | 1 | 1 |
| EspJ | 1 | 1 | 1 | 0 | 1 |
| EspK | 1 | 2 | 1 | 3 | 0 |
| EspL | 1 | 1 | 2 (1) | 2 | 2 (1) |
| EspM | 2 | 2 | 2 | 2 | 0 |
| EspN | 1 | 1 | 1 | 1 | 0 |
| EspO | 2 | 2 | 2 | 2 | 1 (1) |
| EspV | 1 (1) | 1 (1) | 1 (1) | 1 (1) | 0 |
| EspW | 1 | 1 | 1 | 1 | 0 |
| EspX | 1 | 1 | 1 | 1 | 0 |
| EspZa | 1 | 1 | 1 | 1 | 1 |
| Mapa | 1 | 1 | 1 | 1 | 1 |
| NleA/EspI | 1 | 1 (1) | 1 | 1 | 1 |
| NleB | 3 (1) | 1 | 2 | 4 | 3 (1) |
| NleC | 1 | 1 | 2 | 2 (2) | 1 |
| NleD | 1 | 0 | 0 | 0 | 1 |
| NleE | 1 | 1 | 2 | 2 | 2 |
| NleF | 1 | 1 | 1 (1) | 1 | 1 |
| NleG | 14 (6) | 14 | 11 (3) | 8 (2) | 1 |
| NleH | 2 | 2 | 2 | 2 (1) | 3 (1) |
| TccP | 2 (1) | 1 | 1 | 1 | 0 |
| Tira | 1 | 1 | 1 | 1 | 1 |
| Cif | 0 | 1 (1) | 1 (1) | 1 (1) | 1 (1) |
| Ibe | 0 | 2 | 1 | 2 | 0 |
| OspG | 0 | 0 | 1 | 1 (1) | 0 |
| Total | 44 (9) | 44 (3) | 44 (7) | 45 (8) | 26 (5) |
Numbers in parentheses indicate pseudogenes.
aEffectors encoded in the LEE core region.
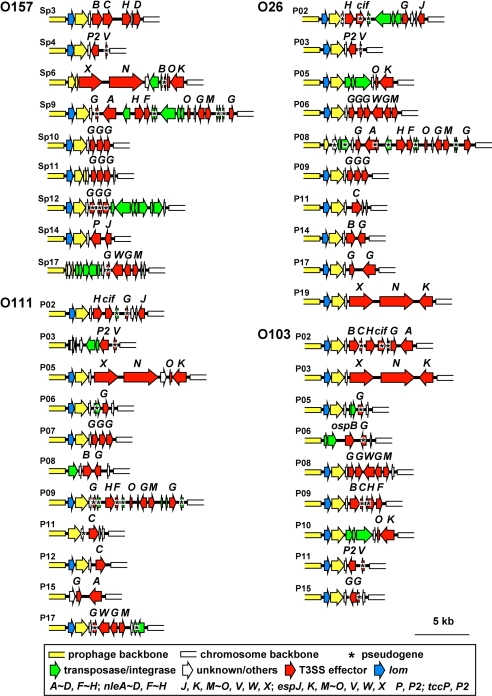
Of the numerous PP/IE-encoded effectors found in the EHECs, which were classified into 28 families (see Table 2), between 10 and 16 were found in the LEEs and SpLE3-like IEs. All other effectors were present in the exchangeable effector loci of lambdoid PPs, which are located just downstream of tail fiber genes (Fig. 5). We found many lambdoid PPs encoding similar, or sometimes identical, effector sets in different EHEC strains. However, such phages had divergent genomic structures and were not always integrated into the same site. This suggests that complex histories of phage infection and subsequent genomic rearrangements existed for each EHEC. O157 contains 5 genes that encode Pch family transcriptional regulators. Three of these genes are on lambdoid PPs, and the others are on a chimeric PP (Sp7) and SpLE1. The first 3 have almost identical sequences (the PchABC subfamily) and positively regulate the expression of many horizontally acquired O157 genes, including the LEE genes and the non-LEE effector genes (32, 33). The non-O157 EHECs also contained genes that encode 3 to 4 PchABC proteins, all of which were on lambdoid PPs. One to 3 genes encoding other subtypes (PchD, PchE, and 2 new subtypes named PchF and PchG) were also found on SpLE1-like IEs or Sp7-like PPs. These genes (excluding pchD, which was present in 4 non-EHEC strains) were present only in EHECs (Table S6).
Fig. 5.
The gene organization of the effector exchangeable loci (of lambdoid PPs identified in the 4 EHEC strains is shown. Effector exchangeable loci are located just downstream of the tail fiber genes and contain various T3SS effector genes. Pseudogenes are indicated by asterisks.
Lambdoid phages also introduced 7 copies of the ileZ-argN-argO operon, encoding 3 extra tRNAs for isoleucine and arginine codons into O157. These codons are rarely used for the backbone genes, but are more frequently used for the foreign genes, including the stx genes and the LEE genes (11). Thus, the 3 tRNAs are likely required for efficient expression of horizontally acquired genes, such as the LEE and stx genes, which may in turn result in their stable retention in the O157 genome. The same may be true for the non-O157 EHECs because they have also acquired 5 to 7 copies of the ileZ-argN-argO operon by way of lambdoid phages. Many non-EHEC strains contained these tRNA genes, but only with a limited number of copies (see Table S6).
Virulence Plasmids.
Conservation of several pEHEC-encoded virulence genes among different serotypes of EHEC has been suggested by previous PCR-based analyses (34, 35). The full sequences of pEHEC plasmids from the 3 non-O157 EHECs confirmed this notion (Fig. S4). Two operons responsible for enterohemolysin production (ehx) and lipid A modification (ecf) were found in all plasmids. Other factors, such as catalase/peroxidase and proteases, were also encoded by various combinations of 2 or 3 pEHEC plasmids. Unexpectedly, however, the locations of these genes differed significantly, the backbone sequences of the plasmids were highly divergent, and their replication systems exhibited notable variations (see Table S3 and Fig. S4). These data indicate that the pEHEC plasmids also possess different and complex evolutionary histories.
Variation in Other Virulence Factors.
In contrast to the virulence genes present in PPs, IEs, and pEHEC plasmids, potential virulence-related genes present on chromosomal backbones exhibited variable conservation patterns among the EHECs. For example, we identified 10 iron utilization systems and 19 fimbrial biosynthesis loci in the 4 EHECs, but many of these loci were also found in other strains and appeared to exhibit distribution patterns associated with the phylogenies of each strain (see Table S6). These genomic differences, together with the minor differences observed in the repertoire of T3SS effectors, may affect both the potential virulence and the host specificities of each EHEC. However, experimental evidence will be required to determine whether this is in fact the case.
Discussion
O26, O111, and O103 EHECs, which we sequenced in this study, are the non-O157 EHECs of the highest clinical importance in many countries. Thus, their genome sequences provide critical genetic information for developing efficient strategies to control non-O157 EHEC infections. Importantly, our genomic comparison of these non-O157 EHECs with O157 EHEC and other fully sequenced E. coli/Shigella strains revealed a genetic mechanism underlying the parallel evolution of EHECs.
Despite their different phylogenies, all of the 4 EHECs have much larger genomes (5.5 to 5.9 Mb) than the other strains (see Table 1 and Fig. S1) and contain surprisingly large numbers of PPs and IEs (21 to 30) (see Table 1 and Table S2). Furthermore, they exhibit a remarkable similarity with regard to their whole gene repertoire and share many genes that are specific to EHEC or are rarely present in other pathotypes (see Fig. 3 and Fig. S3). These genes include not only the stx genes, but also many other genes that are directly or indirectly related to virulence (see Tables S5 and S6), thus conferring a similar virulence potential to each EHEC. The independent acquisition of very similar virulence gene sets is predominantly attributable to mobile elements that are commonly present in the EHECs: multiple lambdoid PPs, several types of IEs, and virulence plasmids. Thus, these mobile elements can be regarded as the primary driving force for the parallel evolution of EHECs. Importantly, despite carrying the same or similar virulence gene sets, these elements exhibit remarkably divergent genomic structures. This property is an indication of their complex and independent evolutionary pathways.
Among the virulence genes shared by EHECs, those associated with the LEE-related T3SS are particularly interesting. The LEE encodes a central part of the T3SS, but SpLE3-like IEs and many lambdoid PPs encode numerous T3SS effectors. Thus, we suggest that the LEE-related T3SS of EHECs has been constructed by genes introduced by these 3 types of mobile elements. Abundance of non-LEE effectors in EHECs is mainly attributable to the acquisition of a large number of lambdoid PPs in each EHEC, although it is not known whether EHECs have the requisite genetic background to allow such accumulation of lambdoid PPs in a single cell. The lambdoid phages have also introduced multiple copies of the PchABC transcriptional regulator and ileZ-argN-argO tRNA genes, which are required for efficient expression of foreign genes, including those required for the T3SS. Acquisition of these genes may be a prerequisite for the development of this highly complex but efficient virulence system in each EHEC.
It is also interesting that virulence plasmids of the 4 EHECs that have apparently different evolutionary histories encode a very similar set of virulence-related genes. All EHECs contain SpLE1-like elements, which also encode many genes potentially related to virulence or dissemination of EHEC. Although the roles of these genes on the virulence plasmids and the SpLE1-like elements in EHEC infection are not fully understood, their specific distribution in EHEC strains suggests that they may play important roles in EHEC pathogenicity or survival and dissemination in environments.
In conclusion, although the evolutionary processes of pathogenic bacteria are often discussed in the context of lineage-associated acquisition of a specific virulence gene set, the present study clearly demonstrates how E. coli strains belonging to different phylogenies can independently evolve into EHEC. The selective forces and special genetic factors (or background) promoting such parallel evolution have yet to be identified (36), but our results yield unique insights into the dynamic evolution of bacterial complex virulence systems.
Materials and Methods
Bacterial Strains.
O26:H11 strain 11368 (stx1+), O111:H- strain 11128 (stx1+/stx2+), and O103:H2 strain 12009 (stx1+/stx2+) were isolated in Japan in 2001 (17). Strain 11368 was isolated from a patient with diarrhea during a diffuse outbreak, and strains 11128 and 12009 were from patients with sporadic cases of diarrhea and bloody stool.
Genome Sequencing and Gene Prediction and Annotation.
The genome sequences of the O26, O111, and O103 strains were determined by a whole-genome shotgun strategy, as described in ref. 18. We constructed 2 plasmid-based shotgun libraries containing shorter (approximately 2 kb) and longer (10 kb) inserts and a fosmid library for each strain. We generated 131,328 (for O26), 119,040 (O103), and 84,480 (O111) sequences from both ends of the clones by using ABI 3730xl sequencers (Applied Biosystems), resulting in 13.5-, 12.8-, and 9.2-fold coverage, respectively. Sequence reads were assembled with the Phred-Phrap-Consed program (37), and gaps were closed as described in ref. 18.
CDSs were identified by using GeneHacker (38), followed by manual inspection of the start codons and ribosome binding sequences of each CDS. Intergenic regions of >150 bp were further reviewed for the presence of small CDSs that encode proteins with significant sequence similarity to known proteins. Functional annotation of the CDSs was performed based on the results of homology searches against the public nonredundant protein database (http://www.ncbi.nlm.nih.gov/) by using BLASTP. RNA genes were identified by using the Rfam database (39) at the Rfam Web site.
Genome-Wide Phylogenetic Analyses.
Selecting the bidirectional best-hits from an all-to-all BLASTP search of the CDSs from the 25 fully sequenced strains (pseudogenes were excluded in this analysis), we identified orthologous CDSs that were conserved in all 25 strains. Among these, 926 groups of orthologous CDSs in which all group members were of the same length were selected. Their concatenated DNA sequences from each of the 25 strains were used for split decomposition analysis, conducted by using SplitsTree4 (22).
From the 926 groups, we identified 345 with a low probability of recombination, based on the PHI-test (23) (cutoff value: P ≥ 0.05). Their concatenated sequences were aligned by using the MAFFT program (40), and the distance matrix was calculated with the DNADIST program in the PHYLIP package (ver. 3.68) (41) by using the Kimura 2-parameter model. Phylogenetic trees were constructed by the neighbor joining and maximum parsimony methods by using the MEGA4 software package (42).
Clustering Analysis of E. coli/Shigella Strains Based on Their Gene Repertoires.
All CDSs of the 25 strains were classified into 12,940 CDS groups (defined by ≥90% sequence identity and ≥60% aligned length coverage of a query sequence) based on the results of the all-to-all BLASTP analysis, and the dataset was converted into binary scores (present = 1 or absent = 0). A cluster analysis of the 25 strains was then performed in Cluster 3.0 based on the conservation patterns of these CDS groups in each strain, and the results were visualized with Treeview (43).
Supplementary Material
Acknowledgments.
We thank A. Yamashita, A. Yoshida, Y. Takeshita, N. Kanemaru, K. Furuya, C. Yoshino, H. Inaba, K. Motomura, Y. Hattori, A. Tamura, and N. Itoh for technical assistance. This work was funded by Grant-in-Aids for Scientific Research on Priority Areas Applied Genomics (to T.H.) and Comprehensive Genomics (to M.H.) from the Ministry of Education, Science and Technology of Japan.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition. The sequences reported in this paper have been deposited in the GeneBank database (accession nos. AP010953–AP010965).
This article contains supporting information online at www.pnas.org/cgi/content/full/0903585106/DCSupplemental.
References
- 1.Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 2.Escobar-Paramo P, et al. A specific genetic background is required for acquisition and expression of virulence factors in Escherichia coli. Mol Biol Evol. 2004;21:1085–1094. doi: 10.1093/molbev/msh118. [DOI] [PubMed] [Google Scholar]
- 3.Reid SD, Herbelin CJ, Bumbaugh AC, Selander RK, Whittam TS. Parallel evolution of virulence in pathogenic Escherichia coli. Nature. 2000;406:64–67. doi: 10.1038/35017546. [DOI] [PubMed] [Google Scholar]
- 4.Bielaszewska M, et al. Detection and characterization of the fimbrial sfp cluster in enterohemorrhagic Escherichia coli O165:H25/NM isolates from humans and cattle. Appl Environ Microbiol. 2009;75:64–71. doi: 10.1128/AEM.01815-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 6.Caprioli A, Morabito S, Brugere H, Oswald E. Enterohaemorrhagic Escherichia coli: Emerging issues on virulence and modes of transmission. Vet Res. 2005;36:289–311. doi: 10.1051/vetres:2005002. [DOI] [PubMed] [Google Scholar]
- 7.Jores J, Rumer L, Wieler LH. Impact of the locus of enterocyte effacement pathogenicity island on the evolution of pathogenic Escherichia coli. Int J Med Microbiol. 2004;294:103–113. doi: 10.1016/j.ijmm.2004.06.024. [DOI] [PubMed] [Google Scholar]
- 8.Wales AD, Woodward MJ, Pearson GR. Attaching-effacing bacteria in animals. J Comp Pathol. 2005;132:1–26. doi: 10.1016/j.jcpa.2004.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coburn B, Sekirov I, Finlay BB. Type III secretion systems and disease. Clin Microbiol Rev. 2007;20:535–549. doi: 10.1128/CMR.00013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dean P, Kenny B. The effector repertoire of enteropathogenic E. coli: Ganging up on the host cell. Curr Opin Microbiol. 2009;12(1):101–109. doi: 10.1016/j.mib.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayashi T, et al. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 2001;8:11–22. doi: 10.1093/dnares/8.1.11. [DOI] [PubMed] [Google Scholar]
- 12.Perna NT, et al. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001;409:529–533. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- 13.Tobe T, et al. An extensive repertoire of type III secretion effectors in Escherichia coli O157 and the role of lambdoid phages in their dissemination. Proc Natl Acad Sci USA. 2006;103:14941–14946. doi: 10.1073/pnas.0604891103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brooks JT, et al. Non-O157 Shiga toxin-producing Escherichia coli infections in the United States, 1983–2002. J Infect Dis. 2005;192:1422–1429. doi: 10.1086/466536. [DOI] [PubMed] [Google Scholar]
- 15.Eklund M, Scheutz F, Siitonen A. Clinical isolates of non-O157 Shiga toxin-producing Escherichia coli: Serotypes, virulence characteristics, and molecular profiles of strains of the same serotype. J Clin Microbiol. 2001;39:2829–2834. doi: 10.1128/JCM.39.8.2829-2834.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ogura Y, et al. Complexity of the genomic diversity in enterohemorrhagic Escherichia coli O157 revealed by the combinational use of the O157 Sakai OligoDNA microarray and the Whole Genome PCR scanning. DNA Res. 2006;13:3–14. doi: 10.1093/dnares/dsi026. [DOI] [PubMed] [Google Scholar]
- 17.Ogura Y, et al. Extensive genomic diversity and selective conservation of virulence-determinants in enterohemorrhagic Escherichia coli strains of O157 and non-O157 serotypes. Genome Biol. 2007;8:R138. doi: 10.1186/gb-2007-8-7-r138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oshima K, et al. Complete genome sequence and comparative analysis of the wild-type commensal Escherichia coli strain SE11 isolated from a healthy adult. DNA Res. 2008;15:375–386. doi: 10.1093/dnares/dsn026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rasko DA, et al. The pangenome structure of Escherichia coli: Comparative genomic analysis of E. coli commensal and pathogenic isolates. J Bacteriol. 2008;190:6881–6893. doi: 10.1128/JB.00619-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Touchon M, et al. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 2009;5:e1000344. doi: 10.1371/journal.pgen.1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dobrindt U. (Patho-)Genomics of Escherichia coli. Int J Med Microbiol. 2005;295:357–371. doi: 10.1016/j.ijmm.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 22.Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 23.Bruen TC, Philippe H, Bryant D. A simple and robust statistical test for detecting the presence of recombination. Genetics. 2006;172:2665–2681. doi: 10.1534/genetics.105.048975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iguchi A, et al. The complete genome sequence and comparative genome analysis of enteropathogenic E. coli O127:H6 strain E2348/69. J Bacteriol. 2008;191:347–354. doi: 10.1128/JB.01238-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ogura Y, et al. Systematic identification and sequence analysis of the genomic islands of the enteropathogenic Escherichia coli strain B171–8 by the combined use of whole-genome PCR scanning and fosmid mapping. J Bacteriol. 2008;190:6948–6960. doi: 10.1128/JB.00625-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tauschek M, Strugnell RA, Robins-Browne RM. Characterization and evidence of mobilization of the LEE pathogenicity island of rabbit-specific strains of enteropathogenic Escherichia coli. Mol Microbiol. 2002;44:1533–1550. doi: 10.1046/j.1365-2958.2002.02968.x. [DOI] [PubMed] [Google Scholar]
- 27.Konczy P, et al. Genomic O island 122, locus for enterocyte effacement, and the evolution of virulent verocytotoxin-producing Escherichia coli. J Bacteriol. 2008;190:5832–5840. doi: 10.1128/JB.00480-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karmali MA, et al. Association of genomic O island 122 of Escherichia coli EDL 933 with verocytotoxin-producing Escherichia coli seropathotypes that are linked to epidemic and/or serious disease. J Clin Microbiol. 2003;41:4930–4940. doi: 10.1128/JCM.41.11.4930-4940.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakano M, et al. Association of the urease gene with enterohemorrhagic Escherichia coli strains irrespective of their serogroups. J Clin Microbiol. 2001;39:4541–4543. doi: 10.1128/JCM.39.12.4541-4543.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Orth D, Grif K, Dierich MP, Wurzner R. Variability in tellurite resistance and the ter gene cluster among Shiga toxin-producing Escherichia coli isolated from humans, animals and food. Res Microbiol. 2007;158:105–111. doi: 10.1016/j.resmic.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 31.Unkmeir A, Schmidt H. Structural analysis of phage-borne stx genes and their flanking sequences in Shiga toxin-producing Escherichia coli and Shigella dysenteriae type 1 strains. Infect Immun. 2000;68:4856–4864. doi: 10.1128/iai.68.9.4856-4864.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iyoda S, Watanabe H. Positive effects of multiple pch genes on expression of the locus of enterocyte effacement genes and adherence of enterohaemorrhagic Escherichia coli O157:H7 to HEp-2 cells. Microbiology. 2004;150:2357–2571. doi: 10.1099/mic.0.27100-0. [DOI] [PubMed] [Google Scholar]
- 33.Abe H, et al. Global regulation by horizontally transferred regulators establishes the pathogenicity of Escherichia coli. DNA Res. 2008;15:25–38. doi: 10.1093/dnares/dsm033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leomil L, Pestana de Castro AF, Krause G, Schmidt H, Beutin L. Characterization of two major groups of diarrheagenic Escherichia coli O26 strains which are globally spread in human patients and domestic animals of different species. FEMS Microbiol Lett. 2005;249:335–342. doi: 10.1016/j.femsle.2005.06.030. [DOI] [PubMed] [Google Scholar]
- 35.Brunder W, Schmidt H, Frosch M, Karch H. The large plasmids of Shiga-toxin-producing Escherichia coli (STEC) are highly variable genetic elements. Microbiology. 1999;145:1005–1014. doi: 10.1099/13500872-145-5-1005. [DOI] [PubMed] [Google Scholar]
- 36.Pallen MJ, Wren BW. Bacterial pathogenomics. Nature. 2007;449:835–842. doi: 10.1038/nature06248. [DOI] [PubMed] [Google Scholar]
- 37.Gordon D, Desmarais C, Green P. Automated finishing with autofinish. Genome Res. 2001;11:614–625. doi: 10.1101/gr.171401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yada T, Hirosawa M. Detection of short protein coding regions within the cyanobacterium genome: Application of the hidden Markov model. DNA Res. 1996;3:355–361. doi: 10.1093/dnares/3.6.355. [DOI] [PubMed] [Google Scholar]
- 39.Griffiths-Jones S, Bateman A, Marshall M, Khanna A, Eddy SR. Rfam: An RNA family database. Nucleic Acids Res. 2003;31:439–441. doi: 10.1093/nar/gkg006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Felsenstein J. Seattle: Department of Genome Sciences, University of Washington; 2005. PHYLIP (Phylogeny Inference Package) version 3.6. Distributed by the author. [Google Scholar]
- 42.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 43.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.