Abstract
Vertebrate vision is maintained by the retinoid (visual) cycle, a complex enzymatic pathway that operates in the retina to regenerate the visual chromophore, 11-cis-retinal. A key enzyme in this pathway is the microsomal membrane protein RPE65. This enzyme catalyzes the conversion of all-trans-retinyl esters to 11-cis-retinol in the retinal pigment epithelium (RPE). Mutations in RPE65 are known to be responsible for a subset of cases of the most common form of childhood blindness, Leber congenital amaurosis (LCA). Although retinoid isomerase activity has been attributed to RPE65, its catalytic mechanism remains a matter of debate. Also, the manner in which RPE65 binds to membranes and extracts retinoid substrates is unclear. To gain insight into these questions, we determined the crystal structure of native bovine RPE65 at 2.14-Å resolution. The structural, biophysical, and biochemical data presented here provide the framework needed for an in-depth understanding of the mechanism of catalytic isomerization and membrane association, in addition to the role mutations that cause LCA have in disrupting protein function.
Keywords: isomerization, metalloprotein, monotopic membrane protein
Vision begins when the 11-cis-retinylidene chromophore of rhodopsin is photoisomerized to all-trans-retinylidene, a process resulting in receptor activation and transduction of the light signal (1). After rhodopsin is photoactivated, it is no longer responsive to light, so for vision to continue, a trans-to-cis isomerization mechanism must be present to regenerate light-sensitive visual pigments. In vertebrates, after photoisomerization, all-trans-retinylidene is hydrolyzed from rhodopsin, reduced to all-trans-retinol, and transported to a tissue adjacent to the photoreceptor layer known as the retinal pigment epithelium (RPE), where enzymatic isomerization occurs (2). An RPE-specific, microsomal membrane protein with an apparent molecular mass of 65 kDa, known as RPE65, was determined to be responsible for trans-to-cis retinoid isomerase activity in the RPE (3–5). The importance of this protein in visual function is also evident from the observation that certain RPE65 mutations cause a form of the hereditary childhood blinding disease known as Leber congenital amaurosis (LCA) or the less severe, later-onset disease, retinitis pigmentosa (RP) (6–8).
Based on sequence homology, RPE65 belongs to a family of carotenoid cleavage oxygenase (CCO) enzymes that oxidatively cleave β-carotene or apocarotenoids (9–11). However, RPE65 is distinct from all other members of this family in that it simultaneously cleaves and isomerizes all-trans-retinyl esters to 11-cis-retinol and a fatty acid rather than oxidatively cleaving carotenoids (3–5, 11, 12). Unlike the reactions catalyzed by other CCO family members, there is no obvious role for molecular oxygen in RPE65 enzymology. The only family member with a known crystal structure is an apocarotenoid oxygenase from Synechocystis that is 25% identical and 42% homologous to human RPE65 (13). All members of this family have a set of four absolutely conserved His residues that bind a required iron cofactor (14).
RPE65 has been shown to associate with microsomal membranes and undergo post-translational modification (15–17). RPE65 was initially reported to exist in both membrane bound and soluble forms (15), and to be reversibly S-palmitoylated on Cys residues 231, 329, and 330 (18, 19). Palmitoylation of these residues in response to light was hypothesized to increase the ratio of membrane-bound to soluble RPE65 leading to greater activity (18, 19). Subsequent findings challenged this hypothesis (20, 21), and currently, the roles of palmitoylation in RPE65 structure and function require clarification.
Here, we present crystallographic, biophysical, and biochemical data that clarify critical issues regarding RPE65 structure and function. Together, these data provide essential information needed to advance the understanding of RPE65 enzymology and membrane association and formulate a structural explanation for how LCA-associated RPE65 amino acid substitutions lead to loss of protein function.
Results and Discussion
Overall Structure.
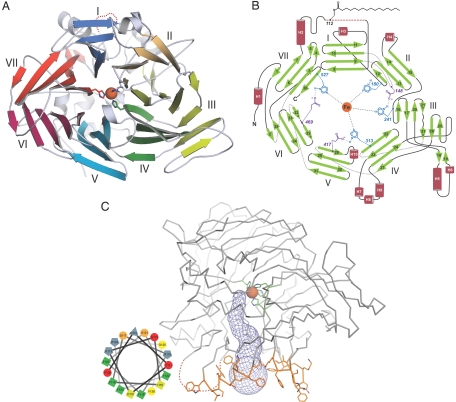
The crystal structure of bovine RPE65 was solved by the single-wavelength anomalous dispersion (SAD) technique using the natively bound iron ion and refined to an Rfree value of 21.6% at 2.14-Å resolution (Tables S1 and S2). The basic RPE65 structural motif is a seven-bladed β-propeller with single-strand extensions on blades VI and VII and a two-strand extension on blade III (Fig. 1 A and B). A seven-bladed β-propeller motif is also observed for the retinal-forming ACO (13), and the two structures superimpose with a rmsd of 2.5 Å for 443 matched Cα positions (Fig. S1) (22). The top face of the β-propeller, defined by the positions of the segments connecting the outer strand of one sheet with the inner strand of the next sheet, is covered by a helical cap that houses the active site. The iron cofactor is located near the top face on the propeller axis and is coordinated by four His residues and three second shell Glu residues. Notably, each blade of the propeller contributes a single residue to the iron ion coordination system. A hydrophobic tunnel leads from the protein exterior to the active site defined by the iron ion and may act as a conduit for passage of retinoids from the membrane to the RPE65 catalytic center (Fig. 1C). Similar tunnels have been observed in other monotopic membrane protein structures (23). The mouth of this tunnel is surrounded by three groups of residues (196–202, 234–236, and 261–271) that are likely to integrate into the lipid bilayer owing to their overall hydrophobicity. Several aromatic residues are found in these segments, suggesting that the depth of RPE65 membrane interaction is restricted to the most proximal portions of phospholipid acyl chains with respect to the polar head groups (24). A hydrophobic interaction with the membrane is consistent with the observation that detergent is required for quantitative solubilization of RPE65. There are also a number of Arg and Lys residues found in these segments that may interact with negatively charged phospholipid head groups. A fourth region of the protein consisting of residues 109–126 is also very likely to contribute to RPE65 membrane affinity, but the electron density for this region is exceptionally weak and insufficient for model building. A helical wheel plot of this segment suggests a structure that is amphipathic with positively charged residues separating the hydrophobic face from the hydrophilic face (Fig. 1C). We note that this segment was predicted to anchor RPE65 to membranes in the study describing the cloning of bovine RPE65 (17). It was recently reported that Cys112, contained within this disordered segment, is S-palmitoylated in native and recombinantly expressed RPE65 (16), and we confirmed this finding via MS in the RPE65 preparation used in this study (Fig. S2). The presence of a palmitoylated residue at this position would strengthen the protein–bilayer interaction and provides further support for our positioning of residues 109–126 relative to the membrane. The causes of the disorder observed for residues 109–126 are not clear, but may be related to delipidation of the protein during purification or destabilization of this region by detergent.
Fig. 1.
RPE65 structure and topology. (A) Structure of an RPE65 monomer viewed from the bottom face of the seven-bladed β-propeller. Blades are numbered I through VII with the blade where “velcro” closure of the core propeller fold occurs labeled as blade VII in accord with established convention. Absolutely conserved His180, His241, His313, and His527 residues are shown as sticks coordinating the natively bound iron ion, shown as an orange sphere. (B) An RPE65 topology diagram showing the positions of residues involved in iron ion-coordination, as well as the position of the palmitoylated Cys residue. Numbers beside these residues indicate their position in the RPE65 amino acid sequence. (C) The proposed membrane-binding surface of RPE65. The mouth of the presumed substrate entry/product exit tunnel (shown as blue mesh) is surrounded by three groups of mainly hydrophobic residues, colored orange, that are likely responsible for anchoring RPE65 to RPE membranes. The maroon dashed line represents the approximate position of a disordered segment. A helical wheel plot of this segment (Inset) indicates that it may form an amphipathic α-helix under appropriate conditions.
Previous reports suggested that RPE65 membrane association and activity are regulated by reversible S-palmitoylation at Cys231, Cys329, and Cys330 (18, 19). Because RPE65 purified from native membranes was used for our structure determination, we would be able to directly observe palmitoyl groups connected to these residues if this hypothesis is correct. Consistent with subsequent mass spectrometric and mutagenesis studies (4, 20, 21), the electron density maps conclusively demonstrate that none of these residues are palmitoylated in purified native RPE65 (Fig. S3). Also, none of these Cys residues are surface exposed or located on the predicted membrane-binding face of the protein, indicating that palmitoylation at these positions would be unlikely to occur.
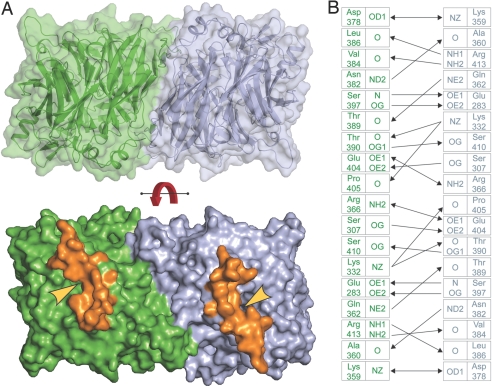
An RPE65 dimer with ≈2-fold rotational symmetry is contained in the crystallographic asymmetric unit (Fig. 2A). The dimer has a parallel arrangement such that entrances to the active sites of each RPE65 protomer face the same side of the dimer. The extensive dimer interface buries a total of ≈3,100 Å2 of surface area. Many of the interacting residues between monomers are part of an extension to blade III of the β-propeller. Interestingly, this extension is absent in Synechocystis ACO, and as a possible consequence, a symmetric dimer similar to that seen in the RPE65 structure is not observed in either of the two reported crystal forms (13). The dimer appears to be held together primarily by a number of hydrogen bonding and ion–ion interactions (Fig. 2B). Analysis of the dimer with the PISA server (25) shows that the P value of the observed solvation free energy gain on dimer formation is 0.33, where values can range from 0 to 1 with values < 0.5 indicating a high probability of the interaction surface being specific. For comparison, this statistic was calculated for known dimeric monotopic membrane proteins prostaglandin H2 synthase-1 [Protein Data Bank (PDB) ID code 1PRH], squalene-hopene cyclase (PDB ID code 1SQC), fatty acid amide hydrolase (PDB ID code 1MT5), and glycerol-3-phosphate dehydrogenase (PDB ID code 2QCU), which yielded values of 0.17, 0.16, 0.30, and 0.75, respectively. Both the absolute value of this statistic for RPE65 and its similarity to a known dimeric monotopic membrane protein, fatty acid amide hydrolase, indicate that the interface is sufficiently specific that it could represent a physiological interaction. To determine whether an RPE65 dimer preforms in detergent-containing solution before crystallization, we performed gel filtration chromatography on purified RPE65 in the presence of 150 mM NaCl. RPE65 eluted at a molecular mass of 83 kDa relative to standards consistent with an RPE65 monomer complexed with a n-octyltetraoxyethylene (C8E4) micelle (Fig. S4). The large buried surface area and symmetric arrangement of the RPE65 dimer observed in the crystal suggest it may be functionally relevant, and although RPE65 is monomeric in solution, it may form dimers on native RPE membranes where it is found at high concentration. Alternatively, the dimer interaction surface could possibly be a binding surface for another microsomal protein such as RDH5, which RPE65 has been shown to interact with (26).
Fig. 2.
RPE65 dimer found in the asymmetric unit. (A) Extensive and complementary nature of the dimer interface. Approximately 3,100 Å2 of surface area is buried between protomers (colored green and blue) (Upper). Positions of the predicted membrane-binding residues (colored orange) in each protomer. The yellow arrowheads indicate the mouths of the predicted substrate entry/product exit tunnels. The observed parallel orientation suggests the observed dimer interaction could be physiological. The red curved arrow indicates a 180° rotation (Lower). (B) Residues mediating the dimer interaction. Single-headed and double-headed arrows indicate hydrogen bonding and ionic interactions, respectively. The inner and outer columns indicate the atom identifiers and residues, respectively. The cut-off distances used for determining these interactions were < 3.4 Å for ionic and van der Waals interactions and < 3.2 Å for hydrogen-bonding interactions.
Iron Ion-Binding Site and Active-Site Cavity.
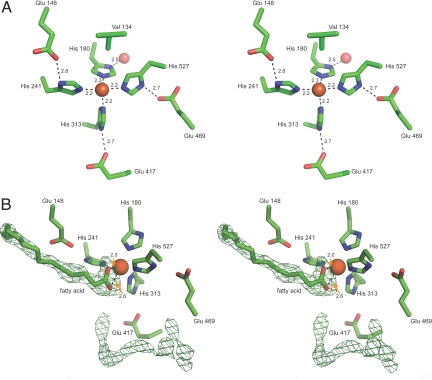
The RPE65 iron cofactor is found near the top face of the propeller axis and is covered by the helical cap. The iron ion is directly coordinated by the Nε atoms of His180, His241, His313, and His527 with average bond lengths of 2.2 Å (Fig. 3A). The geometry of iron ion coordination is approximately octahedral with two open coordination sites. With the exception of His180, the Nδ-H groups are hydrogen-bonded to the Oε atoms of Glu148, Glu417, and Glu469. The Nδ-H group of His180 forms a hydrogen bond with a water molecule found in the lower axial, water-filled cavity of the propeller. These hydrogen-bonding arrangements likely stabilize the octahedral coordination of the iron ion to facilitate catalysis. One of the two open coordination sites in the octahedral geometry is partially blocked by a Cγ atom from Val134 located 4.9 Å away.
Fig. 3.
The RPE65 iron ion-binding site and residual electron density found in the active site cavity. (A) Stereoview of the iron cofactor and its ligands. The iron ion is shown as an orange sphere and the second shell water molecule is shown as a red sphere. The geometry of iron ion coordination is approximately octahedral. The second shell Glu residues most likely help to stabilize this geometry. The side chain of Val134 is 4.9 Å away from the iron ion. Numbers indicate bond distances in angstroms. (B) Stereoview of the residual electron density found in the RPE65 active site. The green mesh represents an unbiased, σ A-weighted Fo − Fc electron density map contoured at 3.5 σ. The appearance of the electron density next to the iron ion is highly suggestive of a bound fatty acid molecule which has been modeled. The lack of features in the second region of electron density precludes molecular assignment. Numbers next to dashed lines indicate bond distances in angstroms.
As mentioned above, the iron ion is accessible through a tunnel (tunnel A) that enters the helical cap of the protein at an angle slightly off 90° relative to the propeller axis (Fig. 1C and Fig. S5A). This tunnel continues past the metal ion forming a bent cavity within the protein. A second tunnel (tunnel B) also leads from the protein exterior to the active-site cavity, but it contains a narrow segment that would most likely occlude the passage of retinoid substrates or products of the isomerization reaction. However, it may permit the passage of water or other small molecules/ions into the active site. Therefore, we hypothesize that retinoid substrate entry and product exit occur through the same tunnel. The putative substrate entry/product exit tunnel and the interior cavity are lined by hydrophobic residues, including several Phe and Tyr residues and a Trp side chain, which confer rigidity and may stabilize intermediates of the isomerization reaction (Fig. S5B). This hydrophobicity would provide an ideal environment to promote the partitioning of lipophilic retinoids from the membrane into the active site.
Tunnel A and the interior cavity contain two regions of strong residual electron density that are not accounted for by protein atoms (Fig. 3B). The first of these two densities is linear with a triangular shape on one end, suggesting the presence of a linear molecule containing a terminal functional group with trigonal planar geometry. The triangular portion is located next to the iron ion and positioned correctly to fulfill one or both of the open coordination sites in the octahedral geometry while the linear portion occupies a portion of tunnel A. The shape of this density suggests it may represent a bound free fatty acid molecule possibly derived from copurified retinyl esters (see below). Notably, in all other reported crystal structures containing 4-His iron coordination motifs similar to that seen in RPE65, besides that of ACO, a carboxylate or bicarbonate ligand is observed in a position similar to the carboxylate moiety of the putative fatty acid bound in the RPE65 active site, indicating that the interaction is chemically favorable. The second region of electron density is linear and bent in shape, and is found in the interior cavity of the protein. This density could represent a bound PEG 200 molecule or a string of partially ordered water molecules, but it is too short to accommodate a C8E4 molecule.
Comparison of the substrate entry/exit tunnels and active sites of RPE65 and ACO reveals a number of similarities, but also many relevant differences. The iron ion-binding site and arrangement of the first and second shell ligands is essentially identical between the two proteins, a somewhat surprising finding given that the two proteins catalyze fundamentally different reactions (Fig. S6). The orientations of the fatty acid in RPE65 and the apocarotenol substrate in ACO are similar except that the latter does not directly interact with the iron ion. There are a number of differences in the residues that line the active-site cavities resulting in different shapes. Differences in side-chain type and position and the different packing of helices 7, 8, and 9 in RPE65 compared with the corresponding residues in ACO (residues 325–342) result in significant differences in the respective active-site cavities. Also, the loosely packed nature of residues 325–342 in ACO results in the presence of a proposed hydrophilic product exit tunnel (13). A constriction in the hydrophilic tunnel of RPE65 would likely prevent exit of the products of isomerization via this route consistent with their hydrophobic nature.
RPE65 Enzymology.
RPE65, like its carotenoid-cleaving relatives, requires ferrous iron for its catalytic activity (27). Consistent with the previous report, we found that the chelating reagent, 1,10-phenanthroline, reduces microsomal retinoid isomerase activity in a dose-dependent fashion, and that this activity is partially restored by addition of ferrous, but not ferric iron (Fig. S7 A and B). Microsomal isomerase activity also was sensitive to the presence of oxidizing agents, suggesting that iron oxidation to the ferric form is responsible for the loss of such activity (Fig. S7C). Because the reaction catalyzed by RPE65 is non-redox, a different property of the iron ion, such as its Lewis acidity, may be used to help catalyze the isomerization reaction.
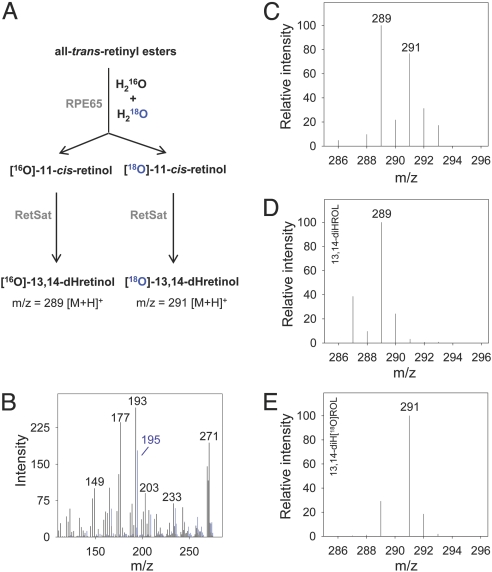
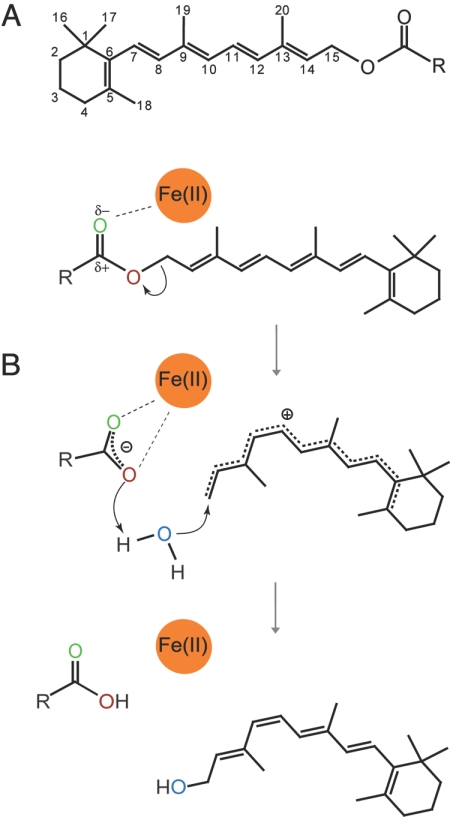
Previously, it was demonstrated that the retinoid isomerization reaction occurs at the alcohol level of oxidation (28), and that esterified retinol is the physiological substrate of the retinoid isomerase (12, 29). Studies using isotopically labeled substrate demonstrate that the stereochemical configuration of C15 inverts during isomerization (30), and that the hydroxyl oxygen in the reactant is lost during the reaction (31, 32). Both observations provide strong evidence that an O-alkyl cleavage event rather than acyl cleavage occurs during the reaction. To determine the origin of the oxygen atom found in the 11-cis-retinol product, we performed isomerization reactions in the presence of H218O, and analyzed the products by using liquid chromatography (LC)-MS (Fig. 4). Our results demonstrate that the oxygen atom found in the 11-cis-retinol product of the isomerization reaction is derived from bulk water and provides additional evidence that the isomerization reaction occurs via an O-alkyl cleavage mechanism.
Fig. 4.
MS analysis of 11-cis-retinol produced in the enzymatic reaction catalyzed by RPE65 in the presence of isotopically labeled water. (A) Experimental scheme: RPE microsomes were incubated with all-trans-retinol in the presence of a 1:1 mixture of H216O to H218O. The product of the enzymatic reaction (11-cis-retinol) was isolated and enzymatically converted into 13,14-dihydroretinol (dHretinol) to allow LC-MS detection of intact retinol molecules. (B) The MS/MS fragmentation pattern of 13,14-dHretinol and [18O]-13,14-dHretinol (blue) with characteristic 2-Da shift of the daughter ion at m/z = 195 owing to the presence of 18O. (C) Incorporation of 18O on the formation of 11-cis-retinol in the presence of H218O (50% vol/vol) as indicated by appearance of the parent ion at m/z = 291 corresponding to [18O]-13,14-dHretinol. (D) The same set of experiments performed in the absence of H218O reveals a product at m/z = 289 matching the mass of unlabeled 13,14-dHretinol. (E) Incubation of synthetic [18O]-retinol with retinol saturase produces exclusively isotopically labeled 13,14-dHretinol (m/z = 291 [M+H]+).
Temporary reduction in π bond order of the polyene chain is a fundamental requirement for retinoid isomerization chemistry to occur. Based on the observation of a putative fatty acid bound to the iron ion in the RPE65 crystal structure, we hypothesize that iron-promoted ester dissociation could generate a retinyl carbocation with reduced π bond order that would allow isomerization about the 11–12 σ bond to occur (Fig. 5). Resolution of the critical issue concerning which residues in RPE65 promote the actual isomerization event and which are responsible for 11-cis specificity cannot be addressed in this study and must await a structure containing a bound retinoid.
Fig. 5.
Proposed mechanism for π bond order reduction of the retinoid polyene chain by RPE65. (A) Chemical structure of an all-trans-retinyl ester with conventional numbering. R is typically palmitate (C16) in vivo. (B) Proposed RPE65 isomerization reaction mechanism. The iron-coordinating His residues are omitted for clarity. The initial interaction of the ester moiety with the iron ion polarizes the C15-O σ bond and makes the fatty ester a good leaving group. Formation of a resonance-stabilized carbocation reduces the π bond order of the system allowing rotation about the C11-C12 σ bond. A water molecule derived from the bulk water attacks the C15 atom quenching the carbocation intermediate. Protonation of the fatty acid carboxylate group might promote its dissociation from the iron cofactor.
From an evolutionary standpoint, it might be expected that the RPE65 catalytic mechanism would resemble that of the carotenoid oxygenases (33, 34). Mechanisms that involve participation of molecular oxygen in the isomerization reaction cannot be definitively ruled out, but would require especially complex chemistry that is not justified by the experimental evidence available to date. Recently, a member of the CCO family, called NinaB, from the Galleria mellonella moth with isomerooxygenase activity was described, indicating that members of this family originally developed isomerase activity while maintaining carotenoid oxygenase activity (35). This finding suggests that the iron cofactor is not directly involved in double bond isomerase activity of NinaB or RPE65.
RPE65 in Retinal Diseases.
A number of RPE65 mutations have been shown to cause LCA or a milder retinal dystropy RP (36). Because human and bovine RPE65 amino acid sequences are 99% identical, it can be assumed that they adopt virtually identical folds. This close relationship allowed us to map human LCA and RP-associated RPE65 amino acid substitutions onto the RPE65 structure to evaluate possible mechanisms by which these substitutions induce changes in function (Fig. S8). It is apparent that a great majority of the amino acid substitutions found in LCA and RP patients are located within or adjacent to the blades of the β-propeller rather than in the connecting loops and helices. Blade VII, where the main β-propeller closure occurs, is frequently affected. Mutations in this region could result in disruption of the β-propeller fold owing to improper “sealing” of the propeller structure. Because each blade of the propeller structure contributes a residue to the iron ion coordination system, substitutions in any sheet could displace the respective critical residue leading to disruption of iron ion binding. Missense mutations that result in substitutions of either the first or second shell iron ion ligands have been found in LCA patients (37–39), consistent with biochemical studies that demonstrated both first and second shell iron ion ligands are essential for RPE65 isomerase activity (4, 40).
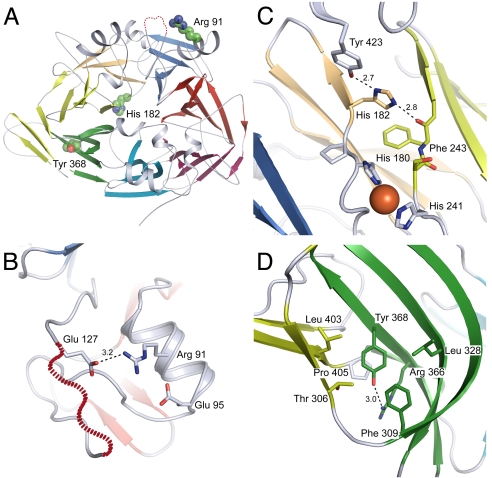
Arg91, Tyr368, and His182 are among the most frequently affected positions in patients with RPE65-associated LCA or RP (Fig. 6A) (7, 36, 39, 41, 42). An analysis of these positions in the RPE65 structure reveals that each amino acid side chain makes specific interactions with surrounding residues that cannot be mimicked by other side chains (Fig. 6 B–D). In many cases, such as His182Arg and Tyr368His substitutions, significant structural distortions would be required to accommodate the volume of the substituted side chain. Arg91 is especially interesting, because it is located outside of the β-propeller core in an α-helix. In the crystal structure, this helix is adjacent to the proposed RPE65 membrane-binding motif. Arg91 forms a salt bridge with Glu127, which is located on the C-terminal side of the sequence of residues that is disordered in the crystal structure. This salt bridge could be critical for the proper positioning of RPE65 membrane-binding elements. Indeed, Arg91 substitutions have been reported to affect RPE65 subcellular localization (43).
Fig. 6.
Locations and local interactions of residues commonly substituted in RPE65-associated LCA or RP. (A) A view of RPE65 from the top face of the β propeller showing the locations of the commonly substituted residues in LCA or RP shown in more detail in B–D. The maroon dashed line indicates the disordered segment (residues 109–126). (B) Arg91 and Glu127 form a salt bridge interaction. Arg91 is located on the surface of the protein in the H2 α-helix forming a salt bridge with Glu127. Glu127 is just upstream of one of the putative RPE65 membrane binding regions. Substitutions in the nearby Glu95 residue have also been found in LCA patients indicating that the structure of this region is critical for the proper function of RPE65. (C) Interaction of His182 with surrounding residues of RPE65. His182 forms hydrogen-bond interactions with the hydroxyl moiety of Tyr423 and the backbone carbonyl oxygen of Phe243. Loss of these specific interactions may destabilize the protein and displace the critical His180 and His241 iron ion-binding residues. (D) Interaction of Tyr368 with surrounding residues. The Tyr side chain forms an electrostatic interaction with Arg366 and several hydrophobic interactions with surrounding side chains. Numbers next to dashed lines indicate bond distances in angstroms.
Methods
RPE65 was purified from bovine RPE by differential centrifugation and anion exchange chromatography. RPE65 crystals were obtained by the hanging-drop vapor-diffusion method. The structure was solved by the SAD technique using the anomalous signal from the natively bound iron ion and refined to Rwork and Rfree values of 18 and 21.6%, respectively. Isotope labeling experiments were carried out by performing the standard retinoid isomerization reaction in the presence of 50% vol/vol H218O. The retinol products were converted to 13,14-dihydroretinol to facilitate MS analysis. The products of the reaction were analyzed by LC-MS/MS. For full methods, see SI Methods.
Supplementary Material
Acknowledgments.
We thank Dr. Wuxian Shi for assistance with diffraction data collection at the NSLS X29 beamline; the staff at the APS 23-ID-D beamline for technical assistance; Drs. Leslie T. Webster, Jr., Johannes von Lintig, and the two anonymous reviewers for excellent comments on the manuscript; and Dr. Alexander Moise (Case Western Reserve University, Cleveland, OH) for providing retinol saturase-expressing cells. This work was supported in part by NIH grants R01-EY009339 (to K.P.), P03-EB09998 and U54-GM074945 (to M.R.C.) and the Visual Sciences Training Program Grant T32-EY007157 (to P.D.K. and D.T.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3FSN).
This article contains supporting information online at www.pnas.org/cgi/content/full/0906600106/DCSupplemental.
References
- 1.Palczewski K. G protein-coupled receptor rhodopsin. Annu Rev Biochem. 2006;75:743–767. doi: 10.1146/annurev.biochem.75.103004.142743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Travis GH, Golczak M, Moise AR, Palczewski K. Diseases caused by defects in the visual cycle: Retinoids as potential therapeutic agents. Annu Rev Pharmacol Toxicol. 2007;47:469–512. doi: 10.1146/annurev.pharmtox.47.120505.105225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jin M, et al. Rpe65 is the retinoid isomerase in bovine retinal pigment epithelium. Cell. 2005;122:449–459. doi: 10.1016/j.cell.2005.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Redmond TM, et al. Mutation of key residues of RPE65 abolishes its enzymatic role as isomerohydrolase in the visual cycle. Proc Natl Acad Sci USA. 2005;102:13658–13663. doi: 10.1073/pnas.0504167102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moiseyev G, Chen Y, Takahashi Y, Wu BX, Ma JX. RPE65 is the isomerohydrolase in the retinoid visual cycle. Proc Natl Acad Sci USA. 2005;102:12413–12418. doi: 10.1073/pnas.0503460102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marlhens F, et al. Mutations in RPE65 cause Leber's congenital amaurosis. Nat Genet. 1997;17:139–141. doi: 10.1038/ng1097-139. [DOI] [PubMed] [Google Scholar]
- 7.Morimura H, et al. Mutations in the RPE65 gene in patients with autosomal recessive retinitis pigmentosa or leber congenital amaurosis. Proc Natl Acad Sci USA. 1998;95:3088–3093. doi: 10.1073/pnas.95.6.3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gu SM, et al. Mutations in RPE65 cause autosomal recessive childhood-onset severe retinal dystrophy. Nat Genet. 1997;17:194–197. doi: 10.1038/ng1097-194. [DOI] [PubMed] [Google Scholar]
- 9.Yan W, et al. Cloning and characterization of a human β, β-carotene-15,15′-dioxygenase that is highly expressed in the retinal pigment epithelium. Genomics. 2001;72:193–202. doi: 10.1006/geno.2000.6476. [DOI] [PubMed] [Google Scholar]
- 10.Redmond TM, et al. Identification, expression, and substrate specificity of a mammalian β-carotene 15,15′-dioxygenase. J Biol Chem. 2001;276:6560–6565. doi: 10.1074/jbc.M009030200. [DOI] [PubMed] [Google Scholar]
- 11.von Lintig J, Vogt K. Filling the gap in vitamin A research. Molecular identification of an enzyme cleaving β-carotene to retinal. J Biol Chem. 2000;275:11915–11920. doi: 10.1074/jbc.275.16.11915. [DOI] [PubMed] [Google Scholar]
- 12.Nikolaeva O, Takahashi Y, Moiseyev G, Ma JX. Purified RPE65 shows isomerohydrolase activity after reassociation with a phospholipid membrane. FEBS J. 2009;276:3020–3030. doi: 10.1111/j.1742-4658.2009.07021.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kloer DP, Ruch S, Al-Babili S, Beyer P, Schulz GE. The structure of a retinal-forming carotenoid oxygenase. Science. 2005;308:267–269. doi: 10.1126/science.1108965. [DOI] [PubMed] [Google Scholar]
- 14.Schwartz SH, Tan BC, Gage DA, Zeevaart JA, McCarty DR. Specific oxidative cleavage of carotenoids by VP14 of maize. Science. 1997;276:1872–1874. doi: 10.1126/science.276.5320.1872. [DOI] [PubMed] [Google Scholar]
- 15.Ma J, et al. Expression, purification, and MALDI analysis of RPE65. Invest Ophthalmol Visual Sci. 2001;42:1429–1435. [PubMed] [Google Scholar]
- 16.Takahashi Y, et al. Identification of a novel palmitylation site essential for membrane association and isomerohydrolase activity of RPE65. J Biol Chem. 2009;284:3211–3218. doi: 10.1074/jbc.M807248200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamel CP, et al. Molecular cloning and expression of RPE65, a novel retinal pigment epithelium-specific microsomal protein that is posttranscriptionally regulated in vitro. J Biol Chem. 1993;268:15751–15757. [PubMed] [Google Scholar]
- 18.Xue L, Gollapalli DR, Maiti P, Jahng WJ, Rando RR. A palmitoylation switch mechanism in the regulation of the visual cycle. Cell. 2004;117:761–771. doi: 10.1016/j.cell.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 19.Xue L, Jahng WJ, Gollapalli D, Rando RR. Palmitoyl transferase activity of lecithin retinol acyl transferase. Biochemistry. 2006;45:10710–10718. doi: 10.1021/bi060897y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin M, Yuan Q, Li S, Travis GH. Role of LRAT on the retinoid isomerase activity and membrane association of Rpe65. J Biol Chem. 2007;282:20915–20924. doi: 10.1074/jbc.M701432200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takahashi Y, Moiseyev G, Chen Y, Ma JX. The roles of three palmitoylation sites of RPE65 in its membrane association and isomerohydrolase activity. Invest Ophthalmol Visual Sci. 2006;47:5191–5196. doi: 10.1167/iovs.06-0614. [DOI] [PubMed] [Google Scholar]
- 22.Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v. 3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Forneris F, Mattevi A. Enzymes without borders: Mobilizing substrates, delivering products. Science. 2008;321:213–216. doi: 10.1126/science.1151118. [DOI] [PubMed] [Google Scholar]
- 24.Ulmschneider MB, Sansom MS, Di Nola A. Properties of integral membrane protein structures: Derivation of an implicit membrane potential. Proteins. 2005;59:252–265. doi: 10.1002/prot.20334. [DOI] [PubMed] [Google Scholar]
- 25.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 26.Hemati N, et al. RPE65 surface epitopes, protein interactions, and expression in rod- and cone-dominant species. Mol Vis. 2005;11:1151–1165. [PubMed] [Google Scholar]
- 27.Moiseyev G, et al. RPE65 is an iron(II)-dependent isomerohydrolase in the retinoid visual cycle. J Biol Chem. 2006;281:2835–2840. doi: 10.1074/jbc.M508903200. [DOI] [PubMed] [Google Scholar]
- 28.Bernstein PS, Rando RR. In vivo isomerization of all-trans- to 11-cis-retinoids in the eye occurs at the alcohol oxidation state. Biochemistry. 1986;25:6473–6478. doi: 10.1021/bi00369a020. [DOI] [PubMed] [Google Scholar]
- 29.Moiseyev G, et al. Retinyl esters are the substrate for isomerohydrolase. Biochemistry. 2003;42:2229–2238. doi: 10.1021/bi026911y. [DOI] [PubMed] [Google Scholar]
- 30.Law WC, Rando RR. Stereochemical inversion at C-15 accompanies the enzymatic isomerization of all-trans- to 11-cis-retinoids. Biochemistry. 1988;27:4147–4152. doi: 10.1021/bi00411a037. [DOI] [PubMed] [Google Scholar]
- 31.McBee JK, et al. Isomerization of all-trans-retinol to cis-retinols in bovine retinal pigment epithelial cells: Dependence on the specificity of retinoid-binding proteins. Biochemistry. 2000;39:11370–11380. doi: 10.1021/bi001061c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deigner PS, Law WC, Canada FJ, Rando RR. Membranes as the energy source in the endergonic transformation of vitamin A to 11-cis-retinol. Science. 1989;244:968–971. doi: 10.1126/science.2727688. [DOI] [PubMed] [Google Scholar]
- 33.Borowski T, Blomberg MR, Siegbahn PE. Reaction mechanism of apocarotenoid oxygenase (ACO): A DFT study. Chemistry. 2008;14:2264–2276. doi: 10.1002/chem.200701344. [DOI] [PubMed] [Google Scholar]
- 34.Kloer DP, Schulz GE. Structural and biological aspects of carotenoid cleavage. Cell Mol Life Sci. 2006;63:2291–2303. doi: 10.1007/s00018-006-6176-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oberhauser V, Voolstra O, Bangert A, von Lintig J, Vogt K. NinaB combines carotenoid oxygenase and retinoid isomerase activity in a single polypeptide. Proc Natl Acad Sci USA. 2008;105:19000–19005. doi: 10.1073/pnas.0807805105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bereta G, et al. Impact of retinal disease-associated RPE65 mutations on retinoid isomerization. Biochemistry. 2008;47:9856–9865. doi: 10.1021/bi800905v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simovich MJ, et al. Four novel mutations in the RPE65 gene in patients with Leber congenital amaurosis. Hum Mutat. 2001;18:164. doi: 10.1002/humu.1168. [DOI] [PubMed] [Google Scholar]
- 38.Simonelli F, et al. Clinical and molecular genetics of Leber's congenital amaurosis: A multicenter study of Italian patients. Invest Ophthalmol Visual Sci. 2007;48:4284–4290. doi: 10.1167/iovs.07-0068. [DOI] [PubMed] [Google Scholar]
- 39.Hanein S, et al. Leber congenital amaurosis: Comprehensive survey of the genetic heterogeneity, refinement of the clinical definition, and genotype-phenotype correlations as a strategy for molecular diagnosis. Hum Mutat. 2004;23:306–317. doi: 10.1002/humu.20010. [DOI] [PubMed] [Google Scholar]
- 40.Takahashi Y, Moiseyev G, Chen Y, Ma JX. Identification of conserved histidines and glutamic acid as key residues for isomerohydrolase activity of RPE65, an enzyme of the visual cycle in the retinal pigment epithelium. FEBS Lett. 2005;579:5414–5418. doi: 10.1016/j.febslet.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 41.Thompson DA, et al. Genetics and phenotypes of RPE65 mutations in inherited retinal degeneration. Invest Ophthalmol Visual Sci. 2000;41:4293–4299. [PubMed] [Google Scholar]
- 42.Lorenz B, et al. Early-onset severe rod-cone dystrophy in young children with RPE65 mutations. Invest Ophthalmol Visual Sci. 2000;41:2735–2742. [PubMed] [Google Scholar]
- 43.Takahashi Y, Chen Y, Moiseyev G, Ma JX. Two point mutations of RPE65 from patients with retinal dystrophies decrease the stability of RPE65 protein and abolish its isomerohydrolase activity. J Biol Chem. 2006;281:21820–21826. doi: 10.1074/jbc.M603725200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.