Abstract
Repeat proteins contain short, tandem arrays of simple structural motifs (20−40 aa). These stack together to form nonglobular structures that are stabilized by short-range interactions from residues close in primary sequence. Unlike globular proteins, they have few, if any, long-range nonlocal stabilizing interactions. One ubiquitous repeat is the tetratricopeptide motif (TPR), a 34-aa helix-turn-helix motif. In this article we describe the folding kinetics of a series of 7 designed TPR proteins that are assembled from arraying identical designed consensus repeats (CTPRan). These range from the smallest 2-repeat protein to a large 10-repeat protein (≈350 aa). In particular, we describe how the energy landscape changes with the addition of repeat units. The data reveal that although the CTPRa proteins have low local frustration, their highly symmetric, modular native structure is reflected in their multistate kinetics of unfolding and folding. Moreover, although the initial folding of all CTPRan proteins involves a nucleus with similar solvent accessibility, their subsequent folding to the native structure depends directly on repeat number. This corresponds to an increasingly complex landscape that culminates in CTPRa10 populating a misfolded, off-pathway intermediate. These results extend our current understanding of the malleable folding pathways of repeat proteins and highlight the consequences of adding identical repeats to the energy landscape.
Keywords: design protein, kinetic traps, misfolding, protein folding
Repeat proteins consist of tandem arrays of simple structural motifs that consist of 20–40 residues. These modules stack together to form elongated, nonglobular protein folds. They are highly abundant, widespread in nature and, second only to immunoglobulins, are the most common protein class to specialize in protein–protein interactions (1). Examples of repeat proteins include tetratricopeptide (TPR), ankyrin, and leucine-rich (LRR) repeats (2, 3). Unusually, unlike globular proteins, these repeat proteins do not rely on complex long-range stabilizing interactions. Instead their recurring modular architectures are dominated by regularized short-range interactions (both inter- and intrarepeat). This distinctive feature, which results in a quasi–1-dimensional structure, has made them extremely attractive targets as models for protein folding and design studies.
The keen interest in repeat proteins has led to successful protein design of consensus TPR (4), ankyrin (5, 6), and LRR proteins (7) and extensive stability and folding studies on natural ankyrin repeat proteins (a 33-residue repeat that forms a β-turn followed by 2 antiparallel α-helices) (8–14). These studies have begun to dissect the equilibrium cooperativity and kinetic folding pathways of repeat proteins. They show that folding is initiated in the most thermodynamically stable unit, with the subsequent route through the energy landscape to the native state being governed by a competition between the stability of individual repeat units and the interactions between repeats. For example, if repeats within a protein have similar stabilities and weak inter-repeat interactions the protein is more likely to have equilibrium intermediates and to fold via a very malleable pathway (more than 1 folding pathway can be accessed) (8, 9, 15, 16). Recently, these studies have been complemented by folding simulations that show and predict that the cooperativity of repeat protein folding decouples on increasing repeat number or where high local frustration occurs (17–19).
Designed consensus repeat proteins provide an excellent system for investigating the fundamental properties of repeat proteins, because each repeat has identical intra- and inter-repeat interactions. Thus, designed repeat proteins are more structurally symmetric than natural repeat proteins and can easily be extended or shortened by adding or removing whole repeats. This ability provides a unique and exquisitely tuneable perturbation that differs radically from normal amino acid mutations and enables a wider exploration of repeat protein folding energy landscapes. For example, dependence of stability, folding rate, cooperativity, and thus folding pathway on repeat number can be explored by engineering a series of proteins that increase in size through addition of identical consensus motifs. In particular, we and the Regan laboratory have used 2 series of designed consensus TPRs (a 34-aa, helix-turn-helix motif), called CTPR and CTPRa proteins, to investigate the dependence of thermodynamic characteristics and denatured state on increasing repeat number (Fig. 1; the 2 series only differ by a 2-aa substitution per repeat) (20–23). These studies have highlighted the 1-dimensionality and changing equilibrium properties of increasing repeat number by showing that the thermodynamic unfolding transition can be described and, importantly, predicted by an Ising model that uses nearest-neighbor interactions within a 1-dimensional lattice (22). Such a description implies that thermodynamically, individual repeats unfold independently of each other and thus can populate partially folded configurations. This observation was confirmed when Cortajarena et al. used equilibrium, NMR-detected hydrogen/deuterium exchange to observe sequential unfolding transitions for 2 CTPR proteins containing 2 and 3 repeats, respectively (20). However, we have also previously shown that kinetic folding of these 2 proteins at 20 °C was 2-state over the limited denaturant concentration range measured (23). These studies have been further complemented by folding simulations showing that although the CTPRs possess very low inter- and intrarepeat frustration, their cooperativity of folding as interpreted by correlation length is roughly 3 repeats (18).
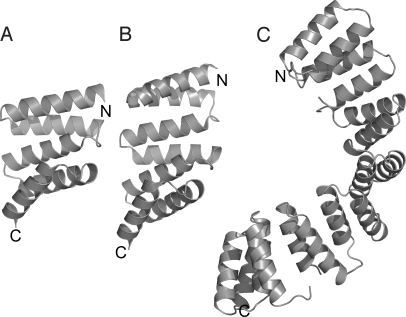
Fig. 1.
Ribbon representation of the crystal structures of (A) CTPR2 [Protein Data Bank (PDB) entry: 1NA3], (B) CTPR3 (PDB entry: 1NA0), and (C) CTPR8 (PDB entry: 2AVP). The figure was prepared using PYMOL.
In this study we investigated the kinetic folding cooperativity and folding energy landscapes on increasing repeat protein length by comparing the folding kinetics of the largest series of highly symmetric designed repeat proteins to date. This consists of 7 designed CTPRa proteins that range from 2.5 to 10.5 repeats (86–358 aa). Our results show that as repeat number increases the landscape becomes more complex, with intermediates increasingly populated, and interestingly, suggest that off-pathway intermediates become populated for the largest protein. Thus, although the CTPRa constructs have low intra- and inter-repeat frustration, on increasing repeat number their highly symmetric and modular native topology causes their cooperative kinetic folding to uncouple and eventually misfold to form kinetic traps. Finally, the folding pathway of this unique TPR system is compared with recently published work on the folding of other types of repeat protein (8, 17, 18).
Results
Structure of CTPRa Proteins.
The consensus TPR proteins (CTPRan) were built from arraying multiple copies (n) of a 34-aa idealized sequence with a C-terminal single “solvating” helix (22) [Fig. 1 and supporting information (SI) Fig. S1A]. All proteins are stable (Fig. 2) and adopt the distinctive TPR fold with the unique feature of possessing identical modular structures.
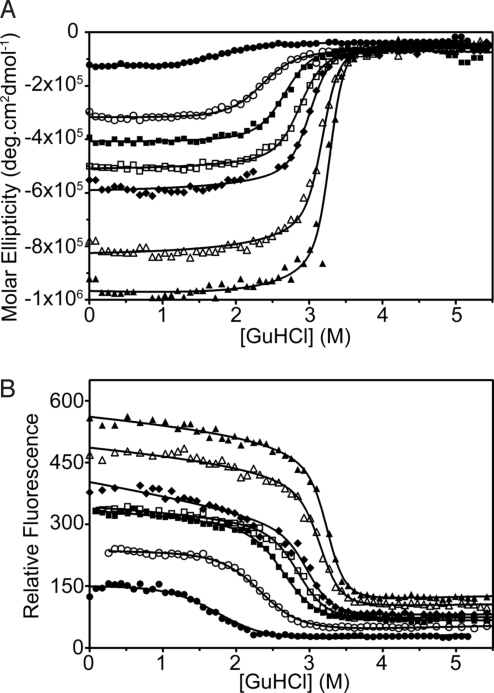
Fig. 2.
GuHCl-induced equilibrium unfolding experiments of the CTPRa proteins at 10 °C monitored by (A) ellipticity and (B) fluorescence [CTPRa2 (3 μM) (•), CTPRa3 (3 μM) (○), CTPRa4 (3 μM) (■), CTPRa5 (1 μM) (□), CTPRa6 (1 μM) (♦), CTPRa8 (1 μM) (▵), and CTPRa10 (1 μM) (▴)]. Solid lines correspond to the global best fits to a description based on the 1-dimensional Ising model (see SI Appendix for details on analysis) (22).
Equilibrium Stability of CTPRa Proteins at 10 °C.
Equilibrium chemical denaturations using guanidine hydrochloride (GuHCl) and urea were performed at 10 °C and at pH 7.0. Ellipticity at 222 nm and fluorescence at 340 nm was monitored as a function of denaturant concentration to follow each structural transition (Fig. 2 and Fig. S1D). All of the CTPRa proteins underwent a single reversible transition that corresponded to the concurrent loss of native secondary [far-UV circular dichroism (CD)–α-helical] and tertiary (fluorescence) structure, to a denatured state that lacked both. This was consistent with previously reported data at 20 °C and 25 °C (22, 23).
Data from these denaturations were initially analyzed by individually fitting each protein to a 2-state model (24). This yielded [D]50% (midpoint of unfolding) and mD-N (change in solvent-accessible surface area upon protein unfolding) from which ΔGD-NH2O/ΔG0→jH2O (free energy of unfolding in water) was calculated (Table S1). However, previous NMR and equilibrium denaturation studies on such designed TPRs have shown that, despite the apparent cooperative equilibrium unfolding, intermediate states are populated through the denaturation transition (21–23). Our data support this because our mD-N values do not increase in direct proportion to chain length of the CTPRa proteins. Moreover, differential scanning calorimetry was performed that showed the larger proteins to be no longer thermally unfolded in a 2-state manner (25).
Recently a number of studies have analyzed multistate equilibrium repeat unfolding using an Ising model (22, 26). This has been used, as the repetitive and modular repeat structure of repeat proteins coupled with the systematic variation of transition midpoint and mD-N mimic the nearest-neighbor coupling of the Ising model. Therefore the data presented here were globally fit using the same Ising model as used when the data were fit at 25 °C (22). This gave xc (midpoint of unfolding of a single α-helix in the protein), m1 (denaturant dependence of a single α-helix in the protein), J (the coupling energy between α-helices), from which H (half of the difference in free energy between the folded and denatured states of a single helix in the absence of coupling to its neighbors) (Table S2). From these data ΔG0→jH2O (the free energy of folding in water for a protein with j α-helices) was calculated for a helix, a repeat (2 helices), 1.5 repeats (3 helices) (−2.4 kcal mol−1, −1.0 kcal mol−1, and 0.4 kcal mol−1, respectively), and each CTPRa protein (Table 1 and Table S2). All of the values of ΔGD-NH2O/ΔG0→jH2O calculated are within error for urea and GuHCl denaturations. As can be seen, the Ising analysis correctly shows that individual helices or repeats are not stable/unfolded at 10 °C. To obtain a folded and stable protein at least 1.5 consecutive repeats are required [as shown previously (23, 27)]. It is interesting to note that for CTPRa2 and CTPRa3 the ΔGD-NH2O/ΔG0→jH2O from 2-state and Ising models are within error, but as the proteins increase in size there is a disparity between results, as one might expect when intermediates are present.
Table 1.
Comparison of kinetic ΔG with equilibrium ΔG and β-Tanford values
| CTPRan | ΔG0→jH2O (kcal mol−1)* | ΔGI-UH2O (kcal mol−1)† | ΔGI-NH2O (kcal mol−1)‡ | ΔGU-NH2O (kcal mol−1)§ | βT TS1 | βT Intermediate | βT TS2 |
|---|---|---|---|---|---|---|---|
| 2 | 3.3 ± 0.9 | - | - | 2.8 ± 0.4 | 0.56 | - | - |
| 3 | 6.1 ± 1.2 | 4.0 ± 1.5 | 5.1 ± 1.5 | 9.1 ± 2.1 | 0.35 | 0.49 | 0.58 |
| 4 | 9.0 ± 1.5 | 3.8 ± 1.4 | 5.9 ± 1.7 | 9.8 ± 2.2 | 0.37 | 0.47 | 0.49 |
| 5 | 11.8 ± 1.9 | 4.2 ± 1.2 | 6.4 ± 2.4 | 10.6 ± 2.7 | 0.30 | 0.45 | 0.46 |
| 6 | 14.7 ± 2.2 | 4.3 ± 0.6 | 10.7 ± 2.5 | 15.0 ± 2.6 | 0.24 | 0.34 | 0.36 |
| 8 | 20.4 ± 2.8 | 4.0 ± 0.3 | 14.6 ± 2.6 | 18.6 ± 2.6 | 0.16 | 0.25 | 0.26 |
| 10 | 26.1 ± 3.5 | 4.3 ± 0.4 | 17.8 ± 3.7 | 22.2 ± 3.7 | 0.15 | 0.23 | 0.23 |
All errors from kinetic data are obtained from propagation of errors obtained from the fitting of the data (shown in Table S3).
*Values averaged from the Ising fit of all equilibrium data and using Eq. 5 (SI Appendix); errors obtained from the propagation of a standard deviation of 3 averaged data sets (shown in Table S2).
†Calculated from kinetic data using ΔGU-1H2O = −RTln(kIUH2O/kUIH2O).
‡Calculated from kinetic data using ΔGI-NH2O = − RTln(kNIH2O/kINH2O).
§Calculated from kinetic data using ΔGU-NH2O = − RTln(kIUH2OkNIH2O/kUIH2OkINH2O). βT values were obtained using Σxmx/Σimi, where Σxmx is the sum of kinetic m-values between the unfolded state and state X on the reaction coordinate, and Σimi is the sum of all kinetic m-values along the reaction coordinate.
Unfolding/Refolding Kinetics of CTPRa Proteins.
The unfolding and refolding kinetics of CTPRa2 to CTPRa10 were measured using stopped-flow fluorescence spectroscopy as a function of GuHCl concentration. This follows the changes in the fluorescence of the tryptophan and tyrosine residues found in each repeat. The unfolding and refolding kinetics of the repeat protein constructs were observed to be very rapid, therefore experiments were performed at 10 °C to expand the range of GuHCl concentration over which data could be collected. This permitted the refolding of CTPRa4 to CTPRa10 to be followed down to 0 M GuHCl, with CTPRa2 and CTPRa3 measured down to 0.7 M and 0.54 M GuHCl, respectively. Within these ranges all of the CTPRa protein kinetics were best described by a monophasic process that fit well to single exponential equation (Fig. S2 A and B show typical kinetic traces for unfolding and refolding, respectively, overlaid with the unfolded or folded baselines). The natural logarithms of the observed rate constants measured as a function of GuHCl concentration for each CTPRa protein are shown as chevrons plots in Fig. 3 and Fig. S3. A number of striking features and trends are immediately apparent from studying the chevrons. These can be broken down into the effects that the addition of TPR motifs have on the rates of refolding and unfolding and the relationship of ln kobs as a function of GuHCl concentration.
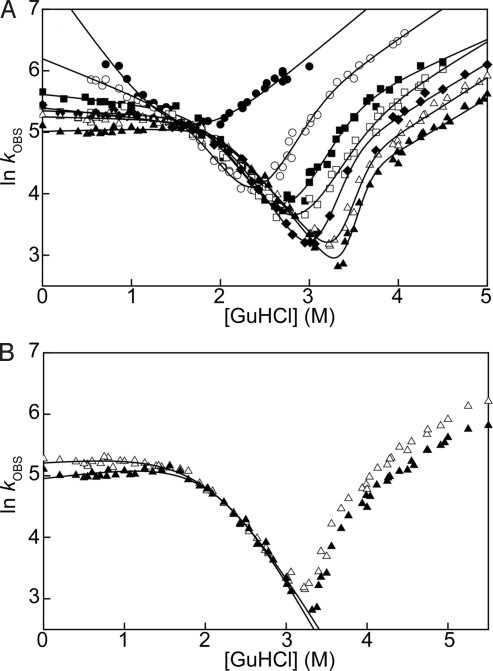
Fig. 3.
(A) Chevron plots for all of the CTPRa proteins in the series. CTPRa2 is fitted to a linear 2-state model of folding (SI Appendix Eq. 6). CTPRa3 to CTPRa10 are fitted to a sequential 3-state on-pathway model (Fig. S1B using SI Appendix Eqs. 8–10). (B) Chevron plots for CTPRa8 and CTPRa10. Here the refolding kinetics are fitted to a minimal dead-end scheme, whereby a compact off-pathway intermediate species I equilibrates with the denatured state (Fig. S1C and SI Appendix Eq. 12). In A and B: CTPRa2 (•), CTPRa3 (○), CTPRa4 (■), CTPRa5 (□), CTPRa6 (♦), CTPRa8 (▵), and CTPRa10 (▴).
Rates of refolding and unfolding.
It is obvious that the main effect of adding TPR motifs is a considerable decrease in the unfolding rate. Thus, the increase in stability of each protein on increasing repeat number can be attributed to a decrease in the rate of unfolding. This was consistent with previously reported data for 2 designed TPRs at 20 °C (23) and agrees well with studies on the effects of adding of ankyrin repeats to ankyrin-containing proteins (8, 28). In comparison, the refolding rates, when measured before any rollover, increase slightly before becoming equally fast.
Nonlinear relationship of ln kobs as a function of [GuHCl].
The most striking feature and trend of the CTPRa chevrons is the changing nonlinearity of both ln kF and ln kU as a function of GuHCl concentration. Although the smallest protein in the series, CTPRa2, exhibits a linear dependence for both refolding and unfolding over the denatured concentration range measured, which is indicative of 2-state behavior, all of the other proteins in the series do not.
When rollover in the refolding arms of the chevron plots are compared, the nonlinearity observed for each progressively bigger CTPRa construct becomes more pronounced until the rollover is no longer a smooth curve but is kinked (Fig. 3 and Fig. S3). The slope of the refolding arm after the kink for the smaller proteins is negative (as one would expect), yet as the repeats become larger the slope becomes flat and for the largest protein (CTPRa10) positive (Fig. 3, Fig. S3, and Table S3). It is interesting to note that the point at which the refolding limb for each protein kinks is essentially the same and coincides with the linear refolding arm of CTPRa2. In a similar manner to the refolding kinetics, the unfolding arms of CTPRa3 to CTPRa10 chevrons have a downward curvature that becomes more curved with increasing repeat number. However, the scale of the rollover and the kinks is less severe.
Whereas previous studies have shown that such nonlinearities can be caused by transient aggregation, ionic effects, and instrumental dead-time (29–31), here they are not. The refolding rate constants for each CTPRa protein were found to be independent of protein concentration over a 100-fold range (measured at 0 M and 0.54 M GuHCl by both pH and [GuHCl] jump experiments, 0.1 μM to 10 μM, and shown in Fig. S3), independent of ionic effects (urea denaturations gave similar rollover to GuHCl denaturations; Fig. S1E) and not limited by instrumental dead-time (folding rates recorded for CTPRa2 are faster than for the curved chevrons of larger CTPRa proteins). Thus, all observed changes in slope of the chevrons can be related to denaturant-dependent changes in either structure or population of differing states in the folding energy landscape.
Confirmation of Significantly Populated Intermediate States During Refolding.
To confirm that the curvature observed in the refolding plots of the larger CTPRa proteins is due to the population of intermediate states, refolding experiments were performed with the hydrophobic dye 1-anilinonaphthalene-8-sulfonate (ANS). ANS is known to bind to exposed clusters of hydrophobic residues frequently found in transient folding intermediates (32). On binding to such a hydrophobic surface, ANS undergoes a large change in fluorescence and so is a sensitive probe of the formation of transient species during folding (32). Therefore, refolding experiments in the presence of ANS at a final concentration of 0.54 M GuHCl were performed. Under these conditions, if the curvature observed in Fig. 3 is due to the population of intermediate states, the intermediates should bind ANS and show a change in fluorescence. This is observed for all of the larger CTPRa proteins with significant curvature (CTPRa4 to CTPRa10; Fig. S2 C and D and Table S4). In contrast, CTPRa2 and CTPRa3 do not. This is expected for CTPRa2, because the refolding arm of its chevron is linear over the denatured concentration range measured. The lack of signal from CTPRa3 might be caused by its rapid folding kinetics at 0.54 M GuHCl (350 s−1 ± 40). When a higher final GuHCl concentrations was used on each CTPRa protein (corresponding to the linear refolding region of each chevron) no signal was observed (Fig. S2 E and F), as expected. The lack of signal at higher GuHCl concentration reflects the fact that the intermediate state is destabilized relative to the denatured state with increasing concentrations of GuHCl.
Characterization of Folding Energy Landscapes.
To analyze the energy landscapes of the CTPRa proteins the kinetic data were fit in 2 ways: (i) CTPRa2: a 2-state folding scheme because no ANS binding was observed; and (ii) CTPRa3 to CTPRa10: a 3-state folding scheme because curved chevron plots and/or ANS binding was observed (thus populating refolding intermediates).
Two-state folding of CTPRa2.
The data were fitted to a linear 2-state model of folding (SI Appendix Eq. 6) (24). The extrapolated kFH2O and kUH2O from the fit were then used to calculate a kinetic ΔGU-NH2O (Table 1 and Table S3). This was consistent with ΔGD-NH2O/ΔG0→5H2O calculated from equilibrium data, showing that in the range measured CTPRa2 folds kinetically in a 2-state manner.
Three-state folding of CTPRa3 to CTPRa10.
Because of the kinked nature of the chevron plots, the data could not be fitted accurately to the sum of 2 quadratic equations (SI Appendix Eq. 7). Therefore, each dataset was fitted to a simple sequential, on-pathway 3-state model whereby the same intermediate is populated in both refolding and unfolding experiments (SI Appendix Eqs. 8–10 and Fig. S1B) (33).
This model is consistent with our ANS results, because it assumes an intermediate is populated in the dead-time of the stopped-flow instrument, causing the rate to be limited (i.e., where the chevron deviates from “2-state” linearity, the refolding kUI + kIU are much faster than kIN and for unfolding the kIN and kNI are much faster than kIU). Fig. 3A shows that our data fit well to this model, and Table S3 shows the rate constants obtained for each CTPRa protein.
Energetics of the Folding Landscape on Addition of Repeats.
Importantly, the results shown in Table S3 enable a direct comparison of the folding pathways for each protein through the calculation of the relative stabilities of each populated state on it. These are shown in Table 1 and consist of the stability of the intermediate relative to the denatured or native state (ΔGU-IH2O and ΔGI-NH2O, respectively) and the stability of the native state relative to the denatured state (ΔGU-NH2O). These highlight 3 important points: (i) when repeat number is increased the overall stability of each protein (ΔGU-NH2O) increases (consistent with equilibrium data and within error of the equilibrium stability, ΔG0→jH2O, calculated from fitting to the Ising model); (ii) the populated intermediates of each CTPRa protein have the same stability relative to the denatured state (ΔGU-NH2O ≈ 4 kcal mol−1) and are therefore independent of repeat number; and (iii) consequently, as the repeat proteins become larger the change in stability between the intermediate and native state increases (ΔGI-NH2O).
Interestingly, the fact that the stability of each CTPRa's intermediate relative to the denatured state (ΔGU-IH2O) does not change with increasing repeat number shows that all CTPRa proteins form a nucleus of comparable stability. Moreover, if the stabilities of helices/repeats obtained from the Ising model are used, the stability of each CTPRa's intermediate (ΔGU-IH2O ≈ 4 kcal mol−1) would correspond to the formation of a nucleus of ≈2.5 repeats (5 helices).
Comparison of Compactness of Transition States and Intermediates of the CTPRa Proteins.
To characterize and compare the compactness of the transition states and intermediates for each CTPRa protein, βT values were calculated from the kinetic data (Table 1). A βT value is a measure of the average degree of exposure in state X on the reaction coordinate relative to that of the denatured state from the native state (34, 35). It is defined as the ratio Σxmx/Σimi, where Σxmx is the sum of kinetic m-values between the unfolded state and state X on the reaction coordinate, and Σimi is the sum of all kinetic m-values along the reaction coordinate. A value of 1 corresponds to a state X that is as solvent exposed as the native state, whereas a value of 0 suggests that a state X is as solvent exposed as the denatured state. It is obvious from Table 1 that as repeats are added the amount of surface area buried relative to their native state of both transition and intermediates states decreases with increasing number of repeats. However, as each protein is increasing in size, this actually corresponds to a similar burial of surface area of transition and intermediates states that is constant throughout all of the CTPRa proteins. This is consistent with the invariant stability of intermediates across the CTPRa proteins (ΔGU-IH2O ≈ 4 kcal mol−1) and implies that the addition of repeats does not greatly change the initial folding process. If the βT values are related to the formation of complete repeats and helices (thus native burial of side chains), the values obtained would equate to the formation of between 1 and 1.5 repeats (2 to 3 helices) in the first transition state, between 2 and 2.5 (4 to 5 helices) in the intermediate, and between 2 and 2.5 (4 to 5 helices) in the second transition state.
Role of Each CTPRa Protein Intermediate in the Folding Landscape.
Further information on the nature of the intermediate states can be obtained by analyzing the slope of the rollover of the refolding arm of each chevron (mI-N; Table S3). In this region of the chevron plot, the slope reports on the transition from the intermediate to the native state, such that the slope depends on the changing solvent-accessible surface area between the intermediate (I) relative to the folding transition state (TS). A negative slope shows that the TS has less solvent-accessible surface area than I, no slope shows that the I and TS have the same solvent-accessible surface area, and a positive slope shows that the TS has more solvent-accessible surface area than I. This would mean a negative dependence on denaturant corresponds to a more collapsed TS relative to I, no slope corresponds to no change in compactness of the TS relative to I, and positive dependence shows that I is more compact relative to the TS. As described above, when repeats are added the rollover turns from negative (CTPRa3 to CTPRa4) to flat (CTPRa5 to CTPRa8) and finally positive (CTPRa10) (Fig. 3 and Table S3). In general, on-pathway folding proceeds through each progressive state being at least as compact if not more compact as the last. Thus, although an off-pathway intermediate cannot be ruled out, the compactness of the intermediates of CTPRa3 to CTPRa8 as judged by the slopes of the rollover observed are consistent with being on pathway (Fig. S1B). In contrast, the positive slope of the rollover observed for CTPRa10 is consistent with an off-pathway intermediate that has to partially unfold through a less-compact TS to fold to the native state (Fig. S1C).
To a first approximation the data are consistent with a minimal dead-end scheme whereby a compact off-pathway intermediate species equilibrates with the denatured state in the dead-time of the experiment (Fig. 3B, Fig. S1C, and Table S5). Thus, although positive slopes have been observed for carbonic anhydrase in the absence of salt (36) and S6 in the presence of salt (37); here, we have explicitly shown that, without any additives, repeat proteins can also exhibit this behavior.
Discussion
Multistate Kinetic Folding Pathways.
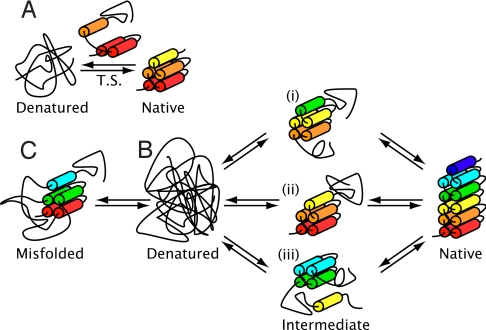
In this study we have performed a comprehensive characterization of the folding pathway of a series of designed CTPRa proteins by investigating their un/folding kinetics and equilibrium denaturations. In particular, our system allowed for full denaturation and thus complete kinetic analysis of 7 proteins (CTPRa2 to CTPRa10) that ranged from 86 aa to 358 aa. The results on these constructs, which differ greatly in size and stability, has enabled us to shed light on the systematic changes and similarities in folding pathway on increasing repeat number. In particular we have been able to show that when the CTPRa proteins' identical modular structure and their modular equilibrium folding thermodynamics are taken into account, our kinetic data are consistent with the following scheme (Fig. 4):
Fig. 4.
Schematic illustration of the proposed folding pathways of CTPRa proteins as they increase in repeat number. Cylinders are coloured from the N terminus in red and correspond to 1 helix. (A) Folding of CTPRa2: 2-state folding over the conditions studied with a transition state (T.S.) that is ≈50% solvent exposed as the native state. The transition state is drawn, for illustrative purposes, as 3 formed helices arranged as a formed repeat and 1 partially formed helix. (B) Folding of CTPRa proteins greater than 3 repeats: multistate folding through a stable intermediate. Although there is no evidence to support a structure for the intermediate state, the proposed structures (i, ii, and iii) are shown and correspond to the ΔGU-I ≈ 4 kcal mol−1. If the stabilities of helices/repeats obtained from the Ising model are used, this would correspond to the formation of a unit equal to ≈2.5 repeats. Folding from the intermediate requires a rearrangement that has no change in compaction when passing through the final transition state on route to the native state. This could consist of the docking of preformed modules. (C) Folding of CTPRa proteins of at least 10 repeats: folding is hampered by the population of a misfolded intermediate (drawn as wrongly docked repeats). The protein has to unfold from this state to continue to fold productively to the final native structure.
(i) All CTPRa proteins begin folding with the formation of a nucleus that has approximately similar burial of surface area (as judged by comparing βT values).
(ii) In the case of a CTPRa protein that contains at least 3 repeats and is more stable than 4 kcal mol−1, stable substructures form to produce metastable intermediates. For CTPRa3, folding from this intermediate still requires further collapse of the protein chain through the final transition state to the native state.
(iii) When the CTPRa protein has more than 4 repeats the intermediate folds to the native state through a transition state that has no change in compaction. This could consist of the docking of preformed modules.
(iv) If the protein has at least 10 repeats it causes the population of a misfolded intermediate. The protein has to unfold from this state to continue to fold productively to the final native structure.
This is an explicit demonstration of misfolding for a repeat protein. In particular, these results provide insight into the kinetic traps that are a direct result of constructing a protein from identical structural and energetic repeated motifs. It is interesting to speculate on the structure of the misfolded intermediate. Two schemes are probable: the misfolded intermediate could be caused by the docking of folded repeats/helices units that are not in the correct native topologic sequence, or the misfolded state could arise from the fact that the CTPRa constructs have unusually compact denatured species (21). However, because the compact denatured structures are found in all of the CTPRa proteins' denatured states and this does not seem to hamper or be prevalent in the kinetic folding of the smaller proteins, we believe the former explanation of wrongly docked modules to be more probable.
Comparison of Kinetics with Equilibrium and Computational Studies.
Our analysis shows that the modular multistate equilibrium folding of the CTPRa proteins (20, 23, 38) is also observed in their kinetic folding and unfolding. Excitingly, our results are also completely consistent with the 2 computational studies on the folding CTPR proteins. First, they mirror the simulations that predict that the kinetic folding pathway of CTPR proteins should be multistate (17, 18). Second, they are consistent with the sequential process obtained from computational simulations by Hagai and Levy (17). These show that as CTPRa proteins increase in repeat number, the occurrence of independently folding intermediates (formed from consecutive folded repeats) increases. Third, our results show that multistate kinetics are only observed when a CTPRa construct contains at least 3 CTPR motifs, and Ferreiro et al. (18) showed that the folding cooperativity of the larger CTPRa proteins, defined by correlation length, is roughly 3 repeats.
Comparison with Ankyrin Repeat Folding Studies.
Although there have been a number published studies on the folding of ankyrin proteins (11, 13), there are few that have characterized such large repeat proteins (9, 10, 15, 16) or compared such a range of repeat protein sizes (8, 28). However, these studies coupled with studies on smaller ankyrin proteins have elegantly shown that folding is controlled through the thermodynamic interplay between the stability of individual repeats and the interactions between repeats. In general, each natural ankyrin protein seems to have a stable core of a repeat or a few repeats that initiates folding and around which the rest of the structure condenses. Whether folding proceeds with no intermediates being populated, a populated kinetic intermediate, or an intermediate populated even under equilibrium conditions seems to be determined by the stability and size of the core compared with the rest of the protein. The results for CTPRa presented here agree well with this scheme and highlight how the difference in structure between natural repeats and our designed constructs affects their folding (i.e., the CTPRa proteins are built from identical repeats and therefore have no repeat/unit that is more stable or less stable than the rest of the protein). This difference manifests itself in the ready accumulation of intermediates along the folding and unfolding pathway and, when the protein is large enough, an off-pathway intermediate. It is interesting to speculate that Nature may have evolved away from complete consensus proteins, not only for binding, but also to avoid such kinetic traps.
Conclusions
Designed TPR motifs have unique properties: identical structural modularity, sequence simplicity, and modular linear structure. We have exploited these properties to explore their folding landscapes by elegantly using the perturbation of adding and subtracting whole repeat motifs. This facilitates a wider exploration of repeat protein folding energy landscapes. Initially on increasing the repeat number, on-pathway intermediates are kinetically populated. Subsequent addition of modular repeats causes off-pathway kinetic traps to predominate. Thus, although proteins constructed of identical repeats have low local frustration, their modular and symmetric structures produce energy landscapes that are prone to kinetic traps. These results complement the existing notion of repeat proteins having very malleable folding pathways and highlight how the structure of consensus repeats affects their folding pathway.
Materials and Methods
Cloning, Protein Production, and Purification.
The designed CTPRa proteins were cloned, expressed, and purified as previously described (22).
Equilibrium Experiments.
Fluorescence and far-UV CD equilibrium unfolding measurements were performed and analyzed as described in detail in SI Appendix.
Kinetic Experiments.
All experiments were performed as described in detail in SI Appendix. In brief, both unfolding and folding phases fitted well to a single-exponential process. No slow, proline isomerization phases were observed in the refolding experiments over a 200-s time scale. It is quite possible that slower phases exist but are difficult to detect owing to instrumental drift.
Data Analysis.
The dependence of ln kobs on [denaturant] for each CTPRa protein was fitted to either a 2-state model or a sequential 3-state model whereby an intermediate is either on pathway (Fig. S1B) or off pathway (Fig. S1C). For full details of equations used see SI Appendix.
Supplementary Material
Acknowledgments.
We thank Dr. Sophie Jackson, Dr. Alan Lowe, and Prof. Lynne Regan for insightful discussion, helpful comments, and suggestions; and members of the Department of Chemistry and Biochemistry (University of Sussex), Dr. Lowe, Dr. Jackson, and Dr. Phillips, for critical reading of the manuscript. Y.J. was supported by an Engineering and Physical Sciences Research Council studentship. This work was partly funded by Biotechnology and Biological Sciences Research Council Grant E005187/1.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0907455106/DCSupplemental.
References
- 1.Grove TZ, Cortajarena AL, Regan L. Ligand binding by repeat proteins: Natural and designed. Curr Opin Struct Biol. 2008;18:507–515. doi: 10.1016/j.sbi.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrade MA, Perez-Iratxeta C, Ponting CP. Protein repeats: Structures, functions, and evolution. J Struct Biol. 2001;134:117–131. doi: 10.1006/jsbi.2001.4392. [DOI] [PubMed] [Google Scholar]
- 3.Groves MR, Barford D. Topological characteristics of helical repeat proteins. Curr Opin Struct Biol. 1999;9:383–389. doi: 10.1016/s0959-440x(99)80052-9. [DOI] [PubMed] [Google Scholar]
- 4.Main ERG, Jackson SE, Regan L. The folding and design of repeat proteins: Reaching a consensus. Curr Opin Struct Biol. 2003;13:482–489. doi: 10.1016/s0959-440x(03)00105-2. [DOI] [PubMed] [Google Scholar]
- 5.Kohl A, et al. Designed to be stable: Crystal structure of a consensus ankyrin repeat protein. Proc Natl Acad Sci USA. 2003;100:1700–1705. doi: 10.1073/pnas.0337680100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mosavi LK, Minor DL, Peng ZY. Consensus-derived structural determinants of the ankyrin repeat motif. Proc Natl Acad Sci USA. 2002;99:16029–16034. doi: 10.1073/pnas.252537899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stumpp MT, Forrer P, Binz HK, Pluckthun A. Designing repeat proteins: Modular leucine-rich repeat protein libraries based on the mammalian ribonuclease inhibitor family. J Mol Biol. 2003;332:471–487. doi: 10.1016/s0022-2836(03)00897-0. [DOI] [PubMed] [Google Scholar]
- 8.Wetzel SK, Settanni G, Kenig M, Binz HK, Pluckthun A. Folding and unfolding mechanism of highly stable full-consensus ankyrin repeat proteins. J Mol Biol. 2008;376:241–257. doi: 10.1016/j.jmb.2007.11.046. [DOI] [PubMed] [Google Scholar]
- 9.Werbeck ND, Rowling PJE, Chellamuthu VR, Itzhaki LS. Shifting transition states in the unfolding of a large ankyrin repeat protein. Proc Natl Acad Sci USA. 2008;105:9982–9987. doi: 10.1073/pnas.0705300105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tripp KW, Barrick D. Rerouting the folding pathway of the notch ankyrin domain by reshaping the energy landscape. J Am Chem Soc. 2008;130:5681–5688. doi: 10.1021/ja0763201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kloss E, Courtemanche N, Barrick D. Repeat-protein folding: New insights into origins of cooperativity, stability, and topology. Arch Biochem Biophys. 2008;469:83–99. doi: 10.1016/j.abb.2007.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Courtemanche N, Barrick D. Folding thermodynamics and kinetics of the leucine-rich repeat domain of the virulence factor Internalin B. Prot Sci. 2008;17:43–53. doi: 10.1110/ps.073166608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barrick D, Ferreiro DU, Komives EA. Folding landscapes of ankyrin repeat proteins: Experiments meet theory. Curr Opin Struct Biol. 2008;18:27–34. doi: 10.1016/j.sbi.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lowe AR, Itzhaki LS. Rational redesign of the folding pathway of a modular protein. Proc Natl Acad Sci USA. 2007;104:2679–2684. doi: 10.1073/pnas.0604653104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Low C, et al. Structural insights into an equilibrium folding intermediate of an archaeal ankyrin repeat protein. Proc Natl Acad Sci USA. 2008;105:3779–3784. doi: 10.1073/pnas.0710657105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Werbeck ND, Itzhaki LS. Probing a moving target with a plastic unfolding intermediate of an ankyrin-repeat protein. Proc Natl Acad Sci USA. 2007;104:7863–7868. doi: 10.1073/pnas.0610315104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hagai T, Levy Y. Folding of elongated proteins: Conventional or anomalous? J Am Chem Soc. 2008;130:14253–14262. doi: 10.1021/ja804280p. [DOI] [PubMed] [Google Scholar]
- 18.Ferreiro DU, Walczak AM, Komives EA, Wolynes PG. The energy landscapes of repeat-containing proteins: Topology, cooperativity, and the folding funnels of one-dimensional architectures. PLoS Comput Biol. 2008;4:e1000070. doi: 10.1371/journal.pcbi.1000070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferreiro DU, Cho SS, Komives EA, Wolynes PG. The energy landscape of modular repeat proteins: Topology determines folding mechanism in the ankyrin family. J Mol Biol. 2005;354:679–692. doi: 10.1016/j.jmb.2005.09.078. [DOI] [PubMed] [Google Scholar]
- 20.Cortajarena AL, Mochrie SGJ, Regan L. Mapping the energy landscape of repeat proteins using NMR-detected hydrogen exchange. J Mol Biol. 2008;379:617–626. doi: 10.1016/j.jmb.2008.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cortajarena AL, et al. Non-random-coil behaviour as a consequence of extensive PPII structure in the denatured state. J Mol Biol. 2008;382:203–212. doi: 10.1016/j.jmb.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kajander T, Cortajarena AL, Main ERG, Mochrie SGJ, Regan L. A new folding paradigm for repeat proteins. J Am Chem Soc. 2005;127:10188–10190. doi: 10.1021/ja0524494. [DOI] [PubMed] [Google Scholar]
- 23.Main ERG, Stott K, Jackson SE, Regan L. Local and long-range stability in tandemly arrayed tetratricopeptide repeats. Proc Natl Acad Sci USA. 2005;102:5721–5726. doi: 10.1073/pnas.0404530102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fersht AR. Structure and Mechanism in Protein Science: Guide to Enzyme Catalysis and Protein Folding. 3rd Ed. New York: W. H. Freeman; 1999. p. 650. [Google Scholar]
- 25.Javadi Y. Falmer near Brighton, UK: University of Sussex; 2009. Structural stability and the folding pathway of a series of consensus tetratricopeptide repeat proteins. Ph.D. thesis. [Google Scholar]
- 26.Aksel T, Barrick D. Analysis of repeat-protein folding using nearest-neighbor statistical mechanical models. Methods Enzymol. 2009;455:95–125. doi: 10.1016/S0076-6879(08)04204-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Main ERG, Xiong Y, Cocco MJ, D'Andrea L, Regan L. Design of stable alpha-helical arrays from an idealized TPR motif. Structure. 2003;11:497–508. doi: 10.1016/s0969-2126(03)00076-5. [DOI] [PubMed] [Google Scholar]
- 28.Tripp KW, Barrick D. Enhancing the stability and folding rate of a repeat protein through the addition of consensus repeats. J Mol Biol. 2007;365:1187–1200. doi: 10.1016/j.jmb.2006.09.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Silow M, Oliveberg M. Transient aggregates in protein folding are easily mistaken for folding intermediates. Proc Natl Acad Sci USA. 1997;94:6084–6086. doi: 10.1073/pnas.94.12.6084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krantz BA, Sosnick TR. Distinguishing between two-state and three-state models for ubiquitin folding. Biochemistry. 2000;39:11696–11701. doi: 10.1021/bi000792+. [DOI] [PubMed] [Google Scholar]
- 31.Went HM, Benitez-Cardoza CG, Jackson SE. Is an intermediate state populated on the folding pathway of ubiquitin? FEBS Lett. 2004;567:333–338. doi: 10.1016/j.febslet.2004.04.089. [DOI] [PubMed] [Google Scholar]
- 32.Jones BE, Jennings PA, Pierre RA, Matthews CR. Development of nonpolar surfaces in the folding of Escherichia coli dihydrofolate reductase detected by 1-anilinonaphthalene-8-sulfonate binding. Biochemistry. 1994;33:15250–15258. doi: 10.1021/bi00255a005. [DOI] [PubMed] [Google Scholar]
- 33.Sanchez IE, Kiefhaber T. Evidence for sequential barriers and obligatory intermediates in apparent two-state protein folding. J Mol Biol. 2003;325:367–376. doi: 10.1016/s0022-2836(02)01230-5. [DOI] [PubMed] [Google Scholar]
- 34.Tanford C. Protein denaturation. Adv Protein Chem. 1968;23:121–282. doi: 10.1016/s0065-3233(08)60401-5. [DOI] [PubMed] [Google Scholar]
- 35.Tanford C. Protein denaturation. C. Theoretical models for the mechanism of denaturation. Adv Protein Chem. 1970;24:1–95. [PubMed] [Google Scholar]
- 36.McCoy LF, Jr, Rowe ES, Wong KP. Multiparameter kinetic study on the unfolding and refolding of bovine carbonic anhydrase B. Biochemistry. 1980;19:4738–4743. doi: 10.1021/bi00562a003. [DOI] [PubMed] [Google Scholar]
- 37.Otzen DE, Oliveberg M. Salt-induced detour through compact regions of the protein folding landscape. Proc Natl Acad Sci USA. 1999;96:11746–11751. doi: 10.1073/pnas.96.21.11746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Main ERG, Lowe AR, Mochrie SGJ, Jackson SE, Regan L. A recurring theme in protein engineering: The design, stability and folding of repeat proteins. Curr Opin Struct Biol. 2005;15:464–471. doi: 10.1016/j.sbi.2005.07.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.