Abstract
Commercial wine yeast strains of the species Saccharomyces cerevisiae have been selected to satisfy many different, and sometimes highly specific, oenological requirements. As a consequence, more than 200 different strains with significantly diverging phenotypic traits are produced globally. This genetic resource has been rather neglected by the scientific community because industrial strains are less easily manipulated than the limited number of laboratory strains that have been successfully employed to investigate fundamental aspects of cellular biology. However, laboratory strains are unsuitable for the study of many phenotypes that are of significant scientific and industrial interest. Here, we investigate whether a comparative transcriptomics and phenomics approach, based on the analysis of five phenotypically diverging industrial wine yeast strains, can provide insights into the molecular networks that are responsible for the expression of such phenotypes. For this purpose, some oenologically relevant phenotypes, including resistance to various stresses, cell wall properties, and metabolite production of these strains were evaluated and aligned with transcriptomic data collected during alcoholic fermentation. The data reveal significant differences in gene regulation between the five strains. While the genetic complexity underlying the various successive stress responses in a dynamic system such as wine fermentation reveals the limits of the approach, many of the relevant differences in gene expression can be linked to specific phenotypic differences between the strains. This is, in particular, the case for many aspects of metabolic regulation. The comparative approach therefore opens new possibilities to investigate complex phenotypic traits on a molecular level.
Saccharomyces cerevisiae is a preferred model organism for studying eukaryotic cells. The haploid yeast genome is compact (12 to 13.5 megabases) and contains only around 6,000 protein-encoding genes (18). However, the functional analysis of the yeast genome remains a challenge, predominantly because many of the putative protein-encoding genes appear not to be amenable to classical genetic approaches. One possible reason is that most studies have been limited to a small number of laboratory strains. While these strains have been selected for their ease of use under laboratory conditions, they lack many of the characteristics that are prominent in industrial isolates of this species. These industrial strains are highly diverse since they have been selected for a large number of different and highly specific tasks. This geno- and phenotypic diversity represents a largely untouched genetic resource. Some of the challenges of large-scale functional genomics can therefore likely be met by including such strains in comparative transcriptomic studies (21, 25).
In commercial wine fermentations, the yeast species S. cerevisiae is the major role player, and wine yeast strains have been isolated and selected for optimized performance in certain key areas of oenological relevance. The specific phenotypic traits selected for include fermentative efficiency, general stress resistance, production of metabolites and in particular aroma compounds, cell wall adhesion properties, and the ability to release enzymes of enological interest (mannoproteins) (35). Various research methodologies have been applied in an attempt to understand crucial aspects of wine yeast physiology. However, many important questions remain unanswered regarding the genetic and molecular regulation of most of these traits, many of which are of a polygenic nature (32) and cannot be fully understood through traditional approaches.
In this paper, we investigate the potential of a comparative functional transcriptomic approach to correlate oenologically relevant phenotypes to specific gene expression patterns. The approach is based on the comparative analysis of physiological data and global gene expression data of five phenotypically diverging commercial wine yeast strains. Phenotypes investigated include general stress resistance, cell wall properties such as adhesion, and metabolic regulation and production.
The ability of fermenting yeast cells to withstand certain stress factors is of extreme importance to the overall fermentation efficiency of the strains (26). During the course of wine fermentation, S. cerevisiae cells are subjected to multiple severe stress conditions that affect growth, viability, and fermentative performance. These stresses include high osmotic pressure, acidity, nutrient deprivation, starvation, and high alcohol concentration (2, 14, 20). Some of these stresses occur sequentially, whereas others occur simultaneously (3). This means that the genetic factors and regulatory networks involved in stress response pathways during fermentation are interwoven into complex regulatory circuits, with notable interplay/overlap between the various response pathways (17). Several gene expression studies of individual wine yeast strains have been undertaken in recent years to shed some light on the adaptation and stress tolerance mechanisms of industrial wine yeast strains. Ivorra et al. (26) and Zuzuarregui et al. (47) both evaluated expression levels of preselected representative genes involved in stress responses in several wine yeast strains during fermentation. Both studies indicated a good correlation between stress resistance and the ability to complete fermentations under suboptimal or optimal conditions.
Large-scale transcriptome monitoring during alcoholic fermentation under conditions mimicking an enological environment has helped to elucidate the coordinated transcriptional reprogramming that takes place during alcoholic fermentation as changes in nutritional, environmental, and physiological conditions occur (31, 39). These approaches have contributed to the identification of regulatory pathways that play a key role in coordinating stress-induced changes in gene expression (39). The data have also pointed to a group of genes designated as fermentation stress response genes that are dramatically induced at various points during fermentation (31). More specific transcriptional profiling of wine yeast strains has focused on cold stress (4) and the response to nitrogen availability (30).
Another important area of yeast performance relates to the ability of yeast cells to adhere to one another and settle out of suspension by the end of fermentation. Adhesion phenotypes such as flocculation and substrate adhesion are directly linked to cell wall properties and can have a major impact in biotechnological processes such as wine making (19). Flocculation (the reversible, calcium-dependent, nonsexual aggregation of yeast cells into flocs), in particular, is a process of great importance to the fermentative characteristics of yeast strains (for a review, see reference 43). Several structurally related lectin-like proteins (flocculins) encoded by FLO genes (12) are responsible for different adhesion phenotypes, suggesting differential programs of pre- and posttranscriptional regulation, a strong likelihood for this gene family (for a review, see reference 44).
The last key area of yeast physiology from an oenological view relates to the general metabolic activity of the cells during fermentation, particularly in terms of the production of volatile flavor and aroma compounds (15, 45). In a related study, using the same transcriptome data set, the gene expression analysis of a selected number of genes involved in metabolism was combined with targeted exo-metabolome analysis (40). This enabled the alignment of a subset of ±200 genes from the transcriptome data with changes in the exo-metabolome and the identification of genes that impact on aroma compound metabolism. This approach helped identify highly correlated gene expression subnetworks that could be linked to specific areas of fermentative metabolism related to the production of organoleptic compounds.
However, not much is known regarding the underlying molecular and biochemical differences between different wine yeast strains in other areas of yeast metabolism. Such differences in wine yeast metabolic activities will have a direct impact on the ability of the cells to tolerate biotic and abiotic stress conditions, as well as influence the likelihood of “stuck” fermentations.
One way to contextualize transcriptomic information is by integrating a priori knowledge of gene interaction and metabolic networks with the gene expression data, as described in Vemuri and Aristidou (42). The combination of comparative microarray data sets with existing models of yeast metabolism and interaction networks offers the potential for in silico evaluation of biologically relevant gene expression changes in the context of key areas of metabolism (16, 34). In this study, we demonstrate that differences in gene expression patterns between strains can be linked to phenotypic differences and to specific reporter metabolites around which significant metabolic and physiological changes can be grouped. Such data provide important clues regarding the key drivers of evolutionary differentiation between industrial strains and will help identify new targets for biotechnological strategies.
MATERIALS AND METHODS
Strains, medium, and culture conditions.
The yeast strains used in this study are the following: VIN13 (Anchor Yeast, South Africa) and EC1118, BM45, 285, and DV10 (all from Lallemand, Inc., Montreal, Canada). All are diploid S. cerevisiae strains used in industrial wine fermentations. Yeast cells were cultivated at 30°C in YPD synthetic medium consisting of 1% yeast extract (Biolab, South Africa), 2% peptone (Fluka, Germany), and 2% glucose (Sigma, Germany). Solid medium was supplemented with 2% agar (Biolab, South Africa).
Fermentation medium.
Fermentation experiments were carried out with synthetic must MS300, which approximates to a natural must as previously described (5). The medium contained 125 g/liter glucose and 125 g/liter fructose, and the pH was buffered at 3.3 with NaOH.
Fermentation conditions.
All fermentations were carried out under microaerobic conditions in 100-ml glass bottles (containing 80 ml of the medium) sealed with rubber stoppers with a CO2 outlet. The fermentation temperature was approximately 22°C, and no continuous stirring was performed during the course of the fermentation. Fermentation bottles were inoculated with YPD cultures in the logarithmic growth phase (approximately an optical density at 600 nm [OD600] of 1) to an OD600 of 0.1 (i.e., a final cell density of approximately 106 CFU·ml−1). The cells from the YPD precultures were briefly centrifuged and resuspended in MS300 to avoid carryover of YPD to the fermentation medium. The fermentations followed a time course of 14 days, and the bottles were weighed daily to assess the progress of fermentation. Samples of the fermentation medium and cells were taken at days 2, 5, and 14 as representative of the exponential, early logarithmic, and late logarithmic growth phases, respectively.
Growth measurement.
Cell proliferation (i.e., growth) was determined spectrophotometrically (Powerwave X; Bio-Tek Instruments) by measuring the OD600 of 200-μl samples of the suspensions over the 14-day experimental period.
Analytical methods: high-performance liquid chromatography.
Culture supernatants were obtained from the cell-free upper layers of the fermentation medium. For the purposes of glucose determination and carbon recovery, culture supernatants and starting medium were analyzed by high-performance liquid chromatography on an Aminex HPX-87H ion exchange column using 5 mM H2SO4 as the mobile phase. An Agilent refractive index detector and UV detector were used in tandem for peak detection and quantification. Analysis was carried out using the HP ChemStation software package.
Enzymatic metabolite assays.
All enzymes and cofactors were obtained from Roche (Germany) or Sigma (Germany). Metabolite concentrations were determined using the enzymatic methods described by Bergmeyer and Bernt (7).
General statistical analysis.
t tests and analysis of variance were conducted using Statistica (version 7). Hierarchical clustering and k-means cluster analysis were carried out using TIGR MeV, version 2.2 (6).
Starvation assays.
Determination of cell viability/survival upon macronutrient starvation was conducted using growth medium limited for key macronutrients. The compositions of the four nutrient-depleted media are summarized in Table 1.
TABLE 1.
Medium composition for carbon, nitrogen, sulfur, and phosphorus starvation assays
| Compound | Amt of compound (g/liter) in medium lackinga:
|
|||
|---|---|---|---|---|
| Carbon | Nitrogen | Sulfur | Phosphorus | |
| (NH4)2SO4 | 5 | NA | NA | 5 |
| KH2PO4 | 3 | 3 | 3 | NA |
| MgSO4·7H2O | 0.5 | 0.5 | NA | 0.5 |
| K2SO4 | NA | 5 | NA | 2 |
| NH4Cl | NA | NA | 5 | NA |
| MgCl2 | NA | NA | 0.5 | NA |
| Glucose | NA | 50 | 50 | 50 |
NA, not added.
Ca2+-dependent flocculation assays.
Yeast colonies for each strain were inoculated (in quadruplicate) in test tubes containing 5 ml of YPD medium and grown to stationary phase. An aqueous solution of EDTA (pH 8.0) was then added to these cultures to a final concentration of 50 mM, and the cultures were agitated vigorously by vortexing at maximum speed setting. The OD600 was determined immediately by mixing 100 μl of the culture with 900 μl of 50 mM EDTA (measurement A). Ca2+-dependent flocculation was then induced by spinning down 1 ml of the liquid cultures in a microcentrifuge, followed by washing in 1 ml of double-distilled H2O (ddH2O) and resuspension in 1 ml of 40 mM CaCl2. The samples were then vigorously agitated as before and left undisturbed for 60 s. A 100-μl sample was then taken from just below the meniscus in the microcentrifuge tube of each sample and mixed thoroughly with 900 μl of a 40 mM CaCl2 solution. A second spectrophotometric measurement was then taken at a wavelength of 600 nm as before (measurement B). For more information, see Bester et al. (8). The extent of Ca2+-dependent flocculation was then calculated by the following formula: flocculation (%) = (A − B)/A × 100 (measurements A and B as described above).
Cell surface hydrophobicity assays.
Yeast cultures grown overnight in YPD medium were diluted to a concentration of 0.5 × 107 cells in 2 ml of ddH2O. After centrifugation and removal of the supernatant, the cells were resuspended in 2 ml of buffer containing 22.2 g·liter−1 K2HPO4, 7.26 g·liter−1 KH2PO4, 1.8 g·liter−1 urea, and 0.2 g·liter−1 MgSO4·7H2O. The absorbance of 1 ml of the cell suspension was determined spectrophotometrically at a wavelength of 660 nm (reading A). To the remaining 1-ml cell suspensions, 100 μl of xylene was added to each sample, and the samples were vortexed vigorously for 30 s and left to stand for 15 min thereafter. The xylene layer was then removed from each tube, and the absorbance of the remaining aqueous layer determined as before at 660 nm (reading B). The modified hydrophobicity index was defined as 1 minus (B/A). High modified hydrophobicity index values are indicative of increased partitioning of cells toward the nonpolar xylene phase and, thus, of a hydrophobic yeast population (22).
Cell surface charge assays.
Yeast cultures grown overnight in YPD medium were diluted to a concentration of 0.5 × 107 cells in 1 ml of 0.2 M acetate buffer (pH 4.0) consisting of 4.1% acetic acid and 0.9% sodium acetate. Cells were washed three times in acetate buffer before final resuspension in acetate buffer containing Alcian blue (0.015 g·liter−1). The cell suspensions were incubated at 120 rpm on a shaker for 5 min at room temperature. Samples were subsequently centrifuged at 4,000 rpm for 5 min, and the absorbance of the supernatant was determined spectrophotometrically at 607 nm. The amount of Alcian blue that remained bound to the cells was calculated using a standard curve set up by diluting the original 0.015 g·liter−1 Alcian blue in acetate buffer. Data were expressed as micrograms of Alcian blue bound per 0.5 × 107 cells (i.e., per 1 ml of OD600 of 0.5) (37).
Mat formation.
The ability of yeast strains to form spreading growth mats (also referred to as biofilm formation) on plates was determined as described previously (38). Ten microliters of a yeast suspension grown overnight in liquid medium was spotted in the center of a YPD plate containing 0.3% (wt/vol) agar and incubated at 23°C.
Heat shock.
Cells were grown continuously at 30°C to an OD600 of 1 before centrifugation and resuspension in an equal volume of ddH2O at a temperature of 55°C. The cell suspensions were then incubated at this temperature for periods of 15, 40, and 45 min; plated out in 10-μl serial dilution ranges on YPD plates; and incubated for 24 to 48 h at 30°C to assess for survival.
Oxidative stress.
Cells were grown to an OD600 of 1 as representative of the mid-exponential growth phase. Samples were plated out in 10-μl serial dilutions on YPD plates containing 1 mM, 2 mM, and 3 mM hydrogen peroxide and incubated at 30°C to detect the growth of tolerant cells.
Osmotic and hypersaline stress.
Cells were grown to an OD600 of 1. Samples were then centrifuged to collect the cells and resuspended in equal volumes of 0.9% NaCl (osmo-neutral). Samples were plated out in 10-μl serial dilutions on YPD plates containing 1 M, 1.5 M, and 2 M sorbitol and incubated at 30°C for 24 to 48 h to assay for osmotic shock. For the hypersaline stress, the same samples were plated out on YPD solid medium containing 1 M, 1.2 M, and 1.5 M sodium chloride (NaCl).
Heavy metal (copper) toxicity.
Mid-logarithmically growing cells were collected by centrifugation, resuspended in slightly buffered ddH2O, and spotted onto YPD plates containing 0.5 mM, 1 mM, 2 mM, and 4 mM copper sulfate (CuSO4), respectively.
Ethanol tolerance.
Yeast cells were grown to the mid-exponential growth phase before centrifugation to collect the cells. The cells were then resuspended in 20%, 25%, and 30% ethanol solutions and incubated at room temperature for 10 min. Serial dilutions of the ethanol-stressed cells were then spotted onto regular YPD plates to determine the relative survival rate of cells of the different strains.
Microarray analysis.
Sampling of cells from fermentations and total RNA extraction were performed as described by Abbott et al. (1). Probe preparation and hybridization to Affymetrix Genechip microarrays were performed according to Affymetrix instructions, starting with 6 μg of total RNA. Results for each strain and time point were derived from three independent culture replicates. The quality of total RNA, cDNA, cRNA, and fragmented cRNA were confirmed using the Agilent Bioanalyzer 2100.
Transcriptomic data acquisition and statistical analysis.
Acquisition and quantification of array images and data filtering were performed using Affymetrix GeneChip operating software, version 1.4. All arrays were scaled to a target value of 500 using the average signal from all gene features using the GeneChip operating software. Genes with expression values below 12 were set to 12 plus the expression value, as previously described (9), in order to eliminate insignificant variations.
Determination of differential gene expression between experimental parameters was conducted using SAM (Significance Analysis of Microarrays) software, version 2 (41). The two-class, unpaired setting was used, and genes with a Q value less than 0.5 (P < 0.0005) were considered differentially expressed. Only genes with a relative change greater than twofold (positive or negative) were taken into consideration.
Random forest analysis was carried out as described by Breiman (11). Genes were differentially ranked according to their ability to discriminate between different time points (clamped strain data) and between different strains (clamped time data). The top 200 open reading frames for each analysis were considered for further in depth analysis and evaluation.
Multivariate data analysis.
The patterns within the different sets of data were investigated by principal-component analysis (PCA; The Unscrambler; Camo Inc., Corvallis, OR). PCA is a bilinear modeling method which gives a visually interpretable overview of the main information in large, multidimensional data sets. By plotting the principal components, it is possible to view statistical relationships between different variables in complex data sets and detect and interpret sample groupings, similarities, or differences, as well as the relationships between the different variables (29).
Reporter metabolite analysis.
Microarray data were analyzed using an algorithm that integrates the topology of the yeast metabolic graph to uncover the transcriptional regulatory architecture of the metabolic pathways. The so-called reporter metabolites are metabolites around which the most significant transcriptional changes can occur. The algorithm allows pairwise and multiple comparisons to be performed using transcriptome data. Thus, multidimensional analysis of gene expression patterns at different times and between different strains is used first to score reporter metabolites. Based on the reconstruction of the metabolic network graphs, it is possible to subsequently uncover highly correlated connected subgraphs (subnetworks) within the enzyme interaction graph. For more information, see the original article by Patil and Nielsen (33).
Microarray data accession number.
Microarray data can be viewed at the Gene Expression Omnibus repository under the accession number GSE11651.
RESULTS
Strain physiology and fermentation.
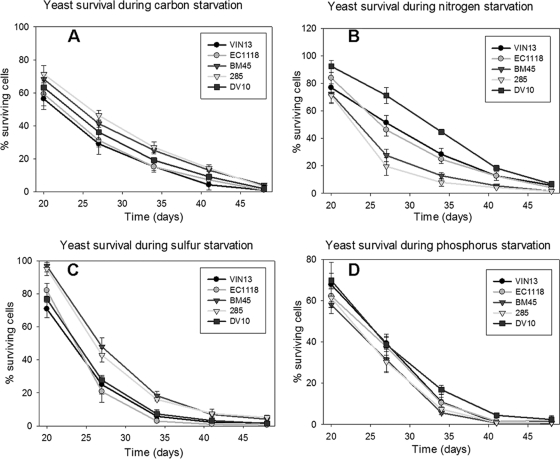
All five commercial strains displayed comparable growth rates and primary fermentation kinetics such as fermentation rate, sugar utilization, and ethanol production while displaying significant differences with regard to the production of extracellular metabolites (40). Additional phenotypic characterization reveals significant differences in the general physiology of these strains. Such differences include the following: (i) phenotypes such as flocculation, invasive growth, and mat formation (Table 2) related to cell surface and adhesion properties; and (ii) the ability of the different strains to tolerate carbon, nitrogen, sulfur, and phosphorous starvation (Fig. 1) as well as key stress conditions such as oxidative, osmotic, and ethanol stress (Fig. 2).
TABLE 2.
Summary of cell wall properties and adhesion phenotypes of the five strains
| Strain | Value for the indicated propertya
|
|||
|---|---|---|---|---|
| Flocculation (%) | MHI | Cell surface charge index | Mat formation | |
| VIN13 | 3.63 ± 0.25 abc | 0.037 ± 0.012 abcd | 25.4 ± 3.7 | No |
| EC1118 | 3.53 ± 0.55 def | 0.193 ± 0.031 aef | 23.3 ± 2.4 a | No |
| BM54 | 12.17 ± 2.08 adg | 0.555 ± 0.074 beg | 26.5 ± 3.2 | Yes |
| 285 | 12.32 ± 2.75 beh | 0.381 ± 0.042 cfg | 23.1 ± 2.2 b | Yes |
| DV10 | 5.85 ± 0.99 cfgh | 0.155 ± 0.027 dg | 28.1 ± 1.2 ab | No |
Lowercase letters indicate values that are significantly different from one another (P < 0.05).
FIG. 1.
Survival response of the strains in this study under carbon (A), nitrogen (B), sulfur (C), and phosphorus (D) starvation conditions.
FIG. 2.
Assays for heat shock (A), oxidative stress (B), osmotic stress (C), hypersaline stress (D), copper toxicity (E), and ethanol (EtOH) tolerance (F) in VIN13, EC1118, BM45, 285, and DV10 strains.
With regard to cell surface properties, a general flocculation test revealed that the different strains showed significant variation in their inherent ability to flocculate in the presence of Ca2+. Flocculation was low in the EC1118 and VIN13 strains, increased in DV10, and was highest for 285 and BM45. The flocculation percentages observed are, however, all much lower than those obtained under similar conditions with laboratory yeast strains (8). Besides flocculation ability, the cell surface hydrophobicity of the various strains also differed significantly from one strain to another. The interstrain trends observed were similar to those reported for the flocculation experiments. Furthermore, only the BM45 and 285 strains showed the ability to form mats or biofilms on semisolid media.
The strains showed various responses to starvation for different macronutrients. Whereas all the strains responded similarly to phosphorous starvation, clear differences were evident for the survival rates of the five strains exposed to carbon-, nitrogen-, and sulfur-depleted media. Once again EC1118, VIN13, and DV10 displayed a close alignment in terms of survival rates while BM45 and 285 exhibited close similarities to one another as well. VIN13 coped best with nitrogen starvation stress, followed by EC1118 and DV10. BM45 and 285 were by far inferior in this regard. However, this pattern was completely reversed in the case of carbon and sulfur starvation, where these two strains displayed a better ability to survive under such conditions.
The strains showed various tolerances to heat shock (Fig. 2A), with the BM45 strain proving to be the least tolerant to this particular stress, followed by the DV10 strain. All three remaining strains showed similar degrees of tolerance to heat stress treatments. At 1 mM concentrations of hydrogen peroxide, the strains 285 and BM45 showed the highest resistance to oxidative stress, followed by the DV10 strain (Fig. 2B). The EC1118 and VIN13 strains were the least tolerant of oxidative stress conditions. Interestingly, the BM45 strain showed the lowest tolerance for high-salt concentrations in the growth medium, with all four of the other strains reacting similarly under these conditions (Fig. 2D). However, all five strains responded with similar growth rates on the medium with a high sorbitol concentration (Fig. 2C). All five strains showed similar levels of tolerance to 20% ethanol, but with 25% ethanol, VIN13 appears to be somewhat more tolerant than the rest (Fig. 2F). Upon exposure to 30% ethanol, however, only the DV10 and VIN13 strains showed observable survival of cells.
Microarray analysis.
As previously reported (40), all facets of the microarray analysis and processing were compliant with the international standards for the minimum information about a microarray experiment. The analysis was reliable and reproducible in terms of the minor variations between independent biological repeats. Changes in gene expression during the course of fermentation also agreed quite well with data from related microarray analysis for the EC1118 (39) and VIN13 strains (30).
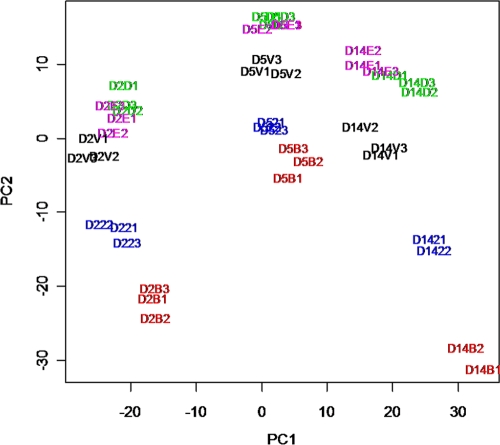
PCA.
The overall structure in the comparative transcriptome data sets of the different strains was analyzed by PCA of differentially expressed genes as identified from two-way analysis of variance (Fig. 3). In terms of design, the samples represent the different fermentations (three independent replicates for each of the five strains) at three different time points. The variables considered are the expression levels of the differentially expressed genes.
FIG. 3.
PCA showing components 1 and 2 (PC1 and PC2, respectively). Time points are indicated by date of sampling (where D2 is day 2, for example), strain, and sample number. Strains can be identified as follows: EC1118, E (purple); VIN13, V (black); BM45, B (red); 285, 2 (blue); DV10, D (green).
It is apparent that the primary experimental factor responsible for the variation in gene expression data is time or, rather, the stage of fermentation. All samples group very well together in time point-specific clusters along the first component or axes of variation. The second component neatly separates the main time point clusters into strain-specific subclusters. The PCA plot thus provides a succinct overview of the overall data structure and the relative relationship between the various strains. The fact that distinct groupings are evident for biological repeats attests to the integrity of the microarray analyses. It is also evident (especially from second component) that certain strains are more similar in terms of their overall gene expression patterns. For instance, BM45 and 285 group closely together at all three time points, while EC1118 and DV10 also appear to group close together, with VIN13 in an intermediate position between the two aforementioned clusters.
Significance and random forest analyses.
A large number of genes were significantly differentially expressed between different strains at the same time point (± 100 to 400) and within a particular strain at different time points during fermentation (± 1,000 to 2,000). The only genes from these detailed results that will be considered in more detail later are those related to the Gene Omnibus process categories of energy and metabolism.
Random forest analysis is a classification and regression tree aggregation technique that can be applied to comparative microarray-type data in order to rank genes in terms of their ability to discriminate between different classes or samples from different experimental conditions. The technique is essentially similar to the concept of biomarker identification and can be applied to discriminate between different experimental factors or conditions. In our research, we used the approach in order to identify genes with a strong discriminatory power, regardless of the absolute magnitude of the relative change in expression (which would otherwise obscure more subtle yet extremely relevant changes in the expression of certain genes).
In the first case, genes are ranked in order of their ability to discriminate between different time points during fermentation (regardless of strain variability). In the second case, genes are ranked based on their ability to discriminate between different yeast strains, regardless of growth cycles or fermentative stage. The top 200 ranked genes for both strain and time point discrimination were subjected to functional categorization. The results of this functional enrichment of the ranked gene lists are summarized in Table 3.
TABLE 3.
Functional categorization of the top 200 genes that discriminate between time points and strains
| Gene discrimination group and MIPS functional categorya | No. of selected genes/ total no. of genes | Genes among the top 200b |
|---|---|---|
| Time point discrimination | ||
| mRNA transcription | 29/556 | PRP45, HTB2, RTG3, ABD1, PRP9, MTF2, SUB2, SNU56, CFT1, SYF1, LRS4, SNU13, PRP22, GLN3, ACA1, FLO8, CEG1, SIP2, DAL81, CTK2, CAF4, PRP16, PRP19, RGR1, HSH155, PFS2, HAL9, RPB10, CET1 |
| mRNA processing | 16/122 | PRP45, ABD1, PRP9, MTF2, SUB2, SNU56, CFT1, SYF1, SNU13, PRP22, CEG1, PRP16, PRP19, CEF1, PFS2, CET1 |
| Cellular transport | 27/494 | CMD1, ARF2, YRB1, CAN1, SBH1, LEU5, VMA10, EPT1, YIL006W, APL1, GAP1, COF1, SED5, VPS34, GSP1, DIC1, ERV41, HXT2, LYP1, PEX15, ITR2, GYP1, SCD5, VMA4, RET3, SAR1, SEC8 |
| Metabolism | 49/1,066 | IMD1, RTG3, RIB7, YBR159W, DUR1,2, DUT1, PHO13, PDC2, EXG2, URH1, CAN1, PMI40, ISC1, GLN3, ILV1, PYC1, ERG1, LAG1, LEU5, EPT1, DCD1, YHR155W, YIL006W, RNR3, LYS12, DAL81, DAL3, URA2, GAP1, RGR1, HMX1, THI7, VPS34, DIC1, NMD4, AMD1, IMD4, HXT2, ADH3, ADE12, LYP1, ITR2, KTR1, TFC7, BTS1, YPL088W, SPT14, PCL8, DPM1 |
| Strain discrimination | ||
| Transport facilitation | 21/312 | SEO1, YBR235W, YBR293W, AUT4, YCL073C, ALR2, YFL054C, AGP3, TPC1, TPO2, MAL11, HXT9, MMP1, SUL2, CTR3, TOM37, PET8, ENB1, PDR5, YOR192C, CTR1 |
| Cell rescue, defense, and virulence | 17/278 | TCM62, SIF2, YBR293W, YCL073C, YER187W, PAU5, ALR2, DAK2, SNO3, TPO2, CPR2, LCB3, HMS2, PAU4, QRI8, ENB1, PDR5 |
| Ion transporters | 7/78 | SEO1, YBR235W, ALR2, SUL2, CTR3, PDR5, CTR1 |
| Metabolism | 43/1,066 | PYC2, AAD3, MDH3, AAD4, THI13, APT2, CHO1, SER3, YFL052W, DAK2, AGP3, AAD6, YFR055W, MIG2, UPF3, TPC1, MAL13, MAL11, YHR033W, DOG2, AAP1, YIL172C, MUC1, YIR035C, LCB3, HXT9, URA8, YJR149W, PGU1, MTD1, MMP1, SUL2, PUT1, PUS5, HMG2, GSF2, TOM37, GLO4, PDR5, YOR192C, CAR1, PPT2, GPH1 |
MIPS, Munich Information Center for Protein Sequences.
Open reading frames were determined based on random forest analysis.
As expected, the time point-discriminatory data contain a large number of genes related to mRNA processing and general cell growth and maintenance. However, the most overrepresented functional categories in the strain-discriminatory sets are related almost entirely to transport facilitation and general metabolism.
Only 12 of the 200 strain-discriminatory ranks are essential genes: RFA1, RSM10, ALG13, BRR6, YGR277c, DSN1, PAM18, CFT2, RSC9, RNA14, YNL260c, and MED4. For the time point-discriminatory rank set a total of 50 genes were essential, which is logical considering the involvement of fermentation stage-specific discriminators in processes such as growth and general cell cycle regulation. There was no overlap between the results of the different ranked lists.
Glycolysis, fermentation, and trehalose metabolism.
Glycolysis, fermentation, and trehalose metabolism were overrepresented in the SAM analysis outputs, which justified further investigation into the various genes coding for enzymes of the key central carbon metabolic pathways. In Fig. 4, the overall change in gene expression over time and between strains is represented as a clustered heat map. The closer the samples aggregate together, the stronger are the statistical relationships between these samples. Accordingly, strains are primarily grouped together in a time-specific manner. Along the vertical plane, genes with similar expression patterns over time and between strains are grouped together. The length of tree branches is inversely related to the strength of the statistical relationship between the genes (i.e., the shorter the branch, the stronger the correlation). It is interesting that for the first two time points, there is the same clustering of the different strains, whereas the strains segregated differently at the last time point. Nevertheless, the three strains EC1118, VIN13, and DV10 cluster closely together at all three time points.
FIG. 4.
Hierarchical clustering of transcripts encoding enzymes involved in glycolysis, fermentation, and trehalose metabolism (data log normalized to the day 2 gene expression average). Red bars denote an increase in expression while green bars indicate a decrease in expression for a given gene.
Reporter metabolite analysis.
The hypothesis-driven approach of reporter metabolite analysis to interpret microarray data aims to uncover the transcriptional regulatory architecture of metabolic networks. The reporter metabolites are those around which the most transcriptional changes occur, which implies that the levels of these metabolites are adjusted in response to the experimental factor(s) in order to maintain metabolic homeostasis within the network.
For the first multiple analysis, all three time points were compared simultaneously for each individual strain. In the second case all strains were simultaneously compared with one another for each of the three time points. For a full list of scored reporter metabolites for all five strains, see Data S1 in the supplemental material. The statistically most significant reporter metabolites from these two analyses are summarized in Tables 4 and 5.
TABLE 4.
Multiple analysis (metabolite reporter analysis) across all strains for days 2, 5, and 14
| Day 2
|
Day 5
|
Day 14
|
|||
|---|---|---|---|---|---|
| Metabolite | No. of neighborsa | Metabolite | No. of neighborsa | Metabolite | No. of neighborsa |
| α,α-Trehalose | 4 | 3-Phospho-d-glyceroyl phosphate | 4 | Hydrogen sulfide | 4 |
| 3-Phospho-d-glyceroyl phosphate | 4 | NADPH | 40 | Sulfite | 3 |
| Biotin | 3 | Dolichyl β-d-mannosyl phosphate | 7 | Intermediate (methylzymosterol) | 2 |
| Orthophosphate | 67 | NADP+ | 43 | Intermediate (zymosterol I) | 2 |
| 3-(4-Hydroxyphenyl)pyruvate | 5 | ATP | 113 | Aminoimidazole ribotide | 2 |
| 3-Dehydrosphinganine | 3 | AMP | 6 | 1-(5-Phospho-d-ribosyl)-5-amino-4-imidazolecarboxylate | 2 |
| Mannose-inositol-P-ceramide | 3 | α,α′-Trehalose 6-phosphate | 4 | Malate | 8 |
| α,α′-Trehalose 6-phosphate | 4 | Mannan | 6 | 3′-Phosphoadenylylsulfate | 2 |
| 7,8-Diaminononanoate | 2 | UMP | 4 | Acetoacetyl-CoA | 2 |
| Sulfite | 3 | d-Mannose 6-phosphate | 5 | O-Acetyl-l-homoserine | 2 |
| (R)-5-Diphosphomevalonate | 2 | GDP | 16 | (R)-3-Hydroxy-3-methyl-2-oxobutanoate M | 2 |
| Dethiobiotin | 2 | Pyrophosphate | 60 | (R)-2,3-Dihydroxy-3-methylbutanoate M | 2 |
| Palmitoyl-CoA | 2 | 3-Methyl-2-oxobutanoate M | 2 | Adenosine 3′,5′-bisphosphate | 3 |
| Oxalosuccinate | 2 | dGDP | 3 | (S)-2,3-Epoxysqualene | 2 |
| NH3xt | 3 | (R)-Pantothenate | 3 | Chitosan | 2 |
| Acetoacetyl-CoA | 2 | Fecosterol | 2 | 3-Phospho-d-glyceroyl phosphate | 4 |
| Inositol phosphorylceramide | 3 | 1-(5′-Phosphoribosyl)-5-amino-4-imidazolecarboxamide | 4 | Citrate M | 5 |
| Pyruvate M | 6 | Malonyl-CoA | 2 | Adenosine | 5 |
| 5-Phospho-alpha-d-ribose 1-diphosphate | 17 | AMP | 38 | γ-l-Glutamyl-l-cysteine | 2 |
| Dolichyl phosphate | 8 | trans,trans-Farnesyl diphosphate | 2 | l-Lysine | 4 |
Number of neighbors refers to the number of reactions in which the reporter metabolite participates.
TABLE 5.
Multiple analysis across all time points within each strain
| Strain | Metabolitea | No. of neighborsb |
|---|---|---|
| EC1118 | Pyrophosphate | 60 |
| NADPH | 40 | |
| dUMP | 4 | |
| Prephenate | 3 | |
| UDP | 15 | |
| Isocitrate | 5 | |
| α,α-Trehalose | 4 | |
| d-Manno 6-phosphate | 5 | |
| (S)-2,3-Epoxysqualene | 2 | |
| CYTS | 5 | |
| NADP+ | 43 | |
| CYTSxt | 4 | |
| ADxt | 4 | |
| GNxt | 4 | |
| α-d-Mannose 1-phosphate | 2 | |
| BM45 | Pyrophosphate | 60 |
| NADPH | 40 | |
| dUMP | 4 | |
| Prephenate | 3 | |
| UDP | 15 | |
| Isocitrate | 5 | |
| α,α-Trehalose | 4 | |
| d-Mannose 6-phosphate | 5 | |
| (S)-2,3-Epoxysqualene | 2 | |
| CYTS | 5 | |
| NADP+ | 43 | |
| CYTSxt | 4 | |
| ADxt | 4 | |
| GNxt | 4 | |
| α-d-Mannose 1-phosphate | 2 | |
| DV10 | NADPH | 40 |
| 5-Phospho-α-d-ribose 1-diphosphate | 17 | |
| Biotin | 3 | |
| d-Fructose 2,6-bisphosphate | 3 | |
| Pyrophosphate | 60 | |
| AMP | 38 | |
| Glutamate M | 7 | |
| dUMP | 4 | |
| NADP+ | 43 | |
| d-Mannose 6-phosphate | 5 | |
| UDP glucose | 12 | |
| UDP | 15 | |
| ASNxt | 4 | |
| GLNxt | 4 | |
| Prephenate | 3 | |
| VIN13 | dUMP | 4 |
| Pyrophosphate | 60 | |
| Glutamate M | 7 | |
| 5-Phospho-α-d-ribose 1-diphosphate | 17 | |
| Dolichyl β-d-mannosyl phosphate | 7 | |
| 2-Oxoglutarate M | 10 | |
| UDP | 15 | |
| AMP | 38 | |
| α-d-Mannose 1-phosphate | 2 | |
| Mannan | 6 | |
| S-Adenosyl-l-methionine | 14 | |
| Prephenate | 3 | |
| 7,8-Diaminononanoate | 2 | |
| dADP | 4 | |
| l-2-Aminoadipate 6-semialdehyde | 2 | |
| 285 | d-Fructose-2,6-bisphosphate | |
| Pyrophosphate | ||
| dUMP | ||
| Prephenate | ||
| NADPH | ||
| Oxaloacetate M | ||
| ASNxt | ||
| GLNxt | ||
| Biotin | ||
| α,α-Trehalose | ||
| GLYxt | ||
| UDP | ||
| NADP+ | ||
| Ergosta-5,7,24(28)-trienol | ||
| ALAxt |
xt, extracellular; AD, adenine; ALA, l-alanine; GN, guanine; GLY, glycine; GLN, l-glutamine; ASN, l-asparagine; CYTS, cytosine.
Number of reactions in which the reporter metabolite participates.
In this case, the analysis identifies the metabolites around which theoretically the most significant changes in gene expression/regulation occur when all the different strains are compared at each specific time point. Several interesting metabolites feature strongly in these interstrain comparisons: trehalose and trehalose-6-P and mannose and mannose-6-P, as well as various reducing equivalents, important cofactors (i.e., biotin and pyrophosphate), and key compounds such as pyruvate and acetyl-coenzyme A (CoA). These results agree quite well with the results of the random forest and significance analyses, as will be discussed shortly.
Table 5 indicates the reporter metabolites for each strain over the three time points. In other words, for each of the strains in the table, the transcriptional changes over time were substantially concerned with regulating the levels of the listed metabolites. In order to validate the assumptions derived from these analyses, several of the high-scoring metabolites were quantified experimentally in the same samples used for transcriptomic analysis.
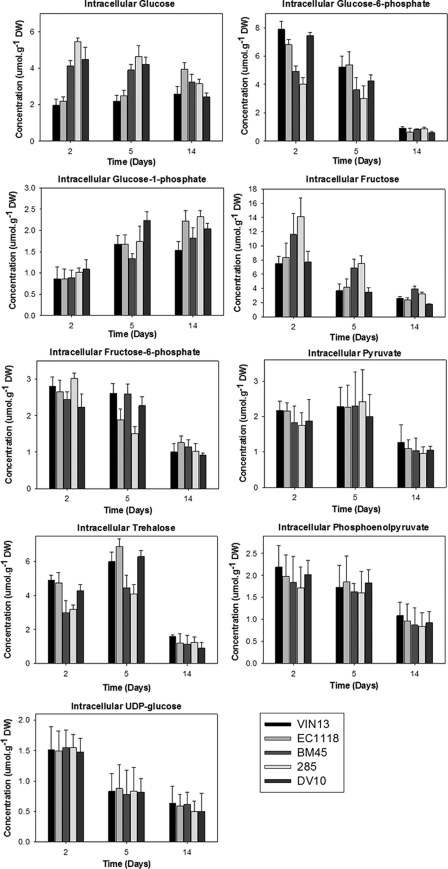
Most metabolite levels followed the same general pattern of decreasing as fermentation progressed (Fig. 5). One notable exception was trehalose, which showed an increase in intracellular concentration between days 2 and 5, with lower levels once again evident at day 14 at the end of fermentation. Intracellular glucose-1-phosphate also showed a definite increase in intracellular levels from the beginning to the end of the fermentation cycle. In terms of interstrain variations, no statistically significant differences were evident across strains for the metabolites phosphoenolpyruvate, pyruvate, and UDP-glucose. Intracellular glucose and fructose concentrations were significantly higher in the BM45 and 285 strains in the early stages of fermentation (days 2 and 5), while glucose-6-phosphate (and to a lesser extent glucose-1-phosphate and fructose-6-phosphate) concentrations were significantly lower in comparison to the EC1118 and VIN13. Intracellular trehalose levels were also consistently higher in EC1118, VIN13, and DV10 than in BM45 and 285, particularly at days 2 and 5.
FIG. 5.
Intracellular metabolite concentrations measured in the five strains at three time points corresponding to the transcriptional analysis. DW, dry weight.
DISCUSSION
Stress responses.
From our comparative transcriptome analysis, it is evident that differences in the expression of genes involved in the various stress response pathways were substantial at all three time points investigated, suggesting that significant differences exist in the manner in which the five strains adapt to the changing fermentation environment at the molecular level. However, from a data analysis perspective, two problems have to be overcome. First, the genomic responses to the different stress conditions share common effectors and regulators, making it difficult to attribute differences in gene expression levels to any specific stress tolerance phenotype. Second, the batch fermentation context does not allow specific control over stresses that apply at specific time points. There are thus inherent complications in terms of connecting differences in stress-related gene expression with stress responses and inherent phenotypic differences between the strains, as revealed by our analysis.
Instead of a haphazard attempt to arbitrarily connect gene expression profiles with stress phenotypes, we sought to explain some of these differences in light of the more concrete (and experimentally validated) metabolic differences between the strains. These links will be explored in more detail in the discussion of the reporter metabolite analysis.
Flocculation.
The flocculation response of yeast cells is a complex process, involving several interrelated signal transduction and stress response pathways as well as numerous structural proteins in the cell wall. Key components of this intricate system include not only the flocculation proteins themselves but also the enzymes responsible for the production and attachment of mannosyl residues to these structural proteins of the cell wall (28, 46).
There is great diversity/variation in the inherent adhesion abilities of different yeast strains used in industrial wine fermentations. In terms of the five strains used in this study, BM45 and 285 are strains with enhanced cell-cell and cell-substrate adhesion properties in comparison to the EC1118 and VIN13 strains (Table 2). In light of the results of our comparative transcriptomic profiling, there may be several genetic factors responsible for these differences.
(i) Flocculin-encoding genes.
Although the ability of S. cerevisiae cells to adhere to other cells and to one another is a component of complex developmental processes, one of the main requirements is the expression of various FLO genes, FLO1, FLO5, FLO10, and FLO11/MUC1 (8, 10, 27, 43). However, these genes are known to present strong strain-dependent variation in size and in their effectiveness in inducing adhesion-related phenotypes (44), and their individual expression levels may not be accurately measured in current arrays because of significant sequence homology. More interesting from a phenotypic perspective should be the expression levels of transcriptional activators that are known to control general adhesion properties such as FLO8 (8, 43, 44). This gene showed significantly higher expression levels during early stationary phase in the BM45 strain than the weaker-flocculating strains VIN13 and EC11118, correlating well with the observed difference in intrinsic flocculation and adhesion ability.
(ii) Cell wall mannoproteins.
Several cell wall mannoproteins of the FIT and DAN families showed significant increases in expression in the BM45 and 285 strains in comparison to the rest. Most notable among these are FIT2, FIT3, DAN1, and DAN4. The FIT genes code for glycosylphosphatidylinositol-anchored cell wall mannoproteins that are involved in the uptake and retention of iron in the cell wall (36). The regulation and roles of the DAN family of mannoproteins are still poorly understood although they are evidently required for anaerobic growth (13). As mannoproteins, the FIT and DAN gene products are cell wall bound and present mannose residues for selective binding by flocculation proteins of adjacent cells. The higher expression levels of genes in both the FIT and DAN families in the two strains with superior cell adhesion properties (BM45 and 285) (Table 2) thus serve as a possible indicator of the potential involvement of these two gene families in establishing different adhesion phenotypes in wine yeast strains.
Central carbon metabolism.
Our data show that a large proportion of differentially expressed transcripts are related to core metabolic activities of the fermenting yeasts (Tables 1 and 2 and Fig. 4). Several enzymes involved in hexose metabolism, glycolysis, trehalose metabolism, and redox balance are differentially expressed between strains at various stages of fermentation. The analysis of the gene expression levels within the framework of enzyme-enzyme and enzyme-metabolite interaction graphs (using the reporter metabolite approach) helped to pinpoint areas of metabolism that could speculatively be related to strain-strain or time point variation.
Trehalose, glucose-6-phosphate, glucose, UDP-glucose, and fructose-6-phosphate scored high on the multiple analyses across time points during fermentation (Table 5). Indeed, these metabolites did show marked differences in concentration between time points (Fig. 5). Trehalose was also a prominent interstrain reporter for days 2 and 5 (Table 4), and once again experimentally determined trehalose levels were found to be significantly different between strains at these time points (Fig. 5). This observation provides confidence that the outputs of the reporter analysis are biologically relevant and useful for an in silico interpretation of metabolic variation between different strains or key time points during fermentation. Several interesting features of these outputs will be discussed briefly in the following section.
Trehalose and trehalose-6-phosphate appear numerous times in the multiple reporter analyses (Tables 4 and 5), indicating that regulation of trehalose levels plays a key role throughout fermentation, both as a stress metabolite and as a key allosteric regulator of several important glycolytic enzymes. Trehalose-6-P restricts sugar influx into glycolysis through inhibition of the hexokinases (23) and thus determines the flux through glycolysis and the provision of energy and intermediates for fermentation, glycerol metabolism, and the oxidative pentose phosphate pathway. Changes in the expression of the genes involved in trehalose metabolism could potentially account for the metabolic restructuring that occurs in carbon metabolism as fermentation progresses. For instance, expression levels of genes encoding TPS1, TPS2, and TSL1 (involved in trehalose synthesis) increase sharply between days 2 and 5 of fermentation. This is mirrored by the experimentally determined increase in trehalose levels between these two time points. Our observation of lower trehalose levels at the end of fermentation can be explained by trehalose mobilization at the final stage of fermentation when sugar exhaustion is imminent. This correlates well with the increasing expression levels of transcripts encoding intracellular trehalose-degrading enzymes (NTH1 and NTH2) as well as with a decrease in the expression of TPS3, which encodes a positive regulator of the TPS1 and TPS3 encoded subunits.
Trehalose concentrations are significantly higher in the EC1118, VIN13, and DV10 strains than in the BM45 and 285 strains during exponential and early logarithmic growth. Trehalose metabolism could thus potentially represent a type of overarching regulator with implications for the general fermentation phenotypes of different wine yeast strains. The differences in trehalose accumulation could be an important contributing factor for the increased thermotolerance and ethanol and osmotic shock tolerance (Fig. 2) exhibited by the three strains that produced trehalose at levels higher other strains and than BM45, in particular (24).
The BM45 and 285 strains were characterized by significantly lower levels of intracellular hexose phosphates and higher levels of fructose. This suggests a possible discrepancy in the phosphorylation efficiency of these strains and may explain their generally lower fermentative rate, higher residual sugars at the end of fermentation, and propensity for stuck or sluggish fermentations.
Several other reporter metabolites are of interest in terms of their likely connection with fermentative phenotypes or relevant physiological traits. The presence of mannose in most of the top 10 reporter metabolite lists for the intrastrain analysis (i.e., the time point differential data) (Table 5) suggests a shift in the provision and availability of mannose residues for mannosylation of cell wall proteins as fermentation progresses. This could again point to protein mannosylation as an important area in terms of determining cell-cell adherence and aggregation during fermentation. Mannose residues or their precursors also feature heavily on the day 2 and day 5 interstrain lists, suggesting that regulation of mannose metabolism differs between the strains in this study and at least partially accounting for their different cell wall properties and flocculation responses.
Reducing equivalents like NAD/P and NADPH as well as related metabolites like glutamate and oxoglutarate (interconversion of these two metabolites is involved in regulating the intercellular NADP/NADH ratios) also stand out in the multiple analyses across time points and strains (Tables 4 and 5). Changes in/maintenance of the ratios of these important reducing equivalents is a major area of metabolic adjustment during anaerobic fermentation due to the decreased availability of sugars (glucose) and the buildup of glycolytic intermediates and other potentially toxic products, e.g., acetaldehyde, as fermentation progresses. The ratio of NAD/P and NADPH will impact on the production of a number of higher alcohols produced by the different strains, and interestingly many of these alcohols do indeed differ significantly between strains (40). Differences in the directionality and flux through dehydrogenase-catalyzed pathways in the different strains affect the overall regulation of NAD/NADH and NADP/NADPH levels in these strains, leading to differences in the cellular balance of reducing equivalents.
Prephenate (which is present in the top 15 reporters for each strain) is an intermediate in the biosynthesis of the aromatic amino acids phenylalanine and tyrosine. This suggests that large-scale changes in the expression of genes involved in this branch of amino acid metabolism occur during the course of fermentation. These aromatic amino acids are starting points for pathways that produce important aroma compounds in wine, namely 2-phenylethanol and 2-phenylethyl acetate (40). The rate at which these volatile compounds appear in the must is rapid during the first few days of fermentation and decreases substantially toward the end. This could reflect a reduced availability of the necessary precursors in the later stages of fermentation, as suggested by the reporter analysis.
Fructose-2,6-biphosphate also features in the DV10 and 285 strains for the intrastrain analysis. This metabolite is not just an intermediary metabolite of glycolysis, but like trehalose it is a very potent regulator of glycolysis by inhibiting Pfkp (phosphofructokinase, platelet) activity. This could potentially be related to the slowing in the rate of glycolysis as fermentation progresses. Biotin (also on the list for DV10 and 285) also exerts a marked effect upon the hexokinase activity of yeast, leading to a stimulation of the rate of glucose and fructose utilization. Once again, this could contribute to the regulation of flux through glycolysis and related pathways, e.g., during the course of fermentation.
The regulatory changes surrounding glycolytic metabolites like 3-phospho-d-glyceroyl phosphate (just below the branch point between the glycerol pathway and the lower glycolysis) are also well represented in the analysis at days 2 and 5. This could indicate a difference in the way that the strains partition carbon toward glycerol production (to maintain NAD/NADH ratios) versus pyruvate production, leading ultimately to ethanol formation. In support of this concept, glycerol production was compared in the five strains, and there were indeed significant differences (40). Such inherent differences in glycerol production by the different strains could account for their various tolerance levels of osmotic stress conditions.
(S)-2,3-Epoxysqualene, methylzymosterol, and zymosterol (Table 4) are uncommon sterols that have proposed to have roles in membrane stabilization under high ethanol levels, which makes sense in light of the high ethanol concentrations at the end of fermentation. The reporter analysis identifies reactions related to the metabolism of these sterols as areas of variability between strains. It is tempting to speculate that this is one of the contributory factors for the differences in ethanol tolerance that was observed in our stress assays.
Conclusions.
This analysis provides a base for further hypothesis-driven investigations into the role of various genetic systems in oenologically relevant phenotypes. By analyzing large comparative transcriptomic data sets of five industrial wine yeast strains, we were able to identify various genes/gene sets that could be linked to relevant aspects of yeast performance in key areas related to flocculation, stress tolerance, and metabolism. The study sheds light on some of the underlying molecular factors related to common interstrain variations between strains and also increased our understanding of metabolic changes that occur during fermentation under wine-making conditions. By using five yeast strains and three time points, we were able to eliminate “noise” and clearly distinguish between differences in gene expression that are related to strain identity alone, as opposed to the specific stage of fermentation. Strains such as BM45 and 285, which were similar in terms of their overall gene expression patterns (as can be seen from the PCA analysis in Fig. 3) showed similar cell adhesion properties and stress tolerance properties (Table 2 and Fig. 1 and 2). The same is true for EC1118, DV10, and VIN13 (to a lesser extent). These strain groupings also hold for the profiles of exo-metabolites produced (40), as well as for the concentrations of the nine key intercellular metabolites measured (Fig. 5). By contextualizing the comparative transcriptome data sets within existing metabolic maps and applying the reporter metabolite algorithm, we were able to pinpoint some of the underlying molecular and genetic factors responsible for these important physiological trait differences between strains. Ultimately, the research presented in this paper provides new insights for targeted engineering strategies aimed at improving the performance of wine yeast strains.
Supplementary Material
Acknowledgments
Funding for the research presented in this paper was provided by the NRF and Winetech and personal sponsorship by the Wilhelm Frank Trust. Funding for travel and collaboration was provided by a Swedish-South African bilateral grant.
We also thank Jo McBride and the Cape Town Centre for Proteomic and Genomic Research for the microarray analysis and the staff and students at the IWBT for their support and assistance in numerous areas.
Footnotes
Published ahead of print on 21 August 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abbott, D. A., T. A. Knijnenburg, L. M. de Poorter, M. J. Reinders, J. T. Pronk, and A. J. van Maris. 2007. Generic and specific transcriptional responses to different weak organic acids in anaerobic chemostat cultures of Saccharomyces cerevisiae. FEMS Yeast Res. 7:819-833. [DOI] [PubMed] [Google Scholar]
- 2.Attfield, P. V. 1997. Stress tolerance: the key to effective strains of baker's yeast. Nat. Biotechnol. 15:1351-1357. [DOI] [PubMed] [Google Scholar]
- 3.Bauer, F. F., and I. S. Pretorius. 2000. Yeast stress response and fermentation efficiency: how to survive the making of wine—a review. S. Afr. J. Enol. 21:27-51. [Google Scholar]
- 4.Beltran, G., M. Novo, V. Leberre, S. Sokol, D. Labourdette, J. M. Guillamon, A. Mas, J. François, and N. Rozes. 2006. Integration of transcriptomic and metabolic analyses for understanding the global responses of low-temperature winemaking fermentations. FEMS Yeast Res. 6:1167-1183. [DOI] [PubMed] [Google Scholar]
- 5.Bely, L., J. Sablayrolles, and P. Barre. 1990. Description of alcoholic fermentation kinetics: its variability and significance. Am. J. Enol. Viticult. 41:319-324. [Google Scholar]
- 6.Ben-Dor, A., R. Shamir, and Z. Yakhini. 1999. Clustering gene expression patterns. J. Comput. Biol. 6:281-297. [DOI] [PubMed] [Google Scholar]
- 7.Bergmeyer, H. U., and E. Bernt. Methods of enzymatic analysis, vol. 3. Verlag Chemie, Weinheim, Germany.
- 8.Bester, M. C., I. S. Pretorius, and F. F. Bauer. 2006. The regulation of Saccharomyces cerevisiae FLO gene expression and Ca2+-dependent flocculation by Flo8p and Mss11p. Curr. Genet. 49:375-383. [DOI] [PubMed] [Google Scholar]
- 9.Boer, V. M., J. H. de Winde, J. T. Pronk, and M. D. Piper. 2003. The genome-wide transcriptional responses of Saccharomyces cerevisiae grown on glucose in aerobic chemostat cultures limited for carbon, nitrogen, phosphorus or sulfur. J. Biol. Chem. 278:3265-3274. [DOI] [PubMed] [Google Scholar]
- 10.Braus, G. H., O. Grundmann, S. Brückner, and H. U. Mösch. 2003. Amino acid starvation and Gcn4p regulate adhesive growth and FLO11 gene expression in Saccharomyces cerevisiae. Mol. Biol. Cell 14:4272-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Breiman, L. 2001. Random forests. Mach. Learn. 45:5-22. [Google Scholar]
- 12.Caro, L. H. P., H. Tettelin, J. H. Vossen, A. F. J. Ram, H. van den Ende, and F. M. Klis. 1997. In silico identification of glycosyl-phosphatidylinositol-anchored plasma-membrane and cell wall proteins of Saccharomyces cerevisiae. Yeast 13:1477-1489. [DOI] [PubMed] [Google Scholar]
- 13.Cohen, B. D., O. Sertil, N. E. Abramova, K. J. A. Davies, and C. V. Lowry. 2001. Induction and repression of DAN1 and the family of anaerobic mannoprotein genes in Saccharomyces cerevisiae occurs through a complex array of regulatory sites. Nucleic Acids Res. 29:799-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.d'Amore, T., and G. G. Stewart. 1987. Ethanol tolerance of yeast. Enzyme Microb. Technol. 9:322-330. [Google Scholar]
- 15.Dickinson, J. R., L. Eshantha, J. Salgado, and M. J. E. Hewlins. 2003. The catabolism of amino acids to long chain and complex alcohols in Saccharomyces cerevisiae. J. Biol. Chem. 278:8028-8034. [DOI] [PubMed] [Google Scholar]
- 16.Förster, J., I. Famili, P. Fu, B. Palsson, and J. Nielsen. 2003. Genome-scale reconstruction of the Saccharomyces cerevisiae metabolic network. Genome Res. 13:244-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gasch, A. P., P. T. Spellman, C. M. Kao, O. Carmel-Harel, M. B. Eisen, G. Storz, D. Botstein, and P. O. Brown. 2000. Genomic expression changes in the response of yeast cells to environmental changes. Mol. Biol. Cell 11:4241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goffeau, A., B. G. Barrell, H. Bussey, R. W. Davis, B. Dujon, H. Feldmann, F. Galibert, J. D. Hoheisel, C. Jacq, M. Johnston, E. J. Louis, H. W. Mewes, Y. Murakami, P. Philippsen, H. Tettelin, and S. G. Oliver. 1996. Life with 6000 genes. Science 274:563-567. [DOI] [PubMed] [Google Scholar]
- 19.Govender, P., J. L. Domingo, M. C. Bester, I. S. Pretorius, and F. F. Bauer. 2008. Controlled expression of the dominant flocculation genes FLO1, FLO5, and FLO11 in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 74:6041-6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hallsworth, J. E. 1998. Ethanol-induced water stress in yeast. J. Ferment. Bioeng. 85:125-137. [Google Scholar]
- 21.Hartwell, L. H., J. J. Hopfield, S. Leibler, and A. W. Murray. 1999. From molecular to modular cell biology. Nature 402:C47-C52. [DOI] [PubMed] [Google Scholar]
- 22.Hinchcliffe, E., W. G. Box, E. F. Walton, and M. Appleby. 1985. The influences of cell wall hydrophobicity on the top fermenting properties of brewing yeast. Proc. Eur. Brew. Congr. 20:323-330. [Google Scholar]
- 23.Hohmann, S., W. Bell, M. J. Neves, D. Valckx, and J. M. Thevelein. 1996. Evidence for trehalose-6-phosphate-dependent and -independent mechanisms in the control of sugar influx into yeast glycolysis. Mol. Microbiol. 20:981-991. [DOI] [PubMed] [Google Scholar]
- 24.Hohmann, S. 1997. Shaping up: the response of yeast to osmotic stress, p. 101-134. In S. Hohmann and W. H. Mager (ed.), Stress responses. Springer, Heidelberg, Germany.
- 25.Ideker, T., T. Galitski, and L. Hood. 2001. A new approach to decoding life: systems biology. Annu. Rev. Genomics Hum. Genet. 2:343-372. [DOI] [PubMed] [Google Scholar]
- 26.Ivorra, C., J. E. Pérez-Ortín, and M. del Olmo. 1999. An inverse correlation between stress resistance and stuck fermentations in wine yeasts. A molecular study. Biotechnol. Bioeng. 54:698-708. [DOI] [PubMed] [Google Scholar]
- 27.Lo, W. S., and A. M. Dranginis. 1998. The cell surface flocculin Flo11 is required for pseudohyphae formation and invasion by Saccharomyces cerevisiae. Mol. Biol. Cell 9:161-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maneesri, J., M. Azuma, Y. Sakai, K. Igarashi, T. Matsumoto, H. Fukuda, A. Kondo, and H. Ooshima. 2005. Deletion of MCD4 involved in glycosylphosphatidylinositol (GPI) anchor synthesis leads to an increase in β-1,6-glucan level and a decrease in GPI-anchored protein and mannan levels in the cell wall of Saccharomyces cerevisiae. J. Biosci. Bioeng. 99:354-360. [DOI] [PubMed] [Google Scholar]
- 29.Mardia, K. V., J. T. Kent, and J. H. Bibby. 1979. Multivariate analysis. Academic Press, London, United Kingdom.
- 30.Marks, V. D., G. K. van der Merwe, and H. J. J. van Vuuren. 2003. Transcriptional profiling of wine yeast in fermenting grape juice: regulatory effect of diammonium phosphate. FEMS Yeast Res. 3:269-287. [DOI] [PubMed] [Google Scholar]
- 31.Marks, V. D., S. J. Ho Sui, D. Erasmus, G. K. van den Merwe, J. Brumm, W. W. Wasserman, J. Bryan, and H. J. J. van Vuuren. 2008. Dynamics of the yeast transcriptome during wine fermentation reveals a novel fermentation stress response. FEMS Yeast Res. 8:35-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marullo, P., M. Bely, I. Masneuf-Pomarede, M. Aigle, and D. Dubourdieu. 2004. Inheritable nature of enological quantitative traits is demonstrated by meiotic segregation of industrial wine yeast strains. FEMS Yeast Res. 4:711-719. [DOI] [PubMed] [Google Scholar]
- 33.Patil, K. R., and J. Nielsen. 2005. Uncovering transcriptional regulation of metabolism by using metabolic network topology. Proc. Natl. Acad. Sci. USA 102:2685-2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pizarro, F. J., M. C. Jewett, J. Nielsen, and E. Agosin. 2008. Growth temperature exerts differential physiological and transcriptional responses in laboratory and wine yeast strains of Saccharomyces cerevisiae. Appl. Environ. Microbiol. 74:6358-6368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pretorius, I. S., and F. F. Bauer. 2002. Meeting the consumer challenge through genetically customized wine-yeast strains. Trends Biotechnol. 20:426-432. [DOI] [PubMed] [Google Scholar]
- 36.Protchenko, O., T. Ferea, J. Rashford, J. Tiedeman, P. O. Brown, D. Botstein, and C. C. Philpott. 2001. Three cell wall mannoproteins facilitate the uptake of iron in Saccharomyces cerevisiae. J. Biol. Chem. 276:49244-49250. [DOI] [PubMed] [Google Scholar]
- 37.Rapoport, A., and M. Becker. 1985. Changes in the surface charge of yeast cells during their dehydration and rehydration. Mikrobiologiya 54:450-453. [Google Scholar]
- 38.Reynolds, T. B., and G. R. Fink. 2001. Bakers' yeast, a model for fungal biofilm formation. Science 291:878-881. [DOI] [PubMed] [Google Scholar]
- 39.Rossignol, T., L. Dulau, A. Julien, and B. Blondin. 2003. Genome-wide monitoring of wine yeast gene expression during alcoholic fermentation. Yeast 20:1369-1385. [DOI] [PubMed] [Google Scholar]
- 40.Rossouw, D., T. Naes, and F. F. Bauer. 2008. Linking gene regulation and the exo-metabolome: a comparative transcriptomics approach to identify genes that impact on the production of volatile aroma compounds in yeast. BMC Genomics 9:530-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tusher, C. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vemuri, G. N., and A. A. Aristidou. 2005. Metabolic engineering in the —omics era: elucidating and modulating regulatory networks. Microbiol. Mol. Biol. Rev. 69:197-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Verstrepen, K. J., G. Derdelinckx, H. Verachtert, and F. R. Delvaux. 2003. Yeast flocculation: what brewers should know. Appl. Microbiol. Biotechnol. 61:197-205. [DOI] [PubMed] [Google Scholar]
- 44.Verstrepen, K. J., T. B. Reynolds, and G. R. Fink. 2004. Origins of variation in the fungal cell surface. Nat. Rev. Microbiol. 2:533-540. [DOI] [PubMed] [Google Scholar]
- 45.Webb, A. D., and J. L. Ingraham. 1963. Fusel oil. Adv. Appl. Microbiol. 5:317-353. [Google Scholar]
- 46.Wiedman, J. M., A.-L. Fabre, B. W. Taron, C. H. Taron, and P. Orlean. 2007. In vivo characterization of the GPI assembly defect in yeast mcd4-174 mutants and bypass of the Mcd4p-dependent step in mcd4Δ cells. FEMS Yeast Res. 7:78-83. [DOI] [PubMed] [Google Scholar]
- 47.Zuzuarregui, A., P. Carrasco, A. Palacios, A. Julien, and M. del Olmo. 2005. Analysis of the expression of some stress induced genes in several commercial wine yeast strains at the beginning of vinification. J. Appl. Microbiol. 98:299-307. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.