Abstract
We have evaluated the interaction that bacterial genotypes and plant hosts have with the loss of pathogenicity in tumors, using seven Agrobacterium tumefaciens strains inoculated on 12 herbaceous and woody hosts. We performed a screening of the agrobacteria present inside the tumors, looking for nonpathogenic strains, and found a high variability of those strains in this niche. To verify the origin of the putative nonpathogenic mutant bacteria, we applied an efficient, reproducible, and specific randomly amplified polymorphic DNA analysis method. In contrast with previous studies, we recovered a very small percentage (0.01%) of nonpathogenic strains that can be considered true mutants. Of 5,419 agrobacterial isolates examined, 662 were nonpathogenic in tomato, although only 7 (from pepper and tomato tumors induced by two A. tumefaciens strains) could be considered to derive from the inoculated strain. Six mutants were affected in the transferred DNA (T-DNA) region; one of them contained IS426 inserted into the iaaM gene, whereas the whole T-DNA region was apparently deleted in three other mutants, and the virulence of the remaining two mutants was fully restored with the T-DNA genes as well. The plasmid profile was altered in six of the mutants, with changes in the size of the Ti plasmid or other plasmids and/or the acquisition of new plasmids. Our results also suggest that the frequent occurrence of nonpathogenic clones in the tumors is probably due to the preferential growth of nonpathogenic agrobacteria, of either endophytic or environmental origin, but different from the bacterial strain inducing the tumor.
Agrobacterium tumefaciens is the causal agent of crown gall, a neoplastic disease induced in a large number of dicotyledonous and some monocotyledonous plants, as well as in gymnosperms, in this case artificially (9, 32, 34, 35). Many studies have focused on the molecular understanding of the process that involves the tumorous growth of plant tissue by means of the transfer of specific genes from a plasmid harbored by the bacterium, called tumor-inducing plasmid (pTi) (7, 18, 30, 43). This transfer system has significant importance for pathogenicity and epidemiology studies, as well as for biotechnology, because this unique mechanism of pathogenicity has allowed the development of a diverse and efficient methodology for the transformation of eukaryotic cells. Other studies have concentrated on the ecology and epidemiology of Agrobacterium spp. to obtain a better knowledge of the spread and stability of the bacterium as a pathogen in tumors, hosts, and soil habitats (5, 11, 36).
The pathogenicity of A. tumefaciens seems to be somewhat unstable both in association with plants and under free-living conditions. Several studies demonstrated that A. tumefaciens populations from tumors induced under natural conditions in several plants consisted mostly of nonpathogenic strains (2, 33, 38), and loss of pathogenicity has been proposed as a likely cause to explain the predominance of nonpathogens over pathogenic strains. Likewise, loss of pathogenicity has also been demonstrated in tumors developed after inoculation of apple plants with A. tumefaciens biovar 2 (3, 4), although the appearance of mutant bacteria was only observed after inoculation of plants grown in vitro, not with those grown in the greenhouse. This, together with the well-known difficulty of isolating pathogenic strains from apple tumors, fostered the hypothesis of a plant-mediated effect on the genetic instability of A. tumefaciens populations (3, 4, 10, 13). The appearance of nonpathogenic mutant bacteria was also observed when Agrobacterium strains were cultured in media supplemented with substances that are naturally present in plant wounds and that favor the activation of pathogenicity genes (13). The molecular bases of the spontaneous loss of pathogenicity in A. tumefaciens are not well understood, although previous work reported changes in the Ti plasmid, in either the regions involved in virulence, the transferred DNA (T-DNA) region, or the vir regions, due to point mutations or to the insertion of mobile elements (10, 13, 14, 21). However, it was not entirely clear that the nonpathogenic isolates were necessarily mutants derived from the inoculated strain rather than naturally occurring nonpathogenic A. tumefaciens strains.
A precise knowledge of the factors that induce the putative loss of pathogenicity in A. tumefaciens, and the role that the plant might play in selecting against virulent bacteria, is pivotal for the development of effective control measures for this important plant pathogen. In the present work, we wanted to expand the preceding studies of the appearance of nonpathogenic variants and the influence of the host and of the bacterial genetic background. To confirm the identity of the nonpathogenic isolates, and to ensure that they were true variants of the inoculated strain, we combined biochemical tests with a highly discriminative randomly amplified polymorphic DNA (RAPD) procedure (24). The analysis of more than 5,400 clones allowed us to show, in contrast with previous studies, that the generation of nonpathogenic variants occurred with a very low frequency, and it appeared to be independent of the plants evaluated; we also show that the mutant bacteria were affected in large areas of the Ti plasmid and in the number of plasmids harbored.
MATERIALS AND METHODS
Bacteria, plasmids and hosts.
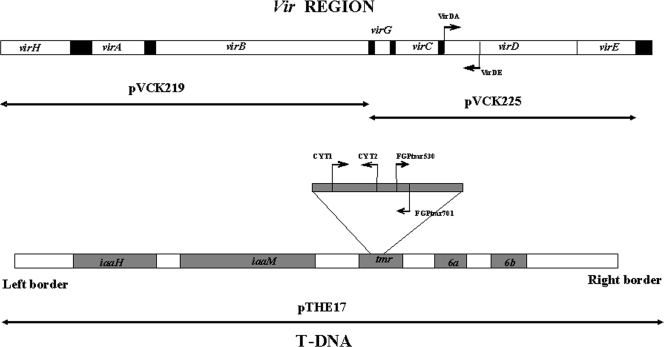
The bacterial strains and host origins are listed in Table 1, and the plasmids employed in the different analyses are shown in Fig. 1. Bacteria were grown on the general medium yeast extract-peptone-glucose agar (YPGA) (22) at 26°C for 48 h. For all the inoculation experiments, the bacterial strains were cultivated using suspensions from single colonies purified three consecutive times in the same medium from water suspensions, to ensure their purity previous to the inoculations. Unless otherwise indicated, antibiotics were used at the following final concentrations (μg/ml): gentamicin, 25; kanamycin, 25; streptomycin, 100; tetracycline, 15.
TABLE 1.
Strains of Agrobacterium tumefaciens employed in the inoculation experiments
| Strain | Host of isolation | Opine type | Biovar | Experiment | Source or reference |
|---|---|---|---|---|---|
| C58 | Cherry | Nopaline | 1 | A | M. Ridé |
| A281a | Agropine | 1 | A | X. Nesme | |
| Ach5 | Prunus sp. | Octopine | 1 | A | M. D. Chilton |
| CFBP42 | Tomato | NDb | 1 | A | CFBPc |
| IVIA325-4 | GF677 | Nopaline | 1 | B | 24 |
| IVIA678-2 | Peach | Nopaline | 1 | B | 24 |
| IVIA1102 | Chrysanthemum | Chrysopine | 1 | A/C | 24 |
| C58Rd | Nopaline | 1 | C | S. K. Farrand |
A281 is strain C58 with pTiBo542.
ND, not determined.
CFBP, Collection Française de Bactéries Phytopathogènes.
C58R is strain C58 resistant to gentamicin.
FIG. 1.
Genes carried by the different plasmids used in complementation analyses and positions of primers employed to check for the presence of the T-DNA regions in the mutants analyzed. For details, see Table 2.
Inoculation experiments and identification analysis.
Three experiments were performed to evaluate the loss of pathogenicity in A. tumefaciens strains inoculated in different hosts in a greenhouse (experiments A and B) and under in vitro conditions (experiment C). Inoculation of A. tumefaciens was done either by direct inoculation of the stems of plant hosts (experiment A) or by growth of the plants in an inoculated sterilized substrate (experiment B).
In experiment A, tomato (cultivar Roma) and pepper (cv. Toledo) plants were separately inoculated with strains A281, Ach5, C58, CFBP42, and IVIA1102, all biovar 1 strains. For the inoculation, a wound was made in the stem of each plant using a sterile scalpel, and a piece of cotton imbibed with 30 μl of a bacterial suspension (optical density at 600 nm [OD600] of 0.3 in water) was placed on the wound and held in place with Parafilm. After 5 weeks, once the tumors had developed, bacteria were isolated from tumors, as follows: the external part of the tumors was peeled, then an internal piece of 100 mg of tumor tissue was comminuted in 9 ml of sterile distilled water, and 30 μl of the suspension was plated on a semiselective medium for Agrobacterium biovar 1 (42). From each tumor, 100 colonies were selected, and a suspension in sterile distilled water was streaked out onto a general medium (YPGA) and purified by single colony transfer. Two tumors, coming from different plants, were analyzed for each combination of host and bacterial strains, and the experiment was performed twice, resulting in the examination of the pathogenicity of 200 different colonies from each inoculated strain used in each experiment (400 in total).
In experiment B, sterilized substrate (Vriezenveen Potgrond BV, Holland) in pots was inoculated with 15 ml of a similar bacterial suspension (OD600 of 0.3 in water) previously cultured on YPGA and distributed evenly over the soil surface, and 1 plant per plot was immediately planted, with 10 plants per host and strain. A wound was inflicted in the crown with a scalpel. Cherry, pear, and apple and hybrid rootstocks of peach × almond Adafuel and GF677 were planted in sterilized substrate inoculated with strain IVIA325-4, whereas peach × almond rootstocks GF677, GN15, and GN22, plum Myrobalan, and peaches Montclar and Nemaguard were planted in substrate inoculated with strain IVIA678-2. When possible, five tumors were selected from five different plants (one tumor per plant), and 50 Agrobacterium colonies were isolated from each tumor as described in experiment A, resulting in a total of 250 colonies analyzed for each combination of host and bacterial strains. Otherwise, when there were not enough tumors or less than 50 colonies per tumor, we increased the number of colonies retained per tumor up to a total of 250 colonies. The plants were maintained for 9 months in a greenhouse at 20 to 25°C and 40 to 60% rH, with a 16-h per day/8-h per night photoperiod and irrigation twice a week.
In experiment C, we evaluated the loss of pathogenicity following repeated culture in liquid medium. Strains IVIA1102 (Kmr) and C58R (Gmr) were analyzed separately in yeast extract-peptone-glucose with and without the corresponding antibiotic to evaluate its possible influence on the appearance of nonpathogenic variants. First, each strain was plated on YPGA medium plus the antibiotic, and 50 colonies each that were well separated were suspended in 500 μl of water. Then, 10 μl of each of these suspensions was inoculated in tomato plants to confirm their pathogenicity, and 20 μl was incubated in 5 ml of yeast extract-peptone-glucose with and without the corresponding antibiotic at 26°C, with shaking for 48 h, resulting in 200 initial clones (50 colonies of each strain in medium with and without antibiotic). After 48 h of incubation, an aliquot of 20 μl was reinoculated in 5 ml of fresh medium, and the procedure was repeated. After four incubation steps (8 days), 10 μl of the last suspension was diluted 1/10 with water and plated on YPGA and YPGA plus antibiotic by triple streaking to obtain isolated colonies. After 48 h at 26°C, one colony was taken from the plates and suspended in 0.2 ml water. Ten microliters was inoculated in tomato plants to check the pathogenicity, and 20 μl of the suspension was incubated into new liquid medium and followed the incubation process. The remaining volume was kept at 4°C for further assays, in case there was a loss in pathogenicity. This process was repeated to perform a total of five inoculation steps, with 100 colonies inoculated in each series of incubations per strain. The experiment was repeated twice, giving a total of 1,000 colonies analyzed with each strain (500 colonies per experiment/strain).
All the bacterial colonies recovered from tumors in experiments A and B were considered putative Agrobacterium when they were showing the typical morphology on a semiselective medium and were positive for the production of urease and hydrolysis of esculin. The pathogenicity assays were performed by inoculating a fresh culture of each bacterium in tomato plants in duplicate and examining for the presence of tumors after 1 month. The inoculation experiment was repeated with those clones that did not induce tumors, which were also inoculated in the original host from which the parental strain was isolated. The clones were considered nonpathogenic when they were not able to produce tumors in any of the two hosts, whereas they were considered to show a loss in virulence if they produced tumors in only one host or if the resulting tumors in at least one host were significantly smaller than those induced by the wild-type strains. Statistical analyses of tumor development were done using a separate experiment, as follows: for each strain analyzed, 15 tomato plants of the same size and age (2 weeks old) were inoculated in the stem with 10 μl of a bacterial suspension (108 CFU/ml, OD600 of ca. 0.65) essentially as described above for experiment A. After 30 days, tumors were carefully collected, avoiding carrying over any healthy tissue, and the cumulative tumor weights per each of the 15 plant batches were compared using one-way analysis of variance, employing the software SPSS v. 10.1.3 (Chicago, IL).
Molecular identification and analysis of mutant bacteria.
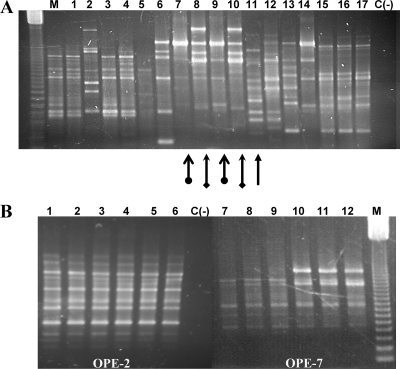
For verification of the origin of bacteria isolated from tumors, we examined their RAPD patterns with primer OPE-2 (5′-GGTGCGGGAA) by following the method of Llop et al. (24), which demonstrated that, under our conditions, the patterns were stable, repetitive, and reproducible. If the resulting patterns were substantially different than those of the inoculated strains (Fig. 2), we considered that the isolates had a different origin. However, if their patterns were similar, we also analyzed their RAPD fingerprints with RAPD primers OPE-7 (5′-AGATGCAGCC), OPE-14 (5′-TGCGGCTGAG), and OPE-20 (5′-AACGGTGACC). We considered that the colonies isolated from tumors were derived from the inoculated strain only when they yielded identical fingerprints with those of these four RAPD primers. The PCRs were performed in a total volume of 25 μl with ca. 70 ng of DNA, 1× PCR buffer (20 mM Tris-HCl [pH 8.0], 50 mM KCl), 1.5 mM MgCl2, 60 μM deoxynucleoside triphosphate, 5 pmol of primer, and 1 U of Taq polymerase (Invitrogen). In all the combinations of host-bacterium strains, and as a positive control, we also analyzed the RAPD fingerprints of 10 randomly selected colonies that retained pathogenicity, which in all cases were identical to those of the inoculated strain. Arbitrarily primed PCR analysis using primers ERIC, BOX, and REP was done with some of the clones to ensure their identity as previously described (8, 25).
FIG. 2.
RAPD pattern diversity obtained with colonies isolated from tumors of different hosts inoculated with strain IVIA325-4 and patterns obtained with mutants of strain IVIA1102. (A) RAPD analysis with primer OPE-2 of colonies obtained with strain IVIA325-4. Lane 1, strain isolated from Adafuel in tumor 1; lanes 2 to 4, isolates from the same host but from tumor 3; lane 5, isolate from Adafuel in tumor 4; lane 6, inoculated strain IVIA325-4; lanes 7 to 17, isolates from host cherry; lanes 7 to 11, isolates from tumor 1; lane 12, isolate from tumor 2; lanes 13 to 14, isolates from tumor 3; lanes 15 to 17, isolates from tumor 5. M, 123-bp molecular mass marker (Life Technologies); C(−), negative control. The arrows indicate the different patterns obtained with isolates from tumor 1 in cherry, the same shape indicates the same pattern, and the different arrows show the colonies that are different. (B) RAPD analysis of colonies obtained with strain IVIA1102 isolated from tomato with primers OPE-2 and OPE-7. Lanes 1 and 7, mutant T22; lanes 2 and 8, mutant T38; lanes 3 and 9, mutant T67; lanes 4 and 10, mutant T32; lanes 5 and 11, mutant T76; lanes 6 and 12, strain IVIA1102. M, 123-bp molecular mass marker (Life Technologies).
Identification of the regions altered in the mutant bacteria.
We analyzed the causes for the loss of pathogenicity by PCR and complementation tests. First, we evaluated the presence of the Ti plasmid in each of the nonpathogenic mutants by PCR, using the following primers directed against the T-DNA and the vir regions (Table 2; Fig. 1): primers FGPtmr530-FGPtmr701 (39) amplify a 172-nucleotide (nt) fragment of the tmr (or ipt) gene; primers CYT1-CYT2 (16) amplify 427 nt of the same tmr gene, but several nucleotides downstream the previous primers, and VirDA-VirDE (16) amplify a 338-nt fragment of the virD gene. Primers CYT1-CYT2 and VirDA-VirDE were employed in a multiplex reaction.
TABLE 2.
Primers employed to examine possible indels responsible for the loss of pathogenicity in the mutants and for sequencing the insertion of mutant T1
| Primer | Sequence (5′-3′) | Size (bp) | Positiona | Reference |
|---|---|---|---|---|
| FGPtmr530 | CCA TGT TGT TTG CTA GCC AG | 172 | ||
| FGPtmr701 | CCT TCG AAT CCG TCG AAA GC | 39 | ||
| VirDA | ATG CCC GAT CGA GCT CAA GT | 338 | ||
| VirDE | CCT GAC CCA AAC ATC TCG GCT GCC CA | 16 | ||
| CYT1 | GAT CGG GTC CAA TGC TGT | 427 | ||
| CYT2b | GAT ATC CAT CGA TCC CTT | 19860 | 16 | |
| CYT21b | TCC ATC GCG TTT ACA GC | 2,449 | 17411 | This work |
| INS-1c | CGA GCA TCT CTC TGA CAA T | This work | ||
| INS-2c | ATG CTC GGT CGC AAG AC | This work | ||
| INS-3b | ACT GGC TTT ACC GTC TCC | 19234 | This work |
Position of each of the primers employed for sequencing the insertion of mutant T1, according to the sequence of pTi of strain C58 from Wood et al. (54), with GenBank accession number AE008690.
Primers employed for the sequencing of the insertion in mutant T1.
Primers designed from the insertion sequence of mutant T1. There is no correspondence with the sequences of T-DNA.
Complementation analyses were performed with plasmids containing either the whole T-DNA region, contained in plasmid pTHE17 (17), or different genes of the vir region, as follows: the virH, virA, and virB genes carried by plasmid pVCK219 (45) and the virG, virC, virD, and virE genes carried by plasmid pVCK225 (45) (Fig. 1). The plasmids were transferred to the nonpathogenic mutants by electroporation using a Gene Pulser II system (Bio-Rad, CA), and the acquisition of each plasmid by the transformants was tested by PCR using the three primer pairs described above. For each combination of mutant and complementing plasmid analyzed, six independent transformants were inoculated on tomato plants. Once the complementation analyses identified a particular DNA region that restored pathogenicity, we examined possible changes in the diverse mutants by PCR using different primers, looking for insertions or deletions that could be distinguished in the amplification (Table 2). All the PCRs were performed in a 50-μl final volume with 3 mM MgCl2, 100 μM deoxynucleoside triphosphate, 3% (vol/vol) formamide, 10 pmol of each primer, and 1 U of Taq DNA polymerase.
Plasmid profiles of the wild-type and mutant strains were visualized by agarose gel electrophoresis using a modified Eckhardt protocol (13) or the method of Zhou et al. (56) to evaluate the existence of changes in the size or number of the native plasmids and, in particular, of the Ti plasmid. Plasmid DNA was digested with EcoRI, HindIII, or PvuI and transferred to nylon membranes (Roche Diagnostics, Mannheim, Germany) using a rapid alkaline transfer protocol (41). As DNA probes, we employed the entire T-DNA region in plasmid pTHE17 (17) (Fig. 1) and the complete pTi from strain IVIA1102. Preparation of labeled probes with digoxigenin, Southern hybridization at 65°C, and detection of hybridization signals using a LAS-3000 system (Fuji, Japan) were carried out with a DIG DNA labeling and detection kit (Roche Diagnostics).
An insertion element (IS) inserted into the iaaM gene in one of the mutants was detected by PCR and sequenced using primers flanking or internal to the IS (Table 2; Fig. 3). The resulting amplicons were separated in agarose gels and purified from the gel using the Mo Bio kit (CA) before sequencing them completely. The sequences were compared against the GenBank databases using the BLAST algorithms (1), and sequences were aligned using CLUSTALW (51).
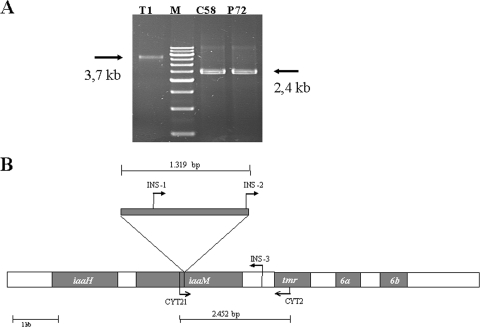
FIG. 3.
Mutant T1 contains an insertion of IS426 in a gene essential for the biosynthesis of 3-indoleacetic acid, iaaM. (A) Amplicons obtained with primer pair CYT21-CYT2. Nonpathogenic mutant bacteria T1 and P72 were isolated from tumors induced in tomato and pepper, respectively, by the wild-type strain C58. M, marker XVII (Life Technologies). (B) Position of IS426 in the T-DNA of mutant T1. The annealing sites of the different primers used for PCR and sequencing are indicated with arrows.
Nucleotide sequence accession number.
Sequences determined in this study have been deposited in the GenBank database under accession no. FJ004947.
RESULTS
Isolation of variants with altered pathogenicity.
Two inoculation experiments (A and B) were carried out using seven A. tumefaciens strains of biovar 1 and 12 different plant hosts in order to evaluate the possible appearance of nonpathogenic variants. In agreement with the difficulties previously found (33), we were unable to recover agrobacteria from any of the tumors analyzed from apple plants. From the remaining tumors, 5,419 typical Agrobacterium-like colonies were recovered on semiselective medium (Table 3); all of them were urease positive, hydrolyzed esculin, and were in consequence preliminarily identified as A. tumefaciens. Their pathogenicity was evaluated by inoculation in tomato plants (Table 3), which allowed for the identification of 662 colonies (12.2%) that were nonpathogenic. In experiment A, 19 tumors were analyzed; 6 of them yielded 184 nonpathogenic isolates from a total of 3,800 colonies examined. In experiment B, 1,619 colonies were recovered from 33 tumors induced by two A. tumefaciens strains in 10 plant hosts. From these, 14 tumors yielded a total of 478 colonies that were nonpathogenic.
TABLE 3.
Occurrence and characteristics of clones with altered pathogenicity isolated from tumors induced by different strains of A. tumefaciens in herbaceous and woody hosts
| Strain | Host | Characteristics of clones with altered pathogenicityd
|
||||
|---|---|---|---|---|---|---|
| No. of clones containing virulence genesa
|
RAPD patternsb
|
|||||
| Proportionc | ipt/tmr | virD | No. of patterns | No. of strains with same pattern as that of the WT | ||
| Experiment A | ||||||
| A281 | Pepper | 0/200 | NA | NA | NA | NA |
| 0/200 | NA | NA | NA | NA | ||
| Tomato | 0/200 | NA | NA | NA | NA | |
| 0/200 | NA | NA | NA | NA | ||
| Ach5 | Pepper | 0/200 | NA | NA | NA | NA |
| 0/200 | NA | NA | NA | NA | ||
| Tomato | 170/200 | 0 | 0 | 2 | 0 | |
| 0/200 | NA | NA | NA | NA | ||
| C58 | Pepper | 1/200 | 1 | 1 | 1 | 1 |
| 0/200 | NA | NA | NA | NA | ||
| Tomato | 1/200 | 1 | 1 | 1 | 1 | |
| 0/200 | NA | NA | NA | NA | ||
| CFBP42 | Pepper | 0/200 | NA | NA | NA | NA |
| Tomato | 0/200 | NA | NA | NA | NA | |
| 0/200 | NA | NA | NA | NA | ||
| IVIA1102e | Pepper | 3/200 | 3 | 3 | 1 | 0 |
| 0/200 | NA | NA | NA | NA | ||
| Tomato | 5/200 | 2 | 2 | 2 | 2 | |
| 4/200 | 0 | 3 | 2f | 3 | ||
| Experiment B | ||||||
| IVIA325-4 | Adafuelg | 200/250 | 0 | 0 | 5 | 0 |
| Appleh | NA | NA | NA | NA | NA | |
| Cherry | 105/250 | 0 | 0 | 4 | 0 | |
| GF677g | 84/250 | 0 | 0 | 3 | 0 | |
| Pear | 0/250 | NA | NA | NA | NA | |
| IVIA678-2 | Peach Nemaguard | 65/200 | 0 | 0 | 4 | 0 |
| GF677g | 0/100 | NA | NA | NA | NA | |
| GxN15g | 4/50 | 0 | 0 | 1 | 0 | |
| GxN22g | 0/100 | NA | NA | NA | NA | |
| Plum Myrobalan | 15/100 | 0 | 0 | 2 | 0 | |
| Peach Montclar | 5/69 | 0 | 0 | 1 | 0 | |
Presence/absence of each gene was examined by PCR with specific primers (see Fig. 1).
Numbers refer only to clones with altered pathogenicity. WT, wild-type strain used for inoculation.
Number of clones with altered pathogenicity/total number of clones analyzed. In experiment A, two tumors per plant were selected from two different plants (four tumors per experiment and strain in total); the experiment was done twice, resulting in a total of 400 colonies examined (except in the case of CFBP42 in pepper, where colonies could be isolated only in one experiment). In experiment B, colonies were isolated from five tumors from different plants to reach a total of 250 isolates per host-strain combination.
NA, not applicable.
Strain IVIA1102 is resistant to kanamycin and, although isolation was carried out on unsupplemented media, all the nonpathogenic isolates were also resistant to kanamycin. Isolates from tumors induced by the remaining A. tumefaciens strains were not tested for resistance to this antibiotic.
Strains showing differences in one band using primer OPE7.
Hybrids of peach × almond.
No Agrobacterium colonies were isolated from the tumors obtained in apple plants.
To ascertain the origin of the 662 nonpathogenic isolates of experiments A and B, we analyzed and compared their fingerprints using a RAPD analysis (24). We could differentiate a total of 26 different RAPD patterns, with one to five RAPD patterns among the nonpathogenic isolates coming from the same combination of host-bacterial strains (Table 3; Fig. 2A). In general, patterns were different for agrobacteria isolated from different hosts, although in a few cases, isolates sharing the same pattern were sometimes found in different tumors of the same plant, as can be observed in Fig. 2A. Only seven of the nonpathogenic agrobacteria analyzed, originating from pepper (one) and tomato (six) plants and produced by two strains (C58 and IVIA1102), showed RAPD patterns that were identical to those of the parental strain (Table 3). These results potentially suggest a different and unknown origin for the remaining 655 nonpathogenic isolates. The seven mutants also displayed patterns identical to those of the inoculated strains using three other RAPD primers, although we detected a variation in one of the fingerprints with three of the nonpathogenic mutants (T22, T38, and T67). They each lacked one prominent band in the RAPD fingerprint using primer OPE-7, which was otherwise identical to the fingerprint of the parental strain IVIA1102 (Fig. 2B). These three mutants suffered plasmid reorganizations and the deletion of at least the complete T-DNA region (see below), which could be responsible for the alterations in this RAPD fingerprint. Additionally, the seven nonpathogenic mutants also displayed identical fingerprints to those of the respective inoculated strains with primers ERIC, BOX, and REP, whereas another 20 randomly selected nonpathogenic clones showed different profiles (data not shown). Together, these results show that only 7 of the 662 nonpathogenic clones analyzed were identical to the inoculated strains of A. tumefaciens.
Strain IVIA1102 employed in experiment A was isolated in Valencia, Spain, from a chrysanthemum tumor and was naturally resistant to high concentrations of kanamycin. Bacteria from tumors induced by this strain were isolated in semiselective medium (42) devoid of kanamycin. Surprisingly, replica plating of the 12 nonpathogenic clones isolated from these tumors showed that although all were resistant to 100 μg/ml kanamycin, 7 of them displayed a RAPD pattern different from that of the parental strain (Table 3 and data not shown), indicating that antibiotic selection was not sufficient to guarantee the recovery of the inoculated strain.
In experiment C, the stability of virulence was examined in 100 individual clones each of two A. tumefaciens strains after five serial inoculation passages in liquid culture medium with or without antibiotic selection. After each passage and inoculation in tomato plants, no nonpathogenic mutant bacteria were found in the 2,000 colonies examined.
Characterization of mutant bacteria with altered pathogenicity.
We analyzed the molecular basis for the loss of pathogenicity in the seven nonpathogenic mutants obtained from tumors induced by strains C58 and IVIA1102 (Table 3). With this purpose, we examined the presence of the Ti plasmid on each clone, the existence of T-DNA and the vir genes, and the complementation of the mutant phenotype using plasmid constructions with overlapping inserts that covered the complete vir region or the complete T-DNA region. These analyses showed that the loss of pathogenicity was due to independent molecular events (Table 4), at least for mutant bacteria isolated from different tumors.
TABLE 4.
Pathogenicity of diverse mutants in tomato using complementation analyses with plasmids covering the vir (pVCK219 and pVCK225) or the T-DNA (pTHE17) region
| Mutant | Origin (strain, host) | Results of complementation analyses witha:
|
|||
|---|---|---|---|---|---|
| No plasmid | pVCK219b | pVCK225c | pTHE17d | ||
| P72 | C58, pepper | − | − | − | − |
| T1 | C58, tomato | − | − | − | + |
| T22, T38, T67 | IVIA1102, tomato | − | − | − | + |
| T32, T76 | IVIA1102, tomato | +d | +d | +d | + |
Results of the inoculation of transformed colonies in tomato. +, tumor; −, no tumor; +d, tumor smaller than that caused by the parental strain. The same results were obtained with six independent transformants per strain for each plasmid tested.
Plasmid containing virH, virA, and virB genes; see Fig. 1.
Plasmid containing virG, virC, virD, and virE genes; see Fig. 1. This plasmid plus pVCK219 covers the whole vir region of strain A6 (see Fig. 1).
Plasmid containing the entire T-DNA region from strain C58.
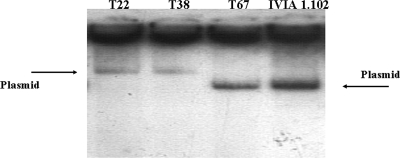
With strain C58, only two true mutants were recovered from tumors, one from a pepper plant (mutant P72) and one from a tomato plant (mutant T1). Mutant P72 did not induce tumors in either tomato or cherry, the host from which its parental strain was isolated. This mutant possessed a Ti plasmid apparently of wild-type size (Fig. 4A) and produced the expected amplicons with primers specific for genes tmr and virD (data not shown); moreover, in complementation analyses, this strain did not recover pathogenicity with any of the plasmid constructions used (Table 4). These results suggest that mutant P72 might contain a mutation in one or more chromosomal genes that renders it nonpathogenic. Mutant T1 presented a reduction in virulence, because it was nonpathogenic in tomato but still produced tumors in cherry. The pathogenicity of this mutant in tomato was completely recovered in complementation analyses with plasmid pTHE17, a construction containing the entire T-DNA region, but not with two other plasmids that covered the vir region (Table 4; Fig. 1). The analysis of the T-DNA region in mutant T1 by PCR amplification with several primers (Table 2) yielded an amplicon covering genes iaaM and tmr that was around 1.3 kb larger than expected (Fig. 3A). Sequencing of this amplicon showed that it contained a 1,319-bp insertion in position 17760 of gene iaaM (accession no. AE007871) that was 99.7% identical to the insertion sequence IS426 (52) present in the genome of A. tumefaciens C58 (accession no. X56562) (Fig. 3B).
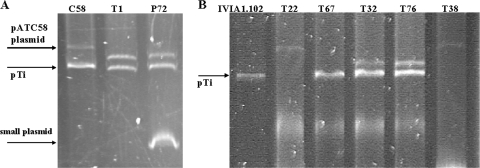
FIG. 4.
Plasmid profiles of the nonpathogenic mutants derived from tumors induced by strains C58 (A) and IVIA1102 (B). The positions of the pTi for C58 (210 kb) and IVIA1102 (120 kb) are indicated with arrows.
Strain C58 harbors two plasmids, pTi and a larger one (pATC58) that has been shown to carry att (attachment) and other genes, which are not essential for pathogenicity but have a quantitative effect on virulence (37). Both P72 and T1 showed no changes in the size of pTi but, instead of pAtC58, contained a smaller plasmid (300 kb rather than 450 kb) (Fig. 4A), which likely is a deletion derivative of pATC58. Additionally, we repetitively observed an intense and distorted band in the plasmid profile of mutant P72 that probably corresponds to a small, high-copy-number plasmid, whose origin is unclear.
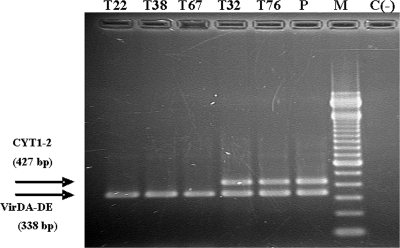
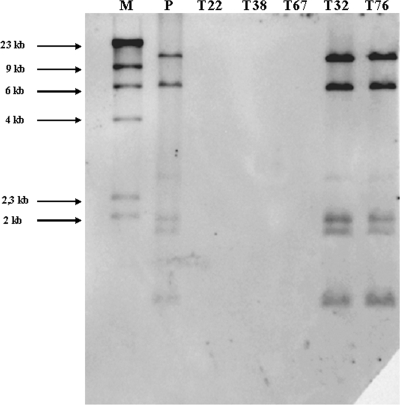
The five remaining mutants were derived from tumors produced in tomato plants by strain IVIA1102. Three of the mutants (T22, T38, and T67) produced no tumors in either tomato or chrysanthemum, the host from which IVIA1102 was isolated. The remaining two mutants (T32, T76), however, showed a reduction in virulence, because they were pathogenic in chrysanthemum but produced tumors in tomato that were significantly smaller than those produced by the parental strain (data not shown). All five mutants produced the expected amplicons using the primer pairs for the virD gene, but only mutants T32 and T76 produced the expected products with the primer pairs specific for the T-DNA region (Fig. 5). Nevertheless, in complementation analyses, the pathogenicity of the five mutants was restored by the construction containing the T-DNA region (plasmid pTHE17) (Table 4), indicating that, independently of the PCR results, all of them were affected in this area. Four of these five mutants (T22, T32, T38, and T76) showed an obviously altered plasmid profile (Fig. 4B). Strain IVIA1102 contains a single plasmid of approximately 120 kb; mutants T22 and T38 also contained a single plasmid, although it was approximately twice that size. On the other hand, mutants T32 and T76 contained a plasmid identical in size to that of the parental strain, but they also harbored an additional plasmid of approximately 300 kb. Mutant T67 was the only one containing a plasmid of a size similar to that of the wild-type strain (Fig. 4B); however, hybridization analyses using T-DNA as a probe clearly showed that this mutant lacks the entire T-DNA region (Fig. 6). Likewise, the hybridization results indicate that the T-DNA region was also absent in mutants T22 and T38, whereas mutants T32 and T76 showed the same hybridization pattern as that of the parental strain (Fig. 6). When the complete plasmid of IVIA1102 was used as a hybridization probe against the plasmids of mutants T22, T38, and T67, these provided a positive hybridization signal, indicating that all these plasmids have sequences similar to that of strain IVIA1102, despite their differences in size (Fig. 7).
FIG. 5.
Presence of genes virD and tmr in nonpathogenic mutant bacteria. Mutants isolated from tumors induced in tomato by strain IVIA1102 were analyzed by multiplex PCR using primer pairs CYT1-CYT2 (CYT1-2) and VirDA-VirDE (VirDA-DE) (see the legend to Fig. 1 for details on amplified fragments). P, parental strain IVIA1102; M, 100-bp DNA size marker (Life Technologies); C(−), negative control. The sizes of the amplicons are detailed on the left.
FIG. 6.
Presence of the T-DNA region in nonpathogenic mutants derived from IVIA1102. Plasmid DNA was digested with HindIII and separated on an agarose gel before Southern hybridization; the probe used was the complete T-DNA region from strain C58, obtained from plasmid pTHE17. P, parental strain IVIA1102; M, marker λHindIII (Invitrogen).
FIG. 7.
Hybridization of uncut plasmids from strain IVIA1102 and mutants T22, T38, and T67 derived from it, using the single plasmid from IVIA1102 as a probe; plasmids were separated in an Eckhardt gel.
DISCUSSION
The population dynamics of Agrobacterium in soil and in the ecological niches that it colonizes (roots, crown of plants) have been investigated in previous works (6, 19, 20, 31, 33, 42). One of the most striking aspects revealed by these works is the predominant number of nonpathogenic isolates in tumors, both natural and derived from inoculations, or in infested soil. In the present work, we examined carefully the frequency of appearance of nonpathogenic variants, both in vitro and in planta, using a larger number of Agrobacterium strains and hosts than that used in previous studies. Additionally, a fundamental aspect of our work was the use of efficient, reproducible, and highly specific RAPD fingerprinting analysis (24) to verify the origin of the putative nonpathogenic clones. In agreement with previous results (33), we observed a relatively high proportion of nonpathogenic clones from different tumors, which generally were of a unique type when originating from a given tumor but differed from those of other tumors and lacked the vir and T-DNA genes (Table 3). However, the analysis of 5,419 colonies recovered from tumors indicates that, under our conditions, the rate of the appearance of bona fide nonpathogenic mutants was very low. Indeed, although 662 of the analyzed clones (12.2%) were nonpathogenic in tomato plants, only 7 of them (0.1%) can be considered to be truly derived from the inoculated strains, as shown by the coincidence of their RAPD fingerprints (Table 3); in fact, this figure is probably an overestimation because mutants T22 and T38 are likely isolates of the same clone, as they were isolated from the same tumor and showed identical phenotypes and plasmid profiles, as well as mutants T32 and T76, which could reduce the total number of mutants found to only 5. These results do not necessarily imply that the nonpathogenic clones isolated from tumors in other works derive from strains that are different from those originating the tumor, although they clearly demonstrate the need to use accurate and reliable methods for verification of the origin of putative nonpathogenic mutant bacteria. For instance, antibiotic resistance has been traditionally used for strain identification; however, we observed that inoculation of a spontaneous kanamycin-resistant strain (Table 3, see strain IVIA1102) did not prevent the isolation of other agrobacteria, which were also resistant to kanamycin but different from the inoculated strain. The inclusion of two or more antibiotics in the selection medium (4) should potentially circumvent this problem, although it would still be necessary to reliably identify the origin of the putative nonpathogenic mutant bacteria, because it is common to find bacteria in plants or soil that are resistant to one or several antibiotics (29).
Other authors found that some strains of A. tumefaciens originated a large proportion of nonpathogenic mutant bacteria in vitro in the presence of acetosyringone (13, 33), a phenolic plant compound produced in wounds that induces the expression of vir genes. This phenomenon was interpreted as an explanation for the putative generation of nonpathogenic mutant bacteria in tumors and their unexplained selective advantage in these tissues. In contrast, our results indicate that the frequent occurrence of nonpathogenic clones in tumors was not the consequence of the repeated generation and strong selection of mutant bacteria but was due mainly to the growth of clones of nonpathogenic agrobacteria different from the strain that produced the tumor. Due to the inoculation of axenic cultures in healthy plants with sterilized substrates, these nonpathogenic environmental agrobacteria that might be colonizing the plant tumors could be at a great numerical disadvantage compared to the bacteria used as inoculum. Additionally, and depending on their origin, these environmental agrobacteria will predictably have to gain access to the interior of the plant and reach the tumor tissues. In spite of this, nonpathogens appeared in just over one-third of the tumors analyzed, suggesting a very strong selective pressure that would favor them rather than the pathogenic bacteria. The coinoculation of wild-type and nonpathogenic strains in competition experiments would be of great interest to test this hypothesis.
We can foresee the following three possible origins for these nonpathogenic agrobacteria that exhibited RAPD patterns different from those of the parental strains: (i) they were present as undetected contaminants in the original bacterial culture used as inoculum; (ii) they are indeed mutant bacteria derived from the inoculated strain; or (iii) they were present as endophytes in the inoculated plants or reached the plants from external sources, such as the irrigation water.
In the first option, it is highly unlikely that the nonpathogenic agrobacteria appear in tumors as a result of the contamination of the culture used as inoculum, because not all the tumors contained nonpathogens and because we found different bacteria in different tumors. Besides, and most importantly, all the bacterial strains used for inoculations were purified several times, were cryoconserved as axenic suspensions, and as an additional precaution, were purified again through three successive single-colony passages before inoculum preparation. Furthermore, we checked the stock of strain C58 for the presence of possible contaminants by PCR targeting the insertion sequence observed in mutant T1, and the results were negative (data not shown). These procedures will practically eliminate the possibility that inocula can be contaminated with as many as five different types of nonpathogenic agrobacteria.
The second option, that all of the nonpathogens derive from the inoculated strain, is also difficult to explain. Three of the true mutants identified here showed the disappearance of one band in the RAPD pattern with one of the four primers used (Fig. 2B), which could be caused by the plasmid reorganizations that they suffered (Fig. 4B). However, it is highly improbable that other nonpathogenic agrobacteria derive from the inoculated strains because their RAPD patterns were highly dissimilar, which would entail the occurrence of frequent and massive genome reorganizations if they had a clonal origin. Nevertheless, spontaneous elevated mutation rates (up to 100-fold), or hypermutation, occur in several bacterial pathogens of animals when exposed to stressful or changing environments or to extreme nutrient limitation (48). Although this phenomenon has not been reported among bacterial plant pathogens, we cannot entirely discard the possibility that part or all of the nonpathogens encountered in tumor tissues are the result of a boom in mutation rates and genome shuffling.
The third possible origin of the nonpathogenic clones whose RAPD patterns differ from those of the inoculated strain (option iii) is that they were present as endophytes in the inoculated plants or reached the plant from external sources, which is, in our opinion, the likeliest explanation. The occurrence of endophytic agrobacteria in otherwise healthy herbaceous and woody plants has been repeatedly shown by other authors (27, 49, 50, 53, 57). Conversely, it was reported that Agrobacterium populations apparently decline rapidly in water, although they can enter a viable but nonculturable state (26, 46), and it is possible that they could recover from this state and colonize plants. Therefore, our results strongly support that the majority of nonpathogenic agrobacteria found in plant tumors are endophytes, or come from other environmental sources, as indicated by the isolation of agrobacteria from the crowns of uninfected tomato plants (data not shown). They coexist in the tumors in a higher number than the pathogenic ones, and surprisingly, they seem to be strongly selected in the tumor environment in detriment of the pathogenic A. tumefaciens originating the tumor, although such a habitat should theoretically favor the pathogenic bacteria and the pTi conjugation (18, 19).
Several authors suggested that the occurrence of nonpathogenic agrobacteria is more common in certain plants than others (4, 33). In this work, we analyzed agrobacteria from tumors induced by seven A. tumefaciens strains in 12 different hosts, and we found only seven nonpathogenic mutants that are clearly derived from the inoculated strain in pepper and tomato (Table 3), although it would be premature to extract conclusions from such a low number of mutants. Additionally, we also found a differential occurrence of nonpathogenic environmental agrobacteria in tumors from different plant hosts (Table 3); for instance, they were very common in tumors from woody plants (experiment B) but less frequent in those from herbaceous hosts (experiment A). Again, we believe that it is not possible to conclude whether or not the host influences the occurrence of nonpathogenic environmental agrobacteria in tumors, because their origin is as yet unknown and the frequency of their occurrence can be subjected to many factors unrelated to the experiment variables. Indeed, plants of experiment A were inoculated in the stem, which would predictably reduce competition and antagonism and probably would result in a larger number of tumors colonized by pathogenic bacteria, as it occurred experimentally. Conversely, pathogenic agrobacteria were added to sterile soil in experiment B, where environmental agrobacteria reaching the plant, for instance, with irrigation water, could invade the same wounds as the pathogens and quickly colonize the tumor. Based on our results, it shall then be necessary to design appropriate new experiments to test the putative differences between herbaceous and woody hosts to promote the growth of nonpathogenic agrobacteria.
Six of the seven true nonpathogenic mutants identified in this work, mutant P72 being the exception, were affected in the T-DNA region, which is responsible for the biosynthesis of phytohormones, and their pathogenicity was fully complemented with plasmid pTHE17, containing this region in its entirety. Mutant T1 contained an insertion of IS426 in the iaaM gene, which is essential for the biosynthesis of the plant phytohormone 3-indoleacetic acid, and was nonpathogenic in tomato but could produce tumors in cherry. This result therefore suggests that the biosynthesis of 3-indoleacetic acid is dispensable for the production of tumors in cherry but essential for the production of tumors in tomato in this strain. A similar effect on host range was observed in Agrobacterium vitis strains, which suffered a reduction in host range capabilities when this gene was removed, due to the insertion of an IS (40). The insertion sequence IS426 has been previously found to be associated with spontaneous mutations in the pTi of strain C58 that lead to nonpathogenic variants (14, 52). Remarkably, we detected alterations in the plasmid profiles of both mutants obtained from strain C58, P72 and T1, although it is unclear if they are in any way related to the loss of pathogenicity. Likewise, mutants T32 and T76 produced smaller tumors than the wild-type strain in tomato but normal tumors in chrysanthemum, suggesting that the production of phytohormones negatively impacts the tumor production process in a host-dependent manner; nevertheless, we cannot discard the possibility that these mutants are also affected in other genes of the T-DNA region. The remaining three mutants, T22, T38, and T67, lacked the complete T-DNA region (ca. 23 kb) and, in consequence, could not induce tumors in either tomato or chrysanthemum. These results strongly suggest that the T-DNA region is a hot spot for the occurrence of diverse mutations that abolish pathogenicity.
All of the seven bona fide mutants suffered genetic changes in their plasmids that led to conspicuous alterations of their native plasmid profiles, except for mutant T67, even though this mutant had suffered the deletion of the T-DNA region. The existence of reorganizations in Agrobacterium is probably favored by the numerous regions of homology among the plasmids and the chromosomes (15, 54). Horizontal plasmid transfer, plasmid cointegration, and rescue of chromosomal DNA occur with a high level of frequency in related bacteria (12, 28, 47, 55), including Agrobacterium (23). Rearrangement between plasmids that did not produce changes in their plasmid profiles despite the interchange of genes they have suffered (55) or structural changes producing the loss of a specific phenotype have also been described (44), which could explain the changes observed in the plasmids of the mutant bacteria obtained. Thus, in three of the mutants (P72, T32, and T76), we observed the appearance of an extra plasmid band, whose origin is unclear, that could be explained if the extra plasmid had been horizontally acquired, probably from other agrobacteria present in the tumor, or due to events of decointegrations, as part of the several rearrangement outcomes already observed in some plasmids in Rhizobium (12).
In conclusion, the presence of true nonpathogenic mutants of Agrobacterium from tumors induced in 11 host plants was found to occur at a very low level of frequency and was apparently nondependent on the host. Our data do not allow us to determine whether the mutations leading to the loss of pathogenicity occurred before inoculation or were favored by the tumor environment. The causes of the total or reduced loss of virulence were variable and generally related to genetic modifications in the Ti plasmid. Also, new plasmids of unclear origin were found colonizing derivates of the inoculated strain, which suggests a high rate of horizontal transfer among bacteria coexisting in the tumor.
Acknowledgments
This research was supported partially by project INIA (grant SC93-117) of the Spanish Ministry of Agriculture and by COST Action 873 (European Union). Pablo Llop has a contract from the Ministry of Education and INIA, with funding from the European Union (Social European Funding).
We are grateful to Robert W. Jackson and Theresa Osinga for critical reading of the manuscript and helpful suggestions.
Footnotes
Published ahead of print on 21 August 2009.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, A. R., and L. W. Moore. 1979. Host specificity in the genus Agrobacterium. Phytopathology 69:320-323. [Google Scholar]
- 3.Bélanger, C., M. L. Canfield, L. W. Moore, and P. Dion. 1993. Detection of avirulent mutants of Agrobacterium tumefaciens in crown-gall tumors produced in vitro. p. 97-101. In E. W. Nester and D. P. S. Verma (ed.), Advances in molecular genetics of plant microbe interactions, vol. 2. Kluwer Academic Publishers, Dordrecht, The Netherlands. [Google Scholar]
- 4.Bélanger, C., M. L. Canfield, L. W. Moore, and P. Dion. 1995. Genetic analysis of nonpathogenic Agrobacterium tumefaciens mutants arising in crown gall tumors. J. Bacteriol. 177:3752-3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouzar, H., D. Ouadah, Z. Krimi, J. B. Jones, M. Trovato, A. Petit, and Y. Dessaux. 1993. Correlative association between resident plasmids and the host chromosome in a diverse Agrobacterium soil population. Appl. Environ. Microbiol. 59:1310-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Canfield, M. L., and L. W. Moore. 1991. Isolation and characterization of opine-utilizing strains of Agrobacterium tumefaciens and fluorescent strains of Pseudomonas spp. from rootstocks of Malus. Phytopathology 81:440-443. [Google Scholar]
- 7.Christie, P. J. 1997. Agrobacterium tumefaciens T-complex apparatus: a paradigm for a new family of multifunctional transporters in eubacteria. J. Bacteriol. 179:3085-3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Bruijn, F. J. 1992. Use of repetitive (repetitive extragenic palindromic and enterobacterial repetitive intergeneric consensus) sequences and the polymerase chain reaction to fingerprint the genomes of Rhizobium meliloti isolates and other soil bacteria. Appl. Environ. Microbiol. 58:2180-2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Cleene, M., and J. Ley. 1976. The host range of crown gall. Bot. Rev. 42:389-466. [Google Scholar]
- 10.Dion, P., C. Belanger, C. Marquis, W. Ream, and S. B. Gelvin. 1996. Ecological significance of avirulence in Agrobacterium, p. 44-58. In W. Ream and S. B. Gelvin (ed.), Crown gall: advances in understanding interkingdom gene transfer. APS Press, St. Paul, MN.
- 11.Duncan, A. V., H. W. Stokes, and G. Daggard. 1992. Genetic exchange in natural microbial communities, p. 383-429. In K. C. Marshall (ed.), Advances in microbial ecology, vol. 12. Plenum Press, New York, NY. [Google Scholar]
- 12.Flores, M., P. Mavingui, X. Perret, W. J. Broughton, D. Romero, G. Hernández, G. Dávila, and R. Palacios. 2000. Prediction, identification, and artificial selection of DNA rearrangements in Rhizobium: toward a natural genomic design. Proc. Natl. Acad. Sci. USA 97:9138-9143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fortin, C., E. W. Nester, and P. Dion. 1992. Growth inhibition and loss of virulence in cultures of Agrobacterium tumefaciens treated with acetosyringone. J. Bacteriol. 174:5676-5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fortin, C., C. Marquis, E. W. Nester, and P. Dion. 1993. Dynamic structure of Agrobacterium tumefaciens Ti plasmids. J. Bacteriol. 175:4790-4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodner, B., G. Hinkle, S. Gattung, N. Miller, M. Blanchard, B. Qurollo, B. S. Goldman, Y. Cao, M. Askenazi, C. Halling, L. Mullin, K. Houmiel, J. Gordon, M. Vaudin, O. Iartchouk, A. Epp, F. Liu, C. Wollam, M. Allinger, D. Doughty, C. Scott, C. Lappas, B. Markelz, C. Flanagan, C. Crowell, J. Gurson, C. Lomo, C. Sear, G. Strub, C. Cielo, and S. Slater. 2001. Genome sequence of the plant pathogen and biotechnology agent Agrobacterium tumefaciens C58. Science 294:2323-2328. [DOI] [PubMed] [Google Scholar]
- 16.Haas, J. H., L. W. Moore, W. Ream, and S. Manulis. 1995. Universal PCR primers for detection of phytopathogenic Agrobacterium strains. Appl. Environ. Microbiol. 61:2879-2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayman, G. T., and S. K. Farrand. 1988. Characterization and mapping of the agrocinopine-agrocin 84 locus on the nopaline Ti plasmid pTiC58. J. Bacteriol. 170:1759-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kado, C. I. 1991. Molecular mechanisms of crown gall tumorigenesis. Crit. Rev. Plant Sci. 10:1-32. [Google Scholar]
- 19.Kerr, A. 1969. Transfer of virulence between isolates of Agrobacterium. Nature 223:1175-1176. [Google Scholar]
- 20.Krimi, Z., A. Petit, C. Mougel, Y. Dessaux, and X. Nesme. 2002. Seasonal fluctuations and long-term persistence of pathogenic populations of Agrobacterium spp. in soils. Appl. Environ. Microbiol. 68:3358-3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LaPointe, G., C. S. Nautiyal, W. S. Chilton, S. K. Farrand, and P. Dion. 1992. Spontaneous mutation conferring the ability to catabolize mannopine in Agrobacterium tumefaciens. J. Bacteriol. 174:2631-2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lelliot, R. A., and D. E. Stead. 1987. Methods for the diagnosis of bacterial diseases of plants, p. 216. In T. F. Preece (ed.), Methods in plant pathology, vol. 2. Blackwell Scientific Publications, Oxford, United Kingdom. [Google Scholar]
- 23.Leloup, L., E. M. Lai, and C. I. Kado. 2002. Identification of a chromosomal tra-like region in Agrobacterium tumefaciens. Mol. Genet. Genomics 267:115-123. [DOI] [PubMed] [Google Scholar]
- 24.Llop, P., B. Lastra, H. Marsal, J. Murillo, and M. M. López. 2003. Tracking Agrobacterium strains by a RAPD system to identify single colonies from plant tumors. Eur. J. Plant Pathol. 109:381-389. [Google Scholar]
- 25.Louws, F. J., D. W. Fulbright, C. T. Stephens, and F. J. De Bruijn. 1994. Specific genome fingerprints of phytopathogenic Xanthomonas and Pseudomonas pathovars and strains generated with repetitive sequences and PCR. Appl. Environ. Microbiol. 60:2286-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manahan, S. H., and T. R. Steck. 1997. The viable but nonculturable state in Agrobacterium tumefaciens and Rhizobium meliloti. FEMS Microbiol. Ecol. 22:29-37. [Google Scholar]
- 27.Martí, R., J. Cubero, A. Daza, J. Piquer, C. I. Salcedo, C. Morente, and M. M. López. 1999. Evidence of migration and endophytic presence of Agrobacterium tumefaciens in rose plants. Eur. J. Plant Pathol. 105:39-50. [Google Scholar]
- 28.Mavingui, P., M. Flores, X. Guo, G. Dávila, X. Perret, W. Broughton, and R. Palacios. 2002. Dynamics of genome architecture in Rhizobium sp. strain NGR234. J. Bacteriol. 184:171-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McManus, P. S., V. O. Stockwell, G. W. Sundin, and A. L. Jones. 2002. Antibiotic use in plant agriculture. Annu. Rev. Phytopathol. 40:443-465. [DOI] [PubMed] [Google Scholar]
- 30.Melchers, L. S., and P. J. J. Hooykaas. 1987. Virulence of Agrobacterium. Oxf. Surv. Plant Mol. Cell Biol. 4:167. [Google Scholar]
- 31.Michel, M. F., A. C. Miranda, C. Depierreux, L. Otten, F. Delmotte, and L. Jouanin. 1990. Identification of different Agrobacterium strains isolated from the same forest nursery. Appl. Environ. Microbiol. 56:3537-3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moore, L. W., and D. A. Cooksey. 1981. Biology of Agrobacterium tumefaciens: plant interactions, p. 15-46. In K. L. Giles and A. C. Atherly (ed.), Biology of Rhizobiaceae. Academic Press, New York, NY.
- 33.Moore, L. W., and M. Canfield. 1996. Biology of Agrobacterium and management of crown gall disease, p. 153-191. In R. Hall (ed.), Principles and practice of managing soilborne plant pathogens. APS Press, St. Paul, MN.
- 34.Morris, J. W., and R. O. Morris. 1990. Identification of an Agrobacterium tumefaciens virulence gene inducer from the pinaceous gymnosperm Pseudotsuga menziesii. Proc. Natl. Acad. Sci. USA 87:3614-3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mougel, C. 2000. Structure génétique des populations d'Agrobacterium spp.: effet sélectif de la plante et implication dans la diffusion conjugative du plasmide Ti. Ph.D. thesis. University of Lyon, France.
- 36.Mougel, C., B. Cournoyer, and X. Nesme. 2001. Novel tellurite-amended media and specific chromosomal and Ti plasmid probes for direct analysis of soil populations of Agrobacterium biovars 1 and 2. Appl. Environ. Microbiol. 67:65-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nair, G. R., L. Zhenying, and A. N. Binns. 2003. Reexamining the role of the accessory plasmid pAtC58 in the virulence of Agrobacterium tumefaciens strain C58. Plant Physiol. 133:989-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nesme, X., F. Michel, and B. Digat. 1987. Population heterogeneity of Agrobacterium tumefaciens in galls of Populus L. from a single nursery. Appl. Environ. Microbiol. 53:655-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nesme, X., M. C. Leclerc, and R. Bardin. 1989. PCR detection of an original endosymbiont: the Ti plasmid of Agrobacterium tumefaciens, p. 47-50. In P. Nardon, V. Gianinazzi-Peason, A. M. Greines, L. Margulis, and D. C. Smith (ed.), Endocytobiology IV. Institut National de la Recherche Agronomique, Paris, France.
- 40.Paulus, F., J. Canaday, F. Vincent, G. Bonnard, C. Kares, and L. Otten. 1991. Sequence of the iaa and ipt region of different Agrobacterium tumefaciens biotype III octopine strains. Reconstruction of octopine Ti plasmid evolution. Plant Mol. Biol. 16:601-614. [DOI] [PubMed] [Google Scholar]
- 41.Reed, K. C., and D. A. Mann. 1985. Rapid transfer of DNA from agarose gels to nylon membranes. Nucleic Acids Res. 25:7207-7221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schroth, M. N., J. P. Thompson, and D. C. Hildebrand. 1965. Isolation of A. tumefaciens-A. radiobacter group from the soil. Phytopathology 55:645-647. [Google Scholar]
- 43.Sheng, J., and V. Citovsky. 1996. Agrobacterium-plant cell DNA transport: have virulence proteins, will travel. Plant Cell 8:1699-1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soberón-Chávez, G., R. Nájera, H. Olivera, and L. Segovia. 1986. Genetic rearrangements of a Rhizobium phaseoli symbiotic plasmid. J. Bacteriol. 167:487-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stachel, S. E., and P. C. Zambryski. 1986. virA and virG control the plant-induced activation of the T-DNA transfer process of A. tumefaciens. Cell 46:325-333. [DOI] [PubMed] [Google Scholar]
- 46.Stockwell, V. O., L. W. Moore, and J. E. Loper. 1993. Fate of Agrobacterium radiobacter K84 in the environment. Appl. Environ. Microbiol. 59:2112-2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sullivan, J. T., and C. W. Ronson. 1998. Evolution of rhizobia by acquisition of a 500-kb symbiosis island that integrates into a phe-tRNA gene. Proc. Natl. Acad. Sci. USA 95:5145-5149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sundin, G. W., and M. R. Weigand. 2007. The microbiology of mutability. FEMS Microbiol. Lett. 277:11-20. [DOI] [PubMed] [Google Scholar]
- 49.Surette, M. A., A. V. Sturz, R. R. Lada, and J. Nowak. 2003. Bacterial endophytes in processing carrots (Daucus carota L. var. sativus): their localization, population density, biodiversity and their effects on plant growth. Plant Soil 253:381-390. [Google Scholar]
- 50.Thomas, P., S. Kumari, G. K. Swarna, and T. K. S. Gowda. 2007. Papaya shoot tip associated endophytic bacteria isolated from in vitro cultures and host-endophyte interaction in vitro and in vivo. Can. J. Microbiol. 53:380-390. [DOI] [PubMed] [Google Scholar]
- 51.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vanderleyden, J., J. Desair, C. De Meirsman, K. Michiels, A. P. Van Gool, M. D. Chilton, and G. C. Jen. 1986. Nucleotide sequence of an insertion sequence (IS) element identified in the T-DNA region of a spontaneous variant of the Ti-plasmid pTiT37. Nucleic Acids Res. 14:6699-6709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang, L. L., E. T. Wang, J. Liu, Y. Li, and W. X. Chen. 2006. Endophytic occupation of root nodules and roots of Melilotus dentatus by Agrobacterium tumefaciens. Microb. Ecol. 52:436-443. [DOI] [PubMed] [Google Scholar]
- 54.Wood, D. W., J. C. Setubal, R. Kaul, D. E. Monks, J. P. Kitajima, V. K. Okura, Y. Zhou, L. Chen, G. E. Wood, N. F. Almeida, Jr., L. Woo, Y. Chen, I. T. Paulsen, J. A. Eisen, P. D. Karp, D. Bovee, Sr., P. Chapman, J. Clendenning, G. Deatherage, W. Gillet, C. Grant, T. Kutyavin, R. Levy, M. J. Li, E. McClelland, A. Palmieri, C. Raymond, G. Rouse, C. Saenphimmachak, Z. Wu, P. Romero, D. Gordon, S. Zhang, H. Yoo, Y. Tao, P. Biddle, M. Jung, W. Krespan, M. Perry, B. Gordon-Kamm, L. Liao, S. Kim, C. Hendrick, Z. Y. Zhao, M. Dolan, F. Chumley, S. V. Tingey, J. F. Tomb, M. P. Gordon, M. V. Olson, and E. W. Nester. 2001. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 294:2317-2323. [DOI] [PubMed] [Google Scholar]
- 55.Zhang, X., B. Kosier, and U. Priefer. 2001. Symbiotic plasmid rearrangement in Rhizobium leguminosarum bv. viciae VF39SM. J. Bacteriol. 183:2141-2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou, C., Y. Yang, and A. Y. Jong. 1990. Miniprep in ten minutes. BioTechniques 8:172-173. [PubMed] [Google Scholar]
- 57.Zoina, A., A. Raio, R. Peluso, and A. Spasiano. 2001. Characterization of agrobacteria from weeping fig (Ficus benjamina). Plant Pathol. 50:620-627. [Google Scholar]