Abstract
Loss of genomic integrity is a defining feature of many human malignancies, including human papillomavirus (HPV)-associated preinvasive and invasive genital squamous lesions. Here we show that aberrant mitotic spindle pole formation caused by abnormal centrosome numbers represents an important mechanism in accounting for numeric chromosomal alterations in HPV-associated carcinogenesis. Similar to what we found in histopathological specimens, HPV-16 E6 and E7 oncoproteins cooperate to induce abnormal centrosome numbers, aberrant mitotic spindle pole formation, and genomic instability. The low-risk HPV-6 E6 and E7 proteins did not induce such abnormalities. Whereas the HPV-16 E6 oncoprotein has no immediate effects on centrosome numbers, HPV-16 E7 rapidly induces abnormal centrosome duplication. Thus our results suggest a model whereby HPV-16 E7 induces centrosome-related mitotic disturbances that are potentiated by HPV-16 E6.
Human papillomaviruses (HPVs) are small epitheliotropic DNA viruses involved in the etiology of several human malignancies. At least 90% of all cervical carcinomas are associated with infections by “high-risk” HPV types such as HPV-16 and -18. The majority of these cancers contain HPV DNA integrated into the host cell genome and express only two viral genes, E6 and E7, both of which encode oncoproteins (1). Both HPV-immortalized cells and high-risk HPV-associated cervical neoplasias, including early precursor lesions, display genomic instability, which is absent in lesions caused by low-risk HPVs (2–6). Induction of genomic plasticity, therefore, constitutes an early and central event in HPV-associated carcinogenesis and may contribute to the integration of HPV DNA into the host genome (7). However, it is not known in detail how HPV E6 and E7 interfere with genomic integrity. HPV E6 and E7 play distinct roles in this process by targeting different pathways (5). Whereas E6 may promote genetic instability by inactivating the tumor suppressor and cell cycle checkpoint protein p53 (5, 8), the mechanism by which E7 subverts the integrity of the host cell genome (5, 9) and whether this function depends on its ability to inactivate the pRB tumor suppressor protein (10, 11) have not been determined.
The centrosome is a cytoplasmic organelle consisting of a pair of centrioles surrounded by a pericentriolar matrix. Each cell contains one or, before a cell division, two centrosomes. During mitosis, the two centrosomes form the poles of a bipolar mitotic spindle, a function that is essential for accurate chromosome segregation. Centrosomes undergo duplication precisely once before cell division. Recent reports have revealed that this process is linked to the cell division cycle via cyclin-dependent kinase 2 (cdk2) activity that couples centriole duplication to the onset of DNA replication at the G1/S transition (12–14).
Various human malignancies exhibit centrosome abnormalities that contribute to defective mitotic spindle pole formation, thus causing chromosome missegregation and genetic instability (15). Here we report that preinvasive and invasive HPV-associated genital lesions contain abnormal centrosome numbers that are associated with mitotic abnormalities. We show that such aberrant centrosomes arise in primary human cells upon expression of HPV-16 E6, E7, or E6 and E7. In contrast, cells expressing low-risk HPV-6 E6 or E7 genes showed no such abnormalities. Whereas acute expression of HPV-16 E6 does not affect centrosome numbers, the HPV-16 E7 oncoprotein rapidly induces abnormal centrosome duplication. Cells expressing both the HPV-16 E6 and E7 oncoproteins showed the most pronounced alterations of centrosome numbers, mitotic spindle poles, and genomic integrity. Our results therefore suggest that the high-risk HPV E6 and E7 oncoproteins cooperate to induce centrosome-related mitotic aberrations, resulting in aneuploidy.
Materials and Methods
Cell Lines and Culture.
Normal human keratinocytes (NHKs) from neonatal foreskins were isolated and cultured as described previously (16). The human osteosarcoma cell line U2OS was obtained from the American Type Culture Collection (ATCC) and cultured in DMEM supplemented with 10% FBS, penicillin (50 units/ml), and streptomycin (50 μg/ml).
Inhibition of cdk2 was performed by treatment with 5 μg/ml roscovitine (Calbiochem) for 24 h (17).
Retroviral Infections.
For retroviral infection of normal human keratinocytes, recombinant LXSN- or pBABE-based retroviral constructs expressing HPV-16 or HPV-6 E6 or E7 were used (16, 18, 19).
Cell Transfections.
pCMVneo-based plasmids (20) containing HPV-16 E6, HPV-16 E7, or mutant HPV-16 E7Δ21–24 were used for transient transfections by calcium phosphate coprecipitation (U2OS) (21) or Lipofectamine Plus (Life Technologies, Grand Island, NY) (NHKs) (22). Cells were cotransfected with a vector encoding farnesylatable green fluorescent protein (pEGFP-F; CLONTECH), and GFP-positive cells were analyzed.
Dominant-negative cdk2 (dn-cdk2) (23) or hemagglutinin epitope-tagged dominant-negative DP1 (dn-DP1) (24) was cotransfected with HPV-16 E7 as indicated. Transfection was monitored by immunoblot detection of the expressed proteins.
For stable transfection, U2OS cells were transfected with a pCMVneo-based plasmid containing the HPV-16 E7 gene or empty plasmid, and the recipients were subjected to G418 (Life Technologies) selection. Expression of HPV-16 E7 protein was monitored in individual clones by Western blotting.
Immunological Methods.
Cell lysates were made and analyzed as previously described (16). The antibodies used were p53 (Ab-6, Calbiochem), HPV-16 E7 (ED17, Santa Cruz Biotechnology), cdk2 (M2, Santa Cruz Biotechnology), actin (Chemicon), and hemagglutinin (HA; Roche Molecular Biochemicals).
For immunofluorescence analysis, cells were grown on coverslips, fixed in 4% paraformaldehyde for 20 min, and permeabilized with 0.5% Triton X-100 (Sigma) in PBS for 15 min, both at room temperature. Sections (5 μm) of formalin-fixed, paraffin-embedded tissue samples were deparaffinized in xylene for 15 min, rehydrated through a graded ethanol series, and exposed to microwave radiation three times for 5 min each in 10 mM citrate buffer (pH 6.0). Normal donkey serum (Jackson ImmunoResearch) was used for blocking at a 1:10 dilution. Monoclonal antibody against γ-tubulin (Sigma) was incubated at a dilution of 1:2000 in PBS overnight at 4°C, followed by a rhodamine red-labeled donkey anti-mouse IgG secondary antibody (Jackson ImmunoResearch) at a dilution of 1:100, for 1 h at 37°C. Alternatively, centrosomes were visualized by staining for pericentrin as described previously by Pihan et al. (15), using a polyclonal antibody against pericentrin (Babco, Richmond, CA) at a 1:50 dilution, followed by a rhodamine red-labeled donkey anti-rabbit IgG antibody (Jackson ImmunoResearch) at a 1:2000 dilution for 1 h at 37°C. Nuclei were visualized by using Hoechst 33258 DNA dye. Cells were analyzed by using a Leica DMLB epifluorescence microscope equipped with a multiband filter set (Omega Optical, Brattleboro, VT) and a Sony DKC5000 digital camera, and images were transferred to Adobe PhotoShop for printout.
Electron Microscopy.
Cells were fixed for 1 h in 2.5% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.4), followed by staining with 1% osmium tetroxide and 1.5% potassium ferrocyanide for 1 h and with 1% uranyl acetate in maleate buffer for 30 min; processed for Epon-Araldite embedding; and analyzed with a JEOL 1200EX transmission electron microscope.
Fluorescence in Situ Hybridization.
For interphase fluorescence in situ hybridization analysis, a Spectrum Green-labeled chromosome 11 α-satellite probe (Vysis, Downers Grove, IL) was used according to the manufacturer's protocol. Slides were washed and counterstained with propidium iodide. Six hundred nuclei were evaluated for each cell population studied.
Statistical Methods.
Student's two-tailed t test for independent samples and the χ2 test were used. Mean percentage and standard error of at least three independent experiments and at least 100 cells evaluated per experiment are given unless indicated otherwise.
Results
Centrosome-Associated Defects of Mitotic Spindle Pole Formation in Cervical Lesions.
To investigate whether abnormal mitoses in HPV-associated genital squamous lesions may be associated with centrosome abnormalities, we analyzed tissue specimens obtained from patients with HPV-associated squamous intraepithelial lesions of the cervix, and a specimen from a vulvar squamous cell carcinoma by immunofluorescence staining, using the pericentriolar marker pericentrin (25). The number of mitotic spindle poles of mitotic cells was evaluated. In both preinvasive and invasive genital squamous lesions, mitoses with abnormal centrosomes were detected (Fig. 1A). The most prevalent alteration was a tripolar spindle pole arrangement. Additional abnormalities were individual spindle poles that contained more than one centrosome. Similarly, the HPV-16-positive cervical cancer cell lines SiHa and Caski also showed centrosome abnormalities (data not shown).
Figure 1.
(A) Mitotic figures with abnormal numbers of centrosomes in both preinvasive (squamous intraepithelial lesions of the cervix, SIL) and invasive (vulvar squamous cell carcinoma, SCC) HPV-associated genital squamous lesions. Centrosomes were visualized by immunofluorescence staining for pericentrin. Nuclei were visualized with Hoechst 33258 DNA dye. Pictures were merged to show the arrangement of centrosomes and mitotic figures. Each of the examples shows a tripolar arrangement of spindle poles. In addition, focusing up and down revealed individual spindle poles consisting of more than one centrosome (arrowheads). (B) (Left) Immunoblot analysis revealed decreased levels of p53 in HPV-16 E6-expressing NHKs. (Right) Immunoblot detection of HPV-16 E7 protein in NHKs with stable expression of this protein. (C) Centrosome abnormalities in HPV-16 E6-, E7-, and E6/E7-expressing NHKs. Centrosomes were visualized by immunofluorescence staining for γ-tubulin (arrowheads), and nuclei were visualized with Hoechst 33258 DNA dye. LXSN-vector-infected keratinocytes (NHK) are shown as the control. Pictures were acquired with a multiband filter set. (D) Quantitation of centrosomal abnormalities in high-risk HPV-16 E6-, E7-, E6/E7-, and low-risk HPV-6 E6 or E7-expressing NHK clones. LXSN-vector infected keratinocytes are shown as the control. Bar graphs show the mean ± SEM of at least triplicate centrosome quantitations.
Normal Human Keratinocytes Expressing HPV-16 E6 and E7 Oncoproteins Display Centrosome Abnormalities.
Next we determined whether NHK populations that express the high-risk HPV-16 E6 and E7 oncoproteins (Fig. 1B) show abnormal centrosome numbers. Centrosome numbers were evaluated in NHKs expressing the HPV-16 E6 or E7 oncoproteins either individually or in combination and compared with matched control keratinocytes, using the pericentriolar marker γ-tubulin (26). In these experiments, only interphase cells were evaluated. Centrosome numbers were considered abnormal when more than two centrosomes per cell (Fig. 1C) were present. The proportion of control NHKs that contained abnormal centrosome numbers was 2.5% (Fig. 1D). In NHK populations expressing the HPV-16 E6 or E7 oncogene individually, the proportion of cells showing abnormal centrosome numbers was increased to 8.3% (3.3-fold) and 8.8% (3.5-fold), respectively. This number was even higher in NHK populations expressing both E6 and E7, where 12.0% of the cells displayed abnormal centrosome numbers (4.8-fold increase; Fig. 1D). Abnormal centrosomes were frequently clustered in a juxtanuclear position (Fig. 1C). Many cells with abnormal centrosome numbers also showed an increased nuclear size or were bi- or multinucleated (Fig. 1C).
In contrast, in low-risk HPV-6 E6- or E7-expressing NHKs, the proportion of cells with centrosome abnormalities was similar to that of control cells (2.6% and 3.3%, respectively; Fig. 1D).
Rapid Induction of Abnormal Centrosome Numbers by Expression of HPV-16 E7 but Not the HPV-16 E6 Oncoprotein.
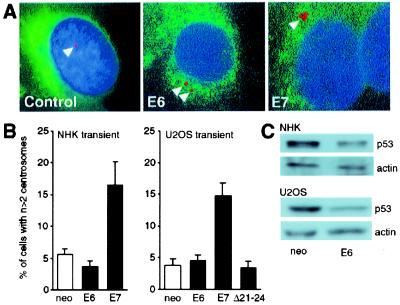
To determine whether abnormal centrosome duplication is an immediate effect of viral oncogene expression, we transiently transfected NHKs with HPV-16 E6 or E7 and analyzed the number of centrosomes in interphase cells at 48 h after transfection (Fig. 2A).
Figure 2.
(A) NHKs were transiently transfected with HPV-16 E6 or E7 and a vector encoding farnesylatable GFP. Only GFP-positive cells were evaluated. Keratinocytes transfected with the parental plasmid were used as the control. Centrosomes were visualized by immunofluorescence staining for γ-tubulin (arrowheads). Nuclei were counterstained with the DNA dye Hoechst 33258. Pictures were obtained with a multiband filter set. (B) Quantitation of centrosomal abnormalities in NHKs and U2OS cells transiently transfected with HPV-16 E6 or E7. Cells transfected with the parental plasmid (neo) were used as the control. Bar graphs show the mean ± SEM of three independent experiments. (C) Decreased p53 levels in response to transient HPV-16 E6 expression in NHKs (Upper) and U2OS cells (Lower). An actin blot is shown to demonstrate equal loading.
In these experiments, 5.6% of NHKs transfected with the parental plasmid used as control and the GFP marker showed abnormal centrosome numbers (Fig. 2B). Acute expression of E6 did not increase the proportion of cells with abnormal centrosome numbers (3.7%), whereas expression of E7 increased the proportion of cells with abnormal centrosome numbers by 2.9-fold to 16.5% (Fig. 2B). Similar results were obtained upon transient transfection of the human osteosarcoma cell line U2OS. Expression of E7 resulted in a 3.9-fold increase (from 3.8% to 14.8%) in cells with centrosome abnormalities, in contrast to E6, which had no significant effect (4.6%) (Fig. 2B). Expression of functional E6 upon transient transfection was documented by decreased steady-state levels of p53 (Fig. 2C). Because both transfected and untransfected cells were analyzed, the differences in the decreases in p53 between U2OS and NHKs likely reflects the differences in transfection efficiencies between the two cell types.
We also studied centrosome numbers in U2OS cells transiently transfected with a transformation-deficient mutant, HPV-16 E7Δ21–24. This mutant is defective for inactivation of pRB (27, 28) and the related pocket proteins p107 and p130. Moreover, it has a severely impaired ability to interact with and inactivate the cdk inhibitor p21Cip1 (16). Centrosome numbers in cells transfected with this E7 mutant were not increased (3.4%) and were similar to those of U2OS cells transfected with the control vector (3.8%) (Fig. 2B). These results indicate that expression of E7 but not E6 directly affects centrosome numbers.
HPV-16 E7 Induces Abnormal Centrosome Duplication.
To investigate whether expression of the HPV E7 oncoprotein induced increased centrosome synthesis and duplication, or caused a defect in centrosome separation, we generated clones of U2OS cells stably expressing HPV-16 E7. In an E7-expressing U2OS clone (Fig. 3A), the proportion of cells with abnormal centrosome numbers was 21.6%. This figure represents a 2.8-fold increase from 7.7% in matched controls (Fig. 3B). Similar results were obtained with a second, independent E7-expressing U2OS clone (data not shown). The E7-expressing U2OS clone and matched controls were then used for ultrastructural analysis of centrosomes by transmission electron microscopy (Fig. 3C). A total of 32 centrioles were examined in control and E7-expressing cells. Procentriole formation was evaluated based on typical morphological characteristics, including a smaller size and diameter, and the position toward the parental centriole (Fig. 3C). In E7-expressing U2OS cells, 7 of the 32 centrioles (21.9%) were procentrioles, compared with 2 procentrioles in a population of 32 (6.3%) in controls. This observation indicates that active duplication of centrioles may be involved in the generation of aberrant centrosome numbers in E7-expressing cells.
Figure 3.
(A) Immunoblot detection of E7 in HPV-16 E7-expressing U2OS cells. (B) Quantitation of centrosomal abnormalities in HPV-16 E7-expressing U2OS cells. Empty vector (neo) transfected cells are shown as the control. Bar graphs show the mean ± SEM of three independent experiments. (C) Ultrastructural analysis of centrosomes in HPV-16 E7-expressing U2OS cells, using transmission electron microscopy. Increased procentriole formation (arrowhead) was observed in E7-expressing U2OS cells compared with matched controls. See text for details. (D) (Left) Treatment of E7-expressing U2OS cells with the cdk2 inhibitor roscovitine decreases the proportion of cells with abnormal centrosome numbers within 24 h. Controls denote E7-expressing U2OS cells treated with solvent (DMSO). (Center) Coexpression of HPV-16 E7 and dominant-negative mutant cdk2 (dn-cdk2) in U2OS cells. (Right) Coexpression of HPV-16 E7 and dominant-negative mutant DP1 (dn-DP1) in U2OS cells. Cell populations transfected with the parental plasmid (neo) are shown as controls. Bar graphs show the mean ± SEM of four independent experiments.
To further support the notion that expression of HPV-16 E7 induces abnormal centrosome synthesis and duplication, we investigated the possibility that interfering with pathways that control centrosome duplication would abrogate the effect of E7. Both the E2F family of transcription factors and cyclin-cdk2 complexes have been implicated in the control of centrosome duplication (12–14, 29). We first determined whether treatment of E7-expressing U2OS cells with the cdk2 inhibitor roscovitine (17) could interfere with abnormal centrosome duplication in E7-expressing cells (Fig. 3D). The proportion of E7-expressing cells with abnormal centrosome numbers was decreased compared with mock-treated cells within 24 h. Expression of a dominant-negative mutant of cdk2 (dn-cdk2) (23) also abrogated the ability of E7 to induce centrosome abnormalities from 11.7% to 3.9%, a value similar to that for mock-transfected cells (4.9%) (Fig. 3D). Cotransfection of U2OS cells with HPV-16 E7 and a dominant-negative mutant of DP1 (dn-DP1) (24), a heterodimerization partner required for the function of E2F, also reduced the number of cells with abnormal centrosomes from 11.7% to 6.3%, close to levels (4.9%) found in mock-transfected U2OS cells (Fig. 3D). Together these results indicate that E7 may affect centrosome synthesis and duplication.
HPV-16 E6 and E7 Oncoproteins Cooperate to Induce Centrosome-Associated Defects of Mitotic Spindle Formation.
Abnormal centrosomes in tumors often retain their ability to nucleate microtubules and assemble abnormal multipolar mitotic spindles (30). To determine whether HPV-16 E6- and E7-induced centrosome abnormalities are associated with aberrant multipolar spindle formation, as we observed in genital squamous lesions (Fig. 1A), we examined metaphases in high-risk HPV oncoprotein-expressing NHKs for the number of participating centrosomes (Fig. 4A). In NHKs expressing the E6 and E7 oncogenes individually, 18% and 9% of metaphase cells, respectively, displayed abnormal numbers of mitotic spindle poles, compared with 6% in control cells. The proportion of metaphase cells with multipolar spindle formation was increased by 8.3-fold to 50% in NHKs expressing both oncogenes, HPV-16 E6 and E7 (Fig. 4B). Many metaphases showed additional alterations, including asymmetrical arrangement of the condensed chromosomes and/or unaligned chromosomal material (Fig. 4A). These changes were most pronounced in E6/E7-expressing cells.
Figure 4.
(A) Abnormal metaphases with multiple participating centrosomes in HPV-16 E6-, E7-, or E6/E7-expressing NHKs. LXSN vector-infected cells are shown as the control. Centrosomes were visualized by immunofluorescence staining for γ-tubulin, and nuclei were visualized with the DNA dye Hoechst 33258. (B) Quantitation of metaphase cells deviant from a bipolar centrosome arrangement in HPV-16 E6-, E7-, or E6/E7-expressing stable clones of NHKs (Left) and U2OS cells transiently transfected with HPV-16 E6, E7, or mutant HPV-16 E7Δ21–24 (Right). LXSN-vector-infected keratinocytes and U2OS cells transfected with the parental plasmid (neo) are shown as controls. The bar graph denotes the result of one representative experiment; at least 100 metaphases were analyzed for each sample. (C) Control anaphase and anaphase of a HPV-16 E6/E7-expressing NHK cell displaying asymmetrical distribution of the condensed chromosomes, a tripolar centrosome arrangement, and one spindle pole with two centrosomes (arrowhead).
To assess whether the formation of aberrant, multipolar metaphase cells reflects an immediate effect of HPV oncogene expression, U2OS cells were transiently transfected with E6, E7, or the transformation-deficient E7Δ21–24 mutant, and the proportion of abnormal metaphases was evaluated 48 h after transfection. In these experiments, expression of E7 induced a 2.9-fold increase from 10% to 29% of multipolar metaphase cells. In contrast, neither E6 nor the transformation-deficient E7Δ21–24 mutant, both of which are unable to affect centrosome numbers in transient assays, had an effect on mitotic spindle pole formation (Fig. 4B).
Similar results were obtained upon transient transfection of NHKs. No centrosome-associated mitotic abnormalities were observed in control and E6-transfected cells, whereas 2.9% of E7-transfected NHKs showed such abnormalities. The lower number most likely reflects the decreased transfection efficiency of these cells.
We then determined the proportion of HPV-16 E6/E7-expressing NHKs progressing into anaphase with aberrant arrangement of the condensed chromosomes (Fig. 4C). A total of 3.2% of cells in anaphase showed abnormalities. Fig. 4C shows a tripolar chromosome arrangement with centrosomes, including one doublet of centrosomes (arrowhead). This observation indicates that some cells with abnormal metaphases can progress through mitosis, thus increasing the risk of daughter cells with chromosomal abnormalities.
Abnormal Centrosome Duplication Induced by HPV Oncoproteins Is Associated with Genomic Instability.
Abnormal centrosome numbers and multipolar spindle formation together with relaxed mitotic checkpoint control increase the propensity for chromosome missegregation and the development of genomic instability (31). To assess the fidelity of chromosome segregation in NHKs stably expressing HPV-16 E6 and/or E7, we used fluorescence in situ hybridization to analyze copy number variations of chromosome 11, which we used as a marker chromosome (Fig. 5A). In NHKs expressing both the E6 and E7 oncoproteins, we found a 4.8-fold increase in cells with chromosome 11 copy numbers deviating from the expected two copies per cell, whereas stable expression of E6 or E7 resulted in 1.8-fold and 2.5-fold increases, respectively (Fig. 5B). Although tri- and tetrasomy were the most prevalent aberrant phenotypes, NHKs expressing both HPV-16 E6 and E7 showed an increased proportion of cells with more than four copies of chromosome 11 (4.8%) compared with E6- or E7-expressing cells (0.6% and 1.8%, respectively).
Figure 5.
(A) Fluorescence in situ hybridization analysis of chromosome 11 in HPV-16 E7-expressing NHKs. Nuclei were counterstained with propidium iodide. Control cells represent matched cultures infected with the parental LXSN vector. (B) Quantitation of chromosome 11 copy numbers in NHKs stably expressing HPV-16 E6, E7, or both viral oncoproteins. LXSN vector-infected keratinocytes are shown as controls.
Discussion
Genomic instability in HPV-associated genital squamous lesions has been reported previously (1, 3). Histopathological analysis of such specimens typically reveals abnormal mitoses, reflecting an increased risk for chromosome missegregation. Here we show that these mitotic abnormalities are associated with abnormal centrosome numbers and that these are caused by the expression of the high-risk HPV E6 and E7 oncoproteins. Expression of high-risk HPV E6 and/or E7 rapidly induces genomic instability in preimmortal normal human cells (5). It has been shown that the integration of the viral genome into the host chromosome, a hallmark of malignant progression, may reflect E6/E7-mediated genomic instability (7). In keratinocytes but not in fibroblasts, expression of E7 was shown to induce aneuploidy (4, 5), which is similar to what we observed in this study (Fig. 5). The molecular basis for the disparity between fibroblasts and keratinocytes is not known, but it has been reported that E7 can overcome G2/M checkpoints in keratinocytes (9), but to a much lesser extent in fibroblasts (32). Our experiments suggest that E6 and E7 cooperate to induce centrosome abnormalities and genomic instability through different mechanisms. Both HPV oncoproteins subvert cell cycle regulatory checkpoints to reactivate and maintain DNA replication competence in differentiating epithelial cells. The HPV-16 E6 oncoprotein inactivates the p53 tumor suppressor protein (8), thus abrogating the arrest of cells at the G1/S transition in response to various cellular insults. In addition, E6 also subverts multiple mitotic checkpoints (9, 32). It is known from previous experiments that loss of p53 or p21Cip1 function results in numeric centrosome abnormalities (33, 34), and it has been proposed that repeated S phase entry and failure to undergo cytokinesis may be involved (34). In our experiments, stable expression of E6, which targets p53 (Fig. 1B), results in centrosome abnormalities (Fig. 1 C and D), phenocopying the loss of p53. Interestingly, however, we did not detect centrosome abnormalities upon acute expression of E6 (Fig. 2B). We interpret this observation to indicate that, at least in our experimental system, abrogation of p53 function through E6 expression does not directly affect centrosome duplication, but allows centrosome abnormalities to develop and accumulate. In contrast, expression of the HPV-16 E7 oncoprotein rapidly induces abnormal centrosome duplication (Figs. 2B and 3). Ultrastructural analysis of centrioles showed evidence for increased procentriole formation in E7-expressing cells. Inhibition of cdk2 and E2F activities, both of which have been implicated in the regulation of centrosome duplication (12–14, 29), abrogates the ability of E7 to induce numeric centrosome abnormalities (Fig. 3D). It is tempting to speculate that the ability of E7 to induce centrosome abnormalities may be related to the ability of this oncoprotein to dysregulate cdk2 and/or E2F activities (16, 35).
Clearly, induction of increased S phase entry by E7 (18) cannot be sufficient to induce centrosome abnormalities as long as centrosome duplication remains coupled to cell division. Possibly relevant for our findings, however, is a report demonstrating that in E7-expressing keratinocytes cyclin E and cyclin A levels are expressed at increased levels throughout the cell division cycle. Cyclin E-associated kinase activity was found to be similarly dysregulated in these cells (35). Hence the centrosome duplication cycle may become effectively uncoupled from the cell division cycle in E7-expressing cells by aberrant expression of normally S phase-specific genes (35, 36). This uncoupling will result in aberrant centrosome numbers. Although our experiments do not directly prove this notion, it would be consistent with our finding that the E7Δ21–24 mutant, which is unable to activate E2F (27), to inactivate the cdk inhibitor p21Cip1 (16), or to increase expression of cyclin E (35), does not induce centrosome abnormalities (Fig. 2B) or multipolar mitoses (Fig. 4B). This model, however, does not rule out the possibility that HPV E7 also targets other components critical for the regulation of the centrosome duplication cycle. Interestingly, an amplification of the chromosomal region encompassing the centrosome-associated kinase, STK15/BTAK, which is involved in centrosome duplication, was reported in HPV E7-expressing cells (6, 37).
In our experiments, coexpression of both HPV-encoded oncogenes, E6 and E7, resulted in an increased proportion of cells with centrosome abnormalities compared with expression of each oncoprotein alone (Fig. 1D). An even more dramatic increase was observed when mitotic spindle poles in metaphases were analyzed with 50% abnormal mitoses in E6/E7-expressing cells (Fig. 4B). This increase shows that coexpression of HPV E6 and E7 as observed in HPV-associated lesions and cancer potentiates mitotic disturbances. However, the proportion of cells with abnormal centrosomes remained relatively constant over time, suggesting that most cells with abnormal metaphases are unable to complete mitosis. Although the fraction of cells that progress into anaphase with a tri- or multipolar chromosome arrangement was relatively small, we were able to detect such abnormalities in 3.2% of HPV-16 E6/E7-expressing keratinocytes. This finding shows that mitotic checkpoint control is sufficiently relaxed in HPV oncogene-expressing cells (9, 32) for some cells with abnormal metaphases to progress through mitosis. Daughter cells derived from abnormal mitoses are likely to have acquired genomic changes that do not confer a growth advantage or may not even be compatible with cellular growth. Regardless, the centrosome-associated mitotic abnormalities induced by E6 and E7 expression in lesions caused by high-risk HPV infections result in the emergence and maintenance of a pool of cells that are prone to malignant progression (38).
Aberrant mitoses with abnormal centrosomes occur already in preinvasive high-risk HPV-associated genital squamous lesions (Fig. 1A). This study strongly suggests that these centrosomal and mitotic abnormalities represent sequelae of HPV E6 and E7 oncogene expression and contribute to genomic instability at a very early stage of high-risk HPV infection. In agreement with this model, expression of low-risk HPV-6 and E7 proteins did not markedly disturb centrosome homeostasis (Fig. 1C). Many cells infected with high-risk HPVs remain clinically unrecognized, and there is often a long latency period after the onset of clinical symptoms and progression to invasive carcinoma (1).
Our results suggest a model whereby the HPV E7 oncoprotein rapidly uncouples centrosome duplication from the cell division cycle and induces increased centrosome duplication and synthesis. The HPV E6 oncoprotein, which does not directly dysregulate centrosome duplication, cooperates with E7 by allowing for cells with abnormal centrosome numbers to accumulate, possibly by relaxing G2/M checkpoint control. The constant presence of a pool of cells that can undergo and complete aberrant mitosis enhances genomic plasticity and increases the likelihood for the emergence of abnormal cells capable of carcinogenic progression.
Acknowledgments
We thank M. Ericsson for performing electron microscopy, R. Kent for valuable assistance with the statistical analysis, D. Galloway and V. Band for retroviral vectors, and P. Hinds and D. Sinclair for critical reading of the manuscript. We are grateful to P. Hinds, N. Dyson, E. Harlow, J. S. Lee, and R. Mulligan for sharing reagents, and E. Kieff for valuable suggestions. This work was supported by National Institutes of Health Grant CA66980 (to K.M.). S.D. is supported by a postdoctoral fellowship from the Deutsche Forschungsgemeinschaft (Du 343/1-1). A.D. is supported by a postdoctoral fellowship from the Dr. Mildred Scheel Stiftung. K.M. is a Ludwig Scholar.
Abbreviations
- HPV
human papillomavirus
- cdk2
cyclin-dependent kinase 2
- NHKs
normal human keratinocytes
- GFP
green fluorescent protein
- dn
dominant-negative
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.170093297.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.170093297
References
- 1.zur Hausen H. Science. 1991;254:1167–1173. doi: 10.1126/science.1659743. [DOI] [PubMed] [Google Scholar]
- 2.Solinas-Toldo S, Dürst M, Lichter P. Proc Natl Acad Sci USA. 1997;94:3854–3859. doi: 10.1073/pnas.94.8.3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heselmeyer K, Schröck E, Du Manoir S, Blegen H, Shah K, Steinbeck R, Auer G, Ried T. Proc Natl Acad Sci USA. 1996;93:479–484. doi: 10.1073/pnas.93.1.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hashida T, Yasumoto S. J Gen Virol. 1991;72:1569–1577. doi: 10.1099/0022-1317-72-7-1569. [DOI] [PubMed] [Google Scholar]
- 5.White A E, Livanos E M, Tlsty T D. Genes Dev. 1994;8:666–677. doi: 10.1101/gad.8.6.666. [DOI] [PubMed] [Google Scholar]
- 6.Reznikoff C A, Belair C, Savelieva E, Zhai Y, Pfeifer K, Yeager T, Thompson K J, DeVries S, Bindley C, Newton M A, et al. Genes Dev. 1994;8:2227–2240. doi: 10.1101/gad.8.18.2227. [DOI] [PubMed] [Google Scholar]
- 7.Kessis T D, Conolly D C, Hedrick L, Cho K R. Oncogene. 1996;13:427–431. [PubMed] [Google Scholar]
- 8.Scheffner M, Werness B A, Huibregtse J M, Levine A J, Howley P M. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 9.Thomas J T, Laimins L A. J Virol. 1998;72:1131–1137. doi: 10.1128/jvi.72.2.1131-1137.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dyson N, Howley P M, Münger K, Harlow E. Science. 1989;243:934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- 11.Boyer S N, Wazer D E, Band V. Cancer Res. 1996;56:4620–4624. [PubMed] [Google Scholar]
- 12.Hinchcliffe E H, Li C, Thompson E A, Maller J L, Sluder G. Science. 1999;283:851–854. doi: 10.1126/science.283.5403.851. [DOI] [PubMed] [Google Scholar]
- 13.Matsumoto Y, Hayashi K, Nishida E. Curr Biol. 1999;9:429–432. doi: 10.1016/s0960-9822(99)80191-2. [DOI] [PubMed] [Google Scholar]
- 14.Lacey K R, Jackson P K, Stearns T. Proc Natl Acad Sci USA. 1999;96:2817–2822. doi: 10.1073/pnas.96.6.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pihan G A, Purohit A, Wallace J, Knecht H, Woda B, Quesenberry P, Doxsey S J. Cancer Res. 1998;58:3974–3985. [PubMed] [Google Scholar]
- 16.Jones D L, Alani R M, Münger K. Genes Dev. 1997;11:2101–2111. doi: 10.1101/gad.11.16.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meijer L, Kim S. Methods Enzymol. 1997;283:113–129. doi: 10.1016/s0076-6879(97)83011-x. [DOI] [PubMed] [Google Scholar]
- 18.Halbert C L, Demers G W, Galloway D A. J Virol. 1991;65:473–478. doi: 10.1128/jvi.65.1.473-478.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soneoka Y, Cannon P M, Ramsdale E E, Griffiths J C, Romano G, Kingsman S M, Kingsman A J. Nucleic Acids Res. 1995;23:628–633. doi: 10.1093/nar/23.4.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baker S J, Markowitz S, Fearon E R, Willson J K, Vogelstein B. Science. 1990;249:912–915. doi: 10.1126/science.2144057. [DOI] [PubMed] [Google Scholar]
- 21.Chen C, Okayama H. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alani R M, Hasskarl J, Grace M, Hernandez M-C, Israel M A, Münger K. Proc Natl Acad Sci USA. 1999;96:9637–9641. doi: 10.1073/pnas.96.17.9637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van den Heuvel S, Harlow E. Science. 1993;262:2050–2054. doi: 10.1126/science.8266103. [DOI] [PubMed] [Google Scholar]
- 24.Wu C L, Classon M, Dyson N, Harlow E. Mol Cell Biol. 1996;16:3698–3706. doi: 10.1128/mcb.16.7.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doxsey S J, Stein P, Evans L, Calarco P D, Kirschner M. Cell. 1994;76:639–650. doi: 10.1016/0092-8674(94)90504-5. [DOI] [PubMed] [Google Scholar]
- 26.Stearns T, Evans L, Kirschner M. Cell. 1991;65:825–836. doi: 10.1016/0092-8674(91)90390-k. [DOI] [PubMed] [Google Scholar]
- 27.Phelps W C, Münger K, Yee C L, Barnes J A, Howley P M. J Virol. 1992;66:2418–2427. doi: 10.1128/jvi.66.4.2418-2427.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones D L, Thompson D A, Münger K. Virology. 1997;239:97–101. doi: 10.1006/viro.1997.8851. [DOI] [PubMed] [Google Scholar]
- 29.Meraldi P, Lukas J, Fry A M, Bartek J, Nigg E A. Nat Cell Biol. 1999;1:88–93. doi: 10.1038/10054. [DOI] [PubMed] [Google Scholar]
- 30.Lingle W L, Lutz W H, Ingle J N, Maihle N J, Salisbury J L. Proc Natl Acad Sci USA. 1998;95:2950–2955. doi: 10.1073/pnas.95.6.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lengauer G, Kinzler K W, Vogelstein B. Nature (London) 1997;386:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 32.Thompson D A, Belinsky G, Chang T H T, Jones D L, Schlegel R, Münger K. Oncogene. 1997;15:3025–3035. doi: 10.1038/sj.onc.1201495. [DOI] [PubMed] [Google Scholar]
- 33.Fukasawa K, Choi T, Kuriyama R, Rulong S, Vande Woude G F. Science. 1996;271:1744–1747. doi: 10.1126/science.271.5256.1744. [DOI] [PubMed] [Google Scholar]
- 34.Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown J P, Sedivy J M, Kinzler K W, Vogelstein B. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 35.Martin L G, Demers G W, Galloway D A. J Virol. 1998;72:975–985. doi: 10.1128/jvi.72.2.975-985.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chellappan S, Kraus V B, Kroger B, Münger K, Howley P M, Phelps W C, Nevins J R. Proc Natl Acad Sci USA. 1992;89:4549–4553. doi: 10.1073/pnas.89.10.4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou H, Kuang J, Zhong L, Kuo W, Gray J W, Sahin A, Brinkley B R, Sen S. Nat Genet. 1998;20:189–193. doi: 10.1038/2496. [DOI] [PubMed] [Google Scholar]
- 38.Cahill D P, Kinzler K W, Vogelstein B, Lengauer C. Trends Cell Biol. 1999;9:M57–M60. [PubMed] [Google Scholar]