Abstract
CIITA is a master transactivator of the major histocompatibility complex class II genes, which are involved in antigen presentation. Defects in CIITA result in fatal immunodeficiencies. CIITA activation is also the control point for the induction of major histocompatibility complex class II and associated genes by interferon-γ, but CIITA does not bind directly to DNA. Expression of CIITA in G3A cells, which lack endogenous CIITA, followed by in vivo genomic footprinting, now reveals that CIITA is required for the assembly of transcription factor complexes on the promoters of this gene family, including DRA, Ii, and DMB. CIITA-dependent promoter assembly occurs in interferon-γ-inducible cell types, but not in B lymphocytes. Dissection of the CIITA protein indicates that transactivation and promoter loading are inseparable and reveal a requirement for a GTP binding motif. These findings suggest that CIITA may be a new class of transactivator.
The major histocompatibility complex (MHC) class II proteins DR, DP, DQ, and the associated molecules DM and invariant chain (Ii) play a central role in the immune response. These proteins present antigenic peptides on the surface of cells for recognition by the T cell receptors of class II-restricted T cells (1). Recently, a novel factor, CIITA, was isolated and shown to be defective in patients afflicted with one class of bare lymphocyte syndrome (BLS) (2). BLS is an often fatal genetic defect, characterized by severe immunodeficiency as a result of failure to express MHC class II genes. The defects fall into at least five complementation groups, all of which act at the level of transcription. Expression of MHC class II molecules in cells deficient in CIITA can be restored by introducing normal CIITA (2–4). In normal cells, CIITA expression is induced by treatment with interferon-γ (IFN-γ) before induction of MHC class II, and CIITA alone is sufficient to induce class II DR expression on the cell surface (3–5). Efficient loading and presentation of peptides also requires coexpression of the DM and Ii proteins. These genes are also regulated by CIITA and, like the MHC class II genes, contain a highly conserved DNA motif S–X–Y (6–8). Furthermore, when B cells terminally differentiate into plasma cells, MHC class II expression is lost concomitantly with CIITA expression (9, 10). CIITA-null mice fail to express MHC class II on their lymphoid cells (11). Thus, CIITA appears to be the key switch for turning MHC class II antigen presentation on or off. Recent evidence has indicated that CIITA may interact with the basal transcription machinery (12), the coactivator BOB (13), and/or the X-box factor RFX5 (14). However, the mode of action of CIITA and its in vivo target have remained elusive.
Changes in promoter assembly or protein/DNA interactions can be detected in intact cells by in vivo genomic footprinting (15). Analysis of the DRA promoter by this technique has revealed interactions at promoter-proximal transcription factor binding sites X and Y in both B lymphocytes and IFN-γ-treated cells (16, 17). The Ii and DMB promoters show similar interactions at the their X and Y sites (18–20). Promoter assembly has a primary dependence on the Y box to initiate factor binding in IFN-γ responsive cells and an interdependence between the X1 and X2 binding activities (21, 22). However, IFN-γ does not induce the X and Y box binding factors, but rather induces expression of CIITA, which does not bind to DNA directly. Importantly, CIITA can serve as a transactivator when artificially tethered to a basal promoter (23, 24). In this report, we demonstrate that CIITA is required for the assembly of transcription factors specifically on the MHC class II promoters and that the promoter assembly function and transactivation are inseparable.
EXPERIMENTAL PROCEDURES
Cell Culture and Transient Transfection.
U373-MG is a glioblastoma multiforme cell line that expresses low basal levels and high IFN-γ-induced levels of MHC class II antigens (25). These cells were maintained in McCoys 5A medium with 10% fetal calf serum (FCS), 2 mM glutamine, 100 units of penicillin, and 100 mg/ml streptomycin. The 2fTGH cell line is a derivative of human HT1080 fibrosarcoma cells (American Type Culture Collection). G3A is a chemically derived mutant of 2fTGH selected for loss of MHC class II expression (26) and it is defective in CIITA expression (3). 2fTGH and G3A cells were grown in DMEM containing 10% FCS as described (26).
Stable Cell Line Transfection and Selection.
Each CIITA expression construct (20 μg) was introduced independently into 5 × 105 G3A cells by standard calcium phosphate precipitation method (3), and the cells were allowed to proliferate for 3 weeks. The cells were then positively selected for MHC class II DR surface expression by using the DR-specific antibody L243 attached to magnetic beads (Dynal, Lake Success, NY). The polyclonal cell population was occasionally monitored for continued DR expression and reselected with magnetic beads. The CIITA expression constructs were positively selected with 250 units/ml hygromycin, beginning 24–36 hr after transfection. Individual resistant colonies were visible after 2–3 weeks, at which time the cells were pooled and analyzed. Cell lines were always maintained in the appropriate drug except when harvested for analysis of chloramphenicol acetyltransferase activity.
Constructs.
The CIITA expression construct pcDNA3.FLAG.CIITA8 contains the CIITA coding sequence driven by a cytomegalovirus promoter in the vector pcDNA3 (Invitrogen) and includes a neomycin resistance marker. An eight amino acid FLAG epitope (DYKDDDDK) is inserted before the first methionine in CIITA. The CIITA expression construct, pREP4.FLAG.CIITA8 is the same as above, except that CIITA is carried in the pREP4 episomal vector (Invitrogen) and, thus, has a hygromycin resistance marker. All of the mutant forms of CIITA used in this report are derivatives of the pREP4.FLAG.CIITA8 construct. These plasmids and the construction of the mutants are described in detail elsewhere (27), except for the RJ-CIITA expression construct, which was described previously (28).
In Vivo Footprinting.
In vivo methylation by dimethyl sulfate of cells and the DNA preparation were as described (29). The ligation-mediated PCR-amplified in vivo genomic footprinting was described originally by Mueller and Wold (15) and modified by Wright and Ting (17). The primers used to reveal the endogenous DRA promoter were as described previously (17). The primer set used to reveal the Ii proximal promoter upper strand have been described (18, 22). The upper strand of the DMB promoter was revealed with the following primer set: DMBIp1, 5′-ATGATCTCCAGACACTGAG-3′; DMBIp2, 5′-ACTGAGCAGAATACTATATTGCCCGGGTC-3′; DMBIp3, 5′-AGCAGAATACTATATTGCCCGGGTCCCTTGAC-3′. The TAP1 promoter region was analyzed in vivo with the R124D primer set as described (30).
RESULTS
Transactivation by CIITA of the MHC class II DRA promoter requires the S–X–Y motifs in B lymphocytes (23). These motifs are also required for activation by CIITA in IFN-γ inducible cell types (K.-C.C., unpublished observation). Because CIITA does not have a recognizable DNA binding motif, it is possible that CIITA interacts directly with transcription factors bound to the S–X–Y motif and not with the DNA. However, these interactions have been difficult to demonstrate in intact cells. Recent evidence from experiments using a yeast one-hybrid system has suggested an interaction between CIITA and recombinant X box factor RFX5 (14). We now address the role of CIITA in a physiologically normal environment, where chromatin may help to regulate the access of factors to the DNA. By using in vivo genomic footprinting, we have evaluated the importance of CIITA for the assembly of factors on the promoters in intact cells.
CIITA Is Required for Promoter Complex Assembly in Vivo.
The fibrosarcoma cell line 2fTGH expresses very low levels of MHC class II on the cell surface, but expression is strongly induced by IFN-γ (3, 26). In vivo genomic footprinting of the MHC class II promoter DRA reveals protein/DNA interactions at the X1, X2, and Y box domains (Fig. 1, lanes 1 and 2). Both guanine residues of the Y box are protected by 50–70%, whereas single guanine residues in the X1 and X2 boxes are protected nearly completely (open arrows). Increased sensitivity of three guanine residues is also revealed (solid arrows). Residues in the lower strand of the X box region are also protected (data not shown). The S box does not show any changes. The same pattern of interactions was detected previously in HeLa and the glioblastoma U373-MG cells and the strength of the interaction is increased upon IFN-γ treatment (17, 32).
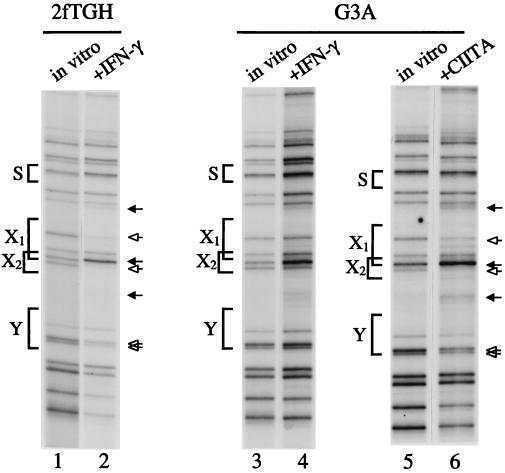
Figure 1.
CIITA mediates in vivo promoter occupancy of MHC class II and associated genes. In vivo footprint of the DRA upper strand in 2fTGH and CIITA-deficient G3A cells. As indicated above each lane, the cells were induced with IFN-γ or stably express CIITA from an integrated transgene. The functional promoter elements are marked on the left. Solid arrows indicate enhancements; open arrows indicate protections; in vitro indicates control, deproteinized methylated genomic DNA in vitro. IFN-γ-induced 2fTGH cells display strong protections and enhancements at the X1 and X2 boxes and substantial protections at the Y box (lanes 1 and 2). Before IFN-γ treatment, the same, but less intense, interactions are observed [data not shown and Wright and Ting (17)]. G3A cells have a bare DRA promoter unless CIITA is introduced (lanes 3–6). In vivo genomic footprinting was done as described on at least three different DNA preparations, analyzed at least twice each. Cells were treated with recombinant IFN-γ at 500 units per ml for 48 hr before harvest.
The in vivo footprint pattern of DNA from a CIITA-negative cell line was analyzed to decipher the effect of CIITA on the assembly of the DRA promoter. G3A is a CIITA-negative mutant cell line derived from 2fTGH (3), and it did not show significant protection over the X1, X2, and Y elements on either the upper strand (Fig. 1, lanes 3 and 4) or the lower strand (data not shown) even after IFN-γ treatment. Stable expression of CIITA into G3A cells restored promoter occupancy in vivo, providing direct evidence that CIITA results in promoter loading in vivo (Fig. 1, lanes 5 and 6). The contacts observed were similar to those in IFN-γ-treated 2fTGH cells and were not increased further by treatment with IFN-γ (data not shown). The site of CIITA integration into the genome did not artifactually cause DRA promoter loading, because identical results were obtained when CIITA was introduced as an episome (see below and Fig. 2C). These findings demonstrate that CIITA is required for transcription factor assembly on MHC class II promoters in response to IFN-γ.
Figure 2.
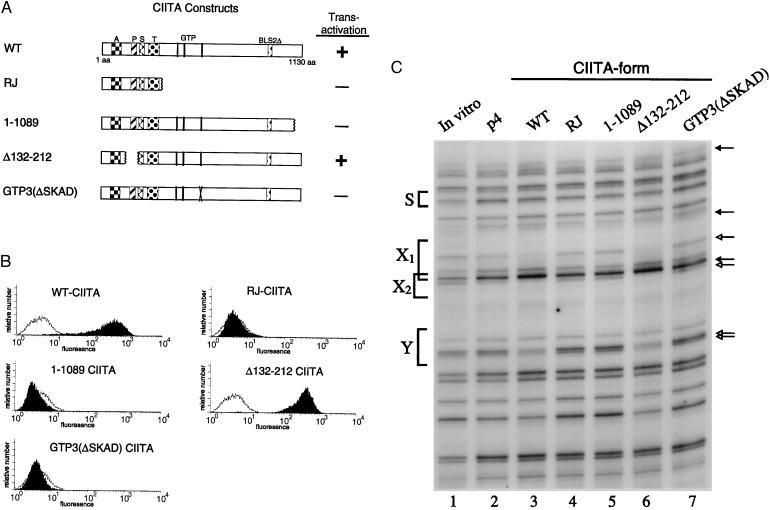
DRA transactivation and promoter assembly in cells expressing mutant forms of CIITA. (A) CIITA mutants. A, acidic domain; P, proline-rich domain; S, serine-rich domain; T, threonine-rich domain; GTP indicates the three homologies to a GTP binding motif; BLS2Δ indicates the region deleted in BLS patient line BLS2. The RJ protein was isolated from the class II-negative cell line RJ2.2.5. Transactivation indicates whether that form of CIITA activated DRA transcription in a cotransient transfection assay. +, Full activation compared with wild-type CIITA; −, absolutely no activity. (B) Cell surface expression of DR in G3A cells stably expressing the indicated form of CIITA was detected by fluorescence-activated cell sorter. Solid profile shows DR expression in the test cells; open profile is the DR expression in G3A cells stably transfected with empty vector. Only the wild-type and Δ132–212 CIITA proteins were able to transactivate. Similar levels of CIITA proteins were expressed in each stably transfected cell line, as detected by Western analysis with the FLAG epitope tag antibody (IBI/Kodak). (C) In vivo footprint analysis of DRA promoter occupancy. Cell lines analyzed are as in B and p4 indicates the cell line with the empty vector. Footprinting and lane markings are as in Fig. 1. Only in the wild-type (lane 3) and Δ132–212 (lane 6) cell lines are protections and enhancements clearly found within the X1, X2, and Y boxes. The other mutant CIITA proteins do not promote factor binding.
In Vivo Promoter Assembly and Transactivation Functions of CIITA Map to the Same Domains of the Protein.
To map the domains of CIITA required for promoter assembly, a series of CIITA mutant proteins were produced. Each was introduced stably into the CIITA-negative G3A cell line and both in vivo promoter assembly and the activation of endogenous MHC class II genes were analyzed. In the initial experiments, four mutant forms of CIITA were examined (Fig. 2A). RJ, isolated from the B lymphoblastoid cell line RJ2.2.5, codes for only the first 336 residues (28). RJ2.2.5 does not express MHC class II because of the defective CIITA, but the DRA promoter is fully occupied in vivo (16). Mutant 1–1089, with a deletion of only the carboxyl-terminal 41 residues, partially disrupts a leucine repeat motif. Mutant Δ132–212 contains an internal deletion of the proline-rich domain and mutant GTP3(ΔSKAD) has a four residue change in a GTP binding motif (27). The four mutant forms of CIITA, the wild-type control, and the empty vector were stably introduced as episomes into G3A cells. Cell surface expression of the MHC class II DR protein was measured in each stable cell line by fluorescence-activated cell sorting. Untransfected G3A cells or G3A cells transfected with the empty vector did not express detectable levels of DR (Fig. 2B). Wild-type CIITA induced an increase of more than 100-fold in cell surface DR expression. Deletion of either the carboxyl-terminal two-thirds of the protein (RJ-CIITA) or just the 41 carboxyl-terminal residues (1-1089) abolished cell surface expression. The proline-rich domain is not required to activate MHC class II expression because Δ132–212 is functional, with a level of cell surface DR similar to that induced by wild-type CIITA. Interestingly, the relatively minor four amino acid change in GTP3(ΔSKAD) completely abolished transactivation by CIITA. The ability of these mutant forms to transactivate the DRA-CAT reporter gene construct in transient assays was also examined. Each form of CIITA that does not activate endogenous gene expression also does not induce transcription in a transient expression assay, whereas both the wild-type and Δ132–212 proteins do activate transcription (ref. 27, and summarized in Fig. 2A). Similar levels of CIITA protein were expressed in each stably transfected cell line, as detected by Western blot analysis (data not shown).
The in vivo promoter occupancy of the DRA gene was examined to determine whether the promoter assembly function of CIITA could be separated from gene activation and DR cell surface expression. Fig. 2C shows the in vivo analysis of the same stable lines as in Fig. 2B. Wild-type and Δ132–212 CIITA proteins, when stably expressed from an episome, induce DRA promoter occupancy at the X1, X2, and Y boxes (compare lanes 3 and 6 with lane 2). This result is consistent with the normal levels of cell surface DR present in these lines. The three CIITA forms that fail to induce cell surface DR are not functional for DRA promoter assembly: none of the protections or enhancements are detected in these lines (lanes 4, 5, and 7). These findings indicate that the requirement for CIITA in transactivation and its requirement for promoter assembly are encoded by overlapping regions within CIITA and are potentially identical.
CIITA also Promotes the Assembly of the Ii and DMB Promoters, but Not the TAP1 Promoter.
The ability of CIITA mutant forms to drive the assembly of factors on the Ii and DMB promoters was examined next. The Ii promoter consists of a distal domain containing the S–X–Y conserved elements and a proximal domain containing a GC box and second Y box. The lower strand of the distal Ii domain is displayed in Fig. 3A. Strong protections were observed within the X box when wild-type CIITA was introduced into CIITA-deficient G3A cells (lanes 2 and 3). The two guanine residues in the Y box were also substantially protected. Similar to the activity of CIITA on the DRA promoter, the RJ, 1–1089, and GTP3(ΔSKAD) mutant proteins do not promote factor binding to the S–X–Y domain of Ii (lanes 4, 5, and 7). The proline-rich deletion Δ132–212, however, was competent to activate Ii promoter loading in the distal region (lane 6). Ii also uses a second promoter-proximal domain for full transcriptional activation. A direct correlation was observed between occupancy of the distal region and the proximal region. The proximal region GC and second Y boxes were only occupied when either the wild-type or Δ132–212 CIITA proteins were present (data not shown). The three transcriptionally inactive forms of CIITA failed to promote factor binding. These findings indicate that CIITA is required for the assembly of factors on the Ii promoter, not only on the conserved S–X–Y domain but also across the entire 270 bp promoter.
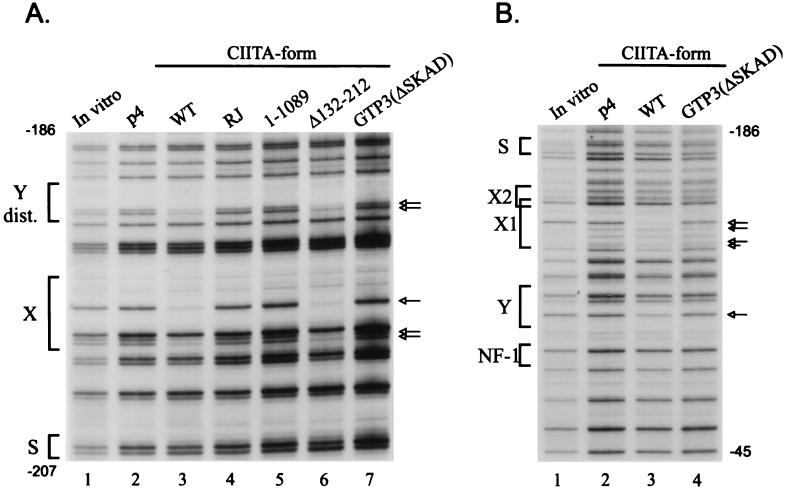
Figure 3.
Invariant chain and DMB promoter occupancy in cells expressing mutant CIITA proteins. (A) Invariant chain distal promoter region. Strong protections of the X and Y elements are seen in the cells expressing wild-type and Δ132–212 CIITA proteins (lanes 3 and 6). The contacts are the same as shown previously in B cells (18, 19) and were confirmed at least four times. (B) DMB promoter region. G3A cells expressing wild-type or GTP3(ΔSKAD) CIITA or the empty vector p4 were examined by in vivo footprinting of the DMB promoter upper strand. The strongest protections are on the X1 box at nucleotide −136 and on the Y box at nucleotide −99. The lane markings are as described in Fig. 2C.
The DMB promoter was also examined to test the importance of CIITA for promoter assembly of nonclassical MHC class II genes. This promoter has a similar S–X–Y domain (20, 33). In vivo footprinting of the upper strand of the DMB promoter revealed that expression of either wild-type or Δ132–212 CIITA were necessary to observe occupancy of the X and Y domains in G3A cells (Fig. 3B, lanes 2 and 3). Similar to the DRA and Ii promoters, the transcriptionally inactive forms of CIITA (lane 4) did not induce assembly of transcription factors on the DMB promoter. These findings indicate that CIITA is required for transcription factor assembly on all of the MHC class II and associated promoters.
To exclude the possibility that CIITA serves as a general chromatin derepressor or nonspecifically disrupts chromatin structure at the MHC class II locus, thus allowing sequence-specific transcription factors to bind, we examined the TAP1 promoter. TAP1, an unrelated gene located within the MHC class II locus, is not induced by CIITA, but is activated by IFN-γ (31). Studies in our laboratory have characterized this promoter and demonstrated both functional GC boxes and NF-κB sites (30). The NF-κB site is occupied only after activation of NF-κB, for example by tumor necrosis factor-α. In addition, the TAP1 promoter contains an interferon response factor element, which binds to IRF-1 in response to IFN-γ (34–36). In the absence of CIITA, the GC box of the TAP1 promoter was clearly occupied (Fig. 4, lanes 1 and 2). Neither the NF-κB site nor the interferon response factor element site were occupied, because the cells were not treated with either IFN-γ or tumor necrosis factor-α. CIITA does not alter TAP1 promoter occupancy (lane 3). These observations demonstrate that the ability of CIITA to activate promoter assembly is specific for MHC class II and associated genes.
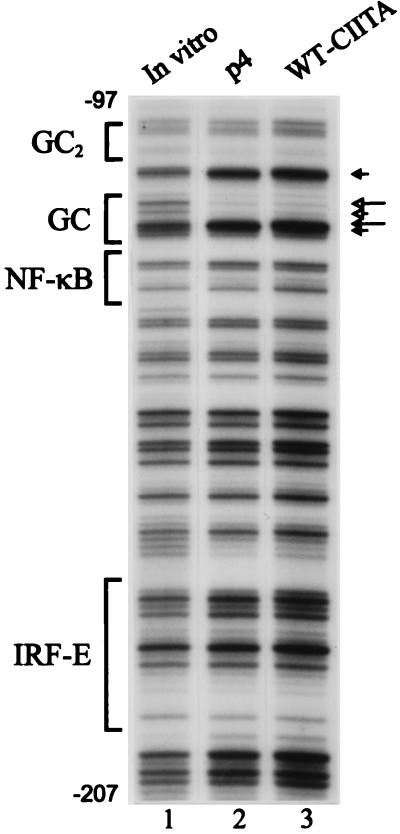
Figure 4.
CIITA does not effect promoter occupancy of the MHC-encoded, nonclass II gene TAP1. The same cell lines analyzed in Fig. 2 were examined for TAP1 promoter binding in vivo. Only the GC box is occupied in these cells, and the in vivo footprint is not altered by CIITA (lanes 2 and 3). The interferon response factor element is not occupied before or after introduction of CIITA. The contacts shown here are similar to those previously reported in HeLa cells (30).
DISCUSSION
The MHC class II gene family is coordinately regulated, and expression is restricted to a small group of antigen-presenting cells. Each of the promoters contains the highly conserved trimeric motif S–X–Y, which is required for expression. Extensive studies of these promoter elements has led to the characterization and/or cloning of the transcription factors bound to them (6, 8, 37). However, these factors do not account for the cell type-specific control and cytokine induction of MHC class II expression. The novel transactivator CIITA mediates MHC class II activation (2–4). Although it has been clearly demonstrated that CIITA is required for MHC class II gene transcription, the mode of its action has remained elusive. The findings presented here now demonstrate that CIITA is absolutely required for the specific assembly of MHC class II promoter complexes in response to IFN-γ, but not on other promoters in the region. This agrees with the suggestion of a previous study that showed a correlation between the lack of CIITA in thymic epithelial cells and a closed promoter (38). In vivo footprinting has been invaluable for understanding gene regulation in living cells. We and others have used it to examine the MHC class II promoters (16, 17). In B cells, which express the MHC class II genes constitutively, strong occupancy of the X1, X2, Y, and octamer sites is observed. In untreated IFN-γ-inducible cells, the X1 and X2 boxes are occupied only weakly, but then become fully occupied after IFN-γ treatment (17). In vivo binding at the Ii promoter is also activated by IFN-γ, but before induction the Ii promoter is completely bare (18).
The role of CIITA in activating MHC class II promoter loading was analyzed in the IFN-γ-inducible cell line 2fTGH and in the mutant cell line G3A in which CIITA is not expressed. In the parental cell, CIITA expression alone was sufficient to promote strong occupancy of the X1, X2, and Y boxes. In the CIITA-deficient G3A cells, the DRA, Ii, and DMB promoters are bare and introduction of CIITA reconstituted normal promoter loading. This result is surprising and contrasts with findings where MHC class II-negative B cells with defective CIITA expression have occupied MHC class II promoters in vivo. Based on the B cell data, it is the general belief that CIITA does not alter promoter assembly. Indeed, examination of the B cell lines RJ2.2.5, RM3, and BLS-2, each members of the CIITA-defective complementation group, reveals a fully occupied DRA promoter (16) with only a minimal alteration in affinity (39). The Ii promoter is also fully loaded with its transcription factors in RJ2.2.5 cells (M.L., unpublished observations). RJ2.2.5 is now known to have a severe deletion of CIITA that prevents transactivation. When the deleted form of CIITA from RJ2.2.5 was expressed stably in G3A cells, there was no enhancement of DRA, Ii, or DMB promoter loading and no transcriptional activation. This demonstrates that the DRA promoter assembly differences observed between G3A and RJ2.2.5 are not due to expression of this deleted CIITA protein in RJ2.2.5. Thus, in cells that require IFN-γ to induce MHC class II transcription, CIITA is necessary to assemble the promoter complex. However, in B cells with constitutive MHC class II transcription, loss of CIITA does not lead to a loss of promoter assembly, only to a loss of transcription. This finding clearly indicates a difference between how B cells and IFN-γ-inducible cells assemble the MHC class II promoter. Importantly, it also defines a previously undescribed function of CIITA in promoter complex assembly.
Previous reports from our laboratory and others have also shown a difference between promoter assembly in B cells and IFN-γ-inducible cells (21, 22, 38, 40). Our findings with the inducible cells demonstrated a complete dependence on the NF–Y/Y box interaction for any of the other interactions to occur in vivo. However, by using the same methodology, B cells displayed no interdependence for binding among the DRA promoter factors. One possibility is that binding of the B cell factor OTF-2 on the DRA promoter modifies the need for CIITA to drive promoter occupancy. This would be consistent with a recent report showing that the OTF coactivator BOB functions synergistically with CIITA, even in the absence of an OTF-2 DNA binding site (13). Alternatively, the X or Y box factors may be modified differently in the two cell types and thus may have different requirements for CIITA. Recent results have shown that the X box factor RFX5 is phosphorylated. However, CIITA does not appear to alter the overall phosphorylation state of RFX5 (41). Thus, CIITA has a dual role in MHC class II-inducible cells in assembling factors on the promoters and in transactivating, whereas in B lymphocytes only transactivation is required.
Homology studies of the CIITA protein have yielded few clues to its activity. However, a putative GTP binding domain consisting of three motifs has been identified and mutated, and each motif has been found to be critical for transactivation (27). This is an intriguing observation because only a few transactivators have been shown to bind ATP and none bind GTP. The Swi/Snf complex, which is functionally and physically associated with the polymerase II holoenzyme, requires ATP, disrupts chromatin, and activates transcription (42). Swi/Snf appears to be recruited to all functional promoters through polymerase II, but affects only some promoters. Conversely, CIITA represents a highly specific regulator only of genes in the MHC class II pathway. A few other transactivators, such as GAGA, the glucocorticoid receptor, and HNF-3-like factors have been shown to disrupt the local structure of chromatin, allowing binding of additional transcription factors (43). However, in contrast to CIITA, each of these factors has an intrinsic DNA binding activity and functions by direct association with the DNA of the promoter.
This report shows that CIITA is required in vivo for promoter assembly in IFN-γ-inducible cells, but not in B cells. In both cell types, CIITA is absolutely required for transactivation. The structure and mode of action of CIITA suggest that it may define a new class of transcriptional coactivators.
ABBREVIATIONS
- MHC
major histocompatibility complex
- BLS
bare lymphocyte syndrome
- IFN-γ
interferon-γ
References
- 1.Schwartz R H. Annu Rev Immunol. 1985;3:237–261. doi: 10.1146/annurev.iy.03.040185.001321. [DOI] [PubMed] [Google Scholar]
- 2.Steimle V, Otten L A, Zufferey M, Mach B. Cell. 1993;75:135–146. [PubMed] [Google Scholar]
- 3.Chin K-C, Mao C, Skinner C, Riley J L, Wright K L, Moreno C S, Stark G R, Boss J M, Ting J P-Y. Immunity. 1994;1:687–697. doi: 10.1016/1074-7613(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 4.Chang C H, Fontes J D, Peterlin M, Flavell R A. J Exp Med. 1994;180:1367–1374. doi: 10.1084/jem.180.4.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steimle V, Siegrist C A, Mottet A, Lisowska-Grospierre B, Mach B. Science. 1994;265:106–109. doi: 10.1126/science.8016643. [DOI] [PubMed] [Google Scholar]
- 6.Glimcher L H, Kara C J. Annu Rev Immunol. 1992;10:13–49. doi: 10.1146/annurev.iy.10.040192.000305. [DOI] [PubMed] [Google Scholar]
- 7.Cogswell J P, Austin J, Ting J P. J Immunol. 1991;146:1361–1367. [PubMed] [Google Scholar]
- 8.Benoist C, Mathis D. Annu Rev Immunol. 1990;8:681–715. doi: 10.1146/annurev.iy.08.040190.003341. [DOI] [PubMed] [Google Scholar]
- 9.Dellabona P, Latron F, Maffei A, Scarpellino L, Accolla R S. J Immunol. 1989;142:2902–2910. [PubMed] [Google Scholar]
- 10.Silacci P, Mottet A, Steimle V, Reith W, Mach B. J Exp Med. 1994;180:1329–1336. doi: 10.1084/jem.180.4.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang C-H, Guerder S, Hong S-C, van Ewijk W, Flavell R A. Immunity. 1996;4:167–178. doi: 10.1016/s1074-7613(00)80681-0. [DOI] [PubMed] [Google Scholar]
- 12.Fontes J D, Jiang B, Peterlin B M. Nucleic Acids Res. 1997;25:2522–2528. doi: 10.1093/nar/25.12.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fontes J D, Jabrane-Ferrat N, Toth C R, Peterlin B M. J Exp Med. 1996;183:2517–2521. doi: 10.1084/jem.183.6.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scholl T, Mahanta S K, Strominger J L. Proc Natl Acad Sci USA. 1997;94:6330–6334. doi: 10.1073/pnas.94.12.6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mueller P R, Wold B. Science. 1989;246:780–786. doi: 10.1126/science.2814500. [DOI] [PubMed] [Google Scholar]
- 16.Kara C J, Glimcher L H. Science. 1991;252:709–712. doi: 10.1126/science.1902592. [DOI] [PubMed] [Google Scholar]
- 17.Wright K L, Ting J P. Proc Natl Acad Sci USA. 1992;89:7601–7605. doi: 10.1073/pnas.89.16.7601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wright K L, Moore T L, Vilen B J, Brown A M, Ting J P-Y. J Biol Chem. 1995;270:20978–20986. doi: 10.1074/jbc.270.36.20978. [DOI] [PubMed] [Google Scholar]
- 19.Brown A M, Wright K L, Ting J P. J Biol Chem. 1993;268:26328–26333. [PubMed] [Google Scholar]
- 20.Ting J P-Y, Wright K L, Chin K-C, Brickey W J, Li G. J Immunol. 1997;159:5457–5462. [PubMed] [Google Scholar]
- 21.Wright K L, Vilen B J, Itoh-Lindstrom Y, Moore T L, Li G, Criscitiello M, Cogswell P, Clarke J B, Ting J P-Y. EMBO J. 1994;13:4042–4053. doi: 10.1002/j.1460-2075.1994.tb06721.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Linhoff M W, Wright K L, Ting J P-Y. Mol Cell Biol. 1997;17:4589–4596. doi: 10.1128/mcb.17.8.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou H, Glimcher L H. Immunity. 1995;2:545–553. doi: 10.1016/1074-7613(95)90034-9. [DOI] [PubMed] [Google Scholar]
- 24.Riley J L, Westerheide S D, Price J A, Brown J A, Boss J M. Immunity. 1995;2:533–543. doi: 10.1016/1074-7613(95)90033-0. [DOI] [PubMed] [Google Scholar]
- 25.Basta P V, Sherman P A, Ting J P. J Immunol. 1987;138:1275–1280. [PubMed] [Google Scholar]
- 26.Mao C, Davies D, Kerr I M, Stark G R. Proc Natl Acad Sci USA. 1993;90:2880–2884. doi: 10.1073/pnas.90.7.2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chin K-C, Li G X, Ting J P-Y. Proc Natl Acad Sci USA. 1997;94:2501–2506. doi: 10.1073/pnas.94.6.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown J A, He X-F, Westerheide S D, Boss J M. Immunogenetics. 1996;43:88–91. doi: 10.1007/BF00186611. [DOI] [PubMed] [Google Scholar]
- 29.Pfeifer G P, Tanguay R L, Steigerwald S D, Riggs A D. Genes Dev. 1990;4:1277–1287. doi: 10.1101/gad.4.8.1277. [DOI] [PubMed] [Google Scholar]
- 30.Wright K L, White L C, Kelly A, Beck S, Trowsdale J, Ting J P-Y. J Exp Med. 1995;181:1459–1471. doi: 10.1084/jem.181.4.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin B K, Chin K-C, Skinner C A, Olsen J C, Dey A, Ozato K, Ting J P-Y. Immunity. 1997;6:591–600. doi: 10.1016/s1074-7613(00)80347-7. [DOI] [PubMed] [Google Scholar]
- 32.Kara C J, Glimcher L H. J Immunol. 1993;150:4934–4942. [PubMed] [Google Scholar]
- 33.Boss J M. Curr Opin Immunol. 1997;9:107–113. doi: 10.1016/s0952-7915(97)80166-5. [DOI] [PubMed] [Google Scholar]
- 34.Pine R, Decker T, Kessler D S, Levy D E, Darnell J E J. Mol Cell Biol. 1990;10:2448–2457. doi: 10.1128/mcb.10.6.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miyamoto M, Fujita T, Kimura Y, Maruyama M, Harada H, Sudo Y, Miyata T, Taniguchi T. Cell. 1988;54:903–913. doi: 10.1016/s0092-8674(88)91307-4. [DOI] [PubMed] [Google Scholar]
- 36.White L C, Wright K L, Felix N J, Ruffner H, Reis L F L, Pine R, Ting JP-Y. Immunity. 1996;5:365–376. doi: 10.1016/s1074-7613(00)80262-9. [DOI] [PubMed] [Google Scholar]
- 37.Ting J P-Y, Baldwin A S. Curr Opin Immunol. 1993;5:8–16. doi: 10.1016/0952-7915(93)90074-3. [DOI] [PubMed] [Google Scholar]
- 38.Rigaud G, Barbaro A D, Nicolis M, Cestari T, Ramarli D, Riviera A-P, Accolla R S. J Immunol. 1996;156:4254–4258. [PubMed] [Google Scholar]
- 39.Rigaud G, Paiola F, Accolla R S. Eur J Immunol. 1994;24:2415–2420. doi: 10.1002/eji.1830241023. [DOI] [PubMed] [Google Scholar]
- 40.Kara C J, Glimcher L H. EMBO J. 1993;12:187–193. doi: 10.1002/j.1460-2075.1993.tb05644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moreno C S, Rogers E M, Brown J A, Boss J M. J Immunol. 1997;158:5841–5848. [PubMed] [Google Scholar]
- 42.Kwon H, Imbalzano A N, Khavari P A, Kingston R E, Green M R. Nature (London) 1994;370:477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- 43.Pina B, Bruggemeier U, Beato M. Cell. 1990;60:719–731. doi: 10.1016/0092-8674(90)90087-u. [DOI] [PubMed] [Google Scholar]