Abstract
The correlates for protection against influenza infection are incompletely characterized. We have applied an ELISA strategy that distinguishes antibodies against native viral surface antigens (potentially neutralizing) from antibodies directed against internal and denatured viral proteins (not neutralizing) to three groups of vaccinated subjects: (1) participants in a study of repeated annual vaccination (2) elderly subjects and (3) patients with Systemic Lupus Erythematosus compared to control subjects. Antibody increase after vaccination was inversely related to the level of pre-existing antibodies in all groups; most subjects had significant initial antibody levels and showed little increase in amount of antibody after vaccination, but the avidity of their serum antibodies tended to increase. Antibodies against denatured virus proteins varied with vaccine formulation; vaccines that are more recent have less total protein for the same amount of native hemagglutinin. We propose an index consisting of rank order of antibody level plus antibody avidity, both measured against native virus, plus hemagglutination-inhibition antibody titer, as a useful measure of immunity against influenza.
1. Introduction
Protection against influenza virus infection is primarily mediated by neutralizing antibodies directed against the major surface antigen, hemagglutinin (HA). Most protective antibodies inhibit binding of the viral HA to sialic acid receptors on host cells, or on red blood cells as measured by the hemagglutination-inhibition (HAI) assay. Antibodies that inhibit NA or M2 (ion channel) activity are also protective, but are of low abundance. Antibodies against internal viral proteins do not neutralize virus and so have no capacity to protect against infection. Antibodies against denatured HA or NA have been reported to neutralize virus, but there is no structural evidence that any of them bind to the native protein [1],
HAI titer is often used as a surrogate marker of protection and increase in HAI titer after immunization has been considered an approximate measure for predicting influenza vaccine efficacy. However, there is not a clear correlation between HAI titer and virus-specific antibodies as measured by ELISA [2], or between HAI titer and protection [3, 4]. Subjects with significant pre-existing HAI titers showed little increase in HAI titer with repeated annual vaccination but appeared to be protected against infection [4, 5].
The levels of virus-specific antibodies measured by ELISA are always greater than those measured either by HAI tests or in neutralization assays. It has been proposed that the difference lies in low avidity antibodies, which bind well enough to be measured in ELISA but not enough to inhibit hemagglutination [6]. However, many "binding but not neutralizing" antibodies may be directed to denatured or other inactive forms such as monomers of glycoprotein. Such antibodies do not offer protection since they do not bind native virions and so cannot inhibit functions [7]. Other non-neutralizing antibodies are against the internal components of the virus, inaccessible to antibody in the infectious particle or infected cell and so not protective.
Non-neutralizing antibodies can be considered to fall into three classes: (i) a memory response against glycoproteins (primarily HA) of earlier strains of influenza, (ii) a response against denatured viral proteins or against the conserved but not protective internal proteins of the virus, or (iii) a response specific for HA, but of too low avidity for neutralization [8]. Non-neutralizing antibodies may help clear viral components via Fc receptors but do not prevent infection.
Influenza vaccines are formulated to contain 15 µg of native HA of each of the three components (H3N2, H1N1 and type B) as measured by single radial immunodiffusion, but the amount of denatured HA is unknown. We have used a sandwich ELISA that separately measures antibodies against native surface antigens and antibodies against denatured viral protein epitopes, referred to as unfoldons [7], in three groups of vaccinated subjects [8, 9]. We analyzed serum samples from a longitudinal trial of inactivated whole-virus influenza vaccine efficacy carried out at Baylor College of Medicine (BCM) from 1983 to 1987 [4, 5], a study of subunit vaccine in the elderly, also carried out at BCM [10], and a study of lupus patients and controls, being conducted at Oklahoma Medical Research Foundation that is still ongoing. We have measured antibodies against the H3N2 component of the vaccine and against an older H3N2 virus for each of these studies.
2. Materials and Methods
2.1 Viruses
Stocks of high-growth, egg-adapted vaccine strains of influenza H3N2 reassorted with PR8 were obtained from the Centers for Disease Control and Prevention, Atlanta, GA. Viruses were propagated in 11-day-old embryonated chicken eggs and purified by sedimentation out of the allantoic fluid followed by 10–40% sucrose density gradient centrifugation. Hemagglutinin titrations were done at 4°C, using human red blood cells. Viral protein was determined by the Bradford method (BioRad Protein Assay kit).
2.2 ELISA to quantitate antibodies against native glycoproteins
Red cell ghosts [11] were solubilized with β-octylpyranoside and used to coat wells of ELISA plates for the sandwich assay described previously [9]. Virus (~64 HAU per 50 µl in CaMg-saline) was captured on the coated wells overnight at 4°C (to inhibit NA activity) and non-specific binding sites were blocked with 20% calf serum in PBS. The human serum samples were diluted 1:16 in 20% calf serum in PBS, and serial two-fold dilutions added to the wells and incubated at room temperature for 1 h. After washing in PBS, alkaline-phosphatase-conjugated goat anti-human polyvalent immunoglobulin (α, γ and μ-chain specific) secondary antibody (Sigma) was bound for an hour, the wells washed free of unbound conjugate and p-nitrophenyl phosphate substrate (Sigma104) added. The color was developed at room temperature for 30 min and absorbance was read at 405 nm.
2.3 ELISA to measure antibodies against denatured proteins (unfoldons)
On a duplicate plate, virus was adsorbed to the red cell membrane-coated wells as above so that the starting amount of virus was the same as for the native protein ELISA. After washing, the wells were treated with methanol for 45 min at 37°C to denature the virus particles and dried before adding serum samples to quantify antibodies against unfoldons. We previously confirmed the native and denatured states of virus in these two assays using a conformation-specific monoclonal antibody that binds only native NA and an anti-peptide antiserum that binds only denatured NA [8]. NP is only detected after methanol treatment.
2.4 Data analysis
Binding curves (absorbance at 405 nm versus µl serum in 50 µl binding mix) were subjected to curve fitting using Kaleidagraph or Prism software as described earlier [8]. The equation used for curve fitting is:
where A405 is the absorbance reading, Amax is the relative amount of anti-influenza antibody in the serum, P0 is the total antibody paratope concentration, P1 is non-specific binding, and Ka(app) is the apparent overall association constant of the serum antibodies. In measuring antibodies in human serum, the “antibody paratope concentration” is defined as volume of serum in the 50 ul binding mix at each point of the binding curve.
3. Results and Discussion
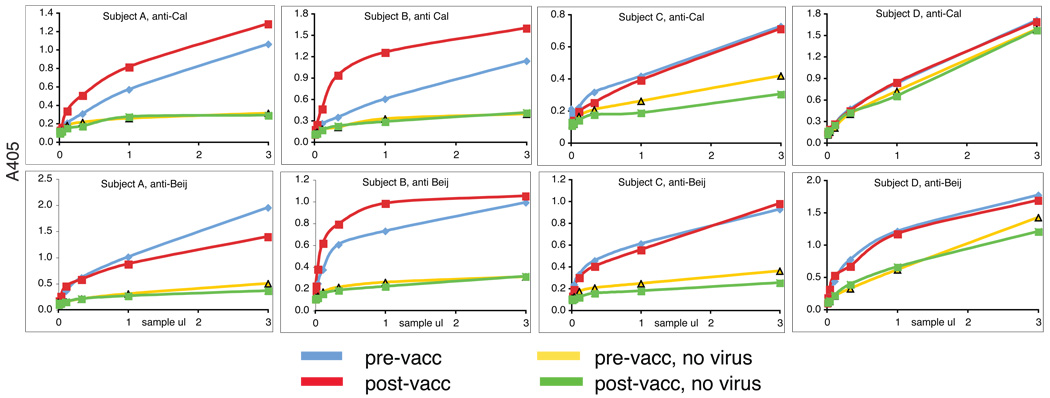
Purified HA or NA is unfolded by adsorption to wells on an ELISA plate, losing biological activity and conformational, neutralizing epitopes, and exposing internal proteins such as matrix (M1) and nucleoprotein (NP). Adsorbed whole virus is partially denatured, so titration of serum samples on virus that has been coated directly onto plastic will detect all anti-influenza antibodies, against external, internal, native and denatured viral proteins, but in unknown proportions [7, 12, 13]. Since only those antibodies, which recognize native, external proteins are neutralizing, we have used a sandwich ELISA technique to measure antibodies against native antigen structure, capturing virus by binding it to sialylated glycans. Only virions containing native HA will attach, and multivalent binding allows the resulting complex to withstand the washing procedures. For studies with the 1980s vaccines [5] we used the glycoprotein fetuin to coat the wells [8], but recent H3N2 viruses have low affinity for fetuin and are washed off. We replaced fetuin with a detergent extract of human erythrocyte ghosts for studies with later viruses [9]. Captured virus, either native, or denatured with methanol, was titrated with serum dilutions to generate a binding curve, and curve-fitting with Kaleidagraph or Prism software yielded the amount of influenza-specific antibody (Amax) and the overall apparent affinity constant, Ka (app), hereafter called Ka. Figure 1 shows some examples of binding curves for antibodies from 4 subjects in the OMRF study, measured with the vaccine virus (A/California/7/2004) and with an older H3N2 virus A/Beijing/89, showing the different levels of pre-existing antibodies and different responses to vaccination. In addition to the patterns of reactivity seen in Figure 1, there is an occasional subject with insignificant pre-vaccination antibody levels and no significant response to the vaccine.
Figure 1. Binding patterns seen among vaccinated subjects.
These results are native ELISAs for four control subjects in the lupus study. The vaccine used contained A/California/7/2004 as the H3N2 component. For an older virus, we used A/Beijing/353/89 because all the subjects were born before 1989.
Top panel: antibodies against California/04. Lower panel: antibodies against Beijing/89. Subject A shows moderate increase of anti-Calif/04 antibodies after vaccination, with no increase against “old” virus Beijing/89.
Subject B shows moderate levels of pre-existing antibodies against California/04 but they are of low affinity. Good response to vaccination in both amount and avidity. Some increase in affinity of antibodies against Beijing/89 after vaccination.
Subject C shows high level of pre-existing antibodies. No increase after vaccination
Subject D shows high levels of non-specific antibodies. No increase after vaccination
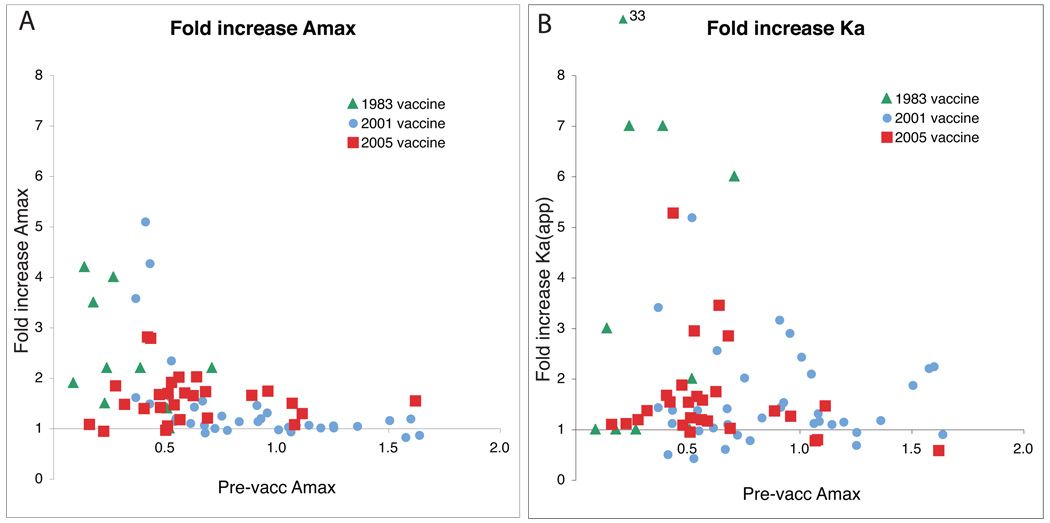
Table 1 shows the average fold increase in Amax and Ka after vaccination for the three studies. The average increase is less than 2-fold in all but the 1980’s study. The reason is that many of the subjects had significant levels of antibodies against the H3N2 vaccine virus before vaccination. When the fold increases for the individuals in the three studies are plotted against the pre-vaccination level of antibody, a clear inverse relationship is seen (Figure 2). Subjects who already have high levels of antibody rarely make any more; there is little increase in antibody amount (Amax, Figure 1A) although the avidity of their antibodies (Ka) may increase (Fig 1B).
Table 1.
Fold increase in antibody amount (Amax) or antibody avidity (Ka) after vaccination
| Study | Virus in vaccine and assay |
Type of vaccine |
Dose of H3 (µg) |
Antibody fold increase | |
|---|---|---|---|---|---|
| Amax | Ka(app) | ||||
| BCM Five year study, yr 1 [8] |
Philippines/82 | Whole virus |
15 | 2.4 ± 1.1 | 6.2 ± 9.8 |
| BCM Elderly [9] | Panama/99 | subunit | 15 | 1.4 ± 0.8 | 1.4 ± 0.6 |
| BCM Elderly [9] | Panama/99 | subunit | 30 | 1.4 ± 0.4 | 1.9 ± 1.4 |
| BCM Elderly [9] | Panama/99 | subunit | 60 | 1.8 ± 1.5 | 1.7 ± 0.9 |
| OMRF yr1 controls | California/04 | subunit | 15 | 1.6 ± 0.5 | 1.6 ± 1.0 |
| OMRF yr1 cases | California/04 | subunit | 15 | 1.4 ± 0.4 | 1.2 ± 0.4 |
| OMRF yr2 controls | Wisconsin/05 | subunit | 15 | 1.2 ± 0.2 | 1.4 ± 0.5 |
| OMRF yr2 cases | Wisconsin/05 | subunit | 15 | 1.2 ± 0.2 | 1.1 ± 0.4 |
Figure 2. Fold increase in Amax is inversely related to pre-vaccination levels of antibodies.
Panel A plots fold increase in Amax against pre-vaccination Amax for the three studies.
Panel B is the same plot for fold increase in Ka. It shows that subjects with higher pre-vaccination levels of antibody may show an increase in avidity after vaccination, even though there is no increase in antibody amount.
We wondered if the lack of increase of antibodies when pre-vaccination levels were high was due to an exaggerated response against denatured antigens. Table 2 shows Amax for antibodies against unfoldons as a percentage of antibodies against native protein. There is considerable variation in the proportion of anti-unfoldon antibodies between the different groups. The serum samples from the oldest study (Philippines/82 and Leningrad/86 H3N2 vaccine components) show very low levels of anti-unfoldon antibodies compared to the later groups. There are two possible reasons for this difference. Firstly, the vaccine in the 1980s was inactivated whole virus and since its manufacture involves fewer steps of processing, the whole virus vaccine might be less denatured during production than the subunit vaccines. Secondly, the initial antibody levels were lower in this group (clustered on the left of Fig 2A), perhaps because vaccination was much less common in the 1980s and the only source of pre-vaccination antibodies may have been previous infection.
Table 2.
Antibodies against unfoldons compared to antibodies against native surface antigens after immunization (unfoldons/native, percent ± standard deviation)
| Study | Virus in vaccine and assay |
Type of vaccine |
Dose of H3 (µg) |
Unfoldons/Native (percent±SD) |
|---|---|---|---|---|
| BCM Five year study, yr 1 |
Philippines/82 | Whole virus | 15 | 23 ± 12 |
| Leningrad/86 | Whole virus | 15 | 24 ± 9 | |
| BCM Five year study, yr 5 |
Philippines/82 | 24 ± 6 | ||
| Leningrad/86 | Whole virus | 15 | 23 ± 9 | |
| BCM Elderly | Panama/99 | subunit | 15 | 83 ± 24 |
| Panama/99 | subunit | 30 | 68 ± 25 | |
| Panama/99 | subunit | 60 | 69 ± 28 | |
| OMRF yr1 controls | California/04 | subunit | 15 | 39 ± 14 |
| OMRF yr1 cases | California/04 | subunit | 15 | 63 ± 19 |
| OMRF yr2 controls | Wisconsin/05 | subunit | 15 | 68 ± 23 |
| OMRF yr2 cases | Wisconsin/05 | subunit | 15 | 82 ± 18 |
We made direct measurements of total protein on different formulations of vaccines to see if there were obvious differences in the amount of protein that was not native HA. We did not have any of the inactivated whole virus vaccines from the 1980s, but we had samples of subunit vaccines going back to 2001. We also measured neuraminidase activity and titered the HA using human red blood cells. The results are shown in Figure 3. The minimal amount of HA per vaccine dose is 45 µg (15 µg of each of the H3N2, H1N1 and type B HAs), measured as native HA by single radial diffusion. There is a marked downward trend in these vaccines, from about 10-fold higher than the minimum HA content in 2001–02 to about 3-fold in 2007–08. The progressive fall in excess protein content from 2001 to 2007 may reflect better manufacturing methods; the excess protein in earlier years may be denatured HA rather than contaminating proteins. In particular, many companies have switched from formalin inactivation to β-propiolactone over the past few seasons, and this could well explain the differences since formalin causes much more extensive cross-linking and denaturation than β-propiolactone. Therefore, although we cannot answer the question of whether whole virus vaccine had less denatured protein than subunit vaccines, we can say that the subunit vaccines now contain less non-native hemagglutinin or other proteins than a few years ago. The high levels of antibodies against unfoldons after vaccination with Panama/99 (Table 2) may have been due to excess denatured protein in the vaccine, but it must be noted that this was an elderly population and we cannot distinguish between age or vaccine composition as the reason for the high anti-unfoldon antibody levels. The HA content of the vaccine is required to be measured as native HA, determined by single radial immunodiffusion. HA titer depends on the number of particles, and the approximately 2 orders of magnitude higher HA titer of vaccine compared to purified whole virus suggests that the rosettes of HA in vaccine contain about 5 HA trimers compared to ~500 in virus particles. The NA content of the vaccine is not reported, so we carried out NA activity assays of each vaccine and found them to be variable but lower than NA of purified viruses (Figure 3C and 3E). The very low NA activity in recent H3N2 viruses California/04 and Wisconsin/05 (Figure 3E) may reflect deletions in the NA gene during egg passage as we have reported for these viruses when grown in MDCK cells [14].
Figure 3. Protein and NA content of vaccines and purified viruses.
Panels A, B and C show the total protein, HA titer and NA activity (fluorescence units) of a dose in five years of vaccines. The results in panel B reflect varying sizes and avidities of HA rosettes in the vaccine, since each vaccine contains 15 µg of each of the three HAs (total 45 µg).
Panels D and E show HA titer and NA activity of some of the viruses that were contained in the vaccines used in A–C. Results were calculated per 45 µg of HA for comparison with A–C.
4. Conclusions
Most of the subjects we have studied have significant levels of anti-influenza antibodies before vaccination, and the fold increase in antibodies after vaccination is inversely proportional to the pre-existing levels of antibodies. The level of protection is determined by amount and avidity of neutralizing antibodies, whether these were pre-existing or induced by the vaccine is irrelevant. Virus neutralization titers in theory should reflect protection but they have such high variability they are not considered reliable measures of protection [15]. The hemagglutination-inhibition (HAI) test, long used as a surrogate measure of protection, has been found to be unreliable with H5N1 viruses [15, 16] and also with recent H3N2 human viruses, apparently due to lower avidity or higher specificity for particular receptors on red blood cells [14]. Only one subject in the OMRF study has been diagnosed with influenza infection, so we have no statistical measure of protection to compare with our native antibody ELISA assay. The level of antibodies against denatured or internal proteins in the three studies is variable. It seems likely that it would be better to keep anti-unfoldon antibodies to a minimum but at this time we have no evidence that high levels of antibodies against unfoldons have a negative effect on antibodies against native proteins that are potentially protective.
For our continuing OMRF study on responses of lupus patients and their matched controls to influenza vaccine, we are using the following parameters to assess likelihood of protection:
The post-vaccination antibody amount against native surface proteins (Amax), i.e. the final titer, whether or not it had been increased after vaccination
The post-vaccination overall avidity of serum antibodies against native surface proteins (Ka)
The post-vaccination HAI titer
In practice we rank subjects from low to high separately for each of these three parameters so that differences in numerical values are removed and extreme outliers do not skew the results. Then the Index of anti-Native Antibodies (INA) for each individual is:
Acknowledgments
The vaccine studies at Baylor College of Medicine were supported by contracts N01-AI65298 and N01-AI25465 from NIAID. Initial studies with the native ELISA assay were supported by R01-AI50933 from NIAID. The current studies of lupus patients and control subjects are supported by contract #HHSN266200500026C (N01-AI-50026) from NIAID.
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Churchill ME, Stura EA, Pinilla C, Appel JR, Houghten RA, Kono DH, et al. Crystal structure of a peptide complex of anti-influenza peptide antibody Fab 26/9. Comparison of two different antibodies bound to the same peptide antigen. J Mol Biol. 1994 Aug 26;241(4):534–556. doi: 10.1006/jmbi.1994.1530. [DOI] [PubMed] [Google Scholar]
- 2.Burlington DB, Wright PF, van Wyke KL, Phelan MA, Mayner RE, Murphy BR. Development of subtype-specific and heterosubtypic antibodies to the influenza A virus hemagglutinin after primary infection in children. J Clin Microbiol. 1985;21(5):847–849. doi: 10.1128/jcm.21.5.847-849.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beyer WE, Palache AM, Sprenger MJ, Hendriksen E, Tukker JJ, Darioli R, et al. Effects of repeated annual influenza vaccination on vaccine sero-response in young and elderly adults. Vaccine. 1996 Oct;14(14):1331–1339. doi: 10.1016/s0264-410x(96)00058-8. [DOI] [PubMed] [Google Scholar]
- 4.Keitel WA, Cate TR, Couch RB, Huggins LL, Hess KR. Efficacy of repeated annual immunization with inactivated influenza virus vaccines over a five year period. Vaccine. 1997;15(10):1114–1122. doi: 10.1016/s0264-410x(97)00003-0. [DOI] [PubMed] [Google Scholar]
- 5.Keitel WA, Cate TR, Couch RB. Efficacy of sequential annual vaccination with inactivated influenza virus vaccine. Am J Epidemiol. 1988;127(2):353–364. doi: 10.1093/oxfordjournals.aje.a114809. [DOI] [PubMed] [Google Scholar]
- 6.de Bruijn IA, Remarque EJ, Jol-van der Zijde CM, van Tol MJ, Westendorp RG, Knook DL. Quality and quantity of the humoral immune response in healthy elderly and young subjects after annually repeated influenza vaccination. J Infect Dis. 1999;179(1):31–36. doi: 10.1086/314540. [DOI] [PubMed] [Google Scholar]
- 7.Laver WG, Air GM, Webster RG, Smith-Gill SJ. Epitopes on protein antigens: Misconceptions and realities. Cell. 1990;61:553–556. doi: 10.1016/0092-8674(90)90464-p. [DOI] [PubMed] [Google Scholar]
- 8.Gulati U, Kumari K, Wu W, Keitel WA, Air GM. Amount and avidity of serum antibodies against native glycoproteins and denatured virus after repeated influenza whole-virus vaccination. Vaccine. 2005;23:1414–1425. doi: 10.1016/j.vaccine.2004.08.053. [DOI] [PubMed] [Google Scholar]
- 9.Gulati U, Keitel WA, Air GM. Increased antibodies against unfolded viral antigens in the elderly after influenza vaccination. Influenza Other Respir Viruses. 2007;1(4):147–156. doi: 10.1111/j.1750-2659.2007.00017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keitel WA, Cate TR, Atmar RL, Turner CS, Nino D, Dukes CM, et al. Increasing doses of purified influenza virus hemagglutinin and subvirion vaccines enhance antibody responses in the elderly. Clin Diagn Lab Immunol. 1996;3(5):507–510. doi: 10.1128/cdli.3.5.507-510.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steck TL, Kant JA. Preparation of impermeable ghosts and inside-out vesicles from human erythrocyte membranes. Methods Enzymol 1974. 1974;31(Pt A):172–180. doi: 10.1016/0076-6879(74)31019-1. [DOI] [PubMed] [Google Scholar]
- 12.Pruett PS, Air GM. Critical interactions in binding antibody NC41 to influenza N9 neuraminidase: amino acid contacts on the antibody heavy chain. Biochemistry. 1998;37:10660–10670. doi: 10.1021/bi9802059. [DOI] [PubMed] [Google Scholar]
- 13.White JM, Wilson IA. Anti-peptide antibodies detect steps in a protein conformational change: low-pH activation of the influenza virus hemagglutinin. J Cell Biol. 1987;105(6 Pt 2):2887–2896. doi: 10.1083/jcb.105.6.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gulati S, Smith DF, Air GM. Deletions of neuraminidase and resistance to oseltamivir may be a consequence of restricted receptor specificity in recent H3N2 influenza viruses. Virology J. 2009;6(22) doi: 10.1186/1743-422X-6-22. epub Feb 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stephenson I, Das RG, Wood JM, Katz JM. Comparison of neutralising antibody assays for detection of antibody to influenza A/H3N2 viruses: an international collaborative study. Vaccine 2007. 2007 May;25(20):4056–4063. doi: 10.1016/j.vaccine.2007.02.039. [DOI] [PubMed] [Google Scholar]
- 16.Rowe T, Abernathy RA, Hu-Primmer J, Thompson WW, Lu X, Lim W, et al. Detection of antibody to avian influenza A (H5N1) virus in human serum by using a combination of serologic assays. J Clin Microbiol. 1999;37(4):937–943. doi: 10.1128/jcm.37.4.937-943.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]