Abstract
The metabolism and efficacy of 5-fluorouracil (FUra) and other fluorinated pyrimidine (FP) derivatives have been intensively investigated for over fifty years. FUra and its antimetabolites can be incorporated at RNA- and DNA-levels, with RNA level incorporation provoking toxic responses in human normal tissue, and DNA-level antimetabolite formation and incorporation believed primarily responsible for tumour-selective responses. Attempts to direct FUra into DNA-level antimetabolites, based on mechanism-of-action studies, have led to gradual improvements in tumour therapy. These include the use of leukovorin to stabilize the inhibitory thymidylate synthase-5-fluoro-2′-deoxyuridine 5′ monophoshate (FdUMP)-5,10-methylene tetrahydrofolate (5,10-CH2FH4) trimeric complex. FUra incorporated into DNA also contributes to antitumour activity in preclinical and clinical studies. This review examines our current state of knowledge regarding the mechanistic aspects of FUra:Gua lesion detection by DNA mismatch repair (MMR) machinery that ultimately results in lethality. MMR-dependent direct cell death signalling or futile cycle responses will be discussed. As 10–30% of sporadic colon and endometrial tumours display MMR defects as a result of human MutL homologue-1 (hMLH1) promoter hypermethylation, we discuss the use and manipulation of the hypomethylating agent, 5-fluorodeoxycytidine (FdCyd), and our ability to manipulate its metabolism using the cytidine or deoxycytidylate (dCMP) deaminase inhibitors, tetrahydrouridine or deoxytetrahydrouridine, respectively, as a method for re-expression of hMLH1 and re-sensitization of tumours to FP therapy.
Keywords: 5-fluorouracil, DNA mismatch repair, thymidylate synthase, hypermethylation, hMLH1, MMR/c-Abl/p73α/GADD45α signalling
The fluorinated pyrimidines (FPs) and their metabolism to DNA-level antimetabolites
5-Fluorouracil (FUra) was developed in 1957 as a potential drug for the treatment of advanced cancers (Heidelberger et al., 1983). Investigation of its antimetabolites resulted in the development of an entire class of fluorinated pyrimidines (FPs). This class of drugs, driven by the work of Dr. Charles Heidelberger (Heidelberger et al., 1983) among many others, represented the first ‘mechanistically designed’ drugs for the treatment of cancer. As enhanced utilization of uracil (Ura) as a precursor of DNA pyrimidines was observed in a series of transplantable tumours, an antimetabolite that resembled uracil was devised. A fluorine atom was substituted for hydrogen at the 5-position of Ura, creating FUra. As theorized, the resulting carbon-fluorine bond was far stronger than the carbon-hydrogen bond, and was insensitive to thymidylate synthase (TS) cleavage following the formation of the TS-5-fluoro-2′-deoxyuridine 5′-monophosphate (FdUMP)-5,10-methylene tetrahydrofolate (5,10-CH2FH4) trimeric inhibitory complex. Because FUra had significant antitumour activity, many related nucleosides were synthesized. One derivative, 5-fluoro-2′-deoxyuridine (FdUrd), also showed considerable antitumour activity. In fact, FdUrd appeared more cytotoxic than FUra in many cancer cell lines in vitro (Willmore and Durkacz, 1993). Moreover, FPs remain the drugs of choice for the treatment of advanced colorectal cancer (Johnston et al., 1996; Sobrero et al., 1997; van Laar et al., 1998). FUra and FdUrd are inactive per se and must be metabolized to nucleotide forms to be cytotoxic (reviewed in Santi, 1980; Heidelberger et al., 1983; Boothman et al., 1989); salient features of this activation pathway are discussed below and demonstrated in Figure 1. Another FP-related antimetabolite, 5-fluoro-2′-deoxycytidine (FdCyd) received much less attention and was simultaneously developed by Greer et al., (Mekras et al., 1984). This fluorodeoxycytidine derivative depends on tumour-selective deamination for activation to FdU-related antimetabolites (Boothman et al., 1985; 1987a,b;). Importantly, the metabolism of deoxycytidine, and therefore 5-fluorodeoxycytidine antimetabolites, can be manipulated for improved cancer-selective uptake and anabolism using specific cytidine and dCMP deaminase inhibitors, tetrahydrouridine (H4Urd) and deoxytetrahydrouridine (dH4Urd) respectively (Boothman et al., 1985; 1987a,b; 1989;). Its use for the treatment of well-defined sporadic MMR-deficient cancers will be discussed below.
Figure 1.
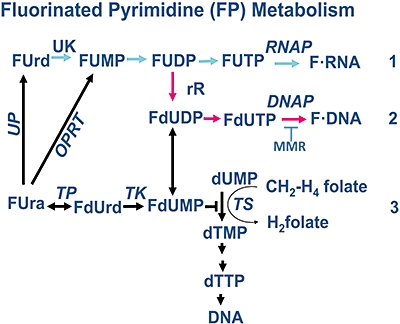
Metabolism of 5-fluorouracil (FUra) to DNA- and RNA-level metabolites. Developed in the late 1950s and studied intensively over the next 40 years, FUra is still a key chemotherapeutic agent used in the treatment of colon cancer, as well as in adjuvant therapies for a variety of other cancers. Upon entering the cell, FUra is rapidly converted to both 5-fluorouridine (FUrd) and 5-fluoro-2'deoxyuridine (FdUrd) antimetabolites by phosphorylases that add on deoxyribose or ribose units, depending on available substrate ribo- or deoxyribo-nucleosides. Once formed, FUrd or FdUrd are phosphorylated by uridine or thymidine kinases (UK or TK), respectively, to retain the antimetabolites in the cell. Basically, all of the FP-antimetabolites are better substrates than the normal metabolites for each enzymatic step. In general, metabolism of FUra to RNA-level antimetabolites (level 1) leads to less antitumour activity and more general toxicity to normal tissue, as the levels of enzymes that metabolize these RNA-level antimetabolites are not elevated in tumour versus normal tissue. In contrast, enzymes [e.g. thymidine kinase (TK) and thymidylate synthase (TS)] that metabolize DNA-level FUra antimetabolites are elevated in tumour above normal tissue (levels 2, 3). The contribution of FdUrd incorporated into DNA to antitumour activity has been misunderstood and greatly under-estimated. Enzyme abbreviations: UP, uridine phosphorylase; UK, uridine kinase; RNAP, RNA polymerase; rR, ribonucleotide reductase; DNAP, DNA polymerase; TP, thymidine phosphorylase; OPRT, orotic acid phosphoribosyl transferase. Adapted from Meyers et al. (2003).
FUra and FdUrd can be converted to common mono-, di-, and tri-phosphate metabolites (Figure 1). FUra may be converted to FdUrd by enzymatic sugar (deoxyribose-1-phosphate) exchange via thymidine phosphorylase (TP). Likewise, TP can convert FdUrd to FUra, depending on the intracellular availability of ribo- or deoxyribo-nucleotide donor pools.
In general, there are three major determinants of the cellular response to FPs. FP exposure can lead to RNA-directed cytotoxicity via incorporation of 5-fluorouridine-5′-triphosphate (FUTP) into RNAs. FUra is converted to FUMP by pyrimidine phosphoribosyl transferase (or converted to fluorouridine (FUrd) by uridine phosphorylase, then to FUMP by uridine kinase), which can then be converted to 5-fluorouridine 5′-diphosphate (FUDP) and ultimately, FUTP. FUTP is an excellent substrate for RNA polymerase, and its incorporation can: (i) interfere with mRNA metabolism and expression (van Laar et al., 1996; 1998;); (ii) inhibit rRNA maturation (Dolnick and Pink, 1983; 1985;); (iii) interfere with tRNA function (Parker and Cheng, 1990); and (iv) possibly lead to the production of a non-functional RNA primer (Spiegelman et al., 1980a,b;). Unfortunately, none of these enzymes are typically elevated in tumour compared with normal tissue. Therefore, such metabolism of FUra derivatives to RNA level antimetabolites leads to normal tissue cytotoxic complications, and not the more desirable efficacious antitumour activity.
Fluorinated pyrimidine exposure can also cause DNA-directed cytotoxicity via incorporation into DNA, and formation of antimetabolites at this level elicits potent antitumour activity. In cell culture, FdUrd at low doses (100–10 000 nM) is primarily metabolized by thymidine kinase (TK) to FdUMP, resulting in DNA-directed cytotoxicity with little or no RNA-directed effects (Parker and Cheng, 1990; Parker and Marinus, 1992). FdUrd can also be converted to FUrd or FUra by TP (Willmore and Durkacz, 1993), causing effects on RNA metabolism (Figure 1). In many cell lines, very high doses (100–1000 nM) of FdU must be given before significant levels of FUrd in DNA are noted (Boothman et al., 1985). FUDP can be converted to 5-fluoro-2′-deoxyuridine 5′-diphosphate (FdUDP) by ribonucleotide reductase (rR), which can then be converted to 5-fluoro-2′-deoxyuridine 5′-triphosphate (FdUTP), a substrate for DNA polymerases-alpha and -beta. Most importantly, incorporation of FdUTP into DNA can result in nucleotide mis-incorporation during replication (Aebersold, 1979). The more traditional form of DNA-directed cytotoxicity in response to FUra exposure is mediated by inhibition of thmidylate synthase (TS), brought on by accumulated FdUMP pools. TS can normally catalyse conversion of dUMP to dTMP, involving transfer of a methyl group from 5, 10-methylene tetrahydrofolate (5,10-CH2FH4) to the number five carbon of uracil (Ura). FdUrd can be converted by TK to FdUMP. FdUMP then forms an inhibitory ternary complex with TS and tetrahydrofolate because of the inability of TS to break the carbon-fluorine bond of FdUMP. Inhibition of TS ultimately results in decreased intracellular dTTP pools and subsequent inhibition of DNA synthesis (Santi, 1980). dNTP pool imbalance can have profound effects on the accuracy of DNA replication, and pool imbalances can greatly increase mutation rates (Caradonna and Cheng, 1980;Hopkins and Goodman, 1980;Ingraham et al., 1980; Prem veer Reddy and Pardee, 1980;veer Reddy and Pardee, 1982; Das et al., 1983; Newman and Miller, 1983). Pool imbalances have been demonstrated to cause infrequent, but potent, formation of FUra:Gua mismatched nucleotides (Meyers et al., 2005). The action of MMR mediates cytotoxicity following recognition of FUra:Gua lesions in several mutually exclusive ways. First, MMR detects FdU in DNA causing its exclusion and stalling replication forks because of the extensive excision repair patch. Second, MMR is required to detect increased mutations caused by pool imbalances resulting from FdUMP-mediated TS inhibition. As a result, MMR stimulates cell cycle checkpoint responses by two proposed pathways: (i) futile cycling, which seems unlikely for a number of reason (discussed below); or (ii) direct signalling via c-Abl/p73α/GADD45α activation, which we have recently implicated in both cell cycle checkpoint and apoptotic responses (Li et al., 2008; Wagner et al., 2008).
Repair pathways known to detect and resolve FUra-induced DNA lesions
There are several instances when uracil (Ura) can be incorporated into genomic DNA, which is highly mutagenic. In response, several DNA repair systems have evolved in mammalian cells for the successful elimination of this moiety. Uracil moieties can form in DNA by either direct incorporation of dUTP during DNA synthesis (Friedberg et al., 2006) or through deamination of cytosine, whose rate has been estimated at 6.9 × 10−8 deaminated moieties per day in double strand DNA (Lindahl and Nyberg, 1974;Shapiro, 1981). A third mechanism for incorporation of Ura into genomic DNA is the action of the activation-induced cytosine deaminase (CD), that can also directly introduce Ura within genomic DNA, but under normal circumstances this is limited to mature B-cells that have been stimulated to undergo the process of class switch recombination or somatic hypermutation (Rada et al., 2002; Imai et al., 2003).
Deamination of cytosine moieties in DNA by chemical or enzymatic activity leads to the formation of Ura:Gua mispairs, while direct incorporation of uracil into DNA leads to Ura:Ade pairs. Both lesions are extremely mutagenic. Uracil incorporated into DNA as Ura:Ade is mainly removed by the action of DNA glycosylases [e.g. uracil-DNA-glycosylases (UDGs)] that activate base excision repair (BER) pathways (Krokan et al., 2002). The major UDGs are UNG1, which is mitochondrial (Otterlei et al., 1998) and UNG2 and SMUG1, which are nuclear (Haug et al., 1998). These enzymes cleave and release Ura from genomic DNA, resulting in apyrimidinic (AP) base damage that, in turn, initiates BER responses.
Activation of AP endonuclease (APE) as a downstream BER response causes a DNA strand break consisting of a 3′-OH group and a 5′-deoxyribose phosphate (5′-dRP) group. 5′-dRP lyase removes the 5′-dRP group and leaves a 5′-phosphate. This is then followed by DNA polymerase activity to fill in the 1–2 base pair gap within the DNA. DNA polymerase β (Polβ) can also perform both removal of the 5′-dRP group and gap filling functions. Once synthesis has been completed, DNA ligase seals the nicks left in DNA. Until recently, BER-mediated DNA repair was believed to be the only mechanism by which Ura incorporated into DNA was removed (Krokan et al., 2002).
The enzymes responsible for preventing FP incorporation into DNA are those, that under normal condition, prevent Ura incorporation into DNA (Caradonna and Cheng, 1980; Ingraham et al., 1980). Both FdUTP and dUTP are substrates for alpha- and beta-DNA polymerases. dUTPase, which dephosphorylates dUTP to dUMP thereby lowering available nuclear dUTP pools (which are normally very low relative to dTTP), also acts on FdUTP formed from FUra exposure (Figure 1). Like most enzymes, dUTPase has a higher affinity for, and can better utilize, FdUTP compared with dUTP. Nevertheless, both Ura and FUra moieties have been detected in the DNA of cells exposed to FPs (Boothman et al., 1985; 1987a,b; 1989; Nio et al., 1991). UDGs recognize both FUra and Ura in DNA, and its activity results in AP sites in DNA. AP sites are, in turn, recognized by APE (Sancar, 1995), which produces a strand scission at the site. Nucleases, including flap-endonuclease 1 (FEN1), recognize these strand scissions, and DNA polymerase (usually Polβ) fills the gap, which is followed by strand resealing by DNA ligase. FUra itself has been reported to competitively inhibit UDG (Wurzer et al., 1994). Aside from UDG, the only other BER enzyme that may recognize FUra moieties in DNA is the MBD4 glycosylase, which has been reported to interact with human MutL homologue-1 (hMLH1) (Bellacosa et al., 1999). The exact pathway(s) important for the repair of FUra moieties is (are) most likely dependent on repair capacity, DNA synthesis status, cell cycle regulation, appropriate balanced deoxyribonucleotide and ribonucleotide pools, as well as other possible tumour microenvironment factors. The contribution of MMR in detecting FUra moieties in DNA, and the resulting cytotoxic responses of exposed cells are described below.
Treatment of mammalian cells with FPs can lead to dNTP pool imbalances. Decreases in dTTP pools because of FdUMP inhibition of TS removes negative feedback inhibition on rR and TK that result in greater levels of FdUTP, which then leads to elevated incorporation of FUra into DNA. Additionally, TS inhibition will cause a build-up of both dUTP and FdUTP pools and eventually exhaust dUTPase. As dUTP and FdUTP accumulate and dTTP levels fall, dUTP and FdUTP pools replace dTTP as substrates for DNA polymerase, resulting in ever-increasing levels of FdUTP or dUTP incorporated into DNA. Given these metabolic changes, it has been puzzling why such low levels of FUra moieties in DNA have been detected in most cancer cells after FP exposure.
DNA mismatch repair and DNA damage signalling
Primary tumours and tumour cell lines containing MMR defects are resistant to a wide variety of commonly used therapeutic agents (see review: Irving and Hall, 2001). These include methylating agents [Temozolomide™; procabazine; N-methyl-N'-nitro-N-nitrosoguanidine (MNNG); N-methyl-N-nitrosourea (MNU)], antimetabolites (6-thioguanine; mercaptopurine), platinum compounds (cisplatin; carboplatin) and perhaps Topoisomerase II inhibitors that have additional effects on cellular redox reactions (doxorubicin; epirubicin). In the last several years, it has become apparent that the drug resistance in MMR-deficient cells was tied to reduced or absent damage-induced G2 arrest and ultimately cell death responses (e.g. apoptosis) (Meyers et al., 1997; Davis et al., 1998; Gong et al., 1999; Hickman and Samson, 1999;Zhang et al., 1999).
Initiation of cellular responses to DNA damage caused by FP exposure requires DNA damage sensors (DS), adaptors/mediators (AM), as well as amplification responses involving MMR-dependent c-Abl responses (Gong et al., 1999; Wagner et al., 2008), or MMR-independent PI-3-like kinases (PIKKs). For simplicity, only G2 arrest and apoptotic responses will be considered here, as these appear to be the primary cellular responses to FP damage. A MMR-independent DS/AM/PIKK complex appears to activate at least two pathways that lead to G2 arrest by cascade phosphorylation of p53 mediated by Chk1. Activation of Chk1, by phosphorylation, leads to the regulation of the Cdc25C phosphatase, by protein modification. During a normal cell cycle, Cdc25C dephosphorylates Cdc2, prior to entry into G2. Thus, inactivation of Cdc25C results in a de facto G2 arrest. Conversely, the phosphorylation-activation of p53 leads to significant up-regulation of 14-3-3σ that, in turn, sequesters Cdc2/cyclinB leading to G2 arrest. These responses can be stimulated in MMR-deficient cells only by high doses of FPs and the responses are more delayed (at least in response to alkylation damage) than are MMR-dependent responses (Wagner et al., 2008).
In MMR-competent cells, another more potent and rapid G2 arrest and apoptotic stimulatory pathway is activated. These responses are seen at >10-fold less doses of FPs or alkylating agents in MMR-competent cells, compared with MMR-independent responses that are noted only after high doses of damaging agents. MMR-dependent sensing of alkylation or FP damage stimulates the activation of c-Abl kinase, which can be suppressed by Gleevec™ or by specific siRNA-c-Abl knockdown (Wagner et al., 2008). These ‘immediate-early’ (i.e., in minutes) G2 arrest responses provide the cellular framework for the MMR signalling pathway. MMR-dependent persistent DNA lesion damage recognition, processing and signalling leads to combined ‘early’ (h) and ‘late’ (h/days) responses that may result in premature senescence, necrosis, or apoptotic cell death. MMR-dependent apoptosis is mediated by induction and stimulated levels of GADD45α and p73α, but not by p53 (Meyers et al., 2001; 2003; 2005; Li et al., 2008). MMR-dependent apoptosis and G2 arrest were p53-independent, as loss of p53 because of E6 expression, somatic knockout, or stable siRNA-p53 knockdown had no affect on apoptotic responses in MMR-dependent cell death (Li et al., 2008). Interestingly, our studies were able to separate MMR-dependent signalling of G2 arrest from apoptotic responses, as siRNA-specific p73α knockdown resulted in loss of apoptosis but not G2 arrest. In contrast, specific knockdown of c-Abl or GADD45α prevented both apoptosis and G2 arrest responses. Loss of p53 in these cells did not affect MMR-dependent responses (Li et al., 2008; Wagner et al., 2008).
Two opposing models have been proposed to account for the MMR-dependent G2 arrest and apoptosis: (i) futile cycling of repair; and (ii) direct MMR-dependent signalling. ‘Futile cycling’ of repair, was originally proposed to explain a similar MMR-dependent cell death effect in bacteria that contained a dam, DNA adenine methylase, mutation (Karran and Marinus, 1982;Fram et al., 1985). In the absence of Dam methylation, the MutH-dependent incision that initiates MMR may occur on either DNA strand on either side of the lesion. Some of these incision events were proposed to lead to bidirectional degradation towards the lesion. This unregulated degradation could then lead to DNA double-strand breaks (DSBs). Multiple DSBs induced by multiple MMR reactions were proposed to result in genetic catastrophe and cell death. As DSBs are a well-known cause of G2 arrest and p53-induced apoptosis in mammalian cells (Kastan et al., 1991), such a mechanism seemed plausible in mammalian cells (Davis et al., 1998). However, there appears to be multiple problems in adapting MMR-dependent ‘futile cycling’ mechanism to mammalian systems, among them the apparent lack of any requirement for p53 (for review see: Meyers et al., 2003; Wang and Edelmann, 2006).
The ‘direct signalling’ model was originally proposed (Davis et al., 1998; Fishel, 1999; 2001;) to explain the MMR-dependent activation of G2 arrest and apoptosis, an apoptotic pathway that later appeared to include c-Abl (Gong et al., 1999), as well as the rapid induction of apoptosis following over-expression of MMR genes (Zhang et al., 1999). More recent evidence suggests that a subset of the MMR proteins (i.e. hMSH2, hMSH6, hMLH1 and hPMS2) serve as sensors of DNA damage (Wang and Edelmann, 2006). For example, Hsieh and colleagues (Yoshioka et al., 2006) have demonstrated that hMSH2-hMSH6 and hMLH1-hPMS2 bound to a methylation-damaged O6-MeG/T mismatched DNA specifically interact with ataxia telangiectasia-and-rad3-related (ATR)-ATRIP (a PIKK family member) and Chk1. These studies suggested the ATR/Chk1 pathway in at least part of the ‘immediate early’ G2 arrest. However, most recently, our group found that MMR-dependent G2 arrest responses triggered by MNNG are dependent on a hMLH1/c-Abl/GADD45α signalling pathway, and that ataxia telangiectasia mutated (ATM)/Chk2, as well as ATR/Chk1, was clearly not involved in the MMR-dependent G2 arrest responses in response to alkylation (Wagner et al., 2008) or FP damage (Meyers et al., 2001; 2003;). The activation of apoptosis following persistent DNA damage was induced by hMLH1/c-Abl/p73α/GADD45α retrograde-signalling pathway, where ATM and p53 were not involved (Li et al., 2008). We also noted that MMR triggers apoptosis in response to MNNG-induced DNA lesions, which along with long-term survival, was completely abrogated by the c-Abl kinase inhibitor, STI571 (Gleevec™). As a result, our data strongly suggest that Gleevec™ may be ill-suited in conjunction with temozolomide or cisplatin, or other clinically used alkylating agents, for efficacious cancer therapy in tumours that are proficient in the MMR pathway (Li et al., 2008).
Remarkably, the introduction of the Msh2 (G674A) or Msh6 (T1217D) mutation into mice resulted in an absence of MMR activity but normal damage-induced apoptosis (Lin et al., 2004; Yang et al., 2004). Thus, dissociation of MMR-dependent multiple excision tracts required for ‘futile cycling’ from a damage-induced apoptotic response would appear to significantly reduce the likelihood of the ‘futile cycling’ mechanism. These functional dissociation mutations are located in separate, but proximal, highly conserved ATP/ADP processing domains of the Msh2-Msh6 heterodimer.
The mechanics of DNA mismatch repair and lesion recognition
Much of our understanding of MMR arose from studies using E. coli (Modrich, 1989; Modrich, 1997). E. coli MMR corrects polymerase mis-incorporation errors by promoting a ‘long patch’, DNA excision reaction that is genetically dependent on MutS, MutL, MutH and MutU (UvrD) gene products. The MutSLH pathway both increases the fidelity of DNA replication (Rydberg, 1978), as well as acts on recombination intermediates containing mispaired bases (Wildenberg and Meselson, 1975; Wagner and Meselson, 1976; Fishel and Kolodner, 1983;Fishel et al., 1986). Strand discrimination for error-free post-replication MMR relies on transient under-methylation of the adenine nucleotide within a GATC DAM sequence. E. coli MMR has been reconstituted in vitro and requires MutS, MutL, MutH and UvrD (helicase II) proteins along with DNA polymerase III holoenzyme, DNA ligase, single-stranded DNA binding protein (SSB) and one of four single-stranded DNA exonucleases (Exo I, Exo VII, RecJ or ExoX); (Lu et al., 1983; Su and Modrich, 1986;Lahue et al., 1987; 1989; Welsh et al., 1987; Grilley et al., 1989; Cooper et al., 1993).
The MutS homodimer has long been known to bind mismatched DNA (Su and Modrich, 1986). In the presence of the MutL homodimer and ATP, the MutS protein footprints around a mismatch (Grilley et al., 1989) and a MutH-dependent endonuclease activity at a hemi-methylated GATC site was enhanced (Welsh et al., 1987). The MutH endonuclease scission was found to direct unwinding and degradation of the unmethylated DNA strand by the coordinated action of Helicase II (UvrD) and one of four single-stranded DNA (ssDNA) exonucleases (RecJ, ExoI, ExoVII, ExoX) (Matson, 1986; Viswanathan and Lovett, 1998;Yamaguchi et al., 1998). Depending on the relative location of the MutH endonuclease in-scission to the mismatch, the resulting excision gap may occur 5′→3′ or 3′→5′ but invariably traverses only the interval between a Dam-site (nick) to just past the mismatch (Figure 2) (Cooper et al., 1993; Grilley et al., 1993). Re-synthesis of the single-stranded gap may be performed by the Pol III holoenzyme (Lahue et al., 1989) or virtually any other polymerase.
Figure 2.
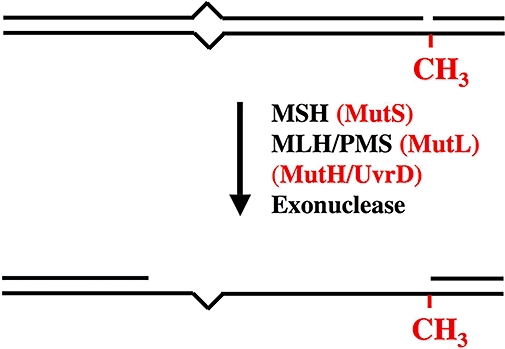
Minimal mismatch repair uniquely requires MutS homologue(s) (MSH), MutL homologue(s) (MLH/PMS) and an Exonuclease (Exo). E. coli (gram-negative enteric bacteria) MMR (red) uniquely requires a hemi-methylated Dam site (-CH3), the MutH endonuclease, and the MutU (UvrD) helicase. The excision tract (250–1000 bp) extends uniquely from the strand scission to just past the DNA mismatch lesion.
Conserved genes and function
The complete human MMR reaction has been reconstituted using cellular extracts (Glazer et al., 1987; Holmes et al., 1990; Thomas et al., 1991) and purified proteins (Genschel and Modrich, 2003;Zhang et al., 2005). As with bacteria, the mismatch recognition requires MutS homologues (MSH) (Drummond et al., 1995; Genschel et al., 1998). Single-nucleotide and small insertion mismatches are recognized by the hMSH2-hMSH6 heterodimer, while insertion/deletion loop-type (IDL) mismatched DNA lesions are recognized by the hMSH2-hMSH3 heterodimer (Acharya et al., 1996; Palombo et al., 1996). Although their detailed role(s) remain enigmatic, the MutL homologue (MLH/PMS) heterodimers function downstream of MSH recognition. Some studies suggest that the hMLH1-hPMS2 and hMLH1-hMLH3 heterodimers may substitute for one another during MMR – although the efficiency of this substitution is controversial (Li and Modrich, 1995;Raschle et al., 1999). Importantly, the mismatched DNA substrate must contain a pre-introduced single-stranded scission (nick), either 5′- or 3′- of the DNA mismatch. Nuclease activity appears to be accomplished by a combination of a 5′-exonuclease (ExoI) and an intrinsic ssDNA endonuclease activity found in some MLH family members (Kadyrov et al., 2006). The minimal 5′→3′ and 3′→5′ excision reaction requires hMSH2-hMSH6 (or hMSH2-hMSH3), hMLH1-hPMS2, ExoI, RPA, PCNA, and RFC (Kadyrov et al., 2006). Re-synthesis of the single-stranded gap requires Polδ, and ligase I, and may be modestly enhanced with HMGB1 (Zhang et al., 2005).
Models for MMR
A detailed biophysical mechanism for MMR remains controversial and incomplete. Modrich and colleagues have proposed a Hydrolysis-Dependent Translocation Motor Model (Figure 3I). It posits the assembly of a MutS-MutL complex at the mismatch (Figure 3IA and IB) (Modrich, 1989; Modrich and Lahue, 1996), which then uses ATP-hydrolysis to motor bi-directionally creating a looped structure (Figure 3IC) (Allen et al., 1997). This DNA tracking process was envisioned to link MutS mismatch recognition to the MutH endonuclease incision, as well as to provide the necessary directionality for subsequent loading of Helicase II (UvrD) and one of the ssDNA exonucleases (Figure 3ID). However, not all predictions arising from this model agree with the genetic or biochemical data (Gradia et al., 1997; 1999; 2000; Fishel, 1998; 1999; Guerrette et al., 1998; 1999; Schmutte et al., 1998; Wilson et al., 1999; Berardini et al., 2000; Schmutte et al., 2001; Mazurek et al., 2002).
Figure 3.
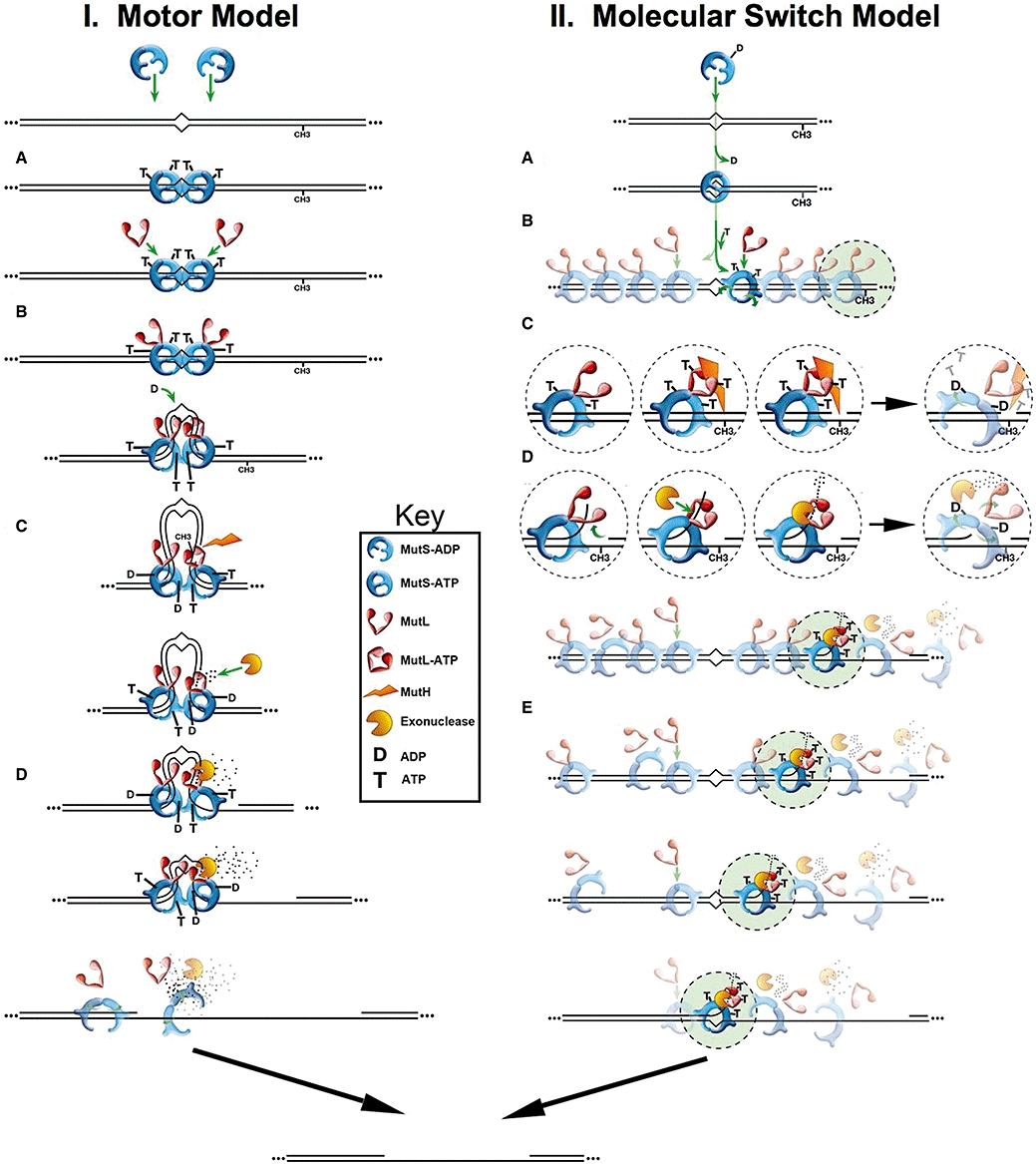
Models for Mismatch Repair. See text for description.
Our work with human MSH proteins led to the Molecular Switch Model[Figure 3II; for review see: (Berardini et al., 2000)]. It is based on the observation that mismatched DNA stimulated the exchange bound ADP for ATP (ADP→ATP exchange) by the human MSH proteins (hMSH2-hMSH6 or hMSH2-hMSH3 (Figure 3IIA) (Gradia et al., 1997; Wilson et al., 1999). ATP binding resulted in the formation of a MSH sliding clamp capable of hydrolysis-independent diffusion/tracking for several thousand nucleotides along the adjoining DNA backbone (Gradia et al., 1999). Iterative loading of multiple sliding clamps was suggested to provide a ‘threshold signal’ that distinguished the mismatch region and provided a ‘gradient’ of MSH proteins that proffered a directionality along the DNA duplex surrounding the mismatch site (Figure 3IIB) (Gradia et al., 1999; Heinen et al., 2002). ATP hydrolysis only occurred when the human MSH proteins dissociated from the DNA ends (Gradia et al., 1999); a rare condition in normal cells. These observations accounted for the low ATPase activity (Haber and Walker, 1991;Gradia et al., 1997; Gradia et al., 2000; Hess et al., 2002) and identified ADP→ATP exchange at the mismatch site as the rate-limiting step. The process appeared remarkably similar to the control of G protein molecular switches by GDP→GTP exchange (Sprang, 1997). Although their exact role in MMR remains enigmatic, it appeared that the MLH/PMS proteins formed a stable ternary complex with ATP-bound MSH sliding clamps (Figure 3IIB) (Acharya et al., 2003). The MSH-MLH/PMS ternary complex was shown to interact with downstream MMR components, such as MutH (Figure 3IIC), MutU (UvrD) helicase (not shown), or a ssDNA end (Figure 3IID). Incremental rearward diffusion, made irreversible by endo- or exo-nuclease digestion, was proposed to create a dynamic and redundant process as multiple MSH-MLH/PMS sliding clamps ‘hand-off’ the excision reaction until it covers the mismatch (Figure 3IIE). At that point no further sliding clamps may be loaded and the minimal MMR reaction is complete. In this model, ATP hydrolysis is only required to recycle MSH sliding clamps. These studies led to a modification of the Motor Model by Modrich and colleagues; although it still required ATP hydrolysis to move the MSH protein along the DNA (Blackwell et al., 1998).
A third mechanism, the Transactivation Model, arose from bacterial MutS, MutL, and MutH structures (Ban and Yang, 1998;Ban et al., 1999; Lamers et al., 2000; Obmolova et al., 2000). However, recent experiments have shown that physically blocking the intervening region between the mismatch and Dam site results in a near complete impairment of MMR (Pluciennik and Modrich, 2007). These observations appear to rule-out this and any similar ‘trans’ models in favour of ‘cis’ models similar to those considered above (Kolodner et al., 2007).
Recognition of mismatches and lesions by MSH proteins
MutS homologue proteins recognize a plethora of DNA mismatches, lesions and structures. This is unusual compared with glycosylases, which often have overlapping recognition with MSH proteins but are highly lesion specific. Comparison(s) of numerous mispair-bound MSH structures appear remarkably similar (Lamers et al., 2000; Obmolova et al., 2000; Natrajan et al., 2003; Warren et al., 2007). An important consequence of mismatch recognition by MSH proteins is the insertion of a highly conserved phenylalanine residue 3′ of the mismatch (Lamers et al., 2000; Obmolova et al., 2000). Using nearest neighbor sequence contexts as a model, we recently found that poorly recognized mismatches display an increased stability of base pairs 3′ to the mismatch (Mazurek et al., 2009). This observation suggests that on at least one strand surrounding a poorly recognized mismatch, the interrogation by MSH proteins may be significantly more difficult. NOE data also suggests that well-recognized mismatches have increased localized dynamic flexibility (Mazurek et al., 2009). Together, these observations suggest that MSH proteins do not recognize the mismatch or lesion but instead recognize the flexibility of the DNA that is induced by the mismatch or lesion. These results have several important implications when considering DNA metabolites that might be useful in provoking the MMR damage signalling response for efficacy in cancer therapeutics.
Role of MMR in FP responses and antitumour activity: a historical perspective
The MMR pathway recognizes all eight single nucleotide mismatches, as well as small insertion/deletion loop-type mismatches. The majority of mismatched nucleotides arise as a result of polymerase mis-incorporation errors. As FPs (especially thymidine analogs) could also be incorporated into DNA across from Gua as a result of deoxynucleotide pool imbalances (e.g. after FUra or FdUrd exposures) and/or because of transition state alterations (e.g. BrdUrd), we investigated the role of MMR in cellular responses to FUra and FdUrd (Meyers et al., 2001). Our group was the first to report that MMR cells were resistant to FUra and FdUrd in an abstract submitted in 1996 (Meyers et al., 1996). We demonstrated that hMLH1-deficient HCT116 colon cancer cells were 20-fold more resistant to FUra (continuous treatment for 72 h) and 17-fold more resistant to FdUrd in clonogenic survival assays compared with genetically matched hMLH1-proficient HCT116 3-6 cells. Likewise, murine MLH1-deficient CT-5 cells were threefold more resistant to a 2-h pulse of FdUrd than their MLH1-proficient ME-10 counterparts. Synchronized MMR-proficient HCT116 3-6 cells treated with low doses of FPs had a twofold greater G2 cell cycle arrest response compared with MMR-deficient HCT116 cells. Asynchronous ME-10 cells demonstrated a fourfold greater G2 arrest after FdUrd treatment compared with CT-5 cells. G2 cell cycle arrest was not a result of mitotic arrest, but rather a true G2 arrest as indicated by elevated cyclin B1 levels and a lack of staining with mitotic protein monoclonal antibody 2. Although p53 levels were induced in FdUrd-treated HCT116 3-6 cells, cell death and G2 arrest responses were not dependent on the function of this tumour suppressor. FdUrd-mediated cytotoxicity was caused by DNA-directed and not RNA-directed effects, as administration of excess dThyd (and not Urd) prevented cytotoxicity, cell cycle arrest and DSB formation. hMLH1-dependent responses to FP treatment were, therefore, predicted to have clinical relevance for the use of DNA-directed FPs in the treatment of tumours with MMR deficiencies.
Clinical data suggest that patients with MMR-deficient cancers do not benefit from FP therapies
Around 10∼15% of sporadic colorectal cancers exhibit mismatch repair deficiencies because of hypermethylation of hMLH1 (Li et al., 2003; Ribic et al., 2003; Jover et al., 2009). FUra has been used in cancer chemotherapy for more than 40 years, and remains the standard of care as an adjuvant chemotherapeutic regimen for the treatment of colorectal cancer. Early studies reported that stages II and III colorectal cancer patients had improved overall survival from FUra adjuvant chemotherapy regardless of MSI status (Elsaleh et al., 2000; Hemminki et al., 2000; Watanabe et al., 2001). However, these studies did not take into account patients with MMR deficiencies that did not receive adjuvant chemotherapy. These considerations decreased the accuracy of the study, as intrinsic overall survival of MMR-deficient colorectal cancer patients related to better prognosis than MMR competent patients. The other study from Elsaleh et al. (2000) was limited by non-randomized sample selection. Recently, both retrospective and prospective studies have demonstrated that colorectal cancer patients with MMR deficiencies do not receive significant benefit from FUra-based adjuvant chemotherapy (Ribic et al., 2003; Carethers et al., 2004; Jover et al., 2006; 2009;). The Ribic et al.'s investigation (Ribic et al., 2003), a retrospective study based on large sample size and appropriate control groups, demonstrated that patients with stages II and III colon cancer benefited from FUra-based adjuvant chemotherapy only when their tumours were MMR-competent. Patients in the same study with tumours resulting from the lack of MMR activity, in contrast, received no benefit from FUra adjuvant therapies (Ribic et al., 2003). The most recent prospective studies further confirmed the retrospective reports suggesting that adjuvant FUra-based chemotherapy may not be useful in stages II and III microsatellite-instable colorectal cancers (Jover et al., 2006; Jover et al., 2009). These clinical data further confirmed our previous findings that MMR-deficient cell lines were less responsive than MMR stable cell lines to FUra treatments (Meyers et al., 2001; 2005;).
MSH2-deficient cells were resistant to FdUrd, but not Tomudex™
We examined human colon cancer cells deficient in hMLH1 expression, as well as both human and mouse cell lines deficient in MSH2 for resistance/sensitivity to FUra, FdUrd or Tomudex™, a non-pyrimidine TS inhibitor. Whereas FdUrd has two major DNA-directed mechanisms of cell killing (i.e. DNA incorporation and inhibition of TS), Tomudex™ specifically inhibits TS. Thus, treatment with Tomudex allowed us to discriminate the relative contributions of DNA incorporation versus TS inhibition in MMR-dependent, FdUrd-mediated cell killing. When corrected for differential TS levels (the isogenic cells used were not different in TS activities) near identical dose-response survival curves for HCT116 versus HCT116 3-6 cells were noted in response to Tomudex, suggesting that incorporation of FUra into DNA accounted for the differential survival noted between these cells (Meyers et al., 2005).
MSH2- cells have reduced G2 arrest after FdUrd or FUra
Restoring hMLH1 expression in HCT116 cells caused significantly more prolonged G2 arrest in response to 6-TG or FdUrd, as we reported (Davis et al., 1998; Meyers et al., 2001). A similar response was noted when examining MSH2−/− and MSH2+/+ murine embryonic stem (ES) cells (Meyers et al., 2005). For example, transient and prolonged G2 arrest responses occurred at drug concentrations (e.g. 1.5 nM FdUrd in MSH2- ES cells) that caused no significant loss of survival. Similar to MLH1-deficient cells, MSH2-deficient cells showed an abrogated G2 arrest response to FdUrd or FUra treatments. Thus, G2 arrest in response to FP exposure also relied on an intact MMR system and was not merely dependent on MLH1 expression. As noted above, no differences in G2 arrest responses were noted after Tomudex™ exposure in isogenic cell lines expressing or lacking MLH1 or MSH2. Thymidine (dThyd) depletion in both cell systems as a consequence of the inhibition of TS activity, S-phase arrest independent of MMR status was found (Meyers et al., 2005) as described (Orlandi et al., 1999; Yin et al., 1999).
In its role in post-replicative DNA repair, MMR detects DNA mispairs/lesions in the context of a newly synthesized DNA strand. It identifies the incorrect base in a mispair (placed there by a DNA polymerase error) because of its presence in the daughter strand (Karran, 2001). FdUrd, formed after FUra exposure, relies on DNA replication for its incorporation into DNA, whereby this pyrimidine analog is incorporated (as the base FU) across from Ade or Gua. We examined the cell cycle arrest responses of HCT116 (hMLH1-, MMR-) and HCT116 3-6 (hMLH1+, MMR+) cells within the first cell cycle after treatment. While both cell lines responded with a strong G2 arrest by 20 h after FdUrd addition, only MMR+ HCT116 3-6 cells responded with a prolonged G2 arrest caused by MMR-dependent proof-reading. Identical G2 arrest responses were noted in the first cell division in MSH2+ cells, whereas MSH2- cells did not arrest.
hMSH2-hMSH6 recognizes FUra:Gua lesions
To assess the ability of MMR to directly recognize FP-induced lesions in DNA, we tested the ability of purified hMSH2-hMSH6 or hMSH2-hMSH3 heterodimers to recognize FP lesions (specifically FU base-paired with Ade or Gua) using 41-mer oligonucleotide substrates. MMR activities using these DNA substrates were assessed by ATPase activities (Mazurek et al., 2002). A Thy:Gua base pair (as a positive control), but not a Thy:Ade base pair (as a negative control), was able to activate the ATPase (i.e. the ATPase velocity) of hMSH2-hMSH6. Interestingly, FUra:Gua and Ura:Gua, but not FUra:Ade or Ura:Ade base pairs, were able to activate MMR activity. Importantly, MMR was not capable of recognizing the dThyd analogs, Ura or FUra, when directly base-paired with Ade. Instead, MMR only detected FUra or Ura when mispaired with Gua. Ade is the expected base-pairing partner for the dThyd/Urd analogs, Ura and FUra. We also examined the ability of hMSH2-hMSH3 complexes to recognize various FUra or Ura substrates. The hMSH2-hMSH3 complex is primarily responsible for recognizing small insertion and deletion loops in DNA (Fishel, 1999). MMR ATPase activity from the hMSH2-hMSH3 complex was observed with the positive control (the Cyt-Ade loop), but not the negative control (Thy:Ade). As expected, neither FUra:Ade nor FUra:Gua were substrates for MMR and, therefore, did not activate the ATPase of the hMSH2-hMSH3 complex.
MMR-deficient cells incorporated higher FUra levels in their DNA
To determine if MMR status influenced the overall amount of radio-labeled FP incorporated into DNA, MMR-deficient and MMR-proficient cells, lacking either MLH1 (HCT116) or MSH2 (ES), were treated with various doses of FdUrd spiked with 20 to 50 mCi·mL−1[3H]FdUrd for 3 days, and genomic DNA purified and assayed for antimetabolite-related, incorporated radioactivity (Meyers et al., 2005). DNA of MLH1-deficient HCT116 or MSH2−/− ES cells treated with 2.5 mM FdUrd contained 2.6- and 2.3-fold greater incorporated radioactive FdUrd, respectively, than their MMR-competent counterparts. Addition of excess dThyd, but not Urd, prevented incorporation of FdUrd into DNA, consistent with the ability of dThyd to rescue FdUrd-induced toxicity in these cells, as previously reported for MMR+ cells (Meyers et al., 2001).
FdUrd-treated MMR- cells selectively incorporated low, but significant levels of FUra:Gua in DNA
As the hMSH2-hMSH6 heterodimer selectively recognized FUra:Gua lesions, we examined the frequency of these mismatch bases in the DNA of FdUrd- or FUra-treated HCT116 cells relative to FUra:Ade base pairs. To distinguish FUra:Ade from FUra:Gua radiolabeled lesions, specific BER enzymes were used. UDG removes Ura and FUra from DNA regardless of their base pairing partners; it can recognize Ura:Ade, FUra:Ade, Ura:Gua, and FUra:Gua, as well as other base pairings (Krokan et al., 2000; Meyers et al., 2003). In contrast, MBD4 (also known as methyl-CpG-binding endonuclease I) only recognizes Ura:Gua or FUra:Gua, but not Ura:Ade or FUra:Ade, in DNA (Petronzelli et al., 2000; Meyers et al., 2003). Indeed, UDG recognized both FUra:Ade and FUra:Gua in the same 41-mer oligomer substrates, indicated by the generation of a cleavage product following hot alkali treatment. In contrast, MBD4 only recognized DNA substrates containing FUra:Gua lesions. In fact, MBD4 recognized FUra:Gua regardless of Cyt methylation status (methylation in human DNA occurs on Cyt in a CpG context). This was interesting as MBD4 was once thought to be the human homologue of the bacterial MutH endonuclease (Bellacosa et al., 1999; Bellacosa, 2001) that allowed MMR to direct repair to the newly synthesized strand based on its methylation status. In bacteria, the daughter strand (which of course would contain the newly incorporated incorrect base) is transiently unmethylated at Ade in a d(GATC) context immediately following replication (Modrich, 1991).
Using UDG and MBD4, MMR-deficient and -proficient cells were analysed for FUra base-pair incorporation analyses. HCT116 and HCT116 3-6 cells were incubated with [3H]FdUrd for 3–10 days and genomic DNA isolated (Meyers et al., 2005). DNA was then treated with UDG or MBD4 (both of which detect Urd or FUrd in DNA and release the free bases, Ura or FUra respectively) to investigate total FUra incorporation compared with FUra incorporated into FUra:Gua lesions. Whereas levels of FUra incorporated across from Gua (as determined by the amount of [3H] released following MBD4 treatment) were equivalent in MMR- HCT116 cells compared with MMR+ HCT116 3-6 cells after 3 days of treatment, there was threefold more FUra:Gua in HCT116 DNA at day 10. This incorporation difference correlated well with lethality, where a difference in cell survival was noted only following longer (i.e. 10 day) exposures to FdUrd (Meyers et al., 2001). In addition, the fact that the amount of [3H] released from DNA after UDG treatment was only modestly higher (in three of four instances) than that released after MBD4 treatment indicates that at least half of the FUra in DNA was paired with Gua. To rule out the possibility that MBD4 levels may be different between MMR- and MMR+ cells, levels of MBD4 protein were examined and found equivalent in HCT116 and HCT116 3-6 cells, as well as in the other cell systems used.
Can FPs be used to treat MSI+, hMLH1- sporadic cancers?
Given that many colon and ovarian cancers are microsatellite instable due to the lack of hMLH1expression caused by hypermethylation of the hMLH1 promoter, is there any hope of treating these cancers using FPs? Recently, one FP, 5-fluoro-2′-deoxycytidine (FdCyd), has been shown to be a hypomethylating agent when incorporated into the DNA of exposed cells (Smith et al., 1992; Chuang et al., 2005). The fluorine group in the 5-position of FdCyd resists methylation because of its carbon-fluorine bond. Indeed, exposing cells to FdCyd alters the methylation pattern of exposed cells (Chuang et al., 2005).
Importantly, prior research from our laboratories has shown that the metabolism of FdCyd can be manipulated to: (i) protect FdCyd from deamination using the dCMP and/or cytidine deaminase inhibitors, deoxytetrahydrouridine (dH4Urd) or tetrahydrouridine (H4Urd) respectively (Boothman et al., 1985; 1987a; 1987b;) (Figure 4); and (ii) direct FdCyd to dramatically increase (∼20-fold) incorporation into the DNA of cells co-treated with dH4Urd, which results in the simultaneous inhibition of both cytidine deaminase (as dH4Urd) and dCMP deaminaase (as dH4UMP) (Figure 4). Indeed, we have recently demonstrated that hMLH1- RKO6 cells, which are normally resistant to FdUrd alone, become sensitive to FdCyd (Figure 5) through the re-expression of hMLH1 (not shown). A dramatic increase in G2 arrest responses were noted when RKO6 cells were exposed to FdCyd. In contrast, RKO6 cells were completely resistant to FdUrd (Figure 5). Recently, Beumer et al. (2006; 2008;) have extended our prior mouse data (Boothman et al., 1987a,b;) by examining pharmacokinetics, metabolism, oral bioavailability, and cytotoxic metabolites of FdCyd in mice and patients. As we previously demonstrated, potential toxic metabolites generated by CD were avoided by combing FdCyd in treatment with 3,4,5,6-tetrahydrouridine (H4Urd). Accompanying plasma 5-FU and FdUrd concentrations were <10% of these observed after therapeutic infusions of 5-FU or FdUrd, while FdCyd levels were well above those required for the inhibition of methylation in vitro. (Beumer et al., 2006; 2008;).
Figure 4.
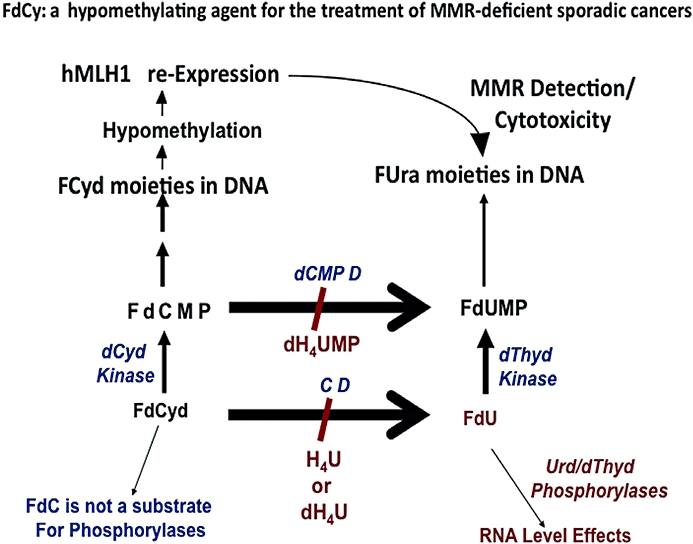
Potential use of 5-fluoro-2′-deoxycytidine (FdCyd) for treatment of hMLH1-, MMR-deficient sporadic cancers. FdCyd, which is not a substrate for RNA-level metabolism (e.g. dThyd or Urd phosphorylases), can be manipulated for increased incorporation into DNA by using the dCyd or dCMP deaminase (CD or dCMPD respectively) inhibitors, tetrahydrouridine (H4Urd) or deoxytetrahydrouridine (dH4Urd) respectively. Once incorporated, FdCyd can lead to hypomethylation of DNA and re-expression of hMLH1 in MSI+ sporadic cancers that are genetically wild-type for hMLH1, but lack hMLH1 expression because of hypermethylation of its promoter, as described by Veigl et al., 1998. Adapted from Meyers et al. (2003). hMLH1, human MutL homologue-1.
Figure 5.

Sensitization of MSI+, hMLH1- RKO6 cells using FdCyd. RKO6 cells are wild-type for the hMLH1 gene, yet are MSI+, and lack expression of hMLH1 protein because of hypermethylation of its promoter. RKO6 cells were synchronized by low serum medium and released by re-plating into 10% fetal calf serum-containing medium. Cells were then mock-treated (untreated cells) or exposed to 1 µM FdUrd or FdCyd for 62 h, past the p53 checkpoint as described (Meyers et al., 2001; 2005;). Cells were then monitored for cell cycle checkpoint responses and changes in the G2 cell population were graphed over time (h) after release. Note that FdCyd treatment caused G2 arrest with corresponding hMLH1 expression (not shown). Dashed line, mock-treated synchronized cells; Open squares, FdUrd (1.0 µM, 4 h); Closed circles, FdCyd (1.0 µM, 4 h). Western blotting demonstrated hMLH1 expression and hPMS2 stabilization after FdCyd, but not after FdUrd, exposures. hMLH1, human MutL homologue-1.
We propose that FdCyd exposures can be used to cause the re-expression of hMLH1, and thereby, convert resistant hypermethylated hMLH1- colon or ovarian cancer cells to sensitive cells that re-express functional hMLH1, and therefore MMR. This is particularly true for colon cancer cells that have elevated levels of dCyd kinase, whereby dH4Urd [that can inhibit both CD and deoxycytidylate deaminase (dCMPD), Figure 4)] can be effectively used to channel FdCyd into DNA (Boothman et al., 1985). After approximately 2–4 days or several cell divisions, reversal of cytidine and dCMP deaminase inhibition (by removal of the reversible inhibitor, dH4Urd) followed by continuous exposure with FdCyd would allow its incorporation into DNA, hypomethlyation of the hMLH1 promoter, stimulated re-expression of hMLH1 protein and restored MMR activity. Restoration of functional MMR would, in turn, result in increased sensitivity of cells to FPs. Once dH4Urd is removed, the accumulated pools of FdCyd and FdCMP then would be converted by deamination to FdUrd and FdUMP by CD and dCMPD (which are also elevated in colon cancers), resulting in enhanced incorporation of FdUrd into DNA and formation of elevated levels of FdUMP (Figure 4). Re-expression of hMLH1 then would increase sensitivity of cells to the now converted FdUrd that incorporates into DNA because of elevated levels of dThyd kinase (TK). Thus, instead of trying to expose cells to aza-cytidine, a typical hypomethylating agent used to re-express genes for increased sensitivity (Compere and Palmiter, 1981), with FUra or FdUrd, one agent (FdCyd) can be used for hMLH1 re-expression and enhanced drug sensitivity of otherwise resistant cells.
Conclusion
Our studies have revealed that DNA mismatch repair (MMR), whose activities greatly affect the sensitivities of cells exposed to FPs. Understanding the mechanisms by which MMR mediates lethality to FPs has revealed several targets that could be exploited for enhanced sensitivity of cancer cells (particularly colon and endometrial cancers that more commonly have MMR deficiencies) to FPs. For example, our studies strongly suggest that c-Abl inhibitors, such as Gleevec™, should not be used in conjunction with regimen that use cisplatin or Temozolomide™ for the treatment of MMR proficient cells. Overcoming hypomethylation of hMLH1 is one example, as noted above. Other mechanisms include the signalling mechanisms that arise after FUra:Gua moieties are detected and responded to by MMR. Although incorporation of FUra:Gua moieties are formed rarely, unlike FUra:Ade lesions that are mutagenic, FUra:Gua (simulating G:T mismatches) become lethal in one cell division by MMR recognition and signalling. Although increased DSBs in FdUrd-treated, MMR-competent cells were noted (Meyers et al., 2001), it is clear that MMR-directed signalling, through the c-Abl/p73α/GADD45α pathway of G2 arrest and apoptosis, plays a critical role in lethality responses to FPs. Thus, activating the c-Abl kinase pathway independent of MMR function in cells devoid of this repair capacity might allow given therapies to overcome this particular resistance mechanism to FPs.
Acknowledgments
This work was supported by Department of Energy (DE-FG-022179) and NIH/NCI (R01-CA-83196) grants to DAB, as well as by NIH grants GM080176 and CA067007 to RF. This publication is CSCN 021 from the Program in Cell Stress and Cancer Nanomedicine, Simmons Comprehensive Cancer Center, UT Southwestern.
Glossary
Abbreviations:
- 5,10-CH2FH4
5,10-methylene tetrahydrofolate
- 5′-dRP
5′-deoxyribose phosphate
- AM
adaptors/mediators
- AP
apyrimidinic/apurinic site
- APE
apurinic/apyrimidinic endonuclease
- ATM
ataxia telangiectasia mutated
- ATR
ataxia telangiectasia-and-rad3-related
- BER
base excision repair
- CD
cytosine deaminase
- dCMP
deoxycytidylate
- dCMPD
deoxycytidylate deaminase
- dH4Urd
deoxytetrahydrouridine
- DS
damage sensors
- DSBs
DNA double-strand breaks
- dThyd
thymidine
- ES
embryonic stem
- FdCyd
5-fluoro-2′-deoxycyticine
- FdUDP
5-fluoro-2′-deoxyuridine 5′-diphosphate
- FdUMP
5-fluoro-2′-deoxyuridine 5′-monophosphate
- FdUTP
5-fluoro-2′-deoxyuridine-5′-triphosphate
- FdUrd
5-fluoro-2′-deoxyuridine
- FEN1
flap-endonuclease 1
- FPs
fluorinated pyrimidines
- FUDP
5-fluorouridine 5′-diphosphate
- FUra (or 5-FU)
5-fluorouracil
- FUrd
fluorouridine
- FUTP
5-fluorouridine-5′-triphosphate
- GADD45
growth arrest and DNA damage-inducible-45 gene/protein
- H4Urd
3,4,5,6-tetrahydrouridine
- hMLH1
human MutL homologue-1
- hMSH2
human MutS homologue-2
- hMSH3
human MutS homologue-3
- hMSH6
human MutS homologue-6
- hPMS2
postmeiotic segregation increased 2
- IDL
insertion/deletion loop-type
- MBD4
methyl-CpG binding domain protein 4
- MMR
mismatch repair
- MNNG
N-methyl-N'-nitro-N-nitrosoguanidine
- MNU
N-methyl-N-nitrosourea
- PIKK
phosphotidylinositol-3′-kinase; Polß; polymerase-beta
- rR
ribonucleotide reductase
- SSB
single-stranded DNA binding protein
- ssDNA
single-stranded DNA
- STI571
Gleevec™
- Thy
thymine
- TK
thymidine kinase
- TP
thymidine phosphorylase
- TS
thymidylate synthase
- UDGs
Uracil-DNA glycosylases
- Ura
uracil
References
- Acharya S, Wilson T, Gradia S, Kane MF, Guerrette S, Marsischky GT, et al. hMSH2 forms specific mispair-binding complexes with hMSH3 and hMSH6. Proc Natl Acad Sci USA. 1996;93:13629–13634. doi: 10.1073/pnas.93.24.13629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acharya S, Foster PL, Brooks P, Fishel R. The coordinated functions of the E. coli MutS and MutL proteins in mismatch repair. Mol Cell. 2003;12:233–246. doi: 10.1016/s1097-2765(03)00219-3. [DOI] [PubMed] [Google Scholar]
- Aebersold PM. Mutation induction by 5-fluorodeoxyuridine in synchronous Chinese hamster cells. Cancer Res. 1979;39:808–810. [PubMed] [Google Scholar]
- Allen DJ, Makhov A, Grilley M, Taylor J, Thresher R, Modrich P, et al. MutS mediates heteroduplex loop formation by a translocation mechanism. EMBO J. 1997;16:4467–4476. doi: 10.1093/emboj/16.14.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban C, Yang W. Structural basis for MutH activation in E. coli mismatch repair and relationship of MutH to restriction endonucleases. EMBO J. 1998;17:1526–1534. doi: 10.1093/emboj/17.5.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban C, Junop M, Yang W. Transformation of MutL by ATP binding and hydrolysis: a switch in DNA mismatch repair. Cell. 1999;97:85–97. doi: 10.1016/s0092-8674(00)80717-5. [DOI] [PubMed] [Google Scholar]
- Bellacosa A. Role of MED1 (MBD4) gene in DNA repair and human cancer. J Cell Physiol. 2001;187:137–144. doi: 10.1002/jcp.1064. [DOI] [PubMed] [Google Scholar]
- Bellacosa A, Cicchillitti L, Schepis F, Riccio A, Yeung AT, Matsumoto Y, et al. MED1, a novel human methyl-CpG-binding endonuclease, interacts with DNA mismatch repair protein MLH1. Proc Natl Acad Sci USA. 1999;96:3969–3974. doi: 10.1073/pnas.96.7.3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berardini M, Mazurek A, Fishel R. The effect of O6-methylguanine DNA adducts on the adenosine nucleotide switch functions of hMSH2-hMSH6 and hMSH2-hMSH3. J Biol Chem. 2000;275:27851–27857. doi: 10.1074/jbc.M003589200. [DOI] [PubMed] [Google Scholar]
- Beumer JH, Eiseman JL, Parise RA, Joseph E, Holleran JL, Covey JM, et al. Pharmacokinetics, metabolism, and oral bioavailability of the DNA methyltransferase inhibitor 5-fluoro-2′-deoxycytidine in mice. Clin Cancer Res. 2006;12:7483–7491. doi: 10.1158/1078-0432.CCR-06-1250. [DOI] [PubMed] [Google Scholar]
- Beumer JH, Parise RA, Newman EM, Doroshow JH, Synold TW, Lenz HJ, et al. Concentrations of the DNA methyltransferase inhibitor 5-fluoro-2′-deoxycytidine (FdCyd) and its cytotoxic metabolites in plasma of patients treated with FdCyd and tetrahydrouridine (THU) Cancer Chemother Pharmacol. 2008;62:363–368. doi: 10.1007/s00280-007-0603-8. [DOI] [PubMed] [Google Scholar]
- Blackwell LJ, Bjornson KP, Modrich P. DNA-dependent activation of the hMutSalpha ATPase. J Biol Chem. 1998;273:32049–32054. doi: 10.1074/jbc.273.48.32049. [DOI] [PubMed] [Google Scholar]
- Boothman DA, Briggle TV, Greer S. Metabolic channeling of 5-fluoro-2′-deoxycytidine utilizing inhibitors of its deamination in cell culture. Mol Pharmacol. 1985;27:584–594. [PubMed] [Google Scholar]
- Boothman DA, Briggle TV, Greer S. Protective, tumor-selective dual pathway activation of 5-fluoro-2′-deoxycytidine provided by tetrahydrouridine in mice bearing mammary adenocarcinoma-755. Cancer Res. 1987a;47:2344–2353. [PubMed] [Google Scholar]
- Boothman DA, Briggle TV, Greer S. Tumor-selective metabolism of 5-fluoro-2′-deoxycytidine coadministered with tetrahydrouridine compared to 5-fluorouracil in mice bearing Lewis lung carcinoma. Cancer Res. 1987b;47:2354–2362. [PubMed] [Google Scholar]
- Boothman DA, Briggle TV, Greer S. Exploitation of elevated pyrimidine deaminating enzymes for selective chemotherapy. Pharmacol Ther. 1989;42:65–88. doi: 10.1016/0163-7258(89)90022-3. [DOI] [PubMed] [Google Scholar]
- Caradonna SJ, Cheng YC. The role of deoxyuridine triphosphate nucleotidohydrolase, uracil-DNA glycosylase, and DNA polymerase alpha in the metabolism of FUdR in human tumor cells. Mol Pharmacol. 1980;18:513–520. [PubMed] [Google Scholar]
- Carethers JM, Smith EJ, Behling CA, Nguyen L, Tajima A, Doctolero RT, et al. Use of 5-fluorouracil and survival in patients with microsatellite-unstable colorectal cancer. Gastroenterology. 2004;126:394–401. doi: 10.1053/j.gastro.2003.12.023. [DOI] [PubMed] [Google Scholar]
- Chuang JC, Yoo CB, Kwan JM, Li TW, Liang G, Yang AS, et al. Comparison of biological effects of non-nucleoside DNA methylation inhibitors versus 5-aza-2′-deoxycytidine. Mol Cancer Ther. 2005;4:1515–1520. doi: 10.1158/1535-7163.MCT-05-0172. [DOI] [PubMed] [Google Scholar]
- Compere SJ, Palmiter RD. DNA methylation controls the inducibility of the mouse metallothionein-I gene lymphoid cells. Cell. 1981;25:233–240. doi: 10.1016/0092-8674(81)90248-8. [DOI] [PubMed] [Google Scholar]
- Cooper DL, Lahue RS, Modrich P. Methyl-directed mismatch repair is bidirectional. J Biol Chem. 1993;268:11823–11829. [PubMed] [Google Scholar]
- Das SK, Benditt EP, Loeb LA. Rapid changes in deoxynucleoside triphosphate pools in mammalian cells treated with mutagens. Biochem Biophys Res Commun. 1983;114:458–464. doi: 10.1016/0006-291x(83)90802-1. [DOI] [PubMed] [Google Scholar]
- Davis TW, Wilson-Van Patten C, Meyers M, Kunugi KA, Cuthill S, Reznikoff C, et al. Defective expression of the DNA mismatch repair protein, MLH1, alters G2-M cell cycle checkpoint arrest following ionizing radiation. Cancer Res. 1998;58:767–778. [PubMed] [Google Scholar]
- Dolnick BJ, Pink JJ. 5-fluorouracil modulation of dihydrofolate reductase RNA levels in methotrexate-resistant KB cells. J Biol Chem. 1983;258:13299–13306. [PubMed] [Google Scholar]
- Dolnick BJ, Pink JJ. Effects of 5-fluorouracil on dihydrofolate reductase and dihydrofolate reductase mRNA from methotrexate-resistant KB cells. J Biol Chem. 1985;260:3006–3014. [PubMed] [Google Scholar]
- Drummond JT, Li GM, Longley MJ, Modrich P. Isolation of an hMSH2-p160 heterodimer that restores DNA mismatch repair to tumor cells. Science. 1995;268:1909–1912. doi: 10.1126/science.7604264. [DOI] [PubMed] [Google Scholar]
- Elsaleh H, Powell B, Soontrapornchai P, Joseph D, Goria F, Spry N, et al. p53 gene mutation, microsatellite instability and adjuvant chemotherapy: impact on survival of 388 patients with Dukes' C colon carcinoma. Oncology. 2000;58:52–59. doi: 10.1159/000012079. [DOI] [PubMed] [Google Scholar]
- Fishel R. Mismatch repair, molecular switches, and signal transduction. Genes Dev. 1998;12:2096–2101. doi: 10.1101/gad.12.14.2096. [DOI] [PubMed] [Google Scholar]
- Fishel R. Signaling mismatch repair in cancer. Nat Med. 1999;5:1239–1241. doi: 10.1038/15191. [DOI] [PubMed] [Google Scholar]
- Fishel R. The selection for mismatch repair defects in hereditary nonpolyposis colorectal cancer: revising the mutator hypothesis. Cancer Res. 2001;61:7369–7374. [PubMed] [Google Scholar]
- Fishel RA, Kolodner R. Gene conversion in Escherichia coli: the identification of two repair pathways for mismatched nucleotides. UCLA Symp Mol Cell Biol. 1983;11:309–324. New Series. [Google Scholar]
- Fishel RA, Siegel EC, Kolodner R. Gene conversion in Escherichia coli. Resolution of heteroallelic mismatched nucleotides by co-repair. J Mol Biol. 1986;188:147–157. doi: 10.1016/0022-2836(86)90300-1. [DOI] [PubMed] [Google Scholar]
- Fram RJ, Cusick PS, Wilson JM, Marinus MG. Mismatch repair of cis-diamminedichloroplatinum(II)-induced DNA damage. Mol Pharmacol. 1985;28:51–55. [PubMed] [Google Scholar]
- Friedberg E, Walker G, Siede W, Wood R, Schultz R, Ellenberger T. DNA Repair and Mutagenesis. Washington DC: ASM press; 2006. [Google Scholar]
- Genschel J, Modrich P. Mechanism of 5′-directed excision in human mismatch repair. Mol Cell. 2003;12:1077–1086. doi: 10.1016/s1097-2765(03)00428-3. [DOI] [PubMed] [Google Scholar]
- Genschel J, Littman SJ, Drummond JT, Modrich P. Isolation of MutSbeta from human cells and comparison of the mismatch repair specificities of MutSbeta and MutSalpha. J Biol Chem. 1998;273:19895–19901. doi: 10.1074/jbc.273.31.19895. [DOI] [PubMed] [Google Scholar]
- Glazer PM, Sarkar SN, Chisholm GE, Summers WC. DNA mismatch repair detected in human cell extracts. Mol Cell Biol. 1987;7:218–224. doi: 10.1128/mcb.7.1.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong JG, Costanzo A, Yang HQ, Melino G, Kaelin WG, Jr, Levrero JY, et al. The tyrosine kinase c-Abl regulates p73 in apoptotic response to cisplatin-induced DNA damage. Nature. 1999;399:806–809. doi: 10.1038/21690. [DOI] [PubMed] [Google Scholar]
- Gradia S, Acharya S, Fishel R. The human mismatch recognition complex hMSH2-hMSH6 functions as a novel molecular switch. Cell. 1997;91:995–1005. doi: 10.1016/s0092-8674(00)80490-0. [DOI] [PubMed] [Google Scholar]
- Gradia S, Subramanian D, Wilson T, Acharya S, Makhov A, Griffith J, et al. hMSH2-hMSH6 forms a hydrolysis-independent sliding clamp on mismatched DNA. Mol Cell. 1999;3:255–261. doi: 10.1016/s1097-2765(00)80316-0. [DOI] [PubMed] [Google Scholar]
- Gradia S, Acharya S, Fishel R. The role of mismatched nucleotides in activating the hMSH2-hMSH6 molecular switch. J Biol Chem. 2000;275:3922–3930. doi: 10.1074/jbc.275.6.3922. [DOI] [PubMed] [Google Scholar]
- Grilley M, Welsh KM, Su SS, Modrich P. Isolation and characterization of the Escherichia coli mutL gene product. J Biol Chem. 1989;264:1000–1004. [PubMed] [Google Scholar]
- Grilley M, Griffith J, Modrich P. Bidirectional excision in methyl-directed mismatch repair. J Biol Chem. 1993;268:11830–11837. [PubMed] [Google Scholar]
- Guerrette S, Wilson T, Gradia S, Fishel R. Interactions of human hMSH2 with hMSH3 and hMSH2 with hMSH6: examination of mutations found in hereditary nonpolyposis colorectal cancer. Mol Cell Biol. 1998;18:6616–6623. doi: 10.1128/mcb.18.11.6616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrette S, Acharya S, Fishel R. The interaction of the human MutL homologues in hereditary nonpolyposis colon cancer. J Biol Chem. 1999;274:6336–6341. doi: 10.1074/jbc.274.10.6336. [DOI] [PubMed] [Google Scholar]
- Haber LT, Walker GC. Altering the conserved nucleotide binding motif in the Salmonella typhimurium MutS mismatch repair protein affects both its ATPase and mismatch binding activities. EMBO J. 1991;10:2707–2715. doi: 10.1002/j.1460-2075.1991.tb07815.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haug T, Skorpen F, Aas PA, Malm V, Skjelbred C, Krokan HE. Regulation of expression of nuclear and mitochondrial form of human uracil-DNA glycosylase. Nucleic Acids Res. 1998;26:1449–1457. doi: 10.1093/nar/26.6.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidelberger C, Danenberg PV, Moran RG. Fluorinated pyrimidines and their nucleosides. Adv Enzymol Relat Areas Mol Biol. 1983;54:58–119. [PubMed] [Google Scholar]
- Heinen CD, Wilson T, Mazurek A, Berardini M, Butz C, Fishel R. HNPCC mutations in hMSH2 result in reduced hMSH2-hMSH6 molecular switch functions. Cancer Cell. 2002;1:469–478. doi: 10.1016/s1535-6108(02)00073-9. [DOI] [PubMed] [Google Scholar]
- Hemminki A, Mecklin JP, Jarvinen H, Aaltonen LA, Joensuu H. Microsatellite instability is a favorable prognostic indicator in patients with colorectal cancer receiving chemotherapy. Gastroenterology. 2000;119:921–928. doi: 10.1053/gast.2000.18161. [DOI] [PubMed] [Google Scholar]
- Hess MT, Gupta RD, Kolodner RD. Dominant Saccharomyces cerevisiae msh6 mutations cause increased mispair binding and decreased dissociation from mispairs by Msh2-Msh6 in the presence of ATP. J Biol Chem. 2002;277:25545–25553. doi: 10.1074/jbc.M202282200. [DOI] [PubMed] [Google Scholar]
- Hickman MJ, Samson LD. Role of DNA mismatch repair and p53 in signaling induction of apoptosis by alkylating agents. Proc Natl Acad Sci USA. 1999;96:10764–10769. doi: 10.1073/pnas.96.19.10764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes J, Jr, Clark S, Modrich P. Strand-specific mismatch correction in nuclear extracts of human and Drosophila melanogaster cell lines. Proc Natl Acad Sci USA. 1990;87:5837–5841. doi: 10.1073/pnas.87.15.5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins RL, Goodman MF. Deoxyribonucleotide pools, base pairing, and sequence configuration affecting bromodeoxyuridine- and 2-aminopurine-induced mutagenesis. Proc Natl Acad Sci USA. 1980;77:1801–1805. doi: 10.1073/pnas.77.4.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai K, Slupphaug G, Lee W-I, Revy P, Nonoyama S, Catalan N, et al. Human uracil-DNA glycosylase deficiency associated with profoundly impaired immunoglobulin class-switch recombination. Nat Immunol. 2003;4:1023–1028. doi: 10.1038/ni974. [DOI] [PubMed] [Google Scholar]
- Ingraham HA, Tseng BY, Goulian M. Mechanism for exclusion of 5-fluorouracil from DNA. Cancer Res. 1980;40:998–1001. [PubMed] [Google Scholar]
- Irving JA, Hall AG. Mismatch repair defects as a cause of resistance to cytotoxic drugs. Expert Rev Anticancer Ther. 2001;1:149–158. doi: 10.1586/14737140.1.1.149. [DOI] [PubMed] [Google Scholar]
- Johnston PG, Takimoto CH, Grem JL, Chabner BA, Allegra CJ, Chu E. Antimetabolites. Cancer Chemother Biol Response Modif. 1996;16:1–27. [PubMed] [Google Scholar]
- Jover R, Zapater P, Castells A, Llor X, Andreu M, Cubiella J, et al. Mismatch repair status in the prediction of benefit from adjuvant fluorouracil chemotherapy in colorectal cancer. Gut. 2006;55:848–855. doi: 10.1136/gut.2005.073015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jover R, Zapater P, Castells A, Llor X, Andreu M, Cubiella J, et al. The efficacy of adjuvant chemotherapy with 5-fluorouracil in colorectal cancer depends on the mismatch repair status. Eur J Cancer. 2009;45:365–373. doi: 10.1016/j.ejca.2008.07.016. [DOI] [PubMed] [Google Scholar]
- Kadyrov FA, Dzantiev L, Constantin N, Modrich P. Endonucleolytic function of MutLalpha in human mismatch repair. Cell. 2006;126:297–308. doi: 10.1016/j.cell.2006.05.039. [DOI] [PubMed] [Google Scholar]
- Karran P. Mechanisms of tolerance to DNA damaging therapeutic drugs. Carcinogenesis. 2001;22:1931–1937. doi: 10.1093/carcin/22.12.1931. [DOI] [PubMed] [Google Scholar]
- Karran P, Marinus MG. Mismatch correction at O6-methylguanine residues in E. coli DNA. Nature. 1982;296:868–869. doi: 10.1038/296868a0. [DOI] [PubMed] [Google Scholar]
- Kastan MB, Onyekwere O, Sidransky D, Vogelstein B, Craig RW. Participation of p53 protein in the cellular response to DNA damage. Cancer Res. 1991;51:6304–6311. [PubMed] [Google Scholar]
- Kolodner RD, Mendillo ML, Putnam CD. Coupling distant sites in DNA during DNA mismatch repair. Proc Natl Acad Sci USA. 2007;104:12953–12954. doi: 10.1073/pnas.0705698104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krokan HE, Nilsen H, Skorpen F, Otterlei M, Slupphaug G. Base excision repair of DNA in mammalian cells. FEBS Lett. 2000;476:73–77. doi: 10.1016/s0014-5793(00)01674-4. [DOI] [PubMed] [Google Scholar]
- Krokan HE, Drablos F, Slupphaug G. Uracil in DNA – occurrence, consequences and repair. Oncogene. 2002;21:8935–8948. doi: 10.1038/sj.onc.1205996. [DOI] [PubMed] [Google Scholar]
- van Laar JA, van der Wilt CL, Rustum YM, Noordhuis P, Smid K, Pinedo HM, et al. Therapeutic efficacy of fluoropyrimidines depends on the duration of thymidylate synthase inhibition in the murine colon 26-B carcinoma tumor model. Clin Cancer Res. 1996;2:1327–1333. [PubMed] [Google Scholar]
- van Laar JA, Rustum YM, Ackland SP, van Groeningen CJ, Peters GJ. Comparison of 5-fluoro-2′-deoxyuridine with 5-fluorouracil and their role in the treatment of colorectal cancer. Eur J Cancer. 1998;34:296–306. doi: 10.1016/s0959-8049(97)00366-3. [DOI] [PubMed] [Google Scholar]
- Lahue RS, Su SS, Modrich P. Requirement for d(GATC) sequences in Escherichia coli mutHLS mismatch correction. Proc Natl Acad Sci USA. 1987;84:1482–1486. doi: 10.1073/pnas.84.6.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahue RS, Au KG, Modrich P. DNA mismatch correction in a defined system. Science. 1989;245:160–164. doi: 10.1126/science.2665076. [DOI] [PubMed] [Google Scholar]
- Lamers MH, Perrakis A, Enzlin JH, Winterwerp HH, de Wind N, Sixma TK. The crystal structure of DNA mismatch repair protein MutS binding to a G x T mismatch. Nature. 2000;407:711–717. doi: 10.1038/35037523. [DOI] [PubMed] [Google Scholar]
- Li GM, Modrich P. Restoration of mismatch repair to nuclear extracts of H6 colorectal tumor cells by a heterodimer of human MutL homologs. Proc Natl Acad Sci USA. 1995;92:1950–1954. doi: 10.1073/pnas.92.6.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LS, Kim NG, Kim SH, Park C, Kim H, Kang HJ, et al. Chromosomal imbalances in the colorectal carcinomas with microsatellite instability. Am J Pathol. 2003;163:1429–1436. doi: 10.1016/S0002-9440(10)63500-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LS, Morales JC, Hwang A, Wagner MW, Boothman DA. DNA mismatch repair-dependent activation of c-Abl/p73alpha/GADD45alpha-mediated apoptosis. J Biol Chem. 2008;283:21394–21403. doi: 10.1074/jbc.M709954200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DP, Wang Y, Scherer SJ, Clark AB, Yang K, Avdievich E, et al. An Msh2 point mutation uncouples DNA mismatch repair and apoptosis. Cancer Res. 2004;64:517–522. doi: 10.1158/0008-5472.can-03-2957. [DOI] [PubMed] [Google Scholar]
- Lindahl T, Nyberg B. Heat-induced deanimation of cytosine residues in deoxyribonucleic acid. Biochemistry. 1974;13:3405–3410. doi: 10.1021/bi00713a035. [DOI] [PubMed] [Google Scholar]
- Lu AL, Clark S, Modrich P. Methyl-directed repair of DNA base-pair mismatches in vitro. Proc Natl Acad Sci USA. 1983;80:4639–4643. doi: 10.1073/pnas.80.15.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson SW. Escherichia coli helicase II (urvD gene product) translocates unidirectionally in a 3′ to 5′ direction. J Biol Chem. 1986;261:10169–10175. [PubMed] [Google Scholar]
- Mazurek A, Berardini M, Fishel R. Activation of human MutS homologs by 8-oxo-guanine DNA damage. J Biol Chem. 2002;277:8260–8266. doi: 10.1074/jbc.M111269200. [DOI] [PubMed] [Google Scholar]
- Mazurek A, Johnson CN, Germann MW, Fishel R. Sequence context effect for hMSH2-hMSH6 mismatch-dependent activation. Proc Natl Acad Sci USA. 2009;106:4177–4182. doi: 10.1073/pnas.0808572106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekras JA, Boothman DA, Perez LM, Greer S. Use of 5-fluorodeoxycytidine and tetrahydrouridine to exploit high levels of deoxycytidylate deaminase in tumors to achieve DNA- and target-directed therapies. Cancer Res. 1984;44:2551–2560. [PubMed] [Google Scholar]
- Meyers M, et al. Loss of DNA mismatch repair reveals resistance to 5-fluorouracil. AACR. 1996 Abstract. [Google Scholar]
- Meyers M, Theodosiou M, Acharya S, Odegaard E, Wilson T, Lewis JE, et al. Cell cycle regulation of the human DNA mismatch repair genes hMSH2, hMLH1, and hPMS2. Cancer Res. 1997;57:206–208. [PubMed] [Google Scholar]
- Meyers M, Wagner MW, Hwang HS, Kinsella TJ, Boothman DA. Role of the hMLH1 DNA mismatch repair protein in fluoropyrimidine-mediated cell death and cell cycle responses. Cancer Res. 2001;61:5193–5201. [PubMed] [Google Scholar]
- Meyers M, Hwang A, Wagner MW, Bruening AJ, Veigl ML, Sedwick WD, et al. A role for DNA mismatch repair in sensing and responding to fluoropyrimidine damage. Oncogene. 2003;22:7376–7388. doi: 10.1038/sj.onc.1206941. [DOI] [PubMed] [Google Scholar]
- Meyers M, Wagner MW, Mazurek A, Schmutte C, Fishel R, Boothman DA. DNA mismatch repair-dependent response to fluoropyrimidine-generated damage. J Biol Chem. 2005;280:5516–5526. doi: 10.1074/jbc.M412105200. [DOI] [PubMed] [Google Scholar]
- Modrich P. Methyl-directed DNA mismatch correction. J Biol Chem. 1989;264:6597–6600. [PubMed] [Google Scholar]
- Modrich P. Mechanisms and biological effects of mismatch repair. Annu Rev Genet. 1991;25:229–253. doi: 10.1146/annurev.ge.25.120191.001305. [DOI] [PubMed] [Google Scholar]
- Modrich P. Strand-specific mismatch repair in mammalian cells. J Biol Chem. 1997;272:24727–24730. doi: 10.1074/jbc.272.40.24727. [DOI] [PubMed] [Google Scholar]
- Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- Natrajan G, Lamers MH, Enzlin JH, Winterwerp HHK, Perrakis A, Sixma TK. Structures of Escherichia coli DNA mismatch repair enzyme MutS in complex with different mismatches: a common recognition mode for diverse substrates. Nucleic Acids Res. 2003;31:4814–4821. doi: 10.1093/nar/gkg677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman CN, Miller JH. Mutagen-induced changes in cellular deoxycytidine triphosphate and thymidine triphosphate in Chinese hamster ovary cells. Biochem Biophys Res Commun. 1983;114:34–40. doi: 10.1016/0006-291x(83)91590-5. [DOI] [PubMed] [Google Scholar]
- Nio Y, Shiraishi T, Tsubono M, Morimoto H, Tseng CC, Kawabata K, et al. Relationship of in vivo antitumor activities of fluorinated pyrimidines to thymidylate synthase activity and intratumoral concentrations of 5-fluorouracil and uracil. Anticancer Res. 1991;11:607–612. [PubMed] [Google Scholar]
- Obmolova G, Ban C, Hsieh P, Yang W. Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature. 2000;407:703–710. doi: 10.1038/35037509. [DOI] [PubMed] [Google Scholar]
- Orlandi L, Bearzatto A, Abolafio G, De Marco C, Daidone MG, Zaffaroni N. Involvement of bcl-2 and p21waf1 proteins in response of human breast cancer cell clones to Tomudex. Br J Cancer. 1999;81:252–260. doi: 10.1038/sj.bjc.6690685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otterlei M, Haug T, Nagelhus TA, Slupphaug G, Lindmo T, Krokan HE. Nuclear and mitochondrial forms of human uracil-DNA glycosylase contains complex nuclear localisation and a strong classical mitochondrial localisation signal, respecticely. Nucleic Acids Res. 1998;26:4611–4617. doi: 10.1093/nar/26.20.4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palombo F, Iaccarino I, Nakajima E, Ikejima M, Shimada T, Jiricny J. hMutSbeta, a heterodimer of hMSH2 and hMSH3, binds to insertion/deletion loops in DNA. Curr Biol. 1996;6:1181–1184. doi: 10.1016/s0960-9822(02)70685-4. [DOI] [PubMed] [Google Scholar]
- Parker BO, Marinus MG. Repair of DNA heteroduplexes containing small heterologous sequences in Escherichia coli. Proc Natl Acad Sci USA. 1992;89:1730–1734. doi: 10.1073/pnas.89.5.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker WB, Cheng YC. Metabolism and mechanism of action of 5-fluorouracil. Pharmacol Ther. 1990;48:381–395. doi: 10.1016/0163-7258(90)90056-8. [DOI] [PubMed] [Google Scholar]
- Petronzelli F, Riccio A, Markham GD, Seeholzer SH, Stoerker J, Genuardi M, et al. Biphasic kinetics of the human DNA repair protein MED1 (MBD4), a mismatch-specific DNA N-glycosylase. J Biol Chem. 2000;275:32422–32429. doi: 10.1074/jbc.M004535200. [DOI] [PubMed] [Google Scholar]
- Pluciennik A, Modrich P. Protein roadblocks and helix discontinuities are barriers to the initiation of mismatch repair. Proc Natl Acad Sci USA. 2007;104:12709–12713. doi: 10.1073/pnas.0705129104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prem veer Reddy G, Pardee AB. Multienzyme complex for metabolic channeling in mammalian DNA replication. Proc Natl Acad Sci USA. 1980;77:3312–3316. doi: 10.1073/pnas.77.6.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada C, Williams GT, Nilsen H, Barnes DE, Lindahl T, Neuberger MS. Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr Biol. 2002;12:1748–1755. doi: 10.1016/s0960-9822(02)01215-0. [DOI] [PubMed] [Google Scholar]
- Raschle M, Marra G, Nystrom-Lahti M, Schar P, Jiricny J. Identification of hMutLbeta, a heterodimer of hMLH1 and hPMS1. J Biol Chem. 1999;274:32368–32375. doi: 10.1074/jbc.274.45.32368. [DOI] [PubMed] [Google Scholar]
- veer Reddy GP, Pardee AB. Coupled ribonucleoside diphosphate reduction, channeling, and incorporation into DNA of mammalian cells. J Biol Chem. 1982;257:12526–12531. [PubMed] [Google Scholar]
- Ribic CM, Sargent DJ, Moore MJ, Thibodeau SN, French AJ, Goldberg RM, et al. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med. 2003;349:247–257. doi: 10.1056/NEJMoa022289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydberg B. Bromouracil mutagenesis and mismatch repair in mutator strains of Escherichia coli. Mutat Res. 1978;52:11–24. doi: 10.1016/0027-5107(78)90091-x. [DOI] [PubMed] [Google Scholar]
- Sancar A. DNA repair in humans. Annu Rev Genet. 1995;29:69–105. doi: 10.1146/annurev.ge.29.120195.000441. [DOI] [PubMed] [Google Scholar]
- Santi DV. Perspective on the design and biochemical pharmacology of inhibitors of thymidylate synthetase. J Med Chem. 1980;23:103–111. doi: 10.1021/jm00176a001. [DOI] [PubMed] [Google Scholar]
- Schmutte C, Marinescu RC, Sadoff MM, Guerrette S, Overhauser J, Fishel R. Human exonuclease I interacts with the mismatch repair protein hMSH2. Cancer Res. 1998;58:4537–4542. [PubMed] [Google Scholar]
- Schmutte C, Sadoff MM, Shim KS, Acharya S, Fishel R. The interaction of DNA mismatch repair proteins with human exonuclease I. J Biol Chem. 2001;276:33011–33018. doi: 10.1074/jbc.M102670200. [DOI] [PubMed] [Google Scholar]
- Shapiro R. Damage to DNA Caused by Hydrolysis. New York: Platium Press; 1981. [Google Scholar]
- Smith SS, Kaplan BE, Sowers LC, Newman EM. Mechanism of human methyl-directed DNA methyltransferase and the fidelity of cytosine methylation. Proc Natl Acad Sci USA. 1992;89:4744–4748. doi: 10.1073/pnas.89.10.4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobrero AF, Aschele C, Bertino JR. Fluorouracil in colorectal cancer – a tale of two drugs: implications for biochemical modulation. J Clin Oncol. 1997;15:368–381. doi: 10.1200/JCO.1997.15.1.368. [DOI] [PubMed] [Google Scholar]
- Spiegelman S, Nayak R, Sawyer R, Stolfi R, Martin D. Potentiation of the anti-tumor activity of 5FU by thymidine and its correlation with the formation of (5FU)RNA. Cancer. 1980a;45:1129–1134. doi: 10.1002/1097-0142(19800315)45:5+<1129::aid-cncr2820451317>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- Spiegelman S, Sawyer R, Nayak R, Ritzi E, Stolfi R, Martin D. Improving the anti-tumor activity of 5-fluorouracil by increasing its incorporation into RNA via metabolic modulation. Proc Natl Acad Sci USA. 1980b;77:4966–4970. doi: 10.1073/pnas.77.8.4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprang SR. G protein mechanisms: insights from structural analysis. Annu Rev Biochem. 1997;66:639–678. doi: 10.1146/annurev.biochem.66.1.639. [DOI] [PubMed] [Google Scholar]
- Su SS, Modrich P. Escherichia coli mutS-encoded protein binds to mismatched DNA base pairs. Proc Natl Acad Sci USA. 1986;83:5057–5061. doi: 10.1073/pnas.83.14.5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas DC, Roberts JD, Kunkel TA. Heteroduplex repair in extracts of human HeLa cells. J Biol Chem. 1991;266:3744–3751. [PubMed] [Google Scholar]
- Veigl ML, Kasturi L, Olechnowicz J, Ma AH, Lutterbaugh JD, Periyasamy S, et al. Biallelic inactivation of hMLH1 by epigenetic gene silencing, a novel mechanism causing human MSI cancers. Proc Natl Acad Sci USA. 1998;95:8698–8702. doi: 10.1073/pnas.95.15.8698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan M, Lovett ST. Single-strand DNA-specific exonucleases in Escherichia coli. Roles in repair and mutation avoidance. Genetics. 1998;149:7–16. doi: 10.1093/genetics/149.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner MW, Li LS, Morales JC, Galindo CL, Garner HR, Bornmann WG, et al. Role of c-Abl kinase in DNA mismatch repair-dependent G2 cell cycle checkpoint arrest responses. J Biol Chem. 2008;283:21382–21393. doi: 10.1074/jbc.M709953200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner R, Jr, Meselson M. Repair tracts in mismatched DNA heteroduplexes. Proc Natl Acad Sci USA. 1976;73:4135–4139. doi: 10.1073/pnas.73.11.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JY, Edelmann W. Mismatch repair proteins as sensors of alkylation DNA damage. Cancer Cell. 2006;9:417–418. doi: 10.1016/j.ccr.2006.05.013. [DOI] [PubMed] [Google Scholar]
- Warren JJ, Pohlhaus TJ, Changela A, Iyer RR, Modrich PL, Beese LS. Structure of the human MutSalpha DNA lesion recognition complex. Mol Cell. 2007;26:579–592. doi: 10.1016/j.molcel.2007.04.018. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Wu TT, Catalano PJ, Ueki T, Satriano R, Haller DG, et al. Molecular predictors of survival after adjuvant chemotherapy for colon cancer. N Engl J Med. 2001;344:1196–1206. doi: 10.1056/NEJM200104193441603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh KM, Lu AL, Clark S, Modrich P. Isolation and characterization of the Escherichia coli mutH gene product. J Biol Chem. 1987;262:15624–15629. [PubMed] [Google Scholar]
- Wildenberg J, Meselson M. Mismatch repair in heteroduplex DNA. Proc Natl Acad Sci USA. 1975;72:2202–2206. doi: 10.1073/pnas.72.6.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmore E, Durkacz BW. Cytotoxic mechanisms of 5-fluoropyrimidines. Relationships with poly(ADP-ribose) polymerase activity, DNA strand breakage and incorporation into nucleic acids. Biochem Pharmacol. 1993;46:205–211. doi: 10.1016/0006-2952(93)90405-l. [DOI] [PubMed] [Google Scholar]
- Wilson T, Guerrette S, Fishel R. Dissociation of mismatch recognition and ATPase activity by hMSH2-hMSH3. J Biol Chem. 1999;274:21659–21664. doi: 10.1074/jbc.274.31.21659. [DOI] [PubMed] [Google Scholar]
- Wurzer JC, Tallarida RJ, Sirover MA. New mechanism of action of the cancer chemotherapeutic agent 5-fluorouracil in human cells. J Pharmacol Exp Ther. 1994;269:39–43. [PubMed] [Google Scholar]
- Yamaguchi M, Dao V, Modrich P. MutS and MutL activate DNA helicase II in a mismatch-dependent manner. J Biol Chem. 1998;273:9197–9201. doi: 10.1074/jbc.273.15.9197. [DOI] [PubMed] [Google Scholar]
- Yang G, Scherer SJ, Shell SS, Yang K, Kim M, Lipkin M, et al. Dominant effects of an Msh6 missense mutation on DNA repair and cancer susceptibility. Cancer Cell. 2004;6:139–150. doi: 10.1016/j.ccr.2004.06.024. [DOI] [PubMed] [Google Scholar]
- Yin MB, Guo B, Panadero A, Frank C, Wrzosek C, Slocum HK, et al. Cyclin E-cdk2 activation is associated with cell cycle arrest and inhibition of DNA replication induced by the thymidylate synthase inhibitor Tomudex. Exp Cell Res. 1999;247:189–199. doi: 10.1006/excr.1998.4346. [DOI] [PubMed] [Google Scholar]
- Yoshioka K, Yoshioka Y, Hsieh P. ATR kinase activation mediated by MutSalpha and MutLalpha in response to cytotoxic O6-methylguanine adducts. Mol Cell. 2006;22:501–510. doi: 10.1016/j.molcel.2006.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Richards B, Wilson T, Lloyd M, Cranston A, Thorburn A, et al. Apoptosis induced by overexpression of hMSH2 or hMLH1. Cancer Res. 1999;59:3021–3027. [PubMed] [Google Scholar]
- Zhang Y, Yuan F, Presnell SR, Tian K, Gao Y, Tomkinson AE, et al. Reconstitution of 5′-directed human mismatch repair in a purified system. Cell. 2005;122:693–705. doi: 10.1016/j.cell.2005.06.027. [DOI] [PubMed] [Google Scholar]


