Abstract
CD1d presents lipid-based antigens (Ag) that are recognised by the semi-invariant T cell receptor (TCR) expressed on Natural Killer T (NKT) cells. While the TCR α-chain is typically invariant, the TCR β-chain expression is more diverse, particularly in mice where at least three different Vβ chains are commonly expressed. We report the structures of Vα14-Vβ8.2 and Vα14-Vβ7 NKT TCRs in complex with CD1d-α-galactosylceramide (α-GalCer), as well as a 2.5 Å structure of the human NKT TCR-CD1d-α-GalCer complex. Both Vβ8.2 and Vβ7 NKT TCRs, as well as the human NKT TCR, ligated CD1d-α-GalCer in a broadly similar manner, thereby highlighting the evolutionarily-conserved nature of this interaction. However, differences within the Vβ domains of the Vβ8.2 and Vβ7 NKT TCR-CD1d complexes not only resulted in altered TCR-β-CD1d-mediated contacts, but also surprisingly modulated recognition mediated by the invariant α-chain. Mutagenesis studies revealed the differing contributions of Vβ8.2 and Vβ7 residues within the CDR2β loop in mediating contacts with CD1d. Collectively we provide a structural basis for the differential NKT TCR Vβ usage in NKT cells.
Introduction
Natural Killer T (NKT) cells are a unique lymphocytic sub-lineage that recognise lipid-based antigens presented by CD1d, a Major Histocompatibility Complex (MHC) class I-like antigen (Ag) presenting molecule (Bendelac et al., 2007). NKT cells are implicated in a broad range of diseases, including microbial immunity, tumour immunity, autoimmunity and allergy (Bendelac et al., 2007; Godfrey and Kronenberg, 2004; Matsuda et al., 2008). NKT cells are present in mice and humans, and typically express a semi-invariant T cell receptor (NKT TCR) consisting of an invariant TCR α-chain (Vα24Jα18 in humans; Vα14Jα18 in mice), paired with a limited selection of TCR β-chains (Vβ11 in humans; Vβ8.2, Vβ7 or Vβ2 in mice)(Burdin et al., 1998; Godfrey et al., 2004; Porcelli et al., 1993). The restricted NKT TCR repertoire is considered to reflect their recognition of the monomorphic CD1d molecule presenting glycolipid antigens. The crystal structure of a human NKT TCR-CD1d-glycolipid (α-galactosylceramide; α-GalCer) complex provided a snapshot into the basis of NKT recognition, and revealed a markedly different mode of TCR recognition in comparison to that observed for TCR-MHC-peptide complexes (Borg et al., 2007). In contrast to the emerging generalities of the TCR-MHC-peptide interaction (Godfrey et al., 2008; Rudolph et al., 2006), the NKT TCR docked parallel to, and at the extreme end of, the CD1d-Ag binding cleft. Within this unusual NKT TCR-CD1d docking framework, interactions with CD1d were dominated by the Complementarity Determining Region (CDR) 3α loop encoded by Jα18 and Vβ11-encoded CDR2β loop, while the CDR1α and CDR3α loops contacted the α-GalCer (Borg et al., 2007). Alanine-scanning mutagenesis studies in the human Vβ11 NKT TCR and mouse Vβ8.2 NKT TCR were consistent with this NKT TCR-CD1d-α-GalCer docking footprint (Scott-Browne et al., 2007; Wun et al., 2008) suggesting a remarkable conservation of this immune recognition event across the 70 million years of evolution that separate mice and humans. For instance, two tyrosine residues (Tyr 48β & Tyr 50β) conserved in the human Vβ11 and mouse Vβ8.2 CDR2β loop were critical for NKT TCR-CD1d-binding (Scott-Browne et al., 2007; Wun et al., 2008), suggesting that the Vβ8.2 NKT TCR docked in a very similar manner to that of the human NKT TCR, which was consistent with the reciprocal cross-species reactivity of these NKT TCRs (Brossay et al., 1998). Structural studies of α-GalCer bound to human and mouse CD1d also revealed a broadly comparable landscape for NKT TCR binding (Koch et al., 2005; Zajonc et al., 2005), but nevertheless differences were apparent in the orientation of the α-galactose head group presented by CD1d from the two different species (Godfrey et al., 2005). It is unclear how the NKT TCR would accommodate such differences when mediating cross-species reactivity. Moreover, it is just as unclear how different NKT TCRs might afford differential reactivity to the same or different glycolipid antigens.
It is established that NKT cells can see an array of different lipid-based antigens (reviewed in (Bendelac et al., 2007; Brutkiewicz, 2006; Godfrey et al., 2008)), including bacteria derived lipid antigens (Fischer et al., 2004; Kinjo et al., 2006; Kinjo et al., 2005; Mattner et al., 2005) and mammalian (self)-glycolipid antigens that include isoglobotrihexosylceramide (iGb3)(Zhou et al., 2004) and GD3 (Wu et al., 2003). Notably, with the exception of α-GalCer, most other glycolipid antigens only seem to be recognised with high affinity by a subset of NKT cells (Brigl et al., 2006; Kinjo et al., 2008; Kinjo et al., 2006; Wu et al., 2003). For example, CD1d tetramers loaded with α-diacylglycerol (Kinjo et al., 2006), α-galacturonosylceramide (Kinjo et al., 2005), or GD3 (Wu et al., 2003), provided a spectrum of staining of NKT cells from negative to bright positive, whereas α-GalCer loaded CD1d tetramers stained the same population with uniformly high intensity (Kinjo et al., 2006; Kinjo et al., 2005; Wu et al., 2003). Similarly, iGb3 seems only to be able to stimulate a subset of α-GalCer reactive NKT cells (Brigl et al., 2006; Zhou et al., 2004). While this suggests that antigen specific subsets of NKT cells may exist, some NKT TCRs are nevertheless capable of recognising several distinct glycolipid antigens similarly ((Scott-Browne et al., 2007); Mallevaey.2009, Immunity submitted), albeit with varying affinity. Given that the NKT TCR α-chain is invariant, this suggests that NKT TCR β-chain plays a role in determining thresholds of antigen reactivity, and that this effectively enables some NKT TCRs to differentiate between antigens. This issue is particularly relevant to mouse NKT cells, which possess a more diverse TCR-β repertoire than humans, due to the frequent use of three Vβ genes (Vβ8.2, Vβ7 and Vβ2), in which Vβ8.2- and Vβ7-containing NKT TCRs represent up to 80% of the mouse NKT cell repertoire. Although human NKT cells also exhibit some TCR β-chain diversity only a small subset lack Vβ11 (Gadola et al., 2002), and furthermore, both human and mouse NKT cells have diverse CDR3β regions (Gadola et al., 2002; Matsuda et al., 2001). In the mouse NKT system, several studies support the differential contribution of TCR β-chains to recognition of different lipid based antigens: α-GalCer is preferentially recognised by NKT cells bearing Vβ8.2 (Schumann et al., 2003), whereas iGb3 is preferentially recognised by NKT cells bearing Vβ7 (Schumann et al., 2006; Wei et al., 2006). Furthermore, while mutations in the CDR3β region of the mouse NKT TCR did not substantially affect α-GalCer mediated activation of NKT TCR expressing hybridomas, they markedly influenced activation by other antigens, including iGb3 and GSL-1(Scott-Browne et al., 2007). Thus, in order to understand how NKT cells can recognise glycolipid antigens, and the selective Vβ gene usage by NKT cells, we need to gain a more complete picture of the different NKT TCRs in complex with CD1d-Ag.
Here we have determined the structures of the Vβ8.2 and Vβ7 NKT TCRs in complex with mouse CD1d-α-GalCer and compared them to a new 2.5 Å resolution structure of the human Vβ11 NKT TCR-CD1d-α-GalCer complex. Our findings, together with associated mutagenesis studies, provide insight into how the Vβ repertoire of NKT TCRs impacts on CD1d-glycolpid recognition.
Results
Vβ8.2-NKT TCR CD1d-α-GalCer complex
To begin to address the varied Vβ usage in mouse NKT TCRs, we expressed and refolded the Vα14Jα18-Vβ8.2 and Vα14Jα18-Vβ7 NKT TCRs (Supplementary Figure 1A & B), then formed and crystallized the complex with the mouse (m)CD1d-α-GalCer. The structure of the Vα14Jα18-Vβ8.2 NKT TCR-mCD1d-α-GalCer complex was subsequently determined to 2.9 Å resolution to an Rfac and Rfree of 23.4 % and 29.8 % respectively (Supplementary Table 1). The initial experimental phases clearly showed unbiased electron density for the α-GalCer and moreover, apart from a small disordered (on account of mobility of the loop) region within the CDR3β loop (residues Gly 98β to Glu 105β), the electron density at the Vα14-Vβ8.2-NKT TCR-mCD1d-α-GalCer interface was unambiguous.
Both the Vβ8.2- and Vβ7-NKT TCRs adopted an acute docking mode, binding approximately parallel to, and above, the F’-pocket of the CD1d-Ag binding cleft (Figure 1A and C). The Vβ8.2-NKT TCR will be discussed first. This TCR interacted with mCD1d residues spanning 76 – 87 and 149 – 153 of the α1-helix and the α2-helix respectively. The buried surface area (BSA) upon ligation was ≈ 760 Å2, in which the TCR α-chain contributes nearly three times more BSA than the TCR β-chain (74 % versus 26 % respectively) (Figure 1B), which is consistent with the α-chain dominating contacts with CD1d-α-GalCer in comparison to the β-chain (Figure 2A, Table 1). The Vβ8.2 chain usage was dictated by the CDR2β loop interacting with mCD1d, as the CDR1β loop does not mediate any contacts with the Ag and the CDR3β loop was mobile (Table 1). The CDR2β loop formed a stretch of interactions exclusively with the α1-helix (residues 83–87) of mCD1d (Table 1 and Figure 1B). Specifically, Tyr 48β and Tyr 50β formed H-bonds and Van der Waals (vdw) contacts with Glu 83 and Lys 86 of CD1d, the latter of which formed a salt bridge with Glu 56β (Figure 2B).
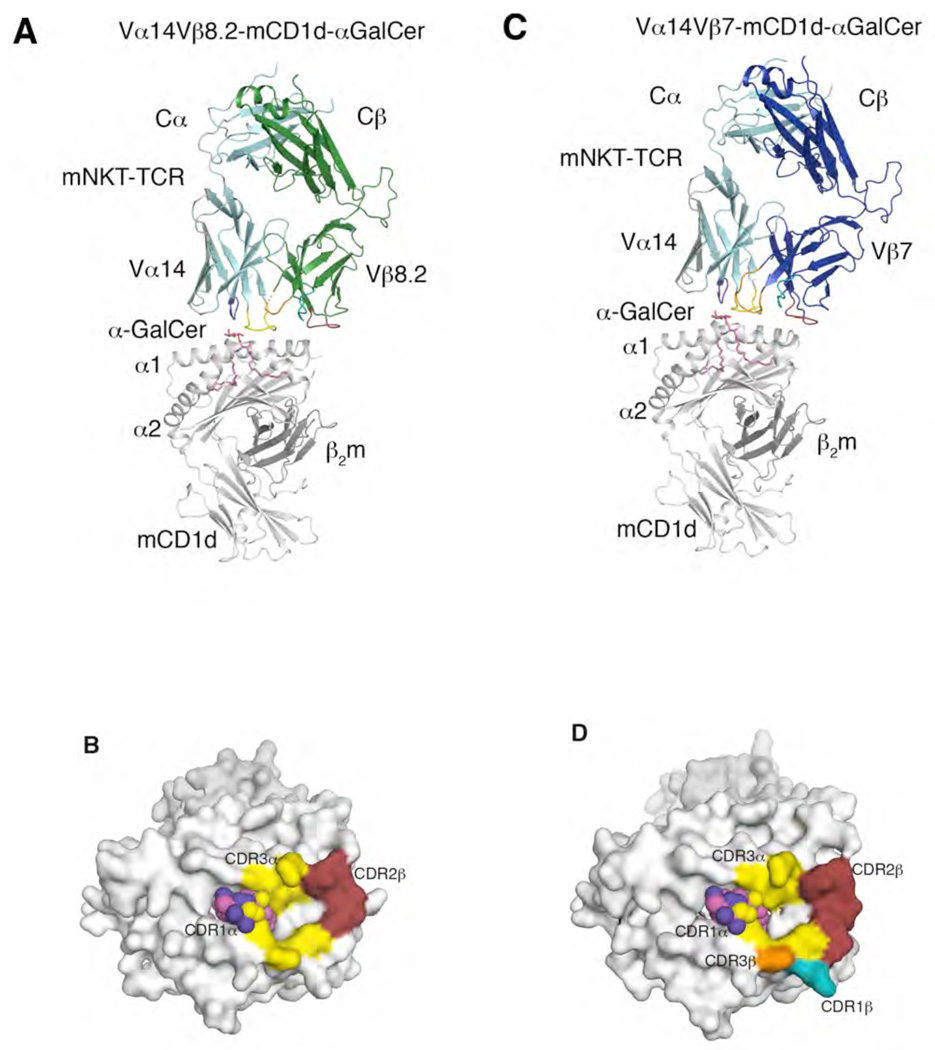
Figure 1.
Structure of mouse NKT TCRs in complex with mouse CD1d-α-GalCer (A) Vα14-Vβ8.2 NKT TCR in complex with mCD1d-α-GalCer. α-GalCer, magenta; mCD1d heterodimer, grey; TCR α-chain, cyan; Vβ8.2 NKT TCR β-chain, green; CDR1α, purple; CDR3α, yellow; CDR1β, teal; CDR2β, ruby; CDR3β, orange; mobile CDR3β region, dashed orange. (B) Footprint of the Vα14-Vβ8.2 NKT TCR on the surface of mouse CD1d-α-GalCer. α-GalCer is shown in spheres. mCD1d, α-GalCer and CDR loops colour coding as in A. (C) Vα14-Vβ7 NKT TCR in complex with mouse CD1d-α-GalCer. Vβ7 NKT TCR β-chain, blue. TCR α-chain, mCD1d, CDR loops and α-GalCer colour coding as in A. (D) Footprint of the Vα14-Vβ7 NKT TCR on the surface of mCD1d-α-GalCer. α-GalCer is shown in spheres. mCD1d, α-GalCer and CDR loops colour coding as in A.
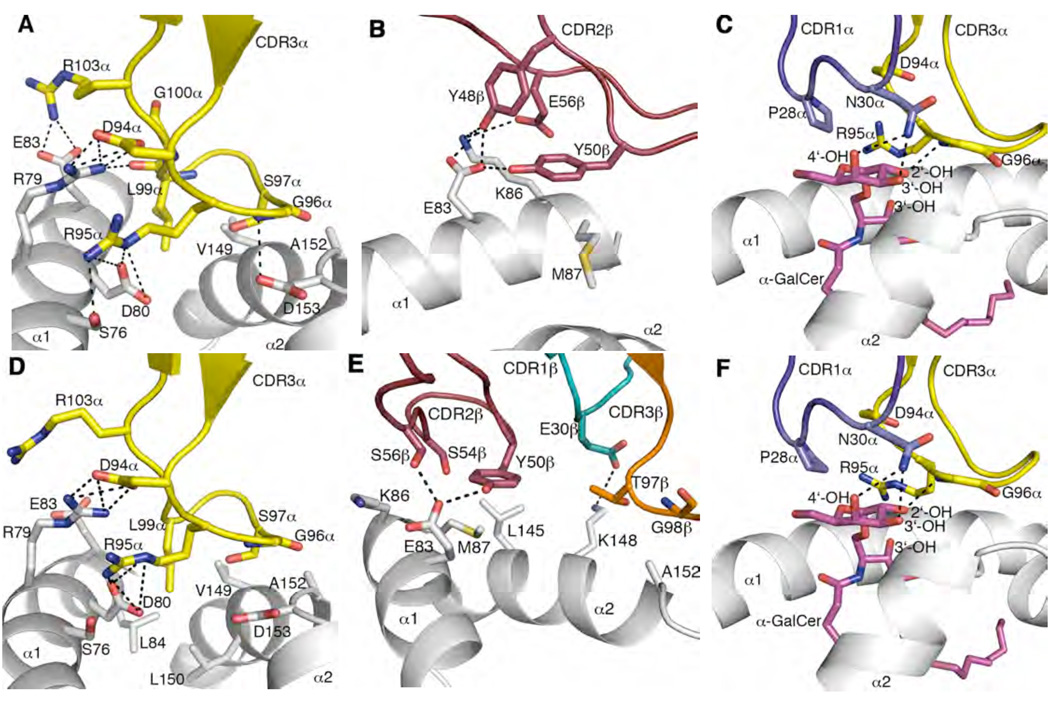
Figure 2.
Mouse CD1d and α-GalCer mediated interactions with mouse NKT TCRs CDR3α mediates multiple contacts between mCD1d α-helices and α-GalCer. CDR2β contacts α1-helix of mCD1d. CDR1α interacts solely with α-GalCer galactose head group. CDR1β mediates polar interactions with the α2-helix only in Vβ7 NKT TCR-mCD1d-α-GalCer. (A) Vβ8.2 NKT TCR CDR3α contacts with mCD1d. (B) Vβ8.2 NKT TCR CDR2β contacts with mCD1d. (C) Vβ8.2 NKT TCR CDR1α and CDR3α contacts with α-GalCer. (D) Vβ7 NKT TCR CDR3α contacts with mCD1d. (E) Vβ7 NKT TCR CDR1β, CDR2β and CDR3β contacts with mCD1d. (F) Vβ7 NKT TCR CDR1α and CDR3α contacts with α-GalCer. CDR1α, purple; CDR3α, yellow; CDR1β, teal; CDR2β, ruby; CDR3β, orange; α-GalCer, magenta; mCD1d, grey. H-bond or salt-bridge interactions are shown in black dashed lines.
Table 1.
Contacts at the mNKT TCR-mCD1d interface
| CDR | Vβ8.2 NKT | mCD1d | Bond | Vβ7 NKT | mCD1d | Bond |
|---|---|---|---|---|---|---|
| CDR3α | Asp94Oδ1 | Arg79Nη1, Arg79Nη2 | Salt-bridge | Asp94Oδ1 | Same as Vβ8.2 | |
| Asp94Oδ2 | Arg79Nη1, Arg79Nη2 | Salt-bridge | Asp94Oδ2 | Same as Vβ8.2 | ||
| Asp94 | Arg79 | VDW | Asp94 | Same as Vβ8.2 | ||
| Arg95Nε | Asp80Oδ1, Asp80Oδ2 | Salt-bridge | Arg95Nε | Same as Vβ8.2 | ||
| Arg95Nη1 | Asp80Oδ1 | Salt-bridge | Arg95Nη1 | Asp80Oδ1, Asp80Oδ2 | Salt-bridge | |
| Ser76Oγ | H-bond | |||||
| Arg95 | Asp80, Arg79, Ser76 | VDW | Arg95 | Same as Vβ8.2 | ||
| Gly96N | Asp153Oδ2 | H-bond | Gly96N | - | ||
| Gly96 | Ala152, Asp153 | VDW | Gly96 | Same as Vβ8.2 | ||
| Ser97 | Val149 | VDW | Ser97 | Val149, Ala152, Asp153 | VDW | |
| Leu99 | Arg79, Val149 | VDW | Leu99 | Arg79, Asp80, Glu83, | VDW | |
| Leu84, Val149, Leu150 | ||||||
| Leu99O | Arg79Nη2 | H-bond | Leu99O | - | ||
| Gly100 | Arg79 | VDW | Gly100 | - | ||
| Arg103 | Arg79, Glu83 | VDW | Arg103 | Arg79 | VDW | |
| Arg103Nη1 | Glu83Oε2, Glu83Oε1 | Salt-bridge | Arg103Nη1 | - | ||
| CDR1β | - | Glu30Oε2 | Lys148Nζ | Salt-bridge | ||
| - | Glu30 | Lys148 | VDW | |||
| CDR2β | Tyr48Oη | Glu83Oε1, Glu83Oε2, | H-bond | - | ||
| Lys86Nζ | ||||||
| Tyr48 | Glu83, Lys86 | VDW | - | |||
| Tyr50Oη | Glu83Oε1 | H-bond | Tyr50Oη | Same as Vβ8.2 | ||
| Tyr50 | Glu83, Met87 | VDW | Tyr50 | Same as Vβ8.2 | ||
| - | Ser54 | Met87, Leu145 | VDW | |||
| Glu56Oε1 | Lys86Nζ | Salt-bridge | Ser56Oγ | Glu83Oε1 | H-bond | |
| Glu56 | Lys86 | VDW | Ser56 | Glu83, Lys86 | VDW | |
| CDR3β | - | Thr97 | Ala152 | VDW | ||
| - | Gly98 | Ala152 | VDW | |||
| CDR | Vβ8.2 NKT | α-GalCer | Bond | Vβ7 NKT | α-GalCer | Bond |
| CDR1α | Pro28 | 6’-OHG, 5’-OG, C-1G | VDW | Pro28 | Same as Vβ8.2 | |
| Asn30 | C-2G, C-3G, C-4G, | VDW | Asn30 | - | ||
| 3’-OHG, 4’-OHG | ||||||
| Asn30Nδ2 | 3’-OHG, 4’-OHG | H-bond | Asn30Nδ2 | Same as Vβ8.2 | ||
| CDR3α | Asp94O | C-1G | VDW | Asp94O | Same as Vβ8.2 | |
| Arg95 | 2’-OHG, C-2G, 3’- | VDW | Arg95 | Same as Vβ8.2 | ||
| OHS | ||||||
| Gly96N | 2’-OHG | H-bond | Gly96N | Same as Vβ8.2 | ||
| Gly96 | C-2G, 3’-OHG | VDW | Gly96 | C-2G, 3’-OHG, 2’-OHG | VDW | |
- Atomic contacts determined using the CCP4i implementation of CONTACT and a cutoff of 4.5 Å.
- Van der Waals interactions defined as non-hydrogen bond contact distances of 4 Å or less.
- Hydrogen bond interactions are defined as contact distances of 3.3 Å or less.
- Salt bridge is defined as contact distance of 4.5 Å or less.
- G = contacts with Galactose head group
- S = contacts with Sphingosine chain
The Vα14-Jα18 α-chain interactions were mediated via the CDR3α and CDR1α loops (57 % and 17 % BSA respectively) (Figure 2A & C, and Table 2). The CDR1α loop interacted with α-GalCer, whereas the Jα18-encoded CDR3α loop interacted with mCD1d and α-GalCer. The importance of the Jα18-encoded region is consistent with the lack of NKT cells in TCR Jα18 gene-inactivated mice (Cui et al., 1997). The CDR3α-mediated interactions were largely electrostatic in nature, but also included some vdw-mediated contacts, including Leu 99α that sat in a small hydrophobic niche, formed by Leu 84, Leu 150, Val 149 of mCD1d, but only made contacts with the latter residue (Figure 2A). There was an inter-digitation of arginine residues at the CDR3α-mCD1d interface, in which Arg 79 from CD1d was flanked by Arg 103α and Arg 95α. This cluster of positively-charged residues were dissipated by neighbouring acidic groups, including Asp 94α, which salt-bridged to Arg 79; Arg 103α that salt-bridged to Glu 83; Arg 95α that salt-bridged to Asp 80 (Figure 2A). Additionally, the main chain amide of Gly 96α H-bonded to Asp 153 of mCD1d and as such, all Jα18 residues at the tip of the CDR3α loop, with the exception of Ala 98α, mediated contacts with mCD1d-α-GalCer (Table 1).
Only the galactose head group of α-GalCer is exposed for recognition by the NKT TCR, and interacted solely with the CDR1α and CDR3α loops (Figure 2C, Table 1). The galactose ring sat below the CDR1α loop and adjacent to the CDR3α loop, forming vdw contacts on one face of the sugar ring with Arg 95α, Gly 96α and Pro 28α. Arg 95α also made vdw contacts with the 3’ hydroxyl of the sphingosine chain (Figure 2C). Gly 96α H-bonds to the 2’ hydroxyl, whereas Asn 30α H-bonds to both the 3’ and 4’ hydroxyl groups of the galactose ring. As such, the galactose ring is sequestered closely by the invariant α-chain of the Vβ8.2 NKT TCR.
The crystal structure of the Vβ8.2 NKT TCR-mCD1d-α-GalCer complex also allowed us to undertake precise structural correlates of the alanine-scanning mutagenesis study previously conducted in this system (Scott-Browne et al., 2007). We can confirm that the effect of some of the Vβ8.2 NKT TCR mutants in interacting with mCD1d-α-GalCer are due to indirect local effects (namely, CDR1α: Val26Ala, Pro28Ala, Asn30Ala, His31Ala, Arg33Ala; CDR1β: Asn31Ala; CDR2β: Ser49Ala; Gly51Ala). On the other hand, the mutational data are in accord with the crystal structure: namely the CDR1α, CDR3α and CDR2α loops represent the energetic footprint of the interaction with mCD1d-α-GalCer. Accordingly, the structure of the Vβ8.2 NKT TCR-mCD1d-α-GalCer complex provided a basis for understanding the biased gene usage of the semi-invariant Vα14Jα18-Vβ8.2 NKT TCR.
Conformational changes upon ligation
The mCD1d-PBS-25 (an analogue of α-GalCer with modifications in the lipids chains) and an engineered variant of Vα14Jα18-Vβ8.2 NKT TCR (which included mutations at the Vα/Vβ interface and within the invariant CDR3α loop) have been solved in the non-liganded state (Zajonc et al., 2005; Zajonc et al., 2008). Hence we evaluated the degree of plasticity in this Vβ8.2 NKT TCR-mCD1d-α-GalCer interaction by comparing the elements of the complex in their liganded and unliganded state. The CDR loops of the Vβ8.2 NKT TCR did not change conformation appreciably upon ligation to mCD1d-α-GalCer, although movements in some side chains (Asn 30α, Tyr 48β and Tyr 50β) were observed (data not shown). Interestingly, upon Vβ8.2 NKT TCR ligation, the α-GalCer head group was observed to be shifted by approximately 1 Å (Supplementary Figure 2). In comparison to the non-liganded Vβ8.2 NKT TCR (Zajonc et al., 2008), there was a slight change in the juxta-positioning (9.5 °) of the Vα14 and Vβ8.2 domains upon ligation. Notably, such movements have been observed previously in TCR-pMHC interactions, where they are thought to relate to signal transmission (Ishizuka et al., 2008).
In addition, there was minimal movement in the mCD1d upon ligation, with some reorientation of side-chain conformations observed including: Arg 79, Glu 83, Lys 86, Lys 148 from mCD1d (data not shown). Overall, the lack of conformational change upon Vβ8.2 NKT TCR-mCD1d-α-GalCer ligation, which was also observed in the human NKT TCR-CD1d-α-GalCer interaction (Borg et al., 2007; Kjer-Nielsen et al., 2006), typifies the innate characteristics of this interaction mediated by a relatively ‘rigid’ receptor-ligand binding, whereas TCRs typically show a greater degree of plasticity upon ligation with pMHC.
Vβ7-NKT TCR CD1d-α-GalCer complex
The Vα14Jα18-Vβ7 NKT TCR is expressed by approximately 15–20% of the mouse NKT T-cell repertoire (Benlagha et al., 2000; Matsuda et al., 2000), and the Vβ7 and Vβ8.2 chains share 54 % sequence identity, with sequence differences located in the CDR1β and CDR2β loops (Supplementary Figure 3). Accordingly, we also aimed to understand how the Vβ7 NKT TCR interacted with mCD1d-α-GalCer. Hence, the refolded Vα14-Vβ7 NKT TCR (Supplementary Figure 1) complexed to mCD1d-α-GalCer was crystallised and the structure determined to 2.8 Å resolution with an Rfac and Rfree of 22.4 % and 27.1 % respectively (Supplementary Table 1). The initial experimental phases clearly showed unbiased electron density for α-GalCer and the electron density at the Vα14-Vβ7 NKT TCR-mCD1d-α-GalCer interface was unambiguous.
Like the Vβ8.2 NKT TCR-CD1d-α-GalCer complex, the Vα14-Vβ7 NKT TCR was perched above the F’-pocket of the CD1d-Ag binding cleft, interacting with a similar stretch of residues on mCD1d (76 – 87 and 145 – 153) (Figure 1C, Figure 2D–F). The footprint of the two TCRs were very similar with the Vα contacts being dictated by the CDR1α and CDR3α loops, the former exclusively contacting α-GalCer, while the latter making substantial contacts with both mCD1d and α-GalCer. However, within these common footprints, there were notable differences in the contacts made between the Vβ7 and Vβ8.2 NKT TCRs and CD1d-α-GalCer, attributable to sequence differences between the NKT TCRs and differing relative juxta-positioning of the Vβ8.2/Vβ7 and Vα14 domains (Figure 3A & B). In turn, these differences also altered the nature of some of the Vα14-Jα18-mCD1d-α-GalCer interactions (Figure 3B and C, Table 1) despite the commonality of the invariant α-chain between the two NKT TCRs.
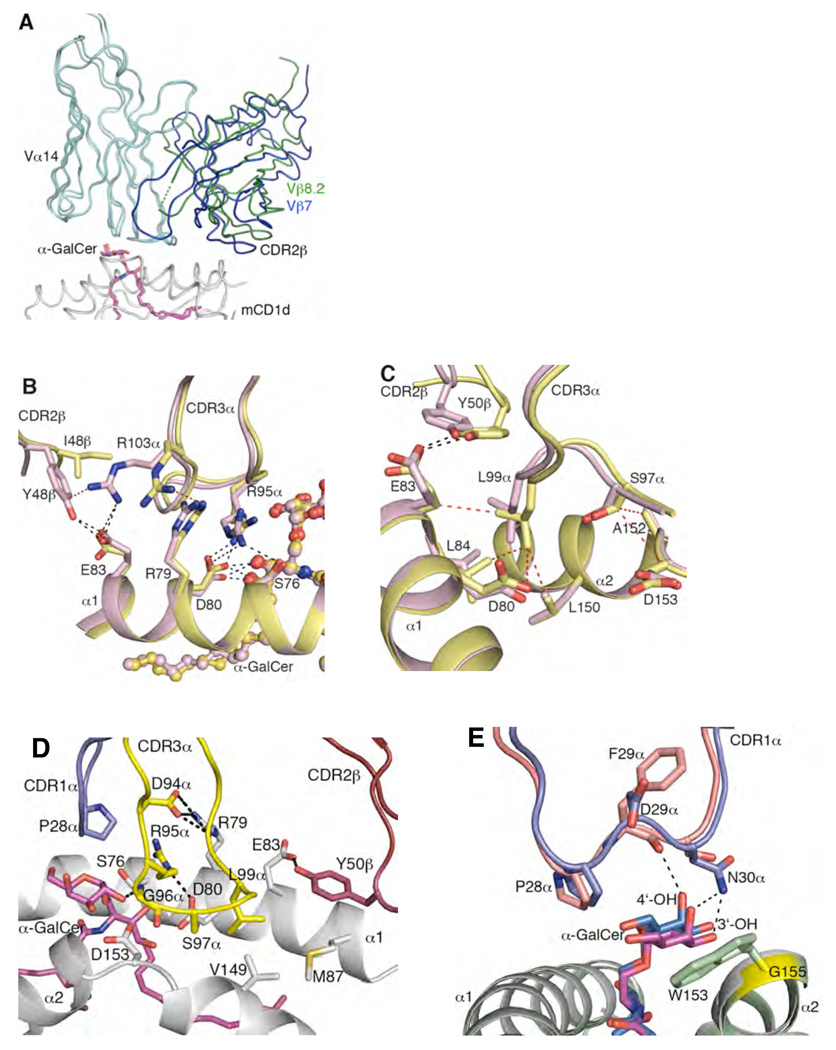
Figure 3.
Comparison of Vα14-Vβ8.2, Vα14-Vβ7 and Vα24-Vβ11 NKT TCR-mCD1d-α-GalCer complexes (A) Superposition of Vα14-Vβ8.2 NKT TCR-mCD1d-α-GalCer and Vα14-Vβ7 NKT TCR-mCD1d-α-GalCer. Differences in the relative juxta-positioning of the Vβ8.2-Vβ7 and Vα14 domains. TCR α-chain, cyan; Vβ8.2 NKT TCR β-chain, green; Vβ7 NKT TCR β-chain, blue; α-GalCer, magenta; mCD1d, grey (B) Differences in the sequence of CDR2β in Vβ8.2 and Vβ7 NKT TCR affected the position of Arg 103α in the CDR3α loop and subsequently altered positions and contacts of Arg 79, Asp 80, Ser 76 and Arg 95α. Vα14-Vβ8.2 NKT TCR-mCD1d-α-GalCer, pink; Vα14-Vβ7 NKT TCR-mCD1d-α-GalCer, yellow. α-GalCer is shown in ball and stick. H-bond or salt-bridge interactions are shown in black dashed lines and vdw interactions are shown in red dashed lines. (C) Altered position of Tyr 50β in Vβ7 NKT TCR affected contacts made by Ser 97α and Leu 99α at the tip of CDR3α with mCD1d. Colour coding as in B. H-bonds are shown in black dashed lines and vdw interactions are shown in red dashed lines. (D) Conserved interactions mediated by CDR1α, CDR3α and CDR2β loops of the human and mouse NKT TCRs on the surface of CD1d and α-GalCer. CDR1α, purple; CDR2β, ruby; CDR3α, yellow; α-GalCer, magenta; CD1d, grey. The numbering shown on CD1d is according to the mouse CD1d. H-bonds or salt-bridge interactions are shown in black dashed lines. (E) The shift in the position of the galactose head group of α-GalCer between mouse and the human NKT TCR-CD1d-α-GalCer structures is due to the presence of a bulky tryptophan side chain in human CD1d (Trp 153) in contrast to glycine (Gly 155, shown in yellow) in mouse CD1d. Human CDR1α, salmon; mouse CDR1α, purple; α-GalCer in human, marine; α-GalCer in mouse, magenta; hCD1d, pale green; mCD1d, grey.
These differences manifested in a larger BSA between the Vβ7-NKT TCR and mCD1d (≈ 860 Å2, Figure 1D) than the corresponding Vβ8.2-NKT TCR-mCD1d-α-GalCer footprint (Figure 1B). This was attributable to a differing juxta-positioning of Vα14 and Vβ7 (9°) compared with that of Vα14-Vβ8.2 NKT TCR, which resulted in the Vβ7 domain being positioned closer to mCD1d (Figure 3A), and hence resulted in more contacts with CD1d when compared to the Vβ8.2 NKT TCR (Table 1). Consequently, the Vα14-chain contributes 59 % BSA and the Vβ7-chain 41 % BSA at the Vβ7-NKT TCR-CD1d-α-GalCer interface.
The Vβ7 chain only interacted with mCD1d, and while the majority of contacts were mediated via the CDR2β loop (27% BSA), the CDR3β (9% BSA) loop and surprisingly the CDR1β loop (3% BSA) also mediated contacts with mCD1d (Figure 1D). Specifically, as a result of the Vβ7 chain leaning more towards the mCD1d in comparison to Vβ8.2, this permitted a salt bridge to be formed between Glu 30β (Asn 30β in Vβ8.2) and Lys 148 (Figure 2E). The interactions with the CDR3β loop were via Thr 97β and Gly 98β, both of which abutted against Ala 152 of mCD1d, and this interaction presumably aided in stabilising the CDR3β loop. The interactions with the Vβ7 CDR2β loop were featured by Tyr 50β, which lay flat against Met 87 of mCD1d, forming a H-bond with Glu 83, the latter of which also formed a H-bond with Ser 56β (Figure 2E). While Tyr 50β, and its interactions with CD1d are conserved between Vβ7 and Vβ8.2, the relative position of Tyr 50β varies in the two complexes (Figure 3C). In addition, Ser 54β formed vdw interactions with Met 87 and Leu 145. The small side chains of Ser 54β and Ser 56β were able to contact mCD1d as a result of the CDR2β chain being closer to mCD1d when compared to the corresponding CDR2β loop of the Vβ8.2 chain (Figure 3A).
Structural differences at the Vα14-Vβ7 and Vα14-Vβ8.2 interfaces also “transmitted” to alterations in some of the Vα14-Jα18-mCD1d contacts (Figure 3B). Specifically, whilst many of the α-chain mediated contacts were similar (Table 1), differences in Jα18-mediated contacts were observed at the tip of the CDR3α loop, namely residues Asp 94α, Arg 95α, Gly 96α, Ser 97α, Leu 99α and Arg 103α. Firstly, these changes appeared to emanate from an altered contact between Arg 103α and the respective Vβ chains. Thus, in the Vβ8.2 NKT TCR, Arg 103α stretches between the Vα/Vβ interface, with its guanadinium group being tethered by Tyr 48β, which enabled Arg 103α to salt bridge to Glu 83 of mCD1d. In Vβ7 however, position 48 is occupied by an Ile residue, which does not contact mCD1d, and moreover results in a loss of interaction with Arg 103α, causing Arg 103α to swing away from the Vα/Vβ interface and pack against Arg 79 of mCD1d, Figure 3B. This in turn affects the conformation of Arg 95α and Asp 80 of mCD1d, the latter of which H-bonds to α-GalCer in Vβ8.2 and Vβ7. The altered conformation of Arg 95α in Vβ7 NKT TCR resulted in loss of an H-bond interaction with Ser 76 of mCD1d (Table 1). Secondly, the altered position of Tyr 50β in Vβ8.2 and Vβ7 NKT TCRs (Figure 3C) also pushed the tip of the CDR3α loop away from CD1d, thereby altering the interactions this region made with mCD1d-α-GalCer (Table 1). For example, this change caused Leu 99α to sit differently within the hydrophobic mCD1d niche, which in turn results in Leu 99α and Ser 97α forming more vdw contacts in the Vβ7 NKT TCR when compared to the Vβ8.2 NKT TCR-mCD1d-α-GalCer interface (Figure 3C, Table 1).
Accordingly, whilst the overall Vβ8.2- and Vβ7- NKT TCR-mCD1d-α-GalCer structures were similar, the Vβ7 domain played a more prominent role at the interface and also affected the invariant α-chain-CD1d contacts.
Comparison to the human NKT TCR-CD1d-αGalCer complex
Previously, we had determined the structure of the human NKT TCR-CD1d-α-GalCer complex to 3.2 Å resolution, making it challenging to accurately assign subtle structural changes that may be present between differing NKT TCR-CD1d-Ag complexes. Accordingly, the human NKT TCR-CD1d-α-GalCer complex was crystallised in a different space group to that originally reported (Borg et al., 2007), and the structure determined to 2.5 Å resolution with an Rfac and Rfree of 21.6 % and 27.9 % respectively (Supplementary Table 1). We then compared the structure of the high resolution human NKT TCR-CD1d-α-GalCer complex to the two mouse NKT TCR-CD1d-α-GalCer complexes to ascertain the evolutionarily conserved characteristics of the NKT TCR-CD1d innate interaction (Figure 3D, Supplementary Figure 4A & B). Overall, the footprints of the human and mouse NKT TCRs were similar, especially between the homologous Vβ domains (Vβ11 in humans and Vβ8.2 in mouse) were compared. Within this common footprint, the following interactions were conserved between the three complexes: Pro 28α to α-GalCer; Asp 94α to α-GalCer; Asp 94α to Arg 79; Arg 95α to α-GalCer; Arg 95α to Asp 80, Arg 79 & Ser 76; Gly 96α to α-GalCer; Gly 96α to Asp153 (Asp 151 in human CD1d); Ser 97α to Val 149 (Val 147 in human CD1d); Leu 99α to Val 149; and Tyr 50β to Glu 83 & Met 87 (Figure 3D).
Nonetheless, a number of differences at the NKT TCR-CD1d interfaces between the three complexes were present, and these were attributable to (i) altered Vα-Vβ juxta-positioning (approximately 13 to 15° rotation between human and mouse NKT TCR-CD1d complexes) (ii) sequence differences between the respective CDR loops that mediated CD1d and α-GalCer recognition (iii) structural differences between hCD1d and mCD1d, which included an altered positioning of the α-GalCer galactose head group due to the presence of a neighbouring Trp 153 (Gly 155 in mCD1d) in hCD1d (Godfrey et al., 2005). Nevertheless, the α-galactose head group was sequestered to a similar extent in all three NKT TCR-CD1d-α-GalCer structures (Figure 3D & E), with the preservation of the H-bond interactions to the 2’, 3’ and 4’ hydroxyl groups.
Accordingly, the NKT TCR footprint on CD1d is broadly comparable across species, highlighting the evolutionarily-conserved nature of this interaction. Nevertheless, differences at the NKT TCR-CD1d interfaces were observed suggest that a degree of malleability in NKT TCR-CD1d recognition plays a role in the reciprocal cross-species reactivity between the human and mouse NKT TCRs.
Vβ7 and Vβ8.2 NKT TCR Binding Affinity
Given the differences in the binding of the human NKT TCR and the mouse Vβ8.2 and Vβ7 NKT TCRs, we determined the affinity and relative avidity of the interactions using surface plasmon resonance (SPR) and CD1d-α-GalCer tetramer inhibition studies respectively.
The affinity (equilibrium dissociation constant, KD) of Vβ8.2 NKT TCR for mCD1d-α-GalCer was 70 nM, whereas the KD for the Vβ7 NKT TCR-mCD1d-α-GalCer interaction was 4-fold lower at 0.3 µM (Supplementary Table 2, Figure 4A and B). These differing affinities were attributable to much longer half life of the Vβ8.2 NKT TCR (t1/2 = 17.3s) compared to the Vβ7 NKT TCR (t1/2 = 6.9s), and were consistent with previous affinity measurements determined via CD1d-multimer staining (Schumann et al., 2003) (Mallevaey.2009, Immunity submitted). Consistent with the reciprocal cross species reactivity of NKT cells, the Vβ8.2 NKT TCR also bound human (h)CD1d-α-GalCer (KD ≈ 94 nM) with similar affinity to mCD1d-α-GalCer, although the respective on and off rates were distinct (Supplementary Table 2, Figure 4C and D). In contrast the Vβ7 NKT TCR bound hCD1d-α-GalCer with approximately 12-fold lower affinity (KD ≈ 3.4 µM), which was consistent with hCD1d-α-GalCer dimer preferentially detecting Vβ8.2 NKT cells (Schumann et al., 2003). Human NKT TCR affinity for hCD1d-α-GalCer was lower (KD ≈ 0.2 µM) than that for the Vβ8.2 NKT TCR-mCD1d-α-GalCer interaction (Supplementary Table 2), and human NKT TCR bound with moderately lower affinity to mCD1d-α-GalCer (KD ≈ 1 µM), as observed previously (Wun et al., 2008).
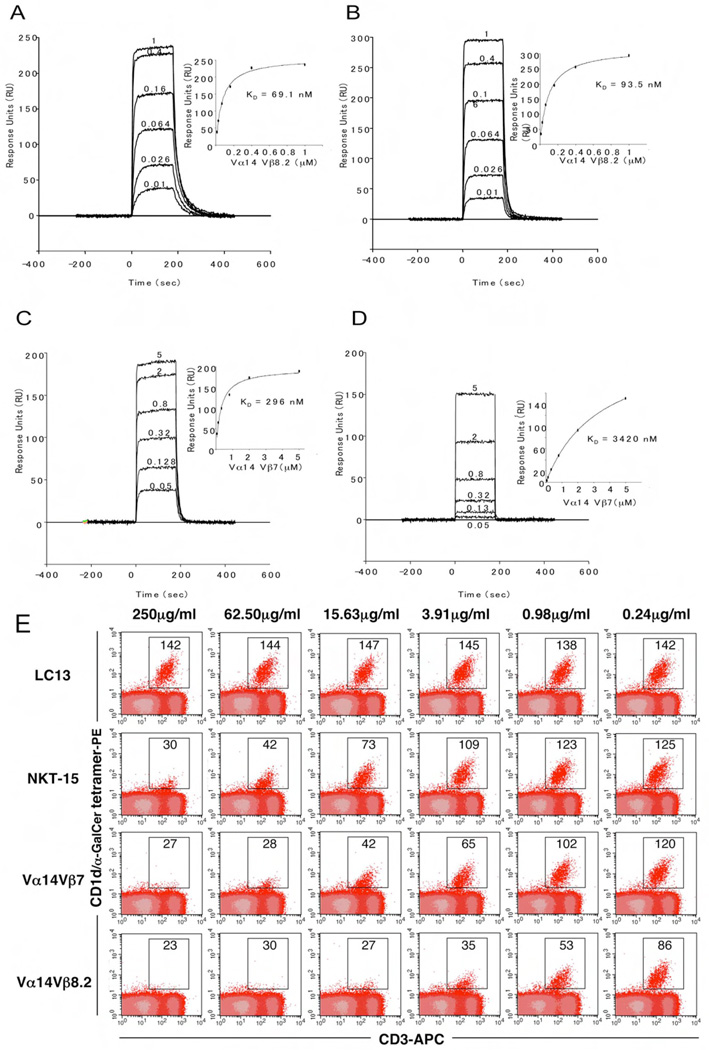
Figure 4.
Differential binding affinities of NKT TCRs to CD1d-α-GalCer. Vα14Jα18-Vβ8.2 (A and B) and Vα14Jα18-Vβ7 (C and D) NKT TCR were injected over streptavidin immobilised mouse (A and C) and human (B and D) CD1d-α–GalCer and simultaneously over a control cell coated with unloaded CD1d. Sensograms show the binding (response units, RU) of increasing concentrations of TCR (0.01 to 1µM for Vα14Jα18-Vβ8.2 and 0.05 to 5 µM for Vα14Jα18-Vβ7) to mouse and human CD1d-α–GalCer following baseline subtraction. Insets show saturation plots demonstrating equilibrium binding of NKT TCR to immobilised CD1d-α–GalCer. The equilibrium dissociation constants (KD) derived by equilibrium analysis were equivalent to those derived by kinetic analysis. E) CD1d-α–GalCer tetramer inhibition. Recombinant soluble NKT TCRs were examined for their ability to block binding of mCD1d/αGC tetramers to mouse NKT cells. PE labelled CD1d-α–GalCer tetramers were pre-incubated with titrating amounts of soluble NKT TCRs or an irrelevant TCR control, LC13, before staining of mouse thymocytes. Cells were analysed by flow cytometry showing mCD1d-α–GalCer tetramer-PE on the vertical axis and anti-CD3 APC on the horizontal axis. CD3+ mCD1d-α–GalCer tetramer+ thymic NKT cells are indicated within the square with the MFI (mean fluorescence intensity) indicated. All measurements were taken in duplicate.
To cross-validate the binding affinity for the three NKT TCRs we used a CD1d-α-GalCer tetramer binding inhibition assay (Kjer-Nielsen et al., 2006) (Figure 4E). The Vβ8.2 NKT TCR blocked tetramer binding with the highest efficiency, still readily detectable to a concentration of 0.98 µg/ml. In contrast, the Vβ7 NKT TCR was approximately 3–4-fold less effective and human NKT TCR was 3–4 fold less effective again. This hierarchy is consistent with the affinities of the different TCRs as determined by SPR analysis. The negative control TCR, LC13 (anti-HLA-B8-FLR) (Kjer-Nielsen et al., 2003), did not inhibit tetramer staining at any of the doses tested.
Accordingly, these data are consistent with our SPR affinity measurements, showing that Vβ8.2 NKT TCR preferentially interacts with CD1d-α-GalCer when compared to Vβ7 NKT TCR.
NKT TCR mutagenesis
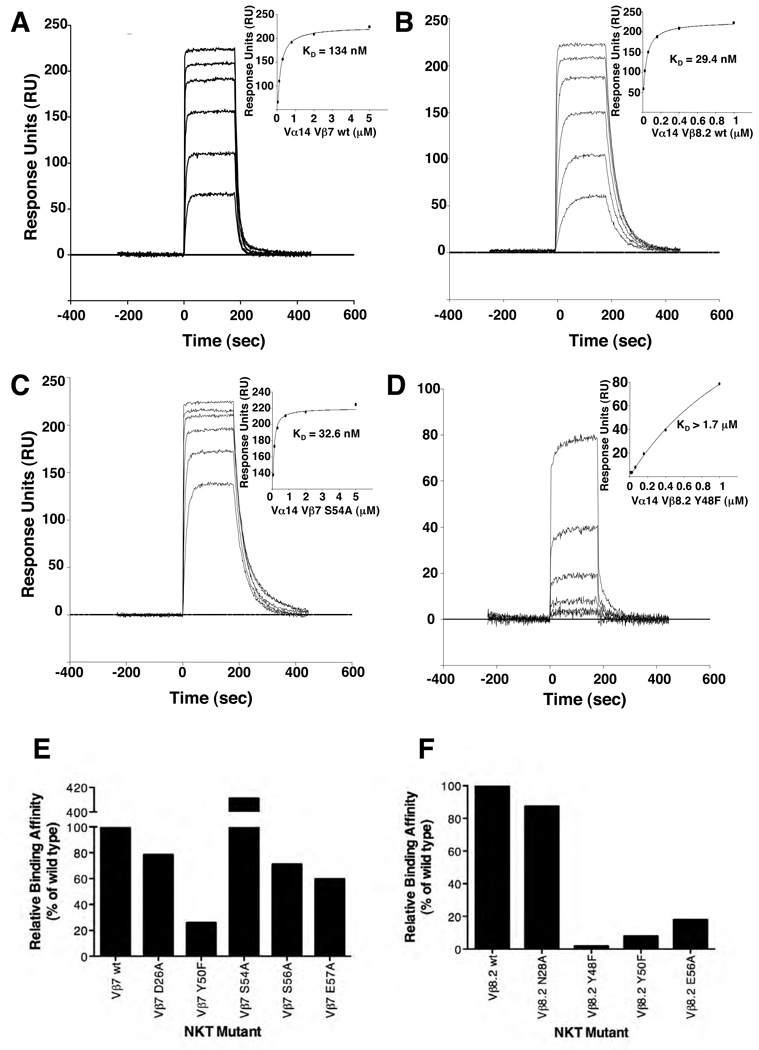
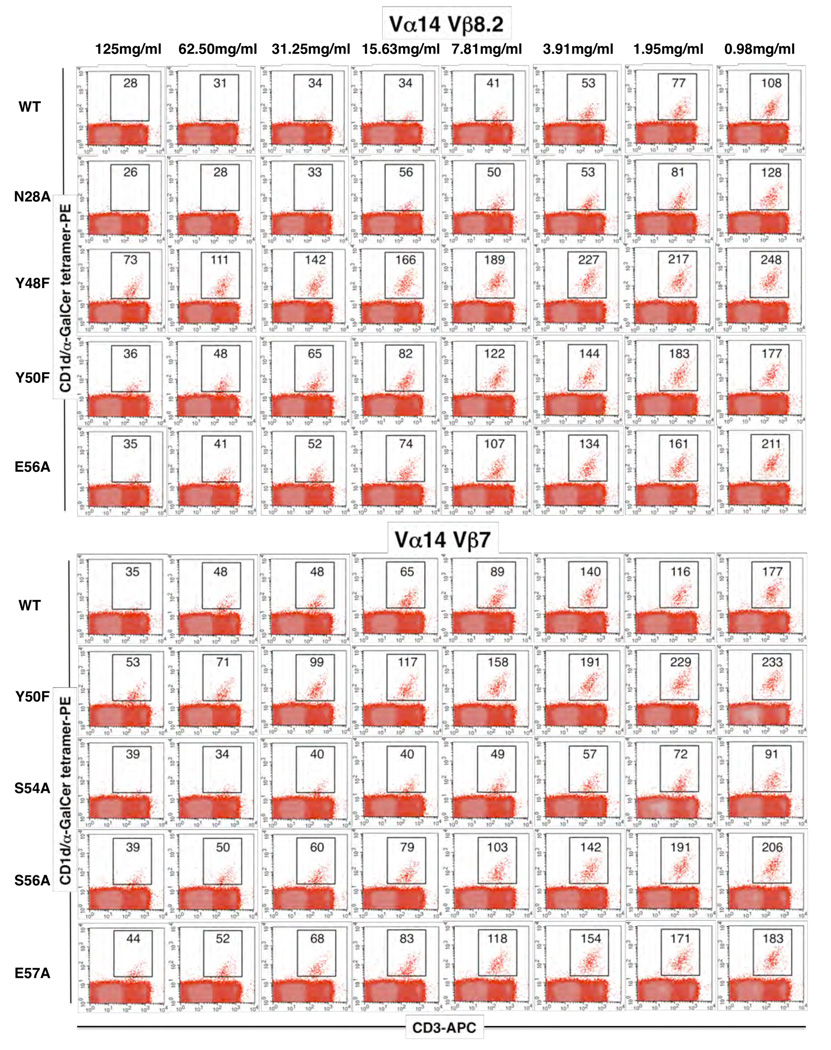
Given the differing contacts made by the CDR2β loop of the Vβ8.2 and Vβ7 NKT TCRs, we next established the importance of the residues within these loops of the respective mouse NKT TCRs. Using the structures as a guide, we mutated CDR2β residues that were observed to contact CD1d, and examined the effect of the mutants via SPR (Figure 5A–F, Supplementary Table 3) and CD1d-α-GalCer tetramer inhibition studies (Figure 6). For the Vβ8.2 and Vβ7 NKT TCRs, three (Tyr48βPhe, Tyr50βPhe, Glu56βAla) and four (Tyr50βPhe, Ser54βAla, Ser56βAla, Glu57βAla) mutants from the CDR2β loop were examined respectively and one residue from the CDR1β loop to serve as a negative control (Vβ8.2, Asn28βAla; Vβ7, Asp26βAla). All of the mutant NKT TCR proteins behaved similarly to the wt NKT TCR in gel filtration and analysis under reducing and non-reducing SDS-PAGE. Furthermore, an ELISA study showed that the NKT TCR mutant proteins were as reactive as the wt NKT TCR to conformation sensitive mAb reactive against the NKT TCR (data not shown). NKT TCR substitutions that caused < 50% change in the affinity of the interaction with CD1d-α-GalCer compared to wt NKT TCR were considered to have no major effect. Conversely, NKT TCR substitutions that caused > 50% change in binding affinity were considered to be energetically important to the interaction. As expected, the control mutations within the CDR1β loop did not affect CD1d-α-GalCer binding (Figure 5 E, F and Figure 6). In the Vβ8.2 NKT TCR, the Tyr48βPhe, Tyr50βPhe and Glu56βAla mutants impacted significantly on CD1d-α-GalCer recognition, consistent with previous alanine-scanning mutagenesis at positions 48 and 50 in the human Vβ11 NKT TCR, although the corresponding Glu 56β position was shown not to be essential for the human NKT TCR-CD1d-α-GalCer interaction. The conservative Tyr48βPhe and Tyr50βPhe mutations revealed the importance of their hydroxyl groups in mediating a series of polar contacts with CD1d. In the Vβ7 NKT TCR, Ser 56β and Glu 57β were shown not to be critical for the interaction, whereas Tyr 50β was shown to play a major role in the interaction with CD1d, again highlighting the importance of the aromatic residues in the CDR2β loop in mediating CD1d contacts. Interestingly, the Ser54βAla mutation in the Vβ7 TCR was observed to markedly improve the affinity of the interaction from 134 nM to 32.6 nM, making the affinity of this mutant comparable to that of the wild type Vβ8.2 NKT TCR. A similar result was observed in another study where CD1d tetramer binding was used to determine TCR affinity (Mallevaey.2009, Immunity submitted). The effect of the Ser54βAla mutant can be attributable to the Ser 54Oγ group being uncompensated within a hydrophobic pocket (Table 1), and accordingly, the Ala mutant forms more favourable vdw interactions when compared to the wild type counterpart. Thus, not all residues within the CDR2β loop of the Vβ7 NKT TCR are of the optimal chemistry and composition to interact with CD1d, and this, in part, may explain why the Vβ8.2 NKT TCR interacts with CD1d-α-GalCer with a higher affinity when compared to the Vβ7 NKT TCR.
Figure 5.
Binding of mutant NKT TCRs to mouse CD1d-α-GalCer as assessed by surface plasmon resonance. Wild type Vβ7 NKT TCR (A) and Vβ8.2 NKT TCR (B) and mutant Vβ7 NKT TCR S54A (C) and mutant Vβ8.2 (Y48F) (D) NKT TCR were injected over streptavidin immobilised mouse CD1d-α-GalCer and over a control cell containing unloaded CD1d. Sensorgrams show the binding (response units, RU) of decreasing concentrations of TCR (5, 2, 0.8, 0.32, 0.13 and 0.05 for Vα14Jα18-Vβ7 TCRs and 1, 0.4, 0.16, 0.064, 0.026 and 0.01µM for Vα14Jα18-Vβ8.2 TCRs) to mouse CD1d-α-GalCer following subtraction of the control flow cell. Insets show saturation plots demonstrating equilibrium binding of NKT TCR to immobilised CD1d-α-GalCer. (E and F) Binding of mutant NKT TCR to mouse CD1d α-GalCer. Site directed mutants of individual Vβ7 or Vβ8.2 residues were refolded with the invariant α-chain. The data is presented as a percentage binding of wild-type NKT TCRTCR.
Figure 6.
Binding of mutant NKT TCRs as assessed by CD1d-α-GalCer tetramer inhibition. Recombinant soluble NKT TCRs, and mutants thereof, were examined for their ability to block binding of mCD1d-α-GalCer tetramers to mouse NKT cells. PE labelled CD1d-α-GalCer tetramers were pre-incubated with titrating amounts of soluble wild type and mutant NKT TCRs before staining of mouse thymocytes. Cells were analysed by flow cytometry showing mCD1d-α–GalCer tetramer-PE on the vertical axis and anti-CD3 APC on the horizontal axis. CD3+ mCD1d-α–GalCer tetramer+ thymic NKT cells are indicated within the square with the MFI indicated.
Discussion
The Vβ8.2 and Vβ7 NKT TCR-mCD1d-α-GalCer structures, in conjunction with the human NKT TCR-CD1d-α-GalCer structure, broadly supports earlier suggestions that the NKT TCR exhibits characteristics of a pattern recognition receptor (Scott-Browne et al., 2007). Namely, the relatively rigid semi-invariant NKT TCR interacts with a monomorphic Ag-presenting molecule in an approximately conserved manner. Within this conserved docking framework that was situated above the CD1d F’ pocket, the invariant CDR1α loop and CDR3α loop contact α-GalCer and CD1d-α-GalCer respectively, while the Vβ domain interacted exclusively with CD1d. Although the NKT TCR is “innate-like”, the NKT TCR Vβ chain nevertheless exhibits diversity in the CDR3β loop and the mouse NKT cell repertoire uses Vβ8.2, Vβ7 and to a lesser extent Vβ2 (Benlagha et al., 2000; Matsuda et al., 2000). Our study addressed the basis and impact of this differential Vβ usage when recognising the prototypic NKT cell Ag, α-GalCer. The Vβ8.2 and Vβ7 NKT TCRs interacted with CD1d in a different manner as a result of sequence and structural differences between these Vβ domains. These differences resulted in a greater involvement of the Vβ7 chain interacting with mCD1d when compared to the Vβ8.2 chain, which included the CDR1β loop of Vβ7 mediating contacts with CD1d. In addition, the importance of specific residues, as directly judged by mutagenesis data, within the respective CDR2β loops varied: in Vβ8.2 NKT TCR, Tyr 48β and Tyr 50β and Glu 56β were essential; whereas only Tyr 50β was critical in Vβ7-mediated recognition of CD1d. Surprisingly, the Ser54βAla mutant improved the affinity of the Vβ7 NKT TCR-CD1d-α-GalCer interaction markedly, thereby simultaneously revealing that the sequence and composition of the NKT TCR Vβ7 CDR2β loop is non-ideal for interacting with CD1d, thereby providing insight into why the Vβ8.2 NKT TCR interacts with CD1d-α-GalCer with higher affinity than Vβ7 NKT TCR. Furthermore, despite the invariant nature of Vα14-Jα18, differences in the Vβ domains were “transferred” to the Jα18 chain, which impacted on Jα18-mediated CD1d-α-GalCer recognition. Collectively, these differences manifested in the Vβ8.2 NKT TCR interacting with mCD1d-α-GalCer with a moderately higher affinity compared to the Vβ7 NKT TCR.
The two mouse NKT TCR-CD1d-α-GalCer complexes, together with the 2.5 Å human NKT TCR-CD1d-α-GalCer structure reported here, allowed us to evaluate the residues in this interaction that are evolutionarily conserved. Namely, the conserved NKT TCR residues that contact identical residues in CD1d are Pro 28α of the CDR1α loop; Asp 94α, Arg 95α, Gly 96α, Ser 97α and Leu 99α of the CDR3α loop; and Tyr 50β of the CDR2β loop. However, given that the CDR2β loop of Vβ2 does not contain any tyrosine residues, this indicates the evolutionary-conserved “interaction codon” that underlies CD1d-restricted α-GalCer recognition by the NKT TCR is encoded by the Vα14-Jα18 domain (Vα24-Jα18 in humans). Recently, conserved binding residues or “interaction codons” have been defined for Vβ8.2 TCRs ligating pMHC (Dai et al., 2008; Feng et al., 2007). This observation allows us to assess whether these residues adopt similar roles in MHC-restricted and CD1d restricted recognition by Vβ8.2 TCRs. While the two Tyr residues forming “interaction codons” within the CDR2β loop of Vβ8.2 TCRs are used in recognising both MHC (Dai et al., 2008; Feng et al., 2007) and CD1d (Scott-Browne et al., 2007), they interact in markedly different regions of these Ag-presenting molecules (Godfrey et al., 2008), thereby highlighting the contrasting characteristics of peptide- and glycolipid-mediated recognition.
Despite the restricted NKT TCR repertoire, NKT cells can recognise a large array of CD1d-restricted lipid antigens (reviewed in (Bendelac et al., 2007; Brutkiewicz, 2006)). Given that the Vβ domains can modulate the affinity for CD1d-Ags (Mallevaey.2009, Immunity submitted), this suggests that any given Vβ chain will be an important factor in determining the range of Ags in which any given NKT TCR can interact with. The structural differences observed in the Vβ8.2 and Vβ7 NKT TCR recognition of mCD1d-α-GalCer may contribute to modulating the affinity towards other CD1d-restricted Ags. Moreover, the differences in CDR3β sequence, structure and their potential for interaction with CD1d in the NKT TCR-CD1d-α-GalCer complexes indicate that they may also play a role in fine-tuning the response to CD1d-Ag. Consistent with this, Vβ8.2+ NKT TCRs were shown to have higher affinity for α-GalCer bound to mCD1d, whereas Vβ7+ NKT TCRs have been reported to preferentially recognise iGb3 when compared to Vβ8.2 NKT TCRs (Schumann et al., 2006; Wei et al., 2006). However, the recent study (Mallevaey.2009, Immunity submitted) found that hybridomas expressing Vβ8.2 NKT TCRs recognise CD1d-iGb3 with higher affinity than those expressing Vβ7 NKT TCRs. This discrepancy might, at least in part, indicate a key contribution by CDR3β regions to the recognition of iGb3 by Vβ7+ NKT cells selected in vivo25, 26. Of potential interest, the sphingosine chain of α-GalCer interacts with Leu 84 of mCD1d, a residue that packs against Leu 99α of the CDR3α loop, the position of which and extent of mCD1d contacts was differentially influenced by the Vβ8.2 and Vβ7 usage. Accordingly, it suggests that different length lipid tails may alter the Leu 84-CDR3α loop contact, thereby providing a subtle mechanism for NKT TCRs to “sense” various ligands differently (McCarthy et al., 2007). In this regard, improving the resolution of the human NKT TCR-CD1d-α-Galcer complex from 3.2 Å to 2.5Å resolution is important, as it will serve as a more accurate benchmark to evaluate the subtle effects differing ligands may impart on the conformation of the CD1d Ag-binding cleft, which in turn may effect NKT TCR recognition. Plasticity in the NKT TCR-CD1d interaction is also suggested by the altered positioning of the α-GalCer head group between the human and mouse CD1d structures (Godfrey et al., 2005). While reciprocal cross species reactivity is observed between the Vβ8.2 and Vβ11 NKT TCR, this is partly diminished in the Vβ7 NKT TCR. In addition, the α-GalCer head group bound to mCD1d was observed to be shifted when ligated to the mouse NKT TCRs, which suggests flexibility in the sugar head group may play a role in the NKT TCR recognition of other Ags, such as the bulky iGb3. Flexibility in the peptide Ag has also been observed in a TCR-pMHC interaction (Tynan et al., 2007). Indeed, a basic superposition of the mCD1d-iGb3 structure (Zajonc et al., 2008) onto our Vβ8.2 NKT TCR-CD1d-α-GalCer structure indicates that such flexibility may occur, unless a significantly different binding mode exists between Vβ8.2 NKT TCR-mCD1d-α-GalCer and Vβ8.2 NKT TCR-mCD1d-iGb3, which seems less likely with the mutagenesis data that suggests a similar docking mode for iGb3 and α-GalCer (Scott-Browne et al., 2007)(Mallevaey.2009, Immunity submitted).
The different NKT TCR β-chains reported here converge on a common CD1d-antigen footprint, yet differences within these footprints were evident. Thus, while the NKT TCR could be considered as a pattern recognition receptor, our study reveals the potential for greater diversity at the NKT TCR-CD1d interface, thus providing greater scope for the differential recognition of a broad variety of CD1d restricted antigens.
Methods
Cloning and expression of genes encoding the mouse NKT Vα14, Vβ8.2 and Vβ7 TCRs
RNA was extracted from NKT-expressing mouse thymocytes (purified by flow cytometric sorting of thymocytes stained with CD1d–α–GalCer tetramers) and reverse transcribed. cDNAs encoding each the mouse NKT Vα14, Vβ8.2 and Vβ7 NKT TCRs were amplified by PCR and cloned into P-GEM Easy (Promega). We were unable to refold the intact ectodomains of mouse NKT TCRs (data not shown) and employed the use of the human constant domains of the NKT TCR to aid in refolding. In brief, soluble chimeric mouse-human TCR gene segments were then PCR-generated by splicing by overlap extension and transferred into the expression vector PET30 (Novagen). Stop codons were inserted immediately before the codons encoding the cysteines naturally forming alpha-beta interchain disulfide bonds. Instead, interchain disulfide pairing was achieved through Thr48Cys and Ser57Cys mutations introduced into the human alpha and beta constant domains respectively. Each chimeric gene was thus predicted to encode a soluble hybrid mouse-human NKT TCR consisting of a mouse variable and a human constant domain, lacking a transmembrane and cytoplasmic domain.
Soluble Vα14-Vβ8.2 and Vα14-Vβ7 NKT TCRs were expressed in BL21 E. coli, and inclusion body protein was prepared, refolded and purified as per the protocol of Garboczi et al. (Garboczi et al., 1996), except protein was refolded in the presence of either 1M (Vα14 and Vβ8.2) or 5 M (Vα14 and Vβ7) urea. The functional integrity of the NKT TCRs was confirmed by gel filtration, gel shift experiments and anti-TCR mAb ELISA reactivity (Supplementary Figure 1).
Expression and purification of CD1d
mCD1d was produced in house as described previously (Matsuda et al., 2000). In brief, mCD1d was made using a dual promoter baculovirus transfer vector, pBacp10pH, kindly provided by Dr Mitchell Kronenberg, La Jolla Institute for Allergy and Immunology, CA, USA. Recombinant mCD1d was produced with a BirA tag followed by a 6 amino acid histidine tag and expressed using HI5 insect cells. Soluble mCD1d protein was purified using Ni-Agarose affinity purification and subsequently passed over a Superdex 200 16/60 gel filtration column to remove aggregated material. For making mCD1d tetramers or for coating Bio-Rad ProteOn chips, purified mCD1d was biotinylated with BirA enzyme (Avidity) as per manufacturer’s protocol. The BirA and His-tag were not removed prior to crystallisation. Human CD1d was made in an analogous manner, with the exception that it lacked a BirA tag. Human CD1d was biotinylated by biotin-malemide treatment of a free cysteine residue at the C-terminal end.
Loading of CD1d
Loading of CD1d was carried out by incubating with α-GalCer (provided by Kirin Brewery Co. and Alexis Biochemicals) in a 3:1 (lipid:protein) molar ratio at room temperature overnight. Excess α-GalCer was removed from CD1d using Superdex 200 10/300 gel filtration.
Complexation of NKT TCRs with CD1d-α̃GalCer
Purified NKT TCR and α–GalCer-loaded CD1d were mixed, and the ternary complex was isolated by gel filtration on a Superdex 200 16/60 column (GE Healthcare), concentrated to 10mg/ml, and used in crystal trials.
Flow cytometry and CD1d-α-GalCer tetramer inhibition assay
Anti mouse TcRβ-allophycocyanin (APC) (clone H57-597) and CD3-APC (clone 145-2C11) were purchased from BD Biosciences. The CD1d/α-GalCer tetramer inhibition assay was carried out as described previously (Kjer-Nielsen et al., 2006). Thymocytes were prepared by gently grinding the organ between frosted glass slides. Stained cells were analysed by flow cytometry using a FACScalibur flow cytometer (Becton Dickinson).
Crystallisation, structure determination and refinement
The Vβ8.2 NKT TCR-CD1d-α-GalCer (7 mg/ml in 10 mM Tris pH 8.0 and 150 mM NaCl) and Vβ7 NKT TCR-CD1d-α-GalCer (6 mg/ml in 10 mM Tris pH 8.0 and 150 mM NaCl) complex crystallised at room temperature in 17% polyethylene glycol 10K, 0.1M ammonium acetate, 0.1M BisTris, pH 5.5 using the hanging drop vapour diffusion technique. Equal ratio of the protein to mother liquor resulted in plate-like crystals after 2–3 days. The crystals were flash frozen prior to data collection in mother liquor containing 20% glycerol as the cryoprotectant. The crystals of Vβ8.2 NKT TCR-CD1d-α-GalCer and Vβ7 NKT TCR-CD1d-α-GalCer complex diffracted to 2.9Å and 2.8Å respectively and belong to the space group P212121, with one ternary complex in the asymmetric unit. The human NKT TCR-CD1d-α-GalCer (10 mg/ml in 10 mM Tris pH 8.0 and 150 mM NaCl) complex crystallised at room temperature in 10–12% PEG 10K, 0.2 M magnesium chloride, 0.1M Tris, pH 9.0. The crystals were equilibrated in the precipitation solution with increasing concentrations of PEG 10K to 35% for at least a few days. The dehydrated crystals were flash frozen in the dehydration solution prior to data collection. The human NKT TCR-CD1d-α-GalCer diffracted to 2.5 Å resolution and belong to the space group P2 with two ternary complexes in the asymmetric unit.
Data for the two mouse NKT TCR complexes were collected at the Australian Synchrotron Facility in Melbourne, Australia, and processed using programs from the CCP4 suite (1994). The data for the human NKT TCR-CD1d-α-GalCer was collected at the Advanced Photon Source synchrotron facility in Chicago and processed using HKL2000 and programs from the CCP4 suite. The crystal structure of the Vβ8.2 NKT TCR-CD1d-α-GalCer was solved by the molecular replacement method, using the program Phaser from the CCP4 Suite. The structure of mouse CD1d-glycosphingolipid complex (Protein Data Bank ID code 2FIK) minus the lipid and the structure of unliganded semi-invariant Vα14 TCR (Protein Data Bank ID code 2Q86) were used as the search models for solving Vβ8.2 NKT TCR-CD1d-α-GalCer. Refmac in CCP4 suite was used for the initial round of rigid body refinement and subsequently restrained refinement interspersed with rounds of model building using Coot (Emsley and Cowtan, 2004). At a later stage of refinement, restrained refinement included translation libration screw parameters. The progress of refinement was monitored by the Rfree value. The Vβ7 NKT TCR-CD1d-α-GalCer was solved by the molecular replacement method in Phaser, using Vβ8.2 NKT TCR-CD1d-α-GalCer minus the lipid as the search model. Initially the structure was refined using rigid body refinement in Refmac followed by the simulated annealing protocol implemented in Phenix (Zwart et al., 2008). The model was improved using iterative rounds of refinement and model building. Translation libration screw parameters were included at a later stage of refinement and the progress of refinement was monitored by the Rfree value. The human NKT TCR-CD1d-α-GalCer was solved by molecular replacement method in Phaser, using the 3.2Å structure solved previously (Protein Data Bank ID code 2PO6) minus the lipid as the search model. The quality of the three structures was assessed with the programs within CCP4. The residues that could not be modelled in the Vβ8.2 NKT TCR-CD1d-α-GalCer were: CD1d, residues 1–7, 90–93 and 110; TCR α-chain, residues 134–135 and 209–210; TCR β-chain, residues 1–2 and 98–105 (CDR3β). The residues that could not be modelled in the Vβ7 NKT TCR-CD1d-α-GalCer were: CD1d, residues 1–7, 88–94 and 108–110; TCR α-chain, residues 130–135, 186–187 and 208–210; TCR β-chain, residues 1 and 122. The residues that could not be modelled in the human NKT TCR-CD1d-α-GalCer were: CD1d, residues 1–5 for chain A and 1–4 for chain C; β2m, residues 98–99 for chain B; TCR α-chain, residues 131–132, 137, 154 for chain E and 136 for chain G; TCR β-chain, residues 1, 100–101 for chain F and 1 for chain H. For data collection and refinement statistics see Supplementary Table 1. All molecular graphics representations were created using PyMol (DeLano, 2002).
Surface plasmon resonance
The interaction between soluble, recombinant CD1d and wild type and mutant NKT TCRs were analysed by SPR using a Bio-Rad ProteOn XPR36 instrument (Hercules, CA). All experiments were performed at 25°C in a buffer containing 10 mM HEPES, pH 7.4, 150 mM NaCl, and 0.005% Tween-20 (HBS-T). Streptavidin was diluted into 10 mM sodium acetate, pH 4.5 and ~3000 RU was immobilised on all 6 flow cells of a GLC Sensorchip (Bio-Rad) by amine coupling. Biotinylated CD1d was passed over the surface of the chip and ~700RU was captured by the streptavidin. Flow cell 1 and 2 contained α–GalCer loaded mouse and human CD1d, respectively, whereas flow cells 3 and 4 contained empty mouse and human CD1d and served as control cells. Recombinant wild type and mutant NKT TCR was subjected to size exclusion chromatography within 24 hours of analysis and the concentration of purified protein estimated by OD280. Wild type and mutant NKT TCRs were then serially-diluted from 5µM to 0.05µM or 1µM to 0.01µM in HBS-T and injected simultaneously over the test and control surfaces at a flow rate of 30 µL/minute. Following subtraction of data from control flow cells, the interactions were analysed using the ProteOn Manager software version 2.1 (Bio-Rad) and steady-state KD values were derived from the equilibrium option of the software package. Kinetic data was derived using the kinetic fit option of the software and data analysis was fitted using the 1:1 Langmuir binding model.
Probing conformational integrity of the wild type and mutant NKT TCRs
100 µl of soluble NKT TCR (5 µg/ml) was added to a 96-well ELISA plate (U96 Maxisorp, Nunc) at 4°C for 16 hours. Plates were then blocked with 200 µl of PBS/1%BSA at 37°C for one hour. Titrated amounts of the conformationally-dependent, constant domain-reactive mAb 12H8 were then added, following which HRP-conjugated anti-mouse Ig was added. O-phenylenediamine substrate (Sigma) was added next and the reaction was terminated with HCl, and ELISA plates were read at 492 nm on a Labsystems Multiscan ELISA plate reader.
Supplementary Material
Acknowledgements
The Australian Research Council (ARC), the National Health and Medical Research Council of Australia (NHMRC) and the Cancer Council of Victoria supported this research. LG was supported by an NIH grant (AI057485). DIG and MJS are supported by NHMRC Research Fellowships, JR is supported by an ARC Federation Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bendelac A, Savage PB, Teyton L. The Biology of NKT Cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- Benlagha K, Weiss A, Beavis A, Teyton L, Bendelac A. In vivo identification of glycolipid antigen-specific T cells using fluorescent CD1d tetramers. Journal of Experimental Medicine. 2000;191:1895–1903. doi: 10.1084/jem.191.11.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borg NA, Wun KS, Kjer-Nielsen L, Wilce MC, Pellicci DG, Koh R, Besra GS, Bharadwaj M, Godfrey DI, McCluskey J, Rossjohn J. CD1d-lipid-antigen recognition by the semi-invariant NKT T-cell receptor. Nature. 2007;448:44–49. doi: 10.1038/nature05907. [DOI] [PubMed] [Google Scholar]
- Brigl M, van den Elzen P, Chen X, Meyers JH, Wu D, Wong CH, Reddington F, Illarianov PA, Besra GS, Brenner MB, Gumperz JE. Conserved and Heterogeneous Lipid Antigen Specificities of CD1d-Restricted NKT Cell Receptors. J Immunol. 2006;176:3625–3634. doi: 10.4049/jimmunol.176.6.3625. [DOI] [PubMed] [Google Scholar]
- Brossay L, Chioda M, Burdin N, Koezuka Y, Casorati G, Dellabona P, Kronenberg M. Cd1d-Mediated Recognition of an Alpha-Galactosylceramide by Natural Killer T Cells Is Highly Conserved through Mammalian Evolution. Journal of Experimental Medicine. 1998;188:1521–1528. doi: 10.1084/jem.188.8.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutkiewicz RR. CD1d ligands: the good, the bad, and the ugly. J Immunol. 2006;177:769–775. doi: 10.4049/jimmunol.177.2.769. [DOI] [PubMed] [Google Scholar]
- Burdin N, Brossay L, Koezuka Y, Smiley ST, Grusby MJ, Gui M, Taniguchi M, Hayakawa K, Kronenberg M. Selective Ability of Mouse CD1 to Present Glycolipids: {alpha}-Galactosylceramide Specifically Stimulates V{alpha}14+ NK T Lymphocytes. The Journal of Immunology. 1998;161:3271–3281. [PubMed] [Google Scholar]
- CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Cui JQ, Shin T, Kawano T, Sato H, Kondo E, Toura I, Kaneko Y, Koseki H, Kanno M, Taniguchi M. Requirement for V(Alpha)14 Nkt Cells in Il-12-Mediated Rejection of Tumors. Science. 1997;278:1623–1626. doi: 10.1126/science.278.5343.1623. [DOI] [PubMed] [Google Scholar]
- Dai S, Huseby ES, Rubtsova K, Scott-Browne J, Crawford F, Macdonald WA, Marrack P, Kappler JW. Crossreactive T Cells Spotlight the Germline Rules for [alpha][beta] T Cell-Receptor Interactions with MHC Molecules. Immunity. 2008;28:324–334. doi: 10.1016/j.immuni.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System. San Carlos, CA, USA: DeLano Scientific; 2002. [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Feng D, Bond CJ, Ely LK, Maynard J, Garcia KC. Structural evidence for a germline-encoded T cell receptor-major histocompatibility complex interaction 'codon'. Nat Immunol. 2007;8:975–983. doi: 10.1038/ni1502. [DOI] [PubMed] [Google Scholar]
- Fischer K, Scotet E, Niemeyer M, Koebernick H, Zerrahn J, Maillet S, Hurwitz R, Kursar M, Bonneville M, Kaufmann SH, Schaible UE. Mycobacterial phosphatidylinositol mannoside is a natural antigen for CD1d-restricted T cells. Proc Natl Acad Sci U S A. 2004;101:10685–10690. doi: 10.1073/pnas.0403787101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadola SD, Dulphy N, Salio M, Cerundolo V. V alpha 24-J alpha Q-independent, CD1d-restricted recognition of alpha-galactosylceramide by human CD4(+) and CD8 alpha beta(+) T lymphocytes. Journal of Immunology. 2002;168:5514–5520. doi: 10.4049/jimmunol.168.11.5514. [DOI] [PubMed] [Google Scholar]
- Garboczi DN, Utz U, Ghosh P, Seth A, Kim J, VanTienhoven EA, Biddison WE, Wiley DC. Assembly, specific binding, and crystallization of a human TCR-alphabeta with an antigenic Tax peptide from human T lymphotropic virus type 1 and the class I MHC molecule HLA-A2. J Immunol. 1996;157:5403–5410. [PubMed] [Google Scholar]
- Godfrey DI, Kronenberg M. Going both ways: immune regulation via CD1d-dependent NKT cells. J Clin Invest. 2004;114:1379–1388. doi: 10.1172/JCI23594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey DI, MacDonald HR, Kronenberg M, Smyth MJ, Van Kaer L. NKT cells: what's in a name? Nat Rev Immunol. 2004;4:231–237. doi: 10.1038/nri1309. [DOI] [PubMed] [Google Scholar]
- Godfrey DI, McCluskey J, Rossjohn J. CD1d antigen presentation: treats for NKT cells. Nat Immunol. 2005;6:754–756. doi: 10.1038/ni0805-754. [DOI] [PubMed] [Google Scholar]
- Godfrey DI, Rossjohn J, McCluskey J. The Fidelity, Occasional Promiscuity, and Versatility of T Cell Receptor Recognition. Immunity. 2008;28:304–314. doi: 10.1016/j.immuni.2008.02.004. [DOI] [PubMed] [Google Scholar]
- Ishizuka J, Stewart-Jones GBE, van der Merwe A, Bell JI, McMichael AJ, Jones EY. The Structural Dynamics and Energetics of an Immunodominant T Cell Receptor Are Programmed by Its V[beta] Domain. Immunity. 2008;28:171–182. doi: 10.1016/j.immuni.2007.12.018. [DOI] [PubMed] [Google Scholar]
- Kinjo Y, Pei B, Bufali S, Raju R, Richardson SK, Imamura M, Fujio M, Wu D, Khurana A, Kawahara K, et al. Natural sphingomonas glycolipids vary greatly in their ability to activate natural killer T cells. Chem Biol. 2008;15:654–664. doi: 10.1016/j.chembiol.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinjo Y, Tupin E, Wu D, Fujio M, Garcia-Navarro R, Benhnia MR, Zajonc DM, Ben-Menachem G, Ainge GD, Painter GF, et al. Natural killer T cells recognize diacylglycerol antigens from pathogenic bacteria. Nat Immunol. 2006;7:978–986. doi: 10.1038/ni1380. [DOI] [PubMed] [Google Scholar]
- Kinjo Y, Wu D, Kim G, Xing GW, Poles MA, Ho DD, Tsuji M, Kawahara K, Wong CH, Kronenberg M. Recognition of bacterial glycosphingolipids by natural killer T cells. Nature. 2005;434:520–525. doi: 10.1038/nature03407. [DOI] [PubMed] [Google Scholar]
- Kjer-Nielsen L, Borg NA, Pellicci DG, Beddoe T, Kostenko L, Clements CS, Williamson NA, Smyth MJ, Besra GS, Reid HH, et al. A structural basis for selection and cross-species reactivity of the semi-invariant NKT cell receptor in CD1d/glycolipid recognition. J Exp Med. 2006;203:661–673. doi: 10.1084/jem.20051777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjer-Nielsen L, Clements CS, Purcell AW, Brooks AG, Whisstock JC, Burrows SR, McCluskey J, Rossjohn J. A structural basis for the selection of dominant alphabeta T cell receptors in antiviral immunity. Immunity. 2003;18:53–64. doi: 10.1016/s1074-7613(02)00513-7. [DOI] [PubMed] [Google Scholar]
- Koch M, Stronge VS, Shepherd D, Gadola SD, Mathew B, Ritter G, Fersht AR, Besra GS, Schmidt RR, Jones EY, Cerundolo V. The crystal structure of human CD1d with and without alpha-galactosylceramide. Nat Immunol. 2005;6:819–826. doi: 10.1038/ni1225. [DOI] [PubMed] [Google Scholar]
- Matsuda JL, Gapin L, Fazilleau N, Warren K, Naidenko OV, Kronenberg M. Natural killer T cells reactive to a single glycolipid exhibit a highly diverse T cell receptor beta repertoire and small clone size. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:12636–12641. doi: 10.1073/pnas.221445298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda JL, Mallevaey T, Scott-Browne J, Gapin L. CD1d-restricted iNKT cells, the 'Swiss-Army knife' of the immune system. Curr Opin Immunol. 2008 doi: 10.1016/j.coi.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda JL, Naidenko OV, Gapin L, Nakayama T, Taniguchi M, Wang C-R, Koezuka Y, Kronenberg M. Tracking the Response of Natural Killer T Cells to a Glycolipid Antigen Using CD1d Tetramers. The Journal of Experimental Medicine. 2000;192:741–754. doi: 10.1084/jem.192.5.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattner J, Debord KL, Ismail N, Goff RD, Cantu C, 3rd, Zhou D, Saint-Mezard P, Wang V, Gao Y, Yin N, et al. Exogenous and endogenous glycolipid antigens activate NKT cells during microbial infections. Nature. 2005;434:525–529. doi: 10.1038/nature03408. [DOI] [PubMed] [Google Scholar]
- McCarthy C, Shepherd D, Fleire S, Stronge VS, Koch M, Illarionov PA, Bossi G, Salio M, Denkberg G, Reddington F, et al. The length of lipids bound to human CD1d molecules modulates the affinity of NKT cell TCR and the threshold of NKT cell activation. J Exp Med. 2007;204:1131–1144. doi: 10.1084/jem.20062342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porcelli S, Yockey CE, Brenner MB, Balk SP. Analysis of T cell antigen receptor (TCR) expression by human peripheral blood CD4-8- alpha/beta T cells demonstrates preferential use of several V beta genes and an invariant TCR alpha chain. The Journal of Experimental Medicine. 1993;178:1–16. doi: 10.1084/jem.178.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph MG, Stanfield RL, Wilson IA. How TCRs bind MHCs, peptides, and coreceptors. Annu Rev Immunol. 2006;24:419–466. doi: 10.1146/annurev.immunol.23.021704.115658. [DOI] [PubMed] [Google Scholar]
- Schumann J, Mycko MP, Dellabona P, Casorati G, Macdonald HR. Cutting Edge: Influence of the TCR Vbeta Domain on the Selection of Semi-Invariant NKT Cells by Endogenous Ligands. J Immunol. 2006;176:2064–2068. doi: 10.4049/jimmunol.176.4.2064. [DOI] [PubMed] [Google Scholar]
- Schumann J, Voyle RB, Wei BY, MacDonald HR. Cutting Edge: Influence of the TCR Vbeta Domain on the Avidity of CD1d:alpha-Galactosylceramide Binding by Invariant Valpha14 NKT Cells. J Immunol. 2003;170:5815–5819. doi: 10.4049/jimmunol.170.12.5815. [DOI] [PubMed] [Google Scholar]
- Scott-Browne JP, Matsuda JL, Mallevaey T, White J, Borg NA, McCluskey J, Rossjohn J, Kappler J, Marrack P, Gapin L. Germline-encoded recognition of diverse glycolipids by natural killer T cells. Nat Immunol. 2007;8:1105–1113. doi: 10.1038/ni1510. [DOI] [PubMed] [Google Scholar]
- Tynan FE, Reid HH, Kjer-Nielsen L, Miles JJ, Wilce MC, Kostenko L, Borg NA, Williamson NA, Beddoe T, Purcell AW, et al. A T cell receptor flattens a bulged antigenic peptide presented by a major histocompatibility complex class I molecule. Nat Immunol. 2007;8:268–276. doi: 10.1038/ni1432. [DOI] [PubMed] [Google Scholar]
- Wei DG, Curran SA, Savage PB, Teyton L, Bendelac A. Mechanisms imposing the V{beta} bias of V{alpha}14 natural killer T cells and consequences for microbial glycolipid recognition. The Journal of Experimental Medicine. 2006;203:1197–1207. doi: 10.1084/jem.20060418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu DY, Segal NH, Sidobre S, Kronenberg M, Chapman PB. Cross-presentation of Disialoganglioside GD3 to Natural Killer T Cells. J Exp Med. 2003;198:173–181. doi: 10.1084/jem.20030446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wun KS, Borg NA, Kjer-Nielsen L, Beddoe T, Koh R, Richardson SK, Thakur M, Howell AR, Scott-Browne JP, Gapin L, et al. A minimal binding footprint on CD1d-glycolipid is a basis for selection of the unique human NKT TCR. The Journal of Experimental Medicine. 2008;205:939–949. doi: 10.1084/jem.20072141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zajonc DM, Cantu C, 3rd, Mattner J, Zhou D, Savage PB, Bendelac A, Wilson IA, Teyton L. Structure and function of a potent agonist for the semi-invariant natural killer T cell receptor. Nat Immunol. 2005;6:810–818. doi: 10.1038/ni1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zajonc DM, Savage PB, Bendelac A, Wilson IA, Teyton L. Crystal structures of mouse CD1d-iGb3 complex and its cognate V[alpha]14 T cell receptor suggest a model for dual recognition of foreign and self glycolipids. Journal of Molecular Biology. 2008;377:1104–1116. doi: 10.1016/j.jmb.2008.01.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Mattner J, Cantu C, 3rd, Schrantz N, Yin N, Gao Y, Sagiv Y, Hudspeth K, Wu YP, Yamashita T, et al. Lysosomal glycosphingolipid recognition by NKT cells. Science. 2004;306:1786–1789. doi: 10.1126/science.1103440. [DOI] [PubMed] [Google Scholar]
- Zwart PH, Afonine PV, Grosse-Kunstleve RW, Hung L-W, Ioerger TR, McCoy AJ, McKee E, Moriarty NW, Read RJ, Sacchettini JC, et al. Automated Structure Solution with the PHENIX Suite. In Structural Proteomics. 2008:419–435. doi: 10.1007/978-1-60327-058-8_28. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.