Abstract
Background and Aims
AtSUC2 encodes a sucrose/proton symporter that localizes throughout the collection and transport phloem and is necessary for efficient transport of sucrose from source to sink tissues in Arabidopsis thaliana. Plants harbouring homozygous AtSUC2 null alleles accumulate sugar, starch, and anthocyanin in mature leaves, have severely delayed development and stunted growth and, in previous studies, failed to complete their life cycle by producing viable seed.
Methods
An AtSUC2 allele with a T-DNA insertion in the second intron was analysed. Full-length transcript from this allele is not produced, and a truncated protein translated from sequences upstream of the insertion site did not catalyse sucrose uptake into yeast, supporting the contention that this is a null allele. Mutant plants were grown in a growth chamber with a diurnal light/dark cycle, and growth patterns recorded.
Key Results
This allele (SALK_038124, designated AtSUC2-4) has the hallmarks of previously described null alleles but, despite compromised carbon partitioning and growth, produces viable seeds. The onset of flowering was chronologically delayed but occurred at the same point in the plastochron index as wild type.
Conclusions
AtSUC2 is important for phloem loading and is therefore fundamental to phloem transport and plant productivity, but plants can complete their life cycle and produce viable seed in its absence. Arabidopsis appears to have mechanisms for mobilizing reduced carbon from the phloem into developing seeds independent of AtSUC2.
Keywords: Arabidopsis thaliana, AtSUC2, sucrose, transporter, symporter, phloem, phloem loading, plant productivity
INTRODUCTION
Photoassimilated carbon is osmotically driven from source tissues to sinks via the phloem. To accentuate the pressure gradient between source and sinks, most plants expend energy to accumulate sugars in the minor-vein phloem of mature leaves. This thermodynamically unfavourable process is known as phloem loading, and two energized mechanisms are well described (Turgeon and Ayre, 2005). In the polymer trap mechanism, sucrose (Suc) diffuses into intermediary cells (specialized phloem companion cells) through highly modified plasmodesmata, and is converted to raffinose family oligosaccharides (RFOs) (Turgeon, 1996). These RFOs are thought to accumulate in the phloem because they are too large to diffuse back toward the mesophyll. The committed step for RFO synthesis is catalysed by galactinol synthase (inositol 1-α-galactosyltransferase; EC 2·4·1·123), which synthesizes galactinol (αd-galactose-[1-3]-1d-myo-inositol), the galactosyl donor for RFO synthesis, from myo-inositol and the energized intermediate, UDP-galactose (Keller and Pharr, 1996). In the second phloem-loading mechanism, sugar, most commonly Suc but also alcohol sugars, is pumped across the plasma membrane from the apoplast into the phloem by sugar/proton symporters and is energized by the proton motive force (Giaquinta, 1977; Lalonde et al., 2003; Hafke et al., 2005; Sauer, 2007; Braun and Slewinski, 2009). Recently, reducing hexose sugars were identified as prominent transport sugars but the mode of entry into the phloem has not been addressed (van Bel and Hess, 2008). Significantly, some plants forego phloem loading and instead have the highest concentration of solute in the mesophyll cells to drive phloem transport (Turgeon and Medville, 1998; Turgeon and Ayre, 2005; Reidel et al., 2009; Rennie and Turgeon, 2009).
Arabidopsis thaliana phloem loads Suc from the apoplast via Suc/proton (Suc/H+) symporters, and, of the nine family members, AtSUC2 is the predominant, if not exclusive, member involved in phloem loading and Suc transport (Lalonde et al., 2004; Sauer, 2007). AtSUC2 is expressed in companion cells of collection and transport phloem (Truernit and Sauer, 1995; Martens et al., 2006), as are the SUT1 orthologues in Solanaceae species (Schmitt et al., 2008). In mutant plants harbouring a T-DNA insertion in each of the functional Suc/H+ symporter-family members, only Atsuc2 mutants demonstrate overtly debilitated phloem transport (Gottwald et al., 2000, Srivastava et al., 2008). Three T-DNA-insertion alleles of AtSUC2 were previously characterized: one has an insert in the first exon downstream of sequences encoding the fourth transmembrane domain and the other two have inserts in the second intron that truncate the protein after the tenth transmembrane domain (Gottwald et al., 2000). The three alleles displayed identical phenotypes: plants were dramatically stunted; accumulated high levels of soluble sugar, starch and anthocyanin; had delayed flowering; and failed to produce viable seeds (Gottwald et al., 2000). Repression of the Solanaceae SUT1 orthologues by antisense RNA resulted in similar characteristics stemming from compromised Suc transport, as well as chlorosis and necrosis in affected tissues (Kuhn et al., 1996; Burkle et al., 1998; Schulz et al., 1998).
In this study, an additional mutant allele of AtSUC2 with a T-DNA insertion in the second intron was analysed, and the designation AtSUC2-4 proposed. Transcripts of this allele were previously demonstrated to be missing the third and forth exons (Srivastava et al., 2008) but transcripts corresponding to exons one and two were present at greatly reduced levels relative to wild type, leaving the possibility that a truncated protein with residual Suc/H+ activity might be produced. However, mutants harbouring this allele did not accumulate [14C]Suc in the minor veins of mature leaves (Srivastava et al., 2009). In this study, the truncated cDNA was expressed in yeast cultures and Suc uptake measured relative to controls to establish further that this allele is a null allele. The phenotype of this allele is identical to the previously described mutants, with the important exception that viable seeds are produced. These results argue that AtSUC2-catalysed phloem loading is not essential for the plant to complete its life cycle, and alternative mechanisms for transporting photoassimilated carbon into maturing seeds are discussed.
MATERIALS AND METHODS
Plasmid constructions and transport assays in yeast
Plasmids pRS424::ADH-MCS and pRS424::ADH-cSUC2 were previously described (Srivastava et al., 2008) and are the empty vector control and full-length AtSUC2 cDNA, respectively, used for [14C]Suc uptake studies in Saccharomyces cerevisiae (yeast). pRS424::ADH-Ex1-2 is an equivalent plasmid harbouring AtSUC2 cDNA truncated to include only the first two exons. The truncated cDNA was created by PCR using pRS424::ADH-cSUC2 as a template and forward oligonucleotide AtSUC25 (5′-TTCAAGGTACCAAATATGGTCAGCCATCCAATG-3′) and reverse oligonucleotide AtSUC2-Ex2Rstop (5′-GAATTCGAGCTCATTGGCCGGCACCGGAATTGGTTG-3′), which incorporates a stop codon (italics) downstream of the amino acid encoding sequences in the second exon of AtSUC2. Phusion Hotstart polymerase (NEB, Beverly, MA, USA) was used according to the manufacturer's instructions. The PCR product was digested with restriction endonucleases KpnI and SacI (NEB; the recognition sequences are underlined in the oligonucleotide sequences), and ligated into the same sites of pRS424::ADH-MCS to create pRS424::ADH-Ex1-2. Transformation of yeast strain SuSy7-URA, growth of cultures, and [14C]Suc uptake assays were as previously described (Srivastava et al., 2008).
Plant material and microscopy
Growth of plants and PCR genotyping were as previously described (Srivastava et al., 2008, 2009). For staining and microscopy to visualize starch and cellulose, plants were kept in the dark for 24 h. Leaves were excised and vacuum infiltrated with 3 % glutaraldehyde in 25 mm phosphate buffer, pH 7·2, and fixed overnight at 4 °C. Tissues were subsequently fixed in ethanol and embedded in paraffin (Paraplast Plus; Kendall Labware, Mansfield, MA, USA) using a Milestone RHS microwave histoprocessor embedding system (Milestone Medical, Kalamazoo, MI, USA) according to the manufacturer's instructions. Transverse 7-μm-thick sections were cut with a rotary microtome, floated in a water bath, and melted onto Probe-on-Plus slides (Fisher Scientific, Houston, TX, USA). Equal volumes of I-KI staining solution (0·2 % iodine, 2 % potassium iodide) and calcofluor white solution (Eng Scientific Inc., Clifton, NJ, USA) were mixed and 200 µL spotted onto deparaffinized and rehydrated sections. Staining proceeded for 10 min; slides were dipped twice in water for 30 s to remove excess stain and mounted in 50 % glycerol for viewing. Sections were viewed under bright field for starch staining and with epifluorescence for calcofluor white staining using excitation and emission filter sets at 340–380 nm and 435–485 nm, respectively. For digital photography, exposure times and settings were equivalent for all samples.
To measure epidermal and palisade cells, leaf tissues were fixed with ethanol:acetic acid (3:1) and washed sequentially in 10 % and 15 % sucrose in phosphate buffered saline (137 mm NaCl, 10 mm phosphate, 2·7 mm KCl, pH 7·4) before staining with 0·1 % propidium iodide solution for 15 min. Stained leaves were visualized by laser-scanning confocal microscopy (Leica TCS SP2 AOBS; Leica Microsystems Inc., Bannockburn, IL, USA). Propidium iodide was detected by exciting with the 488-nm line of the argon laser and emission detected at 620 nm. Cross-sectional area of the cells was calculated with ImageJ version 1·38X (Rasband, 2007).
RESULTS
Atsuc2::T-DNA mutations
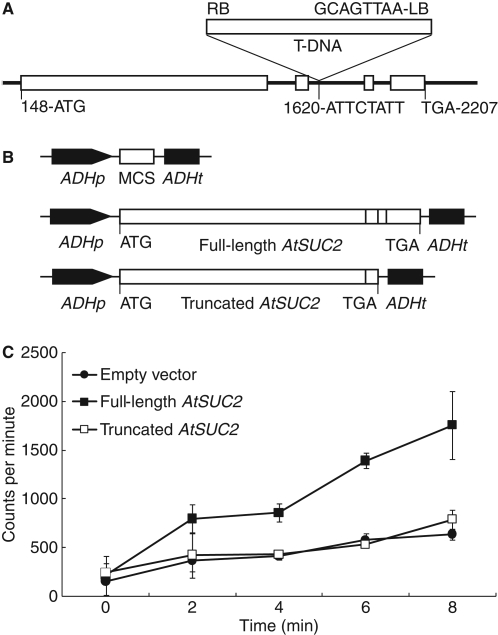
Three lines with T-DNA insertions at the AtSUC2 locus (At1g22710) were obtained through the Arabidopsis Biological Resource Center: SALK_087046, SALK_001331 and SALK_038124 (Alonso et al., 2003). Plant genotyping and molecular characterization of the insertions are detailed elsewhere (Srivastava et al., 2008) but, in brief, an insert was not identified among the 12 SALK_087046 seedlings sampled and the insert in SALK_001331 was localized downstream of the AtSUC2 open reading frame and did not have a visible phenotype. The SALK_038124 T-DNA insertion is in the second intron, 1473 nucleotides downstream of the start codon in the genomic sequence (Fig. 1A). Plants segregating homozygous for the T-DNA (Atsuc2::T-DNA/Atsuc2::T-DNA, designated Atsuc2-4) were severely stunted, and accumulated anthocyanin and starch in mature leaves (Srivastava et al., 2008) as described previously for three other mutants with T-DNA insertions at the AtSUC2 locus (Gottwald et al., 2000). AtSUC2 transcript analysis showed that low levels of transcript corresponding to sequences upstream of the T-DNA insert were present in Atsuc2-4 plants but that transcript corresponding to sequences downstream of the T-DNA insert were not (Srivastava et al., 2008). AtSUC2 cDNA expressed from 2 kb of AtSUC2 promoter was sufficient to restore wild-type growth and carbon partitioning to Atsuc2-4 mutant plants, showing that the observed phenotype was the result of disrupted AtSUC2 activity (Srivastava et al., 2008).
Fig. 1.
Site of T-DNA insertion in Atsuc2-4 (SALK_038124) and expression cassettes used for, and results of, [14C]Suc uptake into yeast. (A) T-DNA insertion site in the second intron of AtSUC2 (At1g22710) in Atsuc2-4 with flanking nucleotides indicated; exons are indicated as white boxes and intergenic regions are indicated by the solid black line; numbering is relative to the start of the 5′ UTR, based on the gene model AT1G22710·1 at http://www.arabidopsis.org; T-DNA is indicated as labelled above the gene model. (B) The three expression cassettes used for [14C]Suc uptake into yeast cells; full-length and truncated AtSUC2 cDNA as labelled with fused exons indicated as white boxes; ATG start and TGA stop codons as labelled; MCS, multiple-cloning site in the empty-vector control, pRS424::ADH-MCS; ADHp, yeast ALCOHOL DEHYDROGENASE promoter; ADHt yeast ADH terminator. (C) Uptake of [14C]Suc uptake into yeast cultures harbouring the indicated plasmids.
Based on the location of the T-DNA inserts, transcript analysis and phenotype, it was argued that the T-DNA insertions identified are true null alleles (Gottwald et al., 2000; Srivastava et al., 2008). However, since small amounts of truncated mRNA were identified in Atsuc2-4 plants (Srivastava et al., 2008), there was a possibility that a truncated protein could be produced and have residual Suc/H+ symporter activity. Any protein resulting from a gene truncated after the second exon would be missing 72 carboxyl-terminal amino acids, corresponding to two membrane-spanning domains, an extracellular domain and the carboxy-terminal cytoplasmic domain (Bush, 1999; Gottwald et al., 2000). To test if such a protein may have residual symporter activity, a cDNA representing AtSUC2 truncated after the second exon was constructed (Fig. 1B), expressed in yeast from a strong ADH1 promoter in a multicopy yeast vector, and [14C]Suc uptake measured (Fig. 1C). Yeast cultures harbouring this truncated AtSUC2 cDNA had Suc uptake rates identical to empty-vector controls, whereas uptake by cells harbouring full-length cDNA was significantly greater. This analysis supports the contention that the T-DNA insertions in SALK_038124 (Srivastava et al., 2008) and the previously identified lines (Gottwald et al., 2000) created true null alleles.
Analysis of growth and carbohydrates in Atsuc2-4 plants
Growth of Atsuc2-4 plants during the vegetative phase of development was previously compared with wild type (AtSUC2/AtSUC2) and to Atsuc2-4 plants complemented with 2 kb of AtSUC2 promoter driving the expression of AtSUC2 cDNA (designated SUC2p::cSUC2; seed stock KD1039) (Srivastava et al., 2008). Vegetative growth was measured as rosette area 21 d after germination, prior to the formation of visible floral structures. Wild-type and KD1039 plants were not statistically different in rosette size (11·21 ± 1·98 cm2 and 9·34 ± 1·26 cm2, respectively) whereas Atsuc2-4 plants were dramatically stunted (0·28 ± 0·08 cm2) (Srivastava et al., 2008).
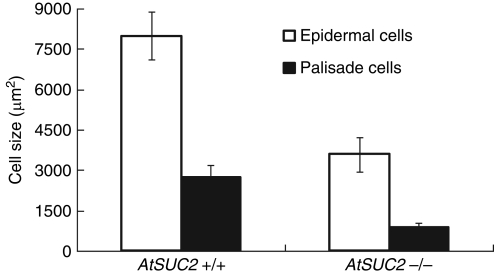
To establish if reduced cell expansion contributed to the extreme stunting of Atsuc2-4 plants, epidermal and palisade cells were examined by staining the cells with propidium iodide and measuring the cross-sectional area with laser-scanning confocal microscopy. Both cell types were smaller (Fig. 2), with Atsuc2-4 epidermal cells having less than half the cross-sectional area of wild type cells, and Atsuc2-4 palisade cells measuring less than one-third of their wild-type counterparts. These results indicate that reduced cell expansion contributed to reduced growth. Despite the smaller cells, however, the number of stomata in a given area was not altered (wild type, 6·0 ± 1·6 per 5·5 × 103 μm2 versus Atsuc2-4, 6·7 ± 1·0 per 5·5 × 103 μm2). In addition, Atsuc2-4 leaves were roughly half as thick as wild-type leaves, and had smaller air spaces among the spongy mesophyll cells (compare Fig. 3A and C with B and D).
Fig. 2.
Relative size of upper-epidermal and palisade cells in wild type (AtSUC2) and Atsuc2-4 plants. Cross-sectional area (μm2) of upper-epidermal and palisade cells as labelled. Measurements are averaged from cells within a single optical field obtained from four different wild-type and Atsuc2-4 plants. Wild-type epidermal cells, n = 14; wild-type palisade cells, n = 110; Atuc2-4 epidermal cells, n = 46; Atsuc2-4 palisade cells, n = 276; where n = total number of cells used to calculate averages and standard deviations.
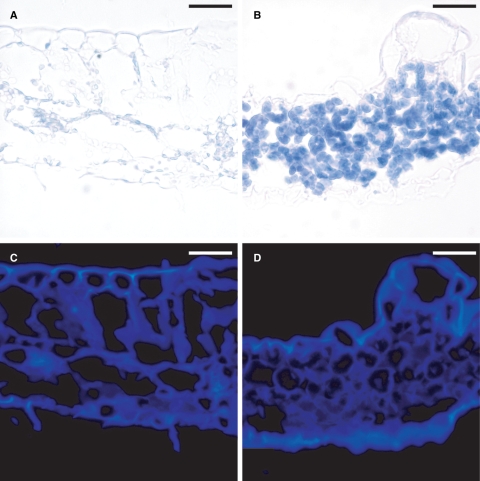
Fig. 3.
Visualization of starch and cellulose in wild-type and Atsuc2-4 plants. Leaf sections stained for starch with iodine from (A) wild-type and (B) Atsuc2-4 plants that were left in darkness for 24 h prior to fixation. (C) and D) Same sections as in (A) and (B), respectively, stained with calcofluor white for cellulose. Scale bars = 40 µm.
Soluble sugars and starch accumulate in Atsuc2-4 plants as a result of compromised transport out of leaves (Srivastava et al., 2008). The accumulation of starch in Atsuc2-4 lamina, relative to wild-type plants was clearly evident by iodine staining (Fig. 3A, B). Recently, Braun and colleagues demonstrated that leaf sectors in the TIE-DYED mutants of Zea mays (Zmtdy1 and Zmtdy2) accumulate sugar, starch and anthocyanin in a manner similar to Atsuc2 plants, most likely because of compromised photoassimilate transport (Braun et al., 2006; Baker and Braun, 2007, 2008). These sectors also develop thicker cell walls with greater deposition of cellulose, as visualized with calcofluor white staining and epifluorescence microscopy (Baker and Braun, 2008). Cellulose was therefore analysed in Atsuc2-4 plants by staining with calcofluor white, but increased levels of cellulose were not observed relative to wild-type plants (Fig. 3C, D).
In addition to being stunted, Atsuc2-4 plants are developmentally delayed (Fig. 4 and Table 1). Twenty-two days after germination, the vegetative rosettes of wild-type plants and KD1039 plants had equivalent numbers of leaves, whereas Atsuc2-4 plants had roughly half as many (Table 1). This developmental delay was also evident in the onset of reproductive growth. Under long-day, inductive conditions, wild-type and KD1039 plants had visible floral organs 30 d after germination, while Atsuc2-4 plants generally required 60 d or longer to form an inflorescence (Fig. 4A–C and Table 1). Variation in the number of days to the onset of flowering was substantial among Atsuc2-4 plants, and one-half had not formed floral buds or an inflorescence after 90 d when the experiment was terminated (Table 1). It is noteworthy, however, that the number of rosette leaves produced between germination and flowering was not significantly different between lines (Table 1), indicating that, although Atsuc2 plants are chronologically delayed, they flower at the same plastochron age (Erickson and Michelini, 1957). Those not flowering by 90 d had <15 leaves.
Fig. 4.
Rosette, inflorescence and siliques from Atsuc2-4 plants. (A) Rosettes of four Atsuc2-4 plants, relative to a single plant segregating either AtSUC2/AtSUC2 or AtSUC2/Atsuc2-4, 30 d after germination. (B) Rosette and nascent inflorescence of a typical Atsuc2-4 plant 60 d after germination. (C) Rosette and inflorescence of a 90-d-old Atsuc2-4 plant. In (A), (B) and (C), white arrows point to mature leaves (cotyledons in A) of Atsuc2-4 plants showing anthocyanin accumulation and necrosis at the distal tips and, in (C), prominent fungal infection. White arrowheads point to floral buds in the centre of the rosettes. Scale bar = 2 mm. (D) Several Atsuc2-4 plants in a single pot, 90 d after germination. Three plants have inflorescences with maturing siliques (white arrows) and one plant has a rosette without an inflorescence (white arrowhead); rosette leaves have visibly evident anthocyanin (white circle) and cauline leaves have less (black circle). Scale bar = 10 mm. (E) Atsuc2-4 plants have shorter siliques (top three) than wild type (bottom three), and do not accumulate anthocyanin. Scale bars = 2 mm.
Table 1.
Atsuc2-4 plants are delayed chronologically, but flower at the same developmental stage
| AtSUC2 | KD1039 | Atsuc2-4 | |
|---|---|---|---|
| Number of leaves 22 d after germination | 15·1 ± 1·7† | n.d.‡ | 7·8 ± 1·8†,* |
| Days from germination to flowering | 30·0 ± 2·8† | 28·5 ± 1·2† | 60·9 ± 13·7§,* |
| Number of leaves at flowering | 15·4 ± 1·4† | 15·9 ± 1·4† | 17·9 ± 3·0§ |
*Student's t-test, relative to wild-type plants (AtSUC2/AtSUC2), P < 0·05.
† n = 12 plants
‡ Not determined.
§ n = 8 flowering plants. An additional 8 plants were not flowering by 90 d after germination, had fewer than 15 leaves, and are not included in this analysis.
The inflorescence that did form on flowering Atsuc2-4 plants was thinner and shorter relative to wild type, which is not surprising considering the limited rosette area available to support inflorescence development (Fig. 4D). Siliques from fertilized flowers developed seeds but were shorter than wild-type siliques and much fewer in number (Fig. 4E and Table 2). Seed yields were also dramatically reduced, but seed size was not affected (Table 2). Germination rates varied substantially among seed batches from individual Atsuc2-4 plants, whereas they were consistently high among wild type plants (Table 2). Progeny of Atsuc2-4 plants grew with the same morphology and had similar fecundity as Atsuc2-4 plants segregating from heterozygous AtSUC2/Atsuc2-4 parents (not shown).
Table 2.
Atsuc2-4 plants produce viable seeds and seedlings to complete their life cycle
| Seed line* | Siliques with seeds | Length of siliques with seeds (mm)† | Seeds per plant‡ | Seed size (mm2)§ | Germination (%) |
|---|---|---|---|---|---|
| KO1 | 11 | 9·2 ± 2·1 | 211 | 0·086 ± 0·022 | 89 |
| KO2 | 13 | 10·3 ± 1·8 | 207 | 0·082 ± 0·019 | 92 |
| KO3 | 8 | 9·4 ± 2·3 | 192 | 0·078 ± 0·026 | 41 |
| KO4 | 11 | 8·8 ± 1·8 | 160 | 0·086 ± 0·023 | 29 |
| KO5 | 10 | 8·7 ± 2·0 | 153 | 0·088 ± 0·025 | 79 |
| KO6 | 10 | 9·3 ± 1·8 | 146 | 0·088 ± 0·022 | 6 |
| KO7 | 9 | 10·5 ± 1·4 | 124 | 0·060 ± 0·024 | 98 |
| KO8 | 4 | 9·0 ± 2·7 | 77 | 0·102 ± 0·027 | 86 |
| KO9 | 2 | 6·5 ± 0·7 | 22 | 0·090 ± 0·013 | 100 |
| KO average | 9·8 ± 3·7 | 9·4 ± 2·0 | 143 ± 62 | 0·084 ± 0·011 | 69 ± 34 |
| WT1 | 172 | 11·8 ± 1·4 | 7500 | 0·085 ± 0·016 | 99 |
| WT2 | 206 | 12·2 ± 1·2 | 9800 | 0·089 ± 0·011 | 97 |
| WT3 | 157 | 12·3 ± 1·4 | 9500 | 0·091 ± 0·010 | 96 |
| WT4 | 336 | 11·9 ± 1·4 | 8500 | 0·092 ± 0·010 | 99 |
| WT5 | 321 | 14·7 ± 1·1 | 7700 | 0·093 ± 0·010 | 98 |
| WT6 | 152 | 12·3 ± 1·4 | 10750 | 0·110 ± 0·010 | 100 |
| WT average | 224 ± 83 | 12·3 ± 1·7 | 8950 ± 1300 | 0·092 ± 0·013 | 98 ± 1 |
*An additional three Atsuc2-4 plants (KO10–KO12) flowered but did not produce seeds and are not included in this analysis. Note that plants used to generate this data are not the same as those in Table 1.
†For calculating variation in silique length, n = number of siliques with seeds for each Atsuc2-4 plant and n = 20 for each wild-type plant.
‡Seeds from Atsuc2-4 plants were counted individually; seeds from wild-type plants were calculated by mass assuming 50 seeds per milligram (Weigel and Glazebrook, 2002).
§For calculating variation in seed size, n = total number of seeds from each Atsuc2-4 plant and n > 50 seeds from each wild-type plant.
Anthocyanin accumulation in rosette leave was prominent, as described previously for other Atsuc2 mutants (Gottwald et al., 2000; Srivastava et al., 2008), but was less evident in cauline leaves and inflorescence stems (Fig. 4D). Levels of anthocyanin in siliques from mutant plants were not visibly different from wild-type siliques (Fig. 4E). Consistent with findings that antisense RNA repression of the AtSUC2 orthologues in Solanaceae species (SUT1) results in chlorosis and necrosis of leaves accumulating starch and soluble sugars, Atsuc2-4 plants showed necrosis in the distal tips of older leaves (Fig. 4A–C). These were also more susceptible to infection by a naturally occurring fungus (most likely a Botrytis species) and developed readily observed hyphae and conidia on older leaves while adjacent and similarly aged wild-type plants did not (Fig. 4C).
DISCUSSION
Earlier work described three alleles of AtSUC2 with T-DNA insertions that blocked formation of a functional protein (Atsuc2-1, -2 and -3), and each conferred identical phenotypes of severely stunted growth, accumulation of sugar and starch in mature leaves from an inability to efficiently transport photoassimilate, and the failure to produce viable seed (Gottwald et al., 2000). In previous work (Srivastava et al., 2008, 2009) and the work presented here, a fourth allele with a T-DNA insert, Atsuc2-4 (SALK_038124), was analysed for growth habit, carbohydrate transport and accumulation, transcript truncation and abundance, and protein activity, with all evidence supporting the assertion that it is a null allele. However, the T-DNA insert of Atsuc2-4 is in the second intron, as are the inserts of Atsuc2-1 and Atsuc2-3, and, in the absence of technology for homologous recombination in plants, the remote possibility that correct splicing may occur at a particular stage of development cannot be formally excluded.
Viable seeds were obtained from Atsuc2-4 plants, demonstrating that AtSUC2 is not essential for arabidopsis to complete its life cycle in the laboratory, but obviously does confer an essential survival advantage: mutant plants were extremely debilitated and required additional care relative to wild type; not all plants produced viable seed, and among those that did, yield was profoundly reduced. It is unlikely that these mutants would produce seed in a natural environment. Since all Atsuc2 T-DNA insertion mutants identified to date harbour apparent null alleles, it is not clear why Atsuc2-4 is able to complete its life cycle while Atsuc2-1, -2 and -3 were not. The SALK lines are in the Columbia Col-0 background (Alonso et al., 2003) while the others are in Ws-2 (Gottwald et al., 2000), but it is unlikely that the genetic background accounts for the observed differences. Different growth conditions are more likely to explain the dissimilar results: Gottwald and colleagues grew their plants with 24 h illumination (Gottwald et al., 2000) while, in the present study, 14 h light and 10 h dark cycles were generally used. Furthermore, Atsuc2-4 rosettes tended to be larger and have less anthocyanin when grown under shorter days and lower light, compared with when grown under longer days with more intense light (not shown). Plants grown under the latter conditions were more difficult to maintain and did not produce viable seed as routinely. Lower irradiance and longer dark periods may be more germane to Atsuc2 null mutants completing their life cycle because stress associated with excessive starch accumulation is minimized (Burkle et al., 1998).
The fact that Atsuc2-4 mutants produce viable seed supports previous hypotheses on the role of AtSUC2 and other members of Suc/H+ symporter gene family in controlling carbon partitioning in different organs during the life cycle of arabidopsis. The microgametophyte, megagametophyte and developing embryo are entirely dependent on the parent sporophyte, and each is symplastically isolated from maternal tissues such that nutrients must be mobilized across membranes (Lalonde et al., 2003; Zhang et al., 2007). Detailed analysis has shown that AtSUC2 expression is limited to the collection phloem of minor veins in mature leaves and the transport phloem of larger vascular bundles, and SUT1 orthologues in Solanaceae species also express in xylem parenchyma cells (Truernit and Sauer, 1995; Stadler et al., 2005a, b; Martens et al., 2006; Schmitt et al., 2008). AtSUC2 and orthologues have not been implicated in supplying nutrients to gametophytes or embryos, and the fact that viable seeds are formed in Atsuc2-4 plants supports this. AtSUC1 is the principal Suc transporter contributing to pollen-tube germination and growth (Stadler et al., 1999; Sivitz et al., 2008) whereas AtSUC5 is implicated in seed filling (Baud et al., 2005).
Considering the importance of AtSUC2 to phloem loading, and the importance of phloem loading to nutrient transport, it is perhaps surprising that Atsuc2 mutants show any growth. Gottwold et al. (2000) noted this and discussed possibilities for how plants may continue to mobilize photoassimilate to sink tissues in the absence of AtSUC2 activity, including overlapping function of other Suc transporters or compensatory activity of hexose transporters. Arabidopsis has nine Suc/H+ symporters, seven of which are functional (Sauer, 2007), and the monosaccharide transporter superfamily has 66 putative members (Lalonde et al., 2004). However, none of the other Suc/H+ symporters appear to have characteristics that could substitute for AtSUC2 (Sivitz et al., 2007, 2008; Sauer, 2007). SUT4 orthologues express in leaf veins, but may have their major role in the release phloem (Chincinska et al., 2008). In addition, although the SUT4 protein from potato was localized to the plasma membrane (Chincinska et al., 2008), orthologous proteins from arabidopsis, barley and Lotus japonicus localized to the tonoplast, which is a location incompatible with a direct role in Suc/H+ symport from the apoplast (Endler et al., 2006; Reinders et al., 2008). Other sugars such as RFOs (Turgeon and Gowan, 1990), hexoses (van Bel and Hess, 2008) and sugar alcohols (Noiraud et al., 2001) all function as transport sugars in various species, and in arabidopsis, specifically, raffinose is found in phloem sap (Haritatos et al., 2000) and a sugar alcohol transporter, AtPLT5, expresses in phloem cells (Klepek et al., 2005). The contribution of these sugars to phloem transport may be elevated in Atsuc2 plants and, in support of this, Atsuc2-4 plants show enhanced uptake of [14C]sorbitol relative to wild type (Srivastava et al., 2009). In addition, the hydrostatic pressure gradient required for phloem transport in Atsuc2 plants may rely more on other solutes, such as K+. K+ channels localize to the phloem (Deeken et al., 2000; Lacombe et al., 2000) and earlier work showed increases in phloem K+ levels associated with decreased sucrose availability (Smith and Milburn, 1980).
Another possibility is that phloem loading – literally the expenditure of energy to load solute against a thermodynamic gradient – is beneficial but not essential for transport in arabidopsis. Turgeon and colleagues showed that willow does not phloem load, but instead has its highest solute concentration in mesophyll cells, and that this correlates with a relatively high number of plasmodesmata connecting mesophyll and minor veins to permit diffusion down a concentration gradient to the phloem for long-distance transport (Turgeon and Medville, 1998). A similar model is proposed for apple (Malus domestica) which transports high levels of sorbitol (Reidel et al., 2009) and a survey of 45 diverse dicotyledonous species suggests that ‘passive’ phloem loading may be more widespread than previously appreciated (Rennie and Turgeon, 2009). A common feature of passive loading is high concentrations of transport compounds in leaves, presumably to generate the hydrostatic pressure necessary to drive phloem transport to sinks. Atsuc2-4 leaves accumulate approx. 18-fold higher levels of Suc in mature leaves than wild type (Srivastava et al., 2008), which would facilitate passive loading and long-distance transport without an energized-concentrating step. The companion cells of arabidopsis minor veins have more plasmodesmatal connections with mesophyll cells than tobacco, which is an archetype for loading sucrose from the apoplast, but fewer than the abundant connections of true passive loaders like willow or apple (Turgeon and Medville, 1998; Haritatos et al., 2000; Reidel et al., 2009). Arabidopsis leaf-vein morphology is therefore compatible with – but not optimized for –source-to-sink transport driven by hydrostatic pressure gradients initiating in mesophyll cells.
It was also observed that cauline leaves accumulated less anthocyanin than rosette leaves, and that inflorescence stems and siliques rarely had visible anthocyanin. These tissues can accumulate anthocyanin as is commonly observed when, for example, the inflorescence grows closer to the growth-chamber lights. Anthocyanin accumulation is a common response to high light and elevated sucrose levels (Solfanelli et al., 2006; Cominelli et al., 2008), and the role of sugar signalling in anthocyanin accumulation is mediated in part by Suc/H+ symporters (Sivitz et al., 2008). The lower levels of anthocyanin in inflorescence stems and siliques of Atsuc2-4 plants, relative to rosette leaves, may be because photoassimilated Suc produced in chlorenchyma cells of these tissues is more efficiently transported to local heterotrophic tissues (e.g. expanding stems, seeds) by AtSUC2-independent transport mechanisms. For example, photoassimilate and phloem sap from source tissues in close proximity to strong sinks could, in principle, flow more readily toward sinks because the smaller pressures generated in the mesophyll cells may be sufficient to overcome the resistance of the sieve-plate pores and sieve elements over short distances (Thompson, 2006).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Elison B. Blancaflor, Samuel Roberts Noble Foundation, Ardmore, OK, USA for confocal laser scanning microscopy equipment and expertise (NSF DBI 0400580), and two anonymous reviewers who helped improve the manuscript. This work was supported by the National Science Foundation (NSF IOS 0344088 to B.G.A.).
LITERATURE CITED
- Alonso JM, Stepanova AN, Leisse TJ, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Baker RF, Braun DM. tie-dyed1 functions non-cell autonomously to control carbohydrate accumulation in maize leaves. Plant Physiology. 2007;144:867–878. doi: 10.1104/pp.107.098814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker RF, Braun DM. tie-dyed2 functions with tie-dyed1 to promote carbohydrate export from maize leaves. Plant Physiology. 2008;146:1085–1097. doi: 10.1104/pp.107.111476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S, Wuilleme S, Lemoine R, et al. The AtSUC5 sucrose transporter specifically expressed in the endosperm is involved in early seed development in Arabidopsis. The Plant Journal. 2005;43:824–836. doi: 10.1111/j.1365-313X.2005.02496.x. [DOI] [PubMed] [Google Scholar]
- van Bel AJE, Hess PH. Hexoses as phloem transport sugars: the end of a dogma? Journal of Experimental Botany. 2008;59:261–272. doi: 10.1093/jxb/erm294. [DOI] [PubMed] [Google Scholar]
- Braun DM, Slewinski TL. Genetic control of carbon partitioning in grasses: roles of sucrose transporters and tie-dyed loci in phloem loading. Plant Physiology. 2009;149:71–81. doi: 10.1104/pp.108.129049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun DM, Ma Y, Inada N, Muszynski MG, Baker RF. tie-dyed1 regulates carbohydrate accumulation in maize leaves. Plant Physiology. 2006;142:1511–1522. doi: 10.1104/pp.106.090381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkle L, Hibberd JM, Quick WP, Kuhn C, Hirner B, Frommer WB. The H+-sucrose cotransporter NtSUT1 is essential for sugar export from tobacco leaves. Plant Physiology. 1998;118:59–68. doi: 10.1104/pp.118.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush DR. Sugar transporters in plant biology. Current Opinion in Plant Biology. 1999;2:187–191. doi: 10.1016/S1369-5266(99)80034-X. [DOI] [PubMed] [Google Scholar]
- Chincinska IA, Liesche J, Krugel U, et al. Sucrose transporter StSUT4 from potato affects flowering, tuberization, and shade avoidance response. Plant Physiology. 2008;146:515–528. doi: 10.1104/pp.107.112334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cominelli E, Gusmaroli G, Allegra D, et al. Expression analysis of anthocyanin regulatory genes in response to different light qualities in Arabidopsis thaliana. Journal of Plant Physiology. 2008;165:886–894. doi: 10.1016/j.jplph.2007.06.010. [DOI] [PubMed] [Google Scholar]
- Deeken R, Sanders C, Ache P, Hedrich R. Developmental and light-dependent regulation of a phloem-localised K+ channel of Arabidopsis thaliana. The Plant Journal. 2000;23:285–290. doi: 10.1046/j.1365-313x.2000.00791.x. [DOI] [PubMed] [Google Scholar]
- Endler A, Meyer S, Schelbert S, et al. Identification of a vacuolar sucrose transporter in barley and Arabidopsis mesophyll cells by a tonoplast proteomic approach. Plant Physiology. 2006;141:196–207. doi: 10.1104/pp.106.079533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson RO, Michelini FJ. The plastochron index. American Journal of Botany. 1957;44:297–305. [Google Scholar]
- Giaquinta RT. Phloem loading of sucrose: pH dependence and selectivity. Plant Physiology. 1977;59:750–753. doi: 10.1104/pp.59.4.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottwald JR, Krysan PJ, Young JC, Evert RF, Sussman MR. Genetic evidence for the in planta role of phloem-specific plasma membrane sucrose transporters. Proceedings of the National Academy of Science of the USA. 2000;97:13979–13984. doi: 10.1073/pnas.250473797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafke JB, van Amerongen J-K, Kelling F, Furch ACU, Gaupels F, van Bel AJE. Thermodynamic battle for photosynthate acquisition between sieve tubes and adjoining parenchyma in transport phloem. Plant Physiology. 2005;138:1527–1537. doi: 10.1104/pp.104.058511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haritatos E, Medville R, Turgeon R. Minor vein structure and sugar transport in Arabidopsis thaliana. Planta. 2000;211:105–111. doi: 10.1007/s004250000268. [DOI] [PubMed] [Google Scholar]
- Keller F, Pharr DM. Metabolism of carbohydrates in sinks and sources: galactosyl-sucrose oligosaccharides. In: Zamski E, Schaffer AA, editors. Photoassimilate distribution in plants and crops: source-sink relationships. New York, NY: Marcel Dekker; 1996. [Google Scholar]
- Klepek Y-S, Geiger D, Stadler R, et al. Arabidopsis POLYOL TRANSPORTER5, a new member of the monosaccharide transporter-like superfamily, mediates H+-symport of numerous substrates, including myo-inositol, glycerol, and ribosele. The Plant Cell. 2005;17:204–218. doi: 10.1105/tpc.104.026641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn C, Quick WP, Schulz A, Riesmeier JW, Sonnewald U, Frommer WB. Companion cell-specific inhibition of the potato sucrose transporter SUT1. Plant, Cell & Environment. 1996;19:1115–1123. [Google Scholar]
- Lacombe B, Pilot G, Michard E, Gaymard F, Sentenac H, Thibaud J-B. A shaker-like K+ channel with weak rectification is expressed in both source and sink phloem tissues of Arabidopsis. The Plant Cell. 2000;12:837–851. doi: 10.1105/tpc.12.6.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalonde S, Tegeder M, Throne-Holst M, Frommer WB, Patrick JW. Phloem loading and unloading of sugars and amino acids. Plant, Cell & Environment. 2003;26:37–56. [Google Scholar]
- Lalonde S, Wipf D, Frommer WB. Transport mechanisms for organic forms of carbon and nitrogen between source and sink. Annual Review of Plant Biology. 2004;55:341–372. doi: 10.1146/annurev.arplant.55.031903.141758. [DOI] [PubMed] [Google Scholar]
- Martens HJ, Roberts AG, Oparka KJ, Schulz A. Quantification of plasmodesmatal endoplasmic reticulum coupling between sieve elements and companion cells using fluorescence redistribution after photobleaching. Plant Physiology. 2006;142:471–480. doi: 10.1104/pp.106.085803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noiraud N, Maurousset L, Lemoine R. Identification of a mannitol transporter, AgMaT1, in celery phloem. The Plant Cell. 2001;13:695–705. doi: 10.1105/tpc.13.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasband WS. ImageJ. Image processing and analysis in Java. 2007. http://rsb.info.nih.gov/ij/
- Reidel EJ, Rennie EA, Amiard V, Cheng L, Turgeon R. Phloem loading strategies in three plant species that transport sugar alcohols. Plant Physiology. 2009;149:1601–1608. doi: 10.1104/pp.108.134791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinders A, Sivitz AB, Starker CG, Gantt JS, Ward JM. Functional analysis of LjSUT4, a vacuolar sucrose transporter from Lotus japonicus. Plant Molecular Biology. 2008;68:289–299. doi: 10.1007/s11103-008-9370-0. [DOI] [PubMed] [Google Scholar]
- Rennie E, Turgeon R. A comprehensive picture of phloem loading strategies; Proceedings of the National Academy of Science of the USA; 2009. pp. 14162–14167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer N. Molecular physiology of higher plant sucrose transporters. FEBS Letters. 2007;581:2309–2317. doi: 10.1016/j.febslet.2007.03.048. [DOI] [PubMed] [Google Scholar]
- Schmitt B, Stadler R, Sauer N. Immunolocalization of Solanaceous SUT1 proteins in companion cells and xylem parenchyma: new perspectives for phloem loading and transport. Plant Physiology. 2008;148:187–199. doi: 10.1104/pp.108.120410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz A, Kuhn C, Riesmeier JW, Frommer WR. Ultrastructural effects in potato leaves due to antisense-inhibition of the sucrose transporter indicate an apoplasmic mode of phloem loading. Planta. 1998;206:533–543. [Google Scholar]
- Sivitz AB, Reinders A, Johnson ME, et al. Arabidopsis sucrose transporter AtSUC9: high-affinity transport activity, intragenic control of expression, and early flowering mutant phenotype. Plant Physiology. 2007;143:188–198. doi: 10.1104/pp.106.089003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivitz AB, Reinders A, Ward JM. Arabidopsis sucrose transporter AtSUC1 is important for pollen germination and sucrose-induced anthocyanin accumulation. Plant Physiology. 2008;147:92–100. doi: 10.1104/pp.108.118992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JAC, Milburn JA. Osmoregulation and the control of phloem-sap composition in Ricinus communis L. Planta. 1980;148:28–34. doi: 10.1007/BF00385438. [DOI] [PubMed] [Google Scholar]
- Solfanelli C, Poggi A, Loreti E, Alpi A, Perata P. Sucrose-specific induction of the anthocyanin biosynthetic pathway in Arabidopsis. Plant Physiology. 2006;140:637–646. doi: 10.1104/pp.105.072579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava AC, Ganesan S, Ismail IO, Ayre BG. Functional characterization of the Arabidopsis AtSUC2 sucrose/H+ symporter by tissue-specific complementation reveals an essential role in phloem loading but not in long-distance transport. Plant Physiology. 2008;148:200–211. doi: 10.1104/pp.108.124776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava AC, Ganesan S, Ismail IO, Ayre BG. Effective carbon partitioning driven by exotic phloem-specific regulatory elements fused to the Arabidopsis thaliana AtSUC2 sucrose-proton symporter gene. BMC Plant Biology. 2009;9:7. doi: 10.1186/1471-2229-9-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler R, Truernit E, Gahrtz M, Sauer N. The AtSUC1 sucrose carrier may represent the osmotic driving force for anther dehiscence and pollen tube growth in Arabidopsis. The Plant Journal. 1999;19:269–278. doi: 10.1046/j.1365-313x.1999.00527.x. [DOI] [PubMed] [Google Scholar]
- Stadler R, Lauterbach C, Sauer N. Cell-to-cell movement of green fluorescent protein reveals post-phloem transport in the outer integument and identifies symplastic domains in Arabidopsis seeds and embryos. Plant Physiology. 2005;a 139:701–712. doi: 10.1104/pp.105.065607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler R, Wright KM, Lauterbach C, et al. Expression of GFP-fusions in Arabidopsis companion cells reveals non-specific protein trafficking into sieve elements and identifies a novel post-phloem domain in roots. The Plant Journal. 2005;b 41:319–331. doi: 10.1111/j.1365-313X.2004.02298.x. [DOI] [PubMed] [Google Scholar]
- Thompson MV. Phloem: the long and the short of it. Trends in Plant Science. 2006;11:26–32. doi: 10.1016/j.tplants.2005.11.009. [DOI] [PubMed] [Google Scholar]
- Truernit E, Sauer N. The promoter of the Arabidopsis thaliana SUC2 sucrose-H+ symporter gene directs expression of β-glucuronidase to the phloem: evidence for phloem loading and unloading by SUC2. Planta. 1995;196:564–570. doi: 10.1007/BF00203657. [DOI] [PubMed] [Google Scholar]
- Turgeon R. Phloem loading and plasmodesmata. Trends in Plant Science. 1996;1:418–423. [Google Scholar]
- Turgeon R, Ayre BG. Pathways and mechanisms of phloem loading. In: Holbrook NM, Zwieniecki MA, editors. Vascular transport in plants. New York, NY: Elsevier Academic Press; 2005. [Google Scholar]
- Turgeon R, Gowan E. Phloem loading in Coleus blumei in the absence of carrier-mediated uptake of export sugar from the apoplast. Plant Physiology. 1990;94:1244–1249. doi: 10.1104/pp.94.3.1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turgeon R, Medville R. The absence of phloem loading in willow leaves. Proceedings of the National Academy of Science of the USA. 1998;95:12055–12060. doi: 10.1073/pnas.95.20.12055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J. Arabidopsis: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2002. [Google Scholar]
- Zhang W-H, Zhou Y, Dibley KE, Tyerman SD, Furbank RT, Patrick JW. Nutrient loading of developing seeds. Functional Plant Biology. 2007;34:314–331. doi: 10.1071/FP06271. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.