Abstract
Imprinting, non-coding RNA and chromatin organization are modes of epigenetic regulation that modulate gene expression and are necessary for mammalian neurodevelopment. The only two known mammalian clusters of genes encoding small nucleolar RNAs (snoRNAs), SNRPN through UBE3A(15q11–q13/7qC) and GTL2(14q32.2/12qF1), are neuronally expressed, localized to imprinted loci and involved in at least five neurodevelopmental disorders. Deficiency of the paternal 15q11–q13 snoRNA HBII-85 locus is necessary to cause the neurodevelopmental disorder Prader–Willi syndrome (PWS). Here we show epigenetically regulated chromatin decondensation at snoRNA clusters in human and mouse brain. An 8-fold allele-specific decondensation of snoRNA chromatin was developmentally regulated specifically in maturing neurons, correlating with HBII-85 nucleolar accumulation and increased nucleolar size. Reciprocal mouse models revealed a genetic and epigenetic requirement of the 35 kb imprinting center (IC) at the Snrpn–Ube3a locus for transcriptionally regulated chromatin decondensation. PWS human brain and IC deletion mouse Purkinje neurons showed significantly decreased nucleolar size, demonstrating the essential role of the 15q11–q13 HBII-85 locus in neuronal nucleolar maturation. These results are relevant to understanding the molecular pathogenesis of multiple human neurodevelopmental disorders, including PWS and some causes of autism.
INTRODUCTION
Epigenetics encompasses multiple mechanisms that temporally and spatially control gene expression throughout development without alteration to the DNA sequence. Non-coding RNAs (ncRNAs), parental imprinting and chromatin organization are among epigenetic mechanisms that contribute to the complexity of gene regulation critical for proper mammalian neurodevelopment. ncRNAs modulate transcription and translation, are found abundantly and sometimes specifically in brain (1) and play pivotal roles in neurodevelopment and long-term memory formation (2,3); 30% of parentally imprinted transcripts are ncRNA (4). Chromatin organization contributes to the spatial context in which genes are developmentally regulated (5), particularly in the highly ordered chromatin of post-mitotic neuronal nuclei (6).
Chromosomal loci 15q11–q13 (murine 7qC) and 14q32.2 (murine 12qF1) are imprinted regions directly involved in at least five neurodevelopmental disorders. Prader–Willi syndrome (PWS) is caused by paternal 15q11–q13 deficiency, whereas Angelman syndrome (AS) is caused by 15q11–q13 maternal deficiency (7,8); and 1–3% of autism cases are caused by 15q11–q13 duplication (9–11). Paternal and maternal uniparental disomy (UPD) for chromosome 14 causes two distinct neurodevelopmental disorders, the latter of which phenotypically resembles PWS (12,13). Both loci contain developmentally regulated neuron-specific transcripts regulated by an imprinting center (IC) (14–16).
Paternal 15q11–q13/7qC expresses a neuron-specific polycistronic transcript containing at least 148 exons over several hundred kilobases that includes the coding sequence for a splicesomal protein (SNRPN), two clusters of small nucleolar RNAs (snoRNAs) (HB/MBII-85,52), four single copy snoRNAs (HBII-436,13, 438a and 438b) and the antisense transcript to a maternally expressed ubiquitin ligase (UBE3A-AS) (17–20) diagrammed in Figure 1D. Maternal 15q11–q13/7qC is comparatively inactive, but expresses UBE3A and ATP10A in the opposite orientation (21–24). 15q11–q13 transcriptional regulation is highly complex, involving multiple allele-specific epigenetic marks, including DNA methylation, histone modification patterns and DNase hypersensitive sites (25–27). Maternal 14q32.2/12qF1 expresses two clusters of snoRNAs (14qI and 14qII) and over forty microRNAs (miRNAs) (14,16). Interestingly, the 15q and 14q tandemly repeated C/D box snoRNA gene clusters are subject to parental imprinting and are unique to eutherian mammals (28).
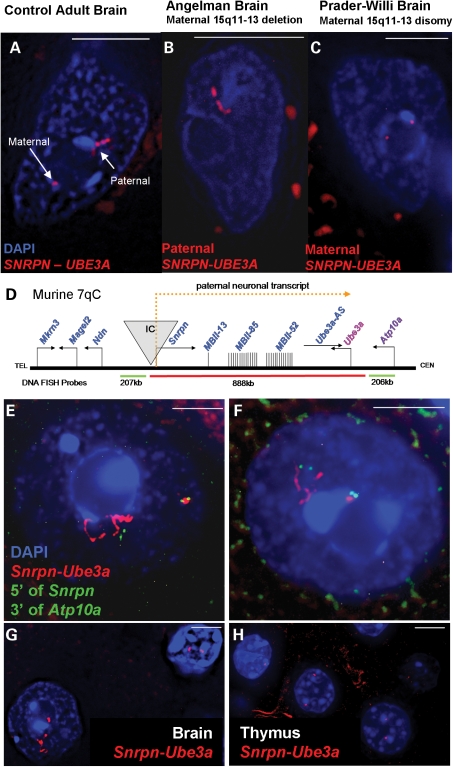
Figure 1.
DNA FISH using a probe contig spanning SNRPN–UBE3A (red fluorescence) on post-mortem human and mouse brain samples reveals two distinct parental chromatin structures in neuronal nuclei, counterstained with DAPI (blue fluorescence). White bar represents 5 µm. (A) Representative control adult neuronal nucleus containing a small maternal SNRPN–UBE3A signal and an extended paternal SNRPN–UBE3A signal. (B) Representative Angelman (maternal 15q11–q13 deletion) neuronal nucleus containing a single extended paternal SNRPN–UBE3A signal. (C) Representative Prader–Willi (maternal disomy) neuronal nucleus with two small maternal SNRPN–UBE3A signals. (D) Neuron-specificity and locus boundaries of chromatin decondensation for murine 7qC (syntenic to human 15q11–q13). A diagram of the Snrpn–Ube3a locus showing paternally expressed genes in blue, maternally expressed genes in red, components of the paternal neuronal transcript in yellow and the imprinting center (IC) in grey. DNA FISH probes are diagramed below and colored to match probe fluorescence. Not drawn to scale. (E and F) Representative images of the Snrpn–Ube3a signals seen in mouse adult neuronal nuclei throughout the brain. Red fluorescence shows a decondensed Snrpn–Ube3a paternal signal and a small compact Snrpn–Ube3a maternal signal corresponding to the transcriptional activity of the alleles. The small green signals are the combined flanking probes 5′ of Snrpn and 3′ of Atp10a. While signal length was variable between individual nuclei, paternal signals averaged ∼4 µm and maternal signals averaged ∼0.5 µm in Purkinje, hindbrain and cortical neurons in adult murine brain (Supplementary Material, Fig. S3). Distances between the flanking probes on each Snrpn–Ube3a allele were utilized to determine the percentage of paternal signals which loop back on to the chromosomal backbone by using the average distance between the flanking probes on the maternal allele plus one standard deviation (1.13 µm + 0.58 = 1.71 µm) as a threshold for looping for the paternal allele. Over 43% of the paternal Snrpn–Ube3a alleles looped back to the IC by this definition as shown by these representative nuclei. (G) Paternally extended Snprn–Ube3a signals are neuron-specific as can be seen in this representative image in which the adult neuronal nucleus shows the decondensed allele (bottom left), whereas only two small signals are seen in the glial nucleus (upper right). (H) No decondensed signals were seen in any other adult tissue (thymus, kidney, spleen, liver), thymus nuclei shown.
SnoRNAs are ncRNAs localized to the nucleolus and generally involved in guiding rRNA modifications (29). The tandemly repeated C/D Box snoRNA clusters within 15q11–q13 and 14q32.2 have greater homology to one another than to any other mammalian snoRNAs and have minimal rRNA homology; however, three snoRNAs from the 15q11–q13 locus have recently been shown to have rRNA homology (30,31). HBII-52 has been shown to regulate splicing of a serotonin receptor gene (32), whereas HBII-85 deficiency appears to be the primary cause of PWS (33,42). Mouse models of MBII-85 deficiency also recapitulate some features of PWS, including hyperphagia and post-natal growth retardation (34,35). The mechanism by which HBII-85 deficiency causes neurodevelopmental abnormalities in PWS and PWS-like abnormalities in mouse models is currently unknown.
Here we provide evidence in both human and mouse brain that post-natal neurons undergo orchestrated chromatin decondensation specifically at two imprinted snoRNA clusters but not other highly transcribed, imprinted or ncRNA containing loci. Chromatin decondensation of imprinted snoRNAs was neuron-specific, developmentally regulated and transcriptionally controlled through the IC. Importantly, deficiency of HB/MBII-85,52 snoRNAs significantly impacted nucleolar size, suggesting that one essential function of the neuronally transcribed snoRNAs is to modify rRNA, thereby increasing nucleolar size during neuronal maturation.
RESULTS
DNA FISH analysis of human and mouse brain sections reveals paternal-specific and neuron-specific chromatin decondensation at SNRPN through UBE3A
To visualize the organization of an imprinted snoRNA cluster in brain, a ∼660 kb DNA FISH probe corresponding to the large paternal neuron-specific transcript from SNRPN through UBE3A within 15q11–q13 was hybridized to human brain, revealing dramatically different signals within neuronal nuclei. Individual neuronal nuclei showed one small compact signal and another strikingly extended (Fig. 1A), indicating differential chromatin organization for the two parental alleles. PWS and AS brain samples revealed that the extended signal was of paternal origin, appearing consistently in AS neurons but never in PWS neurons (Fig. 1B and C).
To further investigate the allele-specific chromatin differences in an experimentally tractable system, a series of murine 7qC DNA FISH probes (Fig. 1D, Supplementary Material, Table S1) were hybridized to adult mouse sagittal brain sections. Probes from Snrpn through Ube3a (∼888 kb) revealed allele-specific extended signals throughout the brain (Fig. 1E and H, red fluorescence). Extended paternal signals were specific to neurons, as glia always exhibited two compact, relatively equivalent signals (Fig. 1G), a FISH pattern that was consistently observed in nuclei within adult liver, thymus, spleen and kidney (Fig. 1H and data not shown). The possibility of a brain specific copy number change was ruled out by qPCR analyses of brain versus liver genomic DNA (Supplementary Material, Fig. S1). Furthermore, extensive RNase A treatment did not change the hybridization signals (Supplementary Material, Fig. S2A), indicating that the extended signals were due to DNA:DNA rather than DNA:RNA hybridization. Tissue digestion with DNase I resulted in degradation of the extended paternal signal more rapidly than the compact maternal signal, indicating that the differential signals seen were due to DNA decondensation and decreased nucleosome coverage (Supplementary Material, Fig. S2B and C).
Over 90% of post-natal day 70 (P70) cortical neuronal nuclei presented an extended paternal Snrpn–Ube3a signal. The paternal Snrpn–Ube3a signal averaged 4.08 µm in length, approximately eight times greater in length than their compact maternal signal counterpart which averaged 0.49 µm. These lengths correspond to a ∼65:1 packing ratio for the paternal allele, only ∼1.6× more compact than a 30 nm fiber (∼40:1 packing ratio) and a ∼540:1 packing ratio for the maternal allele, ∼13.5× more compact than a 30 nm fiber. The largest paternal signals were found in large cortical, Purkinje and hindbrain neuronal nuclei in which paternal signals averaged ∼4 µm in length (Supplementary Material, Fig. S3); the largest signal being 8.94 µm in length corresponding to a ∼30:1 packing ratio. For comparison, a ∼202 kb probe for a gene biallelically expressed and highly transcribed in brain (Stmn4) averaged ∼0.54 µm in adult Purkinje neurons giving a ∼112:1 packing ratio, ∼2.8× more compact than a 30 nm fiber.
No extended signals were seen for probes flanking the Snrpn–Ube3a locus, 5′ of Snrpn and at or 3′ of Atp10a (Fig. 1E and F, green fluorescence), suggesting that this region's highly differential chromatin organization exclusively encompasses the neuron specific poly-cistronic transcript produced only from the paternally inherited allele (17). Paternal Snrpn–Ube3a signals exhibited varied conformations, but 43% of neuronal nuclei showed evidence of paternal allele looping, determined by the distance between flanking probes (Fig. 1E and F, green signals).
Chromatin decondensation is developmentally regulated and correlates to nucleolar enlargement during neuronal maturation
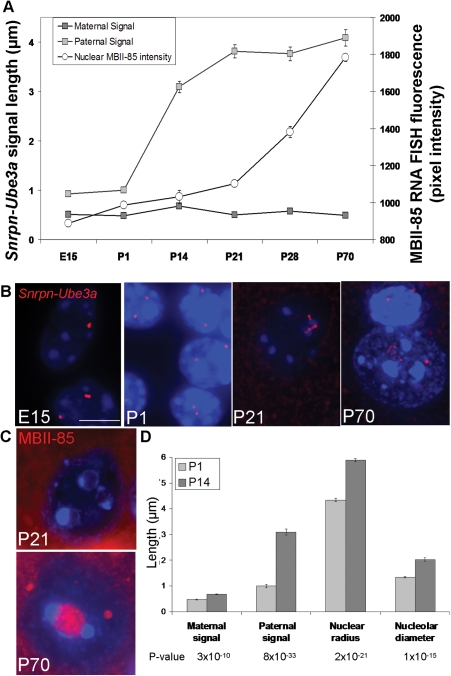
To determine the relationship between Snrpn–Ube3a chromatin organization, paternal snoRNA transcription and nucleolar changes during neuronal maturation, a series of wild-type (wt) murine cortex samples E15, P1, P14, P21, P28 and P70, were separately hybridized with the Snrpn–Ube3a DNA FISH probe, hybridized with a RNA FISH probe for MBII-85, or immunofluorescently stained for nucleolin. Snrpn–Ube3a signals revealed dynamic changes in the decondensation of the paternal allele from birth through adulthood, with the most dramatic changes occurring within the first 2 weeks of life (Fig. 2A and B). Chromatin decondensation of Snrpn–Ube3a preceded increased MBII-85 nuclear accumulation concentrated within neuronal nucleoli (Fig. 2C) and correlated with increased nucleolar and nuclear area, a measure of neuronal maturation (Fig. 2D and Supplementary Material, Fig. S4). The decondensed Snrpn–Ube3a allele did not colocalize with the nucleolus or the major deposition of MBII-85 signal, suggesting that the snoRNA transcripts were transported to the nucleolus following transcription and processing.
Figure 2.
Decondensation of the paternal Snrpn–Ube3a allele is developmentally regulated and correlates to nucleolar enlargement during neuronal maturation. (A) Snrpn–Ube3a DNA FISH probe was hybridized to samples of cerebral cortex of mice aged embryonic day 15 (E15) through post-natal day 70 (P70). Decondensation of the paternal Snrpn–Ube3a allele increases after birth (P1) and precedes increased MBII-85 accumulation (quantified from MBII-85 RNA FISH fluorescence from neuronal nuclei), whereas the maternal allele remains relatively compact throughout development. By P70 the paternal allele is only slightly more compact than a 30 nm fiber and 8-fold longer than the maternal allele (Supplementary Material, Fig. S7). Results represent the mean ± SEM for 100 nuclei per time point. (B) Representative images of Snrpn–Ube3a DNA FISH signals in neuronal nuclei through development: E15, P1, P21 and P70. P70 includes a neuronal nucleus (below) and a glial nucleus (above). (C) Representative images of increased MBII-85 RNA FISH in neuronal nuclei from a P21 nucleus with three small nucleoli and a P70 nucleus with a single large nucleolus in which higher levels of MBII-85 accumulate. (D) Decondensation of the paternal Snrpn–Ube3a allele at P14 corresponds with increases in nuclear (nuclear radius) and nucleolar size (nucleolar diameter), which continue to increase through development (Supplementary Material, Fig. S4). Results represent the mean ± SEM for 100 nuclei/time point. Student's t-test, two tailed. P-values listed below.
A similar developmental change was seen for a smaller Snrpn-MBII-85 (∼140 kb) probe (Supplementary Material, Fig. S5A). Measurements from the Snrpn-MBII-85 probe to the boundaries of heterochromatic foci (determined by dense DAPI fluorescence) revealed that the maternal allele remained substantially closer to heterochromatin (within ∼0.5 µm) throughout development, while the paternal signal remained at least twice that distance away from heterochromatin, in accordance with the parental alleles' transcriptional activity (Supplementary Material, Fig. S5B).
Transcription from the IC is required for paternal Snrpn–Ube3a decondensation
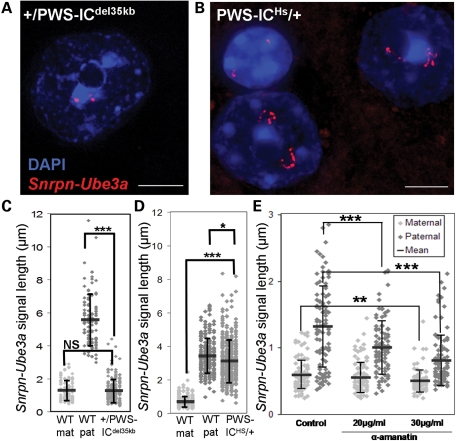
Noting the strong correlation between neuronal maturation, nucleolar snoRNA accumulation and paternal Snrpn–Ube3a allele decondensation, we sought to further understand the link between transcription and chromatin decondensation for Snrpn–Ube3a through genetic mutation. Snrpn–Ube3a DNA FISH to the transcriptionally silent paternally inherited PWS-IC deletion mouse (+/PWS-ICdel35 kb) (36) revealed an absence of decondensed alleles compared with wt littermate neuronal nuclei (Fig. 3A and C), similar to PWS UPD (Fig. 1C), confirming the parental origin of the decondensed allele in murine neurons. In a reverse transgenic mouse line, in which the maternal mouse PWS-IC has been replaced by the human PWS-IC (PWS-ICHs/+) (37), the maternal imprint is lost and therefore the normally silent maternal allele expresses Snrpn, snoRNAs and Ube3a-AS. Snrpn–Ube3a DNA FISH in PWS-ICHs/+ brain samples revealed only decondensed alleles within neuronal nuclei (Fig. 3B and D). These results indicate that an IC capable of activating downstream transcription is necessary for chromatin decondensation of the Snrpn–Ube3a locus. In addition, the transcriptional inhibitor α-amanitin caused a significant decrease in the length of the paternal Snrpn–Ube3a allele in primary cortical neurons (Fig. 3E), further demonstrating the requirement of transcription for chromatin decondensation of the imprinted snoRNA locus. In order to test the genomic neighborhood effect on transcription and decondensation of the Snrpn–Ube3a locus, a transgenic mouse line (+/tg380a) with a transcriptionally active double BAC insertion including PWS-IC through MBII-85 was analyzed. +/tg380a neurons revealed slight decondensation of the BAC transgene allele that was significantly smaller than the endogenous paternal allele, suggesting that transcription alone is not sufficient for complete paternal chromatin decondensation and that genomic neighborhood may play a role in chromatin decondensation of imprinted snoRNAs (Supplementary Material, Fig. S6).
Figure 3.
Transcription is required for paternal Snrpn–Ube3a chromatin decondensation. (A) Representative image of a paternally inherited PWS-IC deletion (+/PWS-ICdel35 kb) neuronal nucleus with two small Snrpn–Ube3a DNA FISH signals due to lack of paternal transcript expression (36). (B) Representative image of two maternally inherited human PWS-IC (PWS-ICHs/+) neurons with two decondensed Snrpn–Ube3a DNA FISH signals due to lack of maternal IC methylation (37). Glial nucleus (upper left) with two small signals. (C) Snrpn–Ube3a signal measurements from +/PWS-ICdel35 kb mouse brain and wild-type (WT mat, WT pat) littermate. Results represent the mean (black bars) ± SD for 100 nuclei/genotype. Student's t-test, two tailed. ***P = 0.000071, NS=not significant. (D) Snrpn–Ube3a signal measurements from PWS-ICHs/+ mouse brain and wild-type littermates; 200 nuclei/genotype. Student's t-test. *P = 0.0027, ***P = 5 × 10−137. +/PWS-ICdel35 kb and PWS-ICHs/+ mice were not directly compared with one another as they are bred on different genetic backgrounds. (E) Murine primary neurons treated for 4 h with the transcriptional inhibitor, α-amanitin, were hybridized with the Snrpn–Ube3a DNA FISH probe and signals measured. Transcriptional inhibition specifically decreased the size of the paternal signal with 20 µg/ml α-amanitin treatment, whereas both signals decreased significantly with 30 µg/ml α-amanitin; 50 nuclei per treatment. Student's t-test. **P < 0.005, ***P < 0.0001.
Neuron-specific chromatin decondensation is uniquely observed at imprinted snoRNA clusters and impacts nucleolar size
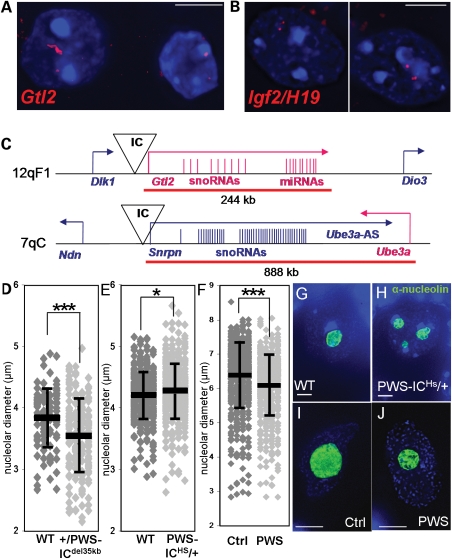
To investigate whether this highly differential, evolutionarily conserved, chromatin decondensation was specific to the Snrpn–Ube3a locus or to a larger group of mammalian loci in neurons, we created a panel of DNA FISH probes for murine loci with similar characteristics to Snrpn–Ube3a (Table 1). This included several imprinted loci (Dlx5/Dlx6, Igf2/H19, Ndn/Mkrn3/Magel2), an imprinted snoRNA cluster (Gtl2), a non-imprinted locus containing a single snoRNA (Htr2c), two loci containing large ncRNAs (Xic and Igf2r/Air), and one containing a large coding transcript (Dmd). Probes were hybridized to adult mouse brain sections and all showed small relatively equally sized signals throughout the brain (Fig. 4B), except Gtl2 (Fig. 4A). The ∼244 kb Gtl2 DNA FISH probe encompassed the maternally expressed snoRNA clusters and multiple miRNAs (Fig. 4C) and also displayed signals similar to Snrpn–Ube3a in mature neuronal nuclei with one small compact signal (presumably the inactive paternal allele) and one decondensed signal (presumably the snoRNA expressing maternal allele). As with Snrpn–Ube3a, the decondensed signal was specific to neuronal nuclei, as it was not observed in glia (Fig. 4A, right nucleus). DNA FISH probes flanking Gtl2 revealed only two small equally sized signals (data not shown). These results implicate transcription of the snoRNA clusters as the driving factor for this high level of chromatin decondensation and not merely imprinting, transcription of ncRNA, nor transcription of a long transcript.
Table 1.
Genetic loci with similarities to Snrpn–Ube3a for which DNA FISH probes were created for chromatin decondensation analyses in adult mouse whole brain sections
| Gene locus | Imprinted | snoRNAsa | ncRNA | Brain expressionb | Transcript size (kb) | DNA FISH probe (kb) | Decondensed chromatin |
|---|---|---|---|---|---|---|---|
| Snrpn–Ube3a | Yes | 80 | Yes | High | 1111 | 888c | Yes |
| Igf2/H19 | Yes | None | Yes | High/low | 8.7/2.7 | 244 | No |
| Dlx5/Dlx6 | Yes | None | No | Med/med | 4.5/4.0 | 203 | No |
| Igf2r | Yes | None | Yes | Med | 8.7 | 204 | No |
| Ndn/Magel2/Mkrn3 | Yes | None | No | High/med/med | 1.7/2.2/2.5 | 169 | No |
| Gtl2 | Yes | 9 | Yes | High | 208 | 244 | Yes |
| Htr2c | No | 1 | Yes | High | 235 | 190 | No |
| Tsix | No | None | Yes | Med | 53 | 224 | No |
| Dmdd | No | None | No | Med–highe | 2256 | 245 | No |
aBased on UCSC Genome Browser Mouse 2007 assembly alignment of homologous human snoRNAs at these loci and current published numbers (13,14,28).
bBased on UCSC Genome Browser heat maps from GNF1M Mouse Chip for all except Snrpn–Ube3a (based on previously published results (17) and Allen Brain Atlas).
cBAC DNA FISH probes (ranging from 127 to 191 kb each) individually showed chromatin decondensation over this region, the combined BAC DNA FISH probe covered the PWS-IC through Ube3a (complete length of the transcript varies by publication).
dDmd was the only single gene locus examined for which the BAC DNA FISH probe did not cover the majority of the transcript.
eWhile Dmd shows only medium expression in brain overall (by UCSC genome browser heat map), it has high expression in adult mouse Purkinje neurons (Allen Brain Atlas) where it was examined for chromatin decondensation.
Figure 4.
Allele-specific chromatin decondensation also seen at another imprinted cluster of snoRNAs. (A–C) The Gtl2 locus, containing imprinted clusters of snoRNAs, reveals similar levels of allele-specific and neuron-specific chromatin decondensation. (A) Representative image of an extended Gtl2 FISH signal (red) for one allele (presumed maternal) in adult murine neuronal nuclei (left nucleus), but not glial nuclei (right nucleus). (B) All other loci tested showed only small signals in neuronal nuclei. Representative image of Igf2/H19 (red), an imprinted locus expressed in brain but without extended chromatin decondensation. (C) Diagram of the imprinted regions of murine 12qF1 and 7qC. Bars and arrows in blue indicate paternally expressed transcripts, whereas pink indicates maternally expressed transcripts. Red lines indicate the location of the DNA FISH probe used. Not to scale. (D) +/PWS-ICdel35 kb Purkinje neurons, lacking snoRNA expression, display significantly smaller nucleoli than their wild-type littermate's Purkinje neurons, correlating to the lack of MBII-85 expression localized to nucleoli. Results represent the mean (black bars) ± SD for 100 nuclei per genotype. Student's t-test, two tailed. ***P < 0.0001. (E) PWS-ICHs/+ Purkinje neurons, with twice the levels of snoRNA expression, display significantly larger nucleoli than their wild-type littermate's Purkinje neurons; 350–400 nuclei/genotype. *P < 0.05. PWS-ICHs/+ Purkinje neurons also had significantly more nucleoli (12.5% had more than one nucleolus) than wild-type littermates (4.45% containing more then one nucleolus), P < 0.00001. (F) Purkinje neurons from PWS individuals also display significantly smaller nucleoli than age-matched controls; 100 nuclei/individual (six controls, five PWS samples). ***P < 0.0001. (G) Representative image of a murine wild-type Purkinje nucleus (DAPI counterstain, blue) with a single large nucleolus (α-nucleolin staining, green). (H) Representative image of a murine PWS-ICHs/+ Purkinje nucleus with multiple nucleoli. (I) Representative image of a human control Purkinje nucleus with a single large nucleolus. (J) Representative image of a PWS Purkinje nucleus with a single smaller nucleolus. White bars represents 5 µm.
To understand the importance of snoRNA cluster chromatin decondensation and transcription to nucleolar maturation of post-natal neurons, we tested the requirement for Snrpn–Ube3a snoRNAs in nucleolar maturation in +/PWS-ICdel35 kb and PWS-ICHs/+ mouse cerebellum. The nucleoli of Purkinje neurons were measured as they are an easily identifiable subset of neurons within the cerebellum that predominantly contain only a single nucleolus in adult mice. +/PWS-ICdel35 kb Purkinje nuclei, deficient for MBII-85/52 expression, had significantly smaller nucleoli than those of wt littermates (Fig. 4D), whereas PWS-ICHs/+ Purkinje nuclei, which express MBII-85/52 from both alleles, had significantly larger and more numerous nucleoli than those of its wt littermates (Fig. 4E, G and H) indicating a role for MBII-85/52 in nucleolar maturation. These alterations in nucleolar size were recapitulated in human Purkinje nucleoli from PWS individuals which were significantly smaller than age-matched controls (Fig. 4F, I and J). These results demonstrate that transcription from the IC and chromatin decondensation of MBII/HBII snoRNAs plays a significant role in regulating nucleolar size during neuronal maturation.
DISCUSSION
Neuronal chromatin undergoes dynamic changes in compaction and organization during post-natal neuronal maturation (38,39), but little is known about chromatin changes to specific loci. Here we have shown visual evidence for a highly orchestrated chromatin decondensation of essential clusters of snoRNAs during neuronal maturation. Although previous research has shown visual evidence of transcriptionally induced changes to chromatin at specific loci in non-neuronal cells (5,40), our results appear to represent the most dramatic chromatin decondensation of an endogenous locus currently reported in interphase mammalian nuclei.
We have shown that allele- and neuron-specific chromatin decondensation of imprinted snoRNA loci correlates with and is required for increased nucleolar size during neuronal maturation. Why this high level of allele-specific chromatin decondensation appears limited to the only two imprinted snoRNA cluster loci is presumably due to high expression of large clusters of snoRNAs and specific events required for snoRNA processing and biogenesis. Two proteins known to associate with biogenesis of C/D box snoRNAs, Tip49a and Tip49b, are involved in an ATP-dependent chromatin remodeling complex which catalyzes ATP-dependent nucleosome sliding (41). Perhaps these or other snoRNA processing factors are brought to the site of transcription to process snoRNAs and remodel the surrounding chromatin. Lack of one of these snoRNA clusters (HBII-85) is necessary to cause PWS (33,42), yet little is known of the function of these snoRNAs within neuronal nuclei. Our results suggest that one function of neuron-specific snoRNAs may be to modify rRNA, thereby increasing nucleolar size during neuronal maturation.
Recent publications point to the HBII-85/52 snoRNA clusters as being central to the PWS phenotype and 15q duplication syndrome. Two individuals have now been found with partial overlapping HBII-85 deletions and PWS-like phenotypes (33,42), whereas overexpression of MBII-52 in a Snprn-Ube3a locus paternal duplication mouse appears to contribute to social and behavioral abnormalities (43). In addition, recent human patients with and without PWS have suggested an exclusion of upstream genes MKRN3, MAGEL2 and NDN from a causal role in the PWS phenotype (44). Our results showing imprinted snoRNA chromatin decondensation impacting nucleolar size are consistent with the essential role of the HBII-85/52 locus in post-natal maturation of mammalian neurons.
While individual snoRNA genes are found in all eukaryotic genomes, snoRNA gene organization and regulation has changed considerably throughout evolution, favoring the colonization of introns to exploit host gene promoters to transcribe multiple snoRNAs from a single promoter (45). The presence of two eutherian-specific C/D snoRNA gene clusters at two evolutionarily distinct chromosomal locations (human 15q and 14q) has been suggested to be a potential evolutionary link to the recent emergence of parental imprinting of these loci (28). Furthermore, human chromosomes 14 and 15 are both acrocentric in humans, encoding repetitive rRNA genes at their p arms, suggesting a potential evolutionary link to localizing snoRNA genes near their target genes for optimal perinucleolar organization. In a recent comparison of gene neighborhood differences between human and chimpanzee, the 15q11–q13 locus was among the most highly divergent in gene neighborhood, suggesting a recent evolutionary advantage in the primate lineage to altered chromosome neighborhood particularly for genes specifically expressed in brain (46). The neuron specificity of the snoRNA chromatin decondensation observed in our study is consistent with a role for the 15q11–q13 snoRNAs in post-natal brain development, a mammalian occurrence under positive selection in the primate lineage.
Epigenetic regulation of both mammalian snoRNA loci is highly complex, requiring imprinting, ncRNA and chromatin remodeling for properly modulated allele- and neuron-specific expression. Homologous trans interactions between maternal and paternal alleles have been reported for both of these loci adding to the complex epigenetic mechanisms required for proper temporal and spatial transcription of these snoRNA loci (47,48). Our DNA FISH results in a transgenic mouse line harboring two additional transcriptionally active copies of the PWS-IC through MBII-85 at another genetic locus revealed that transcriptional activity alone was insufficient to induce full chromatin decondensation, further implicating the importance of snoRNA cluster genomic neighborhood in chromatin remodeling and decondensation.
In the primate lineage, chromosome 15q11–q13 is enriched in segmental low copy repeats or duplicons (49) that predispose it to a series of common breakpoints (50,51). These multiple breakpoints give rise not only to the large deletions and duplications seen in AS, PWS and 15q duplication, but also smaller 15q deletions and duplications distal to the snoRNA cluster found in other neurodevelopmental disorders, including epilepsy (52–54), intellectual disability (54,55), schizophrenia (56) and autism (53). Our results suggest the possibility that imprinted mammalian snoRNA chromatin decondensation during neuronal maturation could exert neighborhood effects on the phenotypic manifestations of copy number variations common to 15q11–q13 in humans.
MATERIALS AND METHODS
Post-mortem human tissue
Tissues of frontal cortex, Broadman Area 9 and cerebellum (<30 h post-mortem) were received frozen and were fixed in 10% formalin, embedded and sectioned (5 µm). Five PWS and six control cerebellum tissue samples were from adults ranging from 39 to 56 years of age. Tissues were obtained through the NICHD Brain and Tissue Bank for Developmental Disorders.
Mouse tissue
Wt male C57BL/6J whole brains for multiple age time points were obtained, fixed and embedded in paraffin and were sectioned (10 µm) for embryonic day 15 (E15), post-natal day 1 (P1), P14, P28 and P70, liver, thymus, kidney and spleen were also taken from a P70 mouse. Whole brains were obtained, fixed, embedded and sectioned for two P8.5wk FVB/PWS-IC 35kb deletion (+/PWS-ICdel35kb) mice and one wt littermate (36). Whole brains were obtained, fixed, embedded and sectioned for two p70 human PWS-IC/C57 (HsIC/+) mice and two wt littermates (37). Whole brains were obtained, fixed, embedded and sectioned for two P15.5wk +/tg380a mice and one littermate control.
Primary neurons
E15 C57BL/6J primary cortical mouse neurons (Lonza, http://www.lonza.com) were cultured according to supplier's protocol for 22 days on four chamber glass slides. Primary neurons were treated with 20 or 30 µg/ml α-amanitin for 4 h. Slides were fixed in Histochoice for 30 min, washed one time in PBS/0.5% Tween and then stored in 70% EtOH. DNA FISH for primary neurons followed the same as for paraffin-embedded tissue starting with the EtOH dehydration.
DNA FISH
DNA isolated from BACs (listed in Supplementary Material, Table S1) ordered from BacPacResources (Oakland, CA, USA) were confirmed by end sequencing. Extracted BAC DNA was labeled with biotin or digoxigenin by nick translation to create DNA FISH probes. Paraffin-embedded tissue was sectioned at 5 µm for human tissue and 10 µm for murine tissue onto glass slides. Slides were baked overnight at 56°C, then placed in four 5 min washes with xylene, then two 5 min washes with 100% ethanol and then 1 h at 95°C in antigen retrieval solution (DAKO). Slides were then washed in 0.2× SSC, post-fixed in Histochoice for 90 min and then washed 5 min in 1× PBS. Slides were dehydrated in 70, 90 and 100% ethanol (10 min each) and then dried at 50°C. A probe mixture containing 3 µl of each probe and 7 µl LSI/WCP buffer (Vysis, Inc.) was warmed to 37°C, then added to the slide, coverslipped and sealed with rubber cement. Probe and tissue were simultaneously denatured at 85°C for 3 min on a slide cycler (Hybaid). Slides were incubated overnight at 37°C, then washed in 50% formamide/50% 2× SSC thrice for 5 min, 2× SSC for 5 min and 2× SSC/0.1% IGEPAL for 5 min, all at 46°C. Slides were dried and 200 µl of blocking solution (warmed to 37°C) was added to each slide, coverslipped and incubated for 30 min at 37°C; and 200 µL of detection solution with α-digoxigenin Rhodamine (1:100) and Avidin-FITC (1:100) was next added to each slide, coverslipped and incubated for 2 h at 37°C. Slides were then washed three times in 1×PBS/0.5% Tween. Slides were air dried and mounted with 5 µg/ml DAPI in Vectrashield (Vector Laboratories) and coverslipped. To test for RNA:DNA hybridization, 250 µg/ml RNase A was added either before Histochoice fixation or after overnight hybridization for 30 min at 37°C. For DNase I digestion slides were incubated in 1 U/ml of DNase I for 10 min at 37°C, then chilled on ice before Histochoice fixation.
RNA FISH
A custom linked nucleic acid MBII-85 digoxigenin labeled probe (5′-DIG-ttccgatgagagtggcggtacaga-3′) was ordered from Exiqon. Paraffin-embedded tissue was sectioned at 5 µm for human tissue and 10 µm for murine tissue onto glass slides. Slides were treated as for DNA FISH up until probe addition with the exceptions that containers were treated with RNase Zap (http://www.ambion.com/) and DEPC water used to dilute solutions. A probe mixture containing 0.2 µl of the MBII-85 probe and 200 µl of hybridization buffer (50% formamide, 5× SSC, 0.1% Tween, 50 µg/ml heparin, 500 µg/ml yeast RNA) was warmed to 37°C, then added to the slide, coverslipped and sealed with rubber cement. Slides were incubated overnight at 57°C, then washed in 50% formamide/50% 2× SSC thrice for 30 m, 2× SSC for 5 min and 2× SSC/0.1% IGEPAL for 5 m, all at 57°C. Slides were dried and 200 µl of blocking solution (warmed to 37°C) was added to each slide, coverslipped and incubated for 30 min at 37°C; and 200 µl of detection solution with α-digoxigenin Rhodamine (1:100) was next added to each slide, coverslipped and incubated for 2 h at 37°C. Slides were then washed three times in 1× PBS/0.5% Tween. Slides were air dried and mounted with 5 µg/ml DAPI in Vectrashield (Vector Laboratories) and coverslipped.
Immunofluorescence
Paraffin embedded tissue slides were baked overnight at 56°C, then placed in four 5 min washes with xylene, then two 5 min washes with 100% ethanol and then 1 h at 95°C in antigen retrieval solution (DAKO). Slides were then washed in 0.2× SSC and then 1:100 dilution of α-nucleolin antibody (Abcam) was added and incubated at 37°C overnight 200 µl of detection solution with α-digoxigenin Rhodamine (1:100) was next added to each slide, coverslipped and incubated for 2 h at 37°C. Slides were air dried and mounted with 5 µg/ml DAPI in Vectrashield (Vector Laboratories) and coverslipped.
Microscopy
Slides were analyzed on an Axioplan 2 fluorescence microscope (Carl Zeiss, Inc., NY, USA) equipped with a Qimaging Retiga EXi high-speed uncooled digital camera, appropriate fluorescent filter sets and automated xyz stage controls. The microscope and peripherals were controlled by a Macintosh running iVision (Scanalytics, Vienna, VA, USA) software. Images were captured for blue, green and red filters at one edge of the specimen, and then repeated at 0.5–1 µm sections through the depth of the tissue. Each image stack was digitally deconvolved to remove out-of-focus light using HazeBuster software (Vaytek, Fairfield, IA, USA). Following haze removal, image stacks for each fluorophore were merged and stacked to create a two-dimensional image representing all of fluorescence within the section. Measurements were taken in preview mode using a 100× oil objective with a 1× zoom, using a drawing tool set to define each pixel with the appropriate number of microns. All measurements for a given experiment were taken with the same exposure times and microscope settings as was appropriate for the fluorescence intensity. For experiments comparing nucleolar size, the investigator making the measurements was blinded to the tissue identity.
Statistical tests
Paired Student t-test were performed for all statistical comparisons between of measurements from individual nuclei.
Packing ratio calculation
Number of base pairs of the BAC probe×the size of a single bp of naked DNA (Packing Ratio for Naked DNA 1:1, 30 nm fiber ∼40:1, Naked DNA = 0.0003 µm/bp). So for the Snrpn–Ube3a combination probe ∼888 kb: 888 × 103 bp × (0.0003 µm/bp) = 266.4 µm (length if naked DNA).
Packing ratio example: paternal signal ∼4 µm: 266.4 µm/4 µmin = 66.6:1 packing ratio (1.7×more compact then a 30 nm fiber) (40).
qPCR
DNA was isolated from adult C57/Blk6 mouse brain and liver using Gentra Puregene Tissue Kit (Qiagen). DNA levels were measured using Nanodrop spectrophotometer and diluted to 50 ng/µl. Reagents from Express SYBR GreenEr qPCR Supermix Universal (Invitrogen) were used for qRT–PCR with the following conditions: 5 µl 2× SYBR Green Buffer (Invitrogen), 1.8 µl 1uMINPrF + PrR (mixed), 2.2 µl nuclease free H2O, 1 µl cDNA. Eppendorf Mastercycle RealPlex was used to perform qPCR with for 40 cycles at 61°C annealing.
qRT–PCR
RNA was isolated from frozen adult mouse brain using 3 ml of TriZol reagent (Invitrogen). cDNA was made using SuperScript™ First-Strand Synthesis System (Invitrogen). cDNA reagents from Express SYBR GreenEr qPCR Supermix Universal (Invitrogen) were used for qRT–PCR with the following conditions: 5 µl 2× SYBR Green Buffer, 1.8 µl 1 µM PrF + PrR (mixed), 2.2 µl nuclease free H2O, 1 µl cDNA. Eppendorf Mastercycle RealPlex was used to perform qRT–PCR for 40 cycles at 62°C annealing.
AUTHOR CONTRIBUTIONS
K.N.L. and J.M.L. designed experiments and wrote manuscript. K.N.L. performed all FISH, immunofluorescence and microscopy. R.O.V. dissected wt mouse tissues and performed qPCR and qRT–PCR. A.J.D. and J.L.R. provided genotyped and dissected transgenic mouse tissues.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
FUNDING
This work was supported by National Institute of Health (1R01HD048799 to J.M.L., HD037872 and C06 RR-12088-01 to J.L.R), National Science Foundation (predoctoral fellowship to K.N.L.). Human tissue was obtained from the National Institute of Child Health and Human Development Brain and Tissue Bank for Developmental Disorders at the University of Maryland, and the Harvard Brain Tissue Resource Center (supported in part by R24MH068855). Funding to pay the Open Access publication charges for this article was provided by the Eunice Kennedy Shriver National Institute of Child Health and Human Development.
ACKNOWLEDGEMENTS
The authors thank Stormy Chamberlain, Karen Johnstone, Joanne K. Suarez, Weston Powell, Frederick Chedin and Mary Delaney.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Mehler M.F., Mattick J.S. Noncoding RNAs and RNA editing in brain development, functional diversification, and neurological disease. Physiol. Rev. 2007;87:799–823. doi: 10.1152/physrev.00036.2006. [DOI] [PubMed] [Google Scholar]
- 2.Wilkinson L.S., Davies W., Isles A.R. Genomic imprinting effects on brain development and function. Nat. Rev. Neurosci. 2007;8:832–843. doi: 10.1038/nrn2235. [DOI] [PubMed] [Google Scholar]
- 3.Mercer T.R., Dinger M.E., Mariani J., Kosik K.S., Mehler M.F., Mattick J.S. Noncoding RNAs in long-term memory formation. Neuroscientist. 2008;14:434–445. doi: 10.1177/1073858408319187. [DOI] [PubMed] [Google Scholar]
- 4.Morison I.M., Ramsay J.P., Spencer H.G. A census of mammalian imprinting. Trends Genet. 2005;21:457–465. doi: 10.1016/j.tig.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 5.Chambeyron S., Da Silva N.R., Lawson K.A., Bickmore W.A. Nuclear re-organisation of the Hoxb complex during mouse embryonic development. Development. 2005;132:2215–2223. doi: 10.1242/dev.01813. [DOI] [PubMed] [Google Scholar]
- 6.Manuelidis L. A view of interphase chromosomes. Science. 1990;250:1533–1540. doi: 10.1126/science.2274784. [DOI] [PubMed] [Google Scholar]
- 7.Knoll J.H., Nicholls R.D., Magenis R.E., Graham J.M., Jr, Lalande M., Latt S.A. Angelman and Prader–Willi syndromes share a common chromosome 15 deletion but differ in parental origin of the deletion. Am. J. Med. Genet. 1989;32:285–290. doi: 10.1002/ajmg.1320320235. [DOI] [PubMed] [Google Scholar]
- 8.Wagstaff J., Knoll J.H., Glatt K.A., Shugart Y.Y., Sommer A., Lalande M. Maternal but not paternal transmission of 15q11–13-linked nondeletion Angelman syndrome leads to phenotypic expression. Nat. Genet. 1992;1:291–294. doi: 10.1038/ng0792-291. [DOI] [PubMed] [Google Scholar]
- 9.Wolpert C.M., Menold M.M., Bass M.P., Qumsiyeh M.B., Donnelly S.L., Ravan S.A., Vance J.M., Gilbert J.R., Abramson R.K., Wright H.H., et al. Three probands with autistic disorder and isodicentric chromosome 15. Am. J. Med. Genet. 2000;96:365–372. doi: 10.1002/1096-8628(20000612)96:3<365::aid-ajmg25>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 10.Baker P., Piven J., Schwartz S., Patil S. Brief report: duplication of chromosome 15q11–13 in two individuals with autistic disorder. J. Autism Dev. Disord. 1994;24:529–535. doi: 10.1007/BF02172133. [DOI] [PubMed] [Google Scholar]
- 11.Wassink T.H., Piven J. The molecular genetics of autism. Curr. Psychiatry Rep. 2000;2:170–175. doi: 10.1007/s11920-000-0063-x. [DOI] [PubMed] [Google Scholar]
- 12.Buiting K., Kanber D., Martin-Subero J.I., Lieb W., Terhal P., Albrecht B., Purmann S., Gross S., Lich C., Siebert R., et al. Clinical features of maternal uniparental disomy 14 in patients with an epimutation and a deletion of the imprinted DLK1/GTL2 gene cluster. Hum. Mutat. 2008;29:1141–1146. doi: 10.1002/humu.20771. [DOI] [PubMed] [Google Scholar]
- 13.Cavaille J., Buiting K., Kiefmann M., Lalande M., Brannan C.I., Horsthemke B., Bachellerie J.P., Brosius J., Huttenhofer A. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc. Natl Acad. Sci. USA. 2000;97:14311–14316. doi: 10.1073/pnas.250426397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cavaille J., Seitz H., Paulsen M., Ferguson-Smith A.C., Bachellerie J.P. Identification of tandemly-repeated C/D snoRNA genes at the imprinted human 14q32 domain reminiscent of those at the Prader–Willi/Angelman syndrome region. Hum. Mol. Genet. 2002;11:1527–1538. doi: 10.1093/hmg/11.13.1527. [DOI] [PubMed] [Google Scholar]
- 15.Sutcliffe J.S., Nakao M., Christian S., Orstavik K.H., Tommerup N., Ledbetter D.H., Beaudet A.L. Deletions of a differentially methylated CpG island at the SNRPN gene define a putative imprinting control region. Nat. Genet. 1994;8:52–58. doi: 10.1038/ng0994-52. [DOI] [PubMed] [Google Scholar]
- 16.Kagami M., Sekita Y., Nishimura G., Irie M., Kato F., Okada M., Yamamori S., Kishimoto H., Nakayama M., Tanaka Y., et al. Deletions and epimutations affecting the human 14q32.2 imprinted region in individuals with paternal and maternal upd(14)-like phenotypes. Nat. Genet. 2008;40:237–242. doi: 10.1038/ng.2007.56. [DOI] [PubMed] [Google Scholar]
- 17.Le Meur E., Watrin F., Landers M., Sturny R., Lalande M., Muscatelli F. Dynamic developmental regulation of the large non-coding RNA associated with the mouse 7C imprinted chromosomal region. Dev. Biol. 2005;286:587–600. doi: 10.1016/j.ydbio.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 18.Runte M., Huttenhofer A., Gross S., Kiefmann M., Horsthemke B., Buiting K. The IC-SNURF-SNRPN transcript serves as a host for multiple small nucleolar RNA species and as an antisense RNA for UBE3A. Hum. Mol. Genet. 2001;10:2687–2700. doi: 10.1093/hmg/10.23.2687. [DOI] [PubMed] [Google Scholar]
- 19.de los Santos T., Schweizer J., Rees C.A., Francke U. Small evolutionarily conserved RNA, resembling C/D box small nucleolar RNA, is transcribed from PWCR1, a novel imprinted gene in the Prader-Willi deletion region, which Is highly expressed in brain. Am. J. Hum. Genet. 2000;67:1067–1082. doi: 10.1086/303106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rougeulle C., Cardoso C., Fontes M., Colleaux L., Lalande M. An imprinted antisense RNA overlaps UBE3A and a second maternally expressed transcript. Nat. Genet. 1998;19:15–16. doi: 10.1038/ng0598-15. [DOI] [PubMed] [Google Scholar]
- 21.Rougeulle C., Glatt H., Lalande M. The Angelman syndrome candidate gene, UBE3A/E6-AP, is imprinted in brain [letter] [In Process Citation] Nat. Genet. 1997;17:14–15. doi: 10.1038/ng0997-14. [DOI] [PubMed] [Google Scholar]
- 22.Vu T., Hoffman A. Imprinting of the Angelman syndrome gene, UBE3A, is restricted to brain. Nat. Genet. 1997;17:12–13. doi: 10.1038/ng0997-12. [DOI] [PubMed] [Google Scholar]
- 23.Herzing L.B.K., Kim S.-J., Cook E.H., Ledbetter D.H. The human aminophospholipid-transporting ATPase gene ATP10C maps adjacent to UBE3A and exhibits similar imprinted expression. Am. J. Hum. Genet. 2001;68:1501–1505. doi: 10.1086/320616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kashiwagi A., Meguro M., Hoshiya H., Haruta M., Ishino F., Shibahara T., Oshimura M. Predominant maternal expression of the mouse Atp10c in hippocampus and olfactory bulb. J. Hum. Genet. 2003;48:194–198. doi: 10.1007/s10038-003-0009-3. [DOI] [PubMed] [Google Scholar]
- 25.Dittrich B., Robinson W.P., Knoblauch H., Buiting K., Schmidt K., Gillessen-Kaesbach G., Horsthemke B. Molecular diagnosis of the Prader–Willi and Angelman syndromes by detection of parent-of-origin specific DNA methylation in 15q11-13. Hum. Genet. 1992;90:313–315. doi: 10.1007/BF00220089. [DOI] [PubMed] [Google Scholar]
- 26.Rodriguez-Jato S., Nicholls R.D., Driscoll D.J., Yang T.P. Characterization of cis- and trans-acting elements in the imprinted human SNURF-SNRPN locus. Nucleic Acids Res. 2005;33:4740–4753. doi: 10.1093/nar/gki786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu M.Y., Tsai T.F., Beaudet A.L. Deficiency of Rbbp1/Arid4a and Rbbp1l1/Arid4b alters epigenetic modifications and suppresses an imprinting defect in the PWS/AS domain. Genes Dev. 2006;20:2859–2870. doi: 10.1101/gad.1452206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Royo H., Cavaille J. Non-coding RNAs in imprinted gene clusters. Biol. Cell. 2008;100:149–166. doi: 10.1042/BC20070126. [DOI] [PubMed] [Google Scholar]
- 29.Boisvert F.M., van Koningsbruggen S., Navascues J., Lamond A.I. The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 2007;8:574–585. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- 30.Cavaille J., Vitali P., Basyuk E., Huttenhofer A., Bachellerie J.P. A novel brain-specific box C/D small nucleolar RNA processed from tandemly repeated introns of a noncoding RNA gene in rats. J. Biol. Chem. 2001;276:26374–26383. doi: 10.1074/jbc.M103544200. [DOI] [PubMed] [Google Scholar]
- 31.Sridhar P., Gan H.H., Schlick T. A computational screen for C/D box snoRNAs in the human genomic region associated with Prader–Willi and Angelman syndromes. J. Biomed. Sci. 2008;15:697–705. doi: 10.1007/s11373-008-9271-x. [DOI] [PubMed] [Google Scholar]
- 32.Kishore S., Stamm S. The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science. 2006;311:230–232. doi: 10.1126/science.1118265. [DOI] [PubMed] [Google Scholar]
- 33.Sahoo T., del Gaudio D., German J.R., Shinawi M., Peters S.U., Person R.E., Garnica A., Cheung S.W., Beaudet A.L. Prader–Willi phenotype caused by paternal deficiency for the HBII-85 C/D box small nucleolar RNA cluster. Nat. Genet. 2008;40:719–721. doi: 10.1038/ng.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ding F., Li H.H., Zhang S., Solomon N.M., Camper S.A., Cohen P., Francke U. SnoRNA Snord116 (Pwcr1/MBII-85) deletion causes growth deficiency and hyperphagia in mice. PLoS ONE. 2008;3:e1709. doi: 10.1371/journal.pone.0001709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Skryabin B.V., Gubar L.V., Seeger B., Pfeiffer J., Handel S., Robeck T., Karpova E., Rozhdestvensky T.S., Brosius J. Deletion of the MBII-85 snoRNA gene cluster in mice results in postnatal growth retardation. PLoS Genet. 2007;3:e235. doi: 10.1371/journal.pgen.0030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chamberlain S.J., Johnstone K.A., DuBose A.J., Simon T.A., Bartolomei M.S., Resnick J.L., Brannan C.I. Evidence for genetic modifiers of postnatal lethality in PWS-IC deletion mice. Hum. Mol. Genet. 2004;13:2971–2977. doi: 10.1093/hmg/ddh314. [DOI] [PubMed] [Google Scholar]
- 37.Johnstone K.A., Dubose A.J., Futtner C.R., Elmore M.D., Brannan C.I., Resnick J.L. A human imprinting centre demonstrates conserved acquisition but diverged maintenance of imprinting in a mouse model for Angelman syndrome imprinting defects. Hum. Mol. Genet. 2005;15:393–404. doi: 10.1093/hmg/ddi456. [DOI] [PubMed] [Google Scholar]
- 38.Martou G., De Boni U. Nuclear topology of murine, cerebellar Purkinje neurons: changes as a function of development. Exp. Cell Res. 2000;256:131–139. doi: 10.1006/excr.1999.4793. [DOI] [PubMed] [Google Scholar]
- 39.Thatcher K., LaSalle J.M. Dynamic changes in Histone H3 lysine 9 acetylation localization patterns during neuronal maturation require MeCP2. Epigenetics. 2006;1:24–31. doi: 10.4161/epi.1.1.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muller W.G., Rieder D., Kreth G., Cremer C., Trajanoski Z., McNally J.G. Generic features of tertiary chromatin structure as detected in natural chromosomes. Mol. Cell Biol. 2004;24:9359–9370. doi: 10.1128/MCB.24.21.9359-9370.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.King T.H., Decatur W.A., Bertrand E., Maxwell E.S., Fournier M.J. A well-connected and conserved nucleoplasmic helicase is required for production of box C/D and H/ACA snoRNAs and localization of snoRNP proteins. Mol. Cell Biol. 2001;21:7731–7746. doi: 10.1128/MCB.21.22.7731-7746.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Smith A.J., Purmann C., Walters R.G., Ellis R.J., Holder S.E., Van Haelst M.M., Brady A.F., Fairbrother U.L., Dattani M., Keogh J.M., et al. A deletion of the HBII-85 class of small nucleolar RNAs (snoRNAs) is associated with hyperphagia, obesity and hypogonadism. Hum. Mol. Genet. 2009;18:3257–3265. doi: 10.1093/hmg/ddp263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakatani J., Tamada K., Hatanaka F., Ise S., Ohta H., Inoue K., Tomonaga S., Watanabe Y., Chung Y.J., Banerjee R., et al. Abnormal behavior in a chromosome-engineered mouse model for human 15q11–13 duplication seen in autism. Cell. 2009;137:1235–1246. doi: 10.1016/j.cell.2009.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanber D., Giltay J., Wieczorek D., Zogel C., Hochstenbach R., Caliebe A., Kuechler A., Horsthemke B., Buiting K. A paternal deletion of MKRN3, MAGEL2 and NDN does not result in Prader–Willi syndrome. Eur. J. Hum. Genet. 2009;17:582–590. doi: 10.1038/ejhg.2008.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dieci G., Preti M., Montanini B. Eukaryotic snoRNAs: a paradigm for gene expression flexibility. Genomics. 2009;94:83–88. doi: 10.1016/j.ygeno.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 46.De S., Teichmann S.A., Babu M.M. The impact of genomic neighborhood on the evolution of human and chimpanzee transcriptome. Genome Res. 2009;19:785–794. doi: 10.1101/gr.086165.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Georges M., Charlier C., Cockett N. The callipyge locus: evidence for the trans interaction of reciprocally imprinted genes. Trends Genet. 2003;19:248–252. doi: 10.1016/S0168-9525(03)00082-9. [DOI] [PubMed] [Google Scholar]
- 48.Thatcher K., Peddada S., Yasui D., LaSalle J.M. Homologous pairing of 15q11-13 imprinted domains in brain is developmentally regulated but deficient in Rett and autism samples. Hum. Mol. Genet. 2005;14:785–797. doi: 10.1093/hmg/ddi073. [DOI] [PubMed] [Google Scholar]
- 49.Bailey J.A., Gu Z., Clark R.A., Reinert K., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- 50.Christian S.L., Fantes J.A., Mewborn S.K., Huang B., Ledbetter D.H. Large genomic duplicons map to sites of instability in the Prader–Willi/Angelman syndrome chromosome region (15q11–q13) Hum. Mol. Genet. 1999;8:1025–1037. doi: 10.1093/hmg/8.6.1025. [DOI] [PubMed] [Google Scholar]
- 51.Robinson W.P., Kuchinka B.D., Bernasconi F., Petersen M.B., Schulze A., Brondum-Nielsen K., Christian S.L., Ledbetter D.H., Schinzel A.A., Horsthemke B., et al. Maternal meiosis I non-disjunction of chromosome 15: dependence of the maternal age effect on level of recombination. Hum. Mol. Genet. 1998;7:1011–1019. doi: 10.1093/hmg/7.6.1011. [DOI] [PubMed] [Google Scholar]
- 52.Sharp A.J., Mefford H.C., Li K., Baker C., Skinner C., Stevenson R.E., Schroer R.J., Novara F., De Gregori M., Ciccone R., et al. A recurrent 15q13.3 microdeletion syndrome associated with mental retardation and seizures. Nat. Genet. 2008;40:322–328. doi: 10.1038/ng.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pagnamenta A.T., Wing K., Akha E.S., Knight S.J., Bolte S., Schmotzer G., Duketis E., Poustka F., Klauck S.M., Poustka A., et al. A 15q13.3 microdeletion segregating with autism. Eur. J. Hum. Genet. 2008;17:687–692. doi: 10.1038/ejhg.2008.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Helbig I., Mefford H.C., Sharp A.J., Guipponi M., Fichera M., Franke A., Muhle H., de Kovel C., Baker C., von Spiczak S., et al. 15q13.3 Microdeletions increase risk of idiopathic generalized epilepsy. Nat. Genet. 2009;41:160–162. doi: 10.1038/ng.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ben-Shachar S., Lanpher B., German J.R., Qasaymeh M., Potocki L., Nagamani S., Franco L.M., Malphrus A., Bottenfield G.W., Spence J.E., et al. Microdeletion 15q13.3: a locus with incomplete penetrance for autism, mental retardation, and psychiatric disorders. J. Med. Genet. 2009;46:382–388. doi: 10.1136/jmg.2008.064378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stefansson H., Rujescu D., Cichon S., Pietilainen O.P., Ingason A., Steinberg S., Fossdal R., Sigurdsson E., Sigmundsson T., Buizer-Voskamp J.E., et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.