Abstract
Genetic instability at palindromes and spaced inverted repeats (IRs) leads to chromosome rearrangements. Perfect palindromes and IRs with short spacers can extrude as cruciforms or fold into hairpins on the lagging strand during replication. Cruciform resolution produces double-strand breaks (DSBs) with hairpin-capped ends, and Mre11p and Sae2p are required to cleave the hairpin tips to facilitate homologous recombination. Fragile site 2 (FS2) is a naturally occurring IR in Saccharomyces cerevisiae composed of a pair of Ty1 elements separated by ∼280 bp. Our results suggest that FS2 forms a hairpin, rather than a cruciform, during replication in cells with low levels of DNA polymerase. Cleavage of this hairpin results in a recombinogenic DSB. We show that DSB formation at FS2 does not require Mre11p, Sae2p, Rad1p, Slx4p, Pso2p, Exo1p, Mus81p, Yen1p, or Rad27p. Also, repair of DSBs by homologous recombination is efficient in mre11 and sae2 mutants. Homologous recombination is impaired at FS2 in rad52 mutants and most aberrations reflect either joining of two broken chromosomes in a “half crossover” or telomere capping of the break. In support of hairpin formation precipitating DSBs at FS2, two telomere-capped deletions had a breakpoint near the center of the IR. In summary, Mre11p and Sae2p are not required for DSB formation at FS2 or the subsequent repair of these DSBs.
PALINDROMES (inverted repeats with no spacer between the repeats) and inverted repeats separated by a short spacer (“IRs”) are hotspots for genetic instability. In bacteria and yeast, palindromes and IRs are frequently deleted (Collins et al. 1982; Dasgupta et al. 1987; Gordenin et al. 1993; Ruskin and Fink 1993), and double-strand breaks (DSBs) and recombination are stimulated by these sequences (Farah et al. 2002, 2005; Lobachev et al. 2002; Lemoine et al. 2005; Cote and Lewis 2008; Eykelenboom et al. 2008). A palindrome introduced as a mouse transgene is a target for deletions and rearrangements and simulates gene conversion (Collick et al. 1996; Akgun et al. 1997). In human cells, the center of a large palindromic AT-rich repeat (PATRR) at 22q11.2 is a hotspot for breaks, translocations, and deletions and drives the most commonly observed non-Robertsonian translocation to 11q23, which also has a PATRR (Kurahashi et al. 2006, 2007; Kogo et al. 2007). IRs are also associated with gene amplification in human cancer cells (Tanaka et al. 2005, 2007) and in yeast (Narayanan et al. 2006).
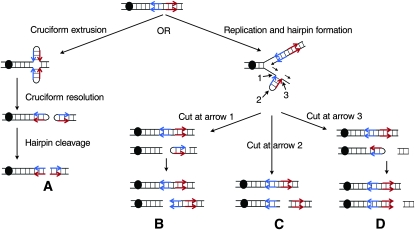
The formation of cruciform or hairpin secondary structures at DNA palindromes and spaced IRs is believed to precipitate the DSBs and genetic instability at these regions. The likelihood that a perfect palindrome will extrude in a cruciform (Figure 1, left-hand side) is affected by base composition at the center of the palindrome and by arm length. Centers with AT base pairs are more likely to extrude than GC centers, presumably due to the easier melting of AT base pairs (Courey and Wang 1988; Zheng and Sinden 1988). Longer arm lengths increase the propensity for stable cruciform formation in both perfect palindromes and IRs (Sinden et al. 1991; Kogo et al. 2007). In plasmids or phage maintained in Escherichia coli, IRs with short spacers of 10 bp often adopt a cruciform secondary structure, but IRs with spacers >20 bp rarely extrude as cruciforms (Sinden et al. 1991; Allers and Leach 1995; Kogo et al. 2007). However, IRs with large spacers can form hairpins on single-stranded DNA (Figure 1, right-hand side), such as within the Okazaki fragment initiation zone on the lagging strand during DNA replication (Trinh and Sinden 1991; Voineagu et al. 2008).
Figure 1.—
Mechanisms of producing a recombinogenic DSB at an IR. The inverted repeat is shown as blue and red arrows, and each line represents a single DNA strand rather than duplicated chromatids. Labeled arrows show the positions of nuclease cleavage at the hairpin structure. The centromere is shown as a black oval. Only those broken DNA molecules containing a centromere are likely to produce a recoverable chromosome rearrangement. (A) Cruciform formation in a nonreplicating DNA molecule. Processing of the resulting structure by a resolvase would be expected to yield two hairpin-capped products that could be subsequently processed to yield uncapped broken DNA molecules. (B–D) DSBs produced by different positions of cleavage of the hairpin intermediate. We show hairpin formation associated with replication of the lagging strand. Cleavage at arrow 1 produces a capped hairpin in the acentric fragment or a centromere-containing fragment with a DSB proximal to FS2. Cleavage at arrow 2 results in a product in which the DSB is between the two elements of the inverted repeat. Cleavage at arrow 3 produces a capped hairpin or, if replication proceeds through the hairpin, results in a centromere-containing fragment with a DSB near the distal Ty element of FS2.
We previously identified a naturally occurring fragile site in Saccharomyces cerevisiae (fragile site 2, FS2) that is a spaced IR on chromosome III. FS2 consists of two 6-kb Ty1 elements in a head-to-head orientation separated by ∼280 bp. Although this site is relatively inactive in wild-type cells, it is a hotspot for DSBs and translocations in cells with low levels of DNA polymerase α or δ (Lemoine et al. 2005, 2008). Polymerases α and δ, respectively, are the primase and replicative polymerase on the lagging strand. FS2-dependent DSBs have been physically observed by separating yeast chromosomes with clamped homogeneous electric field (CHEF) gel electrophoresis and using Southern blotting with chromosome III-specific probes. When cells have low levels of DNA polymerase α, a chromosome fragment of the size expected from a DSB at FS2 is observed. No such fragment is present in cells with wild-type levels of DNA polymerase α or in cells in which the centromere-proximal Ty1 of FS2 has been deleted, disrupting the potential for forming a secondary structure (Lemoine et al. 2005).
Our interpretation of these results is that the reduced level of polymerase α results in accumulation of single-stranded DNA on the lagging strand that permits the inverted Ty1 elements of FS2 to fold into a hairpin (Figure 1, right-hand side). Consistent with this suggestion, Voineagu et al. (2008) argue that the size of the Okazaki fragment initiation zone (OIZ) is a limiting factor in hairpin formation. Since the OIZ in eukaryotic cells is ∼290 nucleotides (DePamphilis and Wassarman 1980; DePamphilis 2002), during normal DNA replication, the pairing of the FS2 Ty elements (separated by an ∼280-bp spacer) will be very infrequent. The enlarged OIZ expected in cells with reduced polymerase α, however, could expose the IR regions flanking the spacer, allowing the formation of a DSB.
The DSB associated with FS2 had several different fates (Lemoine et al. 2005). Failure to repair the break resulted in loss of the broken chromosome. At low frequency, we observed strains with a stable terminal deletion, presumably representing “capping” of the broken chromosome by telomere repeats. A more frequent event was break-induced replication (BIR) between a Ty element of FS2 and a Ty or δ-element located on nonhomologous chromosomes, generating nonreciprocal translocations. δ-Elements are long terminal direct repeats ∼330 bp in length located at the ends of Ty elements and are additionally present as “solo” elements scattered throughout the genome.
In addition to translocations that involve the Ty elements of FS2, we found translocations involving two directly repeated Ty elements located centromere-proximal to FS2; we termed this pair of elements FS1 (Lemoine et al. 2005). Most of the translocations that occur at FS1 are likely to be initiated by a DSB at FS2, since a deletion of one of the two FS2 Ty elements reduces the frequency of both FS2- and FS1-mediated events. Thus, we suggested that DSBs at FS2 are sometimes processed to generate a recombinogenic end in one of the Ty1 elements of FS1.
It has been proposed that cruciform structures are recognized and cleaved in the cell by a Holliday junction resolvase, resulting in two broken ends each capped with a hairpin. Cote and Lewis (2008) demonstrated that Mus81p was required for the resolution of a cruciform formed by a perfect palindrome carried on a plasmid in S. cerevisiae. In a study of an IR consisting of a pair of inverted human Alu elements separated by a 12-bp center spacer that was integrated on a yeast chromosome, however, Lobachev et al. (2002) found that Mus81p was not required for the formation of DSBs at the IR. In both of these studies, hairpin-capped breaks were demonstrated to be present at the center of symmetry, and Mre11p and Sae2p were required for repair of these breaks. In the absence of these proteins, DNA replication across the hairpin-capped sequence generated an extended inverted duplication, suggesting that the essential function of Mre11p and Sae2p is to cleave the hairpin tip. Other studies have also implicated Mre11p and Sae2p in facilitating repair at perfect palindromes and at IRs with very short spacers in yeast (Rattray 2004; Farah et al. 2005; Rattray et al. 2005). Biochemical studies indicate that Mre11p, together with Sae2p, can cleave open small single-stranded DNA loops, such as those at the tips of hairpin-capped DSBs (Trujillo and Sung 2001; Lengsfeld et al. 2007). The previous studies of the effects of various mutants on the stability of inverted repeats have focused on sequences with the potential to extrude as a cruciform. Consequently, we investigated the roles of nucleases and recombination proteins on cleavage and DSB repair at FS2, which is likely to be extruded as a hairpin on the lagging strand rather than as a cruciform.
MATERIALS AND METHODS
Strain construction:
All GAL-POL1 strains in this study are isogenic with MS71, a LEU2 derivative of AMY125 (MATα ade5-1 leu2-3 trp1-289 ura3-52 his7-2) (Kokoska et al. 2000), except for changes introduced by transformation. All mating-type tester strains are isogenic with 1225 (his4-15 leu2 thr4 ura3-52 trp1 Lys−), except for changes introduced by transformation. Strain constructions and genotypes for all strains are in supporting information, Table S1.
Genetic methods and media:
Transformation and mating methods were standard and all strains were grown at 30°. High-galactose medium contained 0.05% galactose and low-galactose medium contained 0.005% galactose, as well as 3% raffinose, plus the standard supplements of yeast extract and peptone; dextrose was omitted. Selective media were standard except for the addition of high or low galactose and the substitution of dextrose with raffinose (Guthrie and Fink 1991).
Quantitation of frequency of illegitimate mating:
For each strain, we examined illegitimate mating in eight independent cultures. Each haploid GAL-POL1 MATα experimental strain was grown overnight in 5 ml low galactose cultures. The mating tester strains (1225a and derivatives of 1225α) were grown overnight in rich growth medium (YPD). Cells were plated onto high galactose to assess viability, and ∼1 × 106 cells of the experimental strains were mixed with a fivefold excess of the tester. These mixtures were concentrated onto a sterile nitrocellulose filter and incubated on high galactose plates for 6 hr at 30°. The cells were rinsed from the filter with water and replated on diploid-selective medium. For legitimate mating, we plated a dilution of the mated cells. For illegitimate mating, the undiluted mixture was plated. After colony formation, we compared the number of diploids to the number of viable cells. Under these conditions, legitimate mating was very efficient, ≥90% in all strains except those with the rad52 or sae2 mutations. In these strains, the efficiency of legitimate mating was ∼60%; the frequency of illegitimate mating was normalized to account for this decreased frequency of legitimate mating.
CHEF analysis, Southern blot analysis of illegitimate diploids, and analysis of DSBs on chromosome III:
Genomic DNA was extracted in agarose plugs to avoid shearing, using the methods described by Lobachev et al. (2002). For CHEF analysis, electrophoresis was performed at 14° in a 1.0% gel, 0.5× TBE buffer in a Bio-Rad (Hercules, CA) CHEF Mapper XA. For analysis of chromosome III translocations in illegitimate diploids, yeast chromosomes were separated with switch times starting at 47 sec and extending to 2 min 49 sec at 5 V/cm for 33 hr. For analysis of the broken chromosome III in haploids with low levels of α-DNA polymerase, separation was done with switch times starting at 9.8 sec and extending to 34.92 sec at 6 V/cm for 18 hr 30 min.
For Southern blot analysis, we used a CHA1 probe to the left arm of chromosome III (sequences 15,838–16,800) produced by PCR amplification of yeast genomic DNA. Probes were labeled by random-priming labeling, using Ready-To-Go DNA Labeling Beads (GE Healthcare). Southern hybridization and washing were standard. Membranes were exposed to a PhosphoImager screen for 1–3 days. Images were captured with a Typhoon imager (GE Healthcare) and quantification was performed using Quantity One analysis software (Bio-Rad).
All illegitimate diploids were initially characterized by CHEF gel separation of chromosomes followed by Southern blotting using the CHA1 probe described above. Several illegitimate diploids were further analyzed using genomic microarrays and additional Southern blots as described by Lemoine et al. (2005). Details of the analysis of each illegitimate diploid are in File S1.
Telomere PCR:
In several of the illegitimate diploids, we detected a chromosome III with terminal deletions. We expected that these chromosomes would be capped with telomeric repeats. From the CHEF gel and microarray analysis, two of these strains (PG297 and PG301) had deleted chromosomes with a breakpoint near the Ty elements of FS2. Using one primer that contained Ty1 sequences (Ty1-f: 5′-AAACGAATTCAGAGTTATTAGATGTGGATACATTGTGA) and one primer with telomere-related sequences (Telo-1-r: 5′-TAAAGCGGCCGCCGCGTCGACTAGTACCACCACACCCAC), we performed PCR using 50 ng of genomic DNA from PG297 or PG301, 35 pmol of each primer, 2.5 units of Taq DNA polymerase (Bioline), 200 μm each dNTP, 1.5 mm MgCl2, and 5 μl of 10× buffer. The PCR conditions were 94° for 2 min followed by 35 cycles at 94° for 30 sec, 60° for 30 sec, and 72° for 4 min. The resulting PCR products were separated by gel electrophoresis and telomeric bands were excised, purified, and sequenced using the primers Ty1-seq1 (5′-GACCAACCAGATGGATTGGC), Ty1-seq2 (5′-CCTGACTCAGGTGATGGAGTG), Ty1-seq3 (5′-GACCCAGGTAGGTAGGAATTGAG), or ARB2-nr (5′-GGCCACGCGTCGACTAGTAC). This strategy for determining the site of telomere addition was based partly on primers designed by Schmidt et al. (2006).
RESULTS
Description of the experimental system:
We previously showed that low levels of DNA polymerase α or δ result in elevated levels of genetic stability as monitored by the frequency of illegitimate mating (Lemoine et al. 2005, 2008). Mating between two MATα strains is usually the consequence of loss of function of the MATα locus from one of the two strains (Strathern et al. 1981). To analyze the various classes of genomic changes that lead to illegitimate mating, we used the system shown in Figure 2. The mating-type locus is located on the right arm of chromosome III. In one MATα haploid strain (the experimental strain), the level of α-DNA polymerase is regulated by a galactose-inducible promoter (GAL-POL1) and the left and right arms of chromosome III contain the wild-type HIS4 and THR4 alleles, respectively. The tester MATα strain has the his4 and thr4 mutant alleles. In our previous study (Lemoine et al. 2005), we showed that growth of the GAL-POL1 strain in medium containing 0.005% galactose and 2% raffinose (low galactose) resulted in very elevated (∼200-fold) levels of illegitimate mating compared to the same strain grown in medium with 0.05% galactose and 2% raffinose (high galactose); the levels of α-DNA polymerase in these two types of medium are ∼10% the wild-type level for the low galactose medium and threefold higher than the wild-type level for the high galactose medium (Lemoine et al. 2005).
Figure 2.—
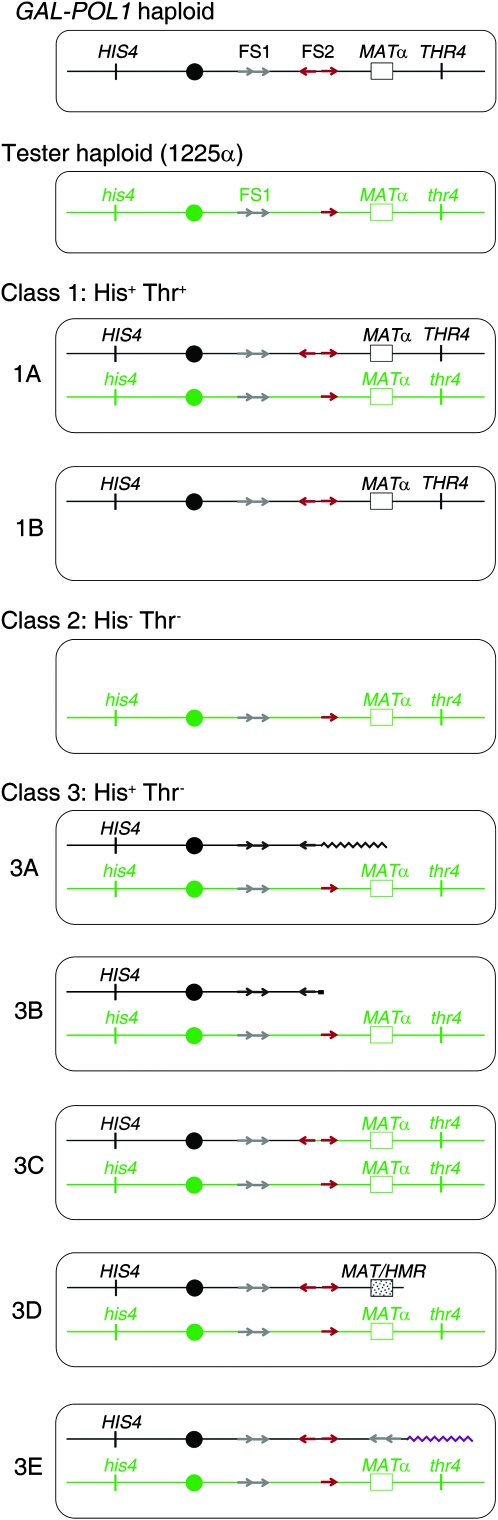
Classes of illegitimate diploids induced by low levels of DNA polymerase α. In our experiments, a GAL-POL1 MATα HIS4 THR4 haploid experimental strain was grown under conditions that result in low α-DNA polymerase. The strain was then mated to a tester strain (1225α) with the genotype MATα his4 thr4. Ty elements are shown as red (FS2) or gray (FS1) arrows, with the orientation of the arrow representing the orientation of the Ty element. On the basis of the phenotypes of the resulting diploids, they were classified as class 1 (His+ Thr+), class 2 (His− Thr−), or class 3 (His+ Thr−). Subsequent analysis showed that there were two types of class 1 events. Class 1A events were a consequence of fusions between two MATα strains without observed genomic changes; class 1B events were a consequence of loss of chromosome III from the tester strain. Class 2 events reflected loss of chromosome III from the experimental strain. The subclasses of class 3 were 3A (translocations with a breakpoint at FS1 or FS2 and at a Ty or δ-element on a nonhomologous chromosome), 3B (telomere-capped terminal deletion on the right arm of III), 3C (DSB on the right arm of III of the experimental strain, followed by repair from the homolog in the tester strain), 3D (deletion fusing MAT and HMR), and 3E (complex rearrangement with the FS2-centered palindrome described further in the text).
In our previous study and in our present study, we observed diploids that were His+ Thr+ (class 1), His− Thr− (class 2), and His+ Thr− (class 3). Class 2 and 3 events are clearly the result of genetic instability in the low polymerase haploid because these events involve loss of markers from chromosome III in the GAL-POL1 strain, but class 1 events may result from instability in either the GAL-POL1 strain or the tester strain. On the basis of further analysis of the diploids by CHEF gels, microarrays, and other physical methods, some of these phenotypic classes can be subdivided. In class 1A diploids, the two chromosomes appear to be identical to the chromosomes of the two MATα haploids. These diploids could represent rare fusions of two haploids of the same mating type without inactivation of MATα information in either haploid, a point mutation within the MATα locus, or a DSB in the tester strain repaired by a BIR event using the homolog derived from the GAL-POL1 strain. Class 1B strains have only a single copy of chromosome III (by CHEF analysis) and, therefore, represent loss of a homolog from the tester strain. Class 2 strains have a single copy of chromosome III derived from the tester and thus represent loss of III from the experimental strain.
Class 3 diploids represent a more diverse class of chromosome rearrangements. Class 3A strains contain translocations that have the left arm of III, the centromere of III, and a portion of the right arm of III fused to sequences of a nonhomologous chromosome. The breakpoint of the translocation on III is usually within the centromere-proximal Ty element of FS2 and the breakpoint on the nonhomologous chromosome is also within a Ty element. We interpret these events as reflecting a DSB at FS2 that was repaired by a BIR event involving an ectopically located Ty element. In class 3B strains, one chromosome has a deletion of the right arm of III that removes the MAT locus and distal sequences. In class 3C strains, the two chromosomes appear to be similar to the chromosomes of the parental haploid strains except the mutant thr4 marker is homozygous. Such strains likely reflect a DSB on III centromere-proximal to the mating-type locus of the experimental strain that was repaired by a BIR event using chromosome III of the tester strain after the mating. In class 3D strains, the chromosome derived from the experimental strain has an interstitial deletion that removes the MAT locus and the THR4 gene as a consequence of recombination between the MAT locus and the silent mating-type information at HMR; deletions of this type were first observed by Hawthorne (1963). In class 3E strains, there is a duplication of the region located between FS1 and FS2 and deletion of the region distal to FS2 with a translocation at the breakpoint to a nonhomologous chromosome arm. As is discussed in detail later, the class 3E rearrangements result from DNA replication across a persistent “hairpin” structure at FS2, generating an extended inverted duplication centered at FS2.
We previously showed that elevated levels of classes 1–3 required both of the Ty elements composing FS2 and low levels of α-DNA polymerase (Lemoine et al. 2005). We inferred that all of these events, therefore, were likely to reflect a structure-specific DSB formed at FS2 in strains with low levels of DNA polymerase, and different modes of repair of this DSB result in the different classes.
The frequency of illegitimate mating of cells with low levels of DNA polymerase α is not affected by mre11-H125N or mre11Δ:
In our previous studies, the GAL-POL1 experimental strain and the test strain were wild type for DNA repair/recombination functions. To determine what genes are required for the generation of DSBs at FS2 or their repair, we constructed GAL-POL1 strains with mutations in various repair/recombination genes, beginning with MRE11. Mre11p is required for processing of hairpins associated with palindromic and spaced IR sequences (Lobachev et al. 2002; Rattray 2004; Rattray et al. 2005; Farah et al. 2005; Lengsfeld et al. 2007; Cote and Lewis 2008). It is generally thought that cruciform extrusion at these sequences, followed by symmetrical cleavage by a Holliday junction resolvase, results in a pair of hairpin-capped DSBs (reviewed in Lewis and Cote 2006). Mre11p, which can cut small DNA loops (Trujillo and Sung 2001; Lengsfeld et al. 2007), then cleaves the tips of these hairpins to produce a free 3′ end available for repair. The mre11-H125N mutant is deficient specifically in Mre11p endonuclease activity (Moreau et al. 1999), and this mutant is phenotypically identical to the mre11 deletion in its failure to repair breaks at cruciforms and spaced IRs (Lobachev et al. 2002). Although the IR at FS2 is unlikely to extrude as a cruciform, given its large central spacer, it could form a hairpin on the lagging strand during DNA synthesis under conditions of low polymerase α, and the tip of this hairpin would be expected to be a substrate for Mre11p.
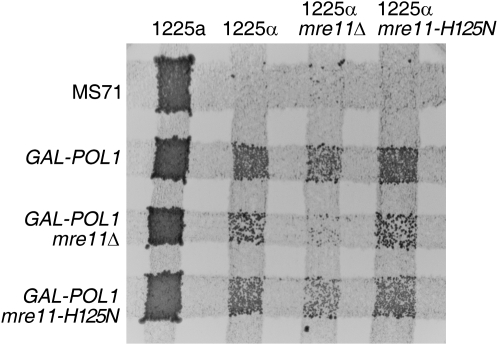
We created both mre11Δ and mre11-H125N mutants in our GAL-POL1 MATα haploid strain and examined instability at FS2 in these mutants under low DNA polymerase conditions. Illegitimate mating was used as a general test of genetic instability at FS2 in these mutants as shown in Figure 3. After pregrowth on low galactose, illegitimate mating by GAL-POL1 mre11-H125N haploids was not substantially different from that of GAL-POL1 haploids. Illegitimate mating was reduced in GAL-POL1 mre11Δ cells, however, particularly when the MATα tester strain also carries an mre11Δ mutation. Using a tester strain carrying the same mutation as the GAL-POL1 strain allows us to study not only the effect of the mutation on DSB formation at FS2, but also the effect of the mutation on repair of these breaks that can occur either before or after mating. We also observed that the viability of mre11Δ mutants was reduced in strains with low DNA polymerase α. After overnight growth in liquid medium with low galactose, only 9% of the GAL-POL1 mre11Δ haploids formed colonies, compared to 40 and 38% of GAL-POL1 mre11-H125N and GAL-POL1 haploids, respectively.
Figure 3.—
Illegitimate matings in strains with mre11 mutations: plate tests of legitimate and illegitimate mating. The MATα wild-type parent strain (MS71) and isogenic GAL-POL1, GAL-POL1 mre11Δ, and GAL-POL1 mre11-H125N (a mutation eliminating the endonuclease activity of Mre11p; Moreau et al. 1999) strains were streaked on medium containing low levels of galactose (0.005%) (resulting in low levels of DNA polymerase α) and grown overnight. These strains were then mated by replica plating to four tester strains: 1225 MATa, 1225 MATα, mre11Δ MATα, and mre11-H125N MATα. After the strains were allowed to mate overnight, they were replica plated to medium on which only diploids were capable of growth.
Since this loss of viability would be expected to affect the efficiency of illegitimate mating, we normalized the frequency of illegitimate mating to the efficiency of legitimate mating between cells of the same genotype. Cells were pregrown in low galactose and then mated to a MATα tester carrying the same mre11 mutation as the GAL-POL1 experimental strain. When corrected for viability and normalized to the frequency of legitimate mating, the average frequencies of illegitimate mating were similar in strains with and without Mre11p in the experimental strain (Table 1). We note, however, that the mre11Δ mutation in the MATα tester strain reduced illegitimate mating to approximately half that of the GAL-POL1 cells mated to a wild-type tester (Lemoine et al. 2005). Since cells lacking the Mre11p complex have increased sensitivity to DNA damaging agents, shortened telomeres, impaired DSB repair and checkpoint signaling, and increased chromosome loss (Tavassoli et al. 1995; Bressan et al. 1998; D'Amours and Jackson 2002; Krishna et al. 2007), it is likely that this inherent genetic instability in the mre11Δ tester strain may impair the ability of these cells to mate or to thrive after mating.
TABLE 1.
Illegitimate (α × α) mating of strains with low levels of α-DNA polymerase
| Experimental genotypea | Test mater genotypea | Frequency of illegitimate mating (×10−5)b | Frequency of class 1 (×10−5)c | Frequency of class 2 (×10−5)c | Frequency of class 3 (×10−5)c |
|---|---|---|---|---|---|
| GAL-POL1 | Wild type | 360 (270–680)d | 14 | 184 | 162 |
| GAL-POL1 | mre11Δ | 148 (132–165) | 73e | 31 | 44 |
| GAL-POL1 mre11Δ | mre11Δ | 156 (138–174) | 64f | 39 | 53 |
| GAL-POL1 | mre11-H125N | 223 (178–268) | 13 | 95 | 115 |
| GAL-POL1 mre11-H125N | mre11-H125N | 140 (115–165) | 6 | 60 | 74 |
| GAL-POL1 | sae2Δ | 164 (137–191) | 10 | 87 | 67 |
| GAL-POL1 sae2Δ | sae2Δ | 79 (59–99) | 8 | 17 | 54 |
| GAL-POL1 | rad52Δ | 170 (132–208) | 29 | 39 | 102 |
| GAL-POL1 rad52Δ | rad52Δ | 40 (35–46) | 14 | 25.5 | 0.5 |
All experimental strains were isogenic with MS71, a LEU2 derivative of AMY125 (MATα ade5-1 leu2-3 trp1-289 ura3-52 his7-2) (Kokoska et al. 2000), except for the GAL-POL1 gene and the indicated mutation. The mating-type tester strains are isogenic with 1225 (MATα his4-15 leu2 thr4 ura3-52 trp1 lys) except for the indicated mutation.
The frequency of illegitimate mating is corrected for viability and normalized to the level of legitimate mating. Numbers in parentheses indicate the 95% confidence interval from 8–10 different cultures.
Fifty to 100 independent illegitimate diploids were examined for each mating to determine the relative frequencies of classes 1, 2, and 3.
Values reported by Lemoine et al. (2005).
Includes class 1B events (defined in Figure 2), which occurred at a frequency of 30 × 10−5.
Includes class 1B events (defined in Figure 2), which occurred at a frequency of 11 × 10−5.
As described above, the phenotypes of the illegitimate diploids (His+/His−, Thr+/Thr−) can be used to divide them into three classes (Figure 2). Of 195 illegitimate diploids from mating of GAL-POL1 mre11-H125N cells to an mre11-H125N tester, classes 1, 2, and 3 were 4, 43, and 53%, respectively, which is a similar distribution to that seen for GAL-POL1 cells mated to a wild-type tester (Lemoine et al. 2005). These percentages were multiplied by the frequency of illegitimate mating to generate the data for classes 1, 2, and 3 shown in Table 1. When either the GAL-POL1 haploid or the GAL-POL1 mre11Δ haploid was mated to the mre11Δ tester, the frequency of class 1 events was substantially elevated relative to the other classes (Table 1).
In our previous studies mating the GAL-POL1 haploid to a wild-type tester, most class 1 diploids (His+ Thr+) had two normal-sized copies of chromosome III and could represent rare fusions between MATα strains, point mutations in MATα, or a DSB centromere-proximal to MAT in the tester strain that is repaired by BIR off the GAL-POL1 chromosome III homolog. Since class 1 diploids do not sporulate and mate as MATα strains (Lemoine et al. 2005), these diploids are not formed by mating-type switching. We sequenced the MAT locus in six class 1 strains derived from illegitimate mating between GAL-POL1 haploids and a wild-type tester (DAMC590 to DAMC595). Sequencing results indicated six polymorphisms in this region between the two parent haploids (File S1 and Table S2). Of the six illegitimate diploids sequenced, five had both sequences derived from the parental haploids. Thus, these diploids appear to reflect rare fusions of MATα haploids rather than point mutations inactivating the MATα locus. One illegitimate diploid, DAMC593, was homozygous for all polymorphisms within the MATα locus that were derived from the GAL-POL1 haploid. Since CHEF gel analysis indicated that this illegitimate diploid contains two normal-sized chromosome IIIs, it is likely that the DAMC593 strain was the result of a DSB centromere-proximal to the MAT locus in the tester that was repaired by BIR using the GAL-POL1 chromosome III as a template.
Although class 1 events after mating to a wild-type tester are primarily rare fusions of MATα haploids, our use of tester strains carrying nuclease mutations could potentially increase class 1 events resulting from instability of chromosome III in the tester haploid. For example, mre11Δ strains have been previously reported to have elevated chromosome loss (Bressan et al. 1998; Krishna et al. 2007). As noted above, the frequency of class 1 events was elevated in our analyses using the mre11Δ tester (Table 1). By quantitating the level of chromosome III vs. other chromosomes in CHEF gels, we found that 12 of 15 class 1 illegitimate diploids derived from a cross of GAL-POL1 mre11Δ cells to the mre11Δ tester had only a single copy of III, consistent with a high rate of chromosome loss in the tester (class 1B). Similarly, 10 of 17 class 1 illegitimate diploids from GAL-POL1 cells mated to the mre11Δ were class 1B strains. In contrast, of 17 class 1 illegitimate diploids from GAL-POL1 haploids mated to a wild-type tester strain, only one was haploid for III. The relative increase in class 1 diploids was observed only in matings using the mre11Δ tester and not the mre11-H125N tester. Thus, loss of chromosomes from the mre11Δ tester strain is not a consequence of loss of the nuclease activity of Mre11p.
DSB formation at FS2 resulting from low polymerase α is not significantly reduced in mre11 mutants and in several other nuclease-deficient mutants:
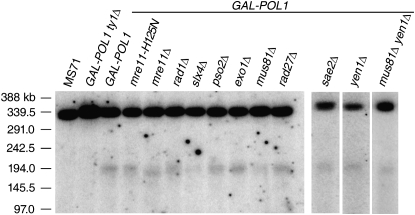
Since the majority of the DSBs on chromosome III in cells with low DNA polymerase α are at FS2 (Lemoine et al. 2005), we measured DSBs at FS2 in mre11 mutant strains. Each strain was grown in high-galactose medium overnight and then incubated in medium with no galactose for 6 hr. We subsequently isolated genomic DNA and separated the chromosomes by CHEF gel electrophoresis, followed by Southern blotting with a probe on the left arm of chromosome III. We observed a DNA molecule of ∼180 kb, the expected size for chromosome III broken at FS2, in the GAL-POL1, GAL-POL1 mre11-H125N, and GAL-POL1 mre11Δ cells (Figure 4). In all three of these strains, ∼7% of the cells had a DSB at FS2 under these conditions. This fragment was not present in an isogenic wild-type strain (MS71) or in GAL-POL1 cells with a deletion of the centromere-proximal Ty1 of FS2. The observation that Mre11p is not required for creating the DSB at FS2 is consistent with our observation that the frequency of illegitimate mating is relatively unaffected in mre11 strains. We note that the broken III molecule in the mre11Δ strain appears smaller in comparison to the other strains (Figure 4). This altered migration could potentially be due to a difference in either telomere length or end resection of the DSB at FS2. Strains with the mre11Δ mutation have short telomeres (Moreau et al. 1999). Also, it has been shown that Mre11p and Sae2p initiate end resection at induced DSBs, and although DSBs can still be resected in the absence of these proteins, this resection occurs more slowly (reviewed in Mimitou and Symington 2009). The mre11-H125N mutation does not affect either telomere length or DSB end resection (Moreau et al. 1999).
Figure 4.—
Physical analysis of DSB formation at FS2 in strains deficient for various nucleases. All strains were grown overnight in high galactose medium and then washed in water and resuspended in medium lacking galactose for 6 hr. DNA was extracted and chromosomal DNA molecules were separated by gel electrophoresis as described in materials and methods. The separated molecules were examined by Southern analysis, using a probe derived from the left end of III. The ratio of chromosome III molecules broken at FS2 (180-kb fragment) vs. the intact III (330 kb) was quantitated using a PhosphoImager. MS71 is a wild-type haploid strain, and GAL-POL1 ty1Δ is isogenic with the GAL-POL1 strain except that it lacks one of the two Ty1 elements that compose FS2 (Lemoine et al. 2005).
We also examined DSBs at FS2 in strains lacking various other nucleases (reviewed by Friedberg et al. 2006; Mimitou and Symington 2009; Rouse 2009) in the GAL-POL1 background, including Exo1p (5′–3′ exonuclease and flap endonuclease), Mus81p (one subunit of a heterodimeric structure-specific nuclease), Pso2p (5′–3′ exonuclease), Rad1p (single-stranded endonuclease), Rad27p (5′–3′ exonuclease, 5′ flap endonuclease), Sae2p (single-stranded exonuclease), Slx4 (5′ flap endonuclease), and Yen1p (a Holliday junction-cleaving enzyme). Each mutant strain was analyzed for FS2-associated DSBs as described above. None of these nuclease mutants eliminated DSB formation at FS2. The ratio of DSB in each mutant strain to DSB in the GAL-POL1 strain (normalized to the amount of intact chromosome III) and the 95% confidence limits, based on at least four measurements for each strain (except rad52), were as follows: mre11-H125N, 0.95 ± 0.16; mre11Δ, 1.37 ± 0.31; sae2, 1.07 ± 0.38; rad1, 0.58 ± 0.42; slx4, 0.57 ± 0.44; pso2, 1.12 ± 0.61; exo1, 1.15 ± 0.8; mus81, 1.04 ± 0.76; rad27, 0.94 ± 0.32; yen1, 1.58 ± 2.13; mus81 yen1, 0.77 ± 0.24; and rad52, 1.14. The ratio of DSBs in the rad52 mutant was measured only once on a gel containing the GAL-POL1 strain for comparison, but FS2-associated DSBs were also observed for this mutant in three other gels.
Mre11p is not required for the formation of FS2-associated translocations:
We next investigated the various types of class 3 events (His+ Thr−) in GAL-POL1-H125N and GAL-POL1 mre11Δ cells. In our previous analysis of GAL-POL1 cells, the most common types of class 3 events are class 3A (BIR resulting in nonreciprocal translocations), class 3B (terminal deletions), and class 3C (BIR events involving homologous chromosomes) (Lemoine et al. 2005). To subdivide the class 3 events in our present study, we first examined the sizes of chromosome III by CHEF gel electrophoresis, followed by Southern analysis using CHA1 (a gene located on the left arm of III) as a hybridization probe. Illegitimate His+ Thr− diploids with two normal-sized copies of III were classified as 3C. Those strains with one normal III and one III of altered size were classified as either 3B or 3A, depending on the size of the alteration. The strains with an altered III of either 150 or 180 kb were considered class 3B, since they are the size expected for a telomere-capped break at FS1 or FS2, respectively; chromosome IIIs of any other size class were considered class 3A.
In GAL1-POL1 illegitimate diploids, we previously reported a ratio of 3:1:4 for subclasses 3A:3B:3C (Lemoine et al. 2005). We did not observe any substantial deviations from this ratio in our analysis of GAL-POL1 cells containing mutations in Mre11p. For GAL-POL1 mre11-H125N cells mated to the mre11-H125N tester, the ratio was 17:1:8 (P = 0.31). For GAL-POL1 mre11Δ cells mated to a wild-type tester, the ratio was 7:0:8 (P = 0.59), and when they were mated to an mre11Δ mutant tester, the ratio was 10:3:12 (P = 0.99). These data indicate that in all cases, the predominant classes are those that result from repair by BIR (classes 3A and 3C).
Chromosome rearrangements in illegitimate diploids generated by mating GAL-POL1 mre11Δ cells to the mre11Δ tester strain:
We confirmed our classifications using DNA microarrays. Previously, we showed that class 3A events result in nonreciprocal translocations in which one breakpoint is in the centromere-proximal Ty of FS2 or one of the two Ty elements of FS1 and the other breakpoint is in a Ty element or a δ-element of a nonhomologous chromosome. Since the frequencies of translocations involving FS1 and FS2 are dependent on the FS2 pair of Ty elements (Lemoine et al. 2005), we suggested that these recombination events are initiated by a DSB that occurs between the Ty elements of FS2 as a consequence of hairpin formation. If the broken molecule is processed to a very limited extent, the Ty of FS2 can undergo a BIR event with a Ty element located on a nonhomologous chromosome, producing the translocation. If the broken chromosome is processed more extensively, then one of the Ty elements of FS1 can initiate the BIR event. The exposed Ty element then initiates a BIR event with a Ty or a δ-element on a nonhomologous chromosome (Figure 5). Almost all of the observed translocations involve Ty elements oriented in such a way that BIR produces a monocentric translocation. Presumably, BIR events that produce dicentric chromosomes or acentric fragments also occur but are selected against during the growth of cells containing the rearrangement. Class 3A events can be diagnosed by DNA microarrays because they result in a deletion of sequences from the right arm of III with a breakpoint in FS1 or FS2 and a duplication of sequences on a nonhomologous chromosome with a breakpoint at a mapped Ty or δ-element (Figure 5A). For some chromosome rearrangements, we used other techniques (Southern analysis or PCR) to confirm breakpoints (Figure 5C).
Figure 5.—
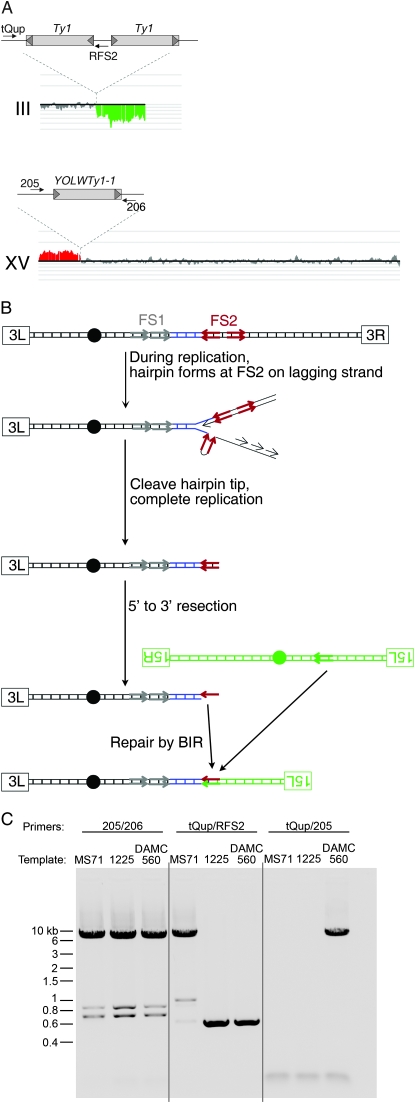
Physical analysis of a translocation produced by a BIR event between nonallelic Ty elements in an mre11Δ/ mre11Δ illegitimate diploid. (A) Microarray analysis. DNA was isolated from a class 3 illegitimate diploid (DAMC560) resulting from the mating of two MATα mre11Δ strains. This sample was labeled with a Cy5 fluorescent nucleotide and mixed with a control DNA sample labeled with a Cy3 fluorescent nucleotide, and this mixture was used a hybridization probe of a microarray containing all of the yeast ORFs and intergenic regions. The ratios of hybridization are indicated as vertical lines with deletions and additions in the experimental strain shown in green and red, respectively (analysis by the CGH-Miner program). No changes were observed on chromosomes other than III and XV. The deletion breakpoint on III is at FS2, and the amplification breakpoint on XV is at YOLWTy1-1. Large gray rectangles represent Ty elements, short gray arrowheads show δ-elements, and small black arrows represent PCR primers. (B) Mechanism for generating the III–XV translocation by BIR. Centromeres are indicated by black circles, left and right telomeres are identified by labeled rectangles, and Ty elements are indicated by arrows. (C) Confirmation of translocation by PCR. The positions of the primers are shown in A. MS71 is the wild-type parental haploid from which all GAL-POL1 experimental strains are derived, and 1225 is the wild-type parental haploid from which all mating-type tester strains are derived. MS71 has the centromere-distal Ty at FS2 that 1225 lacks. As expected, PCR using primers from III (tQup) and XV (205) generates a product when DNA from the strain with the III–XV translocation is used as a template.
In four of the five class 3A illegitimate diploids we examined resulting from mating MATα GAL-POL1 mre11Δ cells to a MATα mre11Δ tester, there was a deletion of chromosome III with a breakpoint at FS1 or FS2 as well as amplification of another chromosome arm with a breakpoint at a Ty1 or a Ty2 element tester (Table 2 and File S1). In these four strains, the altered chromosome had the size expected for a BIR-mediated translocation (File S1). The complete analysis of one of these diploids (DAMC560) is shown in Figure 5. In one diploid (DAMC553), we observed a deletion of sequences distal to FS1, but no amplification. The observed chromosome size in this strain was ∼240 kb, considerably larger that that expected for a simple deletion (class 3B). We did not attempt to characterize this rearrangement further.
TABLE 2.
Chromosome III rearrangements in class 3 illegitimate diploids
| Illegitimate diploid | Relevant genotypea | Subclass | Altered chromosome size (kb)b | Chromosome III alteration | Other chromosome alteration | Interpretationc |
|---|---|---|---|---|---|---|
| DAMC473 |
mre11-H125N mre11-H125N |
3A | 436 | Deletion of sequences distal to FS1 | Amplification of right arm of X distal to YJRWTy1-2 | BIR event |
| DAMC474 |
mre11-H125N mre11-H125N |
3A | 300 | Deletion of sequences distal to FS2 | Amplification of right arm of V distal to YERCTy1-1 | BIR event |
| DAMC475 |
mre11-H125N mre11-H125N |
3A | 700 | Deletion of sequences distal to FS1 | Amplification of right arm of II distal toYBRWTy1-2 | BIR event |
| DAMC476 |
mre11-H125N mre11-H125N |
3E | 776 | Amplification between FS1 and FS2; deletion of sequences distal to FS2 | Amplification of left arm of XIV distal to YNLWTy1-2 | Palindromic amplification between FS1 and FS2 along with BIR event |
| DAMC479 |
mre11-H125N mre11-H125N |
3A | 250 | Deletion of sequences distal to FS1 | Amplification of right arm of I distal to YARWdelta7 | BIR event |
| DAMC483 |
mre11-H125N mre11-H125N |
3A | 850 | Deletion of sequences distal to YCLWTy2-1 | Deletion of left arm of II distal to YBLWTy2-1 | Half crossover; repair by SSA of breaks at or near the repetitive elements involved |
| DAMC484 |
mre11-H125N mre11-H125N |
3A | 813 | Deletion of sequences distal to FS2 | Amplification of right arm of IV distal to YDRCTy1-2 | BIR event |
| DAMC485 |
mre11-H125N mre11-H125N |
3B | 194 | Deletion of sequences distal to FS2 | None detected by genomic microarray | Telomere-capped break |
| DAMC495 |
mre11-H125N mre11-H125N |
3A | 194 | Deletion of sequences distal to FS1 | Amplification of left arm of II distal to YBLWTy2-1 | III/II translocation confirmed; wrong orientation of Ty elements for BIR |
| DAMC551 |
mre11Δ mre11Δ |
3A | 2000 | Deletion of sequences distal to FS2 | Amplification of right arm of XII distal to rDNA array | BIR event, assuming Ty element present within rDNA array |
| DAMC552 |
mre11Δ mre11Δ |
3A | 388 | Deletion of sequences distal to FS2 | Amplification of left arm of II distal to Watson Ty element near YBLCdelta7 (not in sequenced strain) | BIR event |
| DAMC553 |
mre11Δ mre11Δ |
3A | 242 | Deletion of sequences distal to FS1 | None detected by genomic microarray | Uncharacterized rearrangement; chromosome size inconsistent with telomere capping at FS1 |
| DAMC555 |
mre11Δ mre11Δ |
3B | 194 | Deletion of sequences distal to FS2 | None detected by genomic microarray | Telomere-capped break |
| DAMC560 |
mre11Δ mre11Δ |
3A | 300 | Deletion of sequences distal to FS2 | Amplification of left arm of XV distal to YOLWTy1-1 | BIR event |
| DAMC561 |
mre11Δ mre11Δ |
3A | 700 | Deletion of sequences distal to FS1 | Amplification of right arm of II distal to YBRWTy1-2 | BIR event |
| DAMC536 |
sae2Δ sae2Δ |
3A | 300 | Deletion of sequences distal to FS2 | Amplification of right arm of V distal to YERCTy1-1 | BIR event |
| DAMC539 |
sae2Δ sae2Δ |
3A | 400 | Deletion of sequences distal to FS2 | Amplification of left arm of II distal to Watson Ty element near YBLCdelta7 (not in sequenced strain) | BIR event |
| DAMC547 |
sae2Δ sae2Δ |
3D | 250 | Deletion of sequences between MAT and HMR | None | Unequal crossover or SSA (Hawthorne deletion) |
| DAMC549 |
sae2Δ sae2Δ |
3A | 250 | Deletion of sequences distal to FS1 | Amplification of left arm of VII distal to Crick Ty element near YGRWdelta3 (not in sequenced strain) | BIR event |
| DAMC550 |
sae2Δ sae2Δ |
3A | 440 | Amplification between YCRCdelta6 and FS1; deletion of sequences distal to FS2 | Amplification of right arm of X distal to YJRWTy1-1 | Palindromic amplification between YCRCdelta6 and FS1 plus a BIR event |
| PG270 |
rad52Δ rad52Δ |
3A | 654 | Deletion of all III sequences except those centromere-distal to YCLWTy2-1 | Deletion of right arm of VII distal to YGRCTy2-1 | Half crossover, repair by SSA of breaks at or near the repetitive elements involved |
| PG271 |
rad52Δ rad52Δ |
3D | 243 | Deletion between MAT and HMR | None | Unequal crossover or SSA (Hawthorne deletion) |
| PG272 |
rad52Δ rad52Δ |
3A | 218 | Deletion of sequences distal to YCR034W | None | Size is consistent with repair by telomere addition |
| PG273 |
rad52Δ rad52Δ |
3A | 218 | Deletion of sequences distal to FS1 | Deletion of left arm, centromere, and right arm of I up to YARWdelta6 | Half crossover, repair by SSA of breaks at or near the repetitive elements involved |
| PG282 |
rad52Δ rad52Δ |
3A | 218 | Deletion of sequences distal to FS2 | By genomic array, no other changes. By band array, left arm of II at YBLWTy2-1 is attached to the broken III | Half crossover, repair by SSA of breaks at or near the repetitive elements involved |
| PG283 |
rad52Δ rad52Δ |
3A | 267 | Amplification of sequences distal to YCLWTy2-1 and deletion of sequences distal to FS2 | None | BIR |
| PG284 |
rad52Δ rad52Δ |
3A | 230 | Deletion of sequences distal to FS2 | Deletion of left arm, centromere, and right arm of I up to YARCdelta8 | Half crossover, repair by SSA of breaks at or near the repetitive elements involved |
| PG286 |
rad52Δ rad52Δ |
3A | n/a | Monosomic for chromosome III | None | Repair by SSA of a break on III (centromere-proximal to FS2) in both the GAL-POL1 haploid and the tester strain |
| PG297 |
rad52Δ rad52Δ |
3B | 194 | Deletion of sequences distal to FS2 | None detected by genomic microarray | Telomere-capped break |
| PG298 |
rad52Δ rad52Δ |
3A | 218 | Deletion of all III sequences except those centromere-distal to YCLWTy2-1 | Deletion of left arm of VI distal to YFLWTy2-1 | Half crossover, repair by SSA of breaks at or near the repetitive elements involved |
| PG300 |
rad52Δ rad52Δ |
3A | 557 | Deletion of sequences distal to FS2 | Deletion of left arm, centromere, and right arm of XII up to YLRCdelta18 | Half crossover, repair by SSA of breaks at or near the repetitive elements involved |
| PG301 |
rad52Δ rad52Δ |
3B | 194 | Deletion of sequences distal to FS2 | None detected by genomic microarray | Telomere-capped break |
Only the mutations that differ from the progenitor GAL-POL1 strain and the mating-type tester are noted. All strains were derived from MS71 or 1225, as described in the Table 1 legend.
The approximate sizes of the altered chromosome were estimated from a CHEF gel separation of chromosomes, followed by Southern blotting with a probe on the left arm of chromosome III.
Details of the analysis of each illegitimate diploid are in File S1. All of the listed illegitimate diploids were analyzed by CHEF gel and microarray analysis; some were further examined by PCR and Southern analysis.
Chromosome rearrangements in illegitimate diploids generated by mating GAL-POL1 mre11-H125N cells to the mre11-H125N tester strain:
Eight independent class 3A illegitimate diploids were examined. The summary of this analysis is in Table 2 and the details of the analysis for each strain are in File S1. Five of the eight strains had the most common pattern observed in previous studies (Lemoine et al. 2005, 2008), deletion of chromosome III sequences beginning at FS1 or FS2 and amplification of sequences from a different homolog with a breakpoint in a Ty element. The three illegitimate diploids that did not fit this pattern were DAMC495, DAMC483, and DAMC476. The DAMC495 strain had a deletion of sequences distal to FS1 on chromosome III and amplification of sequences distal to YBLWTy2-1 on chromosome II. The Ty elements were, however, in the wrong orientation to produce a monocentric translocation by BIR. One explanation of this result is that the yeast strains used in this study contained an unannotated Ty or δ; this rearrangement was not further analyzed. The DAMC483 diploid had two deletions. One deletion removed all of the sequences of chromosome III except those distal to YCLWTy2-1; this region of III has a cluster of transposable elements and was called the left arm hotspot (LAHS) by Warmington et al. (1986). The second deletion removed the sequences on chromosome II distal to YBLWTy2-1. Southern analysis of this illegitimate diploid was consistent with the repair of two DSBs within these two Ty2 elements by single-strand annealing, resulting in a II–III translocation (File S1).
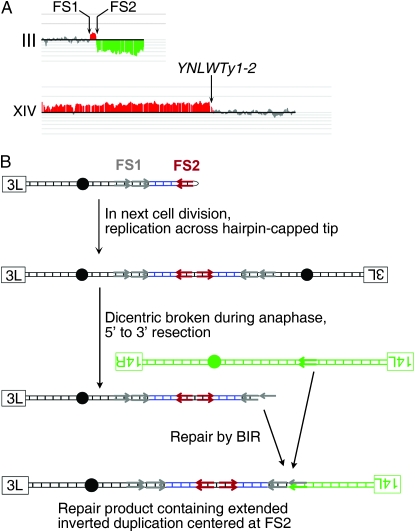
If the endonuclease function of Mre11p is required to cleave a hairpin-capped break at FS2, then we would expect that, in the absence of this protein, replication across hairpin-capped DSBs at FS2 would result in the formation of repair products with an extended inverted repeat centered at FS2, in a mechanism similar to that at the inverted Alu elements described by Narayanan et al. (2006). One of the mre11-H125N/mre11-H125N illegitimate diploids (DMAC476) had this pattern. This strain had a deletion on chromosome III of sequences distal to FS2, a duplication of chromosome III sequences between FS1 and FS2, and a duplication of chromosome XIV sequences distal to YNLWTy1-2 (Figure 6A). Restriction digest mapping and Southern blot analysis of this illegitimate diploid determined that it had a palindromic amplification centered at FS2 and a translocation between the duplicated copy of FS1 centromere-distal to FS2 and YNLWTy1-2 on chromosome XIV. This rearrangement is consistent with the formation of a dicentric chromosome centered at FS2 as a repair intermediate that is broken in or near FS1, followed by BIR-mediated repair of the broken end using a Ty element on chromosome XIV (Figure 6B and File S1). Although this chromosome is consistent with what we expect if the nuclease activity of Mre11p was required to process the spacer of the FS2-associated hairpin, several points should be emphasized. First, we found only one such rearrangement among 13 class 3A events in diploids homozygous for mre11-H125N or mre11Δ, indicating the cleavage of the associated hairpin is efficient in the absence of the Mre11p nuclease. Second, we previously observed a similar chromosome rearrangement in a wild-type GAL-POL1 strain (Lemoine et al. 2005). In summary, the Mre11p endonuclease activity is not required to form the same types of translocations as observed in the wild-type strain.
Figure 6.—
Analysis of a chromosome rearrangement with a 20-kb palindrome centered on FS2 in an mre11−H125N/mre11−H125N illegitimate diploid (DAMC476). (A) Microarray analysis. The analysis was performed as described in Figure 5. On chromosome III, there was a deletion distal to FS2 and a duplication of the sequences between FS1 and FS2. The region of chromosome XIV distal to YNLWTy1-2 was duplicated. Southern analysis (described in File S1) indicated the presence of a chromosome with a large palindrome centered on FS2. (B) Mechanism for generating a rearranged chromosome with an extended palindrome. Centromeres are indicated by black circles, left and right telomeres by labeled rectangles, and Ty elements by arrows.
We also used microarrays to analyze two illegitimate mre11-H125N/mre11-H125N diploids (DAMC485 and DAMC555) assigned to class 3B by CHEF and Southern analysis. This analysis confirmed that these diploids had a deletion of the sequences distal to FS2 on chromosome III with no additional changes (File S1).
Chromosome rearrangements in illegitimate diploids generated by mating GAL-POL1 mre11-H125N or GAL-POL1 mre11Δ cells to a MRE11 tester strain:
We also analyzed 12 class 3A illegitimate diploids derived from mating GAL-POL1 mre11-H125N cells to a wild-type tester. All but one of these diploids had chromosome rearrangements consistent with the BIR event illustrated in Figure 5 (File S1). The exceptional diploid (DAMC461) had a chromosome III with a deletion of sequences distal to FS1 and an amplification of a 190-kb internal segment of VII. The size of the translocation was not consistent with a simple addition of the chromosome VII segment to the truncated chromosome III, and the rearranged chromosome was not further characterized (File S1). We also analyzed four class 3A illegitimate diploids resulting from mating GAL-POL1 mre11Δ cells to a wild-type tester. Three had the typical type of translocation of class 3A diploids, and one (DAMC461) had a complex chromosome rearrangement (File S1).
Sae2p is not required for the formation of FS2-associated translocations:
Sae2p is a nuclease that cooperates with Mre11p in processing of short palindromes in vitro (Lengsfeld et al. 2007). This protein is also involved in the repair of breaks at palindromes and spaced IRs (Lobachev et al. 2002; Rattray 2004; Rattray et al. 2005; Cote and Lewis 2008). Recombinational repair at the Alu-IR generated by Lobachev et al. (2002) is equally defective in mre11Δ, mre11-H125N, and sae2Δ cells, and extended inverted duplications centered at the IR were observed in all of these mutants. We examined illegitimate mating, DSB formation at FS2, and chromosome rearrangements in illegitimate diploids in sae2Δ strains. As noted above, the amount of FS2-associated DSBs in GAL-POL1 sae2Δ cells after 6 hr growth in medium with no galactose was not appreciably different from that in cells with Sae2p (Figure 4).
The frequency of illegitimate diploids was reduced about twofold when both the experimental and the tester strain had the sae2Δ mutation (Table 1). Most of this reduction was in class 2 diploids, representing chromosome loss. Although the reason for this reduction is not clear, it is possible that sae2Δ haploids lacking chromosome III are less capable of being rescued by mating than SAE2 strains. Unlike the mre11Δ tester strain, the sae2Δ mutation in the tester strain did not elevate the frequency of class 1 illegitimate diploids compared to crosses with a wild-type tester, indicating that genome instability in the sae2Δ tester does not substantially contribute to illegitimate mating. As before, we subdivided the class 3 illegitimate diploids by CHEF gel analysis followed by Southern blotting. Of 18 strains examined, the ratio of class 3A:3B:3C was 9:1:8, similar to that observed for the wild-type and mre11 strains.
We analyzed five class 3A strains by microarrays (Table 2 and File S1). Three strains (DAMC536, DAMC539, and DAMC549) had nonreciprocal translocations with one breakpoint at FS1 and FS2 and a second within a Ty element on a nonhomologous chromosome. The strain DAMC547 had a deletion on the right arm of chromosome III between the MAT locus and HMR, likely as a result of unequal crossing over or single-strand annealing between these loci. The last illegitimate diploid, DAMC550, had a deletion of chromosome III distal to FS2, a duplication of chromosome III between YCRCdelta6 and FS1, and a duplication on chromosome X distal to the tandem pair of Ty1 elements YJRWTy1-1 and YJRWTy1-2. This set of alterations is consistent with a mechanism described by Vanhulle et al. (2007) in which a DSB at or distal to FS2 is extensively processed, allowing for pairing between the Ty elements of FS2 and FS1. DNA replication across this intermediate leads to the formation of a dicentric chromosome with an inverted duplication around FS1. This dicentric is broken during anaphase and repaired by BIR (Figure S1). Using restriction digest mapping, we confirmed there is an extended inverted duplication around FS1 in this illegitimate diploid (File S1). Although the frequency of illegitimate mating is slightly reduced in sae2Δ mutants, classes 3A and 3C remain the largest category of illegitimate diploids, indicating that repair of FS2-associated DSBs by BIR is independent of Sae2p.
Most chromosome rearrangements requiring BIR-mediated repair of a DSB at FS2 or FS1 are dependent on Rad52p:
We monitored the frequencies of various classes of illegitimate diploids generated by mating GAL-POL1 strains to a rad52Δ tester and by mating GAL-POL1 rad52Δ strains to a rad52Δ tester (Table 1). The frequencies of class 1 and class 2 diploids were not substantially affected by the rad52Δ mutation (Table 1). This result is expected since the formation of these classes does not require homologous recombination. In contrast, the frequency of class 3 events was reduced ∼100-fold in the diploids formed by mating GAL-POL1 rad52Δ strains to a rad52Δ tester. Since most class 3 events reflect BIR (class 3A) or repair by recombination with the homolog (class 3B), events that require Rad52p (Symington 2002; Davis and Symington 2004), this result is also not surprising.
CHEF gel separation of chromosomes, Southern blotting, and microarrays were used to analyze 12 class 3 illegitimate diploids from rad52Δ strains. Five of these illegitimate diploids contained unusual rearrangements in which the repair product is consistent with single-strand annealing between the broken chromosome III and another broken chromosome, resulting in a “half-crossover” translocation (Haber and Hearn 1985; Smith et al. 2009). File S1 and Figure S2 contain a detailed discussion of the repair events in these illegitimate diploids. We also found two illegitimate diploids that contained deletions that appeared to reflect a DSB near FS2, followed by telomere addition. We used PCR to amplify the deletion breakpoint and determined the exact location of telomere additions in these two strains. Both breakpoints were located within the centromere-proximal Ty1 of FS2 near the spacer that separates the inverted pair of Ty elements (File S1 and Figure S3).
DISCUSSION
As described in the Introduction, palindromic sequences are associated with genetic instability in bacteria, yeast, and mammalian cells. We previously described a naturally occurring fragile site (FS2) composed of an inverted pair of Ty elements separated by a 280-bp spacer (Lemoine et al. 2005, 2008). In the current study, we contrast the regulation of genetic instability of FS2 from that reported for perfect palindromes or IRs with short spacers.
Formation of cruciform and hairpin secondary structures:
Palindromes and IRs have been proposed to form two types of structures, cruciforms (Figure 1, left) and hairpins (Figure 1, right). Physical evidence for the existence of cruciforms has been obtained from in vitro and in vivo studies of plasmids and phage carrying perfect palindromes or IRs separated by <10 bp, but not from plasmids carrying IRs separated by ≥20 bp (Sinden et al. 1983, 1991; Zheng et al. 1991; Allers and Leach 1995; Kogo et al. 2007). However, IRs with large spacers can form hairpins on single-stranded DNA (Figure 1, right), such as within the Okazaki fragment initiation zone on the lagging strand during DNA replication (Trinh and Sinden 1991; Voineagu et al. 2008). The long central spacer at FS2 makes it likely that the recombinogenic structure formed by this sequence is a hairpin. In addition, the elevated rate of instability of spaced IRs (including FS2) under conditions of perturbed DNA replication is more consistent with hairpin formation than with cruciform formation (Gordenin et al. 1992; Lemoine et al. 2005, 2008). It should be noted that hairpin formation, rather than cruciform formation, has also been observed in E. coli for a 111-bp IR interrupted by a 24-bp central spacer (Eykelenboom et al. 2008).
DSB formation and homologous recombination at cruciform and hairpin structures:
Cleavage of a cruciform by a Holliday junction resolvase would be predicted to yield two broken molecules with hairpin-capped ends (Figure 1A). In certain mutant yeast strains, Cote and Lewis (2008) reported such products in plasmids containing perfect palindromes, and Lobachev et al. (2002) observed these products at an IR consisting of a pair of inverted Alu elements separated by 12 bp. Thus, it has been proposed that there are two steps involved in the processing of cruciforms (Lobachev et al. 2002; Cote and Lewis 2008): (1) cleavage by a Holliday junction-like resolvase resulting in two hairpin-capped DSBs and (2) cleavage of the hairpin tips to generate a free 3′ end for repair by homologous recombination (Figure 1A).
Cote and Lewis (2008) reported that the first step, DSB formation, was independent of Mre11p and Sae2p but dependent on Mus81p. Mus81p acting with Mms4p has the ability to cleave certain types of branched DNA structures, although its activity in vitro is different from that of a classic resolvase (reviewed by Mimitou and Symington 2009). In contrast to the results of the Lewis lab, DSB formation at the Alu-IR was independent of Mus81p, Mre11p, Sae2p, and also Yen1p (Lobachev et al. 2002; K. Lobachev, personal communication). Yen1p has the in vitro enzymatic activity expected for a Holliday junction resolvase (Ip et al. 2008). In agreement with the Lobachev lab, we found that DSB formation at FS2 is independent of all tested nucleases including Mre11p, Sae2p, Rad1p, Slx4p, Pso2p, Exo1p, Mus81p, Rad27p, and Yen1p. Thus, either the DSBs at FS2 are generated by an as-yet untested nuclease or these enzymes might act on the FS2-associated secondary structure in a functionally redundant manner.
Both Lobachev et al. (2002) and Cote and Lewis (2008) report that the second step of cruciform processing, cleavage of the hairpin tips to facilitate homologous recombination, is dependent on Sae2p and Mre11p. These results are consistent with other in vivo and in vitro studies (Rattray et al. 2001; Trujillo and Sung 2001; Farah et al. 2002; Lengsfeld et al. 2007). In mre11Δ and sae2Δ mutants, the hairpin tips persist and subsequent replication across the tip results in the formation of an extended inverted duplication centered at the site of the original palindrome or IR (Narayanan et al. 2006; Cote and Lewis 2008). A dicentric chromosome formed by this process could lead to chromosome rearrangements such as that shown in Figure 6. In yeast strains with a palindrome with a short spacer, such rearrangements are common (Narayanan et al. 2006). However, in the current study, we found only a single example of this type of repair product from mre11 or sae2 nuclease mutants. These results indicate that the two-step pathway of DSB formation and processing described for perfect palindromes and IRs with small spacers is unlikely to be the major pathway of DSB formation at FS2.
A hairpin structure at FS2 could be processed by endonucleases that cleave at the base of the hairpin (Figure 1, B and D) or by an endonuclease that cleaves the 280-bp single-stranded loop at the hairpin tip (Figure 1C). Processing by endonucleases as shown in Figure 1B is inconsistent with the observation that most of the chromosome rearrangements have a breakpoint in the centromere-proximal Ty of FS2. Processing of the FS2-associated hairpin as shown in Figure 1D followed by replication through the hairpin following cleavage would generate a centromere-containing chromosome with a DSB near the centromere-distal Ty of FS2. This Ty has not been observed at the breakpoint of rearrangements in our studies. We suggest that FS2-associated DSBs are generated by a single-step pathway as shown in Figure 1C, independent of Mre11p and Sae2p. This process would result in a centromere-containing broken chromosome with the centromere-proximal Ty element of FS2 at the end. Consistent with this model, most of the chromosome rearrangements that we mapped have this Ty element at the breakpoint, and the two terminal deletions that we analyzed were within this Ty element.
Chromosome rearrangements associated with the DSB at FS2:
In this study, as in previous studies (Lemoine et al. 2005, 2008), the most common class of chromosome rearrangements was translocations reflecting BIR events in which the Ty elements of FS1 or FS2 recombined with a Ty or a δ-element on a nonhomologous chromosome. For the Ty elements on the broken chromosome to pair with nonallelic Ty elements, the broken end needs to be processed by nucleases that degrade the duplex 5′–3′, exposing the 3′ end required to initiate the exchange. In S. cerevisiae, this processing is carried out in two steps (Mimitou and Symington 2009), with one step leading to very short single-stranded “tails” (carried out by Sae2p and the MRX complex) and a second reaction leading to longer (several hundred bases) single-stranded ends carried out by Exo1p, Sgs1p complexed with Dna2p, or (possibly) Sgs1p complexed with Exo1p (Mimitou and Symington 2009). Loss of Sae2p or the MRX complex delays, but does not prevent, resection (Mimitou and Symington 2009), consistent with our finding that chromosome rearrangements were not prevented by mutations of MRE11 or SAE2.
Some Ty elements appear to interact more frequently than others in generating translocations, although with the small number of events examined, these biases are not statistically significant. There are 31 Ty1 and Ty2 elements that are in the correct orientation to produce a monocentric translocation in a BIR event involving the centromere-proximal Ty element of FS2. Of 20 translocations involving this Ty element of FS2, 3 interacted with Ty1-1 on chromosome V and 3 interacted with Ty1-2 on IV. Interestingly, the Ty1-1 element on V was shown to be a hotspot for gamma ray-induced translocations (Argueso et al. 2008). There are 21 Ty1 and Ty2 elements in the correct orientation to produce a monocentric translocation by a BIR event with the FS1 Ty elements. Of 14 translocations examined, 3 involved Ty1-2 on chromosome II, and 4 involved a tandem pair of Ty elements (Ty1-1 and Ty1-2) on chromosome IV. If additional data support the preferential use of certain Ty elements in interacting with the FS1 and FS2 Ty's, there are a number of explanations: (1) certain elements have a more open chromatin configuration and are more susceptible to strand invasion, (2) the arrangement of chromosomes within the nucleus affects the frequency of interaction, and (3) the sequences of the Ty elements that interact frequently with the Ty elements of FS1 and FS2 are more similar than those elements that do not interact frequently. Unfortunately, since our genetic background is not identical to that of the sequenced yeast strains, investigation of the last possibility would require extensive resequencing.
Rad52p-independent chromosome rearrangements: half crossovers:
Since the frequency of BIR events is greatly reduced in rad52Δ strains (reviewed by Paques and Haber 1999; Llorente et al. 2008), the low frequency of class 3 events was expected. As described above, the most common type of rearrangement observed in the rad52Δ diploids was likely the result of a half crossover, the production of a single recombinant from two broken chromosomes (Haber and Hearn 1985). Since Ty or δ-elements were observed at the breakpoints of these chromosome rearrangements, it is likely that the recombinant chromosome was formed by single-strand annealing (SSA) between nonallelic Ty or δ-elements (Figure S2). Although the rad52Δ mutation reduces the frequency of SSA for small repeats, the effect on recombination between repeats >2 kb is minor (Paques and Haber 1999). For example, the rate of recombination in the tandem array of 9-kb rRNA gene repeats is unaffected by the rad52Δ mutation (Zamb and Petes 1981; Ozenberger and Roeder 1991).
Conclusion:
On the basis of the data described above, we suggest that the processing of IRs with large spacers differs from the processing of perfect palindromes or IRs with short spacers. Perfect palindromes and IRs with short spacers can extrude as cruciforms and the processing of such structures to form recombinogenic DSBs is hypothesized to involve a two-step mechanism that is dependent on Mre11p and Sae2p (Rattray et al. 2001; Farah et al. 2002; Lobachev et al. 2002; Cote and Lewis 2008). In contrast, FS2 is an IR interrupted by a large spacer and likely does not extrude as a cruciform, but can form a hairpin on the lagging strand during replication. We propose that processing of the hairpin at FS2 to form a recombinogenic DSB occurs by a single-step mechanism that is independent of Mre11p and Sae2p. We suggest that the 280-base loop at the tip of the hairpin at FS2 is cleaved by a single-strand endonuclease; none of the nucleases examined in the current study (Mre11p, Sae2p, Rad1p, Slx4p, Pso2p, Exo1p, Mus81p, Yen1p, and Rad27p) are essential for this processing. The structure of the chromosome rearrangements associated with FS2-generated DSBs suggests that the broken chromosomes are repaired in a Rad52p-dependent BIR pathway involving nonallelic Ty elements.
Acknowledgments
We thank all members of the Petes lab for useful discussions and suggestions and Yiyong Liu for help in mapping chromosome deletion breakpoints. We thank M. Kupiec, K. Lobachev, and S. Jinks-Robertson for their comments on the manuscript. This study was supported by National Institutes of Health grant GM52319 to T.D.P.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.106385/DC1.
The microarray data discussed in this article have been deposited in the NCBI Gene Expression Omnibus (Edgar et al. 2002) and are accessible through GEO Series accession no. GSE16502 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE16502).
References
- Akgun, E., J. Zahn, S. Baumes, G. Brown, F. Liang et al., 1997. Palindrome resolution and recombination in the mammalian germ line. Mol. Cell. Biol. 17: 5559–5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allers, T., and D. R. Leach, 1995. DNA palindromes adopt a methylation-resistant conformation that is consistent with DNA cruciform or hairpin formation in vivo. J. Mol. Biol. 252: 70–85. [DOI] [PubMed] [Google Scholar]
- Argueso, J. L., J. Westmoreland, P. A. Mieczkowski, M. Gawel, T. D. Petes et al., 2008. Double-strand breaks associated with repetitive DNA can reshape the genome. Proc. Natl. Acad. Sci. USA 105: 11845–11850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressan, D. A., H. A. Olivares, B. E. Nelms and J. H. Petrini, 1998. Alteration of N-terminal phosphoesterase signature motifs inactivates Saccharomyces cerevisiae Mre11. Genetics 150: 591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collick, A., J. Drew, J. Penberth, P. Bois, J. Luckett et al., 1996. Instability of long inverted repeats within mouse transgenes. EMBO J. 15: 1163–1171. [PMC free article] [PubMed] [Google Scholar]
- Collins, J., G. Volckaert and P. Nevers, 1982. Precise and nearly-precise excision of the symmetrical inverted repeats of Tn5; common features of recA-independent deletion events in Escherichia coli. Gene 19: 139–146. [DOI] [PubMed] [Google Scholar]
- Cote, A. G., and S. M. Lewis, 2008. Mus81-dependent double-strand DNA breaks at in vivo-generated cruciform structures in S. cerevisiae. Mol. Cell 31: 800–812. [DOI] [PubMed] [Google Scholar]
- Courey, A. J., and J. C. Wang, 1988. Influence of DNA sequence and supercoiling on the process of cruciform formation. J. Mol. Biol. 202: 35–43. [DOI] [PubMed] [Google Scholar]
- D'Amours, D., and S. P. Jackson, 2002. The Mre11 complex: at the crossroads of DNA repair and checkpoint signalling. Nat. Rev. Mol. Cell. Biol. 3: 317–327. [DOI] [PubMed] [Google Scholar]
- DasGupta, U., K. Weston-Hafer and D. E. Berg, 1987. Local DNA sequence control of deletion formation in Escherichia coli plasmid pBR322. Genetics 115: 41–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, A. P., and L. S. Symington, 2004. RAD51-dependent break-induced replication in yeast. Mol. Cell. Biol. 24: 2344–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePamphilis, M. L., 2002. Eukaryotic DNA replication forks. Chemtracts. Biochem. Mol. Biol. 15: 313–325. [Google Scholar]
- DePamphilis, M. L., and P. M. Wassarman, 1980. Replication of eukaryotic chromosomes: a close-up of the replication fork. Annu. Rev. Biochem. 49: 627–666. [DOI] [PubMed] [Google Scholar]
- Edgar, R., M. Domrachev and A. E. Lash, 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30: 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eykelenboom, J. K., J. K. Blackwood, E. Okely and D. R. Leach, 2008. SbcCD causes a double-strand break at a DNA palindrome in the Escherichia coli chromosome. Mol. Cell 29: 644–651. [DOI] [PubMed] [Google Scholar]
- Farah, J. A., E. Hartsuiker, K. Mizuno, K. Ohta and G. R. Smith, 2002. A 160-bp palindrome is a Rad50.Rad32-dependent mitotic recombination hotspot in Schizosaccharomyces pombe. Genetics 161: 461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farah, J. A., G. Cromie, W. W. Steiner and G. R. Smith, 2005. A novel recombination pathway initiated by the Mre11/Rad50/Nbs1 complex eliminates palindromes during meiosis in Schizosaccharomyces pombe. Genetics 169: 1261–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg, E. C., G. C. Walker, W. Siede, R. D. Wood, R. A. Schultz et al., 2006. DNA Repair and Mutagenesis. ASM Press, Washington, DC.
- Gordenin, D. A., A. L. Malkova, A. Peterzen, V. N. Kulikov, Y. I. Pavlov et al., 1992. Transposon Tn5 excision in yeast: influence of DNA polymerases α, δ, and ɛ and DNA repair genes. Proc. Natl. Acad. Sci. USA 89: 3785–3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordenin, D. A., K. S. Lobachev, N. P. Degtyareva, A. L. Malkova, E. Perkins et al., 1993. Inverted DNA repeats: a source of eukaryotic genomic instability. Mol. Cell. Biol. 13: 5315–5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie, C., and G. R. Fink, 1991. Guide to Yeast Genetics and Molecular Biology. Academic Press, San Diego.
- Haber, J. E., and M. Hearn, 1985. RAD52-independent mitotic gene conversion in Saccharomyces cerevisiae frequently results in chromosomal loss. Genetics 111: 7–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawthorne, D. C., 1963. A deletion in yeast and its bearing on the structure of the mating type locus. Genetics 48: 1727–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip, S. C., U. Rass, M. G. Blanco, H. R. Flynn, J. M. Skehel et al., 2008. Identification of Holliday junction resolvases from humans and yeast. Nature 456: 357–361. [DOI] [PubMed] [Google Scholar]
- Kogo, H., H. Inagaki, T. Ohye, T. Kato, B. S. Emanuel et al., 2007. Cruciform extrusion propensity of human translocation-mediating palindromic AT-rich repeats. Nucleic Acids Res. 35: 1198–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokoska, R. J., L. Stefanovic, J. DeMai and T. D. Petes, 2000. Increased rates of genomic deletions generated by mutations in the yeast gene encoding DNA polymerase delta or by decreases in the cellular levels of DNA polymerase delta. Mol. Cell. Biol. 20: 7490–7504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna, S., B. M. Wagener, H. P. Liu, Y. C. Lo, R. Sterk et al., 2007. Mre11 and Ku regulation of double-strand break repair by gene conversion and break-induced replication. DNA Repair (Amst.) 6: 797–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurahashi, H., H. Inagaki, T. Ohye, H. Kogo, T. Kato et al., 2006. Palindrome-mediated chromosomal translocations in humans. DNA Repair (Amst.) 5: 1136–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurahashi, H., H. Inagaki, E. Hosoba, T. Kato, T. Ohye et al., 2007. Molecular cloning of a translocation breakpoint hotspot in 22q11. Genome Res. 17: 461–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemoine, F. J., N. P. Degtyareva, K. Lobachev and T. D. Petes, 2005. Chromosomal translocations in yeast induced by low levels of DNA polymerase a model for chromosome fragile sites. Cell 120: 587–598. [DOI] [PubMed] [Google Scholar]
- Lemoine, F. J., N. P. Degtyareva, R. J. Kokoska and T. D. Petes, 2008. Reduced levels of DNA polymerase delta induce chromosome fragile site instability in yeast. Mol. Cell. Biol. 28: 5359–5368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengsfeld, B. M., A. J. Rattray, V. Bhaskara, R. Ghirlando and T. T. Paull, 2007. Sae2 is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/Xrs2 complex. Mol. Cell 28: 638–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, S. M., and A. G. Cote, 2006. Palindromes and genomic stress fractures: bracing and repairing the damage. DNA Repair (Amst.) 5: 1146–1160. [DOI] [PubMed] [Google Scholar]
- Llorente, B., C. E. Smith and L. S. Symington, 2008. Break-induced replication: What is it and what is it for? Cell Cycle 7: 859–864. [DOI] [PubMed] [Google Scholar]
- Lobachev, K. S., D. A. Gordenin and M. A. Resnick, 2002. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell 108: 183–193. [DOI] [PubMed] [Google Scholar]
- Mimitou, E. P., and L. S. Symington, 2009. Nucleases and helicases take center stage in homologous recombination. Trends Biochem. Sci. 34: 264–272. [DOI] [PubMed] [Google Scholar]
- Moreau, S., J. R. Ferguson and L. S. Symington, 1999. The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol. Cell. Biol. 19: 556–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan, V., P. A. Mieczkowski, H. M. Kim, T. D. Petes and K. S. Lobachev, 2006. The pattern of gene amplification is determined by the chromosomal location of hairpin-capped breaks. Cell 125: 1283–1296. [DOI] [PubMed] [Google Scholar]
- Ozenberger, B. A., and G. S. Roeder, 1991. A unique pathway of double-strand break repair operates in tandemly repeated genes. Mol. Cell. Biol. 11: 1222–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paques, F., and J. E. Haber, 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63: 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattray, A. J., 2004. A method for cloning and sequencing long palindromic DNA junctions. Nucleic Acids Res. 32: e155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattray, A. J., C. B. McGill, B. K. Shafer and J. N. Strathern, 2001. Fidelity of mitotic, double-strand-break repair in Saccharomyces cerevisiae: a role for SAE2/COM1. Genetics 158: 109–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattray, A. J., B. K. Shafer, B. Neelam and J. N. Strathern, 2005. A mechanism of palindromic gene amplification in Saccharomyces cerevisiae. Genes Dev. 19: 1390–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse, J., 2009. Control of genome stability by Slx protein complexes. Biochem. Soc. Trans. 37: 495–510. [DOI] [PubMed] [Google Scholar]
- Ruskin, B., and G. R. Fink, 1993. Mutations in POL1 increase the mitotic instability of tandem inverted repeats in Saccharomyces cerevisiae. Genetics 134: 43–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, K. H., V. Pennaneach, C. P. Putnam and R. D. Kolodner, 2006. Analysis of gross-chromosomal rearrangements in Saccharomyces cerevisiae. Methods Enzymol. 409: 462–476. [DOI] [PubMed] [Google Scholar]
- Sinden, R. R., S. S. Broyles and D. E. Pettijohn, 1983. Perfect palindromic lac operator DNA sequence exists as a stable cruciform structure in supercoiled DNA in vitro but not in vivo. Proc. Natl. Acad. Sci. USA 80: 1797–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinden, R. R., G. X. Zheng, R. G. Brankamp and K. N. Allen, 1991. On the deletion of inverted repeated DNA in Escherichia coli: effects of length, thermal stability, and cruciform formation in vivo. Genetics 129: 991–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, C. E., A. F. Lam and L. S. Symington, 2009. Aberrant double-strand break repair resulting in half crossovers in mutants defective for Rad51 or the DNA polymerase delta complex. Mol. Cell. Biol. 29: 1432–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathern, J., J. Hicks and I. Herskowitz, 1981. Control of cell type in yeast by the mating type. The alpha1-alpha2 hypothesis. J. Mol. Biol. 147: 239–244. [DOI] [PubMed] [Google Scholar]
- Symington, L. S., 2002. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 66: 630–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, H., D. A. Bergstrom, M. C. Yao and S. J. Tapscott, 2005. Widespread and nonrandom distribution of DNA palindromes in cancer cells provides a structural platform for subsequent gene amplification. Nat. Genet. 37: 320–327. [DOI] [PubMed] [Google Scholar]
- Tanaka, H., Y. Cao, D. A. Bergstrom, C. Kooperberg, S. J. Tapscott et al., 2007. Intrastrand annealing leads to the formation of a large DNA palindrome and determines the boundaries of genomic amplification in human cancer. Mol. Cell. Biol. 27: 1993–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavassoli, M., M. Shayeghi, A. Nasim and F. Z. Watts, 1995. Cloning and characterisation of the Schizosaccharomyces pombe rad32 gene: a gene required for repair of double strand breaks and recombination. Nucleic Acids Res. 23: 383–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh, T. Q., and R. R. Sinden, 1991. Preferential DNA secondary structure mutagenesis in the lagging strand of replication in E. coli. Nature 352: 544–547. [DOI] [PubMed] [Google Scholar]
- Trujillo, K. M., and P. Sung, 2001. DNA structure-specific nuclease activities in the Saccharomyces cerevisiae Rad50*Mre11 complex. J. Biol. Chem. 276: 35458–35464. [DOI] [PubMed] [Google Scholar]
- VanHulle, K., F. J. Lemoine, V. Narayanan, B. Downing, K. Hull et al., 2007. Inverted DNA repeats channel repair of distant double-strand breaks into chromatid fusions and chromosomal rearrangements. Mol. Cell. Biol. 27: 2601–2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voineagu, I., V. Narayanan, K. S. Lobachev and S. M. Mirkin, 2008. Replication stalling at unstable inverted repeats: interplay between DNA hairpins and fork stabilizing proteins. Proc. Natl. Acad. Sci. USA 105: 9936–9941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warmington, J. R., R. Anwar, C. S. Newlon, R. B. Waring, R. W. Davies et al., 1986. A ‘hot-spot’ for Ty transposition on the left arm of yeast chromosome III. Nucleic Acids Res. 14: 3475–3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamb, T. J., and T. D. Petes, 1981. Unequal sister-strand recombination within the yeast ribosomal DNA does not require the RAD52 gene product. Curr. Genet. 3: 125–132. [DOI] [PubMed] [Google Scholar]
- Zheng, G. X., and R. R. Sinden, 1988. Effect of base composition at the center of inverted repeated DNA sequences on cruciform transitions in DNA. J. Biol. Chem. 263: 5356–5361. [PubMed] [Google Scholar]
- Zheng, G. X., T. Kochel, R. W. Hoepfner, S. E. Timmons and R. R. Sinden, 1991. Torsionally tuned cruciform and Z-DNA probes for measuring unrestrained supercoiling at specific sites in DNA of living cells. J. Mol. Biol. 221: 107–122. [DOI] [PubMed] [Google Scholar]