Abstract
Ovarioles are the functional unit of the female insect reproductive organs and the number of ovarioles per ovary strongly influences egg-laying rate and fecundity. Social evolution in the honeybee (Apis mellifera) has resulted in queens with 200–360 total ovarioles and workers with usually 20 or less. In addition, variation in ovariole number among workers relates to worker sensory tuning, foraging behavior, and the ability to lay unfertilized male-destined eggs. To study the genetic architecture of worker ovariole number, we performed a series of crosses between Africanized and European bees that differ in worker ovariole number. Unexpectedly, these crosses produced transgressive worker phenotypes with extreme ovariole numbers that were sensitive to the social environment. We used a new selective pooled DNA interval mapping approach with two Africanized backcrosses to identify quantitative trait loci (QTL) underlying the transgressive ovary phenotype. We identified one QTL on chromosome 11 and found some evidence for another QTL on chromosome 2. Both QTL regions contain plausible functional candidate genes. The ovariole number of foragers was correlated with the sugar concentration of collected nectar, supporting previous studies showing a link between worker physiology and foraging behavior. We discuss how the phenotype of extreme worker ovariole numbers and the underlying genetic factors we identified could be linked to the development of queen traits.
THE number of ovariole filaments per ovary is an important female reproductive character that affects fecundity across insect taxa (Richard et al. 2005; Makert et al. 2006). Social insect lineages have evolved a strong dimorphism in ovariole number between reproductive and nonreproductive castes. For example, while most families of bees consistently have 6 total ovarioles, and most species in the family Apidae have 8, the highly social species in the genus Apis (the honeybees) have queens that can have >360 total ovarioles and workers that often have <10 (Winston 1987; Michener 2003). This queen–worker dimorphism is of primary importance because it translates into differential reproductive potential that defines the social roles of these female castes (Winston 1987) and classifies social species in general (Sherman et al. 1995). Furthermore, ovary size (i.e., ovariole number) is the most sensitive indicator of caste-specific development in honeybees (Dedej et al. 1998). The extreme increase in ovariole number for queen honeybees enables high egg-laying rates (>1500 per day) and is apparently a result of selection for increased colony reproduction (growth and fission by swarming) (Seeley 1997). Honeybee queens are thus highly specialized for egg laying, similar to queens of several other social insect taxa, such as army ants or higher termites (Hölldobler and Wilson 1990). Honeybee workers in contrast, do not normally reproduce but perform all other essential activities including foraging for nectar, pollen, and water; caring for brood; and building, maintaining, and defending the colony (Winston 1987; Seeley 1997).
While worker honeybees have drastically reduced ovariole numbers relative to queens, they have retained functional ovaries and can produce unfertilized (haploid) male-destined eggs in the absence of queen pheromonal inhibition (Velthuis 1970; Page and Robinson 1994). In the absence of a queen, variation in worker ovariole number translates into differential reproductive success (Makert et al. 2006), but in the presence of a queen this variation is correlated with several other worker attributes. Variation in worker ovariole number may underlie the pollen hoarding syndrome of honeybees, a set of correlated behavioral and physiological traits associated with biases in pollen vs. nectar foraging within honeybee colonies (Amdam et al. 2004, 2006). Ovariole number is thus an important phenotype associated with queen–worker dimorphism but also worker reproduction and division of labor.
In honeybees, adult ovariole number is determined during larval development by nutrition. Nurse workers feed queen-destined larvae an overabundance of food while the diet of worker-destined larvae is restricted in the fourth and fifth larval instar (Beetsma 1985). Nurse feeding behavior and thus indirect genetic effects of the colony environment can strongly influence larval developmental trajectory (Beekman et al. 2000; Allsopp et al. 2003; Linksvayer et al. 2009). The differential feeding affects larval gene networks sensitive to nutritional status (the target of rapamycin (TOR) pathway; Patel et al. 2007) to change DNA methylation (Kucharski et al. 2008) and juvenile hormone (JH) titers, with titers higher in queen- than in worker-destined larvae (Hartfelder and Engels 1998). Until the fourth instar, queen- and worker-destined larvae have the same number of ovariole primordia (Reginato and Cruz-Landim 2001). Lower JH titer in workers coincides with disintegration of parts of the cytoskeleton in the germ cells and apoptosis, which decreases ovariole number by the fifth instar (Capella and Hartfelder 1998, 2002). In accord, worker ovarioles can be rescued by JH application during the fourth and early fifth instar (Capella and Hartfelder 1998, 2002).
Worker ovariole number and the extent of queen–worker dimorphism for ovariole number vary between species of Apis and between recognized races and strains of Apis mellifera. Both A. cerana and A. mellifera workers typically have <20 total ovarioles (Kapil 1962; Michener and Brothers 1974), but A. cerana queens have only ∼140 ovarioles (Velthuis et al. 1971) while A. mellifera have 200–360 (Michener 1974). In contrast, A. dorsata queens have 248–274 ovarioles and workers have 22–106 (Velthuis et al. 1971). Ruttner and Hesse (1981) studied seven races of A. mellifera and found mean total worker ovariole numbers ranging from 6.4 in A. mellifera mellifera to 18.8 in A. mellifera capensis. Several studies provide evidence that variation in worker ovariole number within populations and between strains has a strong genetic component (Diniz et al. 1993; Thuller et al. 1996, 1998; Jordan et al. 2008).
Here, we compared the distribution of worker ovariole number in colonies from a population of feral Africanized bees in Arizona with commercial European bees. Africanized and European bees are derived from lineages separated for ∼1 million years (Whitfield et al. 2006a) and differ in a variety of traits including body size, development time, defensiveness, and behavioral traits associated with the pollen hoarding syndrome (Winston et al. 1983, 1987; Pankiw 2003). Genome scans have identified a number of loci that differ between Africanized and European lineages, and at least some of these genetic differences seem to be the result of divergent selection (Pankiw 2003; Whitfield et al. 2006a; Zayed and Whitfield 2008). In addition, QTL mapping studies for body size and defensive behavior (Hunt et al. 1998, 2007) have suggested few genes with major effect underlying some of these lineage differences.
We first describe crosses between Africanized and European bees that revealed segregating variation for extreme ovariole number in workers that were sensitive to the social environment. Next, we describe the results of selective pooled DNA QTL mapping of worker ovariole number in two Africanized backcrosses with transgressive worker ovariole phenotypes, and we list potential candidate genes in the regions of the detected QTL. Finally, we demonstrate that variation in ovariole number, albeit unusual, correlates with differences in worker foraging behavior that have previously been shown to be linked to normal variation in ovariole number.
METHODS
Initial survey of ovary variation and mating scheme:
In November 2005, we surveyed 12 Africanized (AHB) and 12 European (EHB) honeybee colonies for worker ovariole number. AHB colonies were collected as feral swarms near Mesa, Arizona. EHB colonies were derived from commercial sources. For each colony, we dissected 20 newly emerged workers and counted the ovarioles in both ovaries. Workers were immobilized by chilling, the abdomen opened dorsally, and both ovaries transferred onto a microscope slide with cover slip to count the ovarioles under a compound microscope. We analyzed ovariole number differences between strains with a general linear mixed model with strain as a fixed effect and colony (strain) as a random effect, using Statistica 7.1 software (StatSoft, Tulsa, OK).
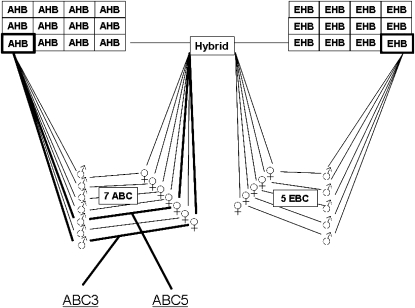
The highest AHB and lowest EHB colonies were used as sources for drone and queen parents, respectively, to produce a hybrid colony (Figure 1). Daughter queens from this mating (i.e., with one set of chromosomes from each, the AHB and EHB parent) were then either backcrossed to drones of the EHB or AHB parent colony to produce EHB backcrosses (EBC) and AHB backcrosses (ABC), respectively (Figure 1). In addition, queens and drones from the original parent AHB and EHB colonies were mated in sibcrosses to produce inbred AHB or EHB worker offspring. Further crosses (e.g., the reciprocal hybrid with an AHB queen and EHB male) were not made due to time, space, and financial constraints.
Figure 1.—
Crossing scheme used to analyze the genetic architecture of ovary size differences between Africanized and European honeybees. Inbred crosses are not shown for clarity.
All parental, hybrid, backcross, and inbred colonies were surveyed for worker ovariole number in May 2006 as described above. We used a similar general linear mixed model described above for the initial ovariole number survey to analyze differences in ovary size between cross types and colonies nested within cross types. We used a post hoc LSD test for differences between specific colonies. On the basis of these initial results, two colonies with workers with extraordinarily high numbers of ovarioles were selected to collect mapping populations of workers (ABC3, n = 94; ABC5, n = 400). In July 2006, the extraordinary worker ovary size in population ABC3 was reconfirmed in an additional series of dissections. Workers were also assessed for queen characteristics other than ovaries (notched mandibles, absence of pollen baskets on hind legs, spermatheca) but no evidence for queen characteristics other than large ovaries was found. Thus, the workers did not represent intercastes described previously in the literature (Weaver 1957; Dedej et al. 1998; Degrandi-Hoffman et al. 2004).
Selective pooled DNA QTL mapping:
Genomic DNA was isolated by a modified CTAB-lysis, combined with a single phenol-chloroform extraction (Hunt and Page 1995). The DNA was resuspended in 100 μl TE and quality and quantity was assessed with a Nanodrop spectrophotometer. DNA samples of sufficient purity (A260/280 ≥ 1.8) and quantity (>100 ng/μl) were diluted to 100 ng/μl and used for subsequent analyses.
We used a selective pooled DNA QTL approach (Darvasi and Soller 1994; Dekkers 2000; Korol et al. 2007; Wang et al. 2007) to map the genomic location in the two AHB backcross populations with the most extreme worker ovary size (ABC3 and ABC5, Figure 1). DNA from individuals in the high and low tails of the phenotypic distribution of ovariole number was pooled into high and low pools for both ABC3 and ABC5. From each cross and each phenotypic tail, three DNA subpools were created as biological replicates with 8 individuals each (fractioned DNA pool design; Korol et al. 2007). Workers composing the high ovariole pools had a median total ovariole number of 37.5 (range, 31–54) (ABC3) and 42 (39–58) (ABC5), workers in the low ovariole pools 14 (5–17) and 13.5 (8–15), respectively. The DNA pools were made up by combining an equal amount of DNA from each individual and genotyped at 1136 verified SNPs on an Illumina (San Diego, CA) array platform according to previous studies (Whitfield et al. 2006a). These SNP markers vary between Arizona AHB and EHB (estimated FST = 0.406; supporting information, Table S2 Whitfield et al. 2006a) and adequately cover the ∼4000 cm (Solignac et al. 2007) or 260 Mb (Honeybee Genome Sequencing Consortium 2006) large honeybee genome.
For each locus we estimated the allele frequency in each DNA pool from the normalized signal intensity of the alternative alleles. To correct for inherent allelic biases, we computed a k-correction factor (Hoogendoorn et al. 2000) on the basis of differences in the allelic normalized signal intensity of samples of individuals that had been previously genotyped as heterozygous, and were kindly provided by C. Whitfield (University of Illinois at Urbana-Champaign). Allele frequencies in the pooled DNA samples were then estimated using the specific k-correction factor for each locus.
As all genotyped workers were offspring of a queen inseminated by a single drone, only a small number of genotypic distributions were possible for each locus. Only the following two mating types produce genetically variable offspring: A1A2 × A2, and A1A2 × A1, both producing half heterozygote and half homozygote offspring, with an overall expected minor allele frequency (i.e., frequency of the rarer allele) of 0.25. If these variable loci segregate perfectly in the high and low tails, one tail will have a minor allele frequency of 0.0 and the other 0.5. We used two different approaches to exclude uninformative marker loci:
(1) Under the more liberal approach, we only excluded loci with minor allele frequencies that were much lower than expected for segregating loci (i.e., these loci were assumed to be fixed for one allele). We excluded all loci with minor allele frequency estimates for both the high and low phenotypic pools that were <0.132, which is the lower 99% confidence interval for a frequency of 0.25 given binomial sampling and the observed technical error variance (see next paragraph; the distribution and confidence intervals were estimated using 100,000 simulated samples). This approach resulted in 486 informative loci.
(2) For the more conservative approach, we excluded loci that had an estimated minor allele frequency averaged across the high and low pools that was outside the interval (0.155, 0.345), the estimated 99% confidence interval for a frequency of 0.25 given binomial sampling and the observed technical error variance. Allele frequencies could be outside this interval if one allele is fixed or if the k-correction factor does not adequately correct for allele frequency estimate biases of the genotyping platform. This second approach resulted in 257 informative loci.
Differences in estimated allele frequencies between upper and lower ovariole pools were calculated for each marker and for each of the AHB BC mapping populations (Darvasi and Soller 1994; Dekkers 2000; Wang et al. 2007). From these differences, a Z statistic was calculated, Z = D/(1/2*0.25/n + VTE)^1/2, where D is the estimated difference in allele frequency between high and low tails, n = 24, the number of individuals per tail, and VTE is the estimate of technical error variance (Darvasi and Soller 1994; Wang et al. 2007). We estimated studywide VTE by comparing allele frequency estimates from the overall pool for ABC3 with the mean of the three replicate subpools for ABC3, and then averaging the VTE estimate for each of the high and low pools to obtain an overall estimate VTE = 0.000424. Z scores from the two backcross populations were combined to give a χ2 statistic for each marker (Wang et al. 2007). In the resulting single marker analysis, the marker with the highest χ2 statistic on a given chromosome or chromosomal region is assumed to be closest to a putative causative locus (Darvasi and Soller 1994; Wang et al. 2007).
Furthermore, both crosses were analyzed for QTL with a least squares interval mapping selective pooled DNA approach developed by Wang et al. (2007). As traditional interval mapping, this analysis combines the information from two adjacent markers (allelic differences between high and low pools) with the estimated recombination rates between markers and hypothetical QTL positions to calculate a χ2 statistic for each genomic position (Wang et al. 2007). χ2 statistics are summed over the two populations at each putative QTL position, and the position with the highest statistic is taken as the estimate of QTL position (Wang et al. 2007). χ2 statistics were subsequently converted to LOD scores. To determine SNP order and recombination distances, we located the SNPs on the physical map of the bee genome by BLAST and assigned recombination distances between adjacent markers by inference from a saturated microsatellite map (Solignac et al. 2007). Specifically, for each chromosome, we created a function relating recombination distance to physical distance using a least squares fit of a six-term polynomial of physical distance to recombination distance; R2 > 0.99 for all chromosomes (Beye et al. 2006; Solignac et al. 2007). After estimating QTL position, QTL allele frequencies were estimated and used to estimate QTL substitution effects, 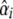 , for each mapping population (following Dekkers 2000; Wang et al. 2007).
, for each mapping population (following Dekkers 2000; Wang et al. 2007).
While standard permutation tests (between subpools, ignoring whether subpools come from the high or low phenotypic pools) can be used to establish genomewide thresholds in the fractioned pool design (Korol et al. 2007), our low number of subpools (3) was not sufficient (only 10 total permutations per backcross). Instead, as suggested by Wang et al. (2007), we simulated allele frequencies of subpools with a binomial distribution using the observed allele frequency estimates and the technical error variance estimates for each locus. These simulated subpools were then randomly distributed between the two phenotypic pools. As for permutation tests (Churchill and Doerge 1994), we established a genomewide threshold as the 95th and 99th quantile of maximum statistic taken from each of 10,000 such simulated data sets. We subsequently used these simulated data sets to estimate P-values for QTL above the genomewide threshold. We also compared our simulated genomewide threshold with a LOD-3 genomewide threshold previously established for QTL mapping in A. mellifera (Hunt et al. 1995; Rueppell et al. 2004). Bootstrap resampling (10,000 random samples) from within high and low subpools was used to estimate the 95% confidence intervals of QTL position (Korol et al. 2007). Because our sample size was very limited we also calculated 1.5-LOD support intervals for QTL position (Lander and Botstein 1989; Dupuis and Siegmund 1999). For single cross analysis, we only report results of the more conservative 1.5-LOD support intervals. All calculations, permutation tests, and bootstrap resampling were done with Mathematica 6.0 (Wolfram Research).
Within the 95% confidence intervals and 1.5-LOD support intervals of the QTL, we searched the NCBI Mapviewer database for the A. mellifera 4.0 genome version (http://www.ncbi.nlm.nih.gov/mapview/map_search.cgi?taxid=7460) for genes whose functional prediction or homology to known genes suggested that they could be involved in ovariole number determination. For this putative functional annotation we used the FlyBase (http://flybase.bio.indiana.edu/), IHOP (http://www.ihop-net.org/UniPub/iHOP/), NCBI (http://www.ncbi.nlm.nih.gov/), and the Geneontology (http://www.geneontology.org/) databases, as well as primary articles. Furthermore, we considered adult and embryonic gene expression of putative candidate gene homologs in Drosophila using FlyAtlas (http://www.flyatlas.org/) (Chintapalli et al. 2007) and the BDGP database (http://www.fruitfly.org/cgi-bin/ex/insitu.pl) (Tomancak et al. 2002). The selection of functional candidates from the list of positional candidate genes is necessarily incomplete and somewhat arbitrary to the extent that our knowledge of the molecular cascade leading to ovary size determination in workers is incomplete.
Foraging behavior of workers with extreme variation in ovariole number:
To produce a sampling population of backcross workers that maximized variation in ovariole number, two backcross colonies that differed greatly in ovariole number were used as sources in the same behavioral experiment in June 2006 (ABC5 and ABC8, see also Figure 3). Over 2 consecutive days, workers emerged in an incubator (50% RH, 34°) from brood combs retrieved from ABC5 and ABC8. Upon emergence, each bee was uniquely tagged by gluing a small numbered plastic tag (BeeWorks) onto her thorax before introduction into a four-frame observation hive that contained an unrelated commercial colony. In total, 200 bees were tagged from each backcross source.
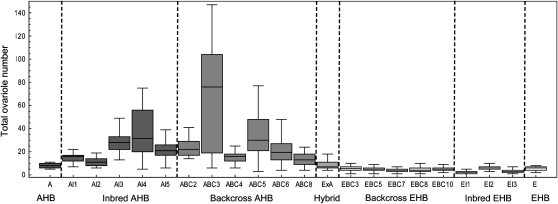
Figure 3.—
Box and whisker plots of total worker ovariole number for the crosses between AHB and EHB that revealed the extreme ovariole number phenotype. From left to right is the AHB parent (A; shown as colony A2 in Figure 2), inbred AHB (AI 1–4), backcross AHB (ABC 1–8), hybrid (ExA), backcross EHB (EBC 3–10), inbred EHB (EI 1–3), and the EHB parent (E; shown as colony E10 in Figure 2). AHB backcross colonies ABC3 and ABC5 with extreme ovariole number phenotype were used for selective pooled DNA QTL mapping.
The observation hive was monitored for 1 hr daily from June 10 to June 30, 2006. To avoid afternoon orientation flights that are not for foraging, worker activities were observed between 6:00 am and 8:30 am. As tagged workers returned after morning foraging trips, they were collected into wire cages at the hive entrance. The bees were anesthetized with CO2 within 30 min. Nectar loads were expelled from the crop into preweighed glass capillary tubes by gently squeezing the bee from the tip of the abdomen to the base of the thorax. Sugar content was estimated using a digital refractometer as described before (Amdam et al. 2006). Thereafter, workers were kept immobile at 4° until ovarioles were counted (see above). We analyzed the effect of source (i.e., ABC5 or ABC8; categorical predictor) and total ovariole number (continuous predictor) on the sugar concentration of nectar collected using stepwise regression.
RESULTS
Results of initial AHB and EHB survey:
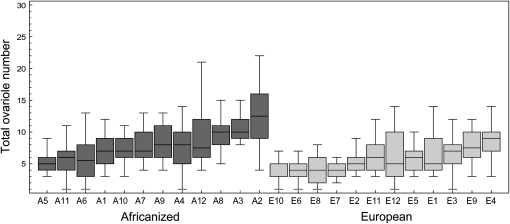
The distribution of total worker ovariole number in the surveyed 12 AHB and 12 EHB colonies is shown in Figure 2. As expected, there was an effect of strain (strain effect, F1,22.1 = 8.89, P = 0.0069) and the total ovariole number of AHB workers was higher (median 8, range 1–30) than the ovariole number of EHB workers (median 5, range 1–14). There was also significant variation among colonies within the strains [colony (strain) effect, F22,438 = 7.89, P < 0.0001], and we chose the AHB and EHB colonies with highest and lowest mean ovariole counts, respectively, as parents for the subsequent crosses (Figure 2).
Figure 2.—
Box and whisker plots of total worker ovariole number for the initial survey of 12 AHB and 12 EHB colonies. The AHB colony with the highest ovariole counts (A2) was used as the AHB parent and the EHB colony with the lowest ovariole counts (E10) was used as the EHB parent in subsequent crosses.
Results of crosses:
The distribution of total worker ovariole number in the parental AHB and EHB colonies, inbred AHB and inbred EHB colonies, hybrid EHB × AHB colony, and backcross AHB and backcross EHB colonies are shown in Figure 3. The ovariole number of hybrid workers (median 7, range 4–20) was statistically indistinguishable from the ovariole number of EHB parent workers (median 6, range 2–8) and AHB parent workers (median 8, range 5–18; LSD test, all P > 0.3, d.f. = 624). The EHB backcross and inbred crosses resulted in phenotypes that were not significantly different from the EHB parental phenotype (LSD test, all P > 0.5, d.f. = 624). In contrast, five out of seven AHB backcross (ABC 1, 2, 3, 5, 6) and two out of four AHB inbred (AI 3, 4) colonies resulted in workers with significantly more ovarioles than the AHB parent colony (LSD test, all P < 0.01, d.f. = 624; Figures 3 and 4). Three crosses (AI4, ABC3, ABC5) contained workers with at least 75 total ovarioles (e.g., Figure 5).
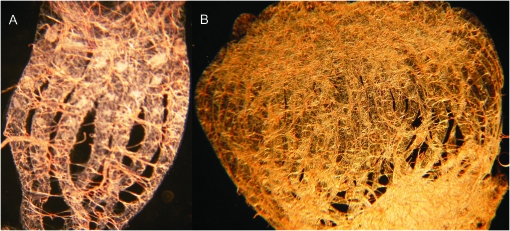
Figure 4.—
Normal-sized worker ovary from the Africanized parent colony A (A) and large worker ovary from Africanized backcross colony ABC3 (B). Each visible filament is an ovariole (e.g., there are 7 ovarioles in A and 46 ovarioles in B).
Figure 5.—
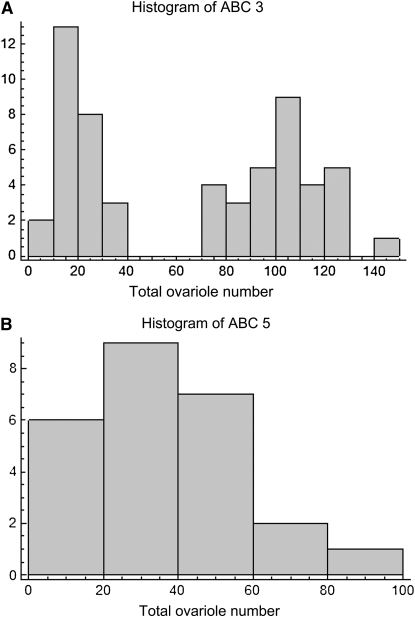
(A) Histogram of ABC3 worker total ovariole number sampled in May and July, raised in natal colony; median 76, range 6–147. (B) Histogram of ABC5 worker total ovariole number sampled in May, raised in natal colony; median 30, range 3–94.
Importance of the social environment for the extreme ovary size phenotype:
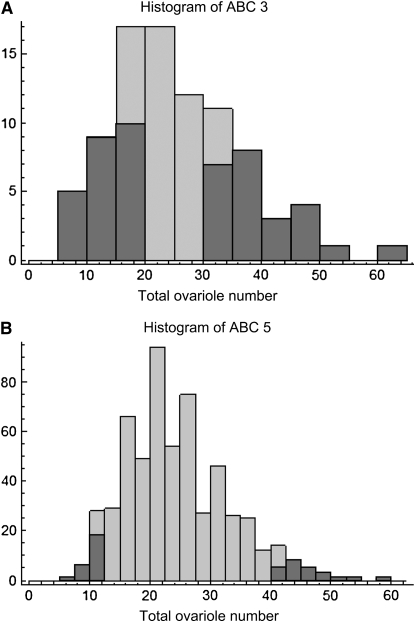
Both times (in May and July) that colony ABC3 was sampled when larvae were reared in their natal colony, the distribution of total ovariole number was strongly bimodal (May, median 77, range 6–110, Figure 3; July, median 32, range 8–147, Figure 5A). In contrast, for the QTL mapping study, larvae from ABC3 were reared in an unrelated EHB nursery colony, resulting in a decrease in ovariole number (median 23.5, range 5–62; Mann-Whitney U-test, P = 0.000030) and a unimodal distribution (Figure 6A). None of the other colonies sampled showed a notable bimodal distribution or such high numbers of ovarioles. However, the second mapping cross (ABC5) was phenotyped under similar conditions and also showed a reduction in worker ovariole number when workers were raised in unrelated nursery colonies (median 23, range 7–58; Figure 5B) compared to their own colony (median 29.5, range 3–94, Figure 6B; Mann-Whitney U-test, P = 0.0072).
Figure 6.—
(A) Histogram of ABC3 worker total ovariole number used for selective pooled QTL mapping, raised in an unrelated nurse colony; median 23.5, range 5–62; N = 88. Shading indicates samples included in the high and low pools. (B) Histogram of ABC5 worker total ovariole number used for selective pooled QTL mapping, raised in an unrelated nurse colony; median 23.0, range 7–58; N = 571. Shading indicates samples included in the high and low pools.
Detailed genetic analysis of two BC colonies:
The results for single marker analysis are summarized in Table 1, listing the 25 loci with the highest score combined from both backcrosses, based on the liberal marker selection. Their relative ranks for populations ABC3 and ABC5 are also listed separately because the backcrosses displayed considerable differences. In addition, their genomic position and information on their overall score under conservative marker selection are given. Many of the most significant markers were excluded under more conservative marker selection criteria.
TABLE 1.
Summary of single marker analysis
| SNP marker | Rank (liberal) | ABC3 rank | ABC5 rank | Chromosome | Position (Mb) | Rank (conservative)a |
|---|---|---|---|---|---|---|
| est6265 | 1 | 30 | 1 | 8 | 6.5 | — |
| est8456 | 2 | 1 | 442 | 11 | 9.9 | 1 |
| ahb2567 | 3 | 12 | 4 | 11 | 10.8 | 2 |
| ahb2105 | 4 | 2 | 239 | 10 | 6.3 | — |
| ahb9731 | 5 | 217 | 2 | 8 | 8.1 | — |
| est7418 | 6 | 3 | 58 | 10 | 8.8 | — |
| ahb12014 | 7 | 157 | 3 | 8 | 6.5 | — |
| est10300 | 8 | 7 | 23 | 14 | 8.2 | — |
| est2045 | 9 | 20 | 10 | 2 | 9.8 | — |
| ahb8451 | 10 | 49 | 7 | 6 | 5.7 | — |
| est8460 | 11 | 4 | 246 | 11 | 9.7 | — |
| ahb8075 | 12 | 5 | 219 | 6 | 1.5 | 3 |
| ahb4562 | 13 | 18 | 18 | 14 | 7.7 | 4 |
| ahb8307 | 14 | 199 | 6 | 6 | 12.7 | — |
| est8926 | 15 | 486 | 5 | 11 | 3.4 | 5 |
| ahb1464 | 16 | 94 | 9 | 1 | 23.0 | 6 |
| ahb8287 | 17 | 272 | 8 | 6 | 12.6 | — |
| ahb2788 | 18 | 40 | 17 | 11 | 12.0 | — |
| ahb7938 | 19 | 32 | 25 | 5 | 6.7 | 7 |
| ahb2458 | 20 | 177 | 11 | 11 | 13.8 | 8 |
| ahb5409 | 21 | 271 | 12 | 2 | 13.8 | — |
| est1736 | 22 | 35 | 30 | 2 | 9.4 | 9 |
| est3715 | 23 | 179 | 13 | 4 | 6.5 | 10 |
| est9592 | 24 | 159 | 14 | 12 | 7.8 | — |
| ahb2783 | 25 | 58 | 29 | 11 | 12.0 | 11 |
Of 486 markers used in the liberal analysis, the top 25 based on combined Z scores for ABC3 and ABC5 mapping populations are shown. Also listed are the ranks in the separate backcross analysis, the genome location, and the markers' overall rank in the conservative analysis.
–, Indicates no rank because the marker was excluded from the analysis.
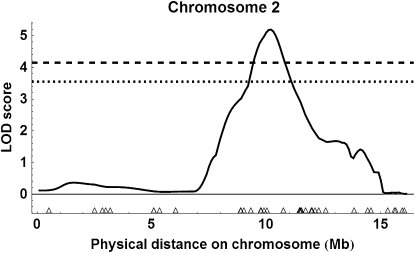
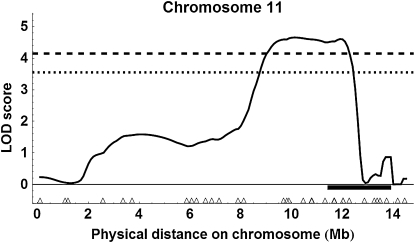
Using the more liberal marker selection criteria, interval mapping for the two populations together revealed two QTL above the 99% genomewide threshold (Figure S1 for all results): on chromosome 2 with an estimated location of 10.1 Mb or 153 cM (Figure 7); peak LOD score = 5.19, P = 0.001; 95% C.I. of location, 10.1–10.5 Mb (= 153–160 cM), 1.5-LOD support interval, 9.2–10.9 Mb (= 146–177 cM)] and an estimated substitution effect of 2.46 ovarioles for ABC3 and 1.78 ovarioles for ABC5; and on chromosome 11, location 10.0 Mb or 160 cM (Figure 8); peak LOD score = 4.66, P = 0.001; 95% C.I. of location, 8.9–12.2 Mb (= 135–205 cM), 1.5-LOD support interval, 8.5–12.3 Mb (=125–206 cM)], and an estimated substitution effect of 3.67 ovarioles for ABC3 and 1.27 ovarioles for ABC5. There was also a QTL on chromosome 4 above the estimated 95% genomewide threshold, location 6.5 Mb or 144 cM [see Figure S1, peak LOD score = 3.66, P = 0.039; 1.5-LOD support interval of location, 4.5–7.8 Mb (= 92–172 cM)].
Figure 7.—
LOD score values for combined ABC3 and ABC5 populations showing the location of a QTL on chromosome 2 with liberal marker selection. Triangles show marker location. The dotted line shows the 95% genomewide threshold for QTL detection and the dashed line the 99% genomewide threshold.
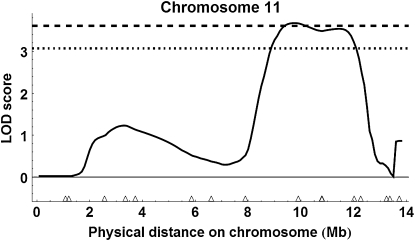
Figure 8.—
QTL on chromosome 11 detected with liberal marker selection. The horizontal black bar shows the 95% C.I. for a QTL affecting age at first foraging described in Rueppell (2009).
We also analyzed the two crosses separately (Figure S2 and Figure S3): For ABC3, the QTL on chromosome 11 was above the 99% genomewide threshold, location 9.6 Mb or 150 cM [peak LOD score = 2.98, P = 0.0065; 1.5-LOD support interval of location, 8.5–12.2 Mb (= 125–205 cM)], and a QTL on chromosome 10 was above the 95% genomewide threshold, location 6.5 Mb or 87 cM [peak LOD score = 2.41, P = 0.0445; 1.5-LOD support interval of location, 5.2–10.9 Mb (= 50–188 cM)]. For ABC5, the QTL on chromosome 2 was above the 99% genomewide threshold, location 10.2 Mb or 154 cM [peak LOD score 3.82, P = 0.0005, 1.5-LOD support interval of location, 9.3–11.4 Mb (= 146–187 cM)], and there were two QTL above the 95% genomewide threshold; on chromosomes 4, location 6.2 Mb or 129 cM [peak LOD score = 2.93, P = 0.0155; 1.5-LOD support interval of location, 4.3–7.7 Mb (= 87–169 cM)], and on chromosome 8, location 7.8 Mb or 129 cM [peak LOD score = 2.61, P = 0.0345; 1.5-LOD support interval of location, 3.8–8.6 Mb (= 68–147 cM)].
Using the more conservative marker selection criteria with 257 marker loci with the two populations combined (see Figure S4), we detected a single QTL above the 99% genomewide threshold on chromosome 11, location 9.6 Mb = 150 cM (Figure 9); peak LOD score = 3.68, P = 0.009; 95% C.I. of location, 8.9–12.0 Mb (= 135–204 cM)], 1.5-LOD support interval, 8.4–12.3 Mb (= 124–206 cM)] and an estimated substitution effect of 4.88 ovarioles for ABC3 and 1.73 ovarioles for ABC5. For single cross analysis with ABC3 (Figure S5), the QTL on chromosome 11 was above the 95% genomewide threshold, location 9.6 Mb or 150 cM [peak LOD score = 2.33, P = 0.049; 1.5-LOD support interval of location, 8.2–12.2 Mb (= 122–205 cM)]. For ABC5, there were no QTL above the 95% genomewide threshold (Figure S6).
Figure 9.—
QTL on chromosome 11 detected with conservative marker selection.
For the combined analyses, both the simulated 95 and 99% genomewide thresholds were more conservative than a LOD 3.0 threshold (Figure S1 and Figure S4), while for the backcross-specific analyses, only the simulated 99% threshold approached or surpassed the LOD 3.0 threshold (Figure S2, Figure S3, Figure S5, and Figure S6). We therefore focused on QTL that were consistently above the 95% threshold in the combined analyses and also were above the 99% cutoff in one of the backcrosses (i.e., the QTL on chromosomes 2 and 11).
Candidate genes:
We searched for potential candidate genes within the confidence intervals for our strongest supported QTL on chromosomes 2 and 11. The confidence interval of the first QTL on chromosome 2 (Figure 7), detected with liberal marker selection, contains only a single gene, which is homologous to the geko gene in Drosophila (CG13695). This gene is classified as an olfactory receptor involved in ethanol response (Shiraiwa et al. 2000). However, the wider genome region indicated by the 1.5-LOD support interval additionally suggested 53 potential candidate genes. The most plausible functional candidates among these are two genes that are involved in actin regulation, the genghis khan homolog LOC412132 and the fimbrin homolog LOC408694, as well as LOC726899, related to the Drosophila gene thread, which is involved in apoptosis (Lisi et al. 2000) and has a degenerate ovary phenotype (Rodriguez et al. 2002).
The second QTL interval on chromosome 11 (Figures 8 and 9), detected with both conservative and liberal marker selection, contained 135 genes, several of which can be regarded as good functional candidates. The most notable ones are quail (LOC410324), which is involved in actin filament-dependent apoptosis in nurse cells in the Drosophila ovary (Matova et al. 1999); cabut (LOC410326), which is an ecdysteroid-responsive transcriptional activator associated with cell death, germ-band shortening, and the JNK cascade (Munoz-Descalzo et al. 2005; Zhao et al. 2008); delta (LOC410351), which affects notch signaling and has been reported to influence the number of germline stem cells in the Drosophila ovary (Ward et al. 2006); and miro (LOC725244), which is involved in mitochondrial homeostasis, apoptotic signal transduction and cytoskeleton organization (Fransson et al. 2003). In addition, several other genes could also be involved in determining ovariole number (Table S1). The wider 1.5-LOD support interval additionally suggested 33 potential candidate genes with the following 3 noteworthy functional candidates: SHC-adaptor protein (LOC412172) affects ovary development in Drosophila via intracellular signaling cascades (Luschnig et al. 2000), FL(2)D (LOC552833) is involved in the female germline sex determination through alternative splicing (Granadino et al. 1992), and the carmer homolog (LOC411458), a methyltransferase that is involved in induction of programmed cell death by ecdysone (Cakouros et al. 2004).
Foraging behavior of workers:
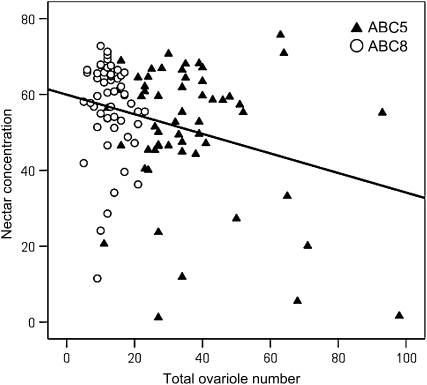
Across the extreme range of ovariole numbers generated by ABC5 and ABC8 (Figure 3), stepwise regression rejected source, and identified forager ovariole number as being correlated with the sugar concentration of nectar collected (R2 = 0.084, F1,107 = 9.82, P = 0.0022; Figure 10).
Figure 10.—
The sugar concentration of nectar collected by foragers is negatively correlated with forager total ovariole number (R2 = 0.084). Foragers from two Africanized backcross colonies (ABC5 and ABC8) were used.
DISCUSSION
Our study reveals genetic variation that strongly affects worker ovariole number in the Africanized honeybee population. Worker ovariole number was sensitive to the social environment and correlated with worker foraging behavior. Interval mapping based on selective DNA pooling revealed one, possibly two, strongly supported QTL, as well as three potential additional backcross-specific QTL. Below we discuss what our crossing design revealed about the genetic architecture of the transgressive worker ovariole number; we discuss candidate genes located in the regions of the two strongest supported QTL; we discuss the relationship between ovariole number and forager behavior; and we consider how the large worker ovary phenotype is related to caste development.
Genetic architecture of honeybee worker ovariole number:
Heritable variation for ovariole number has been found in a variety of insects (Grenier and Nardon 1994; Wayne et al. 1997) and in some cases ovariole number has been shown to be phenotypically plastic, depending on larval nutrition or other environmental factors (Hodin and Riddiford 2000; Tu and Tatar 2003; Bergland et al. 2008). Further study of the genetic architecture of ovariole number in Drosophila spp. has revealed genotype-by-environment interaction (Starmer et al. 1998; Bergland et al. 2008) and several loci of large effect that interact epistatically (Wayne et al. 2001; Richard et al. 2005; Orgogozo et al. 2006).
In accordance with these previous studies, the results of our crosses (Figure 3) suggest a complex genetic architecture for worker honeybee ovariole number. While inbreeding and backcrossing of the EHBs produced offspring that uniformly resembled the parental sources, inbred and backcrossed AHBs showed a wide phenotypic distribution among and within colonies and significantly exceeded the parental phenotypes. Transgressive phenotypes observed in backcrosses are often explained as being due to novel combinations of additive small-effect alleles contributed by each parent that are brought together by hybridization (Rieseberg et al. 1999). However, this model cannot explain our results given that some inbred AHBs also showed the transgressive ovary phenotype: the distribution of ovariole number for the inbred AI4 was approximately the same as the distribution in ABC5, one of the backcrosses with transgressive ovary phenotypes that we mapped (Figure 3). Thus, the AHB parent was the source of alleles causing the transgressive phenotype.
The simplest genetic model that could explain the occurrence of the transgressive phenotype in only some backcross and inbred AHB is that there is a single rare recessive gene of major effect on worker ovariole number with incomplete penetrance; i.e., workers expressing the extreme ovary phenotype are hypothesized to have genotype OO and workers expressing the normal ovary phenotype are hypothesized to have genotype Oo or oo. In this case, if both the AHB parent queen and the hybrid queen were heterozygotes (Oo), half of the AHB BC colonies would be expected to contain workers expressing the extreme ovary phenotype (i.e., these colonies would be composed of OO and Oo workers) and half of the colonies would contain only normal-ovary workers (with genotypes Oo and oo).
However, a more complex model is required to explain the pattern of large variation within and between AHB inbred and backcross colonies. The distribution of ovariole number for the inbred and backcross AHB fell into somewhat discrete categories (see Figure 3, for example ABC3 had the largest ovaries, AI4 and ABC5 had the next largest, etc.). This suggests there may be two (or possibly more) recessive genes of major effect on ovariole number that could interact synergistically. For example, the most extreme individuals may be homozygous recessive at two loci (O1O1O2O2), individuals with moderately sized ovaries homozygous may be recessive at one locus and heterozygous at the second locus (O1o1O2O2 or O1O1O2o2), and all other genotypes produce the normal ovary size. Colonies typically showed a normal distribution of worker ovariole number (but see Figure 5A, discussed in the next paragraph). This together with the fact that we detected mean differences between AHB and EHB workers in our initial survey (Figure 2) suggests that there are additional genetic (or environmental) factors of small effect contributing quantitative variation to ovariole number. One possibility is that the recessive gene(s) of major effect makes the developing ovary more sensitive to other genetic and environmental factors, so that ovariole number is less canalized (see below).
A bimodal distribution of worker ovary size was observed in ABC3 (Figure 5A). Such a distribution could be caused by a single genetic factor (e.g., a recessive major effect allele as discussed above). Alternatively, multiple genetic and environmental factors could affect a normally distributed variable, such as a hormone titer, which is translated via a threshold function into two discrete phenotypic classes (i.e., a threshold character; note that a multiple threshold model may also help to explain the occurrence of multiple more-or-less discrete phenotypic classes; Falconer and Mackay 1996). However, this bimodality occurred only when ABC3 larvae were reared in their natal colony and not when the larvae were reared in an unrelated host colony (compare Figures 5A and 6A). Furthermore, rearing of ABC3 and ABC5 larvae in their natal colonies vs. an unrelated host colony resulted in workers with more ovarioles on average (compare Figures 5 and 6). The conditionality on the social environment suggests additional complexity in the genetic architecture involving genotype-by-environment interaction and indirect genetic effects (Linksvayer 2006, 2007; Linksvayer et al. 2009). The hypothetical loci discussed above with direct effects on ovariole number may also have indirect social effects on ovariole number, or alternatively genetic components of the social environment may arise from distinct loci. Our findings of conditionality of worker ovariole number on the social environment are consistent with previous studies demonstrating that different genotypic combinations of nurse workers, older larvae, and young larvae produce new workers with different ovariole numbers, indicating that a worker's ovariole number is a property of its own genotype as well as the genotypic composition of the colony (Beekman et al. 2000; Allsopp et al. 2003; Linksvayer et al. 2009).
Our QTL analysis also supports the interpretation of the transmission genetics of a complex genetic architecture with loci of varying effects. With our combined mapping effort we detected one, possibly two well-supported QTL with variable effects in the two investigated mapping populations, as well as additional, less well-supported backcross-specific QTL (Figure S2, Figure S3, Figure S5, and Figure S6), despite a high overall genetic similarity between the two mapping populations (r = 0.6875). This is also true for most of the single marker effects (Table 1). The predominance of backcross-specific effects is best explained by multilocus interactions that distinguish the genetic architecture even between the two AHB backcrosses with the highest ovariole number. However, more detailed genetic analyses including individual QTL mapping analyses and characterization of the parental genotypes are needed to synthesize a more detailed genetic model that accommodates specific roles for individual loci.
QTL and candidate genes for worker ovariole number:
In combination, the QTL mapping based on split pools from the phenotypic extremes detected only one or two strongly supported QTL, depending on the stringency of marker exclusion from the analyses. The QTL on chromosome 11 was detected regardless of the marker selection stringency, and several of the highest ranking markers in the single marker analysis were on chromosome 11 (Table 1). Interestingly, this QTL overlapped with a previously mapped behavioral QTL influencing another component of the pollen hoarding syndrome, the age of first foraging (Figure 8; Rueppell 2009), indicating a potential overlap in genetic architecture underlying this suite of correlated traits. In contrast, the QTL on chromosome 2 was not detected with the more conservative marker selection criteria, and markers on chromosome 2 were not among the highest ranking in the single marker analysis (Table 1). Conversely, many of the highest-ranking single markers under liberal marker selection that were excluded in the conservative analysis did not translate into a well-supported QTL in our interval mapping analysis (e.g., on chromosomes 8 and 10). Combined, these findings favor the results of the conservative marker selection.
The detection of one or two QTL supports our interpretation of the phenotypic results of the crossings and it is common to detect only a few QTL of major effect (Mackay 2001). However, QTL studies are biased to detect only QTL with the largest phenotypic effects, and furthermore estimates of the effects of detected QTL are upwardly biased, particularly with small samples size (i.e., the Beavis effect; Xu 2003). Wang et al. (2007) used simulation to evaluate the power of the interval mapping selective DNA pooling approach they developed. They found that selective DNA pooling had similar power as individual genotyping to detect QTL in the middle of a chromosome but less power to detect distally located QTL. Furthermore, they found that estimates of location for distally located QTL were more centrally biased with selective DNA pooling than individual genotyping (Wang et al. 2007). Thus, our study was biased to detect centrally located QTL; our estimates of QTL location may also be centrally biased; and given our small overall sample size, our estimates of QTL effects may be upwardly biased.
The center of the QTL on chromosome 2 is gene poor with only one gene, identified in an olfactory response screen using ethanol (Shiraiwa et al. 2000). However, this locus cannot be ruled out to influence ovary size because it shows expression in many tissues of the adult fly, with highest expression levels in testis (FlyAtlas), and it is expressed during development in the ring gland (BDGP Gene Expression Report). However in the QTL vicinity, within the LOD-1.5 support interval, several candidate genes are located, whose functional annotation indicates a possible role in ovary size determination. In addition, two of these positional candidates are in the top 10 list of genes upregulated in worker-destined fourth instar larvae relative to queen-destined fourth instar larvae [CRMP-PB = LOC408692 and CG30105-PA = LOC726711; see Table 2 in Barchuk et al. (2007)]. These genes are compelling because the fourth instar is the developmental stage that presumably initiates the large differences in worker and queen ovaries (Capella and Hartfelder 1998; Capella and Hartfelder 2002).
The second QTL on chromosome 11 was not affected by our method of marker exclusion, substantiating its significance. It falls into a very gene-rich genome region, containing several genes that are plausible functional candidates, on the basis of their molecular functions in Drosophila and our understanding of the developmental mechanism of the ovary development in honeybees. Probably the most compelling candidate gene for affecting ovary size is quail. In Drosophila, it is mostly expressed in pole cells and later germ cells through early embryonic development (BDGP Gene Expression Report) and also in adult flies, its expression is highest in the testis and ovaries (FlyAtlas). The molecular functions of quail that were identified in Drosophila (Matova et al. 1999) suggest that it could be involved in a JH-titer-dependent disintegration of the actin cytoskeleton in the germ cells leading to the apoptotic reduction of ovariole anlagen in honeybee workers.
Another well-characterized, good candidate is the delta gene, a ligand of notch. It plays a role in cell fate specification throughout Drosophila development and it is involved in many reproductive functions from establishing cell polarity in the oocyte to preventing fusion of egg chambers and necrosis of germ cells (Bender et al. 1993). It is expressed almost ubiquitously during embryonic development and has the highest levels of adult expression in the hindgut and ovary. Overexpression of delta in the germline results in extra germline stem cells (Ward et al. 2006), which could be responsible for extra ovarioles. Cabut was selected as another top candidate because it is expressed in the pole plasm early in development, it is responsive to endocrine stimuli (Zhao et al. 2008), and it is connected to cellular apoptosis via the JNK pathway (Mazars et al. 2001). The last top candidate was miro, selected for its involvement in apoptosis and interaction with the cytoskeleton (Fransson et al. 2003). It is also suggested to be an integrator of intracellular signaling (K.-A. Nilsen, K. E. Ihle, K. Frederick, M. K. Fondrk, B. Smedal, K. Hartfelder, G. V. Amdam, unpublished results) and is expressed in the pole cells during early development and CNS and ovary in adult Drosophila. Finally, the carmer arginine methyltransferase is intriguing because epigenetic transcriptome regulation is a key regulatory step in honeybee caste development (Kucharski et al. 2008), ecdysone titers between workers and queens are different during larval development (Hartfelder and Engels 1998), and worker ovary size determination is largely governed by apoptosis (Capella and Hartfelder 1998; Capella and Hartfelder 2002).
The QTL region in chromosome 11 is broad and we listed many other, second-tier candidate genes. Expression patterns in Drosophila did not provide evidence for the involvement of any of these second-tier candidate genes. However, one candidate gene, the transcription factor NFAT (GB15645) was found among the 100 most differentially expressed genes between developing worker and queen larvae (Barchuk et al. 2007). Another candidate gene was considered secondary on the basis of its molecular and functional characterization, CG16708-homolog GB19002, but it was the only gene among all genes in the QTL intervals whose expression level was significantly affected (P = 2.5 × 10−6) by the JH-analog methoprene (Whitfield et al. 2006b).
Association of ovariole number with foraging behavior:
Our data show a weak but significant negative correlation between ovary size and the concentration of the nectar collected by foragers. These results extend the findings of an earlier study on wild-type European honeybees (Amdam et al. 2006) to the extreme range of ovary sizes observed here. The concentration of the workers' nectar loads is linked to the sucrose sensitivity (Pankiw and Page 2000) and thus a correlation between ovary size and sucrose sensitivity (Tsuruda et al. 2008) can explain the association between ovary size, sensory tuning, and foraging behavior in worker honeybees (Amdam et al. 2009). The association between behavior and ovary size was not very strong (R2 = 0.084; Figure 10), similar to earlier studies (Amdam et al. 2006; Tsuruda et al. 2008). However, this is not surprising because worker ovariole number is a physiological trait fixed during development, while the concentration of collected nectar is a flexible behavioral trait influenced by a variety of dynamic environmental factors such as current nectar availability, individual foraging experience, and current colony conditions (Seeley 1997; Pankiw et al. 2001, 2004).
Link to the development and evolution of caste differences:
Whether a female social insect larva develops into a reproductive queen or a facultatively sterile worker (i.e., reproductive caste determination and differentiation), has traditionally been considered to be caused by environmental factors such as the quantity and quality of nutrition fed to larvae by nurse workers (Wheeler 1986). However, recently a number of examples of strong genetic influences on caste development, i.e., “genetic caste determination,” have been found in various social insect taxa (reviewed by Anderson et al. 2008).
We did not examine the phenotypes of newly produced queens in our study colonies, and thus it is unclear whether there is a direct mechanistic link between the extreme worker ovariole phenotype we discovered and queen development. However, extreme ovariole number is clearly a queen characteristic. The observed phenotype could arise by a mutation in the specific pathway, such as JH-conditional apoptosis, that affects only ovariole number and none of the other queen traits. Alternatively, the ABC3 and ABC5 genotypes that express the extreme worker ovariole number phenotype may have “leaky,” less-canalized queen–worker caste development. Their general developmental physiology may be biased toward the queen trajectory, for example through an increased JH level, even when fed a worker diet. Consequently, workers of these genotypes surpass the queen threshold for one queen trait, ovariole number, but not others, even under conditions that normally produce canalized worker phenotypes with low ovariole number.
Studies in which worker larvae are transferred into queen cells at different ages support our argument above by suggesting that ovary size is the most plastic trait that characterizes the honeybee caste dimorphism and is determined gradually over the course of larval development (Weaver 1957; Woyke 1971; Dedej et al. 1998). In contrast, the determination of external morphological caste characters is less sensitive to the rearing environment early in development (Dedej et al. 1998). External characters may depend on an increase in ecdysteroid titers, which are directly affected by the preceding JH titer (Rachinsky and Hartfelder 1995) and may canalize quantitative JH titer differences into two qualitatively distinct trajectories. This could explain why the differentiation of external morphological traits that occurs later in development is less sensitive to rearing environment than ovariole number.
Nurse honeybee workers strictly control the quantity and quality of food provisioned to brood so that only discrete queen and worker phenotypes are normally produced. This canalization of queen and worker phenotypes at the colony level may usually suppress the effect and detection of genetic variation for caste determination. Rearing effects also clearly affected ovary size in our experimental colonies because workers of ABC3 and ABC5 had larger ovaries when reared in their natal colony, indicating the importance of indirect genetic effects, as described above. However, our crosses have revealed genetic variation that permeated the developmental canalization.
Presumably in the ancestral state, caste differences were less extreme, developmental trajectories less canalized, and large ovary workers more common. Therefore, the transgressive worker ovary phenotype may be regarded as an atavism, and the genetic factors we identified in our crosses may have been involved in the ancestral caste development system and in the evolution of queen–worker dimorphism. Thus, this study reveals potential components involved in queen–worker development and provides a complimentary approach to functional genomic studies (Evans and Wheeler 2001; Wheeler et al. 2006; Barchuk et al. 2007; Patel et al. 2007; Kucharski et al. 2008) that can serve as the basis to understand the molecular mechanisms of caste development, as well as worker behavior, in honeybees.
It is currently unknown how commonly the large worker ovary phenotype that we observed occurs under natural colony conditions. However, previous studies have found evidence for segregating variation for high worker ovariole number (Chaud-netto and Bueno 1979; Thuller et al. 1996, 1998). In a substantial set of crosses within and between AHB and EHB colonies, Thuller et al. (1996, 1998) observed worker ovaries in backcross colonies that exceeded the values of either parent in a similar fashion as we have demonstrated here. These previous studies, together with our results, suggest that alleles associated with extreme worker ovariole number may be somewhat common in honeybee populations, and in some cases the extreme ovariole phenotype may only be expressed following hybridization. These observations bear marked similarities with recent discoveries of genetic components to caste development in several social insects (reviewed by Anderson et al. 2008). Hybridization and cross-fostering may disrupt coevolved components of the honeybee developmental program expressed in larvae and nurses (Linksvayer 2007; Linksvayer et al. 2009), resulting in the production of females with less strictly canalized queen and worker phenotypes.
One possible explanation for the maintenance of alleles causing the large worker ovary phenotype is that workers with more ovarioles are more likely to lay unfertilized male-destined eggs, and successfully mother haploid males, in the event of queen death (Makert et al. 2006); however, at some point, increasing worker ovariole number may actually decrease the probability of ovary activation and egg laying, because worker bees cannot selectively activate a subset of ovarioles and competition among ovarioles for limited circulating resources may lead to no single ovariole receiving enough resources to complete egg development (Velthuis 1970). Selection may have eroded additive genetic variance for worker ovariole number so that most segregating variation is nonadditive, due to dominance or epistasis. We lack the data to analyze these two phenomena at the individual QTL level but the phenotypic distributions of the AHB inbred and backcrossed colonies support this interpretation, as discussed above.
Acknowledgments
T.A.L. was supported by a National Science Foundation (NSF) postdoctoral fellowship. The NSF (0615502) also supported O.R. and G.V.A and funded the genotyping. Additional support to G.V.A. was provided by the Norwegian Research Council (175413, 180504, and 185306), and the PEW Foundation. R.E.P. was supported by the National Institute of Aging (NIA P01 AG22500). Colin Brent and Lauren Groves helped with ovary dissections in the mapping populations. Two anonymous reviewers provided helpful suggestions on an earlier version of this manuscript.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.105452/DC1.
References
- Allsopp, M. H., J. N. M. Calis and W. J. Boot, 2003. Differential feeding of worker larvae affects caste characters in the Cape honeybee, Apis mellifera capensis. Behav. Ecol. Sociobiol. 54: 555–561. [Google Scholar]
- Amdam, G. V., K. Norberg, M. K. Fondrk and R. E. Page, 2004. Reproductive ground plan may mediate colony-level selection effects on individual foraging behavior in honey bees. Proc. Natl. Acad. Sci. USA 101: 11350–11355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amdam, G. V., A. Csondes, M. K. Fondrk and R. E. Page, 2006. Complex social behaviour derived from maternal reproductive traits. Nature 439: 76–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amdam, G. V., K. E. Ihle and R. E. Page, 2009. Regulation of honey bee (Apis mellifera) life histories by vitellogenin, in Hormones, Brain and Behavior, Ed 2, edited by S. Fahrbach. Elsevier, San Diego, CA (in press).
- Anderson, K. E., T. A. Linksvayer and C. R. Smith, 2008. The causes and consequences of genetic caste determination in ants (Hymenoptera: Formicidae). Myrmecol. News 11: 119–132. [Google Scholar]
- Barchuk, A. R., A. S. Cristino, R. Kucharski, L. F. Costa, Z. L. Simoes et al., 2007. Molecular determinants of caste differentiation in the highly eusocial honeybee Apis mellifera. BMC Dev. Biol. 7: 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beekman, M., J. N. M. Calis and W. J. Boot, 2000. Parasitic honeybees get royal treatment. Nature 404: 723. [DOI] [PubMed] [Google Scholar]
- Beetsma, J., 1985. Feeding-behavior of nurse bees, larval food composition and caste differentiation in the honey bee (Apis mellifera L). Fortschritte der Zoologie 31: 407–410. [Google Scholar]
- Bender, L. B., P. J. Kooh and M. A. Muskavitch, 1993. Complex function and expression of Delta during Drosophila oogenesis. Genetics 133: 967–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergland, A. O., A. Genissel, S. V. Nuzhdin and M. Tatar, 2008. Quantitative trait loci affecting phenotypic plasticity and the allometric relationship of ovariole number and thorax length in Drosophila melanogaster. Genetics 180: 567–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beye, M., I. Gattermeier, M. Hasselmann, T. Gempe, M. Schioett et al., 2006. Exceptionally high levels of recombination across the honey bee genome. Genome Res. 16: 1339–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cakouros, D., T. J. Daish, K. Mills and S. Kumar, 2004. An arginine-histone methyltransferase, CARMER, coordinates ecdysone-mediated apoptosis in Drosophila cells. J. Biol. Chem. 279: 18467–18471. [DOI] [PubMed] [Google Scholar]
- Capella, I. C. S., and K. Hartfelder, 1998. Juvenile hormone effect on DNA synthesis and apoptosis in caste-specific differentiation of the larval honey bee (Apis mellifera L.) ovary. J. Insect Physiol. 44: 385–391. [DOI] [PubMed] [Google Scholar]
- Capella, I. C. S., and K. Hartfelder, 2002. Juvenile-hormone-dependent interaction of actin and spectrin is crucial for polymorphic differentiation of the larval honey bee ovary. Cell. Tissue Res. 307: 265–272. [DOI] [PubMed] [Google Scholar]
- Chaud-netto, J., and O. C. Bueno, 1979. Number of ovarioles in workers of Apis mellifera adansonii and Apis mellifera ligustica: a comparative study. J. Apicultural Res. 18: 260–263. [Google Scholar]
- Chintapalli, V. R., J. Wang and J. A. T. Dow, 2007. Using FlyAtlas to identify better Drosophila models of human disease. Nat. Genet. 39: 715–720. [DOI] [PubMed] [Google Scholar]
- Churchill, G. A., and R. W. Doerge, 1994. Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darvasi, A., and M. Soller, 1994. Selective DNA pooling for determination of linkage between a molecular marker and a quantitative trait locus. Genetics 138: 1365–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedej, S., K. Hartfelder, P. Aumeier, P. Rosenkranz and W. Engels, 1998. Caste determination is a sequential process: effect of larval age at grafting on ovariole number, hind leg size and cephalic volatiles in the honey bee (Apis mellifera carnica). J. Apicultural Res. 37: 183–190. [Google Scholar]
- DeGrandi-Hoffman, G., M. Chambers, J. E. Hooper and S. S. Schneider, 2004. Description of an intermorph between a worker and queen in African honey bees Apis mellifera scutellata (Hymenoptera: Apidae). Ann. Entomol. Soc. Am. 97: 1299–1305. [Google Scholar]
- Dekkers, J. C. M., 2000. Quantitative trait locus mapping based on selective DNA pooling. J. Anim. Breed. Genet. 117: 1–16. [Google Scholar]
- Diniz, J. A. F., O. C. Bueno, J. Chaudnetto and O. Malaspina, 1993. Heritability of the number of ovarioles in honey bee workers (Apis mellifera L) (Hym, Apidae). Revista Brasileira De Genetica 16: 917–921. [Google Scholar]
- Dupuis, J., and D. Siegmund, 1999. Statistical methods for mapping quantitative trait loci from a dense set of markers. Genetics 151: 373–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, J. D., and D. E. Wheeler, 2001. Gene expression and the evolution of insect polyphenisms. Bioessays 23: 62–68. [DOI] [PubMed] [Google Scholar]
- Falconer, D. S., and T. F. C. Mackay, 1996. Introduction to Quantitative Genetics, Ed. 4. Prentice Hall, New York.
- Fransson, A., A. Ruusala and P. Aspenstroem, 2003. Atypical Rho GTPases have roles in mitochondrial homeostasis and apoptosis. J. Biol. Chem. 278: 6495–6502. [DOI] [PubMed] [Google Scholar]
- Granadino, B., A. Sanjuan, P. Santamaria and L. Sanchez, 1992. Evidence of a dual function in Fl(2)D, a gene needed for sex-lethal expression in Drosophila melanogaster. Genetics 130: 597–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenier, A. M., and P. Nardon, 1994. The genetic control of ovariole number in Sitophilus oryzae L (Coleoptera, Curculionidae) is temperature sensitive. Genet. Select. Evol. 26: 413–430. [Google Scholar]
- Hartfelder, K., and W. Engels, 1998. Social insect polymorphism: hormonal regulation of plasticity in development and reproduction in the honeybee. Curr. Top. Dev. Biol. 40: 45–77. [DOI] [PubMed] [Google Scholar]
- Hodin, J., and L. M. Riddiford, 2000. Different mechanisms underlie phenotypic plasticity and interspecific variation for a reproductive character in drosophilids (Insecta: Diptera). Evolution 54: 1638–1653. [DOI] [PubMed] [Google Scholar]
- Hölldobler, B., and E. O. Wilson, 1990. The Ants. Harvard University Press, Cambridge, MA.
- Honeybee Genome Sequencing Consortium, 2006. Insights into social insects from the genome of the honeybee Apis mellifera. Nature 443: 931–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogendoorn, B., N. Norton, G. Kirov, N. Williams, M. L. Hamshere et al., 2000. Cheap, accurate and rapid allele frequency estimation of single nucleotide polymorphisms by primer extension and DHPLC in DNA pools. Hum. Genet. 107: 488–493. [DOI] [PubMed] [Google Scholar]
- Hunt, G. J., and R. E. Page, 1995. Linkage map of the honey bee, Apis mellifera, based on RAPD markers. Genetics 139: 1371–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt, G. J., G. V. Amdam, D. Schlipalius, C. Emore, N. Sardesai et al., 2007. Behavioral genomics of honeybee foraging and nest defense. Naturwissenschaften 94: 247–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt, G. J., E. Guzman-Novoa, M. K. Fondrk and R. E. Page, 1998. Quantitative trait loci for honey bee stinging behavior and body size. Genetics 148: 1203–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt, G. J., R. E. Page, M. K. Fondrk and C. J. Dullum, 1995. Major quantitative trait loci affecting honey bee foraging behavior. Genetics 141: 1537–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan, L. A., M. H. Allsopp, B. P. Oldroyd, T. C. Wossler and M. Beekman, 2008. Cheating honeybee workers produce royal offspring. Proc. Biol. Sci. 275: 345–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapil, R. P., 1962. Anatomy and histology of the female reproductive system of Apis indica F. (Hymenoptera, Apidae). Insectes Sociaux 9: 145–163. [Google Scholar]
- Korol, A., Z. Frenkel, L. Cohen, E. Lipkin and M. Soller, 2007. Fractioned DNA pooling: a new cost-effective strategy for fine mapping of quantitative trait loci. Genetics 176: 2611–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucharski, R., J. Maleszka, S. Foret and R. Maleszka, 2008. Nutritional control of reproductive status in honeybees via DNA methylation. Science 319: 1827–1830. [DOI] [PubMed] [Google Scholar]
- Lander, E. S., and D. Botstein, 1989. Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121: 185–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linksvayer, T. A., 2006. Direct, maternal, and sibsocial genetic effects on individual and colony traits in an ant. Evolution 60: 2552–2561. [DOI] [PubMed] [Google Scholar]
- Linksvayer, T. A., 2007. Ant species differences determined by epistasis between brood and worker genomes. PLoS ONE 2: e994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linksvayer, T. A., M. K. Fondrk and R. E. Page, Jr., 2009. Honey bee social regulatory networks are shaped by colony-level selection. Am. Nat. 173: E99–E107. [DOI] [PubMed] [Google Scholar]
- Lisi, S., I. Mazzon and K. White, 2000. Diverse domains of THREAD/DIAP1 are required to inhibit apoptosis induced by REAPER and HID in Drosophila. Genetics 154: 669–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luschnig, S., J. Krauss, K. Bohmann, I. Desjeux and C. Nusslein-Volhard, 2000. The Drosophila SHC adaptor protein is required for signaling by a subset of receptor tyrosine kinases. Mol. Cell 5: 231–241. [DOI] [PubMed] [Google Scholar]
- Mackay, T. F. C., 2001. The genetic architecture of quantitative traits. Annu. Rev. Genet. 35: 303–339. [DOI] [PubMed] [Google Scholar]
- Makert, G. R., R. J. Paxton and K. Hartfelder, 2006. Ovariole number: a predictor of differential reproductive success among worker subfamilies in queenless honeybee (Apis mellifera L.) colonies. Behav. Ecol. Sociobiol. 60: 815–825. [Google Scholar]
- Matova, N., S. Mahajan-Miklos, M. S. Mooseker and L. Cooley, 1999. Drosophila quail, a villin-related protein, bundles actin filaments in apoptotic nurse cells. Development 126: 5645–5657. [DOI] [PubMed] [Google Scholar]
- Mazars, A., F. Lallemand, C. Prunier, J. Marais, N. Ferrand et al., 2001. Evidence for a role of the JNK cascade in Smad7-mediated apoptosis. J. Biol. Chem. 276: 36797–36803. [DOI] [PubMed] [Google Scholar]
- Michener, C. D., 1974. The Social Behavior of the Bees. Harvard University Press, Cambridge, MA.
- Michener, C. D., 2003. The Bees of the World. Johns Hopkins University Press, Baltimore, MD.
- Michener, C. D., and D. J. Brothers, 1974. Were workers of eusocial Hymenoptera initially altruistic or oppressed? Proc. Natl. Acad, Sci. USA 71: 671–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Descalzo, S., J. Terol and N. Paricio, 2005. Cabut, a C2H2 zinc finger transcription factor, is required during Drosophila dorsal closure downstream of JNK signaling. Dev. Biol. 287: 168–179. [DOI] [PubMed] [Google Scholar]
- Orgogozo, V., K. W. Broman and D. L. Stern, 2006. High-resolution quantitative trait locus mapping reveals sign epistasis controlling ovariole number between two Drosophila species. Genetics 173: 197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page, R. E., and G. E. Robinson, 1994. Reproductive competition in queenless honey bee colonies (Apis mellifera L). Behav. Ecol. Sociobiol. 35: 99–107. [Google Scholar]
- Pankiw, T., 2003. Directional change in a suite of foraging behaviors in tropical and temperate evolved honey bees (Apis mellifera L.). Behav. Ecol. Sociobiol. 54: 458–464. [Google Scholar]
- Pankiw, T., and R. E. Page, 2000. Response thresholds to sucrose predict foraging division of labor in honeybees. Behav. Ecol. Sociobiol. 47: 265–267. [Google Scholar]
- Pankiw, T., K. D. Waddington and R.E. Page, 2001. Modulation of sucrose response thresholds in honey bees (Apis mellifera L.): influence of genotype, feeding, and foraging experience. J. Comp. Physiol. A 187: 293–301. [DOI] [PubMed] [Google Scholar]
- Pankiw, T., M. Nelson, R.E. Page and M. K. Fondrk, 2004. The communal crop: modulation of sucrose response thresholds of pre-foraging honey bees with incoming nectar quality. Behav. Ecol. Sociobiol. 55: 286–292. [Google Scholar]
- Patel, A., M. K. Fondrk, O. Kaftanoglu, C. Emore, G. Hunt et al., 2007. The making of a queen: TOR pathway is a key player in diphenic caste development. PLoS ONE 2: e509.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachinsky, A., and K. Hartfelder, 1995. Caste development of honeybees (Apis mellifera): juvenile hormone turns on ecdysteroids. Naturwissenschaften 82: 378–379. [Google Scholar]
- Reginato, R. D., and C. Cruz-Landim, 2001. Differentiation of the worker's ovary in Apis mellifera L. (Hymenoptera, Apidae) during life of the larvae. Invert. Reprod. Dev. 39: 127–134. [Google Scholar]
- Richard, D. S., R. Rybczynski, T. G. Wilson, Y. Wang, M. L. Wayne et al., 2005. Insulin signaling is necessary for vitellogenesis in Drosophila melanogaster independent of the roles of juvenile hormone and ecdysteroids: female sterility of the chico(1) insulin signaling mutation is autonomous to the ovary. J. Insect Physiol. 51: 455–464. [DOI] [PubMed] [Google Scholar]
- Rieseberg, L. H., M. A. Archer and R. K. Wayne, 1999. Transgressive segregation, adaptation and speciation. Heredity 83: 363–372. [DOI] [PubMed] [Google Scholar]
- Rodriguez, A., P. Chen, H. Oliver and J. M. Abrams, 2002. Unrestrained caspace-dependent cell death caused by loss of Diap1 function requires the Drosophila Apaf-1 homolog, Dark. EMBO J. 21: 2189–2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueppell, O., 2009. Characterization of quantitative trait loci for the age of first foraging in honey bee workers. Behav. Genet. 39: 541–553. [DOI] [PubMed] [Google Scholar]
- Rueppell, O., T. Pankiw and R. E. Page, Jr., 2004. Pleiotropy, epistasis and new QTL: the genetic architecture of honey bee foraging behavior. J. Hered. 95: 481–491. [DOI] [PubMed] [Google Scholar]
- Ruttner, F., and B. Hesse, 1981. Specific differences in the development of ovaries and egg-laying of queenless workers of several races of the honeybee, Apis mellifera L. Apidologie 12: 159–183. [Google Scholar]
- Seeley, T. D., 1997. Honey bee colonies are group-level adaptive units. Am. Nat. 150: S22–S41. [DOI] [PubMed] [Google Scholar]
- Sherman, P. W., E. A. Lacey, H. K. Reeve and L. Keller, 1995. The eusociality continuum. Behav. Ecol. 6: 102–108. [Google Scholar]
- Shiraiwa, T., E. Nitasaka and T. Yamazaki, 2000. Geko, a novel gene involved in olfaction in Drosophila melanogaster. J. Neurogenet. 14: 145–164. [DOI] [PubMed] [Google Scholar]
- Solignac, M., F. Mougel, D. Vautrin, M. Monnerot and J. M. Cornuet, 2007. A third-generation microsatellite-based linkage map of the honey bee, Apis mellifera, and its comparison with the sequence-based physical map. Genome Biol. 8: R66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starmer, W. T., M. Polak, L. L. Wolf and J. S. F. Barker, 1998. Reproductive characteristics of the flower breeding Drosophila hibisci bock (Drosophilidae) in eastern Australia: genetic and environmental determinants of ovariole number. Evolution 52: 806–815. [DOI] [PubMed] [Google Scholar]
- Thuller, R. H. C., O. Malaspina, O. C. Bueno and J. Chaud-Netto, 1996. Number of ovarioles in workers descendent from crossings between Africanized and Italian honeybees (Apis mellifera L.): comparing stock, inbred and F1 colonies. Anais da Sociedade Entomologica do Brasil 25: 501–506. [Google Scholar]
- Thuller, R. H. C., O. Malaspina, O. C. Bueno and J. Chaud-Netto, 1998. Number of ovarioles in workers descendent from crossings between Africanized and Italian honeybees, Apis mellifera L.: comparison among backcrosses and ancestors colonies. Anais da Sociedade Entomologica do Brasil 27: 237–243. [Google Scholar]
- Tomancak, P., A. Beaton, R. Weiszmann, E. Kwan, S. Shu et al., 2002. Systematic determination of patterns of gene expression during Drosophila embryogenesis. Genome Biology 3: research0088.0081–0088.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuruda, J., G. V. Amdam and R. E. Page, 2008. Sensory response system of social behavior tied to female reproductive traits. PLoS ONE 3: e3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu, M. P., and M. Tatar, 2003. Juvenile diet restriction and the aging and reproduction of adult Drosophila melanogaster. Aging Cell 2: 327–333. [DOI] [PubMed] [Google Scholar]
- Velthuis, H. H., 1970. Ovarian development in Apis mellifera worker bees. Entomologia Experimentalis et Applicata 13: 377–394. [Google Scholar]
- Velthuis, H. H. W., J. Clement, R. A. Morse and F. M. Laigo, 1971. The ovaries of Apis dorsata from the Philippines. J. Apicultural Res. 10: 63–66. [Google Scholar]
- Wang, J., K. J. Koehler and J. C. M. Dekkers, 2007. Interval mapping of quantitative trait loci with selective DNA pooling data. Genet. Select. Evol. 39: 685–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, E. J., H. R. Shcherbata, S. H. Reynolds, K. A. Fischer, S. D. Hatfield et al., 2006. Stem cells signal to the niche through the Notch pathway in the Drosophila ovary. Curr. Biol. 16: 2352–2358. [DOI] [PubMed] [Google Scholar]
- Wayne, M. L., J. B. Hackett, C. L. Dilda, S. V. Nuzhdin, E. G. Pasyukova et al., 2001. Quantitative trait locus mapping of fitness-related traits in Drosophila melanogaster. Genet. Res. 77: 107–116. [DOI] [PubMed] [Google Scholar]
- Wayne, M. L., J. B. Hackett and T. F. C. MacKay, 1997. Quantitative genetics of ovariole number in Drosophila melanogaster. 1. Segregating variation and fitness. Evolution 51: 1156–1163. [DOI] [PubMed] [Google Scholar]
- Weaver, N., 1957. Experiments on dimorphism in the female honey bee. J. Econ. Entomol. 50: 759–761. [Google Scholar]
- Wheeler, D. E., 1986. Developmental and physiological determinants of caste in social Hymenoptera: evolutionary implications. Am. Nat. 128: 13–34. [Google Scholar]
- Wheeler, D. E., N. Buck and J. D. Evans, 2006. Expression of insulin pathway genes during the period of caste determination in the honey bee, Apis mellifera. Insect Mol. Biol. 15: 597–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield, C. W., S. K. Behura, S. H. Berlocher, A. G. Clark, J. S. Johnston et al., 2006. a Thrice out of Africa: ancient and recent expansions of the honey bee, Apis mellifera. Science 314: 642–645. [DOI] [PubMed] [Google Scholar]
- Whitfield, C. W., Y. Ben-Shahar, C. Brillet, I. Leoncini, D. Crauser et al., 2006. b Genomic dissection of behavioral maturation in the honey bee. Proc. Natl. Acad. Sci. USA 103: 16068–16075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston, M. L., 1987. The Biology of the Honey Bee. Harvard University Press, Cambridge, MA.
- Winston, M. L., O. R. Taylor and G. W. Otis, 1983. Some differences between temperate European and tropical African and South American honeybees. Bee World 64: 12–21. [Google Scholar]
- Woyke, J., 1971. Correlations between the age at which honeybee brood was grafted, characteristics of the resultant queens and result of insemination. J. Apicultural Res. 10: 45–55. [Google Scholar]
- Xu, S. Z., 2003. Theoretical basis of the Beavis effect. Genetics 165: 2259–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zayed, A., and C. W. Whitfield, 2008. A genome-wide signature of positive selection in ancient and recent invasive expansions of the honey bee Apis mellifera. Proc. Natl. Acad. Sci. USA 105: 3421–3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, T., T. Gu, H. C. Rice, K. L. McAdams, K. M. Roark et al., 2008. A Drosophila gain-of-function screen for candidate genes involved in steroid-dependent neuroendocrine cell remodeling. Genetics 178: 883–901. [DOI] [PMC free article] [PubMed] [Google Scholar]