Abstract
The structural and functional properties of human prolactin (hPRL), a 23 kDa protein hormone and cytokine, are pH dependent. The dissociation rate constant for binding to the extracellular domain of the hPRL receptor increases nearly 500-fold over the relatively narrow and physiologic range from pH 8 to 6. As the apparent midpoint for this transition occurs around pH 6.5, we have looked towards histidine residues as a potential biophysical origin of the behavior. hPRL has a surprising number of nine histidines, nearly all of which are present on the protein surface. Using NMR spectroscopy, we have monitored site-specific proton binding to eight of these nine residues and derived equilibrium dissociation constants. During this analysis, a thermodynamic interaction between a localized triplet of three histidines (H27, H30 and H180) became apparent, which was subsequently confirmed by site-directed mutagenesis. After consideration of multiple potential models, we present statistical support for the existence of two negative cooperativity constants, one linking protonation of residues H30 and H180 with a magnitude of approximately 0.1, and the other weaker interaction between residues H27 and H30. Additionally, mutation of any of these three histidines to alanine stabilizes the folded protein relative to the chemically denatured state. Detailed understanding of these complex protonation reactions will aid in elucidating the biophysical mechanism for pH dependent regulation of hPRL’s structural and functional properties.
Keywords: NMR spectroscopy, cooperativity, protonation
Prolactin (PRL) is a 23 kDa protein hormone and a member of the family of hematopoietic cytokines, which includes erythropoietin, granulocyte colony-stimulating factor, interleukin-6, and many others, but is most closely related both evolutionarily and functionally to human growth hormone (hGH) and placental lactogen (hPL) (1). Proteins in this family share a common four α-helical bundle structural topology and recognize a conserved family of type 1 cytokine receptors (2). In humans, PRL is secreted by pituitary lactotrophs under hypothalamic regulation, where it circulates in the bloodstream and acts distally as an endocrine hormone. Human PRL (hPRL) is also synthesized in many extrapituitary tissues, including breast, prostate, and the female reproductive tract, where it appears to act locally to regulate cellular growth and differentiation (3;4). Closely associated with this autocrine/paracrine function as a local growth factor or cytokine, hPRL has been implicated in the growth and development of human cancers arising from the same tissue sites (5-8). Interestingly, a number of post-translational modifications to PRL have been identified, including phosphorylated (9-13) and proteolytically-cleaved (14-17) variants, which appear to largely counter the pro-tumorigenic activities of the wild-type (WT) hormone.
We recently described a dependence of the structural and functional properties of hPRL on solution pH (18). The structural stability of the recombinant protein decreases from 7.6 kcal/mol at pH 8 to 5.6 kcal/mol at pH 6. More striking is the greater than 500-fold decrease in the equilibrium association constant for the extracellular domain of the PRL receptor over this same relatively narrow and physiologic pH range. The biologic consequences of these effects are currently undescribed, but many possibilities exist. A pH dependent conformational change and subsequent aggregation reaction have been postulated to drive packaging of the newly expressed hormone into the secretory granules of pituitary lactotrophs (19;20). The mechanisms for generating the phosphorylated and proteolytically-cleaved variants of hPRL are not well understood, but could be regulated by pH dependent destabilization of the protein. Proteolytic cleavage of hPRL by the endocytic enzyme cathepsin D has been proposed (21-25). As cathepsin D is activated by acidic environments around pH 5 - 6, perhaps concomitant structural destabilization of hPRL increases its susceptibility to proteolytic cleavage in a synergistic manner. Similarly, as phosphorylation of hPRL at S179 is complicated by the complete burial of its side chain within the helical bundle, acid-induced destabilization of the protein may allow phosphorylating enzymes access to the residue. Lastly, the biology of hPRL must be impacted by the dramatic decrease in receptor-binding affinity occurring between the physiologic pH range of around 7.3 - 7.5 to slightly more acidic values between 6.0 - 6.5. A similar shift in pH can be seen in extracellular fluid of malignant versus surrounding tissues (26), implying decreased receptor occupancy of hPRL in hypoxic tumors. As well, after endocytosis of hPRL-receptor complexes, the subsequent decrease in endosomal pH should induce dissociation of the hormone from the receptor.
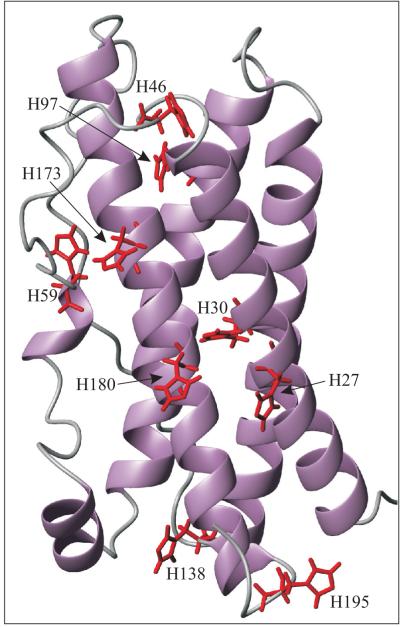
We have begun to explore the biophysical basis for the above pH dependent regulation of the structural and functional properties of hPRL. As the observed perturbations occur largely over a pH range from 6 - 8, with an apparent midpoint to the transition around pH 6.5, protonation of the imidazole side chains of His residues are the most likely origin of the behavior. hPRL contains a surprising number of nine histidines, comprising 4.5% of its amino acid composition, in comparison to the lower 2% incidence of histidines in a survey of modern proteins (27). The closely related proteins hGH and hPL contain three and seven His residues, respectively. Displayed on the tertiary structure of hPRL (28;29) in Figure 1, His imidazoles are largely dispersed throughout the protein surface, with the exception of a single localized cluster of H27, H30 and H180, which forms part of the expected high affinity receptor-binding site (29). To better understand the protonation reactions occurring in hPRL around pH 6.5, we have measured site-specific pH titrations for each of the observable histidines using NMR spectroscopy at near physiologic solution conditions. Although a majority of His residues titrated according to a classic “Henderson-Hasselbach” model, the localized triplet of H27, H30 and H180 displayed more complex behavior. Given the likely importance of these residues in regulating pH dependent receptor-binding, a detailed analysis of their protonation reactions is warranted and presented here.
Figure 1.
Backbone ribbon diagram for the tertiary structure of hPRL (PDB code 1RW5, first member of the ensemble shown) with histidine side chains highlighted in red and labeled with their respective residue number.
Experimental Methods
Preparation of isotope-labeled recombinant proteins for NMR spectroscopy
Wild-type (WT) human prolactin (hPRL) was recombinantly expressed in BL21 DE3 Escherichia coli and purified from inclusion bodies as previously described (30). Expression vectors for the H27A, H30A and H180A hPRL site-directed mutants of hPRL were obtained from Dr. Charles Clevenger (Northwestern University) and protein was similarly prepared. Uniform 13C and 15N protein-labeling was achieved using growth in M9 minimal media, appropriately enriched with 1 g of 15N-NH4Cl and 3 g of U-13C6-glucose (Cambridge Isotope Laboratories, Inc., Andover, MA) per liter of bacterial culture. Selective 15N histidine-labeled WT and H180A hPRL were prepared by transformation of their expression vectors into a histidine auxotroph strain of BL21(DE3) E. coli developed (31) and generously provided by Dr. David Waugh (National Cancer Institute, Bethesda, MD). Similar M9 minimal media was supplemented with 15N histidine (Cambridge Isotope Laboratories, Inc.) and all other unlabelled amino acids (Sigma-Aldrich, Inc., St. Louis, MO) based on a previous protocol (32). After transformation, the auxotrophic bacteria were plated onto LB-enriched agar containing 0.2 g/L ampicillin and incubated overnight at 37°C. A single isolated colony was used to inoculate 50 ml of the minimal media, also with 0.2 g/L ampicillin, and again incubated overnight at 37°C. An aliquot of this culture was used to inoculate one liter of minimal media supplemented with 0.1 g of each unlabeled amino acid and 0.02 g of unlabeled histidine. After growth of this culture at 37°C until its absorbance at 600 nm reached ∼0.8, 0.1 g of 15N-histidine was added and allowed to grow for another ten minutes before inducing recombinant protein expression by the addition of IPTG to a final concentration of 200 mM. Finally, 15N-histidine hPRL was refolded and purified from inclusion bodies as described above with yields between 50 and 100 mg of purified protein.
For NMR spectroscopy, hPRL was concentrated to 0.6 mM (unless otherwise noted) and exchanged into 25 mM potassium-phosphate buffer at pH 7.5, 25 mM NaCl., 5% 2H2O, 1 mM NaN3 and 1 μM of the protease inhibitors leupeptin, pepstatin and PMSF. A series of samples at different pH values, typically within a range of 4 - 8, were prepared for 13C15N-enriched WT, H27A, H30A, H173A and H180A-hPRL and for 15N histidine-labeled WT and H180A-hPRL. The samples were put in susceptibility-matched NMR tubes (Shigemi, Inc., Allison Park, PA) for low volume samples and the pH was adjusted to the desired values with 10 - 50 μl of 0.1 M HCl or 0.1 M NaOH. The pH adjustment was performed adding HCl or NaOH drop by drop, waiting two minutes after each addition to let the pH reach equilibrium. Special care was taken in the preparation of lower pH samples to avoid protein precipitation and the sample was centrifuged if precipitation did occur. The resulting pH of each sample was determined at room temperature before and after NMR data collection. The pH was measured in the NMR tube with an Orion glass micro-pH meter combination electrode (Ag/AgCl) (Thermo Electron Corporation, Waltham, MA, model 9826BN) using an Accumet AP61 pH meter (Fisher Scientific, Pittsburgh, PA).
NMR spectroscopy
All NMR experiments were collected on a Varian INOVA 600 MHz spectrometer using a 5 mm triple resonance probe equipped with triple-axis pulsed magnetic field gradients and utilized pulse sequences from the Varian BioPack User Library (Varian Inc., Palo Alto, CA). Histidine side-chain resonance assignments were determined using lH-13C HSQC and 1H-15N HSQC NMR experiments acquired at 35 °C. The lH-13C HSQC spectra were recorded with a spectral width of 9000.9 Hz and 2048 total (real plus imaginary) points in the 1H dimension, a spectral width of 4100 Hz and 256 quadrature (real plus imaginary) increments in the 13C dimension, signal-averaged over 128 transients, and with a relaxation delay of 1 sec. For the His-tautomeric 1H-15N HSQC NMR experiments, the BioPack library “gNhsqc” pulse-sequence was modified as previously described (33) to allow GARP 13C decoupling during the evolution of the 15N chemical shift and spectra were recorded with a spectral width of 8000 Hz and 1024 total (real plus imaginary) points in the 1H dimension, a spectral width of 2500 Hz and 128 quadrature (real plus imaginary) increments in the 15N dimension, signal-averaged over 256 transients, and with a relaxation delay of 1 sec. In order to detect long-range couplings between imidazole-ring 15N nuclei and the 1Hδ1 and 1Hε2 nuclei, the INEPT relaxation delay was adjusted to correspond to an effective 2JNH coupling constant of 22 Hz, which represents a compromise between the theoretical value of 5 - 10 Hz (absolute value) and losses in signal-to-noise due to transverse (R2) relaxation (33). The spectra were recorded for the WT and mutant hPRL at pH values ranging from 3.97 to 8.30. All NMR spectra were processed using NMRPipe (34), with subsequent display and analysis in Sparky (35). Chemical shifts were referenced indirectly to external DSS at 0.00 ppm, with heteronuclear dimensions referenced using their relative gyromagnetic ratios (36).
Curve fitting and statistical analysis
NMR spectra were recorded at many pH values between pH 4.0 and 8.5 for WT and mutant variants of hPRL. Signs of protein precipitation were noticeable below pH 4.0. Equilibrium thermodynamic parameters were derived for eight of the nine histidines in hPRL from the pH dependencies of their NMR chemical shifts. Using the Scientist 3.0 Software Package (Micromath Research Inc., St. Louis, MO), the observed, pH-dependent chemical shifts (δobs) were fitted to a simple linear combination of the fully protonated (δAH+) and deprotonated (δA) states weighted by their relative populations:
| (Eq. 1) |
For each of the independently titrating residues (H46, H59, H138, H173 and H195),
| (Eq. 2) |
| (Eq. 3) |
For the triplet of coupled titration sites (H27, H30 and H180) modeled in Figure 2, a binding polynomial (37) was written as
| (Eq. 4) |
where, H = 10-pH and Kx = 10pKa,X. The fractions of protonated and unprotonated states with respect to any single residue “X” were expressed as
| (Eq. 5) |
| (Eq. 6) |
Figure 2.
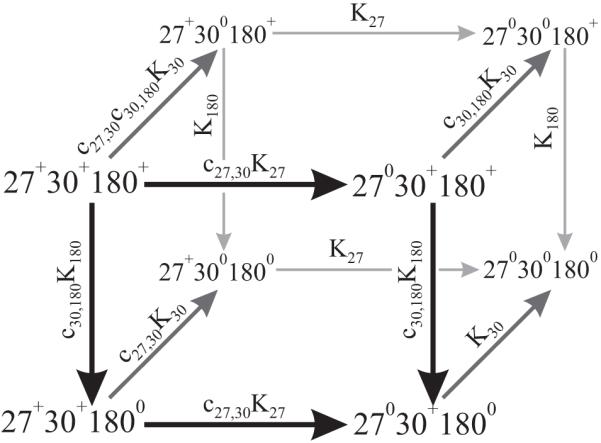
Thermodynamic model for protonation of the imidazole rings of H27, H30, and H180 in hPRL, where the superscript denotes the state of each residue. The linkage coefficients and acid dissociation constants describing each deprotonation reaction are shown above their respective arrows.
A variety of possible thermodynamic linkages were considered for the triplet of H27, H30 and H180, detailed in Results section below. Fixing the two cooperativity constants (c27,30 and c30,180) at unity during global fitting of the data simulated an independent titration model for these three residues. Alternatively, allowing either or both of the cooperativity constants to float during fitting evaluated possible thermodynamic linkages between either residues H27 and H30 alone, residues H30 and H180 alone, or both pairs of residues simultaneously.
For the statistical analysis presented in Tables 2 and 3 of the Results, F-statistics were calculated as follows;
| (Eq. 7) |
where SSE1 and SSE2 represented the sum-squared errors (or residuals) between the experimental data and their best fit values for the two models under comparison. The more simplistic model #1 was considered nested within (or a special case of) the more complex model #2. The number of fitting parameters for each model was represented by P2 and P1, where P2 must be greater than P1, and n was the number of independent experimental data points used to fit both models. Note that by definition the more complex model provides a better fit to the data (i.e. SSE1 > SSE2) and the F-statistic tests whether this improvement in the fit was statistically significant by comparison to corresponding “critical” values derived from the F-distribution, based on a desired probability threshold and the appropriate degrees of freedom in the comparison, in this case (P2 - P1) and (n - P2). Threshold probability values in Table 3 for each calculated F-statistic were calculated in Excel (Microsoft Corporation, Seattle, WA) using the FDIST command.
Table 2.
Summary of the “summed squared errors” (SSEs) and corresponding statistical parameters for each of the binding models considered. SSEs are separately calculated for each independent variable along with the total error
| (1) Independent | (2) c27,30 | (3) c30,180 | (4) c27,30 & c30,180 | N (data pts.) | |
|---|---|---|---|---|---|
| H27 1Hε1 SSEs | 0.024742 | 0.008757 | 0.024742 | 0.016949 | 34 |
| H30 1Hε1 SSEs | 0.087695 | 0.014108 | 0.015538 | 0.012907 | 34 |
| H180 1Hε1 SSEs | 0.058582 | 0.058582 | 0.014959 | 0.018397 | 34 |
| Total SSEs | 0.17102 | 0.081448 | 0.05524 | 0.048253 | 102 |
| No. of Params | 9 | 10 | 10 | 11 |
Table 3.
F-statistic calculations comparing each binding model with corresponding probability values in parentheses.1
| H27 Hε1 | (2) H27-H30 Interacting | (3) H30-H180 Interacting | (4) H27-H30 & H30-H180 Interacting |
|---|---|---|---|
| (1) All Independent | 43.8 (7.6 × 10-7) | Equivalent2 | 5.3 (0.013) |
| (2) H27-H30 Interacting | Rejected3 | ||
| (3) H30-H180 Interacting | 10.6 (0.004) |
| H180 Hε1 | (2) H27-H30 Interacting | (3) H30-H180 Interacting | (4) H27-H30 & H30-H180 Interacting |
|---|---|---|---|
| (1) All Independent | Equivalent2 | 70.0 (1.4 × 10-8) | 25.2 (1.6 × 10-6) |
| (2) H27-H30 Interacting | 50.2 (3.2 × 10-7) | ||
| (3) H30-H180 Interacting | Rejected3 |
| H30 Hε1 | (2) H27-H30 Interacting | (3) H30-H180 Interacting | (4) H27-H30 & H30-H180 Interacting |
|---|---|---|---|
| (1) All Independent | 125.2 (5.2 × 10-11) | 111.5 (1.7 × 10-10) | 66.6 (2.7 × 10-10) |
| (2) H27-H30 Interacting | 2.1(0.16) | ||
| (3) H30-H180 Interacting | 4.7 (0.041) |
| All Data | (2) H27-H30 Interacting | (3) H30-H180 Interacting | (4) H27-H30 & H30-H180 Interacting |
|---|---|---|---|
| (1) All Independent | 101.2 (1.7 × 10-16) | 192.8 (2.66 × 10-24) | 115.8 (9.9 × 10-26) |
| (2) H27-H30 Interacting | 62.6 (5.8 × 10-12) | ||
| (3) H30-H180 Interacting | 13.2 (4.7 × 10-4) |
The first three boxes represent each individual dependent variable used during fitting and the last considers all data.
No improvement in fitbetween models.
More complex model has poorer fit.
Monitoring urea unfolding of hPRL using intrinsic Trp fluorescence
A 9.8 M urea (American Bioanalytical, Natick, MA, ultra-pure urea) stock solution was prepared and its concentration confirmed from refractive index measurements using a hand-held refractometer (Atago U.S.A., Bellevue, WA, model R5000). A series of 24 buffered urea solutions were prepared in quadruplicate in a 96 deep-well tray using an automated reagent dispenser (Thermo Electron Corporation, Waltham, MA, model Multidrop DW) controlled via the serial port of a personal computer. All urea solutions contained 25 mM NaCl and were buffered to their reported pH with 25mM potassium phosphate buffer. All pH measurements were taken with an Accumet AP61 hand-held pH meter (Fisher Scientific, Pittsburgh, PA) and Orion glass micro pH combination electrode (Ag/AgCl) (Thermo Electron Corporation, Waltham, MA, model 9826BN). Protein solutions were manually transferred into the deep-well plates containing the buffered urea solutions using a multi-channel pipette to a typical final concentration of 6 μM, after which the deep-well plates were sealed with adhesive aluminum film, manually tumbled to stir and allowed to equilibrate for a minimum of two hours at room temperature (23 °C) prior to fluorescence readings. Protein/urea solutions were manually transferred to quartz cuvettes in a Cary Eclipse fluorescence spectrophotometer (Varian Instruments, Walnut Creek, CA) employing both temperature control and stirring with micro stir bars. Fluorescence measurements were carried out with an excitation wavelength of 280 nm with a 20 nm slit width and detection at 325 nm with a 5 nm slit width.
Results
Assignment of Histidine imidazole ring chemical shifts
lH-13C HSQC and 1H-15N HSQC spectra of uniformly 13C15N- and 15N-His labeled hPRL recorded at several pH values provided the data for the titration curves of the 1H, 13C and 15N nuclei in the histidine imidazole rings. The assignment of each histidine 1H(13C)ε imidazole chemical shifts signals in the lH-13C HSQC spectra was achieved using uniformly 13C,15N-labeled His to Ala hPRL mutants and pre-existing assignments available in the BMRB (accession codes: 5599 and 6643). These 1Hε chemical shift values from lH-13C HSQC spectra allow us to assign unequivocally the I5Nε I5Nδ and 1Hδ histidine chemical shifts in the 1H-15N HSQC hPRL spectra. The assigned chemical shifts are given as Supporting Information and also have been deposited in the BioMagResBank (http://www.bmrb.wisc.edu), BMRB accession number 15773.
Identification of the histidine tautomeric states
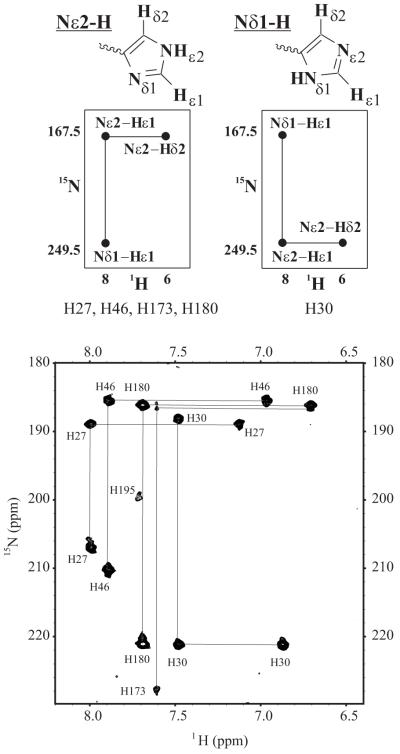
Previous investigators have recognized how histidine side-chains have distinctive 15N NMR chemical shifts depending on the protonation state of the imidazole ring (33;38). In the neutral state, the unprotonated nitrogen is located at higher NMR chemical shifts (up to 80 ppm) compared to its protonated counterpart. As the neutral imidazole ring becomes protonated with lowering of solution pH, the chemical shift of the originally unprotonated nitrogen migrates to progressively lower values. Figure 3 displays how these differences in chemical shifts, along with the relative intensities of the 2JNH-coupled 15Nδ1-1Hε1 and 15Nε2-1Hδ2 crosspeaks in a 1H-15N HSQC NMR spectrum, allow the identification of the His tautomeric state and correct chemical shift assignment of these four nuclei. In a neutral imidazole ring, depending on the location of the nitrogen-bonded hydrogen, two tautomeric states are possible. The most common tautomeric state (Nε2-H) has a pattern as shown in the upper left of Figure 3. The 1Hε1 correlates with two 15N frequencies, one at ∼167.5 ppm corresponding to the protonated nitrogen and one at ∼249.5 ppm corresponding to the unprotonated nitrogen. The 1Hδ2 correlates only with a nitrogen frequency corresponding to one protonated nitrogen as the 3J(15Nδ1-1Hδ2) coupling is too small (1 - 2 Hz) to result in an observable cross-peak. This pattern enables the unambiguous assignment of the nuclei and at the same time specifies which nitrogen is protonated. In the less common Nδ1-H tautomeric state shown in the upper right of Figure 3, the 15Nδ1 is protonated and the 15Nε2-1Hδ2 cross-peak is located at 249.5 ppm for the 15N chemical shift.
Figure 3.
Top: Schematic diagram showing the nomenclature used to describe the two possible tautomeric states of the neutral imidazole ring of histidines, termed Nε2-H and Nδ1-H, and the expected pattern of long-range (2JNH) correlations in the 1H-15N HSQC NMR spectrum [adapted from Pelton et al., 1993]. Bottom: 1H-15N HSQC spectrum of 15N His-labeled hPRL in 25 mM potassium phosphate buffer at pH 7.5 and 25 mM NaCl collected at 35°C. Histidine cross peak patterns are indicated for each residue. Tautomeric assignments for each residue are listed under the representative spectra. The tautomeric states of H57, H97, H138 and H195 are not assigned.
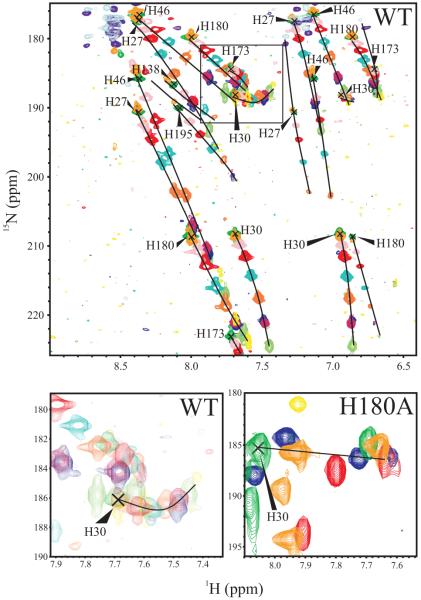
Shown at the bottom of Figure 3 is the 1H-15N HSQC spectrum collected on 15N His-labeled WT-hPRL at pH 7.5, with individual histidine spin systems noted. By simple inspection of their patterns we conclude that, at pH 7.5, H27, H46, H59, H173 and H180 are in the more common Nε2-H tautomeric state; whereas, H30 adopts the less common Nδ1-H tautomeric state. Spin systems for H59, H97, H138 and H195 were either missing or incomplete, precluding assignment of their tautomeric states. Although the 1Hδ2 and 1Hε1 nuclei detected in this experiment do not undergo rapid exchange with solvent, they are susceptible to line broadening from rapid amide hydrogen exchange elsewhere in the imidazole ring or potential conformational exchange processes, which we presume are responsible for the missing resonances. Lastly, Figure 4 shows superposed 1H-15N HSQC spectra of 15N His-labeled WT-hPRL ranging from pH 4.5 to 8.0. The changes in 1H and 15N chemical shifts versus pH are generally linear, consistent with perturbations due to only a single pH-dependent reaction, with the dramatic exception of the 15Nδ1 nucleus of H30. In the spectral expansion shown for WT hPRL, the H30 1Hε1-15Nδ1 crosspeak follows a highly curved trajectory, indicating a dependence on more than one pH-dependent effect. We hypothesize that, given the close relationship between the H30 and H180 imidazole rings in the tertiary structure of hPRL, the H30 15Nδ1 nucleus must detect protonation of both residues approximately equally. This was confirmed by reversion to a linear trajectory in 15N His-labeled H180A hPRL, which can be seen in the second expansion at the bottom right of Figure 4.
Figure 4.
Overlaid 1H-15N HSQC NMR spectra of 15N-his labeled WT and H180A PRL from pH 4.5 to 8.0. The 1H-15N chemical shift changes with pH generally follow linear trajectories as illustrated by the black trace, with low pH peaks at the start of the trajectories in the upper left. Spectral expansions are shown at the bottom depicting the curved trajectory for the H30 1Hε1-15Nδ1 crosspeak in WT hPRL (left), which reverts to a linear trajectory in H180A hPRL (right).
pH-dependent titration of His NMR chemical shifts for WT, H27A, H30A, H173A and H180A-hPRL
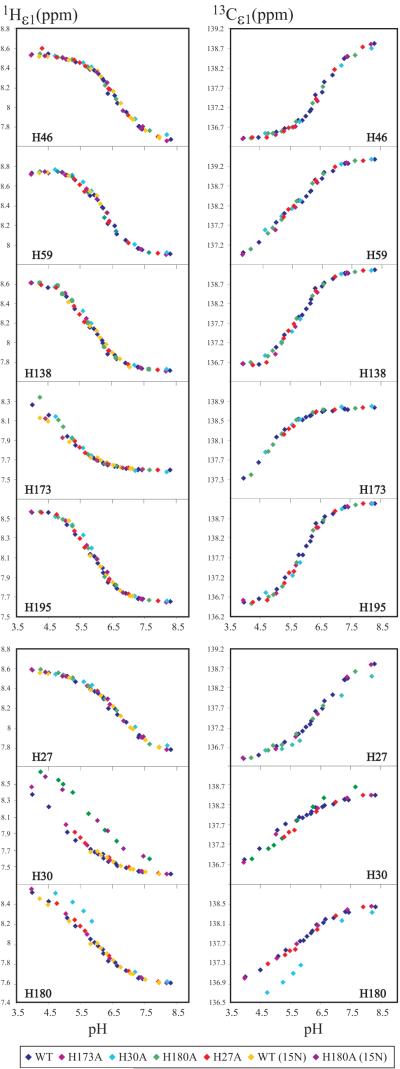
Figure 5 plots the 1Hε1 and 13Cε1 NMR chemical shifts against solution pH for eight of the nine His residues in hPRL. Titrations are superposed for both the WT protein and multiple His to Ala mutants in order to assess the effect of the mutations on the other residues. Not shown are the analogous pH titration curves for the five additional imidazole ring nuclei (i.e., 15Nε2, 15Nδ1, 1Hδ2, and 1Hε2), which were also collected for a majority of the residues. The 1Hε1 and 13Cε1 nuclei provided the largest number of data points and the most complete titration curves. For the five residues shown at the top of the figure, there is very good agreement of the titration data across the large number of independent NMR samples, which represent different protein preparations, labeling schemes and even mutants of hPRL. We compared simultaneous fitting of all six nuclei together, versus 1Hε1 and 13Cε1 together, and versus 1Hε1 chemical shifts on their own and, for all residues, there were no significant differences in the pKa values obtained.
Figure 5.
The 1Hε1 and 13Cε1 NMR chemical shifts are plotted against solution pH for eight of the nine His residues in hPRL. Titrations are superposed for both WT and multiple His to Ala mutants of hPRL.
Four of the nine His residues in hPRL fit well to the simplest model of a single, non-interacting pH titration site. As well, in all of the His to Ala mutations investigated, pH titrations for these residues overlapped well with the WT protein. Hence, we believe that H46, H138, H173 and H195 titrate in classical manner, independently from other residues. Representative, fitted titration curves for the 1Hε1 and 13Cε1 nuclei of H195 are shown at the top of Figure 6. Summarized in Table 4, the fitted pKa values for H46, H138, and H195 fall within the typical range of 6.0 - 6.5; whereas, the pKa for H173 is unusually low at 5.0. This residue is buried in tertiary structure of hPRL. Protonation of H173 would result in the highly unfavorable burial of an uncompensated positive charge, most likely resulting in disruption of the local, favorable structural interactions in order to allow hydration of the charged residue. Therefore, the neutral state of H173 is effectively stabilized, evidenced by its more acidic pKa.
Figure 6.
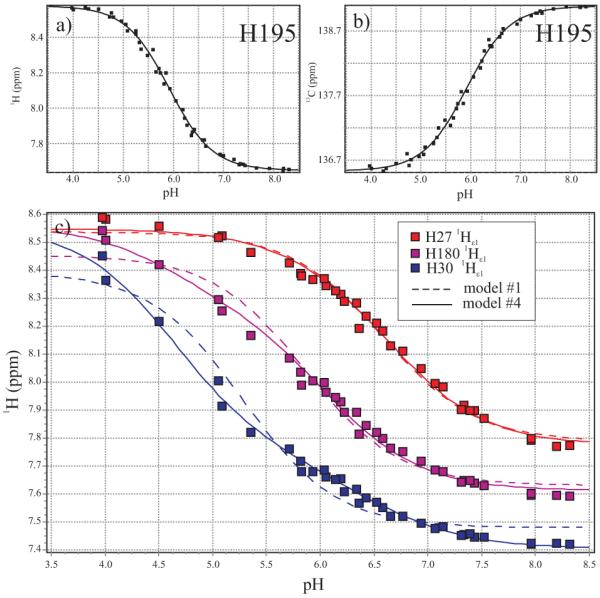
Experimental NMR chemical shifts and best-fit theoretical curves for histidine imidazole ring nuclei in WT hPRL as a function of pH. Panels (a) and (b), respectively, display 1Hε1 and 13Cε1 NMR chemical shifts for H195 fitted to a classic, non-interacting model of protonation. In panel (c), 1Hε1 chemical shifts for residues H30 (blue), H180 (mauve) and H27 (red) are shown with the best-fit curves for both the fully independent model (dashed line, c27,180 = c30,180 = 1, model #1 in Table 1) and the fully interacting model (solid line, c27,30 = 0.42 and c30,180 = 0.14, model #4 in Table 1) for each residue, colored to match the experimental data.
Table 4.
Summary of the best fit binding parameters and the corresponding 95% confidence intervals
| Value | Best Fit | 95% Confidence Interval |
|---|---|---|
| H46 pKa | 6.56 | 6.51-6.58 |
| H59 pKa | 5.84 | 5.71-5.97 |
| H138 pKa | 5.82 | 5.77-5.87 |
| H173 pKa | 4.99 | 4.91-5.07 |
| H195 pKa | 5.91 | 5.87-5.96 |
| H27 pKa | 6.64 | 6.49-6.79 |
| H30 pKa | 6.00 | 5.67-6.33 |
| H180 pKa | 5.96 | 5.83-6.09 |
| c27, 30 | 0.42 | 0.03-0.82 |
| c30, 180 | 0.14 | 0.02-0.27 |
In contrast to the above four independently titrating residues, our data suggest a strong interaction between H27, H30 and H180. Firstly, inspection of Figure 5 shows how mutation of either H30 or H180 strongly perturbs the pH titration curve of the other. Note that an interaction was already suspected based upon the unusual pH dependent trajectory of H30’s 15Nδ1 resonance in Figure 4. Similarly, titration curves for H27 and H30 in Figure 5 also show small, but significant, perturbations when the other is mutated; in contrast, H27 and H180 do not appear to have a significant direct interaction. Additional, independent evidence for these direct interactions can be derived from an analysis of their individual pH titration curves. In Table 1, we compare the results of simultaneous fitting the 1Hε1 titration curves for all three residues according to the thermodynamic model depicted in Figure 2, but considering four separate situations: (1) independent titration of all three residues, (2) linkage between H27 and H30 only, (3) linkage between H30 and H180 only, and (4) simultaneous linkage between both pairs of residues. At the bottom of Figure 6, best fit curves for the fully independent (1) and the fully interacting (4) models are compared to the experimental data.
Table 1.
Summary of fitting the 1Hε1 NMR chemical shifts for the triplet of H27, H30 and H180 according to the model of linked protonation sites in Figure 2
| Model | H27 pKa | H30 pKa | H180 pKa | c27,30 | c30,180 |
|---|---|---|---|---|---|
| (1) All Independent | 6.58 (6.36 - 6.80) |
5.28 (5.06 - 5.51) |
5.86 (5.67 - 6.05) |
1 (fixed) | 1 (fixed) |
| (2) H27-H30 Interacting | 6.66 (6.48 - 6.84) |
6.10 (5.87 - 6.34) |
5.85 (5.73 - 6.00) |
0.07 (0.02 - 0.13) |
1 (fixed) |
| (3) H30-H180 Interacting |
6.58 (6.45 - 6.71) |
5.70 (5.54 - 5.85) |
5.96 (5.82 - 6.10) |
1 (fixed) | 0.10 (0.01 - 0.20) |
| (4) H27-H30 & H30-H180 Interacting | 6.64 (6.49 - 6.79) |
6.00 (5.67 - 6.33) |
5.96 (5.83 - 6.09) |
0.42 (0.03 - 0.82) |
0.14 (0.02 - 0.27) |
Visually, it is evident from Figure 6 that the 1Hε1 titration curves for H30 and H180 do not fit well to the simplest model of independent protonation. The need for a more complex model to describe the titration of H27, H30 and H180 can be more rigorously supported by the use of the F-statistic. Calculation of F-statistics requires the residual sum-squared errors (SSEs) between the experimental data and their best fit values along with the degrees of freedom during fitting, which is derived from both the number of fitting parameters as well as the number of independent pieces of data. In order to more accurately describe the true degrees of freedom during the comparison of our three titration models, we have chosen to include only the 1Hε1 NMR chemical shift data, as the data from this nucleus is the most complete for this triplet of residues. Elsewhere, we have simultaneously fit multiple NMR chemical shifts derived from a single sample (e.g. 1Hε1 and 13Cε1) to derive individual pKa values for each residue. Although inclusion of multiple chemical shifts for each residue should not degrade the accuracy of the best-fit value, it does distort estimation of the statistical errors by over-estimating the number of truly independent pieces of data. In these experiments, the largest source of random error likely derives from the preparation of the individual NMR samples and, to a lesser degree, the separate acquisition of each NMR spectrum. These errors are contained equally in each of the chemical shift values acquired from each NMR spectrum collected on a single sample and, thus, cannot be considered truly independent measurements. This lack of independence is evidenced by a strong correlation between the residual errors (i.e. the difference between the experimental and fitted values) for each of the six nuclei in a single residue (not shown). By restricting our analysis to a single NMR chemical shift per residue, we are largely able to avoid this over-estimation of the statistical degrees of freedom and be more confident in the applicability of the F-statistic to identify and reject overly complex and statistically non-significant models. However, because we have chosen to globally fit the NMR chemical shifts for all three residues simultaneously (necessary for consideration of thermodynamic linkage between residues), we cannot avoid utilizing three independent variables from each NMR spectrum. Therefore, we have chosen to determine SSEs (Table 2) and F-statistics (Table 3) for each of the individual, dependent variables as well as their total. Neither approach is ideal, as the statistics for the isolated dependent variables ignore the compromises occurring during global fitting of the three residues simultaneously and, as well, the statistics for the overall (total) fit likely overestimates the number of independent data points.
Beginning at the top of Table 3, F-statistics for the isolated H27 1Hε1 dependent variable support the inclusion of thermodynamic linkage between H27 and H30, despite the small visual difference between the fitted curves for H27 in Figure 4. Note that the additional inclusion of linkage between H30 and H180 did not improve the fit for H27 when considered on its own, which should be expected. Similarly, when considering only the quality of fit for the H180 1Hε1 dependent variable, the F-statistics clearly support the presence of negative cooperativity between H30 and H180, but consideration of linkage between H27 and H30 does not improve agreement for H180. The situation for the H30 1Hε1 dependent variable is more complex. The quality of fit for H30 is improved in all cases by the addition of thermodynamic linkage between either pairs of residues (see Table 2) and the corresponding F-statistics are significant at a probability >95% for all comparisons except for one (see Table 3). Lastly, when all of the experimental data is considered together inclusion of both thermodynamic linkage parameters greatly improves the quality of fit and the corresponding F-statistics are highly significant in all cases, although the degree of statistical confidence is likely overestimated by the lack of complete independence between the data for each residue. Regardless, when all of the evidence is considered in aggregate, the existence of negative cooperativity between H27 and H30 and also between H30 and H180 is clear. The quality of fit for H27 and H30 depend on their linkage and similarly the quality of fit for H30 and H180 depend on their linkage. This statistical analysis is supported by the deviation in the NMR chemical shift titration curves for each residue in the His to Ala mutants shown in Figure 5.
We present in Table 4 our current, best assessment of the thermodynamic parameters describing the protonation reactions of 8 of the 9 His residues in hPRL. Note that although there can be no doubt about the existence of negative cooperativity between residues H27, H30 and H180, we cannot define the individual cooperativity constants with precision. In our various attempts to globally fit the selected NMR data to various models, these constants displayed the greatest variability in their best fit values. However, interestingly, their product (c27,30*c30,180) remained roughly constrained to the order of ∼ 0.1. This conservation was likely required to properly account for the reduced slope of the transition seen in the H30 1Hε1 titration curve compared to the non-cooperative case. Although our data appears to lack statistical power to precisely define their individual values, there is sufficient evidence to conclude that the negative cooperativity between H30 and H180 is stronger than that for H27 and H30. First, the H30 15Nδ1 NMR chemical shift in Figure 4 appears to depend on titration of both the H30 and H180 imidazole rings. Secondly, a more dramatic change is seen in the titration curves of both H30 and H180 when either is mutated as compared to H27 and H30. This conclusion is further supported by the closer relationship between H30 and H180 in the tertiary structure of hPRL compared to H27 and H30. Therefore, we have chosen to include the best fit values for these cooperativity constants in Table 4 as their relative magnitudes are in agreement with our assessment. However, uncertainty in their precise values is appropriately reflected in their relatively large 95% confidence limits.
Throughout the above description, we have purposely avoided discussion of the results for residues H59 and H97. In all of the above described NMR spectra of WT hPRL, NMR signals for the imidazole ring of H97 are either extremely weak or unobservable. Interestingly, when H59 was mutated to Ala (in order to confirm its assignment) a new strong NMR signal was observed, which was assigned as belonging to H97. Although not in direct contact, H59 and H97 are nearby structurally and both are part of a small cluster of aromatic residues between the two long loops and the 2nd and 4th helices. The most likely explanation for our results is that in WT hPRL the H97 imidazole NMR signals undergo chemical exchange on a timescale similar to their difference in NMR chemical shifts, most likely due to conformational mobility within this aromatic cluster. Mutation of H59 to Ala would be expected to structurally loosen these interactions and, thereby, increase the mobility of H97 to a faster exchange regime where it becomes observable. We also notice that the titration curve for H59 displays slight, systematic deviations from a model of independent titration similar to H30 and H180. We are currently investigating the structural and thermodynamic linkage between H59 and H97, which will be detailed in a future publication.
Contribution of selected histidines to global unfolding of hPRL
The individual contribution of each histidine within the interacting triplet to the global stability of hPRL was investigated using chemical denaturation. Figure 7 plots normalized, intrinsic Trp fluorescence across a wide range of urea concentrations for WT, H27A, H30A and H180A-hPRL. Approximately two-state global unfolding of the proteins is indicated by the sudden and sharp transitions between native-like and denatured protein fluorescence. Shifts in the global unfolding transitions to lower or higher urea concentrations indicate decreases or increases in the global unfolding free energies (ΔGunf), respectively. The experimental data have been fit to a theoretical description of unfolding as previously described (18) to derive quantitative descriptions of [urea]mid, the urea concentration at the midpoint of the transition, and the ΔGunf for each protein. The data clearly indicate that the presence of each individual imidazole ring for residues H27, H30 and H180 is overall destabilizing towards protein folding.
Figure 7.
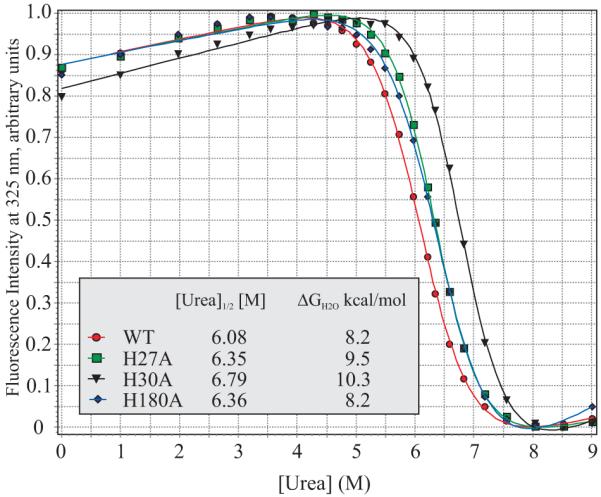
Chemical denaturation curves of WT, H27A, H30A and H180A-hPRL are presented in order to compare their global stabilities. Normalized, intrinsic Trp fluorescence is plotted as a function of urea concentration and the data fitted to theoretical curves, from which are derived the displayed [urea]med and ΔGunf values.
Discussion
Proteins often display pH dependence to their structural stability and functional behavior. Such pH dependence is ultimately mediated by the thermodynamics of protonation reactions of ionizable groups, which are located primarily in the side-chains of Asp, Glu, His, Arg and Lys residues. The pH dependence of any equilibrium between protein structural states (and thus functional states) can be completely described by detailing all the changes in protonation equilibrium constants (i.e. the pKa values) for all the individual ionizable groups. NMR spectroscopy is uniquely suited to describing site-specific protonation and has been routinely utilized to derive residue-specific pKa values (39-49), during which the identification of thermodynamic linkage between protonation sites is not uncommon. We recently discovered dramatic pH dependence to both the structural stability and receptor-binding affinity of hPRL, over the relatively narrow physiologic pH range of 6 - 8 (18). As the initial step to detailing the molecular basis of this pH dependence, we have analyzed the site-specific (or microscopic) titration curves for 8 of the 9 His residues in hPRL. In doing so, we describe a novel approach for combining a statistical analysis of NMR titration curves with site-directed mutagenesis to identify thermodynamic linkage, or cooperativity, between the protonation reactions of individual residues.
The protonation equilibrium constants (pKa values) for the histidines in hPRL all fall in a relatively typical range from 5.8 to 6.6, with the exception of H173 whose unusually acidic pKa of 5.0 is likely the result of the complete burial of its imidazole ring. The unusual and somewhat novel finding is the presence of thermodynamic linkage between the protonation reactions of H27, H30 and H180, corresponding to an overall perturbation in the free energy of protonation of approximately 1.4 kcal/mole. We considered the possibility of hydrogen bonding between these coupled imidazole rings, but this was not supported by their positioning in either the NMR (29) or the X-ray (28) structure of hPRL. Shown in Figure 8, H27 and H30 are not close enough to support hydrogen-bonding (> 3 Å) and H30 and H180 are stacked vertically in a staggered manner consistent with favorable aromatic stacking interactions. Therefore, based upon this positioning, it appears more likely for the negative cooperativity between protonation reactions to be governed primarily by unfavorable electrostatics or, possibly, interference with aromatic stacking.
Figure 8.
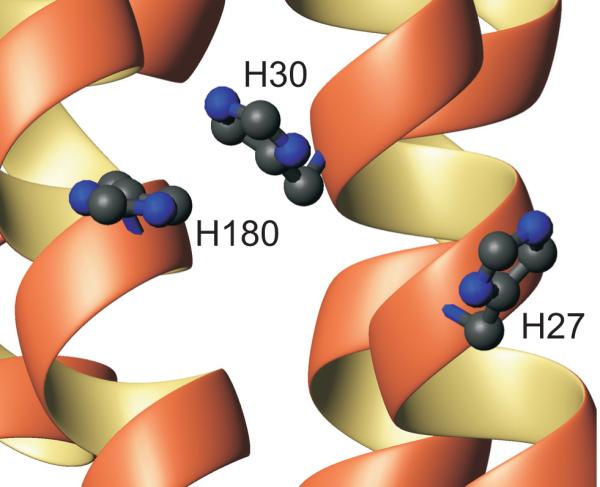
Positions of the H27, H30 and H180 imidazole rings in the X-ray crystallographic structure of a hPRL variant (PDB code: 2Q98).
The thermodynamic interactions between the H27, H30 and H180 described here are likely to contribute towards the biological function of hPRL, based on both their presumed structural location within the high affinity receptor-binding site (29) and an evolutionary argument regarding their unfavorable contribution to protein stability. First, although the structure of hPRL bound to its receptor has not been described, the related protein hGH also binds to the hPRL receptor (but only in the presence of Zn2+) and a high resolution structure of this complex has been reported (50). H27 and H30 from hPRL are conserved in hGH. Structurally, these homologous residues are located within the binding interface and directly coordinate the Zn2+ cation required for receptor recognition. H180 is homologous to an Asp in hGH that is also located within the interface but does not interact directly with the Zn2+ cation. Site-specific mutation of H27, H30 or H180 has been shown to decrease either the receptor-binding affinity or the mitogenic effects of hPRL (51), further supporting their expected location within the high affinity receptor-binding site. We also report here that single-site mutation of either H27, H30 or H180 in hPRL stabilizes the folded protein relative to its chemically denatured state. This is an unusual finding as a majority of amino acids in proteins appear to be evolutionarily optimized towards their contribution to global stability (52). Conformational strain associated with conserved residues, such as seen here, is often an indicator of functional importance. Although the current results do not clearly define the structural basis of the previously reported pH dependent behavior (18), they do form a foundation for ongoing studies on the contribution of specific His residues to the structural stability and receptor-binding properties of hPRL. In the future, we hope that by directly measuring site-specific changes in pKa values associated with receptor-binding and protein unfolding, a better understanding of the biophysical basis of hPRL’s pH dependent behavior will ultimately result.
Supplementary Material
Acknowledgements
We thank Drs. Jay Emerson and John Hartigan in the Department of Statistics, Yale University for useful discussions.
The abbreviations used are
- hPRL
human prolactin
- hGH
human growth hormone
- hPL
human placental lactogen
- WT
wild type
- NMR
nuclear magnetic resonance
- Asp
aspartate
- Glu
glutamate
- Lys
lysine
- Arg
arginine
- His
histidine
- Ala
alanine
- DSS
2,2-Dimethyl-2-silapentane-5-sulfonate sodium salt
- PPM
part per million
- HSQC
heteronuclear single quantum coherence
- 2D
two-dimensional
- INEPT
insensitive nuclei enhanced (by) polarization transfer
- SSE
sum-squared error
- IPTG
Isopropyl β-D-1-thiogalactopyranoside
- PMSF
phenylmethylsulphonyl fluoride
- GARP
Globally Optimized Alternating Phase Rectangular Pulse
Footnotes
This work was supported by the National Institutes of Health R01 grant: CA108992.
Chemical shift data have been deposited in BioMagResBank (BMRB), entry code 15773.
References
- 1.Goffin V, Binart N, Touraine P, Kelly PA. Prolactin: The new biology of an old hormone. Annual Review of Physiology. 2002;64:47–67. doi: 10.1146/annurev.physiol.64.081501.131049. [DOI] [PubMed] [Google Scholar]
- 2.Wells JA, de Vos AM. Hematopoietic receptor complexes. Annu. Rev. Biochem. 1996;65:609–634. doi: 10.1146/annurev.bi.65.070196.003141. [DOI] [PubMed] [Google Scholar]
- 3.Ben-Jonathan N, Mershon JL, Allen DL, Steinmetz RW. Extrapituitary prolactin: distribution, regulation, functions, and clinical aspects. Endocr. Rev. 1996;17:639–669. doi: 10.1210/edrv-17-6-639. [DOI] [PubMed] [Google Scholar]
- 4.Ben-Jonathan N, Liby K, McFarland M, Zinger M. Prolactin as an autocrine/paracrine growth factor in human cancer. Trends in Endocrinology & Metabolism. 2002;13:245–250. doi: 10.1016/s1043-2760(02)00603-3. [DOI] [PubMed] [Google Scholar]
- 5.Clevenger CV, Furth PA, Hankinson SE, Schuler LA. The role of prolactin in mammary carcinoma. Endocrine Reviews. 2003;24:1–27. doi: 10.1210/er.2001-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goffin V, Touraine P, Pichard C, Bernichtein S, Kelly PA. Should prolactin be reconsidered as a therapeutic target in human breast cancer? Mol. Cell. Endocrinol. 1999;151:79–87. doi: 10.1016/s0303-7207(99)00023-4. [DOI] [PubMed] [Google Scholar]
- 7.Leav I, Merk FB, Lee KF, Loda M, Mandoki M, McNeal JE, Ho SM. Prolactin receptor expression in the developing human prostate and in hyperplastic, dysplastic, and neoplastic lesions. Am. J. Pathol. 1999;154:863–870. doi: 10.1016/S0002-9440(10)65333-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu X, Kreye E, Kuo CB, Walker AM. A molecular mimic of phosphorylated prolactin markedly reduced tumor incidence and size when DU145 human prostate cancer cells were grown in nude mice. Cancer Res. 2001;61:6098–6104. [PubMed] [Google Scholar]
- 9.Bernichtein S, Kinet S, Jeay S, Llovera M, Madern D, Martial JA, Kelly PA, Goffin V. S179D-human PRL, a pseudophosphorylated human PRL analog, is an agonist and not an antagonist. Endocrinology. 2001;142:3950–3963. doi: 10.1210/endo.142.9.8369. [DOI] [PubMed] [Google Scholar]
- 10.Xu X, Wu W, Williams V, Khong A, Chen YH, Deng C, Walker AM. Opposite effects of unmodified prolactin and a molecular mimic of phosphorylated prolactin on morphology and the expression of prostate specific genes in the normal rat prostate. Prostate. 2003;54:25–33. doi: 10.1002/pros.10168. [DOI] [PubMed] [Google Scholar]
- 11.Ueda E, Ozerdem U, Chen YH, Yao M, Huang KT, Sun H, Martins-Green M, Bartolini P, Walker AM. A molecular mimic demonstrates that phosphorylated human prolactin is a potent anti-angiogenic hormone. Endocr. Relat. Cancer. 2006;13:95–111. doi: 10.1677/erc.1.01076. [DOI] [PubMed] [Google Scholar]
- 12.Maciejewski PM, Peterson FC, Anderson PJ, Brooks CL. Mutation of serine 90 to glutamic acid mimics phosphorylation of bovine prolactin. J. Biol. Chem. 1995;270:27661–27665. doi: 10.1074/jbc.270.46.27661. [DOI] [PubMed] [Google Scholar]
- 13.Schenck EJ, Canfield JM, Brooks CL. Functional relationship of serine 90 phosphorylation and the surrounding putative salt bridge in bovine prolactin. Mol Cell Endocrinol. 2003;204:117–125. doi: 10.1016/s0303-7207(03)00123-0. [DOI] [PubMed] [Google Scholar]
- 14.Aranda J, Rivera JC, Jeziorski MC, Riesgo-Escovar J, Nava G, Lopez-Barrera F, Quiroz-Mercado H, Berger P, Martinez de la EG, Clapp C. Prolactins are natural inhibitors of angiogenesis in the retina. Invest. Ophthalmol. Vis. Sci. 2005;46:2947–2953. doi: 10.1167/iovs.05-0173. [DOI] [PubMed] [Google Scholar]
- 15.Piwnica D, Touraine P, Struman I, Tabruyn S, Bolbach G, Clapp C, Martial JA, Kelly PA, Goffin V. Cathepsin D processes human prolactin into multiple 16K-like N-terminal fragments: study of their antiangiogenic properties and physiological relevance. Mol. Endocrinol. 2004;18:2522–2542. doi: 10.1210/me.2004-0200. [DOI] [PubMed] [Google Scholar]
- 16.Corbacho AM, Nava G, Eiserich JP, Noris G, Macotela Y, Struman I, Martinez De La Escalera G, Freeman BA, Clapp C. Proteolytic cleavage confers nitric oxide synthase inducing activity upon prolactin. J. Biol. Chem. 2000;275:13183–13186. doi: 10.1074/jbc.275.18.13183. [DOI] [PubMed] [Google Scholar]
- 17.Clapp C, Martial JA, Guzman RC, Rentierdelrue F, Weiner RI. The 16-Kilodalton N-Terminal Fragment of Human Prolactin Is a Potent Inhibitor of Angiogenesis. Endocrinology. 1993;133:1292–1299. doi: 10.1210/endo.133.3.7689950. [DOI] [PubMed] [Google Scholar]
- 18.Keeler C, Jablonski EM, Albert YB, Taylor BD, Myszka DG, Clevenger CV, Hodsdon ME. The kinetics of binding human prolactin, but not growth hormone, to the prolactin receptor vary over a physiologic pH range. Biochemistry. 2007;46:2398–2410. doi: 10.1021/bi061958v. [DOI] [PubMed] [Google Scholar]
- 19.Dannies PS. Mechanisms for storage of prolactin and growth hormone in secretory granules. Mol. Genet. Metab. 2002;76:6–13. doi: 10.1016/s1096-7192(02)00011-2. [DOI] [PubMed] [Google Scholar]
- 20.Sankoorikal BJ, Zhu YL, Hodsdon ME, Lolis E, Dannies PS. Aggregation of human wild-type and H27A-prolactin in cells and in solution: Roles of Zn2+, Cu2+, and pH. Endocrinology. 2002;143:1302–1309. doi: 10.1210/endo.143.4.8732. [DOI] [PubMed] [Google Scholar]
- 21.Hilfiker-Kleiner D, Kaminski K, Podewski E, Bonda T, Schaefer A, Sliwa K, Forster O, Quint A, Landmesser U, Doerries C, Luchtefeld M, Poli V, Schneider MD, Balligand JL, Desjardins F, Ansari A, Struman I, Nguyen NQ, Zschemisch NH, Klein G, Heusch G, Schulz R, Hilfiker A, Drexler H. A cathepsin D-cleaved 16 kDa form of prolactin mediates postpartum cardiomyopathy. Cell. 2007;128:589–600. doi: 10.1016/j.cell.2006.12.036. [DOI] [PubMed] [Google Scholar]
- 22.Leinwand LA. Molecular events underlying pregnancy-induced cardiomyopathy. Cell. 2007;128:437–438. doi: 10.1016/j.cell.2007.01.020. [DOI] [PubMed] [Google Scholar]
- 23.Piwnica D, Fernandez I, Binart N, Touraine P, Kelly PA, Goffin V. A new mechanism for prolactin processing into 16K PRL by secreted cathepsin D. Mol. Endocrinol. 2006;20:3263–3278. doi: 10.1210/me.2006-0044. [DOI] [PubMed] [Google Scholar]
- 24.Clapp C, Gonzalez C, Macotela Y, Aranda J, Rivera JC, Garcia C, Guzman J, Zamorano M, Vega C, Martin C, Jeziorski MC, de la Escalera GM. Vasoinhibins: a family of N-terminal prolactin fragments that inhibit angiogenesis and vascular function. Front. Horm. Res. 2006;35:64–73. doi: 10.1159/000094309. [DOI] [PubMed] [Google Scholar]
- 25.Lkhider M, Castino R, Bouguyon E, Isidoro C, Ollivier-Bousquet M. Cathepsin D released by lactating rat mammary epithelial cells is involved in prolactin cleavage under physiological conditions. J. Cell Sci. 2004;117:5155–5164. doi: 10.1242/jcs.01396. [DOI] [PubMed] [Google Scholar]
- 26.Gerweck LE. The pH difference between tumor and normal tissue offers a tumor specific target for the treatment of cancer. Drug Resist. Updat. 2000;3:49–50. doi: 10.1054/drup.2000.0122. [DOI] [PubMed] [Google Scholar]
- 27.Brooks DJ, Fresco JR, Lesk AM, Singh M. Evolution of amino acid frequencies in proteins over deep time: inferred order of introduction of amino acids into the genetic code. Mol. Biol. Evol. 2002;19:1645–1655. doi: 10.1093/oxfordjournals.molbev.a003988. [DOI] [PubMed] [Google Scholar]
- 28.Jomain JB, Tallet E, Broutin I, Hoos S, van AJ, Ducruix A, Kelly PA, Kragelund BB, England P, Goffin V. Structural and thermodynamical bases for the design of pure prolactin receptor antagonists. X-ray structure of Del1-9-G129R-hPRL. J. Biol. Chem. 2007 doi: 10.1074/jbc.M704364200. [DOI] [PubMed] [Google Scholar]
- 29.Teilum K, Hoch JC, Goffin V, Kinet S, Martial JA, Kragelund BB. Solution structure of human prolactin. J. Mol. Biol. 2005;351:810–823. doi: 10.1016/j.jmb.2005.06.042. [DOI] [PubMed] [Google Scholar]
- 30.Keeler C, Dannies PS, Hodsdon ME. The tertiary structure and backbone dynamics of human prolactin. Journal of Molecular Biology. 2003;328:1105–1121. doi: 10.1016/s0022-2836(03)00367-x. [DOI] [PubMed] [Google Scholar]
- 31.Waugh DS. Genetic tools for selective labeling of proteins with alpha-15N-amino acids. J. Biomol. NMR. 1996;8:184–192. doi: 10.1007/BF00211164. [DOI] [PubMed] [Google Scholar]
- 32.Peterson FC, Gordon NC, Gettins PG. High-level bacterial expression and 15N-alanine-labeling of bovine trypsin. Application to the study of trypsin-inhibitor complexes and trypsinogen activation by NMR spectroscopy. Biochemistry. 2001;40:6275–6283. doi: 10.1021/bi0100992. [DOI] [PubMed] [Google Scholar]
- 33.Pelton JG, Torchia DA, Meadow ND, Roseman S. Tautomeric states of the active-site histidines of phosphorylated and unphosphorylated IIIGlc, a signal-transducing protein from Escherichia coli, using two-dimensional heteronuclear NMR techniques. Protein Sci. 1993;2:543–558. doi: 10.1002/pro.5560020406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. Nmrpipe - a Multidimensional Spectral Processing System Based on Unix Pipes. Journal of Biomolecular NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 35.Kneller DG, Goddard TD. SPARKY. University of California; San Francisco: 1997. [Google Scholar]
- 36.Cavanagh J. Protein NMR spectroscopy: principles and practice. Academic Press; San Diego: 1996. Referencing; pp. 175–176. [Google Scholar]
- 37.Gill SJ, Richey B, Bishop G, Wyman J. Generalized binding phenomena in an allosteric macromolecule. Biophys. Chem. 1985;21:1–14. doi: 10.1016/0301-4622(85)85001-8. [DOI] [PubMed] [Google Scholar]
- 38.Bachovchin WW. 15N NMR spectroscopy of hydrogen-bonding interactions in the active site of serine proteases: evidence for a moving histidine mechanism. Biochemistry. 1986;25:7751–7759. doi: 10.1021/bi00371a070. [DOI] [PubMed] [Google Scholar]
- 39.Andre I, Linse S, Mulder FA. Residue-specific pKa determination of lysine and arginine side chains by indirect 15N and 13C NMR spectroscopy: application to apo calmodulin. J. Am. Chem. Soc. 2007;129:15805–15813. doi: 10.1021/ja0721824. [DOI] [PubMed] [Google Scholar]
- 40.Chivers PT, Prehoda KE, Volkman BF, Kim BM, Markley JL, Raines RT. Microscopic pKa values of Escherichia coli thioredoxin. Biochemistry. 1997;36:14985–14991. doi: 10.1021/bi970071j. [DOI] [PubMed] [Google Scholar]
- 41.Gao G, DeRose EF, Kirby TW, London RE. NMR determination of lysine pKa values in the Pol lambda lyase domain: mechanistic implications. Biochemistry. 2006;45:1785–1794. doi: 10.1021/bi051856p. [DOI] [PubMed] [Google Scholar]
- 42.Joshi MD, Hedberg A, McIntosh LP. Complete measurement of the pKa values of the carboxyl and imidazole groups in Bacillus circulans xylanase. Protein Sci. 1997;6:2667–2670. doi: 10.1002/pro.5560061224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kesvatera T, Jonsson B, Thulin E, Linse S. Measurement and modelling of sequence-specific pKa values of lysine residues in calbindin D9k. J. Mol. Biol. 1996;259:828–839. doi: 10.1006/jmbi.1996.0361. [DOI] [PubMed] [Google Scholar]
- 44.Khare D, Alexander P, Antosiewicz J, Bryan P, Gilson M, Orban J. pKa measurements from nuclear magnetic resonance for the B1 and B2 immunoglobulin G-binding domains of protein G: comparison with calculated values for nuclear magnetic resonance and X-ray structures. Biochemistry. 1997;36:3580–3589. doi: 10.1021/bi9630927. [DOI] [PubMed] [Google Scholar]
- 45.Lindman S, Linse S, Mulder FA, Andre I. Electrostatic contributions to residue-specific protonation equilibria and proton binding capacitance for a small protein. Biochemistry. 2006;45:13993–14002. doi: 10.1021/bi061555v. [DOI] [PubMed] [Google Scholar]
- 46.McIntosh LP, Hand G, Johnson PE, Joshi MD, Korner M, Plesniak LA, Ziser L, Wakarchuk WW, Withers SG. The pKa of the general acid/base carboxyl group of a glycosidase cycles during catalysis: a 13C-NMR study of bacillus circulans xylanase. Biochemistry. 1996;35:9958–9966. doi: 10.1021/bi9613234. [DOI] [PubMed] [Google Scholar]
- 47.Oliveberg M, Arcus VL, Fersht AR. pKA values of carboxyl groups in the native and denatured states of barnase: the pKA values of the denatured state are on average 0.4 units lower than those of model compounds. Biochemistry. 1995;34:9424–9433. doi: 10.1021/bi00029a018. [DOI] [PubMed] [Google Scholar]
- 48.Schubert M, Poon DK, Wicki J, Tarling CA, Kwan EM, Nielsen JE, Withers SG, McIntosh LP. Probing electrostatic interactions along the reaction pathway of a glycoside hydrolase: histidine characterization by NMR spectroscopy. Biochemistry. 2007;46:7383–7395. doi: 10.1021/bi700249m. [DOI] [PubMed] [Google Scholar]
- 49.Tan YJ, Oliveberg M, Davis B, Fersht AR. Perturbed pKA-values in the denatured states of proteins. J. Mol. Biol. 1995;254:980–992. doi: 10.1006/jmbi.1995.0670. [DOI] [PubMed] [Google Scholar]
- 50.Somers W, Ultsch M, DeVos AM, Kossiakoff AA. The X-Ray Structure of a Growth-Hormone Prolactin Receptor Complex. Nature. 1994;372:478–481. doi: 10.1038/372478a0. [DOI] [PubMed] [Google Scholar]
- 51.Goffin V, Shiverick KT, Kelly PA, Martial JA. Sequence-function relationships within the expanding family of prolactin, growth hormone, placental lactogen, and related proteins in mammals. Endocrine Reviews. 1996;17:385–410. doi: 10.1210/edrv-17-4-385. [DOI] [PubMed] [Google Scholar]
- 52.Richards FM. Protein stability: still an unsolved problem. Cell Mol. Life Sci. 1997;53:790–802. doi: 10.1007/s000180050100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.