Abstract
We hypothesize that TCDD induced developmental neurotoxicity is modulated through an AhR dependent interaction with key regulatory neuronal differentiation pathways during telencephalon development. To test this hypothesis we examined global gene expression in both dorsal and ventral telencephalon tissues in E13.5 AhR -/- and wildtype mice exposed to TCDD or vehicle. Consistent with previous biochemical, pathological and behavioral studies, our results suggest TCDD initiated changes in gene expression in the developing telencephalon are primarily AhR dependent, as no statistically significant gene expression changes are evident after TCDD exposure in AhR -/- mice. Based on a gene regulatory network for neuronal specification in the developing telencephalon, the present analysis suggests differentiation of GABAergic neurons in the ventral telencephalon is compromised in TCDD exposed and AhR-/- mice. In addition, our analysis suggests Sox11 may be directly regulated by AhR based on gene expression and comparative genomics analyses. In conclusion, this analysis supports the hypothesis that AhR has a specific role in the normal development of the telencephalon and provides a mechanistic framework for neurodevelopmental toxicity of chemicals that perturb AhR signaling.
Keywords: neurodevelopment, gene expression, bioinformatics, TCDD, dioxin, neurotoxicity
Introduction
Polychlorinated biphenyls (PCBs) and polychlorinated dibenzodioxins (PCDDs) are persistent environmental contaminants that accumulate in fat and are transferred from mother to offspring during gestation and lactation [1, 2]. 2,3,7,8-Tetrachlorodibenzo-pdioxin (TCDD) is considered the most toxic PCDD and exposure results in well studied adverse effects on the immune system, the lung, the liver, and the kidneys [3, 4]. TCDD exposure affects brain development, manifesting as long-term cognitive deficits in rodents [5, 6] and monkeys [7, 8]. Human exposure to PCB mixtures is also correlated with adverse neurodevelopmental outcomes, such as learning, memory, attention and IQ deficits, in numerous studies [9-14].
Most of the known biological effects of TCDD, the PCDDs and some PCBs are mediated through its binding to the aryl hydrocarbon receptor (AhR), a member of the PAS family of bHLH transcription factors [15]. This step is crucial for developmental toxicity endpoints since exposure to high levels of TCDD (40 ug/kg) does not cause any gross teratogenic response in mice lacking the AhR [16]. A more recent study looking at subtle neurodevelopmental toxicity endpoints such as spatial memory tasks in AhR -/-, AhR +/-, and AhR +/+ mice suggests these endpoints are mediated through the AhR as well [17].
In vertebrates, the AhR is found in the cytosol in association with HSP90 chaperones, several accessory proteins and immunophilin-like proteins (XAP2/ARA9/AIP and p23) [15]. Upon ligand binding, the AhR translocates to the nucleus where it complexes with several co-factors to stimulate transcription of Phase 1 detoxification genes, such as cytochrome P450 CYP1A1, as well as several Phase II detoxification genes [18]. This signaling pathway responds to several exogenous compounds not limited to halogenated aromatic hydrocarbons, but also including several classes of plant-derived chemicals [19]. Therefore, one hypothesis is that this detoxification signaling system originally evolved to safely and efficiently metabolize exogenous compounds found in our plant-based diet [19].
Activation of the AhR by a ligand results in the rapid transcriptional activation of a large number of genes whose products control a broad spectrum of cellular functions seemingly unrelated to detoxification pathways, most notably cell division and cell fate [20-24]. Furthermore, several studies have shown activation of mammalian AhR does not need application of an exogenous ligand to be functional [25]. This has lead researchers to hypothesize about the presence of an as yet unknown endogenous ligand for AhR [15, 26-29]. Furthermore, recent studies in Caenorhabditis elegans have shown that the worm AhR protein does not respond to activation by classical xenobiotic ligands of the vertebrate AhR, but that this ancestral form functions to regulate neuronal differentiation, specifically by directing the fate and gene expression phenotype of two of the worm's 26 GABAergic neurons [30-32].
These novel roles of AhR may have important implications for mechanisitic studies of PCB/PCDD toxicity. Although induction of oxidative stress through the classic detoxification pathways may be important for several PCB/PCDD associated toxicity endpoints [33], recent research suggests other AhR-induced pathways may be responsible for other toxicity endpoints including neurodevelopmental endpoints [34]. For example, a study in zebrafish using a morpholino construct to block TCDD-induced Cyp1A expression suggests that TCDD-induced developmental toxicity is not mediated by Cyp1A upregulation [35].
Several important developmentally regulated pathways have been implicated as possible mediators of TCDD-induced developmental toxicity, such as TGF-β retinoic acid signaling, and GABAergic neuronal fate differentiation programs [34]. Furthermore, two independent studies have recently shown AhR-mediated induction of Hes-1, an important regulator of neuronal differentiation via the Notch pathway in the developing telencephalon [36, 37]. In zebrafish, exposure to a relatively low level of TCDD (100 ppt) causes reduced transcription of a key bHLH regulatory proneural differentiation gene, Ngn1 [38]. The products of the proneural bHLH genes, including Ngn1, Ngn2, and Mash1, have been shown to be both necessary and sufficient to direct the differentiation of glutamatergic and GABAergic phenotypes in the dorsal and ventral telencephalon of the mouse [39, 40]. Furthermore, AhR mRNA is colocalized with GAD67, an enzyme specific for the production of GABA, and a well known marker of GABAergic neurons, and TCDD exposure reduces Gad67 mRNA expression in the mouse brain [41].
TCDD exposure also causes reduced expression of Sonic Hedgehog (Shh) in zebrafish [42], an important extracellular morphogen that is key to the initiation of neuronal differentiation in all vertebrates, including rodents and humans [43]. Other studies have shown increased activation of Sp1, AP1 and NF-kB transcription factors in the developing brain after TCDD exposure [44, 45].
All in all, these studies suggest there is much to be discovered about the gene regulatory pathways perturbed in TCDD-induced neurodevelopmental toxicity. In particular, is GABAergic neuronal differentiation preferentially affected by AhR and/or TCDD in the developing mammalian telencephalon, as has been implicated in lower vertebrate and invertebrate species? If so, what are the potential genetic regulatory mechanisms behind this impact? To answer this question, we undertook global gene expression analyses in TCDD-exposed and AhR perturbed developing mouse dorsal and ventral telencephalon. These brain regions are specifically analyzed during peak neurogenesis (E13.5), when GABAergic neurons are being produced in the ventral telencephalon and glutamatergic neurons are being produced in the dorsal telencephalon.
Methods
Animals
Seven adult AhR (+/-) male and female mice generated on 129 × C57BL/6 backgrounds, as described in [46], were provided by Dr. Richard Peterson from the University of Wisconsin to establish a breeding colony. AhR wild type C57BL/6 albino (C57BL/6J-Tyrc-2J/J) animals were also purchased from Jackson Laboratories (Bar Harbor, MA, USA) and mated to AhR (-/-) animals from the initial mating of AhR (+/-) to supplement the original colony. Tail samples were collected for genotyping.
Prior to mating, all animals were singly-housed in polycarbonate cages measuring 17 cm wide, 28 cm long (476 cm2 area), and 13 cm height. When breeding, animals were housed in pairs overnight, one male to one female. Females were singly housed during the day, after morning plug check. All cages contained absorbent heat-treated hardwood bedding and cotton fiber nestlet. The mice were provided Purina Pico Chow No. 5001 (Ralston Purina Co., St. Louis, MO) ad libitum.
Exposure
Breeding age AhR (+/-) male (N=6) and female mice (N=6) were time-mated and two AhR (+/+) breeding pairs were time-mated.. Visual check for breeding plug occurred each morning after overnight matings; plugged females were singly housed, with day of plug counted as day 0.5 of gestation. Body weights of mated females were recorded daily in order to determine correct dosing.
On GD 11.5, three pregnant females were administered a dose of 5 micrograms/kg BW of 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD), CAS No. 1746-01-6 (Battelle, Inc.) in a solution of 95% corn oil/5% acetone, 5 ml/kg BW orally by gavage. Five controls were administered a solution of 95% corn oil/5% acetone, 5 ml/kg BW orally by gavage. This dosing regimen has been previously used to delineate AhR-mediated neurodevelopmental effects of TCDD exposure [17, 47]. Dosing occurred between the hours of 0830 and 1030.
Forty-eight hours after dosing (GD 13.5) all mice were euthanized by CO2 inhalation and exsanguinated by cardiac puncture. Under sterile conditions, the abdominal wall was opened and the uterine horn was removed intact and placed in cold (4°C) PBS. Using a stereoscope and #5 ultra fine forceps, each embryo was dissected from the uterus and transferred with a pipette to a small, clean Petri dish containing fresh, cold PBS where the yolk sac and umbilical vessels, the amniotic sac, and the tail were removed. The tails were placed in a labeled cryovial and stored at or below -70°C for genotyping. The embryos were decapitated at the neck and the heads were embedded in Tissue-Tek O.C.T. (optimum cutting temperature) resin (Sakura Finetek USA, Inc., Torrance, CA) with the superior aspect down, frozen in liquid nitrogen and stored at or below -70°C.
Laser Microdissection (LM)
Transverse sections were acquired per the NIEHS Laser Microdissection (LM) protocol using the Tissue Tek® system for frozen sections (Sakura, Torrance, CA). A total of 22 slides with two serial sections per slide were cut for the purpose of tissue mapping and laser microdissection from each embryo (N=3 per dose and genotype group). Slides numbered 1, 11 and 22 were cut at 6 μm on positive-charged slides (Daigger, Vernon Hills, IL) and stained with HemaCen® (American Master Tech Sci Inc., Lodi, CA) for tissue mapping. Slides numbered 2-10 and 12-21 were cut at 8 μm on PET foil slides (Molecular Machines and Industries, Inc. Haslett, MI), and immediately placed in a sterile slide box partially buried in dry ice to preserve RNA integrity and stored at -80°C for up to one week prior to LM. Following identification of the target areas by the investigator from digital images of the HemaCen®-stained map slides, frozen sections were pulled from the -80°C freezer, air dried for 30 seconds and stained with 1% aqueous Cresyl Violet acetate (Sigma-Aldrich, St. Louis, MO). Laser microdissection was completed within 20 minutes of cresyl violet staining on the UV laser Cellcut® microscope instrument (Molecular Machines and Industries) using the following instrument parameters. Laser speed and laser power ranged from 25-29% and 72-75% respectively while the laser focus was set at 45%.
Dorsal areas were microdissected into a single tube at an average of 1.5 million μm2 for each sample. Ventral areas were microdissected into a second tube at an average of 2.3 million μm2 per sample. To confirm RNA integrity, the entire tissue section from slide 21was lysed and placed into a third tube and designated as a whole lyse positive control. Detailed tissue handling/collection, staining and laser microdissection protocols are available at the NIEHS web site (http://www.niehs.nih.gov/research/atniehs/labs/lep/special/laser/index.cfm).
RNA Isolation and Microarray Hybridization
The PicoPure™ RNA Isolation Kit, Arcturus # KIT0202 / KIT0204 was used to lyse samples and isolate RNA from dorsal and ventral telencephalon samples following manufacturer's instructions. The quality of RNA samples was verified using the PicoChip for Agilent BioAnalyzer. Gene expression analysis was conducted using Affymetrix Mouse Genome 2.0 GeneChip® arrays (Mouse 430 v2, Affymetrix, Santa Clara, CA). Total RNA (3-20 ng) was amplified as directed in the Affymetrix Two-Cycle cDNA Synthesis protocol. Fifteen μg of amplified biotin-cRNAs were fragmented and 10 ug hybridized to each array for 16 hours at 45°C in a rotating hybridization oven using the Affymetrix Eukaryotic Target Hybridization Controls and protocol. Array slides were stained with streptavidin/phycoerythrin utilizing a double-antibody staining procedure and then washed using the EukGE-WS2v5 protocol of the Affymetrix Fluidics Station FS450 for antibody amplification. Arrays were scanned in an Affymetrix Scanner 3000 and data was obtained using the GeneChip® Operating Software (GCOS; Version 1.4.0.036).
Microarray Data Analysis
Initial data preprocessing, normalization, and Principal Component Analysis (PCA) was performed on all 24 samples (N=3 for each dose and genotype group) and all probes to characterize the variability present in the data. This analysis was performed using the Rosetta Resolver system (Version 7.0). Subsequent GC-RMA data normalization, t-test, and FDR calculations were performed in R downloaded from Bioconductor on April 15, 2008.
Quantification of Gene Regulatory Network
The Gene Regulatory Network (GRN) of telencephalon neuronal specification is based on work described in a previous publication [48]. Here, the present data is tested against this network to delineate likely points at which AhR and/or TCDD may perturb this process using the PathScope software originally described elsewhere [49]. Briefly, the strength of the relationships in the GRN were quantified to calculate the posterior probability distribution for the strength of the linkages based on the intensity values seen in the control, AhR -/-, or TCDD treatment gene expression datasets. A log-linear function was used to describe relationships between genes:
| (1) |
where αi is the level of gene expression independent of the network, Iji is an indicator function (0, 1, -1) if a linkage exists from gene j to gene i, βji is the degree to which change in gene j will affect change in gene i, Gj is a variable associated with the relative expression level of gene j compared with normal level j, εi, is the random error in predicted value for gene i and n is the number of genes in the network.
The posterior distributions for the linkages in each network were derived using Markov Chain Monte Carlo (MCMC) sampling methods [49]. For the current analysis, αi are assumed to have normal priors. The priors for the β are assumed normal with mean 0 and variance σ = 1. Finally, εi is assumed to be normally distributed with mean 0 and variance σ2 where σ2 is assumed to have a uniform prior with support defined by the observed data. The MCMC maximum sampling step sizes are 0.03 for the σ, 0.08 for the β, and 0.05 for the α, and 500,000 iterations were performed with decimation of every 10th value. The last 50,000 iterations were used to establish the mean value of βji and the significance of this value. Statistical significance of the parameter βji is defined by less than 5% of iterations with βji ≤ 0. To determine the impact of TCDD treatment and AhR knockout, the control, TCDD treated, and AhR KO datasets were ran individually. The mean ± STD βji ranges were then compared across these three runs. A difference was considered significant if the ranges of βji estimates from the control versus TCDD or AhR-/- did not overlap (corresponding roughly to a p≤0.1).
Comparative Sequence Analysis
The murine Sox11 gene (mm9 chr12:28018207-28027577) was uploaded into the ECR Browser (www.dcode.org). The Mulan algorithm [50] was used to perform a multiple sequence alignment across Sox 11 genes in human (hg 18 chr2:5749959-5758967), monkey (rheMac2 chr13:5770499-5779504), dog (canFam2 chr17:6556513-6563934), opossum (monDom4 chr1:535907235-535917096), and chicken (galGal3 chr3:97427132-97434301). The MultiTF algorithm [50]was then used to find AhR binding sites using the extended 19 bp long TRANSFAC matrix V$AHRARNT_2 (GCGCTGGCATGCAAACTCT) as described in [51].
Results
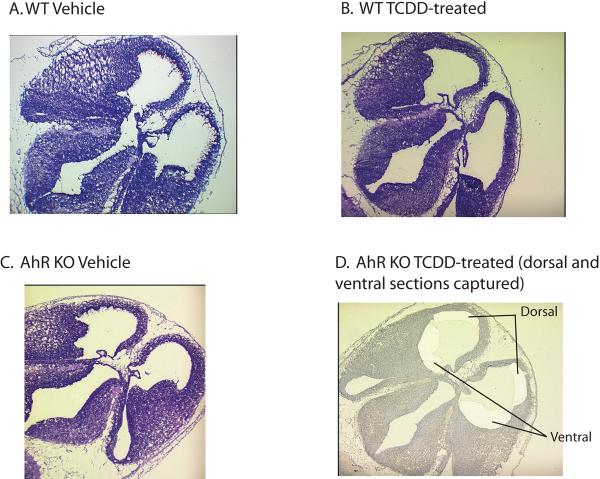
As found in previous analyses of AhR -/- mice [46], no gross morphological changes in the developing telencephalon were seen in the present study when compared with wildtype mice (Figure 1, A. and C.). In addition, a single dose of 5 μg/kg BW of 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) to pregnant females on GD 11.5 did not produce any gross morphological changes in the developing telencephalon on GD 13.5 (Figure 1, B. and D.). Importantly, this dosing regimen has been previously used to delineate AhR-mediated neurodevelopmental effects of TCDD exposure as measured by behavioral test batteries [17, 47] and careful stereological analyses have found evidence of TCDD-induced changes in hippocampal cell number at this dose level [17]. The goal of the present analysis is to determine gene expression changes that may be responsible for these more subtle neurodevelopmental and neurobehavioral impacts.
Figure 1. Frozen sections of developing telencephalon in AhR wild-type (A. and B.) and knockout (C. and D.) E13.5 mouse exposed to vehicle (A. and C.) or TCDD (B. and D.).
A.-C. show sections cut at 6 μm and stained with HemaCen® (American Master Tech Sci Inc., Lodi, CA) for tissue mapping. D. shows a section cut at 8 μm and stained with 1% aqueous Cresyl Violet acetate (Sigma-Aldrich, St. Louis, MO) and highlights the dorsal and ventral regions removed using laser capture microdissection for further gene expression analysis.
We analyzed global gene expression patterns in tissue obtained via laser capture microdissection in the dorsal and ventral developing telencephalon (illustrated in Figure 1, D.) from each treatment group. Between E11 and E15, these areas are undergoing rapid neurogenesis, producing mainly excitatory glutamatergic neurons in the dorsal telencephalon and mainly inhibitory GABAergic neurons in the ventral telencephalon [52]. This process is dependent on precise spatial and temporal expression of several bHLH and Homeobox transcription factors. Therefore our methodology was designed to determine specific AhR- and TCDD- induced impacts in these developing regions.
An initial Principal Component Analysis (PCA) of all probes from each of the 24 samples shows dorsal and ventral samples separate clearly from each other (Supplemental Figure 1). Within ventral samples, genotype and treatment form distinct groups, whereas separation by genotype and treatment are less distinct in dorsal samples (Supplemental Figure 2). This initial global analysis of the dataset supports our hypothesis that AhR-mediated and TCDD- mediated gene expression is preferentially affecting GABAergic inhibitory neuronal generation in the developing ventral telencephalon.
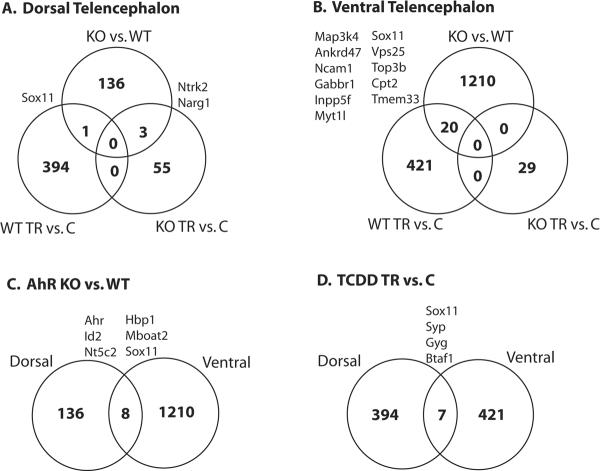
We next determined which probesets showed differential expression according to treatment and genotype within dorsal and ventral tissues. A Venn diagram gives a broad overview of the results (Figure 2). In both dorsal and ventral tissues, TCDD treatment had little or no effect on gene expression in the AhR KO, as only 55 and 29 probesets were considered differentially expressed at a p-value cut-off of 0.01 (FDR = 99%) in either dorsal or ventral tissues, respectively. This result corroborates a substantial body of evidence suggesting TCDD-induced neurodevelopmental toxicity is AhR-dependent [16, 35, 53, 54]. In addition, AhR status has a far greater impact in ventral versus dorsal tissues (1210 versus 136 differentially expressed probesets), suggesting a more prominent endogenous role for AhR in the development of ventral telencephalic structures. Finally, treatment with TCDD produced significant gene expression changes in both dorsal and ventral tissues, some of which are genes that are impacted by AhR status alone. In this Venn diagram, we have highlighted the most likely target genes of TCDD and/or AhR by identifying those genes that showed consistent differential expression across tissue type or in both TCDD- treated and AhR -/- datasets. Full tables of differential expression for each of these comparisons are provided in Supplementary Table 1.
Figure 2. Venn diagram of global gene expression analysis results in dorsal (A.) and ventral (B.) telencephalon.
KO refers to AhR -/-, WT refers to wildtype (AhR +/+), TR refers to treatment of TCDD on GD 11.5, and C refers to vehicle control group. Each circle reports the number of differentially expressed genes (p value < 0.001 and fold change greater than 1.2) within each comparison and the overlap between each comparison.
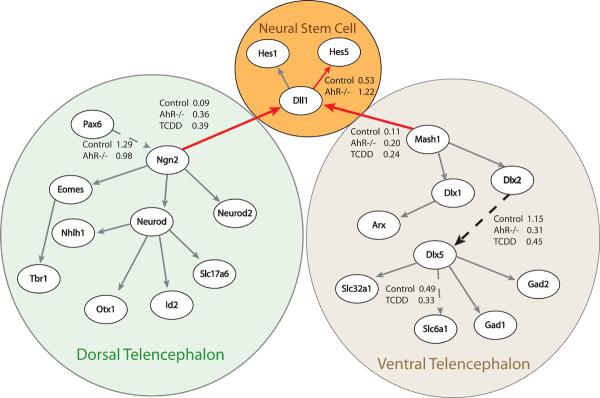
Next, we analyzed the present global gene expression dataset in the context of a gene regulatory network (GRN) describing initiation of neurogenesis in the murine dorsal and ventral telencephalon. A previous study developed this GRN based on a robust body of research describing the differentiation of glutamatergic neurons in the developing dorsal telencephalon via the proneural bHLH transcription factor Ngn2 and differentiation of GABAergic neurons in the developing ventral telencephalon via the proneural bHLH transcription factor Mash1 [48]. Using a previously developed algorithm for the quantification of a GRN [49], we tested whether this GRN is perturbed by AhR status or TCDD exposure, by quantifying the network separately based on either control, TCDD-treated, or AhR -/- datasets. Figure 3 depicts the results of this analysis suggesting that both TCDD exposure and AhR removal results in increased strength of connections between the bHLH proneural proteins (Ngn2 and Mash1) and the Notch signaling pathway (Dll1). Activation of the Notch pathway leads to increased expression of the Hes genes, which maintain the neural progenitor population through direct binding to Ngn2 and Mash1 and inhibition of DNA binding [55]. Consistent with this model, our analysis suggests reduced strength in connections promoting differentiation into neurons, such as Dlx1 to Dlx5, in the TCDD exposed and AhR KO tissues. Overall, these results suggest TCDD binding to AhR or AhR removal may reduce neurogenesis through prolonged Notch activation such that progression of neurogenesis is inhibited. This result is consistent with two other reports suggesting AhR activates Hes1, a downstream target of Notch signaling and a negative regulator of neurogenesis [36, 37].
Figure 3. Impact of TCDD exposure and AhR status on a Gene Regulatory Network (GRN) of neurogenesis in the developing telencephalon.
GRN is based on robust experimental evidence as detailed in [48]. Connection strengths were tested using a network quantification algorithm described in detail previously [49]. Increased thickness of an arrow identifies a significant increase (p< 0.10) in connection strengths when the algorithm is run using TCDD-exposed or AhR-/- datasets. A dotted arrow identifies a significant decrease (p< 0.10) in connection strength when the algorithm is run using TCDD-exposed or AhR -/- datasets.
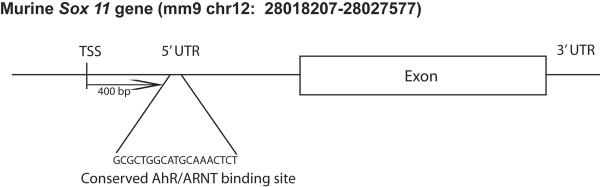
Finally, we annotated the predicted novel target genes of AhR and TCDD identified in Figure 2 to delineate putative relationships with the known GRN described above (Table 1). Corroborating our network analysis, several of the predicted target genes that are down-regulated in AhR-/- or TCDD exposed tissues are markers of neuronal differentiation (Ncam1, Gabbr1, Syp) or are known to play a critical role in neuronal differentiation (Sox11, Myt1l, Hbp1). In particular, Sox11 is differentially expressed based on AhR status and TCDD treatments in both dorsal and ventral tissues, making it a particularly compelling target gene. Therefore, we performed a comparative genomics analysis of Sox11 to determine if conserved binding sites for AhR are present in regions surrounding this gene. Figure 4 shows the results of this analysis depicting a previously unidentified AhR/ARNT binding site (extended 19 bp site developed by Sun et al. 2004) in the 5' untranslated region 400 bp upstream of the transcription start site (TSS). This site is conserved across human, monkey, dog, opossum, mouse, and chicken. Furthermore, we previously predicted Sox11 to be a key player in the neuronal specification process based on network analysis of microarray datasets from gain and loss of function studies for the proneural bHLH proteins [48]. In agreement with this bioinformatics prediction, a recent study showed Sox11 is a direct target for the proneural bHLH transcription factors and plays a critical role in supporting the differentiation cascade into GABAergic neurons in the developing ventral cortex [56].
Table 1.
Putative novel targets for TCDD and/or AhR based on microarray results for developing dorsal and ventral telencephalon tissue in AhR -/- and wildtype animals either treated with TCDD or vehicle.
| Direction of ChangeD/V* | |||||
|---|---|---|---|---|---|
| Gene Symbol | RefSeq ID | Gene Title | GO biological process term | TCDD exposure | AhR -/- genotype |
| Nt5c2 | NM_029810 | 5'-nucleotidase, cytosolic II | nucleotide metabolic process | -- | decreaseD/V |
| Vps25 | NM_026776 | vacuolar protein sorting 25 | regulation of transcription, DNA-dependent | increaseV | increaseV |
| Id2 | NM_010496 | inhibitor of DNA binding 2 | negative regulation of transcription | -- | increaseD/V |
| Mboat2 | NM_026037 | membrane O-acyltransferase domain containing 2 | --- | -- | decreaseD/V |
| Tmem33 | NM_028975 | transmembrane protein 33 | --- | increaseV | increaseV |
| Ncam1 | NM_010875 | neural cell adhesion molecule 1 | positive regulation of calcium-mediated signaling | decreaseV | decreaseV |
| Myt1l | NM_008666 | myelin transcription factor 1-like | nervous system development cell differentiation | decreaseV | decreaseV |
| Inpp5f | NM_178641 | inositol polyphosphate-5-phosphatase F | --- | decreaseV | decreaseV |
| Btaf1 | NM_001080706 | BTAF1 RNA polymerase II | --- | increaseD/V | -- |
| Hbp1 | NM_153198 | high mobility group box transcription factor 1 | Wnt receptor signaling pathway | -- | decreaseD/V |
| Map3k4 | NM_011948 | mitogen-activated | protein kinase kinase kinase 4 protein amino acid phosphorylation | increaseV | increaseV |
| Cpt2 | NM_009949 | carnitine palmitoyltransferase 2 | lipid metabolic process | increaseV | increaseV |
| Syp | NM_009305 | synaptophysin synaptic | transmission | decreaseD/V | -- |
| Gyg | NM_013755 | glycogenin | carbohydrate biosynthetic process | increaseD/V | -- |
| Sox11 | NM_009234 | SRY-box containing gene 11 | regulation of transcription, DNA-dependent | decreaseD/V | decreaseD/V |
| Gabbr1 | NM_019439 | gamma-aminobutyric acid (GABA-B) receptor, 1 | negative regulation of adenylate cyclase activity | decreaseV | decreaseV |
| Top3b | NM_011624 | topoisomerase (DNA) III beta | chromosome segregation | increaseV | increaseV |
D/V signifies whether the significant increase or decrease is found in Dorsal (D) or Ventral(V) tissue samples
Figure 4. Comparative genomics analysis of the Sox11 gene.
Using the ECRbrowser, Mulan, and MultiTF tools on the www.dcode.org site [70], an extended AhR/ARNT binding site (GCGCTGGCATGCAAACTCT) as described in [51] was identified in the 5' untranslated region (UTR) 400 bp upstream of the transcription start site (TSS). This site is conserved across Sox 11 genes in human (hg 18 chr2:5749959-5758967), monkey (rheMac2 chr13:5770499-5779504), dog (canFam2 chr17:6556513-6563934), opossum (monDom4 chr1:535907235-535917096), and chicken (galGal3 chr3:97427132-97434301).
In addition, the putative targets that show increased expression in TCDD-exposed or AhR -/- tissues have known functions in the negative regulation of neuronal differentiation (Id2) [57]or the positive regulation of proliferation (Vps25, Map3k4, Top3b) [58-60]. In the case of Map3k4 (often referred to as Mekk4 in the literature), recent studies show an important role in the regulation of neuronal migration in the developing telencephalon [61].
Discussion
This project elucidates AhR and non-AhR mediated gene expression perturbations by TCDD to key gene regulatory pathways during neurogenesis in the developing telencephalon. Previously, we had analyzed several wildtype and transgenic microarray datasets from developing telencephalon tissues [48], which offers a robust basis for analysis of results from the current project. Specifically, the downstream targets of several key proneural bHLH (Ngn1, Ngn2, Mash1) and Homeobox (Dlx1 and Dlx2) transcription factors are known to be critical for the initiation of neuronal differentiation and for the proper maintenance of the neural progenitor population through regulation of the Notch pathway. Precise balance of this regulatory network defines progression of neuronal generation in the cerebral cortex [62]. In fact, perturbation of this regulatory network has been implicated in the etiology of several neuropsychiatric and neurodegenerative diseases such as Schizophrenia, Bipolar Disorder, ADHD, Alzheimer's, and Parkinson's disease [63-67].
In short, our analysis determined AhR status and TCDD exposure preferentially impacted gene expression in the developing ventral telencephalon. Further analysis suggested loss of AhR either through gene deletion or TCDD binding decreased or delayed progression of neuronal differentiation in the ventral telencephalon, most likely through increased activation of the Notch signaling pathway, which maintains the neural progenitor population through activation of Hes1 and Hes5, both of which negatively regulate DNA binding of the proneural bHLH TFs Ngn2 and Mash1. This finding is consistent with previous reports suggesting Hes1 as a direct target of AhR [36, 37]. Finally, we have identified Sox11, a key regulator of neuronal differentiation in the telencephalon [56], as a novel putative target of AhR. Interestingly, our finding that gene expression changes after TCDD exposure largely replicate gene expression changes seen upon loss of AhR function suggests TCDD binding to the AhR may act as a competitive inhibitor of an endogenous function for AhR.
From a broadened perspective, this analysis provides a mechanistic hypothesis relating AhR perturbation to a specific neurodevelopmental gene regulatory program that could explain the long-term behavioral and intelligence deficits characteristic of prenatal and perinatal PCDD and coplanar PCB exposures [6, 13, 68, 69]. Furthermore, this project has illustrated the utility of a pathway quantification algorithm in identifying critical points at which a gene regulatory network is perturbed by the experimental condition. Future use of such algorithms will greatly increase the efficiency in interpretation of global gene expression datasets.
Supplementary Material
ACKNOWLEDGEMENTS
This research was supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INTEREST STATMENT The authors declare there are no conflicts of interest
REFERENCES
- 1.Hurst CH, DeVito MJ, Birnbaum LS. Tissue disposition of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) in maternal and developing Long-Evans rats following subchronic exposure. Toxicological Sciences. 2000;57(2):275–283. doi: 10.1093/toxsci/57.2.275. [DOI] [PubMed] [Google Scholar]
- 2.Li XL, Weber LWD, Rozman KK. Toxicokinetics of 2,3,7,8-Tetrachlorodibenzo-P-Dioxin in Female Sprague-Dawley Rats Including Placental and Lactational Transfer to Fetuses and Neonates. Fundamental and Applied Toxicology. 1995;27(1):70–76. doi: 10.1006/faat.1995.1109. [DOI] [PubMed] [Google Scholar]
- 3.Birnbaum LS. Endocrine Effects of Prenatal Exposure to Pcbs, Dioxins, and Other Xenobiotics - Implications for Policy and Future-Research. Environmental Health Perspectives. 1994;102(8):676–679. doi: 10.1289/ehp.94102676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walker NJ, et al. Dose-additive carcinogenicity of a defined mixture of “dioxin-like compounds”. Environmental Health Perspectives. 2005;113(1):43–48. doi: 10.1289/ehp.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seo BW, et al. Learning and memory in rats gestationally and lactationally exposed to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) Neurotoxicology and Teratology. 1999;21(3):231–239. doi: 10.1016/s0892-0362(98)00049-x. [DOI] [PubMed] [Google Scholar]
- 6.Widholm JJ, et al. Effects of perinatal exposure to 2,3,7,8-tetrachlorodibenzo-p-dioxin on spatial and visual reversal learning in rats. Neurotoxicology and Teratology. 2003;25(4):459–471. doi: 10.1016/s0892-0362(03)00014-x. [DOI] [PubMed] [Google Scholar]
- 7.Schantz SL, Bowman RE. Learning in Monkeys Exposed Perinatally to 2,3,7,8-Tetrachlorodibenzo-Para-Dioxin (Tcdd) Neurotoxicology and Teratology. 1989;11(1):13–19. doi: 10.1016/0892-0362(89)90080-9. [DOI] [PubMed] [Google Scholar]
- 8.Seegal R, Schantz SL. Neurochemical and behavioral sequelae of exposure to dioxins and PCBs. In: Schecter A, editor. Dioxins and Health. Plenum Press; New York: 1994. pp. 409–447. [Google Scholar]
- 9.Dahlgren J, et al. Health effects on nearby residents of a wood treatment plant. Environmental Research. 2003;92(2):92–98. doi: 10.1016/s0013-9351(02)00065-8. [DOI] [PubMed] [Google Scholar]
- 10.Darvill T, et al. Prenatal exposure to PCBs and infant performance on the Fagan Test of Infant Intelligence. Neurotoxicology. 2000;21(6):1029–1038. [PubMed] [Google Scholar]
- 11.Jacobson JL, Jacobson SW. Evidence for PCBs as neurodevelopmental toxicants in humans. Neurotoxicology. 1997;18(2):415–424. [PubMed] [Google Scholar]
- 12.Jacobson JL, Jacobson SW. Intellectual impairment in children exposed to polychlorinated biphenyls in utero. New England Journal of Medicine. 1996;335(11):783–789. doi: 10.1056/NEJM199609123351104. [DOI] [PubMed] [Google Scholar]
- 13.Huisman M, et al. Neurological condition in 18-month-old children perinatally exposed to polychlorinated biphenyls and dioxins. Early Human Development. 1995;43(2):165–176. doi: 10.1016/0378-3782(95)01674-0. [DOI] [PubMed] [Google Scholar]
- 14.Huisman M, et al. Perinatal Exposure to Polychlorinated-Biphenyls and Dioxins and Its Effect on Neonatal Neurological Development. Early Human Development. 1995;41(2):111–127. doi: 10.1016/0378-3782(94)01611-r. [DOI] [PubMed] [Google Scholar]
- 15.Gu YZ, Hogenesch JB, Bradfield CA. The PAS superfamily: Sensors of environmental and developmental signals. Annual Review of Pharmacology and Toxicology. 2000;40:519–561. doi: 10.1146/annurev.pharmtox.40.1.519. [DOI] [PubMed] [Google Scholar]
- 16.Mimura J, et al. Loss of teratogenic response to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) in mice lacking the Ah (dioxin) receptor. Genes to Cells. 1997;2(10):645–654. doi: 10.1046/j.1365-2443.1997.1490345.x. [DOI] [PubMed] [Google Scholar]
- 17.Powers BE, et al. Tetrachlorodibenzo-p-dioxin exposure alters radial arm maze performance and hippocampal morphology in female AhR(+/-) mice. Genes Brain and Behavior. 2005;4(1):51–59. doi: 10.1111/j.1601-183X.2004.00098.x. [DOI] [PubMed] [Google Scholar]
- 18.Hankinson O. The Aryl-Hydrocarbon Receptor Complex. Annual Review of Pharmacology and Toxicology. 1995;35:307–340. doi: 10.1146/annurev.pa.35.040195.001515. [DOI] [PubMed] [Google Scholar]
- 19.Gonzalez FJ, Nebert DW. Evolution of the P450-Gene Superfamily - Animal Plant Warfare, Molecular Drive and Human Genetic-Differences in Drug Oxidation. Trends in Genetics. 1990;6(6):182–186. doi: 10.1016/0168-9525(90)90174-5. [DOI] [PubMed] [Google Scholar]
- 20.Kolluri SK, et al. p27(Kip1) induction and inhibition of proliferation by the intracellular Ah receptor in developing thymus and hepatoma cells. Genes & Development. 1999;13(13):1742–1753. doi: 10.1101/gad.13.13.1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Puga A, et al. Activation of transcription factors activator protein-1 and nuclear factor-kappa B by 2,3,7,8-tetrachlorodibenzo-p-dioxin. Biochemical Pharmacology. 2000;59(8):997–1005. doi: 10.1016/s0006-2952(99)00406-2. [DOI] [PubMed] [Google Scholar]
- 22.Elferink CJ, Ge NL, Levine A. Maximal aryl hydrocarbon receptor activity depends on an interaction with the retinoblastoma protein. Molecular Pharmacology. 2001;59(4):664–673. doi: 10.1124/mol.59.4.664. [DOI] [PubMed] [Google Scholar]
- 23.Huang GM, Elferink CJ. Multiple mechanisms are involved in Ah receptor-mediated cell cycle arrest. Molecular Pharmacology. 2005;67(1):88–96. doi: 10.1124/mol.104.002410. [DOI] [PubMed] [Google Scholar]
- 24.Martinez JM, et al. Differential toxicogenomic responses to 2,3,7,8-tetrachlorodibenzo-p-dioxin in malignant and nonmalignant human airway epithelial cells. Toxicological Sciences. 2002;69(2):409–423. doi: 10.1093/toxsci/69.2.409. [DOI] [PubMed] [Google Scholar]
- 25.Chang CY, Puga A. Constitutive activation of the aromatic hydrocarbon receptor. Molecular and Cellular Biology. 1998;18(1):525–535. doi: 10.1128/mcb.18.1.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuramoto N, et al. Xenobiotic response element binding enriched in both nuclear and microsomal fractions of rat cerebellum. Journal of Neurochemistry. 2003;85(1):264–273. doi: 10.1046/j.1471-4159.2003.01679.x. [DOI] [PubMed] [Google Scholar]
- 27.Song JS, et al. A ligand for the aryl hydrocarbon receptor isolated from lung. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(23):14694–14699. doi: 10.1073/pnas.232562899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Phelan D, et al. Activation of the Ah receptor signal transduction pathway by bilirubin and biliverdin. Archives of Biochemistry and Biophysics. 1998;357(1):155–163. doi: 10.1006/abbi.1998.0814. [DOI] [PubMed] [Google Scholar]
- 29.McMillan B, Bradfield C. Searching for endogenous activators of the AH receptor. Chemical Research in Toxicology. 2007;20(12):26. [Google Scholar]
- 30.Powell-Coffman JA, Bradfield CA, Wood WB. Caenorhabditis elegans orthologs of the aryl hydrocarbon receptor and its heterodimerization partner the aryl hydrocarbon receptor nuclear translocator. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(6):2844–2849. doi: 10.1073/pnas.95.6.2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang X, Powell-Coffman JA, Jin YS. The AHR-1 aryl hydrocarbon receptor and its co-factor the AHA-1 aryl hydrocarbon receptor nuclear translocator specify GABAergic neuron cell fate in C-elegans. Development. 2004;131(4):819–828. doi: 10.1242/dev.00959. [DOI] [PubMed] [Google Scholar]
- 32.Qin HT, Powell-Coffman JA. The Caenorhabditis elegans aryl hydrocarbon receptor, AHR-1, regulates neuronal development. Developmental Biology. 2004;270(1):64–75. doi: 10.1016/j.ydbio.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 33.Dalton TP, Puga A, Shertzer HG. Induction of cellular oxidative stress by aryl hydrocarbon receptor activation. Chemico-Biological Interactions. 2002;141(12):77–95. doi: 10.1016/s0009-2797(02)00067-4. [DOI] [PubMed] [Google Scholar]
- 34.Puga A, Tomlinson CR, Xia Y. Ah receptor signals cross-talk with multiple developmental pathways. Biochemical Pharmacology. 2005;69(2):199–207. doi: 10.1016/j.bcp.2004.06.043. [DOI] [PubMed] [Google Scholar]
- 35.Carney SA, Peterson RE, Heideman W. 2,3,7,8-tetrachlorodibenzo-p-dioxin activation of the aryl hydrocarbon receptor/aryl hydrocarbon receptor nuclear translocator pathway causes developmental toxicity through a CYP1A-independent mechanism in zebrafish. Molecular Pharmacology. 2004;66(3):512–521. doi: 10.1124/mol.66.3.. [DOI] [PubMed] [Google Scholar]
- 36.Thomsen JS, et al. HES-1, a novel target gene for the aryl hydrocarbon receptor. Molecular Pharmacology. 2004;65(1):165–171. doi: 10.1124/mol.65.1.165. [DOI] [PubMed] [Google Scholar]
- 37.Bansal R, et al. Maternal thyroid hormone increases HES expression in the fetal rat brain: An effect mimicked by exposure to a mixture of polychlorinated biphenyls (PCBs) Developmental Brain Research. 2005;156(1):13–22. doi: 10.1016/j.devbrainres.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 38.Hill A, et al. Effect of 2,3,7,8-Tetrachlorodibenzo-p-dioxin on Early Development and the Neurogenesis Pathway in the Zebrafish (Danio rerio) Neurotoxicology. 2001;53(207210) [Google Scholar]
- 39.Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nature Reviews Neuroscience. 2002;3(7):517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- 40.Schuurmans C, et al. Sequential phases of cortical specification involve Neurogenin-dependent and -independent pathways. Embo Journal. 2004;23(14):2892–2902. doi: 10.1038/sj.emboj.7600278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laroche J, et al. Evidence that GABAergic neruons of the preoptic area of the brain are targets of TCDD reproductive toxicity in adult and perinatal rats. Toxicologist. 2002;66:642. [Google Scholar]
- 42.Hill A, et al. Neurodevelopmental defects in zebrafish (Danio rerio) at environmentally relevant dioxin (TCDD) concentrations. Toxicological Sciences. 2003;76(2):392–399. doi: 10.1093/toxsci/kfg241. [DOI] [PubMed] [Google Scholar]
- 43.Altaba ARI, Palma V, Dahmane N. Hedgehog-Gli signalling and the growth of the brain. Nature Reviews Neuroscience. 2002;3(1):24–33. doi: 10.1038/nrn704. [DOI] [PubMed] [Google Scholar]
- 44.Nayyar T, Zawia NH, Hood DB. Transplacental effects of 2,3,7,8-tetrachlorodibenzo-p-dioxin on the temporal modulation of Sp1 DNA binding in the developing cerebral cortex and cerebellum. Experimental and Toxicologic Pathology. 2002;53(6):461–468. doi: 10.1078/0940-2993-00219. [DOI] [PubMed] [Google Scholar]
- 45.Basha MR, et al. Ontogenetic alterations in prototypical transcription factors in the rat cerebellum and hippocampus following perinatal exposure to a commercial PCB mixture. Neurotoxicology. 2006;27(1):118–124. doi: 10.1016/j.neuro.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 46.Goldschmidt L, et al. Prenatal alcohol exposure and academic achievement at age six: A nonlinear fit. Alcoholism-Clinical and Experimental Research. 1996;20(4):763–770. doi: 10.1111/j.1530-0277.1996.tb01684.x. [DOI] [PubMed] [Google Scholar]
- 47.Lin TM, et al. Role of the aryl hydrocarbon receptor in the development of control and 2,3,7,8-tetrachlorodibenzo-p-dioxin-exposed male mice. Journal of Toxicology and Environmental Health-Part A. 2001;64(4):327–342. doi: 10.1080/152873901316981312. [DOI] [PubMed] [Google Scholar]
- 48.Gohlke JM, et al. Characterization of the proneural gene regulatory network during mouse telencephalon development. Bmc Biology. 2008;6 doi: 10.1186/1741-7007-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Toyoshiba H, et al. Gene interaction network suggests dioxin induces a significant linkage between aryl hydrocarbon receptor and retinoic acid receptor beta. Environmental Health Perspectives. 2004;112(12):1217–1224. doi: 10.1289/txg.7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ovcharenko I, et al. Mulan: Multiple-sequence local alignment and visualization for studying function and evolution. Genome Research. 2005;15(1):184–194. doi: 10.1101/gr.3007205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun YV, et al. Comparative analysis of dioxin response elements in human, mouse and rat genomic sequences. Nucleic Acids Research. 2004;32(15):4512–4523. doi: 10.1093/nar/gkh782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guillemot F. Spatial and temporal specification of neural fates by transcription factor codes. Development. 2007;134(21):3771–3780. doi: 10.1242/dev.006379. [DOI] [PubMed] [Google Scholar]
- 53.Bunger MK, et al. Abnormal liver development and resistance to 2,3,7,8-tetrachlorodibenzo-p-dioxin toxicity in mice carrying a mutation in the DNA-binding domain of the aryl hydrocarbon receptor. Toxicological Sciences. 2008;106(1):83–92. doi: 10.1093/toxsci/kfn149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bunger MK, et al. Resistance to 2,3,7,8-tetrachlorodibenzo-p-dioxin toxicity and abnormal liver development in mice carrying a mutation in the nuclear localization sequence of the aryl hydrocarbon receptor. Journal of Biological Chemistry. 2003;278(20):17767–17774. doi: 10.1074/jbc.M209594200. [DOI] [PubMed] [Google Scholar]
- 55.Hatakeyama J, et al. Hes genes regulate size, shape and histogenesis of the nervous system by control of the timing of neural stem cell differentiation. Development. 2004;131(22):5539–5550. doi: 10.1242/dev.01436. [DOI] [PubMed] [Google Scholar]
- 56.Bergsland M, et al. The establishment of neuronal properties is controlled by Sox4 and Sox11. Genes & Development. 2006;20(24):3475–3486. doi: 10.1101/gad.403406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bai G, et al. Id proteins sustain Hes1 expression to maintain neural stem cells. Cell Research. 2008;18 [Google Scholar]
- 58.Abell AN, Granger DA, Johnson GL. MEKK4 stimulation of p38 and JNK activity is negatively regulated by GSK3 beta. Journal of Biological Chemistry. 2007;282(42):30476–30484. doi: 10.1074/jbc.M705783200. [DOI] [PubMed] [Google Scholar]
- 59.Yamauchi J, et al. Gadd45a, the gene induced by the mood stabilizer valproic acid, regulates neurite outgrowth through JNK and the substrate paxillin in N1E-115 neuroblastoma cells. Experimental Cell Research. 2007;313(9):1886–1896. doi: 10.1016/j.yexcr.2007.02.019. [DOI] [PubMed] [Google Scholar]
- 60.Herz HM, et al. vps25 mosaics display non-autonomous cell survival and overgrowth, and autonomous apoptosis. Development. 2006;133(10):1871–1880. doi: 10.1242/dev.02356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.LoTurco JJ, et al. Citron kinase is a regulator of mitosis and neurogenic cytokinesis in the neocortical ventricular zone. Cerebral Cortex. 2003;13(6):588–591. doi: 10.1093/cercor/13.6.588. [DOI] [PubMed] [Google Scholar]
- 62.Guillemot F. Cell fate specification in the mammalian telencephalon. Progress in Neurobiology. 2007;83(1):37–52. doi: 10.1016/j.pneurobio.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 63.LaMantia AS. Forebrain induction, retinoic acid, and vulnerability to schizophrenia: Insights from molecular and generic analysis in developing mice. Biological Psychiatry. 1999;46(1):19–30. doi: 10.1016/s0006-3223(99)00002-5. [DOI] [PubMed] [Google Scholar]
- 64.Iwamoto K, et al. Molecular characterization of bipolar disorder by comparing gene expression profiles of postmortem brains of major mental disorders. Mol Psychiatry. 2004;9(4):406–16. doi: 10.1038/sj.mp.4001437. [DOI] [PubMed] [Google Scholar]
- 65.Schrimsher GW, et al. Caudate nucleus volume asymmetry predicts attention-deficit hyperactivity disorder (ADHD) symptomatology in children. Journal of Child Neurology. 2002;17(12):877–884. doi: 10.1177/08830738020170122001. [DOI] [PubMed] [Google Scholar]
- 66.Bertram L, et al. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet. 2007;39(1):17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- 67.Bagade S, et al. The PDGene Database. Alzheimer Research Forum. 2008 [Google Scholar]
- 68.Nakajima S, et al. Effects of prenatal exposure to polychlorinated biphenyls and dioxins on mental and motor development in Japanese children at 6 months of age. Environmental Health Perspectives. 2006;114(5):773–778. doi: 10.1289/ehp.8614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vreugdenhil HJI, et al. Effects of prenatal PCB and dioxin background exposure on cognitive and motor abilities in Dutch children at school age. Journal of Pediatrics. 2002;140(1):48–56. doi: 10.1067/mpd.2002.119625. [DOI] [PubMed] [Google Scholar]
- 70.Loots GG, Ovcharenko I. Dcode.org anthology of comparative genomic tools. Nucleic Acids Research. 2005;33:W56–W64. doi: 10.1093/nar/gki355. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.