Abstract
Regulators of G protein signaling (RGS1) are a diverse family primarily known as GTPase-activating proteins (GAPs) for heterotrimeric G proteins. In addition to the RGS domain, which is responsible for GAP activity, most RGS proteins contain other distinct structural motifs. For example, members of the R7 family of RGS proteins contain a DEP, GGL and novel DHEX domain, and are obligatory dimers with the G protein beta subunit Gβ5. Here we show that the Gβ5-RGS7 complex can inhibit Ca2+ mobilization elicited by the muscarinic acetylcholine receptor type 3 (M3R), but not by other Gq-coupled receptors such as M1, M5, histamine H1 and GNRH receptors. Isolated DEP domain of RGS7 is sufficient for the inhibition of M3R signaling, whereas the deletion of the DEP domain renders Gβ5-RGS7 ineffective. Deletion of a portion of the 3rd intracellular loop allowed the receptor (M3R-short) to signal, but rendered it insensitive to the effect of Gβ5-RGS7. Accordingly, recombinant DEP domain bound in vitro to the GST-fused i3 loop of the M3R. These results identify a novel molecular mechanism that can impart receptor-subtype selectivity on signal transduction via Gq-coupled muscarinic receptors.
G protein-coupled receptors (GPCRs) regulate numerous physiological functions in eukaryotes. Agonist-bound GPCRs catalyze the exchange of GDP bound to the G protein α subunits for GTP, which allows the G proteins to modulate the activity of their effector enzymes and ion channels. For example, heterotrimeric G proteins that belong to the Gq class stimulate phospholipase C, which leads to inositol triphosphate-mediated release of Ca2+ from intracellular stores. The duration and amplitude of the activated state of a G protein cascade depends largely on the lifetime of the GTP-bound form of the G protein. For most G proteins, the rate of GTP hydrolysis is increased by a distinct class of approximately thirty diverse proteins known as regulators of G protein signaling (RGS). Their interaction with the G proteins is mediated by a ~120 amino acid RGS domain, which serves as a GTP-ase activating protein (GAP) for Gα subunits [1, 2]. Most RGS proteins also contain other structural motifs that are implicated in a variety of functions [3, 4].
The R7 subfamily of RGS proteins is comprised of four gene products, RGS6, RGS7, RGS9 and RGS11 [5–7]. In addition to the RGS domain, they have three other domains, GGL, DEP, and DHEX. The function of the DHEX (DEP helical extension) domain, which was recently identified by crystallography [8], has not been determined. The GGL (G gamma like) domain is responsible for the interaction with the unique neuro-specific G protein β subunit, Gβ5 [9, 10]. It was shown that Gβ5 and the R7 family of RGS proteins form obligatory dimers in vivo [6, 7]. The DEP domain (first identified in Disheveled, EGL-10 and pleckstrin) was found in a variety of signaling proteins [11]. The function of the DEP domains in the R7 family remained unknown until it was demonstrated that they could bind to R9AP and R7BP, novel proteins that anchor R7 family proteins to the membranes [12–15]. It is interesting to note that a large pool of the Gβ5-RGS7 complex in the native tissue is present in the cytosol apart from the membrane-bound R7BP [16]. Furthermore, the knockout of R7BP produced no apparent phenotype in mice and only slightly affected membrane association of Gβ5-RGS7 [17]. Thus, it appears that Gβ5-RGS7 in the native tissues can exist both as the dimer or trimer with R7BP.
Certain functions of RGS proteins cannot be explained solely by their GAP activity. For example, RGS4 inhibited muscarinic acetylcholine M3 receptor (M3R) with a much higher potency than the cholecystokinin receptor, another Gq-coupled GPCR [18]. This selectivity was dependent on the presence of the N-terminal region of RGS4, but not on the RGS domain. Likewise, another study showed that RGS8 was more potent toward M1R compared to M3R [19]. One of the suggested explanations for the receptor selectivity of RGS action was their direct interaction with GPCRs. Indeed, it was later shown that RGS8 could directly bind to M1R [20].
All GPCRs share the same overall architecture with 7 transmembrane domains, but the difference in their intracellular loops and the C-termini allows them to couple to distinct G proteins and other signaling molecules. For example, muscarinic receptor subtypes M1, M3 and M5 couple to Gq family of G proteins, whereas M2 and M4 receptors couple to Gi. The intracellular regions of GPCRs also contain sites for phosphorylation and arrestin binding, the processes involved in GPCR desensitization [21]. The sites for the interaction of the G protein subunits and arrestins were mapped to the 3rd intracellular (i3) loops of M3 and M2 receptors [22–24]. Studies have also shown that the i3 loops directly bind to proteins such as calmodulin [25], RGS2 [26], casein kinase α [27] and SET, a putative oncogene and protein phosphatase 2A inhibitor [28].
In this paper, we show that the DEP domain of RGS7 can directly bind to the 3rd intracellular loop of the M3R and attenuate receptor-induced Ca2+ mobilization in a M3 subtype-selective manner.
MATERIALS AND METHODS
Reagents and antibodies
The cDNA clones for human Muscarinic M3, M1 and M5, Histamine H1, Gonadotropin Releasing Hormone (GNRH) and Serotonin 2c (5HT2c) receptors were obtained from the Missouri S&T cDNA Resource Center (www.cdna.org). The GFP antibody was from Clontech. RGS7, Gβ5, and Gβ1 antibodies have been described earlier [29]. The rabbit polyclonal antibody against R7BP was provided by Dr. Ken Blumer (Washington University). Carbamoylcholine chloride (carbachol), histamine dihydrochloride, serotonin hydrochloride and human luteinizing hormone releasing hormone acetate were all obtained from Sigma.
GST-MR i3 constructs
The GST-fusion constructs encoding the third intracellular loops (i3) of M1-5 muscarinic receptors were kindly provided by Dr. John Hepler (Emory University) and have been previously described [26, 30]. The DNA fragment encoding the M3R i3 loop region (Asn304-Gln390) was amplified from full length human M3R using the forward primer 5’-TCCGGATCCAACAGGAGGAAGTAT-3’ and the reverse primer 5’-CACGAATTCCTGCAGGTTGTCCGA-3’. The shorter fragment encoding the Ser345-Gln390 part of the i3 loop was amplified using the forward primer 5’-GCCGGTCCTCCCTGGAGAACTCC-3’ and the reverse primer used for Asn304-Gln390. The fragments were cloned into the pGEX-2T vector at the BamH1 and EcoR1 sites. The GST fusion constructs encoding rat M3R i3 Arg252-Gln490 and Val390-Gln490 [22, 31] were kindly provided by Dr. Lanier (Medical University of South Carolina).
Preparation of Brain Homogenates
Mouse brains were homogenized in lysis buffer (20mM Tris-HCl pH 7.5, 1mM EDTA, 50mM NaCl and 2mM β-mercaptoethanol) and centrifuged at 150,000g for 1.5 hrs at 4°C. The supernatant fraction was collected (cytosolic extract) and the pellet containing the membranes was washed twice and resuspended in the same buffer containing 1% sodium cholate. This suspension was left on ice for 15 min then centrifuged at 150,000g for 45 min at 4°C and the supernatant was retained as the membrane extract.
Purification of GST fusion proteins
The purification of the GST fusion proteins was done as previously described [32]. Briefly, bacterial cultures were grown at 37°C and protein expression was induced with the addition of 0.4mM IPTG for 1–1.5 hours at 37°C. The cultures were harvested and centrifuged, and pellets were resuspended in STE (150mM NaCl, 100mM Tris pH 8.0, 1mM EDTA) buffer containing lysozyme, 5mM DTT and protease inhibitors. The cell suspension was briefly sonicated on ice and Sarkosyl (final concentration of 1.5%) and Triton X-100 (final concentration of 2%) were added to the lysate. After gentle rotation at 4°C for 1 hour, the lysate was centrifuged at 19,000 rpm at 4°C for 30 minutes. The clarified lysate was batch processed using GST-Sepharose 4B beads (GE) (0.5 ml of packed beads per 10 ml of lysate, which contained 2–5 mg/ml of total protein) overnight at 4°C. The beads washed and eluted with 20 mM glutathione. The purified GST-fusion protein was desalted on Sephadex G-25 pre-equilibrated with buffer containing 100mM Tris pH 8.0, 150mM NaCl and 15% glycerol and stored frozen in aliquots at −80°C.
Constructs for expression in mammalian cells
The plasmid harboring the M3R-short receptor, which has a 196 amino acids deletion from the i3 loop (amino acids Ala274-Lys469) was previously described [33], and was kindly provided by Dr. Jurgen Wess (NIH). To generate the construct corresponding to amino acids 1–124 of the full length bovine RGS7 (YFP-DEP) nucleotides 1–372 were amplified using the forward primer 5’-TCCGGACTCAGATCTATGGCCCAGGGG-3’ and the reverse primer 5’-GTCTGTGTTAAGCTTTTCCGGCTCCCA-3’. The RGS7 construct that lacks the RGS domain and the C terminus (ΔRGS) was generated by PCR amplification of nucleotides 1–963 corresponding to amino acids 1–321 using the forward primer 5’-TCCGGACTCAGATCTATGGCCCAGGGG-3’ and the reverse primer 5’-CCCAAGCTTTTCTTTGCTTGC-3’. These fragments were cloned into the pEYFP-C1 vector at BglII and Hind III sites. The constructs encoding the C-terminal part of RGS7 (RGS7249–469), YFP-RGS7 and CFP-fused Gβ5 were described previously [32, 34].
Cell culture and transfection
CHO-K1 cells were cultured in F-12K Nutrient Mixture (Kaighn’s modification, Gibco) with 10% fetal bovine serum and penicillin/streptomycin. CHO-R7BP cells [32] were cultured similarly to the CHO-K1 cells with the addition of 400µg/ml of geneticin. 24 hours prior to transfection, the cells were plated to achieve a density of 0.8×106– 1.0×106 cells per 100 mm plate. Transfection was carried out using Lipofectamine 2000 (Invitrogen) as per manufacturer’s instructions. The DNA ratio of RGS7 to Gβ5 was maintained at 5:1, with a total of 8.0 µg of DNA per plate. LacZ DNA was used as a control to ensure that the total DNA per plate used in the CHO-K1 co-transfection assays remained constant. 48 hours after transfection, cells were washed with HBSS, harvested, lysed in hypotonic buffer and used for pull-down assays.
GST pull down
Glutathione Sepharose 4B beads were pre-washed with PBS + 0.1% CHAPS and incubated at 4°C with purified recombinant GST or the GST fusion proteins for 1–2 hours, and then washed three times with PBS + 0.1% CHAPS to remove excess protein. The slurry was incubated overnight at 4°C on a rotary mixer with the investigated CHO cell lysates, as determined by the experiment. At the end of the incubation, the beads were settled by gravity and the supernatant was collected as the unbound fraction. In a typical assay, the packed volume of the resin was 30 µl, and the volume of the protein lysate was 300 µl. The beads were washed three times with 600 µl of PBS + 0.1% CHAPS buffer, and eluted with 30 µl of 2xSDS sample loading buffer. The unbound and eluted fractions were resolved by gel electrophoresis and analyzed by western blotting.
Ca2+-mobilization assay
CHO-K1 cells were transiently transfected with cDNAs for M1, M3, M5, GNRH, H1 or 5HT2c receptors, RGS7 and Gβ5, or Lac Z, as required by the experiment. Transfected cells were grown on 12 mm glass cover slips (Electron Microscopy Sciences). 48 hours after transfection they were washed with 2% FBS in HBSS and then incubated in 2% FBS in HBSS containing 1 µM fura-2AM for 45 minutes at ambient temperature in the dark. This was followed by a 30 min incubation in Locke’s buffer to permit de-esterification of fura-2AM. The cover slips were then secured in a flow chamber and mounted on the stage of a Nikon TE2000 inverted fluorescence microscope. The cells were continuously perfused with Locke’s buffer and stimulated with varying agonist concentrations in the same buffer as required by the experiment. The images were collected in real time every two seconds using a 20x UV objective lens and recorded using Metafluor software. The excitation wavelengths were 340 and 380 nm and the emission was set at 510 nm. Free Ca2+ concentration was determined from the fluorescence measurements using the fura-2 Ca2+ imaging calibration kit (Molecular Probes) according to manufacturer’s instructions.
[3H]NMS binding assay
Muscarinic receptor density was determined by the ligand binding assay using the muscarinic antagonist N-methyl scopolamine chloride ([3H]NMS; 70 Ci/mmol; Perkin-Elmer) essentially as previously described in [35]. Briefly, CHO-K1 cells were transfected in 24 well plates with the M1, M3 or M5 muscarinic receptors alone or together with the Gβ5-RGS7 complex, as required by the experiment. Twenty four hours after transfection, cells were washed and incubated with 1ml of [3H]NMS in a buffer also containing 10 mM HEPES pH 7.4, 4.2 mM NaHCO3, 11.7 mM glucose, 1.2 mM MgSO4, 1.2 mM KH2PO4, 4.7 mM KCl, 118 mM NaCl and 1.3 mM CaCl2. The [3H]NMS concentrations ranged from 20 pM to 14 nM, with seven points used for the saturation curve and Scatchard analysis. Each [3H]NMS concentration was used in duplicate (two wells of cells). Nonspecific binding was determined in the presence of 20 µM atropine. Following 1 hr incubation at 37°C, cells were rapidly washed twice with 1ml of ice-cold buffer and then lysed with 0.5ml of 0.1M NaOH added to the wells. This lysate was neutralized with 0.5ml of 0.1M HCL and the mixture was transferred to the vials for liquid scintillation counting.
RESULTS
Gβ5-RGS7 inhibits M3R-mediated Ca2+ mobilization
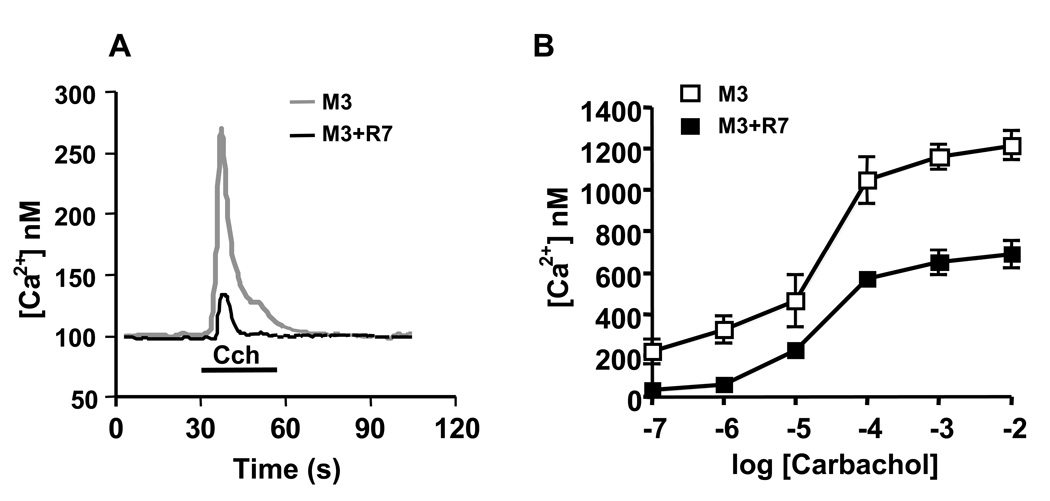
Previous studies showed that Gβ5-RGS7 attenuated Ca2+ mobilization elicited by the muscarinic acetylcholine M3 receptor (M3R) by approximately 50% [29, 32, 34]. Here, we tested if this inhibition could occur to a greater extent at a lower agonist concentration. We reasoned that given the high expression level of the receptor in the transiently transfected cells, the amount of Gβ5-RGS7 was insufficient to quench the receptor-mediated activation of Gq, particularly at saturating agonist concentrations. To reduce the amount of activated receptor, we stimulated cells with a range of carbachol concentrations. We found that at carbachol concentrations of 1 µM and 0.1 µM (below the EC50), the Gβ5-RGS7-mediated inhibition of Ca2+ responses was nearly complete (Fig. 1).
Figure 1. The Gβ5-RGS7 complex inhibits Ca2+ mobilization elicited by the muscarinic M3 receptor.
CHO-K1 cells were transiently transfected with the M3R and Lac Z or the M3R, Gβ5 and RGS7. Cells were plated on glass cover slips, loaded with fura-2AM and the changes in free intracellular Ca2+ concentrations in response to the application of carbachol were recorded in real time as described in Materials and Methods. (A) Representative traces from cells transfected with plasmids encoding M3R and Lac Z (grey) and cells expressing M3R together with Gβ5-RGS7 (black). The application of 100 nM carbachol (Cch) is denoted with the black bar. (B) Dose-response curve obtained by treating cells expressing M3R alone (white squares) M3R plus Gβ5-RGS7 complex (black squares) with increasing concentrations of carbachol. The peak Ca2+ response was recorded from the entire visual field that included 30–60 individual cells. The data represent the mean ± S.D. of six independent experiments.
Receptor selectivity of Gβ5-RGS7
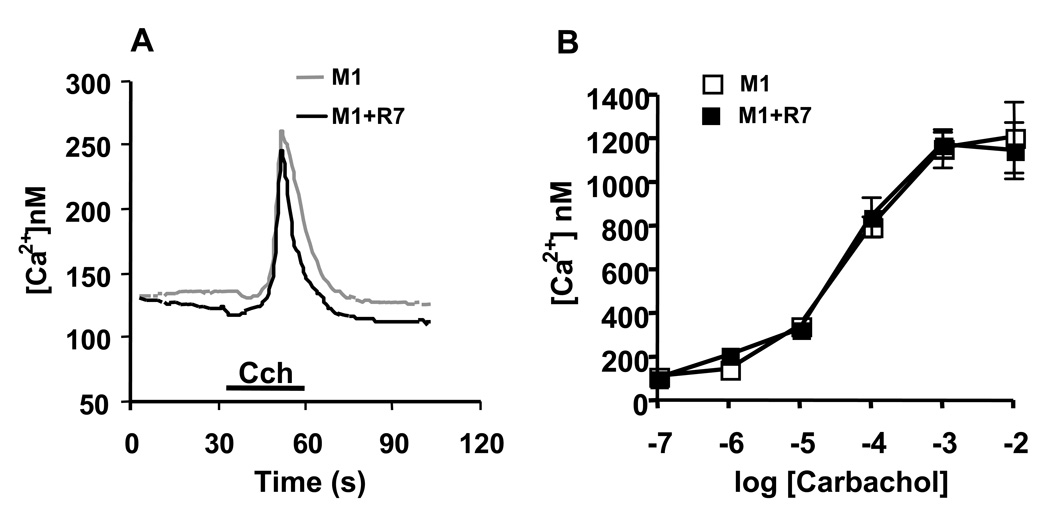
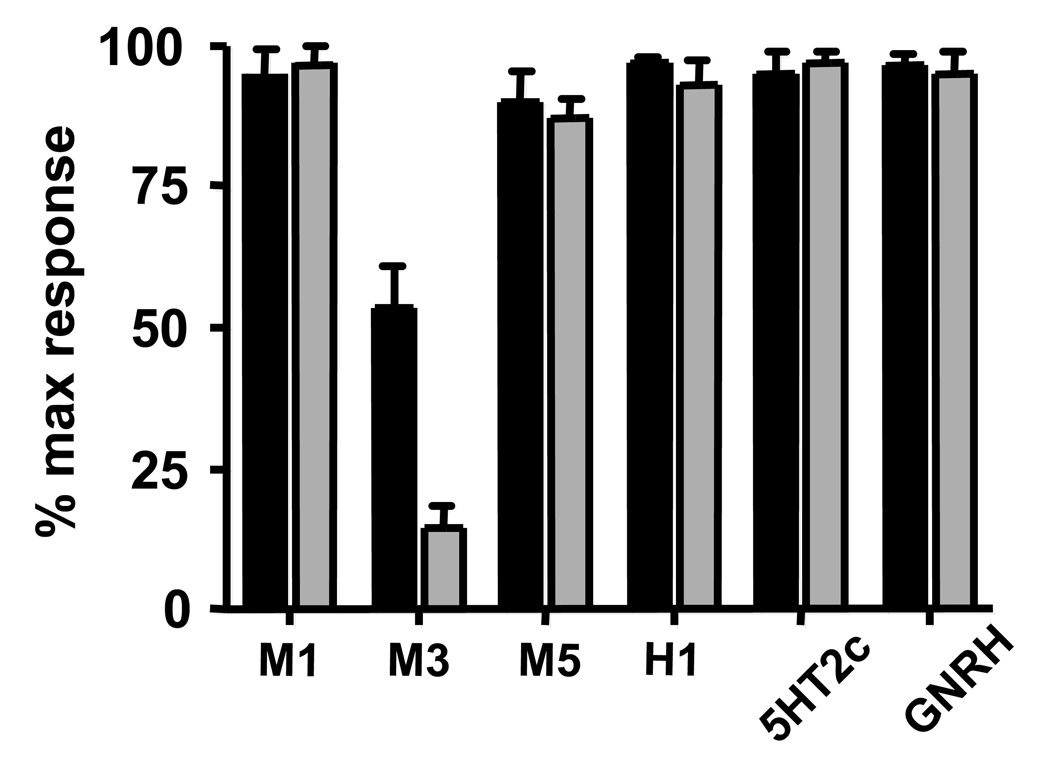
While Gβ5-RGS7 displayed robust inhibitory effect on M3R, it had no effect on other tested Gq-coupled receptors. For example, Figure 2 shows that M1R was insensitive to Gβ5-RGS7 over the wide range of tested agonist concentrations. Likewise, the receptors for histamine (H1), serotonin (5HT2c), GNRH and muscarinic acetylcholine receptor M5R were also not affected by Gβ5-RGS7 (Fig. 3). We determined the number of binding sites on live CHO-K1 cells expressing M1, M3 and M5 receptors using the non-selective muscarinic antagonist [3H]-NMS. The binding of [3H]-NMS was saturable and dose-dependent, and the Bmax in M1R, M3R and M5R transfected cells were as follows: 856 ± 181.4 (n=3), 1066 ± 187.9 (n=4) and 1024 ± 81.5 (n=2) fmol/mg of total protein. The determined Kd for [3H]-NMS were 0.310 ± 0.06, 0.228 ± 0.09, 0.401 ± 0.02 nM respectively. We found that co-expression of Gβ5-RGS7 slightly increased the Bmax of all three tested receptors: 985 ± 118.0 (n=3), 1310 ± 135.2 (n=4), 1304 ± 153.0 (n=2) fmol/mg, which is likely attributable to a minor positive effect on transfection efficiency of the CHO cells. The Kd values in the presence of Gβ5-RGS7 were 0.277 ± 0.08, 0.216 ± 0.04, 0.392 ± 0.06 nM respectively, not appreciably different from the values in the absence of Gβ5-RGS7. Thus, all three muscarinic receptors were expressed at approximately the same level with or without Gβ5-RGS7, however but Gβ5-RGS7 only inhibited signaling elicited by the M3 receptor subtype.
Figure 2. M1R-mediated Ca2+ release is not affected by the Gβ5-RGS7 complex.
CHO-K1 cells were transfected with the M1R and Lac Z or the M1R, Gβ5 and RGS7, and the calcium responses were recorded and analyzed as described in the legend to Fig 1. (A)Representative traces from single cells. Grey trace represents cells with M1R alone, and black shows cells expressing M1R together with Gβ5-RGS7. (B) Dose-response from increasing carbachol concentration. White symbols, M1R alone, black symbols, M1R plus Gβ5-RGS7. The data represent the mean ± S.D. of three independent experiments from measurements of the amplitude of free Ca2+.
Figure 3. Gβ5-RGS7 complex inhibits Gq signaling in receptor selective-manner.
CHO-K1 cells were transiently transfected with cDNAs encoding Gq-couple receptors: M1, M3 and M5 muscarinic, 5HT2c serotonin, H1 histamine and GNRH. Lac Z or the Gβ5 plus RGS7 plasmids were co-transfected with the receptor-encoding plasmids. In each experiment, changes in free intracellular Ca2+ were measured upon treatment of the cells with two agonist concentrations. The highest agonist concentration (black bars) used for the M1, M3 and M5 muscarinic receptors was 10mM and the lowest concentration (grey bars) was 100nM. The highest and the lowest concentrations of 5HT2c, histamine and GNRH were 10µM and 10nM, respectively. The determined response in the presence of Gβ5-RGS7 was expressed as the percent of the signal recorded from cells transfected only with the indicated receptor. Shown is the mean amplitude ± the S.D. of the Ca2+ response from at least three independent experiments.
Since all tested receptors act through Gq, such receptor selectivity indicated that inhibition of M3R signal transduction occurs upstream of the G protein. Therefore, we hypothesized that the Gβ5-RGS7 complex inhibited M3R signal transduction via a mechanism that does not utilize its GAP activity.
The DEP domain of RGS7 is responsible for M3R inhibition
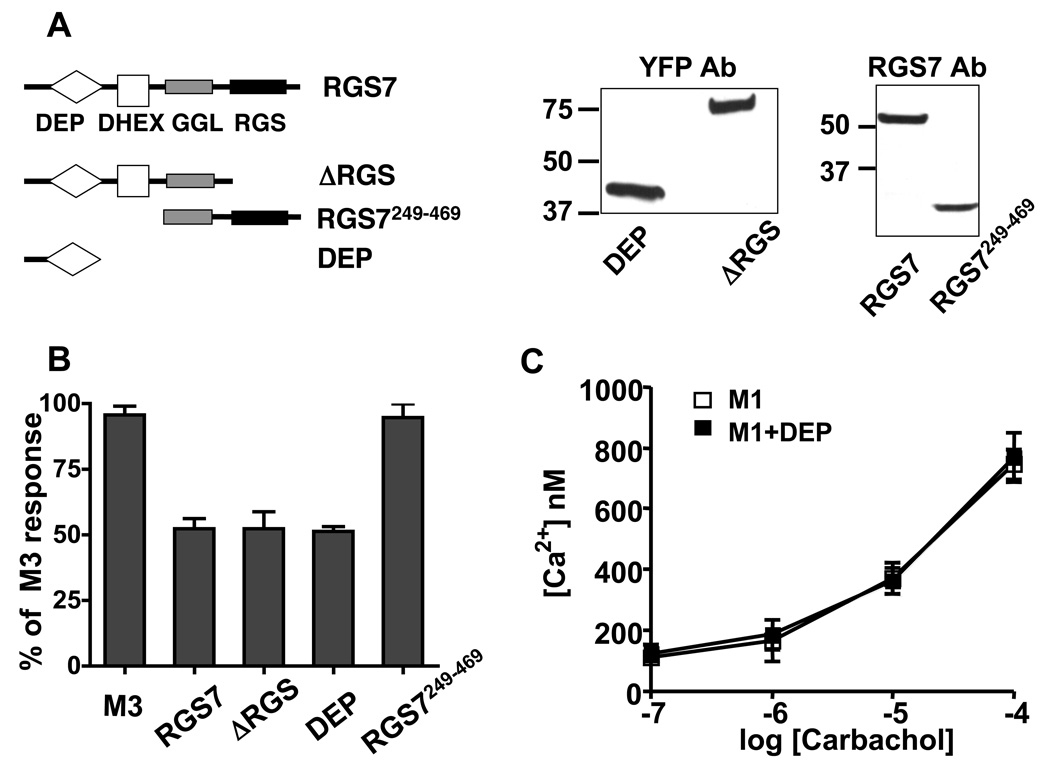
To determine the mechanism by which Gβ5-RGS7 inhibits M3R signaling, we investigated which domain of RGS7 was responsible for this effect. We prepared the following three constructs (Fig 4A): (1) the N-terminal portion of RGS7, which lacked the RGS domain and the C-terminus, termed ΔRGS, (2) the C-terminal portion that lacked the N-terminus, DEP and DHEX domains, termed RGS7249–469, and (3) the N-terminal portion (amino acids 1–124) termed DEP domain. The ΔRGS and the DEP domain constructs were fused to the C-terminus of YFP to enhance their expression and detection. The ΔRGS and RGS7249–469 truncations of RGS7 were co-expressed in CHO cells together with Gβ5. Our results showed that the ΔRGS and DEP constructs inhibited M3R-induced Ca2+ mobilization similarly to the full-length RGS7 protein (Fig. 4B). In contrast, the RGS7249–469, which lacked the DEP domain, had no effect on M3R signaling. These results indicated that the RGS domain is not essential for this inhibition, supporting the hypothesis that Gβ5-RGS7 inhibits M3R-mediated signal transduction via a non-GAP mechanism. Instead, Gβ5-RGS7 complex inhibits M3R signaling via the DEP domain. Similar to the full-length Gβ5-RGS7 (Fig 2), the DEP domain did not inhibit signaling from the M1R, showing the selectivity of the DEP domain of RGS7 toward the M3 receptor subtype (Fig. 4C).
Figure 4. The DEP domain of RGS7 is sufficient for the inhibition of M3R signaling.
(A) CHO-K1 cells were co-transfected with the M3R and RGS7 constructs encoding full-length RGS7, ΔRGS, RGS7249–469 or the DEP domain. The schematic illustrates the location of the DEP (diamond), DHEX (white square), GGL (grey rectangle) and RGS (black rectangle) domains within the RGS7 molecule. The RGS7 constructs containing the GGL domain were cotransfected together with Gβ5 cDNA. The western blot panels show the expression of these constructs in CHO-K1 lysates (12µg of total protein) that were used in these experiments. ΔRGS and DEP YFP fusion proteins were probed with the YFP antibody. Full length RGS7 and RGS7249–469 were probed with the RGS7 antibody. (B) The effects of the constructs described in (A) on M3R-mediated Ca2+ response. The average response in the presence of the constructs was expressed as the percent of the signal from cells transfected with M3R alone. Data show the mean amplitude ± the S.D. from at least three independent experiments. (C) RGS7 DEP does not affect the M1 receptor. Ca2+ responses in the M1R-transfected cells were recorded in the absence (white squares) or the presence of the DEP domain (black squares), over the range of indicated carbachol concentrations. The data represent the mean ± S.D. for two independent experiments.
The DEP domain of RGS7 directly binds to the 3rd loop of M3R
The selectivity of Gβ5-RGS7 toward M3R indicated that it acts upstream of the G protein, suggesting that it may directly bind to the receptor. The i1 and i2 loops are very short and well-conserved among all five muscarinic receptors. In contrast, the i3 loops of muscarinic receptors are longer, much more diverse, and were previously shown to interact with multiple proteins such as Gαq [24], Gβγ[31], β-arrestin [23], RGS2 [26], calmodulin [25] and SET [28]. Therefore, we reasoned that the likely binding site for Gβ5-RGS7 could be located within the 3rd intracellular loop. To test this hypothesis we first investigated if Gβ5-RGS7 had an effect on M3R-short, the deletion mutant of M3R that lacked a large portion of the i3 loop (Fig. 5A). Our results showed that Gβ5-RGS7 had no effect on Ca2+ mobilization elicited by this M3R mutant. We further tested the hypothesis that the DEP binding site is localized within the i3 loop using a pull-down assay with the i3 loop of M3R fused to the C-terminus of GST (Fig. 5B). We found that the DEP domain of RGS7 bound to the M3R i3 loop, but not to GST. The DEP domain also bound to the i3 loop of M5R and also displayed very weak binding to the loops of M1 and M2 receptors.
Figure 5. Direct protein-protein interaction between the third intracellular loop of M3R and the DEP domain of RGS7.
(A) The M3 receptor deletion mutant lacking the 3rd intracellular loop (M3R-short) was transfected into CHO-K1 cells together with Lac Z or Gβ5-RGS7. Changes in Ca2+ were recorded upon the application of 100nM Cch (black bar). The grey line represents a Ca2+ transient recorded from cells transfected with M3R-short alone. Black line represents the M3R-short transfected together with Gβ5-RGS7. (B) GST fusions of the i3 loops of the M1, M2, M3 and M5 receptors or GST were immobilized on glutathione agarose beads. The beads were incubated with the extract from CHO-K1 cells transfected with YFP DEP, as described in Materials and Methods. After the slurry was spun down and unbound material was collected, the resin was washed and eluted with SDS sample buffer. The unbound (U) and eluted (E) material was analyzed by western blot using the anti-GFP antibody. (C) Schematic representation of the sequence of the entire M3R i3 loop. The approximate location of the binding sites for Gαq, Gβγ, SET, regions phosphorylated by GRK2 and casein kinase α (CKα), calmodulin (CaM), and β-arrestin (β-Arr) are indicated according to the literature (see text). The truncated GST-fused M3R i3 loop regions as are shown under the full-length M3R i3 loop. Filled boxes designate constructs that bound the RGS7 DEP domain and the open box shows the construct that did not bind to the DEP domain. The representative results of the pull-down assays obtained with these GST-fusion proteins are shown to the right.
To locate the putative binding site for the DEP domain, we tested its interaction with shorter fragments of the M3R i3 loop. Our results showed that the DEP domain binds to the region encompassing amino acids 345–390 in human M3R, corresponding to the central portion of the i3 loop (Fig. 5C). Thus, it appears that RGS7 DEP binds to the region that is most distant from the membrane, whereas the reported binding sites for the G protein are located at the ends of the 3rd loop [36], presumably near the membrane surface.
The effects of Gβ5 and R7BP on the interaction of RGS7 with M3R
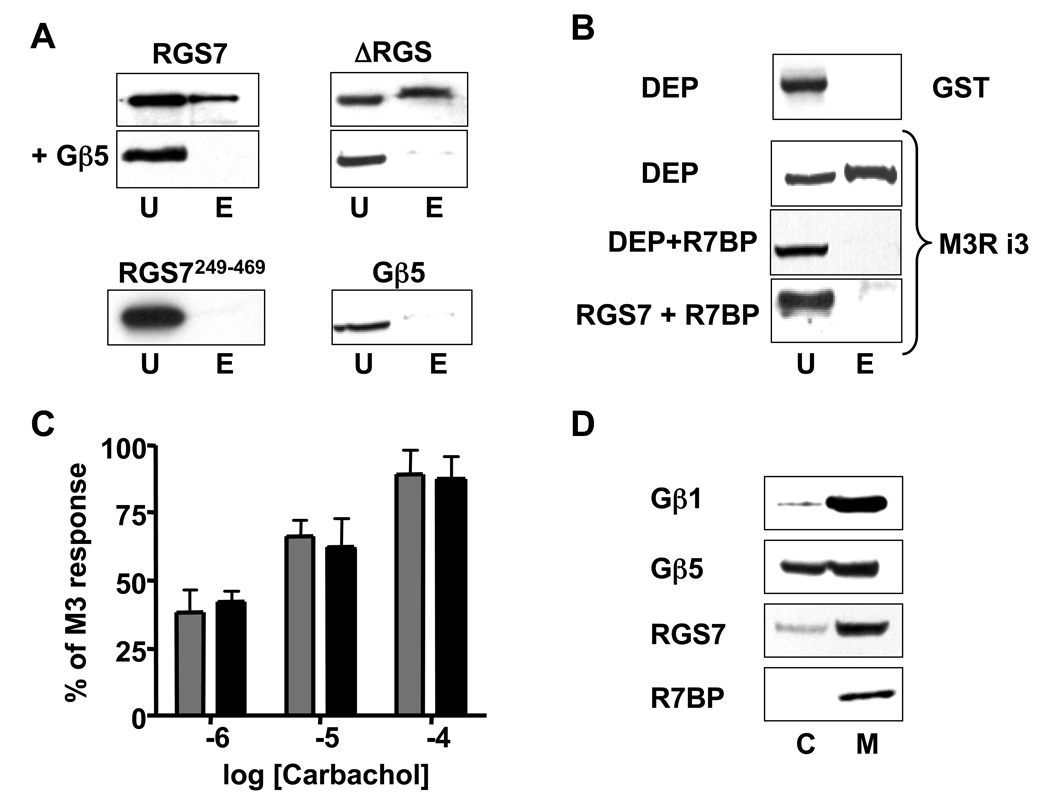
We found that full-length monomeric RGS7 bound to the i3 loop of M3R, but the Gβ5-RGS7 dimer did not (Fig 6A). Likewise, Gβ5 blocked the interaction of the ΔRGS construct with the M3R i3 loop. Neither Gβ5-RGS7249–469 complex, nor monomeric Gβ5 bound to the i3 loop.
Figure 6. Gβ5 and R7BP inhibit the interaction of the DEP domain of RGS7 with the 3rd intracellular loop of M3R.
(A). The N304-I375 region of the i3 loop of M3R fused to GST was used in a series of pull-down assays with the lysates of CHO cells transfected with full-length monomeric RGS7, ΔRGS (with or without Gβ5), Gβ5 alone or RGS7249–469+Gβ5. The unbound (U) and eluted (E) material was analyzed by western blot using the anti-RGS7 (for the full-length RGS7 and the RGS7249–469 construct) or anti-GFP (for ΔRGS or CFP-Gβ5 constructs) antibodies. (B) Wild-type CHO cells or CHO cells stably expressing R7BP were transiently co-transfected with DEP or full-length RGS7. The total cell lysate from the wild-type cells and the membrane extracts from CHO-R7BP cells were compared in the pull-down assay using the GST fusion of M3R i3 (N304-I375) loop. (C) CHO-R7BP cells were transiently transfected with M3R (grey bars) or M3R plus Gβ5-RGS7 (black bars). The Ca2+ responses at three indicated carbachol concentrations were recorded. The data represent the mean amplitude ± S.D. for two independent experiments. (D) Membrane association of R7BP and RGS7 in the brain tissue. Mouse brain was fractionated to obtain cytosolic (C) and membrane (M) fractions as described in Materials and Methods. These fractions were analyzed by western blot for the presence of Gβ1, Gβ5, RGS7 and R7BP. Representative of at least three experiments.
We also investigated the effects of R7BP on the interaction of Gβ5-RGS7 with M3R (Fig. 6 B–D). Our results showed that R7BP prevented the interaction of the DEP domain or the full-length monomeric RGS7 with the i3 loop of M3R (Fig. 6B). This result is consistent with the observation that in CHO cells stably expressing R7BP, Gβ5-RGS7 did not influence M3R-mediated Ca2+ mobilization (Fig. 6C and [32]). We confirmed that Gβ5-RGS7 in the native tissue was present in both membranes and cytosol, whereas R7BP was only found in the membranes (Fig. 6D). These results indicate that binding of DEP to the i3 loop and to R7BP are mutually exclusive and suggest that the regulation of M3R is carried out by the cytosolic form of Gβ5-RGS7.
DISCUSSION
In this paper we investigated the function of a neuronal regulator of G protein signaling, the Gβ5-RGS7 complex. Our study highlighted two novel aspects: the remarkable selectivity of Gβ5-RGS7 toward muscarinic M3 receptors (M3R) and the direct interaction of the DEP domain with the receptor.
Receptor selectivity
Our experiments showed that Gβ5-RGS7 robustly inhibited signaling from M3R, but under the same experimental conditions, it did not influence other Gq-coupled receptors, including M1R (Fig. 1–Fig. 3). This selectivity toward M3R indicated that Gβ5-RGS7 inhibits M3R signaling not via the GAP activity, but upstream of the G protein. We hypothesized that Gβ5-RGS7 interacts directly with the receptor. Since it was shown that M3R contains the binding site for Gβγ subunits [22], we initially thought that the Gβ5/GGL moiety was responsible for this interaction. However, our results showed that the effect of Gβ5-RGS7 was mediated by the DEP domain, whereas neither Gβ5/GGL nor the RGS domain were needed for the inhibition of M3R (Fig 4). Previous studies showed that Gβ5-RGS7 has GAP activity toward Gi but not Gq family G proteins [37, 38]. Therefore, it was not clear why Gβ5-RGS7 inhibited Ca2+ mobilization elicited by the Gq-coupled M3R [29, 32, 34]. Our current findings showed that this inhibition is mediated by the interaction between the receptor and the DEP domain of RGS7. The identification of this novel mechanism resolved the controversy between the lack of the GAP activity toward Gαq and the functional effect of Gβ5-RGS7 on M3R in cells.
The M3R is expressed in a variety of peripheral tissues as well as in the CNS. According to the Allen Brain Atlas (www.brain-map.org), M3R and RGS7 mRNAs co-localize, especially in the cerebral cortex and hippocampus. Interestingly, muscarinic receptors of different subtypes can be found in the same neurons [39, 40]. It is not clear why two Gq-coupled receptors of acetylcholine are needed in one cell. It is reasonable to hypothesize that the interaction with Gβ5-RGS7 differentiates neuronal M3R from the M1R and from M3R expressed in peripheral cells. Mice lacking neuronal M3R have a distinct lean phenotype [41–43], which is different from the phenotypes of other muscarinic receptor knockouts [44]. The Gβ5 knockout mice are born runty [45] and remain lean throughout their lifetime and even on a high fat diet (Slepak lab, unpublished data). The similarity in the phenotypes of Gβ5 and neuronal M3R knockouts may indicate that they participate in the same pathway that is unique for neuronal signaling.
The new role of the DEP domain
We found that the isolated DEP domain of RGS7 mimicked the functional effect of the entire RGS7 complex, supporting the concept that RGS proteins can regulate signal transduction not only via their GAP activity [4]. Our results show that the DEP domain of RGS7 can directly bind to the i3 loop of M3R, which contains binding sites for several other proteins, including Gαq, Gβγ, arrestin, calmodulin and SET [22, 28]. In contrast to the Gαq, RGS7 binds to the middle of the loop, which can potentially protrude far into the cytosol. We speculate that the distance of approximately 100 amino acids between the RGS7 site and juxtamembrane site for Gq could allow cytosolic Gβ5-RGS7 dimer to bind to the receptor at the same time with Gαq. This would be consistent with previous studies that detected FRET between the fluorescently tagged Gβ5-RGS7 and Gαq in cells [34, 46]. The RGS7 binding site partially encompasses the region phosphorylated by GRK2 and casein kinase and a binding site for β-arrestin, which suggests that Gβ5-RGS7 could have a role in the processes of M3R desensitization and β-arrestin-mediated signaling.
Our results showed that binding of the RGS7 DEP domain to the isolated i3 loops of muscarinic receptors (Fig. 5B) had lower sub-type selectivity than the inhibitory effect on the full-length receptors (Fig. 1–Fig. 3). Binding to the M3R i3 loop was the most robust, but the DEP domain also bound well to the M5R i3 loop, and displayed much weaker interaction with the i3 loops of M1 and M2 receptors (Fig. 5B). The reason for the reduced selectivity in the GST pull-down assay compared to the Ca2+ mobilization experiments is not clear at this point. One can speculate that the size of the M3R i3 loop allows the RGS7 complex to remain associated once the G protein binds to the juxtamembrane regions and thus be more effective in inhibiting Gq activation. It is also possible that there is an additional site in the full-length M3R that stabilizes the interaction with the DEP domain and which is absent in other receptors.
It was recently shown that the DEP domain of the yeast RGS protein Sst2 could directly bind to the G protein-coupled receptor, Ste2 [47]. That interaction occurred at the C-terminal tail of Ste2, which is different from the RGS7-M3R interaction. There is no obvious homology between the C-tail of Ste2 and the i3 loop of M3R, however both these regions contain the sites for phosphorylation and play a role in desensitization. Another study showed that the dopamine D2 receptor facilitated the membrane localization of RGS9-2, a member of the R7 family of RGS proteins [48]. Like the interaction of M3R with RGS7 reported in the current paper, the effect of the dopamine receptor was selective for the D2 subtype and was mediated by the DEP domain of RGS9-2. However, it did not require the third loop or the C-terminus of the D2 receptor, indicating that structural elements involved in the interaction with DEP domains can be different for specific receptor subtypes. It is worth noting that another DEP domain-containing signaling protein Disheveled also binds to its cognate seven-pass transmembrane receptor Frizzled. However, this interaction is mediated by its PDZ rather than DEP domain of Disheveled [49]. It will be interesting to find out whether or not the DEP domains found in RGS proteins are unique in their ability to interact with GPCRs.
Potential roles of Gβ5 and R7BP
The fact that the G protein β subunit Gβ5 interacts with RGS proteins of R7 family instead of Gγ subunits was established a decade ago [9, 10, 50, 51]. Gβ5 and the associated RGS protein stabilize each other against rapid proteolysis [29]. This mutual stabilization explained why R7 proteins and Gβ5 have not been found apart from each other in the native tissues [50, 51], and why R7 proteins are absent in Gβ5 knockout animals [45, 52]. The knockout of Gβ5 also causes the disappearance of R7BP [16, 53]. However, it is not clear why such basic function as stabilization of the RGS protein would require association with a G protein β subunit. It appears that there must also be a functional role for Gβ5 within the R7 complex. Early studies suggested that Gβ5 attenuated the interaction of RGS7 with Gαo [10]. This notion was supported by the analyses of RGS9-1, which showed that its GAP activity toward transducin is reduced in the presence of Gβ5 [54, 55]. In this paper, we show that Gβ5 can prevent the protein-protein interaction between the DEP domain and the i3 loop of the M3 receptor (Fig. 6). Thus it appears that Gβ5 can serve as the negative regulator of both the DEP and RGS domains of RGS7.
We found that like Gβ5, R7BP prevents the DEP domain from binding to the i3 loop of M3R. R7BP also blocks the effect of Gβ5-RGS7 complex on M3R signaling [32]. Therefore, R7BP and Gβ5 differ in their effects on the interaction of RGS7 with the full-length M3R in intact cells. Gβ5-RGS7 complex is as effective as the isolated DEP domain of RGS7 in its ability to block M3R signaling under the same experimental conditions (Fig. 4). This indicates that Gβ5 can allow the DEP domain to interact with the i3 loop protruding from the full-length receptor in intact cells. In our previous study we suggested a model where the Gβ5-RGS7 molecule can exist in (at least) two conformations: “closed”, when Gβ5 associates with the DEP domain and “open”, when they do not [32]. Based on the results presented in this paper we now hypothesize that agonist-bound M3R can “open” cytosolic Gβ5-RGS7 dimer so that the DEP domain binds to the i3 loop, which then inhibits M3R-Gq coupling. The interaction with R7BP at the membrane restricts the action of Gβ5-RGS7 to Gi-coupled receptors, such as muscarinic M2 [13], which occurs via the canonical GAP mechanism that involves the RGS domain of RGS7.
ACKNOWLEDGMENTS
We thank Drs. Steve Lanier (Medical University of South Carolina), John Hepler (Emory University), Jurgen Wess (NIH) and Ken Blumer (Washington University) for DNA constructs and antibodies used in this study.
Footnotes
Supported by NIH grant GM 060019.
Abbreviations are: RGS, regulator of G protein signaling; GPCR, G protein-coupled receptor; R7BP, R7 family RGS binding protein; R9AP, RGS9 anchoring protein; GTP, guanosine-triphosphate; GST, glutathione-S-transferase; CHO, Chinese hamster ovary; FRET, fluorescence resonance energy transfer.
REFERENCES
- 1.Berman DM, Gilman AG. Mammalian RGS proteins: barbarians at the gate. J. Biol. Chem. 1998;273(3):1269–1272. doi: 10.1074/jbc.273.3.1269. [DOI] [PubMed] [Google Scholar]
- 2.Abramow-Newerly M, Roy AA, Nunn C, Chidiac P. RGS proteins have a signalling complex: interactions between RGS proteins and GPCRs, effectors,and auxiliary proteins. Cell Signal. 2006;18(5):579–591. doi: 10.1016/j.cellsig.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 3.Hepler JR. RGS protein and G protein interactions: a little help from their friends. Mol. Pharmacol. 2003;64(3):547–549. doi: 10.1124/mol.64.3.547. [DOI] [PubMed] [Google Scholar]
- 4.Willars GB. Mammalian RGS proteins: multifunctional regulators of cellular signalling. Semin. Cell Dev. Biol. 2006;17(3):363–376. doi: 10.1016/j.semcdb.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 5.Siderovski DP, Strockbine B, Behe CI. Whither goest the RGS proteins? Crit. Rev. Biochem. Mo.l Biol. 1999;34(4):215–251. doi: 10.1080/10409239991209273. [DOI] [PubMed] [Google Scholar]
- 6.Witherow DS, Slepak VZ. A Novel Kind of G Protein Heterodimer: The Gbeta5-RGS Complex. Receptors Channels. 2003;9(3):205–212. [PubMed] [Google Scholar]
- 7.Jones MB, Siderovski DP, Hooks SB. The G{beta}{gamma} DIMER as a NOVEL SOURCE of SELECTIVITY in G-Protein Signaling: GGL-ing AT CONVENTION. Mol. Interv. 2004;4(4):200–214. doi: 10.1124/mi.4.4.4. [DOI] [PubMed] [Google Scholar]
- 8.Cheever ML, Snyder JT, Gershburg S, Siderovski DP, Harden TK, Sondek J. Crystal structure of the multifunctional Gbeta5-RGS9 complex. Nat. Struct. Mol. Biol. 2008;15(2):155–162. doi: 10.1038/nsmb.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Snow BE, Krumins AM, Brothers GM, Lee SF, Wall MA, Chung SJ, Mangion S, Gilman AG, Siderovski DP. A G protein gamma subunit-like domain shared between RGS11 and other RGS proteins specifies binding to Gbeta5 subunits. Proc. Natl. Acad. Sci. U. S. A. 1998;95(22):13307–13312. doi: 10.1073/pnas.95.22.13307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levay K, Cabrera JL, Satpaev DK, Slepak VZ. Gbeta5 prevents the RGS7-Galphao interaction through binding to a distinct Ggamma-like domain found in RGS7 and other RGS proteins. Proc. Natl. Acad. Sc.i U. S. A. 1999;96(5):2503–2507. doi: 10.1073/pnas.96.5.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ponting CP, Bork P. Pleckstrin's repeat performance: a novel domain in G-protein signaling? Trends Biochem. Sci. 1996;21(7):245–246. [PubMed] [Google Scholar]
- 12.Hu G, Wensel TG. R9AP, a membrane anchor for the photoreceptor GTPase accelerating protein, RGS9-1. Proc. Natl. Acad. Sci. U. S. A. 2002;99(15):9755–9760. doi: 10.1073/pnas.152094799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drenan RM, Doupnik CA, Boyle MPLJ, Huettner JE, Linder ME, Blumer KJ. Palmitoylation regulates plasma membrane-nuclear shuttling of R7BP, a novel membrane anchor for the RGS7 family. J. Cell Biol. 2005;169(4):623–633. doi: 10.1083/jcb.200502007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martemyanov KA, Yoo PJ, Skiba NP, Arshavsky VY. R7BP, a novel neuronal protein interacting with RGS proteins of the R7 family. J. Biol. Chem. 2005;280(7):5133–5136. doi: 10.1074/jbc.C400596200. [DOI] [PubMed] [Google Scholar]
- 15.Jayaraman M, Zhou H, Jia L, Cain MD, Blumer KJ. R9AP and R7BP: traffic cops for the RGS7 family in phototransduction and neuronal GPCR signaling. Trends Pharmacol. Science. 2008 doi: 10.1016/j.tips.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grabowska D, Jayaraman M, Kaltenbronn KM, Sandiford SL, Wang Q, Jenkins S, Slepak VZ, Smith Y, Blumer KJ. Postnatal induction and localization of R7BP, a membrane-anchoring protein for regulator of G protein signaling 7 family-Gbeta5 complexes in brain. Neuroscience. 2008;151(4):969–982. doi: 10.1016/j.neuroscience.2007.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cao Y, Song H, Okawa H, Sampath AP, Sokolov M, Martemyanov KA. Targeting of RGS7/Gbeta5 to the dendritic tips of ON-bipolar cells is independent of its association with membrane anchor R7BP. J. Neurosci. 2008;28(41):10443–10449. doi: 10.1523/JNEUROSCI.3282-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeng W, Xu X, Popov S, Mukhopadhyay S, Chidiac P, Swistok J, Danho W, Yagaloff KA, Fisher SL, Ross EM, Muallem S, Wilkie TM. The N-terminal domain of RGS4 confers receptor-selective inhibition of G protein signaling. J. Biol. Chem. 1998;273(52):34687–34690. doi: 10.1074/jbc.273.52.34687. [DOI] [PubMed] [Google Scholar]
- 19.Saitoh O, Murata Y, Odagiri M, Itoh M, Itoh H, Misaka T, Kubo Y. Alternative splicing of RGS8 gene determines inhibitory function of receptor type-specific Gq signaling. Proc. Natl. Acad. Sci. U. S. A. 2002;99(15):10138–10143. doi: 10.1073/pnas.152085999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Itoh M, Nagatomo K, Kubo Y, Saitoh O. Alternative splicing of RGS8 gene changes the binding property to the M1 muscarinic receptor to confer receptor type-specific Gq regulation. J. Neurochem. 2006;99(6):1505–1516. doi: 10.1111/j.1471-4159.2006.04220.x. [DOI] [PubMed] [Google Scholar]
- 21.Gesty-Palmer D, Luttrell LM. Heptahelical terpsichory. Who calls the tune? J. Recept. Signal Transduct. Res. 2008;28(1–2):39–58. doi: 10.1080/10799890801941921. [DOI] [PubMed] [Google Scholar]
- 22.Wu G, Benovic JL, Hildebrandt JD, Lanier SM. Receptor docking sites for G-protein betagamma subunits. Implications for signal regulation. J. Biol. Chem. 1998;273(13):7197–7200. doi: 10.1074/jbc.273.13.7197. [DOI] [PubMed] [Google Scholar]
- 23.Wu G, Krupnick JG, Benovic JL, Lanier SM. Interaction of arrestins with intracellular domains of muscarinic and alpha2-adrenergic receptor. J. Biol. Chem. 1997;272(28):17836–17842. doi: 10.1074/jbc.272.28.17836. [DOI] [PubMed] [Google Scholar]
- 24.Wess J, Brann MR, Bonner TI. Identification of a small intracellular region of the muscarinic m3 receptor as a determinant of selective coupling to PI turnover. FEBS Lett. 1989;258(1):133–136. doi: 10.1016/0014-5793(89)81633-3. [DOI] [PubMed] [Google Scholar]
- 25.Lucas JL, Wang D, Sadee W. Calmodulin binding to peptides derived from the i3 loop of muscarinic receptors. Pharm. Res. 2006;23(4):647–653. doi: 10.1007/s11095-006-9784-9. [DOI] [PubMed] [Google Scholar]
- 26.Bernstein LS, Ramineni S, Hague C, Cladman W, Chidiac P, Levey AI, Hepler JR. RGS2 binds directly and selectively to the M1 muscarinic acetylcholine receptor third intracellular loop to modulate Gq/11alpha signaling. J. Biol. Chem. 2004;279(20):21248–21256. doi: 10.1074/jbc.M312407200. [DOI] [PubMed] [Google Scholar]
- 27.Budd DC, McDonald JE, Tobin AB. Phosphorylation and regulation of a Gq/11-coupled receptor by casein kinase 1alpha. J. Biol. Chem. 2000;275(26):19667–19675. doi: 10.1074/jbc.M000492200. [DOI] [PubMed] [Google Scholar]
- 28.Simon V, Guidry J, Gettys TW, Tobin AB, Lanier SM. The proto-oncogene SET interacts with muscarinic receptors and attenuates receptor signaling. J.Biol. Chem. 2006;281(52):40310–40320. doi: 10.1074/jbc.M603858200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Witherow DS, Wang Q, Levay K, Cabrera JL, Chen J, Willars GB, Slepak VZ. Complexes of the G protein subunit gbeta 5 with the regulators of G protein signaling RGS7 and RGS9. Characterization in native tissues and in transfected cells. J. Biol. Chem. 2000;275(32):24872–24880. doi: 10.1074/jbc.M001535200. [DOI] [PubMed] [Google Scholar]
- 30.Levey AI, Kitt CA, Simonds WF, Price DL, Brann MR. Identification and localization of muscarinic acetylcholine receptor proteins in brain with subtype-specific antibodies. J. Neurosci. 1991;11(10):3218–3226. doi: 10.1523/JNEUROSCI.11-10-03218.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu G, Bogatkevich GS, Mukhin YV, Benovic JL, Hildebrandt JD, Lanier SM. Identification of Gbetagamma binding sites in the third intracellular loop of the M(3)-muscarinic receptor and their role in receptor regulation. J. Biol. Chem. 2000;275(12):9026–9034. doi: 10.1074/jbc.275.12.9026. [DOI] [PubMed] [Google Scholar]
- 32.Narayanan V, Sandiford SL, Wang Q, Keren-Raifman T, Levay K, Slepak VZ. Intramolecular Interaction between the DEP Domain of RGS7 and the Gbeta(5) Subunit. Biochemistry. 2007;46(23):6859–6870. doi: 10.1021/bi700524w. [DOI] [PubMed] [Google Scholar]
- 33.Maggio R, Barbier P, Fornai F, Corsini GU. Functional role of the third cytoplasmic loop in muscarinic receptor dimerization. J. Biol. Chem. 1996;271(49):31055–31060. doi: 10.1074/jbc.271.49.31055. [DOI] [PubMed] [Google Scholar]
- 34.Witherow DS, Tovey SC, Wang Q, Willars GB, Slepak VZ. G beta 5.RGS7 inhibits G alpha q-mediated signaling via a direct protein-protein interaction. J. Biol. Chem. 2003;278(23):21307–21313. doi: 10.1074/jbc.M212884200. [DOI] [PubMed] [Google Scholar]
- 35.Willars GB, Muller-Esterl W, Nahorski SR. Receptor phosphorylation does not mediate cross talk between muscarinic M(3) and bradykinin B(2) receptor. Am. J. Physiol. 1999;277(5 Pt 1):C859–C869. doi: 10.1152/ajpcell.1999.277.5.C859. [DOI] [PubMed] [Google Scholar]
- 36.Blin N, Yun J, Wess J. Mapping of single amino acid residues required for selective activation of Gq/11 by the m3 muscarinic acetylcholine receptor. J. Biol.Chem. 1995;270(30):17741–17748. doi: 10.1074/jbc.270.30.17741. [DOI] [PubMed] [Google Scholar]
- 37.Posner BA, Gilman AG, Harris BA. Regulators of G protein signaling 6 and 7. Purification of complexes with gbeta5 and assessment of their effects on g protein-mediated signaling pathway. J. Biol. Chem. 1999;274(43):31087–31093. doi: 10.1074/jbc.274.43.31087. [DOI] [PubMed] [Google Scholar]
- 38.Hooks SB, Waldo GL, Corbitt J, Bodor ET, Krumins AM, Harden TK. RGS6, RGS7, RGS9, and RGS11 stimulate GTPase activity of Gi family G-proteins with differential selectivity and maximal activity. J. Biol. Chem. 2003;278(12):10087–10093. doi: 10.1074/jbc.M211382200. [DOI] [PubMed] [Google Scholar]
- 39.Li GQ, Kevetter GA, Leonard RB, Prusak DJ, Wood TG, Correia MJ. Muscarinic acetylcholine receptor subtype expression in avian vestibular hair cells, nerve terminals and ganglion cells. Neuroscience. 2007;146(1):384–402. doi: 10.1016/j.neuroscience.2007.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Levey AI, Edmunds SM, Koliatsos V, Wiley RG, Heilman CJ. Expression of m1–m4 muscarinic acetylcholine receptor proteins in rat hippocampus and regulation by cholinergic innervation. J. Neurosci. 1995;15(5 Pt 2):4077–4092. doi: 10.1523/JNEUROSCI.15-05-04077.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gautam D, Jeon J, Li JH, Han SJ, Hamdan FF, Cui Y, Lu H, Deng C, Gavrilova O, Wess J. Metabolic roles of the M3 muscarinic acetylcholine receptor studied with M3 receptor mutant mice: a review. J. Recept. Signal Transduct. Res. 2008;28(1–2):93–108. doi: 10.1080/10799890801942002. [DOI] [PubMed] [Google Scholar]
- 42.Yamada M, Miyakawa T, Duttaroy A, Yamanaka A, Moriguchi T, Makita R, Ogawa M, Chou CJ, Xia B, Crawley JN, Felder CC, Deng CX, Wess J. Mice lacking the M3 muscarinic acetylcholine receptor are hypophagic and lean. Nature. 2001;410(6825):207–212. doi: 10.1038/35065604. [DOI] [PubMed] [Google Scholar]
- 43.Gautam D, Gavrilova O, Jeon J, Pack S, Jou W, Cui Y, Li JH, Wess J. Beneficial metabolic effects of M3 muscarinic acetylcholine receptor deficiency. Cell Metab. 2006;4(5):363–375. doi: 10.1016/j.cmet.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 44.Wess J. Muscarinic acetylcholine receptor knockout mice: novel phenotypes and clinical implications. Annu. Rev. Pharmacol. Toxicol. 2004;44:423–450. doi: 10.1146/annurev.pharmtox.44.101802.121622. [DOI] [PubMed] [Google Scholar]
- 45.Chen CK, Eversole-Cire P, Zhang H, Mancino V, Chen YJ, He W, Wensel TG, Simon MI. Instability of GGL domain-containing RGS proteins in mice lacking the G protein beta-subunit Gbeta5. Proc. Natl. Acad. Sci. U. S. A. 2003;100(11):6604–6609. doi: 10.1073/pnas.0631825100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Benians A, Nobles M, Hosny S, Tinker A. Regulators of G-protein signaling form a quaternary complex with the agonist, receptor, and G-protein. A novel explanation for the acceleration of signaling activation kinetics. J. Biol. Chem. 2005;280(14):13383–13394. doi: 10.1074/jbc.M410163200. [DOI] [PubMed] [Google Scholar]
- 47.Ballon DR, Flanary PL, Gladue DP, Konopka JB, Dohlman HG, Thorner J. DEP-domain-mediated regulation of GPCR signaling responses. Cell. 2006;126(6):1079–1093. doi: 10.1016/j.cell.2006.07.030. [DOI] [PubMed] [Google Scholar]
- 48.Kovoor A, Seyffarth P, Ebert J, Barghshoon S, Chen CK, Schwarz S, Axelrod JD, Cheyette BN, Simon MI, Lester HA, Schwarz J. D2 dopamine receptors colocalize regulator of G-protein signaling 9-2 (RGS9-2) via the RGS9 DEP domain, and RGS9 knock-out mice develop dyskinesias associated with dopamine pathways. J. Neurosci. 2005;25(8):2157–2165. doi: 10.1523/JNEUROSCI.2840-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wong HC, Bourdelas A, Krauss A, Lee HJ, Shao Y, Wu D, Mlodzik M, Shi DL, Zheng J. Direct binding of the PDZ domain of Dishevelled to a conserved internal sequence in the C-terminal region of Frizzled. Mol. Cell. 2003;12(5):1251–1260. doi: 10.1016/s1097-2765(03)00427-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cabrera JL, de Freitas F, Satpaev DK, Slepak VZ. Identification of the Gbeta5-RGS7 protein complex in the retina. Biochem. Biophys. Res. Commun. 1998;249(3):898–902. doi: 10.1006/bbrc.1998.9218. [DOI] [PubMed] [Google Scholar]
- 51.Zhang JH, Simonds WF. Copurification of brain G-protein beta5 with RGS6 and RGS7. J. Neurosci. 2000;20(3):RC59. doi: 10.1523/JNEUROSCI.20-03-j0004.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chase DL, Patikoglou GA, Koelle MR. Two RGS proteins that inhibit Galpha(o) and Galpha(q) signaling in C. elegans neurons require a Gbeta(5)-like subunit for function. Curr. Biol. 2001;11(4):222–231. doi: 10.1016/s0960-9822(01)00071-9. [DOI] [PubMed] [Google Scholar]
- 53.Anderson GR, Lujan R, Semenov A, Pravetoni M, Posokhova EN, Song JH, Uversky V, Chen CK, Wickman K, Martemyanov KA. Expression and localization of RGS9-2/Gβ5/R7BP complex in vivo is set by dynamic control of its constitutive degradation by cellular cysteine proteases. J. Neurosci. 2007;27(51):14117–14127. doi: 10.1523/JNEUROSCI.3884-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.He W, Lu L, Zhang X, El-Hodiri HM, Chen CK, Slep KC, Simon MI, Jamrich M, Wensel TG. Modules in the photoreceptor RGS9-1.Gbeta 5L GTPase-accelerating protein complex control effector coupling, GTPase acceleration, protein folding, and stability. J. Biol. Chem. 2000;275(47):37093–37100. doi: 10.1074/jbc.M006982200. [DOI] [PubMed] [Google Scholar]
- 55.Skiba NP, Martemyanov KA, Elfenbein A, Hopp JA, Bohm A, Simonds WF, Arshavsky VY. RGS9-G beta 5 substrate selectivity in photoreceptors. Opposing effects of constituent domains yield high affinity of RGS interaction with the G protein-effector complex. J. Biol. Chem. 2001;276(40):37365–37372. doi: 10.1074/jbc.M106431200. [DOI] [PubMed] [Google Scholar]