Preface
The Gram-negative bacterium Pseudomonas aeruginosa utilizes a complex type III secretion apparatus to inject effector proteins into host cells. The configuration of this secretion machinery, the activities of the proteins that are injected by it, and the consequences of this process to infection are now being elucidated. This Review summarizes our current knowledge of P. aeruginosa type III secretion, including the secretion and translocation machinery, the regulation of this machinery, and the associated chaperones and effector proteins. The features of this interesting secretion system have important implications to the pathogenesis of P. aeruginosa infections and to other type III secretion systems.
Introduction
Pseudomonas aeruginosa is a major cause of health-care associated infections, including pneumonia and infections involving the urinary tract, wounds, burns, and the bloodstream 1. A second population of patients prone to acquire P. aeruginosa infection is those with cystic fibrosis 2. Like many other Gram negative bacteria, P. aeruginosa manipulates eukaryotic host cells by using a type III secretion system (T3SS). This T3SS forms complex needle-like machines on the bacterial surface that function in a highly regulated manner to transport proteins into host cells.
In this review, I will describe the T3SS of P. aeruginosa. Current knowledge of the components of this system will be discussed, with special emphasis on the effector proteins injected by this system. Recent studies in humans and progress in the development of therapeutic agents designed to inhibit the P. aeruginosa T3SS will be summarized.
The P. aeruginosa type III secretion system
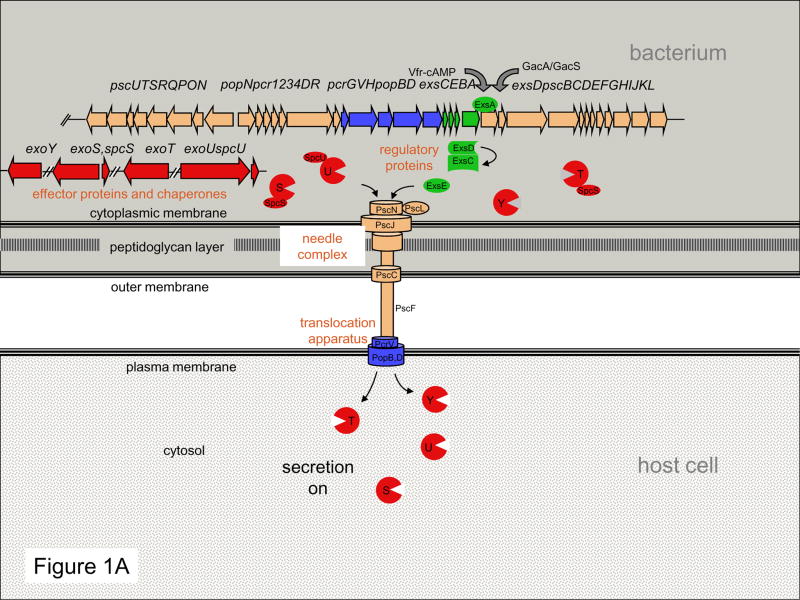
To establish infections, P. aeruginosa employs a broad arsenal of virulence determinants 3, of which its T3SS has been the focus of much recent attention 4. Thirty-six genes encoded in five operons clustered together in the P. aeruginosa chromosome are involved in the biogenesis and regulation of the type III delivery machine. At least six other genes scattered around the chromosome encode effector proteins and their chaperones. This complex regulon can be divided into five parts (Fig. 1A): proteins that comprise the needle complex that transports substrates from the bacterial cytosol to the extracellular environment; proteins that translocate secreted proteins into host cells; proteins that regulate the secretion process; chaperone proteins that facilitate secretion of their cognate partners, and proteins that are injected into host cells, called effector proteins.
Figure 1. Overview of P. aeruginosa type III secretion.
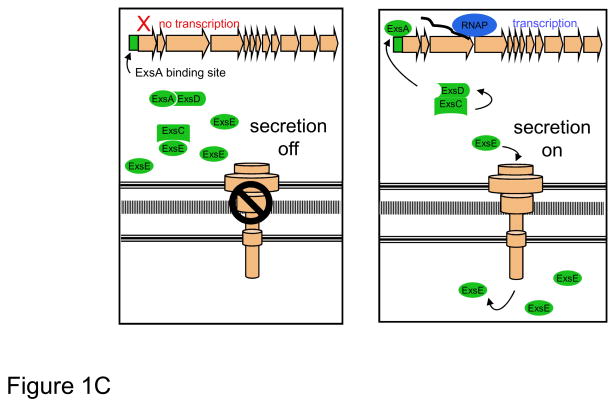
(A) The T3SS may be functionally divided into five components: the needle complex; the translocation apparatus, regulatory proteins, effector proteins, and chaperones. These five parts work together to inject effector proteins into host cells in a highly regulated manner. (C) Linkage of T3SS transcription to protein secretion is achieved through the interactions of four proteins: ExsA, ExsC, ExsD, and ExsE. When secretion is turned off, ExsE accumulates within the bacterium and binds ExsC, allowing ExsD to bind ExsA, and thus preventing transcription of type III secretion genes. When secretion is activated, the regulatory protein ExsE is secreted from the cell, allowing ExsC to bind ExsD. Sequestration of ExsD frees the transcriptional activator ExsA, resulting in unimpeded transcription of type III secretion genes. RNAP, RNA polymerase
The needle complex
The needle complex is the supramolecular structure responsible for transport of specific type III proteins from the bacterial cytosol to the extracellular environment, moving them past the barriers of the bacterial cytoplasmic membrane, the peptidoglycan layer, and the outer membrane. The P. aeruginosa needle complex appears to be similar to the more thoroughly characterized needle complexes of Yersinia spp., Salmonella enterica, and Shigella flexneri and consists of two parts: a multi-ring base and a needle-like filament (Fig. 1A). The needle-like filament, which is comprised of subunits of the protein PscF 5, 6, is thought to be a conduit through which secreted factors move and to also serve as a sensor for host cell contact. These structures are 60–120 nm long and 6–10 nm wide (Fig. 1B), similar in dimensions to the needle-like filaments of Shigella flexneri, Salmonella enterica, and Yersinia enterocolitica 7–10. Relatively little work has been done to characterize the remaining components of the P. aeruginosa needle complex, although some functional assignments can be inferred from the closely related Yersinia type III needle complex (see Box 1). For example, PscN is thought to be an ATPase that powers the P. aeruginosa secretion system and is regulated by PscL 11, 12. PscC is a secretin-like protein that likely oligomerizes with the help of the lipoprotein PscW to form a channel through the bacterial outer membrane 13, 14. Based on studies of its Yersinia homolog, PscP is likely a molecular ruler that dictates the needle length 15. PscJ is predicted to be a lipoprotein component of the basal substructure of the needle complex 16. Although some progress has been made, considerable effort will be required to fully understand the complex mechanism by which the needle complex performs its role in the secretion process.
Box 1. Evolution of the P. aeruginosa type III secretion system.
T3SSs share a common ancestor with flagellar systems 163. But whereas the phylogenies of the flagellar systems follow those of their source organisms, the clusters of genes encoding the T3SSs do not, indicating interspecies horizontal transfer 164. The P. aeruginosa T3SS belongs to the Ysc family of type III systems, which includes the Yop system of Yersinia spp., the Asc system of Aeromonas salmonicida, the Lsc system of Photorhabdus luminescens, the Vcr system of Vibrio parahaemolyticus, the Bsc system of Bordetella spp., and the Dsc system of Desulfovibrio vulgaris 165.
The selective pressures driving the evolution of the P. aeruginosa T3SS are unclear. P. aeruginosa is an opportunistic pathogen in humans, meaning that it usually only infects individuals who are immunocompromised in some way 1. Thus most infections are “iatrogenic,” the consequence of procedures and treatments of modern health care. Even individuals with cystic fibrosis rarely lived long enough to acquire P. aeruginosa prior to the advent of modern antimicrobial therapies 166. For this reason, it is unlikely that the P. aeruginosa T3SS evolved as the result of pressures to survive within humans. What then is the natural target of the P. aeruginosa T3SS? P. aeruginosa is an environmental bacterium that must avoid or survive predation by the many organisms that inhabit the soil and natural bodies of water, and its T3SS may have evolved for this purpose. In support of this notion is that the majority of environmental P. aeruginosa isolates have functional T3SSs 167 and that ExoU, ExoS, ExoT, and ExoY each facilitate killing of environmental amoebae during co-cultivation 168, 169. Thus this system may have evolved to ward off environmental predators, and broad conservation of targeted substrates across eukaryotic organisms resulted in a system also active against human cells.
The translocation apparatus
The translocation apparatus is a proteinaceous membrane pore that accepts effector proteins secreted by the needle complex and delivers them across the host cell plasma membrane (Fig. 1A). The translocation process is quite efficient, with less than 0.1% of secreted effector proteins escaping to the extracellular milieu 17. Like the T3SSs of Yersinia, Salmonella and Shigella spp., the T3SS of P. aeruginosa uses three proteins for translocation: PopB, PopD, and PcrV 18–20.
PopB and PopD are themselves secreted by the type III needle complex and, in a poorly understood manner, interact with each other and the host cell membrane to form the translocation pore 2.8–6.0 nm in diameter 18–22. Recombinant PopB and PopD were sufficient for the formation of oligomers and ring-like structures in lipid vesicles in vitro 21, although PcrV was required for pore formation when these two proteins were injected through the needle complex into the plasma membranes of fibroblasts and red blood cells 23. PcrV is also secreted by the T3SS and is necessary for translocation but does not appear to be itself part of the pore 23, 24. It may instead form a multimeric scaffold at the tip of the type III secretion needle, as it has been shown to do when artificially expressed as part of the related Yersinia enterocolitica T3SS 25. This scaffold may then facilitate the assembly of the PopB/PopD translocation pore within the host cell plasma membrane. Alternatively, PcrV may link the needle complex to preformed PopB/PopD pores, forming part of a continuous conduit through which effector proteins move from the bacterial cytosol to the host cell cytosol 26.
The role of the translocation apparatus may not be limited to transport of effector proteins; the translocation pore by itself may be sufficient to cause the death of host cells 20, 23, 27, 28, either directly through pore-mediated increases in membrane permeability 20, 23, 27, 29 or indirectly through the activation of broad cellular defense responses, as has been observed with the T3SSs of other bacterial pathogens 30. Death of the intoxicated cell in this context may be a protective mechanism of the host rather than a virulence mechanism of the bacterium. Details of such pathways are beginning to emerge. For example, a P. aeruginosa type III secretion/translocation apparatus (in the absence of any of the known effector proteins) is sufficient to trigger the activation of caspase-1 by the inflammasome via Ipaf, a Nod-like receptor (NLR) family member and cytosolic sensor of bacterial products 31–34. As has been shown with Salmonella type III and Legionella type IV secretion, the formation of the translocation pore may facilitate entry of flagellin into the cytosol, where it is recognized by Ipaf, although this remains controversial with P. aeruginosa type III secretion 31–34. Activation of the inflammasome and caspase-1 results in IL-1β and IL-18 production and pyroptosis 20, 32, 33, a form of cell death that is proinflammatory and has features of both apoptosis and necrosis 35. Induction of pyroptosis may explain previous reports of T3SS-dependent but effector-independent death of phagocytes 20, 23, 27, 28. Since bacterial burdens were higher after 12 hr of intraperitoneal infection in Ipaf−/− mice, inflammasome activation appears to be protective and may be an important aspect of the innate immune response against P. aeruginosa 32.
Regulation of type III secretion
Type III secretion in P. aeruginosa is regulated at two levels: transcription of T3SS genes and initiation of secretion itself. These levels are linked in that transcription is induced upon activation of the secretion process, 4, 36 which allows type III secretion components to be produced at high levels when most needed: following contact with host cells.
At its most proximal level, transcription of T3SS genes is controlled by ExsA, a member of the AraC/XylS family of transcriptional regulators. ExsA binds the promoters of T3SS genes, including its own promoter, in a region of DNA that extends through the putative -35 RNA polymerase binding site and 18 nucleotides upstream 37, 38. This region includes an adenine-rich sequence at −55 that is sometimes referred to as an ExsA consensus element 37, 38. An elaborate “catch and release” mechanism involving three additional proteins, ExsC, ExsD, and ExsE, is employed to link ExsA-mediated transcriptional activation to secretion (Fig. 1C). Under conditions that prevent secretion, ExsA is bound by the “anti-activator” ExsD, which inhibits ExsA-dependent transcription 36. ExsC, an “anti-anti-activator,” has the potential to disrupt the ExsD-ExsA interaction by directly binding and sequestering ExsD 39 but under non-secreting conditions is prevented from doing so by its higher binding affinity for ExsE 40, 41. The coupling of transcription with secretion is achieved by the export ExsE through the type III needle complex into either the medium or the cytosol of host cells during secretion 42. Thus under conditions that promote secretion, ExsE levels within the bacterium are depleted. The relative paucity of ExsE frees ExsC to bind ExsD, allowing unimpeded transcriptional activation of type III secretion genes by ExsA. The net result of this regulatory cascade is that production of T3SS proteins, including needle complexes and effector proteins, is maintained at low basal levels until contact with host cells activates secretion, which rapidly increases expression of T3SS genes.
Two global regulatory systems, both of which respond to poorly defined environmental stimuli, are known to work through ExsA to regulate transcription of T3SS genes: intracellular 3′,5′-cyclic AMP (cAMP) and the Gac regulatory system. In the cAMP pathway, the inner membrane protein adenylate cyclase CyaB responds to inducing signals by increasing intracellular cAMP levels 43. cAMP likely allosterically regulates Vfr, a global regulatory protein with homology to the Escherichia coli cAMP receptor protein (CRP) 43. cAMP and Vfr, together with ExsA, regulate expression from T3SS promoters in a poorly understood manner. In the Gac system, the two-component sensors RetS and LadS transduce environmental signals to modulate the two-component GacA/GacS system, which in turn regulates expression of T3SS genes. RetS primes P. aeruginosa for acute infections by inducing T3SS genes and normal piliation but repressing genes that promote biofilm growth, whereas LadS acts in a reciprocal manner, down-regulating type III secretion during chronic infections (Box 2) 44–47. The reader is referred to a recent review for a more detailed discussion of transcriptional regulation of the P. aeruginosa T3SS 48.
Box 2. P. aeruginosa type III secretion in chronic infections.
Whereas P. aeruginosa type III secretion plays a dominant role in acute infections, emerging evidence suggests that it may actually be selected against in chronic infections, such as those afflicting individuals with CF. Many CF patients have antibodies against type III effector proteins 170, suggesting that these factors are expressed at some point during infection, but P. aeruginosa strains gradually lose the ability to secrete type III proteins over time in the CF airways 27, 151, 171. This phenomenon may occur through a number of different mechanisms. P. aeruginosa isolates from chronically infected CF patients often have a mucoid phenotype that results from overexpression of the exopolysaccharide alginate. In these strains, alginate is often overproduced because of mutations within the mucA gene 172, which also result in repression of type III secretion genes 173. A second mechanism is the RetS/LadS/GacA/GacS regulatory system (described in the “Regulation of type III secretion” section), which allows P. aeruginosa to switch from an acute to a chronic infection mode. Finally over a period of years in the CF airways, mutations accumulate in genes essential for type III secretion, such as cyaB, vfr, and exsA 174. Thus multiple mechanisms cause the attenuation of type III secretion during CF infections, underscoring the strength of the selection against this system. It may be that secretion of such destructive toxins is not compatible with bacterial persistence in the airways for years or that T3SS proteins constitute attractive targets for the host immune system.
Triggering of secretion is complex and less well understood, but the physiological stimulus is contact with host cells while the most commonly utilized laboratory signal is exposure to calcium-depleted medium 49, 50. Low calcium concentrations alone are insufficient to trigger secretion but must be accompanied by the presence of certain amino acids, such as glutamate 51. This is likely due to a role played by the citric acid cycle in type III secretion. For example, mutations in the genes encoding citrate synthetases, pyruvate dehydrogenase, and GltR, the regulator of the glucose transporter, all prevented induction of type III secretion 43, 51, 52. In fact, metabolic stress in general modulated the ability of P. aeruginosa to secrete type III proteins 53. Metabolically unfavorable conditions may reduce overall secretion by decreasing the percentage of bacterial cells able to assemble functional secretion machines on their surface 51.
Chaperones
Some but not all type-III-secreted proteins have cognate chaperones to which they bind prior to secretion. These chaperones may facilitate storage of their cognate protein partner in the bacterial cytosol and their appropriate delivery to the secretion apparatus 54. SpcS (formerly called Orf1) serves as a chaperone for both ExoS and ExoT and is required for maximal secretion of these proteins (Fig. 1A, Table 1) 55, 56. Likewise SpcU is a chaperone for ExoU 57. No chaperone has yet been identified for ExoY. Chaperones also exist for type-III-secreted proteins other than effector proteins. For example, PcrH is a chaperone for the translocation proteins PopB and PopD 21, PscE and PscG are chaperones for PscF, the structural subunit of the needle complex 58, and ExsC is a chaperone for the regulatory protein ExsE 40, 41.
Table 1.
Characteristics of P. aeruginosa T3SS effector proteins.
| Effector protein | Size (kDa) | Chaperone | Substrates | Cofactor for activation |
|---|---|---|---|---|
| ExoS | 49 | SpcS | GAP activity: Rho, Rac, and Cdc42 ADPRT activity: ezrin, radixin, moesin, vimentin, cyclophilin A, IgG3, apolipoprotein A1, Ras, Rac1, Cdc42, Rabs 1, 3, 5, 7, 8, and 11, RalA, Rap1, Rap2 |
14-3-3 proteins |
| ExoT | 53 | SpcS | GAP activity: Rho, Rac, and Cdc42 ADPRT activity: Crk-I, Crk-II, phosphoglycerate kinase |
14-3-3 proteins |
| ExoU | 74 | SpcU | phospholipids, lysophospholipids, neutral lipids | SOD1 |
| ExoY | 41 | ? | ATP | ? |
GAP: GTPase activating protein
ADPRT: adenosine diphosphate ribosyl transferase
Crk: CT10-regulator of kinase
SOD1: Cu2+, Zn2+-superoxide dismutase
?: unknown
Please see text for citations
The Effectors
Despite extensive characterization by a number of laboratories, only four effector proteins of the P. aeruginosa T3SS have been identified: ExoS, ExoT, ExoU, and ExoY (Table 1). Thus the P. aeruginosa T3SS appears to have fewer effector proteins than all other well characterized T3SSs. For example, the Yersinia Yop T3SS has 6 effector proteins, the Salmonella SPI1 system ≥ 13, the Salmonella SPI2 system ≥ 10, and the Shigella system ≈ 25. This relative dearth of effector proteins is especially surprising given the wide spectrum of hosts and environments with which P. aeruginosa is associated but may reflect the large number of other types of secretion systems encoded by this bacterium. Alternatively, the small number of effector proteins may have been evolutionarily balanced by their activity against an extremely broad range of eukaryotic organisms.
While virtually all P. aeruginosa strains harbor the genes encoding the type III secretion machinery, most strains do not carry a complete set of effector-encoding genes 59, 60. In isolates from acute infections, the exoS gene is found in 58–72% of isolates, the exoU gene in 28–42%, the exoY gene in 89%, and the exoT gene in 92–100%. For reasons that are not entirely clear, nearly all strains have either the exoS or the exoU gene but not both. As described below, the secreted effector proteins define the phenotype of a strain during infection 60, 61. For example, ExoS-secreting strains cause delayed cell death with features of apoptosis, whereas ExoU secretion is associated with rapid and robust host cell lysis.
ExoS
ExoS is a bi-functional toxin with both GTPase activating protein (GAP) activity and adenosine diphosphate ribosyl transferase (ADPRT) activity (Table 1). This effector protein has a modular domain structure that underscores the complexity of its interactions with host cells (Fig. 2).
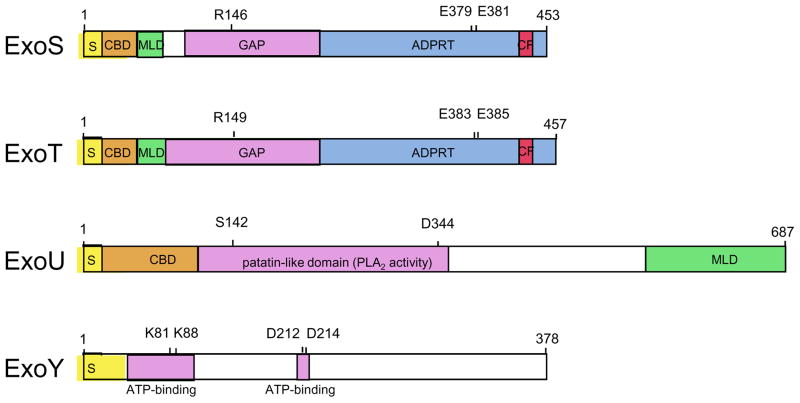
Figure 2. Modular domains of ExoS, ExoT, ExoU, and ExoY.
ExoS is a 453 amino acid bi-functional toxin that has both GAP and ADPRT activity. Arg146 is required for GAP activity and both Glu379 and Glu381 are required for efficient catalytic addition of the ADP-ribose moiety of NAD+ to substrate 175. Within the ADPRT domain, residues 418 to 429 comprise a binding site for the eukaryotic cofactor 14-3-3 that is necessary for activation of the ADPRT activity of ExoS. ExoT is a 457 amino acid protein that is closely related to ExoS. Arg149 is required for the GAP activity of ExoT 76 and residues Glu383 and Glu385 are crucial for its ADPRT activity 175. Residues 422–433 are thought to comprise the site of cofactor 14-3-3 binding. ExoU is a 687 amino acid protein that contains a patatin-like domain necessary for PLA2 activity. Residues Ser142 and Asp344 are required for this activity 117, 118, 121. ExoY is a 378 amino acid adenylate cyclase. Residues Lys81, Lys88, Asp212, and Asp214 are required for its activity and are thought to be necessary for interactions between ExoY and ATP. See text for additional details. S, secretion signal; CBD, chaperone binding site; MLD, membrane localization domain; GAP, GTPase activating protein activity; ADPRT, ADP-ribosyl transferase activity; PLA2, phospholipase A2; CF, cofactor binding site. (Adapted from 176)
As with other type III effector proteins, the extreme amino-terminus of ExoS carries the information for targeting to the type III secretion apparatus. Although no consensus sequence has been identified, the first 15 amino acids of ExoS presumably encode the information necessary for secretion through the type III apparatus 4. Residues 15–51 are believed to comprise a binding site for the ExoS chaperone SpcS 62.
Residues 51–72 contain a symmetrical leucine-rich motif that forms the membrane localization domain (MLD) 63, which is responsible for the initial transient localization of ExoS to the plasma membrane of the host cell (Fig. 3) 64. Subsequent trafficking to the Golgi and endoplasmic reticulum in the perinuclear region of the cell occurs in association with markers for Rab5-containing early endosomes and Rab6- and Rab9-containing late endosomes 64, 65. Of note, localization of ExoS by the MLD to these membrane compartments is essential for efficient modification of its substrates 66, 67.
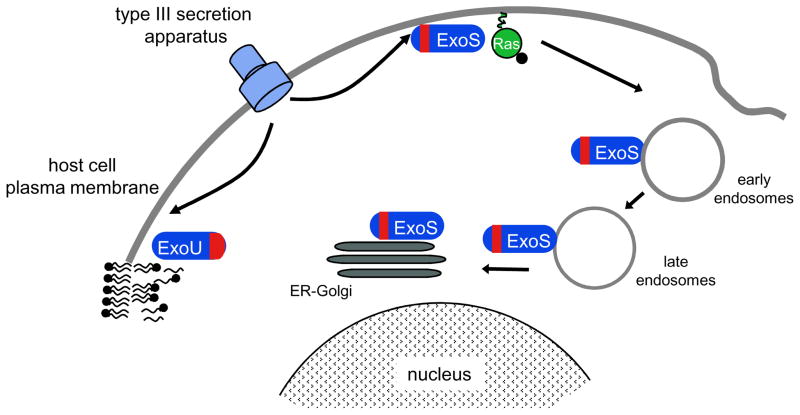
Figure 3. Localization of ExoS and ExoU following injection into mammalian cells.
The N-terminal MLD of ExoS (shown in red) initially targets this effector protein to the plasma membrane and subsequently to early and late endosomes and finally to the endoplasmic reticulum (ER) and Golgi. Membrane localization is necessary for efficient ADP ribosylation of membrane-associated Ras. Following translocation into mammalian cells, ExoU is targeted to the plasma membrane by a C-terminal MLD. At this location, the enzymatic activity of ExoU is thought to cleave phospholipids within the plasma membrane. (Adapted from 65)
Residues 96–233 of ExoS form a GAP domain that targets Rho, Rac, and Cdc42 68, 69, small GTPases that maintain the organization of the actin cytoskeleton. These regulatory GTPases normally switch between an active GTP-bound form and an inactive GDP-bound form, but the ExoS GAP domain biases the switch towards the ADP-bound inactive form, leading to disruption of the actin cytoskeleton 68, 69. This domain has functional and structural similarity with other type III effector proteins such as YopE of Yersinia spp. and StpP of Salmonella enterica 70, 71. The three-dimensional structure of the ExoS GAP domain bound to Rac1 indicates that ExoS uses an arginine finger (Arg146) to stabilize the Rac1 transition state and maintain intrinsic GTPase activity 72. This in turn results in GTP hydrolysis to GDP, causing the GDP-bound form of Rac1 to predominate 69, 73 and leading to inactivation of Rac signaling 74. The transient disruption of the actin cytoskeleton by the GAP activity of ExoS is associated with cell rounding and decreased internalization of P. aeruginosa by certain cell types, suggesting a role in preventing phagocytosis 75–77.
The ADPRT domain of ExoS resides between residues 233 and 453 78. Residues 418 to 429 comprise a binding site for a eukaryotic cofactor that is necessary for activation of ADPRT activity (but not GAP activity 79) 80. This cofactor was initially designated Factor Activating ExoS (FAS) but was later shown to be a 14-3-3 protein 81, 82. These proteins serve as regulatory factors that modulate a wide variety of cellular functions, including cell cycle progression, protein trafficking, and apoptosis. Similar to other 14-3-3 ligands, ExoS binds within a conserved amphipathic groove of 14-3-3 but does so in the opposite orientation of most ligands and utilizes unique intermolecular interactions 83. Deletion of the 14-3-3 binding site renders ExoS unable to interact with its cofactor and results in the loss of ADPRT activity and its consequences to mammalian cells 83. Thus the requirement for a host cell cofactor prevents this potent weapon from being turned against P. aeruginosa itself by precluding activation until after ExoS is injected into the host cell.
Following injection and activation, the ADPRT activity of ExoS has a number of adverse effects on the host cell, including cell death, actin cytoskeletal disruption associated with cell rounding, and inhibition of DNA synthesis, vesicular trafficking and endocytosis 74, 84–86. The irreversible disruption of the cytoskeletal structure may contribute to a reduction in cell-cell adherence, which in turn may facilitate P. aeruginosa penetration through epithelial barriers 74, 84, 86. Cell death has features of apoptosis although loss of cell membrane integrity, more characteristic of necrosis, is also observed 84, 87. It has been postulated that the killing of host immune cells by ExoS may allow P. aeruginosa to persist in the presence of a vigorous host immune response, although the cell types targeted by ExoS in vivo are unclear. Alternatively, this form of cell death may be similar to pyroptosis and may be the culmination of a host cell response to the presence of ExoS aimed at eliminating intoxicated cells.
Defining the substrates modified by ExoS that lead to these pathogenic changes has been the focus of much effort and has been complicated by the promiscuous nature of the ADPRT activity of this toxin. It catalyzes the transfer of ADP-ribose to many different host proteins (Table 1) 85, 88–97, and the challenge has been to link ADP-ribosylation of specific substrates to the observed host cell phenotypes. Two substrates of special interest are Ras and the ezrin, radixin and moesin (ERM) family of proteins. Since Ras signaling affects cellular proliferation, survival, and cytoskeletal structure, this small GTPase has been an attractive candidate for the pathogenically relevant target of ExoS. Modification of Arg41 of Ras results in inhibition of GDP to GTP exchange normally catalyzed by the guanine nucleotide exchange factor (GEF) of Ras 98. This in turn disrupts Ras signaling by preventing association of Ras with its downstream effector, Raf-1 99–101. Over-expression of constitutively active Ras or the protein kinase B/Akt, a downstream component of the Ras signaling pathway, prevented ExoS-mediated cell killing, indicating that ExoS modification of Ras is biologically important 102. ExoS lacking the MLD localized within the cytosol and failed to appropriately ADP-ribosylate membrane-associated Ras, demonstrating the importance of ExoS localization within cells (Fig. 3) 103. The ERM family of proteins 96 bind actin and play an important role in many actin processes, including motility, phagocytosis, adhesion, and the maintenance of cell shape. ADP-ribosylation of ERM proteins prevents their phosphorylation, resulting in inactivation 104. In support of the biological relevance of this modification, dominant active-moesin prevented the cell rounding normally associated with ExoS intoxication 104. Thus the ADPRT activity of ExoS appears to target a number of distinct cell factors and pathways to compromise the host cell.
ExoT
ExoT shares 76% amino acid identity with ExoS, and is also a bifunctional toxin with amino-terminal GAP activity and carboxy-terminal ADPRT activity (Table 1). Although not as thoroughly studied, the domain structure of ExoT appears to be similar to that of ExoS in that residues 1–50 likely encode secretion and chaperone binding information and residues 51–72 are thought to encode a MLD (Fig. 2) 62.
Similar to ExoS, ExoT residues 78–235 contain GAP activity towards Rac, Rho, and Cdc42 105, 106. This activity appears to be very similar to that of the N-terminus of ExoS and causes reversible disruption of the actin cytoskeleton, which manifests as cell rounding, cell detachment, and inhibition of cell migration and phagocytosis 76, 77, 106. The GAP activity of ExoT also contributes to inhibition of cytokinesis through inactivation of Rho 107, 108.
Also similar to ExoS, residues 235–457 of ExoT encode an ADPRT domain that requires binding of the host cell cofactor 14-3-3 for activation 109. However unlike ExoS, ExoT ADP-ribosylates a distinct and limited number of host proteins (Table 1). In particular, ExoT modifies CT10-regulator of kinase (Crk)-I and Crk-II, adaptor proteins 110. Hybrid protein experiments and structural modeling indicated that one small α-helix region (“region A”) present in ExoT but absent in ExoS conferred specificity for Crk 111. ExoT ADP-ribosylates Crk-I at Arg20, which interferes with Crk signaling by preventing association of Crk-I with focal adhesion proteins p130cas and paxillin. This has multiple effects on cells, including disruption of signaling to Rac1 and blockage of cell division at the stage of cytokinesis 107, 108. Thus the GAP and ADPRT activities of ExoT work together to alter the actin cytoskeleton and to inhibit cell migration, adhesion, and proliferation. The net effect of these activities is to block phagocytosis and disrupt epithelial barriers to facilitate bacterial dissemination 27, 28, 76, 112. In addition, the enzymatic activities of ExoT have been linked to delays in wound healing 113, 114, which may augment the opportunistic capacity of P. aeruginosa by allowing it to more fully exploit breaches in mucosal barriers. Although initially reported to have no effect on cell viability, ExoT does cause apoptosis-like cell death predominantly through its ADPRT activity at later time points (10 hours, compared to 2–5 hours for ExoS-mediated killing) 29. Apoptosis may be a consequence of inhibition of Crk signaling, which has been linked to this form of cell death 115.
Multiple studies have demonstrated a pathogenic role for ExoT, but its contribution to disease in mammalian models has been modest relative to ExoU and ExoS 27, 28, 76, 112. ExoT forms a complex with its substrate Crk and a Crk ligand, the E3 ubiquitin ligase Cbl-b 116, leading to polyubiquitination of ExoT by Cbl-b and subsequent proteasomal degradation. Interestingly, Cbl-b deficient mice were much more susceptible to infection with an ExoT+ P. aeruginosa strain than an ExoT− P. aeruginosa strain, although wild-type mice differed only slightly in this regard 116. Thus Cbl-b functions as a host-defense molecule by facilitating the degradation of ExoT.
ExoU
ExoU is a potent phospholipase capable of causing rapid cell death in eukaryotic cells (Table 1). Although not yet demonstrated, the secretion signal directing ExoU to the type III secretion apparatus is assumed to be within the first 15 residues, based on findings from other effector proteins (Fig. 2). A binding domain for the chaperone of ExoU, SpcU, lies within residues 3 through 123 57.
Immediately adjacent to the chaperone binding site, between residues 107 and 357, is a patatin-like domain that encodes phospholipase A2 (PLA2) activity 117, 118. Patatin-like domains are found in several other well-characterized PLA2 enzymes, such as human calcium-independent PLA2 (iPLA2) and human cytosolic phospholipase A2 (cPLA2) 119. PLA2 enzymes hydrolyse the ester bond of the acyl group at the sn-2 position of phospholipids, resulting in release of free fatty acids and lysophospholipids. In actuality, the lipase activity of ExoU has a broad range of substrates, including phospholipids, lysophospholipids, and neutral lipids 117, 118, 120. Sequence alignment with well characterized PLA2 enzymes and mutational analysis suggest that ExoU has a serine-aspartate catalytic dyad involving Ser142 and Asp344 117, 118, 121. Subsequent to the identification of the patatin-like domain in ExoU, similar domains have been identified in a large number of bacterial proteins 122.
Like other P. aeruginosa type III effector proteins, ExoU requires a eukaryotic cofactor for activation 117, 118. Fractionation of eukaryotic cell extracts yielded Cu2+, Zn2+-superoxide dismutase (SOD1) as one such factor 123. Interestingly SOD1 enzymatic activity was not necessary for this effect, suggesting that SOD1 activated ExoU by other mechanisms such as inducing a conformational change or linking ExoU to other factors or its phospholipid substrate. The domain of ExoU that binds SOD1 has not yet been defined.
The C-terminal half of ExoU contains no recognizable homology or motifs but is essential for killing 117, 121, 124, 125. A portion of this part of ExoU, residues 550–683, is now known to contain a MLD that functions to target ExoU to the plasma membrane of the host cell (Fig. 3) 124, 126. The remaining portion of the C-terminal half of ExoU (residues 358 to 549) may also play an important role in the mechanism of this toxin, since 5-amino-acid insertions in this region also destroyed cytotoxicity 121.
Another interesting aspect of ExoU is that it is ubiquitinylated. Once inside the host cell, two ubiquitin molecules are added to lysine 178 of this effector protein. Although ubiquitinylation had a modest effect on ExoU turnover within cells, it did not appreciably alter toxicity or localization 124. Thus the significance of this modification to the pathogenicity of ExoU remains unclear.
The net result of intoxication with ExoU is death of the host cell. Unlike the cell killing caused by ExoS and ExoT, death is characterized by rapid (within 1–2 hours) loss of integrity of the plasma membrane, consistent with necrosis 127, 128. ExoU killing may be directed against phagocytes as well as epithelial barriers, thus promoting bacterial persistence and dissemination 27, 112, 129–131.
ExoY
ExoY is a secreted adenylate cyclase (Table 1) 132. Two domains of ExoY are similar to corresponding domains of the extracellular adenylate cyclases of Bordetella pertussis (CyaA) and Bacillus anthracis (Edema factor) 132 and act together to bind ATP (Fig. 2). ExoY also requires a host cell cofactor for full enzymatic activity, although the identity of this factor is not known 132. Injection of ExoY into mammalian cells results in an elevation of intracellular cAMP concentration and differential expression of multiple genes, including many known to be regulated by cAMP 132, 133. This leads to disruption of the actin cytoskeleton 132, 134, inhibition of bacterial uptake by host cells 135, and increased endothelial permeability 136. Such activities would be predicted to lead to more severe disease, but this has not been uniformly observed 27, 28, and the significance of ExoY in infection remains unclear.
P. aeruginosa type III secretion and disease
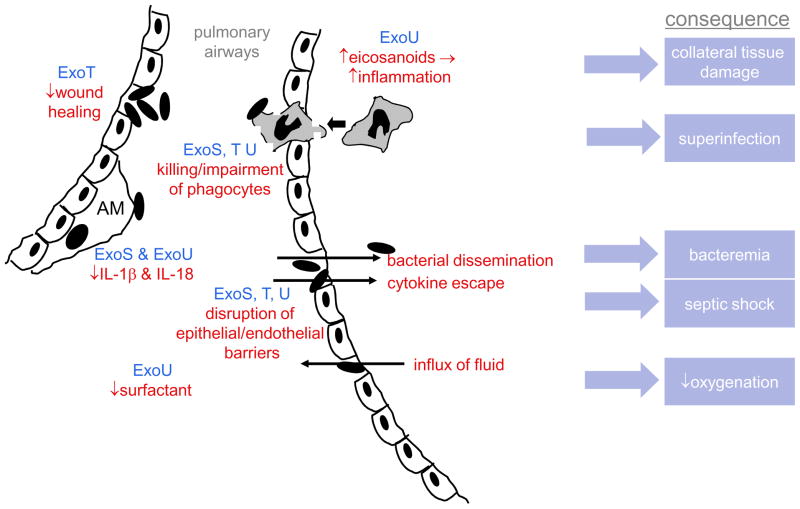
Although not required for infection 61, 137, the P. aeruginosa T3SS enhances disease severity in a number of animal models, including acute pneumonia 27, 127, 128, 138, keratitis 139, bacteremia 28, peritonitis 116, burn infections 140, and gut-derived sepsis in the setting of neutropenia 141. The exact role of type III secretion in the pathogenesis of these infections is less clear, but a model is beginning to emerge. This discussion will focus on acute pneumonia, since the largest amount of data is available for this model system. Following inoculation into the airways, the first lines of defense faced by P. aeruginosa are the bronchial and bronchiolar epithelia. Intact polarized epithelia are relatively resistant to P. aeruginosa adherence and invasion, which may explain the opportunistic nature of this bacterium. In contrast, wounded or disrupted epithelia are quite susceptible to P. aeruginosa binding and injection of type III proteins 142–144. Thus P. aeruginosa may exploit naturally occurring breaches in epithelial barriers or those caused by trauma from endotracheal tubes to gain a foothold in the lungs. The antagonistic effects of ExoT towards wound healing may facilitate the ability of the bacterium to take advantage of these vulnerabilities. Upon contact with sentinel alveolar macrophages, ExoS and ExoU may delay or skew the early phases of the inflammatory response by blocking the inflammasome-mediated production of IL-1β and IL-18 32, 33. Eventually, however, a brisk inflammatory response is induced 130, 131. ExoU, which is homologous to cPLA2, may stoke the inflammatory furnace by generating large amounts of arachidonic acid that in turn leads to increased production of pro-inflammatory eicosanoids 145. The ensuing recruitment of excessive numbers of neutrophils and macrophages to the lungs causes collateral damage to host tissues 145 but fails to eradicate the bacteria, perhaps because these phagocytic cells are killed or otherwise impaired by ExoS, ExoT, and ExoU upon contact with P. aeruginosa bacteria 19, 27, 28, 130, 146. The neutralization of phagocytes also causes a localized immunosuppression, predisposing to superinfection by other pathogens, as has been observed experimentally and clinically 130, 147. Failure to eradicate P. aeruginosa from the lungs allows subsequently secreted type III effector proteins and other toxins to injure pulmonary tissues, which results in the signs and symptoms of pneumonia. Lung compliance is adversely affected by decreased synthesis of the pulmonary surfactant phosphatidylcholine and an influx of protein-rich fluid into the alveoli following disruption of epithelial and endothelial barriers within the lungs by ExoS, ExoT, and ExoU 146, 148–150. Loss of integrity of these barriers also allows bacterial dissemination into the bloodstream 27, 112, 128, 131 and the leakage of proinflammatory cytokines such as TNF-α from the lungs into the systemic circulation, leading to reductions in mean arterial pressure, cardiac output, and base excess, all indicators of septic shock 131. The net result is increased mortality associated with infection by T3SS+ strains 27, 61, 112, 146, 151.
Human studies and therapeutic interventions
In addition to the abundance of evidence in animal models supporting a role for type III secretion in disease, several studies directly link this virulence system to worse outcomes in human infections. Culture of P. aeruginosa isolates with functional T3SSs has been linked to increased mortality, persistence, relapse, and higher bacterial burdens in acutely colonized or infected patients 137, 151–153. Given its important role in pathogenesis, this system is an attractive target for therapeutic intervention. Much effort has been directed towards PcrV, which is accessible at the tip of the needle complex 25 and required for translocation of effector proteins 23, 24. Active or passive immunization using PcrV is protective in animal models of infection 19, 154, apparently by direct blockade of injection of type III proteins into host cells 19. Engineered human anti-PcrV Fab fragments have now been generated and should facilitate the clinical use of this approach 155. Phase I/II clinical studies are ongoing to evaluate the usefulness of PcrV-specific antibodies in the settings of mechanically ventilated patients and individuals with cystic fibrosis (http://www.kalobios.com/kb_pipeline.php).
By residing within cells both before and after secretion, effector proteins remain sheltered from a humoral immune response, and immunization with these proteins is at best minimally effective 19. For this reason, cell-permeable small molecule inhibitors have been sought to neutralize ExoU and ExoS. Inhibitors of the PLA2 activity of ExoU and the ADPRT activity of ExoS have been identified and shown to provide protection in models of infection 156, 157. Likewise inhibitors of ExsA, the activator necessary to express type III secretion genes, are currently under development 158. Targeting patients most likely to benefit from such therapies will require rapid and accurate identification of the effector proteins secreted by P. aeruginosa isolates. For this purpose, several assays capable of rapidly determining the genetic and phenotypic repertoire of effector proteins of P. aeruginosa clinical isolates have been devised 159–161. Thus in a relatively short period of time, remarkable progress has been made in developing interventions to protect patients from the toxic effects of P. aeruginosa type III secretion.
Concluding remarks
The T3SS is a powerful weapon wielded by P. aeruginosa to enhance the pathogenic process. A complex array of environmental signals are integrated to tightly control the injection of ExoS, ExoT, ExoU, and ExoY into host cells, where they are targeted to specific compartments, are activated by host cell cofactors, and manipulate a number of host cell processes. The net result is impairment of the innate immune response and compromise of tissue barriers leading to bacterial dissemination and sepsis.
Despite rapid progress, substantial gaps remain in our understanding of the P. aeruginosa T3SS, and these are the focus of ongoing efforts. Do additional effector proteins exist? Given the extensive variation between P. aeruginosa strains 162, interrogation of a larger number of isolates may be necessary to uncover such proteins or at least exclude their existence. What are the additional regulatory systems that govern type III secretion and how are these systems integrated at the molecular level? What are the cell types targeted for type III injection in vivo, what are the consequences of injection to these cells, and how are these effects orchestrated to bring about enhanced disease severity? Active investigation of these and many other remaining questions promises to reveal novel aspects of bacterial pathogenesis in the years to come.
Online summary
The Pseudomonas aeruginosa type III secretion system consists of five functional parts: (i) proteins that comprise the secretion machine itself, which shall be referred to as the needle complex; (ii) proteins that translocate secreted proteins into host cells; (iii) proteins that regulate the secretion process; (iv) proteins that bind secreted proteins to facilitate the secretion process, called chaperone proteins, and (v) proteins that are actually injected into host cells, called effector proteins.
Only four effector proteins are known to be secreted by the P. aeruginosa type III secretion system: ExoS, ExoT, ExoU, and ExoY.
ExoS is a bifunctional toxin with GTPase activating protein and ADP-ribosyltransferase activities. These activities lead to disruption of the actin cytoskeleton and apoptosis-like cell death.
ExoT is 76% identical to ExoS and also has both GTPase activating protein and ADP-ribosyltransferase activities. However the ADP-ribosyltransferase activity of ExoT is directed against different substrates than that of ExoS. These activities lead to disruption of the actin cytoskeleton and apoptosis-like cell death.
ExoU has phospholipase A2 activity that results in rapid lysis of many mammalian cell types.
ExoY is an adenylate cyclase. Its role in pathogenesis is less clear.
Figure 4. Role of type III effector proteins in the pathogenesis of acute pneumonia.
In early infection, P. aeruginosa exploits breaches in the epithelial mucosa, which is facilitated by ExoT-mediated inhibition of wound healing. ExoU and ExoS block IL-1β and IL-18 production by alveolar macrophages, blunting or biasing the early inflammatory response. When subsequent development of an inflammatory response does occur, it is excessively amplified by ExoU-induced eicosanoid release and causes collateral damage to host tissues. However the recruited phagocytes are unable to eradicate P. aeruginosa because they are killed or impaired by ExoS, ExoT, and ExoU; the resulting paucity of functional phagocytes makes the lungs prone to superinfection by other bacteria. ExoS, ExoT, and ExoU also disrupt epithelial/endothelial barriers, allowing bacteria and proinflammatory cytokines to escape to the bloodstream, leading to bacteremia and septic shock, respectively. These same breaches allow protein-rich fluid to flow into the air spaces of the lung, which together with ExoU-mediated decreases in pulmonary surfactant, causes decreased lung compliance and oxygenation. AM=alveolar macrophage
Acknowledgments
I apologize to colleagues whose work has been omitted due to lack of space and limitations on the allowed number of citations. I thank Nick Cianciotto, Maureen Diaz, Heather Howell, Vanderlene Kung, and Jeffrey Veesenmeyer for reviewing the manuscript and for helpful discussions. This work was supported by the National Institutes of Health (K02 AI065615, R01 AI075191, and R01 AI053674).
A.R.H is a consultant for Microbiotix, Inc.
Glossary
- Inflammasome
a multiprotein cytoplasmic complex that mediates the activation of caspase-1 to promote the maturation of IL 1-β and IL-18. Activation may also lead to a form of cell death termed “pryoptosis.” The inflammasome itself may be activated by NOD-like receptors in response to intracellular microbial triggers
- Flagellin
The protein monomer that forms the hollow filament of the bacterial flagellum
- AraC/XylS transcriptional regulators
A large family of transcriptional activators defined by a 100-amino-acid region of sequence similarity. These proteins regulate diverse bacterial functions including catabolism, stress response, and virulence
- cAMP receptor protein
A regulatory protein that, in the presence of cAMP, binds tightly to a specific DNA sequence in the promoters of a subset of bacterial genes, modulating their transcription. Also known as catabolite gene activator protein (CAP)
- Adenylate cyclase
An enzyme that catalyzes the conversion of ATP to cAMP and pyrophosphate
- Two-component signal transduction systems
Signal transducing proteins that are commonly used by bacteria to couple environmental stimuli to response mechanisms. They consist of sensor and response regulator components that are linked by phosphorelays
- GTPase activating protein
A member of a family of regulatory proteins that stimulate the intrinsic GTPase activity of small signaling G proteins, which results in their inactivation
- ADP ribosyltransferase activity
The enzymatic activity that covalently transfers an ADP-ribose group from nicotinamide adenine dinucleotide (NAD+) to a substrate protein, usually altering the activity of the substrate
- Rab proteins
A family of GTPases that regulate many of the steps in membrane trafficking
- E3 ubiquitin ligase
An enzyme that covalently attaches ubiquitin to lysines in substrate proteins. Single or multiple ubiquitin molecules may be attached to a single lysine and may target the substrate protein for degradation or altered cellular location or function
- Lysophospholipids
Any phospholipid that is missing one of its two acyl chains
- SOD1
A super oxide dismutase that binds copper and zinc ions and converts harmful free superoxide radicals to molecular oxygen and hydrogen peroxide
- Arachidonic acid
A fatty acid that is freed from certain phospholipids by the catalytic activity of PLA2 enzymes. Arachidonic acid is an important inflammatory intermediate that is converted into eicosanoids
Biography
Alan R. Hauser is an Associate Professor in the Departments of Microbiology/Immunology and Medicine at Northwestern University in Chicago, Illinois USA. He received his PhD from the University of Minnesota for studies on the pathogenesis of group A streptococcal infections. He did his postdoctoral studies at the University of California, San Francisco, with Joanne Engel on type III secretion in Pseudomonas aeruginosa. He continues to work on type III secretion as well as genetic determinants for strain-to-strain differences in virulence among P. aeruginosa isolates.
References
- 1.Stryjewski ME, Sexton DJ. In: Severe infections caused by Pseudomonas aeruginosa. Hauser AR, Rello J, editors. Kluwer Academic Publishers; Boston: 2003. pp. 1–15. [Google Scholar]
- 2.Lyczak JB, Cannon CL, Pier GB. Lung infections associated with cystic fibrosis. Clin Microbiol Rev. 2002;15:194–222. doi: 10.1128/CMR.15.2.194-222.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Engel JN. In: Severe infections caused by Pseudomonas aeruginosa. Hauser AR, Rello J, editors. Kluwer Academic Publishers; Boston: 2003. pp. 201–229. [Google Scholar]
- 4.Yahr TL, Goranson J, Frank DW. Exoenzyme S of Pseudomonas aeruginosa is secreted by a type III secretion pathway. Mol Microbiol. 1996;22:991–1003. doi: 10.1046/j.1365-2958.1996.01554.x. This is the first study to show that P. aeruginosa had a type III secretion system. [DOI] [PubMed] [Google Scholar]
- 5.Pastor A, Chabert J, Louwagie M, Garin J, Attree I. PscF is a major component of the Pseudomonas aeruginosa type III secretion needle. FEMS Microbiol Lett. 2005;253:95–101. doi: 10.1016/j.femsle.2005.09.028. This is the first report in which the P. aeruginosa type III secretion needle complexes were visualized. [DOI] [PubMed] [Google Scholar]
- 6.Soscia C, Hachani A, Bernadac A, Filloux A, Bleves S. Cross talk between type III secretion and flagellar assembly systems in Pseudomonas aeruginosa. J Bacteriol. 2007;189:3124–3132. doi: 10.1128/JB.01677-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoiczyk E, Blobel G. Polymerization of a single protein of the pathogen Yersinia enterocolitica into needles punctures eukaryotic cells. Proc Natl Acad Sci U S A. 2001;98:4669–4674. doi: 10.1073/pnas.071065798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cordes FS, et al. Helical structure of the needle of the type III secretion system of Shigella flexneri. J Biol Chem. 2003;278:17103–17107. doi: 10.1074/jbc.M300091200. [DOI] [PubMed] [Google Scholar]
- 9.Kubori T, et al. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science. 1998;280:602–605. doi: 10.1126/science.280.5363.602. [DOI] [PubMed] [Google Scholar]
- 10.Blocker A, et al. The tripartite type III secretion of Shigella flexneri inserts IpaB and IpaC into host membranes. J Cell Biol. 1999;147:683–693. doi: 10.1083/jcb.147.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woestyn S, Allaoui A, Wattiau P, Cornelis GR. YscN, the putative energizer of the Yersinia Yop secretion machinery. J Bacteriol. 1994;176:1561–1569. doi: 10.1128/jb.176.6.1561-1569.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blaylock B, Riordan KE, Missiakas DM, Schneewind O. Characterization of the Yersinia enterocolitica type III secretion ATPase YscN and its regulator, YscL. J Bacteriol. 2006;188:3525–3534. doi: 10.1128/JB.188.10.3525-3534.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koster M, et al. The outer membrane component, YscC, of the Yop secretion machinery of Yersinia enterocolitica forms a ring-shaped multimeric complex. Mol Microbiol. 1997;26:789–797. doi: 10.1046/j.1365-2958.1997.6141981.x. [DOI] [PubMed] [Google Scholar]
- 14.Burghout P, et al. Role of the pilot protein YscW in the biogenesis of the YscC secretin in Yersinia enterocolitica. J Bacteriol. 2004;186:5366–5375. doi: 10.1128/JB.186.16.5366-5375.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Journet L, Agrain C, Broz P, Cornelis GR. The needle length of bacterial injectisomes is determined by a molecular ruler. Science. 2003;302:1757–1760. doi: 10.1126/science.1091422. [DOI] [PubMed] [Google Scholar]
- 16.Burns RE, McDaniel-Craig A, Sukhan A. Site-directed mutagenesis of the Pseudomonas aeruginosa type III secretion system protein PscJ reveals an essential role for surface-localized residues in needle complex function. Microb Pathog. 2008;45:225–230. doi: 10.1016/j.micpath.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 17.Sundin C, Thelaus J, Broms JE, Forsberg A. Polarisation of type III translocation by Pseudomonas aeruginosa requires PcrG, PcrV and PopN. Microb Pathog. 2004;37:313–322. doi: 10.1016/j.micpath.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 18.Sundin C, Wolfgang MC, Lory S, Forsberg A, Frithz-Lindsten E. Type IV pili are not specifically required for contact dependent translocation of exoenzymes by Pseudomonas aeruginosa. Microb Pathog. 2002;33:265–277. doi: 10.1006/mpat.2002.0534. [DOI] [PubMed] [Google Scholar]
- 19.Sawa T, et al. Active and passive immunization with the Pseudomonas V antigen protects against type III intoxication and lung injury. Nature Med. 1999;5:392–398. doi: 10.1038/7391. This is the first study that showed the potential of PcrV as a vaccine candidate. [DOI] [PubMed] [Google Scholar]
- 20.Dacheux D, Goure J, Chabert J, Usson Y, Attree I. Pore-forming activity of type III system-secreted proteins leads to oncosis of Pseudomonas aeruginosa-infected macrophages. Mol Microbiol. 2001;40:76–85. doi: 10.1046/j.1365-2958.2001.02368.x. [DOI] [PubMed] [Google Scholar]
- 21.Schoehn G, et al. Oligomerization of type III secretion proteins PopB and PopD precedes pore formation in Pseudomonas. EMBO J. 2003;22:4957–4967. doi: 10.1093/emboj/cdg499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Faudry E, Vernier G, Neumann E, Forge V, Attree I. Synergistic pore formation by type III toxin translocators of Pseudomonas aeruginosa. Biochemistry. 2006;45:8117–8123. doi: 10.1021/bi060452+. [DOI] [PubMed] [Google Scholar]
- 23.Goure J, et al. The V antigen of Pseudomonas aeruginosa is required for assembly of the functional PopB/PopD translocation pore in host cell membranes. Infect Immun. 2004;72:4741–4750. doi: 10.1128/IAI.72.8.4741-4750.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holmstrom A, et al. LcrV is a channel size-determining component of the Yop effector translocon of Yersinia. Mol Microbiol. 2001;39:620–632. doi: 10.1046/j.1365-2958.2001.02259.x. [DOI] [PubMed] [Google Scholar]
- 25.Mueller CA, et al. The V-antigen of Yersinia forms a distinct structure at the tip of injectisome needles. Science. 2005;310:674–676. doi: 10.1126/science.1118476. [DOI] [PubMed] [Google Scholar]
- 26.Mota LJ. Type III secretion gets an LcrV tip. Trends Microbiol. 2006;14:197–200. doi: 10.1016/j.tim.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 27.Lee VT, Smith RS, Tummler B, Lory S. Activities of Pseudomonas aeruginosa effectors secreted by the type III secretion system in vitro and during infection. Infect Immun. 2005;73:1695–1705. doi: 10.1128/IAI.73.3.1695-1705.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vance RE, Rietsch A, Mekalanos JJ. Role of the type III secreted exoenzymes S, T, and Y in systemic spread of Pseudomonas aeruginosa PAO1 in vivo. Infect Immun. 2005;73:1706–1713. doi: 10.1128/IAI.73.3.1706-1713.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shafikhani SH, Morales C, Engel J. The Pseudomonas aeruginosa type III secreted toxin ExoT is necessary and sufficient to induce apoptosis in epithelial cells. Cell Microbiol. 2008;10:994–1007. doi: 10.1111/j.1462-5822.2007.01102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roy D, et al. A process for controlling intracellular bacterial infections induced by membrane injury. Science. 2004;304:1515–1518. doi: 10.1126/science.1098371. [DOI] [PubMed] [Google Scholar]
- 31.Franchi L, et al. Critical role for Ipaf in Pseudomonas aeruginosa-induced caspase-1 activation. Eur J Immunol. 2007;37:3030–3039. doi: 10.1002/eji.200737532. [DOI] [PubMed] [Google Scholar]
- 32.Sutterwala FS, et al. Immune recognition of Pseudomonas aeruginosa mediated by the IPAF/NLRC4 inflammasome. J Exp Med. 2007;204:3235–3245. doi: 10.1084/jem.20071239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Galle M, et al. The Pseudomonas aeruginosa type III secretion system plays a dual role in the regulation of caspase-1 mediated IL-1beta maturation. J Cell Mol Med. 2007;12:1767–1776. doi: 10.1111/j.1582-4934.2007.00190.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miao EA, Ernst RK, Dors M, Mao DP, Aderem A. Pseudomonas aeruginosa activates caspase 1 through Ipaf. Proc Natl Acad Sci U S A. 2008;105:2562–2567. doi: 10.1073/pnas.0712183105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bergsbaken T, Fink SL, Cookson BT. Pyroptosis: host cell death and inflammation. Nat Rev Microbiol. 2009;7:99–109. doi: 10.1038/nrmicro2070. A recent review summarizing evolving knowledge of the process of pyroptosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCaw ML, Lykken GL, Singh PK, Yahr TL. ExsD is a negative regulator of the Pseudomonas aeruginosa type III secretion regulon. Mol Microbiol. 2002;46:1123–1133. doi: 10.1046/j.1365-2958.2002.03228.x. [DOI] [PubMed] [Google Scholar]
- 37.Hovey AK, Frank DW. Analyses of the DNA-binding and transcriptional activation properties of ExsA, the transcriptional activator of the Pseudomonas aeruginosa exoenzyme S regulon. J Bacteriol. 1995;177:4427–4436. doi: 10.1128/jb.177.15.4427-4436.1995. An early report characterizing the important regulatory activity of ExsA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brutinel ED, Vakulskas CA, Brady KM, Yahr TL. Characterization of ExsA and of ExsA-dependent promoters required for expression of the Pseudomonas aeruginosa type III secretion system. Mol Microbiol. 2008;68:657–671. doi: 10.1111/j.1365-2958.2008.06179.x. [DOI] [PubMed] [Google Scholar]
- 39.Dasgupta N, Lykken GL, Wolfgang MC, Yahr TL. A novel anti-anti-activator mechanism regulates expression of the Pseudomonas aeruginosa type III secretion system. Mol Microbiol. 2004;53:297–308. doi: 10.1111/j.1365-2958.2004.04128.x. [DOI] [PubMed] [Google Scholar]
- 40.Urbanowski ML, Lykken GL, Yahr TL. A secreted regulatory protein couples transcription to the secretory activity of the Pseudomonas aeruginosa type III secretion system. Proc Natl Acad Sci U S A. 2005;102:9930–9935. doi: 10.1073/pnas.0504405102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rietsch A, Vallet-Gely I, Dove SL, Mekalanos JJ. ExsE, a secreted regulator of type III secretion genes in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2005;102:8006–8011. doi: 10.1073/pnas.0503005102. These reports characterized at a molecular level the link between ongoing type III secretion and expression of the type III regulon. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Urbanowski ML, Brutinel ED, Yahr TL. Translocation of ExsE into Chinese hamster ovary cells is required for transcriptional induction of the Pseudomonas aeruginosa type III secretion system. Infect Immun. 2007;75:4432–4439. doi: 10.1128/IAI.00664-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wolfgang MC, Lee VT, Gilmore ME, Lory S. Coordinate regulation of bacterial virulence genes by a novel adenylate cyclase-dependent signaling pathway. Dev Cell. 2003;4:253–263. doi: 10.1016/s1534-5807(03)00019-4. [DOI] [PubMed] [Google Scholar]
- 44.Zolfaghar I, et al. Mutation of retS, encoding a putative hybrid two-component regulatory protein in Pseudomonas aeruginosa, attenuates multiple virulence mechanisms. Microbes Infect. 2005;7:1305–1316. doi: 10.1016/j.micinf.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 45.Laskowski MA, Osborn E, Kazmierczak BI. A novel sensor kinase-response regulator hybrid regulates type III secretion and is required for virulence in Pseudomonas aeruginosa. Mol Microbiol. 2004;54:1090–1103. doi: 10.1111/j.1365-2958.2004.04331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goodman AL, et al. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev Cell. 2004;7:745–754. doi: 10.1016/j.devcel.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 47.Ventre I, et al. Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. Proc Natl Acad Sci U S A. 2006;103:171–176. doi: 10.1073/pnas.0507407103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yahr TL, Wolfgang MC. Transcriptional regulation of the Pseudomonas aeruginosa type III secretion system. Mol Microbiol. 2006;62:631–640. doi: 10.1111/j.1365-2958.2006.05412.x. [DOI] [PubMed] [Google Scholar]
- 49.Vallis AJ, Yahr TL, Barbieri JT, Frank DW. Regulation of ExoS production and secretion by Pseudomonas aeruginosa in response to tissue culture conditions. Infect Immun. 1999;67:914–920. doi: 10.1128/iai.67.2.914-920.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iglewski BH, Sadoff J, Bjorn MJ, Maxwell ES. Pseudomonas aeruginosa exoenzyme S: an adenosine diphoshate ribosyltransferase distinct from toxin A. Proc Natl Acad Sci USA. 1978;75:3211–3215. doi: 10.1073/pnas.75.7.3211. Initial description of ExoS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rietsch A, Mekalanos JJ. Metabolic regulation of type III secretion gene expression in Pseudomonas aeruginosa. Mol Microbiol. 2006;59:807–820. doi: 10.1111/j.1365-2958.2005.04990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dacheux D, et al. Activation of the Pseudomonas aeruginosa type III secretion system requires an intact pyruvate dehydrogenase aceAB operon. Infect Immun. 2002;70:3973–3977. doi: 10.1128/IAI.70.7.3973-3977.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rietsch A, Wolfgang MC, Mekalanos JJ. Effect of metabolic imbalance on expression of type III secretion genes in Pseudomonas aeruginosa. Infect Immun. 2004;72:1383–1390. doi: 10.1128/IAI.72.3.1383-1390.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Parsot C, Hamiaux C, Page AL. The various and varying roles of specific chaperones in type III secretion systems. Curr Opin Microbiol. 2003;6:7–14. doi: 10.1016/s1369-5274(02)00002-4. [DOI] [PubMed] [Google Scholar]
- 55.Yahr TL, Hovey AK, Kulich SM, Frank DW. Transcriptional analysis of the Pseudomonas aeruginosa exoenzyme S structural gene. J Bacteriol. 1995;177:1169–1178. doi: 10.1128/jb.177.5.1169-1178.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shen DK, et al. Orf1/SpcS chaperones ExoS for type three secretion by Pseudomonas aeruginosa. Biomed Environ Sci. 2008;21:103–109. doi: 10.1016/S0895-3988(08)60014-8. [DOI] [PubMed] [Google Scholar]
- 57.Finck-Barbancon V, Yahr TL, Frank DW. Identification and characterization of SpcU, a chaperone required for efficient secretion of the ExoU cytotoxin. J Bacteriol. 1998;180:6224–6231. doi: 10.1128/jb.180.23.6224-6231.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Quinaud M, et al. The PscE-PscF-PscG complex controls type III secretion needle biogenesis in Pseudomonas aeruginosa. J Biol Chem. 2005;280:36293–36300. doi: 10.1074/jbc.M508089200. [DOI] [PubMed] [Google Scholar]
- 59.Feltman H, et al. Prevalence of type III secretion genes in clinical and environmental isolates of Pseudomonas aeruginosa. Microbiology. 2001;147:2659–2669. doi: 10.1099/00221287-147-10-2659. Described of the prevalence of type III secretion effector genes in a large collection of P. aeruginosa clinical and environmental isolates. [DOI] [PubMed] [Google Scholar]
- 60.Fleiszig SMJ, et al. Pseudomonas aeruginosa-mediated cytotoxicity and invasion correlate with distinct genotypes at the loci encoding exoenzyme S. Infect Immun. 1997;65:579–586. doi: 10.1128/iai.65.2.579-586.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schulert GS, et al. Secretion of the toxin ExoU is a marker for highly virulent Pseudomonas aeruginosa isolates obtained from patients with hospital-acquired pneumonia. J Infect Dis. 2003;188:1695–1706. doi: 10.1086/379372. [DOI] [PubMed] [Google Scholar]
- 62.Deng Q, Barbieri JT. Molecular mechanisms of the cytotoxicity of ADP-ribosylating toxins. Annu Rev Microbiol. 2008;62:271–288. doi: 10.1146/annurev.micro.62.081307.162848. [DOI] [PubMed] [Google Scholar]
- 63.Zhang Y, Barbieri JT. A leucine-rich motif targets Pseudomonas aeruginosa ExoS within mammalian cells. Infect Immun. 2005;73:7938–7945. doi: 10.1128/IAI.73.12.7938-7945.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang Y, Deng Q, Barbieri JT. Intracellular localization of type III-delivered Pseudomonas ExoS with endosome vesicles. J Biol Chem. 2007;282:13022–13032. doi: 10.1074/jbc.M606305200. [DOI] [PubMed] [Google Scholar]
- 65.Deng Q, Zhang Y, Barbieri JT. Intracellular trafficking of Pseudomonas ExoS, a type III cytotoxin. Traffic. 2007;8:1331–1345. doi: 10.1111/j.1600-0854.2007.00626.x. [DOI] [PubMed] [Google Scholar]
- 66.Riese MJ, Barbieri JT. Membrane localization contributes to the in vivo ADP-ribosylation of Ras by Pseudomonas aeruginosa ExoS. Infect Immun. 2002;70:2230–2232. doi: 10.1128/IAI.70.4.2230-2232.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang Y, et al. Plasma membrane localization affects the RhoGAP specificity of Pseudomonas ExoS. Cell Microbiol. 2007;9:2192–2201. doi: 10.1111/j.1462-5822.2007.00949.x. [DOI] [PubMed] [Google Scholar]
- 68.Pederson KJ, Vallis AJ, Aktories K, Frank DW, Barbieri JT. The amino-terminal domain of Pseudomonas aeruginosa ExoS disrupts actin filaments via small-molecular-weight GTP-binding proteins. Mol Microbiol. 1999;32:393–401. doi: 10.1046/j.1365-2958.1999.01359.x. [DOI] [PubMed] [Google Scholar]
- 69.Goehring UM, Schmidt G, Pederson KJ, Aktories K, Barbieri JT. The N-terminal domain of Pseudomonas aeruginosa exoenzyme S is a GTPase-activating protein for Rho GTPases. J Biol Chem. 1999;274:36369–36372. doi: 10.1074/jbc.274.51.36369. [DOI] [PubMed] [Google Scholar]
- 70.Evdokimov AG, Tropea JE, Routzahn KM, Waugh DS. Crystal structure of the Yersinia pestis GTPase activator YopE. Protein Sci. 2002;11:401–408. doi: 10.1110/ps.34102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stebbins CE, Galan JE. Modulation of host signaling by a bacterial mimic: structure of the Salmonella effector SptP bound to Rac1. Mol Cell. 2000;6:1449–1460. doi: 10.1016/s1097-2765(00)00141-6. [DOI] [PubMed] [Google Scholar]
- 72.Wurtele M, et al. How the Pseudomonas aeruginosa ExoS toxin downregulates Rac. Nat Struct Biol. 2001;8:23–26. doi: 10.1038/83007. This study reports the three-dimensional structure of ExoS’s GAP domain bound to its substrate Rac. [DOI] [PubMed] [Google Scholar]
- 73.Wurtele M, Renault L, Barbieri JT, Wittinghofer A, Wolf E. Structure of the ExoS GTPase activating domain. FEBS Lett. 2001;491:26–29. doi: 10.1016/s0014-5793(01)02105-6. [DOI] [PubMed] [Google Scholar]
- 74.Rocha CL, Coburn J, Rucks EA, Olson JC. Characterization of Pseudomonas aeruginosa exoenzyme S as a bifunctional enzyme in J774A.1 macrophages. Infect Immun. 2003;71:5296–5305. doi: 10.1128/IAI.71.9.5296-5305.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Frithz-Lindsten E, Du Y, Rosqvist R, Forsberg A. Intracellular targeting of exoenzyme S of Pseudomonas aeruginosa via type III-dependent translocation induces phagocytosis resistance, cytotoxicity and disruption of actin microfilaments. Mol Microbiol. 1997;25:1125–1139. doi: 10.1046/j.1365-2958.1997.5411905.x. [DOI] [PubMed] [Google Scholar]
- 76.Garrity-Ryan L, et al. The arginine finger domain of ExoT contributes to actin cytoskeleton disruption and inhibition of internalization of Pseudomonas aeruginosa by epithelial cells and macrophages. Infect Immun. 2000;68:7100–7113. doi: 10.1128/iai.68.12.7100-7113.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cowell BA, Chen DY, Frank DW, Vallis AJ, Fleiszig SMJ. ExoT of cytotoxic Pseudomonas aeruginosa prevents uptake by corneal epithelial cells. Infect Immun. 2000;68:403–406. doi: 10.1128/iai.68.1.403-406.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Knight DA, Finck-Barbancon V, Kulich SM, Barbieri JT. Functional domains of Pseudomonas aeruginosa exoenzyme S. Infect Immun. 1995;63:3182–3186. doi: 10.1128/iai.63.8.3182-3186.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Riese MJ, et al. Auto-ADP-ribosylation of Pseudomonas aeruginosa ExoS. J Biol Chem. 2002;277:12082–12088. doi: 10.1074/jbc.M109039200. [DOI] [PubMed] [Google Scholar]
- 80.Henriksson ML, et al. A nonphosphorylated 14–3–3 binding motif on exoenzyme S that is functional in vivo. Eur J Biochem. 2002;269:4921–4929. doi: 10.1046/j.1432-1033.2002.03191.x. [DOI] [PubMed] [Google Scholar]
- 81.Fu H, Coburn J, Collier RJ. The eukaryotic host factor that activates exoenzyme S of Pseudomonas aeruginosa is a member of the 14–3–3 protein family. Proc Natl Acad Sci USA. 1993;90:2320–2324. doi: 10.1073/pnas.90.6.2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Coburn J, Kane AV, Feig L, Gill DM. Pseudomonas aeruginosa exoenzyme S requires a eukaryotic protein for ADP-ribosyltransferase activity. J Biol Chem. 1991;266:6438–6446. [PubMed] [Google Scholar]
- 83.Ottman C, et al. Phosphorylation-independent interaction between 14–3–3 and exoenxyme S: from structure to pathogenesis. EMBO J. 2007;26:902–913. doi: 10.1038/sj.emboj.7601530. Reported the structure of 14-3-3 protein bound to a peptide from the ADPRT domain of ExoS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pederson KJ, Barbieri JT. Intracellular expression of the ADP-ribosyltransferase domain of Pseudomonas aeruginosa exoenzyme S is cytotoxic to eukaryotic cells. Mol Microbiol. 1998;30:751–759. doi: 10.1046/j.1365-2958.1998.01106.x. [DOI] [PubMed] [Google Scholar]
- 85.Barbieri AM, Sha Q, Bette-Bobillo P, Stahl PD, Vidal M. ADP-ribosylation of Rab5 by ExoS of Pseudomonas aeruginosa affects endocytosis. Infect Immun. 2001;69:5329–5334. doi: 10.1128/IAI.69.9.5329-5334.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fraylick JE, La Rocque JR, Vincent TS, Olson JC. Independent and coordinate effects of ADP-ribosyltransferase and GTPase-activating activities of exoenzyme S on HT-29 epithelial cell function. Infect Immun. 2001;69:5318–5328. doi: 10.1128/IAI.69.9.5318-5328.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kaufman MR, et al. Pseudomonas aeruginosa mediated apoptosis requires the ADP-ribosylating activity of ExoS. Microbiology. 2000;146:2531–2541. doi: 10.1099/00221287-146-10-2531. [DOI] [PubMed] [Google Scholar]
- 88.Coburn J, Wyatt RT, Iglewski BH, Gill DM. Several GTP-binding proteins, inlcuding p21c-H-ras, are preferred stubstrates of Pseudomonas aeruginosa exoenzyme S. J Biol Chem. 1989;264:9004–9008. [PubMed] [Google Scholar]
- 89.Coburn J, Gill DM. ADP-ribosylation of p21ras and related proteins by Pseudomonas aeruginosa enzoenzyme S. Infect Immun. 1991;59:4259–4262. doi: 10.1128/iai.59.11.4259-4262.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Coburn J, Dillon ST, Iglewski BH, Gill DM. Exoenzyme S of Pseudomonas aeruginosa ADP-ribosylates the intermediate filament vimentin. Infect Immun. 1989;57:996–998. doi: 10.1128/iai.57.3.996-998.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.DiNovo AA, et al. ADP-ribosylation of cyclophilin A by Pseudomonas aeruginosa exoenzyme S. Biochemistry. 2006;45:4664–4673. doi: 10.1021/bi0513554. [DOI] [PubMed] [Google Scholar]
- 92.Henriksson ML, et al. Exoenzyme S shows selective ADP-ribosylation and GTPase-activating protein (GAP) activities towards small GTPases in vivo. Biochem J. 2002;367:617–628. doi: 10.1042/BJ20020714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fraylick JE, Riese MJ, Vincent TS, Barbieri JT, Olson JC. ADP-ribosylation and functional effects of Pseudomonas exoenzyme S on cellular RalA. Biochemistry. 2002;41:9680–9687. doi: 10.1021/bi025826n. [DOI] [PubMed] [Google Scholar]
- 94.Riese MJ, Wittinghofer A, Barbieri JT. ADP ribosylation of Arg41 of Rap by ExoS inhibits the ability of Rap to interact with its guanine nucleotide exchange factor, C3G. Biochemistry. 2001;40:3289–3294. doi: 10.1021/bi002729q. [DOI] [PubMed] [Google Scholar]
- 95.Bette-Bobillo P, Giro P, Sainte-Marie J, Vidal M. Exoenzyme S from P. aeruginosa ADP ribosylates rab4 and inhibits transferrin recycling in SLO-permeabilized reticulocytes. Biochem Biophys Res Commun. 1998;244:336–341. doi: 10.1006/bbrc.1998.8263. [DOI] [PubMed] [Google Scholar]
- 96.Maresso AW, Baldwin MR, Barbieri JT. Ezrin/radixin/moesin proteins are high affinity targets for ADP-ribosylation by Pseudomonas aeruginosa ExoS. J Biol Chem. 2004;279:38402–38408. doi: 10.1074/jbc.M405707200. [DOI] [PubMed] [Google Scholar]
- 97.Rocha CL, Rucks EA, Vincent DM, Olson JC. Examination of the coordinate effects of Pseudomonas aeruginosa ExoS on Rac1. Infect Immun. 2005;73:5458–5467. doi: 10.1128/IAI.73.9.5458-5467.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ganesan AK, Vincent TS, Olson JC, Barbieri JT. Pseudomonas aeruginosa exoenzyme S disrupts Ras-mediated signal transduction by inhibiting guanine nucleotide exchange factor-catalyzed nucleotide exchange. J Biol Chem. 1999;274:21823–21829. doi: 10.1074/jbc.274.31.21823. [DOI] [PubMed] [Google Scholar]
- 99.Henriksson ML, Rosqvist R, Telepnev M, Wolf-Watz H, Hallberg B. Ras effector pathway activation by epidermal growth factor is inhibited in vivo by exoenzyme S ADP-ribosylation of Ras. Biochem J. 2000;347:217–222. [PMC free article] [PubMed] [Google Scholar]
- 100.Vincent TS, Fraylick JE, McGuffie EM, Olson JC. ADP-ribosylation of oncogenic Ras proteins by Pseudomonas aeruginosa exoenzyme S in vivo. Mol Microbiol. 1999;32:1054–1064. doi: 10.1046/j.1365-2958.1999.01420.x. [DOI] [PubMed] [Google Scholar]
- 101.Ganesan AK, Frank DW, Misra RP, Schmidt G, Barbieri JT. Pseudomonas aeruginosa exoenzyme S ADP-ribosylates Ras at multiple sites. J Biol Chem. 1998;273:7332–7337. doi: 10.1074/jbc.273.13.7332. [DOI] [PubMed] [Google Scholar]
- 102.Jansson AL, et al. Exoenzyme S of Pseudomonas aeruginosa is not able to induce apoptosis when cells express activated proteins, such as Ras or protein kinase B/Akt. Cell Microbiol. 2006;8:815–22. doi: 10.1111/j.1462-5822.2005.00668.x. [DOI] [PubMed] [Google Scholar]
- 103.Pederson KJ, Krall R, Riese MJ, Barbieri JT. Intracellular localization modulates targeting of ExoS, a type III cytotoxin, to eukaryotic signalling proteins. Mol Microbiol. 2002;46:1381–1390. doi: 10.1046/j.1365-2958.2002.03256.x. [DOI] [PubMed] [Google Scholar]
- 104.Maresso AW, Deng Q, Pereckas MS, Wakim BT, Barbieri JT. Pseudomonas aeruginosa ExoS ADP-ribosyltransferase inhibits ERM phosphorylation. Cell Microbiol. 2007;9:97–105. doi: 10.1111/j.1462-5822.2006.00770.x. [DOI] [PubMed] [Google Scholar]
- 105.Krall R, Schmidt G, Aktories K, Barbieri JT. Pseudomonas aeruginosa ExoT is a Rho GTPase-activating protein. Infect Immun. 2000;68:6066–6068. doi: 10.1128/iai.68.10.6066-6068.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kazmierczak BI, Engel JN. Pseudomonas aeruginosa ExoT acts in vivo as a GTPase-activating protein for RhoA, Rac1, and Cdc42. Infect Immun. 2002;70:2198–2205. doi: 10.1128/IAI.70.4.2198-2205.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shafikhani SH, Engel J. Pseudomonas aeruginosa type III-secreted toxin ExoT inhibits host-cell division by targeting cytokinesis at multiple steps. Proc Natl Acad Sci U S A. 2006;103:15605–15610. doi: 10.1073/pnas.0605949103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Deng Q, Sun J, Barbieri JT. Uncoupling Crk signal transduction by Pseudomonas exoenzyme T. J Biol Chem. 2005;280:35953–35960. doi: 10.1074/jbc.M504901200. [DOI] [PubMed] [Google Scholar]
- 109.Liu S, Yahr TL, Frank DW, Barbieri JT. Biochemical relationships between the 53-kilodalton (Exo53) and 49-kilodalton (ExoS) forms of exoenzyme S of Pseudomonas aeruginosa. J Bacteriol. 1997;179:1609–1613. doi: 10.1128/jb.179.5.1609-1613.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sun J, Barbieri JT. Pseudomonas aeruginosa ExoT ADP-ribosylates CT10 regulator of kinase (Crk) proteins. J Biol Chem. 2003;278:32794–32800. doi: 10.1074/jbc.M304290200. Although it was initially thought that ExoT had limited ADPRT activity, this report demonstrated that ExoT indeed had robust ADPRT activity but that it was directed toward a unique substrate, Crk proteins. [DOI] [PubMed] [Google Scholar]
- 111.Sun J, Maresso AW, Kim JJ, Barbieri JT. How bacterial ADP-ribosylating toxins recognize substrates. Nat Struct Mol Biol. 2004;11:868–876. doi: 10.1038/nsmb818. [DOI] [PubMed] [Google Scholar]
- 112.Shaver CM, Hauser AR. Relative contributions of Pseudomonas aeruginosa ExoU, ExoS, and ExoT to virulence in the lung. Infect Immun. 2004;72:6969–6977. doi: 10.1128/IAI.72.12.6969-6977.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Garrity-Ryan L, et al. The ADP ribosyltransferase domain of Pseudomonas aeruginosa ExoT contributes to its biological activities. Infect Immun. 2004;72:546–558. doi: 10.1128/IAI.72.1.546-558.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Geiser TK, Kazmierczak BI, Garrity-Ryan LK, Matthay MA, Engel JN. Pseudomonas aeruginosa ExoT inhibits in vitro lung epithelial wound repair. Cell Microbiol. 2001;3:223–236. doi: 10.1046/j.1462-5822.2001.00107.x. [DOI] [PubMed] [Google Scholar]
- 115.Cho SY, Klemke RL. Extracellular-regulated kinase activation and CAS/Crk coupling regulate cell migration and suppress apoptosis during invasion of the extracellular matrix. J Cell Biol. 2000;149:223–236. doi: 10.1083/jcb.149.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Balachandran P, et al. The ubiquitin ligase Cbl-b limits Pseudomonas aeruginosa exotoxin T-mediated virulence. J Clin Invest. 2007;117:419–427. doi: 10.1172/JCI28792. This study demonstrated that ExoT undergoes polyubiquitination by Cbl-b, which limits the virulence associated with this effector protein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sato H, et al. The mechanism of action of the Pseudomonas aeruginosa-encoded type III cytotoxin, ExoU. EMBO J. 2003;22:2959–2969. doi: 10.1093/emboj/cdg290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Phillips RM, Six DA, Dennis EA, Ghosh P. In vivo phospholipase activity of the Pseudomonas aeruginosa cytotoxin ExoU and protection of mammalian cells with phospholipase A2 inhibitors. J Biol Chem. 2003;278:41326–41332. doi: 10.1074/jbc.M302472200. These studies demonstrated that ExoU is a phospholipase. [DOI] [PubMed] [Google Scholar]
- 119.Dessen A, et al. Crystal structure of human cytosolic phospholipase A2 reveals a novel topology and catalytic mechanism. Cell. 1999;97:349–360. doi: 10.1016/s0092-8674(00)80744-8. [DOI] [PubMed] [Google Scholar]
- 120.Tamura M, et al. Lysophospholipase A activity of Pseudomonas aeruginosa type III secretory toxin ExoU. Biochem Biophys Res Commun. 2004;316:323–331. doi: 10.1016/j.bbrc.2004.02.050. [DOI] [PubMed] [Google Scholar]
- 121.Rabin SDP, Hauser AR. Functional regions of the Pseudomonas aeruginosa cytotoxin ExoU. Infect Immun. 2005;73:573–582. doi: 10.1128/IAI.73.1.573-582.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Banerji S, Flieger A. Patatin-like proteins: a new family of lipolytic enzymes present in bacteria? Microbiology. 2004;150:522–525. doi: 10.1099/mic.0.26957-0. [DOI] [PubMed] [Google Scholar]
- 123.Sato H, Feix JB, Frank DW. Identification of superoxide dismutase as a cofactor for the Pseudomonas type III toxin, ExoU. Biochemistry. 2006;45:10368–10375. doi: 10.1021/bi060788j. [DOI] [PubMed] [Google Scholar]
- 124.Stirling FR, Cuzick A, Kelly SM, Oxley D, Evans TJ. Eukaryotic localization, activation and ubiquitinylation of a bacterial type III secreted toxin. Cell Microbiol. 2006;8:1294–1309. doi: 10.1111/j.1462-5822.2006.00710.x. [DOI] [PubMed] [Google Scholar]
- 125.Finck-Barbancon V, Frank DW. Multiple domains are required for the toxic activity of Pseudomonas aeruginosa ExoU. J Bacteriol. 2001;183:4330–4344. doi: 10.1128/JB.183.14.4330-4344.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rabin SD, Veesenmeyer JL, Bieging KT, Hauser AR. A C-terminal domain targets the Pseudomonas aeruginosa cytotoxin ExoU to the plasma membrane of host cells. Infect Immun. 2006;74:2552–2561. doi: 10.1128/IAI.74.5.2552-2561.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hauser AR, Kang PJ, Engel J. PepA, a novel secreted protein of Pseudomonas aeruginosa, is necessary for cytotoxicity and virulence. Mol Microbiol. 1998;27:807–818. doi: 10.1046/j.1365-2958.1998.00727.x. [DOI] [PubMed] [Google Scholar]
- 128.Finck-Barbancon V, et al. ExoU expression by Pseudomonas aeruginosa correlates with acute cytotoxicity and epithelial injury. Mol Microbiol. 1997;25:547–557. doi: 10.1046/j.1365-2958.1997.4891851.x. [DOI] [PubMed] [Google Scholar]
- 129.Zaborina O, et al. Identification of multi-drug resistant Pseudomonas aeruginosa clinical isolates that are highly disruptive to the intestinal epithelial barrier. Ann Clin Microbio Antimicrob. 2006;5:14. doi: 10.1186/1476-0711-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Diaz MH, et al. Pseudomonas aeruginosa induces localized immunosuppression during pneumonia in mammals. Infect Immun. 2008;76:4414–4421. doi: 10.1128/IAI.00012-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kurahashi K, et al. Pathogenesis of septic shock in Pseudomonas aeruginosa pneumonia. J Clin Invest. 1999;104:743–750. doi: 10.1172/JCI7124. In this study, the secretion of ExoU and ExoT was shown to cause septic shock by allowing proinflammatory cytokines to escape from the lungs into the systemic circulation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yahr TL, Vallis AJ, Hancock MK, Barbieri JT, Frank DW. ExoY, an adenylate cyclase secreted by the Pseudomonas aeruginosa type III system. Proc Natl Acad Sci USA. 1998;95:13899–13904. doi: 10.1073/pnas.95.23.13899. This study reported the identification of ExoY and demonstrated its adenylate cyclase activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ichikawa JK, et al. Genome-wide analysis of host responses to the Pseudomonas aeruginosa type III secretion system yields synergistic effects. Cell Microbiol. 2005;7:1635–1646. doi: 10.1111/j.1462-5822.2005.00581.x. [DOI] [PubMed] [Google Scholar]
- 134.Vallis AJ, Finck-Barbancon V, Yahr TL, Frank DW. Biological effects of Pseudomonas aeruginosa type III-secreted proteins on CHO cells. Infect Immun. 1999;67:2040–2044. doi: 10.1128/iai.67.4.2040-2044.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Cowell BA, Evans DJ, Fleiszig SM. Actin cytoskeleton disruption by ExoY and its effects on Pseudomonas aeruginosa invasion. FEMS Microbiol Lett. 2005;250:71–76. doi: 10.1016/j.femsle.2005.06.044. [DOI] [PubMed] [Google Scholar]
- 136.Sayner SL, et al. Paradoxical cAMP-induced lung endothelial hyperpermeability revealed by Pseudomonas aeruginosa ExoY. Circ Res. 2004;95:196–203. doi: 10.1161/01.RES.0000134922.25721.d9. [DOI] [PubMed] [Google Scholar]
- 137.Hauser AR, et al. Type III protein secretion is associated with poor clinical outcomes in patients with ventilator-associated pneumonia caused by Pseudomonas aeruginosa. Crit Care Med. 2002;30:521–528. doi: 10.1097/00003246-200203000-00005. [DOI] [PubMed] [Google Scholar]
- 138.Sawa T, et al. In vitro cellular toxicity predicts Pseudomonas aeruginosa virulence in lung infections. Infect Immun. 1998;66:3242–3249. doi: 10.1128/iai.66.7.3242-3249.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lee EJ, Evans DJ, Fleiszig SM. Role of Pseudomonas aeruginosa ExsA in penetration through corneal epithelium in a novel in vivo model. Invest Ophthalmol Vis Sci. 2003;44:5220–5227. doi: 10.1167/iovs.03-0229. [DOI] [PubMed] [Google Scholar]
- 140.Holder IA, Neely AN, Frank DW. Type III secretion/intoxication system important in virulence of Pseudomonas aeruginosa infections in burns. Burns. 2001;27:129–130. doi: 10.1016/s0305-4179(00)00142-x. [DOI] [PubMed] [Google Scholar]
- 141.Koh AY, Priebe GP, Pier GB. Virulence of Pseudomonas aeruginosa in a murine model of gastrointestinal colonization and dissemination in neutropenia. Infect Immun. 2005;73:2262–2272. doi: 10.1128/IAI.73.4.2262-2272.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Fleiszig SM, et al. Epithelial cell polarity affects susceptibility to P. aeruginosa invasion and cytotoxicity. Infect Immun. 1997;65:2861–2867. doi: 10.1128/iai.65.7.2861-2867.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Saiman L, Prince A. Pseudomonas aeruginosa pili bind to asialoGM1 which is increased on the surface of cystic fibrosis epithelial cells. J Clin Invest. 1993;92:1875–1880. doi: 10.1172/JCI116779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Tsang KW, et al. Interaction of Pseudomonas aeruginosa with human respiratory mucosa in vitro. Eur Respir J. 1994;7:1746–1753. doi: 10.1183/09031936.94.07101746. [DOI] [PubMed] [Google Scholar]
- 145.Saliba AM, et al. Eicosanoid-mediated proinflammatory activity of Pseudomonas aeruginosa ExoU. Cell Microbiol. 2005;7:1811–1822. doi: 10.1111/j.1462-5822.2005.00635.x. [DOI] [PubMed] [Google Scholar]
- 146.Ader F, et al. Alveolar response to Pseudomonas aeruginosa: role of the type III secretion system. Infect Immun. 2005;73:4263–4271. doi: 10.1128/IAI.73.7.4263-4271.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Rello J, Ausina V, Ricart M, Castella J, Prats G. Impact of previous antimicrobial therapy on the etiology and outcome of ventilator-associated pneumonia. Chest. 1993;104:1230–1235. doi: 10.1378/chest.104.4.1230. [DOI] [PubMed] [Google Scholar]
- 148.Kudoh I, Wiener-Kronish JP, Hashimoto S, Pittet JF, Frank D. Exoproduct secretions of P. aeruginosa strains influence severity of alveolar epithelial injury. Am J Physiol. 1994;267(5 Pt 1):L551–L556. doi: 10.1152/ajplung.1994.267.5.L551. [DOI] [PubMed] [Google Scholar]
- 149.Ganter MT, et al. Role of small GPTases and {alpha}v{beta}5 integrin in P. aeruginosa-induced increase in lung permeability. Am J Respir Cell Mol Biol. 2008 doi: 10.1165/rcmb.2007-0454OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Henderson FC, Miakotina OL, Mallampalli RK. Proapoptotic effects of P. aeruginosa involve inhibition of surfactant phosphatidylcholine synthesis. J Lipid Res. 2006;47:2314–2324. doi: 10.1194/jlr.M600284-JLR200. [DOI] [PubMed] [Google Scholar]
- 151.Roy-Burman A, et al. Type III protein secretion is associated with death in lower respiratory and systemic Pseudomonas aeruginosa infections. J Infect Dis. 2001;183:1767–1774. doi: 10.1086/320737. [DOI] [PubMed] [Google Scholar]
- 152.Zhuo H, et al. Increased mortality of ventilated patients with endotracheal Pseudomonas aeruginosa without clinical signs of infection. Crit Care Med. 2008;36:2495–2503. doi: 10.1097/CCM.0b013e318183f3f8. [DOI] [PubMed] [Google Scholar]
- 153.El Solh AA, et al. Persistent infection with Pseudomonas aeruginosa in ventilator-associated pneumonia. Am J Respir Crit Care Med. 2008 doi: 10.1164/rccm.200802-239OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Holder IA, Neely AN, Frank DW. PcrV immunization enhances survival of burned Pseudomonas aeruginosa-infected mice. Infect Immun. 2001;69:5908–5910. doi: 10.1128/IAI.69.9.5908-5910.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Baer M, et al. An engineered human antibody Fab fragment specific for Pseudomonas aeruginosa PcrV antigen has potent anti-bacterial activity. Infect Immun. 2008 doi: 10.1128/IAI.00815-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lee VT, et al. Pseudolipasin A is a specific inhibitor for phospholipase A2 activity of Pseudomonas aeruginosa cytotoxin ExoU. Infect Immun. 2007;75:1089–1098. doi: 10.1128/IAI.01184-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Arnoldo A, et al. Identification of small molecule inhibitors of Pseudomonas aeruginosa exoenzyme S using a yeast phenotypic screen. PLoS Genet. 2008;4:e1000005. doi: 10.1371/journal.pgen.1000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Draper MP, et al. 47th ASM Interscience Conference on Antimicrobial Agents and Chemotherapy Abstract F2–968a; Chicago. 2007. [Google Scholar]
- 159.Ajayi T, Allmond LR, Sawa T, Wiener-Kronish JP. Single-nucleotide-polymorphism mapping of the Pseudomonas aeruginosa type III secretion toxins for development of a diagnostic multiplex PCR system. J Clin Microbiol. 2003;41:3526–3531. doi: 10.1128/JCM.41.8.3526-3531.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Stepinska M, Trafny EA. Diverse type III secretion phenotypes among Pseudomonas aeruginosa strains upon infection of murine macrophage-like and endothelial cell lines. Microb Pathog. 2008;44:448–458. doi: 10.1016/j.micpath.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 161.Li L, Ledizet M, Kar K, Koski RA, Kazmierczak BI. An indirect enzyme-linked immunosorbent assay for rapid and quantitative assessment of Type III virulence phenotypes of Pseudomonas aeruginosa isolates. Ann Clin Microbiol Antimicrob. 2005;4:22. doi: 10.1186/1476-0711-4-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Wolfgang MC, et al. Conservation of genome content and virulence determinants among clinical and environmental isolates of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2003;100:8484–8489. doi: 10.1073/pnas.0832438100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Saier MH., Jr Evolution of bacterial type III protein secretion systems. Trends Microbiol. 2004;12:113–115. doi: 10.1016/j.tim.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 164.Nguyen L, Paulsen IT, Tchieu J, Hueck CJ, Saier MH., Jr Phylogenetic analyses of the constituents of type III protein secretion systems. J Mol Microbiol Biotechnol. 2000;2:125–144. [PubMed] [Google Scholar]
- 165.Troisfontaines P, Cornelis GR. Type III secretion: more systems than you think. Physiology (Bethesda) 2005;20:326–339. doi: 10.1152/physiol.00011.2005. [DOI] [PubMed] [Google Scholar]
- 166.Davis PB. Cystic fibrosis since 1938. Am J Respir Crit Care Med. 2006;173:475–482. doi: 10.1164/rccm.200505-840OE. [DOI] [PubMed] [Google Scholar]
- 167.Ferguson MW, Maxwell JA, Vincent TS, da Silva J, Olson JC. Comparison of the exoS gene and protein expression in soil and clinical isolates of Pseudomonas aeruginosa. Infect Immun. 2001;69:2198–2210. doi: 10.1128/IAI.69.4.2198-2210.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Matz C, et al. Pseudomonas aeruginosa uses type III secretion system to kill biofilm-associated amoebae. ISME J. 2008;2:843–852. doi: 10.1038/ismej.2008.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Abd H, et al. Pseudomonas aeruginosa utilises its type III secretion system to kill the free-living amoeba Acanthamoeba castellanii. J Eukaryot Microbiol. 2008;55:235–243. doi: 10.1111/j.1550-7408.2008.00311.x. [DOI] [PubMed] [Google Scholar]
- 170.Moss J, Ehrmantraut ME, Banwart BD, Frank DW, Barbieri JT. Sera from adult patients with cystic fibrosis contain antibodies to Pseudomonas aeruginosa type III apparatus. Infect Immun. 2001;69:1185–1188. doi: 10.1128/IAI.69.2.1185-1188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Jain M, et al. Type III secretion phenotypes of Pseudomonas aeruginosa strains change during infection of individuals with cystic fibrosis. J Clin Microbiol. 2004;42:5229–5237. doi: 10.1128/JCM.42.11.5229-5237.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Govan JRW, Deretic V. Microbial pathogenesis in cystic fibrosis: Mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev. 1996;60:539–574. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Wu W, Badrane H, Arora S, Baker HV, Jin S. MucA-mediated coordination of type III secretion and alginate synthesis in Pseudomonas aeruginosa. J Bacteriol. 2004;186:7575–7585. doi: 10.1128/JB.186.22.7575-7585.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Smith EE, et al. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci U S A. 2006;103:8487–8492. doi: 10.1073/pnas.0602138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Radke J, Pederson KJ, Barbieri JT. Pseudomonas aeruginosa exoenzyme S is a biglutamic acid ADP-ribosyltransferase. Infect Immun. 1999;67:1508–1510. doi: 10.1128/iai.67.3.1508-1510.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Barbieri JT, Sun J. Pseudomonas aeruginosa ExoS and ExoT. Rev Physiol Biochem Pharmacol. 2004;152:79–92. doi: 10.1007/s10254-004-0031-7. [DOI] [PubMed] [Google Scholar]