Abstract
Barrett’s esophagus (BE) is a metaplastic condition caused by chronic gastroesophageal reflux which represents an early step in the development of esophageal adenocarcinoma (EAC). Single-nucleotide polymorphism microarray (SNP-chip) analysis is a novel, precise, high-throughput approach to examining genomic alterations in neoplasia. Using 250K SNP-chips, we examined the neoplastic progression of BE to EAC, studying 11 matched sample sets: 6 sets of normal esophagus [NE], BE and EAC, 4 of NE and BE, and 1 of NE and EAC. Six (60%) of 10 total BE samples and 4 (57%) of 7 total EAC samples exhibited one or more genomic abnormalities comprising deletions, duplications, amplifications, and copy-number-neutral loss of heterozygosity (CNN-LOH). Several shared abnormalities were identified, including chromosome 9p CNN-LOH (2 BE samples [20%]), deletion of CDKN2A (4 BE samples [40%]), and amplification of 17q12–21.2 involving the ERBB2, RARA and TOP2A genes (3.1Mb, 2 EAC [29%]). Interestingly, one BE sample contained a homozygous deletion spanning 9p22.3-p22.2 (1.2 Mb): this region harbors only one known gene, basonuclin 2 (BNC2). Real-time PCR analysis confirmed deletion of this gene and decreased expression of BNC2 mRNA in the BE sample. Furthermore, transfection and stable expression of BNC2 caused growth arrest of OE33 EAC cells, suggesting that BNC2 functions as a tumor suppressor gene in the esophagus, and that deletion of this gene occurs during the development of EAC. Thus, this SNP-chip analysis has identified several early cytogenetic events and novel candidate cancer-related genes that are potentially involved in the evolution of BE to EAC.
Keywords: esophageal adenocarcinoma, SNP-chip, copy-number-neutral LOH, BNC2
Introduction
Chronic gastroesophageal reflux disease (GERD) is characterized by the retrograde movement of gastric contents into the esophagus, resulting in tissue damage. GERD is the major risk factor for the development of Barrett’s esophagus (BE).1 BE is a premalignant condition, greatly increasing the risk of developing esophageal adenocarcinoma (EAC).2,3
Genomic DNA alterations often contribute to the development of malignant tumors. In BE and EAC, chromosomal aberrations have been discovered by comparative genomic hybridization (CGH) analysis.4–11 CGH analyses have revealed frequent gains of chromosomes 6p (10 – 37%), 7q (17 – 37%), 7p (30 – 60%), 8q (50 – 80%), 10q (20 – 50%), 15q (10 – 40%), 17q (30 – 50%), and 20q (50 – 80%); and frequent losses of chromosomes 4q (20 – 50%), 5q (20 – 50%), 9p (20 – 50%), 14q (30 – 40%), 16q (36 – 40%), 17p (30%), 18q (20 – 60%) and Y (60 – 76%).4–11 These chromosomal alterations have suggested genes associated with esophageal adenocarcinogenesis. For example, the proto-oncogenes MYC (8q), EGFR (7p) and ERBB2 (17q) are often duplicated. The tumor suppressor genes APC, CDKN2A, TP53, and SMAD4 are located on 5q, 9p, 17p and 18q, respectively, and these chromosomal regions are often deleted. Thus, genome-wide analyses of DNA copy-number changes in BE and EAC can identify consensus regions of chromosomal gain and loss, as well as candidate cancer-related genes.
Single-nucleotide polymorphism microarray (SNP-chip) analysis is a novel strategy to examine genomic alterations such as copy-number changes and loss of heterozygosity (LOH).12–14 Importantly, SNP-chip analysis can detect several abnormalities including copy-number-neutral loss of heterozygosity (CNN-LOH) that cannot be detected by either karyotyping or CGH. SNP-chip analysis has been used to study several types of leukemia, including chronic lymphocytic leukemia (CLL),15,16 childhood acute lymphoblastic leukemia (ALL).17,18 and acute myeloid leukemia (AML).19–24
In the current study, we identified chromosomal abnormalities and novel disease-related genomic regions using 250K SNP-chip analysis in matched tissue sample sets that contained normal esophagus (NE), EAC, and/or BE. The use of the CNAG (copy-number analysis for Affymetrix GeneChips) program and the new AsCNAR (allele-specific copy-number analysis using anonymous references) algorithm12,14 provided a highly sensitive technique to detect CNN-LOH as well as copy-number changes in premalignant and malignant esophageal tissue samples.
Materials and methods
Patient samples, isolation of genomic DNA and RNA, and cell culture
Samples of BE and EAC tissues were obtained by endoscopic biopsy. Clinical features of esophageal samples examined in this study are summarized in Table 1. Normal esophageal mucosal (NE) samples were always obtained from the same individual for BE and EAC samples. Tissue samples were snap-frozen in liquid nitrogen. All NE samples were obtained at a minimum of 7 cm proximal to the squamocolumnar junction (the proximal border between BE and NE). All biopsies were examined histopathologically by hematoxylin and eosin staining; and the stromal component averaged approximately 50% and 30% in the BE and EAC samples, respectively. Genomic DNA and total RNA were isolated from tissues using a DNeasy Tissue Kit (Qiagen, Valencia, CA, USA) and Trizol reagent (Invitrogen, Carlsbad, CA, USA), respectively. DNA was determined to be high-MW by agarose gel electrophoresis showing >90% of DNA to be above a length of 20 kb. The EAC cell line, OE33, was maintained in RPMI 1640 medium (Mediatech Inc., Herndon, VA, USA) supplemented with 10% fetal bovine serum (Atlanta Biological, Lawrenceville, GA, USA).
Table 1.
Clinical features of esophageal samples examined by SNP-chip analysis
| Case # | Origin | Procedure | Histology | Age | Gender | Race | EAC Grade | TNM | Stage | Previous Treatment |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | NE | Surgery | Normal epithelium | 54 | M | White | Preoperative chemoradiation (5-FU + cisplatin) | |||
| BE | Metaplasia | |||||||||
| EAC | Adenocarcinoma | Moderately differentiated | T3N1 | III | ||||||
| 2 | NE | Biopsy | Normal epithelium | 68 | M | White | Unknown | |||
| BE | Metaplasia | |||||||||
| EAC | Adenocarcinoma | Moderately differentiated | ||||||||
| 3 | NE | Surgery | Normal epithelium | 52 | M | White | No chemoradiation | |||
| BE | Metaplasia with dysplasia | Moderately differentiated | T1N0 | I | ||||||
| 4 | NE | Surgery | Normal epithelium | 61 | M | White | No chemoradiation | |||
| BE | Metaplasia with focal HGD | |||||||||
| EAC | Adenocarcinoma | Poorly differentiated | T3N1M0 | III | ||||||
| 5 | NE | Biopsy | Normal epithelium | 67 | M | Asian/Pacfic Islander | No chemoradiation | |||
| BE | Metaplasia with LGD and HGD | |||||||||
| EAC | Adenocarcinoma | Well differentiated | ||||||||
| 6 | NE | Biopsy | Normal epithelium | 82 | F | White | Unknown | |||
| EAC | Adenocarcinoma | Poorly differentiated | ||||||||
| 7 | NE | Biopsy | Normal epithelium | 77 | M | White | No chemoradiation | |||
| BE | Metaplasia with LGD and focal HGD | |||||||||
| EAC | Adenocarcinoma | Moderately differentiated | ||||||||
| 8 | NE | Biopsy | Normal epithelium | 80 | M | White | No chemoradiation | |||
| BE | Metaplasia | |||||||||
| 9 | NE | Biopsy | Normal epithelium | 72 | M | White | No chemoradiation | |||
| BE | Metaplasia | |||||||||
| EAC | Adenocarcinoma | Moderately differentiated | T2N0M1* | IV* | ||||||
| 10 | NE | Biopsy | Normal epithelium | 78 | M | White | No chemoradiation | |||
| BE | Metaplasia | |||||||||
| 11 | NE | Biopsy | Normal epithelium | 56 | M | White | No chemoradiation | |||
| BE | Metaplasia | |||||||||
Clinical features of 11 samples examined by SNP-chip analysis are displayed. Abbreviations. NE; normal esophagus, BE; Barrett’s esophagus,
Note. determined by endoscopic ultrasound and CT scans.
High-density SNP-chip analysis
BE or EAC and their matched normal genomic DNA (100 ng) from case #5, #7, #9 and #6, as well as from lung cancer cell line NCI-H2171 and its paired lymphoblastoid cell line NCI-BL2171, were subjected to whole genome amplification using a REPLI-g Midi Kit according to the manufacturer’s protocol (Qiagen). Genomic alterations found in the unamplified aliquots of NCI-H2171 cells were also detected in the amplified aliquots as well. (Supplemental Figure 1). All genomic DNA samples (375 ng) were analyzed on GeneChip Human mapping 250 K microarrays (SNP-chip, Affymetrix, Santa Clara, CA, USA), as described previously.12,14 Hybridization, washing and signal detection were performed on a GeneChip Fluidics Station 400 and a GeneChip scanner 3000, according to the manufacturer’s protocols (Affymetrix). Microarray data were analyzed for the determination of both total and allele-specific copy numbers (AsCNs) using the CNAG program, as previously described,12,14 with minor modifications. All SNPs within a given inferred LOH region were formally analyzed as “heterozygous” SNPs (see reference [14] for mathematical details). For clustering of samples according to copy-number change and CNN-LOH status, GNAGraph softwarewas employed. Size, position and location of genes were identified using the UCSC Genome Browser <http://genome.ucsc.edu/>.
Determination of SNP sequences in cases of CNN-LOH
To validate CNN-LOH, 3 independent SNP sequences (rs2296820, rs668026, and rs2890896) at chromosome 9p were queried in case #11. The genomic region of each SNP site was amplified by genomic PCR using specific primers, and PCR products were purified and sequenced. Primer sequences were as follows: 5′-AAA TGA CCG CAC CTC TGA AG-3′ and 5′-GAG AGC GGC AAA CCA TTA GA-3′ for rs2296820, 5′-TTT GCT AGT CTC ACC ACT TGC-3′ and 5′-CCT TGC ACA TTA TAA ACT CTC GAT-3′ for rs668026, and 5′-GGAAGG GTAGGC TTC CTG AT-3′ and 5′-TCT GTG TCT TTG GTT CTT TTT CA-3′ for rs2890896.
Quantitative genomic and mRNA real-time PCR
Gene dosages of chromosome 9p22.3 in case #11; the ERBB2 gene in cases #5 and #9; the CDKN2A gene in cases #8 and #11; and the basonuclin 2 (BNC2) gene in #11, as well as mRNA expression levels of ERBB2 in case #9 and BNC2 in case #11, were determined by quantitative real-time PCR (iCycler, Bio-Rad, Hercules, CA, USA) using Sybr Green. To determine relative gene dosages and mRNA levels, the chromosome 2p21 region and β-actin were measured as controls, respectively. The delta threshold cycle value (ΔCt) was calculated from the given Ct value by the formula ΔCt = (Ct sample − Ct control). Fold change was calculated as 2−ΔCt. PCR was performed using the following primer pairs: 5′-CCC TCA AAA AGT GGA GAC GA-3′ and 5′-ATT CTT GGG GCA CCT CTC TT-3′ for 9p22.3, 5′-AGT ACC TGG GTC TGG ACG TG-3′ and 5′-CTG GGAACT CAAGCA GGA AG-3′ for ERBB2 genomic DNA, 5′-GTG CCA AAG TGC TCC TGA AGC TG-3′ and 5′-AGC AAA TCT GTT TGG AGG TCTG-3 for the CDKN2A gene, 5′-GGG GAT TCT TCT CGA TGA CA-3′ and 5′-ACT CTC AGG GTC CCC TTG TT-3′ for BNC2 genomic DNA, 5′-GGC AAT CCT GGC TGC GGA TCA AGA-3′ and 5′-ATT TCT GAA CTT CTT GGC TGC C-3′ for the 2p21 region, 5′-GGG ACT ATG TCC GAG GAT AC-3′ and 5′-AGG GTG ATG ATT TCC TCT TC-3′ for BNC2 mRNA, 5′-GTT TGA GTC CAT GCC CAA TC-3′ and 5′-CCC ACG TCC GTA GAA AGG TA-3′ for ERBB2 mRNA, and 5′-CCT GGC ACC CAG CAC AAT-3′ and 5′-GCC GAT CCA CAC GGA GTA CT-3′ for β-actin mRNA
Western blot analysis and colony assays
OE33 cells were transfected with either HA-tagged BNC2 expression vector (kindly provided from Dr. Satrajit Sinha, State University of New York at Buffalo) or empty vector. Cells were harvested, lysed in 2 × sample buffer (12% glycerol, 20 mM Tris-HCl, 4% SDS, 100 mM DTT, 4 mM EDTA, 0.04% Coomassie Brilliant Blue R250), and heat-denatured. Samples were subjected to SDS-PAGE followed by an electro-transfer to polyvinylidene difluoride membrane. The signals were developed with either Supersignal West Pico-Chemiluminescent or -Dura Extended Duration Substrate (Pierce Biotechnology, Rockford, IL, USA). Anti-HA antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA, USA), while anti-GAPDH was from Research Diagnostics (Concord, MA, USA).
For colony formation assays, transfected cells were cultured in the presence of 250 μg/ml G418. After 2 weeks, cells were stained with Crystal Violet Dye (0.1% dissolved in 50% methanol). To quantify the number of surviving colonies, cells were dissolved in 1.0% SDS, and absorbance was measured at 600 nm.
Results
Summary of SNP-chip analysis of esophageal adenocarcinoma samples
SNP chip analysis was performed on genomic DNAs from 11 matched sets (6 of NE, BE, and EAC; 1 of NE and EAC; and 4 of NE and BE) containing in total 10 BE, 7 EAC, and 11 matched NE samples. Because concentrations of genomic DNA in cases #5, #7, #9 and #6 were low, these samples were subjected to whole-genome PCR amplification prior to SNP-chip analysis. In order to confirm the reliability of DNA from whole genome amplification, we compared it to whole DNA. Both were subjected to SNP-chip analysis using the same cell line, NCI-H2171. The genomic changes found in the unamplified DNA from NCI-H2171 cells were also detected in the whole genome amplified samples, suggesting that whole-genome amplification is a useful approach when DNA amounts are limited (Supplemental Figure 1). BE samples from case #3 and from matched BE and EAC samples in cases #1, #4 and #2 revealed no detectable genomic abnormalities (data not shown). In contrast, the remaining specimens (6 BEs and 4 EACs) harbored one or more genomic abnormalities (Table 2 and data not shown). Thus, six (60%) of 10 BE and four (57%) of 7 EAC samples contained genomic alterations, including copy-number changes and CNN-LOH. CNN-LOH is a genomic abnormality that normally cannot be detected by CGH analysis; these regions usually also contain a mutation in a key gene.
Table 2.
Comparison of genomic abnormalities between matched Barrett’s and adenocarcinoma cells
| Status in each tissue |
|||||||
|---|---|---|---|---|---|---|---|
| Case # | Location | Proximal | Distal | Size (Mb) | BE | EAC | Gene(s) in the region |
| 5 | 9p24.3-p13.2 | 30,910 | 37,046,816 | 37 | CNN-LOH | Normal | >10 genes including CDKN2A |
| 17q12-q21.2 | 33,823,479 | 37,542,890 | 3.7 | Normal | Amp | >10 genes including ERBB2, CSF3, RARA, and TOP2A | |
| 7 | 6q22.33-q23.2 | 130,053,359 | 133,756,120 | 3.7 | Normal | Amp | >10 genes including CTGF |
| 6q23.2-q23.3 | 134,406,908- | 136,520,887 | 1.1 | Normal | Amp | 7 genes including MYB | |
| 9p21.3 | 21,455,478 | 22,597,894 | 2.1 | Del | Normal | IFNE1, MTAP, CDKN2A, CDKN2B, and DMRTA1 | |
| 9 | 8p21.3 | 19,243,968 | 20,116,050 | 0.9 | Normal | Amp | SH2D4A, ChGn, LPL, SLC18A1, and ATP6V1B2 |
| 8p12 | 30,973,934 | 33,512,592 | 2.5 | Normal | Amp | PURG, WRN, NRG1, FUT10 and RBM13 | |
| 16p13.13 | 11,095,284 | 11,869,359 | 0.8 | Normal | Amp | 9 genes including SOCS1 | |
| 16p13.12 | 12,744,248 | 13,938,667 | 1.2 | Normal | Amp | ERCC4 | |
| 17q12-q21.2 | 34,176,036 | 37,279,744 | 3.1 | Normal | Amp | >10 genes including ERBB2, CSF3, RARA, and TOP2A | |
| 17q21.32-q22 | 43,910,744 | 48,020,820 | 4.1 | Normal | Amp | >10 genes | |
| 17q23.2-q23.3 | 53,943,071 | 58,922,752 | 5.0 | Normal | Amp | >10 genes | |
| Xp22.13-p22.12 | 17,538,134 | 21,071,174 | 3.5 | Amp | Amp | >10 genes | |
| 6 | 7p11.2 | 54,497,224 | 55,151,950 | 0.7 | - | Amp | SEC61G, and EGFR |
| 11p13-p12 | 33,540,416 | 36,817,806 | 3.3 | - | Amp | >10 genes including LOM2 | |
| 11 | 1p21.3-p11.2 | 97,590,482 | 120,863,833 | 23.3 | Dup | - | >10 genes |
| 3p14.2 | 60,325,749 | 60,953,251 | 0.6 | Del | - | FHIT | |
| 5p15.33-p11 | 81,949 | 46,419,092 | 46.3 | Del | - | >10 genes | |
| 7p21.3 - q11.22 | 8,467,656 | 68,111,884 | 59.6 | Del | - | >10 genes including EGFR | |
| 8p23.3-p12 | 180,568 | 34,774,314 | 34.6 | Dup | - | >10 genes | |
| 8q13.2-q24.3 | 69,209,296 | 146,263,538 | 77.1 | Del | - | >10 genes including MYC | |
| 9p24.3-p21.1 | 30,910 | 31,806,036 | 31.8 | CNN-LOH | - | >10 genes including JAK2 | |
| 9p22.3-p22.2 | 15,773,266 | 16,931,719 | 1.2 | Del | - | BNC2 | |
| 9p21.3 | 21,823,659 | 21,988,733 | 0.2 | Del | - | MTAP and CDKN2A | |
| 10q25.2 - q25.3 | 113,622,691 | 115,912,482 | 2.3 | Del | - | >10 genes | |
| 11p15.3-p15.1 | 10,802,693 | 17,496,138 | 6.7 | Del | - | >10 genes | |
| 11p14.2 | 26,845,102 | 33,281,952 | 6.4 | Del | - | >10 genes including WT1 | |
| 11p12 | 37,649,256 | 39,949,139 | 2.3 | Del | - | no known genes | |
| 14q31.2-q32.33 | 82,684,067 | 106,356,482 | 23.7 | Dup | - | >10 genes including BCL11B, and AKT1 | |
| 16q23.1 | 77,234,871 | 77,398,567 | 0.2 | Del | - | WWOX | |
| 17p13.1-p11.2 | 10,538,736 | 19,971,552 | 9.4 | Dup | - | >10 genes | |
| 17q12-q21.2 | 31,501,499 | 37,358,181 | 5.9 | Del | - | >10 genes including ERBB2, CSF3, RARA, and TOP2A | |
| 17q24.2-q24.3 | 63,302,865 | 65,729,710 | 2.4 | Del | - | >10 genes | |
| 17q24.3-q25.1 | 65,729,710 | 69,223,351 | 3.5 | Del | - | 7 genes | |
| 18q11.2 | 17,918,387 | 20,052,960 | 2.1 | Del | - | >10 genes including GATA6 | |
| 18q11.2-q12.1 | 20,196,461 | 24,997,438 | 4.8 | Dup | - | 9 genes | |
| 19q12-q13.11 | 32,977,359 | 39,248,464 | 6.3 | Del | - | >10 genes including CEBPA and CEBPG | |
| 20q13.13 | 47,968,658 | 48,404,269 | 0.4 | Del | - | ZNF313, SNAI1, Kua-UEV, and CEBPB | |
| 8 | 5q15-q21.1 | 96,914,453 | 98,821,231 | 1.9 | Del | - | RGMB, and CHD1 |
| 7p21.3-p21.1 | 10,332,019 | 13,408,633 | 3.1 | Del | - | NDUFA4, PHF14, SCIN and ARL4A | |
| 7p15.3 | 21,468,520 | 23,844,205 | 2.4 | Del | - | >10 genes | |
| 7p15.1-p14.3 | 27,732,391 | 32,309,160 | 4.6 | Del | - | >10 genes | |
| 7q14.1 | 38,575,532 | 39,469,231 | 0.9 | Del | - | VPS41, POU6F2, and RALA | |
| 7p12.1 | 51,653,697 | 52,398,407 | 0.7 | Del | - | no known gene | |
| 7q21.11 | 79,524,526 | 81,077,769 | 1.6 | Del | - | CD36, SEMA3C, and HGF | |
| 9p24.3 | 182,129 | 1,212,804 | 1.0 | Del | - | DOCK8, ANKRD15, DMRT1, DMRT3, and DMRT2 | |
| 9p24.2-p24.1 | 2,240,370 | 7,362,858 | 5.1 | Del | - | >10 genes including JAK2 | |
| 9p23 | 10,537,886 | 11,701,459 | 1.2 | Del | - | no known gene | |
| 9p21.3 | 20,989,646 | 22,673,792 | 1.7 | Del | - | >10 genes including CDKN2A | |
| 9p21.3-p21.2 | 24,989,702 | 25,954,852 | 1.0 | Del | - | TUSC1 | |
| 9p21.2-p21.1 | 25,954,852 | 31,836,527 | 5.9 | Del | - | 6 genes | |
| 9q21.33-q22.2 | 85,272,210 | 89,347,306 | 4.1 | Del | - | >10 genes | |
| 9p22.2 | 89,347,306 | 90,834,752 | 1.5 | Del | - | GADD45G, DIRAS2, and SYK | |
| 9q22.2-q22.31 | 90,834,752 | 92,063,705 | 1.2 | Del | - | AUH, NFIL3, ROR2, SPTLC1, and IARS | |
| 9q22.33-q31.1 | 97,923,215 | 102,419,896 | 4.5 | Del | - | >10 genes | |
| 9q31.1-q31.2 | 103,707,323 | 107,191,745 | 3.5 | Del | - | >10 genes | |
| 9q33.3 | 123,643,047 | 125,825,598 | 2.2 | Del | - | >10 genes | |
| 21q11.2-q21.1 | 14,892,587 | 16,494,111 | 1.6 | Del | - | NRIP1, and USP25 | |
| 21q21.1 | 17,139,470 | 22,209,634 | 5.1 | Del | - | CXADR, BTG3, CHODL, PRSS7, and NCAM2 | |
| 10 | 1p32.3-p21.2 | 54,223,221 | 100,428,188 | 46.2 | Del | - | >10 genes |
| 3p14.2 | 60,589,486 | 61,002,116 | 0.4 | Del | - | FHIT | |
| 4p16.3-p16.2 | 3,510,571 | 4,842,056 | 1.3 | Del | - | 6 genes | |
| 7p22.3-q22.1 | 141,322 | 102,675,814 | 102.5 | Dup | - | >10 genes including EGFR and HGF | |
| 7q22.1-q36.3 | 102,675,814 | 158,377,134 | 55.7 | CNN-LOH | - | >10 genes including MET and BRAF | |
| 8p23.3-p12 | 180,568 | 30,728,603 | 30.5 | Dup | - | >10 genes | |
| 9p24.3-p13.3 | 30,910 | 35,208,693 | 35.2 | Del | - | >10 genes including JAK2, BNC2, and CDKN2A | |
| 9p13.2 | 36,443,176 | 38,589,297 | 2.1 | Del | - | >10 genes | |
| 10p15.3-p15.1 | 148,946 | 4,785,268 | 4.6 | Del | - | 10 genes | |
| 12q21.1 | 73,327,357 | 73,956,816 | 0.6 | Amp | - | KCNC2 | |
| 12q21.1-q24.33 | 73,979,022 | 132,377,151 | 58.4 | Del | - | >10 genes | |
| 13q (whole) | Dup | - | >10 genes | ||||
| 17p13.3-p11.2 | 18,901 | 22,029,237 | 22.0 | Del | - | >10 genes | |
| 17q25.3 | 76,868,173 | 78,598,059 | 1.7 | CNN-LOH | - | >10 genes | |
| 18p11.32 | 210,071 | 1,738,722 | 1.5 | CNN-LOH | - | 8 genes | |
| 18p11.32-q11.2 | 2,013,693 | 20,887,311 | 18.9 | Dup | - | >10 genes | |
| 18q11.2-q12.3 | 23,055,299 | 39,594,161 | 16.5 | Del | - | >10 genes | |
| 18q12.3-q21.1 | 40,929,681 | 47,747,005 | 6.8 | Dup | - | >10 genes including SMAD2, SMAD7, and SMAD4 | |
| 18q21.2-q21.31 | 51,216,340 | 52,531,252 | 1.3 | Del | - | TCF4, WDR7 and WDR7 | |
| 18q21.31-q21.32 | 52,633,158 | 55,879,013 | 3.2 | Dup | - | >10 genes | |
| 18q21.32-q22.1 | 56,357,316 | 60,406,673 | 4 | Dup | - | >10 genes including BCL2 | |
| 18q22.1-q22.2 | 61,129,618 | 65,324,331 | 4.2 | Del | - | CDH7, CDH19, TXNDC10 and DOK6 | |
| 18q22.2-q23 | 65,568,556 | 73,578,012 | 8 | Dup | - | >10 genes | |
| 18q23 | 73,598,711 | 74,272,671 | 0.7 | Del | - | no known gene | |
| 18q23 | 74,163,515 | 76,082,467 | 1.9 | CNN-LOH | - | 8 genes | |
| 20p12.1-p11.1 | 11,928,996 | 26,236,535 | 14.3 | Del | - | >10 genes including FOXA2 | |
| 21q11.2-q22.11 | 14,986,474 | 32,776,077 | 17.8 | Del | - | >10 genes | |
| 21q22.12-q22.3 | 35,141,436 | 46,885,639 | 11.7 | Del | - | >10 genes | |
Physical localization and size (Mb) are obtained from UCSC Genome Browser. If less than 5 genes are in these regions, all gene names are displayed. Note that amplification of Xp22.13-p22.12 (3.5 Mb) were found in both BE and EAC sample of case #9. Abbreviations: Del; deletion, Dup; duplication (gain of copy number), Amp; amplification, CNN-LOH; copy number neutral loss of heterozygosity; -, not analyzed.
Chromosomal abnormalities in matched BE and EAC specimens
Since BE is believed to constitute an early step in EAC development; we compared the genomic status of BE and EAC with that of NE from the same individuals (Figure 1). Three cases (#1, #4 and #2) contained no genomic changes in either their BE or their EAC samples. BE tissue from case #9 contained one amplification on the X chromosome (Xp22.13-p22.12, 3.5 Mb). Interestingly, EAC tissue from this patient exhibited 7 additional amplified regions, including 8p21.3 (0.9 Mb), 8p12 (2.5 Mb), 16p13.13 (0.8 Mb), 16p13.12 (1.2 Mb), 17q12-q21.2 (3.1 Mb), 17q21.32-q22 (4.1 Mb), and 17q23.2-q23.3 (5.0 Mb), as well as Xp22.13-p22.12 (3.5 Mb). The EAC sample from case #5 contained an amplification of 17q12-q21.2 (3.7 Mb), while the EAC from case #7 had two amplifications at chromosome 6q (6q22.33-q23.2 [3.7 Mb] and 6q23.2-q23.3 [1.1 Mb]). These amplifications were not detected in matching BE samples. Unexpectedly, CNN-LOH of 9p-terminal - p13.2 (37.0 Mb) occurred in BE tissue from case #5, as did deletion of 9p21.3 (2.1 Mb) in BE tissue from case #7; however, these aberrations were not detected in their matched EAC tissues, possibly because contamination with normal cells masked this abnormality in the frank tumors.
Figure 1. Summary of genomic abnormalities in esophageal samples.

Genomic DNAs from 6 matched sets (NE, BE and EAC), 1 matched NE-EAC pair, and 4 matched NE-BE pairs (in total, 10 BE and 7 EAC samples) were subjected to SNP-chip analysis; genomic abnormalities are summarized by color: pink (copy-number-neutral LOH [CNN-LOH]); green (hemizygous deletion); blue (homozygous deletion); red (duplication/amplification). Six (60%) of 10 BE (cases #5, #7, #9, #11, #8 and #10) and four (57%) of 7 EAC (cases #5, #7, #9, #6) samples possessed genomic alterations.
Other CNN-LOH and copy-number changes
All other abnormalities, including CNN-LOH and copy-number changes, are displayed in Table 2. Based on SNP-chip analyses, 3 cases exhibited CNN-LOH. BE samples from cases #5 and #11 had 9p CNN-LOH, while case #10 contained 4 regions of CNN-LOH, including 7q22.1-q36.3 (55.7 Mb), 17q25.3 (1.7 Mb), 18p11.32 (1.5 Mb), and 18q23 (1.9 Mb) (Figure 1 and Table 2). Deletions and duplications were found only in BE samples. Deleted regions involving 7p, 9p, 9q, and 18q were found frequently, and duplicated regions involving 18q were often observed (Figure 1 and Table 2). Of interest, 1 BE sample (case #10) manifested amplification at 12q.21.1 (0.6 Mb); this region contains only one known gene, the Shaw-related voltage-gated potassium channel gene known as KCNC2. Furthermore, EAC samples exhibited amplifications at 6q, 8p, 11p, 16p and 17q, but no duplications or CNN-LOH (Figure 1 and Table 2).
Shared abnormalities found in BE and/or EAC samples
Several chromosomal abnormalities were shared by two or more different patients. As shown in Table 3, several chromosomal abnormalities were present in BE and EAC samples, suggesting that these loci harbored aberrant genes altered early during BE-EAC evolution. Shared abnormalities included two BE cases (#5 and #11) with 9p CNN-LOH; 2 BE cases (#10 and #11) with duplication of 8p23.3-p12; 2 BE cases (#8 and #10) with deletion of 21q11.2-q21.1. Two EAC samples (#5 and #9) exhibited amplification of 17q12-q21.2; this was the only region with gains in more than 1 EAC sample. Furthermore, the CDKN2A gene was deleted in 4 BE (#8, #11, #7 and #10) and the FHIT gene was deleted in 2 BE (#10 and #11) samples.
Table 3.
Shared abnormalities found in either BE and/or EAC samples
| Chromosome | Size | Status | Case # |
|---|---|---|---|
| 9p24.3-p21.1 | 31.7 Mb | CNN-LOH | 5 BE 11 BE |
| 8p23.3-p12 | 30.5 Mb | duplication | 10 BE 11 BE |
| 21q11.2-q21.1 | 1.6 Mb | deletion | 8 BE 10 BE |
| 17q12-q21.2 | 3.1 Mb | amplification | 5 EAC 9 EAC |
| CDKN2A gene | - | deletion | 8 BE* 11 BE* 7 BE 10 BE |
| FHIT gene | - | deletion | 10 BE 11 BE |
Six regions were identified as shared abnormalities including CNN-LOH, duplication, deletion, and amplification.
Abbreviations: CNN-LOH; copy number neutral loss of heterozygosity, BE; Barrett’s esophagus, EAC; esophageal adenocarcinoma,
; homozygous deletion.
Validation of SNP-chip data using nucleotide sequencing quantitative real-time PCR and expression analysis
We validated CNN-LOH detected by SNP-chip using several different techniques including determining SNP sequences, as well as gene-dosage, in the CNN-LOH region. It was presumed that when LOH occurred, SNP sequences in this region should exhibit homozygosity, whereas those of matched normal samples should be heterozygous. Therefore, we examined 3 independent SNP sequences on chromosome 9p in the CNN-LOH region in case #11 (NE and BE) (Figure 2A). In the BE sample, all 3 SNP sites (rs2296820, rs668026, and rs2890896) clearly showed only a single signal (Figure 2B). In contrast, these 3 SNP sites in the matched NE sample showed heterozygosity. These results strongly suggested that LOH occurred in this region.
Figure 2. Representative SNP-chip analysis in Barrett’s esophagus samples.

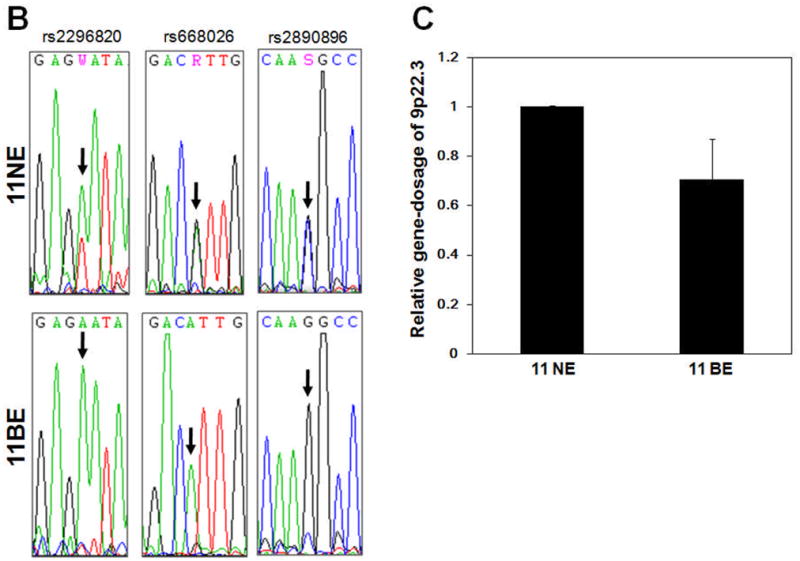
(A) SNP-chip data for chromosome 9 in BE samples (cases #8 and #11). Red dots are SNP sites as probes and indicate total copy number (CN). Blue line is an average of copy number and shows gene dosage. Green bars are heterozygous (hetero) SNP calls. Red and green lines show allele-specific copy number (AsCN). Both cases exhibited deletions of CDKN2A (boxed regions). Case #11 had CNN-LOH at 9p and a deletion of the basonuclin 2 gene (circled). (B) SNP sequences in the 9p CNN-LOH region for validation of CNN-LOH in case #11. Three independent SNP sites (rs2296820, rs668026, and rs2890896) were sequenced. All 3 SNP sites showed heterozygosity in matched NE samples; whereas their corresponding BE samples showed homozygosity, consistent with loss of heterozygosity. (C) Determination of gene dosage in the 9p region. Gene dosage of 9p22.3 (the CNN-LOH region) in case #11 was compared between matched NE and BE genomic DNA, using quantitative genomic real-time PCR. Gene dosage levels were calculated as the ratio between 9p22.3 and the reference genomic DNA, 2p21. Results represent the mean of 3 experiments ± SD.
Next, we determined gene dosage in this region to exclude the possibility of a hemizygous deletion. Gene dosage at 9p22.3 in the region of CNN-LOH in BE case #11 was compared to that in the matched NE using quantitative genomic real-time PCR (Q-PCR). Gene dosage levels were calculated as the ratio between 9p22.3 and the reference genomic DNA at 2p21, which displayed normal gene dosage by SNP-chip analysis. DNA levels at the 9p22.3 region in BE case #11 were almost identical to those in matched NE at this site, indicating that this region possessed normal copy number (Figure 2C). Thus, our SNP sequence and genomic Q-PCR data validated results of SNP-chip analysis, clearly showing CNN-LOH at the 9p region.
We also validated copy-number changes. Four BE samples (#8, #11, #7 and #10) showed deletion of the CDKN2A gene by SNP-chip analysis (Figure 2A and Table 3). Gene dosage of CDKN2A was examined by genomic Q-PCR. Levels of CDKN2A in BE samples from cases #8 and #11 were approximately 10-fold lower relative to matched NE (data not shown). EAC samples from cases #5 and #9 exhibited amplification of 17q12-q21.2 by SNP-chip analysis; and this region contains the ERBB2, CSF3, RARA and TOP2A genes. Genomic Q-PCR revealed that levels of ERBB2 in EAC samples #5 and #9 were approximately 5-fold and 8-fold higher than in matched NE samples, respectively (Figure 3A). Taken together, these results validated the observations of our SNP-chip analysis.
Figure 3. Relationship between copy-number change and gene expression.

(A) Amplification of the ERBB2 gene: Two EAC samples (#5 and #9) had amplification of the 17q12-q21.2 region, which includes the ERBB2 gene. Gene dosage of the ERBB2 gene was compared between matched NE, BE and EAC genomic DNA using genomic Q-PCR. Gene dosage levels were calculated as the ratio between the ERBB2 gene and the reference genomic region, 2p21. Results represent the mean of 3 experiments ± SD. (B) Expression of ERBB2 mRNA: Expression of ERBB2 mRNA in case #9 was compared between matched NE, BE and EAC samples using qRT-PCR. Expression levels were calculated as the ratio between ERBB2 and the reference gene, β-actin. Results represent the mean of 3 experiments ± SD.
Furthermore, we compared copy-number change to gene expression levels. Expression of ERBB2 mRNA was examined with quantitative reverse transcriptase PCR (qRT-PCR) in case #9, which showed amplification of this gene by SNP-chip and genomic Q-PCR. Levels of ERBB2 mRNA in the BE and EAC from case #9 were approximately 6-fold and 14-fold higher than in matched NE, respectively (Figure 3B). Taken together, these data suggested that copy-number changes resulted in aberrant gene expression.
Deletion of basonuclin 2 in Barrett’s sample
As shown in Figure 2A, the BE sample from case #11 clearly showed deletion involving the 9p22.3-p22.2 region (1.2 MB, circled); only one known gene, basonuclin 2 (BNC2), is located here. BNC2, a zinc finger protein, can bind to DNA and behave as a transcription factor.25–27 Genomic Q-PCR revealed that the level of BNC2 in BE sample #11 was approximately 3.5-fold lower than in matched NE (Figure 4A). The level of BNC2 mRNA in this BE sample was also approximately 3.5-fold lower than in matched NE (Figure 4B), consistent with decreased expression of BNC2 in this BE sample due to loss of this gene.
Figure 4. Deletion of basonuclin 2 in Barrett’s esophagus.
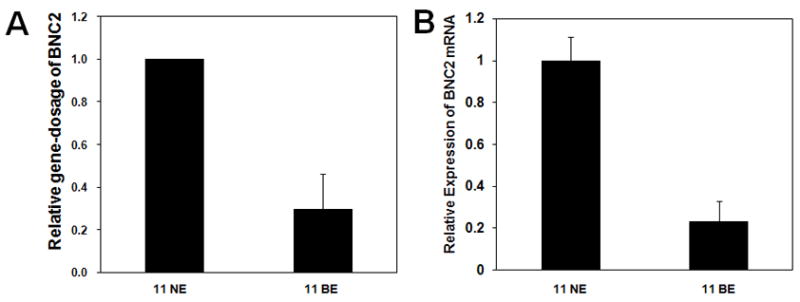
(A) Deletion of the basonuclin 2 (BNC2) gene: BE sample #11 exhibited deletion at 9p22.3-p22.2; the deleted region contained only the BNC2 gene. Gene dosage of BNC2 in the BE sample was compared to matched NE genomic DNA using genomic Q-PCR. Gene dosage levels were calculated as the ratio between the BNC2 gene and the reference genomic region, 2p21. Results represent the mean of 3 experiments ± SD. (B) Expression of BNC2 mRNA. Expression of BNC2 mRNA in case #11 BE was compared to its matched NE sample using qRT-PCR. Expression levels were calculated as the ratio between BNC2 and the reference gene, β-actin. Results represent the mean of 3 experiments ± SD.
These findings prompted us to examine forced expression of BNC2 in the EAC cell line, OE33. Clonogenic assays were used to assess the effect of BNC2 on the growth rate of OE33 cells, since these cells do not express endogenous BNC2 mRNA (data not shown). OE33 cells were transfected with either a BNC2 expression vector or an empty vector as a control. Each vector contained the neomycin resistance gene. Untransfected cells all died in 250 μg/ml G418 after 2 weeks. Therefore, cells were cultured in media containing 250 μg/ml G418 for 2 weeks and then stained to determine the number of surviving colonies. BNC2-transfected cells formed 60% fewer colonies than did empty vector-transfected controls (Figures 5B and 5C). Taken together, these results suggested that BNC2 inhibits clonal proliferation of EAC cells and is a potential EAC tumor suppressor gene.
Figure 5. Inhibition of proliferation of esophageal adenocarcinoma cells by BNC2.
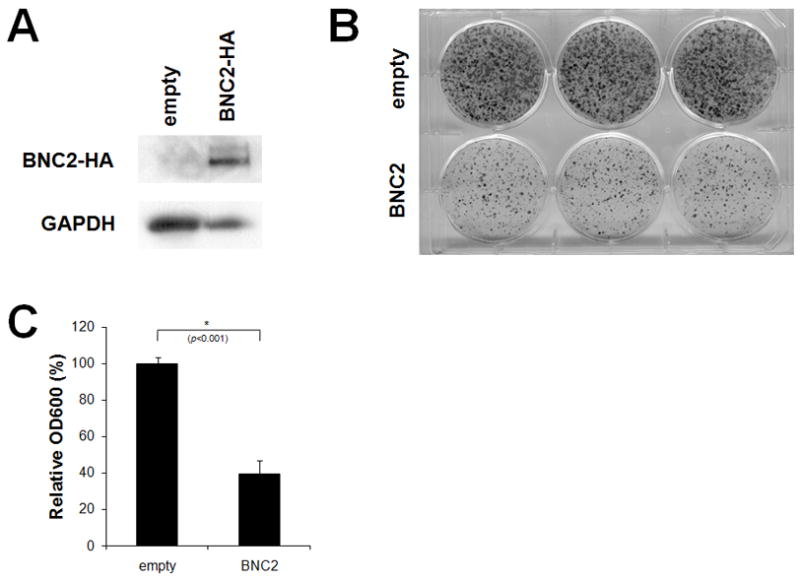
(A) Expression of exogenous HA-tagged BNC2. OE33 esophageal adenocarcinoma cells were transfected with either a BNC2 expression vector or an empty vector as a control. Expression of HA-tagged BNC2 was determined by Western blot analysis. GAPDH was used as an internal control. (B) Colony formation assay. Transfected cells were cultured in the presence of 250 μg/ml G418 for 2 weeks and then stained with 0.1% crystal violet to determine the number of surviving colonies. (C) Quantification of the number of surviving colonies. The colonies were dissolved in 1.0% SDS and absorbance was measured at 600 nm. Results were significantly different between control and BNC2-transfected cells (p<0.001).
Discussion
In the current study, we found that six (60%) of 10 BE and four (57%) of 7 EAC samples contained genomic alterations, including both copy-number changes and CNN-LOH. Analysis of matched NE, BE, and EAC sample sets revealed several chromosomal regions potentially involved in the progression from BE to EAC. Furthermore, we identified a zinc finger protein, BNC2, as a potential EAC tumor suppressor gene.
Our analyses identified deletions of 9p (4 cases) and 18q (2 cases) in BE samples and amplification of 17q (2 cases) in EAC samples. These findings are consistent with previous EAC results obtained by CGH analysis.4–11 We also found that the 17q12-q21.2 region was amplified in two EAC samples; this region includes the ERBB2, CSF3, RARA and TOP2A genes. ERBB2 is a transmembrane glycoprotein with tyrosine kinase activity, for which 10% to 70% of EAC samples show amplification.28–34 An antagonist of ERBB2, Trastuzumab/Herceptin, inhibits growth of the OE19 EAC cell line, which exhibits high expression of ERBB2.35 DNA topoisomerase II alpha (TOP2A) is associated with active cell proliferation, and its overexpression has been reported in a number of tumors, including esophageal squamous cell carcinoma.36,37 An inhibitor of topoisomerase II, etoposide, is used for chemotherapy of solid tumors including small-cell lung cancer. Data suggest that breast cancers with amplification of ERBB2 and TOP2A have a better response when they receive the combination of both Trastuzumab/Herceptin and a TOP2A inhibitor.38 In reference to our observed amplification of RARA, it is notable that retinoic acid, including all-trans retinoic acid (ATRA) and 9-cis retinoic acid (9-cis RA), induces the differentiation of acute promyelocytic leukemia cells and neuroblastoma cells and is used in treatment of these and other cancers.
Our SNP-chip analysis showed that the EAC from case #7 exhibited amplification of 6q22.33-q23.2 including the CTGF gene. This gene is involved in cell adhesion, migration, proliferation and angiogenesis.39 A recent study by our group demonstrated that 75% of esophageal squamous cell carcinomas overexpressed CTGF with accumulation of β-catenin in the nucleus40, supporting the importance of the β-catenin signaling pathway in esophageal tumorigenesis. In case #9, the BE sample contained only one chromosomal amplification (Xp22.13-p22.12), whereas the EAC sample from the same patient had numerous abnormalities including X-chromosomal alteration. This finding suggests that X-chromosomal alteration was acquired as an early event; this alteration may enhance the evolution from BE to EAC.
We also identified several CNN-LOH regions that included the entire 9p arm, 7q22.1-q36.3 (55.7 Mb), 17q25.3 (1.7 Mb), 18p11.32 (1.5 Mb) and 18q23 (1.9 Mb). CNN-LOH is a genomic abnormality that cannot normally be detected by karyotypic analysis; CNN-LOH regions often contain mutated genes. For example, the constitutively active forms of either JAK2 V617F mutant, FLT3-ITD, or an AML1/RUNX1 frameshift were found in a CNN-LOH region in AML cells from our SNP-chip analysis.24 The 7q22.1-q36.3 CNN-LOH region in BE contains the proto-oncogenes MET and BRAF. An activating mutation of MET (Y1253D) was detected in 15 (11%) of 138 oropharyngeal squamous cell carcinoma patients.41 An activating mutation of BRAF (V600E) was found in 66% of malignant melanomas, as well as at lower frequencies in a wide range of human cancers;42 in particular, 11% of Barrett’s EACs have been reported to possess BRAF mutations.43 These findings prompted us to determine exon sequences at mutational hotspots (MET Y1253 and BRAF V600) in sample #10; however, these genes did not contain detectable mutations (data not shown). Thus, further studies are indicated to identify key dysregulated or abnormal gene(s) in these regions.
In cancer cells, tumor suppressor genes are often inactivated by deletion, mutation and/or hypermethylation of their promoter regions. The cyclin-dependent kinase inhibitor gene, CDKN2A, is homozygously or hemizygously deleted, mutationally inactivated, or hypermethylated in approximately 50%, 5%, and 60% of EACs, respectively, and hypermethylated or hemizygously deleted in up to 40% of BEs.44–49 These findings are consistent with our identification of CDKN2A deletions in 4 BE samples (40%) and emphasize that CDKN2A inactivation represents an early event in the BE-EAC carcinogenic cascade. Similarly, the FHIT gene was deleted in 2 BE samples (20%). A prior report observed that 86% of BE and 93% of EAC samples exhibited altered FHIT expression.50 Although aberrant FHIT expression has also been detected in normal tissues,51 FHIT-deficient mice developed tumors in several tissues, suggesting that this gene functions as a tumor suppressor.52,53
The frequency of chromosomal changes in our study differed slightly from previous published CGH results; this discrepancy may have been due to high normal cell contamination of several of our samples or if several were triploidy and tetraploidy. This can be difficult to identify by SNP-chip analysis. Surprisingly, chromosomal alterations sometimes differed between BE and EAC samples. BE samples from cases #5 and #7 contained chromosomal changes in the 9p region, but these alterations disappeared in the corresponding EAC samples. One possible explanation for this conundrum is that the 9p alteration-bearing population was dominant in the BE sample; expansion of this clone altered the cellular microenvironment, thus enhancing the growth of a completely different (i.e., 9p-euploid) cellular population, which then became dominant in the EAC sample. Such clonal evolution has been previously reported in Barrett’s neoplastic evolution.54–58
Interestingly and to our knowledge for the first time, homozygous deletion of the BNC2 gene occurred in one BE sample. BNC2 is a zinc finger protein that is highly expressed in normal keratinocytes, ovary, testis, kidney and lung.25–27 BNC2 can bind to a sequence in the promoter region of the rRNA gene, and its protein is localized to the nucleus, suggesting that it functions as a transcriptional regulator.25–27 Also interestingly, the esophageal adenocarcinoma cell lines OE33 and OE19 did not express this gene (data not shown), and overexpression of BNC2 inhibited clonal proliferation of OE33 cells, suggesting that this gene has potential tumor-suppressive function. Inactivation by methylation and/or deletion of several TSGs, including CDKN2A and TP53, are known to be involved in esophageal tumorigenesis. Expression of BNC2 was low in a BE sample and the EAC cell lines. This inactivation could contribute to transformation to esophageal cancer cells. Recently, Nancarrow et al analyzed 23 EAC samples by SNP-chip.59 These investigators found several chromosomal lesions that were similar to our data, including homozygous deletions of CDKN2A and FHIT, loss of copy number at 17p, and CNN-LOH at 17q25.3, further emphasizing the importance of these regions or genes in the development of EAC.
In summary, we have demonstrated that BE frequently exhibits certain genomic and chromosomal alterations consistent with its precancerous state, often with early timing of these events. Furthermore, we have identified several novel genomic abnormalities in BE or EAC, notably CNN-LOH and inactivation of the zinc-finger gene, BNC2.
Supplementary Material
A lung cancer cell line NCI-H2171 and its paired lymphoblastoid cell line NCI-BL2171 were analyzed by SNP-chip analysis from both genomic and whole-genome amplified DNA. SNP-chip data from DNA with (+) or without (-) whole-genome amplification were compared. Genomic abnormalities are summarized by color: pink (copy-number-neutral LOH [CNN-LOH]); green (hemizygous deletion); red (duplication/amplification). Genomic changes found in the unamplified DNA aliquots were also detected in the amplified DNA aliquots.
Acknowledgments
We thank members of laboratories for helpful discussions. We are grateful to Dr. Satrajit Sinha, State University of New York at Buffalo, for his gift of the BNC2 expression vector. This work was supported by NIH grants CA85069, CA106763, CA 84986, and CA01808, as well as the Parker Hughes Fund. S.J.M. holds the Harry and Betty Myerberg/Thomas R. Hendrix Endowed Professorship in Gastroenterology and is a member of the Sidney Kimmel Comprehensive Cancer Center and the Cellular and Molecular Medicine Graduate Program at JHU. H.P.K. is the holder of the Mark Goodson endowed Chair in Oncology Research and is a member of the Jonsson Cancer Center and the Molecular Biology Institute, UCLA.
Footnotes
Brief statements
SNP-chip analysis revealed chromosomal abnormalities and novel disease-related genomic changes during progression from Barrett’s esophagus to esophageal adenocarcinoma. Zinc finger protein BNC2 is identified as a potential tumor suppressor gene in the esophagus.
References
- 1.Shaheen N, Ransohoff DF. Gastroesophageal reflux, Barrett’s esophagus, and esophageal cancer: scientific review. JAMA. 2002;287:1972–81. doi: 10.1001/jama.287.15.1972. [DOI] [PubMed] [Google Scholar]
- 2.Wijnhoven BP, Tilanus HW, Dinjens WN. Molecular biology of Barrett’s adenocarcinoma. Ann Surg. 2001;233:322–37. doi: 10.1097/00000658-200103000-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McManus DT, Olaru A, Meltzer SJ. Biomarkers of esophageal adenocarcinoma and Barrett’s esophagus. Cancer Res. 2004;64:1561–9. doi: 10.1158/0008-5472.can-03-2438. [DOI] [PubMed] [Google Scholar]
- 4.Moskaluk CA, Hu J, Perlman EJ. Comparative genomic hybridization of esophageal and gastroesophageal adenocarcinomas shows consensus areas of DNA gain and loss. Genes Chromosomes Cancer. 1998;22:305–11. [PubMed] [Google Scholar]
- 5.van Dekken H, Geelen E, Dinjens WN, Wijnhoven BP, Tilanus HW, Tanke HJ, Rosenberg C. Comparative genomic hybridization of cancer of the gastroesophageal junction: deletion of 14Q31–32.1 discriminates between esophageal (Barrett’s) and gastric cardia adenocarcinomas. Cancer Res. 1999;59:748–52. [PubMed] [Google Scholar]
- 6.van Dekken H, Vissers CJ, Tilanus HW, Tanke HJ, Rosenberg C. Clonal analysis of a case of multifocal oesophageal (Barrett’s) adenocarcinoma by comparative genomic hybridization. J Pathol. 1999;188:263–6. doi: 10.1002/(SICI)1096-9896(199907)188:3<263::AID-PATH374>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 7.Walch AK, Zitzelsberger HF, Bruch J, Keller G, Angermeier D, Aubele MM, Mueller J, Stein H, Braselmann H, Siewert JR, Höfler H, Werner M. Chromosomal imbalances in Barrett’s adenocarcinoma and the metaplasia-dysplasia-carcinoma sequence. Am J Pathol. 2000;156:555–66. doi: 10.1016/S0002-9440(10)64760-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Varis A, Puolakkainen P, Savolainen H, Kokkola A, Salo J, Nieminen O, Nordling S, Knuutila S. DNA copy number profiling in esophageal Barrett adenocarcinoma: comparison with gastric adenocarcinoma and esophageal squamous cell carcinoma. Cancer Genet Cytogenet. 2001;127:53–8. doi: 10.1016/s0165-4608(00)00423-4. [DOI] [PubMed] [Google Scholar]
- 9.Riegman PH, Vissers KJ, Alers JC, Geelen E, Hop WC, Tilanus HW, van Dekken H. Genomic alterations in malignant transformation of Barrett’s esophagus. Cancer Res. 2001;61:3164–70. [PubMed] [Google Scholar]
- 10.Croft J, Parry EM, Jenkins GJ, Doak SH, Baxter JN, Griffiths AP, Brown TH, Parry JM. Analysis of the premalignant stages of Barrett’s oesophagus through to adenocarcinoma by comparative genomic hybridization. Eur J Gastroenterol Hepatol. 2002;14:1179–86. doi: 10.1097/00042737-200211000-00004. [DOI] [PubMed] [Google Scholar]
- 11.Su M, Chin SF, Li XY, Edwards P, Caldas C, Fitzgerald RC. Comparative genomic hybridization of esophageal adenocarcinoma and squamous cell carcinoma cell lines. Dis Esophagus. 2006;19:10–4. doi: 10.1111/j.1442-2050.2006.00530.x. [DOI] [PubMed] [Google Scholar]
- 12.Nannya Y, Sanada M, Nakazaki K, Hosoya N, Wang L, Hangaishi A, Kurokawa M, Chiba S, Bailey DK, Kennedy GC, Ogawa S. A robust algorithm for copy number detection using high-density oligonucleotide single nucleotide polymorphism genotyping arrays. Cancer Res. 2005;65:6071–9. doi: 10.1158/0008-5472.CAN-05-0465. [DOI] [PubMed] [Google Scholar]
- 13.Engle LJ, Simpson CL, Landers JE. Using high-throughput SNP technologies to study cancer. Oncogene. 2006;25:1594–601. doi: 10.1038/sj.onc.1209368. [DOI] [PubMed] [Google Scholar]
- 14.Yamamoto G, Nannya Y, Kato M, Sanada M, Levine RL, Kawamata N, Hangaishi A, Kurokawa M, Chiba S, Gilliland DG, Koeffler HP, Ogawa S. Highly sensitive method for genomewide detection of allelic composition in nonpaired, primary tumor specimens by use of affymetrix single-nucleotide-polymorphism genotyping microarrays. Am J Hum Genet. 2007;81:114–26. doi: 10.1086/518809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pfeifer D, Pantic M, Skatulla I, Rawluk J, Kreutz C, Martens UM, Fisch P, Timmer J, Veelken H. Genome-wide analysis of DNA copy number changes and LOH in CLL using high-density SNP arrays. Blood. 2007;109:1202–10. doi: 10.1182/blood-2006-07-034256. [DOI] [PubMed] [Google Scholar]
- 16.Lehmann S, Ogawa S, Raynaud SD, Sanada M, Nannya Y, Ticchioni M, Bastard C, Kawamata N, Koeffler HP. Molecular allelokaryotyping of early-stage, untreated chronic lymphocytic leukemia. Cancer. 2008;112:1296–305. doi: 10.1002/cncr.23270. [DOI] [PubMed] [Google Scholar]
- 17.Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, Girtman K, Mathew S, Ma J, Pounds SB, Su X, Pui CH, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–64. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 18.Kawamata N, Ogawa S, Zimmermann M, Kato M, Sanada M, Hemminki K, Yamatomo G, Nannya Y, Koehler R, Flohr T, Miller CW, Harbott J, et al. Molecular allelokaryotyping of pediatric acute lymphoblastic leukemias by high-resolution single nucleotide polymorphism oligonucleotide genomic microarray. Blood. 2008;111:776–84. doi: 10.1182/blood-2007-05-088310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raghavan M, Lillington DM, Skoulakis S, Debernardi S, Chaplin T, Foot NJ, Lister TA, Young BD. Genome-wide single nucleotide polymorphism analysis reveals frequent partial uniparental disomy due to somatic recombination in acute myeloid leukemias. Cancer Res. 2005;65:375–8. [PubMed] [Google Scholar]
- 20.Fitzgibbon J, Smith LL, Raghavan M, Smith ML, Debernardi S, Skoulakis S, Lillington D, Lister TA, Young BD. Association between acquired uniparental disomy and homozygous gene mutation in acute myeloid leukemias. Cancer Res. 2005;65:9152–4. doi: 10.1158/0008-5472.CAN-05-2017. [DOI] [PubMed] [Google Scholar]
- 21.Tyybäkinoja A, Elonen E, Vauhkonen H, Saarela J, Knuutila S. Single nucleotide polymorphism microarray analysis of karyotypically normal acute myeloid leukemia reveals frequent copy-number-neutral loss of heterozygosity. Haematologica. 2008;93:631–2. doi: 10.3324/haematol.12232. [DOI] [PubMed] [Google Scholar]
- 22.Gorletta TA, Gasparini P, D’Elios MM, Trubia M, Pelicci PG, Di Fiore PP. Frequent loss of heterozygosity without loss of genetic material in acute myeloid leukemia with a normal karyotype. Genes Chromosomes Cancer. 2005;44:334–7. doi: 10.1002/gcc.20234. [DOI] [PubMed] [Google Scholar]
- 23.Fitzgibbon J, Smith LL, Raghavan M, Smith ML, Debernardi S, Skoulakis S, Lillington D, Lister TA, Young BD. Association between acquired uniparental disomy and homozygous gene mutation in acute myeloid leukemias. Cancer Res. 2005;65:9152–4. doi: 10.1158/0008-5472.CAN-05-2017. [DOI] [PubMed] [Google Scholar]
- 24.Akagi T, Ogawa S, Dugas M, Kawamata N, Yamamoto G, Nannya Y, Sanada M, Miller CW, Yung A, Schnittger S, Haferlach T, Haferlach C, et al. Frequent genomic abnormalities in acute myeloid leukemia/myelodysplastic syndrome with normal karyotype. Haematologica. 2009;94:213–23. doi: 10.3324/haematol.13024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vanhoutteghem A, Djian P. Basonuclin 2: an extremely conserved homolog of the zinc finger protein basonuclin. Proc Natl Acad Sci USA. 2004;101:3468–73. doi: 10.1073/pnas.0400268101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Romano RA, Li H, Tummala R, Maul R, Sinha S. Identification of Basonuclin2, a DNA-binding zinc-finger protein expressed in germ tissues and skin keratinocytes. Genomics. 2004;83:821–33. doi: 10.1016/j.ygeno.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 27.Vanhoutteghem A, Djian P. Basonuclins 1 and 2, whose genes share a common origin, are proteins with widely different properties and functions. Proc Natl Acad Sci USA. 2006;103:12423–8. doi: 10.1073/pnas.0605086103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakamura T, Nekarda H, Hoelscher AH, Bollschweiler E, Harbeck N, Becker K, Siewert JR, Harbeck N. Prognostic value of DNA ploidy and c-erbB-2 oncoprotein overexpression in adenocarcinoma of Barrett’s esophagus. Cancer. 1994;73:1785–94. doi: 10.1002/1097-0142(19940401)73:7<1785::aid-cncr2820730703>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 29.Jankowski J, Hopwood D, Wormsley KG. Expression of epidermal growth factor, transforming growth factor alpha and their receptor in gastro-oesophageal diseases. Dig Dis. 1993;11:1–11. doi: 10.1159/000171396. [DOI] [PubMed] [Google Scholar]
- 30.Hardwick RH, Barham CP, Ozua P, Newcomb PV, Savage P, Powell R, Rahamin J, Alderson D. Immunohistochemical detection of p53 and c-erbB-2 in oesophageal carcinoma; no correlation with prognosis. Eur J Surg Oncol. 1997;23:30–5. doi: 10.1016/s0748-7983(97)80139-4. [DOI] [PubMed] [Google Scholar]
- 31.Hardwick RH, Shepherd NA, Moorghen M, Newcomb PV, Alderson D. c-erbB-2 overexpression in the dysplasia/carcinoma sequence of Barrett’s oesophagus. J Clin Pathol. 1995;48:129–32. doi: 10.1136/jcp.48.2.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duhaylongsod FG, Gottfried MR, Iglehart JD, Vaughn AL, Wolfe WG. The significance of c-erb B-2 and p53 immunoreactivity in patients with adenocarcinoma of the esophagus. Ann Surg. 1995;221:677–84. doi: 10.1097/00000658-199506000-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fléjou JF, Paraf F, Muzeau F, Fékété F, Hénin D, Jothy S, Potet F. Expression of c-erbB-2 oncogene product in Barrett’s adenocarcinoma: pathological and prognostic correlations. J Clin Pathol. 1994;47:23–6. doi: 10.1136/jcp.47.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Polkowski W, van Sandick JW, Offerhaus GJ, ten Kate FJ, Mulder J, Obertop H, van Lanschot JJ. Prognostic value of Lauren classification and c-erbB-2 oncogene overexpression in adenocarcinoma of the esophagus and gastroesophageal junction. Ann Surg Oncol. 1999;6:290–7. doi: 10.1007/s10434-999-0290-2. [DOI] [PubMed] [Google Scholar]
- 35.Dahlberg PS, Jacobson BA, Dahal G, Fink JM, Kratzke RA, Maddaus MA, Ferrin LJ. ERBB2 amplifications in esophageal adenocarcinoma. Ann Thorac Surg. 2004;78:1790–800. doi: 10.1016/j.athoracsur.2004.05.037. [DOI] [PubMed] [Google Scholar]
- 36.Kim R, Ohi Y, Inoue H, Toge T. Expression and relationship between topoisomerase I and II alpha genes in tumor and normal tissues in esophageal, gastric and colon cancers. Anticancer Res. 1999;19:5393–8. [PubMed] [Google Scholar]
- 37.Ohashi Y, Sasano H, Yamaki H, Shizawa S, Kikuchi A, Shineha R, Akaishi T, Satomi S, Nagura H. Topoisomerase II alpha expression in esophageal squamous cell carcinoma. Anticancer Res. 1999;19:1873–80. [PubMed] [Google Scholar]
- 38.Pritchard KI, Messersmith H, Elavathil L, Trudeau M, O’Malley F, Dhesy-Thind B. HER-2 and topoisomerase II as predictors of response to chemotherapy. J Clin Oncol. 2008;26:736–44. doi: 10.1200/JCO.2007.15.4716. [DOI] [PubMed] [Google Scholar]
- 39.Igarashi A, Okochi H, Bradham DM, Grotendorst GR. Regulation of connective tissue growth factor gene expression in human skin fibroblasts and during wound repair. Mol Biol Cell. 1993;4:637–45. doi: 10.1091/mbc.4.6.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deng YZ, Chen PP, Wang Y, Yin D, Koeffler HP, Li B, Tong XJ, Xie D. Connective tissue growth factor is overexpressed in esophageal squamous cell carcinoma and promotes tumorigenicity through beta-catenin-T-cell factor/Lef signaling. J Biol Chem. 2007;282:36571–81. doi: 10.1074/jbc.M704141200. [DOI] [PubMed] [Google Scholar]
- 41.Aebersold DM, Landt O, Berthou S, Gruber G, Beer KT, Greiner RH, Zimmer Y. Prevalence and clinical impact of Met Y1253D-activating point mutation in radiotherapy-treated squamous cell cancer of the oropharynx. Oncogene. 2003;22:8519–23. doi: 10.1038/sj.onc.1206968. [DOI] [PubMed] [Google Scholar]
- 42.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, Davis N, Dicks E, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–54. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 43.Sommerer F, Vieth M, Markwarth A, Röhrich K, Vomschloss S, May A, Ell C, Stolte M, Hengge UR, Wittekind C, Tannapfel A. Mutations of BRAF and KRAS2 in the development of Barrett’s adenocarcinoma. Oncogene. 2004;23:554–8. doi: 10.1038/sj.onc.1207189. [DOI] [PubMed] [Google Scholar]
- 44.Esteve A, Martel-Planche G, Sylla BS, Hollstein M, Hainaut P, Montesano R. Low frequency of p16/CDKN2 gene mutations in esophageal carcinomas. Int J Cancer. 1996;66:301–4. doi: 10.1002/(SICI)1097-0215(19960503)66:3<301::AID-IJC5>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 45.Muzeau F, Flejou JF, Thomas G, Hamelin R. Loss of heterozygosity on chromosome 9 and p16 (MTS1, CDKN2) gene mutations in esophageal cancers. Int J Cancer. 1997;72:27–30. doi: 10.1002/(sici)1097-0215(19970703)72:1<27::aid-ijc3>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 46.Suzuki H, Zhou X, Yin J, Lei J, Jiang HY, Suzuki Y, Chan T, Hannon GJ, Mergner WJ, Abraham JM, Meltzer SJ. Intragenic mutations of CDKN2B and CDKN2A in primary human esophageal cancers. Hum Mol Genet. 1995;4:1883–7. doi: 10.1093/hmg/4.10.1883. [DOI] [PubMed] [Google Scholar]
- 47.Zhou X, Suzuki H, Shimada Y, Imamura M, Yin J, Jiang HY, Tarmin L, Abraham JM, Meltzer SJ. Genomic DNA and messenger RNA expression alterations of the CDKN2B and CDKN2 genes in esophageal squamous carcinoma cell lines. Genes Chromosomes Cancer. 1995;13:285–90. doi: 10.1002/gcc.2870130409. [DOI] [PubMed] [Google Scholar]
- 48.Wong DJ, Barrett MT, Stöger R, Emond MJ, Reid BJ. p16INK4a promoter is hypermethylated at a high frequency in esophageal adenocarcinomas. Cancer Res. 1997;57:2619–22. [PubMed] [Google Scholar]
- 49.Klump B, Hsieh CJ, Holzmann K, Gregor M, Porschen R. Hypermethylation of the CDKN2/p16 promoter during neoplastic progression in Barrett’s esophagus. Gastroenterology. 1998;115:1381–6. doi: 10.1016/s0016-5085(98)70016-2. [DOI] [PubMed] [Google Scholar]
- 50.Michael D, Beer DG, Wilke CW, Miller DE, Glover TW. Frequent deletions of FHIT and FRA3B in Barrett’s metaplasia and esophageal adenocarcinomas. Oncogene. 1997;15:1653–9. doi: 10.1038/sj.onc.1201330. [DOI] [PubMed] [Google Scholar]
- 51.Chen YJ, Chen PH, Lee MD, Chang JG. Aberrant FHIT transcripts in cancerous and corresponding non-cancerous lesions of the digestive tract. Int J Cancer. 1997;72:955–8. doi: 10.1002/(sici)1097-0215(19970917)72:6<955::aid-ijc6>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 52.Zanesi N, Fidanza V, Fong LY, Mancini R, Druck T, Valtieri M, Rudiger T, McCue PA, Croce CM, Huebner K. The tumor spectrum in FHIT-deficient mice. Proc Natl Acad Sci U S A. 2001;98:10250–5. doi: 10.1073/pnas.191345898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fujishita T, Doi Y, Sonoshita M, Hiai H, Oshima M, Huebner K, Croce CM, Taketo MM. Development of spontaneous tumours and intestinal lesions in Fhit gene knockout mice. Br J Cancer. 2004;91:1571–4. doi: 10.1038/sj.bjc.6602182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maley CC, Galipeau PC, Finley JC, Wongsurawat VJ, Li X, Sanchez CA, Paulson TG, Blount PL, Risques RA, Rabinovitch PS, Reid BJ. Genetic clonal diversity predicts progression to esophageal adenocarcinoma. Nat Genet. 2006;38:468–73. doi: 10.1038/ng1768. [DOI] [PubMed] [Google Scholar]
- 55.Barrett MT, Sanchez CA, Prevo LJ, Wong DJ, Galipeau PC, Paulson TG, Rabinovitch PS, Reid BJ. Evolution of neoplastic cell lineages in Barrett oesophagus. Nat Genet. 1999;22:106–9. doi: 10.1038/8816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wong DJ, Paulson TG, Prevo LJ, Galipeau PC, Longton G, Blount PL, Reid BJ. p16(INK4a) lesions are common, early abnormalities that undergo clonal expansion in Barrett’s metaplastic epithelium. Cancer Res. 2001;61:8284–9. [PubMed] [Google Scholar]
- 57.Blount PL, Meltzer SJ, Yin J, Huang Y, Krasna MJ, Reid BJ. Clonal ordering of 17p and 5q allelic losses in Barrett dysplasia and adenocarcinoma. Proc Natl Acad Sci USA. 1993;90:3221–5. doi: 10.1073/pnas.90.8.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boynton RF, Huang Y, Blount PL, Reid BJ, Raskind WH, Haggitt RC, Newkirk C, Resau JH, Yin J, McDaniel T, Meltzer SJ. Frequent loss of heterozygosity at the retinoblastoma locus in human esophageal cancers. Cancer Res. 1991;51:5766–9. [PubMed] [Google Scholar]
- 59.Nancarrow DJ, Handoko HY, Smithers BM, Gotley DC, Drew PA, Watson DI, Clouston AD, Hayward NK, Whiteman DC. Genome-wide copy number analysis in esophageal adenocarcinoma using high-density single-nucleotide polymorphism arrays. Cancer Res. 2008;68:4163–72. doi: 10.1158/0008-5472.CAN-07-6710. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A lung cancer cell line NCI-H2171 and its paired lymphoblastoid cell line NCI-BL2171 were analyzed by SNP-chip analysis from both genomic and whole-genome amplified DNA. SNP-chip data from DNA with (+) or without (-) whole-genome amplification were compared. Genomic abnormalities are summarized by color: pink (copy-number-neutral LOH [CNN-LOH]); green (hemizygous deletion); red (duplication/amplification). Genomic changes found in the unamplified DNA aliquots were also detected in the amplified DNA aliquots.


