Summmary
Efficient chromosome segregation during mitosis relies on the coordinated activity of molecular motors with proteins that regulate kinetochore attachments to dynamic spindle microtubules [1]. CLASPs are conserved kinetochore- and microtubule-associated proteins encoded by two paralogue genes, clasp1 and clasp2, and have been previously implicated in the regulation of kinetochore-microtubule dynamics [2–4]. However, it remains unknown how CLASPs work in concert with other proteins to form a functional kinetochore-microtubule interface. Here we have identified mitotic interactors of human CLASP1 using a proteomic approach. Among these, the microtubule plus-end directed motor CENP-E [5] was found to form a complex with CLASP1 that co-localizes to multiple structures of the mitotic apparatus in human cells. We found that CENP-E recruits both CLASP1 and CLASP2 to kinetochores independent of its motor activity or the presence of microtubules. Depletion of CLASPs or CENP-E by RNAi in human cells causes a significant and comparable reduction of kinetochore-microtubule poleward flux and turnover rates, as well as rescues spindle bipolarity in Kif2a–depleted cells. We conclude that CENP-E integrates two critical functions that are important for accurate chromosome movement and spindle architecture: one relying directly on its motor activity and the other involving the targeting of key microtubule regulators to kinetochores.
Keywords: Flux, Kinetochore, Microtubule turnover, Mitosis, Mitotic spindle
Results and Discussion
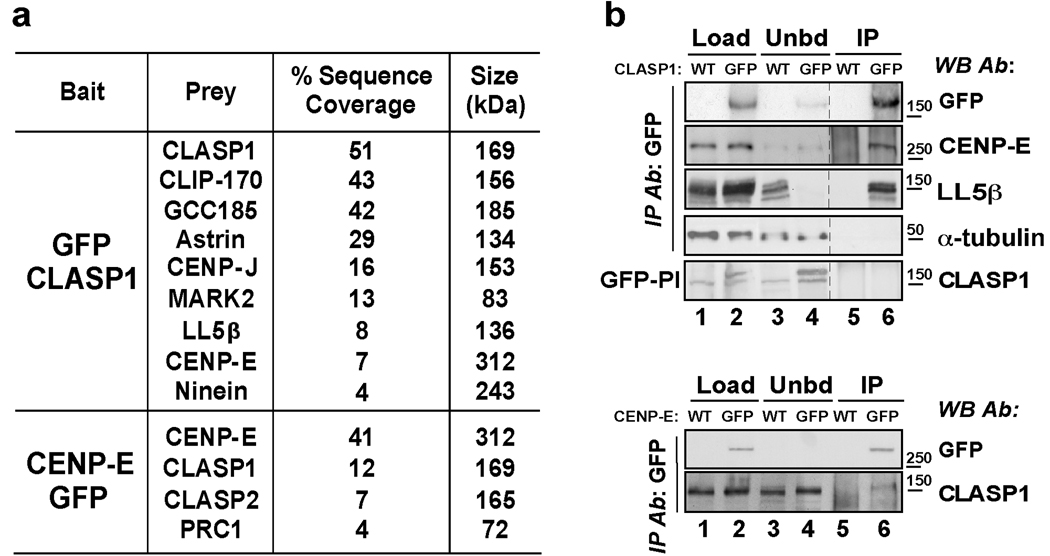
To shed light on the molecular context of human CLASPs during mitosis we identified CLASP1-interacting proteins from nocodazole arrested HeLa cells stably expressing LAP-tagged CLASP1 [4]. LAP purification [6] followed by mass spectrometry analysis recovered known CLASP1 interactors in mammals, such as CLIP-170, LL5β, GCC185 and Astrin [7–9] (Manning et al., submitted). Additionally, we identified CENP-E in our human CLASP1 purification, confirming previous results obtained in Xenopus meiotic egg extracts [10]. This approach also identified novel candidate CLASP1 binding partners, including the centriolar proteins CENP-J/CPAP and Ninein [11, 12], as well as MARK kinases [13] (Figure 1a).
Figure 1. Human CLASP1 interacts with CENP-E.
(a) Mass spectrometry analysis of affinity purified GFP-(LAP)-CLASP1 and CENP-E-GFP identifies novel protein interactions. Polypeptides identified (Prey) and the percentages of the relative sequence coverage are indicated. A complete list of the polypeptides identified during this analysis is given in Table S1. (b) Anti-GFP Immunoprecipitation from mitotic enriched HeLa cells stably expressing GFP-(LAP)-CLASP1 or CENP-E-GFP. Native protein extracts (Load) obtained from the indicated cell lines, unbound proteins (Unbd) and immunoprecipitations (IP) were subjected to western blot analysis with the indicated antibody. Immunoprecipitations were blotted for LL5β and α-tubulin as positive and negative controls. Immunoprecipitations performed using anti-GFP pre-immunization serum (GFP-PI) as precipitating antibody were analyzed by western blotting with rabbit anti-CLASP1 antibody. Quantification of CENP-E levels in the GFP-CLASP1 immunoprecipitation revealed +131% increase relative to control. Quantification of CLASP1 levels in the CENP-E-GFP immunoprecipitation revealed +135% increase relative to control.
From the full list of CLASP1 interactors, (Table S1) CENP-E is the only bona fide kinetochore (KT) protein [14]. Importantly, the functional significance of the CLASP1/CENP-E interaction remains unknown and therefore was selected for an in-depth analysis. We started by using mass spectrometry to confirm that endogenous CLASP1 co-purifies with CENP-E (Figure 1a). Notably, endogenous CLASP2 was also found in the purification, suggesting that CENP-E forms distinct complexes with CLASP1 and CLASP2 (Figure 1a). The reciprocal interaction of human CLASP1 with CENP-E was confirmed by Western blot after immunoprecipitation with anti-GFP antibodies in nocodazole arrested HeLa cells stably expressing GFP-CLASP1 or CENP-E-GFP (Figure 1b). Finally, immunofluorescence analysis showed that endogenous CLASP1 and CENP-E co-localize to multiple structures of the mitotic apparatus throughout mitosis, including centrosomes, KTs, spindle mid-zone and mid-body (Figure S1). Altogether these data suggest that CLASPs and CENP-E may be involved in functionally related aspects of mitosis.
Previous work in C. elegans, an organism lacking CENP-E orthologues, has shown that the CENP-F-like proteins HCP-1 and HCP-2 recruit CLASP to KTs [15]. However, as opposed to C. elegans, CENP-F depletion in human cells apparently does not affect CLASP1 KT-recruitment [16]. In order to test whether in human cells CENP-E could fulfill the task of targeting CLASPs to KTs, we depleted ∼80% of CENP-E from HeLa cells (Figure S2), which led to a significant reduction of CLASP1 (∼80%, n Luciferase RNAi=387 KTs from 18 cells; n CENP-E RNAi=579 KTs from 15 cells; p<0.001, Mann-Whitney test) and CLASP2 (∼65%, n Luciferase RNAi=332 KTs from 10 cells; n CENP-E RNAi=321 KTs from 10 cells; p<0.001, Mann-Whitney test) KT levels, in a microtubule (MT)-independent manner (Figure 2a, b and f, Figures S3 and S4). Importantly, CLASP1 and CLASP2 localization in the spindle and centrosomes was not affected by CENP-E depletion (Figure 2b, Figure 3b, and data not shown). On the contrary, depletion of both CLASPs from HeLa cells by RNAi (∼90% depletion, Figures S2 and S6) caused no measurable change in CENP-E localization at KTs (n Luciferase RNAi =350 KTs from 9 cells; n CLASPs-RNAi=348 KTs from 8 cells; p=0.438, Mann-Whitney test), regardless of the presence of MTs (Figure 2c, and Figures S3 and S6). Under these conditions, CLASP1 was completely removed from KTs, but a detectable RNAi resistant pool of stable protein remained associated with structures at spindle poles resembling centrioles (Figure 2c) [4]. Overall, these results indicate that CENP-E is required to specifically target a very significant pool of CLASP1 and CLASP2 to KTs.
Figure 2. CENP-E targets CLASP1 to KTs in a Motor-independent manner.
(a–c) Interdependency analysis of CLASP1 and CENP-E localization. HeLa cells treated with the indicated specific siRNA were prepared for chromosomes spreads in the presence of 10 µM nocodazole to fully depolymerize MTs. Following fixation, cells were stained for endogenous CLASP1 and CENP-E. ACA was used as an inner-KT marker and DNA was stained with DAPI. (d and e) The dependency of CLASP1 KT targeting from the motor domain of CENP-E was tested in HeLa cells transfected with a construct over-expressing a dominant negative motor-less GFP-CENP-E (GFP-CENP-E NΔ803) (d), or HeLa cells treated with UA62784 (e), an inhibitor of CENP-E ATPase activity. Cells were prepared for chromosomes spreads, fixed and stained for CLASP1 (d) or CLASP1 and CENP-E (e). For each cell, a magnification of an area containing KTs (colored as indicated) is shown in the smaller panels. Arrowheads indicate centrosomes. Scale bar= 5 µm. (f) Quantification of CLASP1/ACA KT fluorescence ratio in control (a), CENP-E RNAi (c), cells transfected with GFP-CENP-E NΔ803 (d) or treated with UA62784 (e).
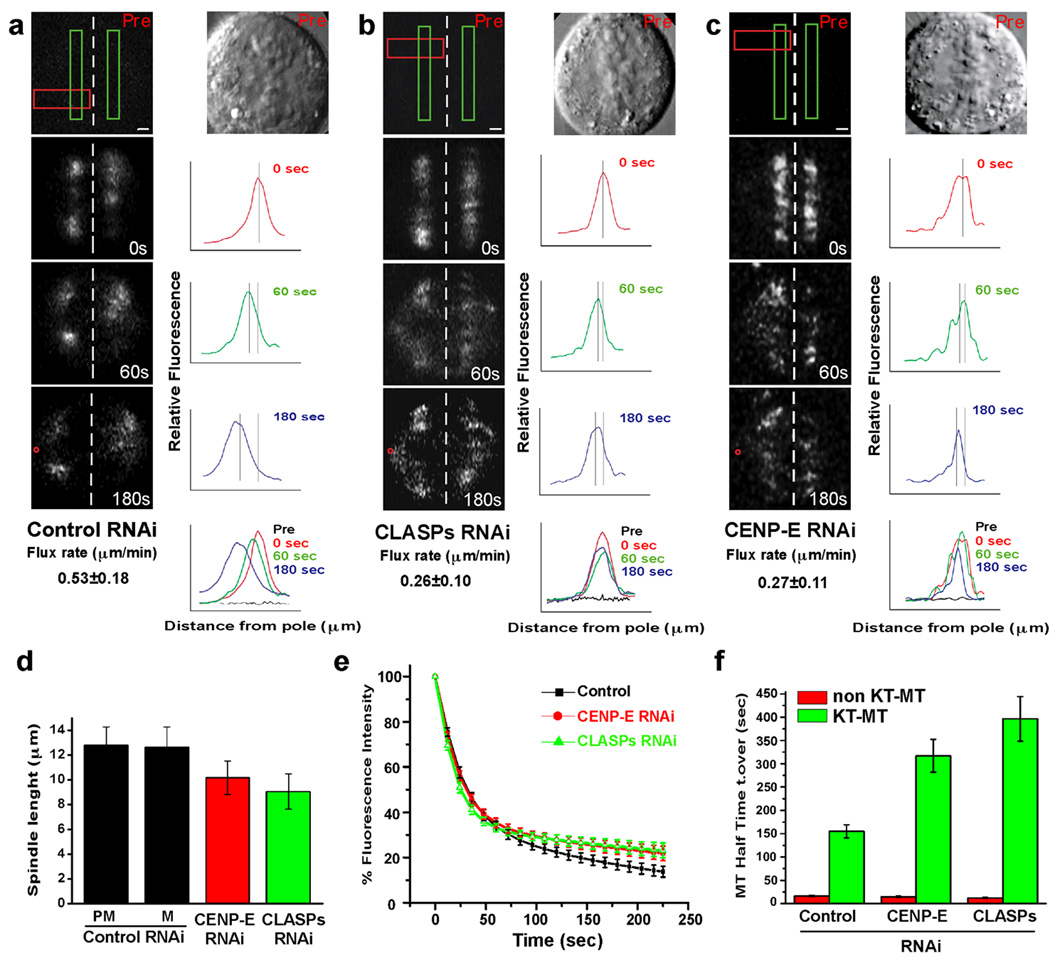
Figure 3. CENP-E targeting of CLASP1 to KTs is required to sustain normal KTMT dynamics.
(a–c) Human U2OS cells in late prometaphase/metaphase depleted for CLASPs or CENP-E display a significant reduction of KT MT poleward flux. Metaphase cells were identified by DIC and imaged (Pre) while flourescence images were captured before (Pre) and at various times (indicated in seconds) after photoactivation of GFP-α-tubulin with pulses from a 405 nm laser in one or two areas of the spindle (green rectangles). Line scans represent the relative fluorescence intensity of individual KT-MTs in a defined area (red rectangle) of the fluorescence images. Lines indicate the position of peak fluorescence intensity and flux rates were determined by plotting the position of peak fluorescence intensity as a function of time. Red circles indicate the position of spindle poles. Scale bar= 1 µm. (d) Cells deficient for CENP-E or CLASPs show shorter spindles (PM=Prometaphase; M=Metaphase). Data represent the mean±standard deviation. (e) Cells depleted for CENP-E or CLASPs show a significantly higher KT MT half time turnover. Normalized fluorescence intensity over time after photoactivation of U20S cells (in late PM/M) following treatment with the indicated siRNA. Data indicate mean±standard error corrected for background subtraction and photobleaching. (f) Calculated MT half time turnover for U2OS cells in e. Error bars represent standard error.
The MT independence of CENP-E-mediated targeting of CLASPs to KTs makes it unlikely to rely on the MT plus-end directed motor activity of CENP-E. To directly test this prediction we quantified CLASP1 KT levels in HeLa cells over-expressing a motor-less CENP-E construct (GFP-CENP-E NΔ803), which causes a dominant-negative effect by preventing endogenous CENP-E from assembling onto KTs [17] and recruits CLASP1 to many cytoplasmic aggregates (Figure S5). Under these conditions, CLASP1 KT levels were similar to non-transfected control cells (Figure 2d and f, n non-transfected= 322 KTs from 8 cells; n transfected=308 KTs from 8 cells). To confirm these results, we used a recently identified fluorenone, UA62784, reported to inhibit CENP-E ATPase activity but not its KT localization [18]. HeLa cells treated with 100 nM of UA62784 for 12 h showed normal CENP-E and CLASP1 localization at KTs (Figure 2e and f and Figure S3; n Control=314 KTs from 8 cells; n UA62784 treated=299 KTs from 8 cells). Overall, these results lead to two conclusions: 1) CENP-E motor domain is not required for interaction with CLASP1; and 2) recruitment of CLASP1 to KTs is a novel motor-independent function of CENP-E.
One remarkable feature of KTs is the capacity of constantly renewing their MT composition (i.e. KT-MT turnover), while allowing the poleward translocation/flux of attached MTs [19]. This is critical to ensure proper chromosome segregation and genomic stability by preventing the formation of incorrect KT-MT attachments [20, 21]. Studies in D. melanogaster culture cells showed that the single CLASP orthologue in this organism is required for the poleward translocation of MT subunits within KT-MTs [3]. To dissect the functional significance of the interaction between CLASPs and CENP-E in this process, we used pulses from a 405 nm laser to photoactivate GFP-α-tubulin stably expressed in human U2OS cells and measured the velocity at which the fluorescent mark activated in the proximity of chromosomes approached the pole. Consistent with previous reports [22], in control cells at late prometaphase/metaphase the fluorescent mark approached the pole with a mean velocity of 0.53 ± 0.18 µm/min (Figure 3a, Table 1 and Movie S1), with cells entering anaphase with normal kinetics after photoactivation (data not shown). In contrast, after RNAi depletion of ∼90% of CLASP1 or both CLASPs (Figure S2), the fluorescent mark approached the pole at 0.36 ± 0.09 µm/min and 0.26±0.10 µm/min, respectively (Figure 3b, Table 1 and Movie S1). Depletion of ∼80% of CENP-E by RNAi (Sup. Figure 2d) phenocopies the simultaneous depletion of both CLASPs, with the fluorescent mark approaching the pole at 0.27 ± 0.11 µm/min (Figure 3c, Table 1 and Movie S1). In a small subset of experiments we were successful in marking both half-spindles and noted a similar reduction in the rates that fluorescent marks on opposing KT-MTs move apart after CLASPs or CENP-E RNAi in comparison with controls (Table 1). Altogether, these results suggest that flux rates in human cells are sensitive to the KT levels of CLASP1 and CLASP2, which are largely determined by CENP-E. Curiously, loss of function of the single CLASP orthologue in Drosophila causes bipolar spindles to gradually collapse into monopolar spindles due to continuous depolymerization of MTs at their minus-ends, while tubulin subunit incorporation at the plus-ends is attenuated [3, 23]. This scenario is somewhat different from our knock-down of CLASPs (or CENP-E) in human cells, where spindles are 20–30% shorter than control cells in prometaphase or metaphase (n Luciferase RNAi=29; n CLASPs RNAi=28; n CENP-E RNAi=13; p<0.001, t-test), but only rarely form monopolar spindles (Figure 3d; [24] and our unpublished observations). However, anti-CLASP1 antibody injections in HeLa cells did cause the formation of monopolar spindles [2], suggesting that some residual function of CLASPs after RNAi is still sufficient to prevent the full collapse of the spindle, while allowing some poleward MT flux.
Table I.
Poleward MT flux rates in Photoactivatable-GFP-α-Tubulin U2OS cells
| Control | CLASP1 RNAi | CLASPs RNAi | CENP-E RNAi | |
|---|---|---|---|---|
| Mark-to-pole (µm/min) | 0.53±0.18 (28) | 0.36±0.09 (8;<0.001) | 0.26±0.10 (19; <0.001) | 0.27±0.11 (13; <0.001) |
| Mark-away-from-mark on opposing KT MTs/2 (µm/min) | 0.53±0.07 (3) | 0.26±0.07 (4; <0.001) | 0.28±0.15 (3; 0.002) | 0.23±0.02 (3; 0.002) |
The values indicate mean ± standard deviation; n and p are given in parentheses.
We also determined how CLASPs and CENP-E affect KT MT turnover by measuring fluorescence loss on the photoactivated area over time, after background subtraction and photobleaching correction, and fitting the results to a double exponential curve [19–21, 25]. The fast-decay component has been interpreted to represent non-KT-MTs that rapidly lose their activated fluorescence, while the slower-decay component likely corresponds to the more stable KT MTs in which the activated fluorescence is more persistent (Figure 3e). Surprisingly, the calculated half-time turnover for KT-MTs in cells depleted for CLASPs or CENP-E was significantly higher (respectively 396.5 ± 48 sec; n=8, p<0.001, and 317.3 ± 35.2 sec; n=13, p<0.025) than in control cells (155.2±13.9 sec, n=14) (Figure 3e and f and Figure S2), suggesting increased KT-MT stability. Previous studies in mammalian cells lacking CENP-E or that were microinjected with function-blocking antibodies have shown 23–50% reduction in MT binding at KTs, which was interpreted as CENP-E being required to stabilize KT-MT attachments [26, 27]. However, depletion of CENP-E or CLASPs in HeLa cells did not prevent the formation of cold-stable KT-MTs (Figure S7), indicating that despite of a reduction in the number of KT-MTs, these are actually more stably attached, possibly through point contacts with core KMN components [28], but the capacity to recruit new MTs might be impaired. Importantly, the half-time turnover observed for non-KT MTs in control (16 ± 1.2 sec), CLASPs-depleted (11.9±0.9 sec) and CENP-E-depleted cells (14.6±1.2 sec) was similar, indicating that the contribution of these proteins to non-KT MT turnover is minor. Thus, CENP-E-mediated targeting of CLASPs to KTs renders attached MTs to exchange more rapidly and overall become less stable. Interestingly, both CLASPs and CENP-E levels at KTs decrease as cells progress from prometaphase to anaphase [2, 4, 29], consistent with the observation that KT MTs become less dynamic at anaphase onset [19]. One plausible explanation for how CLASPs render KT MTs less stable is by evoking their role in the recruitment of the kinesin-13 MT depolymerase Kif2b to KTs, thereby assisting in MT detachment as cells progress into metaphase [21, 30].
Depletion of Kif2a by RNAi in U2OS cells leads to the formation of monopolar spindles and bipolarity can be rescued with treatments that either disrupt the formation of KT-MTs or renders them less dynamic [21, 31]. In order to investigate whether CLASPs and CENP-E at KTs work in concert in the same molecular pathway required for spindle architecture we compared the efficiency of bipolar spindle formation after co-depleting CLASPs or CENP-E with Kif2a by RNAi (Figure S8). CLASP1 or double CLASPs depletion in U2OS cells is sufficient to rescue spindle bipolarity in ∼80% of Kif2a–deficient cells while maintaining KT-MTs (Figure S8 and our unpublished observations). This is likely to be due to a relatively low contribution of CLASP2 during mitosis in U2OS cells, when compared to HeLa cells [24]. Interestingly, a functional cooperation between CLASP and the kinesin-13 protein KLP10A has been previously observed in Drosophila S2 cells [32, 33], suggesting evolutionary conservation of spindle architecture in both systems. Finally, CENP-E depletion rescued spindle bipolarity in Kif2a–deficient cells in 85% of the cases (Figure S8), confirming that CLASPs and CENP-E at KTs work in the same pathway involved in spindle formation and maintenance.
Based on in vitro studies it has been proposed that CENP-E tethers KTs to dynamic MTs, independently of its ATPase activity [34]. Based on our functional data, this property of CENP-E can be explained in vivo by a novel motor-independent role in targeting CLASPs to KTs, thereby contributing to establish/maintain functional KT-MT attachments. Noteworthy, perturbation of CLASPs function does not give persistent mono-oriented chromosomes like many cells following CENP-E inibition [5, 17, 26, 35]. This suggests that there are some unique activities of CENP-E that are not fulfilled by CLASPs. In this context, we propose that CENP-E integrates at least two activities. One is to power (through its kinesin motor activity) chromosome alignment [35], a function that is somewhat independent of CLASPs because knocking down CLASPs does not lead to the same congression problems (i.e. persistent mono-oriented chromosomes). The second function of CENP-E is to recruit CLASPs to KTs to functionally modulate KT-MT dynamics, which explains why KT-MT flux and turnover change if either CLASPs or CENP-E are lost. On the basis of the more complex spindle phenotype associated with CLASPs depletion in HeLa cells there might be other CLASPs-interacting proteins involved in centriole and spindle function found in this study, which do not interact with CENP-E (our unpublished observations). Finally, the results presented in this paper challenge current models for how CENP-E communicates with the spindle-assembly checkpoint (SAC) [36–38]. According to these models, CENP-E has been seen to be important for the capture and stabilization of MTs at KTs, thereby contributing to SAC silencing. In light of the new data presented in this paper supporting that CENP-E destabilizes rather than stabilizes MT attachment, alternative models must be put forward. An attractive one might involve the participation of CENP-E in destabilizing erroneous MT-KT attachments under control of Aurora B [20], while antagonizing the MT-stabilizing role of BubR1 [39, 40]. Since Aurora B controls the targeting of BubR1, CENP-E and Kif2b to KTs [21, 41] it is a likely possibility that CLASPs are also important parameters under regulation by this pathway to make mitosis error-free.
Experimental Procedures
Cell culture, RNAi, Drug treatments, Transfections and Western Blot Analysis
Human HeLa cells, HeLa LAP-CLASP1 [4], HeLa CENP-E-GFP [42] and U2OS-PA-GFP-α-tubulin cells [22] were grown in DMEM at 37°C in the presence of 5% CO2 and supplemented with 10% fetal bovine serum (FBS) and antibiotics. Human CLASP1, CLASP2, CENP-E, Kif2a and Nuf2 levels were reduced using specific siRNA oligonucleotides as previously described [24, 31, 43, 44] (Manning et al., submitted). Phenotypes were analyzed and quantified 48 h and 72 h after RNAi treatment and protein depletion was monitored by western blot using the following antibodies: rabbit anti-CLASP1 1:1000 [45]; rat anti-CLASP1 1:200, rabbit anti-CENP-E 1:500 (Santa Cruz); anti-Kif2a [31]; anti-Eg5 (Manning et al., submitted) and mouse anti-α-tubulin 1:2000 (Sigma). HRP-conjugated secondary antibodies (Amersham) were visualized using the ECL system (Pierce). To enrich HeLa LAP-CLASP1 or HeLa CENP-E-GFP cell cultures for mitotic cells for mass spectrometry and immunoprecipitation analyses, 100 ng/mL or 5ng/mL nocodazole, respectively, was added to the media for 16 or 18 h before harvesting. For inhibition of CENP-E ATPase activity, 100 nM of UA62784 was added to the media for 12 h before the analysis [18]. For photobleaching correction in the MT half time turnover experiments, control cells were treated with 5 µm Taxol 10 min before starting imaging. Transfection of HeLa cells with GFP-CENP-E NΔ803 was performed as described [2] and analysed 48 h after transfection. Wild-type and clasp2 KO MEFs were grown as previously described [4].
Immunofluorescence
HeLa or U20S cells were processed for immunofluorescence as described previously [2, 31]. Primary antibodies used were: rabbit anti-CLASP1 1:300 [2]; sheep anti-CENP-E 1:500 (gift from W. Earnshaw, University of Edinburgh, Edinburgh, UK); rat anti-CLASP1 1:100; rat anti-CLASP2 1:100, rabbit anti-CENP-E 1:300 (Santa Cruz); human anti-ACA 1:1000 (gift from W. Earnshaw) and mouse anti-α-tubulin 1:2000 (Sigma). Immunofluorescence on chromosome spreads was performed as described [2]. Secondary antibodies used were Alexa 488, 568 and 647 1:2000 (Invitrogen, Molecular Probes) and DNA was counterstained with DAPI (1 µg/ml). For cold induced MT-depolymerisation experiments, HeLa cells were incubated 15 min on ice and 1 min with 0.2% Triton X-100 in PHEM buffer (60 mM Pipes, 25 mM Hepes, 10 mM EGTA, 2 mM MgCl2). After a 30 min fixation in 4% paraformaldehyde in PHEM buffer followed by a 5 min methanol post-fixation, cells were processed for immunofluorescence as described above.
Mass spectrometry and Immunoprecipitation
Native protein extracts from HeLa LAP-CLASP1 [4] or HeLa CENP-E-GFP [42] cell cultures enriched for mitotic cells by nocodazole treatment were prepared for mass spectrometry analysis as described [6, 46]. The presented list of CLASP1-interacting proteins was filtered for known common contaminants in LAP-purifications. Immunoprecipitation experiments were performed with aliquots of native protein extracts (3 mg of total protein in a total volume of 500 µl of IP buffer: 150 mM Kcl, 75mM Hepes p.H 7.5, 1.5 mM EGTA, 1.5 mM MgCl2, 10 % glycerol, 0.1 % NP40, protease inhibitors) prepared from HeLa LAP-CLASP1 or HeLa CENP-E-GFP cell cultures enriched for mitotic cells by incubation with nocodazole. Protein extracts were incubated with the precipitating antibody at 4 oC for 4 h on a rotating platform. Precipitating primary antibodies used were: rabbit anti-GFP 1:100 and rabbit anti-GFP pre-immunization serum (GFP-PI) 1:100. These extracts were then incubated with 40 µl of protein A-Sepharose for 2 h at 4 °C on a rotating platform. Samples were centrifuged, the supernatant was retained as unbound sample and the pelleted beads washed 3 times with washing buffer (IP buffer with 250 mM KCl). Precipitated proteins were removed from the beads by boiling 5 min in SDS sample buffer and subjected to electrophoresis followed by western blot analysis with the appropriate antibody: rabbit anti-GFP 1:1000; rabbit anti-CENP-E 1:100 (Santa Cruz); anti-LL5β 1:2000 (gift from Anna Akhmanova, Erasmus Medical Center, Rotterdam, The Netherlands); anti-α-tubulin 1:2000 (Sigma) and rabbit anti-CLASP1 1:1000 [45].
Fluorescence quantification at KTs
Protein accumulation at KTs of HeLa cells prepared for chromosome spreads and immunostained with rat anti-CLASP1 or rat anti-CLASP2, rabbit anti-CENP-E and human anti-ACA, was measured for individual KTs by quantification of the pixel gray levels of the focused z-plane within a region of interest (ROI). Background was measured outside the ROI and was subtracted to the measured fluorescent intensity inside the ROI. Results were normalized against a constitutive KT marker (ACA) using a custom routine written in Matlab.
GFP-α-tubulin photoactivation analysis
For photoactivaton studies, mitotic human U2OS cells stably expressing photoactivatable GFP-α-tubulin [22] were identified by DIC microscopy and after acquisition of a pre-activation frame, 2×0.8 sec pulses from a 405-nm laser were used to activate GFP-α-tubulin in one or two areas of ∼7 µm2 inside the spindle. Imaging was performed with a Leica SP2 spectral confocal with a 63×/1.4 NA objective lens with an additional zoom of 7x and images were acquired every 3 sec in the first 4 min and subsequently every 30 sec. For MT poleward flux experiments the quantification of fluorescence intensity of the activated areas and the quantification of flux rates were performed as described [22]. Quantification of MT half time turnover was performed as previously described [19–21, 25] where the background-subtracted fluorescence values of an activated area were corrected for photobleaching by determining the fluorescence loss in activated spindles from taxol-treated cells. The fluorescent values were normalized to the first time-point following photoactivation and averaged from different cells for each time-point. The kinetics of fluorescence loss after activation were fit to a double exponential curve and the regression analysis was performed as described [19–21, 25] using the Origin 6 software (Originlab, MA).
Spindle structure analysis
Spindle length from prometaphase or metaphase HeLa cells were calculated with ImageJ software by measuring the distance between spindle poles in 3D. CLASP1 or CENP-E staining at the poles was used as reference. Assays for the analysis of bipolar spindle formation after siRNA depletion of the indicated proteins was performed as described [31].
Supplementary Material
Table S1 – Complete list of the names of polypeptides identified during the mass spectrometry analysis of CLASP1 interactors (Figure 1a) and the percentages of sequence coverage that was obtained for each protein.
Figure S1 – Human CLASP1 colocalises with CENP-E at multiple locations of the mitotic apparatus. Immunodetection of endogenous CLASP1 (rabbit anti-CLASP1), CENP-E (sheep anti-CENP-E) and α-tubulin in fixed mitotic HeLa cells. Scale bar= 5 µm.
Figure S2 – Cells treated with specific siRNAs for CLASPs or CENP-E show significantly reduced levels of the target protein. (a–f) Western blot analysis of CLASPs and CENP-E depletion efficiency. Total cell extracts from HeLa cells (a and b) or U20S–PA-GFP-α-tubulin cells (c–f), untreated or treated with the indicated siRNA were run on SDS-PAGE and blotted with either rabbit anti-CLASP1, rabbit anti-CENP-E or mouse anti α-tubulin (loading control).
Figure S3 – CENP-E targets CLASP1 to KTs in a Motor-independent manner. (a–c) HeLa cells treated with the indicated specific siRNA were fixed and stained for endogenous CLASP1 and CENP-E. ACA was used as an inner-KT marker and DNA was stained with DAPI. For each cell, a magnification of an area containing KTs (colored as indicated) is shown in the smaller panels. Arrowheads indicate centrosomes. Scale bar= 5 µm. (d) Correlative quantification of CLASP1 and CENP-E KT levels in chromosome spreads prepared in the presence of 10 µM nocodazole from control, CLASPs RNAi, CENP-E RNAi, or cells treated with the CENP-E inhibitor UA62784.
Figure S4 – CENP-E targets CLASP2 to KTs. (a–b) HeLa cells treated with the indicated specific siRNA were prepared for chromosome spreads in the presence of 10 µM nocodazole to fully depolymerize MTs. Following fixation, cells were stained for endogenous CLASP2 and CENP-E. ACA was used as an inner-KT marker and DNA was stained with DAPI. For each cell, a magnification of an area containing KTs (colored as indicated) is shown in the smaller panels. Arrowheads indicate centrosomes. Scale bar= 5 µm. (c) Correlative quantification of CLASP2 and CENP-E KT levels in control (a) or CENP-E RNAi (b) cells
Figure S5 – Over-expression of GFP-CENP-EΔN803 causes a dominant-negative effect and recruits CLASP1 to cytoplasmic aggregates. (a) Mild overexpression of a dominant negative motor-less GFP-CENP-E (CENP-E NΔ803) in a HeLa cell. (b) Strong overexpression of the same dominant negative construct in a HeLa cell. Endogenous CLASP1 (green), CENP-E NΔ803 (red) and DNA (blue) panels alone are indicated. Scale bar= 5µm.
Figure S6 – Western blot characterization of anti-CLASP1 and anti-CLASP2 rat monoclonal antibodies. (a and b) Total cell protein extracts from HeLa cells either untreated or treated with the indicated siRNA, or transiently transfected with GFP-CLASP1 or GFP-CLASP2, were run on SDS-PAGE and blotted with mouse anti α-tubulin (loading control), rat anti-CLASP1 (a) or rat anti CLASP2 (b). Note that expression of GFP-CLASP1 or GFP-CLASP2 down-regulates the expression levels of endogenous CLASP1 or CLASP2. (b) Total cell protein extracts from WT or clasp2 KO Mouse Embryonic Fibroblasts (MEFs) [15] were run on SDS-PAGE and probed with mouse anti-α-tubulin (loading control) and rat anti-CLASP2.
Figure S7 – Depletion of CENP-E or CLASPs does not prevent the formation of cold-stable KT-MTs. Nuf2 RNAi was used as positive control. ACA was used as an inner-KT marker. For each cell, a magnification of an area containing KTs is shown in the small panels. Scale bar= 5 µm.
Figure S8 – CENP-E and CLASPs cooperate to ensure proper spindle structure. (a–h) CLASPs and CENP-E RNAi rescue spindle bipolarity in Kif2a-depleted cells. U2OS cells treated with the indicated siRNAs were fixed and stained for CLASP1 (rabbit anti-CLASP1; red), CENP-E (rabbit anti-CENP-E; red), MTs (green) and DNA (blue). (i) Quantification of the efficiency of bipolar spindle formation in cells treated with the indicated siRNAs. Numbers in the graph bars represent percentage of cells that form bipolar spindles (mean±standard deviation). (j and k) Cells treated with specific siRNA for Kif2a or CENP-E show significantly reduced levels of the target protein. Total cell extracts from U20S cells untreated or treated with the indicated siRNA were run on SDS-PAGE and either probed with anti-Kif2a antibody, anti-CENP-E antibody or with anti-Eg5 antibody (loading control).
Movie S1 - Human U2OS cells in late prometaphase/metaphase depleted for CLASP1, CLASPs or CENP-E display a significant reduction of KT MT poleward flux. Flourescence images were captured at various times after photoactivation of GFP-α-tubulin with pulses from a 405 nm laser in two areas of the spindle. Time zero corresponds to the first frame after photoactivation. Scale bar= 2µm; Time indicated in seconds. Three representative examples for each treatment are shown.
Acknowledgements
We thank I. Cheeseman for guidance and providing reagents for LAP purifications, A. J. Pereira and S. Bakhoum for the development of Matlab routines used in this paper and advice on the quantification of microtubule dynamics parameters, P. Sampaio for help with photoactivation and A. Akhmanova, T. Yen and W. Earnshaw for the generous gift of reagents. S.M., A.R.R.P., and A.L.P hold fellowships from Fundação para a Ciência e a Tecnologia (FCT) of Portugal (SFRH/BPD/26780/2006; SFRH/BD/32976/2006; SFRH/BD/25084/2005). Work in the lab of D.A.C. is supported by National Institutes of Health grant GM51542. Work in the laboratory of H.M. is supported by grants PTDC/BIA-BCM/66106/2006 and PTDC/SAU-OBD/66113/2006 from FCT and the Gulbenkian Programme on the Frontiers in Life Sciences.
Abbreviations
- KT
kinetochore
- MT
microtubule
- GFP
green fluorescent protein
- LAP
localization affinity purification
- SAC
Spindle-assembly checkpoint.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cheeseman IM, Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol. 2008;9:33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- 2.Maiato H, Fairley EA, Rieder CL, Swedlow JR, Sunkel CE, Earnshaw WC. Human CLASP1 is an outer kinetochore component that regulates spindle microtubule dynamics. Cell. 2003;113:891–904. doi: 10.1016/s0092-8674(03)00465-3. [DOI] [PubMed] [Google Scholar]
- 3.Maiato H, Khodjakov A, Rieder CL. Drosophila CLASP is required for the incorporation of microtubule subunits into fluxing kinetochore fibres. Nat Cell Biol. 2005;7:42–47. doi: 10.1038/ncb1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pereira AL, Pereira AJ, Maia AR, Drabek K, Sayas CL, Hergert PJ, Lince-Faria M, Matos I, Duque C, Stepanova T, et al. Mammalian CLASP1 and CLASP2 cooperate to ensure mitotic fidelity by regulating spindle and kinetochore function. Mol Biol Cell. 2006;17:4526–4542. doi: 10.1091/mbc.E06-07-0579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wood KW, Sakowicz R, Goldstein LS, Cleveland DW. CENP-E is a plus end-directed kinetochore motor required for metaphase chromosome alignment. Cell. 1997;91:357–366. doi: 10.1016/s0092-8674(00)80419-5. [DOI] [PubMed] [Google Scholar]
- 6.Cheeseman IM, Desai A. A combined approach for the localization and tandem affinity purification of protein complexes from metazoans. Sci STKE 2005. 2005:11. doi: 10.1126/stke.2662005pl1. [DOI] [PubMed] [Google Scholar]
- 7.Akhmanova A, Hoogenraad CC, Drabek K, Stepanova T, Dortland B, Verkerk T, Vermeulen W, Burgering BM, De Zeeuw CI, Grosveld F, et al. Clasps are CLIP-115 and −170 associating proteins involved in the regional regulation of microtubule dynamics in motile fibroblasts. Cell. 2001;104:923–935. doi: 10.1016/s0092-8674(01)00288-4. [DOI] [PubMed] [Google Scholar]
- 8.Lansbergen G, Grigoriev I, Mimori-Kiyosue Y, Ohtsuka T, Higa S, Kitajima I, Demmers J, Galjart N, Houtsmuller AB, Grosveld F, et al. CLASPs attach microtubule plus ends to the cell cortex through a complex with LL5beta. Dev Cell. 2006;11:21–32. doi: 10.1016/j.devcel.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 9.Efimov A, Kharitonov A, Efimova N, Loncarek J, Miller PM, Andreyeva N, Gleeson P, Galjart N, Maia AR, McLeod IX, et al. Asymmetric CLASP-dependent nucleation of noncentrosomal microtubules at the trans-Golgi network. Dev Cell. 2007;12:917–930. doi: 10.1016/j.devcel.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hannak E, Heald R. Xorbit/CLASP links dynamic microtubules to chromosomes in the Xenopus meiotic spindle. J Cell Biol. 2006;172:19–25. doi: 10.1083/jcb.200508180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delgehyr N, Sillibourne J, Bornens M. Microtubule nucleation and anchoring at the centrosome are independent processes linked by ninein function. J Cell Sci. 2005;118:1565–1575. doi: 10.1242/jcs.02302. [DOI] [PubMed] [Google Scholar]
- 12.Kleylein-Sohn J, Westendorf J, Le Clech, M., Habedanck R, Stierhof YD, Nigg EA. Plk4-induced centriole biogenesis in human cells. Dev Cell. 2007;13:190–202. doi: 10.1016/j.devcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 13.Drewes G, Ebneth A, Preuss U, Mandelkow EM, Mandelkow E. MARK, a novel family of protein kinases that phosphorylate microtubule-associated proteins and trigger microtubule disruption. Cell. 1997;89:297–308. doi: 10.1016/s0092-8674(00)80208-1. [DOI] [PubMed] [Google Scholar]
- 14.Cooke CA, Schaar B, Yen TJ, Earnshaw WC. Localization of CENP-E in the fibrous corona and outer plate of mammalian kinetochores from prometaphase through anaphase. Chromosoma. 1997;106:446–455. doi: 10.1007/s004120050266. [DOI] [PubMed] [Google Scholar]
- 15.Cheeseman IM, MacLeod I, Yates JR, 3rd, Oegema K, Desai A. The CENP-F-like proteins HCP-1 and HCP-2 target CLASP to kinetochores to mediate chromosome segregation. Curr Biol. 2005;15:771–777. doi: 10.1016/j.cub.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 16.Bomont P, Maddox P, Shah JV, Desai AB, Cleveland DW. Unstable microtubule capture at kinetochores depleted of the centromere-associated protein CENP-F. Embo J. 2005;24:3927–3939. doi: 10.1038/sj.emboj.7600848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schaar BT, Chan GK, Maddox P, Salmon ED, Yen TJ. CENP-E function at kinetochores is essential for chromosome alignment. J Cell Biol. 1997;139:1373–1382. doi: 10.1083/jcb.139.6.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henderson MC, Shaw YJ, Wang H, Han H, Hurley LH, Flynn G, Dorr RT, Von Hoff DD. UA62784, a novel inhibitor of centromere protein E kinesin-like protein. Mol Cancer Ther. 2009;8:36–44. doi: 10.1158/1535-7163.MCT-08-0789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhai Y, Kronebusch PJ, Borisy GG. Kinetochore microtubule dynamics and the metaphase-anaphase transition. J Cell Biol. 1995;131:721–734. doi: 10.1083/jcb.131.3.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cimini D, Wan X, Hirel CB, Salmon ED. Aurora kinase promotes turnover of kinetochore microtubules to reduce chromosome segregation errors. Curr Biol. 2006;16:1711–1718. doi: 10.1016/j.cub.2006.07.022. [DOI] [PubMed] [Google Scholar]
- 21.Bakhoum SF, Thompson SL, Manning AL, Compton DA. Genome stability is ensured by temporal control of kinetochore-microtubule dynamics. Nat Cell Biol. 2009;11:27–35. doi: 10.1038/ncb1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ganem NJ, Upton K, Compton DA. Efficient mitosis in human cells lacking poleward microtubule flux. Curr Biol. 2005;15:1827–1832. doi: 10.1016/j.cub.2005.08.065. [DOI] [PubMed] [Google Scholar]
- 23.Maiato H, Sampaio P, Lemos CL, Findlay J, Carmena M, Earnshaw WC, Sunkel CE. MAST/Orbit has a role in microtubule-kinetochore attachment and is essential for chromosome alignment and maintenance of spindle bipolarity. J Cell Biol. 2002;157:749–760. doi: 10.1083/jcb.200201101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mimori-Kiyosue Y, Grigoriev I, Sasaki H, Matsui C, Akhmanova A, Tsukita S, Vorobjev I. Mammalian CLASPs are required for mitotic spindle organization and kinetochore alignment. Genes Cells. 2006;11:845–857. doi: 10.1111/j.1365-2443.2006.00990.x. [DOI] [PubMed] [Google Scholar]
- 25.DeLuca JG, Gall WE, Ciferri C, Cimini D, Musacchio A, Salmon ED. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127:969–982. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- 26.McEwen BF, Chan GK, Zubrowski B, Savoian MS, Sauer MT, Yen TJ. CENP-E is essential for reliable bioriented spindle attachment, but chromosome alignment can be achieved via redundant mechanisms in mammalian cells. Mol Biol Cell. 2001;12:2776–2789. doi: 10.1091/mbc.12.9.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Putkey FR, Cramer T, Morphew MK, Silk AD, Johnson RS, McIntosh JR, Cleveland DW. Unstable kinetochore-microtubule capture and chromosomal instability following deletion of CENP-E. Dev Cell. 2002;3:351–365. doi: 10.1016/s1534-5807(02)00255-1. [DOI] [PubMed] [Google Scholar]
- 28.Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- 29.Hoffman DB, Pearson CG, Yen TJ, Howell BJ, Salmon ED. Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores. Mol Biol Cell. 2001;12:1995–2009. doi: 10.1091/mbc.12.7.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manning AL, Ganem NJ, Bakhoum SF, Wagenbach M, Wordeman L, Compton DA. The kinesin-13 proteins Kif2a, Kif2b, and Kif2c/MCAK have distinct roles during mitosis in human cells. Mol Biol Cell. 2007;18:2970–2979. doi: 10.1091/mbc.E07-02-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ganem NJ, Compton DA. The KinI kinesin Kif2a is required for bipolar spindle assembly through a functional relationship with MCAK. J Cell Biol. 2004;166:473–478. doi: 10.1083/jcb.200404012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laycock JE, Savoian MS, Glover DM. Antagonistic activities of Klp10A and Orbit regulate spindle length, bipolarity and function in vivo. J Cell Sci. 2006;119:2354–2361. doi: 10.1242/jcs.02957. [DOI] [PubMed] [Google Scholar]
- 33.Buster DW, Zhang D, Sharp DJ. Poleward tubulin flux in spindles: regulation and function in mitotic cells. Mol Biol Cell. 2007;18:3094–3104. doi: 10.1091/mbc.E06-11-0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lombillo VA, Nislow C, Yen TJ, Gelfand VI, McIntosh JR. Antibodies to the kinesin motor domain and CENP-E inhibit microtubule depolymerization-dependent motion of chromosomes in vitro. J Cell Biol. 1995;128:107–115. doi: 10.1083/jcb.128.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kapoor TM, Lampson MA, Hergert P, Cameron L, Cimini D, Salmon ED, McEwen BF, Khodjakov A. Chromosomes can congress to the metaphase plate before biorientation. Science. 2006;311:388–391. doi: 10.1126/science.1122142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abrieu A, Kahana JA, Wood KW, Cleveland DW. CENP-E as an essential component of the mitotic checkpoint in vitro. Cell. 2000;102:817–826. doi: 10.1016/s0092-8674(00)00070-2. [DOI] [PubMed] [Google Scholar]
- 37.Mao Y, Desai A, Cleveland DW. Microtubule capture by CENP-E silences BubR1-dependent mitotic checkpoint signaling. J Cell Biol. 2005;170:873–880. doi: 10.1083/jcb.200505040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yao X, Abrieu A, Zheng Y, Sullivan KF, Cleveland DW. CENP-E forms a link between attachment of spindle microtubules to kinetochores and the mitotic checkpoint. Nat Cell Biol. 2000;2:484–491. doi: 10.1038/35019518. [DOI] [PubMed] [Google Scholar]
- 39.Lampson MA, Kapoor TM. The human mitotic checkpoint protein BubR1 regulates chromosome-spindle attachments. Nat Cell Biol. 2005;7:93–98. doi: 10.1038/ncb1208. [DOI] [PubMed] [Google Scholar]
- 40.Maia AF, Lopes CS, Sunkel CE. BubR1 and CENP-E have antagonistic effects upon the stability of microtubule-kinetochore attachments in Drosophila S2 cell mitosis. Cell Cycle. 2007;6:1367–1378. doi: 10.4161/cc.6.11.4271. [DOI] [PubMed] [Google Scholar]
- 41.Ditchfield C, Johnson VL, Tighe A, Ellston R, Haworth C, Johnson T, Mortlock A, Keen N, Taylor SS. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J Cell Biol. 2003;161:267–280. doi: 10.1083/jcb.200208091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poser I, Sarov M, Hutchins JR, Heriche JK, Toyoda Y, Pozniakovsky A, Weigl D, Nitzsche A, Hegemann B, Bird AW, et al. BAC TransgeneOmics: a high-throughput method for exploration of protein function in mammals. Nat Methods. 2008;5:409–415. doi: 10.1038/nmeth.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harborth J, Elbashir SM, Bechert K, Tuschl T, Weber K. Identification of essential genes in cultured mammalian cells using small interfering RNAs. J Cell Sci. 2001;114:4557–4565. doi: 10.1242/jcs.114.24.4557. [DOI] [PubMed] [Google Scholar]
- 44.DeLuca JG, Moree B, Hickey JM, Kilmartin JV, Salmon ED. hNuf2 inhibition blocks stable kinetochore-microtubule attachment and induces mitotic cell death in HeLa cells. J Cell Biol. 2002;159:549–555. doi: 10.1083/jcb.200208159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mimori-Kiyosue Y, Grigoriev I, Lansbergen G, Sasaki H, Matsui C, Severin F, Galjart N, Grosveld F, Vorobjev I, Tsukita S, et al. CLASP1 and CLASP2 bind to EB1 and regulate microtubule plus-end dynamics at the cell cortex. J Cell Biol. 2005;168:141–153. doi: 10.1083/jcb.200405094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Link AJ, Eng J, Schieltz DM, Carmack E, Mize GJ, Morris DR, Garvik BM, Yates JR., 3rd Direct analysis of protein complexes using mass spectrometry. Nat Biotechnol. 1999;17:676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 – Complete list of the names of polypeptides identified during the mass spectrometry analysis of CLASP1 interactors (Figure 1a) and the percentages of sequence coverage that was obtained for each protein.
Figure S1 – Human CLASP1 colocalises with CENP-E at multiple locations of the mitotic apparatus. Immunodetection of endogenous CLASP1 (rabbit anti-CLASP1), CENP-E (sheep anti-CENP-E) and α-tubulin in fixed mitotic HeLa cells. Scale bar= 5 µm.
Figure S2 – Cells treated with specific siRNAs for CLASPs or CENP-E show significantly reduced levels of the target protein. (a–f) Western blot analysis of CLASPs and CENP-E depletion efficiency. Total cell extracts from HeLa cells (a and b) or U20S–PA-GFP-α-tubulin cells (c–f), untreated or treated with the indicated siRNA were run on SDS-PAGE and blotted with either rabbit anti-CLASP1, rabbit anti-CENP-E or mouse anti α-tubulin (loading control).
Figure S3 – CENP-E targets CLASP1 to KTs in a Motor-independent manner. (a–c) HeLa cells treated with the indicated specific siRNA were fixed and stained for endogenous CLASP1 and CENP-E. ACA was used as an inner-KT marker and DNA was stained with DAPI. For each cell, a magnification of an area containing KTs (colored as indicated) is shown in the smaller panels. Arrowheads indicate centrosomes. Scale bar= 5 µm. (d) Correlative quantification of CLASP1 and CENP-E KT levels in chromosome spreads prepared in the presence of 10 µM nocodazole from control, CLASPs RNAi, CENP-E RNAi, or cells treated with the CENP-E inhibitor UA62784.
Figure S4 – CENP-E targets CLASP2 to KTs. (a–b) HeLa cells treated with the indicated specific siRNA were prepared for chromosome spreads in the presence of 10 µM nocodazole to fully depolymerize MTs. Following fixation, cells were stained for endogenous CLASP2 and CENP-E. ACA was used as an inner-KT marker and DNA was stained with DAPI. For each cell, a magnification of an area containing KTs (colored as indicated) is shown in the smaller panels. Arrowheads indicate centrosomes. Scale bar= 5 µm. (c) Correlative quantification of CLASP2 and CENP-E KT levels in control (a) or CENP-E RNAi (b) cells
Figure S5 – Over-expression of GFP-CENP-EΔN803 causes a dominant-negative effect and recruits CLASP1 to cytoplasmic aggregates. (a) Mild overexpression of a dominant negative motor-less GFP-CENP-E (CENP-E NΔ803) in a HeLa cell. (b) Strong overexpression of the same dominant negative construct in a HeLa cell. Endogenous CLASP1 (green), CENP-E NΔ803 (red) and DNA (blue) panels alone are indicated. Scale bar= 5µm.
Figure S6 – Western blot characterization of anti-CLASP1 and anti-CLASP2 rat monoclonal antibodies. (a and b) Total cell protein extracts from HeLa cells either untreated or treated with the indicated siRNA, or transiently transfected with GFP-CLASP1 or GFP-CLASP2, were run on SDS-PAGE and blotted with mouse anti α-tubulin (loading control), rat anti-CLASP1 (a) or rat anti CLASP2 (b). Note that expression of GFP-CLASP1 or GFP-CLASP2 down-regulates the expression levels of endogenous CLASP1 or CLASP2. (b) Total cell protein extracts from WT or clasp2 KO Mouse Embryonic Fibroblasts (MEFs) [15] were run on SDS-PAGE and probed with mouse anti-α-tubulin (loading control) and rat anti-CLASP2.
Figure S7 – Depletion of CENP-E or CLASPs does not prevent the formation of cold-stable KT-MTs. Nuf2 RNAi was used as positive control. ACA was used as an inner-KT marker. For each cell, a magnification of an area containing KTs is shown in the small panels. Scale bar= 5 µm.
Figure S8 – CENP-E and CLASPs cooperate to ensure proper spindle structure. (a–h) CLASPs and CENP-E RNAi rescue spindle bipolarity in Kif2a-depleted cells. U2OS cells treated with the indicated siRNAs were fixed and stained for CLASP1 (rabbit anti-CLASP1; red), CENP-E (rabbit anti-CENP-E; red), MTs (green) and DNA (blue). (i) Quantification of the efficiency of bipolar spindle formation in cells treated with the indicated siRNAs. Numbers in the graph bars represent percentage of cells that form bipolar spindles (mean±standard deviation). (j and k) Cells treated with specific siRNA for Kif2a or CENP-E show significantly reduced levels of the target protein. Total cell extracts from U20S cells untreated or treated with the indicated siRNA were run on SDS-PAGE and either probed with anti-Kif2a antibody, anti-CENP-E antibody or with anti-Eg5 antibody (loading control).
Movie S1 - Human U2OS cells in late prometaphase/metaphase depleted for CLASP1, CLASPs or CENP-E display a significant reduction of KT MT poleward flux. Flourescence images were captured at various times after photoactivation of GFP-α-tubulin with pulses from a 405 nm laser in two areas of the spindle. Time zero corresponds to the first frame after photoactivation. Scale bar= 2µm; Time indicated in seconds. Three representative examples for each treatment are shown.