Abstract
HuD is a neuronal RNA-binding protein associated with the stabilization of mRNAs for GAP-43 and other neuronal proteins that are important for nervous system development and learning and memory mechanisms. To better understand the function of this protein, we generated transgenic mice expressing human HuD (HuD-Tg) in adult forebrain neurons. We have previously shown that expression of HuD in adult dentate granule cells results in an abnormal accumulation of GAP-43 mRNA via post-transcriptional mechanisms. Here we show that this mRNA accumulation leads to the ectopic expression of GAP-43 protein in mossy fibers. Electrophysiological analyses of the mossy fiber to CA3 synapse of HuD-Tg mice revealed increases in paired-pulse facilitation (PPF) at short inter-pulse intervals and no change in long-term potentiation (LTP). Presynaptic calcium transients at the same synapses exhibited faster time constants of decay, suggesting a decrease in the endogenous Ca2+ buffer capacity of mossy fiber terminals of HuD-Tg mice. Under resting conditions, GAP-43 binds very tightly to calmodulin sequestering it and then releasing it upon PKC-dependent phosphorylation. Therefore subsequent studies examined the extent of GAP-43 phosphorylation and its association to calmodulin. We found that despite the increased GAP-43 expression in HuD-Tg mice, the levels of PKC-phosphorylated GAP-43 were decreased in these animals. Furthermore, in agreement with the increased proportion of non-phosphorylated GAP-43, HuD-Tg mice showed increased binding of calmodulin to this protein. These results suggest that a significant amount of calmodulin may be trapped in an inactive state, unable to bind free calcium and activate downstream signaling pathways. In conclusion, we propose that an unregulated expression of HuD disrupts mossy fiber physiology in adult mice in part by altering the expression and phosphorylation of GAP-43 and the amount of free calmodulin available at the synaptic terminal.
Keywords: HuD, GAP-43, mossy fiber, residual calcium, paired pulse facilitation
INTRODUCTION
Post-transcriptional regulation by RNA splicing, mRNA export, stability, localization, and translation add to the complexity of gene expression. These mechanisms are particularly relevant in neurons, where mRNAs are localized to axons and dendrites and thus segregated from the primary control of transcription (for reviews see, Sossin and DesGroseillers, 2006; Wang et al., 2007 and Schuman et al., 2006, and for specific examples, Smith et al., 2004). Among these post-transcriptional mechanisms, mRNA stability mechanisms are estimated to affect about 10 % of human genes (Bakheet et al., 2006). Hu proteins are homologs of Drosophila ELAV (embryonic lethal abnormal vision) and are the best known mRNA stabilizers in mammalian cells. Although there is only one ELAV protein in Drosophila, four mammalian ELAV-like proteins have been identified. Three of these proteins (HuB, HuC, and HuD) are developmentally regulated and expressed in neurons. The fourth member, HuR (also called HuA), is also expressed in other tissues. Although Hu proteins were initially described as early markers of neuronal differentiation (Barami et al., 1995), significant levels of these proteins persist throughout life, particularly in the neocortex and hippocampus (Okano and Darnell, 1997; Bolognani, 2004; Bolognani et al., 2007b. The recent findings that HuD expression increases during learning and memory (Quattrone et al., 2001; Bolognani, 2004; Pascale et al., 2004) and that overexpression results in cognitive deficits (Bolognani et al., 2007a) suggests that this RNA-binding protein may play an important role in mechanisms of synaptic plasticity in the adult CNS.
One of the best characterized targets of HuD is the mRNA of the growth- and plasticity-associated protein GAP-43. We have shown previously that HuD binds to a highly conserved U-rich motif in the GAP-43 3′UTR stabilizing the transcript (Kohn et al., 1996; Tsai et al., 1997). The HuD-dependent stabilization of the GAP-43 mRNA has significant physiological consequences as it enhances the levels of this protein and promotes neurite outgrowth and neuronal differentiation (Mobarak et al., 2000; Anderson et al., 2000; Anderson et al., 2001). In addition to its role in growth cone function, GAP-43 is known to have many functions in pre-synaptic terminals. In its non-phosphorylated state, GAP-43 is thought to serve as a calmodulin trap, sequestering this calcium-binding protein and keeping it inaccessible (reviewed by Liu and Storm, 1990 and Skene, 1990). GAP-43 is also a primary target of protein kinase C (PKC) (Akers and Routtenberg, 1985) and this activation releases calmodulin (Alexander et al., 1987) and allows for GAP-43 to facilitate neurotransmitter release (Dekker et al., 1989; Ivins et al., 1993) via its interactions with the synaptic core complex (Haruta et al., 1997) and the endocytotic machinery (Neve et al., 1998). Therefore, the pre-synaptic function of GAP-43 depends on the dynamic equilibrium between binding to calmodulin and releasing it upon PKC-dependent phosphorylation (for reviews see Benowitz and Routtenberg, 1997 and Perrone-Bizzozero, 2006).
Dentate granule cells (DGC) of the hippocampus express GAP-43 mRNA and HuD only during the first 2 weeks of postnatal development and these molecules are not detected in mature DGC (Meberg and Routtenberg, 1991; Bendotti et al., 1997; Okano and Darnell, 1997; Clayton, 1998; Bolognani et al., 2006; Bolognani et al., 2007b). This decline in GAP-43 expression is not due to a decrease in transcription as mature DGC continue to transcribe this gene at high levels, but to the post-transcriptional destabilization of the mRNA, presumably due to the absence of HuD (Namgung and Routtenberg, 2000; Bolognani et al., 2006). Supporting this idea, we have shown recently that adult transgenic mice in which HuD was overexpressed via the αCaMKin II promoter exhibited increased stabilization and accumulation of GAP-43 mRNA in dentate granule cells and other neuronal populations (Bolognani et al., 2006). Here we report that the HuD-dependent stabilization of the GAP-43 mRNA in dentate granule cells leads to ectopic GAP-43 expression in mossy fiber terminals, and that these changes are correlated with alterations in GAP-43 phosphorylation and calmodulin binding as well as decreased intracellular calcium buffering and increased paired-pulse facilitation.
METHODS AND MATERIALS
Transgenic mice
HuD transgenic mice were bred and genotyped by PCR as described before (Bolognani et al., 2006). Animals were backcrossed to C57Bl/6 background for at least 7 generations and adult male mice (3 – 5 months of age) were used for all studies. All procedures were approved by the University of New Mexico and Vanderbilt Medical Center Institutional Animal Care and Use Committee (IACUC) in compliance with the Guidelines for the Care and Use of Laboratory Animals established by the National Institutes of Health (NIH).
Western blot analysis
Hippocampal tissue was prepared as previously described (Tanner et al., 2004). Briefly, hippocampi were dissected on ice from fresh brains, frozen on dry ice, and stored at −80°C until further use. Tissues were homogenized in ice-cold buffered sucrose solution (20 mM Tris-HCl, pH 7.4, 0.32 M sucrose, 2 mM EDTA, 2 mM EGTA, 0.3 mM PMSF, 50 mM sodium fluoride, 2 μg/μl leupeptin, and 2 μg/μl aprotinin). Homogenates were centrifuged at 1000 × g for 4 min, and an aliquot of the resulting supernatant (S1) was centrifuged at 4°C at 17,000 × g, yielding a crude synaptosomal fraction (S2) and a membrane-associated pellet (P2). Protein was determined according to the method of Bradford (Bradford, 1976) using bovine serum albumin (Bio-Rad, Hercules, CA). Samples were resolved by 10% SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to PVDF membranes. Levels of total GAP-43, phosphorylated GAP-43, calbindin, and calmodulin were determined in the S1 fractions. The levels of GAP-43-bound calmodulin were measured in P2 fractions. Primary antibodies used were: sheep anti-GAP-43 (1:2000, Benowitz et al., 1988), rabbit anti-GAP-43 phosphoSer41 (1:1000; Chemicon, Temeculah, CA), vitamin D-dependent 28K calbindin (Calbindin-D28K,1:1000, Swant, Bellinzona, Switzerland) and mouse anti-calmodulin (1:1000; Upstate, Lake Placid, NY). Blots were developed with species appropriate HRP-conjugated secondary IgGs followed by Western Lightning enhanced chemiluminescence (Perkin-Elmer, Boston MA) and quantified on Fluor-S MultiImager (BioRad).
Immunohistochemistry
Mice were perfussed with 0.4% paraformaldehyde (PFA) in phosphate buffered saline and brains were dissected and cryoprotected for at least 24 hours in 30% sucrose in PBS. Sections (40 μm) were cut with a sliding microtome and processed free-floating. Hippocampal sections were incubated with sheep anti-GAP-43 (1:2000) (Benowitz et al., 1988), and rabbit anti-Calbindin D-28k (1:2000) (Swant, Bellinzona, Switzerland) followed by incubation with anti-sheep Alexa 633 and anti-rabbit Alexa 568 secondary antibodies (both at 1:100) (Molecular Probes, Eugene, OR). Epifluorescent images were taken on a BioRad BX-60 microscope with either an Optronics MagnaFire or an Olympus DP71 CCD-digital camera and associated image analysis software (Olympus America Inc., Melville, NY).
Calmodulin-GAP-43 Binding studies
The interaction of GAP-43 with calmodulin was measured by immunoprecipation with anti-GAP-43 antibodies followed by western blots of the precipitated complex. Briefly, a 50% slurry of protein G magnetic beads (Dynal beads, Invitrogen Corporation Carlsbad, California) was incubated overnight at 4°C with 10 μl of sheep anti-GAP-43 antiserum (Benowitz et al., 1988). The antibody-bound beads were then incubated for 2 hours with 50 μg of total brain protein extract in 1 ml of NP40 buffer (1%NP-40, 0.5% BSA, 2mM EDTA, 10 mM Tris-HCl pH 7.5, and 0.15 M NaCl). Beads were washed 4 times with NP-40 buffer and twice with high salt NP40 buffer (the same as above but with 0.5 M NaCl). Protein was extracted, resolved in SDS-PAGE, and transferred to PVDF membranes. The amount of precipitated GAP-43 was measured by western blot. The optical density of the GAP-43 band in the first gel was used to correct the loading of a second gel, which contained equal amounts of GAP-43 in each lane. This second gel was used to measure the amount of calmodulin bound to GAP-43 in control and transgenic mice.
Electrophysiology
Transverse hippocampal slices were prepared from adult mice of different genotypes according to standard procedures. Briefly, mice were killed by cervical dislocation and decapitated. The brains were quickly removed and placed in ice-cold high sucrose cutting solution containing (in mM) 110 sucrose, 6 NaCl, 3 KCl, 26 NaHCO3, 1.25 NaH2PO4, 7 MgCl2, 0.5 CaCl2, 0.6 sodium ascorbate, and 10 glucose, pH 7.3–7.4. Horizontal slices (350–400μm) were cut in high sucrose cutting solution using a vibratome. The slices were kept at RT in ACSF containing (in mM): 125 NaCl, 2.5 KCl, 1.25 NaH2PO4, 26 NaHCO3, 1.2 MgCl2, 2.0 CaCl2, and 10 glucose (pH 7.4), saturated with 95% O2/5% CO2. The slices were allowed to recover for at least 1 hour before being transferred to the recording chamber, which was kept at 32 ± 0.5°C with a laminar ACSF flow rate of 2–3 ml/min. To measure mossy fiber-CA3 synapse responses, field excitatory postsynaptic potentials (fEPSP) were recorded in the CA3 stratum radiatum layer. Recording glass micropipettes were filled with 3 M NaCl and had 1–4 MΩ resistance. Bipolar stimulating electrodes were constructed from twisted insulated stainless-steel wires and were driven by biphasic electrical stimulus generated by the Digidata 1322A interface (Molecular Devices, Sunnyvale, CA) and a stimulus isolator (model 2200, A-M Systems, Carlsborg, WA) under control of Clampex 9.0 software (Molecular Devices, Sunnyvale, CA). Recorded electrical signals were amplified using a differential amplifier (model 1800, A-M Systems, Carlsborg, WA), filtered at 1 kHz and digitized at 10 kHz.
Calcium Imaging
Hippocampal slices were cut at 300 μm on a vibratome from the brains of mice whose genotype was unknown to the experimenter. Presynaptic fibers were filled with the Ca2+-Mg2+ fluorophore, Mg-Green AM, using a previously described technique (Atluri and Regehr, 1996 ; Schiess et al., 2006). Briefly, an ejection electrode containing Mg Green-AM in ACSF was lowered into the fiber path between a stimulating electrode and the presynaptic mossy fiber terminal field. While observing the emission image following 490nm excitation, a pressure pulse was applied with a syringe to produce a small bright spot (1 μl) in the fiber pathway. The slice was then maintained with 2 ml/min flow of oxygenated ACSF for ~1 hr until the fluorescence was visible at the presynaptic imaging site 500 μm away from the ejection site. The excitation light source was focused with a diaphragm in the epi-illumination pathway and the emission was then measured with a photomultiplier tube. Single pulses set to produce ~50% maximum fEPSP were delivered orthodromically through the stimulating electrode by a Master 8 pulse generator under control of the imaging system. The bath solution was then changed to one containing 10 μM CNQX, 25 μM D-AP5, and 20 μM bicuculline to block any Ca2+ signal from postsynaptic cells and 5 ΔF/F0 records were averaged. Following this, 600 nM TTX was added to the bath and 5 more records were averaged. The TTX record was then subtracted from that measured in the presence of ionotropic receptor blockers to eliminate any signal from cells filled directly at the ejection site. The decay phase of the difference ΔF/F0 record was fit with a least squares regression to a single exponential and the time constant of this fit used as a measure of the decay of presynaptic residual [Ca2+].
Statistical Analyses
The levels of total and phosphorylated GAP-43 and calcium binding proteins in wild type mice and the two lines of HuD transgenic animals were analyzed by a one-way ANOVA and the electrophysiology properties of these mice were analyzed by a two-way ANOVA, followed by Bonferroni post-hoc comparison tests using Prism 4.0 software for Windows (GraphPad, San Diego, CA).
RESULTS
Increased levels of GAP-43 protein in the hippocampus of HuD-Tg mice
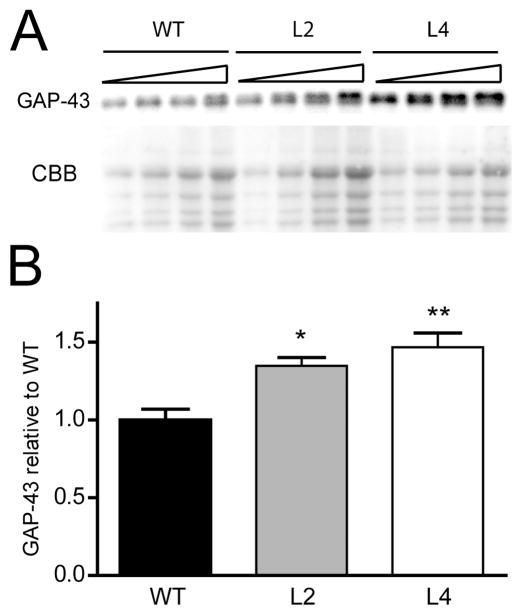
We have previously generated HuD-Tg mice overexpressing human HuD in forebrain neurons under the control of the α CaMKin II promoter (Bolognani et al., 2006). Using these animals, we demonstrated that ectopic expression of this RNA-binding protein in adult dentate granule cells (DGCs) of mice was sufficient to induce the accumulation of GAP-43 mRNA in cells that do not normally contain this mRNA (Bolognani et al., 2006). Since the original studies were based on the analyses of two different transgenic lines (lines 2 and 4), we used these lines of HuD-Tg mice for subsequent protein analyses. Western blots were performed using serial dilutions of hippocampal homogenates containing different amounts of total protein and the GAP-43 signal was normalized using Coomassie brilliant blue (CBB) staining (Fig. 1A). The advantage of loading multiple dilutions of a given sample is that protein levels can be calculated from the slope of the dose response curve rather than from a single determination, thus increasing the sensitivity of the measurement. Using this method, we found that normalized optical densities of GAP-43 were significantly higher in line 2 and line 4 HuD-Tg mice than those from wild type control littermates (Fig. 1B). No significant differences in GAP-43 protein levels were found between the two lines of HuD-Tg mice. This is not surprising since, although the amount of transgenic protein in line 4 are higher than in line 2, the total amount of HuD protein in the hippocampus is not different in these lines (Bolognani et al, 2006). Altogether, these results indicate that increased levels of GAP-43 mRNA in HuD-Tg mice lead to the translation and accumulation of this protein in the hippocampus.
Figure 1. Increased levels of GAP-43 in the hippocampus of HuD-Tg mice.
The levels of GAP-43 were determined in hippocampal homogenates from adult HuD-Tg mice of line 2 and line 4 and from non-transgenic littermates (WT) by western blots. (A) Panels show GAP-43 protein levels and CBB staining of serial dilutions of hippocampal extracts. (B) The optical densities of GAP-43 were corrected by that of CBB. Both lines of HuD-Tg mice show significant increases in GAP-43 expression (N=9 *p<0.05 and **p<0.01, one-way ANOVA).
Abnormal expression of GAP-43 in mossy fiber terminals of HuD-Tg mice
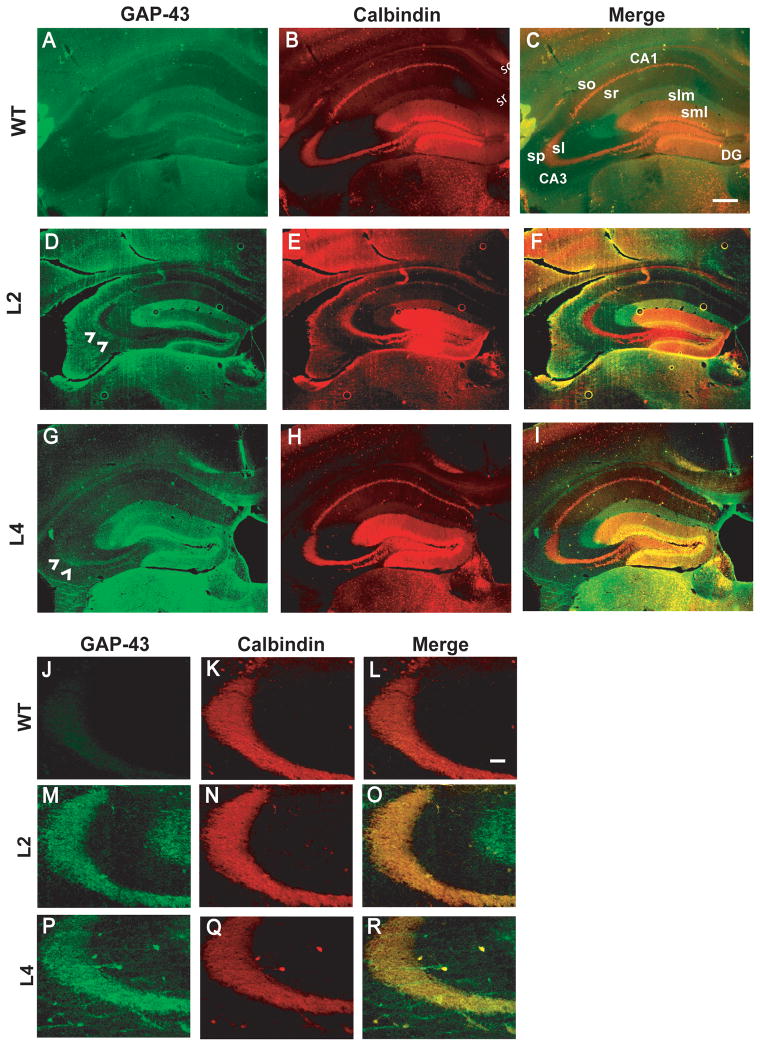
Immunohistochemical studies were performed to identify cellular origins of increased GAP-43 expression in HuD-Tg mice. As shown in Figure 2A, GAP-43 is not detected in mossy fibers of adult wild type mice, however, adult animals from both HuD-Tg lines show robust GAP-43 immunoreactivity at these terminals,(see arrowheads in Panels D and G). In addition, we observed an increased expression of GAP-43 in perforant path axons from the entorhinal cortex and in the inner molecular layer of the dentate gyrus. Consistent with these finding, high levels of the HuD transgene were expressed in layer II and III neurons of the entorhinal cortex (data not shown). As seen in the high magnification images of Panels J-R, GAP-43 immunoreactivity in the CA3 region co-localizes with that of calbindin D28K (CB) in the stratum lucidum of lines 2 and 4 mice. In addition, a few GAP-43 immunostained fibers we found to extend over to the stratum pyramidale (SP) of line 4 mice. These fibers, however, are not stained by calbindin (Panels P–R) or Timm’s (data not shown). Given the role of GAP-43 in sprouting, it is conceivable that the GAP-43 +/CB- fibers seen in Panel P, correspond to collateral sprouts from mossy fibers, which are immature and thus, calbindin negative. Alternatively, these fibers could be part of the recurrent CA3 network or sprouts from commissural/association fibers. While the precise origin of these sprouts remains to be elucidated, it is clear that mossy fibers of HuD transgenic mice are both calbindin and GAP-43 positive. Since mossy fibers normally lack GAP-43, we then sought to examine whether the ectopic expression of this protein had any effects in the short-term and long-term synaptic physiology at these terminals.
Figure 2. GAP-43 protein is present in mossy fiber terminals of HuD transgenic mice but not in wild type mice.
GAP-43 immunofluorescence (green, A, D,G) Calbindin D28K ( red, B,E,H) and merged images (C,F,I) show the increased expression of GAP-43 and its colocalization with calbindin in HuD-Tg mice. Images shown in the top panels were taken using a 4X objective and those in the bottom panels were taken at 20X. Note that although wild type mice only show background GAP-43 staining in mossy fibers (A and J), both lines of HuD-Tg mice (M, P) express high levels of this protein (D, F, K, P). Slm, stratum lacunosum moleculare; sml, stratum moleculare; sl, stratum lucidum; so, stratum oriens; sr, stratum radiatum, and sp, stratum pyramidale. Scale bars = 300 μm (A–I) and 50 μm (J–R).
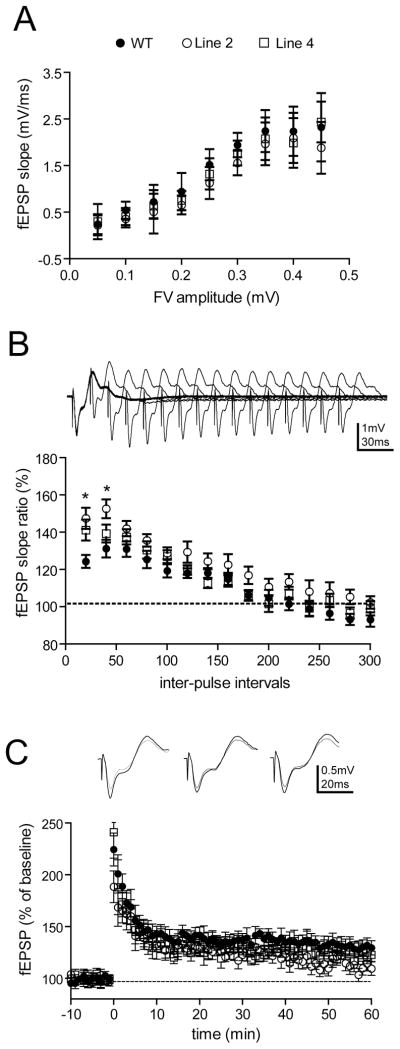
Increased Mossy Fiber-CA3 paired pulse facilitation in HuD-Tg mice
Analysis of the general electrophysiological properties of the hippocampus in HuD-Tg mice revealed no genotypic difference in the basal synaptic transmission in mossy fiber-CA3 (MF-CA3) synapses (Fig. 3A). However, we found a robust effect of HuD overexpression on paired pulse facilitation (PPF) at MF-CA3 synapses, with both lines of HuD-Tg mice showing increased PPF at short interpulse intervals (two-way ANOVA, post hoc Bonferroni tests, *p<0.05, Fig. 3B). We then measured the effect of genotype on LTP in these synapses. Unlike LTP in the perforant path, Schaffer collaterals, and commissural/association synapses, mossy fiber-CA3 LTP (MF-LTP) is NMDA receptor-independent. MF-LTP was induced by four 1-sec trains of 100Hz stimuli that were separated by a 20-sec interval in the presence of 50 μM AP5 to block other forms of LTP. In contrast to the changes in short-term plasticity, we did not find any significant changes in tetanus-induced MF-LTP in either genotype (Fig. 3C. one-way ANOVA of averaged responses at 60 min post tetanus).
Figure 3. Enhanced paired pulse facilitation in mossy fiber terminals of HuD-Tg mice.
A) The input-output relationships of mossy fibers, as represented by the presynaptic fiber volley amplitude (binned in 0.05 mV) vs. fEPSP slopes, did not reveal significant changes in either genotypes (two-way ANOVA, p>0.05 for genotype effects). B) Altered HuD and GAP-43 expression leads to increased paired pulse facilitation in mossy fiber-CA3 synapses at short interpulse intervals (two-way ANOVA, with Bonferroni post hoc tests. *p<0.05). C) No changes of tetanus-induced, NMDAR-independent LTP in either transgenic line. Mossy fiber LTP was induced by four 1-sec trains of 100Hz stimuli that were separated by a 20-sec inter pulse interval. 50 uM AP5 was present during the experimental duration. No significant changes were obtained at 60 min post-tetanus (one-way ANOVA p>0.05). All the electrophysiological measurements used 7–8 mice per group. Filled circles, WT; open circles, line 2; open squares, Line 4
Decreased time constant of decay of residual calcium in mossy fibers of HuD-Tg mice
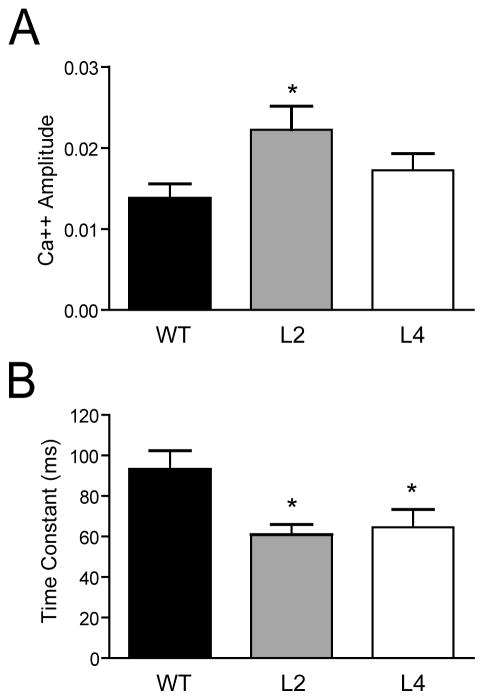
Residual calcium ([Ca2+]res) is known to play an important role in PPF at mossy fibers and other synapses with low basal probability of release (Zucker and Regehr, 2002). Calcium transients were measured in distal mossy fibers after proximal stimulation. The initial [Ca2+]amplitude and the time constant (τ) of decay of presynaptic [Ca2+]res in WT, line 2 and line 4 mice are shown in Figure 4. Both lines of HuD-Tg mice exhibited decreases in the decay τ, with line 2 mice showing also a significant increase in the initial Ca2+ amplitude. Considering that the levels of decay of [Ca2+]res in mossy fiber terminals are primarily controlled by the concentration of endogenous buffers (Blatow et al., 2003; Scott and Rusakov, 2006), one likely interpretation for the faster decay of residual calcium in both lines of HuD-Tg is that these animals have reduced levels of endogenous calcium buffer, leading to increased [Ca2+]res and higher PPF at short interpulse intervals.
Figure 4. Changes in residual calcium levels and dynamics in mossy fibers of HuD Tg-mice.
Calcium transients were measured in distal mossy fibers after proximal stimulation using Mg Green as described in the Materials and Methods. Animals of line 2 showed increased in the initial amplitude of the calcium signal (A) whereas both lines of HuD-Tg mice exhibited decreases in the time constant of decay (B). *p<0.05.
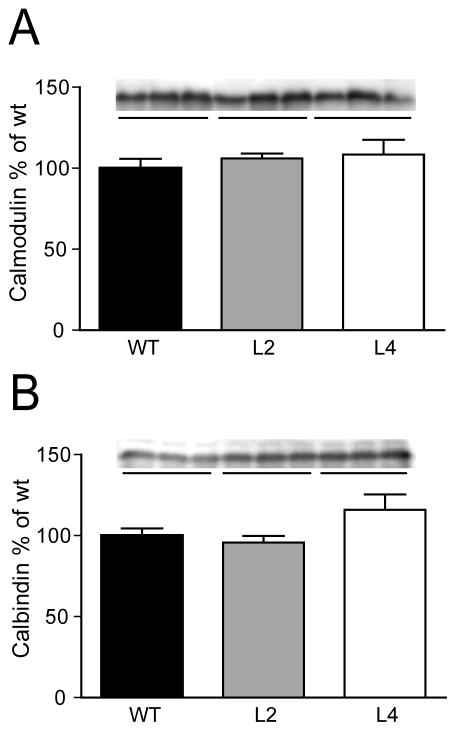
The main calcium binding proteins in mossy fibers are calbindin D28K and calmodulin. Therefore, subsequent studies measured the levels of these proteins in HuD-Tg mice by western blots. As shown in Figure 5, no differences in the total levels of the calcium-binding proteins were observed in any of the transgenic lines.
Figure 5. No differences in the total levels of the calcium-binding proteins calmodulin and calbindin in the hippocampus of HuD-Tg mice.
Protein levels were determined by western blots in the S1 fraction as described in the Methods. Results are representative from three independent experiments using 5 animals per group.
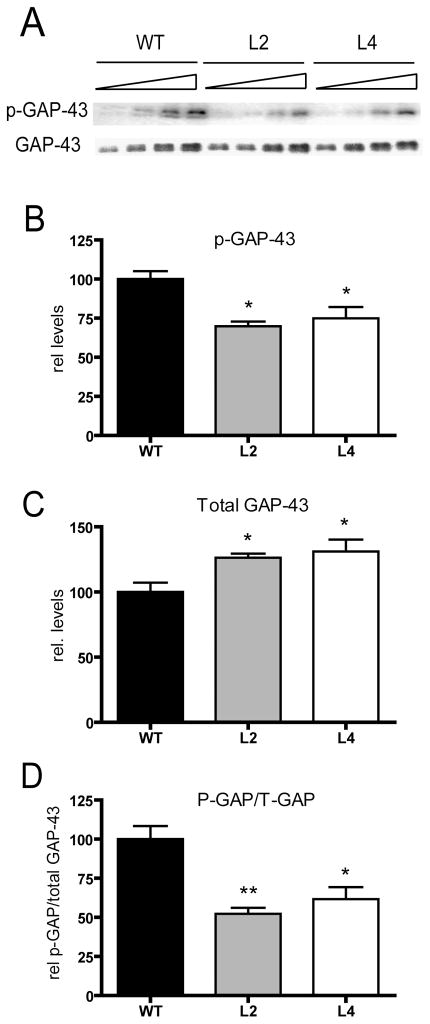
Decreased GAP-43 phosphorylation and increased calmodulin binding in HuD-Tg mice
Given that GAP-43 binds calmodulin with high affinity and this interaction is controlled by the PKC-dependent phosphorylation of GAP-43, subsequent studies examined the levels of phosphorylated GAP-43 (p-GAP-43) and the ratio of phosphorylated to total GAP-43 in HuD-Tg mice. We found that, although GAP-43 expression was increased in HuD-Tg mice, both the levels of phosphorylated protein and the ratio of p-GAP-43 to total GAP-43 (phosphorylated and non-phosphorylated) was significantly decreased in both lines of transgenic animals (Fig. 6).
Figure 6. Decreased levels of PKC-phosphorylated GAP-43 in HuD-Tg mice.
The levels of phosphorylated GAP-43 in hippocampal homogenates were determined using a phospho-specific antibody. A) Western blots showing the levels of phosphorylated and total GAP-43 protein increased in HuD-Tg mice. (B–C). The levels of phosphorylated GAP-43 (p-GAP) and total GAP-43 (T-GAP) were corrected by Coomassie blue staining. (D) The ratios of p-GAP-43 to total GAP-43 were calculated using the intensities of the bands corresponding to these proteins on the same blots. *p<0.05**p<0.01, N=5 animals per group.
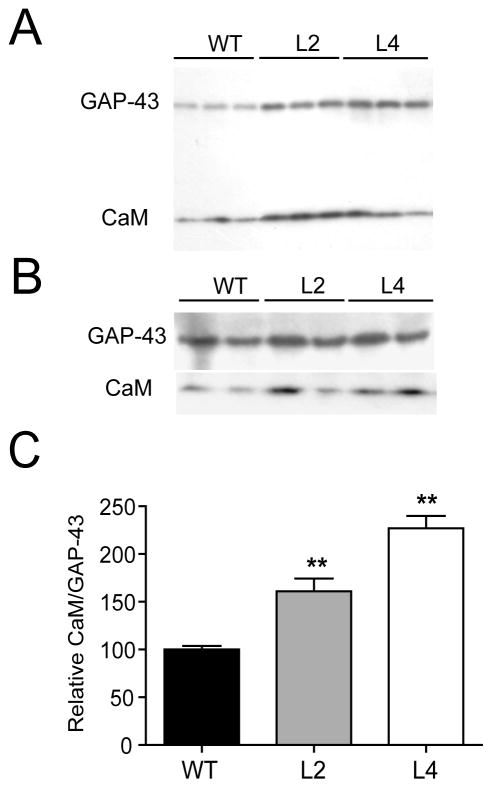
To determine whether an increased proportion of non-phosphorylated GAP-43 molecules in HuD-Tg mice affects the binding of calmodulin to this protein, we performed two types of studies. First, we measured the levels of calmodulin in the membrane fraction, presumably because of its association to GAP-43 (Fig. 7A) and second, we immunoprecitated GAP-43 from hippocampal extracts and measured the relative amount of bound calmodulin (Fig. 7B). Both methods revealed that there was a significant increase in the amount of calmodulin bound to GAP-43 in both lines of HuD-Tg mice (Fig. 7C). These whole hippocampal tissue analyses suggest that mossy fibers of HuD-Tg animals may have lower levels of free calmodulin, which is in agreement with the increased in PPF observed at these synapses.
Figure 7. Increased binding of calmodulin to GAP-43 in HuD-Tg mice.
Western blots show the levels of calmodulin and GAP-43 in the membrane fraction (A) and after immunoprecipitation with GAP-43 specific antibodies (B). The amount of calmodulin bound to GAP-43 increased significantly in both lines of HuD-Tg mice (C). Results are representative of three independent experiments run in duplicates using 3 animals from each group. **p<0.01
DISCUSSION
The neuron-specific RNA binding protein HuD plays an important role in the posttranscriptional regulation of neuronal gene expression during development and synaptic plasticity. This protein is known to stabilize GAP-43 mRNA and other neuronal mRNAs containing AU-rich sequences in their 3′ UTR. In this study, we examined the effect of overexpression of this RNA-binding protein in dentate granule cells on GAP-43 expression, function and on the electrophysiological properties of mossy fibers. Normally, mature dentate granule cells express neither GAP-43 mRNA nor HuD. Our results indicate that ectopic expression of HuD in DGCs of HuD-Tg mice increases the levels of GAP-43 protein in mossy fibers and that this change was correlated with an increase in PPF. This enhancement in short-term plasticity was associated with increases in residual calcium, which in turn correlated with an increased proportion of calmodulin molecules bound to GAP-43. These results highlight the role of HuD in the post-transcriptional control of GAP-43 gene expression in vivo and suggest the potential role of GAP-43 in controlling hippocampal physiology by its interactions with calmodulin.
GAP-43 and HuD expression levels show a strong developmental correlation, particularly in hippocampal dentate granule cells (Bolognani et al., 2007b). During development of the mouse hippocampus, GAP-43 mRNA levels in DGCs gradually decline from P4 to P14 despite ongoing transcription (Cantallops and Routtenberg, 1999). Furthermore, these authors suggested that activity-dependent changes in GAP-43 mRNA levels in DGCs are controlled post-transcriptionally (Cantallops and Routtenberg, 1999), as mRNA levels changed during early DGC development but transcriptional activity did not. This constant level of transcription persists in adult mice even when no mature GAP-43 mRNA is detected in the cells (Namgung and Routtenberg, 2000; Bolognani et al., 2006; Cantallops and Routtenberg, 1999). We recently found that the levels of HuD mRNA and protein also show a similar developmental decline suggesting that, in the absence of this mRNA stabilization factor, the GAP-43 mRNA is quickly degraded. Supporting this idea we recently demonstrated that ectopic expression of HuD in mature DGCs is sufficient to stabilize the GAP-43 mRNA leading to increased mRNA (Bolognani et al., 2006) and protein (Figs. 1 and 2) levels.
GAP-43 binds very tightly to calmodulin sequestering the protein in synaptic terminals and releasing it upon PKC-dependent phosphorylation (Alexander et al., 1987). Given their relative abundance and binding affinity, it has been estimated that under resting conditions most of the calmodulin in neurons is bound to GAP-43 (Liu and Storm, 1990). The levels of free calmodulin in the brain depend not only on the levels of GAP-43, but also on its state of phosphorylation (Alexander et al., 1987). In our HuD transgenic mice, we found that the increases in GAP-43 protein levels were associated with a decrease in both the total amount of phosphorylated GAP-43 and the ratio of phosphorylated to total GAP-43 molecules, which in turn correlated with a higher proportion of calmodulin molecules bound to GAP-43.
Mossy fiber synapses are quite distinct from other CNS synapses in that they exhibit very low probability of release, very robust PPF, and an NMDA-independent form of LTP of presynaptic origin (Nicoll and Schmitz, 2005). PPF at these synapses is known to be controlled by residual calcium levels (Zucker and Regehr, 2002), which in turn is regulated by the saturation of endogenous calcium buffers (Blatow et al., 2003). Even though calbindin D28K is the main calcium-buffering protein in mossy fibers, calmodulin contributes up to 50% of buffer saturation under stimulated conditions (Berggard et al., 2002). Therefore, it is likely that the estimated decreases in free calmodulin in HuD-Tg mice could alter the ability of this calcium-binding protein to buffer calcium in the terminal, leading to increases in both residual calcium and PPF. In contrast with short-term plasticity (PPF), long-term plasticity (LTP) was not affected by the presence of HuD and GAP-43 in mature mossy fibers. Inhibition of LTP at these synapses via pharmacological or genetic manipulation does not affect PPF or other forms of synaptic plasticity, suggesting that short-term plasticity at the MF-CA3 synapse may have greater importance to animal behavior than MF-LTP (Urban et al., 2001). In contrast to mossy fibers, Schaffer collateral and perforant path terminals normally contain GAP-43 and GAP-43 phosphorylation at these terminals increases soon after the establishment of LTP (Lovinger et al., 1986; Ramakers et al., 1995). Consistent with the role of GAP-43 phosphorylation in LTP, mice expressing a mutation in GAP-43 protein that mimics its constitutive phosphorylation by PKC show increased LTP in CA1 and dentate gyrus (DG) (Hulo et al., 2002; Routtenberg et al., 2000). Although we did not observe an effect of HuD overexpression in MF-LTP, we did find that LTP in the PP-DG synapses of line 2 mice was significantly impaired (data not shown). These results suggest that the effect of HuD on LTP may be dependent both on the specific type of LTP and on the levels of GAP-43 normally present in the pre-synaptic terminal.
In addition to the changes in GAP-43 expression and hippocampal electrophysiology, the HuD overexpressor (OE) mice show significant deficits in associative memory. The same two lines of HuD-Tg mice used in these studies were shown to have deficits in both contextual fear conditioning and the Morris water maze (Bolognani et al., 2007b). The changes in behavior of our HuD OE mice appeared to be different from GAP-43 OE mice, which show increased learning of a delayed non-matching to sample task (Routtenberg et al., 2000). Interestingly, a recent study suggests that not all of the GAP-43 OE mice have increased learning abilities and animals that have excessive amounts of GAP-43 in the hippocampus show substantial learning and memory deficits (Holahan et al., 2007). It will therefore be of interest to determine whether the levels of GAP-43 considered excessive in that report are close to those generated in the present study.
CONCLUSIONS
Our results suggest that increases in GAP-43 expression and decreases in GAP-43 phosphorylation in the hippocampus contribute to the electrophysiological and behavioral changes observed in HuD-Tg mice. Since HuD binds to other neuronal mRNAs, it is possible that the dysregulated expression of other HuD targets could play a role in the changes in short-term mossy fiber plasticity in HuD-Tg animals. Nonetheless, we believe that our findings are primarily due to the abnormal expression of GAP-43 in mossy fibers for the following reasons. First, GAP-43 transcription is persistently high in dentate granule cells, making the expression of this gene particularly sensitive to the effect of HuD. Second, given that mature mossy fibers normally do not contain GAP-43, these synapses are quite sensitive to the accumulation of the mRNA and protein in HuD-Tg mice. Finally, mossy fiber PPF is very sensitive to the changes of endogenous concentration of calcium buffer. Given the high affinity of GAP-43 for calmodulin, a higher amount of calmodulin may be sequestered, and thus inactivated, in HuD-Tg, leading to alterations in calcium signaling. In conclusion, we propose that the unregulated expression of HuD disrupts mossy fiber physiology in adult mice in part by altering the expression and phosphorylation of GAP-43 and the amount of free calmodulin available at the synaptic terminal. This report and another recent study from our group (Bolognani, et al., 2007) provide the first evidence that altered mRNA stability in vivo can lead to alterations in hippocampal physiology and behavior.
Acknowledgments
This work was supported by NS30255 to NPB and AG022574 to EJW. DCT was supported in part by NIAAA training grant T32 AA1427. We wish to thank Sayuri Nixon for genotyping the mouse lines used in these studies and Dr. Aryeh Routtenberg for critical reading of the manuscript.
References
- Akers RF, Routtenberg A. Protein kinase C phosphorylates a 47 Mr protein (F1) directly related to synaptic plasticity. Brain Res. 1985;334(1):147–51. doi: 10.1016/0006-8993(85)90576-1. [DOI] [PubMed] [Google Scholar]
- Alexander KA, Cimler BM, Meier KE, Storm DR. Regulation of calmodulin binding to P-57. A neurospecific calmodulin binding protein. J Biol Chem. 1987;262(13):6108–13. [PubMed] [Google Scholar]
- Anderson KD, Morin MA, Beckel-Mitchener A, Mobarak CD, Neve RL, Furneaux HM, Burry R, Perrone-Bizzozero NI. Overexpression of HuD, but not of its truncated form HuD I+II, promotes GAP-43 gene expression and neurite outgrowth in PC12 cells in the absence of nerve growth factor. J Neurochem. 2000;75(3):1103–14. doi: 10.1046/j.1471-4159.2000.0751103.x. [DOI] [PubMed] [Google Scholar]
- Anderson KD, Sengupta J, Morin M, Neve RL, Valenzuela CF, Perrone-Bizzozero NI. Overexpression of HuD accelerates neurite outgrowth and increases GAP-43 mRNA expression in cortical neurons and retinoic acid-induced embryonic stem cells in vitro. Exp Neurol. 2001;168(2):250–8. doi: 10.1006/exnr.2000.7599. [DOI] [PubMed] [Google Scholar]
- Atluri PP, Regehr WG. Determinants of the time course of facilitation at the granule cell to Purkinje cell synapse. J Neurosci. 1996;16(18):5661–71. doi: 10.1523/JNEUROSCI.16-18-05661.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakheet T, Williams BR, Khabar KS. ARED 3.0: the large and diverse AU-rich transcriptome. Nucleic Acids Res. 2006;34(Database issue):D111–4. doi: 10.1093/nar/gkj052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barami K, Iversen K, Furneaux H, Goldman SA. Hu protein as an early marker of neuronal phenotypic differentiation by subependymal zone cells of the adult songbird forebrain. J Neurobiol. 1995;28(1):82–101. doi: 10.1002/neu.480280108. [DOI] [PubMed] [Google Scholar]
- Bendotti C, Baldessari S, Pende M, Southgate T, Guglielmetti F, Samanin R. Relationship between GAP-43 expression in the dentate gyrus and synaptic reorganization of hippocampal mossy fibres in rats treated with kainic acid. Eur J Neurosci. 1997;9(1):93–101. doi: 10.1111/j.1460-9568.1997.tb01357.x. [DOI] [PubMed] [Google Scholar]
- Benowitz LI, Apostolides PJ, Perrone-Bizzozero N, Finklestein SP, Zwiers H. Anatomical distribution of the growth-associated protein GAP-43/B-50 in the adult rat brain. J Neurosci. 1988;8(1):339–52. doi: 10.1523/JNEUROSCI.08-01-00339.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benowitz LI, Routtenberg A. GAP-43: an intrinsic determinant of neuronal development and plasticity. Trends Neurosci. 1997;20(2):84–91. doi: 10.1016/s0166-2236(96)10072-2. [DOI] [PubMed] [Google Scholar]
- Berggard T, Miron S, Onnerfjord P, Thulin E, Akerfeldt KS, Enghild JJ, Akke M, Linse S. Calbindin D28k exhibits properties characteristic of a Ca2+ sensor. J Biol Chem. 2002;277(19):16662–72. doi: 10.1074/jbc.M200415200. [DOI] [PubMed] [Google Scholar]
- Blatow M, Caputi A, Burnashev N, Monyer H, Rozov A. Ca2+ buffer saturation underlies paired pulse facilitation in calbindin-D28k-containing terminals. Neuron. 2003;38(1):79–88. doi: 10.1016/s0896-6273(03)00196-x. [DOI] [PubMed] [Google Scholar]
- Bolognani F, Merhege M, Twiss A, Nora J, Perrone-Bizzozero I. Dendritic localization of the RNA-binding protein HuD in hippocampal neurons: association with polysomes and upregulation during contextual learning. Neurosci Letters. 2004;371:152–157. doi: 10.1016/j.neulet.2004.08.074. [DOI] [PubMed] [Google Scholar]
- Bolognani F, Qiu S, Tanner DC, Paik J, Perrone-Bizzozero NI, Weeber EJ. Associative and spatial learning and memory deficits in transgenic mice overexpressing the RNA-binding protein HuD. Neurobiol Learn Mem. 2007a;87(4):635–43. doi: 10.1016/j.nlm.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Bolognani F, Tanner DC, Merhege M, Deschenes-Furry J, Jasmin B, Perrone-Bizzozero NI. In vivo post-transcriptional regulation of GAP-43 mRNA by overexpression of the RNA-binding protein HuD. J Neurochem. 2006;96(3):790–801. doi: 10.1111/j.1471-4159.2005.03607.x. [DOI] [PubMed] [Google Scholar]
- Bolognani F, Tanner DC, Nixon S, Okano HJ, Okano H, Perrone-Bizzozero NI. Coordinated Expression of HuD and GAP-43 in Hippocampal Dentate Granule Cells During Developmental and Adult Plasticity. Neurochem Res. 2007b Jun 19; doi: 10.1007/s11064-007-9388-8. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Cantallops I, Routtenberg A. Activity-dependent regulation of axonal growth: posttranscriptional control of the GAP-43 gene by the NMDA receptor in developing hippocampus. J Neurobiol. 1999;41(2):208–20. [PubMed] [Google Scholar]
- Clayton GH, Perez GM, Smith RL, Owens GC. Expression of mRNA for the elav-like neural-specific RNA binding protein, HuD, during nervous system development Brain Res. Dev Brain Res. 1998;109:271–280. doi: 10.1016/s0165-3806(98)00074-1. [DOI] [PubMed] [Google Scholar]
- Dekker LV, De Graan PN, Oestreicher AB, Versteeg DH, Gispen WH. Inhibition of noradrenaline release by antibodies to B-50 (GAP-43) Nature. 1989;342(6245):74–6. doi: 10.1038/342074a0. [DOI] [PubMed] [Google Scholar]
- Haruta T, Takami N, Ohmura M, Misumi Y, Ikehara Y. Ca2+-dependent interaction of the growth-associated protein GAP-43 with the synaptic core complex. Biochem J. 1997;325 (Pt 2):455–63. doi: 10.1042/bj3250455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holahan MR, Honegger KS, Tabatadze N, Routtenberg A. GAP-43 gene expression regulates information storage. Learn Mem. 2007;14(6):407–15. doi: 10.1101/lm.581907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulo S, Alberi S, Laux T, Muller D, Caroni P. A point mutant of GAP-43 induces enhanced short-term and long-term hippocampal plasticity. Eur J Neurosci. 2002;15(12):1976–82. doi: 10.1046/j.1460-9568.2002.02026.x. [DOI] [PubMed] [Google Scholar]
- Ivins KJ, Neve KA, Feller DJ, Fidel SA, Neve RL. Antisense GAP-43 inhibits the evoked release of dopamine from PC12 cells. J Neurochem. 1993;60(2):626–33. doi: 10.1111/j.1471-4159.1993.tb03194.x. [DOI] [PubMed] [Google Scholar]
- Kohn DT, Tsai KC, Cansino VV, Neve RL, Perrone-Bizzozero NI. Role of highly conserved pyrimidine-rich sequences in the 3′ untranslated region of the GAP-43 mRNA in mRNA stability and RNA-protein interactions. Brain Res Mol Brain Res. 1996;36(2):240–50. doi: 10.1016/0169-328x(95)00239-o. [DOI] [PubMed] [Google Scholar]
- Liu YC, Storm DR. Regulation of free calmodulin levels by neuromodulin: neuron growth and regeneration. Trends Pharmacol Sci. 1990;11(3):107–11. doi: 10.1016/0165-6147(90)90195-e. [DOI] [PubMed] [Google Scholar]
- Lovinger DM, Colley PA, Akers RF, Nelson RB, Routtenberg A. Direct relation of long-term synaptic potentiation to phosphorylation of membrane protein F1, a substrate for membrane protein kinase C. Brain Res. 1986;399(2):205–11. doi: 10.1016/0006-8993(86)91510-6. [DOI] [PubMed] [Google Scholar]
- Meberg PJ, Routtenberg A. Selective expression of protein F1/(GAP-43) mRNA in pyramidal but not granule cells of the hippocampus. Neuroscience. 1991;45(3):721–33. doi: 10.1016/0306-4522(91)90284-u. [DOI] [PubMed] [Google Scholar]
- Mobarak CD, Anderson KD, Morin M, Beckel-Mitchener A, Rogers SL, Furneaux H, King P, Perrone-Bizzozero NI. The RNA-binding protein HuD is required for GAP-43 mRNA stability, GAP-43 gene expression, and PKC-dependent neurite outgrowth in PC12 cells. Mol Biol Cell. 2000;11(9):3191–203. doi: 10.1091/mbc.11.9.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namgung U, Routtenberg A. Transcriptional and post-transcriptional regulation of a brain growth protein: regional differentiation and regeneration induction of GAP-43. Eur J Neurosci. 2000;12(9):3124–36. doi: 10.1046/j.1460-9568.2000.00196.x. [DOI] [PubMed] [Google Scholar]
- Neve RL, Coopersmith R, McPhie DL, Santeufemio C, Pratt KG, Murphy CJ, Lynn SD. The neuronal growth-associated protein GAP-43 interacts with rabaptin-5 and participates in endocytosis. J Neurosci. 1998;18(19):7757–67. doi: 10.1523/JNEUROSCI.18-19-07757.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoll RA, Schmitz D. Synaptic plasticity at hippocampal mossy fibre synapses. Nat Rev Neurosci. 2005;6(11):863–76. doi: 10.1038/nrn1786. [DOI] [PubMed] [Google Scholar]
- Okano HJ, Darnell RB. A hierarchy of Hu RNA binding proteins in developing and adult neurons. J Neurosci. 1997;17(9):3024–37. doi: 10.1523/JNEUROSCI.17-09-03024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascale A, Gusev PA, Amadio M, Dottorini T, Govoni S, Alkon DL, Quattrone A. Increase of the RNA-binding protein HuD and posttranscriptional up-regulation of the GAP-43 gene during spatial memory. Proc Natl Acad Sci U S A. 2004;101(5):1217–22. doi: 10.1073/pnas.0307674100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrone-Bizzozero NI, Tanner D. GAP-43 in neural development and plasticity. In: Lajitha A, Lim R, editors. The handbook of neurochemistry and molecular neurobiology. 3. Heidelberg, Germany: Springer; 2006. pp. 315–329. [Google Scholar]
- Quattrone A, Pascale A, Nogues X, Zhao W, Gusev P, Pacini A, Alkon DL. Posttranscriptional regulation of gene expression in learning by the neuronal ELAV-like mRNA-stabilizing proteins. Proc Natl Acad Sci U S A. 2001;98(20):11668–73. doi: 10.1073/pnas.191388398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakers GM, De Graan PN, Urban IJ, Kraay D, Tang T, Pasinelli P, Oestreicher AB, Gispen WH. Temporal differences in the phosphorylation state of pre- and postsynaptic protein kinase C substrates B-50/GAP-43 and neurogranin during long-term potentiation. J Biol Chem. 1995;270(23):13892–8. doi: 10.1074/jbc.270.23.13892. [DOI] [PubMed] [Google Scholar]
- Routtenberg A, Cantallops I, Zaffuto S, Serrano P, Namgung U. Enhanced learning after genetic overexpression of a brain growth protein. Proc Natl Acad Sci U S A. 2000;97(13):7657–62. doi: 10.1073/pnas.97.13.7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiess AR, Scullin CS, Partridge LD. Neurosteroid-induced enhancement of short-term facilitation involves a component downstream from presynaptic calcium in hippocampal slices. J Physiol. 2006;576(Pt 3):833–47. doi: 10.1113/jphysiol.2006.118505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuman EM, Dynes JL, Steward O. Synaptic regulation of translation of dendritic mRNAs. J Neurosci. 2006;26(27):7143–6. doi: 10.1523/JNEUROSCI.1796-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott R, Rusakov DA. Main determinants of presynaptic Ca2+ dynamics at individual mossy fiber-CA3 pyramidal cell synapses. J Neurosci. 2006;26(26):7071–81. doi: 10.1523/JNEUROSCI.0946-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene JH. GAP-43 as a ‘calmodulin sponge’ and some implications for calcium signalling in axon terminals. Neurosci Res Suppl. 1990;13:S112–25. doi: 10.1016/0921-8696(90)90040-a. [DOI] [PubMed] [Google Scholar]
- Smith CL, Afroz R, Bassell GJ, Furneaux HM, Perrone-Bizzozero NI, Burry RW. GAP-43 mRNA in growth cones is associated with HuD and ribosomes. J Neurobiol. 2004;61(2):222–35. doi: 10.1002/neu.20038. [DOI] [PubMed] [Google Scholar]
- Sossin WS, DesGroseillers L. Intracellular trafficking of RNA in neurons. Traffic. 2006;7(12):1581–9. doi: 10.1111/j.1600-0854.2006.00500.x. [DOI] [PubMed] [Google Scholar]
- Tsai KC, Cansino VV, Kohn DT, Neve RL, Perrone-Bizzozero NI. Post-transcriptional regulation of the GAP-43 gene by specific sequences in the 3′ untranslated region of the mRNA. J Neurosci. 1997;17(6):1950–8. doi: 10.1523/JNEUROSCI.17-06-01950.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban NN, Henze DA, Barrionuevo G. Revisiting the role of the hippocampal mossy fiber synapse. Hippocampus. 2001;11(4):408–17. doi: 10.1002/hipo.1055. [DOI] [PubMed] [Google Scholar]
- Wang W, van Niekerk E, Willis DE, Twiss JL. RNA transport and localized protein synthesis in neurological disorders and neural repair. Dev Neurobiol. 2007;67(9):1166–82. doi: 10.1002/dneu.20511. [DOI] [PubMed] [Google Scholar]
- Zucker RS, Regehr WG. Short-term synaptic plasticity. Annu Rev Physiol. 2002;64:355–405. doi: 10.1146/annurev.physiol.64.092501.114547. [DOI] [PubMed] [Google Scholar]