Abstract
Muscleblind-like 2 (Mbnl2) is a zinc finger protein first identified in Drosophila. It appears to be essential for photoreceptor development and to be involved in RNA splicing. Here we report that Mbnl2 is strongly expressed in the rat pineal gland. The abundance of pineal Mbnl2 transcripts follows a marked circadian rhythm with peak levels approximately 7-fold higher at night than day levels. Mbnl2 protein exhibits a similar rhythm. In vitro studies indicate that the abundance of Mbnl2 transcripts and protein are controlled by an adrenergic/cAMP mechanism.
Introduction
There is abundant evidence indicating that the pinealocyte and retinal photoreceptor evolved in chordates from a common primitive photodetector, and that this chordate structure is derived from a photosensitive system that subsequently evolved into the various eyes found in all animals (Gehring 2004; Klein 2004). To a large degree, this common evolution is reflected in the expression of a shared set of genes dedicated to development and signal transduction in eyes and in the vertebrate pineal gland. Against this background, we were intrigued by the results of preliminary gene profiling studies of the pineal gland which suggested that this tissue expressed Mbnl2 3, a gene which encodes a little-studied zinc finger protein first identified in the Drosophila eye and thought to be essential for photoreceptor differentiation (Begemann et al. 1997).
Mbnl2 is a member of the muscleblind (Mbnl) family of proteins, which are so named because of their similarity to genes that play a role in muscle and eye development in Drosophila. Members of the Mbnl family exhibit different developmental and tissue distribution patterns (Fardaei et al. 2002; Pascual et al. 2006). These proteins contain zinc fingers and bind to CUG RNA expansions to influence alternative RNA splicing (Miller et al. 2000; Timchenko et al. 2002; Kanadia et al. 2003; Ho et al. 2004; Ho et al. 2005; Paul et al. 2006; Warf and Berglund 2007; Orengo et al. 2008); recent studies have identified RNA binding region in the protein (He et al. 2009).
In cases where there is marked expansion of CTG or CCGT repeats, muscleblind proteins bind to these thereby inhibiting binding of factors required for normal processing, resulting in incorrect splicing (Ho et al. 2004; Dansithong et al. 2005; Paul et al. 2006; Warf and Berglund 2007). Of note in this regard is evidence that human myotonic dystrophy is caused by binding of muscleblind proteins to CTG or CCGT repeats in the gene encoding a chloride channel, resulting in defective transcript splicing (Charlet et al. 2002; Mankodi et al. 2002).
Our interest in Mbnl2 was strengthened by the possibility that it might play a role in the pineal gland similar to that in the Drosophila eye because of the conserved features of eye development and of the pineal gland (Ashery-Padan and Gruss 2001; Klein 2004; Gehring 2005; Klein 2006); in addition, it might control alternative splicing in these tissues (Kim et al. 2005; Kim et al. 2007).
The studies presented here were designed to verify the evidence that Mbnl2 is expressed in the pineal gland and to extend this. Our studies confirm that Mbnl2 is expressed in the pineal gland and also in the retina and several brain regions. We have also found that that pineal Mbnl2 transcripts and protein levels exhibit marked daily rhythms and that both are neurally regulated by an adrenergic/cAMP mechanism, similar to that which controls other functions in the pineal gland (Klein 1985).
Materials and Methods
Materials
32P-αdCTP was obtained from GE Healthcare (Piscataway, NY) and 35S-α-dATP was from Perkin-Elmer (Boston, MA). Norepinephrine, 8-Br cAMP, dibutyryl cAMP, isoproterenol, actinomycin D, puromycin and KT5720 were purchased from Sigma (St. Louis, MO).
Animals and tissue preparations
Rats (Sprague-Dawley, 150 to 200 grams) used for experimental manipulations or for organ culture experiments were obtained from Taconic Farms Inc. (Germantown, NY). All animals used for Northern and Western blot studies were housed for two weeks in light/dark (LD) 14:10 lighting cycles, except for the experiment presented in Fig. 2C, in which some animals were maintained in constant darkness (DD) for 5 days prior tissue removal. Surgically prepared male rats were obtained from Taconic Farms. For surgery, the superior cervical ganglia were approached through the ventral neck and located bilaterally near the carotid trigone, medial to the carotid bifurcation For superior cervical ganglionectomy, (SCGX), the adjacent sympathetic trunk was cut and the ganglion were removed. For decentralization, a caudal portion of the sympathetic turn was removed. Animals for the two point LD night/day studies were killed at zeitgeber time (ZT) 7 or19; for the constant darkness (DD) experiment or at circadian time (CT) 7 or 19. For the studies presented in Fig. 2A and 2B animals were killed at ZT 1,7,13.15.19,23.
Fig. 2. Daily rhythm in rat pineal Mbnl2 mRNA and protein.
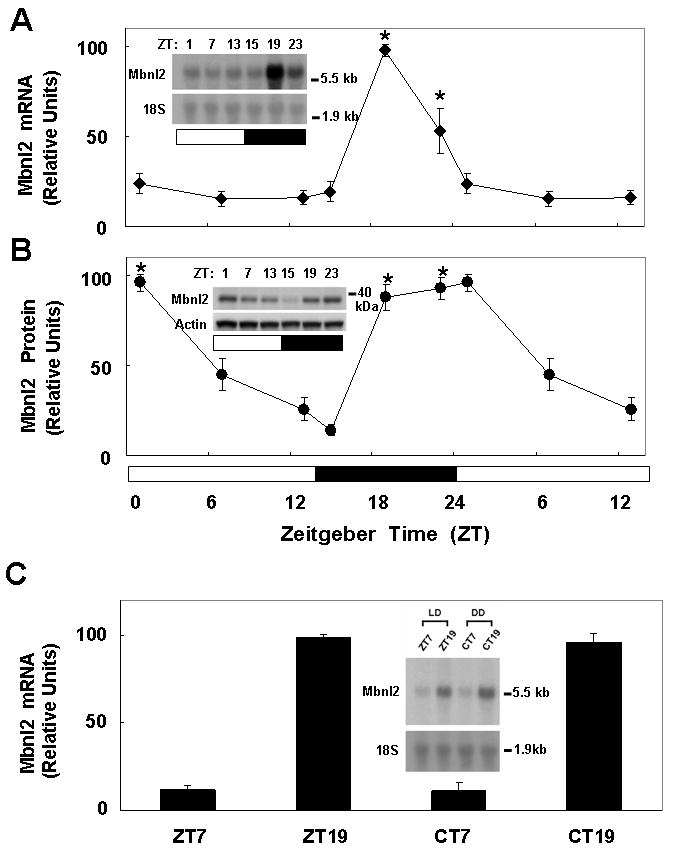
Animals were housed in a controlled lighting environment for two weeks (LD 14:10 for panels A and B). Pineal glands were obtained at the indicated times. A. Mbnl2 mRNA: Each lane was loaded with 8 μg of total RNA. The blot was hybridized with a rat Mbnl2 probe. To normalize for RNA loading, the blot was reprobed for 18S rRNA. The ZT1, 7, and 13 data points are double plotted. Results are presented as the mean ± SE of three replicates. *, p < 0.01 vs. ZT7 group. B. Mbnl2 protein: Each lane was loaded with 60 μg protein from a pineal extract. Immunodetection was performed with a polyclonal rabbit anti-Mbnl2 serum at a dilution of 1:10,000. The Mbnl2 band appears around 40 kDa. The ZT1, 7, and 13 data points are double plotted. Results are presented as the mean ± SE of three replicates. *, p < 0.01 vs. ZT15 group. C. Pineal Mbnl2 mRNA. Animals were housed for two weeks in LD 14:10 (ZT7 and ZT9) or nine days in LD 14:10 followed by 5 days in constant darkness (DD)(Circadian time [CT] 7 and 19). Each lane was loaded with 8 μg of total RNA. The blot was hybridized with a rat Mbnl2 probe. To normalize for RNA loading, the blot was reprobed for 18S rRNA. Results are presented as the mean ± SE of three replicates. *, p < 0.01 vs. ZT7 group. The insert is a typical Northern blot image. For further details see the Materials and Methods section.
Rats (males) used for radioactive in situ hybridization and immunohistochemistry were obtained from Charles River (Sulzfeld, Germany) and were housed in LD 12:12. Animals were sacrificed by CO2 asphyxiation and decapitated. Tissues were prepared as described previously (Kim et al. 2005; Kim et al. 2007). For immunohistochemical analysis, male rats were perfusion-fixed in 4% paraformaldehyde in 0.1 M phosphate buffer at ZT6 and ZT18, respectively; brains were removed, post-fixed in the same fixative and cryoprotected in 25% sucrose.
In some experiments, male rats were injected with either isoproterenol (ISO; 10 mg/kg) or norepinephrine (NE; 1 mg/kg); the compounds were first dissolved in 0.9% NaCl and the appropriate volume was injected subcutaneously in the nape of the neck. Rats were injected with ISO at ZT 4 and killed at ZT 7; NE was injected into SCGX rats at ZT16 and the rats were killed at ZT19. Pineal glands were removed and immediately placed on solid CO2, and then stored at −80°C until use.
Animal use and care protocols were in accordance with NIH guidelines, with Health Sciences Animal Policy and with the guidelines of EU Directive 86/609/EEC (approved by the Danish Council for Animal Experiments).
Organ culture
Pineal glands from female rats were cultured in BGJb medium containing 1 mg/ml BSA as described previously (Kim et al. 2005; Kim et al. 2007). In experiments where mRNA and protein levels are reported, each analysis was performed on separate sets of pineal glands treated in culture on the same day.
Analysis of Mbnl2 mRNA
Northern blot analysis
Total RNA was extracted using the guanidine-HCl/phenol procedure (TRIzol Reagent, Invitrogen, Carlsbad, CA). RNA was separated on a 1.5% agarose/0.7 M formaldehyde gel, transferred to a charged nylon membrane (Nytran, Schleicher and Schuell, Dassel, Germany) by passive capillary transfer and cross-linked to the membrane using ultraviolet light. The Mbnl2 hybridization probes used were based on Mbnl2 sequence (Supp. Fig. 1A). Probes were 32P-labeled by random priming using a DNA Labeling kit (GE Healthcare). Blots were stripped and reprobed for 18S rRNA (nucleotides 484-1080, GenBank accession number M11188) to monitor the quality of the RNA and equal loading. Blots were hybridized at 68 °C for 1.5 h in QuikHyb buffer (Stratagene, La Jolla, CA). The final wash was in 0.1 × SSC/0.1% SDS at 60 °C for 20 min. Blots were visualized and quantitated using a PhosphorImager (Molecular Dynamics, Sunnyvale, CA); values were normalized against the signals generated by 18S rRNA.
Radiochemical in situ hybridization histology
A published method was used (Kim et al. 2005; Kim et al. 2007). Sagittal cryostat sections (12 μm) were thawed and fixed for 5 min in 4% paraformaldehyde in PBS, washed 2 × 1 min in PBS, and acetylated (0.25% acetic anhydride in 0.9% NaCl containing 0.1 M triethanolamine; 10 min). The sections were then dehydrated in a graded series of ethanol and delipidated in 100% chloroform (5 min). They were partially rehydrated in 100% and 95% ethanol (1 min each) and allowed to dry.
For hybridization, a 35S-labeled oligonucleotide probe was prepared based on rat Mbnl2 sequence (5′-TACAAACGGTTACGGTGTTGTCGTTTGTGTCG-3′). The labeled probe was diluted in the hybridization buffer consisting of 50% (v/v) formamide, 4 × SSC, 1 × Denhardt's solution, 10% (w/v) dextran sulfate, 10 mM DTT, 0.5 mg/ml salmon sperm DNA and 0.5 mg/ml yeast tRNA. A 200 μl sample of the labeled probe was placed on each section. The sections were then covered with Parafilm and incubated in a humid chamber overnight at 37 °C. After hybridization, the slides were washed in 1 × SSC for 4 × 15 min at 55 °C, 2 × 30 min at room temperature, and rinsed twice in distilled water. The sections were dried and exposed to X-ray film for 1 to 2 weeks. The pineal hybridization signals on X-ray films were quantified using “Image 1.42” (Wayne Rasband, NIH). Optical density was converted to dpm/mg tissue using simultaneously exposed 14C-standards calibrated by comparison with 35S brain-paste standards. Results are based on the analysis of pineal glands from five animals killed at night and five during the day.
Analysis of Mbnl2 protein
Immunoblots
To prepare samples, 2 glands were homogenized by brief sonication (three 1 second pulses) using a Biosonik sonicator (Bronwill Scientific, Rochester, NY) in 50 μl of 0.1M sodium phosphate buffer, pH 6.8, 4°C; the homogenate was centrifuged (13,000 × g, 15 min, 4 °C) and the supernatant was used for analytical procedures. Protein was measured by a dye binding method using a commercial reagent (Bio-Rad Protein Assay, Hercules, CA) with BSA as the standard. Samples containing 60 μg of protein were resolved on pre-formed 10% Tris/glycine (1 mm) gels using the manufacturer's protocol (Novex, San Diego, CA). Rainbow standards (GE Healthcare) were used to determine the molecular mass of the proteins.
Proteins were electroblotted onto an Immobilon-P (0.45 μM) transfer membrane (Millipore, Bedford, MA) in a semi-dry blotting system (Investigator Graphite Electroblotter System, Genomic Solutions, Chelmsford, MA) according to the manufacturer's protocol. The proteins were equilibrated (5 min) and transferred using 10 mM CAPS (3-cyclohexylamino-1-propanesulfonic acid) buffer, pH 11, containing 20% methanol and 0.01% SDS. The conditions for electrical transfer were 400 μA/cm2 (20 min), 600 μA/cm2 (20 min), 800 μA/cm2 (20 min), followed by 1200 μA/cm2 (45 min).
Membranes were air-dried and then blocked for 2 h in PBS, pH 7.4, containing 10% non-fat dry milk (Bio-Rad), 0.2% Tween-20 (Bio-Rad), and 0.05% thimerosal (Sigma). An anti-Mbnl2 polyclonal serum was raised in New Zealand White rabbits against an internal peptide sequence (rat Mbnl2 137-157) (Supp. Fig. 1B) which detects a single band (∼40 kDa) in the rat pineal gland and retina and in extracts of HEK293 cells transfected with Mbnl2 (Supp. Fig. 1C).
The antiserum was diluted (1:10,000) in PBS containing 1 mg/ml BSA fraction V and 0.05% thimerosal. The membranes were incubated (18 h, room temperature) with antiserum and washed [2 × 5 min in TPBS (PBS containing 0.05% Tween-20) followed by 2 × 5min in PBS] and then incubated with goat anti-rabbit IgG conjugated to horseradish peroxidase (9 ng/ml, 1 h, room temperature, Kirkegaard and Perry, Gaithersburg, MD) prepared in TPBS containing 0.05% thimerosal and 2.5 μl/ml normal goat serum (Pierce, Rockford, IL). The membranes were then washed 3 × 10 min in TPBS followed by 3 × 5 min washes in PBS. The secondary antibody was detected by enhanced chemiluminescence (Lumiglo, Kirkegaard and Perry). Loading was monitored by stripping the blot and reprobing with anti-actin serum (Sigma). The blots were exposed to BIO-MAX MR (Eastman Kodak Company, Rochester, NY) and quantitated using ImageQuant 5.2 software (Molecular Dynamics, Sunnyvale, CA).
Mbnl2 immunohistochemistry
Cryostat sections (14 μm) were washed in PBS and incubated in 5% normal swine serum diluted in PBS for 30 min. This was followed by incubation in antiserum diluted (1:1,000) in PBS containing 1% BSA fraction V and 0.3% Triton X-100 for 16 h at 4°C. Specificity control was performed by incubating parallel sections in pre-immune serum. Sections were washed in PBS containing 0.25% BSA and 0.1% Triton X-100 followed by incubation for 1 h in biotinylated swine anti-rabbit IgG (Dako, Glostrup, Denmark) diluted (1:500) in the same buffer. Sections were washed in PBS with 0.1% Triton X-100 and incubated for 45 min in ABC-Vectastain solution (Vector Laboratories, Burlingame, CA) diluted (1:100) in the same buffer. After washing PBS with 0.1% Triton X-100 and in 0.05 M Tris (pH 7.6), chromogenic development was performed in 1.4 mM diaminobenzidine and 0.01% H2O2 in 0.05 M Tris (pH 7.6) for 20 min. The reaction was terminated by extensive washing of the sections in deionized water. Finally, the sections were dried and embedded in Pertex (Histolab, Gothenburg, Sweden).
Statistical analysis
All data are expressed as means ± SE values for the number of determinations indicated. Statistical analyses were performed using Student's t-test for two groups and one-way analysis of variance (ANOVA) for multiple groups. Asterisk (*), p < 0.01.
Results
Tissue distribution and daily rhythm in Mbnl2 mRNA
Mbnl2 mRNA (∼5.5 kb) was detected as a single band by Northern blot analysis (Fig. 1A). The daytime expression level of pineal Mbnl2 was lower than in the cerebellum, retina and skeletal muscle (Fig. 1A); expression in other tissues examined was nearly undetectable. Expression in the retina confirms the results of studies on embryonic chicken (Huang et al. 2008). Pineal Mbnl2 mRNA levels increased ∼ 7-fold at night; significant night/day differences in the abundance of Mbnl2 transcripts in other tissues were not detected.
Fig. 1. Tissue distribution of Mbnl2 mRNA.
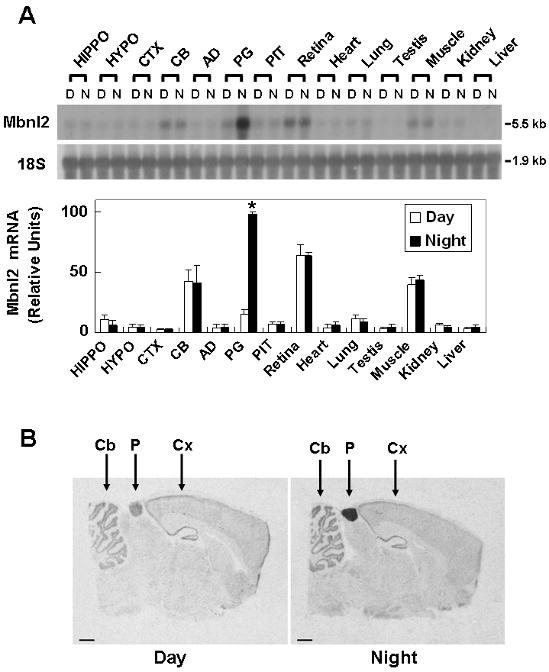
A. Northern blot analysis of Mbnl2 mRNA in selective tissues at night and during day. Rats were housed in a controlled lighting environment (LD 14:10). Total RNA was obtained from day tissues removed at ZT7 and night tissues removed at ZT19 under dim red light. RNA preparation and Northern blot analysis were performed as described in Materials and Methods. The blot was hybridized with a rat Mbnl2 probe. To normalize for RNA loading, blots were stripped and reprobed for 18S rRNA. Abbreviations are HIPPO, hippocampus; HYPO, hypothalamus; CTX cerebral cortex; CB, cerebellum; AD, adrenal gland; PG, pineal gland; PIT, pituitary gland; Muscle, skeletal muscle. Results are presented as the mean ± SE of three replicates. B. Radiochemical in situ hybridization histology of Mbnl2 mRNA in the rat brain. Animals were housed in a controlled lighting environment for two weeks (LD 12:12). The day sample is from an animal killed during daytime (ZT6) and the night one is from an animal killed during nighttime (ZT18). Scale bars, 1 mm. CB, cerebellImmuum; PG, pineal gland; CTX, cortex. For further details see the Materials and Methods section.
In situ
hybridization (Fig. 1B) revealed an ∼17-fold (p < 0.001) night/day difference in Mbnl2 expression in the pineal gland. Distinct expression was also detected in the cerebellum, hippocampus and the ventromedial hypothalamus; however night/day differences were not observed.
The daily rhythm in Mbnl2 transcripts was examined in detail by sampling at six time points (Fig. 2A). This indicated that the peak expression (∼7-fold higher than day values) occurred 4 hours after lights off and was transient.
To determine whether the daily rhythm in Mbnl2 mRNA was translated into a daily rhythm in Mbnl2 protein, a polyclonal antiserum was raised in rabbits against an internal Mbnl2 peptide, corresponding to rat Mbnl2 137-157 (Supp. Fig. 1B). This revealed that there is a > 9-fold difference in Mbnl2 protein (∼40 kDa), with peak levels achieved 4 hours after lights off (Fig. 2B). In contrast to the transient peak in Mbnl2 mRNA levels, peak Mbnl2 protein levels remained sustained for ∼ 7 hours.
The rhythm in Mbnl2 mRNA was found to persist in animals maintained in constant darkness (DD), indicating that it was truly circadian, apparently driven by an endogenous clock and not by environmental lighting (Fig. 2C).
Immunohistochemical analysis revealed a universal staining of all the cells in the pineal parenchyma and no staining in the perivascular spaces, indicating the gene is expressed in pinealocytes. Although all parenchymal cells exhibited immunoreactivity (Fig. 3), the staining pattern was not uniform; a few cells exhibited stronger staining. Similar staining patterns were observed at both ZT6 and ZT18, times at which quantitative analysis (Fig. 2B) indicated that protein levels differed by ∼2-fold; the pre-immune serum did not generate a signal, providing evidence that the anti-Mbnl2 serum was specific.
Fig. 3. Immunohistochemical detection of Mbnl2 protein in the rat pineal gland.
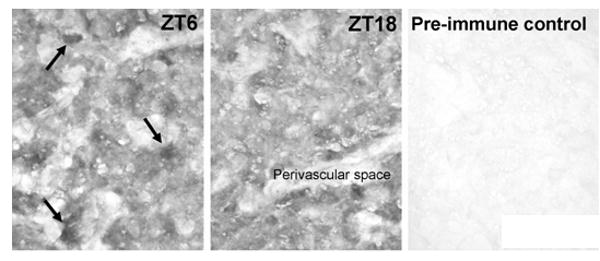
Animals were housed in a controlled lighting environment for two weeks (LD12:12). Pineal glands were obtained at ZT6 and ZT18. Immunohistochemical detection was done with the polyclonal rabbit anti-Mbnl2 used in Fig. 2B. at a dilution of 1:1,000. A pre-immunization negative control (ZT18) is displayed. Note the universal staining of the cells in the pineal parenchyma and a few intensively stained cells (arrows in ZT6). Scale bar, 50 μm. For further details see the Materials and Methods section.
Neural control of Mbnl2 mRNA
In most cases in which expression of genes increases at night in the pineal gland, a neural mechanism is involved. This is controlled by an internal circadian clock located in the suprachiasmatic nucleus of the hypothalamus, which is linked to the pineal gland by a multi-synaptic neural circuit that passes through central (paraventricular nucleus) and peripheral (superior cervical ganglia, SCG) neural structures. Here, we found that interrupting this circuit by surgically removing the SCG (SCGX) or by transection of the sympathetic trunk caudal to the SCG, which removes the afferent nervous input to the ganglion (DCNT), blocked the increase in expression of Mbnl2 at night (Fig. 4A).
Fig. 4. Regulation of pineal Mbnl2 transcripts by a photoneural mechanism.
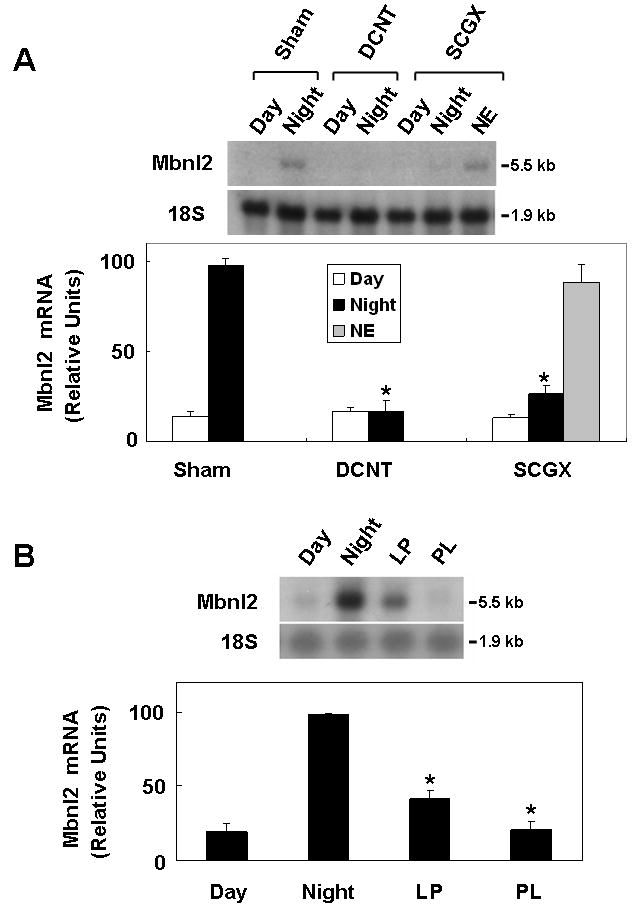
Each lane contains 3 μg total RNA obtained from a pool of two pineal glands. Northern blots were probed with a rat Mbnl2 probe. To normalize for variations in RNA loading, the blot was reprobed for 18S rRNA. Results are presented as the mean ± SE of three replicates. For further details see the Materials and Methods section. A. The nocturnal increase in Mbnl2 mRNA is driven by neural input from the SCG. Pineal RNA was obtained from Sham, DCNT, and SCGX rats killed during the day (ZT7) or at night (ZT19). In addition, one group of SCGX rats was injected with 1 mg/kg NE at ZT16 and killed 3 hours later with the other nighttime experimental group. *, p < 0.01 vs. Sham Night group. B. Effect of light exposure at night on Mbnl2 mRNA level in the rat pineal gland. Rats were housed in a controlled lighting environment (LD 14:10). Pineal glands were obtained during the day (ZT7), at night (ZT19) in the dark, after 1 hr light pulse (LP) or after 5 hr of prolonged light (PL). *, p < 0.01 vs. Night group. For further details see the Materials and Methods section.
Another approach to blocking neural stimulation of the pineal gland is to expose animals to light. This blocks the transmission to the pineal gland of stimulatory signals generated by the suprachiasmatic nucleus at night. In these studies, light exposure during the first 4 hours of the night period (PL) blocked the increase in Mbnl2 transcript level (Fig. 4B). We also found that a 1 hour light pulse (LP) caused a rapid decrease in Mbnl2 mRNA. Accordingly, it appears that the daily rhythm in Mbln2 expression is driven by the well-described neural system which regulates pineal function (Klein 1985).
Adrenergic-cAMP control of Mbnl2 mRNA and protein
The sympathetic neurons in the SCG which innervate the pineal gland act by releasing NE. Here we found that treatment of SCGX animals with NE elevated Mbnl2 expression at ZT4 (Fig. 4A). We also found that treatment of intact animals with the β-adrenergic agonist isoproterenol elevated Mbnl2 transcript abundance (Fig. 5).
Fig. 5. Isoproterenol (ISO) elevates Mbnl2 transcript levels in vivo.
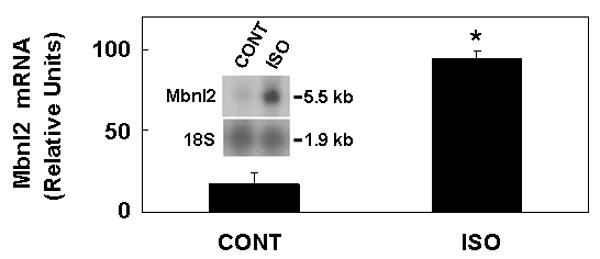
Rats were injected subcutaneously with isoproterenol (20 mg/kg) at ZT4. All pineal glands were obtained at ZT7 and glands were removed and stored on solid CO2. Each lane was loaded with 8 μg total RNA obtained from a pool of two pineal glands. Northern blots were probed with a rat Mbnl2 probe. To normalize for variations in RNA loading, the blot was reprobed for 18S rRNA. The results were confirmed in three experiments. Results are mean ± SE of three replicates. *, p < 0.01 vs. CONT group. For further details see the Materials and Methods section.
To determine if NE was acting directly on the pineal gland, organ culture was used. Treatment of glands in organ culture with NE elevated both Mbnl2 mRNA and Mbnl2 protein (Fig. 6).
Fig. 6. Evidence that NE and cAMP control Mbnl2 mRNA and protein.
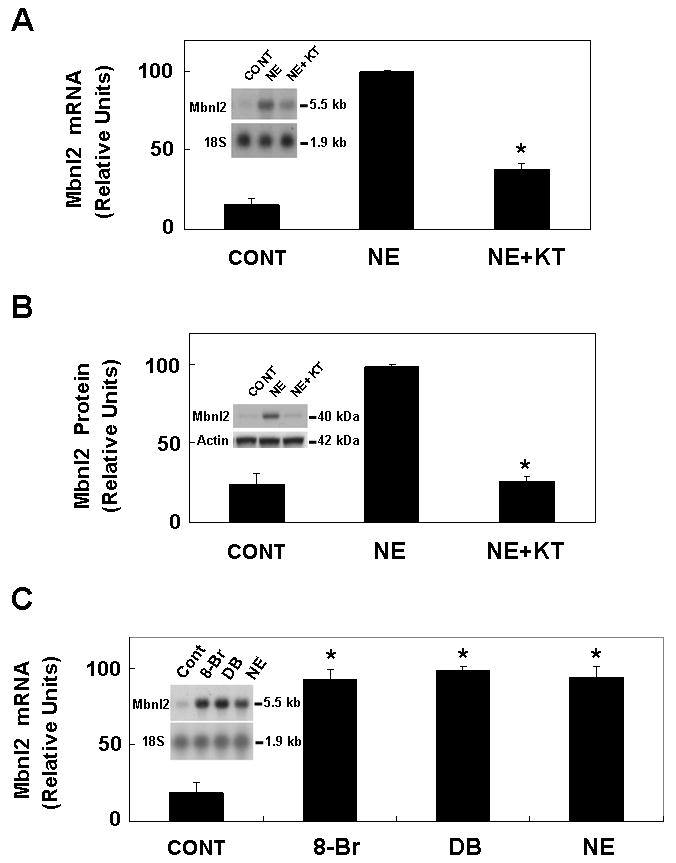
A. Mbnl2 mRNA: Cultured pineal glands were pre-incubated with 3 μM KT5720 alone for 1 hr; NE (1 μM) was then added and incubation was continued for 6 hours. Each lane was loaded 8 μg of total RNA from pools of three or four glands. Northern blots were probed with a rat Mbnl2 probe. To normalize for RNA loading, the blot was reprobed for 18S rRNA. Results are presented as the mean ± SE of three replicates. *, p < 0.01 vs. NE group. B. Mbnl2 protein: Using pineal glands from the same experiment shown in panel A, each lane was loaded with 60 μg protein from a pineal extract. Immunodetection was performed with an anti-Mbnl2 serum at a dilution of 1:10,000. Results are presented as the mean ± SE of three replicates. *, p < 0.01 vs. NE group. C. Pineal glands were cultured for 6 hours with 500 μM 8-Br cAMP (8-Br), 500 μM dibutyryl cAMP (DB) or 1 μM NE; control (CONT). *, p < 0.01 vs. CONT group. For further technical details see 5.A. and the Materials and Methods section.
Many effects of NE are mediated by the second messenger cAMP, acting through cAMP dependent protein kinase (PKA). The possible participation of cAMP in the control of Mbnl2 mRNA was studied in two ways. First, it was found that blocking the action of PKA, using the PKA-selective inhibitor KT5720, blocked stimulation of Mbnl2 mRNA and protein by NE (Fig. 6A, B). Second, it was found that either of two cAMP protagonists, DB cAMP or 8-Br cAMP, increased Mbnl2 mRNA levels (Fig. 6 C).
In contrast to the marked stimulatory effects of PKA activators on Mbnl2, it was found that treatment with an activator of the protein kinase C family, phorbol myrstic acetate (PMA), did not alter Mbnl2 mRNA (Fig. 7).
Fig. 7. NE-induced elevation in Mbnl2 mRNA requires transcription but not translation.
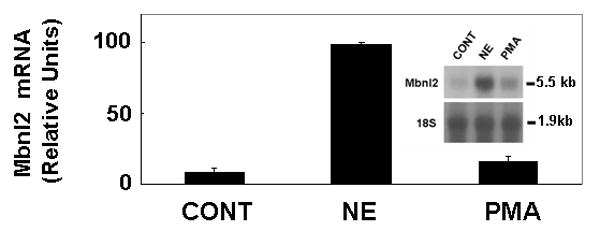
Pineal glands were treated with 30 μg/ml actinomycin D (ACTD) or 50 μg/ml puromycin (PURO) for 1 hour and during the subsequent 6 hours treatment with 1 μM NE. Each lane was loaded 8 μg total RNA obtained from a pool of four pineal glands. After hybridization with the Mbnl2 probe, the blot was stripped and reprobed with an 18S rRNA probes as a control. Results were confirmed in three independent experiments. The values are mean ± SE of three replicates. For further details see the Materials and Methods section.
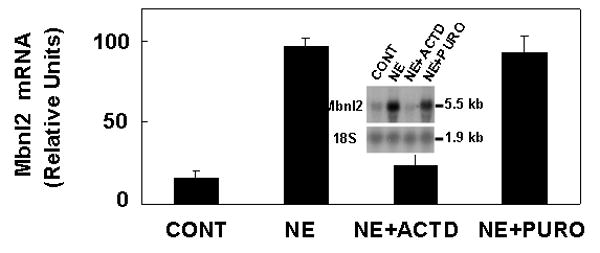
Adrenergic induction of Mbnl2 requires de novo synthesis of mRNA, but not protein
The induction of Mbnl2 by NE was blocked by treatment with a dose of actinomycin D known to inhibit mRNA synthesis in this system by >95%, indicating that de novo synthesis of mRNA was required (Fig. 8). The increase in Mbnl2 mRNA, however, was not blocked by treatment with puromycin, a blocker of protein synthesis, indicating that de novo synthesis of a protein is not required for induction of Mbnl2 expression.
Fig. 8.
Discussion
The studies presented here provide a clear indication that Mbnl2 is expressed at relatively high levels in the rat pineal gland and that expression exhibits a daily rhythm in this tissue. The finding that the rhythm in mRNA is translated into a change in Mbnl2 protein indicates that a major factor controlling the abundance of this protein is translation, rather than control via transcriptional or posttranscriptional mechanisms. Our finding that peak protein levels are maintained for a longer period than those of mRNA suggests that the protein is more stable than the mRNA, which appears to disappear quickly following photic-induced cessation of neural stimulation (Fig. 4B). The relatively more stable nature of Mbnl2 protein, as compared to mRNA may reflect binding to proteins or nucleic acids, or both.
Moreover, several findings made here are consistent with the conclusion that expression of this gene is circadian in nature and controlled by the well described neural system which includes the SCN (Klein 1985), and drives 24-hour patterns in gene expression by PKA activation (Bailey et al. 2009). These include the observations that interruption of the neural input to the gland blocks the nocturnal increase in Mbnl2 expression, that adrenergic/cAMP stimulation elevates expression, and that the Mbnl2 mRNA rhythm persists in constant darkness. Our finding that an activator of PKC does not mimic the effects of NE supports the view that activation of PKC alone is not sufficient for induction of Mbnl2 expression and that PKC signaling does not play an essential regulatory role.
It is of interest to note that the induction of Mbnl2 expression was not blocked by an inhibitor of protein synthesis. Accordingly, induction does not require new synthesis of a transcription factor or coactivator for expression. Rather, cAMP may initiate Mbnl2 expression by stimulating phosphorylation of pre-existing cAMP response element binding protein, which act by binding to cAMP response elements in the Mbnl2 promoter.
The evidence that 24-hour changes in Mbnl2 appear to be part of the global changes in gene expression and function which characterize pineal biology, implies that it plays an important role in this tissue. This role is likely to be related to the correct synthesis and splicing of some of the transcripts that are generated on a differential night/day basis. The task of identifying specific regulated genes may be aided by the both the knowledge that Mbnl2 binds to RNA CUG repeat sequences and the results of recent gene profiling analysis of the rat pineal gland which has identified a large number of rhythmically expressed and highly expressed genes(Bailey et al. 2009). It is known that several genes in the pineal gland are expressed as alternatively spliced transcripts, including Crem, Mat2a, Pde4b2 and Drd44 (Stehle et al. 1993; Kim et al. 2005; Kim et al. 2007). The relative abundance of the isoforms of these genes could be determined by Mbnl2.
The finding that expression of Mbnl2 is regulated by a cAMP mechanism provides reasons to suspect that cAMP might act in this way in other cell types, either in response to first messenger signaling by transmitters or hormones and their agonists; or in response to exogenous agents which elevate cAMP through inhibition of phosphodiesterases, including caffeine and PDE subtype targeted drugs. This hypothesis requires testing, because the regulation of this gene may be strongly influenced by cellular context.
Whereas it is clear that alternative splicing is an essential element in transcript processing, little is known about whether this can be dynamically regulated. The results presented here provide clear evidence that one likely participant in splicing can be rapidly controlled by neural signals. The question of whether this occurs with other proteins involved in splicing remains to be determined.
The finding that Mbnl2 is highly expressed in both the pineal gland and retina places it among a small set of genes that are selectively expressed in both tissues (Klein 2004, 2006). Whereas temporal changes in expression of genes in the pineal gland appears to reflect the action of cAMP, it is thought that the retinal/pineal pattern of expression reflects the action of one or more of a small group of transcription factors which control of cell fate determination and maintenance of phenotype in these tissues. Accordingly, expression is controlled by cAMP acting in concert with one or more of these transcription factors, which includes Pax4 (Rath et al. 2009a; Rath et al. 2009b), Pax6 (Gehring 2005; Philips et al. 2005), Crx (Furukawa et al. 1999; Rath et al. 2006; Rath et al. 2007), Otx2 (Nishida et al. 2003; Rath et al. 2007), and NeuroD (Morrow et al. 1999; Munoz et al. 2007). Pax6 is notable in that it is also found in Drosophila and other animal eyes, where it appears to play a dominant role in development (Matus et al. 2007). Likewise, the fact that Mbnl2 is expressed in the vertebrate pineal gland and retina in addition to Drosophila eyes points to the likelihood that it has an evolutionarily conserved role in the biology of eyes and the vertebrate pineal gland.
Supplementary Material
Acknowledgments
This research was supported in part by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health (J.K., S.L.C., J.L.W., D.C.K.) and grants from the Danish Agency for Science Technology and Innovation, the Lundbeck Foundation, the Danish Medical Research Council (grant numbers 271-07-0412 and 271-06-0754), the Novo Nordisk Foundation, the Carlsberg Foundation, and Simon Foughner Hartmann's foundation (M.M. and M.F.R.). S.B. was a Howard Hughes Medical Institute Fellow of the Life Sciences Research Foundation.
Abbreviations
- ACTD
actinomycin D
- 8-Br
8-bromo cAMP
- CT
circadian time
- DB
dibutyryl cAMP
- DD
constant darkness
- DCNT
decentralization of the superior cervical ganglia
- ISO
isoproterenol
- LD
light dark
- Mbnl2
muscleblind-like 2
- NE
norepinephrine
- PKA
protein kinase A
- PKC
protein kinase C
- PMA
phorbol myrstic acetate
- PURO
puromycin
- SCG
superior cervical ganglia
- SCGX
superior cervical ganglionectomy
- ZT
Zeitgeber time
Footnotes
Mlbn2 mRNA in the rat pineal gland was found to be 5- to 6-fold higher at night relative to day in spotted cDNA array (n = 3) analysis (Blackshaw S, Coon SL, Kim J-S, Cepko CL, and Klein DC, unpublished data). This was supported by subsequent results from an oligonucleotide array (Affymetrix) study (Bailey et al. 2009).
Circadian expression of Drd4 in pineal gland: Adrenergic/cAMP control mechanism requires thyroid hormone. (Kim J-S, Bailey MJ, Weller JL, Sugden D, Rath MF, Møller M and Klein DC, unpublished.)
References
- Ashery-Padan R, Gruss P. Pax6 lights-up the way for eye development. Curr Opin Cell Biol. 2001;13:706–714. doi: 10.1016/s0955-0674(00)00274-x. [DOI] [PubMed] [Google Scholar]
- Bailey MJ, Coon SL, Carter DA, Humphries A, Kim JS, Shi Q, Gaildrat P, Morin F, Ganguly S, Hogenesch JB, Weller JL, Rath MF, Moller M, Baler R, Sugden D, Rangel ZG, Munson PJ, Klein DC. Night/day changes in pineal expression of >600 genes: central role of adrenergic/cAMP signaling. J Biol Chem. 2009;284:7606–7622. doi: 10.1074/jbc.M808394200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begemann G, Paricio N, Artero R, Kiss I, Perez-Alonso M, Mlodzik M. muscleblind, a gene required for photoreceptor differentiation in Drosophila, encodes novel nuclear Cys3His-type zinc-finger-containing proteins. Development. 1997;124:4321–4331. doi: 10.1242/dev.124.21.4321. [DOI] [PubMed] [Google Scholar]
- Charlet BN, Savkur RS, Singh G, Philips AV, Grice EA, Cooper TA. Loss of the muscle-specific chloride channel in type 1 myotonic dystrophy due to misregulated alternative splicing. Mol Cell. 2002;10:45–53. doi: 10.1016/s1097-2765(02)00572-5. [DOI] [PubMed] [Google Scholar]
- Dansithong W, Paul S, Comai L, Reddy S. MBNL1 is the primary determinant of focus formation and aberrant insulin receptor splicing in DM1. J Biol Chem. 2005;280:5773–5780. doi: 10.1074/jbc.M410781200. [DOI] [PubMed] [Google Scholar]
- Fardaei M, Rogers MT, Thorpe HM, Larkin K, Hamshere MG, Harper PS, Brook JD. Three proteins, MBNL, MBLL and MBXL, co-localize in vivo with nuclear foci of expanded-repeat transcripts in DM1 and DM2 cells. Hum Mol Genet. 2002;11:805–814. doi: 10.1093/hmg/11.7.805. [DOI] [PubMed] [Google Scholar]
- Furukawa T, Morrow EM, Li T, Davis FC, Cepko CL. Retinopathy and attenuated circadian entrainment in Crx-deficient mice. Nat Genet. 1999;23:466–470. doi: 10.1038/70591. [DOI] [PubMed] [Google Scholar]
- Gehring WJ. Historical perspective on the development and evolution of eyes and photoreceptors. Int J Dev Biol. 2004;48:707–717. doi: 10.1387/ijdb.041900wg. [DOI] [PubMed] [Google Scholar]
- Gehring WJ. New perspectives on eye development and the evolution of eyes and photoreceptors. J Hered. 2005;96:171–184. doi: 10.1093/jhered/esi027. [DOI] [PubMed] [Google Scholar]
- He F, Dang W, Abe C, Tsuda K, Inoue M, Watanabe S, Kobayashi N, Kigawa T, Matsuda T, Yabuki T, Aoki M, Seki E, Harada T, Tomabechi Y, Terada T, Shirouzu M, Tanaka A, Guntert P, Muto Y, Yokoyama S. Solution structure of the RNA binding domain in the human muscleblind-like protein 2. Protein Sci. 2009;18:80–91. doi: 10.1002/pro.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho TH, Charlet BN, Poulos MG, Singh G, Swanson MS, Cooper TA. Muscleblind proteins regulate alternative splicing. EMBO J. 2004;23:3103–3112. doi: 10.1038/sj.emboj.7600300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho TH, Savkur RS, Poulos MG, Mancini MA, Swanson MS, Cooper TA. Colocalization of muscleblind with RNA foci is separable from mis-regulation of alternative splicing in myotonic dystrophy. J Cell Sci. 2005;118:2923–2933. doi: 10.1242/jcs.02404. [DOI] [PubMed] [Google Scholar]
- Huang H, Wahlin KJ, McNally M, Irving ND, Adler R. Developmental regulation of muscleblind-like (MBNL) gene expression in the chicken embryo retina. Dev Dyn. 2008;237:286–296. doi: 10.1002/dvdy.21408. [DOI] [PubMed] [Google Scholar]
- Kanadia RN, Johnstone KA, Mankodi A, Lungu C, Thornton CA, Esson D, Timmers AM, Hauswirth WW, Swanson MS. A muscleblind knockout model for myotonic dystrophy. Science. 2003;302:1978–1980. doi: 10.1126/science.1088583. [DOI] [PubMed] [Google Scholar]
- Kim JS, Bailey MJ, Ho AK, Moller M, Gaildrat P, Klein DC. Daily rhythm in pineal phosphodiesterase (PDE) activity reflects adrenergic/3′,5′-cyclic adenosine 5′-monophosphate induction of the PDE4B2 variant. Endocrinology. 2007;148:1475–1485. doi: 10.1210/en.2006-1420. [DOI] [PubMed] [Google Scholar]
- Kim JS, Coon SL, Blackshaw S, Cepko CL, Moller M, Mukda S, Zhao WQ, Charlton CG, Klein DC. Methionine adenosyltransferase:adrenergic-cAMP mechanism regulates a daily rhythm in pineal expression. J Biol Chem. 2005;280:677–684. doi: 10.1074/jbc.M408438200. [DOI] [PubMed] [Google Scholar]
- Klein DC. Photoneural regulation of the mammalian pineal gland. Ciba Found Symp. 1985;117:38–56. doi: 10.1002/9780470720981.ch4. [DOI] [PubMed] [Google Scholar]
- Klein DC. The 2004 Aschoff/Pittendrigh lecture: Theory of the origin of the pineal gland--a tale of conflict and resolution. J Biol Rhythms. 2004;19:264–279. doi: 10.1177/0748730404267340. [DOI] [PubMed] [Google Scholar]
- Klein DC. Evolution of the vertebrate pineal gland: the AANAT hypothesis. Chronobiol Int. 2006;23:5–20. doi: 10.1080/07420520500545839. [DOI] [PubMed] [Google Scholar]
- Mankodi A, Takahashi MP, Jiang H, Beck CL, Bowers WJ, Moxley RT, Cannon SC, Thornton CA. Expanded CUG repeats trigger aberrant splicing of ClC-1 chloride channel pre-mRNA and hyperexcitability of skeletal muscle in myotonic dystrophy. Mol Cell. 2002;10:35–44. doi: 10.1016/s1097-2765(02)00563-4. [DOI] [PubMed] [Google Scholar]
- Matus DQ, Pang K, Daly M, Martindale MQ. Expression of Pax gene family members in the anthozoan cnidarian, Nematostella vectensis. Evol Dev. 2007;9:25–38. doi: 10.1111/j.1525-142X.2006.00135.x. [DOI] [PubMed] [Google Scholar]
- Miller JW, Urbinati CR, Teng-Umnuay P, Stenberg MG, Byrne BJ, Thornton CA, Swanson MS. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 2000;19:4439–4448. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow EM, Furukawa T, Lee JE, Cepko CL. NeuroD regulates multiple functions in the developing neural retina in rodent. Development. 1999;126:23–36. doi: 10.1242/dev.126.1.23. [DOI] [PubMed] [Google Scholar]
- Munoz EM, Bailey MJ, Rath MF, Shi Q, Morin F, Coon SL, Moller M, Klein DC. NeuroD1: developmental expression and regulated genes in the rodent pineal gland. J Neurochem. 2007;102:887–899. doi: 10.1111/j.1471-4159.2007.04605.x. [DOI] [PubMed] [Google Scholar]
- Nishida A, Furukawa A, Koike C, Tano Y, Aizawa S, Matsuo I, Furukawa T. Otx2 homeobox gene controls retinal photoreceptor cell fate and pineal gland development. Nat Neurosci. 2003;6:1255–1263. doi: 10.1038/nn1155. [DOI] [PubMed] [Google Scholar]
- Orengo JP, Chambon P, Metzger D, Mosier DR, Snipes GJ, Cooper TA. Expanded CTG repeats within the DMPK 3′ UTR causes severe skeletal muscle wasting in an inducible mouse model for myotonic dystrophy. Proc Natl Acad Sci U S A. 2008;105:2646–2651. doi: 10.1073/pnas.0708519105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual M, Vicente M, Monferrer L, Artero R. The Muscleblind family of proteins: an emerging class of regulators of developmentally programmed alternative splicing. Differentiation. 2006;74:65–80. doi: 10.1111/j.1432-0436.2006.00060.x. [DOI] [PubMed] [Google Scholar]
- Paul S, Dansithong W, Kim D, Rossi J, Webster NJ, Comai L, Reddy S. Interaction of muscleblind, CUG-BP1 and hnRNP H proteins in DM1-associated aberrant IR splicing. EMBO J. 2006;25:4271–4283. doi: 10.1038/sj.emboj.7601296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philips GT, Stair CN, Young Lee H, Wroblewski E, Berberoglu MA, Brown NL, Mastick GS. Precocious retinal neurons: Pax6 controls timing of differentiation and determination of cell type. Dev Biol. 2005;279:308–321. doi: 10.1016/j.ydbio.2004.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath MF, Morin F, Shi Q, Klein DC, Moller M. Ontogenetic expression of the Otx2 and Crx homeobox genes in the retina of the rat. Exp Eye Res. 2007;85:65–73. doi: 10.1016/j.exer.2007.02.016. [DOI] [PubMed] [Google Scholar]
- Rath MF, Bailey MJ, Kim JS, Coon SL, Klein DC, Moller M. Developmental and daily expression of the Pax4 and Pax6 homeobox genes in the rat retina: localization of Pax4 in photoreceptor cells. J Neurochem. 2009a;108:285–294. doi: 10.1111/j.1471-4159.2008.05765.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath MF, Munoz E, Ganguly S, Morin F, Shi Q, Klein DC, Moller M. Expression of the Otx2 homeobox gene in the developing mammalian brain: embryonic and adult expression in the pineal gland. J Neurochem. 2006;97:556–566. doi: 10.1111/j.1471-4159.2006.03773.x. [DOI] [PubMed] [Google Scholar]
- Rath MF, Bailey MJ, Kim JS, Ho AK, Gaildrat P, Coon SL, Moller M, Klein DC. Developmental and diurnal dynamics of Pax4 expression in the mammalian pineal gland: nocturnal down-regulation is mediated by adrenergic-cyclic adenosine 3′,5′-monophosphate signaling. Endocrinology. 2009b;150:803–811. doi: 10.1210/en.2008-0882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehle JH, Foulkes NS, Molina CA, Simonneaux V, Pevet P, Sassone-Corsi P. Adrenergic signals direct rhythmic expression of transcriptional repressor CREM in the pineal gland. Nature. 1993;365:314–320. doi: 10.1038/365314a0. [DOI] [PubMed] [Google Scholar]
- Timchenko LT, Tapscott SJ, Cooper TA, Monckton DG. Myotonic dystrophy: discussion of molecular basis. Adv Exp Med Biol. 2002;516:27–45. doi: 10.1007/978-1-4615-0117-6_2. [DOI] [PubMed] [Google Scholar]
- Warf MB, Berglund JA. MBNL binds similar RNA structures in the CUG repeats of myotonic dystrophy and its pre-mRNA substrate cardiac troponin T. RNA. 2007;13:2238–2251. doi: 10.1261/rna.610607. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


