Abstract
It has been known for a long time that genetic factors affect sleep quantity and quality. Genetic screens identified several mutations that affect sleep across species, pointing to an evolutionary conserved regulation of sleep. Moreover, it has also been recognized that sleep affects the expression of genes. These findings have given valuable clues about the molecular underpinnings of sleep regulation and function that might lead the way to more efficient treatments for sleep disorders.
Introduction
Studies with twins in the 1930’s suggested for the first time that sleep is, to some extent, under genetic control. In 1992 PRNP, which codes for the prion protein PRNP, was the first specific gene linked to a human sleep disorder, fatal familial insomnia 1 and was later on shown to affect sleep regulation in mice 2,3. In 1999 the hypocretin/orexin system was implicated in human narcolepsy through genetic studies in dogs 4 and mice 5. Since then, other genes have been linked to human sleep disorders or have been shown to affect sleep in animals across species, suggesting that some of the core mechanisms underlying sleep and its regulation are conserved. Recently, molecular studies have also identified hundreds of brain transcripts across species that change their expression level between sleep and waking, suggesting that the functional consequences of sleep are also shared across species. Thus, specific genes can profoundly affect sleep and, conversely, sleep can influence brain gene expression.
The recent progress in identifying sleep genes and sleep-dependent expression patterns was driven by multiple experimental approaches, from classical mutagenesis screening in flies, quantitative trait loci (QTL) analysis in mice, genome-wide association studies in humans to extensive transcriptomic analysis in flies and rodents (Box 1).
Box 1. Methods to identify genes affecting sleep phenotypes.
Several strategies can be used. In reverse genetics a candidate gene is mutated first, and then the effects of the mutation on sleep are assessed (from genotype to phenotype). Candidate genes are chosen based on their known function, which makes them likely to be relevant for sleep. In forward genetics the starting point is the phenotype, and a significant part of the work involves going back to the genotype to identify the responsible gene (from phenotype to genotype). This approach is unbiased, and novel genes can be discovered. Forward genetic methods include quantitative trait loci (QTLs) analysis and mutagenesis screening, which can complement each other. Quantitative trait loci (QTLs) are stretches of genome closely linked to the genes that underlie the phenotype under study. QTL analysis starts with the crossing between two inbred mouse strains that differ in the trait under study, and maps a chromosomal region segregating with the phenotype in the progeny. The region may contain either a single “major” gene (that can explain >25% of the genetic variance of a trait) or several genes with small effects. In mutagenesis screenings random small mutations are induced over the entire genome. Insertional mutagenesis uses transposable elements (in flies) to induce mutations, while chemical mutagenesis uses ethylmethane sulfonate (EMS, in flies) or N-ethyl N-nitrosourea (ENU, in mice). In these studies hundreds/thousands of mutated flies or mice are usually screened for the phenotype of interest, and behavioral analysis is preferred because less time consuming and less expensive 120.
This Review will first describe the multiple aspects of sleep that can be grouped in circadian and homeostatic sleep phenotypes, and then discuss some of the genes whose mutations significantly affect sleep in flies, mice, and humans. Overall, these genes can be broadly subdivided into four major functional categories: ion channels, circadian regulation, neurotransmission, and other signaling pathways/hormones. Finally, the effects of sleep on brain gene expression will be reviewed, and the functional categories of waking-related and sleep-related transcripts will be discussed.
Sleep phenotypes
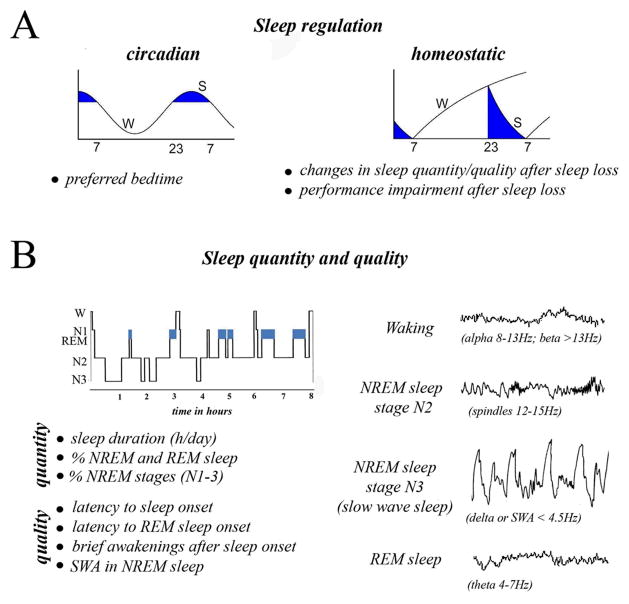
Studies in the 1930’s reported that some sleep phenotypes have higher concordance in monozygotic than in dizygotic twins, suggesting some genetic basis (reviewed in 6). A first requirement for studying this phenomenon is to define and characterize the phenotypes of sleep: how much, how well, and when we sleep (Fig 1). It is noteworthy, that sleep phenotypes can change independently of each other. For instance, sleep can be poorly restorative and yet of “normal” duration. Similarly, in some circadian sleep disorders the preferred bedtime occurs too early or too late, but daily sleep amount and sleep quality are preserved.
Figure 1.
Sleep phenotypes. Sleep phenotypes such as regulation, sleep duration (quantity), or sleep intensity (quality) may reflect different aspects of sleep. A) Sleep regulation. Blue areas indicate time of day most conducive to sleep in humans, due to the combined effect of the circadian system, which consolidates sleep during the dark phase, and the homeostatic system, which increases sleep pressure as a function of waking duration. B) Sleep quantity and quality. Top left, night distribution of sleep stages in adult humans. Right, representative EEG traces in waking, NREM sleep and REM sleep. The waking “activated” EEG is dominated by low voltage fast activity in the beta (>13Hz) and alpha (8–13Hz) range. NREM sleep comprises a transitional stage 1 (N1, not shown), when alpha activity disappears, followed by stage 2 (N2), rich in sleep spindles, and then N3 or slow wave sleep (SWS, also called stages 3+4), when the EEG shows prominent slow waves 119. The higher the number of slow waves, the deeper is NREM sleep, i.e. the more difficult is to wake up. Sleep spindles are waxing and waning oscillations of thalamic origin whose frequency (12–15 Hz) is comprised within the sigma band (12–16Hz), while slow waves are of cortical origin and are comprised within the delta band, also called slow wave activity (SWA, <4.5Hz).
Sleep is tightly regulated by two sets of mechanisms that work partly independent of each other: the circadian and homeostatic mechanism (Fig 1A). The circadian mechanism reflects how sleep propensity changes during the 24 hours, and its function is to restrict sleep to a time of day that is ecologically appropriate. In mammals, the suprachiasmatic nucleus (SCN) of the hypothalamus is responsible for circadian (circa-diem, approximately 24 hours) rhythms. After SCN lesions sleep is no longer consolidated in one major phase (the day in rodents, the night in flies and humans), but occurs in short episodes throughout the 24-hour period. The homeostatic mechanism, instead, reflects how sleep pressure accumulates during wakefulness and is discharged when we sleep. If waking is prolonged beyond its physiological duration (~ 16 hours in humans) sleepiness increases, performance decreases, and when sleep is finally permitted its duration and/or intensity are greater than in baseline conditions. Sleep homeostasis is controlled centrally by the interaction between sleep-promoting neuronal groups in hypothalamus and basal forebrain, and waking-promoting groups in hypothalamus and brainstem. Sleep need is also locally regulated: brain regions that have undergone synaptic potentiation (e.g. after learning) show larger slow waves, reflecting deeper sleep states 7.
Sleep quantity (duration of total sleep and of its various phases) and quality (latency to sleep, brief arousals after sleep onset, amount of large slow waves) also belong to the sleep phenotypes that have to be accounted for when sleep regulation and function are assessed (Fig 1B).
Sleep quantity and quality in humans is assessed by measuring brain electrical activity with the electroencephalogram (EEG). EEG patterns change predictably depending on behavioral state, from the low voltage fast activity of waking and REM sleep to the slow waves and spindles of NREM sleep (Fig. 1B). More refined information about sleep quality comes from the analysis of the EEG power spectrum, which measures the extent to which specific frequency bands are represented in the EEG signal (Fig. 1B). Interestingly, recent studies found strong heritability of the sleep EEG power spectrum 8,9 (Box 2). Evidence for the heritability of other sleep traits is listed in Supplementary Table 1.
Box 2. Heritability of the sleep and waking EEG.
Two recent reports found strong heritability of the sleep EEG power spectrum 8,9, extending previous results in normal sleepers (not twins), which showed that the sleep EEG is very consistent across nights in the same subject, but varies considerably from one individual to another. Other studies found that the trait-like nature of the human sleep EEG is evident at most frequencies below 15–16 Hz121–126. Heritability is probably not as strong for delta activity, which is significantly affected by sleep/waking history. It is worth mentioning that the spectral composition of the human EEG shows striking heritability also during waking, with estimates ranging from 70 to 90% for most frequency bands 127,128. The coherence of the waking EEG is also under strong genetic influence 129,130. In fact, the EEG is among the most hereditable traits in humans, although few specific genes have been identified so far. A linkage disequilibrium between the beta frequency of the human EEG and a GABAA receptor locus has been reported in a rest state with eyes closed, but it is unknown whether this finding is specific for relaxed waking or extends to sleep 131. Thus, the challenge for the future will be not only to identify which specific genes influence the human EEG, but also to determine to which extent their effects are unique to sleep or waking. It is likely that at least some of the interindividual differences in the human EEG are related to genetic factors that are independent of behavioral state.
The most studied is the EEG delta activity, the EEG power in the 0.5–4.5Hz range during NREM sleep, also called slow wave activity (SWA). The higher the SWA, the deeper is NREM sleep, i.e. the more difficult it is to wake up when a stimulus is delivered 10. Moreover, the longer the duration of waking, the higher is the level of SWA at sleep onset (refs in 11). For these reasons delta activity is considered a marker of sleep intensity as well as of sleep need. Sleep in rodents, birds, and other animals is also customarily quantified using EEG analysis, while behavioral criteria are used as indicators of sleep in fish and invertebrates. Indeed, it is worth pointing out that in almost all animals sleep is characterized by quiescence (marine mammals are an exception) and increased arousal threshold. The latter is the single most important feature that distinguishes sleep from “rest” or quiet waking, suggesting that the reduced ability to respond to the environment is a requirement for the restorative function of sleep to occur, and can be captured without the EEG 12. Thus, the EEG is crucial for a detailed analysis of NREM and REM sleep in many vertebrates, but our understanding of sleep regulation and functions can equally benefit from behavioral analysis, as done in several newly introduced invertebrate animal models (Table 1).
Table 1.
The best characterized non-mammalian animal models currently used in sleep research, together with major similarities and differences relative to sleep in mice and other mammals.
| Aspects of sleep documented in mice and other mammals | Fruit fly (Drosophila melanogaster) | Zebrafish (Danio rerio) | Caenorhabditis elegans |
|---|---|---|---|
| Behavioral definition of sleep is met | yes: quiescence and increased arousal threshold 88,132 | yes: quiescence and increased arousal threshold 14–16 | yes: quiescence and increased arousal threshold during development (lethargus) 17; preliminary evidence for quiescence and increased rest after deprivation in adults 133 |
| Documented changes in brain activity | yes: brain electrical activity is reliably correlated with behavioral state 134 | ||
| Homeostatic regulation of sleep is present | yes: increase in sleep time, arousal threshold, duration of sleep episodes and decrease in brief awakenings after sleep deprivation; homeostatic regulation largely independent of the circadian clock 88,132,135; however, sleep is more fragmented in cycle and Clock mutants, and female (but not male) cycle mutants show exaggerated response to sleep deprivation 40,97 | yes: increase in sleep time, arousal threshold, and duration of sleep episodes after sleep deprivation by electrical stimulation or vibration; weak or no homeostatic response after sleep deprivation by light exposure 14,16 | yes: following deprivation quiescence occurs earlier, is more consolidated; arousal threshold increased relative to baseline 17 |
| Circadian regulation of sleep is present | yes: sleep mainly at night in entrained light:dark conditions or constant darkness; arrhythmic sleep after lesions of the circadian clock 88 | yes: sleep mainly at night 14–16 | Lethargus is time locked to the expression of Lin-42, the C. elegans ortholog of the circadian gene Period 136 |
| Changes in brain gene expression associated with sleep and waking | yes: some are similar to those seen in mammals79,84 | ||
| Changes in sleep parameters with aging | yes:sleep fragmentation in old flies 137 | (not tested?) | (sleep-like state well defined only during larval development) |
| Drugs and signaling pathways: similarities with mammals | increase in waking with caffeine, modafinil, amphetamines, octopamine (insect equivalent of norepinephrine), increase in sleep with antihistamines 88,132,138–140
|
increased sleep with melatonin, GABAergic hypnotics, alpha2-adrenergic agonists, histaminergic H1 antagonists 14,145,146 | most major mammalian neurotransmitters present (ACh, glutamate, dopamine, serotonin, GABA) As in mammals, genetic manipulations that increase the EGFR pathway block locomotion and feeding 147, but untested whether this quiescence is a sleep-like state (i.e. with increased arousal threshold) |
| Drugs and signaling pathways: differences with mammals |
|
unclear whether the hypocretin/orexin system is wake-promoting as in mammals 15, or sleep- promoting 16 | |
| Major differences relative to mammals |
|
|
|
| Major strengths and limitations as animal model for sleep |
|
|
|
Sleep studies across species
Studies across in flies, mice and humans have identified genes that affect sleep. These genes can be grouped into four major functional categories (Supplementary Table 2), which will be discussed below after briefly discussing contributions from various species.
Studies in flies
Studies over the past 8 years have demonstrated that the fruit fly shows most of the fundamental features of mammalian sleep (Table 1). Flies lend themselves ideally to forward genetic approaches (Box 1) because their genome is less redundant than mouse or human genomes, which means that a mutation in the fly is more likely to yield a strong phenotype. Recently, the naturally occurring variation in both sleep phenotypes and mRNA expression levels of wild-derived inbred lines has also been used as a promising approach to identify candidate genes 13. The genetic basis of sleep will also benefit from studies in the zebrafish Danio rerio and the roundworm Caenorhabditis elegans (Table 1), both of which have recently been shown to have a sleep-like state 14–17.
Studies in mice
Almost all mouse genes known to affect sleep have been identified by reverse genetics studies, with more than 70 mutant lines tested so far, starting from two pioneering reports in 1996 2,18 (Supplementary Table 2). Quantative trait loci (QTL) analysis (Box 1) has recently succeeded twice, in linking a single mouse gene to a specific frequency of the sleep EEG 19,20, while no mouse gene so far has been identified using mutagenesis screenings. A few general conclusions can be drawn from these studies. First, most mouse mutant lines show effects on at least one sleep phenotype, which is perhaps not surprising since these lines carry mutations in “candidate” genes. Second, the effects on sleep quantity – if any- are usually small, with increases or, in most cases, decreases in total sleep of ~ 20% or less, due to a decline in NREM or REM sleep. Very few mutations affect both sleep phases, and then almost always in the same direction (prokineticin 2-deficient mice 21 and corticotrophin-releasing hormone overexpressing mice 22 are exceptions; Supplementary Table 2). Third, changes in the response to sleep deprivation, when present, are also usually small, although caution is needed to interpret these results, because most studies do not assess sleep homeostasis in a comprehensive manner.
Studies of sleep disorders in human
In several cases the genetic basis of human sleep disorders has been clarified, and the identified candidate genes have been tested in animal models. Examples that will be discussed include the hypocretin/orexin system and its role in narcolepsy, and Period (Per) genes, whose mutations result in abnormal circadian regulation.
Ion Channels
Over the last 3 years, mutagenesis screenings have identified two fly genes with striking effects on fly sleep, namely Shaker and Sleepless. The first gene, Shaker, which was identified with EMS mutagenesis (Box 1), codes for the alpha subunit of a tetrameric potassium channel that passes a voltage-activated fast-inactivating IA current 23. Homologous channels in vertebrates have similar properties and, in both mammals and flies, IA plays a major role in the control of membrane repolarization and transmitter release 23. Flies carrying minisleep, or other Shaker loss of function mutations, sleep only 2–4 hours every day rather than 8–10 hours, but their circadian and homeostatic regulation of sleep are normal 24. Moreover, learning and memory in these flies is impaired, and lifespan is reduced, although it is still unclear whether these deficits can be ascribed to reduced sleep 24,25. Hyperkinetic codes for a beta regulatory subunit that interacts with the alpha pore forming subunits coded by Shaker. Hyperkinetic loss of function mutations also show a short sleeping phenotype, impairment in learning and memory, and reduced life span 25. In Hyperkinetic mutants sleep is not reduced as much as in Shaker mutants (by 30–50%), which is expected since Hyperkinetic null mutations reduce, but do not abolish, the IA current.
Sleepless, was identified using insertional mutagenesis 26 (Box 1). Sleepless flies, like the most extreme Shaker null mutants, sleep only ~ 2 hours a day (~ 85% less than controls), mainly due to a decrease in sleep episode duration. Sleepless codes for a glycosyl-phosphatidylinositol-anchored protein with unknown function, and has no obvious vertebrate homolog. Quiver, however, a previously identified mutation that affects the IA current, is allelic to Sleepless, and Sleepless flies have reduced levels of Shaker. This suggests that the Sleepless short sleeping phenotype is at least in part mediated by the Shaker current. There are nevertheless some interesting differences between Shaker and Sleepless flies, most notably that only the latter show a reduced homeostatic response, as indicated by no changes in sleep duration after sleep deprivation.
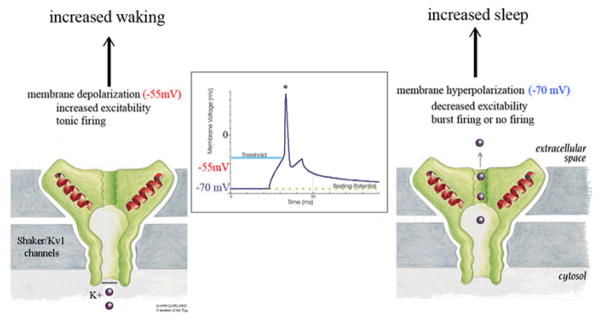
Why do mutations affecting the IA current decrease sleep duration so significantly? In mammals, neurons in the cortex and thalamus are more hyperpolarized in sleep than in waking 27, and one possibility is that these mutations may impinge on the core cellular mechanisms of sleep by changing overall neuronal excitability (Fig. 2). Based on sequence similarity, the closest mammalian homologues of the Drosophila Shaker gene are the alpha subunits of the Kv1 family of potassium channels, while the Kv2, Kv3, and Kv4 families are more distantly related (“Shaker-like”). Kv1 channels activate in the subthreshold voltage range in many neurons, and can act as extremely diverse regulators of neuronal excitability 28 (Figure 2). Mice lacking the closest mammalian homologue of Shaker, Kv1.2 (Kcna2), sleep less. Their short sleeping phenotype, however, is far from being as dramatic as in Shaker flies 29, perhaps because of redundancy - there is one Shaker gene in Drosophila, but at least 16 genes code for alpha subunits of voltage-dependent potassium channels in mammals 30,31.
Figure 2.
Proposed mechanism for the short sleeping phenotype caused by loss of function mutations of Shaker/Kv1 potassium channels. Normally, the opening of these channels allow K+ ions to exit the neuron, bringing the membrane potential to more hyperpolarized (more negative) levels, close to the resting membrane potential. Mutations that reduce the total number of these channels, and/or decrease the time the channel can remain open, tend to bring the membrane potential to more positive (depolarized levels), closer to the threshold for firing an action potential (* in the figure).
A striking sleep phenotype is observed in double knockout (KO) mice lacking the voltage-dependent potassium channels Kv3.1 and Kv3.332, which sleep less (by 40%), have shorter sleep episodes, an overall decrease in the EEG power spectrum more evident in NREM sleep, and no response to sleep deprivation. These mice are also hyperactive and show motor dysfunction, which may be partly responsible for their sleep fragmentation 32,33. The mechanism underlying the effect of these mutations on sleep is most likely different from those involved in Shaker and Sleepless mutants, because Kv3-type channels are mainly expressed in cortical and thalamic GABAergic interneurons, where their presence enables these cells to fire repetitively at high-frequency 34. Whether human extreme short sleepers have mutations in voltage-dependent potassium channels remains unknown, but in one case of Morvan’s syndrome, a rare autoimmune disorder with central symptoms, marked sleeplessness has been associated with the presence of autoantibodies against voltage-dependent potassium channels 35.
Another candidate sleep gene that was identified in mice is Cav3.1, which codes the alpha1G subunit of the T-type calcium channels. These channels play a crucial role in most of the neuronal oscillations displayed by thalamic and cortical neurons during NREM sleep 36. Two independent studies have reported a decrease of ~20% in NREM sleep and more frequent brief awakenings, a sign of disrupted sleep, in mutant mice carrying a Cav3.1 deletion either globally or in most of the thalamus, while cortical deletion had no effect37,38. However, detailed experiments to assess the effects of these mutations on the sleep EEG still need to be carried out.
Circadian regulation
At the molecular level, circadian genes interact and determine circadian rhythmicity. Most of these genes have been tested for their effects on sleep (Supplementary Table 2). Double KO mice of the circadian genes cryptochrome1 and cryptochrome2 sleep more and have higher NREM delta activity, but show little further increase in sleep duration and intensity after sleep deprivation 39. Other mutations of circadian mouse genes (Clock, brain and muscle ARNT-like protein 1, neuronal PAS domain protein 2, prokineticin 2) also result in an abnormal response to sleep deprivation, as do mutations of the fly circadian genes Cycle and Clock (Table 1). Overall, it appears therefore that circadian mutations not only affect the timing of sleep, as expected, but also its homeostatic regulation.
Period (Per) genes
A functional circadian clock seems to require oscillations of Per protein levels and its phosphorylation, and/or its rhythmic nuclear localization (Fig. 3). Flies lacking Per have a disrupted rest/activity cycle and reduced sleep, but whether the short sleeping phenotype is due to the specific loss of Per remains unclear 40. Loss of Per2 in mice (mammals have 3 Per genes) also disrupts locomotor activity rhythms; moreover, cortical Per2 levels change depending on both time of day (high at night) and behavioral state (high in waking) 39,41–43. Since Per2 induction is sensitive to the NAD/NADH ratio 44,45 it may reflect the level of brain activity 46, which varies with sleep and waking. Thus, Per2 could be an essential part of the central clock as well as of the mechanism that regulates sleep need based on waking duration (Fig. 3). However, studies in animals remain inconclusive, because Per1, Per2, Per3, and double Per1/Per2 KO mice have normal sleep duration and sleep homeostasis.
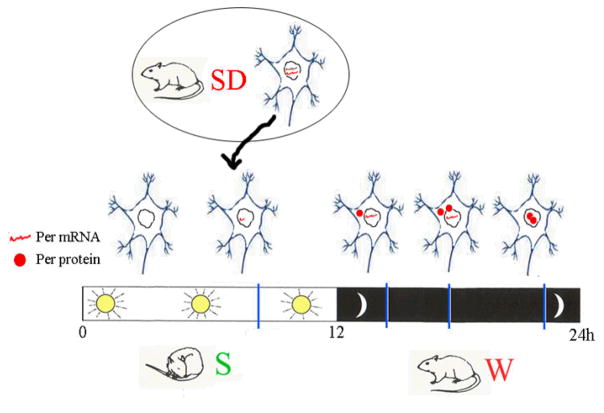
Figure 3.
Model showing changes in the expression of Period genes (mRNA and protein) as a function of the 24-hour cycle and in response to sleep deprivation (SD): mRNA levels grow during the day, while protein levels peak at night, when Per enters the nucleus and blocks its own expression. These circadian changes are similar regardless of whether the animal is diurnal (like flies, which sleep mainly at night) or nocturnal (like rats, which sleep mostly during the day). Rodents forced to stay awake during the day show increased Per mRNA levels in the cerebral cortex. In both flies and mammals, however, sleep deprivation usually does not reset the circadian clock, i.e. does not cause phase shifts.
Studies in humans, instead, point to an important role for PERIOD3, which contains variable-number tandem-repeat polymorphisms in its coding region. Relative to PER34/4 carriers, PER35/5 individuals have longer duration of SWS and higher NREM delta activity, the latter also after sleep deprivation 47. PER35/5 subjects also show higher alpha and theta EEG activity during waking and REM sleep (both signs of increased sleep pressure), and after total sleep deprivation (but not chronic sleep restriction 48) are more impaired in executive functioning tasks in the early morning 47,49. Significant interindividual differences in the cognitive impairment caused by sleep loss have been found before (reviewed in 50). Recent experiments suggest that these individual differences are also associated with different patterns of neural activation as measured using functional MRI 51. Further studies are required to confirm that PER3 plays a crucial role in modulating susceptibility to sleep deprivation.
The human familial advanced sleep phase syndrome (FASPS) is transmitted in a highly penetrant autosomal dominant manner. FASPS subjects have a normal duration of sleep but go to sleep ~ 4-hour earlier than usual (extreme early birds). Some FASPS individuals carry a serine to glycine mutation in PER2, and the mutation in vitro affects the ability of casein kinase I epsilon to phosphorylate PER2 52. The same mutation in mice recapitulates the human FASPS phenotype 53. A missense mutation in CSNK1D (human casein kinase I delta gene) also results in FASPS, and reduces enzymatic activity in vitro 54. These and other studies show that the extent to which PERIOD proteins are phosphorylated greatly affects their nuclear accumulation and thus influence the endogenous circadian period, suggesting that future proteomic studies can help our understanding of how sleep and circadian rhythms are regulated. Genetic association studies have also suggested a link between delayed sleep phase syndrome (DSPS, extreme night owls) and several genes, including PER3 (refs in 55).
Neurotransmission
In general, genetic studies have largely confirmed the arousal-promoting role of the noradrenergic, histaminergic, serotonergic, cholinergic and hypocretin/orexin systems (detailed refs in Supplementary Table 2). In some cases, these studies have also revealed a more specific role for some of these systems in regulating certain sleep phenotypes. For instance hypocretins/orexins have been shown to be essential for stabilizing sleep and wakefulness 56. Indeed narcolepsy, a neurological disorder caused by hypocretins/orexins deficiency, is characterized by the inability to maintain long waking periods, abrupt transitions into NREM sleep, and abnormal intrusions of REM sleep into waking 56. Canine narcolepsy is an autosomal recessive, fully penetrant, disorder due to a mutation in the gene coding for the hypocretin receptor 2 4. In mice, a narcoleptic-like phenotype is present in both the preprohypocretin/orexin KO mice, where the hypocretins/orexins producing cells are spared but the corresponding peptides are lost, and in the hypocretin/orexin-ataxin 3 transgenic rats and mice, where both cells and peptides are missing.
Genetic studies have also shown that the noradrenergic and histaminergic systems are important in maintaining arousal, since mice deficient in histamine (Hdc KO mice) or norepinephrine/epinephrine (Dbh KO mice) tend to sleep more, and have less trouble in going back to sleep after exposure to mild stress. On the other hand, mice lacking the serotonin transporter SERT or serotonin receptor 1A have 40–50% more REM sleep, in line with the role of serotonin in the regulation of this behavioral state.
Reverse genetics has also clarified the mechanisms by which some drugs affect sleep. For instance, adenosinergic transmission, which is blocked by caffeine, plays an important role in the homeostatic regulation of sleep, and adenosine levels increase during waking and decrease during sleep in cortex and basal forebrain of rats and cats (reviewed in 57). Mice lacking the adenosine receptor A2AR are insensitive to the wake-promoting effects of caffeine, consistent with the fact that in humans a polymorphism in A2AR contributes to individual sensitivity in the effects of caffeine on sleep 57. In humans, a functional polymorphism in the gene coding for the catabolic enzyme adenosine deaminase (ADA) results in a ~ 20–30% decrease in enzymatic activity in blood cells, ~ 50% increase in SWS duration, and ~60% increase in NREM delta activity, with no change in total sleep time 58. Whether this genetic variation also affects the response to sleep deprivation remains untested.
Other signaling pathways and hormones
A variety of molecules have been tested for their effects on sleep, but no systematic analysis of all major signaling pathways has been attempted so far. While it is not possible to provide a comprehensive summary of these studies in the main text (but see Supplementary Table 2), it is clear that a few mutations produce large (40–50%) changes in sleep quantity. They include those affecting RIM1alpha, a protein involved in synaptic vesicle release 59, and UBE3A, a ubiquitin protein ligase whose null mutation, however, also causes abnormal EEG discharges 60. Other signaling pathways have been linked to a specific frequency of the sleep EEG. During REM sleep the EEG shows prominent theta activity, which varies greatly in frequency among inbred mouse strains (slow and fast theta strains). By using QTL analysis followed by fine mapping, one study was able to link Acads to theta frequency 19. Acads encodes the short-chain acyl-coenzyme A dehydrogenase, an enzyme involved in fatty acid beta oxidation, but the mechanism by which its deficiency can slow down the peak theta frequency (from ~8 to 6 Hz) remains unclear 19. Another study that used forward, molecular, and reverse genetic approaches showed that Rarb, the gene encoding the retinoid acid receptor beta, is important to determine the contribution of delta activity to the EEG during NREM sleep 20. The mechanism by which this nuclear receptor affects the amount of delta activity, and thus cortical synchrony during sleep, is unclear, but it may depend on its role in development, neural plasticity, and dopaminergic transmission. Rarb does not seem to play a role in sleep homeostasis.
The Dps1 locus
A recent study using 25 recombinant inbred mice strains found that the increase in NREM delta activity after sleep deprivation depends on both duration of sleep and genotype. It was found that Dps1, a QTL locus on mouse chromosome 13 that contains more than 200 genes, accounts for 49% of the genetic variance in this trait 61,62. Two follow-up studies identified Homer1a, an immediate early gene induced by neuronal activity 63,64, as a likely candidate gene within Dps1 65,66. The conclusion was based on the consistent and strong induction of Homer1a after sleep deprivation in the whole brain of several mice strains 65, the higher expression of cortical HOMER1A in the strain with the greatest increase in delta activity, and the presence of single nucleotide polymorphisms in Homer1a upstream regulatory region 66. It remains to be proven conclusively whether Homer1a plays a role in sleep homeostasis. Future studies should show, for instance, that Homer1a KO mice have a significantly impaired response to sleep deprivation. Since Dps1 contains numerous genes, it is also possible that other major candidates will be identified within this locus. Flies lacking the single Drosophila Homer gene are hyperactive and show defects in courtship behavior, but whether their sleep regulation is abnormal is unknown 67.
Studies in humans: genetic control of sleep disorders
Fatal familial insomnia (FFI)
Fatal familial insomnia (FFI) is a rare autosomal dominant disease due to a point mutation at codon 178 of PRNP. The same mutation is also present in patients affected by the familial form of Creutzfeldt-Jakob disease (CJD), another prion disease with extensive cortical, rather than thalamic, degeneration, and in which dementia, rather than insomnia, is the main clinical feature. In FFI patients, codon 129 on the mutated allele codes for a methionine, while in CJD patients it codes for a valine (reviewed in 68,69). FFI is characterized by a decrease in sleep spindles, sleep fragmentation, reduction in total sleep time, loss of the circadian regulation of sleep, disappearance of SWS, and intrusion of a REM-like state into wakefulness. FFI patients show accumulation of abnormal PRNP in the brain, but PRNP levels do neither correlate with the severity of the disease, nor with the extent of neuronal degeneration. It is also unclear to what extent the sleep disturbances contribute to the death of FFI patients. Mice lacking Prnp have fragmented sleep, suggesting that the normal protein may promote sleep consolidation 2,3. Sleep problems also exist in CJD patients, and transgenic mice carrying the mouse homolog of the human D178N/V129 mutation show a large decrease in REM sleep and periods of a “mixed” state that cannot be easily classified as either sleep or waking 70.
Narcolepsy
While in dogs and mice narcolepsy is genetically determined, non-genetic factors play a major role in human narcolepsy, as suggested by its low (~30%) concordance in monozygotic twins. No association has been found between human narcolepsy and polymorphisms in the genes of the hypocretin/orexin system, and so far only one case of narcolepsy with an unusually early onset has been associated with a mutation in preprohypocretin 71. Yet, most patients with narcolepsy-cataplexy have low or undetectable levels of hypocretins, and narcolepsy is strongly associated with human leucocyte antigen (HLA) alleles, in particular HLA DQB1*0602. This suggests that the deficit in hypocretinergic neurotransmission in human narcolepsy could be due to an autoimmune attack 72. Consistent with this, a recent genome-wide association study found that narcolepsy is associated with the T-cell receptor alpha locus73.
Restless leg syndrome (RLS)
RLS is a common sleep disorder often characterized by periodic limb movements during sleep. RLS can be familial (autosomal dominant in up to 1/3 of cases). Although dopaminergic agonists are used to treat primary RLS, no association has been found between this disorder and genes involved in dopaminergic transmission. Instead, recent genome-wide case-control studies identified 4 predisposing loci on chromosomes 2p, 6p, 9p and 15q. One locus is within the homeobox gene MEIS1, which is involved in limb development. The second within BTBD9, whose function may be related to iron storage, the third between the gene coding for the kinase MAP2K5 and LBXCOR1, a homeodomain transcription factor important for the development of GABAergic interneurons in the dorsal horn of the spinal cord, and the fourth within PTPRD (protein tyrosine phosphatase receptor type delta) 74,75. A second genome-wide association study also found an association between BTBD9 and RLS with period limb movements 76. Together, the 4 loci may explain more than 50% of the risk for RLS in individuals with European ancestry.
Obstructive sleep apnea syndrome (OSAS)
OSAS, a common disorder characterized by recurrent episodes of apnea/hypopnea (no or reduced airflow) during sleep, is also to some extent under genetic control. The genetic basis of this disorder, however, is difficult to study, because many of the risks factors for OSAS, including obesity and alterations of the craniofacial morphology, are also under genetic control 77.
Effects of normal sleep and waking on brain gene expression
It is well established that brains of sleeping and awake animals differ in terms of patterns of neural activity, content of many neurotransmitters and neuromodulators, metabolism, and ability to react to external stimuli. Recent whole-genome transcriptomic studies have now revealed that brains of sleeping and awake animals also differ at the molecular level. In rats, mice, flies, and sparrows hundreds of brain transcripts change their level of expression between sleep and wakefulness 41,65,78–86. In the most tested species so far, rats and mice, these changes occur mainly in the cerebral cortex, cerebellum, and hypothalamus, but also other brain areas. For instance, in one study up to ~5% (752 of 15,459) of the transcripts tested in the cerebral cortex was up- or down-regulated in rats that had slept for 8 hours relative to rats that had been spontaneously awake or sleep deprived for 8 hours 41. A similar number of cortical transcripts changed their expression because of time of day independent of behavioral state, suggesting that day/night time and sleep/wakefulness influence cortical gene expression to a similar extent 41. In general, across studies, there are as many “sleep” transcripts (with higher expression during sleep relative to waking) as there are “waking” transcripts. This result suggests that, despite being usually associated with behavioral inactivity, sleep is far from being a quiet state at the cellular level. Most importantly, sleep-related and wakefulness-related transcripts belong to different functional categories, suggesting that the two behavioral states may favor different cellular processes 41. Moreover, several of the molecular correlates of sleep and wakefulness first identified in rats and mice have subsequently been found in fruit flies and/or sparrows (Fig. 4). The overlap in some major gene categories is remarkable considering that these studies differ not only with respect to the animal species that was tested, but also in terms of design (e.g. sleep deprived vs. spontaneously awake animals), duration of wakefulness (3–8 hours), brain region, and statistical approach.
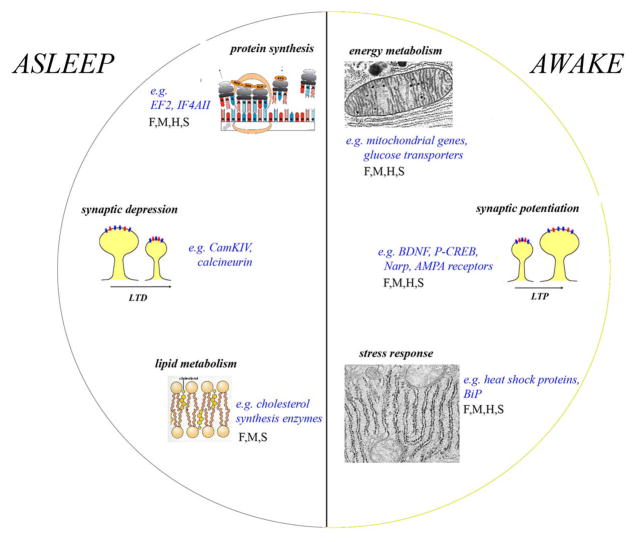
Figure 4.
Schematic representation of the major functional categories of genes whose expression is higher in the rat brain after several hours of wakefulness (including after 3–8 hours of sleep deprivation) or after several hours of sleep 41,78,87. F, M, H, S indicate when changes in the same functional category are also present in the brain of fruit flies, mice, Djungarian hamsters (Tom DeBoer, Irene Tobler, Chiara Cirelli, unpublished results) and sparrows, respectively.
Waking-related transcripts
Three functional categories of transcripts are most consistently increased during waking and short-term sleep deprivation relative to sleep (Fig. 4). The first group comprises genes involved in energy metabolism, including those coding for mitochondrial proteins, glucose transporters, and proteins related to glycogen metabolism 86–89. Their up-regulation may represent a mechanism by which the brain responds to the high energy requirements of wakefulness. This response, however, does not seem to persist when sleep deprivation is prolonged for more than a few hours 80,90. Imaging studies in animals 91, normal human subjects 92, and FFI patients 93 also show either no change or a decrease in cerebral metabolic rate after prolonged sleep loss.
The second group of waking-related transcripts codes for proteins involved in the response to cellular stress, including heat shock proteins and chaperones 41,78,79,82,83,85,86. This suggests that the absence of sleep may indeed represent a cellular stress for brain cells. In support of this hypothesis, in the mouse cerebral cortex a few hours of sleep deprivation induce the “unfolded protein response”, a global stress response that involves the induction of the chaperone BiP in the endoplasmatic reticulum, which promotes the degradation of misfolded proteins, and a decrease in protein synthesis 94. Importantly, there are no major signs of brain cellular damage after prolonged waking (refs in 95,96), an indication that the stress response must be protective. This is consistent with the observation that mutant flies unable to mount a strong stress response during sleep deprivation die of sleep loss earlier than controls 97.
The third group of wakefulness-related transcripts plays a role in synaptic plasticity, and more specifically in synaptic potentiation 41,65,79,86,98. Overall, the induction of these genes suggests that synaptic potentiation is favored during wakefulness relative to sleep. Consistent with this, recent experiments in flies and rats show that molecular and electrophysiological markers of net synaptic potentiation prevail in waking, while markers of net synaptic depression are present in sleep 99–101.
Sleep-related transcripts
Some of the transcripts with increased expression during sleep are involved in protein synthesis 41,84–86(Fig. 4). These findings are in agreement with previous studies that identified a positive correlation between sleep and protein synthesis 102–107. Whether sleep favors protein synthesis globally, or enhances the synthesis of specific classes of proteins, is still unclear.
Another group of sleep-related transcripts includes CamK4 (calmodulin-dependent protein kinase IV), a gene that has recently been involved in synaptic scaling 108, and other genes that have been associated with synaptic depression and depotentiation, such those coding for calcineurin, FK506 binding protein 12, inositol 1,4,5-trisphosphate receptor and amphiphysin II 41. Thus, while wakefulness is the appropriate time for memory acquisition and synaptic potentiation, sleep may favor complementary aspects of plasticity, such as synaptic consolidation and/or downscaling. An involvement of sleep in such processes is suggested by behavioral and physiological experiments showing that sleep improves the performance of different learning tasks 109,110. At this stage, however, the mechanism by which sleep enhances performance is still debated. One idea is that sleep consolidates synapses activated by learning during the previous waking period, perhaps through mechanisms that require cortical spindle activity and the slow waves of NREM sleep 111,112. Another possibility is that, since wakefulness is associated with a diffuse potentiation of synaptic circuits that results in a net increase in synaptic weight, sleep produces a generalized depression of synapses. This downscaling would benefit the brain because it decreases the energetic cost of synaptic activity, eliminates weak and ineffective synapses, reduces cellular stress and increases signal to noise ratios 113.
Finally, a large group of sleep-related transcripts is involved in membrane trafficking and maintenance 41,84–86. Some of these transcripts are involved in exocytosis and neurotransmitter release, others in synaptic vesicle recycling, tethering/docking of vesicles to their target organelles, and cycling between trans-Golgi network and plasma membrane. Other transcripts are important for the synthesis/maintenance of membranes in general and of myelin in particular, including oligodendrocitic genes coding for myelin structural proteins, myelin-related receptors, and enzymes. Also, transcripts with higher expression in sleep code for enzymes involved in the synthesis and transport of cholesterol, a major constituent of myelin and other membranes and an important factor in regulating synaptic efficacy 114,115. Depletion of cholesterol/sphingolipid leads to instability of surface AMPA receptors and gradual loss of synapses and dendritic spines 116. Thus, it may not be by chance that sleep seems to be linked to membrane trafficking and cholesterol synthesis on one hand, and to protein synthesis and synaptic homeostasis on the other hand.
Future directions
There is little doubt that more genes affecting sleep phenotypes will be identified in the near future, and it is reasonable to assume that most mutations will have less striking effects on sleep in mammals than in simpler organisms, since mammalian genomes are redundant. Even in simpler organisms, however, we should not expect single genes to completely abolish sleep or its regulation, because sleep is a complex and highly regulated behavior, present in all the animal species that have been carefully studied so far, suggesting that it has one or more fundamental functions, such as maintaining brain plasticity and saving energy 12. A similar argument applies to the brain regions that are involved in initiating and maintaining sleep and waking: there are several sleep-promoting and arousal-promoting neural systems, and the lesion of one or even a few of them does not result in the complete and permanent lack of either sleep or waking.
Sleep genetic studies are bound to contribute further to our understanding of sleep regulation and function but are also challenging. The identification of the mutated gene in animal models or human disorders is becoming faster and cheaper by the day. One major challenge lies in the choice of the phenotype: Which sleep parameters should we measure when searching for “insomnia genes”? Another issue is the large number of subjects required for genome-wide association studies, which may be a limiting factor unless extensive collaborations across sleep centers around the world are initiated. Finally, large-scale mutagenesis screening remains time-consuming and costly in mice, until – in parallel to flies - automatic, high-throughput and yet sensitive methods to assess sleep in hundreds of animals at a time become available.
Nevertheless, as reviewed in this article, some genes with profound effects on sleep have already been identified even in mammals, and others will be discovered. Neurons are more hyperpolarized during sleep than during waking, and mutations that can significantly change the resting membrane potential, the balance between inhibitory and excitatory neurotransmission, and/or the overall neuronal excitability of large sets of neurons are likely to affect quantity and quality of sleep, as Shaker, Sleepless, and Kv3.1/Kv3.3 mutants suggest. Also, in flies as well as in mammals, sleep homeostasis reflects not only the duration of prior waking, but also its intensity, and sleep need increases when waking is associated with learning 7,117,118. Thus, it is likely that mutations in many plasticity-related genes will affect the homeostatic regulation of sleep. Overall, the identification of mutations that change the need for sleep, or make subjects resistant to the negative effects of sleep deprivation, may prove crucial to further our understanding of the functions of sleep.
Supplementary Material
Acknowledgments
This work was funded by NIH grants P20 MH077967 and R01 GM075315. I thank Drs Daniel Bushey, Stephanie Maret and Giulio Tononi for critical comments on the manuscript, and Drs Daniel Bushey, Emmanuel Mignot and David Raizen for providing the pictures for Table 1.
Glossary terms
- Hypocretin/orexin system
A group of neurons in the posterior hypothalamus that have diffuse projections to the CNS and release hypocretins/orexins. These neuropeptides have been involved in the regulation of sleep and arousal, feeding, and energy metabolism
- Non-rapid-eye-movement sleep
(NREM sleep) One of the two types of sleep observed in mammals and birds. NREM sleep includes slow wave sleep (SWS, also called stages 3+4 or N3), characterized mainly by large slow waves 119
- REM sleep
The second phase of sleep observed in mammals and birds. In REM sleep the muscle tone is reduced or absent, but the EEG is similar to waking. REM theta activity (4–7Hz) is also present
References
- 1.Medori R, et al. Fatal familial insomnia, a prion disease with a mutation at codon 178 of the prion protein gene [see comments] N Engl J Med. 1992;326:444–9. doi: 10.1056/NEJM199202133260704. First study to show that a sleep disorder is caused by a gene mutation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tobler I, et al. Altered circadian activity rhythms and sleep in mice devoid of prion protein. Nature. 1996;380:639–642. doi: 10.1038/380639a0. First study in mice to show that a null mutation affects sleep regulation. [DOI] [PubMed] [Google Scholar]
- 3.Tobler I, Deboer T, Fischer M. Sleep and sleep regulation in normal and prion protein-deficient mice. J Neurosci. 1997;17:1869–79. doi: 10.1523/JNEUROSCI.17-05-01869.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin L, et al. The sleep disorder canine narcolepsy is caused by a mutation in the hypocretin (orexin) receptor 2 gene. Cell. 1999;98:365–376. doi: 10.1016/s0092-8674(00)81965-0. Seminal study that identifies the autosomal recessive mutation responsible for canine narcolepsy. [DOI] [PubMed] [Google Scholar]
- 5.Chemelli RM, et al. Narcolepsy in orexin knockout mice: Molecular genetics of sleep regulation. Cell. 1999;98:437–451. doi: 10.1016/s0092-8674(00)81973-x. This study shows that mice lacking hypocretin/orexin have a narcolepsy-like phenotype. [DOI] [PubMed] [Google Scholar]
- 6.Dauvilliers Y, Maret S, Tafti M. Genetics of normal and pathological sleep in humans. Sleep Med Rev. 2005;9:91–100. doi: 10.1016/j.smrv.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 7.Huber R, Ghilardi MF, Massimini M, Tononi G. Local sleep and learning. Nature. 2004;430:78–81. doi: 10.1038/nature02663. This study shows for the first time in humans that local changes in sleep intensity as measured by slow wave activity are driven by learning. [DOI] [PubMed] [Google Scholar]
- 8.Ambrosius U, et al. Heritability of Sleep Electroencephalogram. Biol Psychiatry. 2008 doi: 10.1016/j.biopsych.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 9.De Gennaro L, et al. The electroencephalographic fingerprint of sleep is genetically determined: a twin study. Ann Neurol. 2008;64:455–60. doi: 10.1002/ana.21434. [DOI] [PubMed] [Google Scholar]
- 10.Blake H, Gerard R. Brain potentials during sleep. Am J Physiol. 1937;119:692–703. [Google Scholar]
- 11.Achermann P, Borbely AA. Mathematical models of sleep regulation. Front Biosci. 2003;8:s683–93. doi: 10.2741/1064. [DOI] [PubMed] [Google Scholar]
- 12.Cirelli C, Tononi G. Is sleep essential? PLoS Biol. 2008;6:e216. doi: 10.1371/journal.pbio.0060216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harbison ST, et al. Co-regulated transcriptional networks contribute to natural genetic variation in Drosophila sleep. Nat Genet. 2009;41:371–5. doi: 10.1038/ng.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhdanova IV, Wang SY, Leclair OU, Danilova NP. Melatonin promotes sleep-like state in zebrafish. Brain Res. 2001;903:263–8. doi: 10.1016/s0006-8993(01)02444-1. [DOI] [PubMed] [Google Scholar]
- 15.Prober DA, Rihel J, Onah AA, Sung RJ, Schier AF. Hypocretin/orexin overexpression induces an insomnia-like phenotype in zebrafish. J Neurosci. 2006;26:13400–10. doi: 10.1523/JNEUROSCI.4332-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yokogawa T, et al. Characterization of Sleep in Zebrafish and Insomnia in Hypocretin Receptor Mutants. PLoS Biol. 2007;5:e277. doi: 10.1371/journal.pbio.0050277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raizen DM, et al. Lethargus is a Caenorhabditis elegans sleep-like state. Nature. 2008;451:569–72. doi: 10.1038/nature06535. [DOI] [PubMed] [Google Scholar]
- 18.Zhang J, Obal F, Jr, Fang J, Collins BJ, Krueger JM. Non-rapid eye movement sleep is suppressed in transgenic mice with a deficiency in the somatotropic system. Neurosci Lett. 1996;220:97–100. doi: 10.1016/s0304-3940(96)13232-8. [DOI] [PubMed] [Google Scholar]
- 19.Tafti M, et al. Deficiency in short-chain fatty acid beta-oxidation affects theta oscillations during sleep. Nat Genet. 2003;34:320–5. doi: 10.1038/ng1174. First study in mice to use QTL analysis to identify a gene that affects a specific feature of the sleep EEG (theta rhythm during REM sleep). [DOI] [PubMed] [Google Scholar]
- 20.Maret S, et al. Retinoic acid signaling affects cortical synchrony during sleep. Science. 2005;310:111–3. doi: 10.1126/science.1117623. [DOI] [PubMed] [Google Scholar]
- 21.Hu WP, et al. Altered circadian and homeostatic sleep regulation in prokineticin 2-deficient mice. Sleep. 2007;30:247–56. [PMC free article] [PubMed] [Google Scholar]
- 22.Kimura M, et al. Conditional corticotropin-releasing hormone overexpression in the mouse forebrain enhances rapid eye movement sleep. Molecular Psychiatry. 2009 doi: 10.1038/mp.2009.46. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwarz TL, Tempel BL, Papazian DM, Jan YN, Jan LY. Multiple potassium-channel components are produced by alternative splicing at the Shaker locus in Drosophila. Nature. 1988;331:137–42. doi: 10.1038/331137a0. [DOI] [PubMed] [Google Scholar]
- 24.Cirelli C, et al. Reduced sleep in Drosophila Shaker mutants. Nature. 2005;434:1087–92. doi: 10.1038/nature03486. First gene with strong effects on sleep duration identified in flies using mutagenesis screening. [DOI] [PubMed] [Google Scholar]
- 25.Bushey D, Huber R, Tononi G, Cirelli C. Drosophila Hyperkinetic mutants have reduced sleep and impaired memory. J Neurosci. 2007;27:5384–93. doi: 10.1523/JNEUROSCI.0108-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koh K, et al. Identification of SLEEPLESS, a sleep-promoting factor. Science. 2008;321:372–6. doi: 10.1126/science.1155942. Second gene with strong effects on sleep duration identified in flies using mutagenesis screening. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steriade M. The corticothalamic system in sleep. Front Biosci. 2003;8:D878–99. doi: 10.2741/1043. [DOI] [PubMed] [Google Scholar]
- 28.Guan D, et al. Expression and biophysical properties of Kv1 channels in supragranular neocortical pyramidal neurones. J Physiol. 2006;571:371–89. doi: 10.1113/jphysiol.2005.097006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Douglas CL, et al. Sleep in Kcna2 knockout mice. BMC Biol. 2007;5:42. doi: 10.1186/1741-7007-5-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Misonou H, Trimmer JS. Determinants of voltage-gated potassium channel surface expression and localization in Mammalian neurons. Crit Rev Biochem Mol Biol. 2004;39:125–45. doi: 10.1080/10409230490475417. [DOI] [PubMed] [Google Scholar]
- 31.Yuan LL, Chen X. Diversity of potassium channels in neuronal dendrites. Prog Neurobiol. 2006;78:374–89. doi: 10.1016/j.pneurobio.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 32.Espinosa F, Marks G, Heintz N, Joho RH. Increased motor drive and sleep loss in mice lacking Kv3-type potassium channels. Genes Brain Behav. 2004;3:90–100. doi: 10.1046/j.1601-183x.2003.00054.x. [DOI] [PubMed] [Google Scholar]
- 33.Espinosa F, Torres-Vega MA, Marks GA, Joho RH. Ablation of Kv3.1 and Kv3.3 potassium channels disrupts thalamocortical oscillations in vitro and in vivo. J Neurosci. 2008;28:5570–81. doi: 10.1523/JNEUROSCI.0747-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rudy B, McBain CJ. Kv3 channels: voltage-gated K+ channels designed for high-frequency repetitive firing. Trends Neurosci. 2001;24:517–26. doi: 10.1016/s0166-2236(00)01892-0. [DOI] [PubMed] [Google Scholar]
- 35.Liguori R, et al. Morvan’s syndrome: peripheral and central nervous system and cardiac involvement with antibodies to voltage-gated potassium channels. Brain. 2001;124:2417–26. doi: 10.1093/brain/124.12.2417. [DOI] [PubMed] [Google Scholar]
- 36.Crunelli V, Cope DW, Hughes SW. Thalamic T-type Ca2+ channels and NREM sleep. Cell Calcium. 2006;40:175–90. doi: 10.1016/j.ceca.2006.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee J, Kim D, Shin HS. Lack of delta waves and sleep disturbances during non-rapid eye movement sleep in mice lacking alpha1G-subunit of T-type calcium channels. Proc Natl Acad Sci U S A. 2004;101:18195–9. doi: 10.1073/pnas.0408089101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anderson MP, et al. Thalamic Cav3.1 T-type Ca2+ channel plays a crucial role in stabilizing sleep. Proc Natl Acad Sci U S A. 2005;102:1743–8. doi: 10.1073/pnas.0409644102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wisor JP, et al. A role for cryptochromes in sleep regulation. BMC Neurosci. 2002;3:20. doi: 10.1186/1471-2202-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hendricks JC, et al. Gender dimorphism in the role of cycle (BMAL1) in rest, rest regulation, and longevity in Drosophila melanogaster. J Biol Rhythms. 2003;18:12–25. doi: 10.1177/0748730402239673. [DOI] [PubMed] [Google Scholar]
- 41.Cirelli C, Gutierrez CM, Tononi G. Extensive and divergent effects of sleep and wakefulness on brain gene expression. Neuron. 2004;41:35–43. doi: 10.1016/s0896-6273(03)00814-6. First genome-wide microarray study to show that hundreds of transcripts in the rat cerebral cortex change their expression because of sleep and waking, independent of circadian time. [DOI] [PubMed] [Google Scholar]
- 42.Franken P, Thomason R, Heller HC, O’Hara BF. A non-circadian role for clock-genes in sleep homeostasis: a strain comparison. BMC Neurosci. 2007;8:87. doi: 10.1186/1471-2202-8-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wisor JP, et al. Sleep deprivation effects on circadian clock gene expression in the cerebral cortex parallel electroencephalographic differences among mouse strains. J Neurosci. 2008;28:7193–201. doi: 10.1523/JNEUROSCI.1150-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rutter J, Reick M, Wu LC, McKnight SL. Regulation of clock and NPAS2 DNA binding by the redox state of NAD cofactors. Science. 2001;293:510–4. doi: 10.1126/science.1060698. [DOI] [PubMed] [Google Scholar]
- 45.Reick M, Garcia JA, Dudley C, McKnight SL. NPAS2: an analog of clock operative in the mammalian forebrain. Science. 2001;293:506–9. doi: 10.1126/science.1060699. [DOI] [PubMed] [Google Scholar]
- 46.Kasischke KA, Vishwasrao HD, Fisher PJ, Zipfel WR, Webb WW. Neural activity triggers neuronal oxidative metabolism followed by astrocytic glycolysis. Science. 2004;305:99–103. doi: 10.1126/science.1096485. [DOI] [PubMed] [Google Scholar]
- 47.Viola AU, et al. PER3 polymorphism predicts sleep structure and waking performance. Curr Biol. 2007;17:613–8. doi: 10.1016/j.cub.2007.01.073. First study in humans to identify a gene polymorphism that affects the response to sleep deprivation. [DOI] [PubMed] [Google Scholar]
- 48.Goel N, Banks S, Mignot E, Dinges DF. PER3 polymorphism predicts cumulative sleep homeostatic but not neurobehavioral changes to chronic partial sleep deprivation. PLoS One. 2009;4:e5874. doi: 10.1371/journal.pone.0005874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Groeger JA, et al. Early morning executive functioning during sleep deprivation is compromised by a PERIOD3 polymorphism. Sleep. 2008;31:1159–67. [PMC free article] [PubMed] [Google Scholar]
- 50.Van Dongen HP, Vitellaro KM, Dinges DF. Individual differences in adult human sleep and wakefulness: Leitmotif for a research agenda. Sleep. 2005;28:479–96. doi: 10.1093/sleep/28.4.479. Review of the data suggesting a genetic basis for the interindividual variability in the response to sleep loss. [DOI] [PubMed] [Google Scholar]
- 51.Lim J, Choo WC, Chee MW. Reproducibility of changes in behaviour and fMRI activation associated with sleep deprivation in a working memory task. Sleep. 2007;30:61–70. doi: 10.1093/sleep/30.1.61. [DOI] [PubMed] [Google Scholar]
- 52.Toh KL, et al. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science. 2001;291:1040–3. doi: 10.1126/science.1057499. First study to identify a mutation responsible for FASPS. [DOI] [PubMed] [Google Scholar]
- 53.Xu Y, et al. Modeling of a human circadian mutation yields insights into clock regulation by PER2. Cell. 2007;128:59–70. doi: 10.1016/j.cell.2006.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu Y, et al. Functional consequences of a CKIdelta mutation causing familial advanced sleep phase syndrome. Nature. 2005;434:640–4. doi: 10.1038/nature03453. [DOI] [PubMed] [Google Scholar]
- 55.Okawa M, Uchiyama M. Circadian rhythm sleep disorders: characteristics and entrainment pathology in delayed sleep phase and non-24-h sleep-wake syndrome. Sleep Med Rev. 2007;11:485–96. doi: 10.1016/j.smrv.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 56.Sakurai T. The neural circuit of orexin (hypocretin): maintaining sleep and wakefulness. Nat Rev Neurosci. 2007;8:171–81. doi: 10.1038/nrn2092. [DOI] [PubMed] [Google Scholar]
- 57.Landolt HP. Sleep homeostasis: a role for adenosine in humans? Biochem Pharmacol. 2008;75:2070–9. doi: 10.1016/j.bcp.2008.02.024. [DOI] [PubMed] [Google Scholar]
- 58.Retey JV, et al. A functional genetic variation of adenosine deaminase affects the duration and intensity of deep sleep in humans. Proc Natl Acad Sci U S A. 2005;102:15676–81. doi: 10.1073/pnas.0505414102. First study in humans to identify a gene polymorphism that affects the duration of NREM sleep and slow wave activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lonart G, Tang X, Simsek-Duran F, Machida M, Sanford LD. The role of active zone protein Rab3 interacting molecule 1 alpha in the regulation of norepinephrine release, response to novelty, and sleep. Neuroscience. 2008;154:821–31. doi: 10.1016/j.neuroscience.2008.03.047. [DOI] [PubMed] [Google Scholar]
- 60.Colas D, Wagstaff J, Fort P, Salvert D, Sarda N. Sleep disturbances in Ube3a maternal-deficient mice modeling Angelman syndrome. Neurobiol Dis. 2005;20:471–8. doi: 10.1016/j.nbd.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 61.Franken P, Chollet D, Tafti M. The homeostatic regulation of sleep need is under genetic control. J Neurosci. 2001;21:2610–21. doi: 10.1523/JNEUROSCI.21-08-02610.2001. This study used QTL analysis to show that sleep need, as measured by the increase in slow wave activity after prolonged waking, is strongly influenced by genetic factors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Andretic R, Franken P, Tafti M. Genetics of sleep. Annu Rev Genet. 2008;42:361–88. doi: 10.1146/annurev.genet.42.110807.091541. [DOI] [PubMed] [Google Scholar]
- 63.Kato A, Ozawa F, Saitoh Y, Hirai K, Inokuchi K. vesl, a gene encoding VASP/Ena family related protein, is upregulated during seizure, long-term potentiation and synaptogenesis. FEBS Lett. 1997;412:183–9. doi: 10.1016/s0014-5793(97)00775-8. [DOI] [PubMed] [Google Scholar]
- 64.Brakeman PR, et al. Homer: a protein that selectively binds metabotropic glutamate receptors. Nature. 1997;386:284–8. doi: 10.1038/386284a0. [DOI] [PubMed] [Google Scholar]
- 65.Maret S, et al. Homer1a is a core brain molecular correlate of sleep loss. Proc Natl Acad Sci U S A. 2007;104:20090–5. doi: 10.1073/pnas.0710131104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mackiewicz M, Paigen B, Naidoo N, Pack AI. Analysis of the QTL for sleep homeostasis in mice: Homer1a is a likely candidate. Physiol Genomics. 2008;33:91–9. doi: 10.1152/physiolgenomics.00189.2007. [DOI] [PubMed] [Google Scholar]
- 67.Diagana TT, et al. Mutation of Drosophila homer disrupts control of locomotor activity and behavioral plasticity. J Neurosci. 2002;22:428–36. doi: 10.1523/JNEUROSCI.22-02-00428.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schenkein J, Montagna P. Self management of fatal familial insomnia. Part 1: what is FFI? MedGenMed. 2006;8:65. [PMC free article] [PubMed] [Google Scholar]
- 69.Lugaresi E, Provini F. Fatal familial insomnia and agrypnia excitata. Rev Neurol Dis. 2007;4:145–52. [PubMed] [Google Scholar]
- 70.Dossena S, et al. Mutant prion protein expression causes motor and memory deficits and abnormal sleep patterns in a transgenic mouse model. Neuron. 2008;60:598–609. doi: 10.1016/j.neuron.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 71.Peyron C, et al. A mutation in a case of early onset narcolepsy and a generalized absence of hypocretin peptides in human narcoleptic brains. Nat Med. 2000;6:991–7. doi: 10.1038/79690. [DOI] [PubMed] [Google Scholar]
- 72.Nishino S. Clinical and neurobiological aspects of narcolepsy. Sleep Med. 2007;8:373–99. doi: 10.1016/j.sleep.2007.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hallmayer J, et al. Narcolepsy is strongly associated with the T-cell receptor alpha locus. Nat Genet. 2009;41:708–11. doi: 10.1038/ng.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Winkelmann J, et al. Genome-wide association study of restless legs syndrome identifies common variants in three genomic regions. Nat Genet. 2007;39:1000–6. doi: 10.1038/ng2099. [DOI] [PubMed] [Google Scholar]
- 75.Schormair B, et al. PTPRD (protein tyrosine phosphatase receptor type delta) is associated with restless legs syndrome. Nat Genet. 2008;40:946–8. doi: 10.1038/ng.190. [DOI] [PubMed] [Google Scholar]
- 76.Stefansson H, et al. A genetic risk factor for periodic limb movements in sleep. N Engl J Med. 2007;357:639–47. doi: 10.1056/NEJMoa072743. [DOI] [PubMed] [Google Scholar]
- 77.Kimura M, Winkelmann J. Genetics of sleep and sleep disorders. Cell Mol Life Sci. 2007;64:1216–26. doi: 10.1007/s00018-007-6532-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cirelli C, Tononi G. Gene expression in the brain across the sleep-waking cycle. Brain Res. 2000;885:303–21. doi: 10.1016/s0006-8993(00)03008-0. [DOI] [PubMed] [Google Scholar]
- 79.Cirelli C, LaVaute TM, Tononi G. Sleep and wakefulness modulate gene expression in Drosophila. J Neurochem. 2005;94:1411–9. doi: 10.1111/j.1471-4159.2005.03291.x. [DOI] [PubMed] [Google Scholar]
- 80.Cirelli C, Faraguna U, Tononi G. Changes in brain gene expression after long-term sleep deprivation. J Neurochem. 2006;98:1632–45. doi: 10.1111/j.1471-4159.2006.04058.x. [DOI] [PubMed] [Google Scholar]
- 81.Terao A, Greco MA, Davis RW, Heller HC, Kilduff TS. Region-specific changes in immediate early gene expression in response to sleep deprivation and recovery sleep in the mouse brain. Neuroscience. 2003;120:1115–24. doi: 10.1016/s0306-4522(03)00395-6. [DOI] [PubMed] [Google Scholar]
- 82.Terao A, et al. Differential increase in the expression of heat shock protein family members during sleep deprivation and during sleep. Neuroscience. 2003;116:187–200. doi: 10.1016/s0306-4522(02)00695-4. [DOI] [PubMed] [Google Scholar]
- 83.Terao A, et al. Gene expression in the rat brain during sleep deprivation and recovery sleep: an Affymetrix GeneChip study. Neuroscience. 2006;137:593–605. doi: 10.1016/j.neuroscience.2005.08.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zimmerman JE, et al. Multiple mechanisms limit the duration of wakefulness in Drosophila brain. Physiol Genomics. 2006;27:337–50. doi: 10.1152/physiolgenomics.00030.2006. [DOI] [PubMed] [Google Scholar]
- 85.Mackiewicz M, et al. Macromolecule biosynthesis - a key function of sleep. Physiol Genomics. 2007 doi: 10.1152/physiolgenomics.00275.2006. [DOI] [PubMed] [Google Scholar]
- 86.Jones S, Pfister-Genskow M, Benca RM, Cirelli C. Molecular correlates of sleep and wakefulness in the brain of the white-crowned sparrow. J Neurochem. 2008;105:46–62. doi: 10.1111/j.1471-4159.2007.05089.x. [DOI] [PubMed] [Google Scholar]
- 87.Cirelli C, Tononi G. Differences in gene expression between sleep and waking as revealed by mRNA differential display. Brain Res Mol Brain Res. 1998;56:293–305. doi: 10.1016/s0169-328x(98)00057-6. [DOI] [PubMed] [Google Scholar]
- 88.Shaw PJ, Cirelli C, Greenspan RJ, Tononi G. Correlates of sleep and waking in Drosophila melanogaster. Science. 2000;287:1834–7. doi: 10.1126/science.287.5459.1834. [DOI] [PubMed] [Google Scholar]
- 89.Petit JM, Tobler I, Allaman I, Borbely AA, Magistretti PJ. Sleep deprivation modulates brain mRNAs encoding genes of glycogen metabolism. Eur J Neurosci. 2002;16:1163–7. doi: 10.1046/j.1460-9568.2002.02145.x. [DOI] [PubMed] [Google Scholar]
- 90.Rhyner TA, Borbely AA, Mallet J. Molecular Cloning of Forebrain mRNAs which are Modulated by Sleep Deprivation. Eur J Neurosci. 1990;2:1063–1073. doi: 10.1111/j.1460-9568.1990.tb00018.x. Pioneer study that used subtractive cDNA cloning to identify rat forebrain transcripts affected by sleep deprivation. [DOI] [PubMed] [Google Scholar]
- 91.Everson CA, Smith CB, Sokoloff L. Effects of prolonged sleep deprivation on local rates of cerebral energy metabolism in freely moving rats. J Neurosci. 1994;14:6769–78. doi: 10.1523/JNEUROSCI.14-11-06769.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wu JC, et al. The effect of sleep deprivation on cerebral glucose metabolic rate in normal humans assessed with positron emission tomography. Sleep. 1991;14:155–162. [PubMed] [Google Scholar]
- 93.Cortelli P, et al. Cerebral metabolism in fatal familial insomnia: relation to duration, neuropathology, and distribution of protease-resistant prion protein. Neurology. 1997;49:126–33. doi: 10.1212/wnl.49.1.126. [DOI] [PubMed] [Google Scholar]
- 94.Naidoo N, Giang W, Galante RJ, Pack AI. Sleep deprivation induces the unfolded protein response in mouse cerebral cortex. J Neurochem. 2005;92:1150–7. doi: 10.1111/j.1471-4159.2004.02952.x. [DOI] [PubMed] [Google Scholar]
- 95.Cirelli C, Shaw PJ, Rechtschaffen A, Tononi G. No evidence of brain cell degeneration after long-term sleep deprivation in rats. Brain Res. 1999;840:184–93. doi: 10.1016/s0006-8993(99)01768-0. [DOI] [PubMed] [Google Scholar]
- 96.Gopalakrishnan A, Ji LL, Cirelli C. Sleep deprivation and cellular responses to oxidative stress. Sleep. 2004;27:27–35. doi: 10.1093/sleep/27.1.27. [DOI] [PubMed] [Google Scholar]
- 97.Shaw PJ, Tononi G, Greenspan RJ, Robinson DF. Stress response genes protect against lethal effects of sleep deprivation in Drosophila. Nature. 2002;417:287–91. doi: 10.1038/417287a. [DOI] [PubMed] [Google Scholar]
- 98.Hendricks JC, et al. A non-circadian role for cAMP signaling and CREB activity in Drosophila rest homeostasis. Nat Neurosci. 2001;4:1108–15. doi: 10.1038/nn743. [DOI] [PubMed] [Google Scholar]
- 99.Vyazovskiy VV, Cirelli C, Pfister-Genskow M, Faraguna U, Tononi G. Molecular and electrophysiological evidence for net synaptic potentiation in wake and depression in sleep. Nat Neurosci. 2008;11:200–8. doi: 10.1038/nn2035. This study shows that the overall synaptic strength in the rat cerebral cortex increases during waking and decreases during sleep. [DOI] [PubMed] [Google Scholar]
- 100.Gilestro G, Tononi G, Cirelli C. Widespread changes in synaptic markers as a function of sleep and waking in Drosophila. Science. 2009;324:109–112. doi: 10.1126/science.1166673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Donlea JM, Ramanan N, Shaw PJ. Use-dependent plasticity in clock neurons regulates sleep need in Drosophila. Science. 2009;324:105–8. doi: 10.1126/science.1166657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Reich P, Driver JK, Karnovsky ML. Sleep: effects on incorporation of inorganic phosphate into brain fractions. Science. 1967;157:336–8. doi: 10.1126/science.157.3786.336. [DOI] [PubMed] [Google Scholar]
- 103.Reich P, Geyer SJ, Steinbaum L, Anchors M, Karnovsky ML. Incorporation of phosphate into rat brain during sleep and wakefulness. J Neurochem. 1973;20:1195–205. doi: 10.1111/j.1471-4159.1973.tb00088.x. [DOI] [PubMed] [Google Scholar]
- 104.Voronka G, Demin NN, Pevzner LZ. Total protein content and quantity of basic proteins in neurons and neuroglia of rat brain supraoptic and red nuclei during natural sleep and deprivation of paradoxical sleep. Dokl Akad Nauk SSSR. 1971;198:974–7. [PubMed] [Google Scholar]
- 105.Drucker-Colin RR, Spanis CW, Cotman CW, McGaugh JL. Changes in protein levels in perfusates of freely moving cats: relation to behavioral state. Science. 1975;187:963–5. doi: 10.1126/science.167436. [DOI] [PubMed] [Google Scholar]
- 106.Ramm P, Smith CT. Rates of cerebral protein synthesis are linked to slow wave sleep in the rat. Physiol Behav. 1990;48:749–53. doi: 10.1016/0031-9384(90)90220-x. [DOI] [PubMed] [Google Scholar]
- 107.Nakanishi H, et al. Positive correlations between cerebral protein synthesis rates and deep sleep in Macaca mulatta. Eur J Neurosci. 1997;9:271–9. doi: 10.1111/j.1460-9568.1997.tb01397.x. [DOI] [PubMed] [Google Scholar]
- 108.Ibata K, Sun Q, Turrigiano GG. Rapid synaptic scaling induced by changes in postsynaptic firing. Neuron. 2008;57:819–26. doi: 10.1016/j.neuron.2008.02.031. [DOI] [PubMed] [Google Scholar]
- 109.Born J, Rasch B, Gais S. Sleep to remember. Neuroscientist. 2006;12:410–24. doi: 10.1177/1073858406292647. [DOI] [PubMed] [Google Scholar]
- 110.Walker MP, Stickgold R. Sleep, memory, and plasticity. Annu Rev Psychol. 2006;57:139–66. doi: 10.1146/annurev.psych.56.091103.070307. [DOI] [PubMed] [Google Scholar]
- 111.Steriade M. Coherent oscillations and short-term plasticity in corticothalamic networks. Trends Neurosci. 1999;22:337–45. doi: 10.1016/s0166-2236(99)01407-1. [DOI] [PubMed] [Google Scholar]
- 112.Sejnowski TJ, Destexhe A. Why do we sleep? Brain Res. 2000;886:208–223. doi: 10.1016/s0006-8993(00)03007-9. [DOI] [PubMed] [Google Scholar]
- 113.Tononi G, Cirelli C. Sleep function and synaptic homeostasis. Sleep Med Rev. 2006;10:49–62. doi: 10.1016/j.smrv.2005.05.002. This review suggests that a major function of sleep is to reduce synaptic strength in large brain areas, and discusses how sleep could allow synaptic downscaling. [DOI] [PubMed] [Google Scholar]
- 114.Mauch DH, et al. CNS synaptogenesis promoted by glia-derived cholesterol. Science. 2001;294:1354–7. doi: 10.1126/science.294.5545.1354. [DOI] [PubMed] [Google Scholar]
- 115.Christopherson KS, et al. Thrombospondins are astrocyte-secreted proteins that promote CNS synaptogenesis. Cell. 2005;120:421–33. doi: 10.1016/j.cell.2004.12.020. [DOI] [PubMed] [Google Scholar]
- 116.Hering H, Lin CC, Sheng M. Lipid rafts in the maintenance of synapses, dendritic spines, and surface AMPA receptor stability. J Neurosci. 2003;23:3262–71. doi: 10.1523/JNEUROSCI.23-08-03262.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ganguly-Fitzgerald I, Donlea J, Shaw PJ. Waking experience affects sleep need in Drosophila. Science. 2006;313:1775–81. doi: 10.1126/science.1130408. [DOI] [PubMed] [Google Scholar]
- 118.Huber R, Tononi G, Cirelli C. Exploratory behavior, cortical BDNF expression, and sleep homeostasis. Sleep. 2007;30:129–39. doi: 10.1093/sleep/30.2.129. [DOI] [PubMed] [Google Scholar]
- 119.Silber MH, et al. The visual scoring of sleep in adults. J Clin Sleep Med. 2007;3:121–31. [PubMed] [Google Scholar]
- 120.Pack AI, et al. Novel method for high-throughput phenotyping of sleep in mice. Physiol Genomics. 2007;28:232–8. doi: 10.1152/physiolgenomics.00139.2006. [DOI] [PubMed] [Google Scholar]
- 121.Werth E, Achermann P, Dijk DJ, Borbely AA. Spindle frequency activity in the sleep EEG: individual differences and topographic distribution. Electroencephalogr Clin Neurophysiol. 1997;103:535–42. doi: 10.1016/s0013-4694(97)00070-9. [DOI] [PubMed] [Google Scholar]
- 122.Tan X, Campbell IG, Palagini L, Feinberg I. High internight reliability of computer-measured NREM delta, sigma, and beta: biological implications. Biol Psychiatry. 2000;48:1010–9. doi: 10.1016/s0006-3223(00)00873-8. [DOI] [PubMed] [Google Scholar]
- 123.Finelli LA, Achermann P, Borbely AA. Individual ‘fingerprints’ in human sleep EEG topography. Neuropsychopharmacol. 2001;25:S57–62. doi: 10.1016/S0893-133X(01)00320-7. [DOI] [PubMed] [Google Scholar]
- 124.De Gennaro L, Ferrara M, Vecchio F, Curcio G, Bertini M. An electroencephalographic fingerprint of human sleep. Neuroimage. 2005;26:114–22. doi: 10.1016/j.neuroimage.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 125.Buckelmuller J, Landolt HP, Stassen HH, Achermann P. Trait-like individual differences in the human sleep electroencephalogram. Neuroscience. 2006;138:351–6. doi: 10.1016/j.neuroscience.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 126.Tucker AM, Dinges DF, Van Dongen HP. Trait interindividual differences in the sleep physiology of healthy young adults. J Sleep Res. 2007;16:170–80. doi: 10.1111/j.1365-2869.2007.00594.x. [DOI] [PubMed] [Google Scholar]
- 127.van Beijsterveldt CE, Molenaar PC, de Geus EJ, Boomsma DI. Heritability of human brain functioning as assessed by electroencephalography. Am J Hum Genet. 1996;58:562–73. [PMC free article] [PubMed] [Google Scholar]
- 128.van Beijsterveldt CE, van Baal GC. Twin and family studies of the human electroencephalogram: a review and a meta-analysis. Biol Psychol. 2002;61:111–38. doi: 10.1016/s0301-0511(02)00055-8. [DOI] [PubMed] [Google Scholar]
- 129.van Beijsterveldt CE, Molenaar PC, de Geus EJ, Boomsma DI. Genetic and environmental influences on EEG coherence. Behav Genet. 1998;28:443–53. doi: 10.1023/a:1021637328512. [DOI] [PubMed] [Google Scholar]
- 130.Chorlian DB, et al. Heritability of EEG coherence in a large sib-pair population. Biol Psychol. 2007;75:260–6. doi: 10.1016/j.biopsycho.2007.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Porjesz B, et al. Linkage disequilibrium between the beta frequency of the human EEG and a GABAA receptor gene locus. Proc Natl Acad Sci U S A. 2002;99:3729–33. doi: 10.1073/pnas.052716399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hendricks JC, et al. Rest in Drosophila is a sleep-like state. Neuron. 2000;25:129–38. doi: 10.1016/s0896-6273(00)80877-6. [DOI] [PubMed] [Google Scholar]
- 133.Tobler I, Wigger E, Durr R, Hajnal A. C.elegans: a model organism to investigate the genetics of sleep and sleep homeostasis. Journal of Sleep Research. 2004;13:1. [Google Scholar]
- 134.Nitz DA, van Swinderen B, Tononi G, Greenspan RJ. Electrophysiological correlates of rest and activity in Drosophila melanogaster. Curr Biol. 2002;12:1934–40. doi: 10.1016/s0960-9822(02)01300-3. [DOI] [PubMed] [Google Scholar]
- 135.Huber R, et al. Sleep homeostasis in Drosophila melanogaster. Sleep. 2004;27:628–39. doi: 10.1093/sleep/27.4.628. [DOI] [PubMed] [Google Scholar]
- 136.Jeon M, Gardner HF, Miller EA, Deshler J, Rougvie AE. Similarity of the C. elegans developmental timing protein LIN-42 to circadian rhythm proteins. Science. 1999;286:1141–6. doi: 10.1126/science.286.5442.1141. [DOI] [PubMed] [Google Scholar]
- 137.Koh K, Evans JM, Hendricks JC, Sehgal A. A Drosophila model for age-associated changes in sleep:wake cycles. Proc Natl Acad Sci U S A. 2006;103:13843–7. doi: 10.1073/pnas.0605903103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hendricks JC, Kirk D, Panckeri K, Miller MS, Pack AI. Modafinil maintains waking in the fruit fly drosophila melanogaster. Sleep. 2003;26:139–46. doi: 10.1093/sleep/26.2.139. [DOI] [PubMed] [Google Scholar]
- 139.Andretic R, van Swinderen B, Greenspan RJ. Dopaminergic modulation of arousal in Drosophila. Curr Biol. 2005;15:1165–75. doi: 10.1016/j.cub.2005.05.025. [DOI] [PubMed] [Google Scholar]
- 140.Crocker A, Sehgal A. Octopamine regulates sleep in drosophila through protein kinase A-dependent mechanisms. J Neurosci. 2008;28:9377–85. doi: 10.1523/JNEUROSCI.3072-08a.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kume K, Kume S, Park SK, Hirsh J, Jackson FR. Dopamine is a regulator of arousal in the fruit fly. J Neurosci. 2005;25:7377–84. doi: 10.1523/JNEUROSCI.2048-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wisor JP, et al. Dopaminergic role in stimulant-induced wakefulness. J Neurosci. 2001;21:1787–94. doi: 10.1523/JNEUROSCI.21-05-01787.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Agosto J, et al. Modulation of GABAA receptor desensitization uncouples sleep onset and maintenance in Drosophila. Nat Neurosci. 2008;11:354–9. doi: 10.1038/nn2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Graves LA, et al. Genetic Evidence for a Role of CREB in Sustained Cortical Arousal. J Neurophysiol. 2003;23:23. doi: 10.1152/jn.00882.2002. [DOI] [PubMed] [Google Scholar]
- 145.Renier C, et al. Genomic and functional conservation of sedative-hypnotic targets in the zebrafish. Pharmacogenet Genomics. 2007;17:237–53. doi: 10.1097/FPC.0b013e3280119d62. [DOI] [PubMed] [Google Scholar]
- 146.Ruuskanen JO, Peitsaro N, Kaslin JV, Panula P, Scheinin M. Expression and function of alpha-adrenoceptors in zebrafish: drug effects, mRNA and receptor distributions. J Neurochem. 2005;94:1559–69. doi: 10.1111/j.1471-4159.2005.03305.x. [DOI] [PubMed] [Google Scholar]
- 147.Van Buskirk C, Sternberg PW. Epidermal growth factor signaling induces behavioral quiescence in Caenorhabditis elegans. Nat Neurosci. 2007;10:1300–7. doi: 10.1038/nn1981. [DOI] [PubMed] [Google Scholar]
- 148.Kramer A, et al. Regulation of daily locomotor activity and sleep by hypothalamic EGF receptor signaling. Science. 2001;294:2511–5. doi: 10.1126/science.1067716. [DOI] [PubMed] [Google Scholar]
- 149.Snodgrass-Belt P, Gilbert JL, Davis FC. Central administration of transforming growth factor-alpha and neuregulin-1 suppress active behaviors and cause weight loss in hamsters. Brain Res. 2005;1038:171–82. doi: 10.1016/j.brainres.2005.01.030. [DOI] [PubMed] [Google Scholar]
- 150.Kushikata T, Fang J, Chen Z, Wang Y, Krueger JM. Epidermal growth factor enhances spontaneous sleep in rabbits. Am J Physiol. 1998;275:R509–14. doi: 10.1152/ajpregu.1998.275.2.R509. [DOI] [PubMed] [Google Scholar]
- 151.Foltenyi K, Greenspan RJ, Newport JW. Activation of EGFR and ERK by rhomboid signaling regulates the consolidation and maintenance of sleep in Drosophila. Nat Neurosci. 2007;10:1160–7. doi: 10.1038/nn1957. [DOI] [PubMed] [Google Scholar]
- 152.Morrow JD, Vikraman S, Imeri L, Opp MR. Effects of serotonergic activation by 5-hydroxytryptophan on sleep and body temperature of C57BL/6J and Interleukin-6-deficient mice are dose and time related. Sleep. 2008;31:21–33. doi: 10.1093/sleep/31.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Yuan Q, Joiner WJ, Sehgal A. A sleep-promoting role for the Drosophila serotonin receptor 1A. Curr Biol. 2006;16:1051–62. doi: 10.1016/j.cub.2006.04.032. [DOI] [PubMed] [Google Scholar]
- 154.You YJ, Kim J, Raizen DM, Avery L. Insulin, cGMP, and TGF-beta signals regulate food intake and quiescence in C. elegans: a model for satiety. Cell Metab. 2008;7:249–57. doi: 10.1016/j.cmet.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.