Abstract
The epithelial-to-mesenchymal transition (EMT) that occurs during embryonic development is recapitulated during tumor metastasis. Important regulators of this process include growth factors, transcription factors, and adhesion molecules. New evidence suggests that microRNA (miRNA) activity contributes to metastatic progression and EMT; however, the mechanisms leading to altered miRNA expression during cancer progression remain poorly understood. Importantly, overexpression of the epidermal growth factor receptor (EGFR) in ovarian cancer correlates with poor disease outcome and induces EMT in ovarian cancer cells. We report that EGFR signaling leads to transcriptional repression of the miRNA miR-125a through the ETS family transcription factor PEA3. Overexpression of miR-125a induces conversion of highly invasive ovarian cancer cells from a mesenchymal to an epithelial morphology, suggesting miR-125a is a negative regulator of EMT. We identify AT-rich interactive domain 3B (ARID3B) as a target of miR-125a and demonstrate that ARID3B is overexpressed in human ovarian cancer. Repression of miR-125a through growth factor signaling represents a novel mechanism for regulating ovarian cancer invasive behavior.
Introduction
MicroRNA (miRNA) are a novel class of noncoding regulatory RNA molecules. They regulate gene expression by associating with the 3′untranslated region (3′UTR) of mRNA. Every tumor type surveyed thus far expresses different miRNA than the corresponding normal tissue, and multiple reports indicate that miRNA are differentially expressed in ovarian cancer [1,2]. These findings suggest that miRNA may be involved in tumor initiation, growth, or progression; however, the mechanisms leading to regulation of miRNA expression in ovarian cancer have not been characterized.
The epidermal growth factor receptor (EGFR) is often mutated or overexpressed in ovarian cancer [3–8], and expression correlates with poor survival. One of the ways that EGFR is thought to promote tumor progression is by induction of the epithelial-to-mesenchymal transition (EMT). Loss of E-cadherin is a critical step in EMT [9]. EGFR induces loss of E-cadherin in ovarian tumor cells, and EGFR expression correlates with E-cadherin loss in ovarian tumors [10]. Activation of EGFR also drives ovarian cancer cell invasion and activation of matrix metalloproteinases [10–13]. Therefore, multiple signaling pathways and gene products that contribute to cancer progression are EGF-regulated.
In Drosophila, EGFR induces transcription of miR-7 during photoreceptor differentiation through degradation of the ETS family transcription factor YAN [14]. Because EGFR-mediated signaling events are conserved between Drosophila and humans, we hypothesized that EGFR may regulate miRNA in human disease. Because YAN negatively regulated miRNA gene expression, we chose to identify miRNA with reduced expression in ovarian cancer. Two separate studies reported that miR-125a was decreased in ovarian cancer, so we focused on EGFR regulation of miR-125a [1,2].
In this study, we examined the expression of miR-125a in response to EGF treatment and showed for the first time in mammalian cells that EGF regulated miRNA expression at the transcriptional level through the ETS factor PEA3. Because miR-125a expression induced a mesenchymal-to-epithelial transition in invasive ovarian cancer cells, we chose to identify targets of miR-125a that may regulate EMT. AT-rich interactive domain 3B (ARID3B) seemed to be a strong target by sequence alignment. ARID3B transforms fibroblasts (with MYCN) and is tumor-promoting in neuroblastoma cells [15]. Interestingly, ARID3B is essential for the development of mesenchymal cells in embryos and loss of ARID3B is embryonic lethal [16]. These studies imply that ARID3B may play an important role in EMT and tumor progression. Our findings demonstrate that miR-125a regulates ARID3B expression and that ARID3B is elevated in ovarian cancer.
Materials and Methods
Cell Culture
The ovarian carcinoma cell lines OVCA433 and DOV13 (and stable cell lines) were grown in minimum essential medium supplemented with 10% FBS, 0.2 mM l-glutamine, 1 mM sodium pyruvate, 50 U/ml penicillin, and 50 µg/ml streptomycin. OVCA433 cells expressing PEA3 were described previously [11]. To generate miR-125a expressing cell lines, pre-miR-125a was cloned into pSilencer (Ambion, Austin, TX), and OVCA433 and DOV13 cells were transfected according to the instructions using Superfect (Qiagen, Valencia, CA). Stable cell lines were maintained in either 1 mg/ml G418 or 200 µg/ml Hygromycin B (Invitrogen, Carlsbad, CA). Epidermal growth factor was purchased from Biomedical Technologies, Inc (Stoughton, MA).
Western Blots
Cells were lysed in RIPA buffer (50 mM Tris pH 7.5, 150 mM NaCl, 1% Triton X-100, 5 mM EDTA). Protein concentration was determined using the BCA kit (Pierce, Rockford, IL). Twenty-five micrograms of protein was separated by SDS-PAGE and transferred to nitrocellulose. Blots were probed with the following antibodies: E-cadherin, N-cadherin, and β-catenin from BD Transduction Laboratories (San Jose, CA); GAPDH from Chemicon (Temecula, CA); and ARID3B from Abcam (Cambridge, MA). Antibodies for Snail and β-tubulin were from Santa Cruz Biotechnologies (Santa Cruz, CA). Densitometry was performed using the Kodak Image Station using Molecular Imaging Software version 4.0 (Carestream Health, New Haven, CT). Statistics were calculated using Student's t test.
Microscopy
Phase-contrast imaging and immunofluorescence microscopy were performed using an Olympus BH-2 inverted microscope or an Olympus 1X70 fluorescence microscope (Olympus, Center Valley, PA), respectively. For immunofluorescence studies cells were fixed in ice-cold acetone, blocked with BSA, and were then incubated with anti-ARID3B (Abcam), anti-E-cadherin, anti-N-cadherin, or anti-β-catenin (all from BD Transduction Laboratories). Fluorescein isothiocyanate-conjugated antirabbit or antimouse secondary antibodies from Chemicon were used. Coverslips were mounted using Vectashield mounting medium with 4β,6-diamidino-2-phenylindole (Vector Laboratories, Burlingame, CA). Bright field (for immunohistochemistry [IHC]) images were collected on a Zeiss Axioskop2 MOT (Carl Zeiss Micro Imaging, Inc, Thornwood, NY) using Slidebook software (Olympus).
Immunohistochemistry
IHC was performed on US Biomax, Inc (Rockville, MD) tissue arrays (OV807, OV1001, OV808) for ARID3B. IHC was performed according to the staining procedure for paraffin sections provided by the VECTASTAIN ABC KIT using DAB as a substrate solution and Hematoxylin QS as a counterstain (all from Vector Laboratories).
RNA Detection
Total RNA was extracted with TRIzol (Invitrogen) according to the manufacturer's instructions. Complementary DNA (cDNA) was generated from 1 µg of RNA using High Capacity cDNA Archive Kit or 5 ng of RNA using a miRNA kit according to the manufacturer's instructions (Applied Biosciences, Foster City, CA). For quantitative PCR (Q-PCR) SYBR Master Mix and TaqMan Master Mix solution was obtained from Applied Biosciences, and primer/probe QuantiTect Primer Assays for GAPDH and ARID3B were obtained from Qiagen and miR-125a Primer Assays were included in the miRNA cDNA kit. All reactions were performed in duplicate, and all experiments performed at least three times using a 7900HT sequence detection system (Applied Biosciences). ΔΔCT calculations were used to normalize signal versus a GAPDH control. Statistics were calculated using Student's t test. Reverse transcription-polymerase chain reaction (RT-PCR) for primiR-125a and 18s RNA was performed using the following primers: Pri-F, 5′-GGGTTCCTTGGGGAGGAG-3′ and Pri-R, 5′-ATTCCCCAGGTGTGTGGTT-3′; 18sF, 5′-AAACGGCTACCACATCCAAG-3′ and 18sR, 5′-CCTCCAATGGATCCTCGTTA-3′.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed as described previously [11]. PCR was performed on the miR-125a promoter using the following primers: 5′-GGGTAGGTAGGAAGCAGGTG-3′ and 5′-CAAGGAACCCAGGAGTCCA-3′. Immunoprecipitation (IP) with anti-β-tubulin (Santa Cruz Biotechnologies) was used as a negative control. ChIP controls were also performed on samples without chromatin or without antibody. As an additional negative control, we performed ChIP to a region upstream of the miR-125a promoter using the following primers: 5′-CTTAGGTCCGGGGAATCTCT-3′ and 5′-GGGTCTACGGCCAGCTCT-3′.
Statistical Analysis
Statistical analysis for immunohistochemistry was performed using a Jonckheere-Terpstra exact test for trend was used to compare the distribution of expression levels (high, medium, low, none) across tissue types (serous tumor, benign tumor, normal epithelium) followed by pairwise comparisons of tissue types. A Bonferroni adjustment was applied to the P values for the pairwise comparisons. All tests were two-sided.
Results
EGF and PEA3 Regulate miR-125a Expression
To identify miRNA down-modulated in ovarian cancer, we compared the published reports from Nam et al. [2] and Iorio et al. [1]. The only miRNA that were decreased in both reports were members of either the miR-125a/b family or the miR-99a/b100 family. Analysis of the genomic regions of DNA 1000 bp upstream of miR-125a, miR-125b, miR-99a, and miR-100 (data not shown) revealed that miR-125a contained five PEA3 binding sites, making it a candidate for EGF regulation [11]. miR-100 had one PEA3 site in the upstream region, whereas miR-99a and miR-125a had no binding sites (data not shown). Therefore, we chose to focus our studies on miR-125a regulation.
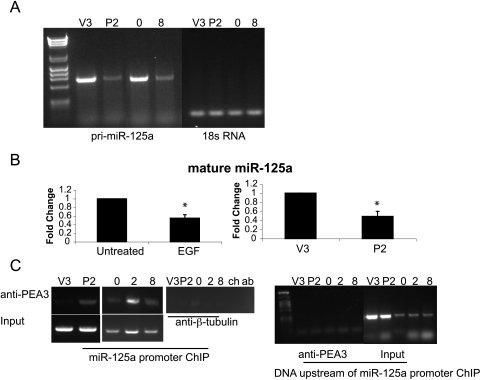
For the studies presented here, we used two different cell lines. The ovarian cancer cell line OVCA433 exhibits an epithelial morphology and is poorly invasive. Importantly, OVCA433 cells respond robustly to EGF treatment by undergoing an EMT and becoming invasive [10,11,17]. DOV13 cells will also be used because they are highly invasive ovarian tumor cells. We have shown that EGFR stimulates the activity of the transcription factor PEA3 [11]. To determine whether PEA3 represses miR-125a, we performed RT-PCR for the pri-miR-125a (the original miRNA transcript that has not undergone processing) on vector control (V3) and PEA3 overexpressing (P2) cells (which were characterized in the study of Cowden Dahl et al. [11] and derived from the OVCA433 cells). Pri-miR-125a expression was reduced in PEA3 overexpressing cells (Figure 1A), and similarly, 20 nM EGF treatment for 8 hours also reduced transcription of miR-125a (Figure 1A). We next assessed if the mature miR-125a was reduced as a result of EGFR and PEA3 activation. miR-125a expression was decreased by 40% (P < .05) and 50% (P < .05) by EGF treatment or PEA3 overexpression, respectively (Figure 1B). We observed a 20% decrease in miR-125a in DOV13 cells and immortalized ovarian epithelial cells (IOSE 398; data not shown). We predict that the smaller change in miR-125a expression in response to EGF in DOV13 cells results from the cells being less responsive to EGF stimulation. EGF stimulates OVCA433 invasion by 12-fold [11] and DOV13 invasion by only 2-fold [13]. Interestingly, the total PEA3 protein expression is similar between DOV13 and OVCA433 cells and is provided in Figure W1. We find that EGFR signaling and PEA3 activity repress transcription of miR-125a leading to decreased mature miR-125a.
Figure 1.
EGFR signaling through PEA3 represses miR-125a expression. (A) RT-PCR for pri-miR-125a (lanes 2–5) and 18s RNA (lanes 6–9) on vector (V3), PEA3 (P2), OVCA433 untreated (0), and OVCA433 + 8 hours of EGF treatment (8). (B) TaqMan PCR for mature miR-125a on untreated OVCA433 cells or treated with EGF for 8 hours and in V3 and P2 cells. Expression was normalized to untreated OVCA433 and V3 cells, respectively. *P < .05. (C) ChIP for PEA3 association with the PEA3 binding sites immediately upstream of miR-125a in V3 and P2 cells. ChIP was also performed on OVCA433 cells untreated or treated with EGF for 2 or 8 hours. IP with anti-β-tubulin was used as a negative control. ChIP performed in the absence of chromatin (ch) or antibody (ab) did not result in PCR products. Input chromatin for each sample is shown. ChIP was also performed on chromosomal DNA upstream of the miR-125a promoter on samples and input DNA.
We next examined if miR-125a repression occurs through the EGF regulated transcription factor PEA3. ChIP demonstrated that treatment of cells with 20 nM EGF enhanced PEA3 association with the miR-125a promoter (Figure 1C) indicating that EGFR activation of PEA3 represses miR-125a transcription. Of note, we see PEA3 associates with the miR-125a promoter after 2 hours of EGF treatment followed by the decrease in mature miR-125a between 4 and 8 hours (data not shown and Figure 1B). We predict that PEA3 associates with the miR-125a promoter within 2 hours of EGF treatment, but the decrease in total miR-125a expression is evident once the existing pool of mature miR-125a is degraded. As controls, we performed ChIP for a region of DNA upstream of the miR-125a promoter. This work represents the first example of EGF leading to transcriptional regulation of a miRNA in human cancer cells.
Overexpression of miR-125a Results in a Mesenchymal-to-Epithelial Transition
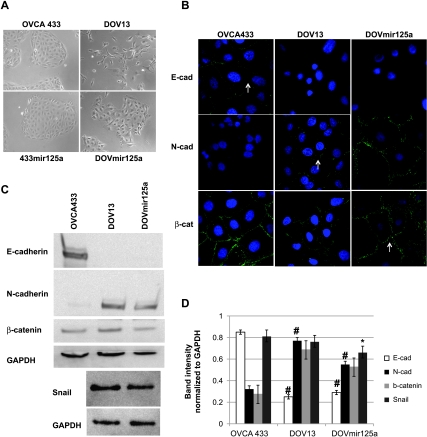
Because EGFR activity represses miR-125a, we generated OVCA433 and DOV13 cells that overexpress miR-125a (433mir125a and DOV-mir125a, respectively). These cell lines enable us to investigate a functional role for this miR-125a in cell types with different invasive potential. OVCA433 cells normally exhibit an epithelial phenotype, whereas DOV13 cells have a mesenchymal phenotype (Figure 2A). Mature miR-125a was increased 2.5-fold in the 433mir125a and Dov13mir125a clones (data not shown). Overexpression of miR-125a does not alter the morphology of the epithelial OVCA433 cells (Figure 2A). The 433mir125a cells appear identical to the parental cells and express E-cadherin and junctional β-catenin, and N-cadherin is not detectable (Figure W2). However, forced expression of miR-125a in DOV13 cells drives the cells toward an epithelial morphology (Figure 2A), suggesting that miR-125a targets are important in the acquisition or maintenance of the mesenchymal morphology.
Figure 2.
miR-125a overexpression induces a mesenchymal-to-epithelial transition. (A) Phase-contrast images were collected for OVCA433 and DOV13 cells stably transfected with miR-125a (433mir125a and DOVmir125a). Original magnification, x20. (B) Immunofluorescence was performed on OVCA433, DOV13, and DOVmir125a cells for E-cadherin, N-cadherin, and β-catenin (in green). Nuclei are stained with DAPI. Original magnification, x63. (C) Western blot for E-cadherin, N-cadherin, β-catenin, and GAPDH on OVCA433, DOV13, and DOVmir125a cells. Western blot for Snail and GAPDH on DOV13 and DOVmir125a cells. (D) Densitometry on Western blots from panel C. #P > .05 for OVCA433 cells compared with DOV13 or DOVmir125a cells. *P > .05 DOV13 cells compared with DOVmir125a.
OVCA433 cells have junctional E-cadherin, which is typical of epithelial cells, and DOV13 and DOVmir125a cells do not express junctional E-cadherin but instead express N-cadherin (Figure 2, B–D). These data suggest that E-cadherin is epigenetically silenced or actively repressed in DOV13 cells and introduction of miR-125a does not reverse the gene silencing. In support of this theory, we find that Snail expression is slightly reduced (by approximately 15%) in the DOVmir125a cells (Figure 2, C and D), but we do not find a concomitant increase in E-cadherin. Others have demonstrated that restoration of E-cadherin to junctions after EMT requires inhibition of MAP kinase signaling and increased E-cadherin expression in cells that overexpress RAS [18]. Interestingly, DOV13 cells naturally express activated RAS [19], suggesting that multiple events would need to occur simultaneously to restore E-cadherin at junctions. Instead, elevated miR-125a in DOVmir125a cells resulted in the distribution of N-cadherin to junctional complexes and localization of β-catenin to cell-cell junctions (Figure 2B). These staining patterns indicate that in cells overexpressing miR-125a epithelial-like adherens junctions are formed with N-cadherin in place of E-cadherin. Therefore, miR-125a expression represses the mesenchymal phenotype.
ARID3B Is a Target of miR-125a Repression
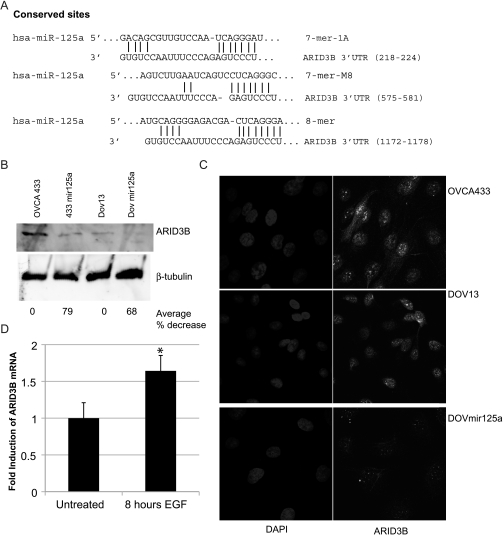
To identify targets of miR-125a that are important in mesenchymal morphology, we used the online algorithm TargetScan. The 3′UTR of ARID3B has three conserved miR-125a sites (Figure 3A). ARID3B null mice die during midgestation with multiple defects including failure to maintain cranial mesenchyme [16]. Because ARID3B is critically important for mesenchymal developmental during embryogenesis, we hypothesized that ARID3B also promotes mesenchymal morphologies or behaviors in cancer. First to assess if miR-125a regulates endogenous ARID3B expression, we performed Western blot analysis on whole-cell lysates from miR-125a overexpressing and parental cell lines. ARID3B is expressed in OVCA433 and DOV13 cells (Figure 3, C and D). However, in 433mir125a and DOVmir125a cells, there is approximately a 70% to 80% decrease in total ARID3B expression (Figure 3B). We confirmed that ARID3B expression was greatly diminished in DOVmir125a cells by immunofluorescence (Figure 3C), indicating that ARID3B is a likely target. Because miR-125a is an EGF-regulated microRNA, we investigated ARID3B regulation by EGFR. By Q-PCR, we find that ARID3B expression is induced by 1.6-fold in response to EGF (P = .0398; Figure 3D). Our data imply that ARID3B may be an important mediator of mesenchymal morphology in response to growth factor signaling.
Figure 3.
miR-125a targets ARID3B for repression. (A) Alignment of human miR-125a with the three conserved sites in the ARID3B 3′UTR. (B) Western blot for ARID3B and β-tubulin on OVCA433, 433mir125a, DOV13 and DOVmir125a cells. The average percent decrease in ARID3B protein (for three experiments) is indicated. (C) Immunofluorescence for ARID3B (in green) and DAPI was performed on OVCA433, DOV13, and DOVmir125a cells. Original magnification, x63. (D) Q-PCR for ARID3B on OVCA433 cells that are untreated or treated with 20 nM EGF for 8 hours. *P > .05.
ARID3B Is Overexpressed in Ovarian Cancer
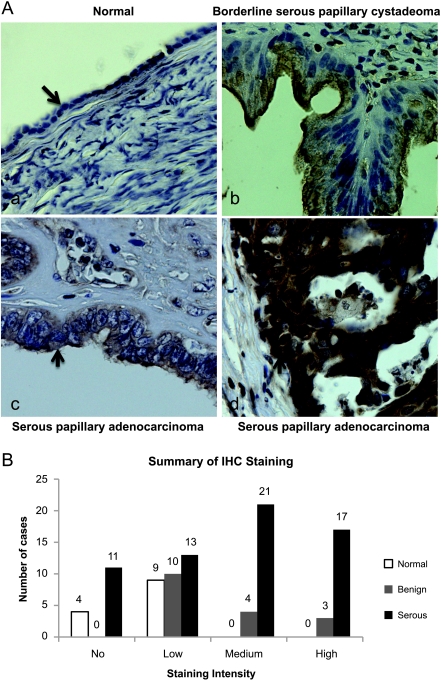
The function of ARID3B is unknown, but its role in development and its ability to promote tumorigenesis suggests that ARID3B may have an important role in tumor progression [15,16]. Furthermore, because PEA3 is overexpressed in ovarian cancer and miR-125a is decreased, we predict that ARID3B will be elevated in ovarian tumors [20,21]. Ovarian cancer tissue arrays were probed for ARID3B protein expression by IHC. Using a combination of US Biomax tissue arrays, 62 serous ovarian tumors, 17 benign ovarian tumors, and 13 normal ovarian epithelia sections were evaluated for ARID3B staining. Examples of a typical normal ovarian epithelium, a benign borderline tumor, and two malignant ovarian tumors (one with low (+) and one with high ARID3B staining (+++)) are shown (Figure 4A). Normal epithelia had either little to no ARID3B (Figure 4B). Benign lesions (borderline serous or mucinous papillary cystadenomas, and serous or mucinous cystadenomas) had generally low staining, although 41% of the tissues had either moderate (23%) or high (18%) staining (Figure 4B) indicating heterogeneity of ARID3B expression in benign lesions. In contrast, 82%of malignant serous tumors (serous papillary cystadenocarcinomas or adenocarcinomas) stained positive for ARID3B. Of these, 75% of the tumors had moderate to high expression. There was an observed trend of higher expression levels in both serous tumors and benign tumors than in normal epithelium (P < .003). The expression level of ARID3B in normal epithelium was significantly lower than serous tumors (P = .002). The difference in ARID3B expression between benign tumors and normal epithelium was not statistically significant (P > .500). We have shown that ARID3B is overexpressed in serous ovarian cancer and therefore may contribute to cancer progression.
Figure 4.
ARID3B expression is elevated in ovarian cancer. (A) Immunohistochemistry for ARID3B in normal ovary (a), benign ovarian tumor (b), and two example of malignant ovarian tumors one with low staining (c) and one with high staining (d). Original magnification, x40. Pathologic diagnosis is indicated. Arrow in panel (a) marks ovarian epithelium. Arrowheads in b to d denote brown ARID3B staining. (B) Summary of ARID3B expression in normal ovarian epithelium, benign ovarian lesions, and serous ovarian malignant tumors. (-) = no staining, (+) = low, (++) = moderate, (+++) = high. Staining was only scored for normal and tumor epithelia (not stroma).
Discussion
We demonstrate that the EGFR pathway described in Drosophila leading to transcriptional repression of a miRNA through an ETS factor is conserved in human ovarian tumor cells. Recently, it was shown that miR-21, a tumor-promoting miRNA, is induced by EGFR in lung cancer; however, the mechanism for this regulation is unknown [22]. Our data provide evidence for a novel mechanism of miRNA regulation downstream of EGFR signaling. This is also the first example of EGFR-mediated miRNA repression in human cells. Therefore, miRNA regulation by growth factor signaling represents an additional way of regulating tumor progression perhaps by fine-tuning oncogenic signaling.
Regulation of the mesenchymal-to-epithelial transition by miR-125a represents the second example of a miRNA regulating this process. Recently, it has been shown that the miR-200 family of miRNA correlates with E-cadherin expression [23]. Forced expression of miR-200 miRNA induces a mesenchymal-to-epithelial transition in mesenchymal cells, and inhibition of miR-200 induces EMT [23]. miR-125a regulation of mesenchymal-to-epithelial transition seems to be through a different mechanism because miR-125a does not regulate cadherin expression. Alternatively, our data suggests that miR-125a regulates N-cadherin localization through an indirect mechanism. We predict that miR-125a regulation of ARID3B contributes to the switch between mesenchymal and epithelial morphologies.
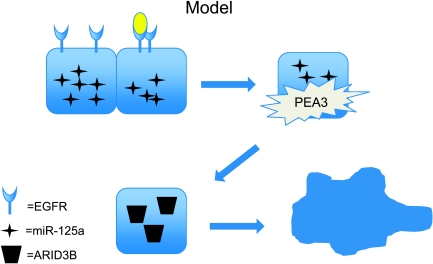
Aberrant expression of miR-125a in ovarian cancer led to the identification of ARID3B as a target of miR-125a. ARID3B is required for mesenchymal development in the mouse [16], and we predict that miR-125a repression of ARID3B plays a role in maintaining an epithelial morphology. We propose a model in Figure 5 where EGFR signaling leads to miR-125a transcriptional repression. The repression of miR-125a releases ARID3B promoting a mesenchymal phenotype and contributing to disease progression.
Figure 5.
Model: EGFR repression of miR-125a leads to ARID3B accumulation. Epithelial ovarian cancer cells express miR-125a. When EGFR is ligand-activated PEA3 activity is induced leading to transcriptional repression of miR-125a. The decrease in miR-125a then allows ARID3B protein to accumulate. ARID3B up-regulation leads to a mesenchymal transformation.
Supplementary Material
Acknowledgments
The authors thank Ed Bedrick for statistical analysis. Images (fluorescence and bright field) were generated in the University of New Mexico Cancer Center Fluorescence Microscopy Facility, supported as detailed on the Web page: http://hsc.unm.edu/crtc/microscopy/Facility.html.
Abbreviations
- ARID3B
AT-rich interactive domain 3B
- ChIP
chromatin immunoprecipitation
- EGFR
epidermal growth factor receptor
- IHC
immunohistochemistry
- miRNA
microRNA
- 3βUTR
3βuntranslated region
Footnotes
K.D.C.D. is supported by K99 CA133190-01 and an American Cancer Society Institutional Research Grant IRG-92-024 and was formerly supported by NRSA F32 1F32CA119729. K.D.C.D. and L.G.H. are faculty in the University of New Mexico Cancer Research and Treatment Center National Institutes of Health grant P20 CA888070.
This article refers to supplementary materials, which are designated by Figures W1 and W2 and are available online at www.neoplasia.com.
References
- 1.Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–8707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- 2.Nam EJ, Yoon H, Kim SW, Kim H, Kim YT, Kim JH, Kim JW, Kim S. MicroRNA expression profiles in serous ovarian carcinoma. Clin Cancer Res. 2008;14:2690–2695. doi: 10.1158/1078-0432.CCR-07-1731. [DOI] [PubMed] [Google Scholar]
- 3.Berns EM, Klijn JG, Henzen-Logmans SC, Rodenburg CJ, van der Burg ME, Foekens JA. Receptors for hormones and growth factors and (onco)-gene amplification in human ovarian cancer. Int J Cancer. 1992;52:218–224. doi: 10.1002/ijc.2910520211. [DOI] [PubMed] [Google Scholar]
- 4.Bartlett JM, Langdon SP, Simpson BJ, Stewart M, Katsaros D, Sismondi P, Love S, Scott WN, Williams AR, Lessells AM, et al. The prognostic value of epidermal growth factor receptor mRNA expression in primary ovarian cancer. Br J Cancer. 1996;73:301–306. doi: 10.1038/bjc.1996.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Niikura H, Sasano H, Sato S, Yajima A. Expression of epidermal growth factor-related proteins and epidermal growth factor receptor in common epithelial ovarian tumors. Int J Gynecol Pathol. 1997;16:60–68. doi: 10.1097/00004347-199701000-00010. [DOI] [PubMed] [Google Scholar]
- 6.Scambia G, Benedetti Panici P, Battaglia F, Ferrandina G, Baiocchi G, Greggi S, De Vincenzo R, Mancuso S. Significance of epidermal growth factor receptor in advanced ovarian cancer. J Clin Oncol. 1992;10:529–535. doi: 10.1200/JCO.1992.10.4.529. [DOI] [PubMed] [Google Scholar]
- 7.Stewart CJ, Owens OJ, Richmond JA, McNicol AM. Expression of epidermal growth factor receptor in normal ovary and in ovarian tumors. Int J Gynecol Pathol. 1992;11:266–272. doi: 10.1097/00004347-199210000-00004. [DOI] [PubMed] [Google Scholar]
- 8.van der Burg ME, Henzen-Logmans SC, Foekens JA, Berns EM, Rodenburg CJ, van Putten WL, Klijn JG. The prognostic value of epidermal growth factor receptors, determined by both immunohistochemistry and ligand binding assays, in primary epithelial ovarian cancer: a pilot study. Eur J Cancer. 1993;29A:1951–1957. doi: 10.1016/0959-8049(93)90451-k. [DOI] [PubMed] [Google Scholar]
- 9.Huber MA, Kraut N, Beug H. Molecular requirements for epithelial-mesenchymal transition during tumor progression. Curr Opin Cell Biol. 2005;17:548–558. doi: 10.1016/j.ceb.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 10.Cowden Dahl KD, Symowicz J, Ning Y, Gutierrez E, Fishman DA, Adley BP, Stack MS, Hudson LG. Matrix metalloproteinase 9 is a mediator of epidermal growth factor-dependent e-cadherin loss in ovarian carcinoma cells. Cancer Res. 2008;68:4606–4613. doi: 10.1158/0008-5472.CAN-07-5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cowden Dahl KD, Zeineldin R, Hudson LG. PEA3 is necessary for optimal epidermal growth factor receptor-stimulated matrix metalloproteinase expression and invasion of ovarian tumor cells. Mol Cancer Res. 2007;5:413–421. doi: 10.1158/1541-7786.MCR-07-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellerbroek SM, Halbleib JM, Benavidez M, Warmka JK, Wattenberg EV, Stack MS, Hudson LG. Phosphatidylinositol 3-kinase activity in epidermal growth factor-stimulated matrix metalloproteinase-9 production and cell surface association. Cancer Res. 2001;61:1855–1861. [PubMed] [Google Scholar]
- 13.Ellerbroek SM, Hudson LG, Stack MS. Proteinase requirements of epidermal growth factor-induced ovarian cancer cell invasion. Int J Cancer. 1998;78:331–337. doi: 10.1002/(SICI)1097-0215(19981029)78:3<331::AID-IJC13>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 14.Li X, Carthew RW. A microRNA mediates EGF receptor signaling and promotes photoreceptor differentiation in the Drosophila eye. Cell. 2005;123:1267–1277. doi: 10.1016/j.cell.2005.10.040. [DOI] [PubMed] [Google Scholar]
- 15.Kobayashi K, Era T, Takebe A, Jakt LM, Nishikawa S. ARID3B induces malignant transformation of mouse embryonic fibroblasts and is strongly associated with malignant neuroblastoma. Cancer Res. 2006;66:8331–8336. doi: 10.1158/0008-5472.CAN-06-0756. [DOI] [PubMed] [Google Scholar]
- 16.Takebe A, Era T, Okada M, Martin Jakt L, Kuroda Y, Nishikawa S. Microarray analysis of PDGFR alpha+ populations in ES cell differentiation culture identifies genes involved in differentiation of mesoderm and mesenchyme including ARID3b that is essential for development of embryonic mesenchymal cells. Dev Biol. 2006;293:25–37. doi: 10.1016/j.ydbio.2005.12.016. [DOI] [PubMed] [Google Scholar]
- 17.Zeineldin R, Rosenberg M, Ortega D, Buhr C, Chavez MG, Stack MS, Kusewitt DF, Hudson LG. Mesenchymal transformation in epithelial ovarian tumor cells expressing epidermal growth factor receptor variant III. Mol Carcinog. 2006;45:851–860. doi: 10.1002/mc.20237. [DOI] [PubMed] [Google Scholar]
- 18.Sangha N, Wu R, Kuick R, Powers S, Mu D, Fiander D, Yuen K, Katabuchi H, Tashiro H, Fearon ER, et al. Neurofibromin 1 (NF1) defects are common in human ovarian serous carcinomas and co-occur with TP53 mutations. Neoplasia. 2008;10:1362–1372. doi: 10.1593/neo.08784. following 1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Q, Mattingly RR. Restoration of E-cadherin cell-cell junctions requires both expression of E-cadherin and suppression of ERK MAP kinase activation in Ras-transformed breast epithelial cells. Neoplasia. 2008;10:1444–1458. doi: 10.1593/neo.08968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davidson B, Goldberg I, Gotlieb WH, Kopolovic J, Ben-Baruch G, Reich R. PEA3 is the second Ets family transcription factor involved in tumor progression in ovarian carcinoma. Clin Cancer Res. 2003;9:1412–1419. [PubMed] [Google Scholar]
- 21.Hibbs K, Skubitz KM, Pambuccian SE, Casey RC, Burleson KM, Oegema TR, Jr, Thiele JJ, Grindle SM, Bliss RL, Skubitz AP. Differential gene expression in ovarian carcinoma: identification of potential biomarkers. Am J Pathol. 2004;165:397–414. doi: 10.1016/S0002-9440(10)63306-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seike M, Goto A, Okano T, Bowman ED, Schetter AJ, Horikawa I, Mathe EA, Jen J, Yang P, Sugimura H, et al. MiR-21 is an EGFR-regulated anti-apoptotic factor in lung cancer in never-smokers. Proc Natl Acad Sci USA. 2009;106:12089–12090. doi: 10.1073/pnas.0905234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.