Abstract
K-RBP is a KRAB-containing zinc finger protein with multiple zinc finger motifs and represses Kaposi’s sarcoma-associated herpesvirus (KSHV) transactivator RTA-mediated transactivation of several viral lytic gene promoters, including the ORF57 promoter. Whether K-RBP binds DNA through its zinc fingers and the role of zinc finger domain in repressing gene expression are unclear. Here we report that K-RBP binds DNA through its zinc finger domain and the target DNA sequences contain high GC content. Furthermore, K-RBP binds to KSHV ORF57 promoter, which contains a GC-rich motif. K-RBP suppresses the basal ORF57 promoter activity as well as RTA-mediated activation. The zinc finger domain of K-RBP is sufficient for the suppression of ORF57 promoter activation mediated by the viral transactivator RTA. Finally, we show that K-RBP inhibits RTA binding to ORF57 promoter. These findings suggest that the DNA-binding activity of K-RBP plays an important role in repressing viral promoter activity.
Keywords: KSHV/HHV-8, zinc finger, DNA binding, transcription repression
Introduction
KSHV is the etiologic agent of Kaposi’s sarcoma (KS) (Chang et al., 1994; Chang and Moore, 1996). Like other herpesviruses, KSHV establishes a latent infection in infected cells but lytic replication can occur spontaneously or upon activation by various stimuli (Jenner et al., 2001; Sarid et al., 1998). Induction of KSHV lytic gene expression occurs via the expression of the replication and transcription activator RTA (Gradoville et al., 2000; Lukac et al., 1998; Sun et al., 1998). RTA activates the transcription of various viral genes, and one of the highly responsive genes to RTA-mediated transactivation is ORF57 (West and Wood, 2003). The transactivation activity of RTA can be modulated by many cellular and viral factors through different mechanisms (West and Wood, 2003). One of these cellular factors is a KRAB-containing zinc finger protein ZnF426 which we have identified previously; it interacts with the KSHV RTA, and was named KSHV-RTA binding protein (K-RBP) (Wang et al., 2001).
KRAB (Kruppel-associated box)-containing zinc finger proteins are the largest family of zinc finger regulatory proteins encoded in the human genome with about 300 members. They play important roles in various cellular processes, such as cell differentiation, apoptosis and neoplastic transformation (Urrutia, 2003). The KRAB-containing zinc finger proteins are characterized by the presence of 1–2 KRAB domains at N-terminus followed by multiple C2H2 zinc finger motifs. Many of the KRAB domains repress transcription when tethered to DNA. The KRAB domains also interact with proteins, such as TIF1β, and their interactions are important for their repression function (Moosmann et al., 1996). The tandem C2H2 zinc fingers in KRAB-containing zinc finger proteins were proposed to recognize specific DNA elements and bind to DNA. However, not all zinc fingers bind DNA and not all zinc fingers contribute equally to the DNA binding specificity in proteins containing multiple zinc finger motifs (Peng et al., 2002). Furthermore, there is at least one zinc finger protein that was found not to bind to DNA (Kwan et al., 2003).
Similar to other KRAB-containing zinc finger proteins, K-RBP contains a Kruppel-associated box (KRAB) at the N-terminus and multiple zinc finger motifs at the C-terminus; and it is expressed in a number of different cell lines tested (Wang et al., 2001). The K-RBP protein represses RTA-mediated transactivation and knockdown of K-RBP expression enhances RTA-mediated transactivation of gene driven by the ORF57 promoter. More interestingly, knockdown of K-RBP expression in KSHV latently infected cells also increases KSHV-RTA mediated lytic gene expression, suggesting that K-RBP plays a role in repressing RTA-mediated KSHV lytic gene expression in latently infected cells (Yang and Wood, 2007). Therefore, it is of interest to characterize the molecular properties of K-RBP to study the mechanism of its function on viral promoters. The KRAB domain of K-RBP is a repression domain but the role of its zinc finger motifs has not yet been defined (Yang and Wood, 2007). Whether the zinc finger motifs of K-RBP bind to DNA, especially the viral promoter, and play a role in repressing KSHV promoter is of interest and needs to be investigated.
In this report, we demonstrate that K-RBP binds DNA through its zinc finger motifs and the binding elements are GC-rich. In addition, we found that the DNA-binding activity of K-RBP significantly contributes to the repression of KSHV ORF57 gene promoter in the absence and presence of RTA. It is possible that K-RBP regulates other viral gene expression and viral replication using a similar mechanism. The characterization of K-RBP DNA-binding activity and identification of K-RBP binding sequences will provide important molecular insights towards understanding the functions of K-RBP and its role in regulating KSHV gene expression and replication at the molecular level.
Results
Expression and purification of recombinant K-RBP protein
K-RBP contains 12 zinc finger motifs, with 11 of them being classic C2H2 motifs and the other one being a non-typical C2H2 motif (Fig. 1A). In addition, 6 of the 11 linkers between the zinc fingers of K-RBP are composed of the sequence TG(E)EKPY which is a common canonical sequence in tandem DNA-binding C2H2 zinc finger proteins (Wolfe et al., 2000). Even though zinc finger motifs are well-known DNA-binding motifs, not all zinc finger proteins bind DNA (Kwan et al., 2003). To test whether K-RBP binds to DNA, recombinant His-tagged K-RBP was over-expressed to a high level in E. coli and could be detected in the insoluble fraction of the bacterial lysate (Fig. 1B). The protein was purified and then refolded by dialyzing against refolding buffer. Coomassie blue staining of the SDS-PAGE and Western blot analysis showed that the refolded protein was pure (Fig. 1C). The purified and refolded K-RBP protein was subsequently used for DNA binding analysis.
FIG. 1.

(A) Schematic representation of K-RBP protein showing the KRAB domain and 12 zinc finger motifs. Amino acid alignment of the zinc finger motifs in K-RBP showing the amino acids involved in Zn2+ binding (in bold). The −1, +3 and +6 indicate the amino acids in the α-helical region of a zinc finger motif that determine its DNA-binding specificity. (B) Coomassie blue staining of cell lysates from E. coli expressing K-RBP protein. Lane 1: protein marker; Lane 2: Lysate from E. coli cells containing K-RBP expression plasmid without IPTG induction; Lane 3: soluble fraction of Lysate from E. coli cells containing K-RBP expression plasmid with IPTG induction; Lane 4: insoluble fraction of the lysate from E. coli cells harboring K-RBP expression plasmid with IPTG induction. Arrow indicates K-RBP protein. (C) Coomassie blue staining and Western blot analysis of His-tagged K-RBP protein after purification, urea-solubilization and refolding. Lane 1: Coomassie blue staining of K-RBP protein after purification; Lane 2: Coomassie blue staining of K-RBP protein after refolding; Lane 3: Western blot analysis of protein using anti-His antibody after refolding. Arrow indicates K-RBP protein.
K-RBP binds DNA via its zinc finger domain
To investigate whether K-RBP binds DNA, we conducted an in vitro DNA-cellulose assay. K-RBP protein was examined for its association with cellulose alone or cellulose conjugated to dsDNA. Consistent with it being a potential DNA-binding protein, K-RBP bound tightly to dsDNA cellulose but not to cellulose alone (Fig. 2A). In addition, the DNA-binding activity of K-RBP was affected by the addition of Zn2+. An increase in ZnSO4 concentration resulted in more K-RBP being bound to DNA, and the addition of metal-chelating reagent EDTA inhibited K-RBP binding (Fig. 2B). This suggests that Zn2+ is required for optimal DNA binding by K-RBP. Similar effect was also observed with another KRAB-containing zinc finger protein (Jheon et al., 2001).
FIG. 2.
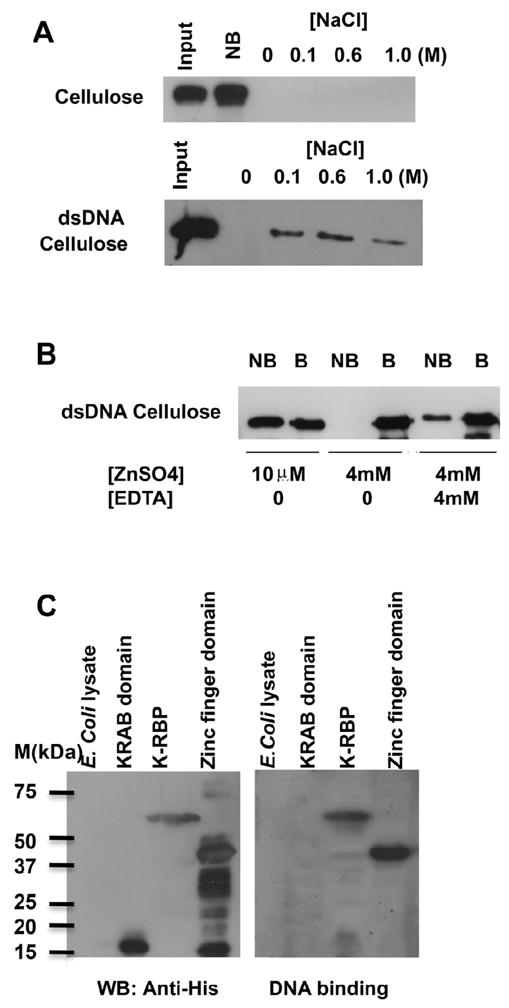
DNA-binding activity of K-RBP protein. (A) The bacterially expressed K-RBP was refolded and incubated with cellulose without DNA-conjugation or dsDNA-cellulose. The bound (B) and non-bound (NB) fractions were separated by centrifugation. The bound protein was washed and eluted by increasing concentrations of NaCl in the binding buffer. The K-RBP protein in different fractions was separated by SDS-PAGE and analyzed by Western blot using anti-K-RBP antibody. (B) K-RBP was incubated with dsDNA cellulose in the presence of ZnSO4 and EDTA. DNA cellulose bound protein was eluted with 1.25% SDS. (C) Zinc finger domain, but not KRAB domain of K-RBP binds DNA. The E. coli expressed His-tagged KRAB domain (1–98), zinc finger domain (141–554), full-length K-RBP and E. coli lysate were resolved on SDS-PAGE, transferred to PVDF membrane and analyzed by Western blot using anti-His antibody (left panel). The second membrane with the same samples was incubated in K-RBP refolding buffer overnight at 4°C and hybridized to the random dsDNA probe labeled with [α-32P] dATP (right panel).
To examine whether K-RBP binds DNA through its zinc finger domain, full-length His-tagged K-RBP and two truncated K-RBP clones, KRAB domain K-RBP1-98 and zinc finger domain K-RBP141-554, were constructed, and the resulting proteins expressed in E. coli were used in DNA filter binding assay. Membrane-immobilized full-length K-RBP and zinc finger domain (K-RBP141-554), but not KRAB domain (K-RBP1-98), were found to bind [α-32P] dATP labeled DNA (Fig. 2C). These results indicate that the zinc finger domain of K-RBP is responsible for the DNA binding activity of K-RBP.
Identification of K-RBP binding sequences
To investigate whether K-RBP binds specific DNA sequences, we incubated the His-tagged K-RBP fusion protein with a pool of double stranded 63-mers containing 27 nucleotides of random sequence, N27. The K-RBP bound oligonucleotides were recovered by Ni-NTA, amplified by PCR, and reselected as described in Materials and Methods and shown in figure 3A. A total of 33 clones were analyzed and 70% of them (23 clones) contain (T/G)CGG or its complementary sequence CCG(A/C) in the randomly synthesized region (data not shown). This result suggests that (T/G)CGG sequence could be a core motif for K-RBP binding.
FIG. 3.
Selection of K-RBP binding sequences. (A) Schematic representation of the procedure used to identify K-RBP binding sequences. (B) Consensus (Con) sequences for K-RBP binding. Numbers indicate the frequencies of the specific nucleotide at each position (Pos). The most abundant nucleotides at each position were used to generate the consensus sequence shown in the bottom. The underlined CGG motif was pre-synthesized in the pool of the second round selection oligomers. (C) K-RBP binds clone #14 probe in EMSA. The probe was labeled with [α-32P]dATP. The clone #14 and Oct-1 probe were used as unlabeled competitors. Unlabeled competitors were added at an 80–100-fold molar excess over the labeled probe. Various amounts of Ni-NTA agarose beads (5 and 15 μl) were used to remove the His-tagged K-RBP protein in EMSA. GST beads (15 μl) were used as a control. Mouse anti-His or Flag antibodies were added in the reaction prior to adding the probe as shown in lanes 11 and 12. The arrow indicates the bound complex. One μl K-RBP protein contains 5 ng purified K-RBP protein approximately.
To better define the K-RBP binding sequences, we performed a second set of selection with oligomers containing the central CGG core identified during the first selection flanked by 14 and 15 nucleotides of random sequences at each end (N14-CGG-N15). After 8 rounds of selection, the selected oligonucleotides were cloned and sequenced. A total of 100 clones were sequenced, of which 82 good quality sequences were analyzed and the frequencies of different nucleotides appearing at each position were calculated. The analysis revealed a GC-rich feature, with a GC-rich element in the consensus sequence: G(G/T)GGG(G/T)G(G/T)NNGCGGG(G/T)GG (Fig. 3B). Notably, several sequences featured multiple times among the selected 82 clones, and all of them were rich in GC content. These results suggest that K-RBP preferably binds to sequences with high GC content.
To confirm the binding of K-RBP with the selected sequences, we performed EMSA using one of the selected DNA target clone #14 as a probe. This sequence was present in 7 out of 82 analyzed sequences (Fig. 3C). The result showed that the K-RBP protein bound the probe specifically, and increasing amounts of protein resulted in stronger binding (Fig. 3C, lanes 2 and 3). The binding was competed efficiently by the unlabeled clone #14 probe but not by Oct-1 binding element probe which is not GC-rich (Fig. 3C, lanes 4 and 5). In addition, when the K-RBP protein was depleted by Ni-NTA beads prior to binding, the intensity of the protein-DNA complex decreased. However, the addition of GST beads had no effect (Fig. 3C, lanes 6 to 8). We also used anti-His antibody to test for a supershift of the K-RBP/DNA complex. However, the addition of this antibody did not generate a supershifted band but the intensity of the K-RBP/DNA complex band decreased significantly. In contrast, the anti-Flag antibody had little effect (Fig. 3C, lanes 9 to 12). These results suggest that binding of K-RBP to this probe is specific.
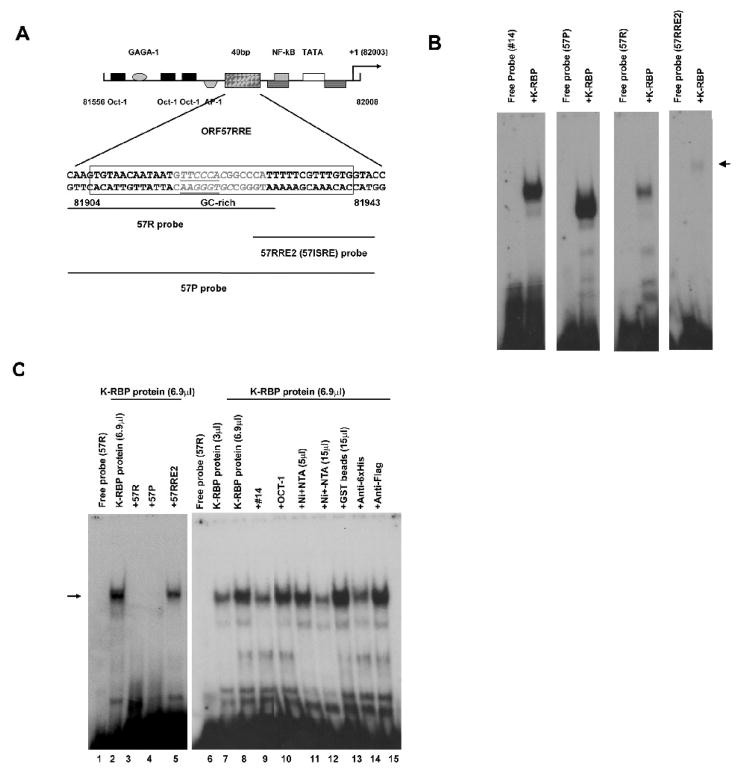
K-RBP binds KSHV ORF57 promoter and suppresses its activity
K-RBP has been shown to suppress KSHV ORF57 and K8 promoter activities activated by RTA (Yang and Wood, 2007). We found a GC-rich element beside RTA response elements in ORF57 promoter. The ORF57 gene is an important early protein required for KSHV lytic replication (Duan et al., 2001; Lukac et al., 2001; Wang et al., 2005) and we tested whether K-RBP actually binds this promoter and whether the binding can lead to the suppression of the promoter activity. Fig. 4A shows the schematic representation of the ORF57 promoter in which we have identified at least two RTA binding elements (RRE1 and RRE2) (Wang et al., 2005). The GC-rich element is located between RRE1 and RRE2. We then performed EMSA using 57R, 57RRE2, the entire promoter element 57P and clone #14 as probes to determine their interaction with K-RBP. K-RBP bound to the 57P and 57R probes, but poorly to 57RRE2. Binding of K-RBP to 57P was stronger than that of 57R (Fig. 4B). These results suggest that even though the AT-rich 57RRE2 is not a preferable binding target of K-RBP, it enhances the binding of K-RBP to 57R since stronger binding of K-RBP to 57P was observed relative to 57R (Fig. 4B). The binding of K-RBP to 57R was specific because increasing protein/DNA complex was observed with an increasing amount of protein added (Fig. 4C, lanes 7 and 8). The addition of the various unlabelled competitors which included clone #14, 57R and 57P significantly decreased binding of K-RBP to 57R, but the addition of 57RRE2 and the unrelated Oct-1 binding element had little or no inhibitory effect (Fig. 4C, lanes 3, 4, 5, 9 and 10). It is interesting to note that although clone #14 cold probe can only partially compete with K-RBP for binding to 57R at an 80-fold excess, it completely inhibited K-RBP binding at a 200-fold excess (data not shown). This suggests that the binding between K-RBP and ORF57 promoter is strong. Depletion of K-RBP protein by Ni-NTA beads but not GST beads reduced the intensity of the protein-DNA complex (Fig. 4C, lane 11 to 13). Also, anti-His antibody diminished K-RBP binding to 57R, but anti-Flag antibody had no effect (Fig. 4C, lane 14 to 15). These results demonstrate that K-RBP can specifically bind to ORF57 promoter.
FIG. 4.
K-RBP binds ORF57 promoter. (A) Schematic representation of RRE in KSHV ORF57 promoter on which the locations of various putative promoter-regulatory elements are indicated. The whole 57RRE region was boxed. The GC-rich region is in gray letters. The RBP-Jkappa binding element is underlined. The bold lines below the sequence indicate the ORF57 promoter probes that were used for EMSA. (B) EMSA of the clone #14, 57R, 57RRE2 and 57P probes with K-RBP protein. Purified His-tagged K-RBP was incubated with 32P labeled probes. Arrow indicates the weak binding between K-RBP and 57RRE2. (C) K-RBP binds 57R probe in EMSA. Experiment was performed as described in Fig. 3C except that different unlabeled competitors were used as indicated. Arrow indicates the binding complex between K-RBP and 57R. One μl K-RBP protein contains about 5 ng purified K-RBP protein.
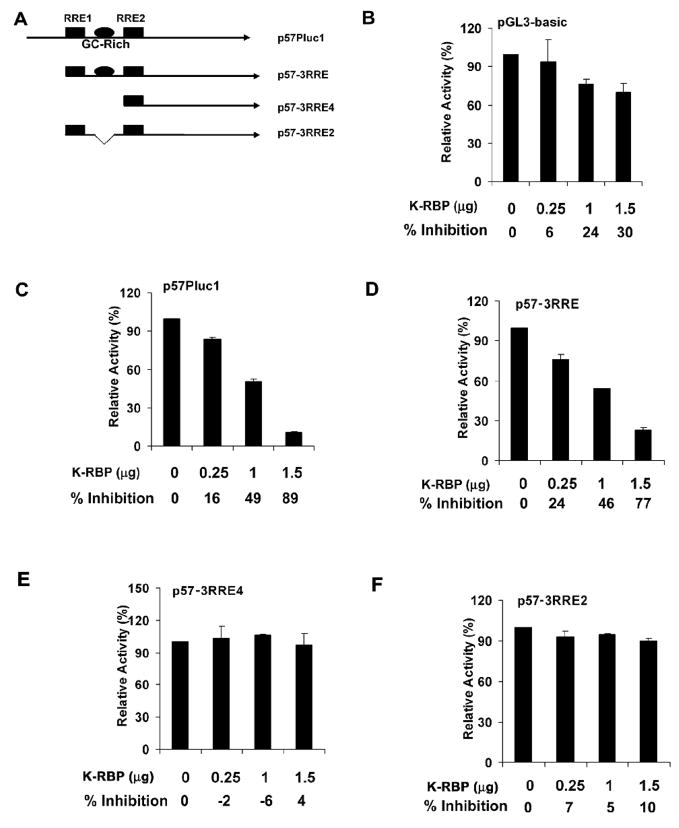
Since K-RBP is a transcriptional repressor, the binding of K-RBP to the ORF57 promoter may play a role in suppression of the promoter activity. Therefore we performed luciferase reporter assay in transfected 293T cells to investigate this possibility. The schematic representations of the luciferase reporter constructs of ORF57 promoter and the mutants used in the study are shown in Fig. 5A. Plasmid pGL3-basic was used for the construction of the luciferase reporters. The p57Pluc1 and p57-3RRE contain the intact ORF57 RREs; the p57-3RRE4 has deletion in RRE1 and does not contain the GC-rich element; p57-3RRE2 has a deletion in the GC-rich element of p57-3RRE. K-RBP had only a marginal suppressive effect on pGL3-basic at high concentration (Fig. 5B), but strongly suppressed p57Pluc1 and p57-3RRE with the intact GC-rich element (Fig. 5C and D). As expected, K-RBP did not suppress p57-3RRE4 and p57-3RRE2, which lack the GC-rich element (Fig. 5E and F). These results together suggest that K-RBP binds the ORF57 promoter via the GC-rich element and functionally suppresses the promoter activity.
FIG. 5.
K-RBP suppresses KSHV ORF57 promoter is dependent on the GC-rich element. (A) Schematic representation of the luciferase promoter reporters used. (B-F) 293T cells were transfected with 100ng of pGL3-basic as shown in (B), p57Pluc1 in (C), p57-3RRE in (D), p57-3RRE4 in (E), p57-3RRE2 in (F) using the indicated amounts of K-RBP plasmid pcDNAK-RBP. The luciferase activities were detected at 24 h post-transfection. For all transfections, total DNA amounts used in each transfection were normalized by the addition of control plasmid. The percentages of inhibition by K-RBP were indicated. The error bars indicate standard deviations of at least 3 independent experiments. Transfection efficiency for each experiment was normalized using a β-Gal expression plasmid as internal control. The basal levels of the promoter activity were normalized to 100%.
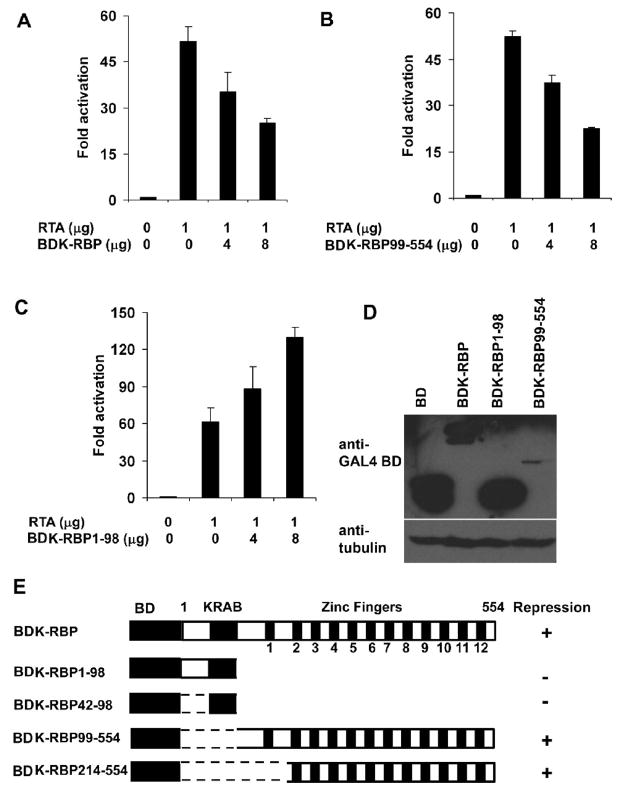
The zinc finger domain of K-RBP alone can repress KSHV RTA-activated ORF57 gene promoter
We have shown that K-RBP can repress RTA-mediated ORF57 gene promoter activation in 293T cells in an earlier study (Yang and Wood, 2007). Similar effect can be observed in another cell line, BJAB cells, and the expression of K-RBP readily suppressed RTA-activated ORF57 gene expression (Fig. 6A). We next sought to determine whether the DNA-binding activity of K-RBP is required to mediate the repression. Several GAL4BD-tagged K-RBP deletion mutants were tested for their effect on RTA-activated ORF57 promoter. The use of GAL4BD tag would provide nuclear localization to the truncated proteins and the ORF57 promoter construct does not contain GAL4 binding site. Plasmids pBDK-RBP42-98 and pBDK-RBP1-98 retain the N-terminal KRAB domain; pBDK-RBP99-554 and pBDK-RBP214-554 retain the full and partial zinc finger domains, respectively (Fig. 6E). Interestingly, as shown in Fig. 6B and C, K-RBP99-554 repressed RTA-activated ORF57 luciferase reporter activity to a similar level as full-length K-RBP, but K-RBP1-98 failed to cause any repression. On the contrary, the addition of deletion mutant K-RBP1-98 enhanced RTA-mediated transcriptional activation by about 2-fold (Fig. 6C). The effect of deletion mutant K-RBP214-554 was similar to K-RBP99-554 which also repressed RTA-mediated transactivation (Fig. 6E). The effect of deletion mutant K-RBP42-98 was similar to K-RBP1-98. It enhanced RTA-mediated transactivation by about 2-fold (Fig. 6E and data not shown). The repression of RTA-mediated transactivation by the K-RBP zinc finger domain was also observed in 293T cells (Data not shown). Since K-RBP99-554 retained its repressor activity on RTA-mediated activation of ORF57 promoter but K-RBP1-98 did not, we examined the expression levels of the two mutants in comparison to the intact K-RBP (Fig. 6D). K-RBP1-98 was expressed at relatively higher level, while the BDK-RBP99-554 was expressed at lower level, suggesting that the inability of K-RBP1-98 to cause repression was not due to inefficient expression of the K-RBP1-98 protein. Together our results suggest that the DNA-binding zinc finger domain of K-RBP plays an important role in repressing RTA-mediated transactivation of the ORF57 promoter.
FIG. 6.
Effect of RTA-mediated transactivation of ORF57 promoter (p57Pluc1) by K-RBP and its mutants in BJAB cells. (A–C) BJAB cells were transfected with 1 μg of p57Pluc1, expression plasmids of RTA and GAL4BD-tagged K-RBP in (A), K-RBP99-554 (zinc fingers) in (B), K-RBP1-98 (KRAB) in (C). The luciferase activities were detected at 48 h post-transfection. In all transfections, the total DNA amounts used in each transfection were normalized by the addition of control expression plasmid. The basic promoter activity was normalized as 1 fold. The error bars indicate standard deviations of at least 3 independent experiments. Transfection efficiency for each experiment was normalized using a β-Gal expression plasmid as internal control. (D) The expression of K-RBP and its mutants. The indicated proteins expressed from plasmids were immunoblotted with rabbit anti-GAL4BD antibody. The higher molecular band from the control plasmid in the first lane is probably a GAL4BD fusion protein translated from a read-through transcript from the vector. Tubulin was used as the loading control. (E) Schematic diagrams of K-RBP deletion mutants used and their effects on RTA-mediated ORF57 transactivation. “+” and “−” indicate the ability of the various deletion clones to repress RTA-mediated transactivation.
Inhibition of RTA binding to ORF57 promoter by K-RBP
The zinc finger domain of K-RBP is sufficient to repress RTA-mediated ORF57 promoter transactivation and this domain mediates DNA binding activity, which suggests that K-RBP may inhibit RTA binding to ORF57 promoter to repress RTA-mediated transactivation. We first showed that the zinc finger domain of K-RBP was sufficient to bind to ORF57 promoter. As shown in Fig. 7A, the membrane-immobilized zinc finger domain of K-RBP was tightly bound to [α-32P] dATP labeled 57P probe as well as to the full-length K-RBP. This is consistent with the result showing that the zinc finger domain was responsible for the DNA-binding activity of K-RBP (Fig. 2C). Since RTA also binds to ORF57 promoter (Lukac et al., 2001), we then examined RTA binding to the promoter in the presence of K-RBP protein using EMSA. The baculovirus expressed RTA bound to 57P probe efficiently and the binding of RTA to 57P was competed efficiently by unlabeled 57P probe, but not by an Oct-1 binding element (Fig. 7B, lanes 3 and 4). In order to determine whether K-RBP affects RTA binding to 57P, EMSA was carried out in the presence of both RTA and K-RBP proteins. As shown in Fig. 7B (lanes 6–8), RTA binding to 57P was decreased significantly as an increasing amount of K-RBP was added. The upper RTA/57P complex disappeared and the intensities of the middle and lower RTA/57P complexes decreased. Our results suggest that K-RBP can inhibit the binding of RTA to ORF57 promoter. The inhibition of RTA binding to ORF57 promoter by K-RBP likely contributes to the repression of RTA-mediated ORF57 promoter transactivation.
FIG. 7.
Inhibition of RTA binding to ORF57 promoter by K-RBP. (A) The zinc finger domain of K-RBP binds ORF57 promoter. The E. coli expressed His-tagged zinc finger domain, full-length K-RBP and E. coli lysate were resolved on SDS-PAGE, transferred to PVDF membrane and analyzed by Coomassie blue staining (left panel). The second membrane with the same samples was refolded and hybridized to the 57P probe labeled with [α-32P] dATP (right panel). (B) Baculovirus expressed RTA, E. coli expressed K-RBP and [α-32P] dATP labeled 57P probe were used in EMSA. The EMSA was performed as described in Fig. 3C except that different unlabeled competitors and proteins were used as indicated in the figure. Open arrows indicate specific binding of RTA to probes. Filled arrows indicate the binding of K-RBP to probes. Numbers below the bands indicate the relative intensities of the bands measured by Image J.
Discussion
In this study, we characterized the DNA-binding activity of K-RBP and demonstrated that the DNA-binding zinc finger domain of K-RBP plays an important role in suppressing RTA-mediated ORF57 promoter transactivation. This study, as well as the earlier findings that KSHV RTA utilizes other cellular factors such as IRF-7 and RBP-Jκ to regulate the transactivation activity (Liang et al., 2002; Wang et al., 2005), supports that the modulation of RTA-DNA binding is an important mechanism to regulate RTA activity.
DNA binding activity of K-RBP
Even though KRAB-containing zinc finger proteins are predicted to function as sequence-specific transcriptional repressors by binding DNA via the zinc fingers and repressing transcription using the KRAB domain (Gebelein and Urrutia, 2001; Urrutia, 2003), recent studies have shown that zinc fingers are more diverse in selecting their binding partners than previously thought. The zinc fingers could be RNA or protein binding motifs (Brayer and Segal, 2008), and not all zinc finger proteins were shown to bind to DNA (Kwan et al., 2003). While some studies have suggested that the −1, +3 and +6 amino acids (Fig. 1A) in the α-helical region of a zinc finger motif determine its DNA-binding specificity (Pavletich and Pabo, 1991; Pavletich and Pabo, 1993; Wolfe et al., 2000), other studies have reported that other amino acids in the α-helix also contribute to the binding specificity of a zinc finger motif (Peng et al., 2002). In addition, in the context of a full-length protein, not all zinc finger motifs bind DNA (Peng et al., 2002; Zheng et al., 2000).
Since K-RBP binds DNA through its zinc fingers, in order to study whether K-RBP regulates RTA-mediated transactivation via DNA-binding, we decided to examine the DNA-binding activity of K-RBP. We chose the random selection approach to identify K-RBP binding DNA sequences because the accurate prediction of zinc finger protein binding sites is unreliable. Among the target sequences that were bound by K-RBP, the majority of them contain GC-rich elements throughout the sequences. This is consistent with the findings that the consensus binding sequences of many zinc finger proteins are also GC rich even though there are a few that are not (Christy and Nathans, 1989; Swirnoff and Milbrandt, 1995). Our results suggest that K-RBP preferentially binds to the consensus sequence but its targets may not be limited to these sequences. The relatively broad binding specificity of K-RBP was also evidenced by its tight binding to ORF57 promoter, which does not show 100% similarity to the selected consensus sequence. The plasticity of K-RBP DNA-binding is not unprecedented since this reflects the complexity of DNA-binding for a protein with multiple zinc fingers. This has also been observed in other zinc finger proteins. For example, the cellular protein Ying Yang 1 (YY1) can bind to several different consensus sequences (Hyde-DeRuyscher et al., 1995; Kim and Kim, 2008). Further studies such as mutageneisis analysis will be needed to dissect the preferential binding sites and the fingers within K-RBP responsible for DNA and protein bindings.
The repression of KSHV ORF57 promoter activity by K-RBP
KSHV establishes latency after primary infection, which is characterized by the repression of a number of viral lytic genes. There are multiple mechanisms involved in suppression of KSHV lytic gene expression but they are not fully understood. A number of cellular factors may contribute to the maintenance of KSHV latency, and K-RBP is one of them (Brown et al., 2003; Gwack et al., 2001; Gwack et al., 2003; Wang et al., 2005). K-RBP negatively regulates RTA-activation of lytic genes such as ORF57 gene (Yang and Wood, 2007). Our results here indicate that one of the mechanisms for repression of ORF57 expression is the direct binding of K-RBP to its promoter. The binding of K-RBP to viral ORF57 promoter may bring co-repressors to silence gene expression. An example is TIF1β, which was found to interact with the KRAB domain of K-RBP and some other KRAB-containing proteins (Abrink et al., 2001; Yang and Wood, 2007). The repression mechanism of KRAB-containing zinc finger proteins has been shown to involve recruitment of repression complex by KRAB domain, including TIF1β (transcription intermediary factor 1β) (Friedman et al., 1996; Kim et al., 1996; Moosmann et al., 1996). The KRAB/TIF1β complex recruits cellular factors heterochromatin protein 1 (HP1) family, a family of non-histone heterochromatin-associated proteins with gene-silencing function, HDAC and SETDB1, a SET domain-containing protein that methylates lysine 9 of histone H3 to the DNA regulatory region (Cammas et al., 2004; Lechner et al., 2000; Nielsen et al., 1999; Peng et al., 2000; Ryan et al., 1999; Schultz et al., 2002; Schultz et al., 2001; Sripathy et al., 2006). This may also explain why the KRAB domain of K-RBP in the absence of the zinc finger motifs can moderately enhance RTA-mediated transactivation (Fig. 6C). It may function as a dominant-negative mutant that interacts with repressors to prevent their targeting to ORF57 promoter to mediate repression.
Our study showed that the zinc finger domain of K-RBP alone can bind to ORF57 promoter, compete with RTA in binding to ORF57 promoter and repress RTA-activated ORF57 promoter. This suggests that the DNA-binding activity of K-RBP plays an important role for inhibiting RTA-mediated transactivation. However, the DNA binding activity of K-RBP may be not the sole mechanism in repressing RTA-mediated transactivation. An example is the KSHV ORFK8, it contains the GGGGGTGG sequence motif and its activation by RTA can be repressed by K-RBP (Yang and Wood, 2007). However, K-RBP cannot repress ORFK8 promoter expression in the absence of RTA (unpublished data). This suggests that the interaction between K-RBP and RTA may also play a role in the repression of RTA-mediated transactivation, in addition to the DNA binding activity of K-RBP. It is also possible that a combination of the above mechanisms could be involved in the repression of RTA-mediated transactivation by K-RBP.
The potential role of K-RBP in KSHV life cycle
We have recently found that RTA could induce K-RBP degradation through a proteasome-dependent pathway leading to lytic reactivation (Yang et al., 2008). It is likely that there is a dynamic relationship between repression by K-RBP and K-RBP degradation induced by RTA in KSHV infected cells, and this balance may change at various stages of KSHV infection. During latency, RTA-mediated transactivation may be blocked by K-RBP and other repressors when RTA expression level is very low. However, during lytic induction, when RTA expression is induced, RTA may overcome the suppression and lead to lytic replication.
The identification of K-RBP as a repressor of KSHV gene expression and as an ORF57 promoter binding protein is interesting because other KRAB-containing zinc finger proteins have also been found to be involved in regulating viral replication. K-RBP is one of a few members of the zinc finger protein family that are known to repress viral replication. The KRAB-containing zinc finger protein OTK18 represses human immunodeficiency virus type 1 (HIV-1) replication (Carlson et al., 2004). Recently, a KRAB containing protein was found to repress lentivirus proviral transcription (Bulliard et al., 2006). Engineered KRAB-containing zinc finger proteins were also used to repress HIV transcription and replication in several studies (Eberhardy et al., 2006; Reynolds et al., 2003; Segal et al., 2004). The DNA-binding activity plays an important role in specifying its effective targets. It is possible that the KRAB-containing zinc finger proteins play a broader role and function as intracellular antiviral molecules to inhibit viral replication. This possibility needs to be further explored.
K-RBP may have other roles in KSHV life cycle in addition to transcriptional regulation. The origin of replication of KSHV is a GC-rich region, whether K-RBP binds this region to regulate viral replication needs to be investigated (Russo et al., 1996). K-RBP may also regulate the expression of other KSHV genes or other viruses such as Epstein-Barr virus (EBV), since co-infection of primary effusion lymphoma by both KSHV and EBV occurs frequently (Drexler et al., 1998), and there are a number of KSHV and EBV viral promoters harboring GC-rich elements. Further study will be needed to map other K-RBP binding promoters on KSHV viral genome. This may include investigating the in vivo binding targets of K-RBP on viral and cellular genome by Chromatin immunoprecipitation followed by new deep pyrosequencing technique. In conclusion, the regulation of KSHV gene expression by K-RBP through its DNA-binding activity suggests a DNA binding-dependent mechanism for K-RBP to regulate KSHV gene expression.
Materials and Methods
Plasmids
Expression plasmids pcDNAK-RBP and pcDNAORF50, which encode full-length K-RBP and KSHV RTA, have been described elsewhere (Wang et al., 2001). The mammalian expression plasmids GAL4 DNA binding domain (GAL4BD)-tagged K-RBP and its mutant clones pBDK-RBP, pBDK-RBP42-98, pBDK-RBP1-98, pBDK-RBP214-554 and pBDK-RBP99-554 encoding full-length, K-RBP fragments from amino acids 42 to 98, 1 to 98, 214 to 554 and 99 to 554 respectively, have also been described elsewhere (Yang and Wood, 2007). The E. coli expression plasmids pET28K-RBP, pET28K-RBP1-98 and pET28K-RBP141-554 were constructed by PCR-amplification of corresponding K-RBP DNA fragments and inserting into pET28 vector. Reporter plasmid p57Pluc1, containing promoter region of KSHV ORF57, has also been described previously (Duan et al., 2001). The luciferase reporter plasmids p57-3RRE, p57-3RRE2 and p57-3RRE4, as shown in Fig. 5, were constructed by amplifying corresponding DNA fragments of ORF57 promoter by PCR and inserting into pGL3-basic vector (Promega, Madison, WI). The β-galactosidase (β-Gal) expression plasmid pCMV-β was purchased from BD Clontech (Mountain View, CA) and used for the normalization of transfection efficiency. All inserts amplified by PCR were confirmed by DNA sequence analysis.
Expression, purification and refolding of recombinant K-RBP protein
E. coli DE3 cells containing His-tagged K-RBP expression plasmid pET28K-RBP were induced by 1 mM isopropyl-beta-D-thiogalactopyranoside (IPTG) at 37°C for 4–5 h to express K-RBP protein. The cells were suspended in 1x PBS containing 1mM Phenylmethylsulphonylfluoride (PMSF) and then lysed by sonication. The insoluble fraction was collected by centrifugation and washed sequentially with 1x PBS containing 2 M urea, 0.1% SDS, 1% NP-40 and 1% TritonX-100. The majority of the insoluble fraction after extensive washing contains K-RBP protein (see Fig. 1B). The purified protein was solubilized in 1x PBS containing 8 M Urea, and was renatured by successive dialysis against refolding buffer (25 mM HEPES, pH 7.9, 50 mM NaCl, 0.1% NP-40, 1 mM PMSF, 0.16 mM dithiothreitol [DTT], 10 μM ZnSO4, 10% glycerol) containing 4, 2, 1 and 0 M Urea at 4°C. The resulting protein was evaluated by coomassie blue staining and Western blot analysis with anti-His antibody following SDS-PAGE. The protein expression of K-RBP1-98 and K-RBP141-554 was carried out in E. coli similar to K-RBP protein.
Expression and purification of recombinant KSHV RTA protein
KSHV RTA was expressed in Sf9 cells using Bac-N-Blue baculovirus expression system (Invitrogen, Carlsbad, CA). Briefly, full-length KSHV RTA was PCR-amplified and cloned into pBlueBacHis2A vector, which was co-transfected into Sf9 cells with the wild-type baculovirus vector. Recombinant virus was plaque purified twice to obtain a high titer stock. To express protein, Sf9 cells were infected at an MOI of 10 with the recombinant virus. The purification of His-tagged RTA protein was carried out using Ni-NTA superflow (Qiagen, Valencia, CA) according to the manufacture’s recommendation.
DNA cellulose binding assay
DNA cellulose binding assay was carried out according to published procedure with modifications (Nishiya et al., 1998). Briefly, approximately 5 μg of K-RBP protein was incubated with 5 mg of cellulose or double-stranded (ds) DNA-cellulose in binding buffer (25 mM HEPES, pH 7.9, 50 mM KCl, 0.1% NP-40, 1 mM PMSF, 0.16 mM DTT, 10 μM ZnSO4, 10% glycerol) in 0.5 ml volume overnight at 4°C. The cellulose was preincubated with 5% bovine serum albumin (BSA) in binding buffer for 1 h prior to use. After incubation with K-RBP, the cellulose/protein complex was washed 4 times with binding buffer. The protein bound to DNA-cellulose was eluted with 1.25% SDS or binding buffer containing increasing concentration of NaCl, resolved by SDS-PAGE, and then detected by Western blot analysis using rabbit anti-K-RBP antibody.
DNA filter binding assay
Filter binding assay was adapted from published procedure with some modifications (Jheon et al., 2001). Cell lysates from E. coli were separated by SDS-PAGE and transferred to Polyvinylidene fluoride (PVDF) membrane. The membrane was renatured overnight at 4°C in binding buffer (25 mM HEPES, pH 7.9, 50 mM KCl, 0.1% NP-40, 1 mM PMSF, 0.16 mM DTT, 10 μM ZnSO4, 10% glycerol, 30–50 μg/ml salmon testes DNA). DNA probes were end-labeled with [α-32P] dATP (PerkinElmer Life Sciences, Waltham, MA) using Klenow DNA polymerase (New England Biolabs, Ipswich, MA). Approximately 1 × 105 cpm/ml of the labeled DNA was hybridized to the membrane in binding buffer overnight at room temperature. The membrane was washed for 6 h (3–4 times) at room temperature in 50 mM KCl, and the protein/DNA binding complexes were analyzed by autoradiography.
Selection of K-RBP binding sequences
The selection of consensus DNA binding sequences of K-RBP was carried out by the Selected and Amplified Binding Sequence (SAAB) method with modifications (Blackwell and Weintraub, 1990). For the first set of binding sequence selection, a pool of double-stranded random oligomers was obtained by PCR amplification using the 63-base random oligonucleotide N27 (5′-ATGGTACCGAAGTCCTCA-N27-GACATCAGTCTCGAGTCT-3′) as template (125 ng), 0.5 μg of primer 1 (5′-ATGGTACCGAAGTCCTCA -3′) and 0.5 μg of primer 2 (5′-AGACTCGAGACTGATGTC-3′). The dsDNA (~0.5 μg) was incubated with 0.5 μg of His-tagged K-RBP protein for 1 h with rotation in a final volume of 150 μl of binding buffer (25 mM HEPES, pH 7.9, 50 mM KCl, 0.1% NP-40, 1mM PMSF, 0.16 mM DTT, 10 μM ZnSO4, 10% glycerol) containing 10 μg/ml poly(dI·dC) at room temperature. The resulting K-RBP/DNA complex was isolated by incubating the solution with Ni-NTA superflow for 1 h, and the beads were washed 4 times by binding buffer. The K-RBP/DNA complex was separated from Ni-NTA by 100 μl of elution buffer (250 mM imidazole, 300 mM NaCl, 50 mM NaH2PO4), heated at 95°C for 10 min and partially purified by Qiagen nucleotide removal kit. Ten μl of the recovered DNA was amplified by PCR with primers 1 and 2. After seven rounds of selection and purification, the final round PCR product was cloned directly into pGEM-T easy vector system (Promega), amplified and sequenced.
The second set of binding sequence selection was performed using a pool of double-stranded random oligomers obtained by PCR, using 100 ng of the 68-base random oligonucleotide N14-CGG-N15 (5′-GACTGACAGATCTGTGAG-N14-CGG-N15-CTGTCATATGACAGTCAC-3′) as template, 0.5 μg of primer 3 (5′-GACTGACAGATCTGTGAG -3′) and 0.5 μg of primer 4 (5′-GTGACTGTCATATGACAG-3′). Binding was carried out as described above but the selection was carried out for eight rounds. The PCR product from the eighth round was inserted into pGEM-T and sequenced.
DNA sequencing and analysis
After multiple rounds of selection, PCR amplification and purification, the affinity-selected oligonucleotides were ligated into the pGEM-T vector. After transforming into E. coli, the DNA inserts from the resulting clones were sequenced. The sequences were analyzed, aligned using the BioEdit sequence analysis program and adjusted manually.
Electrophoretic mobility shift assay (EMSA)
The double-stranded oligonucleotides used for EMSA were end-labeled with [α-32P] dATP using Klenow DNA polymerase. The labeled probes were incubated with the protein for 30 min in binding buffer (10 mM Tris-HCl [pH 7.5], 150 mM KCl, 5.7 mM MgCl2, 1 mM EDTA, 0.2 μg of poly[dI·dC], 5 μg of BSA, 0.67 mM DTT, 0.67 mM PMSF, 5% glycerol) in the presence or absence of unlabeled probes. If antibody was added to detect supershift, the antibody and protein were preincubated for 20 min before the labeled probes were added. The protein/DNA binding samples were loaded onto a 4.5% polyacrylamide gel in 1xTGE buffer (5 mM Tris, 190 mM glycine, and 1 mM EDTA [pH 8.3]), then dried and exposed to X-ray film. Signal intensities were quantified by the NIH Image J software. The probes 57P, 57R and 57RRE2 that were used are indicated in Fig. 4A. The probe clone #14 is indicated in Fig. 3D. One strand of Oct-1 binding sequence is: 5′-TGTCGAATGCAAATCACTAG-3′.
Antibodies
Anti-K-RBP antibody was described elsewhere (Yang and Wood, 2007). The mouse anti-tubulin monoclonal antibody was purchased from Sigma (St. Louis, MO). The mouse anti-Flag M2 monoclonal antibody was purchased from Stratagene (La Jolla, CA) and the mouse anti-His monoclonal antibody was purchased from BD Clontech. Mouse and rabbit anti-GAL4BD antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Cell culture, transfection, and luciferase assay
Human 293T cells were grown in Dulbecco’s Modified Eagle Medium (DMEM, Invitrogen) supplemented with 10% fetal bovine serum (FBS, Invitrogen) and 1% penicillin-streptomycin (Mediatech, Manassas, VA) at 37°C with 5% CO2. Transfection of 293T cells was carried out using Lipofectamine 2000 (Invitrogen) according to manufacturer’s recommendations. Human BJAB cells were grown in RPMI 1640 medium (Gibco BRL) supplemented with 10% FBS and 1% penicillin-streptomycin at 37°C with 5% CO2. Transfection of BJAB cells was conducted using Amaxa Nucleofactor according to manufacturer’s recommendations. For all transfections, the total DNA amounts used in each transfection were normalized by adding control plasmid. Luciferase activities were determined at 48 h post-transfection by the Luciferase Assay System (Promega) according to the manufacturer’s recommendations. Each result was an average of at least three independent transfections. Transfection efficiency for each experiment was normalized by co-transfecting with a β-gal expression plasmid pCMV-β which was used as internal control.
Western blotting analysis
Proteins were separated by SDS-PAGE and transferred to PVDF membrane. The membrane was blocked with 3–5% nonfat dry milk in TBST (50 mM Tris [pH 7.5], 200 mM NaCl, 0.05% Tween 20) at room temperature for 1 h. The membrane was then incubated with primary antibody with the same concentration of milk in TBST for 1 h at room temperature, washed three times with TBST for 15 min, then incubated with the HRP-conjugated secondary antibody at room temperature for 1 h. The membrane was again washed three times with TBST, treated with SuperSignal (Pierce Biotechnology) detection reagents and exposed to Kodak Light film.
Acknowledgments
This study was supported by PHS grant CA76903 and NCRR COBRE grant RR15635 to C.W.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abrink M, Ortiz JA, Mark C, Sanchez C, Looman C, Hellman L, Chambon P, Losson R. Conserved interaction between distinct Kruppel-associated box domains and the transcriptional intermediary factor 1 beta. Proc Natl Acad Sci U S A. 2001;98(4):1422–1426. doi: 10.1073/pnas.041616998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell TK, Weintraub H. Differences and similarities in DNA-binding preferences of MyoD and E2A protein complexes revealed by binding site selection. Science. 1990;250 (4984):1104–1110. doi: 10.1126/science.2174572. [DOI] [PubMed] [Google Scholar]
- Brayer KJ, Segal DJ. Keep your fingers off my DNA: protein-protein interactions mediated by C2H2 zinc finger domains. Cell Biochem Biophys. 2008;50(3):111–131. doi: 10.1007/s12013-008-9008-5. [DOI] [PubMed] [Google Scholar]
- Brown HJ, Song MJ, Deng H, Wu TT, Cheng G, Sun R. NF-kappaB inhibits gammaherpesvirus lytic replication. J Virol. 2003;77 (15):8532–8540. doi: 10.1128/JVI.77.15.8532-8540.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulliard Y, Wiznerowicz M, Barde I, Trono D. KRAB can repress lentivirus proviral transcription independently of integration site. J Biol Chem. 2006;281(47):35742–35746. doi: 10.1074/jbc.M602843200. [DOI] [PubMed] [Google Scholar]
- Cammas F, Herzog M, Lerouge T, Chambon P, Losson R. Association of the transcriptional corepressor TIF1beta with heterochromatin protein 1 (HP1): an essential role for progression through differentiation. Genes Dev. 2004;18(17):2147–2160. doi: 10.1101/gad.302904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson KA, Leisman G, Limoges J, Pohlman GD, Horiba M, Buescher J, Gendelman HE, Ikezu T. Molecular Characterization of a Putative Antiretroviral Transcriptional Factor, OTK18. J Immunol. 2004;172(1):381–391. doi: 10.4049/jimmunol.172.1.381. [DOI] [PubMed] [Google Scholar]
- Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;266 (5192):1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- Chang Y, Moore PS. Kaposi’s Sarcoma (KS)-associated herpesvirus and its role in KS. Infect Agents Dis. 1996;5(4):215–222. [PubMed] [Google Scholar]
- Christy B, Nathans D. DNA Binding Site of the Growth Factor-Inducible Protein Zif268. Proc Natl Acad Sci U S A. 1989;86(22):8737–8741. doi: 10.1073/pnas.86.22.8737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler HG, Uphoff CC, Gaidano G, Carbone A. Lymphoma cell lines: in vitro models for the study of HHV-8+ primary effusion lymphomas (body cavity-based lymphomas) Leukemia. 1998;12 (10):1507–1517. doi: 10.1038/sj.leu.2401160. [DOI] [PubMed] [Google Scholar]
- Duan W, Wang S, Liu S, Wood C. Characterization of Kaposi’s sarcoma-associated herpesvirus/human herpesvirus-8 ORF57 promoter. Arch Virol. 2001;146(2):403–413. doi: 10.1007/s007050170185. [DOI] [PubMed] [Google Scholar]
- Eberhardy SR, Goncalves J, Coelho S, Segal DJ, Berkhout B, Barbas CF., 3rd Inhibition of human immunodeficiency virus type 1 replication with artificial transcription factors targeting the highly conserved primer-binding site. J Virol. 2006;80(6):2873–2883. doi: 10.1128/JVI.80.6.2873-2883.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JR, Fredericks WJ, Jensen DE, Speicher DW, Huang XP, Neilson EG, Rauscher FJ., 3rd KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev. 1996;10(16):2067–2078. doi: 10.1101/gad.10.16.2067. [DOI] [PubMed] [Google Scholar]
- Gebelein B, Urrutia R. Sequence-specific transcriptional repression by KS1, a multiple-zinc-finger-Kruppel-associated box protein. Mol Cell Biol. 2001;21(3):928–939. doi: 10.1128/MCB.21.3.928-939.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradoville L, Gerlach J, Grogan E, Shedd D, Nikiforow S, Metroka C, Miller G. Kaposi’s sarcoma-associated herpesvirus open reading frame 50/Rta protein activates the entire viral lytic cycle in the HH-B2 primary effusion lymphoma cell line. J Virol. 2000;74(13):6207–6212. doi: 10.1128/jvi.74.13.6207-6212.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwack Y, Byun H, Hwang S, Lim C, Choe J. CREB-binding protein and histone deacetylase regulate the transcriptional activity of Kaposi’s sarcoma-associated herpesvirus open reading frame 50. J Virol. 2001;75(4):1909–1917. doi: 10.1128/JVI.75.4.1909-1917.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwack Y, Nakamura H, Lee SH, Souvlis J, Yustein JT, Gygi S, Kung H-J, Jung JU. Poly(ADP-Ribose) polymerase 1 and ste20-like kinase hKFC act as transcriptional repressors for gamma-2 herpesvirus lytic replication. Mol Cell Biol. 2003;23(22):8282–8294. doi: 10.1128/MCB.23.22.8282-8294.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde-DeRuyscher RP, Jennings E, Shenk T. DNA binding sites for the transcriptional activator/repressor YY1. Nucleic Acids Res. 1995;23(21):4457–4465. doi: 10.1093/nar/23.21.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenner RG, Alba MM, Boshoff C, Kellam P. Kaposi’s sarcoma-associated herpesvirus latent and lytic gene expression as revealed by DNA arrays. J Virol. 2001;75(2):891–902. doi: 10.1128/JVI.75.2.891-902.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jheon AH, Ganss B, Cheifetz S, Sodek J. Characterization of a novel KRAB/C2H2 zinc finger transcription factor involved in bone development. J Biol Chem. 2001;276(21):18282–18289. doi: 10.1074/jbc.M010885200. [DOI] [PubMed] [Google Scholar]
- Kim JD, Kim J. YY1’s longer DNA-binding motifs. Genomics. 2008 doi: 10.1016/j.ygeno.2008.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SS, Chen YM, O’Leary E, Witzgall R, Vidal M, Bonventre JV. A novel member of the RING finger family, KRIP-1, associates with the KRAB-A transcriptional repressor domain of zinc finger proteins. Proc Natl Acad Sci U S A. 1996;93(26):15299–15304. doi: 10.1073/pnas.93.26.15299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan AH, Czolij R, Mackay JP, Crossley M. Pentaprobe: a comprehensive sequence for the one-step detection of DNA-binding activities. Nucleic Acids Res. 2003;31(20):e124. doi: 10.1093/nar/gng124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner MS, Begg GE, Speicher DW, Rauscher FJ., 3rd Molecular determinants for targeting heterochromatin protein 1-mediated gene silencing: direct chromoshadow domain-KAP-1 corepressor interaction is essential. Mol Cell Biol. 2000;20(17):6449–6465. doi: 10.1128/mcb.20.17.6449-6465.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Chang J, Lynch SJ, Lukac DM, Ganem D. The lytic switch protein of KSHV activates gene expression via functional interaction with RBP-Jkappa (CSL), the target of the Notch signaling pathway. Genes Dev. 2002;16(15):1977–1989. doi: 10.1101/gad.996502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukac DM, Garibyan L, Kirshner JR, Palmeri D, Ganem D. DNA binding by Kaposi’s sarcoma-associated herpesvirus lytic switch protein is necessary for transcriptional activation of two viral delayed early promoters. J Virol. 2001;75(15):6786–6799. doi: 10.1128/JVI.75.15.6786-6799.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukac DM, Renne R, Kirshner JR, Ganem D. Reactivation of Kaposi’s sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology. 1998;252 (2):304–312. doi: 10.1006/viro.1998.9486. [DOI] [PubMed] [Google Scholar]
- Moosmann P, Georgiev O, Le Douarin B, Bourquin JP, Schaffner W. Transcriptional repression by RING finger protein TIF1 beta that interacts with the KRAB repressor domain of KOX1. Nucleic Acids Res. 1996;24(24):4859–4867. doi: 10.1093/nar/24.24.4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen AL, Ortiz JA, You J, Oulad-Abdelghani M, Khechumian R, Gansmuller A, Chambon P, Losson R. Interaction with members of the heterochromatin protein 1 (HP1) family and histone deacetylation are differentially involved in transcriptional silencing by members of the TIF1 family. Embo J. 1999;18(22):6385–6395. doi: 10.1093/emboj/18.22.6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiya N, Sabe H, Nose K, Shibanuma M. The LIM domains of hic-5 protein recognize specific DNA fragments in a zinc-dependent manner in vitro. Nucleic Acids Res. 1998;26(18):4267–4273. doi: 10.1093/nar/26.18.4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavletich NP, Pabo CO. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991;252 (5007):809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- Pavletich NP, Pabo CO. Crystal structure of a five-finger GLI-DNA complex: new perspectives on zinc fingers. Science. 1993;261 (5129):1701–1707. doi: 10.1126/science.8378770. [DOI] [PubMed] [Google Scholar]
- Peng H, Begg GE, Schultz DC, Friedman JR, Jensen DE, Speicher DW, Rauscher FJ., 3rd Reconstitution of the KRAB-KAP-1 repressor complex: a model system for defining the molecular anatomy of RING-B box-coiled-coil domain-mediated protein-protein interactions. J Mol Biol. 2000;295(5):1139–1162. doi: 10.1006/jmbi.1999.3402. [DOI] [PubMed] [Google Scholar]
- Peng H, Zheng L, Lee WH, Rux JJ, Rauscher FJ., 3rd A common DNA-binding site for SZF1 and the BRCA1-associated zinc finger protein, ZBRK1. Cancer Res. 2002;62(13):3773–3781. [PubMed] [Google Scholar]
- Reynolds L, Ullman C, Moore M, Isalan M, West MJ, Clapham P, Klug A, Choo Y. Repression of the HIV-1 5′ LTR promoter and inhibition of HIV-1 replication by using engineered zinc-finger transcription factors. Proc Natl Acad Sci U S A. 2003;100(4):1615–1620. doi: 10.1073/pnas.252770699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo JJ, Bohenzky RA, Chien MC, Chen J, Yan M, Maddalena D, Parry JP, Peruzzi D, Edelman IS, Chang Y, Moore PS. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci U S A. 1996;93(25):14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan RF, Schultz DC, Ayyanathan K, Singh PB, Friedman JR, Fredericks WJ, Rauscher FJ., 3rd KAP-1 corepressor protein interacts and colocalizes with heterochromatic and euchromatic HP1 proteins: a potential role for Kruppel-associated box-zinc finger proteins in heterochromatin-mediated gene silencing. Mol Cell Biol. 1999;19(6):4366–4378. doi: 10.1128/mcb.19.6.4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarid R, Flore O, Bohenzky RA, Chang Y, Moore PS. Transcription mapping of the Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) genome in a body cavity-based lymphoma cell line (BC-1) J Virol. 1998;72(2):1005–1012. doi: 10.1128/jvi.72.2.1005-1012.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ., 3rd SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16(8):919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DC, Friedman JR, Rauscher FJ., 3rd Targeting histone deacetylase complexes via KRAB-zinc finger proteins: the PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2alpha subunit of NuRD. Genes Dev. 2001;15(4):428–443. doi: 10.1101/gad.869501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal DJ, Goncalves J, Eberhardy S, Swan CH, Torbett BE, Li X, Barbas CF., 3rd Attenuation of HIV-1 replication in primary human cells with a designed zinc finger transcription factor. J Biol Chem. 2004;279(15):14509–14519. doi: 10.1074/jbc.M400349200. [DOI] [PubMed] [Google Scholar]
- Sripathy SP, Stevens J, Schultz DC. The KAP1 corepressor functions to coordinate the assembly of de novo HP1-demarcated microenvironments of heterochromatin required for KRAB zinc finger protein-mediated transcriptional repression. Mol Cell Biol. 2006;26(22):8623–8638. doi: 10.1128/MCB.00487-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun R, Lin SF, Gradoville L, Yuan Y, Zhu F, Miller G. A viral gene that activates lytic cycle expression of Kaposi’s sarcoma-associated herpesvirus. Proc Natl Acad Sci U S A. 1998;95(18):10866–10871. doi: 10.1073/pnas.95.18.10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swirnoff AH, Milbrandt J. DNA-binding specificity of NGFI-A and related zinc finger transcription factors. Mol Cell Biol. 1995;15(4):2275–2287. doi: 10.1128/mcb.15.4.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urrutia R. KRAB-containing zinc-finger repressor proteins. Genome Biol. 2003;4(10):231. doi: 10.1186/gb-2003-4-10-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhang J, Zhang L, Harrington W, Jr, West JT, Wood C. Modulation of human herpesvirus 8/Kaposi’s sarcoma-associated herpesvirusreplication and transcription activator transactivation by interferon regulatory factor 7. J Virol. 2005;79(4):2420–2431. doi: 10.1128/JVI.79.4.2420-2431.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Liu S, Wu MH, Geng Y, Wood C. Identification of a cellular protein that interacts and synergizes with the RTA (ORF50) protein of Kaposi’s sarcoma-associated herpesvirus in transcriptional activation. J Virol. 2001;75(24):11961–11973. doi: 10.1128/JVI.75.24.11961-11973.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West JT, Wood C. The role of Kaposi’s sarcoma-associated herpesvirus/human herpesvirus-8 regulator of transcription activation (RTA) in control of gene expression. Oncogene. 2003;22 (33):5150–5163. doi: 10.1038/sj.onc.1206555. [DOI] [PubMed] [Google Scholar]
- Wolfe SA, Nekludova L, Pabo CO. DNA recognition by Cys2His2 zinc finger proteins. Annu Rev Biophys Biomol Struct. 2000;29(1):183–212. doi: 10.1146/annurev.biophys.29.1.183. [DOI] [PubMed] [Google Scholar]
- Yang Z, Wood C. The transcriptional repressor K-RBP modulates RTA-mediated transactivation and lytic replication of Kaposi’s sarcoma-associated herpesvirus. J Virol. 2007;81(12):6294–6306. doi: 10.1128/JVI.02648-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Yan Z, Wood C. Kaposi’s Sarcoma-Associated Herpesvirus (KSHV) Transactivator RTA Promotes Degradation of the Repressors to Regulate Viral Lytic Replication. J Virol. 2008;82(7):3590–3603. doi: 10.1128/JVI.02229-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Pan H, Li S, Flesken-Nikitin A, Chen PL, Boyer TG, Lee WH. Sequence-specific transcriptional corepressor function for BRCA1 through a novel zinc finger protein, ZBRK1. Mol Cell. 2000;6 (4):757–768. doi: 10.1016/s1097-2765(00)00075-7. [DOI] [PubMed] [Google Scholar]