Abstract
The C protein of human parainfluenza virus type 3 (HPIV3) is a multifunctional accessory protein which inhibits viral transcription and interferon (IFN) signaling. In the present study, we found that removal of N-terminal 25 or 50 amino acid residues from the C protein (CNΔ25 or CNΔ50) totally abolished viral RNA synthesis in the HPIV3 minigenome system. Further N-terminal or C-terminal deletion impaired the inhibitory ability of CNΔ25 and CNΔ50. Subsequent mutagenesis analysis suggested that the N-terminal charged amino acid residues (K3, K6, K12, E16, and R24) contribute to the higher inhibition caused by CNΔ25 than the C protein. Consistent with viral RNA synthesis inhibition, the growth of HPIV3 was significantly decreased by 5 logs in HeLa-derived cell line expressing CNΔ25. Interestingly, replication of respiratory syncytial virus (RSV), another important respiratory tract pathogen, was also strongly inhibited in the presence of CNΔ25. These findings provide a promising potential to use CNΔ25 as an antiviral agent against the clinically important respiratory tract diseases caused by HPIV3 and RSV.
Keywords: HPIV3, C protein, CNΔ25 protein, antivirals
Introduction
Human parainfluenza virus type 3 (HPIV3) is one of the parainfluenza viruses, which is classified in the subfamily paramyxovirinae, family paramyxoviridae, order mononegavirales (the nonsegmented negative-strand RNA virus). Along with another respirovirus (HPIV1), and two rubulaviruses (HPIV2 and HPIV4), the parainfluenza viruses (HPIVs) are the most common cause of respiratory tract diseases, second only to respiratory syncytial virus (RSV), for infections in young children and infants (e.g. pneumonia, bronchitis, and bronchiolitis). Infection of HPIVs and RSV has also been linked to increasing rates of mortality in immuno-compromised patients (Cortez et al., 2001; Madhi et al., 2002). Although some antiviral regents and vaccine candidates have been generated and tested in preclinical and clinical trials (Sato et al., 2008; Mao et al., 2008; Liuzzi et al., 2005; Nichols et al., 2008; Nokes et al., 2008), there is no single industry-approved vaccine or antiviral drug on the market for this group of patients. HPIVs infection is treated symptomatically, and RSV is treated with nonspecific antiviral ribavirin, which is not very effective either in children or in adults (Nokes et al., 2008).
The C protein of HPIV3 is an accessory protein, encoded by the P gene using an alternative open reading frame. The P gene encodes the P protein (phosphoprotein), an important cofactor of the viral RNA polymerase L protein and also the D protein and possibly the V protein. The mRNAs of the latter two proteins are thought to be produced during viral transcription by inserting Gs into an editing site in the P gene (Durbin et al., 1999). These proteins possess the same N-terminal amino acid sequences as the P protein but distinct C-termini. Until now, only the D protein, but not the V protein, has been detected during HPIV3 infection (Durbin et al., 1999; Galinski et al., 1992; Wells et al., 2008).
The accessory C proteins of paramyxoviruses were reported to be involved in many of the viral functions, including replication (Kurotani et al., 1998; Escoffier et al., 1999; Sleeman et al., 2008; Dubin et al., 1999), assembly (Hasan et al., 2000), and budding, (Sakaguchi et al., 2005), as well as suppressing virus-induced apoptosis (Koyama et al., 2003; Toth et al., 2009) and antagonizing host innate immunity (Kato et al., 2001; Kato et al., 2007; Nakatsu et al, 2006; Nakatsu et al, 2008; Malur et al., 2005). Within the paramyxoviruses, only respiroviruses (e.g. HPIV3, Sedai virus/SeV) (Malur et al., 2004; Kato et al., 2007), morbilliviruses (e.g. measles virus/MV)(Nakatsu et al., 2008) and henipaviruses (e.g. Nipah/NiV)(Lo et al., 2009) encode C proteins. Interestingly, HPIV3, MV and NiV encode only one C protein, whereas SeV encodes a set of C proteins (C’, C, Y1 and Y2) through different translation initiation codons. Although there are very limited amino acid sequence similarities among these different sources of C proteins, inhibition of viral replication occurs heterotypically by the C proteins. For example, HPIV3 C protein inhibits replication of minigenomes of NiV and MV, whereas C proteins of SeV, but not C proteins of NiV and MV, inhibit HPIV3 minigenome replication (Malur et al., 2004, Sleeman et al., 2008). As for HPIV3, a coiled-coiled motif within the C protein was implicated to be involved in the viral transcription inhibition (Malur et al., 2004). Although the precise mechanism of C protein mediated replication inhibition is not known, the SeV C protein was reported to inhibit viral replication by binding to the L protein (Horikami et al., 1997; Grogan et al., 2001), and its N-terminal was dispensable for such inhibition (Kato et al., 2002; Kato et al., 2004).
In this study, we identified the domain in HPIV3 C protein responsible for the inhibition of viral transcription. We found that CNΔ25 and CNΔ50, the N-terminal 25 and 50 amino acids truncated mutants of HPIV3 C protein, totally abrogated viral transcription in the HPIV3 minigenome system and the inhibitory effect of CNΔ25 in HPIV3 growth was significant. Interestingly, CNΔ25 also strongly inhibited the replication of RSV. Our findings suggest that CNΔ25 has a potential to be used as an effective antiviral agent against the clinically important respiratory viruses including HPIV3 and RSV.
Results
The domain of HPIV3 C protein that inhibits viral RNA synthesis
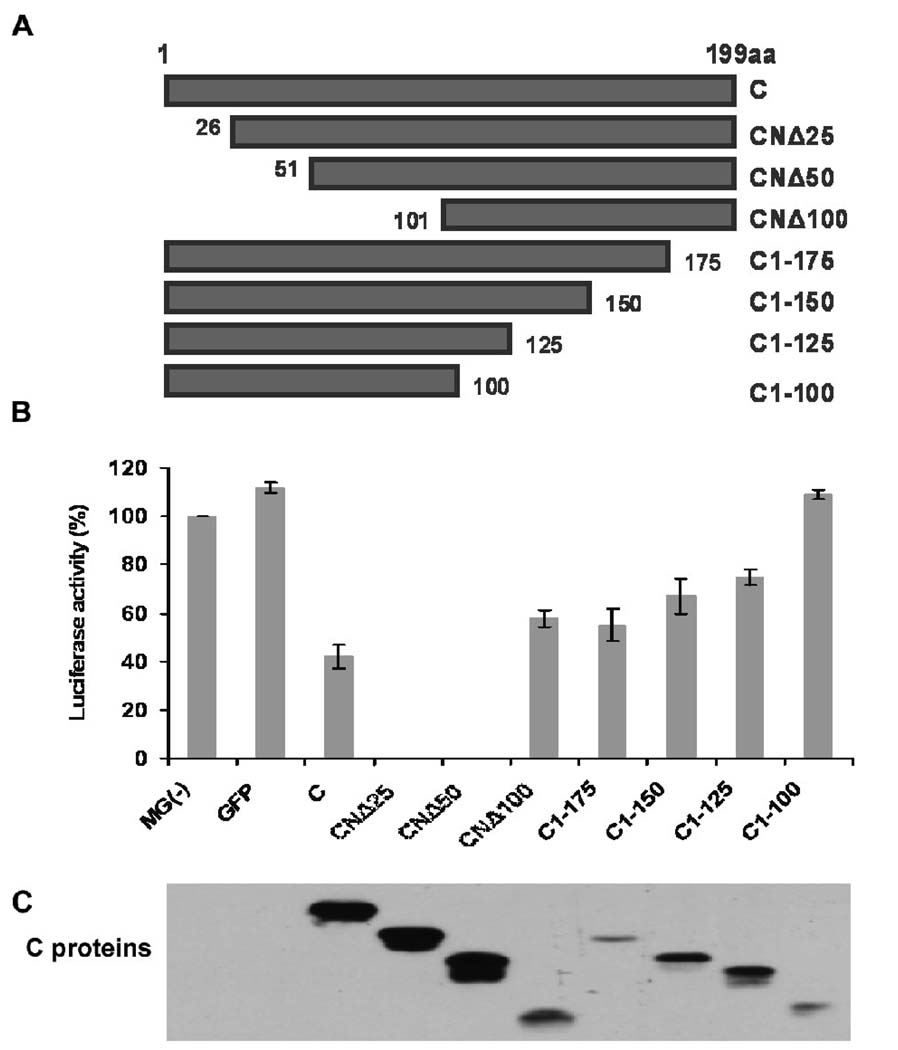
To identify the domain of the C protein involved in viral RNA synthesis inhibition, we constructed a series of plasmids encoding flag-tagged (C-terminal) C proteins, truncated from both N- and C-termini (Fig. 1A). We then tested these C mutants, together with the wild-type (wt) C protein, in the HPIV3 minigenome system as described in Materials and Methods. As previously observed (Malur et al., 2004), the C protein inhibited the luciferase activity by about 60% (Fig. 1B). Surprisingly, N-terminal deletion of 25 amino acids (CNΔ25) or 50 amino acids (CNΔ50) virtually abolished the luciferase activity, whereas further deletion (CNΔ100) reduced the inhibition back to 42%, less than the inhibition caused by the C protein (60%). In contrast, the deletion from the C-terminal end of the C protein resulted in the loss of inhibitory ability from 45% for C1-175 with virtually no inhibition for C1-100. Expression of the C protein and its mutants was confirmed by Western blot analysis using the anti-flag antibody (Fig. 1C). The expression levels of the C and CNΔ25 proteins were comparable, although the CNΔ50 protein expressed about one half-fold higher (Fig. 1C), suggesting the different levels of inhibition of viral RNA synthesis caused by C and CNΔ25 were not due to the amount of proteins expressed. The other C mutants were expressed in much lower levels than the C protein, but increasing their expression levels did not significantly increase their inhibitory activities (data not shown). These results suggest that the C-terminal region of the C protein downstream of residue 25 (CNΔ25) functions in inhibiting viral RNA synthesis, with N-terminal 25 amino acids performing some regulatory role in the inhibition process.
Fig. 1.
The domain of the C protein of HPIV3 that inhibits viral RNA synthesis. (A) Diagram of the N- and C-terminally truncated mutants of the C protein. (B) HPIV3 minigenome assay. The HeLa cells, preinfected with vaccinia virus vTF7-3 to express the T7 RNA polymerase, were transfected with the minigenome plasmid pMG(−), supporting plasmids pGEM4-L/pGEM4-P/pGEM4-N, as well as the C protein/mutants encoding plasmids. At 24 h post-transfection, the cells were lysed and the activity of luciferase was determined using a luminometer. The luciferase activity (%) is a percentage of the positive control MG(−) after subtracting the background level MG(−)/-L (L protein is absent in the system). A plasmid expressing GFP was used as control for the minigenome system. (C) The cell lysates were detected for the expression of the flag-tagged C proteins using anti-flag antibody.
Deletion mapping of CNΔ25
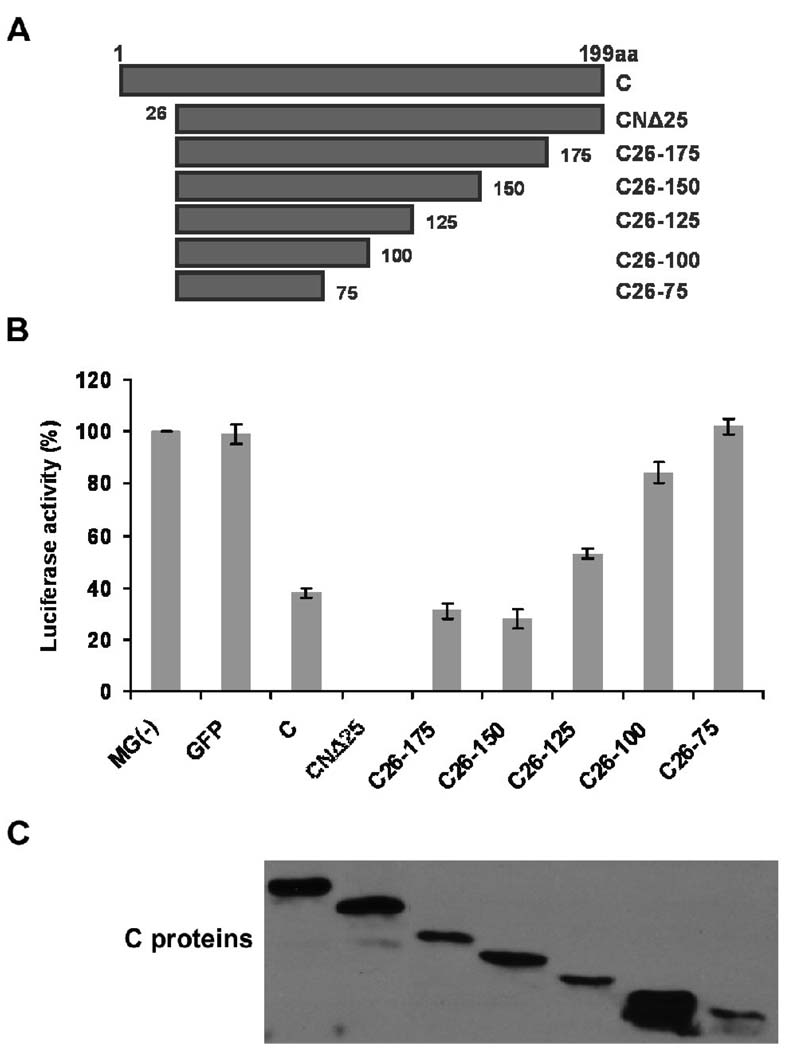
Since the CNΔ25 protein was able to reproducibly inhibit the minigenome RNA synthesis, we attempted to identify the minimum fragment of CNΔ25, without significantly losing its inhibitory ability. We truncated CNΔ25 protein from its C-terminus, and generated the following mutants C26-175, C26-150, C26-125, C26-100 and C26-75 (Fig. 2A). These mutants were tested in HPIV3 minigenome system, along with the wt C and CNΔ25 proteins. As shown in Fig. 2B, with increasing deletion from the C terminus of CNΔ25, the luciferase activity increased and finally no inhibition occurred when the C26-75 mutant was present in the minigenome system. Expression levels of these C proteins were also analyzed by Western blot using the anti-flag antibody (Fig. 2C). By comparing CNΔ25 and C26-150 proteins, which were expressed in similar levels, it was evident that the C-terminal 50 amino acids deletion from CNΔ25 caused the decrease of the inhibitory activity of CNΔ25. When we compared the effects of C26-175, C26-125 and C26-75 proteins, which were also comparably expressed, we found that with the removal of the C-terminal portion, the inhibitory activity of CNΔ25 was progressively decreased. It is noteworthy that, although the expression level of the C26-100 protein was much higher than other C proteins, it only resulted in slight inhibition (about 16%) of luciferase activity. We further investigated the relationship between the amounts of protein and the extent of RNA synthesis inhibition, which showed different does-dependent curves (data not shown). Consequently, we compared the inhibitory effects of proteins with similar expression levels, instead of using percent inhibition per unit amount of proteins for any single experiment. The data were consistent with what was observed for the C terminal truncated mutants of the C protein (Fig. 1), suggesting that the C-terminal region of the C protein is required for the maximum inhibition of viral RNA synthesis and CNΔ25 manifests maximum inhibition of viral RNA synthesis.
Fig. 2.
Deletion mapping of CNΔ25. (A) Diagram of the C-terminal truncated mutants of the CNΔ25 protein. (B) HPIV3 minigenome assay. The HeLa cells, preinfected with vaccinia virus vTF7-3, were transfected with the minigenome plasmid pMG(−), supporting plasmids pGEM4-L/pGEM4-P/pGEM4-N, as well as C, CNΔ25 or CNΔ25 mutants encoding plasmids. At 24 h post-transfection, the cells were lysed and the lysates were used to detect luciferase activity (C) Cell lysates were used for the detection of the flag-tagged C proteins expression using anti-flag antibody.
Interaction of the C protein with the L protein
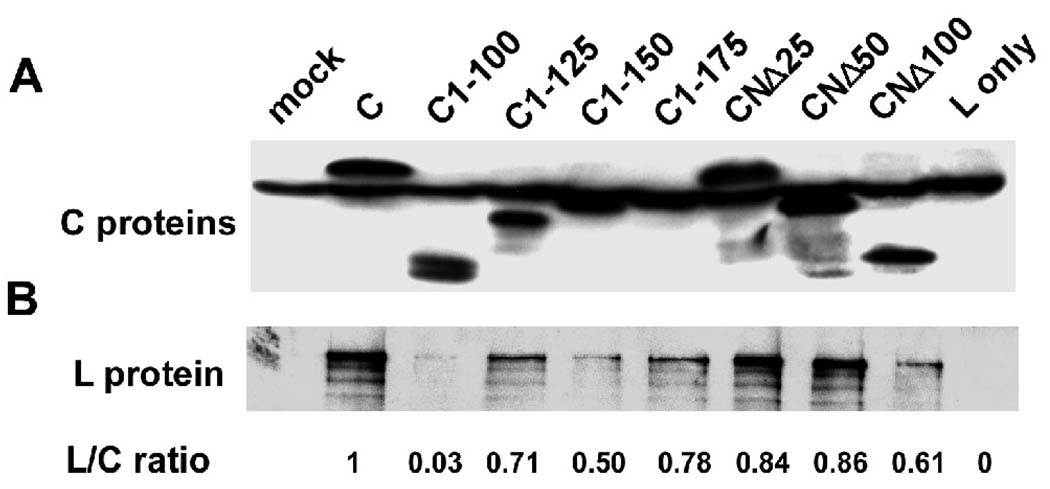
Since SeV C protein was suggested to inhibit viral transcription by binding to the viral RNA polymerase (Horikami et al., 1997; Grogan et al., 2001), we also studied the interaction between HPIV3 C proteins and viral RNA polymerase L protein using the co-immunoprecipitation assay. The C-terminally flag-tagged C encoding plasmids were co-transfected with non-tagged L plasmid into HeLa cells as described in Materials and Methods. Cell lysates harvested at 24 h post-transfection were incubated with the anti-flag affinity gel overnight. We used anti-flag and the anti-L antibodies separately to detect flag-tagged C protein (full length and mutants) and non-tagged L protein, respectively. As shown in Fig. 3A, flag-C proteins were pulled down by the anti-flag affinity gel effectively. Simultaneously, different amounts of L protein precipitated with the C proteins (Fig. 3B). To accurately judge the relative efficiency of C-L interaction from blots shown in Fig. 3, we quantified the intensity of each band using ImageQuant 5.2 software and measured the L/C ratio as indicated below the figure. In this experiment, the L/C ratio represents the interaction affinity between L and C proteins. The L/C ratio for CNΔ25 and CNΔ50, the two C mutants abrogating the minigenome transcription, are 0.84 and 0.86, respectively. The C1-100 mutant, which showed no inhibitory effect on minigenome replication (Fig. 3), only has very weak interaction with L protein (with L/C ratio of 0.03). Other C mutants with inhibitory ability between C and C1-100 mutant interact with L protein with different affinities (L/C ratio: 0.50-0.78). Thus, it seems that the interaction affinity between L and C correlated, albeit partially, with the inhibitory activity of the C protein mutants.
Fig. 3.
The interaction between the C protein/mutants and the L protein detected by co-immunoprecipitation assay. The HeLa cells, preinfected with the vaccinia virus vTF7-3, were transfected with C protein/mutants encoding plasmids and the L protein encoding plasmid. At 24 h post-transfection, the cells were lysed and incubated with the anti-flag M2 affinity gel overnight. The affinity gel was washed and subsequently boiled with SDS-PAGE loading buffer and was subjected to Western blot analyses using anti-flag (A) and anti-L (B) antibodies, respectively. Mock: both L protein and C protein are absent; L only: C protein is absent. The intensity of each band was quantified using ImageQuant 5.2 software and the L/C ratio was measured as indicated below the figure.
Role of charged amino acid residues in the N-terminal 25 amino acids of the C protein in the inhibition of viral RNA synthesis
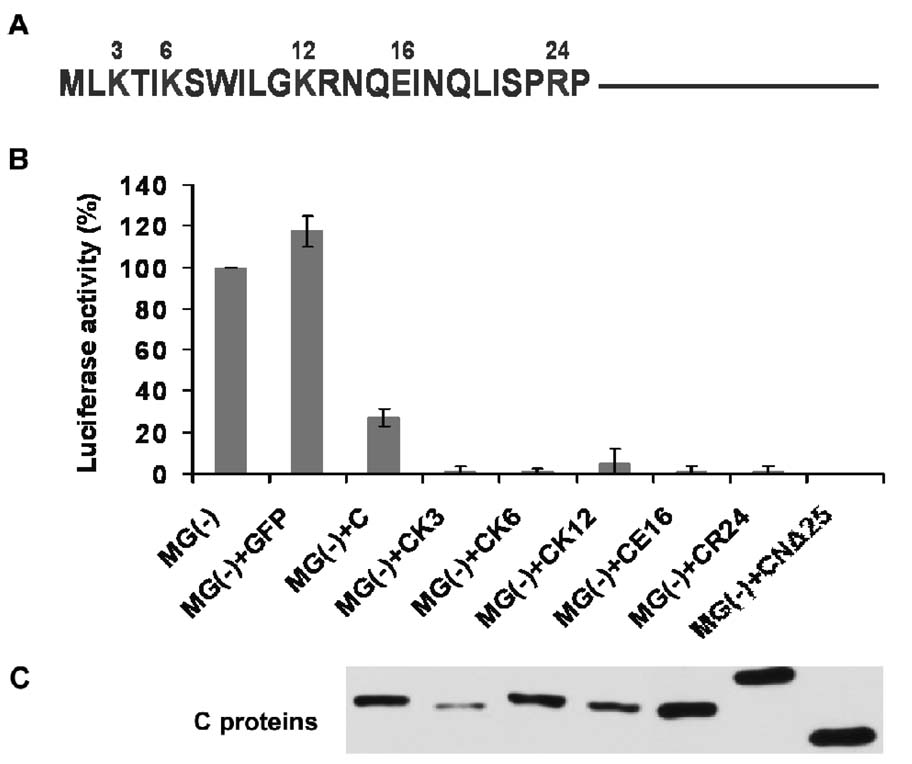
Since we did not observe differential L protein binding affinity between C and CNΔ25 proteins, by the Co-IP assay (Fig. 3A), the question remains as to how the 25 amino acid residues in the N-terminus of HPIV3 C protein (Fig. 4A) affect the inhibitory activity of the C protein on viral RNA synthesis. We initially found that charged amino acid residues in the C terminus of the CNΔ25 protein decreased the inhibition caused by the CNΔ25 protein (data not shown), suggesting that the net charge of the protein somehow changes its conformational structure or its interaction with other viral components. Accordingly, we substituted the charged amino acid residues within the N-terminal 25 amino acids of the C protein into alanine individually, as indicated in the Fig. 4A. The R13 residue was not included due to lack of expression of the corresponding CR13 protein. Note that CR24- a mutant of the C protein in which the arginine at position 24 was substituted with alanine, migrated slower during SDS-PAGE probably due to decreased pI value of CR24 due to substitution of R to A (Fig. 4C). We, then, tested these C protein mutants for transcription inhibition in the HPIV3 minigenome system. We found that all of the C mutants (K3, K6, K12. E16 and R24) increased the inhibitory activity of the C protein to almost 100%, similar to CNΔ25 (Fig. 4B), despite their various expression levels (Fig. 4C), suggesting these N-terminal charged amino acid residues play a critical role in the inhibition of viral RNA synthesis.
Fig. 4.
The role of charged amino acid residues in the N-terminal 25aa of the C protein in inhibiting the viral RNA synthesis. (A) The N-terminal 25 amino acids of the C protein, with the charged amino acid residues indicated. (B) The HPIV3 minigenome assay. The HeLa cells, preinfected with the vaccinia virus vTF 7-3 to express the T7 RNA polymerase, were transfected with the minigenome plasmid pMG(−), the supporting plasmids pGEM4-L/pGEM4-P/pGEM4-N, and the C/CNΔ25/site-directed C mutants expressing plasmids. At 24 h pi, the cells were lysed for the luciferase assay. (C) The expression of C/CNΔ25/site-directed C mutants was detected by Western blot using the anti-flag antibody.
Effects of the C proteins on HPIV3 growth
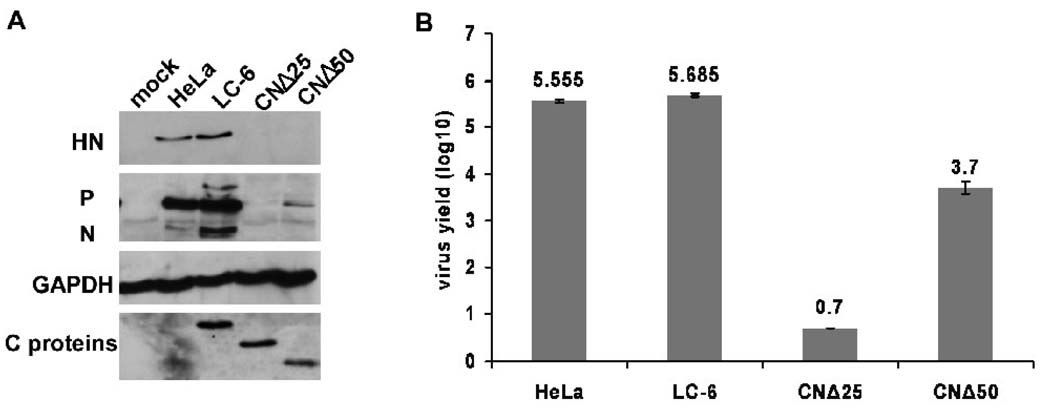
Since CNΔ25 and CNΔ50 virtually abolished viral RNA synthesis, it was of interest to study their effects on viral growth. We established HeLa-derived stable cell lines expressing CNΔ25 or CNΔ50 using lentivirus vector, and infected the cell lines including C expressing LC-6 cells previously described (Malur et al., 2005) with HPIV3 at an MOI of 0.1. At 24 h post-infection, cells were lysed for immunoblotting with anti-HN, anti-RNP or anti-C antibodies, and supernatants were collected and titered by plaque assay. As shown in Fig. 5A, the expression of C, CNΔ25 and CNΔ50 proteins was confirmed in the stable cell lines. Viral glycoprotein HN, as well as P and N proteins, were slightly higher in LC-6 cells than in parent HeLa cells. In contrast, all three viral proteins were drastically decreased in CNΔ25 and CNΔ50 -expressing cells. In fact, in CNΔ25 cells, viral proteins were virtually undetectable by Western blot analysis. Virus titers in the supernatants showed consistent results (Fig. 5B). In the presence of CNΔ25 and CNΔ50 proteins virus titers decreased by 5 logs and 2 logs, respectively, whereas the C protein had no effect on HPIV3 growth. It is noteworthy that the amount of viral protein expression in infected CNΔ50-expressing cells was about 1% of that in infected wt C-expressing cells, which was consistent with the amount of viruses produced in CNΔ50-expressing cells (103.7) compared to that in wt C-expressing cells (105.685, 2 log difference). Thus, it seems that CNΔ25 exerts considerably more inhibitory effect on HPIV3 infection than CNΔ50.
Fig. 5.
The effects of the C, CNΔ25 and CNΔ50 proteins on HPIV3 growth. (A) The HeLa, LC-6, CNΔ25 and CNΔ50 cells were infected with mock or HPIV3 (MOI of 0.1) for 24 h. The cells were lysed for Western blot analysis using the anti-HN and anti-RNP antibodies to detect the viral proteins HN, P and N. GAPDH was used as a loading control. The expression of C/CNΔ25/CNΔ50 proteins was confirmed in the LC-6/CNΔ25CNΔ50 cells using the anti-flag antibody. (B) The supernatants from samples in A were titered by plaque assay. The plaque assay for each sample was performed 3 times. The average and the standard deviation (SD) of the viral yields are indicated.
Effects of the C proteins on the expression of rg-RSV proteins
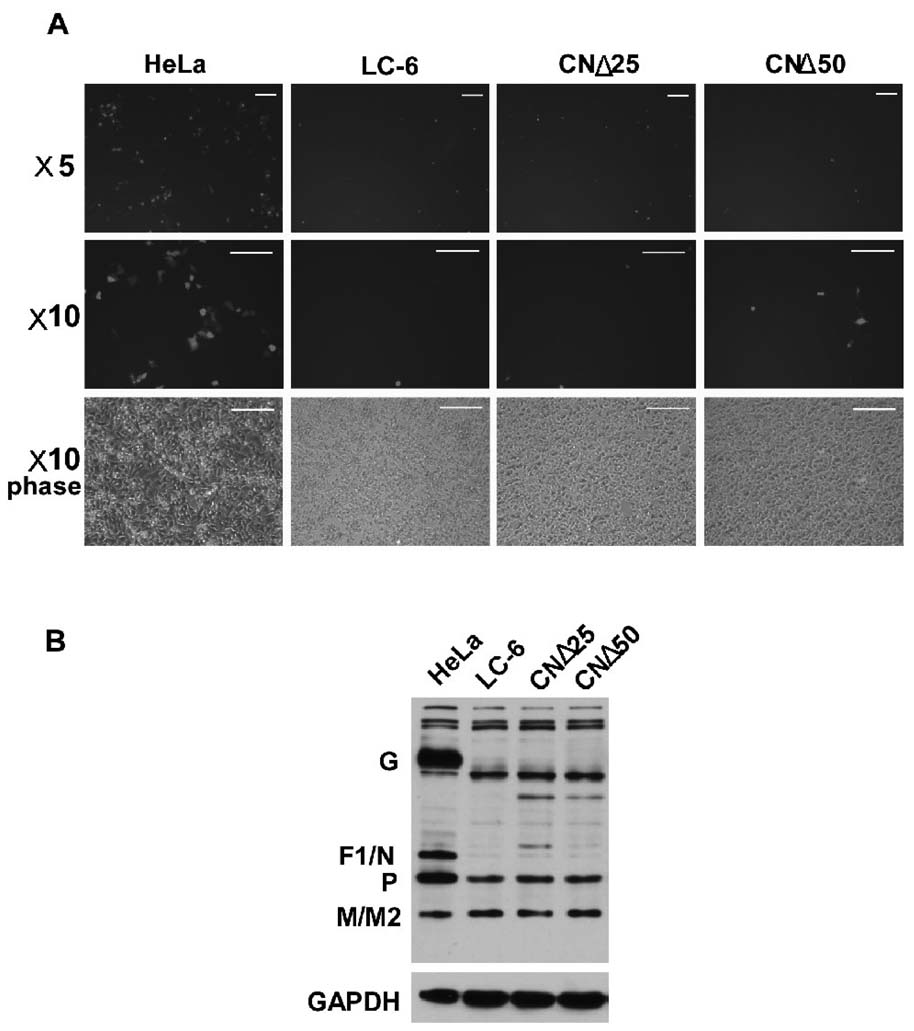
As mentioned above, HPIV3 C protein heterotypically inhibited the transcription of MV and NiV within the paramyxoviruse family. We, thus, wanted to investigate whether CNΔ25 had any effect on the multiplication of RSV, the most common respiratory pathogen belonging to the paramxyovirus family. We infected three cell lines with the GFP tagged recombinant rg-RSV (Hallak et al., 2000) and at 24 h post-infection, we observed GFP expression in cells under the fluorescence microscope. We also harvested cell lysates for immunoblotting with anti-RSV antibody. As shown in Fig. 6A, GFP expression was significantly and equally inhibited in LC-6, CNΔ25 and CNΔ50 cells as compared to that in infected HeLa cells. Similarly, RSV viral proteins G and F1/N were undetectable, and the P protein was also significantly decreased in three infected cell lines (Fig. 6B). Interestingly, viral M/M2 proteins were comparable in all cells. The significant inhibition of viral protein synthesis strongly suggests that the CNΔ25 protein, as well as C and CNΔ50 proteins, also possesses potent antiviral activity against RSV infection.
Fig 6.
The effects of the C, CNΔ25 and CNΔ50 proteins on the RSV infection. (A) HeLa, LC-6, CNΔ25 and CNΔ50 cells were infected with mock or gfp-RSV (MOI of 0.1) for 24 h. The expression of GFP protein in infected cells was observed under the fluorescence microscope. Three sets of pictures are shown, ×5 fluorescence, ×10 fluorescence, and ×10 phase. The white bars represent 100µm. (B) Cell lysates in A were detected by Western blot using the anti-RSV virion antibody for the expression of the viral proteins including the G, F1/N, P, and M/M2. GAPDH was used as a loading control.
Discussion
The paramyxoviral C protein is a multifunctional protein involved in regulating various viral and cellular functions (Kurotani et al., 1998; Escoffier et al., 1999; Sleeman et al., 2008; Dubin et al., 1999; Hasan et al., 2000; Sakaguchi et al., 2005; Koyama et al., 2003; Toth et al., 2009; Kato et al., 2001; Kato et al., 2007; Nakatsu et al, 2006; Nakatsu et al, 2008; Malur et al., 2005). The major studies on the C proteins have been directed towards exploring their inhibitory roles in viral transcription and counteracting IFN signaling pathways (Grogan et al., 2001; Horikami et al., 1997; Kato et al., 2002; Kato et al., 2004; Kato et al., 2001; Kato et al., 2007; Malur et al., 2004; Malur et al., 2005; Nakatsu et al, 2006; Nakatsu et al, 2008; Sleeman et al., 2008). Using a reporter gene carrying minigenome transcription assay (Hoffman et al., 2000) we demonstrated that HPIV3 C protein inhibits viral transcription by 60% and identified a coil-coil motif within the protein, the removal of which resulted in significant abrogation of the inhibitory function (Malur et al., 2004). In the present study, we demonstrated that removal of N-terminal 25 or 50 amino acids of the C proteins (CNΔ25 & CNΔ50) completely inhibited viral transcription in HPIV3 minigenome system (Fig. 1), whereas further N-terminal deletion (CNΔ100), impaired the inhibitory activity of the C protein, suggesting that C50-100 fragment is critical for the inhibitory activity of C protein. These findings are consistent with our previous observation that a coil-coil structure of the C protein, C60-80, played a critical role in the inhibition of viral transcription (Malur et al., 2004). However, the mutants C26-75 and C26-100 (overlapping or containing C60-80) exerted less effect on minigenome transcription (Fig. 2). Moreover, progressive deletion from the C-terminal end of CNΔ25 resulted in gradual loss of the inhibitory effect (Fig. 1 & 2), suggesting that C-terminal region also plays a role in the inhibition process. Altering the charged amino acids within the N-terminal 25 amino acids of the full-length C protein to alanine rendered C protein to strongly inhibit transcription similar to CNΔ25 (Fig. 4). Thus, it seems that the overall structure of the C protein, rather than the single peptide, correlates with the RNA synthesis inhibition phenotype.
To further explore the mechanism by which CNΔ25 inhibits RNA synthesis we studied its interaction with the L protein. Previously, it has been shown for SeV that the inhibition of viral RNA synthesis by the C protein directly correlated with its interaction with the L protein (Horikami et al., 1997; Grogan et al., 2001). Our studies similarly demonstrated that HPIV3 C as well as CNΔ25 and CNΔ50 bound to the L protein to a similar extent (Fig. 3), whereas the C-terminally deleted C, that are less inhibitory to RNA synthesis, interacted to the L protein in significantly lesser degree. The cogent question remains as to why CNΔ25 and CNΔ50, which bind similarly to the L protein as wt C, exert such a different RNA inhibitory phenotype in the minigenome assay. It can be envisaged that these two proteins manifest different functional attributes during RNA synthesis inhibition by their altered structural conformation or interacting differently with the essential cofactor, the P protein, although the later protein also interact with the wt C and CNΔ25 to the same extent (data not shown). Thus, the precise mechanism by which CNΔ25 and CNΔ50 inhibit transcription so dramatically needs further studies.
The interesting and potential clinically important findings are that particularly CNΔ25 appears to function as a potent antiviral protein. We established stable cell lines expressing CNΔ25 and CNΔ50 using lentiviral system similar to that previously established for the wt C protein, LC6 cells (Malur et al., 2005). The growth of HPIV3 was severely restricted in CNΔ25 expressing cells both in viral protein synthesis and in virus yields (Fig. 5). The inhibition of viruses was by 5 logs in CNΔ25 cells, whereas the viral growth was inhibited by 2 logs in CNΔ50 and no inhibition was observed in LC6 cells. These results reinforce the contention that the mode of CNΔ25 mediated inhibition of viral RNA synthesis must be different from the observed inhibition by the wt C protein and possibly CNΔ50. Interestingly, CNΔ25 expressing cells also inhibited replication of GFP expressing RSV (Fig. 6). The viral protein synthesis, specifically G and F1/N was greatly inhibited. However, we did not observe any plaque formed in plaque assay, even by the supernatant harvested from infected HeLa cells. The reason for this might be due to rg-RSV being severely attenuated than wt RSV (data not shown). The precise reason for the inhibitory property of CNΔ25 on RSV replication remains unclear, however, heterotypic inhibition of RNA synthesis by paramyxoviral C proteins have been well documented (Sleeman et al., 2008).
In summary, removal of the N-terminal 25aa of the C protein (CNΔ25) significantly potentiated the inhibitory activity of the C protein on viral transcription. The charged amino acid residues K3, K6, K12, E16, and R24 within the N-terminal 25aa appear to be involved in the regulation of transcription inhibition. Remarkably, CNΔ25 acts as a potent inhibitor against the infection of both of HPIV3 and RSV, raising a possibility that the CNΔ25 can be used as an antiviral agent in future studies.
Materials and Methods
Cells and viruses
HeLa (S) cells were cultured in DMEM (10 % fetal bovine serum, penicillin/streptomycin). HeLa-derived stable cell lines expressing C protein (LC-6) (Malur et al., 2005), CNΔ25 and CNΔ50 cells were established using recombinant lentiviruses, and single cell clones were obtained through blasticidin selection followed by purification through limited dilution. HPIV3 (HA-1; NIH 47885) and vesicular stomatitis virus/VSV (Indiana) were grown in HeLa cells. The recombinant GFP-tagged RSV (rg-RSV) was generously supplied by Dr. Brian R. Murphy in the National Institute of Allergy and Infectious Diseases (NIAI.D).
HPIV3 minigenome assay
HPIV3 minigenome assay was performed as described previously (Mao et al., 2008; Hoffman et al., 2000) with slight modifications. HeLa cell monolayers in 6 well plate, grown to 70–80 % confluence, were infected with recombinant vaccinia virus vTF7-3, which expresses T7 RNA polymerase, at an MOI of 2. At 1 h post-infection, cells were washed with phosphate buffered saline (PBS) and transfected with HPIV3 minigenome plasmid pMG (−) (200 ng) encoding luciferase reporter gene, supporting plasmids encoding N (640 ng), P (700 ng) and L (100 ng) proteins, as well as the C/mutants expressing plasmids (500 ng) using Lipofectin (Invitrogen). MG(−)/-L was used as the negative control in which the L expressing plasmid was absent. At 24 h post-transfection, cells were lysed in 100 µl of lysis buffer, from which 20 µl aliquots were used to determine luciferase activity in a luminometer (VICTOR) according to the manufacturer’s instructions (Luciferase Assay Kit; Roche).
Western Blotting
Western Blotting was performed according to the protocol reported previously (Mao et al., 2008) with slight modifications. In brief, cell extracts were subject to 10 % sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membrane. The membrane was blocked with 5 % nonfat milk in PBST (PBS with 0.1 % Tween 20) for 1h and incubated with primary antibody in 5 % nonfat milk at 4°C for overnight. The anti-flag antibody was purchased from Sigma. The anti-L antibody was prepared from a rabbit by the Hybridoma Core in the Lerner Research Institute (Chattopadhyay et al., 2008). The substrate reaction was performed using ECL™ Western blotting detection kit (GE Healthcare).
Co-immunoprecipitation assay
The Co-immunoprecipitation assay was performed as described previously (Chattopadhyay et al., 2009). In brief, HeLa cells were infected with the recombinant vaccinia virus, vTF7-3 and plasmids encoding L protein (2.5 µg) and C-terminally flag tagged C protein (0.5 µg) or C mutant proteins (0.5 µg) were co-transfected using Lipofectin (Invitrogen). At 24 h post-transfection, the cells were lysed in the lysis buffer (Roche). The cell lysates were incubated with the anti-flag M2 affinity gel (Sigma A2220) at 4°C overnight. The affinity gel was washed with Tris Buffered Saline (TBS) for five times and subsequently boiled in the presence of 5×loading buffer. The immunoprecipitated proteins were resolved in SDS-PAGE gel and transferred to a nitrocellulose membrane by Western blotting. The nitrocellulose membrane containing transferred proteins was incubated with either anti-L polyclonal antibody or anti-flag M2 monoclonal antibody (Sigma F3165), followed by incubation with appropriate secondary antibodies for the detection of L or C proteins by chemiluminescence (GE healthcare).
Mutagenesis
Mutagenesis was performed using Quick-Change® II site-directed kit (Stratagene, #200523). In brief, primer pairs were used to amplify site-mutated C genes by Ultra polymerase with the C protein encoding plasmids as the template. The products were treated with DpnI (1u) at 37°C for 1 h to digest the methylated template. One µl of sample was used to transform XL-1 Blue competent cells. Colonies were identified by restriction enzyme digestion and confirmed by sequencing.
Plaque assay for HPIV3
The virus titer was determined by plaque assay as described previously (Mao et al., 2008). Briefly, the supernatant of the infected cells was serially diluted in 1 ml of OPTI-MEM. Confluent monolayer of CV1 cells in 6 well plates were washed with PBS and incubated with the diluted suspension at 37°C, 5 % CO2 for 1.5 h. The media was then removed and cells were washed with PBS and overlaid with 0.8 % methylcellulose. After 48 h, the methylcellulose was aspirated and cells were stained with 1 % crystal violet (in 50 % methanol).
Acknowledgements
We thank Dr. Ratan K. Maitra for helping us establish the C proteins expressing stable cell lines. We appreciate Dr. Brian R. Murphy and Dr. Peter L. Collins for kindly providing us the rg-RSV and anti-RSV virion antibody. This work was supported by the NIH grant to AKB (AI 32037).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Chattopadhyay S, Banerjee AK. Phosphoprotein, P of human parainfluenza virus type 3 prevents self-association of RNA-dependent RNA polymerase, L. Virology. 2009;383(2):226–236. doi: 10.1016/j.virol.2008.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez KJ, Erdman DD, Peret TC, Gill VJ, Childs R, Barrett AJ, Bennett JE. Outbreak of human parainfluenza virus 3 infections in a hematopoietic stem cell transplant population. J Infect Dis. 2001;184(9):1093–1097. doi: 10.1086/322041. [DOI] [PubMed] [Google Scholar]
- Durbin AP, McAuliffe JM, Collins PL, Murphy BR. Mutations in the C, D and V open reading frames of human parainfluenza virus type 3 attenuate replication in rodents and primates. Virology. 1999;261(2):319–330. doi: 10.1006/viro.1999.9878. [DOI] [PubMed] [Google Scholar]
- Escoffier C, Manié S, Vincent S, Muller CP, Billeter M, Gerlier D. Nonstructural C protein is required for efficient measles virus replication in human peripheral blood cells. J Virol. 1999;73(2):1695–1698. doi: 10.1128/jvi.73.2.1695-1698.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galinski MS, Troy RM, Banerjee AK. RNA editing in the phosphoprotein gene of the human parainfluenza virus type 3. Virology. 1992;186(2):543–550. doi: 10.1016/0042-6822(92)90020-P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grogan CC, Moyer SA. Sendai virus wild-type and mutant C proteins show a direct correlation between L polymerase binding and inhibition of viral RNA synthesis. Virology. 2001;288(1):96–108. doi: 10.1006/viro.2001.1068. [DOI] [PubMed] [Google Scholar]
- Hasan MK, Kato A, Muranaka M, Yamaguchi R, Sakai Y, Hatano I, Tashiro M, Nagai Y. Versatility of the accessory C proteins of Sendai virus: contribution to virus assembly as an additional role. J Virol. 2000;74(12):5619–5628. doi: 10.1128/jvi.74.12.5619-5628.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman MA, Banerjee AK. Precise mapping of the replication and transcription promoters of human parainfluenza virus type 3. Virology. 2000;269(1):201–211. doi: 10.1006/viro.2000.0223. [DOI] [PubMed] [Google Scholar]
- Horikami SM, Hector RE, Smallwood S, Moyer SA. The Sendai virus C protein binds the L polymerase protein to inhibit viral RNA synthesis. Virology. 1997;235(2):261–270. doi: 10.1006/viro.1997.8702. [DOI] [PubMed] [Google Scholar]
- Kato A, Ohnishi Y, Kohase M, Saito S, Tashiro M, Nagai Y. Y2, the smallest of the Sendai virus C proteins, is fully capable of both counteracting the antiviral action of interferons and inhibiting viral RNA synthesis. J Virol. 2001;75(8):3802–3810. doi: 10.1128/JVI.75.8.3802-3810.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato A, Ohnishi Y, Hishiyama M, Kohase M, Saito S, Tashiro M, Nagai Y. The amino-terminal half of Sendai virus C protein is not responsible for either counteracting the antiviral action of interferons or down-regulating viral RNA synthesis. J Virol. 2002;76(14):7114–7124. doi: 10.1128/JVI.76.14.7114-7124.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato A, Cortese-Grogan C, Moyer SA, Sugahara F, Sakaguchi T, Kubota T, Otsuki N, Kohase M, Tashiro M, Nagai Y. Characterization of the amino acid residues of sendai virus C protein that are critically involved in its interferon antagonism and RNA synthesis down-regulation. J Virol. 2004;78(14):7443–7454. doi: 10.1128/JVI.78.14.7443-7454.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato A, Kiyotani K, Kubota T, Yoshida T, Tashiro M, Nagai Y. Importance of the anti-interferon capacity of Sendai virus C protein for pathogenicity in mice. J Virol. 2007;81(7):3264–3271. doi: 10.1128/JVI.02590-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama AH, Irie H, Kato A, Nagai Y, Adachi A. Virus multiplication and induction of apoptosis by Sendai virus: role of the C proteins. Microbes Infect. 2003;5(5):373–378. doi: 10.1016/s1286-4579(03)00043-1. [DOI] [PubMed] [Google Scholar]
- Kurotani A, Kiyotani K, Kato A, Shioda T, Sakai Y, Mizumoto K, Yoshida T, Nagai Y. Sendai virus C proteins are categorically nonessential gene products but silencing their expression severely impairs viral replication and pathogenesis. Genes Cells. 1998;3(2):111–124. doi: 10.1046/j.1365-2443.1998.00170.x. [DOI] [PubMed] [Google Scholar]
- Liuzzi M, Mason SW, Cartier M, Lawetz C, McCollum RS, Dansereau N, Bolger G, Lapeyre N, Gaudette Y, Lagacé L, Massariol MJ, Dô F, Whitehead P, Lamarre L, Scouten E, Bordeleau J, Landry S, Rancourt J, Fazal G, Simoneau B. Inhibitors of respiratory syncytial virus replication target cotranscriptional mRNA guanylylation by viral RNA-dependent RNA polymerase. J Virol. 2005;79(20):13105–13115. doi: 10.1128/JVI.79.20.13105-13115.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo MK, Harcourt BH, Mungall BA, Tamin A, Peeples ME, Bellini WJ, Rota PA. Determination of the henipavirus phophoprotein gene mRNA editing frequencies and detection of the C, V and W proteins of Nipah virus in virus-infected cells. J Gen Virol. 2009;90(Pt 2):398–404. doi: 10.1099/vir.0.007294-0. [DOI] [PubMed] [Google Scholar]
- Madhi SA, Ramasamy N, Petersen K, Madhi A, Klugman KP. Severe lower respiratory tract infections associated with human parainfluenza viruses 1–3 in children infected and noninfected with HIV type 1. Eur J Clin Microbiol Infect Dis. 2002;21(7):499–505. doi: 10.1007/s10096-002-0754-9. [DOI] [PubMed] [Google Scholar]
- Malur AG, Hoffman MA, Banerjee AK. The human parainfluenza virus type 3 (HPIV3) C protein inhibits viral transcription. Virus Res. 2004;99(2):199–204. doi: 10.1016/j.virusres.2003.11.009. [DOI] [PubMed] [Google Scholar]
- Malur AG, Chattopadhyay S, Maitra RK, Banerjee AK. Inhibition of STAT 1 phosphorylation by human parainfluenza virus type 3 C protein. J Virol. 2005;79(12):7877–7882. doi: 10.1128/JVI.79.12.7877-7882.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao H, Thakur CS, Chattopadhyay S, Silverman RH, Gudkov A, Banerjee AK. Inhibition of human parainfluenza virus type 3 infection by novel small molecules. Antiviral Res. 2008;77(2):83–94. doi: 10.1016/j.antiviral.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsu Y, Takeda M, Ohno S, Koga R, Yanagi Y. Translational inhibition and increased interferon induction in cells infected with C protein-deficient measles virus. J Virol. 2006;80(23):11861–11867. doi: 10.1128/JVI.00751-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsu Y, Takeda M, Ohno S, Shirogane Y, Iwasaki M, Yanagi Y. Measles virus circumvents the host interferon response by different actions of the C and V proteins. J Virol. 2008;82(17):8296–8306. doi: 10.1128/JVI.00108-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols WG, Peck Campbell AJ, Boeckh M. Respiratory viruses other than influenza virus: impact and therapeutic advances. Clin Microbiol Rev. 2008;21(2):274–290. doi: 10.1128/CMR.00045-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nokes JD, Cane PA. New strategies for control of respiratory syncytial virus infection. Curr Opin Infect Dis. 2008;21(6):639–643. doi: 10.1097/QCO.0b013e3283184245. [DOI] [PubMed] [Google Scholar]
- Sakaguchi T, Kato A, Sugahara F, Shimazu Y, Inoue M, Kiyotani K, Nagai Y, Yoshida T. AIP1/Alix is a binding partner of Sendai virus C protein and facilitates virus budding. J Virol. 2005;79(14):8933–8941. doi: 10.1128/JVI.79.14.8933-8941.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Wright PF. Current status of vaccines for parainfluenza virus infections. Pediatr Infect Dis J. 2008;27(10 Suppl):S123–S125. doi: 10.1097/INF.0b013e318168b76f. [DOI] [PubMed] [Google Scholar]
- Sleeman K, Bankamp B, Hummel KB, Lo MK, Bellini WJ, Rota PA. The C, V and W proteins of Nipah virus inhibit minigenome replication. J Gen Virol. 2008;89(Pt 5):1300–1308. doi: 10.1099/vir.0.83582-0. [DOI] [PubMed] [Google Scholar]
- Toth AM, Devaux P, Cattaneo R, Samuel CE. Protein kinase PKR mediates the apoptosis induction and growth restriction phenotypes of C protein-deficient measles virus. J Virol. 2009;83(2):961–968. doi: 10.1128/JVI.01669-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells G, Malur A. Expression of human parainfluenza virus type 3 PD protein and intracellular localization in virus infected cells. Virus Genes. 2008;37(3):358–367. doi: 10.1007/s11262-008-0269-2. [DOI] [PubMed] [Google Scholar]