Abstract
BNIP3 and NIX are proteins related to the BH3-only family, which induce both cell death and autophagy. Consistent with their ability to induce cell death, BNIP3 and NIX are implicated in the pathogenesis of cancer and heart disease. In tumor cells, BNIP3 and NIX are regulated by hypoxia, and the deregulation of BNIP3 or NIX expression is associated with tumor growth. In heart muscle, BNIP3 and NIX are regulated by hypoxia and Gαq-dependent signaling, respectively, and their expression is associated with decreased myocardial function. Apart from their role in cell death, BNIP3 and NIX are also implicated in the induction of autophagy. In erythroid cells, NIX is required for a specialized type of autophagy that targets mitochondria for elimination (mitophagy). Similarly, BNIP3 regulates mitophagy in response to hypoxia. In this review, we will discuss possible mechanisms by which BNIP3 and NIX induce cell death and mitophagy. We will also consider the potential relationship between cell death pathways and autophagy in development and homeostasis.
BNIP3 (BCL2 and adenovirus E1B 19 kDa-interacting protein 3) and BNIP3-like (BNIP3L), also known as NIX, are proteins with homology to BCL2 in the BH3 domain, which induce both cell death and autophagy. Although other proteins may induce cell death or autophagy more efficiently than BNIP3 or NIX, the ability of these proteins to do both makes them useful reagents to explore the relationship between these alternate cell fates. With this in mind, here we review the role of BNIP3 and NIX in cell death and autophagy, and discuss potential mechanisms by which they may function. Additionally, we discuss a specialized type of autophagy that is relevant during development, namely mitophagy, in the erythroid lineage.
Discovery and Characterization of BNIP3
BNIP1, BNIP2, and BNIP3, were initially identified in a yeast two-hybrid screen as BCL2 and adenovirus E1B 19 kDa-interacting proteins.1 E1B 19 kDa protein protects cells from death after adenovirus infection,2,3 and mutants of this protein that are defective for binding to BNIP proteins are also defective for cell death suppression.1 The antiapoptotic protein BCL2 can functionally replace E1B 19 kDa protein in this context,4,5 and also directly interacts with BNIP proteins.1 Further, as is the case for E1B 19 kDa protein, mutants of BCL2 that are defective for binding to BNIP proteins are also defective in cell death suppression. These findings suggest that BNIP1, BNIP2, and BNIP3 are proapoptotic proteins that are suppressed by E1B 19 kDa protein or BCL2. BNIP1 and BNIP2 localize to the nuclear envelope and endoplasmic reticulum, whereas BNIP3 localizes to mitochondria when overexpressed.1 Thus, BNIP3 is a proapoptotic protein that may function through a mitochondrial pathway.
BNIP3 has death-inducing activity in cell lines, although its maximum effect is delayed relative to other proapoptotic proteins.6–8 BNIP3 has a putative BH3 domain (Figure 1), and C-terminal transmembrane domain.7 The BNIP3 BH3 domain differs from the consensus BCL2 family sequence, (L/V/M)1XXXG5D6(D/E)7F8E9R10, at two evolutionarily conserved residues, W7 and W11 (ref. 9). This domain, substituted for the corresponding sequence of BAX, confers proapoptotic activity, and the ability to heterodimerize with BCL-XL.7 Further, in stably transfected baby rat kidney cells undergoing p53-dependent cell death, the BNIP3 BH3 domain is required to reverse the antiapoptotic effect of BCL-XL. On the other hand, mutation of the BNIP3 BH3 domain is associated with minimal loss of proapoptotic activity in transiently transfected MCF-7 breast carcinoma cells, and at least partial retention of the ability to bind BCL2 and BCL-XL. 7,8 By comparison, the C-terminal transmembrane domain of BNIP3 is essential for mitochondrial localization and proapoptotic activity.6–8 The BNIP3 transmembrane domain is required for interaction with BCL2, although either the transmembrane domain or the N-terminal 49 amino acids is sufficient for interaction with BCL-XL. Interestingly, a cytochrome b5 transmembrane domain substitution mutant of BNIP3, targeted to the ER, retains its ability to interact with BCL2 and BCL-XL, and its proapoptotic activity.8 Thus, the BNIP3 transmembrane domain plays an important role in the interaction of BNIP3 with BCL2 or BCL-XL, and in BNIP3-induced cell death, and the BNIP3 BH3 domain may mediate cell death in some conditions.
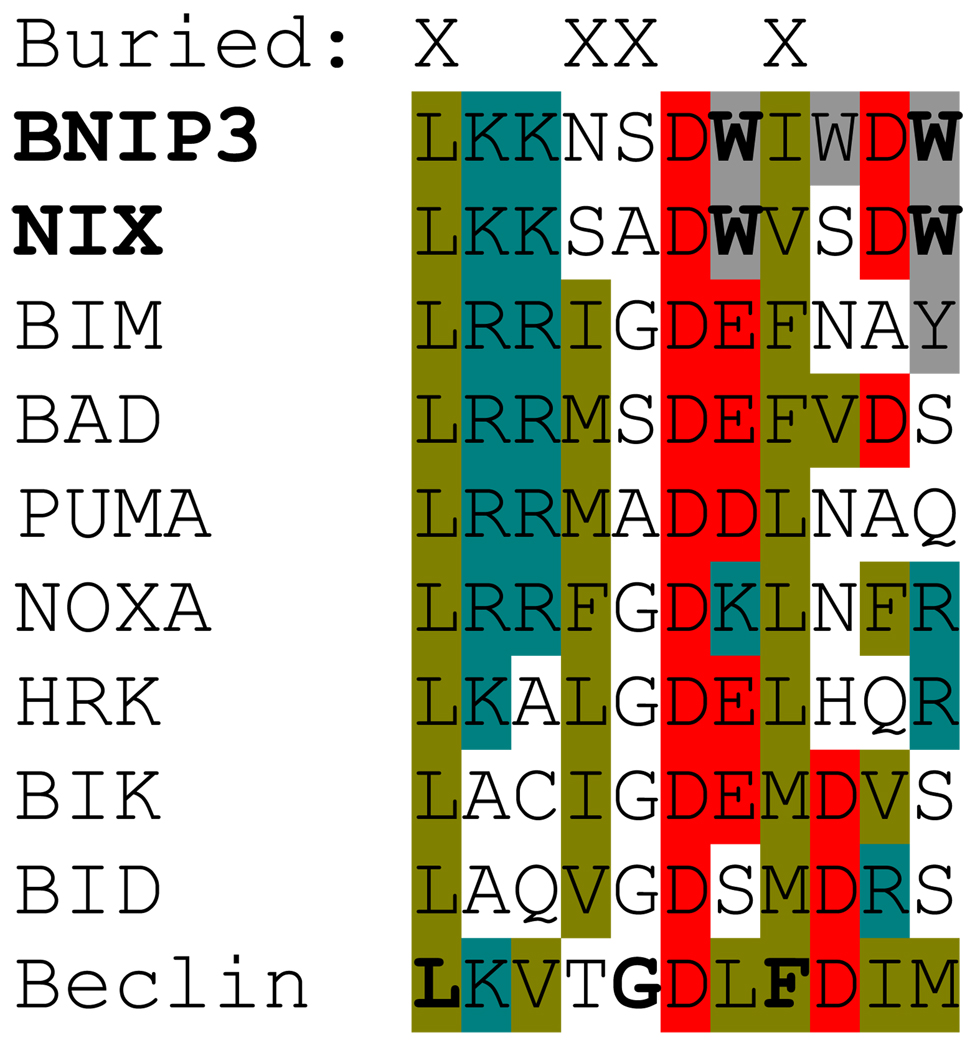
Figure 1.
BH3 domain alignment. An alignment of the BH3 domains of human BNIP3, NIX, other BH3-only proteins, and Beclin-1 is shown. Hydrophobic nonpolar residues are highlighted in olive, hydrophobic polar residues in gray, negatively charged residues in red, and positively charged residues in teal. Conserved tryptophan residues in BNIP3 and NIX are shown in bold. BH3 residues that are partially or fully buried when bound to BCL-XL are indicated by an “X” at the top. Critical helical contact residues in Beclin-1 that disrupt the interaction with BCL-XL (L, G),79 or BCL2 (F),78 when mutated are shown in bold. The first leucine residue in each row corresponds to position 1 in ref. 9.
A putative BNIP3 homolog has been identified in C. elegans that is 21% identical with BNIP3 overall.10,11 Like mammalian BNIP3, ceBNIP3 induces death in cell lines, and interacts with CED-9 and CED-3. Also like BNIP3, the ceBNIP3 BH3 domain is not required for interaction with CED-9 or for induction of cell death, whereas the transmembrane domain is required for both.8,11 Additionally, ceBNIP3-induced cell death is not prevented by caspase inhibitors. These features, and others that will be discussed, suggest that BNIP3 and ceBNIP3 cause an atypical form of cell death.
Discovery and Characterization of NIX
NIX was cloned from a human placenta cDNA library based on its homology (56% identical) to BNIP3.12 Stable NIX expression retards the growth of cancer cell lines, suggesting that NIX may be a tumor suppressor. Like BNIP3, NIX localizes to mitochondria, interacts with BCL2 and BCL-XL, and is proapoptotic.13 Also similar to BNIP3, the C-terminal transmembrane domain of NIX, but not its BH3 domain, is essential for its proapoptotic activity. Further, recombinant NIX protein induces loss of mitochondrial membrane potential (Δψm) and cytochrome c release from isolated mitochondria in vitro. Ohi et al. identified NIX, while investigating adenovirus E1B 19 kDa interacting proteins.14 They found that NIX localizes to the nuclear envelope, endoplasmic reticulum, and mitochondria, but in contrast to other investigators, did not find that NIX induced apoptosis. This lack of effect may be attributed to experimental factors, such as the use of an early time point after transfection, and a late indicator of cell death. By contrast, Chen et. al showed that NIX localizes to mitochondria, and is able to cause apoptosis when transiently expressed.15 Further, they showed that BCL2 and BCL-XL protect cells from NIX-induced death, and that NIX is sensitive to proteosome-dependent degradation. Thus, NIX shares several key features with BNIP3, including an ability to interact with BCL2 and BCL-XL, and to induce apoptosis through its C-terminal transmembrane domain; with regard to its proapoptotic activity, NIX appears to be similar to BNIP3.15
BNIP3, NIX, and Hypoxia-induced Cell Death: Role in Cancer and Heart Disease
BNIP3 and NIX are implicated in hypoxia-induced tumor cell death. BNIP3 was identified in a subtractive hybridization screen in Chinese hamster ovary-K1 cells exposed to hypoxia, and hypoxia strongly induced BNIP3 mRNA.16 Further, BNIP3 protein was induced by hypoxia in these cells, and the kinetics of induction correlated with cell death. The BNIP3 promoter has two HIF-1α binding sites, and the site at −234 relative to the translational start codon is required for transactivation by hypoxia and HIF-1α. In another study, hypoxia induced BNIP3 expression in tumor cell lines, and BNIP3 was expressed in the perinecrotic areas of several epithelial cell carcinomas.17 In this study, BNIP3 was suppressed by Von Hippel-Lindau protein in a renal cell carcinoma cell line, consistent with its regulation through the HIF-1α pathway. Hypoxia in tumors is a negative prognostic indicator; accordingly, deregulation of BNIP3 expression is associated with aggressive disease (reviewed in ref. 18).
Regulation of NIX by hypoxia in tumor cell lines is also reported.16,17,19 In U2OS osteosarcoma cells, NIX is regulated transcriptionally and post-transcriptionally by hypoxia and p53.19 The transcriptional mechanism is postulated to involve HIF-1α-dependent recruitment of CBP to the Nix gene, followed by recruitment of p53, whereas the post-transcriptional mechanism is not known. Experimental downregulation of NIX increases the growth of these cells in a tumor transplant model, suggesting that NIX may inhibit tumor growth in hypoxic conditions. In studies of human cancer, hypermethylation of the BNIP3 promoter was found in pancreatic cancer,20 and the Nix gene was infrequently mutated in a panel of primary breast and ovarian tumors.21 Thus, BNIP3, and NIX are regulated by hypoxia in tumor cells, and their expression is associated with tumor cell death.
In heart muscle, there is evidence that BNIP3 and NIX have a role in pathological cell death. BNIP3 is induced by hypoxia in primary neonatal rat cardiomyocytes, and in adult ventricular myocytes.22–24 Apart from the effect of hypoxia on BNIP3 expression, the combination of hypoxia and acidosis causes alkalai-resistant association of BNIP3 with mitochondria.23 Hypoxia-induced cardiac myocyte cell death exhibits apoptotic features, such as DNA fragmentation and preservation of plasma membrane integrity;23 however, the role of caspases is controversial. It is reported that BNIP3 expression causes cytochrome c release and myocyte death, which is blunted by caspase inhibitors,24 but it is also reported that cytochrome c is not released, caspase-3 is inactive, and caspase inhibitors do not prevent cell death.23 The role of caspases aside, it is generally agreed that BNIP3 causes mitochondrial depolarization in cardiac myocytes, which is blocked by mitochondrial permeability transition pore (MPTP) inhibitors. It is also reported that a C-terminal deletion mutant of BNIP3 lacking its transmembrane domain functions as a dominant negative regulator of BNIP3-induced cell death.23,24 Notably, a role for BNIP3 in cardiac myocyte cell death has been confirmed in vivo; in mice subjected to ischemia-reperfusion, BNIP3 deficiency limits post-myocardial infarct ventricular remodeling by reducing peri-infarct apoptosis.25 Conversely, high-level BNIP3 expression in the heart, driven by α-myosin heavy chain regulatory sequences, significantly increases the frequency of apoptotic cells, and causes cardiomyopathy without ischemia.
NIX also has a role in heart disease, but with an important difference from BNIP3. NIX is regulated by cellular signals associated with cardiac hypertrophy, not hypoxia, in neonatal rat cardiac myocytes.26 Specifically, NIX is regulated by Gαq signaling and protein kinase Cα through an effect on Sp1 binding to the NIX promoter. As is the case for BNIP3, a role for NIX in cardiac myocyte apoptosis has been confirmed in vivo. High-level cardiac expression of NIX causes lethal perinatal cardiomyopathy, whereas expression of a naturally-occurring C-terminally truncated variant of NIX, which is defective in mitochondrial localization, protects Gαq-overexpressing neonates from this condition.27 Additionally, NIX deficiency protects mice from Gαq-mediated and pressure-overload cardiomyopathy.28 Thus, BNIP3 and NIX mediate parallel death pathways, which are regulated by cardiac ischemia and hypertrophy, respectively.
Mechanism of BNIP3 and NIX-induced Cell Death
The molecular mechanism through which BNIP3 and NIX induce cell death is not well understood. Vande Velde et al. first demonstrated that BNIP3 mediates a non-apoptotic type of cell death. Specifically, they showed that overexpression of BNIP3 induces the death of 293T cells, in the absence of apaf-1, caspase-9, or caspase-3, and without cytochrome c release;29 cell death in this setting appears to be necrotic. Similar results were obtained for BNIP3-mediated cell death in cardiac myocytes, but only in the presence of acidosis, and dependent on opening of the MPTP.23 In isolated liver mitochondria, recombinant BNIP3 causes opening of the MPTP, and triggers cytochrome c release, through a transmembrane domain-dependent, BH3 domain-independent, mechanism.30 Similar results were reported for NIX, but only at a 20-fold higher concentration of recombinant protein.13 Another group, using less NIX, failed to see an effect on the MPTP, but did observe cytochrome c release.31 In murine embryonic fibroblasts, BAX or BAK are required for BNIP3-induced loss of Δψm, and cytochrome c release.32 In this study, inhibition of the MPTP was partially effective in preventing BNIP3-induced cell death, but did not prevent BAX or BAK activation.
Collectively, the data suggest that there is not a single mechanism through which BNIP3 and NIX induce cell death. Instead, there appears to be several, and the mechanism likely depends on the cell type and experimental conditions. One model, which is generally accepted, is that upon activation BNIP3 inserts into the mitochondrial outer membrane, which causes opening of the MPTP, loss of Δψm, generation of reactive oxygen species, and necrosis (Figure 2, left panel). BAX and BAK have a role in BNIP3-induced cell death,32 and therefore are included in this model; however, their contribution to opening of the MPTP by this mechanism, if any, is not well defined.
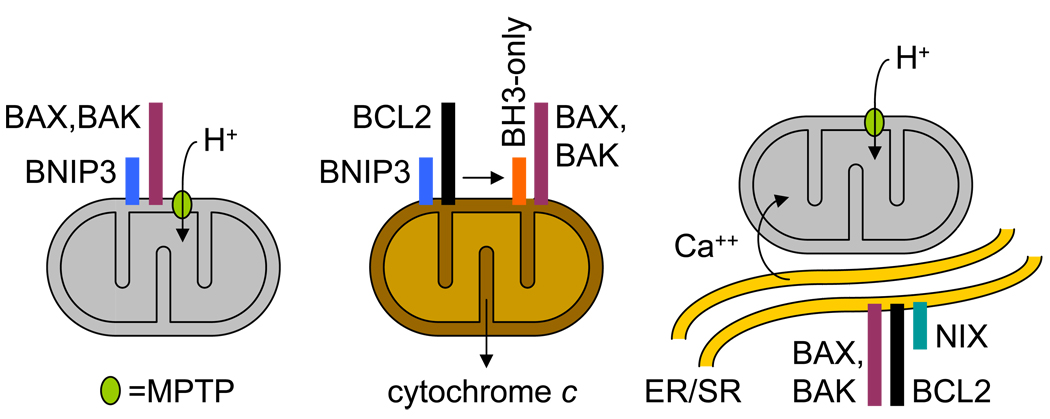
Figure 2.
Potential mechanisms of BNIP3 and NIX-induced cell death. (Left panel) BNIP3 (blue box) or NIX (teal box), in concert with BAX or BAK (plum box), cause opening of the MPTP (lime oval), mitochondrial depolarization (represented as gray color), increased generation of reactive oxygen species, and cell death. (Center panel) BNIP3 or NIX bind BCL2 or BCL-XL (black box), releasing BH3-only protein (orange box) to interact with BAX or BAK, causing cytochrome c release, caspase activation, and cell death. (Right panel) NIX binds to endoplasmic reticulum/sarcoplasmic reticulum (ER/SR), increasing ER/SR calcium (Ca++) stores, causing mitochondrial calcium uptake, opening of the MPTP, and cell death.
A second possibility is that BNIP3 and NIX function as typical BH3-only proteins (Figure 2, center panel); however, several lines of evidence suggest that this is not the major pathway of BNIP3 or NIX-induced cell death. First, compared to typical BH3-only proteins, BNIP3 and NIX are weak inducers of cell death.33 Second, whereas typical BH3-only proteins trigger cell death through cytochrome c release, independent of the MPTP,34 BNIP3 and NIX-induced cell death involves opening of the MPTP. Third, whereas the proapoptotic activities of typical BH3-only proteins are highly dependent on their BH3 domains,35 the proapoptotic activities of BNIP3 and NIX are primarily determined by their transmembrane domains. Fourth, while typical BH3-only proteins cause apoptotic cell death, BNIP3 and NIX-induced cell death has necrotic features. We do not exclude the possibility that this mechanism may contribute to BNIP3 or NIX-induced cell death; however, it may be difficult to evaluate in the presence of the major MPTP-dependent pathway.
A recent study provides evidence for a third potential mechanism of BNIP3 or NIX-induced cell death (Figure 2, right panel).36 BAX or BAK, and BCL2, have opposing effects on cell death, mediated at least in part through their effects on ER/SR calcium stores, mitochondrial calcium uptake, and the MPTP.37–40 In this study, enforced expression of NIX was shown to increase, and loss of NIX was shown to decrease, ER/SR calcium stores in cardiac myocytes. Restoration of calcium stores by genetic ablation of an inhibitor of the ER/SR calcium-uptake pump, reversed the beneficial effect of NIX deficiency on Gαq-mediated cardiomyopathy. Further, ER-targeted NIX caused mitochondrial depolarization and cell death. Whether this mechanism is operative in non-cardiac cells remains to be determined.
Biophysical properties of the BNIP3 transmembrane domain
BNIP3 and NIX could open the MPTP directly through an interaction with BAX or BAK, or a component of the MPTP,41 or indirectly through increased mitochondrial calcium uptake. Alternatively, BNIP3 and NIX could form channels in the mitochondrial outer membrane, which activate the MPTP. BNIP3 and NIX undergo homodimerization in cells, which is resistant to denaturation by sodium dodecyl sulfate (SDS), or reducing conditions, and is mediated by their transmembrane domains.6,13 Given the importance of the transmembrane domains in BNIP3 and NIX-induced cell death, several studies have used biophysical methods to characterize this homotypic interaction. The BNIP3 transmembrane domain strongly self-associates in E. Coli membranes, and SDS micelles.42 Dimers form a right-handed parallel helix-helix structure with a continuous hydrophilic track that spans the lipid bilayer.43,44 Critical interfacial residues that mediate BNIP3 homodimerization are S172H173XXA176XXL179G180XXI183G184 (numbers from human BNIP3). These residues are conserved between BNIP3 and NIX, include an AXXXGXXXG glycine zipper motif,45 and participate in specific interactions, such as intermonomeric hydrogen bonds between polar residues at S172 and H173. It has been proposed that the residues S172 and H173 control an acid-sensitive proton channel in the mitochondrial outer membrane that can initiate cell death.44; however, results with an H173A mutant of BNIP3 are inconsistent,46,47 and other dimerization-defective mutants of BNIP3 (L179S and G180E) fail to prevent BNIP3-induced cell death8. It is possible that these mutants induce cell death, because they can still associate in vivo; regardless, additional studies will be needed to establish a physiological role for BNIP3 or NIX-dependent channel formation.
Autophagy: An Alternate Cell Fate Determined by BNIP3 and NIX
Apart from the role of BNIP3 and NIX in cell death, an intriguing aspect of the biology of these proteins is their role in autophagy. Autophagy was initially described based on its ultrastructural features, in particular double-membraned structures that surrounded cytoplasm and organelles in cells, known as autophagosomes.48,49 Autophagy is a catabolic program present in organisms as primitive as yeast, which is activated in response to starvation or changing nutrient conditions. Genetic studies in yeast have identified a number of proteins, and elucidated several biochemical pathways, that are essential for autophagy.50 One protein is the serine-threonine kinase, autophagy-related 1 (Atg1), which is required for preautophagosomal structure formation, and is negatively regulated by mTOR. Another is Atg6, which is a component of a class III PI3-K-containing complex. The mammalian homolog of Atg6, Beclin-1, has tumor suppressor activity.51,52 Growth of the preautophagosomal structure depends on the activity of two parallel ubiquitin-like conjugation pathways, which are both regulated by an E1-like enzyme Atg7. Apart from its role in the maintenance of cellular homeostasis, autophagy is required for differentiation in lower eukaryotes,53 and there is a growing appreciation of its role in mammalian development.54
Daido et al. first described a role for BNIP3 in autophagy in the ceramide-induced autophagic death of malignant glioma cells.55 In these cells, ceramide induces an increase in BNIP3 expression, and BNIP3 causes mitochondrial depolarization and autophagy. This study was followed by another that showed a role for BNIP3 in arsenic trioxide-induced autophagic cell death.56 In an ischemia-reperfusion model, BNIP3 induced autophagy, which protected HL-1 myocytes from cell death.57 Similarly, hypoxia-induced BNIP3 increased autophagic death, but protected Saos2 cells from necrosis-type cell death.58 Finally, in muscle wasting disorders, where autophagy is implicated in the pathogenesis, BNIP3 and NIX are upregulated, and the expression of either in skeletal muscle induces autophagosome formation.59 Thus, BNIP3 and NIX exhibit a dual nature; they induce cell death, and they also participate in the induction of autophagy. Induction of autophagy by BNIP3 or NIX has a protective effect in some settings, whereas in others it is associated with autophagic cell death. With regard to the latter, it is unclear if death is due to excess autophagy or to an independent death-inducing function of BNIP3 or NIX.
Role of NIX in Erythroid Development
NIX mRNA is expressed in many tissues at a fairly constant level;26 however, along with the mammalian homologs of Atg1 and Atg8 (ULK1 and LC3), NIX is strongly upregulated during terminal erythroid differentiation.60–62 In contrast to the regulation of BNIP3 by hypoxia, or NIX by Gαq-dependent signaling, the cis-acting sequences responsible for upregulation of NIX during erythroid differentiation have not been identified, nor has direct regulation of NIX by erythroid transcription factors been demonstrated. Possibly, NIX is induced by the master regulator of erythroid differentiation, GATA1. Another possibility is the AKT-regulated transcription factor FoxO3. FoxO3 activity is increased by starvation in muscle cells, binds LC3, Bnip3 and Nix, increases their expression, and induces autophagy.59 FoxO3 expression, nuclear localization, and transcriptional activity are induced during erythroid differentiation.63,64 Thus, an intriguing albeit untested model is that NIX and LC3 expression are induced by FoxO3.
Consistent with the erythroid-specific pattern of NIX expression, NIX-deficient mice exhibit defects in erythroid development. Nix−/− mice have mild to moderate anemia, reticulocytosis, and an increase in splenic erythropoiesis.31,61,65 NIX and BCL-XL are coordinately upregulated during erythroid differentiation,60,61 and it is suggested that the pro- and antiapoptotic balance between these proteins regulates erythrocyte production.31 In general agreement with the notion that NIX deficiency confers a survival benefit, Nix−/− erythroid progenitors show improved survival in vitro following erythropoietin deprivation or treatment with a calcium ionophore. On the other hand, the inability of NIX deficiency to rescue BCL-XL deficiency (unpublished results), suggests that there is a proapoptic factor besides NIX that causes cell death in the absence of BCL-XL. Parenthetically, the rescue of BCL-XL deficiency by combined deficiency of BAX and BAK,61 indicates that this factor functions through the intrinsic apoptotic pathway.
Further analysis of NIX-deficient mice revealed a defect in erythroid development that is not directly related to the regulation of death pathways. Nix−/− reticulocytes have a defect in mitochondrial clearance (Figure 3).61,65 Nascent reticulocytes generated by the enucleation of late orthochromatic erythroblasts undergo maturation in the bone marrow and circulation. During this process, they lose surface area and volume,66 undergo remodeling of the plasma membrane and cytoskeleton,67 and eliminate all ribosomes and membrane-bound organelles, such as mitochondria.68,69 In Nix−/− reticulocytes, these processes proceed normally with the exception of mitochondrial clearance. Ultrastructural studies show that mitochondria are cleared from reticulocytes through an autophagy-related process with the major difference that the contents of mature autophagic vacuoles are not recycled but are eliminated by exocytosis.68,70 Like wild-type reticulocytes undergoing maturation, autophagy is induced in Nix−/− reticulocytes; however, the mitochondria are not eliminated. This failure to eliminate mitochondria indicates that they are not properly targeted to autophagosomes in the absence of NIX. There is also evidence that BNIP3 is required for mitophagy in response to hypoxia, suggesting a broader role for these proteins in mitochondrial elimination.71 In the next section, we will consider three models for the mechanism of NIX-dependent mitophagy.
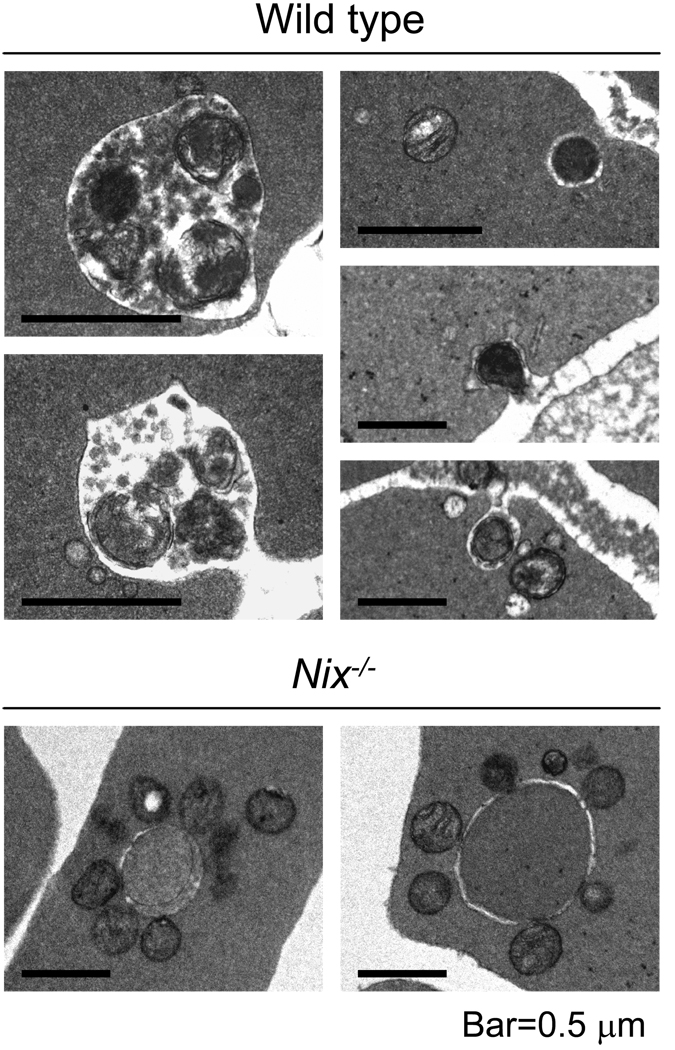
Figure 3.
NIX is required for mitophagy in reticulocytes. (Top panel set) Mitochondria in wild type reticulocytes undergoing exocytosis. Mitochondria in mature autophagic vacuoles (left two panels). Elimination of individual mitochondria (right three panels). (Bottom panel set) Mitochondria in Nix−/− reticulocytes accumulate on the cytoplasmic face of autophagosomes, but are not eliminated. A 0.5 µm bar is shown for scale in all panels. Magnification is 25,000× in all panels, except in the upper panel set the left two panels are 75,000×, and the upper right panel is 50,000×.
Mechanism of NIX-dependent Mitophagy
The first and simplest model of NIX-dependent mitophagy we consider is that NIX triggers mitochondrial depolarization, which in turn causes mitochondrial autophagy and clearance (Figure 4, top row, left). This model is simplest in the sense that there is already considerable evidence, reviewed herein, that BNIP3 and NIX can trigger mitochondrial depolarization, and that mitochondrial depolarization is sufficient to cause mitophagy.72,73 In support of this model, Sandoval et al. show that reticulocyte mitochondria depolarize in culture over a period of 16 hours, and that the mitochondrial clearance defect caused by NIX deficiency can be rescued by compounds that cause mitochondrial depolarization.65 However, one caveat in interpreting these results is the lack of evidence that NIX directly triggers mitochondrial depolarization in reticulocytes. As illustrated in figure 4 (second row, right), it is equally plausible that NIX-dependent mitochondrial depolarization occurs after autophagosome formation,74 but prior to elimination.75 Studies of mitochondrial clearance in autophagy-defective reticulocytes should help resolve this question.
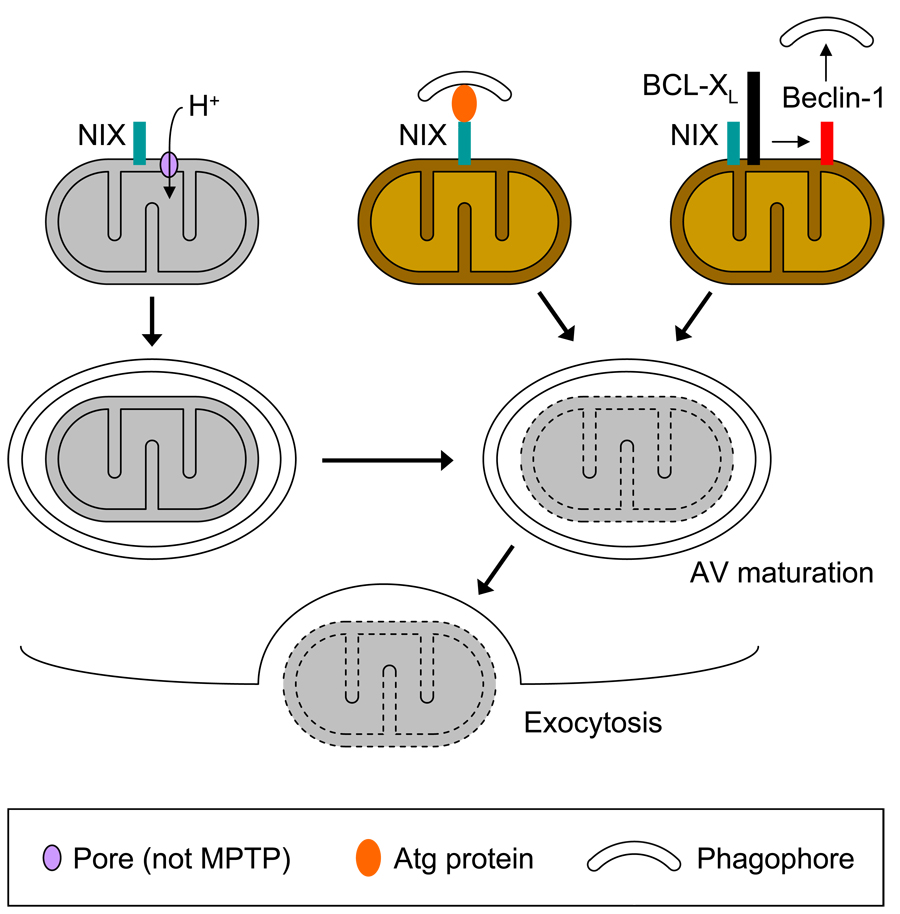
Figure 4.
Potential mechanisms of NIX-dependent mitophagy in reticulocytes. (Top row, left panel) NIX (teal box) opens non-MPTP pore (lavender oval), causing mitochondrial depolarization (represented as gray color), and inducing mitophagy. (Top row, center panel) NIX recruits autophagy protein (orange oval), which supports formation of phagophore. (Top row, right panel) NIX binds BCL-XL (black box), BCL-XL releases Beclin-1 (red box), which activates autophagy, and supports formation of phagophore. (Middle row) Completed autophagosome matures into autophagic vacuole (AV), and mitochondria is degraded. Depending on the mechanism, mitochondrial depolarization either occurs prior to this step or at this step. (Bottom row) A degraded mitochondria is eliminated by exocytosis. A legend is shown at the bottom.
Another problem with the idea that NIX first triggers mitochondrial depolarization is that mitophagy in reticulocytes occurs independent of BAX and BAK.61 In this regard, ABT-737, which rescues the NIX defect,65 requires BAX or BAK to induce mitochondrial depolarization (unpublished results) and cell death;76 therefore, its mechanism of action is likely different from NIX. Arguably, NIX may still cause mitochondrial depolarization through a BAX- or BAK-independent pathway; however, this remains speculative, primarily since mitochondrial clearance also occurs independent of the MPTP.61 This independence from BAX, BAK, and the MPTP, suggests that the mechanism of NIX-dependent mitophagy is distinct from any of the proposed mechanisms of BNIP3 or NIX-induced cell death (Figure 2).
A second model of NIX-dependent mitophagy assumes a novel function of NIX, namely an ability to recruit autophagy components, independent of an ability to trigger mitochondrial depolarization (Figure 4, top row, center). As previously discussed, enforced expression of BNIP3 or NIX induces autophagy in different conditions,55–59 which suggests that the ability to induce autophagy may be an intrinsic property of these proteins. Importantly, BNIP3 and NIX are not homologs of any of the canonical autophagy proteins identified in yeast,50 and NIX deficiency does not generally impair autophagy in erythroid cells, but instead interferes with the targeting of mitochondria to autophagosomes.61,65 A simple model consistent with these findings is that NIX functions as an adaptor protein, and recruits components of the autophagy machinery to mitochondria. Consistent with this suggestion, preliminary studies indicate that NIX directly interacts with LC3 (Ivan Dikic, personal communication).
A third model of NIX-dependent mitophagy is derived from a recently proposed model of crosstalk between autophagy and cell death pathways. Both BNIP3 and Beclin-1 were initially identified as BCL2 or BCL-XL-interacting proteins.1,77 Beclin-1 is part of a class III PI3-K-containing complex that regulates autophagy induction,50 and BCL2 inhibits autophagy by binding to Beclin-1.78 Structural and mutational studies show that this interaction is mediated through a BH3-like domain in Beclin-1 (Figure 1).79,80 BH3-only proteins or mimetics can induce autophagy by competing with Beclin-1 for binding to BCL2 or BCL-XL.81 Thus, it is possible that BNIP3 or NIX compete with Beclin-1 for binding to BCL-XL, and that increased NIX expression during erythroid differentiation disrupts pre-existing BCL-XL-Beclin-1 complexes, thereby releasing Beclin-1 and activating autophagy (Figure 4, top row, right). However, there are two caveats with this model. First, as already noted, autophagy is not generally impaired in NIX-deficient reticulocytes, suggesting that the core autophagy machinery is intact, and that there are other routes of autophagy activation in erythroid cells. In this regard, NIX may only be required to activate autophagy near the mitochondrial outer membrane, where it is located. Second, NIX must exhibit specificity for BCL-XL-Beclin-1 complexes. Otherwise, if NIX were to disrupt BCL-XL interactions with typical BH3-only proteins, it could lead to cell death. The same is true for Beclin-1, and at least for Beclin-1 there is a preliminary report that its overexpression does not induce cell death.82
Conclusions and Perspective
BNIP3 and NIX share certain features with the BH3-only protein subgroup of the BCL2 family, such as sequence homology in the BH3 domain, residence in the mitochondrial outer membrane, and the ability to interact with BCL2 and BCL-XL. Consequently, initial studies of these proteins focused on their role in cell death. Intriguingly, these studies found that BNIP3 and NIX do not function like typical BH3-only proteins. BNIP3 and NIX are weak inducers of cell death that cause loss of mitochondrial Δψm, opening of the MPTP, and, variably, release of cytochrome c. Additionally, the transmembrane domains, but not the BH3 domains, of BNIP3 and NIX, play a major role in the induction of cell death. Subsequent studies revealed that BNIP3 and NIX induce an atypical form of cell death, and that they also induce autophagy.
A central question is whether the ability of BNIP3 and NIX to induce cell death and autophagy are related or independent functions. As discussed in the previous section, they may be mechanistically related; namely, the death-inducing function of BNIP3 and NIX, which is mediated through mitochondrial depolarization, may also serve as the event that initiates autophagy. Alternatively, they may be mechanistically independent, but functionally related. In that case, mitochondrial depolarization and autophagy-inducing functions may localize to different domains of BNIP3 and NIX. Finally, they may be completely independent functions. How might mitochondrial depolarization and autophagy be functionally related? In some respects, they are opposing functions. Mitochondrial depolarization eventually leads to compromise of the mitochondrial outer membrane, and this leads to the release or activation of destructive enzymes. Autophagy-generated membranes serve to compartmentalize these enzymes, and perform a protective function. Together, these properties may allow for limited subcellular destruction, which in turn may be important in cellular remodeling, homeostasis, and development. However, if the destructive process were to exceed autophagy-dependent containment, cell death would result.
Acknowledgements
This research was supported by a grant from the National Institutes of Health to P.A.N. (R21 DK074519), and by the American, Lebanese, and Syrian Associated Charities.
Contributor Information
Ji Zhang, Email: ji.zhang@stjude.org.
Paul A. Ney, Email: paul.ney@stjude.org.
References
- 1.Boyd JM, Malstrom S, Subramanian T, et al. Adenovirus E1B 19 kDa and Bcl-2 proteins interact with a common set of cellular proteins. Cell. 1994;79:341–351. doi: 10.1016/0092-8674(94)90202-x. [DOI] [PubMed] [Google Scholar]
- 2.Takemori N, Riggs JL, Aldrich C. Genetic studies with tumorigenic adenoviruses. I. Isolation of cytocidal (cyt) mutants of adenovirus type 12. Virology. 1968;36:575–586. doi: 10.1016/0042-6822(68)90189-x. [DOI] [PubMed] [Google Scholar]
- 3.Takemori N, Cladaras C, Bhat B, Conley AJ, Wold WS. cyt gene of adenoviruses 2 and 5 is an oncogene for transforming function in early region E1B and encodes the E1B 19,000-molecular-weight polypeptide. J.Virol. 1984;52:793–805. doi: 10.1128/jvi.52.3.793-805.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rao L, Debbas M, Sabbatini P, et al. The adenovirus E1A proteins induce apoptosis, which is inhibited by the E1B 19-kDa and Bcl-2 proteins. Proc.Natl.Acad.Sci.U.S.A. 1992;89:7742–7746. doi: 10.1073/pnas.89.16.7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Subramanian T, Boyd JM, Chinnadurai G. Functional substitution identifies a cell survival promoting domain common to adenovirus E1B 19 kDa and Bcl-2 proteins. Oncogene. 1995;11:2403–2409. [PubMed] [Google Scholar]
- 6.Chen G, Ray R, Dubik D, et al. The E1B 19K/Bcl-2-binding protein Nip3 is a dimeric mitochondrial protein that activates apoptosis. J.Exp.Med. 1997;186:1975–1983. doi: 10.1084/jem.186.12.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yasuda M, Theodorakis P, Subramanian T, Chinnadurai G. Adenovirus E1B-19K/BCL-2 interacting protein BNIP3 contains a BH3 domain and a mitochondrial targeting sequence. J.Biol.Chem. 1998;273:12415–12421. doi: 10.1074/jbc.273.20.12415. [DOI] [PubMed] [Google Scholar]
- 8.Ray R, Chen G, Vande VC, et al. BNIP3 heterodimerizes with Bcl-2/Bcl-X(L) and induces cell death independent of a Bcl-2 homology 3 (BH3) domain at both mitochondrial and nonmitochondrial sites. J.Biol.Chem. 2000;275:1439–1448. doi: 10.1074/jbc.275.2.1439. [DOI] [PubMed] [Google Scholar]
- 9.Aouacheria A, Brunet F, Gouy M. Phylogenomics of life-or-death switches in multicellular animals: Bcl-2, BH3-Only, and BNip families of apoptotic regulators. Mol.Biol.Evol. 2005;22:2395–2416. doi: 10.1093/molbev/msi234. [DOI] [PubMed] [Google Scholar]
- 10.Yasuda M, Sa-Eipper C, Gong XL, Chinnadurai G. Regulation of apoptosis by a Caenorhabditis elegans BNIP3 homolog. Oncogene. 1998;17:2525–2530. doi: 10.1038/sj.onc.1202467. [DOI] [PubMed] [Google Scholar]
- 11.Cizeau J, Ray R, Chen G, Gietz RD, Greenberg AH. The C. elegans orthologue ceBNIP3 interacts with CED-9 and CED-3 but kills through a BH3- and caspase-independent mechanism. Oncogene. 2000;19:5453–5463. doi: 10.1038/sj.onc.1203929. [DOI] [PubMed] [Google Scholar]
- 12.Matsushima M, Fujiwara T, Takahashi E, et al. Isolation, mapping, and functional analysis of a novel human cDNA (BNIP3L) encoding a protein homologous to human NIP3. Genes Chromosomes.Cancer. 1998;21:230–235. [PubMed] [Google Scholar]
- 13.Imazu T, Shimizu S, Tagami S, et al. Bcl-2/E1B 19 kDa-interacting protein 3-like protein (Bnip3L) interacts with bcl-2/Bcl-xL and induces apoptosis by altering mitochondrial membrane permeability. Oncogene. 1999;18:4523–4529. doi: 10.1038/sj.onc.1202722. [DOI] [PubMed] [Google Scholar]
- 14.Ohi N, Tokunaga A, Tsunoda H, et al. A novel adenovirus E1B19K-binding protein B5 inhibits apoptosis induced by Nip3 by forming a heterodimer through the C-terminal hydrophobic region. Cell Death.Differ. 1999;6:314–325. doi: 10.1038/sj.cdd.4400493. [DOI] [PubMed] [Google Scholar]
- 15.Chen G, Cizeau J, Vande VC, et al. Nix and Nip3 form a subfamily of pro-apoptotic mitochondrial proteins. J.Biol.Chem. 1999;274:7–10. doi: 10.1074/jbc.274.1.7. [DOI] [PubMed] [Google Scholar]
- 16.Bruick RK. Expression of the gene encoding the proapoptotic Nip3 protein is induced by hypoxia. Proc.Natl.Acad.Sci.U.S.A. 2000;97:9082–9087. doi: 10.1073/pnas.97.16.9082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sowter HM, Ratcliffe PJ, Watson P, Greenberg AH, Harris AL. HIF-1-dependent regulation of hypoxic induction of the cell death factors BNIP3 and NIX in human tumors. Cancer Res. 2001;61:6669–6673. [PubMed] [Google Scholar]
- 18.Burton TR, Gibson SB. The role of Bcl-2 family member BNIP3 in cell death and disease: NIPping at the heels of cell death. Cell Death.Differ. 2009 doi: 10.1038/cdd.2008.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fei P, Wang W, Kim SH, et al. Bnip3L is induced by p53 under hypoxia, and its knockdown promotes tumor growth. Cancer Cell. 2004;6:597–609. doi: 10.1016/j.ccr.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 20.Okami J, Simeone DM, Logsdon CD. Silencing of the hypoxia-inducible cell death protein BNIP3 in pancreatic cancer. Cancer Res. 2004;64:5338–5346. doi: 10.1158/0008-5472.CAN-04-0089. [DOI] [PubMed] [Google Scholar]
- 21.Lai J, Flanagan J, Phillips WA, Chenevix-Trench G, Arnold J. Analysis of the candidate 8p21 tumour suppressor, BNIP3L, in breast and ovarian cancer. Br.J.Cancer. 2003;88:270–276. doi: 10.1038/sj.bjc.6600674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo K, Searfoss G, Krolikowski D, et al. Hypoxia induces the expression of the pro-apoptotic gene BNIP3. Cell Death.Differ. 2001;8:367–376. doi: 10.1038/sj.cdd.4400810. [DOI] [PubMed] [Google Scholar]
- 23.Kubasiak LA, Hernandez OM, Bishopric NH, Webster KA. Hypoxia and acidosis activate cardiac myocyte death through the Bcl-2 family protein BNIP3. Proc.Natl.Acad.Sci.U.S.A. 2002;99:12825–12830. doi: 10.1073/pnas.202474099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Regula KM, Ens K, Kirshenbaum LA. Inducible expression of BNIP3 provokes mitochondrial defects and hypoxia-mediated cell death of ventricular myocytes. Circ.Res. 2002;91:226–231. doi: 10.1161/01.res.0000029232.42227.16. [DOI] [PubMed] [Google Scholar]
- 25.Diwan A, Krenz M, Syed FM, et al. Inhibition of ischemic cardiomyocyte apoptosis through targeted ablation of Bnip3 restrains postinfarction remodeling in mice. J.Clin.Invest. 2007;117:2825–2833. doi: 10.1172/JCI32490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Galvez AS, Brunskill EW, Marreez Y, et al. Distinct pathways regulate proapoptotic Nix and BNip3 in cardiac stress. J.Biol.Chem. 2006;281:1442–1448. doi: 10.1074/jbc.M509056200. [DOI] [PubMed] [Google Scholar]
- 27.Yussman MG, Toyokawa T, Odley A, et al. Mitochondrial death protein Nix is induced in cardiac hypertrophy and triggers apoptotic cardiomyopathy. Nat.Med. 2002;8:725–730. doi: 10.1038/nm719. [DOI] [PubMed] [Google Scholar]
- 28.Diwan A, Wansapura J, Syed FM, et al. Nix-mediated apoptosis links myocardial fibrosis, cardiac remodeling, and hypertrophy decompensation. Circulation. 2008;117:396–404. doi: 10.1161/CIRCULATIONAHA.107.727073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vande Velde C, Cizeau J, Dubik D, et al. BNIP3 and genetic control of necrosis-like cell death through the mitochondrial permeability transition pore. Mol.Cell Biol. 2000;20:5454–5468. doi: 10.1128/mcb.20.15.5454-5468.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim JY, Cho JJ, Ha J, Park JH. The carboxy terminal C-tail of BNip3 is crucial in induction of mitochondrial permeability transition in isolated mitochondria. Arch.Biochem.Biophys. 2002;398:147–152. doi: 10.1006/abbi.2001.2673. [DOI] [PubMed] [Google Scholar]
- 31.Diwan A, Koesters AG, Odley AM, et al. Unrestrained erythroblast development in Nix−/− mice reveals a mechanism for apoptotic modulation of erythropoiesis. Proc.Natl.Acad.Sci.U.S.A. 2007;104:6794–6799. doi: 10.1073/pnas.0610666104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kubli DA, Ycaza JE, Gustafsson AB. Bnip3 mediates mitochondrial dysfunction and cell death through Bax and Bak. Biochem.J. 2007;405:407–415. doi: 10.1042/BJ20070319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim H, Rafiuddin-Shah M, Tu HC, et al. Hierarchical regulation of mitochondrion-dependent apoptosis by BCL-2 subfamilies. Nat.Cell Biol. 2006;8:1348–1358. doi: 10.1038/ncb1499. [DOI] [PubMed] [Google Scholar]
- 34.Shimizu S, Tsujimoto Y. Proapoptotic BH3-only Bcl-2 family members induce cytochrome c release, but not mitochondrial membrane potential loss, and do not directly modulate voltage-dependent anion channel activity. Proc.Natl.Acad.Sci.U.S.A. 2000;97:577–582. doi: 10.1073/pnas.97.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kelekar A, Thompson CB. Bcl-2-family proteins: the role of the BH3 domain in apoptosis. Trends Cell Biol. 1998;8:324–330. doi: 10.1016/s0962-8924(98)01321-x. [DOI] [PubMed] [Google Scholar]
- 36.Diwan A, Matkovich SJ, Yuan Q, et al. Endoplasmic reticulum-mitochondria crosstalk in NIX-mediated murine cell death. J.Clin.Invest. 2008 doi: 10.1172/JCI36445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Foyouzi-Youssefi R, Arnaudeau S, Borner C, et al. Bcl-2 decreases the free Ca2+ concentration within the endoplasmic reticulum. Proc.Natl.Acad.Sci.U.S.A. 2000;97:5723–5728. doi: 10.1073/pnas.97.11.5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scorrano L, Oakes SA, Opferman JT, et al. BAX and BAK regulation of endoplasmic reticulum Ca2+: a control point for apoptosis. Science. 2003;300:135–139. doi: 10.1126/science.1081208. [DOI] [PubMed] [Google Scholar]
- 39.Nutt LK, Pataer A, Pahler J, et al. Bax and Bak promote apoptosis by modulating endoplasmic reticular and mitochondrial Ca2+ stores. J.Biol.Chem. 2002;277:9219–9225. doi: 10.1074/jbc.M106817200. [DOI] [PubMed] [Google Scholar]
- 40.Baines CP, Kaiser RA, Purcell NH, et al. Loss of cyclophilin D reveals a critical role for mitochondrial permeability transition in cell death. Nature. 2005;434:658–662. doi: 10.1038/nature03434. [DOI] [PubMed] [Google Scholar]
- 41.Marzo I, Brenner C, Zamzami N, et al. Bax and adenine nucleotide translocator cooperate in the mitochondrial control of apoptosis. Science. 1998;281:2027–2031. doi: 10.1126/science.281.5385.2027. [DOI] [PubMed] [Google Scholar]
- 42.Sulistijo ES, Jaszewski TM, MacKenzie KR. Sequence-specific dimerization of the transmembrane domain of the "BH3-only" protein BNIP3 in membranes and detergent. J.Biol.Chem. 2003;278:51950–51956. doi: 10.1074/jbc.M308429200. [DOI] [PubMed] [Google Scholar]
- 43.Sulistijo ES, MacKenzie KR. Sequence dependence of BNIP3 transmembrane domain dimerization implicates side-chain hydrogen bonding and a tandem GxxxG motif in specific helix-helix interactions. J.Mol.Biol. 2006;364:974–990. doi: 10.1016/j.jmb.2006.09.065. [DOI] [PubMed] [Google Scholar]
- 44.Bocharov EV, Pustovalova YE, Pavlov KV, et al. Unique dimeric structure of BNip3 transmembrane domain suggests membrane permeabilization as a cell death trigger. J.Biol.Chem. 2007;282:16256–16266. doi: 10.1074/jbc.M701745200. [DOI] [PubMed] [Google Scholar]
- 45.Kim S, Jeon TJ, Oberai A, et al. Transmembrane glycine zippers: physiological and pathological roles in membrane proteins. Proc.Natl.Acad.Sci.U.S.A. 2005;102:14278–14283. doi: 10.1073/pnas.0501234102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frazier DP, Wilson A, Graham RM, et al. Acidosis regulates the stability, hydrophobicity, and activity of the BH3-only protein Bnip3. Antioxid.Redox.Signal. 2006;8:1625–1634. doi: 10.1089/ars.2006.8.1625. [DOI] [PubMed] [Google Scholar]
- 47.Kubli DA, Quinsay MN, Huang C, Lee Y, Gustafsson AB. Bnip3 functions as a mitochondrial sensor of oxidative stress during myocardial ischemia and reperfusion. Am.J.Physiol Heart Circ.Physiol. 2008;295:H2025–H2031. doi: 10.1152/ajpheart.00552.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clark SL., Jr Cellular differentiation in the kidneys of newborn mice studies with the electron microscope. J.Biophys.Biochem.Cytol. 1957;3:349–362. doi: 10.1083/jcb.3.3.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ashford TP, Porter KR. Cytoplasmic components in hepatic cell lysosomes. J.Cell Biol. 1962;12:198–202. doi: 10.1083/jcb.12.1.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yorimitsu T, Klionsky DJ. Autophagy: molecular machinery for self-eating. Cell Death.Differ. 2005;12 Suppl 2:1542–1552. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qu X, Yu J, Bhagat G, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J.Clin.Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc.Natl.Acad.Sci.U.S.A. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Otto GP, Wu MY, Kazgan N, Anderson OR, Kessin RH. Macroautophagy is required for multicellular development of the social amoeba Dictyostelium discoideum. J.Biol.Chem. 2003;278:17636–17645. doi: 10.1074/jbc.M212467200. [DOI] [PubMed] [Google Scholar]
- 54.Cecconi F, Levine B. The role of autophagy in mammalian development: cell makeover rather than cell death. Dev.Cell. 2008;15:344–357. doi: 10.1016/j.devcel.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Daido S, Kanzawa T, Yamamoto A, et al. Pivotal role of the cell death factor BNIP3 in ceramide-induced autophagic cell death in malignant glioma cells. Cancer Res. 2004;64:4286–4293. doi: 10.1158/0008-5472.CAN-03-3084. [DOI] [PubMed] [Google Scholar]
- 56.Kanzawa T, Zhang L, Xiao L, et al. Arsenic trioxide induces autophagic cell death in malignant glioma cells by upregulation of mitochondrial cell death protein BNIP3. Oncogene. 2005;24:980–991. doi: 10.1038/sj.onc.1208095. [DOI] [PubMed] [Google Scholar]
- 57.Hamacher-Brady A, Brady NR, Logue SE, et al. Response to myocardial ischemia/reperfusion injury involves Bnip3 and autophagy. Cell Death.Differ. 2007;14:146–157. doi: 10.1038/sj.cdd.4401936. [DOI] [PubMed] [Google Scholar]
- 58.Tracy K, Dibling BC, Spike BT, et al. BNIP3 is an RB/E2F target gene required for hypoxia-induced autophagy. Mol.Cell Biol. 2007;27:6229–6242. doi: 10.1128/MCB.02246-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mammucari C, Milan G, Romanello V, et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6:458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 60.Aerbajinai W, Giattina M, Lee YT, Raffeld M, Miller JL. The proapoptotic factor Nix is coexpressed with Bcl-xL during terminal erythroid differentiation. Blood. 2003;102:712–717. doi: 10.1182/blood-2002-11-3324. [DOI] [PubMed] [Google Scholar]
- 61.Schweers RL, Zhang J, Randall MS, et al. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc.Natl.Acad.Sci.U.S.A. 2007;104:19500–19505. doi: 10.1073/pnas.0708818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kundu M, Lindsten T, Yang CY, et al. Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood. 2008;112:1493–1502. doi: 10.1182/blood-2008-02-137398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marinkovic D, Zhang X, Yalcin S, et al. Foxo3 is required for the regulation of oxidative stress in erythropoiesis. J.Clin.Invest. 2007;117:2133–2144. doi: 10.1172/JCI31807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bakker WJ, Blazquez-Domingo M, Kolbus A, et al. FoxO3a regulates erythroid differentiation and induces BTG1, an activator of protein arginine methyl transferase 1. J.Cell Biol. 2004;164:175–184. doi: 10.1083/jcb.200307056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sandoval H, Thiagarajan P, Dasgupta SK, et al. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008;454:232–235. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Waugh RE, McKenney JB, Bauserman RG, et al. Surface area and volume changes during maturation of reticulocytes in the circulation of the baboon. J.Lab Clin.Med. 1997;129:527–535. doi: 10.1016/s0022-2143(97)90007-x. [DOI] [PubMed] [Google Scholar]
- 67.Chasis JA, Prenant M, Leung A, Mohandas N. Membrane assembly and remodeling during reticulocyte maturation. Blood. 1989;74:1112–1120. [PubMed] [Google Scholar]
- 68.Gronowicz G, Swift H, Steck TL. Maturation of the reticulocyte in vitro. J.Cell Sci. 1984;71:177–197. doi: 10.1242/jcs.71.1.177. [DOI] [PubMed] [Google Scholar]
- 69.Koury MJ, Koury ST, Kopsombut P, Bondurant MC. In vitro maturation of nascent reticulocytes to erythrocytes. Blood. 2005;105:2168–2174. doi: 10.1182/blood-2004-02-0616. [DOI] [PubMed] [Google Scholar]
- 70.Heynen MJ, Tricot G, Verwilghen RL. Autophagy of mitochondria in rat bone marrow erythroid cells. Relation to nuclear extrusion. Cell Tissue Res. 1985;239:235–239. doi: 10.1007/BF00214924. [DOI] [PubMed] [Google Scholar]
- 71.Zhang H, Bosch-Marce M, Shimoda LA, et al. Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J.Biol.Chem. 2008;283:10892–10903. doi: 10.1074/jbc.M800102200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 72.Elmore SP, Qian T, Grissom SF, Lemasters JJ. The mitochondrial permeability transition initiates autophagy in rat hepatocytes. FASEB J. 2001;15:2286–2287. doi: 10.1096/fj.01-0206fje. [DOI] [PubMed] [Google Scholar]
- 73.Twig G, Elorza A, Molina AJ, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim I, Rodriguez-Enriquez S, Lemasters JJ. Selective degradation of mitochondria by mitophagy. Arch.Biochem.Biophys. 2007;462:245–253. doi: 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang J, Ney PA. NIX induces mitochondrial autophagy in reticulocytes. Autophagy. 2008;4:354–356. doi: 10.4161/auto.5552. [DOI] [PubMed] [Google Scholar]
- 76.van Delft MF, Wei AH, Mason KD, et al. The BH3 mimetic ABT-737 targets selective Bcl-2 proteins and efficiently induces apoptosis via Bak/Bax if Mcl-1 is neutralized. Cancer Cell. 2006;10:389–399. doi: 10.1016/j.ccr.2006.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liang XH, Kleeman LK, Jiang HH, et al. Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J.Virol. 1998;72:8586–8596. doi: 10.1128/jvi.72.11.8586-8596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pattingre S, Tassa A, Qu X, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 79.Oberstein A, Jeffrey PD, Shi Y. Crystal structure of the Bcl-XL-Beclin 1 peptide complex: Beclin 1 is a novel BH3-only protein. J.Biol.Chem. 2007;282:13123–13132. doi: 10.1074/jbc.M700492200. [DOI] [PubMed] [Google Scholar]
- 80.Feng W, Huang S, Wu H, Zhang M. Molecular basis of Bcl-xL's target recognition versatility revealed by the structure of Bcl-xL in complex with the BH3 domain of Beclin-1. J.Mol.Biol. 2007;372:223–235. doi: 10.1016/j.jmb.2007.06.069. [DOI] [PubMed] [Google Scholar]
- 81.Maiuri MC, Le TG, Criollo A, et al. Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1. EMBO J. 2007;26:2527–2539. doi: 10.1038/sj.emboj.7601689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Maiuri MC, Tasdemir E, Criollo A, et al. Control of autophagy by oncogenes and tumor suppressor genes. Cell Death.Differ. 2009;16:87–93. doi: 10.1038/cdd.2008.131. [DOI] [PubMed] [Google Scholar]