Abstract
The rebinding kinetics of CO to protoheme (FePPIX) in the presence and absence of a proximal imidazole ligand reveals the magnitude of the rebinding barrier associated with proximal histidine ligation. The ligation states of the heme under different solvent conditions are also investigated using both equilibrium and transient spectroscopy. In the absence of imidazole, a weak ligand (probably water) is bound on the proximal side of the FePPIX-CO adduct. When the heme is encapsulated in micelles of cetyltrimethylammonium bromide (CTAB), photolysis of FePPIX-CO induces a complicated set of proximal ligation changes. In contrast, the use of glycerol-water solutions leads to a simple two-state geminate kinetic response with rapid (10–100 ps) CO recombination and a geminate amplitude that can be controlled by adjusting the solvent viscosity. By comparing the rate of CO rebinding to protoheme in glycerol solution with and without a bound proximal imidazole ligand, we find the enthalpic contribution to the proximal rebinding barrier, Hp, to be 11 ± 2 kJ/mol. Further comparison of the CO rebinding rate of the imidazole bound protoheme with the analogous rate in myoglobin (Mb) leads to a determination of the difference in their distal free energy barriers: ΔGD ≈ 12 ± 1 kJ/mol. Estimates of the entropic contributions, due to the ligand accessible volumes in the distal pocket and the xenon-4 cavity of myoglobin (~3 kJ/mol), then lead to a distal pocket enthalpic barrier of HD ≈ 9 ± 2 kJ/mol. These results agree well with the predictions of a simple model and with previous independent room-temperature measurements (Tian et al. Phys. Rev. Lett. 1992, 68, 408) of the enthalpic MbCO rebinding barrier (18 ± 2 kJ/mol).
Introduction
Iron protoporphyrin-IX (FePPIX or heme) is the prosthetic group of a large class of heme proteins and is intimately involved with physiological functions associated with the binding of small diatomic ligands such as CO, O2, and NO. The binding of these diatomic ligands is crucial to the varied catalytic, signaling, and oxygen storage functions of this important class of proteins.1–5 The diatomic ligand binds to the heme iron from the distal side, while an amino acid residue from the protein matrix (often histidine or cysteine) binds from the other (proximal) side. The dynamics associated with ligand binding to the heme group has been studied for many years in both native and mutant proteins3–15 and in heme model compounds.16–24 The binding process for model compounds involves at least two steps: the diffusion of ligand through the surrounding solvent or buffer followed by the bond formation step between the diatomic ligand and the iron. In proteins, the process is complicated by the protein matrix where various entry and exit channels, as well as docking sites and cavities within the protein, lead to a complexity in the overall reaction process that is still not fully understood.12–14,24–30
The specific interactions that control the activation barrier for the Fe-diatomic bond formation in terms of the heme, the proximal ligand, and the distal pocket protein architecture have been studied extensively. For simplicity, the enthalpic part of the ligand rebinding barrier can be separated into two contributions: the proximal term, HP, which is controlled by the proximal ligand and the heme geometry (e.g., the Fe out-of-plane displacement) and the distal term, HD, which is determined by the distal pocket environment around the dissociated ligand. It remains unclear if one or the other of these two terms dominates the overall enthalpic barrier, or if both are equally important, as suggested by a prior analysis of the nonexponential rebinding kinetics of MbCO at low temperatures.31,32 On the other hand, when the entropic part of the free energy rebinding barrier is considered, the distal contribution (associated with the ligand accessible volumes33 of the distal pocket and the Xe4 cavity) is more important than the proximal contribution.
Several studies of Mb under conditions (e.g., low pH, H93G mutation) that remove the proximal histidine (His93) ligand have been carried out in order to determine its contribution to the rebinding barrier.10,15,34,35 Unfortunately, the situation becomes complex because of the increased acidity of the iron atom when CO binds. For example, the complex ligation kinetics observed for Mb at low pH required a ligand switching model involving both His93 and H2O as possible ligands.35 The ligand switch arises because of the presence of the Fe–His bond when CO is bound to the partially unfolded35,36 protein at low pH. In contrast, a transient proximal Fe–OH2 bond is formed35,36 when CO is photolyzed. In the case of the H93G mutant of Mb, where the native proximal His93 ligand is replaced by glycine and no exogenous ligand is added, another histidine (His97) is recruited as the proximal ligand when CO binds and acidifies the heme iron.37,38 As a result of these complexities, the kinetic effects of removing the proximal histidine ligand have not been isolated in Mb using either pH induced unfolding or the H93G mutant system.
One obvious way to probe the proximal contribution to the rebinding barrier, free of complications from the various protein-mediated intermediate states and the distal pocket contribution, is to study the CO rebinding kinetics of heme model compounds with and without the proximal imidazole ligand. Surprisingly, a complete set of such kinetic measurements has not been reported and analyzed. If Fe–PPIX is simply dissolved in aqueous solution at high pH, then dimerization takes place, which significantly perturbs the rebinding kinetics.39 To prevent heme dimerization, it is possible to use micelles formed from materials such as CTAB (cetyltrimethylammonium bromide), which stabilize the hemes in a monodispersed solution.40,41
Model compound kinetic studies have been performed in CTAB under conditions where an imidazole ligand is bound to the heme.18,23 However, for such compounds, the CO ligand escape rate is much larger than its geminate rebinding rate, so the latter process is difficult or impossible to detect.23 Moreover, in the absence of an added imidazole ligand, the CTAB encapsulated heme samples again display complex kinetic evolution, probably due to a base elimination mechanism18,42 (i.e., upon CO photolysis, there is a delayed dissociation of the weak proximal base before CO rebinds). The problems mentioned above have made clear kinetic comparisons between the imidazole ligated and unligated species problematic.
In contrast to FePPIX-CO kinetics in CTAB solutions, early studies16,17,43–45 of CO binding to iron PPIX in highly concentrated glycerol solutions (with no added proximal ligand) revealed a relatively simple kinetic response, suggesting that heme aggregation and spurious proximal ligation was not a problem under these highly viscous solution conditions. Kinetic measurements of diatomic ligand binding to FePPIX in the presence of a proximal imidazole ligand are also feasible under identical conditions of highly concentrated glycerol. For example, Traylor and Magde19 used 1-methlyimidazole (1-MeIm) as a ligand to form a 1-MeImFePPIX-CO complex in 98% glycerol and observed a geminate yield of 0.4 with a ~300 ps time constant. However, there have been no systematic attempts made to isolate and assess the effects of proximal imidazole ligation on the CO geminate rebinding kinetics using a self-consistent set of solution conditions.
The measurements reported here present the first such controlled comparisons of the CO geminate rebinding kinetics, with and without a proximal imidazole ligand. We study heme in its bare form, FePPIX, without the protein matrix and we focus on the effect of the proximal ligand by adding exogenous 2-methylimidazole (2-MeIm) to the heme in highly concentrated (80–95%) glycerol solution. We quantitatively compare the kinetics of the imidazole ligated species to that of the unligated bare heme in the same highly viscous glycerol solution, where it is known to undergo very rapid and large amplitude CO geminate recombination.20,44 Such measurements directly probe Hp, the contribution of the proximal imidazole ligation to the CO rebinding barrier. In addition, we are also able to compare the kinetics of the imidazole ligated FePPIX-CO model compound in glycerol with that of native Mb, to assess the contribution of the distal pocket protein architecture to the rebinding barrier.
Experimental Methods
Hemin (ferric protoporphyrin IX chloride) is purchased from Porphyrin Products Inc., dissolved in 1N NaOH, and then diluted into glycerol or CTAB solution. For typical 80% (v/v) glycerol solutions (glycerol obtained from Acros Organics), the final sample pH is 12. CTAB solutions (1% (w/v) cetyltrimethylammonium bromide obtained from Sigma, Inc.) are adjusted to pH 7 with 0.1 M phosphate buffer. The 2-methylimidazole concentration is 0.5 M when used. The hemin concentrations for kinetics measurements are typically 50–100 μM. At higher heme concentration in glycerol solution aggregation in deoxy state is observed, as is evidenced by the blue-shift of Soret and red-shift of band III in the deoxy sample spectra. After flushing with argon, the sample is reduced by addition of a small amount of degassed sodium dithionite solution. The CO adduct is formed from the reduced sample by flushing with CO for at least 30 min. The equilibrium absorption spectra of the samples are obtained using a Hitachi U-3410 spectrophotometer.
Both cw and pulsed laser sources were used to excite Raman scattering in resonance with the heme Soret band. Raman scattering excited by the 413.1 nm line of a Kr ion laser (Innova 300, Coherent, Inc.) was collected at right angles to the excitation beam and focused into a single grating monochromator (1870B, Spex Industries). A 76 MHz train of 3 ps pulses, obtained by second harmonic generation of the picosecond oscillator (Mira 900P, Coherent, Inc.), excited scattering at 435 nm, which was collected in a backscattering geometry. In both cases, we used interferometric notch filters (Kaiser Instruments) to reject the intense Rayleigh scattered light and a liquid nitrogen cooled CCD detector (Princeton Instruments) to record the spectra. In additional experiments, either the 363.8 nm line from an Ar laser (Coherent) or tunable output from a dye laser, containing stilbene 1 or stilbene 3 and pumped by the UV lines of the Ar laser, was used in conjunction with a triple monochromator (SPEX) equipped with an intensified photodiode array (Princeton Instruments). Infrared data were recorded on a Bio-rad FTS-60A FTIR spectrometer.
Two laser systems are used to measure the kinetics over all time scales. One covers the subpicosecond time scale out to 5 ns, and the other system covers from 10 ns to 100 ms. The ultrafast kinetics measurement setup and experimental procedure are described in detail elsewhere.46,47 An amplified Ti:sapphire laser system (REGA 9050, Coherent, Inc.) generates ~50 fs laser pulses at ~800 nm with an average power of ~ 1 W and repetition rate of 200 kHz. These fundamental pulses are used to pump an optical parametric amplifier (OPA, Coherent 9400) and to generate a white light continuum. The OPA output is tuned at 580 nm and used to photolyze the sample. The white light continuum, which probes the sample, is created by focusing the 800 nm fundamental into a spinning fused silica disk, with a resulting spectrum that extends from 390 to 800 nm. A monochromator (Instruments SA, Triax 320) is used to disperse the probe light after the sample. The time delay for the probe (white light continuum) with respect to the pump (OPA pulse) is controlled using a servo-motor translation stage (Daedal Division, Parker Automation) with a full translation range of 5.6 ns.
The pump and probe beams are focused into a spinning sample cell using an achromatic lens. Polarizations of pump and probe beams form the magic angle to remove rotational relaxation artifacts, which are (in any case) negligible for the heme group on short time scales in a high viscosity (glycerol based) solvent.48 The sample cell has a 2 in. diameter and spins at a rate of over 7000 rpm to ensure that every pump–probe pulse pair detects fresh re-equilibrated sample. Care is also taken to make the pump beam completely overlap the probe beam at the sample. The time resolution of 200 fs is determined by pump–probe cross-correlation.
The kinetics measurements employ synchronous detection, where the pump beam is modulated by a mechanical chopper (3 kHz), and the pump-induced change in the probe signal is detected at each time delay using a PMT and a lock-in amplifier. Transient absorption spectra are taken with a diode array detector (TBY1024 Princeton Instruments, Inc.) by using a shutter (Uniblitz, Inc.) to sequentially block and unblock the pump beam for an equal amount of time (about 0.5 s) so that pump-off and pump-on spectra can be accumulated. The logarithm of the ratio yields the absolute magnitude of the pump induced transient absorption difference spectrum.
The experimental system for the longer time (> 10 ns) kinetics measurement has also been described in detail previously.29 Briefly, a cw beam produced by a universal arc lamp (Oriel Instruments, Model 66021) and a 0.25 m monochromator (Oriel Instruments, Model 77200) probe the kinetic response of the sample at selected wavelengths. The transmitted beam is detected by a high-linearity, low noise, photomultiplier (Hamamatsu, H6780) and recorded by a transient digitizer (LeCroy 9420). A laser pulse (10 ns, 532 nm), produced by a 10 Hz Nd-doped yttrium–aluminum-garnet (YAG) laser (Continuum, Inc.), is used to photolyze the sample with a typical pulse energy of 25 mJ. The polarization of the photolysis beam is scrambled using quartz wedges to eliminate the rotational relaxation artifact.
A 3-state kinetic model49 is used to describe the ligand rebinding process after photolysis of CO from the Fe–PPIX
| (1) |
where the FePPIX:CO represents a contact pair state50 that can undergo geminate recombination and FePPIX + CO represents the state with CO free in solution. Within this simple 3 state model (under the condition [FePPIX] ≪ [CO]), the absorption change as a function of time can be expressed as
| (2) |
with 0 ≤ β ≤ 1, accounting for short time scale relaxation processes. The observed geminate rate kg = kBA + kout is the sum of the intrinsic rate of bond formation and the ligand escape rate. The geminate yield is Ig = {kBA/(kBA + kout)}.
Results and Analysis
Equilibrium Spectra
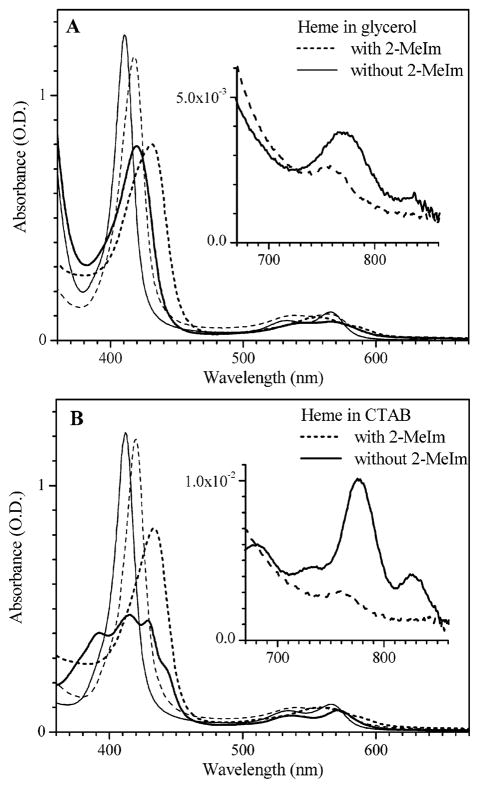
The absorption spectra of the reduced (or “deoxy”) state of FePPIX in 80% glycerol is shown in Figure 1A. The spectra with and without the proximal ligand (2-methylimidazole, 2-MeIm), show the characteristic asymmetric line shape of a five-coordinate heme.51 The thick solid line peaking at 420 nm corresponds to the sample with no added ligand and the thick dashed line peaking at 432 nm corresponds to the 2-MeIm adduct. The other lines in Figure 1A correspond to the samples with (thin dashed) and without (thin solid) 2-MeIm, following the addition of CO, peaking at 418 and 411 nm, respectively. As can be seen, addition of the 2-methylimidazole shifts the Soret peaks of both the deoxy and CO-bound FePPIX species to the red, very near the analogous Soret peaks found in native Mb. Figure 1B shows the corresponding spectra in CTAB solution. The multiple Soret peaks of the deoxy form without external ligand have also been observed in benzene,52 and assigned to a four-coordinated heme. However, the four-coordinate heme in partially unfolded deoxy Mb has a single Soret peak at 383 nm35. Raman vibrational markers provide a more definite test for heme coordination and suggest (see below) a mixture of four- and five-coordinate species for FePPIX in CTAB solution in the absence of imidazole.
Figure 1.
Equilibrium spectra of protoheme in 80% (v/v) glycerol (A) and 1% (w/v) CTAB (B). Dashed and solid lines are samples with and without the 2-MeIm ligand. The thin and thick lines are for CO-bound and deoxy protoheme, respectively. Soret peak positions for heme in glycerol without 2-MeIm are 420 nm (deoxy) and 411 nm (CO bound). When 2-MeIm is bound the Soret peaks are 432 nm (deoxy) and 418 nm (CO bound). For CO bound heme in CTAB in the absence of 2-MeIm, the Soret peak is found at 412 nm. When 2-MeIm is bound to heme in CTAB, the Soret peaks are at 434 nm (deoxy) and 420 nm for CO bound. The multiple peaks seen in the Soret region for FePPIX in CTAB in the absence of 2-MeIm and CO are located at 392, 415, 430, and 444 nm.
Band III, shown in the insert, is found near 760 nm in the presence of 2-MeIm for both glycerol and CTAB solutions and is typical of a five-coordinated heme species.9,31,53–62 This band shifts to 770 nm in the absence of 2-MeIm as indicative of four-coordinate hemes.35,63 Another band near 840 nm also appears when the imidazole ligand is absent and sharpens at low temperature (not shown). Several studies have identified correlations between the band III frequency and diatomic ligand binding rates.9,31,55,56 More specifically, the iron displacement from the mean plane of the heme has been proposed as a common underlying structural parameter that influences both the band III frequency, the Fe–His frequency, and ligand binding rates.31,56,57,62,64 On the other hand, it should be recognized that local electric fields can also influence the band III frequency.59,60 Recently, a strong correlation between the Fe displacement and the oscillator strength of band III has been used61 to explain the unusually strong temperature dependence observed31 for band III in myoglobin. The proposed correlations61 suggest that the blue shift and intensity decrease of band III, which are observed when 2-MeIm binds and displaces the water ligand to Fe(PPIX), reflect an increased Fe displacement from the heme plane and thus a higher barrier to CO binding.31,32 Measurements of geminate CO rebinding to these compounds, described below, allow a direct test of this hypothesized correlation.
Transient Spectra
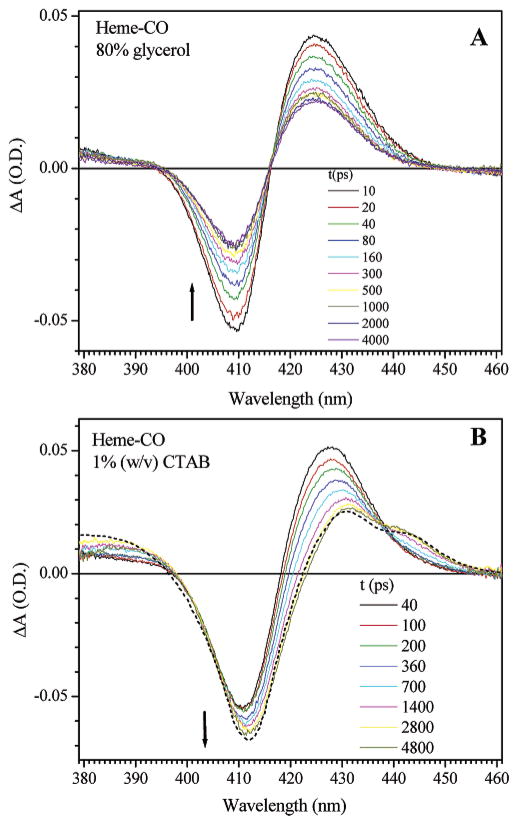
The upper part of Figure 2 shows the transient absorption difference spectra following CO photolysis from the bare heme in glycerol buffer. The transient spectra show the characteristics of a simple two state rebinding dynamics, with fixed isosbestic points and with a photoproduct spectrum that is nearly identical to corresponding equilibrium deoxy species. In contrast, the transient spectra of heme in CTAB solution display a dynamic evolution of the photoproduct in the 420–450 nm region. The initial photoproduct is close to that in glycerol solution, but is different from the equilibrium deoxy Mb species in CTAB. Within the first five ns, the initial photoproduct in CTAB does not rebind with CO, as is shown by the increase in the bleach signal. It instead relaxes (with a time constant of about 1 ns) to a four-coordinate and/or weakly bound five-coordinate equilibrium deoxy state, as is shown by the equilibrium difference spectrum (dashed line) and the characteristic multiple Soret peaks.
Figure 2.
Transient absorption spectra of photolyzed FePPIX-CO. Panel A shows a sample in 80% (v/v) glycerol solution. Panel B shows data for a sample in 1% (w/v) CTAB solution, the dashed line in B is the scaled equilibrium difference spectra between the deoxy and CO bound sample (from Figure 1B without 2-MeIm).
Vibrational Spectra
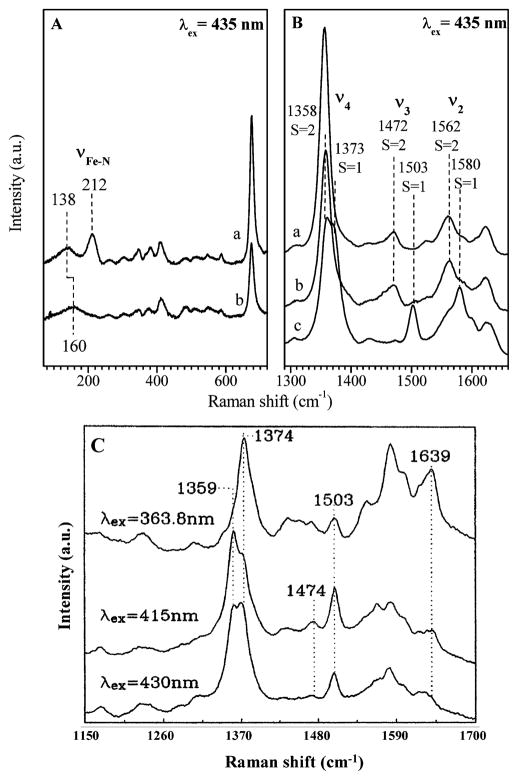
Figure 3A and 3B show the low and high-frequency Raman spectra of the deoxy FePPIX excited at 435 nm. For heme in glycerol, the core marker bands show features characteristic of a high-spin five-coordinated species65 with or without the addition of 2-methylimidazole. The 2-MeIm adduct shows a strong υ(Fe–NIm) mode at 212 cm−1, demonstrating the ligation of the imidazole ligand. When excited at 435 nm, as shown in Figure 3B(c), the resonance Raman spectrum of deoxy FePPIX in CTAB solution without the addition of 2-methylimidazole shows mainly features of intermediate spin, which is characteristic of a four coordinated heme species. The spectra also show minor features of a high spin species, most notably the doublet in the υ4 region indicates a mixture of both four and five coordinate species. A more detailed Raman study of deoxy FePPIX in CTAB solution, which involved varying the excitation wavelength, is shown in Figure 3C. A minority species of five coordinate heme manifests its presence when the laser excitation is tuned to 415 nm and the high-spin (υ3) marker band at 1474 cm−1 is clearly resolved. As the excitation moves further to the blue near 364 nm, the intermediate spin species again dominates the spectra. This behavior is consistent with the multiple peaked nature of the absorption spectrum shown in Figure 1B and suggests that a 5-coordinate heme minority species, absorbing near 420 nm (e.g., see Figure 1A), underlies the multiple peaked 4-coordinate heme absorption spectrum observed in CTAB solution.
Figure 3.
Panels A and B show the resonance Raman spectra of deoxy FePPIX in glycerol with (a) and without (b) 2-MeIm. Deoxy FePPIX in CTAB without 2-MeIm is shown in (c) of panel B and as a function of excitation wavelength in panel C.
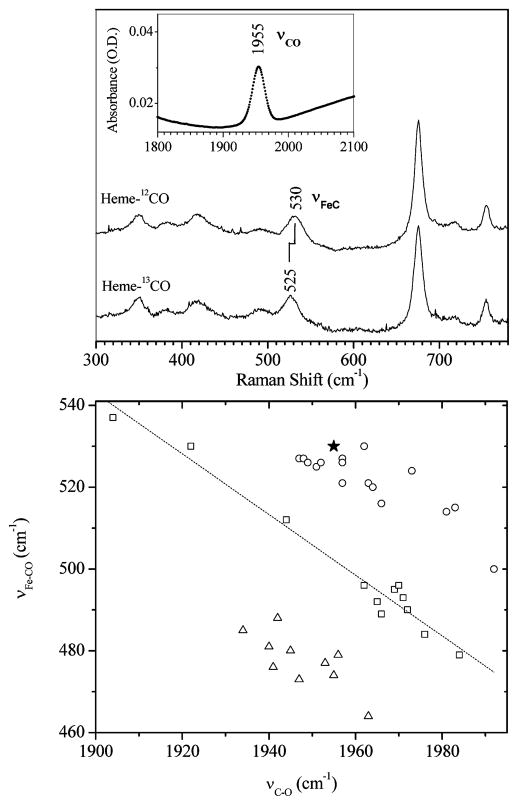
In Figure 4, we show the Raman and infrared data for the CO bound heme species in glycerol (without adding 2-MeIm). The correlation of υ(C–O) and υ(Fe–CO) indicates the proximal ligand is either weak66 or absent.67 The correlation curve probably cannot effectively separate a five coordinate CO bound state from a six coordinate CO species with a weak proximal ligand (e.g., water).
Figure 4.
Upper panel shows measurements of FePPIX-CO in 80% (v/v) glycerol solution. The υ(Fe-CO) assignment is made using isotopic labeling and resonance Raman spectroscopy (excitation at 413 nm). The υ(C–O) mode is detected using infrared absorption (inset). The lower panel is a correlation plot of υ(Fe–CO) and υ(C–O), with all data points except the solid star (present work) from Vogel et al.67 The open squares are data for imidazole (or histidine) ligated heme systems and the dashed line is a fit to the data. The open triangles represent thiolate ligated heme systems and the open circles and the solid star (this work) represent heme systems with weak or absent proximal ligands.
CO Rebinding Kinetics
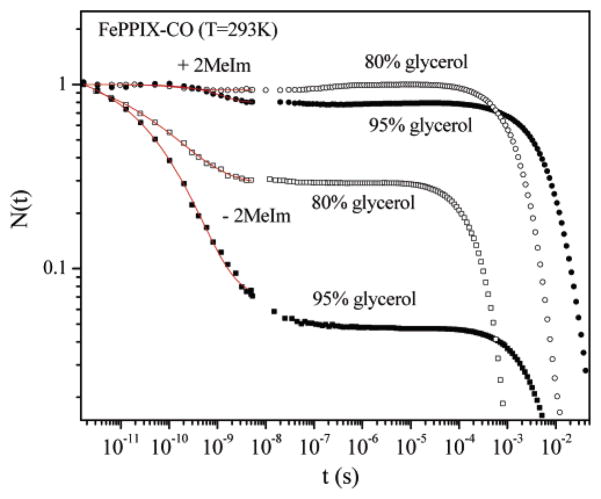
As can be seen from Figure 5 and Table 1, the high viscosity of the glycerol samples mainly affects the CO escape rate, kout, (see eq 1) and only weakly affects the bond formation rate, kBA (see eq 1). This same conclusion was also arrived at in earlier studies.68 Addition of 2-MeIm as a proximal ligand significantly reduces the geminate rebinding rate and the amplitude of the geminate process. For heme without a strong proximal ligand, kBA is about 2 orders of magnitude larger than for the heme with 2-methylimidazole. The CO rebinding barrier difference between the proximally ligated (with 2-MeIm) and unligated (no 2-MeIm added) states can be found from the rates in Table 1:
Figure 5.
FePPIX-CO recombination kinetics: in 80% (v/v) glycerol solution, with (open circle) and without (open rectangle) 2-MeIm; in 95% (v/v) glycerol solution, with (solid circle) and without (solid rectangle) 2-MeIm. The results of fitting the data are given in Table 1.
Table 1.
Kinetics of CO Rebinding in Glycerol Solution
| kBA (s−1) | kout (s−1) | kg (s−1) | Ig | β | ks (s−1) | |||
|---|---|---|---|---|---|---|---|---|
| Heme-CO | 80% glycerol | − 2-MeIm | 1.5 × 1010 | 5 × 109 | 2 × 1010 | 0.75 | 0.33 | 3.3 × 103 |
| + 2-MeIm | 3.3 × 108 | 4.4 × 109 | 4.7 × 109 | 0.07 | 1 | 5 × 102 | ||
| 95% glycerol | − 2-MeIm | 1.9 × 1010 | 1 × 109 | 2 × 1010 | 0.95 | 0.36 | 1.9 × 102 | |
| + 2-MeIm | 1.7 × 108 | 6.7 × 108 | 8.3 × 108 | 0.2 | 1 | 1.1 × 102 | ||
| 99% EG | − 2-MeIm | 1.5 × 1010 | 2.5 × 1010 | 4 × 1010 | 0.38 | 0.37 | ||
| Mb-CO | 75% glycerol | 2.8 × 106 | 2 × 107 | 2.3 × 107 | 0.12 | 0.5 | 6.5 × 102 |
For 80% glycerol
| (3a) |
For 95% glycerol
| (3b) |
where we have assumed that the entropic contribution to the free energy barrier is independent of the proximal ligation state (i.e., the spin entropy changes are common to both reactions as seen by the characteristic spin-sensitive spectroscopic indicators in Figures 1A, 3, and 4). Thus, the proximal contribution to the difference in the enthalpic rebinding barriers at 293 K is found to be ΔHP = 9.5 kJ/mol (80% glycerol) and ΔHP = 11.2 kJ/mol (95% glycerol), leading us to conclude
| (4) |
FePPIX complexed with 2-MeIm in glycerol, in the absence of a distal pocket protein matrix, also shows a faster rebinding rate than when the heme is embedded in the protein matrix (i.e., Mb). In the latter case, barriers associated with the distal pocket will also retard the CO rebinding process. The ratio of the rebinding rates with and without the protein matrix can be used to find the free energy difference in the rebinding barrier due to the presence of the distal pocket and protein matrix.
| (5) |
Thus, the difference in the free energy barrier at 293 K for CO rebinding due to the presence of the distal pocket in Mb is approximately
| (6) |
where we explicitly acknowledge (through ΔSD in eq 5) that the entropic contribution to the distal barrier may differ between the glycerol solution and the protein matrix.
Discussion
Ligation States of Fe-PPIX in Glycerol
Protoheme in glycerol solution without the addition of 2-MeIm has a weak proximal ligand, derived either from water or glycerol.52,63,69,70 For example, the Soret band in Figure 1 displays an asymmetric line shape, characteristic of a five-coordinate heme,51 but blue shifted to 420 nm as might be expected for a weak fifth ligand. In addition, the Raman core-size marker bands71 of PPIX in glycerol are indicative of a high-spin, five coordinated heme and are very similar to the marker bands we observe when 2-MeIm is added (see Figure 3). In contrast, the Raman spectra of CTAB-encapsulated Fe(PPIX) show marker bands characteristic of both 4- and 5-coordinate Fe(II) hemes, indicating that the weak ligand is probably water, which is destabilized in the hydrophobic interior of the CTAB micelle.
The spectral evidence indicates that the CO-bound PPIX species in both the CTAB and the glycerol samples is six-coordinated with a weak proximal ligand from water or OH. Photolysis of CO does not dissociate the weak proximal ligand initially. In glycerol solution, the CO has a much smaller escape rate due to the glycerol viscosity, so that geminate rebinding of CO takes place before any other proximal ligation changes occur. On the other hand, for samples in the aqueous CTAB solution, the viscosity is much lower so that CO can escape more efficiently. When this happens, a dynamic dissociation of the weak proximal ligand can take place in the first few ns because the affinity of the heme iron for the proximal ligand (probably water) in the hydrophobic micelle environment is lowered when the CO bond is broken.
Figure 4 displays the correlation between the bound C–O and the Fe–CO vibrational frequencies in the absence of 2-MeIm and demonstrates that, when the CO complex is formed in glycerol, the proximal ligand bound trans to the CO is either weak or absent. The deoxy Fe–PPIX sample in glycerol also has a red-shifted (and more intense) band III (see Figure 1), which is consistent with a more planar conformation of the heme, as is expected for a high-spin ferrous heme with a weak (or absent) proximal ligand. Finally, the Raman spectra show that the heme is coordinated by a weak fifth ligand, even in the absence of CO and 2-MeIm. This fact, along with the ca. 100-fold increase in heme affinity for water observed following CO binding to deuterioheme,70 suggests that (in the absence of added imidazole) water is likely bound as a weak proximal ligand in the CO bound complex as well as in the deoxy state.
When 2-MeIm is added to Fe-PPIX in glycerol solution, the ligation state remains five coordinate because the steric interactions between the 2-methyl group and the heme prevent the bis (six coordinate) complex from being formed.69 The Soret bands with and without CO (see Figure 1) mimic very closely the spectra observed for native Mb. The low-frequency Raman spectra, shown in Figure 3, locate the Fe–NIm mode at 212 cm−1 and give definitive evidence that the 2-MeIm has bound to form the 5 coordinate complex. Band III of the 2-MeIm FePPIX complex is also located at 760 nm, as observed in deoxy Mb.
In summary, the ligation properties of the model system composed of FePPIX in glycerol is remarkably well-behaved. When 2-MeIm is added, the sample displays optical and Raman spectra that mimic very closely what is observed for native Mb. More importantly, when no exogenous 2-MeIm ligand is added, the spectral evolution observed following CO photolysis remains characteristic of a simple two state rebinding process (Figure 2, upper). This should be contrasted with the spectral evolution found for FePPIX in CTAB micelles (Figure 2, lower), which displays more complicated behavior, probably involving a ligand switching mechanism.36,37
Entropic Contribution to the Distal Barrier
We can estimate the differences in the entropic contributions to the distal barrier between Mb and the 2-MeIm bound FePPIX by assuming that the entropy at the transition state for CO bond formation to the heme, S†D, is independent of the details of the distal architecture for the different samples (i.e., S†D(Mb) ≈ S†D(+Im) in eq 5). Thus, the difference in the entropic contribution to the distal free energy barrier in eq 5 is given by
| (7) |
where SD(Mb) = k ln ΩMb and SD(+Im) = k ln Ω+Im with Ω denoting the number of possible distal pocket states associated with the CO ligand in its initial configuration prior to geminate recombination. Thus, eq 7 leads directly to
| (8) |
We now take the number of accessible initial states for the CO ligand in Mb, ΩMb, to be proportional to the volume of the region from which the CO ligand must return during the geminate rebinding process. Similarly, the number of possible reactant states for geminate rebinding of the CO ligand to 2-MeImFePPIX in glycerol solvent, Ω+Im, is proportional to the volume of the glycerol solvent that is displaced by the photolyzed CO ligand. In the case of Mb, the ligand accessible volume, relevant for geminate rebinding, can be taken as the sum of the distal and Xe4 pocket volumes,72 which has been quantified73a,b and is found to be ~4.6 Å3. For the FePPIX in glycerol, we approximate73b the accessible volume as the cube of the CO bond length, ca. 1.3 Å3. Taking the ratio and combining eqs 5, 6, and 8, we find TΔSD = kT ln(Ω+Im/ΩMb) = −3.2 kJ/mol so that
| (9) |
Thus, given the various approximations, we conclude
| (10) |
Proximal and Distal Contributions to the Rebinding Barrier
The advantage of systematic studies of a well-behaved model system is that we can separately assess the proximal and distal contributions to the enthalpic rebinding barrier. Dlott and co-workers measured the temperature dependence of the CO rebinding to FePPIX in glycerol (with no added ligand) and found that the barrier for rebinding, H0BA, was quite small (~1 kJ/mol).44,73c This allows both the proximal contribution to the barrier (due to the imidazole ligation) and the distal contribution (due to the protein matrix) to be directly assessed by using eqs 4 and 10.
The weak proximal ligand, observed in the absence of the 2-MeIm, is consistent with a more in-plane position of the heme iron and a much lower rebinding barrier. Thus, we denote the barrier for FePPIX-CO in glycerol, associated with kBA(−Im), as H0BA. Then the definition of ΔHP in eq 3 leads to ΔHP = HP − H0BA so that the absolute enthalpic barrier associated with the proximal imidazole ligation can be found using HP = ΔHP + H0BA. Here, we have assigned the magnitude44 of H0BA ≈ 1 kJ/mol to the weak proximal ligation of water, which is an approximation that falls within our margin of error. Given the uncertainty of H0BA as measured by Miers et al.,44 and its assignment to the proximal part of the enthalpic barrier, we use eq 4 with an expanded error margin to find
| (11) |
On the other hand, distal effects have also been implicated in the rebinding process and extensive distal pocket mutation studies5,12,74–76 have led some workers to suggest that the distal pocket is the main source of the ligand binding barrier.77–79 We can estimate the distal contribution to the barrier using the same approach as above because the 2-MeIm ligated FePPIX (which has no distal pocket) can be compared to Mb, which does have a distal pocket. Here, as above, we assume that the very small (~1 kJ/mol) rebinding (enthalpy) barrier observed for FePPIX in glycerol44 is purely proximal in origin (due to the weakly bound proximal water ligand), and that the distal contribution, H0D, can be taken to be approximately zero. Taking HD = ΔHD + H0D, and using the ratio of the CO rebinding rates as outlined in eqs 5–10, yields the distal pocket contribution to the barrier
| (12) |
An early analysis32 of the low-temperature CO rebinding to Mb, which also separated the proximal and distal contributions to the rebinding barrier, found that a fit to the low-temperature data7 required a distal term of ~7 kJ/mol32 and a (distributed) proximal term of ~5 kJ/mol. However, at room temperature the photolyzed heme is no longer “quenched” by the frozen protein (below the glass transition of the glycerol solvent) and it can relax to a fully domed out-of-plane position. Using the simple model of Srajer et al.,32 along with a difference between the average iron out-of-plane position for temperatures below (~0.2 Å) and above (~0.4 Å) the solvent glass transition, the room-temperature proximal barrier HP was predicted32 to increase from 5 to 12 kJ/mol. Thus, the overall barrier at room temperature was estimated32 to be HBA ≈ HP + HD ≈ 12kJ/mol + 7kJ/mol ≈ 19 kJ/mol. These early predictions agree surprisingly well with the MbCO rebinding barrier determined80,81 near room temperature (HBA = 18 ± 2 kJ/mol prior to distal relaxation) as well as with the room-temperature measurements using the FePPIX model system presented here (HP + HD ≈ 11 kJ/mol + 9 kJ/mol = 20 kJ/mol). It is worth emphasizing that this same simple model32,64 also predicts a heme doming force constant of 22 ± 3 kcal mol−1 Å−2, associated with the proximal rebinding barrier, which is consistent with recently detected heme doming oscillations near 1 THz.82–84
In conclusion, we emphasize that a systematic analysis of CO rebinding to a “well behaved” heme model compound has been carried out so that the magnitude of the proximal and distal contributions to the rebinding barrier can be separately determined. The results can be cross-checked by noting that the sum of the two contributions HBA = HP + HD ≈ 20 kJ/mol is very close to independent measurements80,81 of the MbCO rebinding barrier (HBA = 18 ± 2 kJ/mol) and to the room temperature barrier predicted by an early model.32,80
Acknowledgments
This work is supported by NSF 0211816 and NIH DK035090 (PMC) and by NSF0240955 and NIH GM-52002 (JTS).
References
- 1.Reedy CJ, Gibney BR. Chem Rev. 2004;104:617–649. doi: 10.1021/cr0206115. [DOI] [PubMed] [Google Scholar]
- 2.Karow DS, Pan D, Tran R, Pellicena P, Presley A, Mathies RA, Marletta MA. Biochemistry. 2004;43:10203–10211. doi: 10.1021/bi049374l. [DOI] [PubMed] [Google Scholar]
- 3.Sage JT. Hemoglobins: O2 Uptake and Transport. In: Atwood JL, Steed JW, editors. Encyclopedia of Supramolecular Chemistry. Marcel Dekker; New York: 2004. pp. 636–644. [Google Scholar]
- 4.Sage JT, Champion PM. Small Substrate Recognition in Heme Proteins. In: Suslick KS, editor. Comprehensive Supramolecular Chemistry. Pergamon; Oxford, U. K: 1996. pp. 171–218. [Google Scholar]
- 5.Springer BA, Sligar SG, Olson JS, Phillips GN., Jr Chem Rev. 1994;94:699–714. [Google Scholar]
- 6.Antonini E, Brunori M. Hemoglobin and Myoglobin in Their Reactions with Ligands. North-Holland Publishing Co; Amsterdam-London: 1971. [Google Scholar]
- 7.Austin RH, Beeson KW, Eisenstein L, Frauenfelder H, Gunsalus IC. Biochemistry. 1975;14:5355–5373. doi: 10.1021/bi00695a021. [DOI] [PubMed] [Google Scholar]
- 8.Carver TE, Rohlfs RJ, Olson JS, Gibson QH, Blackmore RS, Springer BA, Sligar SG. J Biol Chem. 1990;265:20007–20020. [PubMed] [Google Scholar]
- 9.Campbell BF, Chance MR, Friedman JM. Science. 1987;238:373–376. doi: 10.1126/science.3659921. [DOI] [PubMed] [Google Scholar]
- 10.Franzen S. J Phys Chem B. 2002;106:4533–4542. [Google Scholar]
- 11.Walda KN, Liu XY, Sharma VS, Magde D. Biochemistry. 1994;33:2198–2209. doi: 10.1021/bi00174a029. [DOI] [PubMed] [Google Scholar]
- 12.Olson JS, Phillips GN., Jr J Biol Chem. 1996;271:17593–17596. doi: 10.1074/jbc.271.30.17593. [DOI] [PubMed] [Google Scholar]
- 13.Scott EE, Gibson QH, Olson JS. J Biol Chem. 2001;276:5177–5188. doi: 10.1074/jbc.M008282200. [DOI] [PubMed] [Google Scholar]
- 14.Nienhaus K, Deng P, Olson JS, Warren JJ, Nienhaus GU. J Biol Chem. 2003;278:42532–42544. doi: 10.1074/jbc.M306888200. [DOI] [PubMed] [Google Scholar]
- 15.Cao W, Ye X, Sjodin T, Christian JF, Demidov AA, Berezhna S, Wang W, Barrick D, Sage JT, Champion PM. Biochemistry. 2004;43:11109–11117. doi: 10.1021/bi049077g. [DOI] [PubMed] [Google Scholar]
- 16.Hasinoff BB. Arch Biochem Biophys. 1981;211:396–402. doi: 10.1016/0003-9861(81)90470-7. [DOI] [PubMed] [Google Scholar]
- 17.Hill JR, Cote MJ, Dlott DD, Kauffman JF, Macdonald JD, Steinbach PJ, Berendzen JR, Frauenfelder H. Springer Series in Chemical Physics. 1986;46(Ultrafast Phenom 5):433–435. [Google Scholar]
- 18.White DK, Cannon JB, Traylor TG. J Am Chem Soc. 1979;101:2443–2454. [Google Scholar]
- 19.Traylor TG, Magde D, Taube DJ, Jongeward KA, Bandyopadhyay D, Luo JK, Walda KN. J Am Chem Soc. 1992;114:417–429. [Google Scholar]
- 20.Miers JB, Postlewaite JC, Cowen BR, Roemig GR, Lee IYS, Dlott DD. J Chem Phys. 1991;94:1825–1836. [Google Scholar]
- 21.Huang Y, Marden MC, Lambry JC, Fontaineaupart MP, Pansu R, Martin JL, Poyart C. J Am Chem Soc. 1991;113:9141–9144. [Google Scholar]
- 22.Tetreau C, Lavalette D, Momenteau M, Lhoste JM. Proc Natl Acad Sci USA. 1987;84:2267–2271. doi: 10.1073/pnas.84.8.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cao W, Ye X, Georgiev GY, Berezhna S, Sjodin T, Demidov AA, Wang W, Sage JT, Champion PM. Biochemistry. 2004;43:7017–7027. doi: 10.1021/bi0497291. [DOI] [PubMed] [Google Scholar]
- 24.Tian WD, Sage JT, Champion PM. J Mol Biol. 1993;233:155–166. doi: 10.1006/jmbi.1993.1491. [DOI] [PubMed] [Google Scholar]
- 25.Lim M, Jackson TA, Anfinrud PA. Nat Struct Biol. 1997;4:209–214. doi: 10.1038/nsb0397-209. [DOI] [PubMed] [Google Scholar]
- 26.Brunori M, Vallone B, Cutruzzolà F, Travaglini-Allocatelli C, Berendzen J, Chu K, Sweet RM, Schlichting I. Proc Natl Acad Sci USA. 2000;97:2058–2063. doi: 10.1073/pnas.040459697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tian WD, Sage JT, Champion PM, Chien E, Sligar SG. Biochemistry. 1996;35:3487–3502. doi: 10.1021/bi952474u. [DOI] [PubMed] [Google Scholar]
- 28.Elber R, Karplus M. J Am Chem Soc. 1990;112:9161–9175. [Google Scholar]
- 29.Cao W, Christian JF, Champion PM, Rosca F, Sage JT. Biochemistry. 2001;40:5728–5737. doi: 10.1021/bi010067e. [DOI] [PubMed] [Google Scholar]
- 30.Schotte F, Lim M, Jackson TA, Smirnov AV, Soman J, Olson JS, Phillips GN, Jr, Wulff M, Anfinrud PA. Science. 2003;300:1944–1947. doi: 10.1126/science.1078797. [DOI] [PubMed] [Google Scholar]
- 31.Srajer V, Champion PM. Biochemistry. 1991;30:7390–7402. doi: 10.1021/bi00244a005. [DOI] [PubMed] [Google Scholar]
- 32.Srajer V, Reinisch L, Champion PM. J Am Chem Soc. 1988;110:6656–6670. [Google Scholar]
- 33.Tilton RF, Jr, Kuntz ID, Jr, Petsko GA. Biochemistry. 1984;23:2849–2857. doi: 10.1021/bi00308a002. [DOI] [PubMed] [Google Scholar]
- 34.Decatur SM, DePillis GD, Boxer SG. Biochemistry. 1996;35:3925–3932. doi: 10.1021/bi952625t. [DOI] [PubMed] [Google Scholar]
- 35.Sage JT, Morikis D, Champion PM. Biochemistry. 1991;30:1227–1237. doi: 10.1021/bi00219a010. [DOI] [PubMed] [Google Scholar]
- 36.Sage JT, Li PS, Champion PM. Biochemistry. 1991;30:1237–1247. doi: 10.1021/bi00219a011. [DOI] [PubMed] [Google Scholar]
- 37.Franzen S, Bailey J, Dyer RB, Woodruff WH, Hu RB, Thomas MR, Boxer SG. Biochemistry. 2001;40:5299–5305. doi: 10.1021/bi0023403. [DOI] [PubMed] [Google Scholar]
- 38.Franzen S, Peterson ES, Brown D, Friedman JM, Thomas MR, Boxer SG. Eur J Biochem. 2002;269:4879–4886. doi: 10.1046/j.1432-1033.2002.03193.x. [DOI] [PubMed] [Google Scholar]
- 39.White WI. In: The Porphyrins. Dolphin D, editor. Academic; NewYork: 1979. [Google Scholar]
- 40.Simplicio J, Schwenzer K. Biochemistry. 1973;12:1923–1929. doi: 10.1021/bi00734a014. [DOI] [PubMed] [Google Scholar]
- 41.Medhi OK, Mazumdar S, Mitra S. Inorg Chem. 1989;28:3243–3248. [Google Scholar]
- 42.Larsen RW. Inorg Chim Acta. 1999;288:74–81. [Google Scholar]
- 43.Beece D, Eisenstein L, Frauenfelder H, Good D, Marden MC, Reinisch L, Reynolds AH, Sorensen LB, Yue KT. Biochemistry. 1980;19:5147–5157. doi: 10.1021/bi00564a001. [DOI] [PubMed] [Google Scholar]
- 44.Miers JB, Postlewaite JC, Zyung T, Chen S, Roemig GR, Wen X, Dlott DD, Szabo A. J Chem Phys. 1990;93:8771–8776. [Google Scholar]
- 45.Moore JN, Hansen PA, Hochstrasser RM. Proc Natl Acad Sci USA. 1988;85:5062–5066. doi: 10.1073/pnas.85.14.5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang W, Ye X, Demidov AA, Rosca F, Sjodin T, Cao W, Sheeran M, Champion PM. J Phys Chem B. 2000;104:10789–10801. [Google Scholar]
- 47.Ye X, Demidov AA, Champion PM. J Am Chem Soc. 2002;124:5914–5924. doi: 10.1021/ja017359n. [DOI] [PubMed] [Google Scholar]
- 48.Hansen PA, Moore JN, Hochstrasser RM. Chem Phys. 1989;131:49–62. [Google Scholar]
- 49.Henry ER, Sommer JH, Hofrichter J, Eaton WA. J Mol Biol. 1983;166:443–451. doi: 10.1016/s0022-2836(83)80094-1. [DOI] [PubMed] [Google Scholar]
- 50.Jongeward KA, Magde D, Taube DJ, Marsters JC, Traylor TG, Sharma VS. J Am Chem Soc. 1988;110:380–387. [Google Scholar]
- 51.Srajer V, Schomacker KT, Champion PM. Phys Rev Lett. 1986;57:1267–1270. doi: 10.1103/PhysRevLett.57.1267. [DOI] [PubMed] [Google Scholar]
- 52.Brault D, Rougee M. Biochemistry. 1974;13:4591–4597. doi: 10.1021/bi00719a019. [DOI] [PubMed] [Google Scholar]
- 53.Iizuka T, Yamamoto H, Kotani M, Yonetani T. Biochim Biophys Acta. 1974;371:126–139. doi: 10.1016/0005-2795(74)90161-5. [DOI] [PubMed] [Google Scholar]
- 54.Eaton WA, Hanson LK, Stephens PJ, Sutherland JC, Dunn JBR. J Am Chem Soc. 1978;100:4991–5003. [Google Scholar]
- 55.Agmon N. Biochemistry. 1988;27:3507–3511. doi: 10.1021/bi00409a057. [DOI] [PubMed] [Google Scholar]
- 56.Ahmed AM, Campbell BF, Caruso D, Chance MR, Chavez MD, Courtney SH, Friedman JM, Iben IET, Ondrias MR, Yang M. Chem Phys. 1991;158:329–351. [Google Scholar]
- 57.Rich PR, Moody AJ, Ingledew WJ. FEBS Lett. 1992;305:171–173. doi: 10.1016/0014-5793(92)80659-5. [DOI] [PubMed] [Google Scholar]
- 58.Christian JF, Unno M, Sage JT, Champion PM, Chien E, Sligar SG. Biochemistry. 1997;36:11198–11204. doi: 10.1021/bi9710075. [DOI] [PubMed] [Google Scholar]
- 59.Franzen S, Moore LJ, Woodruff WH, Boxer SG. J Phys Chem B. 1999;103:3070–3072. [Google Scholar]
- 60.Nienhaus K, Lamb DC, Deng P, Nienhaus GU. Biophys J. 2002;82:1059–1067. doi: 10.1016/s0006-3495(02)75465-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stavrov SS. Biopolymers. 2004;74:37–40. doi: 10.1002/bip.20039. [DOI] [PubMed] [Google Scholar]
- 62.Kiger L, Stetzkowski-Marden F, Poyart C, Marden MC. Eur J Biochem. 1995;228:665–668. doi: 10.1111/j.1432-1033.1995.tb20307.x. [DOI] [PubMed] [Google Scholar]
- 63.Brault D, Rougee M. Biochemistry. 1974;13:4598–4602. doi: 10.1021/bi00719a020. [DOI] [PubMed] [Google Scholar]
- 64.Champion PM. J Raman Spectrosc. 1992;23:557–567. [Google Scholar]
- 65.Parthasarathi N, Hansen C, Yamaguchi S, Spiro TG. J Am Chem Soc. 1987;109:3865–3871. [Google Scholar]
- 66.Ray GB, Li XY, Ibers JA, Sessler JL, Spiro TG. J Am Chem Soc. 1994;116:162–176. [Google Scholar]
- 67.Vogel KM, Kozlowski PM, Zgierski MZ, Spiro TG. Inorg Chim Acta. 2000;297:11–17. [Google Scholar]
- 68.Marden MC, Hazard ESI, Gibson QH. Biochemistry. 1986;25:2786–2792. doi: 10.1021/bi00358a008. [DOI] [PubMed] [Google Scholar]
- 69.Brault D, Rougee M. Biochem Biophys Res Commun. 1974;57:654–659. doi: 10.1016/0006-291x(74)90596-8. [DOI] [PubMed] [Google Scholar]
- 70.Rougee M, Brault D. Biochemistry. 1975;14:4100–4106. [Google Scholar]
- 71.Spiro TG, Li HY. Resonance Raman Spectroscopy of Metalloporphyrins. In: Spiro TG, editor. Biological Applications of Raman Spectroscopy. Wiley-Interscience Publication; New York, Chichester, Brisbane, Toronto, Singapore: 1988. pp. 1–37. [Google Scholar]
- 72.Hummer G, Schotte F, Anfinrud PA. Proc Natl Acad Sci U S A. 2004;101:15330–15334. doi: 10.1073/pnas.0405295101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liang J, Edelsbrunner H, Fu P, Sudhakar PV, Subramaniam S. Proteins. 1998;33:18–29.Alternatively, as an estimate of the minimum total volume explored by the dissociated ligand we can take the 45% occupancy factor ( Srajer V, et al. Biochemistry. 40:13802–13815. doi: 10.1021/bi010715u.) found for CO localized in the distal pocket volume of 1.4 Å3. This leads to Vmin(Mb) ~ 1.4 Å3/0.45 = 3.1 Å3. For FePPIX in glycerol, the ligand accessible volume is expected to be smaller than the 1.4 Å3 distal pocket of the protein, so the 1.3 Å3 bond length cube is probably an upper limit. (c) Recent temperature dependent ligand rebinding kinetics experiments performed in our lab suggest that this enthalpy barrier may be as high as 3 kJ/mol.
- 74.Olson JS, Mathews AJ, Rohlfs RJ, Springer BA, Egeberg KD, Sligar SG, Tame J, Renaud JP, Nagai K. Nature. 1988;336:265–266. doi: 10.1038/336265a0. [DOI] [PubMed] [Google Scholar]
- 75.Sugimoto T, Unno M, Shiro Y, Dou Y, Ikeda-Saito M. Biophys J. 1998;75:2188–2194. doi: 10.1016/S0006-3495(98)77662-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Draghi F, Miele AE, Travaglini-Allocatelli C, Vallone B, Brunori M, Gibson QH, Olson JS. J Biol Chem. 2002;277:7509–7519. doi: 10.1074/jbc.M109206200. [DOI] [PubMed] [Google Scholar]
- 77.Lim M, Jackson TA, Anfinrud PA. J Biol Inor Chem. 1997;2:531–536. [Google Scholar]
- 78.Lim M, Jackson TA, Anfinrud PA. Time-Resolved Infrared Studies of Ligand Dynamics in Heme Proteins. In: Fayer MD, editor. Ultrafast Infrared and Raman Spectroscopy. Marcel Dekker; New York: 2001. pp. 191–226. [Google Scholar]
- 79.McMahon BH, Stojkovic BP, Hay PJ, Martin RL, Garcia AE. J Chem Phys. 2000;113:6831–6850. [Google Scholar]
- 80.Tian WD, Sage JT, Srajer V, Champion PM. Phys Rev Lett. 1992;68:408–411. doi: 10.1103/PhysRevLett.68.408. [DOI] [PubMed] [Google Scholar]
- 81.Kumar ATN, Zhu L, Christian JF, Demidov AA, Champion PM. J Phys Chem B. 2001;105:7847–7856. [Google Scholar]
- 82.Zhu LY, Sage JT, Champion PM. Science. 1994;266:629–632. doi: 10.1126/science.7939716. [DOI] [PubMed] [Google Scholar]
- 83.Liebl U, Lipowski G, Negrerie M, Lambry JC, Martin JL, Vos MH. Nature. 1999;401:181–184. doi: 10.1038/43699. [DOI] [PubMed] [Google Scholar]
- 84.Rosca F, Kumar ATN, Ionascu D, Ye X, Demidov AA, Sjodin T, Wharton D, Barrick D, Sligar SG, Yonetani T, Champion PM. J Phys Chem A. 2002;106:3540–3552. [Google Scholar]