Abstract
The well-characterized secretory glycoprotein, rice (Oryza sativa) α-amylase isoform I-1 (AmyI-1), was localized within the plastids and proved to be involved in the degradation of starch granules in the organelles of rice cells. In addition, a large portion of transiently expressed AmyI-1 fused to green fluorescent protein (AmyI-1-GFP) colocalized with a simultaneously expressed fluorescent plastid marker in onion (Allium cepa) epidermal cells. The plastid targeting of AmyI-1 was inhibited by both dominant-negative and constitutively active mutants of Arabidopsis thaliana ARF1 and Arabidopsis SAR1, which arrest endoplasmic reticulum-to-Golgi traffic. In cells expressing fluorescent trans-Golgi and plastid markers, these fluorescent markers frequently colocalized when coexpressed with AmyI-1. Three-dimensional time-lapse imaging and electron microscopy of high-pressure frozen/freeze-substituted cells demonstrated that contact of the Golgi-derived membrane vesicles with cargo and subsequent absorption into plastids occur within the cells. The transient expression of a series of C-terminal-truncated AmyI-1-GFP fusion proteins in the onion cell system showed that the region from Trp-301 to Gln-369 is necessary for plastid targeting of AmyI-1. Furthermore, the results obtained by site-directed mutations of Trp-302 and Gly-354, located on the surface and on opposite sides of the AmyI-1 protein, suggest that multiple surface regions are necessary for plastid targeting. Thus, Golgi-to-plastid traffic appears to be involved in the transport of glycoproteins to plastids and plastid targeting seems to be accomplished in a sorting signal–dependent manner.
INTRODUCTION
Cereal α-amylases (EC 3.2.1.1) are typical secretory proteins found in many plants. In germinating cereal seeds, these enzyme molecules are biosynthesized and secreted from the secretory tissues, the scutellar epithelium and the aleurone, to the starchy endosperm, which has undergone programmed cell death. Numerous α-amylase isoforms have been identified in cereals, but the predominant α-amylase isoform I-1 (AmyI-1) in rice (Oryza sativa) is a unique glycoprotein that bears N-linked oligosaccharide side chains (Hayashi et al., 1990; Terashima et al., 1994). The biosynthesis and secretion of AmyI-1 have been extensively investigated: mRNA translation on endoplasmic reticulum (ER) membrane–bound ribosomes, signal sequence–dependent translocation of the ER, core glycosylation in the ER lumen, vesicular transport to the Golgi apparatus, oligosaccharide modification to the complex type, and exocytosis all proceed according to the canonical secretory mechanism (Palade, 1975; Blobel, 1980; Kornfeld and Kornfeld, 1985).
Protein targeting into plastids is an essential cellular event for maintaining plant function and plant life. Plastids, including chloroplasts in green leaves and amyloplasts in starchy cells, contain the genetic machinery required to synthesize their own proteins, although most plastidial proteins are encoded in the nuclear DNA. Nuclear-encoded plastidial proteins are normally synthesized in the cytosol and posttranslationally imported into the organelle. In most cases, precursor proteins are synthesized with an NH2-terminal presequence called a transit peptide (Bruce, 2000; Zhang and Glaser, 2002; Lee et al., 2008). The transit peptide is necessary for and also sufficient for plastidial targeting and translocation initiation. Upon import, the transit peptide is proteolytically removed by a stromal processing peptidase, and the mature protein attains its correct conformation. The transit peptide is recognized by receptors on the chloroplast's surface, which are integral components of the Toc (translocon at the outer envelope of chloroplast) complex. Import across the inner envelope is facilitated by the Tic (translocon at the inner envelope of chloroplast) complex (Schnell and Hebert, 2003; Soll and Schleiff, 2004). Yu et al. (2005) have revealed that the precursor of Arabidopsis thaliana AMY3 (99.7 kD) has a predicted N-terminal transit peptide for plastidial localization, and a 93.5-kD AMY3 protein is present in chloroplasts isolated from Arabidopsis leaves (Yu et al., 2005). In contrast with secretory α-amylase, plastidial AMY3 from Arabidopsis appears to be synthesized in the cytosol as a precursor with the transit peptide, imported through the Toc/Tic apparatus to the stroma, and then processed to the 93.5-kD mature form.
Recently, however, trafficking of glycoproteins and proteins to the plastid via the endomembrane system has been reported (Jarvis, 2008). Several research groups have identified the presence of glycoproteins within the chloroplasts (Gaikwad et al., 1999; Asatsuma et al., 2005). Proteomic analysis of the Arabidopsis chloroplast has provided supporting evidence that there are many proteins (>8% of the total chloroplast proteins) with predicted signal peptides for ER translocation (Kleffmann et al., 2004). A rice α-amylase (αAmy3) exhibited dual localization to plastids and extracellular compartments in plant cells (Chen et al., 1994, 2004). The αAmy3 protein had an ER signal sequence, but no predicted N-glycosylation site. The αAmy3 mRNA was translated in the presence of canine microsomes, and translation products lacking the N-terminal extrapeptide were detected inside the lumen of the organelle. Moreover, a reporter protein with the signal peptide of αAmy3 was mainly targeted to amyloplasts in rice cells (Chen et al., 2004). These results imply the existence of a signal peptide–dependent plastid protein transport pathway through the endomembrane system. More recently, convincing evidence has been presented for a traffic route from the ER-Golgi system to the plastid (Villarejo et al., 2005; Nanjo et al., 2006). Arabidopsis carbonic anhydrase 1 (CAH1), which catalyzes the reversible reaction between carbon dioxide hydration and bicarbonate dehydration, was found to localize in the chloroplast stroma despite having a signal sequence. The CAH1 protein has been shown to contain five predicted N-glycosylation sites, and the mature chloroplast CAH1 was actually N-glycosylated. The oligosaccharide side chain of CAH1 bore β(1,2)-xylose and α(1,3)-fucose residues, which are conjugated in the Golgi apparatus, and the transport of CAH1 to chloroplast was reversibly prevented by a drug, Brefeldin A, which inhibits ER-to-Golgi traffic (Villarejo et al., 2005). Rice nucleotide pyrophosphatase/phosphodiesterase 1 (NPP1), a recently identified enzyme that exhibits hydrolytic activity toward ADP-glucose, which is the substrate for starch biosynthesis, is a chloroplast glycoprotein conjugated with N-linked oligosaccharide chains that are recognized by concanavalin A. Similarly, the enzyme glycoprotein was shown to be transported and localized to the chloroplasts from the secretory pathway via Brefeldin A–sensitive vesicular transport (Nanjo et al., 2006). These results indicate that ER-to-Golgi traffic is essential for chloroplast targeting of glycoproteins in both monocot and dicot plants.
Our previous studies using transgenic rice plants with suppressed expression or overexpression of AmyI-1 revealed that the enzyme glycoprotein is involved in degradation of plastidial starch in living cells (Asatsuma et al., 2005, 2006). Immunocytochemical analysis with specific anti-AmyI-1 antibodies, the expression and targeting of AmyI-1-GFP in redifferentiated green cells, and cell biochemical analysis of chloroplastic AmyI-1 demonstrated that AmyI-1 is present in the chloroplasts of rice leaves. The predicted precursor sequence of AmyI-1 contains the signal peptide for translocating the ER membrane, but no transit peptide. Here, we report a unique transport system involved in the plastidial targeting of the AmyI-1 glycoprotein and identify a plastid targeting signal of AmyI-1 that is common to rice and onion (Allium cepa) cells.
RESULTS
AmyI-1 Degrades Starch Granules in Living Rice Cells
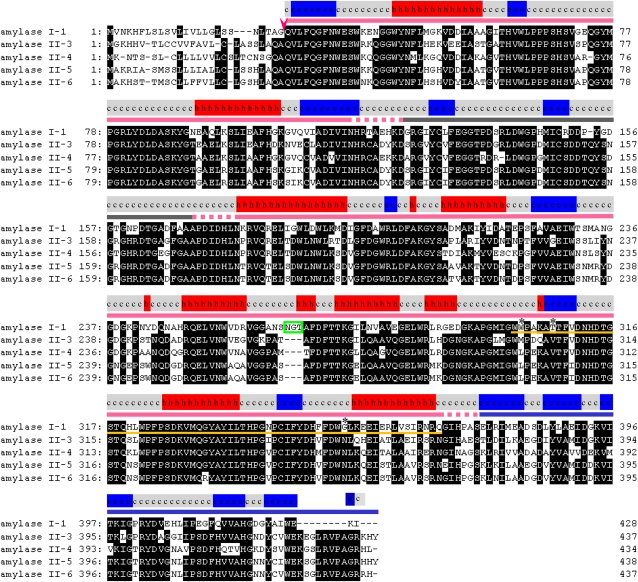
α-Amylases in rice are polymorphic enzymes. The α-amylase isoforms AmyI-1 (RAmy1A, αAmy7), II-3 (RAmy3E, αAmy8), II-4 (RAmy3D, αAmy3), and II-5/6 (RAmy3B/3C, αAmy6) are extensively expressed in rice plants and tissue culture (Mitsui et al., 1996; Nanjo et al., 2004). The amino acid sequence alignment of these isoforms, with a possible secondary structure of AmyI-1 predicted by the Phyre program (Bennett-Lovsey et al., 2008), is shown in Figure 1. The presence of a typical N-terminal signal peptide for ER membrane translocation was confirmed in all precursor forms of these isoforms. The oligosaccharide structure and its conjugating site for AmyI-1 have been determined (Hayashi et al., 1990; Terashima et al., 1994). Thus, there is no doubt that AmyI-1 is a secretory glycoprotein.
Figure 1.
Amino Acid Sequence Alignments of Rice α-Amylase Isoforms.
Arrowhead indicates predicted cleavage sites, and open green box shows a consensus sequence of the N-glycosylation site. Conserved amino acids are boxed in black. A possible secondary structure of AmyI-1, predicted by the Phyre program, is presented on the top of sequence alignment: h, α-helix; e, β-strand; c, coil. Red, gray, and blue underlining represents A, B, and C domains, respectively. Yellow underlining indicates the region important for plastid targeting of AmyI-1. Asterisks represent the three sites of point mutations described in Figure 8.
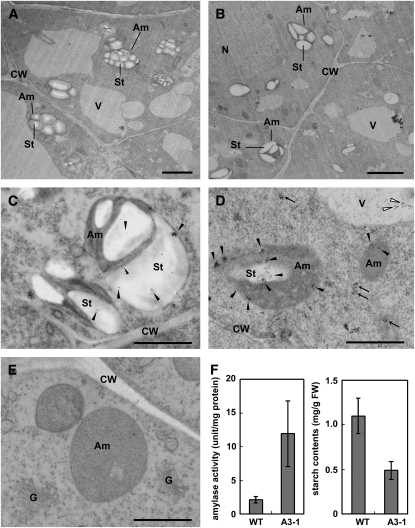
Rice callus cells derived from the embryo portion of the seed actively synthesize and secrete α-amylases under sugar-starved conditions, whereas sucrose supplementation almost entirely prevents the extracellular liberation of AmyI-1 and causes a significant amount of the enzyme molecule to be retained intracellularly (Mitsui et al., 1999). We used callus cells derived from transgenic rice seeds (A3-1 line) with constitutively high levels of AmyI-1 expression to examine the localization and function of AmyI-1 by electron microscopy. In contrast with the ripe starch granules in the amyloplasts of wild-type cells (Figure 2A), the amyloplasts of A3-1 plants had starch granules separated by large spaces under sucrose-supplemented conditions (Figure 2B). Quantitative measurements showed that the starch content in the callus cells overexpressing AmyI-1 was reduced to approximately half of that in the wild type (Figure 2F). To determine the subcellular localization of AmyI-1, we performed immunocytochemical analyses on ultrathin sections of both wild-type and A3-1 cells using anti-AmyI-1 antibodies. Some of the immunogold particles that label AmyI-1 were detected in the amyloplasts of wild-type cells (Figure 2C, black dots). A heavier and more clustered labeling pattern was seen in the amyloplasts and small membrane vesicles of A3-1 cells (Figure 2D). Sections treated with the preimmune sera showed no signal (Figure 2E). Thus, it is evident from these results that AmyI-1 targets and degrades the starch granules in the amyloplasts of living callus cells. The positive signal was also detected in the vacuole (Figure 2D). It has been recently reported that ribulose-1,5-bisphosphate carboxylase/oxygenase and stromal-targeted fluorescent proteins can be mobilized to the vacuole through an autophagic process (Ishida et al., 2008). The AmyI-1 proteins in the vacuole may come from the plastids.
Figure 2.
Electron Microscopy and Biochemical Characterization of Rice A3-1 Callus Cells Constitutively Expressing High Levels of AmyI-1 under Sugar-Supplemented Conditions.
Wild-type ([A] and [C]) and A3-1 ([B], [D], and [E]) cells cultured in Murashige and Skoog (MS) medium containing 3% sucrose at 28°C for 4 d and immediately subjected to fixation. G, Golgi; Am, amyloplast; St, starch; CW, cell wall; V, vacuole; N, nucleus. Bars = 1 μm.
(A) and (B) Morphological observations.
(C) to (E) Immunocytochemical observations. The ultrathin sections were incubated with either an affinity-purified polyclonal antibody against AmyI-1 ([C] and [D]) or preimmune serum (E) and 10-nm colloidal gold-conjugated protein A, sequentially. Arrows and closed and open arrowheads indicate immunogold signals of AmyI-1 in small vesicles, amyloplasts, and vacuoles, respectively.
(F) Quantitative data of amylase activity and starch content in wild-type and A3-1 cells. Values are mean ± sd obtained from three independent experiments.
AmyI-1 Contains a Plastid-Targeting Signal Common to Both Rice and Onion Cells
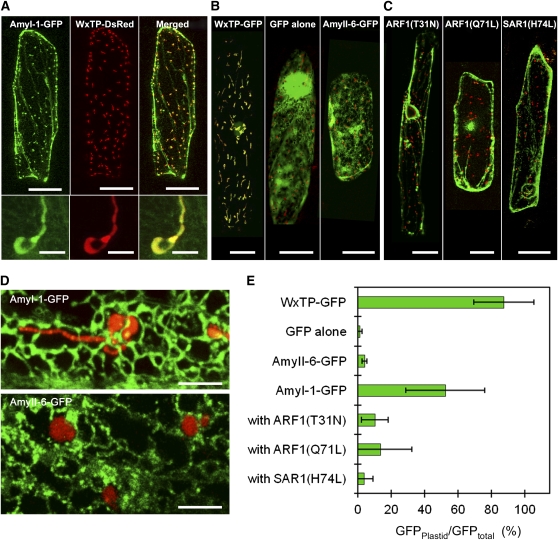
To characterize the nature of plastid localization of AmyI-1 in details, we transiently expressed α-amylase isoforms (I-1, II-3, II-4, II-5, and II-6) fused with green fluorescent protein (GFP) in onion epidermal cells. Since onion cells have no chlorophyllous autofluorescence, it is necessary to visualize the plastids for evaluating the plastid targeting of Amy-GFP. The transit peptide of Waxy (Klösgen and Weil, 1991) fused to either GFP or red fluorescent protein from Discosoma sp (DsRed) was expressed in onion epidermal cells using the particle bombardment procedure. The plastids and stromules extending from their surfaces were visualized with two fluorescence proteins, WxTP-GFP and WxTP-DsRed (see Supplemental Figure 1 online), and their distributions were distinguishable from those of peroxisomes and mitochondria, which were visualized with per-GFP and mt-GFP, respectively (see Supplemental Figure 2 online). Thus, WxTP-GFP and WxTP-DsRed were shown to be suitable plastid markers in onion cells.
AmyI-1-GFP was simultaneously expressed with a plastid marker, WxTP-DsRed. As shown in Figure 3A, a large portion of AmyI-1-GFP fluorescence overlapped with the plastid fluorescence visualized by WxTP-DsRed. Some types of AmyI-1-GFP fluorescence, however, labeled the ER-Golgi network in addition to the inside of the plastids in the cells (Figure 3D, top), suggesting that AmyI-1 is transported to plastids from ER-Golgi in onion cells. The distribution of the other rice amylase isoforms was also tested, but unlike for AmyI-1-GFP, the fluorescence of AmyII-3-, AmyII-4-, AmyII-5-, and AmyII-6-GFP rarely merged with that of the plastid marker (Figure 3B, right panel; see Supplemental Figure 3A online) and was constantly detected in the ER-Golgi network (Figure 3D, bottom panel). Statistical analysis confirmed that the targeting abilities of the other isoforms are much lower than that of AmyI-1 (Figure 3E; see Supplemental Figure 3B online), indicating that the plastid targeting of AmyI-1 is not a result of artificial overexpression of secretory proteins. Thus, AmyI-1 possesses a plastid-targeting signal common to both rice and onion cells.
Figure 3.
Expression and Localization of AmyI-1-GFP in Onion Epidermal Cells.
(A) Onion epidermal cells expressing AmyI-1-GFP and WxTP-DsRed. AmyI-1-GFP, green (left); WxTP-DsRed, red (middle); GFP and DsRed merged (right). AmyI-1-GFP colocalized with the plastid stroma marker WxTP-DsRed. Bars = 100 μm. Bottom panels are close-up views of plastids. Bars = 5 μm.
(B) Control experiments. Labeling with WxTP-DsRed and either WxTP-GFP (left), GFP (middle), or AmyII-6-GFP (right) was performed. Bars = 100 μm.
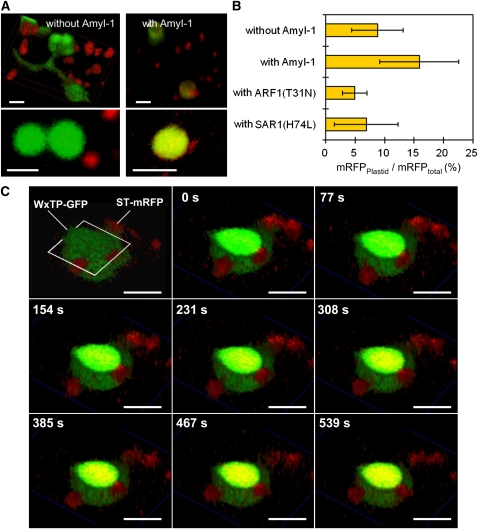
(C) Effects of Arabidopsis ARF1(T31N), ARF1(Q71L), and SAR1(H74L) on the targeting of AmyI-1-GFP into plastids, as visualized with WxTP-DsRed. Bars = 100 μm.
(A) to (C) are projections of stacks of 20 to 30 images per cell, acquired from the top to the middle of the cell, every 1 to 2 μm.
(D) The precise distribution of AmyI-1-GFP and AmyII-6-GFP in onion cells. Top: AmyI-1-GFP and WxTP-DsRed. Extended focus view constructed from 13 sections spanning the entire plastid, every 0.64 μm. Bottom: AmyII-6-GFP and WxTP-DsRed. View constructed from 15 sections, every 0.69 μm. Bars = 5 μm.
(E) Statistical evaluation of the plastidial localization of AmyI-1-GFP. Ratios of the fluorescence intensity of GFP in the plastidial area to GFP in the whole cell (GFPplastid/GFPtotal) (%) were determined. Values are represented as mean ± sd (n = 12).
Membrane Trafficking Is Necessary for Plastid Targeting of AmyI-1
ARF1 and SAR1 GTPases are essential for membrane trafficking between the ER and the Golgi apparatus in tobacco (Nicotiana tabacum) and Arabidopsis cells (Takeuchi et al., 2000, 2002). Expression of dominant-negative or constitutively active mutants of Arabidopsis ARF1 and SAR1, which are defective in GTPase cycling, inhibits the ER-to-Golgi traffic (Takeuchi et al., 2000, 2002). Golgi-resident proteins and secretory and vacuolar proteins are therefore retained in the ER network structure in the presence of such mutants. The effects of Arabidopsis ARF1(T31N), ARF1(Q71L), and SAR1(H74L) mutants on the plastid targeting of AmyI-1-GFP were tested. The transport of AmyI-1-GFP into plastids was severely inhibited by the ARF1 and SAR1 mutants, and the fluorescent proteins remained in the ER network structure (Figure 3C). Punctate structures in addition to the ER network were also fluorescent, probably representing ER exit sites; they never overlapped with plastids.
A statistical analysis showed that the transport activity of AmyI-1-GFP into plastids was significantly reduced in the ARF1 and SAR1 mutants (Figure 3E). As controls, the effects of these mutants on the localization of a trans-Golgi marker (transmembrane domain of sialyltransferase fused to the monomeric red fluorescent protein, ST-mRFP) (Kim et al., 2001; Latijnhouwers et al., 2005) were tested. The trans-Golgi marker in the cells expressing ARF1(T31N), ARF1(Q71L), or SAR1(H74L) was relocated to the ER network (see Supplemental Figure 4A online). By contrast, as would be expected, these mutants exhibited no significant effect on the behavior of plastidial (WxTP-GFP), peroxisomal (per-GFP, the peroxisomal targeting signal 2 fused with GFP), or mitochondrial (mt-GFP, the presequence of the γ subunit of Arabidopsis F1-ATPase fused with GFP) markers (see Supplemental Figures 4B to 4D online).
We previously demonstrated that another rice N-glycosylated glycoprotein, NPP1, is transported from the secretory pathway to the chloroplast (Nanjo et al., 2006). α-Carbonic anhydrase was also reported to be N-glycosylated and transported to chloroplasts in Arabidopsis leaves and suspension-cultured cells (Villarejo et al., 2005). Both glycoproteins were targeted to plastids in a Brefeldin A–sensitive manner (Ritzenthaler et al., 2002). Overall, these results strongly suggest that the membrane trafficking from the ER is necessary for the plastid targeting of these glycoproteins.
Golgi-to-Plastid Traffic
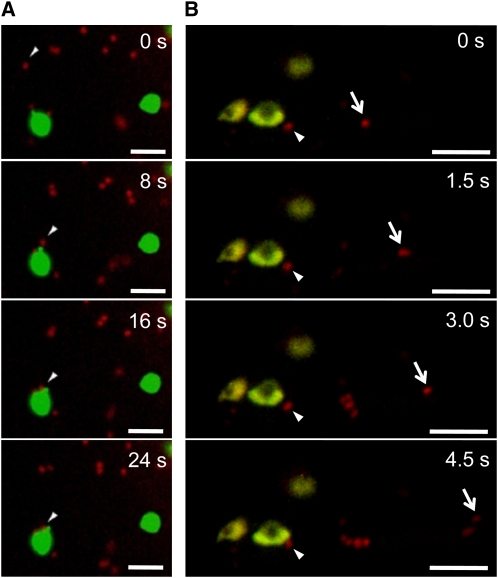
The mammalian Golgi apparatus has been shown to cluster and form a juxta- or perinuclear network in the cells. By contrast, plant Golgi stacks are dispersed singly or in small clusters throughout the cytoplasm and show active and stop-and-go tumbling movements along actin microfilaments (Nebenführ et al., 1999). The active movement of plant Golgi stacks suggests that the Golgi stack itself serves as a cargo container. If a genuine route from the Golgi complex to plastids exists, we consider it possible that these two organelles might be in constant communication. To test this idea, we performed high-speed two-dimensional and three-dimensional time-lapse analyses using confocal laser scanning microscopy. When ST-mRFP and WxTP-GFP were simultaneously expressed together with AmyI-1, which would activate Golgi-to-plastid traffic in onion cells, significant merging of ST-mRFP with the GFP-labeled plastids was observed (Figures 4 and 5). Moreover, the time-lapse scans caught several dramatic scenes: (1) the Golgi bodies made a soft landing on the surface of the plastid (Figure 4A; see Supplemental Movie 1 online). (2) Close contact between the two organelles was occasionally observed (Figure 4B; see Supplemental Movie 2 online). (3) Small membrane vesicles derived from the trans-Golgi were detected on the surface of the plastid (Figure 5A, right panel; see Supplemental Movie 3 online). Intriguingly, the fluorescent membrane vesicles did not stay long on the envelope membranes. They were eventually relocated to the interior of the plastid (Figure 5C; see Supplemental Movie 4 online).
Figure 4.
Two-Dimensional Time-Lapse Imaging of a Communication between Golgi and Plastid.
ST-mRFP and WxTP-GFP dual labeling with AmyI-1 expression was performed in onion cells. ST-mRFP, red; WxTP-GFP, green. Bars = 5 μm.
(A) The images were taken at the rate of 1 frame per 4 s. Cropped frames from the movie (see Supplemental Movie 1 online) are shown here. Arrowhead shows the Golgi body (red) soft-landing on the surface of the plastid (green).
(B) The images were taken at a rate of 1 frame per 0.5 s. Cropped frames from the movie (see Supplemental Movie 2 online) are shown here. Arrows show Golgi bodies with different locomotion characteristics: one remains around the plastids (arrowhead) and the other exhibits active movement (arrow).
Figure 5.
Three-Dimensional Time-Lapse Imaging of Golgi-to-Plastid Traffic.
(A) ST-mRFP and WxTP-GFP labeling without (left) or with (right) AmyI-1. Top panels represent three-dimensional images reconstructed from 10 optical sections spanning the entire plastid, every 1.5 μm. Bottom panels show a magnified view of a representative section of a plastid. ST-mRFP, red; WxTP-GFP, green. Bars = 5 μm.
(B) Statistical evaluation of the plastidial localization of ST-mRFP. To evaluate the incorporation of ST-mRFP into plastids without or with AmyI-1, ARF1(T31N) or SAR1(H74L) expression, the ratio of the fluorescence intensity of RFP in the plastidial area to RFP in the whole cell (RFPplastid/RFPtotal) was determined. Values are represented as means ± sd (n = 5 to 13).
(C) Three-dimensional time-lapse images of cells expressing ST-mRFP, WxTP-GFP, and AmyI-1. Ten optical sections, 1.5 μm apart, were taken at 0.844-s intervals. A representative section, the third section from the top indicated by open white quadrangle and three-dimensional surface images constructed with sections from No. 4 to 10 using the α-blend procedure were reconstructed into movies (see Supplemental Movie 4 online). ST-mRFP, red; WxTP-GFP, green. The change in color from green to yellow in the stroma is due to the uptake of the Golgi marker ST-mRFP. Bars = 5 μm.
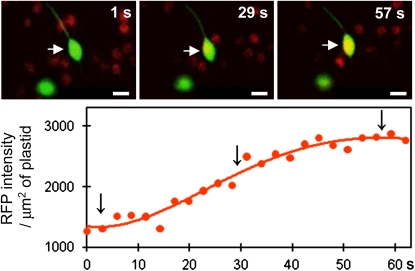
In the cells expressing ST-mRFP and WxTP-GFP without AmyI-1, such overlapping of mRFP and GFP appeared to be less frequent (Figures 5A and 5B). Three-dimensional time-lapse imaging showed that even without AmyI-1 expression, the ST-mRFP–labeled Golgi adheres to the surface of plastids and stromules (Figure 5A, left; see Supplemental Movie 5 online). The incorporation of ST-mRFP into plastids still occurred (Figure 6), albeit at lower frequency. In addition, Golgi-to-plastid trafficking was noticeably prevented by the expression of either ARF1(T31N) or SAR1(Q74L) (Figure 5B). We propose that these results suggest that Golgi-to-plastid basal communication occurs under tranquil physiological conditions and that AmyI-1 coexpression enhances the flow of Golgi-to-plastid traffic and activates the communication between two organelles in the cell.
Figure 6.
Merging of the Trans-Golgi Marker into a Plastid That Lacks AmyI-1 Expression.
ST-mRFP (red) and WxTP-GFP (green) dual labeling without AmyI-1 expression was performed in onion cells. Top panel: two-dimensional time-lapse scanning. The images were taken at a rate of 1 frame per 1.156 s. Three frames at intervals of 28.9 s are shown. A selected plastid, indicated by the white arrow, was subjected to quantitative analysis. Bottom panel: quantitative evaluation of the plastidial localization of ST-mRFP. The values of RFP fluorescence intensity by area unit of plastid (RFP intensity/μm2 of plastid) were determined at intervals of 2.312 s. Bars = 5 μm.
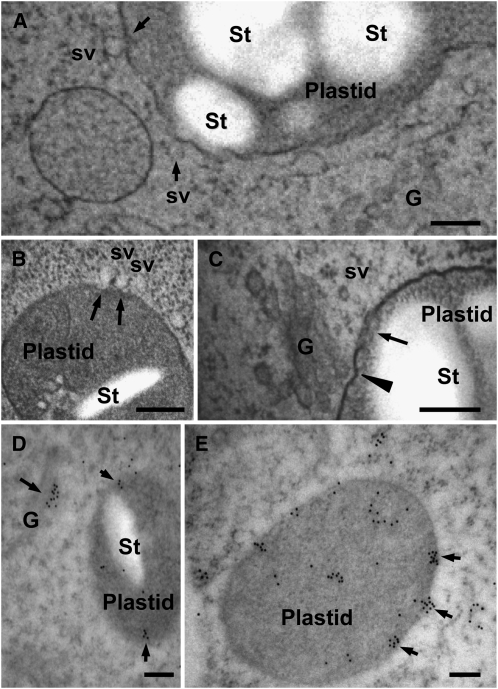
To clarify the precise mechanism of Golgi-to-plastid traffic, electron microscopy studies were performed on suspension-cultured cells derived from transgenic rice seed (A3-1 line) with a constitutively high expression of AmyI-1 using high-pressure frozen/freeze-substituted techniques (Toyooka et al., 2006, 2009). In the electron microscopy images, it was frequently observed that small membrane vesicles, perhaps derived from the Golgi apparatus stayed by plastids, adhered to the envelope membrane of plastids (Figure 7A). Tight association of the Golgi stacks with the envelope membrane was also observed in the same cells (Figure 7C). As shown in Figures 7A to 7C, membrane vesicles are apparently taken up into the plastid, possibly passing through the envelope membranes. Furthermore, immunological staining of ultrathin sections with anti-AmyI-1 antibodies revealed the presence of AmyI-1 in both the Golgi apparatus and the plastids (Figures 7D and 7E) and, moreover, the clustering of AmyI-1 proteins on and beneath the envelope membrane, representing the instant of plastid entry of clustered AmyI-1 proteins (Figure 7E).
Figure 7.
Electron Microscopy Characterization of Golgi-to-Plastid Traffic using High-Pressure Frozen/Freeze-Substituted Cells.
Rice cells (A3-1) cultured in MS medium containing 3% sucrose at 28°C for 4 d were rapidly frozen in a high-pressure freezer. G, Golgi; St, starch; sv, small membrane vesicle. Bars = 200 nm.
(A) to (C) Morphological observations. Arrows in (A) represent obvious adhesions of membrane vesicles to plastids. In (B), arrows show the moment of plastid entry of membrane vesicles. In (C), a tight association between the Golgi stack and a plastid (arrowhead) and vesicles taken up inside the plastid (arrow) are shown.
(D) and (E) Immunocytochemical observations. The ultrathin sections were labeled with anti-AmyI-1 antibodies and 12-nm colloidal gold particles coupled to goat anti-rabbit IgG. Immunogold particles (arrows) were deposited in both Golgi apparatus and plastids. Arrows in (E) indicate the clustered AmyI-1 proteins entering the plastid.
Characterization of the Plastid-Targeting Signal of AmyI-1
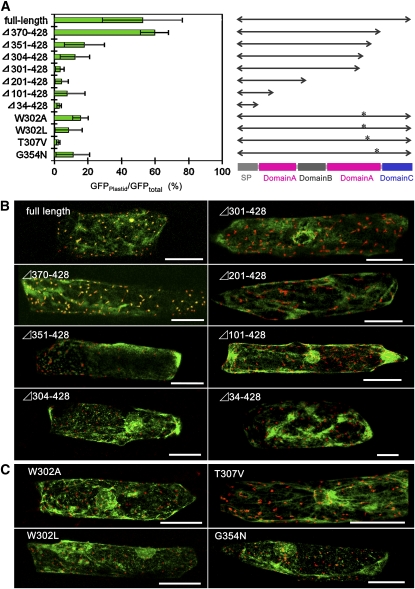
The majority of chloroplast proteins are synthesized in the cytosol as precursors with an N-terminal transit peptide and imported posttranslationally through the Toc/Tic complex into the organelle. As described above, however, the predicted precursor sequence of AmyI-1 contains no transit peptides for plastidial localization (Figure 1). To identify the plastid-targeting signal of AmyI-1, the transient expression and localization of a series of C-terminal truncated AmyI-1-GFP fusion proteins was examined in bombarded onion cells. The predicted signal peptidase cleavage site is the N-terminal side of Gln-26. AmyI-1(Δ34–428) includes only the N-terminal ER signal peptide of AmyI-1. These results indicate that AmyI-1 cannot be targeted to the plastid by the signal peptide alone (Figures 8A and 8B). We thus conclude that the signal peptide of AmyI-1 is necessary for entry into the secretory pathway but is not sufficient for plastid targeting.
Figure 8.
Evaluation of Plastid-Targeting Abilities of Various C-Terminal Truncated and Site-Directed Mutated AmyI-1.
Simultaneous expression of WxTP-DsRed and either a C-terminal truncated AmyI-1 (either full length, Δ370–428, Δ351–428, Δ304–428, Δ301–428, Δ201–428, Δ101–428, or Δ34–428), AmyI-1(W302A), AmyI-1(W302L), AmyI-1(T307V), or AmyI-1(G354N) fused to GFP was performed in onion cells. Bars = 100 μm.
(A) Left: quantitative results. Values show the mean ± sd obtained in four independent experiments. Right: diagram of the AmyI-1 series. Asterisks indicate the relative positions of the substituted residues.
(B) Typical images of plastid localization of the C-terminal truncated AmyI-1 series.
(C) Typical images of plastid localization of the point-mutated AmyI-1 series.
Like the barley (Hordeum vulgare) α-amylase structure (Robert et al., 2005), AmyI-1 appears to be composed of a large central domain of (βα)8-barrels (A domain), a loop domain (B domain), and a C-terminal domain that forms a five-stranded, antiparallel β-sheet (C domain). AmyI-1(Δ370–428) contains the full (βα)8-barrel domain A and the loop domain B, but lacks the C domain. As shown in Figures 8A and 8B, AmyI-1(Δ370–428)-GFP fusion protein was normally targeted into the plastids, like the full-length AmyI-1. Further C-terminal deletion of AmyI-1 gave rise to the gradual loss of plastid targeting ability, and AmyI-1(Δ301–428) exhibited no plastid targeting. This indicates the importance of the peptide region from Trp-301 to Gln-369 for plastid targeting of AmyI-1. A putative surface starch binding site Typ-301–Trp-302 and substrate binding subsites and catalytic residue in the active site cleft (His-313, Asp-314, and Gln-319) were located in this region (Søgaard et al., 1993).
Amino acid sequence alignment of Amy isoforms revealed that the AmyI-1 polypeptide has 12 unique amino acid residues in the region from 301 to 369 (Figure 1). Of these amino acid residues, we selected Trp-302, Thr-307, and Gly-354, which are located in different areas in the three-dimensional structure of the protein, for the next set of site-directed mutagenesis experiments. The substitution of Ala or Leu for Trp-302 arrested the targeting of AmyI-1-GFP into the plastids. The substitution of Val for Thr-307 or Asn for Gly-354 also arrested the plastid targeting of AmyI-1-GFP, as did the W302A and W302L mutations (Figures 8A and 8C). Furthermore, the chimera proteins of AmyII-6(1-266):AmyI-1(256-369) and AmyII-6(1-266):AmyI-1(287-369) failed to localize in the plastids (see Supplemental Figure 5 online). From these results, we infer that conformational changes in the region from Trp-301 to Gln-369 influence the targeting ability of AmyI-1.
DISCUSSION
AmyI-1 Degrades Starch Granules in Living Rice Cells
It is widely accepted that α-amylase is the main enzyme responsible for starch breakdown in germinating cereal seeds. This enzyme is excreted from the secretory tissues to the storage tissue, in which the starch granules are accumulated in dead cells. However, recent investigations have also revealed the involvement of α-amylase in starch degradation in living rice cells (Chen et al., 1994; Asatsuma et al., 2005). Immnocytochemical studies have shown that α-amylases are localized in multiple compartments, such as amyloplasts, vacuoles, and cell walls (Chen et al., 1994). Moreover, a rice α-amylase isoform, αAmy3, occurred in the chloroplasts of transgenic tobacco leaves (Chen et al., 2004). Transgenic rice plants that overexpressed α-amylases produced white immature seeds with reduced starch accumulation (Asatsuma et al., 2006). This study provides further evidence that the AmyI-1 glycoprotein is localized in the amyloplasts and degrades starch granules within the organelles in rice callus cells (Figure 2). Thus, rice α-amylase exhibits dual targeting to endomembranes/extracellular spaces and plastids (Chen et al., 2004). Protein import to plastids through a secretory pathway does not agree with the traditional cell biological context of protein transport and appears to be an inelegant system compared with the canonical plastid import machinery. However, the chloroplast targeting of α-amylase in leaf cells is physiologically relevant, since starch turnover in chloroplasts is a crucial process in leaves, and no starch is present outside of the cells. The plastid-targeting route from the endomembrane system appears to be operated as a physiological event in plants.
AmyI-1 Contains a Plastid-Targeting Signal Common to Both Rice and Onion Cells
Several α-amylase isoforms, such as AmyI-1 (RAmy1A, αAmy7), II-3 (RAmy3E, αAmy8), and II-4 (RAmy3D, αAmy3), have been shown to occur within the plastids in rice cells (Chen et al., 2004; Asatsuma et al., 2005). However, only AmyI-1 exhibited clear and efficient targeting into the plastids in the onion epidermal cells. AmyII-3 and AmyII-4 were rarely transported to the plastids (Figure 3; see Supplemental Figure 3 online). AmyI-1's plastid-targeting signal thus functions in both rice and onion cells. Plastid-targeting signals of two other isoforms are considered to be insufficient in the context of the onion system. Chen et al. (2004) have revealed, employing a series of loss-of-function and gain-of-function analyses, that the ER signal peptides of αAmy3 and αAmy8 direct cargo proteins to plastids and extracellular compartments in rice and tobacco cells. By contrast, the signal peptide of AmyI-1 failed to deliver reporter protein GFP to plastids in onion cells and chloroplasts in transformed rice cells (Figure 8; see Supplemental Figure 6 online). We believe that the AmyI-1 signal peptide contains no information for directing cargo protein, including its own structural protein, to the plastids. The divergence in roles of the signal peptides of rice α-amylases remains unresolved.
Golgi-to-Plastid Traffic
Intracellular compartments in eukaryotic cells can be divided into at least five distinct families. The first and second compartments are the cytosol and the nucleus. The third includes all single-membrane organelles that function in the secretory and endocytic pathways and possibly microbodies (peroxisomes). The fourth and fifth are mitochondria and plastids, and the latter includes chloroplasts in green leaves and amyloplasts in starchy cells. The lumen of the ER, the Golgi apparatus, and lysosomes or vacuoles are topologically equivalent to the extracellular space, and the membranes of the organelles, collectively called endomembrane organelles, may have evolved from the plasma membrane. Cycles of membrane budding and fusion permit communication with the endomembrane and cell exterior via transport vesicles (Rothman and Wieland, 1996). It has been long accepted that mitochondria and plastids are isolated from the traffic between organelles in the third compartment group (Blobel, 1980). However, communications between the intracellular compartments in higher plant cells are much more flexible than previously thought. In this study, we demonstrated that the Golgi stacks were tightly associated with the plastids (Figure 7C) and that a Golgi-resident protein, ST-mRFP, was actually incorporated into the interior of the plastid (Figure 5C), indicating the existence of a protein traffic route from the Golgi complex to plastids. In addition, the incorporation of Golgi-resident proteins into plastids was stimulated by AmyI-1 expression (Figure 5B), and the plastid-targeting of both AmyI-1 and ST-mRFP was similarly inhibited by arresting the ER-to-Golgi traffic (Figures 3E and 5B). Thus, the Golgi apparatus appears to communicate with plastids directly to import glycoprotein cargo into plastids.
In Euglena, a fusion event of the Golgi-derived vesicles to the outermost of the three chloroplast envelope membranes has been observed (van Dooren et al., 2001). The membrane vesicle transport between the Golgi apparatus and the plastids is a potential candidate for the import mechanism of glycoproteins into plastids in higher plant cells. In this model, however, two puzzling problems must be resolved: one is the topology of the targeted space and the other is the recycling of fused membranes. Membrane fusion between the Golgi-derived vesicles and the outer envelope of plastids would result in localization of the cargo in the intermembrane space between the outer and inner envelopes. It has recently been proposed that three routes may operate in the transfer of cargo from the intermembrane space to the stroma after delivering cargo materials to the space: (1) engagement of an unknown transporter in the inner envelope membrane, (2) passage through the Tic transporter operating independently of the Toc machinery, or (3) vesicle budding from the inner membrane itself (Radhamony and Theg, 2006). Our study strongly supports vesicle-mediated import of the AmyI-1 glycoprotein into plastids. Electron microscopy observations have shown that the Golgi-derived vesicles physically bind to plastids, and the clustered AmyI-1 proteins appeared to pass through the envelope membranes and enter the stromal space (Figure 7). However, it should be noted that AmyI-1 importation does not take place by means of the simple membrane fusion described in the Euglena model. Three-dimensional time-lapse imaging shows that the trans-Golgi membranes appear to be taken up and spread inside the plastid (Figure 5C; see Supplemental Movie 4 online). In addition, the moment of plastid entry of membrane vesicles was seen in EM observations using quick-frozen cells (Figures 7A and 7B), suggesting that the vesicles might pass through the plastid envelope membranes by a mechanism other than by microautophagy-like invagination into the organelle. If microautophagy-like invagination were involved in the uptake of membrane vesicles, vesicles encircled by the plastid envelope membranes in addition to small invagination would be frequently detected in the stroma. However, distinctive vesicles such as these were extremely rare, suggesting that an unresolved vesicle uptake mechanism operates in AmyI-1 importation into plastids. The imported vesicles are perhaps subsequently broken up in the organelle.
The Golgi apparatus is known to be a multifunctional organelle, responsible for the biosynthesis of cell wall polysaccharides, the processing and modification of glycoproteins, and the sorting of polysaccharides and proteins destined for different locations (Staehelin and Moore, 1995). During the mitosis of tobacco BY-2 cells, ∼20% of the Golgi bodies remain close to the spindle, whereas another 20% relocate to the equatorial region near the plasma membrane. These regions are devoid of mitochondria and plastids (Nebenführ et al., 2000). Our results revealed that the Golgi bodies in onion cells have at least two different modes of locomotion. One group moved around the plastids, while the other traveled further than those that stayed in the vicinity of the plastids (Figure 4B; see Supplemental Movie 2 online). Thus, the localization of Golgi bodies in the cells depends upon their environment and functions. Recent investigations have demonstrated that Golgi stacks are constructed de novo and mature in yeast (Matsuura-Tokita et al., 2006), and the rate of cisternal maturation is rapid, matching the rate of protein transport through the secretory pathway (Losev et al., 2006). It is conceivable that Golgi bodies destined for plastid delivery are also newly formed for appropriate situations.
Characterization of the Plastid-Targeting Signal of AmyI-1
Secretion is thought to be a default targeting pathway in both plant and animal cells; that is, a secretory protein without any other sorting information within the protein travels through the ER and Golgi network and is finally liberated from the cells. Plastidial glycoproteins may possess certain targeting signals for their localization; however, no details of plastid-targeting signals have so far been revealed. This study demonstrated that the plastid-targeting signal of AmyI-1 is embedded in the protein structure of the enzyme. The results obtained by site-directed mutations of Trp-302 and Gly-354, located on the surfaces of opposite sides of the AmyI-1 protein, suggest that multiple surface regions are necessary for plastid targeting (Figure 8). There was no sequence resemblance among the three glycoproteins (AmyI-1, NPP1, and CAH1), which were all shown to occur in plastids. We thus infer that the plastid-targeting signals of glycoproteins are expressed by as yet uncharacterized three-dimensional structures.
Vacuolar proteins are synthesized and translocated into the ER lumen and then delivered to the vacuole through the secretory pathway. The sorting of vacuolar proteins requires interaction between the protein's sorting signal and a sorting receptor within the endomembranes (Kirsch et al., 1996; Matsuoka and Neuhaus, 1999; Vitale and Hinz, 2005; Fuji et al., 2007). Vacuolar sorting signals are grouped into three categories: sequence-specific signals, C-terminal signals, and physical structure signals (Vitale and Hinz, 2005). Three vacuolar proteins have been reported to possess physical structure vacuolar sorting signals (Saalbach et al., 1991; von Schaewen and Chrispeels, 1993; Törmäkangas et al., 2001). One of these, barley aspartic protease (phytepsin), was shown to reach the vacuoles through the Golgi apparatus in a coat protein complex (COP)II-mediated manner. A plant-specific insert of 104 amino acids present in phytepsin serves as a vacuolar sorting signal. Deletion of the plant-specific insert causes efficient secretion of the truncated phytepsin in a COPII-independent manner. We observed that the dominant-negative and constitutively active mutants of ARF1 and SAR1 effectively prevent the targeting of AmyI-1 into the plastids and keep them in the ER in onion epidermal cells (Figure 3C), indicating that the plastid targeting of AmyI-1 requires COP-mediated membrane traffic. It is apparent that the ER-Golgi system, located in the early secretory pathway, acts as a set of sorting stations for delivering proteins to the plastids in addition to the vacuoles. However, the molecular machinery that recognizes these physical targeting signals remains unknown.
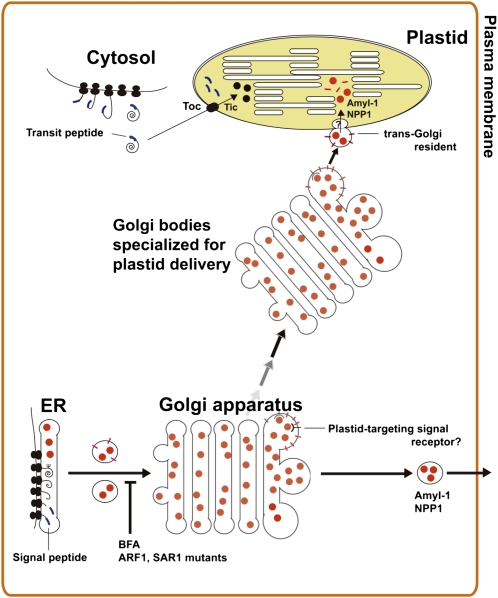
Based on our overall results, we propose a model for glycoprotein targeting to plastids in higher plant cells (Figure 9). The plastidial glycoprotein is synthesized in the ER and transported to the Golgi in a way that is dependent on ARF1 and SAR1 GTPases. A physical structure signal for their plastid targeting is recognized by hypothetical receptors on the early secretory pathway. Golgi bodies with cargo, which are specialized for delivering a glycoprotein cargo to the plastids, might move to the plastid in a targeting signal–dependent manner, after which small membrane vesicles derived from the Golgi adhere to the plastid's envelope membrane. The imported vesicles are eventually broken up within the plastid stroma, allowing the liberated glycoproteins to function inside the organelle.
Figure 9.
A Hypothetical Model for Plastid Targeting of Glycoproteins from the Golgi Apparatus through the Secretory Pathway.
The plastid-destined glycoproteins, such as AmyI-1 and NPP1, are synthesized in the ER lumen and transported to the Golgi apparatus. Plastid targeting of glycoproteins is prevented by the drug Brefeldin A and/or mutants of ARF1 and SAR1, indicating that membrane trafficking is essential to their plastid targeting. The Golgi bodies with cargo, which are specialized for delivering cargo to the plastids, might move to the plastid in a targeting signal–dependent manner, after which small membrane vesicles derived from the Golgi adhere to the envelope membrane of the plastid. Both the membrane component and glycoprotein cargo are eventually imported into the interior of the plastid through the envelope membranes via as yet unknown translocation machinery.
METHODS
Plant Materials
Rice seeds (Oryza sativa cv Nipponbare) were supplied by the Niigata Agricultural Research Institute (Niigata, Japan). The production of transgenic rice seed (A3-1 line) has been described previously (Asatsuma et al., 2005). Rice calli derived from the embryo portions of the seeds were cultured as follows. About 2 g of rice callus cells were grown in a 500-mL Sakaguchi flask holding 120 mL of MS medium containing 3% (w/v) sucrose, 2 mg/L 2,4-D, and 5 mg/L thiamine-HCl, placed on a reciprocal shaker operated at 110 strokes/min with 70-mm amplitude, at 28°C in darkness. The established suspension-cultured cells were subcultured at 7-d intervals. All of the procedures were performed under aseptic conditions. Amylase activity and starch content were determined as described in earlier studies (Mitsui et al., 1996; Asatsuma et al., 2005).
Plasmid Constructs
Primer sequences for PCR amplifications are given in Supplemental Table 1 online. The constructions of pAmyI-1, pGFP, and pAmyI-1-GFP have been described previously (Asatsuma et al., 2005). For DsRed2 expression in onion (Allium cepa) epidermal cells, the BamHI-SacI PCR-amplified fragment from pDsRed2 (Takara Bio) was cloned into the same sites of pGFP to produce pDsRed. To create pWxTP-DsRed, we PCR-amplified the first 1 to 111 amino acid residues, including the transit peptide sequence, from the rice waxy gene (pWCW) (Klösgen and Weil, 1991; Itoh et al., 2003) with two flanking primers and then digested the PCR product with BamHI. The BamHI-digested fragment was inserted into the same site as pDsRed. To construct pWxTP-GFP, we further inserted the PCR-amplified WxTP fragment into the BamHI site of pGFP. We also used pST-mRFP (Kim et al., 2001; Latijnhouwers et al., 2005), pper-GFP (Mano et al., 2002), and pmt-GFP (Niwa et al., 1999) to visualize the trans-Golgi, peroxisomes, and mitochondria, respectively. The dominant-negative mutants of Arabidopsis thaliana ARF1 and constitutive active mutant of SAR1 (pMT121-ARF1 T31N, pMT121-ARF1 Q71L, and pMT121-SAR1 H74L, respectively) were reported previously (Takeuchi et al., 2002). Double entry vector pdEV(WxTP-GFP)(AmyI-1) was created from pWxTP-GFP and pAmyI-I using the Gateway system according to the manufacturer's protocol (Invitrogen).
Each full-length AmyII-3, II-4, II-5, and II-6 was PCR amplified using pAmyII-3, pAmyII-5, pAmyII-6, or pAmyII-4 (Asatsuma et al., 2006) as DNA template and flanking primer sets. To construct pAmy-GFP, the PCR-amplified AmyII-3 (identical to accession number M59352, except that this sequence contained no BamHI site), II-4, II-5, and II-6 were inserted into pGFP.
To construct a set of pAmyI-1(truncated)- and pAmyI-1(point-mutated)-GFPs, the C-terminal truncated AmyI-1 series (Δ34–428, Δ101–428, Δ201–428, Δ301–428, Δ304–428, Δ351–428, or Δ370–428) and the point-mutated AmyI-1 series (W302L, W302A, T307V, or G354N) were PCR amplified using pAmyI-1 as the DNA template and specific primer sets and then inserted into pGFP.
We PCR-amplified AmyI-1(256-369)(Gly)4 and AmyI-1(287-369)(Gly)4 from pAmyI-1 and 35S-AmyII-6(1-266) (no BamHI site) from pAmyII-6-GFP with flanking primer sets. To construct pAmyII-6(1-266):AmyI-1(256-369)-GFP or pAmyII-6(1-266):AmyI-1(287-369)-GFP, the BamHI-KpnI–digested AmyI-1(256-369) or AmyI-1(287-369) and HindIII-BamHI–digested 35S-AmyII-6(1-266) were sequentially inserted into pGFP.
The construction of binary vectors pZH2B-35S-AmyI-1, pZH2B-35S-GFP, and pZH2B-35S-AmyI-1-GFP has been described previously (Asatsuma et al., 2005). To create pZH2B-35S-AmyI-1(Δ101–428)-GFP and pZH2B-35S-AmyI-1(Δ370–428)-GFP, the 35S-AmyI-1(Δ101–428)-GFP and 35S-AmyI-1(Δ370–428)-GFP genes were PCR amplified from pAmyI-1(Δ101–428)-GFP and pAmyI-1(Δ370-428)-GFP. The HindIII-NotI–digested fragment was then inserted into the same site of pZH2B. These vectors were transformed into competent cells of Agrobacterium tumefaciens strain EHA101 (Hood et al., 1986) and treated with 20 mM CaCl2. Agrobacterium-mediated transformation and regeneration of rice plants were performed according to the methods described by Hiei et al. (1994).
Introduction of Plasmids into Onion Cells
To introduce plasmid DNA into onion epidermal cells, the particle bombardment method was adopted using a helium-driven particle accelerator (PDS-1000/He; Bio-Rad) with all basic adjustments set according to the manufacturer's recommendations. Three micrograms of plasmid DNA in 10 μL distilled water was mixed with 10 μL of a 60 mg/mL gold particle (diameter 1.0 μm) solution, 10 μL of 2.5 mM CaCl2, and 4 μL of 0.1 M spermidine, and incubated for 30 min at room temperature. Gold particles coated with plasmid DNA were rinsed with cold ethanol and then gently suspended in 10 μL ethanol. The gold particles were bombarded twice into onion cells using the particle delivery system with 1100 p.s.i. rupture discs. The bombarded onion epidermal cells were cultured on 0.6% Gelrite with 2,4-D-free MS medium at 25°C in darkness.
Microscopy
For fluorescence microscopy, images of GFP, DsRed, and mRFP fluorescence in whole cells were observed using a microscopy setup composed of a BX-61 microscope (Olympus) and a cooled CCD camera (EM-CCD; Hamamatsu). An Hg lamp was used to excite the fluorescent proteins. GFP fluorescence was obtained by excitation at 470 to 490 nm and detection at 510 to 550 nm, and DsRed and mRFP fluorescence by 520- to 550-nm excitation with ≥580-nm detection. Deconvolution was performed using Lumina Vision imaging software (Mitani). Twenty to thirty images per cell, from the top to middle of the cell, every 1 to 2 μm, were taken and combined into one image.
For confocal laser scanning microscopy, three microscopy settings were used for confocal fluorescence imaging of living cells. An Olympus FV300-BX-61 confocal laser scanning microscope (CLSM) was used for two-dimensional time-lapse observations and an Olympus FV1000-IX-71 for three-dimensional time-lapse observations. Ar and green He/Ne lasers were used to excite GFP at 488 nm and mRFP and DsRed at 543 nm. Reconstructions of three-dimensional surface images were performed using an α-blend procedure using FV10-ASW imaging software (Olympus). A custom-made system produced by the Dynamic-Bio Project was used for higher-resolution and higher-speed observations (see Supplemental Movie 2 online). In this system, an Olympus IX-70 microscope was equipped with a special high signal-to-noise ratio color confocal system (Yokogawa Electric), image intensifiers (Hamamatsu Photonics), and high-speed and ultrahigh-sensitivity HARP cameras (NHK Engineering Services and Hitachi Kokusai Electric). An Ar+/Kr+ laser (Melles Griot) was used to excite GFP at 488 nm and mRFP at 568 nm simultaneously. For three-dimensional imaging, the objective lens was oscillated vertically to the sample plane by means of a piezo actuator system (Yokogawa Electric). Collected pictures were analyzed using Volocity software (Improvision) (Matsuura-Tokita et al., 2006).
For quantitative analysis, the fluorescence intensity in plastids and in whole cells was determined using Lumina Vision imaging software. The background was always set at the maximum value of fluorescence intensity for the area in which no structural image was present. The area visualized with either WxTP-GFP or WxTP-DsRed was defined as the plastidial area. In analysis of the whole cell, each individual image from the top to the middle of the cell, 15 to 20 frames every 1 to 2 μm, was evaluated. To evaluate the plastid-targeting abilities of GFP-labeled proteins, we determined the ratio of the fluorescence intensity of GFP in the plastidial area provided with WxTP-DsRed to GFP in the whole cell (GFPplastid/GFPtotal). By contrast, to estimate the incorporation of ST-mRFP into plastids, the ratio of the fluorescence intensity of RFP in the plastidial area provided with WxTP-GFP to RFP in the whole cell (RFPplastid/RFPtotal) was determined and compared. For time-lapse experiments, the values of RFP fluorescence intensity per area unit of plastid visualized with WxTP-GFP (RFP intensity/μm2 of plastid) were determined.
Electron Microscopy Studies
For the high-pressure frozen/freeze-substituted cells: wild-type and A3-1 rice cells constitutively and highly expressing AmyI-1 cultured in MS medium containing 3% sucrose at 28°C for 4 d were immediately placed on a flat specimen carrier and frozen in a high-pressure freezer (EM-PACT; Leica Microsystems). The frozen samples were fixed in anhydrous acetone containing 2% osmic acid (OsO4) for 3 to 4 d at –80°C for morphological observation or fixed with anhydrous acetone containing 1% glutaraldehyde and 1% OsO4 for 3 to 4 d at –80°C for immunocytochemistry. The tubes containing the frozen samples were warmed at 3°C/h to a temperature of –20°C, and at 1°C/h from –20°C to 4°C, and kept for 2 h at 4°C using an automatic freeze substitution system (EM-AFS; Leica Microsystems). For morphological observation, the samples were stained with 1% tannic acid in acetone for 1 h at room temperature. The samples were then washed with 100% acetone and embedded in epoxy resin (TAAB). The fixed samples for immunocytochemistry were embedded in LR White resin (London Resin).
The ultrathin sections for morphological and immunocytochemical observations were mounted on 400-mesh Cu and 600-mesh Ni grids, respectively. Immunocytochemical detection of AmyI-1 was performed according to Toyooka et al. (2009). The sections on Ni grids were treated with 10% BSA in Tris-buffered saline (TBS) for 30 min at room temperature. The sections were incubated with an affinity-purified polyclonal antibody toward AmyI-1 (1:100) in TBS. After washing with TBS, the sections were further incubated with 12-nm colloidal gold particles conjugated to goat anti-rabbit IgG (Jackson ImmunoResearch). The sections were washed with TBS and rinsed in distilled water. Both sections for morphological and immunochemical observations were finally stained with 4% aqueous uranyl acetate for 5 min and examined using a transmission electron microscope (JEM-1011; JEOL) at 80 kV. Images were acquired using a Gatan DualView camera and Digital Micrograph software or transmission electron microscopy films.
For the unfrozen cells, freshly harvested rice cells were fixed with 4% (w/v) paraformaldehyde and 60 mM sucrose in 50 mM cacodylate buffer, pH 7.2, overnight at 4°C. The samples were dehydrated and embedded in either epoxy or LR White resin and then cut into ultrathin sections. For immunocytochemical detection of AmyI-1, the sections on Ni grids were washed with PBST containing 1% BSA for 20 min and then incubated in a 1:100 dilution of anti-AmyI-1 antibodies or nonimmune sera in PBST at 4°C for 1 h. The sections were washed with PBST for 20 min and then incubated with 10-nm colloidal gold-conjugated protein A at 4°C for 30 min. The sections were washed with PBST and distilled water and then finally stained with uranyl acetate for 10 min and lead citrate for 2 min, sequentially. They were examined using a Hitachi H-600 transmission electron microscope at an accelerating voltage of 75 kV.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL database under the following accession numbers: Amy I-1 (AAA33885, M24286), II-3 (AAA33896, M59352), II-4 (AAA33886, M24287), II-5 (CAA39777, X56337), and II-6 (CAA39778, X56338).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Fluorescence Images in Onion Epidermal Cells Expressing Either GFP, DsRed, WxTP-GFP, or WxTP-DsRed.
Supplemental Figure 2. Fluorescence Images in Onion Cells Simultaneously Expressing WxTP-DsRed with per-GFP or mt-GFP.
Supplemental Figure 3. Distinct Localization of Various Amy Isoforms in Onion Epidermal Cells.
Supplemental Figure 4. Effects of ARF1(T31N), ARF1(Q71L), and SAR1(H74L) on the Localization of Various Organelle Markers.
Supplemental Figure 5. Plastid Targeting Abilities of AmyII-6:AmyI-1 Chimera Proteins.
Supplemental Figure 6. Plastid Targeting Abilities of AmyI-1(Δ101–428)- and AmyI-1(Δ370–428)–GFP in Cells of Transgenic Rice Plants.
Supplemental Table 1. Primer Sequences for PCR Amplification.
Supplemental Movie 1. Video of Figure 4A.
Supplemental Movie 2. Video of Figure 4B.
Supplemental Movie 3. Video of Figure 5A, Right.
Supplemental Movie 4. Video of Figure 5C.
Supplemental Movie 5. Video of Figure 5A, Left.
Supplementary Material
Acknowledgments
This research was supported by the Ministry of Agriculture, Forestry, and Fisheries of Japan (Genomics for Agricultural Innovation Grant IPG0021) and by Grants-in-Aid 16658042 and 17051011 from the Ministry of Education, Culture, Sports, Science, and Technology, Japan, to T.M. and 17078009 to K.M. We thank Kimiko Itoh and Hidetaka Hori (Niigata University, Japan) for valuable and helpful comments during preparation of the manuscript, A. Kato (Niigata University, Japan), Y. Niwa (University of Shizuoka, Japan), I. Hwang (Pohang University of Science and Technology, Korea), and R. Tsien (University of California, San Diego, CA) for providing pper-GFP, pmt-GFP, ST, and mRFP, and Susan T. Hamamoto (University of California, Berkeley, CA) for tannic acid. We thank Y. Hayashi (Niigata University, Japan), Yumi Goto, and Mayuko Sato (Plant Science Center, RIKEN) for their valued contributions to the electron microscopy study. We also thank Miyuki Koumura (Olympus, Tokyo, Japan) for technical support with confocal laser scanning microscopy.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Toshiaki Mitsui (t.mitsui@agr.niigata-u.ac.jp).
Online version contains Web-only data.
Open access articles can be viewed online without a subscription.
References
- Asatsuma, S., Sawada, C., Itoh, K., Okito, M., Kitajima, A., and Mitsui, T. (2005). Involvement of α-amylase I-1 in starch degradation in rice chloroplasts. Plant Cell Physiol. 46 858–869. [DOI] [PubMed] [Google Scholar]
- Asatsuma, S., Sawada, C., Kitajima, A., Asakura, T., and Mitsui, T. (2006). α-Amylase affects starch accumulation in the rice grain. J. Appl. Glycosci. 53 187–192. [Google Scholar]
- Bennett-Lovsey, R.M., Herbert, A.D., Sternberg, M.J., and Kelley, L.A. (2008). Exploring the extremes of sequence/structure space with ensemble fold recognition in the program Phyre. Proteins 70 611–625. [DOI] [PubMed] [Google Scholar]
- Blobel, G. (1980). Intracellular protein topogenesis. Proc. Natl. Acad. Sci. USA 77 1496–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce, B.D. (2000). Chloroplast transit peptides: Structure, function and evolution. Trends Cell Biol. 10 440–447. [DOI] [PubMed] [Google Scholar]
- Chen, M.H., Huang, L.F., Li, H.M., Chen, Y.R., and Yu, S.M. (2004). Signal peptide-dependent targeting of a rice α-amylase and cargo proteins to plastids and extracellular compartments of plant cells. Plant Physiol. 135 1367–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, M.H., Liu, L.F., Chen, Y.R., Wu, H.K., and Yu, S.M. (1994). Expression of α-amylase, carbohydrate metabolism, and autophagy in cultured rice cells is coordinately regulated by sugar nutrient. Plant J. 6 625–636. [DOI] [PubMed] [Google Scholar]
- Gaikwad, A., Tewari, K.K., Kumar, D., Chen, W., and Mukherjee, S.K. (1999). Isolation and characterization of the cDNA encoding a glycosylated accessory protein of pea chloroplast DNA polymerase. Nucleic Acids Res. 27 3120–3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuji, K., Shimada, T., Takahashi, H., Tamura, K., Koumoto, Y., Utsumi, S., Nishizawa, K., Maruyama, N., and Hara-Nishimura, I. (2007). Arabidopsis vacuolar sorting mutants (green fluorescent seed) can be identified efficiently by secretion of vacuole-targeted green fluorescent protein in their seeds. Plant Cell 19 597–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi, M., Turu, A., Mitsui, T., Takahashi, N., Hanzawa, H., Arata, Y., and Akazawa, T. (1990). Structure and biosynthesis of the xylose-containing carbohydrate moiety of rice α-amylase. Eur. J. Biochem. 191 287–295. [DOI] [PubMed] [Google Scholar]
- Hiei, Y., Ohta, S., Komari, T., and Kumashiro, T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6 271–282. [DOI] [PubMed] [Google Scholar]
- Hood, E.E., Helmer, G.L., Fraley, R.T., and Chilton, M.D. (1986). The hyper-virulence of Agrobacterium tumefaciens A281 is encoded in a region of pTiBo542 outside of T-DNA. J. Bacteriol. 168 1291–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida, H., Yoshimoto, K., Izumi, M., Reisen, D., Yano, Y., Makino, A., Ohsumi, Y., Hanson, M.R., and Mae, T. (2008). Mobilization of Rubisco and stromal-localized fluorescent proteins of chloroplasts to the vacuole by an ATG gene-dependent autophagic process. Plant Physiol. 148 142–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh, K., Ozaki, H., Okada, K., Hori, H., Takeda, Y., and Mitsui, T. (2003). Introduction of Wx transgene into rice wx mutants leads to both high- and low-amylose rice. Plant Cell Physiol. 44 473–480. [DOI] [PubMed] [Google Scholar]
- Jarvis, P. (2008). Targeting of nucleus-encoded proteins to chloroplasts in plants. New Phytol. 179 257–285. [DOI] [PubMed] [Google Scholar]
- Kim, D.H., Eu, Y.J., Yoo, C.M., Kim, Y.W., Pih, K.T., Jin, J.B., Kim, S.J., Stenmark, H., and Hwang, I. (2001). Trafficking of phosphatidylinositol 3-phosphate from the trans-Golgi network to the lumen of the central vacuole in plant cells. Plant Cell 13 287–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsch, T., Saalbach, G., Raikhel, N.V., and Beevers, L. (1996). Interaction of a potential vacuolar targeting receptor with amino- and carboxyl-terminal targeting determinants. Plant Physiol. 111 469–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleffmann, T., Russenberger, D., von Zychlinski, A., Christopher, W., Sjolander, K., Gruissem, W., and Baginsky, S. (2004). The Arabidopsis thaliana chloroplast proteome reveals pathway abundance and novel protein functions. Curr. Biol. 14 354–362. [DOI] [PubMed] [Google Scholar]
- Klösgen, R.B., and Weil, J.H. (1991). Subcellular location and expression level of a chimeric protein consisting of the maize waxy transit peptide and the beta-glucuronidase of Escherichia coli in transgenic potato plants. Mol. Gen. Genet. 225 297–304. [DOI] [PubMed] [Google Scholar]
- Kornfeld, R., and Kornfeld, S. (1985). Assembly of asparagine-linked oligosaccharides. Annu. Rev. Biochem. 54 631–664. [DOI] [PubMed] [Google Scholar]
- Latijnhouwers, M., Hawes, C., Carvalho, C., Oparka, K., Gillingham, A.K., and Boevink, P. (2005). An Arabidopsis GRIP domain protein locates to the trans-Golgi and binds the small GTPase ARL1. Plant J. 44 459–470. [DOI] [PubMed] [Google Scholar]
- Lee, D.W., Kim, J.K., Lee, S., Choi, S., Kim, S., and Hwang, I. (2008). Arabidopsis nuclear-encoded plastid transit peptides contain multiple sequence subgroups with distinctive chloroplast-targeting sequence motifs. Plant Cell 20 1603–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losev, E., Reinke, C.A., Jellen, J., Strongin, D.E., Bevis, B.J., and Glick, B.S. (2006). Golgi maturation visualized in living yeast. Nature 441 1002–1006. [DOI] [PubMed] [Google Scholar]
- Mano, S., Nakamori, C., Hayashi, M., Kato, A., Kondo, M., and Nishimura, M. (2002). Distribution and characterization of peroxisomes in Arabidopsis by visualization with GFP: Dynamic morphology and actin-dependent movement. Plant Cell Physiol. 43 331–341. [DOI] [PubMed] [Google Scholar]
- Matsuoka, K., and Neuhaus, J. (1999). Cis-elements of protein transport to the plant vacuoles. J. Exp. Bot. 50 165–174. [Google Scholar]
- Matsuura-Tokita, K., Takeuchi, M., Ichihara, A., Mikuriya, K., and Nakano, A. (2006). Live imaging of yeast Golgi cisternal maturation. Nature 441 1007–1010. [DOI] [PubMed] [Google Scholar]
- Mitsui, T., Loboda, T., Kamimura, I., Hori, H., Itoh, K., and Mitsunaga, S. (1999). Sucrose-controlled transport and turnover of α-amylase in rice (Oryza sativa L.) cells. Plant Cell Physiol. 40 884–893. [Google Scholar]
- Mitsui, T., Yamaguchi, J., and Akazawa, T. (1996). Physicochemical and serological characterization of rice α-amylase isoforms and identification of their corresponding genes. Plant Physiol. 110 1395–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanjo, Y., Asatsuma, S., Itoh, K., Hori, H., and Mitsui, T. (2004). Proteomic identification of α-amylase isoforms encoded by RAmy3B/3C from germinating rice seeds. Biosci. Biotechnol. Biochem. 68 112–118. [DOI] [PubMed] [Google Scholar]
- Nanjo, Y., Oka, H., Ikarashi, N., Kaneko, K., Kitajima, A., Mitsui, T., Muñoz, F.J., Rodríguez-López, M., Baroja-Fernández, E., and Pozueta-Romero, J. (2006). Rice plastidial N-glycosylated nucleotide pyrophosphatase/phosphodiesterase is transported from the ER-Golgi to the chloroplast through the secretory pathway. Plant Cell 18 2582–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebenführ, A., Frohlick, J.A., and Staehelin, L.A. (2000). Redistribution of Golgi stacks and other organelles during mitosis and cytokinesis in plant cells. Plant Physiol. 124 135–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebenführ, A., Gallagher, L.A., Dunahay, T.G., Frohlick, J.A., Mazurkiewicz, A.M., Meehl, J.B., and Staehelin, L.A. (1999). Stop-and-go movements of plant Golgi stacks are mediated by the acto-myosin system. Plant Physiol. 121 1127–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa, Y., Hirano, T., Yoshimoto, K., Shimizu, M., and Kobayashi, H. (1999). Non-invasive quantitative detection and applications of non-toxic, S65T-type green fluorescent protein in living plants. Plant J. 18 455–463. [DOI] [PubMed] [Google Scholar]
- Palade, G. (1975). Intracellular aspects of the process of protein synthesis. Science 189 347–358. [DOI] [PubMed] [Google Scholar]
- Ritzenthaler, C., Nebenfuhr, A., Movafeghi, A., Stussi-Garaud, C., Behnia, L., Pimpl, P., Staehelin, L.A., and Robinson, D.G. (2002). Reevaluation of the effects of Brefeldin A on plant cells using tobacco Bright Yellow 2 cells expressing Golgi-targeted green fluorescent protein and COPI antisera. Plant Cell 14 237–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhamony, R.N., and Theg, S.M. (2006). Evidence for an ER to Golgi to chloroplast protein transport pathway. Trends Cell Biol. 16 385–387. [DOI] [PubMed] [Google Scholar]
- Robert, X., Haser, R., Mori, H., Svensson, B., and Aghajari, N. (2005). Oligosaccharide binding to barley α-amylase 1. J. Biol. Chem. 280 32968–32978. [DOI] [PubMed] [Google Scholar]
- Rothman, J.E., and Wieland, F.T. (1996). Protein sorting by transport vesicles. Science 272 227–234. [DOI] [PubMed] [Google Scholar]
- Saalbach, G., Jung, R., Kunze, G., Saalbach, I., Adler, K., and Muntz, K. (1991). Different legumin protein domains act as vacuolar targeting signals. Plant Cell 3 695–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell, D.J., and Hebert, D.N. (2003). Protein translocons: Multifunctional mediators of protein translocation across membranes. Cell 112 491–505. [DOI] [PubMed] [Google Scholar]
- Søgaard, M., Kadziola, A., Haser, R., and Svensson, B. (1993). Site-directed mutagenesis of histidine 93, aspartic acid 180, glutamic acid 205, histidine 290, and aspartic acid 291 at the active site and tryptophan 279 at the raw starch binding site in barley α-amylase 1. J. Biol. Chem. 268 22480–22484. [PubMed] [Google Scholar]
- Soll, J., and Schleiff, E. (2004). Protein import into chloroplasts. Nat. Rev. Mol. Cell Biol. 5 198–208. [DOI] [PubMed] [Google Scholar]
- Staehelin, L.A., and Moore, I. (1995). The plant Golgi apparatus: Structure, functional organization and trafficking mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 46 261–288. [Google Scholar]
- Takeuchi, M., Ueda, T., Sato, K., Abe, H., Nagata, T., and Nakano, A. (2000). A dominant negative mutant of sar1 GTPase inhibits protein transport from the endoplasmic reticulum to the Golgi apparatus in tobacco and Arabidopsis cultured cells. Plant J. 23 517–525. [DOI] [PubMed] [Google Scholar]
- Takeuchi, M., Ueda, T., Yahara, N., and Nakano, A. (2002). Arf1 GTPase plays roles in the protein traffic between the endoplasmic reticulum and the Golgi apparatus in tobacco and Arabidopsis cultured cells. Plant J. 31 499–515. [DOI] [PubMed] [Google Scholar]
- Terashima, M., Kubo, A., Suzawa, M., Itoh, Y., and Katoh, S. (1994). The roles of the N-linked carbohydrate chain of rice α-amylase in thermostability and enzyme kinetics. Eur. J. Biochem. 226 249–254. [DOI] [PubMed] [Google Scholar]
- Törmäkangas, K., Hadlington, J.L., Pimpl, P., Hillmer, S., Brandizzi, F., Teeri, T.H., and Denecke, J. (2001). A vacuolar sorting domain may also influence the way in which proteins leave the endoplasmic reticulum. Plant Cell 13 2021–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyooka, K., Goto, Y., Asatsuma, S., Koizumi, M., Mitsui, T., and Matsuoka, K. (2009). A mobile secretory vesicle cluster involved in mass transport from the Golgi to plant cell exterior. Plant Cell 21 1212–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyooka, K., Moriyasu, Y., Goto, Y., Takeuchi, M., Fukuda, H., and Matsuoka, K. (2006). Protein aggregates are transported to vacuoles by a macroautophagic mechanism in nutrient-starved plant cells. Autophagy 2 96–106. [DOI] [PubMed] [Google Scholar]
- van Dooren, G.G., Schwartzbach, S.D., Osafune, T., and McFadden, G.I. (2001). Translocation of proteins across the multiple membranes of complex plastids. Biochim. Biophys. Acta 1541 34–53. [DOI] [PubMed] [Google Scholar]
- von Schaewen, A., and Chrispeels, M.J. (1993). Identification of vacuolar sorting information in phytohemagglutinin, an unprocessed vacuolar protein. J. Exp. Bot. 44 339–342. [Google Scholar]
- Villarejo, A., et al. (2005). Evidence for a protein transported through the secretory pathway en route to the higher plant chloroplast. Nat. Cell Biol. 7 1224–1231. [DOI] [PubMed] [Google Scholar]
- Vitale, A., and Hinz, G. (2005). Sorting of proteins to storage vacuoles: how many mechanisms? Trends Plant Sci. 10 316–323. [DOI] [PubMed] [Google Scholar]
- Yu, T.S., et al. (2005). α-Amylase is not required for breakdown of transitory starch in Arabidopsis leaves. J. Biol. Chem. 280 9773–9779. [DOI] [PubMed] [Google Scholar]
- Zhang, X.P., and Glaser, E. (2002). Interaction of plant mitochondrial and chloroplast signal peptides with the Hsp70 molecular chaperone. Trends Plant Sci. 7 14–21. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.