Abstract
We have isolated and characterized the cDNA encoding the ornamental tobacco (Nicotiana langsdorffii X N. sanderae) homolog of the antirrhinum (Antirrhinum majus) MYB305. This transcription factor was robustly expressed at Stage 12 of nectary development but was only weakly expressed in the earlier Stage 6 nectaries. The ornamental tobacco MYB305 contains a conserved R2R3 MYB DNA binding domain with 76 amino acids in the activation domain. A green fluorescent protein-MYB305 fusion localized to nucleus of tobacco protoplasts and yeast one-hybrid assays demonstrated that it functions as a transcription activator. A conserved 23–amino acid C-terminal domain is required to activate gene expression. The coding region of the myb305 cDNA was expressed in Escherichia coli as a glutathione S-transferase fusion protein and was purified to homogeneity. This protein shows binding to two consensus MYB binding sites on the ornamental tobacco Nectarin I (nec1) promoter as well as to the single site located on the Nectarin V (nec5) promoter. Deletions of either of the binding sites from the nec1 promoter significantly reduced expression in nectary tissues. Temporally, MYB305 expression precedes that of nec1 and nec5, as would be expected if the MYB305 factor regulates expression of the nec1 and nec5 genes. Ectopic expression of MYB305 in foliage was able to induce expression of both nec1 and nec5, as well as two flavonoid biosynthetic genes in the foliage. Finally, RNA interference knockdown of MYB305 resulted in reduced expression of both nectarins and flavonoid biosynthetic genes. We conclude that expression of MYB305 regulates expression of the major nectarin genes in the floral nectary.
INTRODUCTION
Many species of angiosperms attract animals (insects, birds, and mammals) to visit their reproductive organs and physically transfer pollen from anthers, where it is produced, to the stigma where it begins the fertilization process. On the surface, reliance on other organisms to mediate sexual fertilization seems an unsatisfactory strategy; however, the wide diversity and great success of the angiosperms that have evolved over the past 125 million years punctuates the tremendous success of this strategy. Plants attract these visiting pollinators to the flower by offering a reward of nutritionally rich nectar, which is produced from an unusual floral organ, the nectary, that typically is located at the base of the flower near the ovary. Floral nectar accumulates in the cup-like environment formed at the floral base (Nepi, 2007). Although nectar is an aqueous solution of sugars, it also contains amino acids (Carter et al., 2006) as well as a variety of other components. Krebs cycle intermediates, lipids, vitamins, proteins, and biologically important metal cations have all been identified in nectars from a variety of species (Nicolson and Thornburg, 2007).
Because visiting pollinators are promiscuous and are not sterile, the plant's reproductive tract, which is bathed in this metabolically rich nectar, is at high risk for microbial infection. Teleologically, few places in the plant are as important as the ovary, where the next generation's seeds will develop. Therefore, plants must have mechanisms to defend the ovary from microbial attack. We have identified a number of proteins (termed nectarins) that accumulate in the soluble nectar of ornamental tobacco (Nicotiana langsdorffii X N. sanderae) plants (Carter et al., 1999; Carter and Thornburg, 2000, 2004b, 2004c; Naqvi et al., 2005). Understanding the biochemistry of these proteins has led to the identification of the nectar redox cycle, a complete biochemical pathway expressed in the soluble nectar of ornamental tobacco plants that produces a defense substance. We propose that this pathway has evolved to defend the gynoecium from invading microorganisms (Thornburg et al., 2003; Carter and Thornburg, 2004a; Carter et al., 2007).
The nectar redox cycle involves the production of very high levels of hydrogen peroxide: up to 4 mM (Carter and Thornburg, 2000). This pathway is initiated by a developmentally regulated, nectary-expressed NADPH oxidase that begins producing superoxide just prior to anthesis (Carter et al., 2007). The superoxide is subsequently disproportionated into oxygen and hydrogen peroxide by the major nectar protein, Nectarin I (NEC1; Carter and Thornburg, 2000). Among the other nectar proteins, Nectarin V (NEC5) is a glucose oxidase that also produces hydrogen peroxide but via a different mechanism from NEC1 (Carter and Thornburg, 2004c). Because of the highly oxidative nature of ornamental tobacco nectar, antioxidants (both ascorbate and β-carotene) are important in nectary function (Carter and Thornburg, 2004b; Horner et al., 2007). Other nectarins function to limit fungal invasion of the gynoecium by inhibiting fungal enzymes that degrade plant cell walls (Naqvi et al., 2005).
While nectar proteins have been long reported (Beutler, 1935; Zimmerman, 1954), their study at the molecular level is still in its infancy. The floral nectarins of only two species, ornamental tobacco and leek (Allium porrum), have been completely characterized. In leek, a mannose binding protein and alliinase were identified (Peumans et al., 1997), and both of these proteins are thought to function in defense of nectar. Recently, nectarins have been identified in extrafloral nectar of Acacia spp in Jacaranda mimosafolia floral nectar as well as in the reproductive secretions of gymnosperms. In each of these cases, the defensive proteins found in extrafloral nectar (Heil et al., 2005; González-Teuber et al., 2009), in Jacaranda floral nectar (Kram et al., 2008), and in gymnosperm secretions (Poulis et al., 2005; O'Leary et al., 2007; Wagner et al., 2007) appear to be classical defense proteins, such as pathogenesis-related protein lipase and thaumatin. Thus, nectarins represent a unique and quite varied group of proteins that function to protect plant secretions using a variety of molecular strategies.
To understand better the mechanisms that regulate expression of the nectarins in ornamental tobacco nectar, we analyzed nectarin gene expression and determined that several different transcriptional programs regulate their expression (Carter et al., 1999; Carter and Thornburg, 2004b, 2004c; Naqvi et al., 2005). For NEC1, the major nectar protein, we previously evaluated the expression from the nec1 promoter in transgenic plants (Carter and Thornburg, 2003). From deletion studies, we identified a consensus MYB binding site within the nec1 promoter and predicted that a MYB transcription factor was involved in the temporal expression of NEC1 and possibly NEC5 as well (Carter and Thornburg, 2003). This consensus MYB binding site was previously identified in flavonoid biosynthetic genes, including phenylalanine ammonia lyase (pal) and chalcone isomerase (chi) of antirrhinum (Antirrhinum majus), bean (Phaseolus vulgaris), and parsley (Petroselinum crispum; Sablowski et al., 1994). Two MYB transcription factors, MYB305 and MYB340, were shown to bind to this sequence and to regulate expression of flavonoid biosynthetic genes in antirrhinum flowers. It is interesting to note that the A. majus MYB305 gene was also highly expressed in nectaries (Moyano et al., 1996).
In other gene expression studies, we performed an EST study of gene expression at three different stages of ornamental tobacco nectary development (D.-L. Yin, K. Taylor, and R.W. Thornburg, unpublished results). One outcome of this study was the identification of a highly expressed MYB transcription factor in the nectary. This study was therefore designed to investigate a potential role of this highly expressed MYB transcription factor in regulating NEC1 and NEC5 expression.
RESULTS
The myb305 cDNA from LxS8 ornamental tobacco plants was cloned as part of a nectary EST project. Because we had previously suggested that a MYB transcription factor was responsible for the temporal activation of the nec1 promoter in tobacco nectaries (Carter and Thornburg, 2003), we characterized this factor in detail.
Expression of MYB305
To determine where this myb gene was expressed, we initially examined various whole-plant tissues by RT-PCR to evaluate gene expression. As seen in Figure 1, this myb gene is strongly expressed only in flowers. It was not expressed in stems (lane 6), roots (lane 8), or leaves (lane 12). In reproductive tissues, this myb gene was expressed in the ovary (lane 5), the floral tube (lane 7), and petals (lane 13) but was most strongly expressed in the floral nectary (lanes 1 to 4). This myb gene was not detected in anthers (lane 9), stamens (lane 10), or sepals (lane 11). Based on this analysis, we conclude that in ornamental tobacco, this myb gene was expressed uniquely in flowers with the highest level in the nectary and with lower levels in the ovary, floral tube, and petals.
Figure 1.
Temporal and Spatial Expression of myb305 in LxS8 Plants.
RNAs were isolated from individual tissues and processed for RT-PCR as described in Methods. Lane 1, stage 2 nectary; lane 2, stage 6 nectary; lane 3, stage 9 nectary; lane 4, stage 12 nectary; lane 5, ovary at anthesis; lane 6, stem; lane 7, floral tube at anthesis; lane 8, root; lane 9, anthers at anthesis; lane 10, stamens at anthesis; lane 11, sepals at anthesis; lane 12, leaf; lane 13, petals at anthesis. Cycle number for both myb305 and gapdh is 24 for all tissues tested.
Phylogenetic Analysis
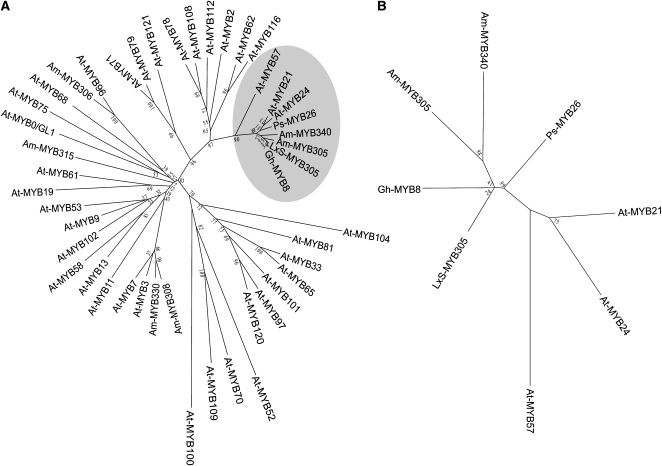
BLAST analysis (Altschul et al., 1990) of the translated protein sequence clearly identifies it as an R2R3 MYB transcription factor (e value = 2e-74) and reveals that this protein is most closely related to a group of MYB proteins that includes the A. majus MYB transcription factors MYB305 and MYB340 (Jackson et al., 1991), the Pisum sativum MYB26 (Uimari and Strommer, 1997), and the Gerbera hybrida MYB8 (Laitinen et al., 2005). Because Am-MYB305 was the first of this group of proteins to be identified (Jackson et al., 1991), we named this protein the N. langsdorffii X N. sanderae (LxS) MYB305.
To place this gene into a phylogenetic context, we analyzed the MYB DNA binding domains of a group of MYB proteins, including a representative subset of 37 of the 132 Arabidopsis thaliana R2R3 MYB proteins and six Antirrhinum MYB proteins using the neighbor joining method. The 37-protein Arabidopsis subset was chosen to cover all phylogenetic space but to simplify the analysis by limiting the number of individual proteins in this analysis (Kranz et al., 1998; Stracke et al., 2001). As seen in Figure 2A, the MYB DNA binding domains identify a distinct clade of proteins containing LxS-MYB305 and other closely related proteins, including one each from G. hybrida and P. sativa and two each from A. majus (Am-MYB305 and Am-MYB340) and Arabidopsis proteins (At-MYB21 and At-MYB24). In addition, At-MYB57 appears near the root of this clade. These three Arabidopsis genes are collectively known as the Family 19 clade of MYB genes (Kranz et al., 1998; Stracke et al., 2001). Based upon MPSS analysis (mpss.udel.edu/at/), both At-MYB21 and At-MYB24 are exclusively expressed in the MPSS inflorescence libraries, while At-MYB57 is expressed in flowers and in a variety of other tissues.
Figure 2.
Phylogenetic Analysis of LxS-MYB305.
(A) Analysis of the LxS-MYB305 DNA binding domain and the MYB DNA binding domains from A. majus, Arabidopsis, G. hybrida cv Terra Regina, and P. sativum MYBs. The alignment used for this analysis and GenBank accessions are provided in Supplemental Figure 5 online, and a text file of the alignment is available as Supplemental Data Set 1 online. The shaded area shows a distinct clade of proteins containing LxS-MYB305 and its closely related proteins.
(B) Analysis of the complete protein sequences of LxS-MYB305 and its closely related MYBs. The alignment used for this analysis and GenBank accessions are provided in Supplemental Figure 6 online, and a text file of the alignment is available as Supplemental Data Set 2 online.
To get an enhanced picture of this clade, the full protein sequences (DNA binding domain plus activation domain) of the eight most closely related genes were phylogenetically analyzed and are displayed in Figure 2B. As can be seen, LxS-MYB305 more closely resembles G. hybrida MYB8 than the A. majus MYBs or P. sativa MYB26. The association of the Arabidopsis proteins with this group suggests that the MYB305 family proteins may share some functions with At-MYB Family 19 proteins; however, the Arabidopsis proteins also appear to form a separate subclade within this group, and this clade separation suggests that the MYB305 family genes may have unique functions.
Nuclear Localization of MYB305
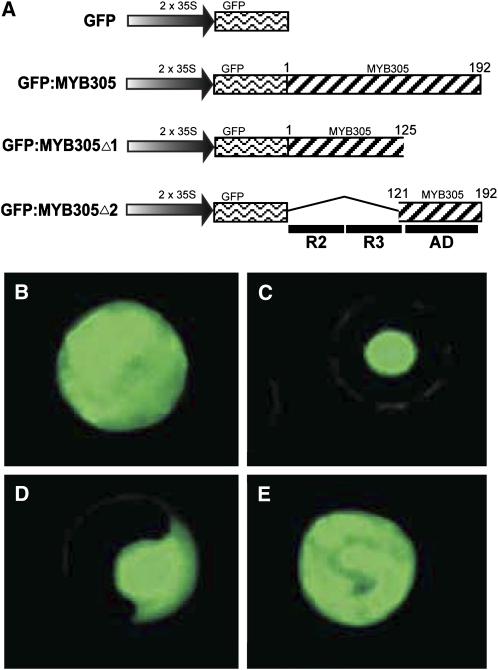
To evaluate the subcellular localization of MYB305, we used a transient transformation assay using tobacco mesophyll protoplasts. For these studies, the green fluorescent protein (GFP) reporter gene was fused in frame to the 5′ end of the LxS-MYB305 open reading frame (Figure 3A). The plasmid pMDC43 encoding GFP alone served as a control. Protoplasts transformed with pMDC43 showed GFP fluorescence throughout the entire cell (Figure 3B). By contrast, cells transformed with the full-length GFP:MYB305 construct showed bright fluorescence localized to the nucleus (Figure 3C). To investigate which part of the MYB protein possessed the nuclear localization signal, LxS-MYB305 was truncated into two parts, MYB305Δ1 and MYB305Δ2 (Figure 3A). MYB305Δ1 contains the R2R3 MYB DNA binding domain but not the transcriptional activation domain, whereas MYB305Δ2 contains the transcriptional activation domain but not the R2R3 DNA binding domain. The two truncated MYB fragments were each fused in frame with GFP. The GFP:MYB305Δ1 fusion protein localized to the nucleus, whereas the GFP:MYB305Δ2 fusion protein did not (Figures 3D and 3E), indicating that the nuclear localization signal resides within the R2R3 domain.
Figure 3.
Nuclear Localization of LxS-MYB305.
(A) The plasmid constructs used for the nuclear localization studies. The amino acid number of the included LxS-MYB305 fragments is shown above the hatched bars. The locations of the R2R3 DNA binding domains and the activation domain (AD) of the LxS-MYB305 protein are shown below the figure.
(B) to (E) Transient expression assay of GFP fluorescence in transformed N. tabacum cv Xanthi mesophyll protoplasts. Typical results of tobacco protoplasts transformed with constructs encoding GFP only (pMDC43; [B]), full-length GFP:MYB305 fusion (pRT656; [C]), truncated GFP:MYB305Δ1 fusion (pRT657; [D]), and truncated GFP:MYB305Δ2 fusion (pRT658; [E]).
MYB305 Is a Transcription Activator in Yeast
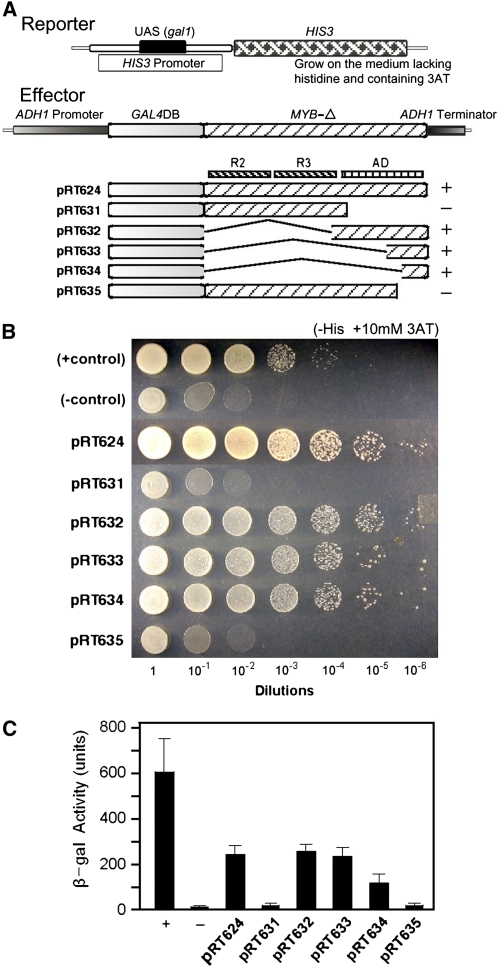
To investigate whether MYB305 can serve as a transcriptional activator, we used yeast one-hybrid assays to drive the expression of marker genes (Shioda et al., 1997). Initially, the full-length LxS-MYB305 coding sequence was fused to the 3′ end of the GAL4 DNA binding domain to generate the effector construct pRT624 (Figure 4A). The reporter construct contains a modified his3 gene in which a gal1 upstream activation sequence has been inserted into the his3 promoter. Positive and negative controls were a reconstructed GAL4 (GAL4DB-AD) and a construct containing only the GAL4 DNA binding domain (pDBleu). All constructs were transformed into the yeast strain MaV203 (Vidal et al., 1996) that harbors his3− reporter systems (Figure 4A) as well as a similar lacZ reporter. All constructs were assayed visually using dilution growth tests, and expression was quantitated using the lacZ reporter system.
Figure 4.
LxS-MYB305 Is a Transcriptional Activator in Yeast and It Activates the Transcription of Reporter Genes via Its C-Terminal Domain.
(A) Constructs used in these studies. The GAL4 DNA binding domain (DB) and LxS-MYB305-Δ fragment (MYB-Δ) fusion genes driven by the ADH1 promoter serve as effectors; the HIS3 and LacZ (data not shown) genes with Gal4 DNA binding site on their promoters serve as reporters. The constructs of each effector are shown with DNA fragments identified above the individual constructs. R2 and R3 represent the R2 and R3 repeat of MYB DNA binding domains, respectively; AD represents the activation domain. The + control contains a recombined Gal4 DNA binding domain and activation domain; the – control contains Gal4 DNA binding domain only.
(B) Serial dilution growth assays of transformed yeast on selective YSM His− + 10 mM 3AT medium. Constructs are identified to the left of the figure. Dilutions are identified below the figure.
(C) Quantitation of β-galactosidase activity in yeast. Constructs are identified below the bars (means ± sd, n = 4 with two replicates for each sample). +, positive control construct; −, negative control construct.
[See online article for color version of this figure.]
As shown in Figure 4B, little growth was observed when the pDBleu strain (negative control) was grown on medium lacking histidine. By contrast, strong growth was observed with the reconstructed GAL4 strain (positive control) at 10−3 and even 10−4 dilutions. The effector construct, pRT624, that encoded the GAL4 DNA binding domain fused with the complete MYB305 protein also resulted in strong growth of transgenic yeast. It also gave ∼20-fold more β-galactosidase activity than the negative control in the quantitative lacZ reporter assay (Figure 4C). Based upon these data, we conclude that the LxS-MYB305 is capable of activating transcription in yeast.
MYB305 Activates Transcription via Its Conserved C Terminus
Because MYB305 is a transcription activator, we sought to identify those portions of the molecule responsible for transcription activation. Therefore, we prepared a series of DNA constructs that lacked various portions of the activation domain. These constructs, also shown in Figure 4A, contain the entire R2R3 domain but no activation domain (amino acids 1 to 125; pRT631), the complete activation domain but no R2R3 domain (amino acids 108 to 192; pRT632), the C-terminal half of the activation domain (amino acids 158 to 192; pRT633), the C-terminal end of the activation domain (amino acids 169 to 192; pRT634), and the entire MYB305 protein lacking only the C-terminal 30 amino acids (amino acids 1 to 162; pRT635). All constructs were transformed into the yeast strain MaV203 and were assayed by dilution growth tests and quantitated with the lacZ reporter system.
As shown in Figure 4B, yeast cells harboring pRT631, which lacks the complete activation domain, do not grow well on media lacking histidine. By contrast, each of the constructs that contains the C-terminal end of the activation domain (pRT632, pRT633, and pRT634) all grow quite well on media lacking histidine, indicating that sequences in the C-terminal region of the MYB305 are responsible for transcriptional activation. To confirm this, we tested pRT635 containing the complete protein sequence except for the final 30 amino acids. This construct did not grow well on the media lacking histidine, confirming that the C terminus of the protein mediates transcriptional activation. Quantitation of the β-galactosidase activity in these yeast strains (Figure 4C) demonstrated that pRT632, pRT633, and pRT634 provide 10- to 20-fold greater transcriptional activity than the control strain, pDBleu. Activity measured from constructs lacking either the entire activation domain (pRT631) or the final 30 amino acids (pRT635) of that domain were not above background level.
Acidic Residues Near the MYB305 C Terminus Mediate Transcriptional Activation
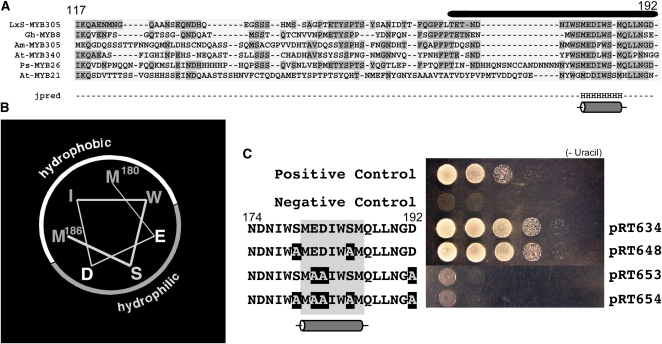
To evaluate further the function of the activation domain, we analyzed its predicted secondary structure (amino acids 117 to 192) using Jpred, a neural network-based program (Cuff and Barton, 1999, 2000) that identifies structural homologs, performs multiple sequence alignment, and classifies each residue as likely to reside in an α-helix, β-sheet, or random coil secondary structure. As shown in Figure 5A, the algorithm identified and aligned the same transcription factors found in our earlier BLAST analyses. This alignment also shows that the C-terminal region required for transcriptional activation identified in construct pRT634 (black bar above the sequences) is the most conserved feature of these transcription factors. The Jpred secondary structure prediction algorithm identified only one feature shared among the aligned activation domains, a single α-helix that is seven amino acids long, residing near the C terminus. The remaining portions of the activation domains of these transcription factors were predicted to be unstructured.
Figure 5.
Analysis of the MYB305 Activation Domain.
(A) Alignment of amino acid sequences of proteins closely related to LxS-MYB305. The amino acid numbers in LxS-MYB305 are indicated above the sequences, and the bar represents the minimal activation domain identified in Figure 4. Regions of identity are shaded. The secondary structural predictor Jpred predicts only one region of secondary structure, a single α-helix located in the center of the minimal activation domain.
(B) Surface analysis of the predicted α-helix showing the hydrophobic and hydrophilic surfaces. The positions of Met-180 and Met-186 are identified.
(C) Site-specific mutagenesis of the minimal activation domain of the LxS-MYB305. Serial dilution growth assays of transformed yeast on selective YSM Ura− medium. Effector and reporter constructs are identical to those in Figure 4. Positive (Gal4DB-AD) and negative (pDBleu) controls are shown. The site-specific mutants are identified to the left of the figure, with construct numbers shown at right. The location of the predicted α-helix is shaded and shown below these sequences. The spots are serial dilutions of each transformed yeast strain.
[See online article for color version of this figure.]
The predicted α-helix lies near the center of the sequence that is required for transcriptional activation. When we mapped amino acid residues from this region of LxS-MYB305 onto a helical structure (Figure 5B), the hydrophobic residues Met-180, Ile-183, Trp-184, and Met-186 lie on one face of the α-helix, while the hydrophilic residues Glu-181, Asp-182, and Ser-185 are located on the opposite face of the helix, suggesting this may be a classical amphipathic α-helix. Because amphipathic α-helices often present important amino acid side chains for biological activity, we hypothesize that the conserved amphipathic nature of this helix contributes to the transcriptional activation function in these proteins.
Short acidic sequences are well known to activate transcription (Ruden et al., 1991). To evaluate the potential role of acidic sequences in the activation activity of the LxS-MYB305 C-terminal domain, we made a series of site-directed mutations designed to alter acidic amino acids. In addition to the Asp and Glu residues that occur in this region, acidic charges can also be generated by phosphorylation of amino acids in this portion of the molecule. Phosphorylation is a common posttranslational modification of transcription factors (Magasanik, 1988; Decker and Kovarik, 1999) and the online analysis tool NetPhos (Blom et al., 1999) predicted that Ser-185 in the C-terminal domain of LxS-MYB305 may be phosphorylated. Therefore, we prepared a series of modified C-terminal domains fused to the GAL4 DNA binding domain. In construct pRT648, both Ser-179 and Ser-185 were changed to Ala. In construct pRT653, the acidic residues Glu-181, Asp-182, and Asp-192 were changed to Ala. In pRT654, all five residues were changed to Ala. As shown in Figure 5C, changing the Ser residues to Ala in construct pRT648 had no effect on the transcriptional activation of the modified protein, suggesting that phosphorylation of these Ser residues may not be important for transcriptional activation. By contrast, changing the acidic residues to Ala in either construct pRT653 or pRT654 completely abolished the transcriptional activation activity of the fusion protein. We conclude that acidic residues located near the MYB305 C terminus mediate transcriptional activity of this transcription factor.
MYB305 Binds the nec1 and nec5 Promoters in Vitro
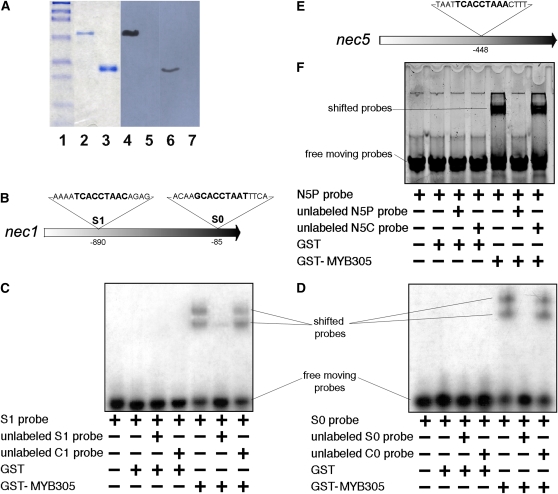
NEC1 is the major protein secreted into nectar of ornamental tobacco (Carter et al., 1999; Carter and Thornburg, 2000). Cloning of the nec1 promoter and subsequent promoter deletion analysis revealed the presence of a consensus MYB DNA binding site located at −899 to −891 of the promoter (Carter and Thornburg, 2003). Deletion of this site resulted in loss of reporter gene expression in the nectary. Based upon these analyses, we previously predicted the involvement of a MYB protein in nec1 transcriptional activation. In this study, we reexamined the nec1 promoter sequence and identified a second potential MYB binding site located at −86 to −78 of the nec1 promoter (Figure 6B). To evaluate whether LxS-MYB305 can bind to these regions of the nec1 promoter, we expressed this transcription factor as a glutathione S-transferase (GST) protein fusion in Escherichia coli and purified it by virtue of the GST tag. The GST-MYB305 fusion protein was purified to a single protein band as visualized on SDS-PAGE (Figure 6A, lane 2). We also developed antisera against the LxS-MYB305 for use in these studies. This antiserum reacts with the GST-MYB305 fusion protein (lane 4) but not against GST alone (lane 5). The antibodies also recognize the native MYB305 in nectary tissues (lane 6) but show no reactivity against any proteins present in the foliage of the ornamental tobacco plants (lane 7).
Figure 6.
MYB305 Binds to the nec1 and nec5 Promoter Fragments.
(A) Expression and purification of LxS-MYB305 protein. Lanes 1 to 3 are Coomassie blue stained: lane 1, protein standards; lane 2, purified GST-MYB305 protein; lane 3, GST protein. Lanes 4 to 7 are immunoblots using purified anti-LxS8-MYB305 antibodies: lane 4, purified GST-MYB305 protein; lane 5, GST protein; lane 6, 100 μg of protein from Stage 12 nectary tissue of wild-type LxS8 plant; lane 7, 100 μg of protein from foliage of wild-type LxS8 plant.
(B) Structure of the N. plumbaginifolia nec1 promoter showing the location and sequences of the two consensus MYB binding sites S1 and S0.
(C) Mobility shift of the radiolabeled S1 probe. Inclusion of individual components is indicated below each lane. S1 probe represents the S1 MYB binding site on the nec1 promoter. The unlabeled S1 probe is identical to the radiolabeled S1 probe and the unlabeled C1 probe differs from S1 probe with MYB consensus sequence replaced by poly A/T as indicated in Supplemental Table 1 online. Film exposure was 2 h.
(D) Mobility shift of the radiolabeled S0 probe. Inclusion of individual components is indicated below each lane. S0 probe represents the S0 MYB binding site of the nec1 promoter. The unlabeled S0 probe is identical to the radiolabeled S0 probe, and the unlabeled C0 probe differs from S1 probe with MYB consensus sequence replaced by poly A/T as indicated in Supplemental Table 1 online. Film exposure was 2 h.
(E) Structure of the N. plumbaginifolia nec5 promoter showing the location of the consensus MYB binding site.
(F) Mobility shift of the fluorescently labeled N5P probe. Inclusion of individual components is indicated below each lane. The unlabeled N5P probe is identical to the fluorescently labeled N5P probe, and the unlabeled N5C probe differs from N5P probe as indicated in Supplemental Table 1 online.
[See online article for color version of this figure.]
This purified GST-MYB305 fusion protein was then used in electrophoretic mobility shift assays to evaluate its interactions with [32P]-labeled DNA probes. Two duplex oligonucleotide probes were used for these analyses, the S1 probe (N1S1; see Supplemental Table 1 online) corresponds to the upstream MYB binding site (−899 to −891) and the S0 probe (N1S0) corresponds to a second binding site (−86 to −78) within the nec1 promoter. The GST protein alone cannot bind either S1 or S0 probes (Figures 6C and 6D), whereas GST-MYB305 can bind both labeled probes. In each case, the addition of GST-MYB305 resulted in a substantial mobility shift of the probe. The addition of 10-fold excess unlabeled S1 or S0 probe successfully competed for the binding of GST-MYB305 to the respective labeled probe. We also tested whether the addition of a 10-fold excess of an unlabeled, modified competitor C1 (N1C1) or C0 (N1C0) probe that lacked the MYB binding site could displace the interaction of GST-MYB305 with the original labeled probes. In each case, the competitor probes, either C1 or C0, were identical to the S1 and S0 probes with the exception that the nine-nucleotide MYB consensus sequence was replaced with a poly A/T track (see Supplemental Table 1 online). As shown in Figure 6, the addition of these unlabeled competitor probes had no effect on the binding of the GST-MYB305 to the labeled S1 and S0 probes. Based on these observations, we conclude that LxS-MYB305 binds specifically to the S1 and S0 MYB binding sites in vitro.
The regulation of the Nectarin V (nec5) promoter is similar to that of the nec1 promoter, and there is a single consensus MYB binding site located at −448 of the nec5 promoter (Figure 6E; Carter and Thornburg, 2003). Therefore, we tested the purified GST-MYB305 for interaction with a synthetic nec5 promoter element. As shown in Figure 6F, GST-MYB305 did indeed interact specifically with the nec5 MYB binding site. This binding could be competed with an excess of unlabeled N5P probe but not with the unlabeled mutant probe (N5C). These observations led us to conclude that in addition to binding the nec1 promoter, LxS-MYB305 also interacts with the MYB binding site in the nec5 promoter.
The patterns of mobility shifts observed in Figures 6C and 6D show multiple interactions of the DNA probes with GST-MYB305, suggesting a possible multimerization of the proteins interacting with the probes. GST is known to dimerize in vitro (Lim et al., 1994), and the observed patterns may be due to GST multimerization. To test whether MYB305 could form a homodimer, we used the yeast two-hybrid method. We fused the first 162 amino acids of LxS-MYB305 lacking the final 30 amino acids of the activation domain with the yeast two-hybrid bait vector, pDEST32 (www.invitrogen.com), and the full-length LxS-MYB305 to the two-hybrid prey vector, pDEST22, respectively. No activity was detected in the two-hybrid interaction (see Supplemental Figure 1 online), and we conclude that the truncated 162–amino acid LxS-MYB305 protein does not homodimerize in the yeast two-hybrid assay and that multimerization is likely due to the presence of the GST in the GST-MYB305 fusion protein. However, because the bait construct lacked the C-terminal 30 amino acids, we cannot unequivocally conclude that the intact protein does not dimerize through this small C-terminal region.
Wild-Type, but Not Mutant, nec1 Promoters Drive β-Glucuronidase Expression
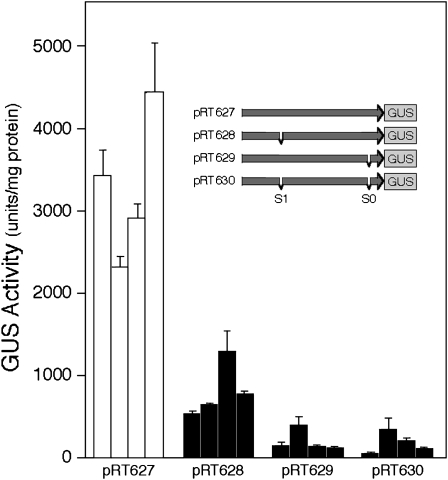
To evaluate the role of the MYB binding sites in regulating transcription from the nec1 promoter, we generated a series of promoter-β-glucuronidase (GUS) constructs that contained the wild-type nec1 promoter or the nec1 promoter with a mutation at the S1 site, at the S0 site, or at both sites (Figure 7, inset). These promoter-GUS constructs were mobilized into Agrobacterium tumefaciens LBA4404 and used to transform N. tabacum cv Xanthi plants. After selection, the plants were grown to floral maturity. Subsequently, nectaries were isolated and tested for nec1 promoter function. As shown in Figure 7, four transgenic plants, each harboring the wild-type nec1 promoter-GUS fusion construct, all expressed between 2300 and 4400 units of GUS activity per mg nectary protein. By contrast, when either the S1 site, the S0 site, or both sites were mutated, there was a dramatic reduction in GUS activity expressed in the nectaries of each of four independent transformants prepared from each construct, suggesting that both the S1 and S0 consensus MYB binding sites are both important for promoter activation. These data are consistent with our earlier observation that deletion of the upstream site results in a dramatic loss of promoter activity (Carter and Thornburg, 2003).
Figure 7.
Analysis of Wild-Type and Mutant nec1 Promoters in Transgenic N. tabacum cv Xanthi Plants.
Different promoter constructs were transformed into tobacco, and mature floral nectaries of transgenic plants were assayed for β-glucuronidase activity. Inset shows the clone number and structure of the various constructs tested. The MYB site mutations were effected by deletions of the binding site: pRT627 contains the full-length unmodified promoter (−1216..+50); pRT628 contains the full nec1 promoter with S1 MYB site deleted (−1216.. −898 + −892..+50); pRT629 contains the full nec1 promoter with the S0 MYB site deleted (−1216.. −84 + −78..+50); pRT630 contains the full nec1 promoter with both MYB sites deleted (−1216.. −898 + −892.. −84 + −78..+50). GUS activity is presented in units/mg of nectary protein for four independent transformants for each of the four constructs (mean ± sd, n = 6).
Temporal Expression of myb305, nec1, and nec5
We have provided evidence that MYB305 may function as a transcriptional regulator of nec1 expression. The LxS-MYB305 protein is nuclear localized, it activates transcription in yeast, it binds to nec1 and nec5 promoter sequences in vitro, and mutations of the binding sites in the nec1 promoter destroy promoter activity in vivo. If MYB305 does indeed regulate nec1 and nec5 expression in plants, it should be expressed prior to either of these genes. Therefore, we used RT-PCR to evaluate when the nectarin genes and MYB305 are expressed during nectary development.
Tobacco flower development previously has been divided into 12 morphologically distinct stages between the mature bud stage (stage S1) and anthesis (S12) (Koltunow et al., 1990). To evaluate when myb305 is expressed, we identified stage-specific flowers, isolated the nectaries, extracted stage-specific mRNA, and performed RT-PCR for myb305 as well as for nec1 and nec5 mRNAs. We used 18S and 26S RNA as loading and PCR controls.
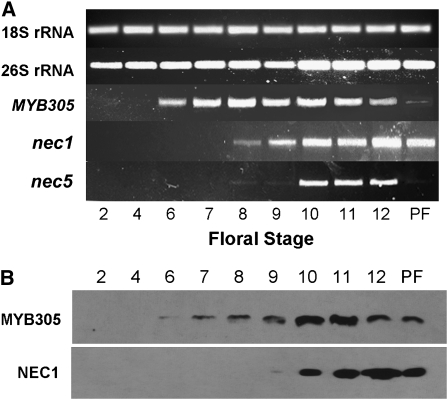
As shown in Figure 8A, myb305 is expressed by floral stage S6 and remains on until floral stage S12 (anthesis). These results are similar to the results shown in Figure 1. After fertilization, myb305 is abruptly downregulated. By contrast, both nec1 and nec5 begin to be expressed after stage S8 (∼36 h later). Although nec1 is more strongly expressed than nec5, they both continue to be expressed, accumulating to high levels at anthesis (stage S12). These results are entirely consistent with our earlier observations on the expression patterns of nec1 and nec5 (Carter et al., 1999; Carter and Thornburg, 2003, 2004c). Figure 8B shows MYB305 and NEC1 protein levels by immunoblot. These protein blots confirm the expression of myb305 and nec1 in Figure 8A. Together, these results demonstrate that myb305 is expressed prior to nec1 and nec5 and that MYB305 protein accumulates in the nectary prior to expression of both the nec1 transcript and the NEC1 protein. Thus, we conclude that expression of myb305 temporally precedes the expression of nec1 and nec5 in developing nectaries.
Figure 8.
Temporal Expression of myb305 in Nectaries of Wild-Type LxS8 Plants.
(A) RT-PCR of myb305, nec1, and nec5 in the nectary glands of LxS8 plants at different floral stages. Both 18S and 26S rRNAs were used as controls. PF = 48 h postfertilization. Amplification cycle number for 18S rRNA, 26S rRNA, and myb305 is 24 for all floral stages. Amplification cycle number for nec1 and nec5 is 26 for all floral stages.
(B) Protein blotting of MYB305 and NEC1 proteins in the nectary glands of LxS8 plants at different floral stages. Film exposure for MYB305 was 20 min. Film exposure for NEC1 was 30 min.
Ectopic Expression of MYB305 Causes Foliar Expression of nec1 and nec5
To test further the functionality of MYB305 in nectarin gene expression, we expressed the LxS-myb305 cDNA in the foliage of N. tabaccum plants and tested whether this ectopic expression was able to drive expression of nectarin genes in leaves. In addition, two genes in the flavonoid biosynthetic pathway, pal and chi, have previously been shown to be regulated by the A. majus MYB305 (Sablowski et al., 1994, 1995), so we also evaluated expression of these genes in the foliage of these plants.
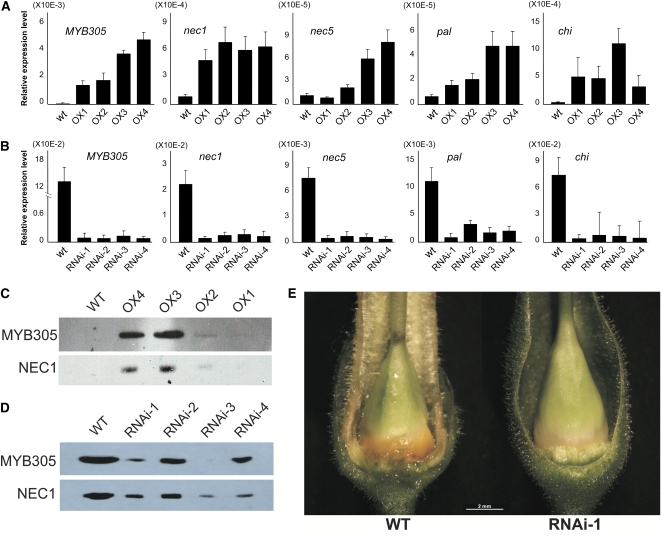
For this study, we developed four independent transgenic overexpressor (OX) plant lines that express the coding region of LxS-myb305 under the control of the strong cauliflower mosaic virus 35S promoter. The phenotype of the OX plants was similar to the wild-type plants, with the exception that they were shorter, growing to ∼50% the height of wild-type plants (see Supplemental Figure 2 online). As shown in Figure 9A, quantitative RT-PCR analysis demonstrates that none of the genes of interest were highly expressed in the foliage of wild-type, untransformed tobacco plants. These results confirm the expression pattern of myb305 shown in Figure 1 as well as the published patterns of expression previously observed for nec1 and nec5 (Carter et al., 1999; Carter and Thornburg, 2003, 2004c). As opposed to the wild-type plants, the myb305 mRNA is detectable in the foliage of each of the overexpression lines. Similarly, nec1 and the nec5 transcripts were found in the overexpression lines. One line, OX1, showed a very weak signal for nec5, while all other lines showed substantial levels of nec1 and nec5 expression in the MYB305 overexpression lines. In addition, we also tested the expression of the flavonoid genes pal and chi. Neither of these flavonoid genes was expressed in the foliage of wild-type tobacco plants (Figure 9A), but both were observed in each of the MYB305 overexpression lines. Based on these results, we conclude that ectopic expression of each of the target genes (nec1, nec5, pal, and chi) in the foliage correlates with the expression of myb305. In contrast with nec1 and nec5, neither Nectarin III (nec3) nor Nectarin IV (nec4) were expressed in the foliage of these plants (see Supplemental Figure 3 online), suggesting that these genes are not regulated by MYB305. These results for MYB305 and NEC1 were confirmed at the protein level by immunoblot analysis. Wild-type plants showed undetectable levels of both MYB305 and NEC1 (Figure 9C). Again, these results are internally consistent with our earlier observations (Figure 7A) and with previously published observations (Carter et al., 1999, 2007). Similarly, MYB305 and NEC1 proteins were readily observed in the OX plants, especially OX3 and OX4. The OX1 and OX2 plants showed low levels of MYB305 and NEC1 and required long exposures to observe the bands. The wild-type plants failed to show any bands, even with these long exposures.
Figure 9.
Analysis of LxS-MYB305 Overexpression and RNAi Plants.
(A) Quantitative RT-PCR analysis of LxS-myb305, nec1, nec5, pal, and chi expression in the foliage of LxS-MYB305 overexpression N. tabacum plants. Means + sd are shown (four biological replicates with two technical replicates each).
(B) Quantitative RT-PCR analysis of LxS-myb305, nec1, nec5, pal, and chi expression in the stage 12 nectary glands of LxS-MYB305 RNAi N. tabacum plants. Means + sd are shown (four biological replicates with two technical replicates each).
(C) Immunoblotting of MYB305 and NEC1 proteins in the foliage of LxS-MYB305 overexpression N. tabacum plants. Film exposure time was 1 h for MYB305 and 12 h for NEC1.
(D) Immunoblotting of MYB305 and NEC1 proteins in the stage 12 nectary glands of LxS-MYB305 RNAi N. tabacum plants. Film exposure for both MYB305 and NEC1 was 30 min.
(E) Phenotype comparison of wild-type N. tabacum nectary and RNAi N. tabacum nectary. Bar = 2 mm.
RNA Interference Knockdowns of MYB305 Also Knocks Down Expression of nec1 and nec5
Finally, we also prepared a series of RNA interference (RNAi) lines that showed reduced expression of myb305 in the nectary. Quantitative RT-PCR analysis of Stage 12 nectaries from wild-type plants (Figure 9B) shows strong expression of myb305, both nec1 and nec5, as well as both of the flavonoid biosynthetic genes (pal and chi) in the wild-type plants. Again, these data are consistent internally with our earlier observations (Figures 1 and 8) as well as with previously published observations (Moyano et al., 1996; Carter et al., 1999; Carter and Thornburg, 2003, 2004c). In each of the four independent RNAi knockdown lines, nectary expression of myb305 is reduced. Similarly, both nec1 and nec5 are also reduced in the nectaries of these knockdown lines, as are both of the flavonoid biosynthetic genes. Thus, we conclude that a reduction in the level of myb305 expression correlates with reduced levels of nec1 and nec5 expression as well as of pal and chi expression. As before, we also examined the expression of MYB305 and NEC1 at the protein level using immunoblots (Figure 9D). While the wild-type plants showed substantial amounts of both proteins, each of the RNAi knockdown lines showed reduced levels of the proteins in Stage 12 nectaries, confirming that the protein levels accumulating in the nectaries of these RNAi plants reflect the levels of nectary-expressed myb305 and nec1 mRNAs.
Additional Phenotypes Associated with the MYB305 RNAi and OX Plants
In addition to the molecular phenotypes observed in the RNAi lines and OX lines (i.e., myb305, nec1, nec5, pal, and chi expression), we also have observed additional phenotypes that affect the nectary, flower, and the whole plant. The phenotypes of RNAi lines include failure of the nectary to accumulate high levels of β-carotene (Figure 9E), a considerably reduced flow of nectar, failure of the flower petals to expand at anthesis, decreased pigmentation in the petals, an elongated style, and an abscission phenotype in which the entire floral bud falls from the stalk. Because these phenotypes are unrelated to the role of MYB305 in regulating the nectarin genes, these additional phenotypes will be the subject of a future, more detailed investigation of these plants. The ectopic expression of MYB305 results in a unique plant shape phenotype: the OX plants grow slower, are dwarf, branch earlier, and have crinkly leaves (see Supplemental Figure 2 online). The broad nature of these additional phenotypes suggests that MYB305 has roles in floral development that go beyond flavonoid metabolism and nectar protein expression.
DISCUSSION
In earlier work, we identified a consensus MYB binding site within the nec1 promoter and hypothesized that a MYB family transcription factor might activate the temporal expression of the nec1 gene in vivo (Carter and Thornburg, 2003). In this study, we identified MYB305 as a major nectary-expressed transcription factor. We have shown that this factor is expressed exclusively in flowers and have presented evidence that this protein is the transcription factor that directly binds to and activates transcription of the both the nec1 and nec5 promoters. Phylogenetically, LxS-MYB305 is closely related to snapdragon MYB305 and homologs from G. hybrida and P. sativa, as well as to a group of MYB proteins from Arabidopsis (MYB21, MYB24, and MYB57).
This group of proteins has long been thought to function in the activation of flavonoid biosynthetic genes. Am-MYB305 was first identified in 1991 in Antirrhinum flowers (Jackson et al., 1991). It was later shown to regulate transcription of the pal, chi, and 4-coumaryl-CoA-ligase genes in snapdragon flowers (Sablowski et al., 1994). Another related MYB protein, MYB340, is functionally redundant in snapdragon flowers (Moyano et al., 1996) and is closely related to this group of proteins (Figure 2). These two proteins, MYB305 and MYB340, appear to be regulated by phosphorylation in Antirrhinum flowers (Moyano et al., 1996). When the Am-MYB305 protein was expressed in foliage, ectopic expression of flavonoids was observed (Sablowski et al., 1995). In pea, MYB26 binds the P-box–like binding sites in the promoter regions of several flavonoid biosynthetic genes (Uimari and Strommer, 1997). Gh-MYB8 interacts with bHLH factor GMYC1 and is required for activation of a late anthocyanin biosynthetic gene promoter PGDFR2 (Laitinen et al., 2005). So activation of genes in the flavonoid and anthocyanin pigmentation pathways may be a general feature of these proteins.
The Arabidopsis MYB21 and MYB24 genes are involved in jasmonate response during stamen development (Mandaokar et al., 2006). In Arabidopsis, they are required for proper anther development (Li et al., 2006; Mandaokar et al., 2006). It has also been shown that overexpression of MYB24 causes aberrant anther development (Yang et al., 2007). MYB21 is a flower-specific gene, but in cop1 mutants, MYB21 is expressed throughout the seedling tissues (Shin et al., 2002), although MYB24 is not (Li et al., 2006). Whether cop1 regulates the myb305 genes in other species is not clear and because cop1 mutants are not available in snapdragon, gerbera, tobacco, or pea, this may remain unclear for some time.
Because the Arabidopsis genes appear to function in anther development, we also carefully examined anther length in the flowers of our wild-type LxS-MYB305 RNAi plants as well as the LxS-MYB305 OX plants. While the anthers all appeared to be morphologically normal, they were affected in their length (see Supplemental Table 2 online). Both the OX and the RNAi lines had shorter anthers than the wild type, with the RNAi lines being more severely affected. Likewise, the style length for each of the mutant lines was also shorter than the wild type; however, style length showed a more severe reduction with the OX lines. Thus, while both anther and style lengths were affected by both overexpression of MYB305 as well as by knockdown of MYB305 expression, there was not a clear direct association of MYB305 expression with the phenotype. Whether the related MYB305 proteins in other species also function in anther development is unknown. None of these earlier papers dealing with MYB305 that have been published over the past 15 years has mentioned the role of these proteins in anther development; however, this does not rule out the possibility that these proteins could have marginal function in anther development as observed in tobacco.
We have also examined nectaries of the Arabidopsis myb21 and myb24 mutant lines as well as the myb21 and myb24 double mutant lines (Mandaokar et al., 2006), but we have not observed any clear nectary phenotypes in these mutants. Thus, although they share high identity, the relationship of the myb305 genes with the Arabidopsis Family 19 MYB genes is far from clear. However, it is interesting to note that the MYB26 protein sequence from P. sativa (a rosid) clusters together with the sequence from ornamental tobacco (an asterid) rather than the sequences from Arabidopsis (another rosid). To confirm the robustness of this phylogeny, we analyzed the ornamental tobacco sequence together with only the rosid sequences (from pea and Arabidopsis). As shown in Supplemental Figure 4 online, the pea sequence still clusters with the ornamental tobacco sequence rather than with the other rosid sequences. This further highlights the differences between the myb305 family genes and the Arabidopsis Family 19 MYB genes and suggests that the myb305 genes may have additional functions. It is noteworthy that the myb305 family genes are identified in species that produce relatively large quantities of nectar as opposed to Arabidopsis, which produces very little nectar (Davis et al., 1998). Whether the additional functions of MYB305 relate to high levels of nectar production is currently under investigation.
We demonstrated that the LxS-MYB305 protein accumulates in nuclei of transiently transformed tobacco protoplasts and that the nuclear localization signal is localized in the R2R3 domain. Arabidopsis MYB24 has also been shown to be nuclear (Yang et al., 2007). Within the nucleus, these proteins interact with the promoters of several flavonoid and nectarin genes. We demonstrated that the LxS-MYB305 protein functions as a transcriptional activator in yeast one-hybrid assays, confirming its potential role as a transcriptional regulator. A 75–amino acid C-terminal domain, when fused with the GAL4 DNA binding domain, strongly activated transcription. A series of deletions within this C-terminal domain more precisely localized the transcriptional activation domain to a 23–amino acid region at the C terminus of the protein.
Alignments of the related MYB proteins indicated that this 23–amino acid region at the C terminus is the most conserved portion of the activation domain. This conservation among MYB proteins has been previously reported (Kranz et al., 1998; Li et al., 2006). However, this study evaluates the structural features of this region and reveals new insights. Secondary structure predictions suggested that the activation domain lacks defined secondary structure, with the single exception of a predicted short α-helix located near the middle of the 23–amino acid activation region. This α-helix was found to be amphipathic with hydrophilic residues exposed on one side of the helix and hydrophobic residues on the other. Such α-helices are often form strong activation regions in transcription factors (Bushman et al., 1989).
This 23–amino acid region was found to contain a number of acidic residues, as well as two Ser residues that could be phosphorylated to provide additional acidic residues in this region. Short acidic peptide sequences are well known to activate transcription when brought into proximity of DNA (Ma and Ptashne, 1987). In the case of LxS-MYB305, we found that a short 23–amino acid peptide was all that was required to activate transcription when fused to the GAL4 DNA binding domain. This identified peptide has a net excess of four acidic residues. Site-specific mutagenesis revealed that Ala substitutions of two Ser residues did not alter transcriptional activation; however, replacement of three acidic residues with Ala residues completely abolished transcription.
To determine whether LxS-MYB305 could bind specific motifs within the nec1 promoter, we expressed the protein as a GST fusion and tested the purified protein for direct interaction with the MYB binding sites in the nec1 promoter. For both of the S1 and S0 MYB binding sites in the nec1 promoter, unlabeled probe competed for binding of the GST-MYB305 protein to the labeled duplex oligonucleotide probes, but unlabeled mutated probes did not compete, indicating that the interaction was specific for the probe sequence. Similarly, GST-MYB305 also bound the MYB binding site in the nec5 promoter. Unlabeled duplex probes also competed for this binding, but unlabeled mutated probes did not.
To evaluate further the role of these MYB binding sites, we prepared a series of GUS reporter constructs that contained either the wild-type or mutagenized nec1 promoters. In transgenic plants, we found that the wild-type nec1 promoter expressed GUS activity at appreciable levels in four independent transformants; however, deletion of either or both of the MYB binding sites from the nec1 promoter resulted in loss of promoter activity. The finding that both sites are required to achieve high levels of expression is somewhat surprising and suggests that multiple MYB-DNA interactions may be required to achieve high-level activation of the nec1 promoter. Further studies along these lines will be needed to reveal the role of these interactions in transcriptional activation of these promoters.
In addition, we have generated transgenic plant lines that show ectopic expression of LxS-MYB305. These plants also showed ectopic expression of the flavonoid genes, pal and chi, in foliage as expected (Sablowski et al., 1994; Moyano et al., 1996; Uimari and Strommer, 1997; Laitinen et al., 2005) as well as ectopic expression of the nectarin genes. Both nec1 and nec5 were coordinately expressed in the foliage of these plants; however, nec3 and nec4 were not (see Supplemental Figure 3A online). Based upon differential patterns of expression, we had previously predicted that nec3 and nec4 were expressed via different mechanisms from nec1 and nec5 (Carter and Thornburg, 2004b; Naqvi et al., 2005), and these studies confirm this.
Finally, we generated RNAi lines that show severely reduced levels of LxS-MYB305. These lines each show reduced accumulation of nec1 and nec5 transcripts in nectaries. These lines also show reduced expression of the flavonoid genes pal and chi. In addition, the nectaries of these RNAi plants do not accumulate high levels of β-carotene as wild-type nectaries do. These plants also show reduced levels of nectar accumulation in the flower. Both of these characteristics (β-carotene accumulation and nectar production) are associated with starch metabolism in nectaries (Horner et al., 2007; Ren et al., 2007a, 2007b). The role of LxS-MYB305 in nectary starch metabolism is currently an active project in our laboratory.
In this manuscript, we provide evidence that MYB305 is a transcriptional regulator that functions in expression of Nectarin I and Nectarin V. LxS-MYB305 is expressed most strongly in the nectary, but it also is expressed in the ovary, floral tube, and petals. The protein is nuclear localized, it activates transcription in yeast, and it binds to the nec1 and nec5 promoters in vitro. Furthermore, mutation of the MYB binding sites within the nec1 promoter destroys promoter activity in vivo, and myb305 is transcribed prior to expression of the target genes nec1 and nec5. In addition, ectopic expression of LxS-MYB305 in foliage results in foliar expression the target genes nec1, nec5, pal, and chi and reduced expression of LxS-myb305 via RNAi results in a coordinated knockdown of these same target genes nec1, nec5, pal, and chi. Based on all of these data, we conclude that in addition to regulating the flavonoid biosynthetic genes, MYB305 also regulates the nec1 and nec5 genes in the nectaries of ornamental tobacco. Furthermore, the additional phenotypes observed in the RNAi knockdown and overexpression plants suggest that LxS-MYB305 has roles in floral development that are far beyond flavonoid metabolism and nectar protein expression.
METHODS
Materials
Unless otherwise noted, the materials used in these studies were obtained from either Fisher Chemical or Sigma-Aldrich Chemical and were of the highest quality available. All radioisotopes were from Perkin-Elmer.
Plants
The LxS8 ornamental tobacco (Nicotiana tabacum) plants used in these studies, growth condition of plants, and methods for isolation of nectar and floral tissues was previously described (Carter et al., 1999). If not used immediately, tissues were frozen at −20°C until use. Flowers were staged as described (Koltunow et al., 1990). N. tabacum cv Xanthi plants were also previously described (Thornburg et al., 1987). The Arabidopsis thaliana myb21, myb24 mutants, and the double mutant (Mandaokar et al., 2006) were kindly supplied by John Browse, Washington State University.
Genes
Complete sequencing was performed on both strands of all clones at the Iowa State University Nucleic Acid Facility. Synthesis of the various cDNA libraries used to produce the LxS-myb305 cDNAs was previously described (Carter and Thornburg, 2004c; Naqvi et al., 2005).
Anti-LxS-MYB305 and Anti-NEC1 Antiserum
Rabbit anti-NEC1 antiserum was previously described (Carter et al., 1999). Rabbit anti-LxS-MYB305 antiserum was produced by GenScript using a synthesized polypeptide, H2N-PFLTETNDNIWSMED-COOH, which corresponds to the C terminus of the LxS-MYB305 protein. The antiserum was affinity purified prior to use. The peptide was coupled to CNBr-activated Sepharose 4 resin (GE Healthcare). Twenty milliliters of crude antiserum were passed through the column and after washes with 100 mL 50 mM Tris-HCl, pH 7.0, 100 mL 10 mM Tris-HCl, pH 7.0, containing 0.5 M NaCl, and finally with 100 mL 50 mM Tris-HCl, pH 7.0, with 0.2 M NaCl, the anti-LxS-MYB305 antibodies were eluted with 100 mM glycine, pH 2.5. The antibodies were neutralized with 1 M Tris-HCl, pH 8, 50% glycerol, 0.25 M NaCl, and 0.1 M KCl prior to storage at −20°C and use in the immunoblot experiments.
Phylogenetic Analysis
All sequences other than LxS-MYB305 were obtained from GenBank. The R2R3 MYB region of LxS-MYB305 was aligned with other MYB proteins' MYB regions using the ClustalW integrated in the MEGA program (Tamura et al., 2007). The aligned sequences (see Supplemental Figure 5 and Supplemental Data Set 1 online) were used to produce phylogenetic trees using MEGA (Tamura et al., 2007). The phylogenetic trees were produced using the neighbor joining method with the following parameters: complete deletion and P-distance. One thousand bootstrap replications of each tree were produced to test the robustness of the phylogenetic tree. Similarly, the full-length LxS-MYB305 protein sequence and closely related protein sequences were aligned (see Supplemental Figures 6 and 7 and Supplemental Data Sets 2 and 3 online) and were analyzed using the same method and parameters as above.
Secondary Structure Prediction
The activation domain of the LxS-MYB305 coding sequence was identified from the one-hybrid studies and input on the Jpred server (www.compbio.dundee.ac.uk/www-jpred/ index.html).
Expression of MYB305 in Escherichia coli and Purification of Recombinant Protein
The full-length cDNA of LxS-myb305 was amplified by PCR using the MYBGST1 and MYBGST2 primers (see Supplemental Table 1 online). The PCR product was digested with BamHI and SalI (Sigma-Aldrich) and ligated into the similarly digested vector pGEX-4T-3 (GE Life Sciences) to generate the expression vector, pRT608. The plasmid was then transferred into E. coli strain BL21 Codon Plus. The E. coli were grown in 2YT media with 1% glucose at 30°C, and 0.1% IP Isopropyl β-D-1- thiogalactopyranoside was used to induce the expression of the soluble fusion protein. The fusion GST-MYB305 protein was purified using Glutathione Sepharose 4B (GE Life Sciences). The protein concentration was determined by the method of Bradford (1976).
Preparation of Labeled Oligonucleotide Probes and Mobility Shift Assays
The probes for analysis of the MYB binding sites in the nec1 promoter were prepared by [32P]-labeling. The duplex N1S1 probe was prepared by annealing two single-stranded oligonucleotides, N1P1-UPPER and N1P1-LOWER (see Supplemental Table 1 online). The two oligonucleotides were mixed in a 0.2-mL tube and heated to 95°C using thermal cycler. Then, the tube was floated on a 250-mL beaker filled with 95°C water sitting at room temperature. As the water temperature decreased and reached room temperature, the paired oligonucleotides annealed. Likewise, the duplex N1S0 probe was similarly prepared using the oligonucleotides N1P2-UPPER and NIP2-LOWER. Both probes were labeled with [γ-32P]ATP by T4 polynucleotide kinase (Promega). The duplex competitor probes N1C1 and N1C0 were similarly prepared. Mobility gel shift assays were performed using the Promega gel shift assay system according to the manufacturer's instructions. The N5P probe for the analysis of the MYB binding site in the nec5 promoter was prepared using a fluorescent HEX tag incorporated during synthesis at the 5′ end of the oligonucleotide (Integrated DNA Technologies). The gel shift assay was performed the same way as with N1S1 and N1S0 probes, except that the gel was imaged with a Typhoon 8600 imaging system (GE Life Sciences) instead of x-ray film.
Transcriptional Activation Assays
Yeast One- and Two-Hybrid Assays
Details of the construct preparation for the yeast one- and two-hybrid studies are presented in the Supplemental Methods online. The constructs were transformed into the Saccharomyces cerevisiae strain MaV203 (Invitrogen) using the method of Gietz et al. (1992). The transformed yeast was cultured in complete synthetic yeast medium (Sherman et al., 1986) without Leu (SYM Leu−) for transactivation assays or in SYM Leu− Trp− for two-hybrid assays. In both cases, cells were grown in the liquid medium until they reached an OD600 of 1.0 before transferring to SYM Ura− or SYM His− plates for serial dilution growth assays.
β-Galactosidase Activity and Yeast Cell Growth Assays
β-Galactosidase activity in the transformed yeast cells was quantified using chlorophenol red-β-d-galactopyranoside (Sigma-Aldrich) as substrate (Serebriiskii and Golemis, 2000). The serial dilution growth of transformed yeast cells was assayed in SYM Leu− according to the manufacturer's protocol (Invitrogen).
Subcellular Localization
Details of the construct preparation for the subcellular localization studies are presented in the Supplemental Methods online.
Protoplast Preparation and Transformation
In vitro shoot cultures of N. tabacum cv Xanthi were used for protoplast isolation. Plants were maintained on Murashige and Skoog (MS) solid medium (Murashige and Skoog, 1962) without growth regulators at 25°C. Sterile leaves from 2-month-old in vitro–grown plants were digested overnight a solution of 1% (w/v) Onozuka Cellulase (Research Products International) and 0.2% (w/v) Macerocyme (Julie and Brent, 1986) dissolved in K3S medium (Nagy and Maliga, 1976). Following digestion, the protoplasts were collected and concentrated by floating on K3S1 medium in Bobcock bottles (Julie and Brent, 1986). The protoplasts were transiently transformed using polyethylene glycol (Locatelli et al., 2003). The transformed protoplasts were grown in K3G1 medium for 24 h without selection and then for a second 24-h period in K3G1 medium + 25 μg/mL Hygromycin B before observation.
Fluorescence Microscopy
Protoplasts selected as described above were imaged using an Olympus BX60 upright fluorescent microscope fitted with a Burner module (U-ULS100HG). Photographs were taken by Nikon Coolpix 950 digital camera using the GFP fluorescence filter. For each transformant, there were ∼5 to 20 observations, and the cells shown are typical of each transformant.
nec1 Promoter Assays
Details of the construct preparation for the nec1 promoter assays are presented in the Supplemental Methods online.
Plant Transformation
N. tabacum cv Xanthi plants were transformed with Agrobacterium tumefaciens LBA 4404 (Ooms and Bakker, 1982), by cocultivation as described (Thornburg et al., 1987). Transgenic progeny lines were selected on MS plates containing 0.1 μg/mL naphthalene acetic acid, 1.0 μg/mL benzylaminopurine, 50 μg/mL Hygromycin B, and 100 μg/mL Carbenicillin. After regeneration, plantlets were transferred to MS media with 50 μg/mL Hygromycin B and 100 μg/mL Carbenicillin, but containing no hormones for rooting. Once the rooted shoots were ∼5 cm tall, they were transferred into soil and grown to floral maturity in the greenhouse.
GUS Activity Assays
Floral nectaries from the transformed tobacco plants were collected and the tissue was homogenized in extraction buffer (50 mM NaHPO4, pH 7.0, containing 10 mM β-mercaptoethanol, 10 mM EDTA, 0.1% [w/v] sodium lauryl sarcosine, and 0.1% [w/v] Triton X-100). The protein concentration was determined using the method of Bradford (1976). The activities of GUS were determined by fluorometric assays using 4-methylumbeliferyl-β-glucuronide as substrate (Jefferson, 1987).
LxS-MYB305 Overexpression and LxS-MYB305 RNAi Plants
The full-length LxS-myb305 cDNA sequence (AF132671) was PCR amplified by attBMYB-1 and attBMYB-2 primers and then recombined (BP reaction) into the pDONRZeo vector (Invitrogen) to obtain the entry clone pRT613. This plasmid was subsequently recombined (LR reaction) into pMDC32 (Curtis and Grossniklaus, 2003) vector to form an overexpression plant transformation vector, pRT621. Transgenic plants were prepared as described above.
The 3′ end of the cDNA sequence of LxS-myb305 excluding MYB DNA binding domains was PCR amplified by attBMYB-13 and attBMYB-2 primers and subsequently recombined (BP reaction) into pDONRZeo to obtain the entry clone, pRT671. This plasmid was then recombined (LR reaction) into pB7GWIWG2 (Karimi et al., 2002) to get an RNAi plant transformation vector, pRT672. Transgenic plants were prepared as described above, except the RNAi plants were selected by 20 μg/mL ammonium glufosinate instead of Hygromycin B in both selection and rooting stages.
RT-PCR
The tobacco tissues were collected and frozen in liquid nitrogen; RNAs were then purified using the TRIzol reagent method (Invitrogen) according to the manufacturer's instructions. The first-strand cDNAs were made using the Eppendorf cMaster RT-PCR system. All primers used in this study are given in Supplemental Table 1 online. Minimal cycles of amplification were used to detect the genes of interest to ensure that analyses were conducted in a linear portion of the amplification spectrum. Initially, RT-PCR reactions were run with even cycle numbers starting from 20 cycles on. After analysis, the cycle number when the bands first became visible was chosen for the final analysis conditions. The gels were stained with ethidium bromide, and individual cycle numbers are indicated in each figure legend.
Quantitative RT-PCR
The leaves of LxS-MYB305 overexpression N. tabacum and nectaries of the LxS-MYB305 RNAi N. tabacum plants were collected and frozen in liquid nitrogen. Total RNA was purified using the TRIzol reagent method according to the manufacturer's instructions. The first-strand cDNAs were made using the Eppendorf cMaster RT-PCR system, and the first-strand cDNAs were used as template in the quantitative PCR reaction. Primers used are given in Supplemental Table 1 online. The PCR reactions were performed on Stratagene mx4000 multiplex quantitative PCR system using the Brilliant II SYBR Green QPCR Master Mix (Stratagene). 26S rRNA was used as an internal reference to normalize the relative level of each transcript. Four to six biological replications were used to calculate each relative expression value.
Immunoblotting
Various tobacco tissues were collected and immediately frozen in liquid nitrogen. Proteins were isolated using a plant total protein extraction kit (PE0230; Sigma-Aldrich), and 100 μg of total protein from leaves or different nectary stages were electrophoresed on 12% SDS-PAGE. After running the gel, the proteins were electrophoretically transferred to polyvinylidene fluoride membranes. The proteins were detected with either 1:2000 rabbit anti-LxS-MYB305 antiserum or 1:5000 rabbit anti-NEC1 antiserum and then with horseradish peroxidase–conjugated goat anti-rabbit antiserum. The SuperSignal West Pico Chemiluminescent Substrate (Pierce Scientific) was used to detect the signals on membranes; the membranes were then exposed to x-ray films. Individual exposure times are indicated in each figure legend.
Accession Numbers
The sequence of the LxS8-MYB305 cDNA used in these studies was deposited in GenBank with accession number EU111679. A second, nearly identical MYB305 cDNA was also cloned and was deposited with accession number EU111678. Other genes used in this study include the promoters of the Nicotiana plumbaginifolia nec1 gene (AF132671) and nec5 gene (AF503441). Sequences used in the phylogenetic analysis are presented in the legends of the supplemental figures online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Yeast Two-Hybrid Test of Homodimerization with LxS-MYB305.
Supplemental Figure 2. Comparison of Wild-Type and MYB305 OX Phenotypes.
Supplemental Figure 3. Gene Expression in Wild Type and Mutant Plants.
Supplemental Figure 4. Phylogenetic Analysis of LxS-MYB305 with Rosid Homologs.
Supplemental Figure 5. Alignment of R2R3 MYB DNA Binding Domains for Phylogenetic Analysis.
Supplemental Figure 6. Alignment of Complete MYB Amino Acid Sequences for Phylogenetic Analysis.
Supplemental Table 1. Oligonucleotides Used in These Studies.
Supplemental Table 2. Length of Various Organs from Flowers at Anthesis.
Supplemental Data Set 1. Alignments Used for the Phylogenetic Analysis in Figure 2A.
Supplemental Data Set 2. Alignment Used for the Phylogenetic Analysis in Figure 2B.
Supplemental Data Set 3. Alignment Used in Supplemental Figure 4.
Supplemental Methods. Preparation of DNA Constructs for Experiments Used in These Studies.
Supplementary Material
Acknowledgments
This work was supported by the National Science Foundation (IBN 0235645), Carver Trust, the Hatch Act, and State of Iowa funds.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Robert W. Thornburg (thorn@iastate.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215 403–410. [DOI] [PubMed] [Google Scholar]
- Beutler, R. (1935). Nectar. Bee World 24 106–116, 128–136, 156–162. [Google Scholar]
- Blom, N., Gammeltoft, S., and Brunak, S. (1999). Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 294 1351–1362. [DOI] [PubMed] [Google Scholar]
- Bradford, M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principles of protein-dye binding. Anal. Biochem. 72 248–254. [DOI] [PubMed] [Google Scholar]
- Bushman, F.D., Shang, C., and Ptashne, M. (1989). A single glutamic acid residue plays a key role in the transcriptional activation function of lambda repressor. Cell 58 1163–1171. [DOI] [PubMed] [Google Scholar]
- Carter, C., Graham, R., and Thornburg, R.W. (1999). Nectarin I is a novel, soluble germin-like protein expressed in the nectar of Nicotiana sp. Plant Mol. Biol. 41 207–216. [DOI] [PubMed] [Google Scholar]
- Carter, C., Healy, R., O'Tool, N.M., Naqvi, S.M.S., Ren, G., Park, S., Beattie, G.A., Horner, H.T., and Thornburg, R.W. (2007). Tobacco nectaries express a novel NADPH oxidase that is implicated in the defense of floral reproductive tissues against microorganisms. Plant Physiol. 143 389–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter, C., Shafir, S., Yehonatan, L., Palmer, R.G., and Thornburg, R. (2006). A novel role for proline in plant floral nectars. Naturwissenschaften 93 72–79. [DOI] [PubMed] [Google Scholar]
- Carter, C., and Thornburg, R.W. (2000). Tobacco Nectarin I: Purification and characterization as a germin-like, manganese superoxide dismutase implicated in the defense of floral reproductive tissues. J. Biol. Chem. 275 36726–36733. [DOI] [PubMed] [Google Scholar]
- Carter, C., and Thornburg, R.W. (2003). The nectary-specific pattern of gene expression is regulated by multiple promoter elements in the tobacco Nectarin I promoter. Plant Mol. Biol. 51 451–457. [DOI] [PubMed] [Google Scholar]
- Carter, C., and Thornburg, R.W. (2004. a). Is the nectar redox cycle a floral defense against microbial attack? Trends Plant Sci. 9 320–324. [DOI] [PubMed] [Google Scholar]
- Carter, C., and Thornburg, R.W. (2004. b). Tobacco Nectarin III is a bifunctional enzyme with monodehydroascorbate reductase and carbonic anhydrase activities. Plant Mol. Biol. 54 415–425. [DOI] [PubMed] [Google Scholar]
- Carter, C., and Thornburg, R.W. (2004. c). Tobacco Nectarin V is a flavin-containing berberine bridge enzyme-like protein with glucose oxidase activity. Plant Physiol. 134 460–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuff, J., and Barton, G. (1999). Evaluation and improvement of multiple sequence methods for protein secondary structure prediction. Proteins 34 508–519. [DOI] [PubMed] [Google Scholar]
- Cuff, J., and Barton, G. (2000). Application of multiple sequence alignment profiles to improve protein secondary structure prediction. Proteins 40 502–511. [DOI] [PubMed] [Google Scholar]
- Curtis, M.D., and Grossniklaus, U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, A., Pylatuik, J., Paradis, J., and Low, N. (1998). Nectar-carbohydrate production and composition vary in relation to nectary anatomy and location within individual flowers of several species of Brassicaceae. Planta 205 305–318. [DOI] [PubMed] [Google Scholar]
- Decker, T., and Kovarik, P. (1999). Transcription factor activity of STAT proteins: Structural requirements and regulation by phosphorylation and interacting proteins. Cell. Mol. Life Sci. 55 1535–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz, D., St. Jean, A., Woods, R.A., and Schiestl, R.H. (1992). Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20 1425–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Teuber, M., Eilmus, S., Muck, A., Svatos, A., and Heil, M. (2009). Pathogenesis-related proteins protect extrafloral nectar from microbial infestation. Plant J. 58 464–473. [DOI] [PubMed] [Google Scholar]
- Heil, M., Rattke, J., and Boland, W. (2005). Postsecretory hydrolysis of nectar sucrose and specialization in ant/plant mutualism. Science 308 560–563. [DOI] [PubMed] [Google Scholar]
- Horner, H.T., Healy, R.A., Ren, G., Fritz, D., Seames, C., and Thornburg, R.W. (2007). Amyloplast to chromoplast conversion in developing ornamental tobacco floral nectaries provides sugar for nectar and antioxidants for protection. Am. J. Bot. 94 12–24. [DOI] [PubMed] [Google Scholar]
- Jackson, D., Culianez-Macia, F., Prescott, A.G., Roberts, K., and Martin, C. (1991). Expression patterns of myb genes from Antirrhinum flowers. Plant Cell 3 115–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson, R.A. (1987). Assaying chimeric genes in plants: The GUS gene fusion system. Plant Mol. Biol. Rep. 5 387–405. [Google Scholar]
- Julie, A., and Brent, H. (1986). Techniques for enhanced release of leaf protoplasts in populus. Plant Cell Rep. 5 284–287. [DOI] [PubMed] [Google Scholar]
- Karimi, M., Inze, D., and Depicker, A. (2002). Gateway vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7 193–195. [DOI] [PubMed] [Google Scholar]
- Koltunow, A.M., Truettner, J., Cox, K.H., Walroth, M., and Goldberg, R.B. (1990). Different temporal and spatial gene expression patterns occur during anther development. Plant Cell 2 1201–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kram, B.W., Bainbridge, E.A., Perera, M.A., and Carter, C. (2008). Identification, cloning and characterization of a GDSL lipase secreted into the nectar of Jacaranda mimosifolia. Plant Mol. Biol. 68 173–183. [DOI] [PubMed] [Google Scholar]
- Kranz, H.D., et al. (1998). Towards functional characterisation of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J. 16 263–276. [DOI] [PubMed] [Google Scholar]
- Laitinen, R.A., Immanen, J., Auvinen, P., Rudd, S., Alatalo, E., Paulin, L., Ainasoja, M., Kotilainen, M., Koskela, S., Teeri, T.H., and Elomaa, P. (2005). Analysis of the floral transcriptome uncovers new regulators of organ determination and gene families related to flower organ differentiation in Gerbera hybrida (Asteraceae). Genome Res. 15 475–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., Yang, X., Wang, Y., Li, X., Gao, Z., Pei, M., Chen, Z., Qu, L.J., and Gu, H. (2006). Two groups of MYB transcription factors share a motif which enhances trans-activation activity. Biochem. Biophys. Res. Commun. 341 1155–1163. [DOI] [PubMed] [Google Scholar]
- Lim, K., Ho, J.X., Keeling, K., Gilliland, G.L., Ji, X., Rüker, F., Carter, D.C. (1994). Three-dimensional structure of Schistosoma japonicum glutathione s-transferase fused with a six-amino acid conserved neutralizing epitope of gp41 from hiv. Protein Sci. 3 2233–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locatelli, F., Vannini, C., Magnani, E., Coraggio, I., and Bracale, M. (2003). Efficiency of transient transformation in tobacco protoplasts is independent of plasmid amount. Plant Cell Rep. 21 865–871. [DOI] [PubMed] [Google Scholar]
- Ma, J., and Ptashne, M. (1987). A new class of yeast transcriptional activators. Cell 51 113–119. [DOI] [PubMed] [Google Scholar]
- Magasanik, B. (1988). Reversible phosphorylation of an enhancer binding protein regulates the transcription of bacterial nitrogen utilization genes. Trends Biochem. Sci. 13 475–479. [DOI] [PubMed] [Google Scholar]
- Mandaokar, A., Thines, B., Shin, B., Lange, B.M., Choi, G., Koo, Y.J., Yoo, Y.J., Choi, Y.D., Choi, G., and Browse, J. (2006). Transcriptional regulators of stamen development in Arabidopsis identified by transcriptional profiling. Plant J. 46 984–1008. [DOI] [PubMed] [Google Scholar]
- Moyano, E., Martinez-Garcia, J.F., and Martin, C. (1996). Apparent redundancy in myb gene function provides gearing for the control of flavonoid biosynthesis in antirrhinum flowers. Plant Cell 8 1519–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 15 473–497. [Google Scholar]
- Nagy, J.I., and Maliga, P. (1976). Callus induction and plant regeneration from mesophyll protoplasts of Nicotiana sylvestris. Z. Pflanzenphysiol. 78 453–455. [Google Scholar]
- Naqvi, S., Harper, A., Carter, C., Ren, G., Guirgis, A., York, W.S., and Thornburg, R.W. (2005). Tobacco Nectarin IV is a specific inhibitor of fungal xylosidases secreted into the nectar of ornamental tobacco plants. Plant Physiol. 139 1389–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nepi, M. (2007). Nectary structure and ultrastructure. In Nectaries and Nectar, S.W. Nicolson, M. Nepi, and E. Pacini, eds (Dordrecht, The Netherlands: Springer), pp. 129–166.
- Nicolson, S., and Thornburg, R.W. (2007). Nectar chemistry. In Nectary and Nectar: A Modern Treatise, S.W. Nicolson, M. Nepi, and E. Pacini, eds (Amsterdam: Springer-Varlig), pp. 215–263.
- O'Leary, S.J.B., Poulis, B.A.D., and von Aderkas, P. (2007). Identification of two thaumatin-like proteins (TLPs) in the pollination drop of hybrid yew that may play a role in pathogen defence during pollen collection. Tree Physiol. 27 1649–1659. [DOI] [PubMed] [Google Scholar]
- Ooms, G., and Bakker, A. (1982). T-DNA organization in homogeneous and heterogeneous octopine-type crown gall tissues of Nicotiana tabacum. Cell 30 589–597. [DOI] [PubMed] [Google Scholar]
- Peumans, W.J., Smeets, K., Van Nerum, K., Van Leuven, F., and Van Damme, E.J.M. (1997). Lectin and alliinase are the predominant proteins in nectar from leek (Allium porrum L.) flowers. Planta 201 298–302. [DOI] [PubMed] [Google Scholar]
- Poulis, B.A.D., O'Leary, S.J.B., Haddow, J.D., and von Aderkas, P. (2005). Identification of proteins present in the Douglas fir ovular secretion: an insight into conifer pollen selection and development. Int. J. Plant Sci. 166 733–739. [Google Scholar]
- Ren, G., Healy, R., Horner, H.T., James, M., and Thornburg, R.W. (2007. b). Expression of starch metabolic genes in the developing nectaries of ornamental tobacco plants. Plant Sci. 173 621–637. [Google Scholar]
- Ren, G., Healy, R., Klyne, A., Horner, H.T., James, M., and Thornburg, R.W. (2007. a). Transient starch metabolism in ornamental tobacco floral nectaries regulates nectar composition and release. Plant Sci. 173 277–290. [Google Scholar]
- Ruden, D.M., Ma, J., Li, Y., Wood, K., and Ptashne, M. (1991). Generating yeast transcriptional activators containing no yeast protein sequences. Nature 350 250–252. [DOI] [PubMed] [Google Scholar]
- Sablowski, R.W., Baulcombe, D.C., and Bevan, M. (1995). Expression of a flower-specific Myb protein in leaf cells using a viral vector causes ectopic activation of a target promoter. Proc. Natl. Acad. Sci. USA 92 6901–6905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablowski, R.W., Moyano, E., Culianez-Macia, F.A., Schuch, W., Martin, C., and Bevan, M. (1994). A flower-specific Myb protein activates transcription of phenylpropanoid biosynthetic genes. EMBO J. 13 128–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serebriiskii, I., and Golemis, E. (2000). Uses of lacZ to study gene function: Evaluation of [beta]-galactosidase assays employed in the yeast two-hybrid system. Anal. Biochem. 285 1–15. [DOI] [PubMed] [Google Scholar]
- Sherman, F., Fink, G.R., and Hicks, J.B. (1986). Laboratory Course Manual for Methods in Yeast Genetics. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory).
- Shin, B., Choi, G., Yi, H., Yang, S., Cho, I., Kim, J., Lee, S., Paek, N.C., Kim, J.H., Song, P.S., and Choi, G. (2002). AtMYB21, a gene encoding a flower-specific transcription factor, is regulated by COP1. Plant J. 30 23–32. [DOI] [PubMed] [Google Scholar]
- Shioda, T., Fenner, M.H., and Isselbacher, K.J. (1997). MSG1 and its related protein MRG1 share a transcription activating domain. Gene 204 235–241. [DOI] [PubMed] [Google Scholar]
- Stracke, R., Werber, M., and Weisshaar, B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4 447–456. [DOI] [PubMed] [Google Scholar]
- Tamura, K., Dudley, J., Nei, M., and Kumar, S. (2007). MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24 1596–1599. [DOI] [PubMed] [Google Scholar]
- Thornburg, R.W., An, G., Cleveland, T.E., Johnson, R., and Ryan, C.A. (1987). Wound-inducible expression of a potato inhibitor II-chloramphenicol acetyltransferase gene fusion in transgenic tobacco plants. Proc. Natl. Acad. Sci. USA 84 744–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornburg, R.W., Carter, C., Powell, A., Mittler, R., Rizhsky, L., and Horner, H.T. (2003). A major function of the tobacco floral nectary is defense against microbial attack. Plant Syst. Evol. 238 211–218. [Google Scholar]
- Uimari, A., and Strommer, J. (1997). Myb26: A MYB-like protein of pea flowers with affinity for promoters of phenylpropanoid genes. Plant J. 12 1273–1284. [DOI] [PubMed] [Google Scholar]
- Vidal, M., Brachmann, R.K., Fattaey, A., Harlow, E., and Boeke, J.D. (1996). Reverse two-hybrid and one-hybrid systems to detect dissociation of protein-protein and DNA-protein interactions. Proc. Natl. Acad. Sci. USA 93 10315–10320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, R., Mugnaini, S., Sniezko, R., Hardie, D., Poulis, B., Nepi, M., Pacini, E., and von Aderkas, P. (2007). Gymnosperm pollination drop proteins are conservative and indicate complex function. Sex. Plant Reprod. 20 181–189. [Google Scholar]
- Yang, X.Y., Li, J.G., Pei, M., Gu, H., Chen, Z.L., and Qu, L.J. (2007). Over-expression of a flower-specific transcription factor gene AtMYB24 causes aberrant anther development. Plant Cell Rep. 26 219–228. [DOI] [PubMed] [Google Scholar]
- Zimmerman, J.G. (1954). Uber die sekretion saccharosespaltender trans-glucosidasen in pflanzlichen nektar. Experientia 10 145–149. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.