Abstract
Successful integration of viral genome into a host chromosome depends on interaction between viral integrase and its recognition sequences. We have used a reconstituted concerted human immunodeficiency virus, type 1 (HIV-1), integration system to analyze the role of integrase (IN) recognition sequences in formation of the IN-viral DNA complex capable of concerted integration. HIV-1 integrase was presented with substrates that contained all 4 bases at 8 mismatched positions that define the inverted repeat relationship between U3 and U5 long terminal repeats (LTR) termini and at positions 17–19, which are conserved in the termini. Evidence presented indicates that positions 17–20 of the IN recognition sequences are needed for a concerted DNA integration mechanism. All 4 bases were found at each randomized position in sequenced concerted DNA integrants, although in some instances there were preferences for specific bases. These results indicate that integrase tolerates a significant amount of plasticity as to what constitutes an IN recognition sequence. By having several positions randomized, the concerted integrants were examined for statistically significant relationships between selections of bases at different positions. The results of this analysis show not only relationships between different positions within the same LTR end but also between different positions belonging to opposite DNA termini.
Integration of viral genome into the host chromosome is essential for stable infection of the retrovirus and occurs in a concerted reaction in which the ends of the viral DNA are brought together and inserted into the acceptor in a coordinated manner (1, 2). In vitro, the viral protein integrase (IN)1 is both necessary and sufficient to catalyze integration of viral sequences into a target DNA. Some cellular proteins were found to stimulate the reaction (3–6). IN forms a homodimer or a higher order multimer structure, which recognizes the terminal sequences in the U3 and U5 LTRs. In most retroviruses these sequences are related to one another in that they are nearly perfect inverted repeats. The number of bases required for specific IN recognition varies with the virus and ranges between 10 and 20 bp (7). In Moloney murine leukemia virus, the IN recognition sequences at U5 and U3 LTRs are identical in sequence. In avian sarcoma virus and HIV-1, they differ at the 3- and 8-positions of 15 and 20 bp, respectively.
Insights into the molecular details of integration reaction have been gained mainly through the use of the systems reconstituted in vitro (3, 5, 6, 8–16). Most of these systems employ duplex oligodeoxyribonucleotides substrates (8–13), which do not exhibit the concerted properties of DNA integration in vivo. To analyze the integration reaction in detail, we have used an in vitro integration system that employs a mini donor DNA containing U3 and U5 IN recognition sequences at each end and demonstrates concerted DNA integration reactions as determined by direct sequencing of integrants (3, 5, 14). To examine sequence preferences by HIV-1 IN, we randomized the “mismatched” positions or the conserved positions 17–19 nucleotides from the ends of unprocessed viral DNA in U5 and/or U3 LTR IN recognition sequences. We then allowed IN to “choose” sequences in the mini donor DNAs that constitute a functional recognition sequence. Results from analysis of enzymatic activity and sequences selected from the population of substrates indicate that in addition to sequences in and around the conserved CA dinucleotide, positions 17–20 are also important for reconstitution of concerted integration in vitro. Moreover, there were statistically significant relationships among the bases selected at different positions such that the selection of a base at one position was found in combination with the selection of a base at another.
EXPERIMENTAL PROCEDURES
Reagents
[α-32P]dCTP (3,000 Ci/mmol) was purchased from Amersham Biosciences. Proteinase K (30 units/mg) and glycogen were from Roche Molecular Biochemicals. HMG-I(Y) was purified as described by Nissen and Reeves (17). IN was purified as described by Yi et al. (18). Vent DNA polymerase (2 units/μl) was from New England Biolabs (Beverly, MA). Oligodeoxyribonucleotides were purchased from Operon (Alameda, CA) and purified by PAGE under denaturing conditions. The following oligodeoxyribonucleotides were used in this study: U5(WT), 5′ACTGCTAGAGATTTTCCACACTGGGCGGAGCCTATG 3′; U5 N8, 5′ACTGNNAGNGNTNNTNCACNCTGGGCGGAGCCTATG 3′; U5 N3G-TG, 5′ACTGCTAGAGATTTTCNNNACTGGGCGGAGCCTATG 3′; U5 18GA19, 5′ACTGCTAGAGATTTTCCCTACTGGGCGGAGCCTATG 3′; U5 17AAT19, 5′ACTGCTAGAGATTTTCTTAACTGGGCGGAGCCTATG 3′; U3(WT), 5′ACTGGAAGGGCTAATTCACTCGTTGCCCGGATCCGG 3′; U3 N8, 5′ACTGNNAGNGNTNNTNCACNCGTTGCCCGGATCCGG 3′; U3 N3GTG, 5′ACTGGAAGGGCTAATTNNNTCGTTGCCCGGATCCGG 3′; U5seq, 5′AGAATTCGGCGTTGCTGGCGTTTTTCCATA 3′; and U3seq, 5′CTGCCGTCATCGACTTCGAAGGTTCGAATC 3′. The U5 N8 and U5 N3GTG oligodeoxyribonucleotides along with U3(WT) were used to prepare HIV-1 donor-concerted DNA integration substrates with randomization in the U5 terminus sequence. The U5 18GA19 and U5 17AAT19 together with U3(WT) oligodeoxyribonucleotides were used to prepare HIV-1-concerted DNA integration substrates with mutations at positions 17–19. In each case, the sequence refers to the 3′-cleaved strand of the U5 LTR IN recognition sequence The U3 N8 and U3 N3GTG oligodeoxyribonucleotides along with U5(WT) were used to prepare comparable donor DNAs with randomization in the U3 IN recognition sequence. The U5seq and U3seq oligodeoxyribonucleotides were used as sequencing primers. The U3seq primer is complementary to plasmid πvx nucleotides 180 to 151, and the U5seq primer is complementary to plasmid πvx nucleotides 312–341.
Bacterial Strains and Growth Conditions
Escherichia coli DH5α (Invitrogen) and MC1061/P3 (Invitrogen) strains were used for these studies. MC1061/P3 is a derivative of MC1061 containing the male episome, P3, which can be selected for the presence of an encoded Kanr gene. In addition, P3 possesses amp (Am) and tet (Am) genes, the expression of which can be rescued by the supF amber suppressor tRNA. Under these conditions, MC1061/P3 can be selected for ampicillin, tetracycline, and kanamycin resistance.
Plasmid Constructions and Preparations
Plasmid pHHIV2 was used in this study as a template to amplify donor DNA and is a variation of pBCSK+ in which a wild type HIV-1 donor DNA PCR product was inserted into pBCSK+ catalyzed by IN, resulting in the loss of 2 bp from the LTR ends. This plasmid was propagated in E. coli MC1061/P3 under the conditions described above. The integration acceptor was plasmid pBCSK+ (Stratagene, La Jolla, CA) and was propagated in E. coli DH5α. Plasmids were purified with Qiaprep columns (Qiagen, Chatsworth, CA) according to the manufacturer’s instructions. The growth of DH5α containing pBCSK+ was selected by addition of chloramphenicol (35 μg/ml).
Preparation of Donor DNAs
Integration donors were amplified by using thermostable Vent DNA polymerase and the primers listed above. Twenty five pmol of each primer and 50 ng of pHHIV2 DNA, as the template, were used for each PCR. Vent DNA polymerase was used according to the manufacturer’s instructions. A total of 20 rounds of amplification was performed in each reaction. The amplification conditions were 94 °C for 2 min, 50 °C for 1 min, and 72 °C for 1 min for three rounds. This was followed by amplification conditions that used 94 °C for 2 min, 57 °C for 1 min, and 72 °C for 45 s for 17 additional rounds. The resultant product donor DNA was isolated after electrophoresis through 2% agarose gels equilibrated with 0.5× Tris borate/EDTA (5). The purified DNA (600 ng) was recovered using QIAquick gel extraction kit (Qiagen). The integration donors were ~300 bp in length and were internally labeled during the PCR by the inclusion of [α-32P]dCTP (3,000 Ci/mmol, 10 mCi/ml). The final concentrations of deoxyribonucleoside triphosphates during amplification reactions were 0.25 mM each of unlabeled dATP, dGTP, and dTTP. The final dCTP concentration was 0.0502 mM (12 Ci/mmol, 0.6 mCi/ml).
Standard Integration Reaction Conditions
The concerted integration reaction conditions were similar to those described previously (5, 14). Briefly, 15 ng (0.15 pmol of ends) of donor DNA was mixed with 50 ng of acceptor DNA (0.02 pmol) and 80 ng of HIV-1 IN (1.25 pmol) in an 8.5-μl preincubation reaction mixture containing, at final concentrations, 25 mM MOPS (pH 7.2), 23 mM NaCl, 10 mM dithiothreitol, 5% polyethylene glycol 8000, 10% dimethyl sulfoxide, 0.05% Nonidet P-40, 1% glycerol, 1.6 mM HEPES (pH 8.0), and 3.3 mM EDTA. The IN was diluted in a buffer containing 30% glycerol, 0.5 M NaCl, 50 mM HEPES (pH 8.0), 1 mM dithiothreitol, and 0.1 mM EDTA. Where specified 100 ng of HMG-I(Y) was added to the reaction mixtures. The preincubation reaction mixtures were placed on ice overnight. The volume of each preincubation mixture was then increased to 10 μl with the addition MgCl2 of to a final concentration of 7.5 mM, and the integration assay mixture was incubated at 37 °C for 2 h. The reactions were stopped by increasing the volume to 150 μl by the addition of EDTA (final concentration of 4.25 mM), SDS (final concentration of 0.44%), and proteinase K (final concentration of 0.06 mg/ml). After digestion for 60 min at 37 °C, the reaction mixtures were extracted with phenol followed by phenol/chloroform/isoamyl alcohol (25:24:1 mixture). Fifteen μl of 3 M sodium acetate (pH 5.2) was added along with 1 μl of glycogen (10 mg/ml stock solution). The reaction products were precipitated by the addition of 450 μl of 100% ethanol and washed twice with 70% ethanol prior to electrophoresis and autoradiography. The reaction products were separated on a 1% agarose gel run in 0.5× Tris borate, EDTA, and ethidium bromide at 10 V/cm for 2 h. Following electrophoresis, gels were submerged in 5% trichloroacetic acid for 20 min or until the bromphenol blue dye turned bright yellow. After washing with water, the gels were dried on DE81 paper (Whatman) in a Bio-Rad slab gel dryer at 80 °C for ~2 h under vacuum. Quantitation of reaction products was carried out using a PhosphorImager and ImageQuant 5.0 software. Experiment with WT donor integrants always accompanied experiment with mutant donor integrants as controls. All experiments were repeated at least 2 times.
Cloning and Sequencing of Integrants
In all experiments, integration products were used directly for transformation of bacteria. The integration products were introduced into E. coli MCI061/P3 by electroporation, using a Bio-Rad electroporator with 0.1-cm electroporation cuvettes, 1.8-kV voltage, 25-microfarad capacitance, and 200-ohm resistance. The P3 episome is maintained at a low copy number. Therefore, only 40 μg/ml ampicillin, 15 μg/ml kanamycin, or 10 μg/ml tetracycline were required for selection. Under these conditions, we detected no colonies after supF selection when the donor, acceptor, or donor and acceptor were electroporated into cells in the absence of IN. Plasmid DNAs were recovered from individual clones, and integration junctions were sequenced by using primers U3seq (for sequencing the U3 junction) and U5seq (for sequencing the U5 junction). Sequencing was performed using the Thermo-Sequenase kit (U. S. Biochemical Corp.).
Statistical Analysis
We used the χ2 test to examine the statistical significance of the influence of base composition at one position on the sequence selection at the other. HIV-1 sequences with the following GenBank™ accession numbers were analyzed for sequence conservation/variability and relationship between bases (Table VIII): AB023804, AB032740, AB032741, AB049811, AB052867, AB052995, AF003887, AF003888, AF004394, AF004885, AF033819, AF042100, AF042101, AF042106, AF049494, AF049495, AF064699, AF069140, AF070521, AF075719, AF084936, AF086817, AF110959, AF110962, AF110963, AF110964, AF110965, AF110966, AF110967, AF110968, AF110969, AF110970, AF110971, AF110972, AF110973, AF110974, AF110975, AF110976, AF110977, AF110978, AF133821, AF164485, AF197338, AF197339, AF197340, AF197341, AF119819, AF119820, AF256205, AF256206, AF256207, AF256208, AF256210, AF256211, AF259954, AF259955, AF286236, AF286365, AF290030, AF321523, AF385934, AF385935, AF385936, AJ006287, AJ237565, AJ245481, AJ271370, AJ271445, AJ288981, AJ288982, AJ291719, AJ302646, AJ302647, D10112, D86068, K02007, K02013, K02083, K03454, K03455, L02317, L20571, L20587, L31963, L39106, M17449, M17451, M19921, M26727, M38429, M62320, M93258, M93259, NC_001802, U12055, U21135, U23487, U34603, U34604, U37270, U43096, U43141, U51189, U54771, U69584, U69585, U39362, U69586, U69587, U69588, U69589, U69592, U88822, X01762, X04415, and Z11530.
Table VIII. Frequency of each base at positions 1–20 of U5 and U3 LTRs.
Data derived from 116 sequences of HIV-1 published in GenBank™ that contain both U3 and U5 LTRs.
| 20 | 19 | 18 | 17 | 16 | 15 | 14 | 13 | 12 | 11 | 10 | 9 | 8 | 7 | 6 | 5 | 4 | 3 | 2 | 1 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| U5 LTRa | ||||||||||||||||||||
| G | 28 | 90 | 23 | 86 | 77 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 114 | 0 | 0 | 115 | 0 |
| A | 3 | 11 | 11 | 9 | 34 | 116 | 115 | 116 | 116 | 0 | 0 | 0 | 0 | 0 | 116 | 0 | 0 | 116 | 1 | 0 |
| T | 85 | 15 | 79 | 21 | 5 | 0 | 0 | 0 | 0 | 116 | 1 | 116 | 0 | 116 | 0 | 0 | 1 | 0 | 0 | 116 |
| C | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 114 | 0 | 116 | 0 | 0 | 2 | 115 | 0 | 0 | 0 |
| U3 LTR | ||||||||||||||||||||
| G | 0 | 96 | 0 | 23 | 0 | 0 | 0 | 0 | 0 | 76 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 116 | 0 |
| A | 116 | 5 | 2 | 93 | 115 | 116 | 1 | 0 | 116 | 31 | 0 | 0 | 0 | 38 | 0 | 0 | 0 | 116 | 0 | 0 |
| T | 0 | 0 | 99 | 0 | 0 | 0 | 91 | 116 | 0 | 9 | 1 | 0 | 0 | 78 | 116 | 0 | 0 | 0 | 0 | 116 |
| C | 0 | 15 | 15 | 0 | 1 | 0 | 24 | 0 | 0 | 0 | 115 | 116 | 116 | 0 | 0 | 116 | 116 | 0 | 0 | 0 |
Numbers shown in boldface correspond to the wild type sequence.
RESULTS
Sequence Selection by IN at Mismatched Positions in the U5 LTR
To examine sequence requirements by HIV-1 IN for the U3 and U5 IN recognition sequences, we constructed a library of mini donor DNA substrates that contained randomized nucleotides at different positions in the IN recognition sequences. These substrates were then used to reconstitute concerted DNA integration in vitro in which IN was allowed to select those sequences from the library that supported concerted DNA integration. The conditions for reconstitution of integration are described under “Experimental Procedures.” The first mini donor DNA library analyzed contained randomization deoxyribonucleotides at positions 5, 6, 9, 11, 13, 14, 16, and 20 (see Fig. 1) of U5 while leaving the U3 IN recognition sequence as wild type. These positions represent the mismatched bases in the nearly perfect inverted repeat in the HIV-1 U3 and U5 RNA termini.
Fig. 1. Mismatched positions in HIV-1 LTR ends and sites of randomization.
Twenty base pairs of the IN recognition sequences derived from the U3 and U5 LTR ends are shown. Bases that differ between the two LTR ends are in bold.
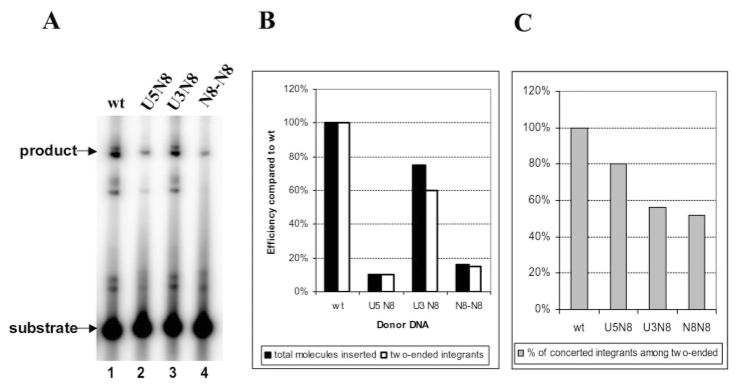
Integration products from reactions with labeled donor DNA were separated by agarose gel electrophoresis and quantified by PhosphorImaging. The yield of integrants obtained with the library of U5 randomized donors was 10% that observed with the wild type donor DNA (Fig. 2A, lanes 1 and 2). This decrease was also reflected in the numbers of colonies recovered after the integration reactions were introduced into bacteria (Fig. 2B) as described under “Experimental Procedures.” These colonies are derived from integrants arising from two-ended insertions of the donor into an acceptor. When individual integrants were sequenced, we found that 80% arose by a concerted mechanism, exhibiting all of the characteristics associated with integration in vivo (Fig. 2, B and C, and Table I in Supplemental Material). The remainder arose by a non-concerted mechanism, which introduced deletions into the acceptor DNA (Table I in Supplemental Material).
Fig. 2. Integration of donors with randomized mismatched positions in U3 and/or U5 LTR.
Integration reactions were reconstituted as described under “Experimental Procedures.” A, gel electrophoresis analysis of integration products from reactions with wild type (wt) donor (lane 1), donor containing randomized positions 5, 6, 9, 11, 13, 14, 16, and 20 in U5 LTR end (U5N8) (lane 2), donor with U3 LTR end randomized at the same positions (U3N8) (lane 3), or donor with both IN recognition sequences containing randomized mismatched positions (N8-N8) (lane 4). B, summary of the percentage of RFII-like products shown in A (closed bars) compared with wild type and the total number of colonies containing two-ended integrants after integration reaction products introduced into bacteria (open bars). Integration efficiency of wild type mini donor DNA was set as 100%. The data shown are an average of two separate experiments with the standard deviation between experiments of 0.5–2%. C, percent of integrants derived from Supplemental Material to Tables I–III formed by a concerted mechanism involving two ends of the same donor DNA.
In examining the bases selected in the different randomized positions of the recovered and sequenced concerted DNA integrants, we found all 4 bases at each randomized position (Table I). The fact that these integrants arose by a concerted DNA integration mechanism indicates that there is considerable plasticity as to what constitutes an IN recognition sequence, especially considering that we have effectively mutated 40% of the U5 IN recognition sequence. However, upon a closer analysis, selection preferences for and against different bases were detected. For instance, only two concerted DNA integrants contained T at position 5. Also G was almost completely excluded from positions 6, 9, and 20 (Table I). In contrast, A was preferred at position 6, whereas T was most often selected at position 9 as found in the U5 wild type viral sequence. At position 16, A and C were preferred even though G is found at this position in the wild type U5 viral sequence. T and C were selected at position 20 where T is the wild type base (Table I). The data presented in Table I were derived from two separate experiments. The distribution of selected bases at each position is presented in Table I, see the numbers in parentheses.
Table I. Selection of bases in the HIV-1 U5 IN recognition sequence in vitro from donor DNAs with eight non-conserved base positions randomized.
Number of each base selected at each randomized position of U5 N8 donor in the sequenced pool of concerted integrants is shown. The numbers in parentheses are derived from the two separate experiments. The sum is shown outside of the parentheses. Individual sequences may be found in the Supplemental Material.
| 5 | 6 | 9 | 11 | 13 | 14 | 16 | 20 | |
|---|---|---|---|---|---|---|---|---|
| G | 17 (8, 9) | 2 (0, 2) | 4 (2, 2) | 7 (4, 3) | 8 (3, 5) | 12 (1, 11) | 5 (2, 3) | 2 (1, 1) |
| A | 16 (10, 6) | 24 (11, 13) | 8 (5, 3) | 13 (7, 6) | 9 (3, 6) | 10 (7, 3) | 17 (9, 8) | 8 (1, 7) |
| T | 2 (1, 1) | 10 (6, 4) | 22 (12, 10) | 15 (7, 8) | 15 (7, 8) | 18 (11, 7) | 7 (4, 3) | 19 (10, 9) |
| C | 13 (5, 8) | 12 (7, 5) | 14 (5, 9) | 13 (6, 7) | 16 (11, 5) | 8 (5, 3) | 19 (9, 10) | 19 (12, 7) |
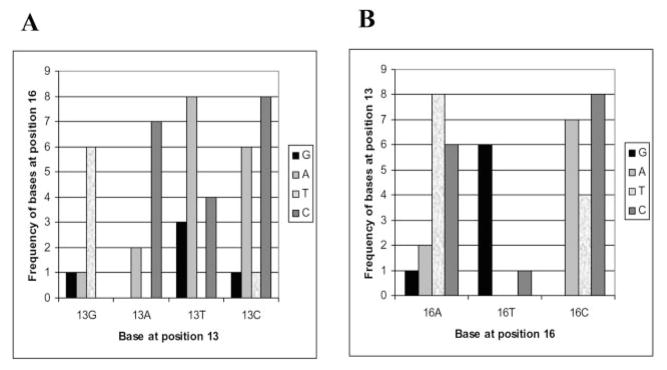
A further analysis of the sequence of the concerted integrants revealed that the bases at certain positions were found in combination with bases at other positions (Fig. 3A). For instance, the strongest statistically significant relationship occurs between positions 13 and 16. The probability (p) of position 13 as a specific base in combination with position 16 is 0.000022. The probability was calculated using χ2 analysis. A p value of less than 0.1 was considered significant. For example, when G is found in position 13, one finds T at position 16; when A is found at position 13, one now finds C at position 16; when T is found at position 13, one finds A at position 16 (Fig. 4). Other examples of preferred combination of bases were at positions 6 and 13, at positions 6 and 16, at positions 14 and 9, and at positions 20 and 6. The presence of the wild type T (as opposed to C) at position 20 resulted in selection of a wild type base A at position 6. Also, even though a wild type A was not a preferred base at position 13, 5 of 9 A nucleotides selected at this position were in donors containing wild type A at position 6. Only five donors were selected with wild type G at position 16. Interestingly, four of five were in donors containing wild type base A at position 6. The presence of wild type base A at position 14 resulted in selection of wild type T at position 9. Other combinations of interactions were not considered statistically significant.
Fig. 3. Relationships between randomized positions in donor DNA-HIV-1 IN complex.
Donor contained IN recognition sequence with randomized mismatched positions at one end and wild type U3 (A) or wild type U5 IN recognition sequence at the other end (B). Data were derived from Supplemental Material to Table I(i) for A and from Supplemental Material to Table II(ii) for B. χ2 test was used to reveal statistically significant relationships between positions.
Fig. 4. Example of relationship between positions in U5 LTR end.
A, relationship of base content at position 13 with base selection at position 16. B, relationship of base content at position 16 with base selection at position 13. DNA integration reactions were carried out with IN, HMG-(I/Y), acceptor DNA, and a mini donor DNA containing randomized bases at mismatched positions in U5 LTR end, whereas U3 LTR end contained wild type terminal U3 sequence. Frequencies of particular base selection at position 16 (A) and position 13 (B) are shown by height of correspondent bars.
Sequence Selection by IN at Mismatched Positions in the U3 LTR
A similar analysis was carried out using a library of donor DNAs in which the same mismatched positions were randomized in the U3 IN recognition sequences while maintaining the U5 LTR IN recognition sequence as wild type. In this instance, the number of integrants from the library as analyzed by agarose gel was 75% that of wild type donor level (Fig. 2A, lanes 1 and 3). As expected, there was a further decrease in the percentage of colonies recovered compared with wild type when introduced into bacteria (Fig. 2B). Previous analysis of specific mutant substrates indicated that a wild type U5 promotes one-ended insertion events when the U3 IN recognition sequence is mutated (14). Of the two-ended donor insertion integrants sequenced, only 56% arose by a concerted integration mechanism (Fig. 2C and Table II in the Supplemental Material). An analysis of concerted integrants indicated that all bases were found in all mutated positions (Table II), similar to what was observed with the randomized U5 substrates. There was a preference for selection of the wild type T at position 6, the U5 G at position 5, and U5 T at position 20. As with the previous data set, sequences were derived from two independent experiments (Table II, see the numbers in parentheses).
Table II. Selection of bases in the HIV-1 U3 IN recognition sequence in vitro from donor DNAs with eight non-conserved base positions randomized.
Number of each base selected at each randomized position of U3 N8 donor in the sequenced pool of concerted integrants is shown. The numbers in parentheses are derived from the two separate experiments. The sum is shown outside of the parentheses. Individual sequences may be found in the Supplemental Material.
| 5 | 6 | 9 | 11 | 13 | 14 | 16 | 20 | |
|---|---|---|---|---|---|---|---|---|
| G | 16 (9, 7) | 4 (2, 2) | 11 (6, 5) | 12 (7, 5) | 9 (6, 3) | 5 (3, 2) | 9 (5, 4) | 4 (1, 3) |
| A | 8 (4, 4) | 9 (5, 4) | 6 (2, 4) | 5 (2, 3) | 7 (5, 2) | 11 (4, 7) | 5 (3, 2) | 8 (4, 4) |
| T | 4 (3, 1) | 15 (9, 6) | 10 (4, 6) | 6 (2, 4) | 8 (2, 6) | 10 (5, 5) | 10 (7, 3) | 14 (8, 6) |
| C | 5 (1, 4) | 5 (1, 4) | 6 (5, 1) | 10 (6, 4) | 9 (4, 5) | 7 (5, 2) | 9 (2, 7) | 7 (4, 3) |
We further analyzed the data for statistically significant biases in selection of bases at the different positions (Fig. 3B). The selection of specific bases at positions 6 and 16 deviated significantly from random. Four of five sequences containing A at position 16 as in wild type U3 also had T at position 6 as in wild type U3. Combination of specific bases at position 13 with positions 5, 6, 11, 14, and 20 were also detected. Choice of base at position 20 affected base selection at position 13. Only the presence of viral base T at position 13 resulted in selection of A at position 20 which is present in wild type U3 LTR and only the presence of an A at position 20 induced selection of T at position 13. The presence of T at position 13 led to selection of G or A at position 5, whereas a wild type C was excluded; C was preferred at position 11 where G is found in viral U3 LTR; at position 14 the wild type base T was also not preferred. Four of the five C nucleotides at position 6 were found in donors containing T at position 13. T was almost exclusively found at position 6 when G was present at position 13. Specific combinations of bases were found at positions 9 and 20, at positions 11 and at positions 13 and 14, and positions 14 and 6. Other combinations were considered not statistically significant.
Randomization of Mismatched Positions in Both LTRs
IN was presented with a library of donor DNAs containing all 4 bases at positions 5, 6, 9, 11, 13, 14, 16, and 20 in both IN recognition sequences. Thus a total of 16 of the 40 specific base pair positions of the IN recognition sequences were randomized in these substrates. The integration reaction with these donor molecules, as analyzed by gel electrophoresis, was similar to that observed when using a donor DNA in which only the U5 IN recognition sequence was mutated (Fig. 2A, lanes 2 and 4, and Fig. 2B). Of the two-ended integrants sequenced, 51% was derived by a concerted DNA integration mechanism (Fig. 2C and Table III in the Supplemental Material). This was similar to what was observed with the substrates containing only randomizations in U3 IN recognition sequence.
The pattern of base selection by HIV-1 IN was analyzed from the pool of sequenced concerted integrants. Overall, T was selected predominantly at position 20, and G was almost excluded from position 6, whereas T was under-represented at position 5 (Table III). These trends are similar to that observed when only one IN recognition sequence was randomized. Because all mismatched positions were randomized in these substrates, there can be no distinction between U3 and U5 ends. Therefore it is difficult to relate intra-relationships of bases selected within a given IN recognition sequence to the data presented in Tables I and II in the Supplemental Material, where one IN recognition sequence remained wild type. However, randomizing both LTR ends permits one to examine the sequenced integrants for inter-LTR relationships in selection of bases. For instance, in most cases A was selected at position 6 in one LTR end, whereas T was selected at position 6 in the other. A statistical analysis of relationships for selection of bases in different positions is presented in Table IV. A significant inter-LTR as well as intra-LTR termini relationship was observed among positions 6 and 16 (Table IV and Fig. 3). Another example is a correlation between the bases at position 5 in one LTR with that at position 20 in the other (Table IV).
Table III. Selection of bases in the HIV-1 U5 and U3 IN recognition sequence in vitro from donor DNAs with eight non-conserved base positions randomized.
Number of each base selected at each randomized position of N8N8 donor in the sequenced pool of concerted integrants is shown. The numbers in parentheses are derived from the two separate experiments. The sum is shown outside of the parentheses. Individual sequences may be found in the Supplemental Material.
| 5 | 6 | 9 | 11 | 13 | 14 | 16 | 20 | 20 | 16 | 14 | 13 | 11 | 9 | 6 | 5 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G | 9 (6, 3) | 1 (1, 0) | 4 (2, 2) | 7 (0, 7) | 7 (3, 4) | 7 (4, 3) | 8 (2, 6) | 5 (3, 2) | 6 (5, 1) | 6 (4, 2) | 5 (1, 4) | 8 (4, 4) | 6 (3, 3) | 5 (3, 2) | 3 (1, 2) | 11 (6, 5) |
| A | 8 (4, 4) | 19 (9, 10) | 5 (2, 3) | 5 (3, 2) | 9 (6, 3) | 11 (5, 6) | 10 (6, 4) | 4 (2, 2) | 6 (3, 3) | 12 (6, 6) | 7 (2, 5) | 8 (5, 3) | 12 (6, 6) | 12 (5, 7) | 7 (4, 3) | 7 (4, 3) |
| T | 2 (0, 2) | 6 (4, 2) | 16 (9, 7) | 11 (8, 3) | 11 (6, 5) | 7 (4, 3) | 9 (5, 4) | 19 (10, 9) | 17 (7, 10) | 8 (5, 3) | 15 (10, 5) | 8 (3, 5) | 11 (7, 4) | 9 (5, 4) | 18 (8, 10) | 7 (4, 3) |
| C | 14 (7, 7) | 7 (3, 4) | 8 (4, 4) | 10 (6, 4) | 6 (2, 4) | 8 (4, 4) | 6 (4, 2) | 5 (2, 3) | 4 (2, 2) | 7 (2, 5) | 6 (4, 2) | 9 (5, 4) | 4 (1, 3) | 7 (4, 3) | 5 (4, 1) | 8 (3, 5) |
Table IV. Relationships between randomized positions of both LTR ends.
Data were derived from Supplemental Material for Table III. Analysis was performed as described under “Experimental Procedures.” Statistically significant probabilities of influence of randomized positions in one LTR on other positions in the opposite LTRs are shown in cells belonging to the column of the position that having an influence on sequence selection and to the row of the position being affected.
| Opposite | Same LTR |
|||||||
|---|---|---|---|---|---|---|---|---|
| 5 | 6 | 9 | 11 | 13 | 14 | 16 | 20 | |
| 5 | 0.0179 | |||||||
| 6 | 0.0117 | 0.0137 | 0.0672 | 0.0142 | 0.0107 | |||
| 9 | ||||||||
| 11 | 0.0035 | 0.0195 | ||||||
| 13 | ||||||||
| 14 | 0.0158 | |||||||
| 16 | 0.0219 | |||||||
| 20 | 0.005 | |||||||
Randomization of Positions 17–19 in U5 LTR
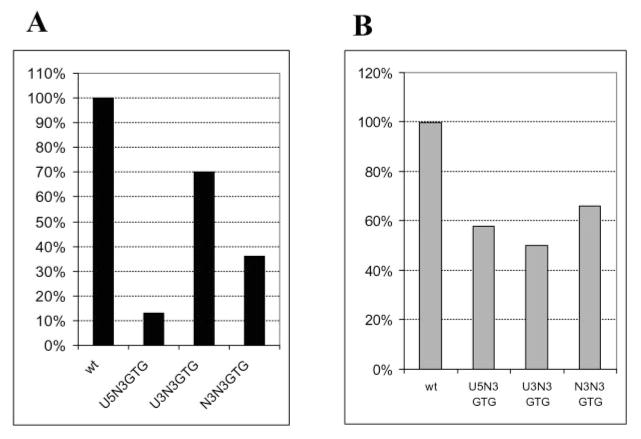
Because we detected a preference for selection of a specific base at position 20 in the HIV-1 U3 and U5 IN recognition sequences, we thought that adjacent regions in the IN recognition sequences might be also important. Therefore, we examined the effect of introducing random sequence into positions of 17–19. These base pairs are conserved in the two LTR ends. When these positions were randomized in U5, there was a decrease in recovered colonies to 13% that observed with a wild type donor DNA (Fig. 5A). This suggests that these positions are important in recognition of the donor substrate. Of the sequenced integrants, 58% were derived by a concerted mechanism (Fig. 5B). Sequence analysis of the concerted integrant pool showed that C and G were preferred at position 19, T and C at position 18, and C at position 17 (Table V). Even though these are conserved bases in the U5 and U3 LTR ends, wild type bases were not preferred at all three positions among sequenced integrants derived by a concerted mechanism (Table V). As in the case of mismatched positions, the selection of a base at one position was correlated with that at another. Combinations were found between positions 17 and 19 (p = 0.047). When a wild type G was at position 19, a wild type G was also at position 17; when C was at position 19, C was also at position 17 and vice versa.
Fig. 5. Randomization of positions 17–19 in U3 and/or U5 LTR.
Concerted DNA integration reactions were carried out with IN, HMG-(I/Y), acceptor DNA, and a wild type (wt) mini donor DNA, donor DNA in which positions 17–19 were randomized in U5 (U5N3GTG), U3 LTR (U3N3GTG), and in both (N3N3GTG). A, total number of colonies containing two-ended integrants. B, percent of integrants derived from Supplemental Material to Tables V–VII formed by a concerted mechanism involving two ends of the same donor DNA.
Table V. Selection of bases in the HIV-1 U5 IN recognition sequence in vitro from donor DNAs with 17–19 conserved base positions randomized.
Number of each base selected at each randomized position of U5 N3 donor in the sequenced pool of concerted integrants is shown. The numbers in parentheses are derived from the two separate experiments. The sum is shown outside of the parentheses. Individual sequences may be found in the Supplemental Material.
| 19 | 18 | 17 | |
|---|---|---|---|
| G | 10 (4, 6) | 4 (2, 2) | 8 (2, 6) |
| A | 4 (3, 1) | 6 (2, 4) | 4 (3, 1) |
| T | 6 (3, 3) | 13 (8, 5) | 8 (6, 2) |
| C | 14 (7, 7) | 11 (5, 6) | 14 (6, 8) |
To confirm that positions 17–19 were indeed important for HIV-1 IN recognition, we analyzed two donor DNAs with specific U5 base pair changes at these positions and compared their relative efficiency to a wild type donor. The first mutant (U5–18GA19) contained G and A substitution at U5 positions 18 and 19, respectively. The second mutant (U5–17AAT19) contained AAT instead of GTG at positions 17–19, respectively. The number of colonies containing U5–18GA19 integrants was reduced to ~5% the number found using a wild type donor. The U5–17AAT19 mutation caused a 50% decrease in the number of colonies compared with wild type.
Randomization of Positions 17–19 in U3 LTR
Positions 17–19 of the U3 IN recognition sequence were also randomized. In this case the yield of colonies was 70% of the number obtained with a wild type donor (Fig. 5A). Among them only 50% were derived by a concerted integration mechanism (Fig. 5B and Table VI in the Supplemental Material). Analysis of the concerted integrants showed that C was preferred at position 19, T at position 18, and T/G/A at position 17 (Table VI). Moreover, base choice at position 19 restricted base selection at position 17 (as in U5 LTR end), p = 0.07.
Table VI. Selection of bases in the HIV-1 U3 IN recognition sequence in vitro from donor DNAs with 17–19 conserved base positions randomized.
Number of each base selected at each randomized position of U5 N8 donor in the sequenced pool of concerted integrants is shown. The numbers in parentheses are derived from the two separate experiments. The sum is shown outside of the parentheses. Individual sequences may be found in the Supplemental Material.
| 19 | 18 | 17 | |
|---|---|---|---|
| G | 6 (4, 2) | 9 (5, 4) | 8 (3, 5) |
| A | 6 (2, 4) | 3 (2, 1) | 7 (3, 4) |
| T | 6 (3, 3) | 13 (5, 8) | 10 (6, 4) |
| C | 11 (5, 6) | 4 (2, 2) | 4 (2, 2) |
Randomization of Positions 17–19 in Both LTRs in the Same Donor
To analyze interaction between the two LTR ends, positions 17–19 in both U5 and U3 IN recognition sequences were randomized. The yield of colonies was 36% of wild type (Fig. 5A), and 65% contained plasmids formed by a concerted mechanism (Fig. 5B and Table VII in Supplemental Material). An analysis of the concerted integrants indicated that C was preferred at positions 17–19 in U5 LTR, G at positions 17 and 19 of U3 LTR, and A at position 18 of U3 LTR (Table VII). No statistically significant interactions were found in this instance.
Table VII. Selection of bases in the HIV-1 U5 and U3 IN recognition sequence in vitro from donors DNAs with 17–19 conserved base positions randomized.
Number of each base selected at each randomized position of U5 N8 donor in the sequenced pool of concerted integrants is shown. The numbers in parentheses are derived from the two separate experiments. The sum is shown outside of the parentheses. Individual sequences may be found in the Supplemental Material.
| 19 | 18 | 17 | 19 | 18 | 17 | |
|---|---|---|---|---|---|---|
| G | 11 (9, 2) | 8 (2, 6) | 10 (6, 4) | 13 (8, 5) | 10 (2, 8) | 15 (8, 7) |
| A | 5 (1, 4) | 7 (5, 2) | 9 (4, 5) | 8 (4, 4) | 13 (6, 7) | 9 (5, 4) |
| T | 8 (3, 5) | 7 (5, 2) | 7 (3, 4) | 10 (5, 5) | 10 (8, 2) | 7 (4, 3) |
| C | 16 (7, 9) | 18 (8, 10) | 14 (7, 7) | 9 (3, 6) | 7 (4, 3) | 9 (3, 6) |
DISCUSSION
We have used a reconstituted HIV-1 integration system capable of concerted DNA integration (5, 14) to examine the requirements for HIV-1 IN recognition sequences. The donor DNA substrates used in these studies contained 20 bp derived from the U3 and U5 LTR termini at each end. Although duplex oligodeoxyribonucleotide substrates are capable of supporting processing and strand transfer reactions in vitro with less than 20 bp (13), we chose to use the larger sequence in our donor DNA substrates because of a high degree of sequence conservation among LTR sequences derived from HIV-1-infected patients which included position 20 of the U3 LTR (Table VIII). The 20-bp length also consists of the inverted repeat in the LTR termini. In RSV, the inverted repeat defines the IN recognition sequence capable of supporting concerted DNA integration in vitro (3, 5, 11).
Sequence analysis of the integrants formed by the concerted integration mechanism using a single donor DNA containing random sequence at the mismatched bases of the inverted repeat indicates that HIV-1 IN tolerates considerable variation in both the U3 and U5 termini. This is also true for the matched sequences at positions 17–19 in both termini. The ability of HIV-1 IN to tolerate sequence variation in vitro was described previously (8, 9) using duplex oligodeoxyribonucleotide substrates. Despite the finding that successful integrants were selected with all 4 bases at randomized positions, some preferences for specific bases were observed, such as at positions 5, 6, and 20. Positions 5 and 6, adjacent to the conserved CA dinucleotide, were found to be important in previous analyses where mutations in these positions caused a decrease in donor DNA integration efficiency and/or change in mechanism of integration (14). Because these are only preferences, a question arises as to whether a sufficient number of sequences were examined to indicate that the observed sequence selection was not biased by sample size. To control for this possibility, each randomized donor DNA substrate was analyzed in two separate reactions. An examination of the bases selected in the randomized positions from the two pools, with some exceptions, were the same suggesting that a sufficient number of sequences were examined.
Preferences observed for specific bases at position 20 in both HIV-1 LTR ends suggested there was a second region in the LTR termini, distal to the CA dinucleotide, important for formation of an HIV-1 IN-DNA complex capable of supporting concerted integration in vitro. This appears to be the case because randomization of the adjacent positions 17–19 in the U3 and/or the U5 LTR resulted in a significant decrease in the total number of two-ended DNA integrants recovered concomitant with an increase in the percentage of non-concerted integrants detected by sequencing. The need for sequences distal to the region around the CA dinucleotide for concerted DNA integration was confirmed by analysis of two mutants that contained specific sequences substituted at positions 18 and 19 and 17–19, respectively. Both substitutions resulted in a significant decrease in the efficiency of the concerted DNA integration reaction compared with a wild type donor. The fact that these distal sequences are not required for processing and strand transfer reactions using duplex oligodeoxyribonucleotide substrates suggests that they are specifically needed for formation of concerted DNA integration complexes and may constitute a second region of contact between multimers of IN and the substrate. Nevertheless, an analysis of 116 HIV-1 GenBank™ sequences containing both U3 and U5 IN recognition sequences indicated that the majority of variation in vivo in the U5 LTR end was found at positions 16–20 (Table VIII), the region for which the above in vitro system highlighted as important for concerted DNA integration. No mutations were found at positions 1, 3, 6–9, 11–13, and 15. In the U3 LTR, the majority of mutations were found at positions 7, 11, 14, and 17–19, and no mutations were found at positions 1–5, 8–9, 12–13, 15, and 20. Because IN tolerates sequence variation in vitro, the in vivo pattern of conservation could be explained by requirements for replication functions other than integration. For instance, U5 terminal sequence is preserved to maintain the structure of the U5 IR stem/loop to support efficient initiation of reverse transcription (19–21). HIV-1 U3 partially encodes the Nef gene (22) so that the U3 LTR sequence variation is likely to be restricted by this coding function.
Although sequence variation is tolerated by HIV-1 IN in vitro, we examined integrants derived by a concerted integration mechanism for correlations among bases selected at different positions within the same IN recognition sequence. For instance, when the U3 IN recognition sequence remained wild type and the mismatched bases in U5 were randomized, we found statistically significant relationships for combinations of bases selected at positions 6, 13, and 16. A different set of preferences was found when the U3 end was randomized and U5 remained wild type. This highlights the asymmetric recognition of these sequences by HIV-1 IN and is consistent with previous analyses (14). It is also interesting to note that the statistical correlation between intra-base selections at different positions in U5 was stronger than found in U3. In addition, U3 randomized donors produced six times more colonies than U5 randomized donors. Therefore, the HIV-1 U3 tolerates much more sequence variation than the U5 termini.
A relationship among positions within the HIV-1 IN recognition sequence was suggested previously using duplex oligodeoxyribonucleotide substrates (9). In this work, 5′ AAGCA 3′ was selected at positions 3–7 from a pool of substrates in which these positions were randomized. However, when positions 3–11 were randomized, 5′ AACACAGCA 3′ was a selected sequence. Selection of the base at position 7 (in bold) depended on the size of the randomized region and therefore is likely to be influenced by the upstream sequence. In either case, the base selected at position 7 was not the one present in either wild type U5 or U3 LTR. Bases selected at positions 8, 10, and 11 were also different from wild type and those selected in the study by Esposito and Craigie (8). In contrast, when single-ended duplex oligodeoxyribonucleotide substrates representing the RSV LTR termini were randomized at positions 3–7 and positions 3–11, sequences selected by RSV IN at positions 3–7 were independent of the randomized region. Thus, specific combinations of sequence selection were not detected in the analyses of these RSV substrates (9).
Besides intra-relationships for selection of bases noted above, we examined the sequence data of concerted integrants for inter-relationships in selection of bases between LTR ends, which might be expected for a concerted reaction which brings both ends of the viral DNA together into a complex with IN. We detected statistically significant preferences for specific combinations of bases at some randomized positions between the two IN recognition sequences (Table IV). This included position 5 of one DNA terminus being correlated with position 20 at the other end and position 16 with positions 5 and 6 of the opposite LTR end. A similar but not identical picture emerges in examining the 116 HIV-1 GenBank™ sequences. By using the χ2 test analysis, it appears that choice of bases in several positions is correlated with the sequence at other positions belonging to the same as well as different LTR ends (Tables IX and X). For example, positions 16–20 (variable region of U5 LTR) have very strong relationships with each other (most likely to preserve the structure of the IR-stem loop) and with position 7 of the U3 LTR in vivo. This correlates with findings in our reconstituted concerted integration system where position 20 of one LTR and position 5 of the opposite LTR are interconnected. Correlation between the two viral DNA termini was observed previously for Moloney murine leukemia virus in vivo (1) and in vitro (2) and for an AMV in reconstituted concerted integration system (15).
Table IX. Probabilities of statistically significant influence of positions 16–20 of U5 LTR on sequence variation at positions 16–20 of U5 LTR and positions 7, 11, 14, 17–19 of U3 LTR.
Patient-derived viral isolates of sequence data published in GenBank™ was analyzed for sequence variation dependence between positions by χ2 test. When U5 positions shown in the first row in boldface influence any position shown in the first column, probability of this event is printed in the cell belonging to the column of U5 position and the row of the position with which it interacts. NA, not applicable.
| Position | U5 LTR |
||||
|---|---|---|---|---|---|
| 20 | 19 | 18 | 17 | 16 | |
| U5 20 | NA | 8.4 × 10−18 | 2.48 × 10−19 | 4.215 × 10−20 | 3.21 × 10−10 |
| U5 19 | 6.525 × 10−19 | NA | 5.01 × 10−19 | 1.49 × 10−18 | 2.96 × 10−13 |
| U5 18 | 3.4 × 10−20 | 3.35 × 10−18 | NA | 1.653 × 10−23 | 1.36 × 10−9 |
| U5 17 | 4.066 × 10−18 | 1.49 × 10−18 | 8.54 × 10−17 | NA | 13.08 × 10−10 |
| U5 16 | 7.447 × 10−10 | 5.1 × 10−12 | 4.15 × 10−15 | 1.451 × 10−9 | NA |
| U3 19 | 3.33 × 10−5 | ||||
| U3 18 | 7.86 × 10−21 | 7.038 × 10−18 | 3.72 × 10−5 | ||
| U3 17 | 3.109 × 10−12 | 0.036 | 0.01 | ||
| U3 14 | 0.087 | 0.052 | |||
| U3 11 | 0.079 | 0.074 | |||
| U3 7 | 4.99 × 10−6 | 2.03 × 10−5 | 7.26 × 10−5 | 2.38 × 10−7 | 0.019 |
Table X. Probabilities of statistically significant influence of positions 7, 11, 14, 17–19 of U3 LTR on sequence variation at positions 7, 11, 14, 17–19 of U3 LTR and positions 16–20 of U5 LTR.
Notations are as in legend to Table IX except U5 is now U3. NA, not applicable.
| U3 LTR |
||||||
|---|---|---|---|---|---|---|
| 19 | 18 | 17 | 14 | 11 | 7 | |
| U3 19 | NA | 1.76 × 10−25 | 0.012 | 0.05 | ||
| U3 18 | 7.88 × 10−25 | NA | 0.085 | 0.079 | 0.064 | |
| U3 17 | 8.42 × 10−10 | 0.037 | NA | 0.044 | 0.006 | |
| U3 14 | NA | |||||
| U3 11 | 0.044 | NA | 0.002 | |||
| U3 7 | 0.018 | 0.022 | 0.006 | 0.002 | NA | |
| U5 20 | 0.089 | 0.066 | 2.99 × 10−5 | |||
| U5 19 | 0.003 | 0.046 | 2.03 × 10−5 | |||
| U5 18 | 0.026 | 0.097 | 0.092 | 1.407 × 10−4 | ||
| U5 17 | 0.071 | 2.38 × 10−7 | ||||
| U5 16 | 1.51 × 10−5 | 2.98 × 10−5 | 0.018 | 0.076 | 0.09 | 0.028 |
Interactions between the U5 and U3 LTR ends most likely reflect restraints imposed on them by their function in the integration reaction. This would be consistent with recent findings of transposase and integrase interactions with their respective substrate DNAs. It was shown that each DNA terminus is specifically bound by the N terminus of one transposase monomer, whereas the terminal nucleotides are bound by the catalytic domain of the other monomer (for review see Ref. 23). In the case of HIV-1 IN, it was shown that the C-terminal domain and the catalytic core domain, both from different IN monomers, bind each end of the viral DNA (16). Thus, the coordination of base selection at the viral DNA terminus and a distal region of the IN recognition sequence at the other end might be due to structural requirements imposed by the correct folding of the full-length IN monomer that binds to the two regions. The interwoven structure of two domains of the same monomer on the two DNA ends brings the two termini together in a protein-DNA complex. Correlation between base selections at proximate positions could be explained by their binding to the same IN domain and structural requirements imposed by this binding. These predictions await a more complete structural analysis of IN-DNA complexes.
Supplementary Material
Acknowledgments
We thank Dr. Ann Skalka, Fox Chase Cancer Center, for purified preparations of HIV-1 IN and reading this manuscript and Ray Reeves, Washington State University, for purified preparations of HMG-I(Y).
Footnotes
This work was supported in part by United States Public Health Service Grants CA38046 and CA52047.
The on-line version of this article (available at http://www.jbc.org) contains Tables I–III and V–VII.
The abbreviations used are: IN, integrase; HIV-1, human immunodeficiency virus, type 1; HMG-I(Y), high mobility group protein I/Y; LTR, long terminal repeat; RSV, Rous sarcoma virus: MOPS, 4-morpholinepropanesulfonic acid; WT, wild type.
References
- 1.Murphy JE, Goff SP. J Virol. 1992;66:5092–5095. doi: 10.1128/jvi.66.8.5092-5095.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wei SO, Mizuuchi K, Craigie R. Proc Natl Acad Sci U S A. 1998;95:10535–10540. doi: 10.1073/pnas.95.18.10535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aiyar A, Hindmarsh P, Skalka AM, Leis J. J Virol. 1996;70:3571–3580. doi: 10.1128/jvi.70.6.3571-3580.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farnet CM, Bushman FD. Cell. 1997;88:483–492. doi: 10.1016/s0092-8674(00)81888-7. [DOI] [PubMed] [Google Scholar]
- 5.Hindmarsh P, Ridky T, Reeves R, Andrake M, Skalka AM, Leis J. J Virol. 1999;73:2994–3003. doi: 10.1128/jvi.73.4.2994-3003.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carteau S, Gorelick RJ, Bushman FD. J Virol. 1999;73:6670–6679. doi: 10.1128/jvi.73.8.6670-6679.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hindmarsh P, Leis J. Microbiol Mol Biol Rev. 1999;63:836–843. doi: 10.1128/mmbr.63.4.836-843.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Esposito D, Craigie R. EMBO J. 1998;17:5832–5843. doi: 10.1093/emboj/17.19.5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou H, Rainey GJ, Wong SK, Coffin JM. J Virol. 2001;75:1359–1370. doi: 10.1128/JVI.75.3.1359-1370.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engelman A, Mizuuchi K, Craigie R. Cell. 1991;67:1211–1221. doi: 10.1016/0092-8674(91)90297-c. [DOI] [PubMed] [Google Scholar]
- 11.Katzman M, Katz RA, Skalka AM, Leis J. J Virol. 1989;63:5319–5327. doi: 10.1128/jvi.63.12.5319-5327.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sherman PA, Dickson ML, Fyfe JA. J Virol. 1992;66:3593–3601. doi: 10.1128/jvi.66.6.3593-3601.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morgan AL, Katzman M. J Gen Virol. 2000;81:839–849. doi: 10.1099/0022-1317-81-3-839. [DOI] [PubMed] [Google Scholar]
- 14.Brin E, Leis J. J Biol Chem. 2002;277:10938–10948. doi: 10.1074/jbc.M108116200. [DOI] [PubMed] [Google Scholar]
- 15.McCord M, Chiu R, Vora AC, Grandgenett DP. Virology. 1999;259:392–401. doi: 10.1006/viro.1999.9782. [DOI] [PubMed] [Google Scholar]
- 16.Gao K, Batler SL, Bushman F. EMBO J. 2001;20:3565–3576. doi: 10.1093/emboj/20.13.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nissen MS, Reeves R. J Biol Chem. 1995;270:4355–4360. doi: 10.1074/jbc.270.9.4355. [DOI] [PubMed] [Google Scholar]
- 18.Yi J, Asante-Appiah E, Skalka AM. Biochemistry. 1999;38:8458–8468. doi: 10.1021/bi982870n. [DOI] [PubMed] [Google Scholar]
- 19.Miller JT, Ge Z, Morris S, Leis J. J Virol. 1997;71:7648–7656. doi: 10.1128/jvi.71.10.7648-7656.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aiyar A, Ge Z, Leis J. J Virol. 1994;68:611–618. doi: 10.1128/jvi.68.2.611-618.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cobrinik D, Aiyar A, Ge Z, Katzman M, Huang H, Leis J. J Virol. 1991;65:3864–3872. doi: 10.1128/jvi.65.7.3864-3872.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muesing MA, Smith DH, Cabradilla CD, Benton CV, Lasky LA, Capon DJ. Nature. 1985;313:450–458. doi: 10.1038/313450a0. [DOI] [PubMed] [Google Scholar]
- 23.Rice PA, Baker TA. Nat Struct Biol. 2001;8:302–307. doi: 10.1038/86166. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.