Abstract
To increase our understanding of imprinted genes in swine, we carried out a comprehensive analysis of this gene family using two complementary approaches: expression and phenotypic profiling of parthenogenetic fetuses, and analysis of imprinting by pyrosequencing. The parthenote placenta and fetus were smaller than those of controls but had no obvious morphological differences at Day 28 of gestation. By Day 30, however, the parthenote placentas had decreased chorioallantoic folding, decreased chorionic ruggae, and reduction of fetal-maternal interface surface in comparison with stage-matched control fetuses. Using Affymetrix Porcine GeneChip microarrays and/or semiquantitative PCR, brain, fibroblast, liver, and placenta of Day 30 fetuses were profiled, and 25 imprinted genes were identified as differentially expressed in at least one of the four tissue types: AMPD3, CDKN1C, COPG2, DHCR7, DIRAS3, IGF2 (isoform specific), IGF2AS, IGF2R, MEG3, MEST, NAP1L5, NDN, NNAT, OSBPL1A, PEG3, APEG3, PEG10, PLAGL1, PON2, PPP1R9A, SGCE, SLC38A4, SNORD107, SNRPN, and TFPI2. For DIRAS3, PLAGL1, SGCE, and SLC38A4, tissue-specific differences were detected. In addition, we examined the imprinting status of candidate genes by quantitative allelic pyrosequencing. Samples were collected from Day 30 pregnancies generated from reciprocal crosses of Meishan and White Composite breeds, and single-nucleotide polymorphisms were identified in candidate genes. Imprinting was confirmed for DIRAS3, DLK1, H19, IGF2AS, NNAT, MEST, PEG10, PHLDA2, PLAGL1, SGCE, and SNORD107. We also found no evidence of imprinting in ASB4, ASCL2, CD81, COMMD1, DCN, DLX5, and H13. Combined, these results represent the most comprehensive survey of imprinted genes in swine to date.
Keywords: assisted reproductive technology, comparative genomic imprinting, epigenetics, gene regulation, genomic imprinting, parthenogenesis, placenta, swine parthenote
Expression profiling of porcine parthenote and biparental fetuses combined with pyrosequencing-based allelic quantification using reciprocal crosses confirms the imprinting status of 36 genes in several fetal tissues at Day 30 of gestation.
INTRODUCTION
In contrast to the majority of genes, where expression is from both alleles, genomic imprinting leads to parent-specific monoallelic expression from either the paternal or maternal chromosome. Currently, there are more than 100 known imprinted genes and, with some notable exceptions [1–3] (Catalogue of Imprinted Genes; http://igc.otago.ac.nz/home.html), their imprinting status is conserved between humans and mice. On a molecular level, imprinted genes are controlled by an epigenetic “stamp” of chromatin markings, including DNA methylation or repressive histone modifications to silence one parental allele, thus resulting in monoallelic expression [4]. The parental conflict or kinship theory proposed by Moore and Haig [5] suggests that in mammals, paternally expressed imprinted genes act on the placenta to promote extraction of resources from the mother to enhance offspring development and fitness, whereas maternally expressed imprinted genes act to restrict fetal growth to conserve maternal resources for long-term reproductive fitness of the mother. Imprinted genes can also act directly on the fetus by influencing cellular proliferation or apoptosis and can also affect fetal growth by influencing the flux of maternal nutrients through the placenta. Recent evidence also suggests a role for imprinted genes in cognitive behaviors, because gene inactivation studies of paternally expressed Peg3 demonstrated a deficiency in maternal care in mice, with females inheriting a null allele from their fathers having impaired milk ejection and inability to rear pups [6]. Work from several laboratories has also shown that incomplete epigenetic reprogramming of animals cloned by somatic cell nuclear transfer leads to aberrant expression of imprinted genes and may contribute to placentomegaly [7, 8]. Our earlier work documented phenotypic variation in cloned livestock [9], with evidence suggesting incomplete epigenetic reprogramming of imprinted genes as one culprit of the phenotypic variation.
To increase our understanding of the role of imprinted genes in porcine reproductive biology and to understand how different mammalian species are regulated by imprinting [10], it is important that a comprehensive analysis of imprinted genes be carried out in swine. Although there have been several reports of imprinted genes in swine [11–18], there is still a considerable amount of information missing. Additionally, the potential role for imprinted dysregulation in placental function is lacking. The feasibility of genome-wide detection of epigenetic asymmetry has been demonstrated previously by using uniparental models (parthenotes [PRTs] and androgenotes) [19–22]. This model is driven by the hypothesis that expression patterns of imprinted genes will differ between PRTs, with two sets of maternal chromosomes and no paternal chromosomes, and biparental (BP) embryos, with one set of maternal and one set of paternal chromosomes. In spite of some recognized weaknesses [23, 24], the parthenogenetic model has been very useful for exploration of genomic imprinting because it can identify known imprinted genes as well as previously unreported imprinted genes [10, 25, 26]. In the present study, we define imprinting as an allelic expression pattern that differs from the expected 50:50 and that maintains a parent-of-origin effect. To confirm imprinting, reciprocal crosses between two breeds of pigs (White Composite [27] and Meishan) were used to clarify the parent-of-origin effects, and quantitative allelic pyrosequencing (QUASEP) was used to quantitate allelic imbalances, followed by a statistical test to determine significance. In cases where we were unable to identify an informative polymorphism, we assigned provisional imprinting status I(PD) based on differential expression between uniparental and BP samples essentially as described by others [10, 25, 26], with the exception that a stringent statistical analysis of the data was added. Although recent studies have identified a large number of genes that are expressed from only one allele (monoallelic) [28, 29], these genes are not expressed in a parent-of-origin nature. In addition to describing for the first time placental defects associated with parthenogenesis in swine, the work described here is the most comprehensive analysis of imprinted genes in swine to date and forms the basis for future studies to elucidate their functional significance in many aspects of reproductive biology, including fetal and placental growth and development, as well as fecundity [30].
MATERIALS AND METHODS
Generation of Swine PRT and BP Fetuses
To create a diploid embryo containing only maternally derived chromosomes, in vitro-matured sow oocytes of occidental origin (Landrace × Yorkshire) were obtained from a local abattoir and were activated by a single DC pulse of 50 V/mm for 100 μsec, and extrusion of the second polar body was inhibited by culture for 6 h in 10 μg/ml cycloheximide [31]. Diploidization was assessed by karyotyping our individual parthenogenetic fibroblast cell lines. One to two hours after removal from cycloheximide, a midventral laparotomy was performed on a synchronized recipient at the first day of standing estrus, and 25–30 PRTs were transferred into the oviduct. Biparental embryos were produced by natural matings from occidental crossbreed of Yorkshire × Landrace × Duroc animals from the Swine Educational Unit at North Carolina State University. Fetal tissues were collected at Day 28 or 30, fetal and placental weights were recorded, and porcine fibroblast lines were established as described previously [31].
Analysis of Morphology and Histology Between Stage-Matched BP and PRT Fetuses
For histological analysis, at least three independent placentas were analyzed from Day 28 and Day 30 PRT placentas and age-matched BP controls. Sections were stained with hematoxylin and eosin and analyzed for vascularity and overall placental/uterine morphology. Three or more representative histological sections prepared from BP or PRT placentas were used 1) to count maternal blood capillaries or 2) to count chorionic protrusion/ruggae, and 3) to measure chorionic surface area. Briefly, photographs of identical dimensions of the representative histological slides were used to outline the nonlinear path of the maternal-fetal interface and the total length corresponding to the fetal-maternal interface length calculated. At least three independent measures per sample type were taken. An unpaired one-tailed Student t-test was used to determine significance of maternal capillaries, chorionic ruggae, or surface area between control and PRT fetuses.
Generation of Reciprocal Crosses and Sample Collection
Reciprocal crosses were generated from natural matings of White Composite (WC) [27] with purebred Meishans (MS) ages 6 mo or older. The WC represents a four-breed composite population of Yorkshire, Landrace, Large White, and Chester White breeds, and a full description of the WC is provided in Cassady et al. [27]. Pregnant gilts from each cross were killed on the morning of Gestational Day 30, and fetuses and their placentae were collected from each uterine horn. Fetuses were dissected, and brain, carcass, liver, and placenta were placed in cryovials, snap frozen in liquid nitrogen, and stored at −80°C until they could be processed further.
All experimental procedures involving animals were approved by the U.S. Meat Animal Research Center Animal Care Committee and/or by the Institutional Animal Care and Use Committee at North Carolina State University.
Identification of Transcript Single-Nucleotide Polymorphisms
Three approaches were taken to identify transcript single-nucleotide polymorphisms (tSNPs) in order to query the parent-of-origin status of the imprinted allele: namely, 1) an SNP resource panel previously established by our group [32] of crossbred White Composite (WC) [27], purebred Meishans, and their hybrid fetuses (total of eight animals) was used to scan for tSNPs in genes of interest and to identify the paternal and maternal allele via direct sequencing of genomic DNA; 2) multiple-sequence alignments of swine genomic traces from the National Center for Biotechnology Information repository [33]; and 3) examination of microarray data from a placental expression study to identify single-feature polymorphisms via modeling probe-by-breed differences [34].
RNA Processing for Microarray and Pyrosequencing Studies
Total RNA was extracted from each fetal tissue using the RNAqueous Kit (Ambion/Applied Biosystems, Austin, TX) following the instructions of the manufacturer, with minor modifications. Briefly, 200–300 mg of placental tissue was pulverized in a mortar and pestle under liquid nitrogen, resuspended in lysis buffer at a ratio of 600 μl of buffer per 50 mg of tissue, vortexed vigorously for 60–90 sec, and precleared by centrifugation for 5 min at 3000 × g. To remove bulk genomic contamination, the precleared supernatant was applied to an Agilent mini-prefilter column and centrifuged at 10 000 × g for 1 min. The filtrate was precipitated with an equal volume of 64% ethanol, and the RNAqueous protocol was continued according to the manual. Total RNA was quantitated by the NanoDrop ND-1000 spectrophotometer, and integrity was assessed by loading 2 μg per well for denaturing agarose gel electrophoresis (NanoDrop Technologies, Wilmington, DE). Total RNA was then stored at −80°C until it could be processed further.
Microarray Experimental Design and Statistical Analysis
Three control (Landrace × Yorkshire × Duroc crossbred; BP) and three diploid PRT (Landrace × Yorkshire crossbred) pregnancies were generated for collection of organs and fibroblast cell lines. For whole organs (brain, liver, placenta), tissues from one pregnancy were pooled and used as a biological replicate, for a total of three BP and three PRT replicates per tissue. For the fibroblast cell lines, each biological replicate consisted of fibroblasts derived from a randomly selected fetus and cultured for two passages. A total of 24 short oligonucleotide microarray arrays (two sources [BP and PRT] times four tissues times three replicates) were hybridized as described previously (Affymetrix Porcine GeneChip; Affymetrix, Santa Clara, CA) [31]. The microarray processing was performed by a commercial service provider (Expression Analysis, Durham, NC), in accordance with methods specified by the manufacturer for target preparation and hybridization. Before target production, the quality and quantity of each RNA sample being used for hybridization were assessed using a 2100 BioAnalyzer and an RNA 6000 Nano LabChip Kit (Agilent Technologies, Palo Alto, CA). Microarray data quality was assessed by examining array-to-array correlations as described by us in Tsai et al. [31] and others [35], as well as by principal component analysis [36].
Expression of all candidate genes was analyzed using MAS 5.0 presence/absence algorithm [37] with a conservative P < 0.001 as a criterion of expression. Expressed genes were identified and analyzed for differences in expression between BP and PRTs using a gene-by-gene linear mixed model. All tissues were analyzed independently. Briefly, log2-transformed perfect-match intensities for all probes on the array were normalized by fitting to a linear mixed model with fixed effects for treatment and probe and a random effect for array using SAS and JMP/Genomics (SAS, Cary, NC). The residuals from this first model were then fit to a gene-specific linear mixed model with fixed effects for treatment and probe and a random array effect. Least-square means were estimated for the differences in treatment effect. In the case of IGF2, probe sets were analyzed exon by exon using the linear mixed model with the same fixed (probe and treatment) and a random (array) effect. The full microarray dataset used in this study is publicly available at Gene Expression Omnibus [38] under accession number GSE10443 and meets requirements established by Minimum Information About a Microarray Experiment consortium [39].
To represent the microarray data, two approaches were used. First, the ratios of expression of the BP sample compared with the PRT samples were used. The expectations are that genes expressed from both alleles will have a ratio of 1 (both maternal and paternal chromosomes expressed equally so BP and PRT are equivalent diploids); genes that are expressed only from the paternal chromosome will have a ratio greater than 1, and the PRT sample should have nondetectable expression (present in the BP with both maternal and paternal chromosomes but absent in the PRT with only maternal chromosomes); genes expressed from the maternal chromosome will have a ratio of less than 1, but there will be expression from both the BP and PRT samples (one copy of maternal chromosome in the BP and two copies in the PRT). Comparison of this ratio allows for quick examination of a large number of genes with a wide range of expression levels. Second, to facilitate interpretation of cases where apparent activation of alternative isoforms was detected in a tissue-specific manner, probe-by-probe plots for the relevant tissues were used. This allows the direct observation of the changes in the profile of hybridization in each tissue, and it is used to illustrate cases of tissue-specific imprinting. Finally, to validate the microarray data and to examine isoform-specific expression, semiquantitative PCR was used. Primers were designed to the Affymetrix probe target sequences, and identity of amplicons was confirmed by restriction mapping and/or sequencing. All RNA pools were normalized using the 60S ribosomal protein L18 housekeeping gene RPL18 as a reference [40, 41]. For each primer set, samples were amplified separately for various PCR cycle intervals (15, 18, 20, 22, 25, 28, 32, 35, etc., and up to 45 cycles if needed), and products were separated in agarose gel electrophoresis, stained with ethidium bromide, and imaged for further analysis. It should be noted each PCR represents an optimized cycle number for each individual gene. Band intensities between figures may differ to keep within the linear amplification range in all semiquantitative RT-PCR gels. Images of RT-PCR cycles showing linear amplification were analyzed using the National Institutes of Health Image densitometric software ImageJ1.5.13 (http://rsbweb.nih.gov/ij/index.html) to determine the intensity of each PCR band, and a ratio of BP:PRT normalized to RPL18 was calculated. Two independent cDNA pools were used to produce the RT-PCR data: the original cDNA pool that was used to generate the microarray data, and an additional pool that was prepared from independent BP and PRT samples. This was done to both confirm that the results seen applied to independent samples and to be able to carry additional RT-PCRs that could not have been completed using the initial cDNA pool because of sample availability limitations.
QUASEP Experiments
Recently, new sequencing technologies have become available to quantitate allele-specific expression patterns. In this study, QUASEP [42] was used to quantify transcript abundance from maternal or paternal alleles to evaluate parent-of-origin effects to confirm the microarray data and extend our observations to putative imprinted genes not represented in the microarrays. Briefly, pyrosequencing of an informative polymorphism will provide a 50:50 ratio of bioluminescence intensity, which is indicative of heterozygous genomic DNA. In the case of an imprinted gene, where one allele is silenced, the ratio of bioluminescence intensity will bias one allele, in some cases completely to a ratio of 100:0. The combination of reciprocal crosses (where a 100:0 allelic ratio in one cross shifts to 0:100 in the reciprocal cross) and quantitative allelic sequencing of an informative polymorphism provide unambiguous interpretation of imprinting status, and thus serve as a rigorous quantitative method to clarify epigenetic asymmetry.
For pyrosequencing analysis, RNA was isolated as described above, and contaminating genomic DNA in the total RNA extractions was removed with 6 units of hypermorphic DNAse I (TURBO DNase; Applied Biosystems/Ambion) per 30 μg of nucleic acid. The first-strand cDNA was synthesized in a final volume of 20 μl from the following reagents: 1) 5 μg of total RNA using 2) a ∼15-μM random pentadecamer priming method [43] and 3) a thermostable, RNase H-negative Moloney murine leukemia virus-reverse transcriptase (SuperScript VILO cDNA Synthesis Kit; Applied Biosystems/Invitrogen, Foster City, CA), with 4) 4 ng/μl thermostable single-stranded DNA-binding protein (ET-SSB; Biohelix, Beverly, MA) and 5) an optimized RNase inhibitor (SUPERase In; Applied Biosystems/Ambion). Reaction conditions were 65°C, 5 min, denaturation; 50°C, 60 min and 55°C, 90 min, reverse transcription; and 85°C, 5 min, heat inactivation. First-strand cDNA product was diluted to 33 ng/μl with nuclease-free H2O. An RT-negative control was used to check for genomic DNA contamination by omission of reverse transcriptase, and these controls were negative on PCR amplification. In addition, all cDNAs were further tested for genomic DNA contamination by using primers that amplify across introns, and only those found to be free of contamination were used for further analysis.
The universal biotinylated primer approach developed by Aydin et al. [44] was used to generate PCR products as input for QUASEP [44]. QUASEP was performed using PCR products from genomic DNA (genotyping) and compared to cDNA (allelic quantitation) from animals of interbreed reciprocal crosses. The PCR conditions to generate biotinylated products for input into QUASEP are summarized below. Polymerase chain reaction was performed with 25-μl reactions containing a final volume/concentration of the following components: 3 μl of reverse transcription product (100 ng of cDNA template), 50 nM each gene-specific primers synthesized with the 5′ universal overhangs, 250 nM each universal primers (with a terminal 5′-biotinylated moiety on one primer based on directionality of PSQ assay design), 3.5 mM MgCl2, and 20.5 μl of 1.1× PCR master mix (Platinum Blue PCR SuperMix; Applied Biosystems/Invitrogen). The PCR thermocycling conditions were: 95°C, 15 sec, denaturation; 60°C, 15 sec, annealing; 72°C, 30 sec, extension (n = 8 cycles), followed by 95°C, 15 sec, denaturation; 58°C, 15 sec, annealing; 72°C, 30 sec, extension (n = 40 cycles), and a final extension at 72°C for 30 sec. Short amplicons (75–200 bp) were designed using the exemplar sequences available from Affymetrix NetAFFX. All primers used for these assays to amplify specific probes were designed using Primer3, BatchPrimer3, or MPrime and were chemically synthesized with only desalting purification (IDT, Coralville, IA; a full list of primers is available in supplemental Table S1, available at www.biolreprod.org). Biotinylated products were visualized to check for quantity and quality of PCR amplification on 8% PAGE or 2% agarose gel electrophoresis prior to pyrosequencing.
Procedures for preparing single-stranded PCR product for QUASEP are summarized below. Streptavidin-coated Sepharose beads (200 μg; GE Biosciences, Piscataway, NJ) were prewashed twice in washing buffer (Qiagen/Biotage, Valencia, CA) using a vacuum pump manifold and were resuspended in 45 μl of binding buffer (Qiagen/Biotage). An equivalent volume (25–45 μl) of biotinylated PCR product was agitated with resuspended beads for efficient immobilization (15 min, 25°C), then washed briefly in 70% ethanol (10 sec, 25°C). To remove the nonbiotinylated DNA strand, immobilized duplexed DNA was melted using a NaOH-containing denaturation buffer (5 sec, 25°C; Qiagen/Biotage) and were subsequently washed once in wash buffer (10 sec, 25°C; Qiagen/Biotage). Sequencing primer was added in 40 μl of annealing buffer at a final concentration of 0.4 μM and was hybridized to template by incubation (95°C for 2 min) in a heat block and being allowed to slowly cool to ambient temperature. The QUASEP reaction was performed using the automated PSQ 96MA machine, which uses a disposable cartridge (PSQ 96 Reagent Cartridge; Qiagen/Biotage) to deliver enzymes, substrates, and each of the four nucleotides, per manufacturer's protocol (PSQ 96 SNP Reagent Kit 5x96; Qiagen/Biotage). Data acquisition of bioluminescence was captured and transcribed into spectra (pyrograms) for analysis. To accurately measure allelic frequency and correct for experimental bias from unequal PCR amplification, genomic DNA allelic frequencies were compared to cDNA allelic frequencies.
To statistically analyze the QUASEP data, we combined the results from all informative fetuses (and reciprocal crosses, if available) and compared the genomic versus the cDNA allelic ratios for each gene by heteroscedastic two-tailed t-test to determine whether the cDNA allelic frequencies differed from the genomic frequencies. Significance was set at P < 0.05.
RESULTS
Phenotypic and Gene Profiling Characteristics of Day 30 BP and PRT Fetuses
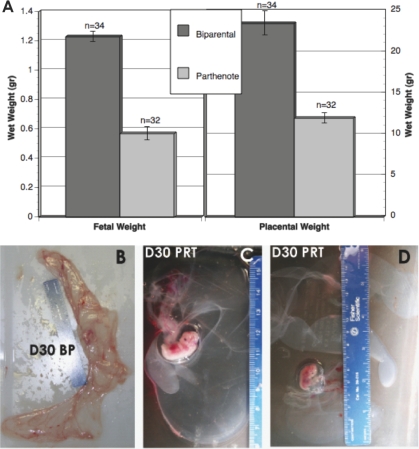
Of 352 PRT embryos transferred into seven recipients, four became pregnant. From the four pregnancies, we were able to collect 52 viable fetuses at Days 28–30 of gestation (15%; based on fetal heart beats), and fetal and placental weights were collected from 32 fetuses. The additional fetuses were used for experiments unrelated to the present study. Fetal and placental weights were compared between PRT and BP fetuses and, as predicted by the parental conflict hypothesis, both were significantly reduced in the PRT fetuses (Fig. 1). Histological analysis of PRT and control placentas at Day 28 revealed no significant differences between these placentae. By Day 30, however, there were placental differences, with the PRT samples having a reduction of branched structures or interdigitation, reduced number of chorionic protrusions or simple villus (P < 0.0136), and reduced chorionic surface area (P < 0.0175; Fig. 1). In addition, maternal-fetal crosstalk seemed to be impaired, because uterine epithelium showed a trend (P < 0.129) toward reduction of the total number of maternal blood vessels at Day 30.
FIG. 1.
A) Fetal and placental weight comparisons between BP and PRT concepti at Day 30 of gestation. Bars indicate mean ± SEM; n indicates the number of observations. B–D) Gross morphology of placentae from Day 30 (D30) naturally mated (B) and PRT (C and D) gestations. Reduced vasculogenesis and angiogenesis are readily apparent in the swine PRT, compared with stage-matched BP controls. The PRT placental tissue often appears translucent, possibly resulting from ischemia, as would be expected from decreased oxygen transport. Ruler is in centimeters.
Additionally, the prediction that expression profiling of uniparental pregnancies could be used to identify conserved imprinted genes was evaluated. To reduce the dimensionality of the transcriptome data into clusters of similar arrays, and as a quality control to determine the quality of the hybridization and identify arrays that did not meet required quality controls, we performed a principal component analysis to clarify which of the four tissue-specific arrays clustered together (Supplemental Fig. S1 illustrates the data after mixed-model normalization [45] and principal components analysis). The first three principal components were used because they explained 86%, 5%, and 5% of the total variation, respectively. Two arrays (LG2 and BG3; liver PRT 2 and brain PRT 3, respectively) fell outside the 95% concentration ellipse and were excluded from downstream analysis (Supplemental Fig. S1).
Because our experimental focus was the study of conservation of the imprinted gene family, we extracted from the microarray data informative probes that detected known or putative imprinted genes (Catalogue of Imprinted Genes). Of the 49 genes analyzed in this manner, eight were identified as not expressed at P < 0.001 in any tissue tested; they included CALCR, DIO3, GABRA5, HTR2A, INS, OSBPL5, SLC22A2, and WT1.
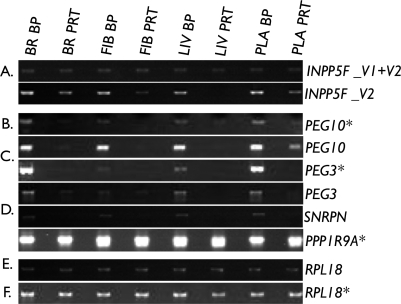
To examine in more detail the remaining expressed genes, we mapped each Affymetrix probe sequence to the known porcine transcript, or in its absence to the human transcript, and examined each gene individually. This identified the probe sets for GNAS, INPP5F, KCNQ1, and PPP1R9A as noninformative because of their inability to discriminate known imprinted and nonimprinted isoforms. To clarify the expression status of these genes, we attempted to design isoform-specific RT-PCR. Unfortunately, we were unable to do so for GNAS or KCNQ1. However, a semiquantitative RT-PCR assay for INPP5F variant 2 (INPP5F_V2) and PPP1R9A were successfully designed. Results shown in Figure 2A indicate that INNP5F_V2 is preferentially expressed in carcass and liver BP tissues but not in brain and placental samples. Similarly, for PPP1R9A, results from the semiquantitative RT-PCR indicated that expressions from BP and PRT samples were similar in brain, fibroblasts, and liver (PRT:BP ratios not different from 1). In contrast, in the placental sample, expression from the PRT sample was higher than the BP sample, with a PRT:BP ratio of 1.7 (Fig. 2E).
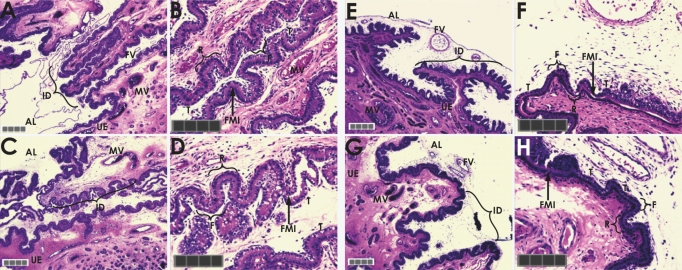
FIG. 2.
Fine structure of fetal-maternal interface from stage-matched, naturally mated controls (Day 28 [A and B] and Day 30 [E and F]) and swine PRTs (Day 28 [C and D] and Day 30 [G and H]) stained with hematoxylin and eosin. Defects become apparent in Day 30 PRT placental tissue and include the number, extent, and reduced complexity of chorionallantoic folding or interdigitation and reduced ruggae. AL, allantois; FV, fetal vessels; T, chorionic trophoblasts; FMI, fetal-maternal interface; UE, uterine epithelium; MV, maternal vessels; R, top of fetal ridge or ruggae; F, base of fetal trough or fossae. Original magnifications ×10 (A, C, E, and G) and ×20 (B, D, F, and H). Gray bar = 200 μm; each square = 50 μm.
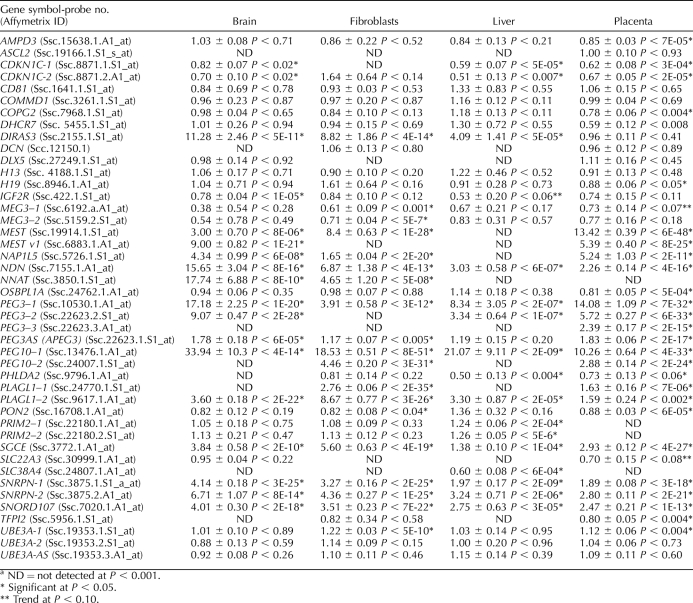
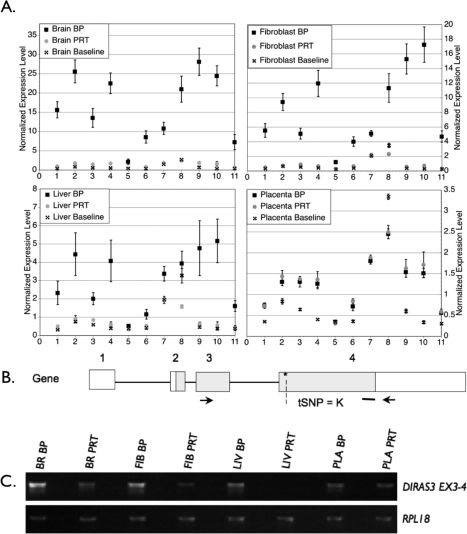
For the remaining genes, we used the Affymetrix array data to determine the ratio of expression of the BP tissues to the PRT tissues and determined whether the ratios differed from 1, an indication of a shift from biallelic expression. As shown in Table 1, DIRAS3, MEST, NNAT, NAP1L5, NDN, PEG3, APEG3, PEG10, PLAGL1, PRIM2A, SGCE, and SNRPN had ratios greater than 1, indicating greater expression from the BP samples, a pattern expected of paternally expressed genes. For MEST, NNAT, NAP1L5, NDN, PEG3, APEG3, PEG10, and SNRPN, increased expression from the BP sample and lack of PRT expression were detected in all samples where the genes were expressed. Results of the semiquantitative RT-PCR for PEG3, PEG10, and SNRPN are shown in Figure 2 and confirm the microarray data. Biparental:PRT expression ratios for each of these three genes ranged from a low of 2.7 (PEG10, liver sample) to a high of 150 (PEG3, brain sample), but in all cases there were clear differences between BP and PRT samples. For PRIM2A, significant expression differences were observed in the paternal direction (greater than 1). However, the microarray data indicated transcript expression in PRTs, and hence do not support complete silencing of the maternal allele. For DIRAS3, PLAGL1, and SGCE, tissue-specific differences were observed. DIRAS3 (ARHI) expression between BP and PRT was significantly different in brain, fibroblasts, and liver but not in the placenta (Table 1). In the placenta, there was significant expression from the PRT sample, something not seen in any of the other tissues. Results in Figure 3A show the detection of a transcript in the placental PRT sample but not in the other tissues. To confirm this expression pattern, an RT-PCR was performed. Results confirmed that the placenta had similar expression levels from the BP and PRT samples (BP:PRT = 1.3), whereas the other tissues showed greater differences between BP and PRT expression (BP:PRT ratios of 3.6, 5.6, and 48.2 for brain, fibroblast, and liver, respectively; Fig. 4).
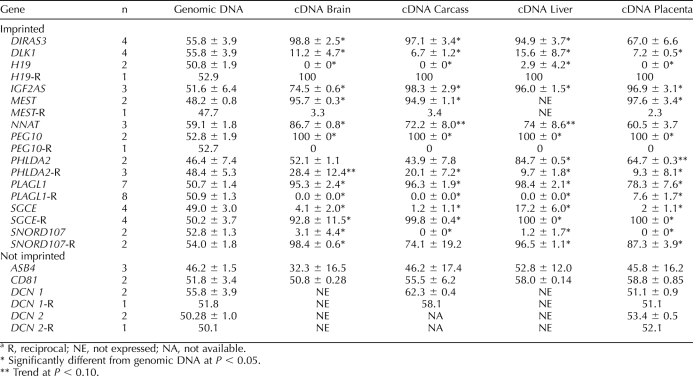
TABLE 1.
Gene expression in biparental (BP) and parthenogenetic (PRT) fetal tissues expressed as BP/PRT ratios (mean ± SE).a
FIG. 3.
Semiquantitative PCR analysis of candidate imprinted genes. Samples were analyzed as described in the text. A) Expression of variant 1 and/or variant 2 for INPP5F. INNP5F_V2 has increased expression in BP compared with PRT samples in all tissues tested. B–D) Combined results for PEG10, PEG3, and SNRPN showing expression from the BP samples but lack of or highly reduced expression from the PRT sample, a pattern supportive of paternal expression. Additionally, the PCR results confirm the microarray data. E) Expression of PPP1R9A showing greater expression in the PRT than the BP placental sample but similar BP and PRT expressions in all other tissues. F) RPL18 was used as the control baseline transcript. Two independent RNA pools were used to generate these results. BR, brain; FIB, fibroblast; LIV, liver; PLA, placenta. *Both cDNA pools behaved similarly.
FIG. 4.
Tissue-specific differences in BP and PRT fetal tissues for DIRAS3. A) A comparison of expression of BP and PRT samples using a probe-by-probe analysis. This allows for the identification of tissue-specific differences. Each Affymetrix probe set contains 11 oligonucleotides with sequence homology to different regions of the gene. These plots show how each probe in the set behaves. The expression value is normalized, taking into account all of the genes in the array, and allows for comparisons of relative expression levels between genes and between tissues. The baseline value refers to background hybridization for each probe. This is calculated using the mismatched hybridization values. As expected of paternally expressed genes, expression was seen in the BP but not the PRT tissues, with the exception of the placenta. In the placenta, expression levels were low relative to other tissues but, in addition, the BP and the PRT samples had similar levels of expression. This supports expression of a different isoform in the placenta or lack of expression of the imprinted isoform. B) Diagram of the DIRAS3 gene and isoform that have been detected in swine and/or humans. Locations of PCR primers are designated by arrows. The heavy bar indicates where array probes bind, and an asterisk/dashed line indicates tSNP for QUASEP. C) The PCR results confirming the exon-by-exon microarray analysis, a pattern supportive of paternal expression. BR, brain; FIB, fibroblast; LIV, liver; PLA, placenta; Iso, mRNA isoform.
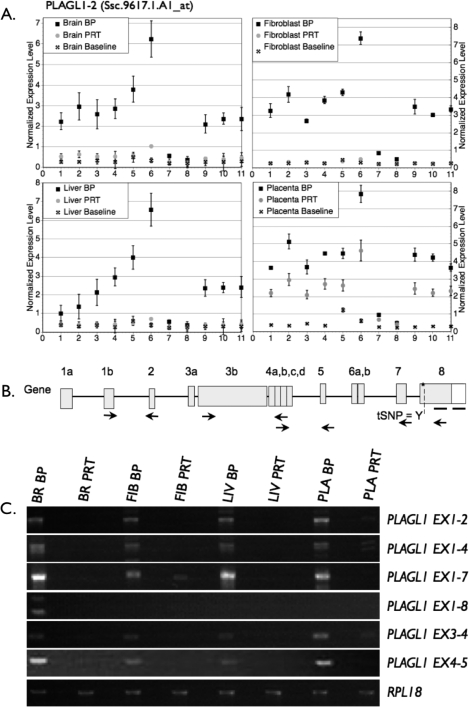
Figure 5A shows the presence of PLAGL1 expression in PRT placental tissues; however, no expression was detected in PRT brain, fibroblast, or liver tissues. In comparison, the expression level was still significantly higher in the BP placenta than the PRT sample, which suggests either coexpression of an imprinted and nonimprinted isoform or partial relaxation of imprinting. A series of RT-PCRs amplifying different exons was developed, and results supported a complex pattern of tissue- and isoform-specific imprinting, with PLAGL1 exon 1–2, exon 1–4, and exon 3–4 showing slight expression from the PRT placenta but not other PRT tissues (Fig. 5). In contrast, PLAGL1 exon 1–7 had partial maternal expression in fibroblasts but lack of PRT expression in other tissues. PLAGL1 exon 1–8 was only detected in the BP brain.
FIG. 5.
Analysis of expression at the PLAGL1 locus by isoform-specific semiquantitative PCR. A) PLAGL1 expression in two representative samples. Although pattern of expression in brain shows lack of expression in the PRT sample, in the placental sample there is significant PRT expression. As for DIRAS3, this supports the presence of a second isoform in the placenta compared with the other tissues. B) Diagram of the PLAGL1 gene and isoforms that have been detected in swine and/or humans. Locations of PCR primers are designated by arrows. The heavy bar indicates where array probes bind, and an asterisk/dashed line indicates tSNP for QUASEP. P1 and P2 represent alternative exon 1 expressed from promoter 1 or promoter 2 isoforms as identified in human studies. More than 50 PLAGL1 isoforms have been annotated by human/mouse expressed sequence tag databanks. The two depicted isoforms are representative of imprinted and nonimprinted isoforms as identified in human studies. C) The PCR results confirming the exon-by-exon microarray analysis, a pattern supportive of paternal expression. BR, brain; FIB, fibroblast; LIV, liver; PLA, placenta; Iso, mRNA isoform.
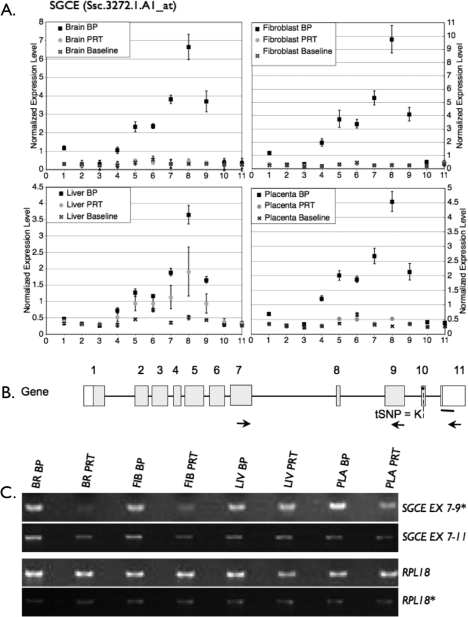
SGCE also had a tissue-specific pattern, but in this case it was the liver that showed expression from the PRT genome when compared to the other three tissues (Fig. 3C). An RT-PCR amplifying exon 7–9 and exon 7–11 confirmed expression from the PRT liver seen in the probe-by-probe analysis, but in addition it indicated SGCE exon 7–9 is also expressed, albeit at a low level compared with the BP, in all PRT samples (Fig. 6). However, BP:PRT ratios were lower in the liver and placenta (0.9 and 1.8, respectively) than in brain and fibroblast (7.8 and 4.0, respectively). The RT-PCR for exon 7–11 had a similar pattern, with respective BP:PRT ratios for liver and placenta of 0.9 and 1.0 versus 3.0 and 4.0 for brain and fibroblasts.
FIG. 6.
Analysis of expression at the SGCE locus by isoform-specific semiquantitative PCR. A) Expression of SGCE in BP and PRT tissues. Although there was expression of SGCE in the PRT liver, no PRT expression could be detected in any other tissue, including the brain. B) Schematic of SGCE gene and transcript isoforms that have been detected in swine and/or humans. Locations of PCR primers are designated by arrows. The heavy bar indicates where array probes bind, and an asterisk/dashed line indicates tSNP for QUASEP. C) The PCR results confirming the exon-by-exon microarray analysis, a pattern supportive of paternal expression. The asterisk is used to depict cDNA pools as described in Figure 2. BR, brain; FIB, fibroblast; LIV, liver; PLA, placenta; Iso, mRNA isoform.
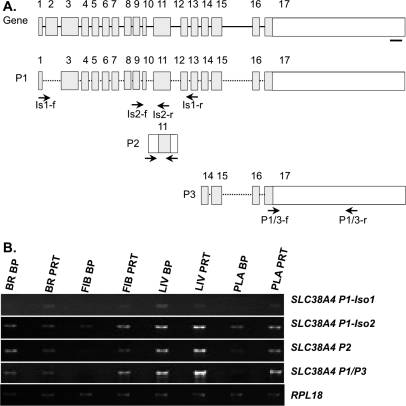
In addition, AMPD3, CDKN1C, COPG2, DHCR7, H19, IGF2R, MEG3, OSBPL1A, PHLDA2, PON2, SLC38A4, and TFPI2 had ratios lower than 1 in at least one tissue type examined, indicating greater expression from the PRT than the BP samples, a pattern expected of maternally expressed genes (Fig. 7). In the case of SLC38A4, the array was capable of detecting expression in liver with a higher level of expression in the PRT than the BP sample. In humans, transcription of SLC38A4 produces eight different mRNAs, six alternatively spliced variants, and two unspliced isoforms. From three alternative SLC38A4 [46], we designed a series of RT-PCRs for different regions of the gene and, as shown in Figure 7B, a complex pattern of expression was seen. For the P1-Iso1 transcript, expression was greater in the PRT than the BP sample in all tissues except the liver, where the opposite was true. In contrast, for P1-Iso2, P2, and P1+P3, ratios of BP:PRT were lower than 1 (range, 0.8–0.03) in all tissues except the brain. In the brain, ratios were 1.4, 2.3, and 1.5 for P1-Iso2, P2, and P1+P3, respectively. For SLC22A3, there was a trend (Table 1) toward overexpression in the PRT placenta, which is suggestive of maternal imprinted gene expression. H19 had an unexpected result, with only the placenta showing a significant allelic imbalance. Consistent with the pattern of a maternally expressed imprinted gene, H19 showed higher expression in the PRT placental tissue. Unexpectedly, wide variation among replicates constrained the detection of significant maternal expression in other tissues (brain, fibroblast, liver) by microarray expression profiling, as would be predicted by the PRT samples. Fortunately, we were able to test imprinting of H19 by QUASEP and confirmed that H19 was imprinted in all tissues tested (Table 2). ASCL2, CD81, COMMD1, DCN, DLX5, H13, and UBE3A-AS were not differentially expressed between PRT and BP embryos in any tissue analyzed (Table 1).
FIG. 7.
Analysis of expression at the SLC38A4 locus by isoform-specific semiquantitative PCR. A) Cartoon of SLC38A4 gene and mRNA isoforms that have been detected in swine and/or humans. Locations of PCR primers are designated by arrows. The heavy bar indicates where array probes bind. B) The PCR results show patterns supportive of maternal expression for some assays but not others, indicating complex locus control. BR, brain; FIB, fibroblast; LIV, liver; PLA, placenta; Iso, mRNA isoform, P, putative promoters 1, 2, or 3.
TABLE 2.
Quantitative allelic pyrosequencing (QUASEP) analysis of reciprocal Meishan × White composite crosses (mean ± SE).a
Analysis of IGF2
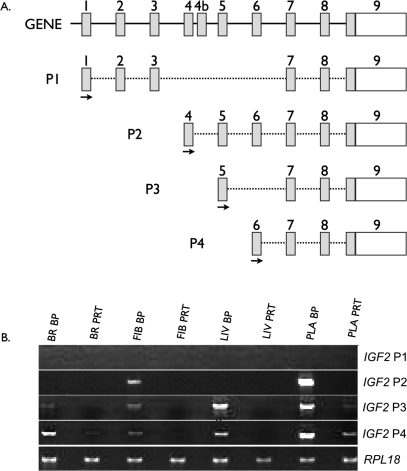
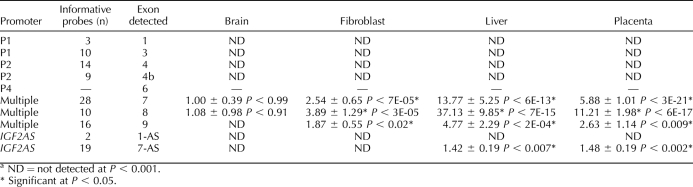
The IGF2 locus is particularly complex because of the presence of several distinct isoforms originating from different promoters, only some of which have been reported to be imprinted [47]. In the arrays used, there were nine independent probe sets capable of detecting different isoforms of IGF2 and two capable of detecting IGF2AS. Each probe set was carefully mapped to the known porcine IGF2 locus to determine which exon each probe set was detecting, and the data were analyzed exon by exon. This allowed us to gather expression information for different exons. As shown in Figure 8A and Table 3, Affymetrix probes targeting transcripts generated from the P1 and P2 promoters were not detectable. In contrast, probes that can detect the P3 and P4 promoters combined showed a high bias toward overexpression in the BP tissues, indicating paternal expression. To identify which of the two promoters, P3 or P4, was active in the different tissues and to confirm lack of expression from P1 and P2 promoters, promoter-specific PCR was used. Results indicated that there was no detectable expression from promoter P1 in any of the tissues tested. To confirm that the primers used could detect P1 transcripts, we isolated cDNA from adult porcine liver, and all primers successfully detected transcripts originating from P1 promoter (data not shown). For the P2 promoter, there was a low level of expression in the BP but not the PRT placenta and fibroblasts (Fig. 8B). The P3 transcript was expressed at high levels in liver and placenta and was barely detectable in brain and fibroblasts (BP:PRT ratios were 13.1, 7.3, 3.3, and 5.3, respectively). The pattern of expression of the P4 transcript was similar to P3 (BP:PRT ratios were 5.6, 2.7, 8.5, and 3.1 for brain, fibroblasts, liver, and placenta, respectively).
FIG. 8.
Analysis of expression at the IGF2 locus by isoform-specific semiquantitative PCR. A) Diagram of the different promoters/isoforms of IGF2 that have been detected in swine and/or humans. Locations of PCR primers are designated by arrows. B) The PCR results confirming the exon-by-exon microarray analysis (Table 3). Both sets of results confirm overexpression of the P2, P3, and P4 isoforms in the BP samples, a pattern supportive of paternal expression. BR, brain; FIB, fibroblast; LIV, liver; PLA, placenta.
TABLE 3.
Expression differences of IGF2 isoforms and IGF2AS between biparental and parthenogenetic fetal tissues.a
Analysis of Imprinting by QUASEP
Although the expression profiling gave an overall view of the conservation of imprinted genes in swine, and it provided a unique set of observations with respect to imprinted gene expression, it was important to both validate the microarray data in a more direct way and to expand the analysis to imprinted genes not represented in the arrays. Thus, we developed hybrid crosses between purebred Meishans and WC (hybrid of Yorkshire, Landrace, Large White, and Chester White breeds) and used a pyrosequencing-based approach to examine monoallelic versus biallelic expression. Using methods described previously, tSNPs were identified in our reference population for all genes described in Figure 9 and Table 2.
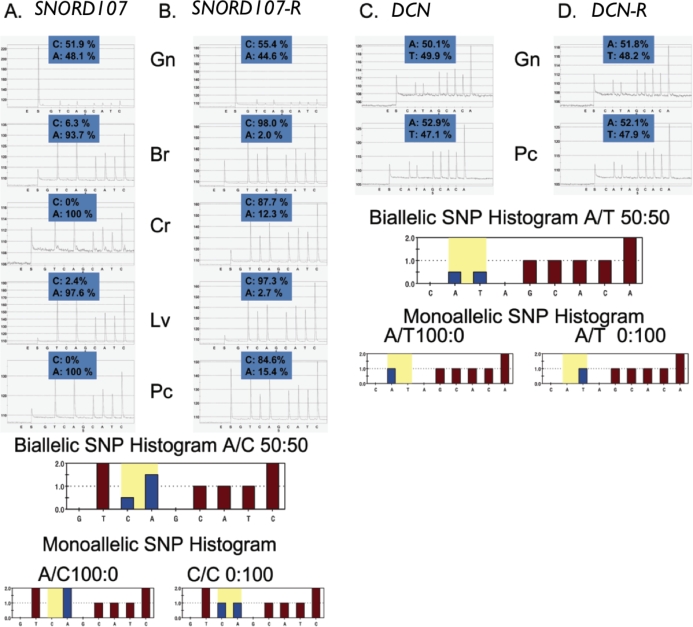
FIG. 9.
QUASEP results of Day 30 fetal samples obtained from reciprocal Meishan × WC matings. A representative set of pyrosequencing results for both (A and B) imprinted (SNORD107) and (C and D) nonimprinted (DCN) genes. Reciprocal matings (R) from WC × Meisan and Meishan × WC for SNORD107 (B) and DCN (D) are included. Allelic quantification of each pyrogram can be calculated on the basis of peak height and the data used to test for bias from the genomic sample. DCN was not expressed on microarrays in brain or liver tissues, and carcass assays were not available. The biallelic and monoallelic histograms represent the ideal expected peak heights that would correspond for each particular tSNP. Neighboring sequence may affect interpretation of expected peak height ratios, as depicted in the SNORD107 pyrograms (i.e., TTcA looks different than TTCA). Additionally, to account for any PCR bias, the genomic and cDNA results are shown for all tissues and each assay. The numbers above the peaks denote the percentage of each allele identified by QUASEP. In all cases, they should add up to 100%. Multiple replications of this QUASEP assay were used to develop the results shown in Table 2. Gn, genomic DNA sample; Br, brain cDNA; Cr, carcass cDNA; Lv, liver cDNA; Pc, placenta cDNA.
The identified tSNPs were analyzed by QUASEP using DNA and cDNA collected from fetal tissues (brain, carcass, liver, and placenta) from both reciprocal interbreed crosses. Each of the 15 interbreed fetuses collected (seven fetuses from WC × MS and eight fetuses from MS × WC) were screened by QUASEP to identify heterozygotes. In general, three to six animals containing the informative polymorphisms were identified from reciprocal matings to clarify the imprinting status for each gene. These informative polymorphisms were identified in both reciprocal crosses, WC × MS and MS × WC, for all genes except ASB4, DLK1, IGF2AS, and NNAT; in these exceptions, tSNPs were identified in only one direction of the litter matings: WC × MS or MS × WC, but not both. A representative set of results is shown in Figure 9 depicting allelic quantification for DNA and cDNA. Analogous pyrograms were developed for each of the genes above and used to generate the results shown in Table 2.
As indicated previously, we define imprinting as a 1) significant allelic imbalance from 50:50 and 2) display of a parent-of-origin effect. In the current study, reciprocal crosses were used to clarify the parent-of-origin effects, and QUASEP was used to quantitate allelic imbalances, followed by a statistical test to determine significance. Although recent studies have identified genes that are expressed monoallelically, these genes are not expressed in a parent-of-origin nature. Taken together, QUASEP identified genes that are imprinted across all tissues tested in a tissue-specific manner or biallelically expressed genes (Table 2). Moreover, reciprocal crosses, if available, confirmed the parent-of-origin effect of the allelic bias as exemplified by SGCE (Fig. 9), with allelic shifts from 100 to 0 versus the expected biallelic 50:50. There were also two cases where the reciprocal crosses differed in the extent of allelic bias. For PHLDA2, the reciprocal crosses differed in the fibroblast and placenta, with one cross having a significant allelic bias and the other not having it. Similarly, for SGCE, the liver tissue showed complete bias in one direction (100% or 0%) but a 17% or 73% in the other direction (Table 2). For ASB4, CD81, and DCN, the allelic bias between genomic DNA and cDNA was not significantly different, indicating that these genes are not imprinted in swine.
DISCUSSION
In spite of the importance of imprinted genes for the development and function of the placenta and the fetus, there is a dearth of knowledge about this gene family in domestic species, including swine. Our goal was to determine the effects of unbalanced imprinting, as represented by the PRT model, on placental and fetal development, and to use gene profiling, the PRT model, and quantitative genotyping tools (QUASEP) to carry out a comprehensive analysis of imprinted genes in swine. It should be noted that some differences in gene expression might occur because of differences in genetic background between swine, epistasis between imprinted and nonimprinted genes, or issues unique to uniparental fetuses independent of imprinting. However, the biological material—both BP fetuses from natural matings and PRTs—was of occidental origin, and multiple biological replicates were used. Additionally, to formally assign imprinting status, candidate genes were analyzed by QUASEP using reciprocal crosses of occidental and Meishan swine fetal tissues (whole brain, carcass, liver, and placentae). Thus, it is highly unlikely that the observed differences are due to genetic effects independent from imprinting.
Phenotypic Placental and Fetal Effects of Unbalanced Imprinting
As shown in Figure 1, both placental and fetal weights were significantly reduced in the PRTs. This supports the parental conflict hypothesis and coincides with results seen in other species [5]. What was somewhat unexpected was the overall normality of the PRT placenta at Day 28, where no significant changes in morphology were noted. Yet, by Day 30, there were changes in chorionic ruggae numbers, maternal-fetal interface surface area, and a trend toward lower vascularization (P = 0.12). These observations suggest that paternally expressed genes, although relevant, are not essential for the initiation of fetal and placental development, but as pregnancy progresses their role becomes more critical. This is supported by the fact that we could not maintain any pregnancies beyond Day 33 of gestation. We are now examining in more detail this transitional Day 30 to Day 33 period to see whether we can identify the factors responsible for the inability of the PRT placenta and/or fetus to survive beyond Day 33. This will be greatly facilitated by the information and resources that we have developed to study imprinted genes in swine as described below.
Identification of Tissue-Specific Imprinting
A series of novel tissue-specific isoforms for DIRAS3, PLAGL1, SLC38A4, and SGCE were identified by expression profiling and/or QUASEP. In addition, others had presented information on tissue-specific imprinting in the IGF2 [47] and PHLDA2 locus [48] in other species, and we confirmed or extended these observations to swine.
DIRAS3 is a known tumor suppressor gene, and small changes in levels of expression could have significant effects on proliferation and differentiation. Recently, it was reported that the porcine DIRAS3 was imprinted in all tissues sampled from five heterozygous 2-mo-old piglets using an occidental and Meishan hybrid line similar to that in our study [11]. Our microarray data support these conclusions of imprinted paternal expression in brain, fibroblast, and liver. In addition, we report an unusual pattern of expression in the placenta, with either expression of a nonimprinted isoform or partial reactivation of the imprinted allele in the PRT samples. Both QUASEP and RT-PCR results confirm these observations and point to a unique mode of regulation of this gene in the placenta (Fig. 3 and Table 1). Although expression levels were low in the placenta (3.5-fold over background) compared with, for instance, brain (35-fold over background; Fig 3), our data convincingly show the presence of placental-specific isoforms in the PRT. Their identification can lead to further studies to clarify their role in porcine placental development and function. At present, there are no reports for any functional role of DIRAS3 in the placenta of any species, yet this unique form of expression regulation suggests an important role for this protein in placental development and function.
PLAGL1 is known to be important for growth regulation, is considered a tumor suppressor gene and, like TP53, can induce cell cycle arrest and apoptosis [49]. Disruption of the PLAGL1 paternal allele in Plagl1+/−pat mice results in intrauterine growth-restricted placentas (IUGRs) but fails to significantly alter placental development and/or function, such as amino acid transport, total placental weight, or extraembryonic morphology [50]. Effects on fetal growth are supported by a report of PLAGL1 being downregulated in human IUGR [51]. In humans, transcription at the PLAGL1 locus produces multiple isoforms [52]. Our data from the microarray (Table 1), confirmed by RT-PCR (Fig. 4), support the existence of multiple isoforms in swine and suggest a complex tissue-specific expression pattern of imprinted and nonimprinted isoforms, a phenomenon that has not been reported previously in any other species. This observation was also supported by QUASEP, which showed a bias in paternal allelic expression of PLAGL1 in the placenta compared with other tissues (Fig. 4 and Table 2), suggesting that different isoforms are expressed in the placenta (Table 2). From a biological perspective, the end result of the presence of nonimprinted isoforms is that there is a double dose of PLAGL1 in the placenta compared with other tissues. This raises several questions: How is the normal imprinted expression overridden? What is the importance of this increased expression in the placenta, and how does it affect fetal growth in the absence of any identifiable placental defect, at least in mice [30]? Moreover, because this is the first report of placental-specific PLAGL1 regulation of imprinting, at this point we cannot determine whether this observation is unique to swine or is also seen in other placental mammals.
SGCE is a component of the sarcoglycan complex and is involved in linking F-actin to the extracellular matrix. Mutations in SGCE are associated with a range of diseases, including myoclonus-dystonia, compulsive disorders, and alcohol dependence, among others. To date, there is no known role for SGCE in placental function other than it is known to be expressed throughout gestation in the human placenta [53]. Our data support imprinting in all tissues tested, consistent with previous observations in mice [26]. In addition, we identified a unique expression pattern in the liver supportive of expression from the usually silent maternal allele (Fig. 5). A similar observation of weak maternal expression had been reported previously for the mouse brain but not the liver [54]. Although there are no known published reports of porcine SGCE isoforms, nine possible isoforms have been predicted by genome annotation in the mouse, and four in humans [46]. Recently, it has been reported that SGCE is upregulated in human hepatocellular carcinoma [55], suggesting that SGCE plays a role in hepatocyte proliferation. Thus, it is plausible that maternal expression of the usually silent allele, leading to a relative increase in SGCE levels, is a compensatory mechanism present at a developmental time of very rapid liver growth. It will be interesting to see whether this pattern of expression is species conserved, and/or present only at the fetal stages or in instances of compensatory hypertrophy.
PHLDA2 is a maternally expressed imprinted gene that has been implicated in placental function in humans and mice. It is expressed in the villous cytotrophoblasts in humans and in type II trophoblasts in the labyrinthine layer in mice [56]. Inactivation of Phlda2 in murine placentae resulted in expansion of spongiotrophoblast layer and placental overgrowth [57], whereas overexpression resulted in placental stunting [58]. In humans, upregulation of PHLDA2 has been implicated in IUGR [51, 59]. Imprinting at the PHLDA2 locus is complex and tissue specific. In both humans and mice, there is predominant maternal expression but significant expression from the supposedly silenced paternal allele [48]. Our QUASEP data indicate that the same occurs in swine with detection of significant levels of expression of the paternal allele (from 10% to 30% in the placenta; Table 2). Moreover, we observed tissue-specific differences, suggesting that these maternal:paternal expression ratios may shift depending on the tissues and stages being analyzed. Both the probe-by-probe analysis (data not shown) and the QUASEP data (Table 2) supported greater expression from the paternal allele in placenta than in liver. Because expression levels of PHLDA2 have a direct effect of trophoblast growth and differentiation in both humans and mice, it will be of great interest to determine whether the same is true in swine, as well as to examine how this protein acts to affect trophoblast function.
The growth factor IGF2 has been reported as imprinted and paternally expressed in all therian and eutherian mammals. Swine have at least four different promoters (P1–P4) that regulate IGF2 expression in an isoform-specific manner [60]. Nezer et al. [14] demonstrated paternal expression in two Day 70 fetal swine tissues: muscle and liver. Equally important, they demonstrated that the transcript from promoter P1 was imprinted in the liver at this stage. This has been confirmed recently in nonfetal tissues, and data have presented for the presence of imprinted transcript from other promoters [47]. Our results show that in the placenta, P2, P3, and P4 were expressed, and they were expressed in a pattern supporting imprinting and paternal expression. However, we could not detect expression of the P1 transcript in any tissue tested (Fig. 7). We postulate that our inability to detect expression from P1 is due to its activation later in fetal development, because it has been detected in postnatal and Day 70 porcine fetuses [14]. This would suggest that growth factor demand increases during gestation lead to activation of the P1 promoter in the liver. How this switch is accomplished, whether it is species specific, and the timing of its activation remain to be determined.
Species-Specific Imprinted Genes
In addition to tissue-specific and isoform-specific imprinting, we also found no evidence of imprinting in seven genes (ASCL2, ASB4, CD81, COMMD1, DCN, DLX5, and H13) that had been reported previously as imprinted in mice. ASCL2 was only detected in the placenta, and there was no evidence of imprinting (Table 1). This is analogous to reports in the early human placenta, where biallelic expression is seen [61], but is different than the maternal expression reported in mice [62]. DLX5 had been reported previously as imprinted in both human [63] and mouse brain [64], but those results have been questioned [54, 65], and at this time it is believed this gene is not imprinted in these two species. There is a previous report of imprinting of DLX5 in swine with unusual results indicating imprinting in some tissues, such as skeletal muscle, spleen, lung, and stomach, but biallelic expression in others, such as heart, liver, and kidney [66]. Neither brain nor placenta was tested. We could only detect expression in brain and placenta and found no evidence for imprinting in either tissue. H13 has been reported as imprinted and preferentially maternally expressed in mice [67]. The data presented by Wood et al. [67], based on a nonquantitative, allele-specific PCR sequencing, show imprinting in the fetal brain, but for other tissues there was considerable presence of the supposedly silenced allele. This was particularly evident in the placental sample where biallelic expression was seen. Thus, even in the only species for which imprinting for this gene has been reported, the data support a complicated and tissue-specific form of imprinting control. In swine, H13 was expressed in all tissues tested, but no evidence of imprinting was found. For ASB4, CD81, COMMD1, and DCN, our data support no imprinting in swine, as has been reported for human and/or bovine, and are discordant to reports in mice (Supplemental Table S2). In general, the imprinting pattern seen in swine was similar to humans but dissimilar to mice. At this point, it is difficult to determine the biological relevance of these apparent differences because of the conflicting evidence in the literature regarding imprinting at this locus in mice. At the very least, we have been able to conclusively demonstrate that these genes are not imprinted in swine.
Additionally, discrepancies from previous reports were observed for COPG2, PRIM2, and SLC38A4. Copg2 is a complex gene, with reported maternal expression in the mouse [68], disputed paternal expression in humans [69], and biallelic expression in sheep and cattle [70]. Our provisional data support imprinting and maternal expression in the swine placenta (Table 1 and Supplemental Table S2). Because artiodactyls (sheep, cow, and pig) have different modes of placentation (e.g., cotyledonary placentae with villi in sheep and cattle, compared with diffuse placentae with folded interdigitation in swine), COPG2 represents a unique case of species-specific genomic imprinting where swine diverge, and it may lend clues to different modes of mammalian placentation. In a functional context, COPG2 facilitates intracellular trafficking of proteins through budding from the Golgi membrane. The locus is implicated in Silver-Russell syndrome, a condition of severe intrauterine and postnatal growth retardation.
PRIM2 has been reported as imprinted and maternally expressed in human lymphoblastoid lines [71]. Our results differ and support paternal expression in liver only, and both probes used gave analogous information. However, unlike other paternally expressed genes, such as PEG10, PRIM2 was also expressed in the PRT sample, suggesting that there is incomplete silencing of the maternal allele. PRIM2 functions in DNA replication as a multimeric protein complex of polymerase and primase. Epigenetic regulation at the PRIM2 locus may lead to delayed DNA replication timing, impaired trophoblast proliferation, and developmental growth retardation in porcine PRTs.
SLC38A4, a gene involved in cell proliferation, tissue growth [49], and the transport of arginine and lysine across the plasma membrane [72], has been reported as imprinted and paternally expressed in all tissues tested in mice [73], and as not imprinted in cattle [74]. Our results conflict with those in cattle and mice [75] and support a complex isoform- and tissue-specific form of imprinting regulation. This is particularly evident for the P1-iso1 isoform, which clearly shows lack of expression in the BP samples in brain, fibroblasts, and placenta, but expresses in the liver (Fig. 6). Because this gene plays a key role in the transport of arginine and lysine, changes in its expression levels can have a significant effect on fetal growth. As to why it would be paternally expressed in mice and maternally expressed in swine, we can only speculate that it may be due to differences in placental types between these two species, or due to the presence of different isoforms, some maternally expressed and some paternally expressed, as our data support (Fig. 6).
Comparison of Array and QUASEP
The parthenogenetic model and expression profiling facilitated rapid screening of parent-of-origin effects for 24 genes (Table 1 and Supplemental Table S2). When the data from the microarrays, semiquantitative RT-PCRs, and the QUASEP analysis were compared, it was reassuring to see that all approaches provided analogous information. Of the 14 genes analyzed by both QUASEP and microarrays, all were correctly identified as imprinted, and with the imprinting being in the same direction. However, differences in the sensitivity of detection between the two methods (expression profiling or QUASEP) resulted in some gene/tissues combinations being identified as imprinted in one assay and not the other (H19, IGF2AS, INPP5F, PHLDA2, PPP1R9A, and NNAT). In the cases of INPP5F and PPP1R9A, Affymetrix probes did not discriminate between nonimprinted isoforms, so semiquantitative RT-PCR was used to demonstrate preferential paternal expression of INPP5F variant 2 (Fig. 2A) and PRT overexpression in the placenta of PPP1R9A, as depicted in Figure 2E. For NNAT, QUASEP detected imprinting in brain, liver, and fibroblasts, whereas the array detected differences only in brain and fibroblasts but no expression in liver and placenta. Other differences between the two assays pointed to the existence of isoforms. For instance, the microarray data indicated a tissue-specific expression pattern for PLAGL1, with expression only in the PRT placental sample, suggestive of either reactivation of the imprinted allele or expression of a nonimprinted allele (Fig. 4 and Table 1). Similarly, the array results helped us identify tissue-specific differences for DIRAS3 and SGCE (Figs. 3, and 5, respectively). Overall, however, the microarray and QUASEP results showed remarkable concordance, and they support the use of uniparental models and expression profiling as a mechanism for studying imprinting in mammalians species.
In summary, the key findings of this paper include the analysis of fetal and placental abnormalities in porcine PRTs, a comprehensive analysis of imprinting, including previously unreported cases of evolutionary divergence, and the identification of tissue-specific isoforms for several imprinted genes, most of which had not been reported previously in any species. Overall, this work represents the most comprehensive study of imprinted genes in swine to date. As we have indicated throughout the text, there is a disappointing lack of information on the role of these genes in swine fetal and placental development and function. It is our hope that this work will stimulate and assist in providing a framework upon which research in this critical area can be expanded.
Supplementary Material
Acknowledgments
We thank Jose L. Estrada for the generation of uniparental pregnancies, Bashir Mir for technical assistance with RNA extraction and sexing of animals used in microarray studies, and well as Abby York, Troy Gramke, Sue Hauver, and Sam Nejezchleb for technical assistance.
Footnotes
Supported by the U.S. Department of Agriculture Cooperative State Research, Education, and Extension Service National Research Initiative grant 2005-35604-15343 to J.A.P. and B.A.F.; by National Institutes of Health/National Institute of Child Health and Human Development grant HD048510 to J.A.P.; by a National Science Foundation (NSF) Integrative Graduate Education and Research Traineeship fellowship to S.R.B. and S.T.; a North Carolina State College of Veterinary Medicine epigenetics doctoral fellowship to S.R.B.; an NSF Graduate Research Fellowship to S.T.; and a National Institute of Environmental Health Sciences training grant to N.H. This work was performed as part of an initiative from the Center for Comparative Medicine and Translational Research at the North Carolina State University College of Veterinary Medicine.
These authors contributed equally to this work.
REFERENCES
- McGrath J, Solter D.Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell 1984; 37: 179–183.. [DOI] [PubMed] [Google Scholar]
- Morison IM, Ramsay JP, Spencer HG.A census of mammalian imprinting. Trends Genet 2005; 21: 457–465.. [DOI] [PubMed] [Google Scholar]
- Monk D, Arnaud P, Apostolidou S, Hills FA, Kelsey G, Stanier P, Feil R, Moore GE.Limited evolutionary conservation of imprinting in the human placenta. Proc Natl Acad Sci U S A 2006; 103: 6623–6628.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis A, Mitsuya K, Umlauf D, Smith P, Dean W, Walter J, Higgins M, Feil R, Reik W.Imprinting on distal chromosome 7 in the placenta involves repressive histone methylation independent of DNA methylation. Nat Genet 2004; 36: 1291–1295.. [DOI] [PubMed] [Google Scholar]
- Moore T, Haig D.Genomic imprinting in mammalian development: a parental tug-of-war. Trends Genet 1991; 7: 45–49.. [DOI] [PubMed] [Google Scholar]
- Li L, Keverne EB, Aparicio SA, Ishino F, Barton SC, Surani MA.Regulation of maternal behavior and offspring growth by paternally expressed Peg3. Science 1999; 284: 330–333.. [DOI] [PubMed] [Google Scholar]
- Mann MR, Chung YG, Nolen LD, Verona RI, Latham KE, Bartolomei MS.Disruption of imprinted gene methylation and expression in cloned preimplantation stage mouse embryos. Biol Reprod 2003; 69: 902–914.. [DOI] [PubMed] [Google Scholar]
- Suemizu H, Aiba K, Yoshikawa T, Sharov AA, Shimozawa N, Tamaoki N, Ko MS.Expression profiling of placentomegaly associated with nuclear transplantation of mouse ES cells. Dev Biol 2003; 253: 36–53.. [DOI] [PubMed] [Google Scholar]
- Archer GS, Dindot S, Friend TH, Walker S, Zaunbrecher G, Lawhorn B, Piedrahita JA.Hierarchical phenotypic and epigenetic variation in cloned swine. Biol Reprod 2003; 69: 430–436.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurston A, Taylor J, Gardner J, Sinclair KD, Young LE.Monoallelic expression of nine imprinted genes in the sheep embryo occurs after the blastocyst stage. Reproduction 2008; 135: 29–40.. [DOI] [PubMed] [Google Scholar]
- Cheng HC, Zhang FW, Deng CY, Jiang CD, Xiong YZ, Li FE, Lei MG.NNAT and DIRAS3 genes are paternally expressed in pigs. Genet Sel Evol 2007; 39: 599–607.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Bergmann A, Lucas S, Stone R, Stubbs L.Lineage-specific imprinting and evolution of the zinc-finger gene ZIM2. Genomics 2004; 84: 47–58.. [DOI] [PubMed] [Google Scholar]
- Zhou QY, Huang JN, Xiong YZ, Zhao SH.Imprinting analyses of the porcine GATM and PEG10 genes in placentas on days 75 and 90 of gestation. Genes Genet Syst 2007; 82: 265–269.. [DOI] [PubMed] [Google Scholar]
- Nezer C, Moreau L, Brouwers B, Coppieters W, Detilleux J, Hanset R, Karim L, Kvasz A, Leroy P, Georges M.An imprinted QTL with major effect on muscle mass and fat deposition maps to the IGF2 locus in pigs. Nat Genet 1999; 21: 155–156.. [DOI] [PubMed] [Google Scholar]
- Braunschweig MH, Van Laere AS, Buys N, Andersson L, Andersson G.IGF2 antisense transcript expression in porcine postnatal muscle is affected by a quantitative trait nucleotide in intron 3. Genomics 2004; 84: 1021–1029.. [DOI] [PubMed] [Google Scholar]
- Killian JK, Nolan CM, Wylie AA, Li T, Vu TH, Hoffman AR, Jirtle RL.Divergent evolution in M6P/IGF2R imprinting from the Jurassic to the Quaternary. Hum Mol Genet 2001; 10: 1721–1728.. [DOI] [PubMed] [Google Scholar]
- Xu C, Su L, Zhou Q, Li C, Zhao S.Imprinting analysis of the porcine MEST gene in 75 and 90 day placentas and prenatal tissues. Acta Biochim Biophys Sin (Shanghai) 2007; 39: 633–639.. [DOI] [PubMed] [Google Scholar]
- Zhang FW, Cheng HC, Jiang CD, Deng CY, Xiong YZ, Li FE, Lei MG.Imprinted status of pleomorphic adenoma gene-like I and paternal expression gene 10 genes in pigs. J Anim Sci 2007; 85: 886–890.. [DOI] [PubMed] [Google Scholar]
- Kuzmin A, Han Z, Golding MC, Mann MR, Latham KE, Varmuza S.The PcG gene Sfmbt2 is paternally expressed in extraembryonic tissues. Gene Expr Patterns 2008; 8: 107–116.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menheniott TR, Woodfine K, Schulz R, Wood AJ, Monk D, Giraud AS, Baldwin HS, Moore GE, Oakey RJ.Genomic imprinting of Dopa decarboxylase in heart and reciprocal allelic expression with neighboring Grb10. Mol Cell Biol 2008; 28: 386–396.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz R, Menheniott TR, Woodfine K, Wood AJ, Choi JD, Oakey RJ.Chromosome-wide identification of novel imprinted genes using microarrays and uniparental disomies. Nucleic Acids Res 2006; 34: e88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido I, Saito C, Mizuno Y, Meguro M, Bono H, Kadomura M, Kono T, Morris GA, Lyons PA, Oshimura M, Hayashizaki Y, Okazaki Y.Discovery of imprinted transcripts in the mouse transcriptome using large-scale expression profiling. Genome Res 2003; 13: 1402–1409.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruf N, Dunzinger U, Brinckmann A, Haaf T, Nurnberg P, Zechner U.Expression profiling of uniparental mouse embryos is inefficient in identifying novel imprinted genes. Genomics 2006; 87: 509–519.. [DOI] [PubMed] [Google Scholar]
- Ruf N, Bahring S, Galetzka D, Pliushch G, Luft FC, Nurnberg P, Haaf T, Kelsey G, Zechner U.Sequence-based bioinformatic prediction and QUASEP identify genomic imprinting of the KCNK9 potassium channel gene in mouse and human. Hum Mol Genet 2007; 16: 2591–2599.. [DOI] [PubMed] [Google Scholar]
- Cruz NT, Wilson KJ, Cooney MA, Tecirlioglu RT, Lagutina I, Galli C, Holland MK, French AJ.Putative imprinted gene expression in uniparental bovine embryo models. Reprod Fertil Dev 2008; 20: 589–597.. [DOI] [PubMed] [Google Scholar]
- Piras G, El Kharroubi A, Kozlov S, Escalante-Alcalde D, Hernandez L, Copeland NG, Gilbert DJ, Jenkins NA, Stewart CL.Zac1 (Lot1), a potential tumor suppressor gene, and the gene for epsilon-sarcoglycan are maternally imprinted genes: identification by a subtractive screen of novel uniparental fibroblast lines. Mol Cell Biol 2000; 20: 3308–3315.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassady JP, Young LD, Leymaster KA.Heterosis and recombination effects on pig reproductive traits. J Anim Sci 2002; 80: 2303–2315.. [PubMed] [Google Scholar]
- Gimelbrant A, Hutchinson JN, Thompson BR, Chess A.Widespread monoallelic expression on human autosomes. Science 2007; 318: 1136–1140.. [DOI] [PubMed] [Google Scholar]
- Krueger C, Morison IM.Random monoallelic expression: making a choice. Trends Genet 2008; 24: 257–259.. [DOI] [PubMed] [Google Scholar]
- Ramanathan P, Martin IC, Gardiner-Garden M, Thomson PC, Taylor RM, Ormandy CJ, Moran C, Williamson P.Transcriptome analysis identifies pathways associated with enhanced maternal performance in QSi5 mice. BMC Genomics 2008; 9: 197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai S, Mir B, Martin AC, Estrada JL, Bischoff SR, Hsieh WP, Cassady JP, Freking BA, Nonneman DJ, Rohrer GA, Piedrahita JA.Detection of transcriptional difference of porcine imprinted genes using different microarray platforms. BMC Genomics 2006; 7: 328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrenkrug SC, Freking BA, Smith TP, Rohrer GA, Keele JW.Single nucleotide polymorphism (SNP) discovery in porcine expressed genes. Anim Genet 2002; 33: 186–195.. [DOI] [PubMed] [Google Scholar]
- Panitz F, Stengaard H, Hornshoj H, Gorodkin J, Hedegaard J, Cirera S, Thomsen B, Madsen LB, Hoj A, Vingborg RK, Zahn B, Wang X, et al. SNP mining porcine ESTs with MAVIANT, a novel tool for SNP evaluation and annotation. Bioinformatics 2007; 23: i387–391.. [DOI] [PubMed] [Google Scholar]
- Bischoff SR, Tsai S, Hardison N, York A, Freking BA, Nonneman D, Rohrer G, Piedrahita JA.Identification of SNPs and INDELS in swine transcribed sequences using short oligonucleotide microarrays. BMC Genomics 2008; 9: 252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Chu TM, Gibson G, Wolfinger RD.A mixed model approach to identify yeast transcriptional regulatory motifs via microarray experiments. Stat Appl Genet Mol Biol 2004; 3: Article 22. [DOI] [PubMed] [Google Scholar]
- Wall ME, Rechtsteiner A, Rocha LM. Singular Value Decomposition and Principal Component Analysis. Norwell, MA:: Kluwer;; 2003. [Google Scholar]
- Liu WM, Mei R, Di X, Ryder TB, Hubbell E, Dee S, Webster TA, Harrington CA, Ho MH, Baid J, Smeekens SP.Analysis of high density expression microarrays with signed-rank call algorithms. Bioinformatics 2002; 18: 1593–1599.. [DOI] [PubMed] [Google Scholar]
- Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, et al. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res 2009; 37: D885–D890.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, Gaasterland T, Glenisson P, et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet 2001; 29: 365–371.. [DOI] [PubMed] [Google Scholar]
- Kullberg M, Nilsson MA, Arnason U, Harley EH, Janke A.Housekeeping genes for phylogenetic analysis of eutherian relationships. Mol Biol Evol 2006; 23: 1493–1503.. [DOI] [PubMed] [Google Scholar]
- Zhu J, He F, Song S, Wang J, Yu J.How many human genes can be defined as housekeeping with current expression data? BMC Genomics 2008; 9: 172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutsch F, Ruf N, Vaingankar S, Toliat MR, Suk A, Hohne W, Schauer G, Lehmann M, Roscioli T, Schnabel D, Epplen JT, Knisely A, et al. Mutations in ENPP1 are associated with ‘idiopathic’ infantile arterial calcification. Nat Genet 2003; 34: 379–381.. [DOI] [PubMed] [Google Scholar]
- Stangegaard M, Dufva IH, Dufva M.Reverse transcription using random pentadecamer primers increases yield and quality of resulting cDNA. Biotechniques 2006; 40: 649–657.. [DOI] [PubMed] [Google Scholar]
- Aydin A, Toliat MR, Bahring S, Becker C, Nurnberg P.New universal primers facilitate pyrosequencing. Electrophoresis 2006; 27: 394–397.. [DOI] [PubMed] [Google Scholar]
- Wolfinger RD, Gibson G, Wolfinger ED, Bennett L, Hamadeh H, Bushel P, Afshari C, Paules RS.Assessing gene significance from cDNA microarray expression data via mixed models. J Comput Biol 2001; 8: 625–637.. [DOI] [PubMed] [Google Scholar]
- Thierry-Mieg D, Thierry-Mieg J.AceView: a comprehensive cDNA-supported gene and transcripts annotation. Genome Biol 2006; 7(suppl 1):S12.1–14.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Bin Y, Curchoe C, Yang L, Feng D, Jiang Q, O'Neill M, Tian XC, Zhang S.Genetic imprinting of H19 and IGF2 in domestic pigs (Sus scrofa). Anim Biotechnol 2008; 19: 22–27.. [DOI] [PubMed] [Google Scholar]
- Qian N, Frank D, O'Keefe D, Dao D, Zhao L, Yuan L, Wang Q, Keating M, Walsh C, Tycko B.The IPL gene on chromosome 11p15.5 is imprinted in humans and mice and is similar to TDAG51, implicated in Fas expression and apoptosis. Hum Mol Genet 1997; 6: 2021–2029.. [DOI] [PubMed] [Google Scholar]
- Lui JC, Finkielstain GP, Barnes KM, Baron J.An imprinted gene network that controls mammalian somatic growth is down-regulated during postnatal growth deceleration in multiple organs. Am J Physiol Regul Integr Comp Physiol 2008; 295: R189–R196.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varrault A, Gueydan C, Delalbre A, Bellmann A, Houssami S, Aknin C, Severac D, Chotard L, Kahli M, Le Digarcher A, Pavlidis P, Journot L.Zac1 regulates an imprinted gene network critically involved in the control of embryonic growth. Dev Cell 2006; 11: 711–722.. [DOI] [PubMed] [Google Scholar]
- McMinn J, Wei M, Schupf N, Cusmai J, Johnson EB, Smith AC, Weksberg R, Thaker HM, Tycko B.Unbalanced placental expression of imprinted genes in human intrauterine growth restriction. Placenta 2006; 27: 540–549.. [DOI] [PubMed] [Google Scholar]
- Valleley EM, Cordery SF, Bonthron DT.Tissue-specific imprinting of the ZAC/PLAGL1 tumour suppressor gene results from variable utilization of monoallelic and biallelic promoters. Hum Mol Genet 2007; 16: 972–981.. [DOI] [PubMed] [Google Scholar]
- Smallwood A, Papageorghiou A, Nicolaides K, Alley MK, Alice J, Nargund G, Ojha K, Campbell S, Banerjee S.Temporal regulation of the expression of syncytin (HERV-W), maternally imprinted PEG10, and SGCE in human placenta. Biol Reprod 2003; 69: 286–293.. [DOI] [PubMed] [Google Scholar]
- Monk D, Wagschal A, Arnaud P, Muller P, Parker-Katiraee L, Bourchis D, Scherer SW, Feil R, Stanier P, Moore GE.Comparative analysis of human chromosome 7q21 and mouse proximal chromosome 6 reveals a placental-specific imprinted gene, TFPI2/Tfpi2, which requires G9a and Eed for allelic-silencing. Genome Res 2008; 18: 1270–1281.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Ge X, Shen Y, Chen L, Kong Y, Zhang H, Man X, Tang L, Yuan H, Wang H, Zhao G, Jin W.Gene expression profile analysis of human hepatocellular carcinoma using SAGE and LongSAGE. BMC Med Genomics 2009; 2: 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena A, Frank D, Panichkul P, Van den Veyver IB, Tycko B, Thaker H.The product of the imprinted gene IPL marks human villous cytotrophoblast and is lost in complete hydatidiform mole. Placenta 2003; 24: 835–842.. [DOI] [PubMed] [Google Scholar]
- Frank D, Fortino W, Clark L, Musalo R, Wang W, Saxena A, Li CM, Reik W, Ludwig T, Tycko B.Placental overgrowth in mice lacking the imprinted gene Ipl. Proc Natl Acad Sci U S A 2002; 99: 7490–7495.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas M, John R, Saxena A, Barton S, Frank D, Fitzpatrick G, Higgins MJ, Tycko B.Placental growth retardation due to loss of imprinting of Phlda2. Mech Dev 2004; 121: 1199–1210.. [DOI] [PubMed] [Google Scholar]
- Apostolidou S, Abu-Amero S, O'Donoghue K, Frost J, Olafsdottir O, Chavele KM, Whittaker JC, Loughna P, Stanier P, Moore GE.Elevated placental expression of the imprinted PHLDA2 gene is associated with low birth weight. J Mol Med 2007; 85: 379–387.. [DOI] [PubMed] [Google Scholar]
- Amarger V, Nguyen M, Van Laere AS, Braunschweig M, Nezer C, Georges M, Andersson L.Comparative sequence analysis of the INS-IGF2-H19 gene cluster in pigs. Mamm Genome 2002; 13: 388–398.. [DOI] [PubMed] [Google Scholar]
- Miyamoto T, Hasuike S, Jinno Y, Soejima H, Yun K, Miura K, Ishikawa M, Niikawa N.The human ASCL2 gene escaping genomic imprinting and its expression pattern. J Assist Reprod Genet 2002; 19: 240–244.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemot F, Caspary T, Tilghman SM, Copeland NG, Gilbert DJ, Jenkins NA, Anderson DJ, Joyner AL, Rossant J, Nagy A.Genomic imprinting of Mash2, a mouse gene required for trophoblast development. Nat Genet 1995; 9: 235–242.. [DOI] [PubMed] [Google Scholar]
- Okita C, Meguro M, Hoshiya H, Haruta M, Sakamoto YK, Oshimura M.A new imprinted cluster on the human chromosome 7q21-q31, identified by human-mouse monochromosomal hybrids. Genomics 2003; 81: 556–559.. [DOI] [PubMed] [Google Scholar]
- Horike S, Cai S, Miyano M, Cheng JF, Kohwi-Shigematsu T.Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome. Nat Genet 2005; 37: 31–40.. [DOI] [PubMed] [Google Scholar]
- Schule B, Li HH, Fisch-Kohl C, Purmann C, Francke U.DLX5 and DLX6 expression is biallelic and not modulated by MeCP2 deficiency. Am J Hum Genet 2007; 81: 492–506.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng HC, Zhang FW, Jiang CD, Li FE, Xiong YZ, Deng CY.Isolation and imprinting analysis of the porcine DLX5 gene and its association with carcass traits. Anim Genet 2008; 39: 395–399.. [DOI] [PubMed] [Google Scholar]
- Wood AJ, Roberts RG, Monk D, Moore GE, Schulz R, Oakey RJ.A screen for retrotransposed imprinted genes reveals an association between X chromosome homology and maternal germ-line methylation. PLoS Genet 2007; 3: e20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Park CW, Hahn Y, Park J, Lee J, Yun JH, Hyun B, Chung JH.Mit1/Lb9 and Copg2, new members of mouse imprinted genes closely linked to Peg1/Mest(1). FEBS Lett 2000; 472: 230–234.. [DOI] [PubMed] [Google Scholar]
- Yamasaki K, Hayashida S, Miura K, Masuzaki H, Ishimaru T, Niikawa N, Kishino T.The novel gene, gamma2-COP (COPG2), in the 7q32 imprinted domain escapes genomic imprinting. Genomics 2000; 68: 330–335.. [DOI] [PubMed] [Google Scholar]
- Khatib H.The COPG2, DCN, and SDHD genes are biallelically expressed in cattle. Mamm Genome 2005; 16: 545–552.. [DOI] [PubMed] [Google Scholar]
- Pant PV, Tao H, Beilharz EJ, Ballinger DG, Cox DR, Frazer KA.Analysis of allelic differential expression in human white blood cells. Genome Res 2006; 16: 331–339.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constancia M, Angiolini E, Sandovici I, Smith P, Smith R, Kelsey G, Dean W, Ferguson-Smith A, Sibley CP, Reik W, Fowden A.Adaptation of nutrient supply to fetal demand in the mouse involves interaction between the Igf2 gene and placental transporter systems. Proc Natl Acad Sci U S A 2005; 102: 19219–19224.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno Y, Sotomaru Y, Katsuzawa Y, Kono T, Meguro M, Oshimura M, Kawai J, Tomaru Y, Kiyosawa H, Nikaido I, Amanuma H, Hayashizaki Y, et al. Asb4, Ata3, and Dcn are novel imprinted genes identified by high-throughput screening using RIKEN cDNA microarray. Biochem Biophys Res Commun 2002; 290: 1499–1505.. [DOI] [PubMed] [Google Scholar]
- Zaitoun I, Khatib H.Assessment of genomic imprinting of SLC38A4, NNAT, NAP1L5, and H19 in cattle. BMC Genet 2006; 7: 49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RJ, Dean W, Konfortova G, Kelsey G.Identification of novel imprinted genes in a genome-wide screen for maternal methylation. Genome Res 2003; 13: 558–569.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.