Summary
Genome-wide studies have identified abundant small, non-coding RNAs including snRNAs, snoRNAs, cryptic unstable transcripts (CUTs), and upstream regulatory RNAs (uRNAs) that are transcribed by RNA polymerase II (pol II) and terminated by a Nrd1-dependent pathway. Here, we show that the prolyl isomerase, Ess1, is required for Nrd1-dependent termination of ncRNAs. Ess1 binds the carboxy terminal domain (CTD) of pol II and is thought to regulate transcription by conformational isomerization of Ser-Pro bonds within the CTD. In ess1 mutants, expression of ∼10% of the genome was altered, due primarily to defects in termination of snoRNAs, CUTs, SUTs and uRNAs. Ess1 promoted dephosphorylation of Ser5 (but not Ser2) within the CTD, most likely by the Ssu72 phosphatase, and we provide evidence for a competition between Nrd1 and Pcf11 for CTD-binding that is regulated by Ess1-dependent isomerization. This is the first example of a prolyl isomerase required for interpreting the “CTD code.”
Introduction
Gene expression in eukaryotes requires distinct co-factor complexes that assemble on the RNA polymerase II (pol II) core enzyme and help it to carry out the sequential steps of the transcription cycle, including initiation, elongation, termination, and RNA 3′-end processing (Cho, 2007; Pandit et al., 2008). The timing of co-factor assembly is regulated, in part, by reversible covalent and non-covalent modification of the carboxy terminal domain (CTD) of Rpb1, the largest subunit of pol II (Egloff and Murphy, 2008; Hirose and Ohkuma, 2007; Phatnani and Greenleaf, 2006).
Covalent modification of the CTD involves phosphorylation and dephosphorylation of Ser2 and Ser5 (Chapman et al., 2007; Palancade and Bensaude, 2003; Patturajan et al., 1998; Phatnani and Greenleaf, 2006), and Ser7 (Chapman et al., 2007; Egloff et al., 2007; Keogh et al., 2003) within the CTD heptad repeat (Tyr1-Ser2-Pro3-Thr4-Ser5-Pro6-Ser7). Phosphorylation of Ser5 is enhanced at the 5′ ends of genes and is associated with initiation complex formation, 5′-capping, and the transition to elongation, while phosphorylation of Ser2 increases toward the 3′ end of genes and is associated with elongation and termination (Bird et al., 2004; Kobor and Greenblatt, 2002; Licatalosi et al., 2002; McCracken et al., 1997; Schroeder et al., 2000). The role of Ser7 phosphorylation in yeast is unknown. A number of conserved Ser2- and Ser5- specific CTD kinases and phosphatases have been identified, and many co-factor proteins required for transcription and chromatin modification specifically associate with either Ser2- or Ser5- phosphorylated forms of the CTD (reviewed in Egloff and Murphy, 2008).
Much less is known about non-covalent changes to the CTD such as prolyl isomerization. The CTD repeat contains two Ser-Pro peptide bonds that are targets of enzymes known as peptidyl prolyl cis/trans isomerases (Schiene and Fischer, 2000). Isomerization is predicted to cause dramatic conformational changes in the CTD that likely affect co-factor binding (Cramer, 2006; Meinhart et al., 2005). The CTD isomerase Ess1 was identified in Saccharomyces cerevisiae (Hanes et al., 1989; Hani et al., 1995), and has a human ortholog, Pin1 (Lu et al., 1996; Lu and Zhou, 2007). Ess1 deletion is lethal in yeast and mutants undergo mitotic arrest (Hanes et al., 1989; Wu et al., 2000). Pin1 knockdowns and knockouts also show cell cycle defects (Fujimori et al., 1999; Liou et al., 2002; Lu et al., 1996), and Pin1 misregulation has been associated with cancer and neurological disorders (Lu and Zhou, 2007; Yeh and Means, 2007). Pin1 has been shown to interact with a wide variety of target proteins (Joseph et al., 2003).
Both Ess1 and Pin1 bind Ser2- and Ser5- phosphorylated forms of the CTD but they do not bind unphosphorylated CTD (Morris et al., 1999; Verdecia et al., 2000; Yaffe et al., 1997; Zhang et al., 2002). Ess1 binds and isomerizes phospho-Ser5-Pro6 (P-Ser5-Pro6) ∼5-fold better than phospho-Ser2-Pro3 (P-Ser2-Pro3) within the CTD (Gemmill et al., 2005), consistent with genetic experiments identifying P-Ser5-Pro6 as a major functional target (Wilcox et al., 2004). Genetic experiments also indicate that ESS1 plays important roles in initiation, elongation, and termination of pol II transcription (Wilcox et al., 2004; Wu et al., 2001; Wu et al., 2003; Wu et al., 2000; Krishnamurthy et al., 2009). Indeed, ESS1 (PTF1) was recovered in a screen for mutations that impair transcription termination in a readthrough reporter assay (Hani et al., 1995).
In yeast, two distinct pathways for transcription termination have been described (Kim et al., 2004; Kim et al., 2006; Lykke-Andersen and Jensen, 2007; Rondon et al., 2008; Steinmetz and Brow, 1996; Steinmetz et al., 2001). The major pathway is the canonical mRNA termination pathway that results in 3′ cleavage and polyadenylation of transcripts derived from protein coding genes. This pathway requires members of the CFI complex including Hrp1, Pcf11, Rna14, and Rna15, and the CPF complex including Cft2, Pap1, Pta1, Pti1, Ref2, Rat1, Ssu72 and Swd2 (Aranda and Proudfoot, 2001; Birse et al., 1998; Buratowski, 2005; Nedea et al., 2003). The products of this pathway are relatively stable mRNAs that carry long (∼70 nt) poly(A) tails and are exported to the cytoplasm for translation (Barabino and Keller, 1999). In contrast, other pol II transcripts that are typically small, nonpolyadenylated, and largely restricted to the nucleus, are terminated by an alternative pathway that is dependent upon Nrd1 (Steinmetz et al., 2001). The Nrd-1 pathway is used to terminate most small nucleolar RNAs (snoRNAs), small nuclear RNAs (snRNAs), cryptic unstable transcripts (CUTs), and some short mRNAs (Arigo et al., 2006; Kim et al., 2006; Steinmetz et al., 2001; Thiebaut et al., 2006). This pathway utilizes a distinct, but partially overlapping set of proteins that includes Glc7, Nrd1, Nab3, Paf1, Pcf11, Rna14, Rna15, Sen1, Ssu72, Swd2 and Trf4 (Lykke-Andersen and Jensen, 2007). It is not yet established which pathway is used for a newly described set of small transcripts called SUTs (stable unannotated transcripts; Xu et al., 2009).
Here we sought to identify Ess1 target genes and study the role of Ess1 in their expression. Using genome-wide approaches, we found that about 10% of the genome is mis-regulated in ess1 mutants, and that the most prominent defect is a failure to correctly terminate snoRNA gene transcription. Readthrough of snoRNAs in ess1 mutants is similar to that observed in nrd1, nab3, pcf11 and paf1-complex mutants, and indeed genetic interactions and chromatin immunoprecipitation (ChIP) data indicate that Ess1 is linked to the Nrd1 pathway. As expected if Ess1 functions in the Nrd1 pathway, ess1 mutants show a failure to process CUTs and upstream regulatory RNAs (uRNAs), whose misregulation leads to constitutive expression (or repression) of downstream ORFs. We also show that Ess1 controls the phosphorylation state of the CTD and present a model for how Ess1-directed isomerization of the CTD plays a critical role in Nrd1-dependent transcription termination.
Results
Genome-wide expression analysis reveals a gene-specific requirement for Ess1
Current evidence does not distinguish whether Ess1 (or Pin1) affects the transcription of all pol II-dependent genes, or only a subset. To resolve this issue, we used whole-genome expression analysis with microarrays containing all the yeast ORFs. First, we compared expression between wild-type and ess1ts mutant yeast, at both permissive (24°C) and non-permissive (34°C) temperatures. Second, we used a highly-regulated activator system (GAL4-ER-VP16) to drive high or low levels of the wild-type or mutant Ess1 proteins (Gemmill et al., 2005). The results using these different methods were remarkably congruent and suggest that in yeast, only a subset of genes are preferentially affected (>2-fold) when Ess1 activity is deficient (Table S1A,C,D). At 24°C, ∼3% of all annotated ORFs in ess1ts cells were affected, while after 3 hr after a shift to 34°C this increased to ∼10% (Fig. S1A). Similarly, using the regulated promoter system, ∼10% of ORFs were affected (Table S1A, Fig. S1B), and 218 of these overlapped with ORFs identified in the ts experiment (P=8.7e-88, P=2.9e-29 for up- and down-regulated genes, respectively; Fig. S1C). Thus, only some ORFs appear to be Ess1-dependent. We also observed a >2 fold change in expresion of 233 non-coding RNAs and intergenic regions (e.g., SAGE tags, snoRNAs, Ty1LTRs; Fig. S1A).
Ess1 is required for termination of snoRNA genes
Although we expected to identify alterations in mRNA levels for a key set of G2/M cell-cycle genes, which could have explained the ess1 mitotic arrest phenotype, this was not the case. However, we did observe a remarkable similarity to microarray data from studies of ssu72 mutants (Ganem et al., 2003; Nedea et al., 2003). SSU72 encodes a Ser5-specific CTD phosphatase originally isolated as a suppressor of TFIIB (SUA7) mutations (Sun and Hampsey, 1996). Our results, based on a composite of the ts and regulated-promoter microarray data, mirror those obtained for ssu72 mutants (Table S1B). For example, eight of the 30 genes that showed the largest increase in expression in ess1 mutants are among the 28 genes that show the largest increase in ssu72 mutants (Fig. S1D). This overlap is highly significant (P=1.7e-12). Equally remarkable is that 11 of the 30 genes that showed the largest increases in ess1 mutants have snoRNAs immediately upstream (Table S1B), a situation similar to that in the ssu72 study (Ganem et al., 2003).
Based on studies done with Ssu72 and Nrd1 (Carroll et al., 2004; Dichtl et al., 2002; Ganem et al., 2003; Steinmetz and Brow, 2003; Steinmetz et al., 2001), we tested whether the increase in expression of selected genes in ess1 mutants was due to readthrough from upstream snoRNAs. Total RNA from wild-type and ess1 mutant yeast was reverse transcribed (RT) and amplified by PCR, using primers to intergenic regions downstream of snoRNAs (Fig. 1A). No intergenic transcription was detected in wild-type cells, whereas strong signals were detected in ess1ts mutants, even at permissive temperature (Fig. 1B). As expected, readthrough was generally more pronounced after a shift to 37°C. Readthrough from SNR5 into HEM4 was confirmed by the identification of fusion transcripts (Fig. 1C).
Figure 1. Readthrough of snoRNA gene transcription in ess1 mutants.
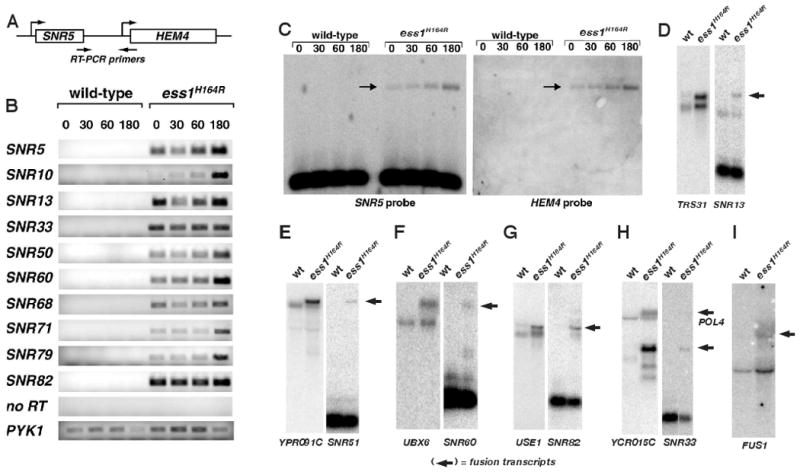
(A) Scheme used to detect intergenic transcription downstream of the snoRNA gene, SNR5. Total RNA was reverse-transcribed, and the resulting cDNA was subjected to PCR (25 cycles). (B) Ethidium-bromide stained gel reveals intergenic transcription in ess1H164R ts-cells but not wild type at 0, 30, 60, 180 min after shift to 37°C. Controls are no reverse transcriptase (no RT), and the PYK1 gene. (C) Northern analysis of total RNA from cells as described in B. Probes are indicated and the arrows identify fusion transcripts containing both SNR5 and HEM4 sequences. The normal HEM4 transcript is not expressed. (D-H) Northern analysis of SNR loci of RNAs harvested from the indicated cells at 180 min. after shifting to 37°C.
Tiling array analysis of ess1 mutants reveals global termination defects
To survey transcription readthrough in ess1 mutants on a genome-wide scale, we used tiling microarrays (David et al., 2006) to compare RNA from wild-type and ess1ts mutant cells grown at 24°C or 34°C. Nearly all snoRNA genes exhibited readthrough transcription in ess1ts mutant cells, as suggested by a composite average (Fig. S2A), and shown for selected individual genes (Fig. 2A-F).
Figure 2. Integrated Genome Browser (IGB) display of tiling array data.
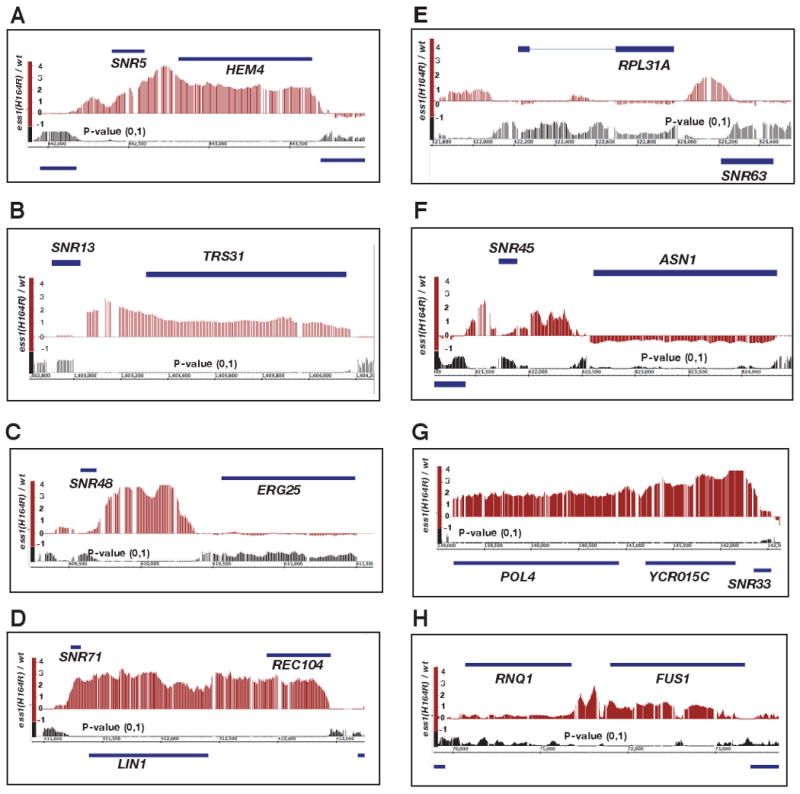
(A-H) Genes diagrammed as solid bars above the signal of the difference plot (e.g. SNR5, HEM4) are transcribed left to right (→), while genes shown below the P-value plot (e.g. LIN1, SNR63) are transcribed right to left (←). Signals above the line of the X-axis represent increased expression in ess1H164R mutant cells relative to wild-type (both at 34°C), while signals below the line represent decreased expression. P-values tended to be significant (typically P < 0.01) across regions where large difference signals are detected. Gaps in the signals are due to absence of probes at those positions on the tiling array.
For individual genes, different patterns of readthrough were observed. At the SNR5 locus, readthrough proceeds into the adjacent mRNA gene, HEM4, before termination occurs (Fig. 2A). A similar pattern was observed for SNR13-TRS31 (Fig. 2B) and SNR82-USE1 (Fig. S3A). At the SNR48 and SNR8 loci, in contrast, readthrough ends abruptly following the intergenic region, prior to the start of the respective downstream genes, ERG25 and YTM1 (Fig. 2C; Fig. S3B). Readthrough into an adjacent ORF, convergently transcribed, can also occur as for SNR71-LIN1-REC104 (Fig. 2D), and the polycistronic SNR41-SNR70-SNR51 genes (Fig. S3C). In other cases, readthrough stops abruptly at the 3′ end of the ORF on the opposite strand (e.g., SNR63-RPL31A, Fig. 2E). We do not know why some ORFs are read through while others are not, but the differences might involve cryptic termination sites, or mechanism(s) of transcriptional occlusion (e.g. colliding polymerases) and/or chromatin structural impediments to elongation. Readthrough from some snoRNAs appears to downregulate the adjacent ORF (e.g. SNR45-ASN1, Fig. 2F). As a control, SNR52, which is transcribed by RNA pol III, did not show significant readthrough in ess1 mutants (Fig. S3D).
Readthrough detected in the tiling array was confirmed by Northern analysis in which fusion transcripts were detected using both SNR and adjacent ORF probes (Fig. 1D-H). These and other data (not shown) indicate that termination of nearly all independently-transcribed snoRNA genes are Ess1-dependent. In contrast, tiling array data did not reveal obvious termination defects for the pol II-transcribed spliceosomal U1, U2, U3, U4 and U5 snRNA genes (data not shown). Thus, Ess1 is important for termination of some, but not all classes of pol II-dependent small non-coding RNA genes.
Finally, some mRNA genes also exhibited readthrough in ess1 mutants. For example, SNR33 reads through into YCR015C, but termination after this ORF also appears defective, given that transcription extends into the downstream POL4 gene (Fig 1H, Fig. 2G). Another example is the readthrough of RNQ1 into FUS1 (Fig. 1I, Fig. 2H).
To determine the extent of mRNA readthrough genome-wide, we compared RNA levels between ess1 mutants and wild-type cells at a defined window (-300 to +300 bp) centered on the predicted 3′ transcription termination site (TTS; Nagalakshmi et al., 2008), for each of 5,074 ORFs. The results suggest that readthrough occurs in ess1 mutant cells, with maximal values obtained at 25-150 bp downstream of the TTS (Fig. S2B). The magnitude of the average signal is small, because only a small subset of the 5,073 genes analyzed show aberrant 3′ transcription (Fig. S4A). Most abundant among the 100 most affected genes in ess1 mutants are ribosomal protein genes and other highly expressed genes (Table S2). An example of probable readthrough is shown in Figure S5A. In many cases, the 3′ signal is probably due to antisense transcription rather than readthrough (Fig. S5B-E), since some of these transcripts have been identified as CUTs or SUTs (Fig. S5B,C; see Table S2). In summary, our tiling array data indicate that termination of nearly all snoRNAs, as well as a small subset of mRNA genes, is Ess1 dependent. In addition, the analysis indicates that both known and newly-identified small non-coding transcripts (CUTs/SUTs) are stabilized in ess1 mutants (see also below).
Ess1 functions in the Nrd1 pathway
The Nrd1 pathway is the primary means by which transcription of snoRNA genes is terminated (Steinmetz et al., 2001; Vasiljeva and Buratowski, 2006). Both Nrd1 and Ess1 mutants show defects in snoRNA termination. To test whether Ess1 functions in the Nrd1 pathway, we first examined readthrough of reporter genes that carry snoRNA gene terminators, which contain Nrd1 and/or Nab3 binding sites (Carrol et al., 2004). As shown previously, nrd1-5 mutants exhibit readthrough (Fig. 3A, upper panels). Likewise, we find that ess1 mutants show readthtrough, which is slightly stronger for the SNR47 reporter than for the SNR13 reporter (Fig. 3A, lower panels). These results demonstrate that the activity of cis-acting termination signals of snoRNA genes depends upon Ess1.
Figure 3. Ess1 is linked to the Nrd1 pathway.
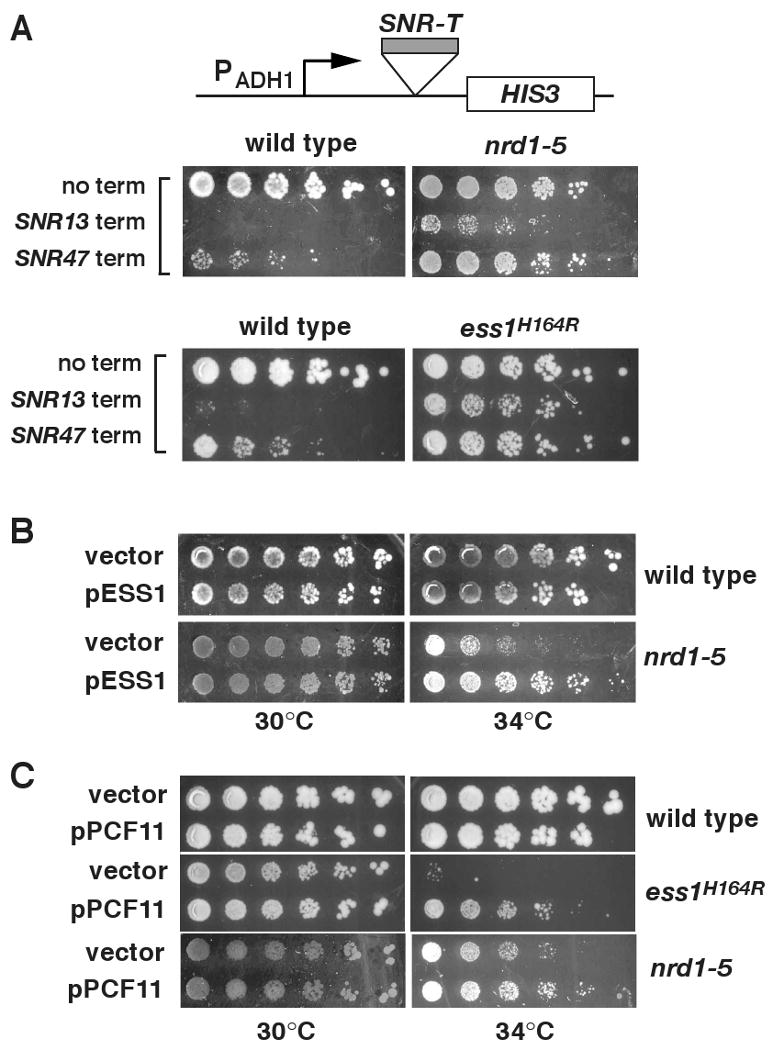
(A) SNR terminators are Ess1-dependent. Reporter plasmids with or without SNR13 or SNR47 terminators upstream of HIS3 were transformed into nrd1-5 or ess1H164R cells or their parent strains and tested for growth on CSM -his medium. Shown are 1:5 dilutions (starting at 1.2 ×106 cells) grown at 30°C for 4 days. (B) Overexpression of ESS1 from a high-copy plasmid suppresses the nrd1-5 ts growth defect at 34°C. Shown are 1:3 dilutions (starting at 1.4 ×106 cells) grown for 4 days. (C) Overexpression of PCF11 from a high-copy plasmid suppresses the ts-growth defects of ess1H164R and nrd1-5 at 34°C. Shown are 1:5 dilutions (starting at 1.2 ×106 cells) grown for 3 days.
We next examined genetic interactions between ESS1 and three genes in the Nrd1 pathway, NRD1, NAB3 and PCF11. Overexpression of Ess1 suppressed the ts-growth defect in a nrd1 mutant (Fig. 3B), whereas little or no suppression was observed in nab3 or pcf11 mutants (data not shown). Reciprocal experiments in which NRD1, NAB3 and PCF11 were overexpressed in ess1 mutant cells, showed that only PCF11 suppressed (Fig. 3C; data not shown). Together, the genetic results and the failure of ess1 mutants to terminate at SNR13 and SNR47 terminators, suggest that Ess1 functions in the Nrd1 pathway.
We next used ChIP to examine the recruitment of RNA pol II, Ess1, and Nrd1 to snoRNA loci in ess1 mutants. As expected, in ess1 mutants, levels of pol II in 3′ regions were elevated, consistent with transcription readthough (Fig. 4B,F,J). Localization of a TAP-tagged Ess1 in wild-type cells suggests a broad distribution of Ess1 along snoRNA genes, although detection levels were modest at best (Fig. 4C,G,K). Most revealing was the localization of Nrd1. In wild type cells, Nrd1 is recruited to the 3′ ends of genes, where it binds both the pol II CTD and the nascent RNA (Carroll et al., 2007; Vasiljeva et al., 2008). If Ess1 is important for Nrd1 recruitment, then Nrd1 localization should be lost in ess1 mutants, as observed in paf1 mutants (Sheldon et al., 2005), thus explaining the transcription readthrough. Surprisingly, we found the opposite, that in ess1 mutants, Nrd1 recruitment increased at the 3′ ends of snoRNA genes (Fig. 4C,G,K). Conversely, in cells that overexpress Ess1, Nrd1 recruitment decreased (Fig. S6B,D). These results suggest that Ess1 promotes release of Nrd1 from the CTD. In contrast, Ess1 promotes the binding of Pcf11 to the CTD, as indicated by ChIP data on a CUT locus that shows Pcf11 recruitment is diminished in ess1 mutants, (Fig. S6E-G; S. Buratowski, pers. comm.).
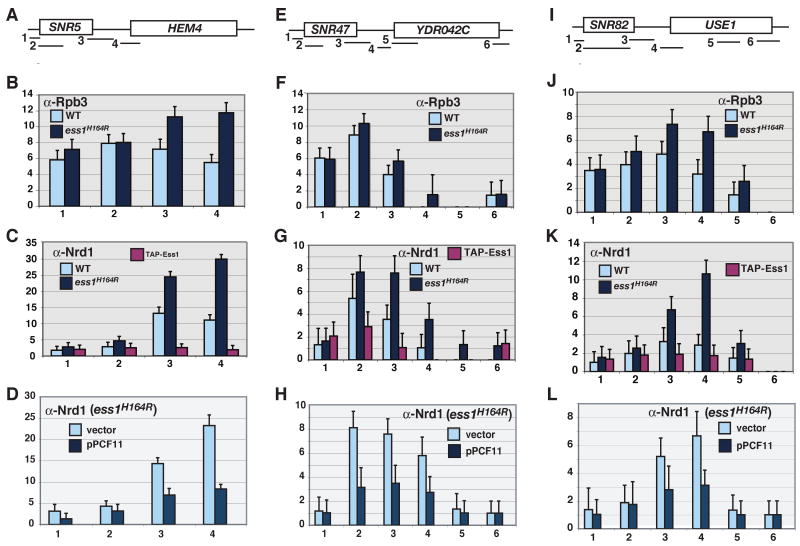
Figure 4. ChIP of Nrd1, Rpb3 and Ess1 at selected SNR loci.
Cells were grown at 30°C. Fold changes are relative to a chromosome V control. (A,E,I) Schematics showing locations of PCR products used for ChIP. (B,F,J) RNA pol II readthrough in ess1H164R mutant cells as detected by localization of the Rpb3 subunit. (C,G,K) Nrd1 localization at the 3′ ends of snoRNA genes is increased in ess1H164R mutant cells. Localization of Ess1 is also shown. (D,H,L) Overexpression of PCF11 in ess1H164R mutants inhibits Nrd1 recruitment. Error bars are standard deviations of 3-4 biological replicates.
Based on these results, we suggest that Ess1 is required for Nrd1-dependent termination; loss of Ess1 results in pol II readthrough, perhaps due to a failure to release Nrd1 and concomitant blocking of Pcf11 recruitment. Consistent with such a competition model, forced overexpression of Pcf11 reversed the effect on Nrd1 observed in ess1 mutants, reducing Nrd1 recruitment at snoRNA terminators (Fig. 4D,H, L). Importantly, the effects on Nrd1 recruitment to terminators were not likely due to changes in overall Nrd1 protein levels in mutant backgrounds. This was a concern since in ess1 mutants at 34°C, standard and tiling arrays showed a ∼1.5 and ∼2-fold increase in Nrd1 mRNA levels, respectively (Fig. S1A, Fig. S7E). However, this change is not likely to account for the larger changes in Nrd1 recruitment (3-4 fold) identified by ChIP (Fig. 4), since both ChIP and Western analysis were done at 30°C, where there is no detectable change in Nrd1 protein levels (Fig. 5A, bottom panel; Fig. 5C).
Figure 5. Ess1 promotes de-phosphorylation of the CTD at Ser5.
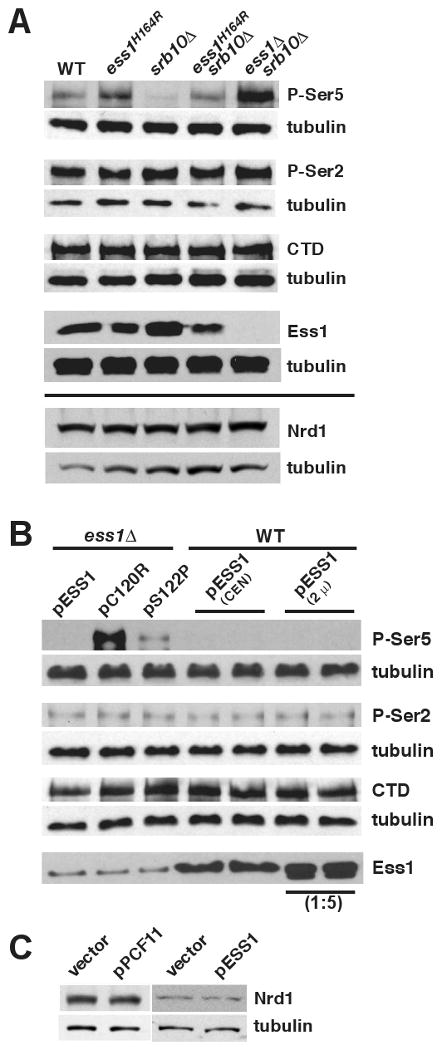
(A) Whole cell extracts from wild-type cells (WT; W303-1a) or the indicated mutant strains (see Table S4) were analyzed by Western blotting using antibodies specific for the P-Ser5 (H14), P-Ser2 (H5), or hypophosphorylated forms of the CTD (8WG16), or a tubulin control (Abcam). Bottom panel, Nrd1 levels in same strains. (B) Extracts from ess1Δ or wild-type cells bearing the indicated plasmids were analyzed by Western blotting as in (A). pC120R and pS122P (CEN) plasmids encode catalytic mutants of Ess1 (see text for details). Ess1 was detected with anti-Ess1 antibodies. A 1:5 dilution of cell extract was used for the 2μ-Ess1 overexpression samples. In both (A) and (B) independent blots were done for each antibody (P-Ser5, P-Ser2, CTD, Ess1) along with an anti-tubulin control, using fractions from the same protein extracts. (C) Nrd1 levels in strains overexpressing PCF11 or ESS1.
If Ess1 functions in the Nrd1 pathway, then we would expect the expression of genes altered in Nrd1-pathway mutants to be altered in ess1 mutants. Indeed, we find a significant number of genes whose expression increases in both ess1 and nab3 mutants (P=2.4e-18; Fig. S1E). Comparable nrd1 array data were not available.
Ess1 controls levels of CTD-Ser5 phosphorylation in vivo
How might Ess1 affect Nrd1 release from the CTD in terminator regions? Our microarray results indicate that Ess1 and Ssu72, a Ser5-specific CTD phosphatase, share common target genes (Table S1B). It is also known that Nrd1 preferentially binds the CTD phosphorylated at Ser5 (Vasiljeva et al., 2008)) and that Nrd1-dependent termination is sensitive to the ratio of P-Ser5 and P-Ser2 levels (Gudipati, et al., 2008). Therefore, one way in which Ess1 might control the Nrd1 pathway is through effects on phosphorylation of the CTD. As shown in Figure 5A, ess1 mutants showed an increase in levels of P-Ser5, but no change in levels of P-Ser2. Two alleles were used in this experiment, a ts-allele (ess1H164R), and a null allele (ess1Δ), where viability is maintained by the presence of a suppressor mutation, srb10Δ. The control strain (srb10Δ) showed no increase in P-Ser5 levels relative to the wild type.
To confirm these results, we carried out the reciprocal experiments. Wild-type Ess1 or catalytic mutants were overexpressed in wild-type or ess1Δ backgrounds (Fig. 5B). Overexpression of wild-type Ess1 reduced levels of P-Ser5, as expected, while overexpression of a catalytic mutant (C120R), whose activity is down ∼10,000-fold (Gemmill et al., 2005), did not. An intermediate allele (S122P) whose catalytic activity is down ∼5-fold showed an intermediate level of P-Ser5. No effects were detected on levels of P-Ser2. These results indicate that Ess1, a prolyl isomerase, has a strong influence on the phosphorylation state of the pol II CTD in vivo. Specifically, Ess1 promotes dephosphorylation at Ser5-Pro6, which can explain its effect on Nrd1 localization.
Ess1 is required for termination of CUTs and upstream regulatory RNAs
CUTs are observed throughout the genome, and are detected most readily when Nrd1-dependent termination and/or nuclear exosome function is compromised, such as in nrd1, nab3, sen1 and rrp6 mutants. To test whether Ess1 is required for CUT processing, we examined the expression of several previously identified CUTs in ess1 mutants. Among these, we selected GRE1, which has a large upstream CUT, and NMR026W. Both of these CUTs showed increased expression in microarray analysis of ess1 mutants. As controls we chose NGR060W and NEL025C, whose expression did not change appreciably in ess1 mutants. Indeed, qRT-PCR shows that levels of GRE1 and NMR026W increased significantly, whereas levels of NGR060W and NEL025C did not (Fig. 6A). GRE1 readthrough transcripts were detected by Northern analysis, as was a stabilized NMR026W transcript (Fig. 6B).
Figure 6. Ess1 mutants reveal potential CUTs and upstream regulatory RNAs.
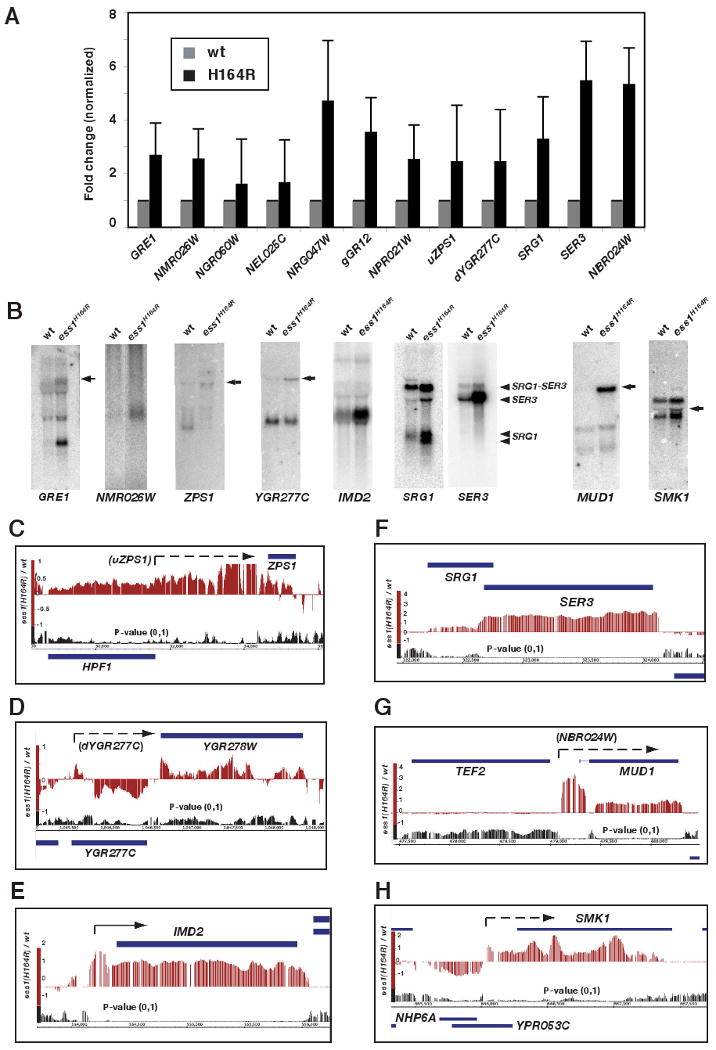
(A) Quantitative real-time PCR analysis of potential CUT and uRNA loci in ess1H164R mutant cells. All data were normalized to expression of the ACT1 gene, and wild-type was set equal to one. Error bars are standard deviations from the mean log values. (B) Northern analysis reveals CUTs and uRNA readthrough products in ess1H164R mutants. Probes are indicated, and arrows identify fusion transcripts. In the case of SRG1, the two transcripts previously identified (Martens and Winston, 2004) are indicated. A SRG1-SER3 fusion transcript is also identified. (C-H) Tiling array data reveal patterns of CUT and uRNA expression and effects on neighboring genes (see text for details). Dashed arrows indicate positions of predicted CUTs, solid arrows represent previously annotated CUTs.
To identify potential new CUTs that are stabilized in ess1 mutants, we used two approaches. First, we chose transcripts whose levels increased in expression arrays of ess1H164R mutants at restrictive temperature, and in ess1H164R shut-off cells (Table S1A). We excluded annotated ORFs, snoRNAs, and regions downstream of snoRNAs, and focused on transcripts that overlapped SAGE tags (∼200 transcripts; Velculescu et al., 1997), which identify potential CUTs and indicate the direction of transcription (Wyers et al., 2005). For the top 50 non-annotated transcripts, we examined tiling array data and found examples of increased signals located 5′, 3′ and within nearby ORFs in both the sense and antisense orientations. We selected SAGE tags, NGR047W, gGR12, and NPR021W for further analysis, and found they are elevated in ess1 mutant cells (at 34°C) as detected by qRT-PCR, with the largest increases in NGR047W and gGR12 (Fig. 6A). Tiling data suggest that these are CUTs, fail to terminate correctly, and become stabilized (Fig. S7A,B).
In a second approach, we analyzed ORFs whose expression increases or decreases in ess1 mutants, but which do not have nearby snoRNAs. For example, tiling array profiles (Fig. 6C,D) and qRT-PCR (Fig. 6A) identify potential CUT readthrough from upstream of ZPS1 (uZPS) and downstream of YGR277C (dYGR277C). As predicted, we detected readthrough transcripts by Northern analysis (Fig. 6B).
Several CUT-like RNAs have been discovered that overlap the transcription start sites (TSS) of protein coding genes, and downregulate the adjacent ORFs (Kuehner and Brow, 2008; Martens et al., 2004; Thiebaut et al., 2008). These RNAs are neither cryptic nor unstable, but are short, non-coding, and exert powerful regulatory effects on neighboring genes (Corden, 2008; Dichtl, 2008). We found that termination of several of these depend upon Ess1. For example, in ess1 mutants, upstream RNAs read through into IMD2 and SER3 (Fig. 6E,F), resulting in their upregulation (SER3 is normally downregulated by SRG1; Martens et al., 2004). Readthrough is also detected into the HRP1, IMD3, NRD1, and URA8 genes (Fig. S7C-F). Increased expression was confirmed for IMD2, SRG1, and SER3 (Fig 6A,B). We discovered additional genes, including MUD1 (NBR024W) and SMK1, that show increased expression in ess1 mutants (Table S1C), have increased 5′ transcription (Fig. 6A,G,H), and show readthrough transcripts by Northern analysis (Fig. 6B), all of which is consistent with regulation by newly-identified upstream RNAs.
To determine how widespread the aberrant transcription of potential CUTs and uRNAs is in ess1 mutants, we analyzed tiling array data for increased signal in the vicinity (+/-300 bp) of the transcription start sites (TSS) for 4,367 genes (Nagalakshmi et al., 2008). Our analysis revealed an overall increase in signal 5′ to the TSS in ess1 mutants (Fig. S2C), which was due to aberrant 5′ transcription in a subset of the 4,367 genes analyzed (Fig. S4B). Among the top 100 genes (Table S3), many encode ribosomal proteins and other highly expressed proteins, and are known to be nucleosome-depleted in their 5′ upstream regions (Lee et al., 2007; Xu et al., 2009). Many of the 5′ transcripts are CUTs or SUTs previously identified in rrp6 mutant backgrounds (Neil et al., 2009; Xu et al., 2009). Bidirectional CUTs (Neil et al., 2009) are also identified (e.g. Fig. S5J). Interestingly, tiling array profiles for each of the top 100 genes (summarized in Table S3) indicate that most contain upstream transcripts not previously identified (e.g. Fig. S5F-I). Thus, tiling analysis of ess1 mutants has revealed a new set of genes that are potentially regulated by upstream regulatory RNAs.
Discussion
Results presented here show that the Ess1 prolyl isomerase is necessary for regulating expression of short non-coding RNAs, including snoRNAs, CUTs, SUTs and potential regulatory RNAs. Regulation occurs via the Nrd1 pathway of transcription termination and likely reflects the activity of Ess1 on its major yeast substrate, the phosphorylated CTD of RNA polymerase II.
Gene-specfic requirement for Ess1 function
Prior to this work, it was not clear whether the Ess1 (or Pin1) prolyl isomerase was necessary for the expression of all pol II-transcribed genes, or just a subset of them (Wu et al., 2000; Xu et al., 2003; Krishnamurthy et al., 2009). Here, using two experimental regimes, we found that expression of only ∼10% of genes are significantly affected in ess1 mutants under standard laboratory conditions. These changes were largely the result of transcription termination defects that resulted in stabilization of small non-coding RNAs, although some mRNAs were also affected. Our results indicate that Ess1 functions mainly in the Nrd1 termination pathway rather than the mRNA termination pathway.
A mechanism for Ess1 function in Nrd1-dependent termination
We propose the following model for Ess1 function in termination of small non-coding RNAs via the Nrd1 pathway (Fig. 7). Ess1 binds and isomerizes P-Ser5-Pro6 bonds in the pol II CTD causing a conformational change (left and middle panels). The trans isomer, favored by some phosphatases (Zhou et al., 2000), would be targeted by Ssu72, resulting in dephosphorylation at Ser5, thus releasing Nrd1 (in complex with Nab3). Nrd1 release would then allow binding by Pcf11 to adjacent P-Ser2 sites (right panel) so that termination can occur. The mechanism suggests an ordered competition between Nrd1 and Pcf11 for binding to the CTD, and that the proper sequence of binding (Nrd1 must bind first) is mediated by Ess1, which is required for termination.
Figure 7. Model for the role of Ess1 in Nrd1-dependent termination.
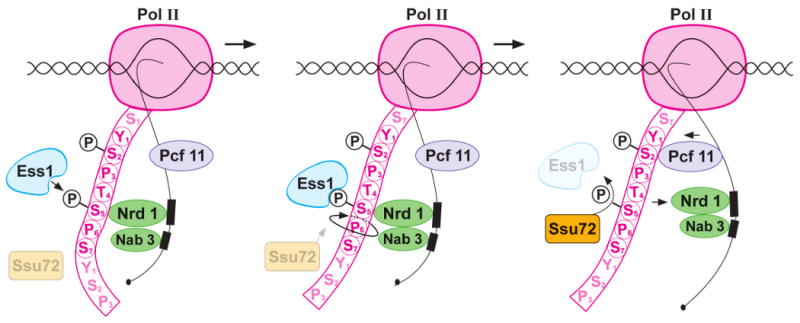
RNA polymerase II is depicted transcribing into a Nrd1-dependent terminator. Only one complete CTD repeat (YSPTSPS) is shown for simplicity. Ess1 targets P-Ser5-Pro6 (left panel), where Nrd1 binds in a complex with Nab3. Ess1 catalyzes a cis/trans isomerization, changing the conformation of the CTD (middle panel), making it a better substrate (e.g. trans form) for the P-Ser5-specific phosphatase, Ssu72. Ssu72 dephosphorylates P-Ser5, dissociating Nrd1/Nab3 from the CTD, thus allowing Pcf11 to bind adjacent P-Ser2 sites (right panel). The model predicts a competition between Nrd1 and Pcf11 for CTD binding that is controlled by Ess1.
A summary of the supporting data is as follows. First, Ess1 binds the CTD and preferentially targets P-Ser5-Pro6 (Wu et al, 2000; Wilcox et al., 2004; Gemmill et al., 2005). Here we showed that Ess1 is required for termination of most small ncRNAs as well as some mRNAs (Figs. 1,2,6). We found that Ess1 was required for the function of Nrd1-dependent terminators (Fig. 3A), and that Ess1 promoted the release of Nrd1 from terminator regions (Fig. 4C,G,K; Fig. S6B,D,F). In contrast, Ess1 promoted the recruitment of Pcf11 to terminators (Fig. S6G; S. Buratowski, unpublished). Consistent with a competition model, forced overexpression of Pcf11 compensated for ess1 defects genetically (Fig. 3C), and it reduced the abnormally high levels of Nrd1 found at terminators in ess1 mutant cells (Fig. 4D,H,L).
Regulation of Nrd1 association with the pol II CTD by Ess1 is probably the result of Ess1-dependent stimulation of de-phosphorylation of P-Ser5-Pro6 (Fig. 5). De-phosphorylation is likely carried out by Ssu72, consistent with our microarray results, which revealed a common set of target genes shared by Ess1 and Ssu72 (Fig. S1, Table S1B), and supported by the finding that overexpression of Ssu72 suppresses ess1 mutations (Krishnamurthy et al., 2009; Wu and Hanes, unpublished data). In the absence of Ess1 activity, P-Ser5 levels increased, whereas P-Ser2 levels were unchanged (Fig. 5). This altered ratio favored Nrd1 binding, which prefers P-Ser5 (Vasiljeva et al., 2008), but reduced the binding of Pcf11, which prefers P-Ser2 (Licatalosi et al., 2002).
Nrd1-dependent termination occurs on short pol II-transcribed genes because they are enriched for the P-Ser5 form of the CTD (Gudipati et al., 2008). On longer genes, P-Ser5 decreases toward the 3′ end while P-Ser2 increases, leading to termination by the standard mRNA pathway. Since Ess1 preferentially targets P-Ser5-Pro6, this would explain why its primary effects are on the Nrd1-dependent termination pathway and why all genes do not have termination defects in ess1 mutants (i.e. they are not Nrd1-dependent).
The identification of additional non-coding RNAs
Our genome-wide analysis of ess1 mutants identified hundreds of small non-coding RNA transcripts that were stabilized as a result transcription readthrough and/or defects in RNA processing (Tables S2 & S3). Although many of these CUTs and other small RNAs were in common with those identified in studies of other genes in the Nrd1 pathway, including nrd1, nab3, ssu72, rrp6, and sen1, the sets were not identical (Fig. S7G). That is, each mutation, including ess1, revealed a different spectrum of small, non-coding RNA transcripts of the so-called hidden transcriptome, perhaps reflecting a differing level of importance of each factor at a given locus. Thus, our comprehensive bioinformatic analysis identified a new set of “ess1-specific” genes that are potentially regulated by 5′ or 3′ non-coding transcripts (sense and anti-sense). Further studies will be needed to determine possible the regulatory mechanisms that function at each locus.
Ess1 is a key player in interpreting the CTD code
Genetic studies suggested roles for Ess1 in transcription initiation, elongation and termination (Hani, et al., 1995; Krishnamurthy et al., 2009; Wilcox et al., 2004; Wu et al., 2003; Wu et al., 2000). Here, we show, for the first time, a role for Ess1 in the termination of non-coding RNAs via the Nrd1 pathway. How can Ess1 function at so many key steps of transcription? An emerging theme is that Ess1 (and Pin1), in its capacity as a prolyl cis/trans isomerase, induces conformational changes in the CTD that regulate binding of multiple cofactors (e.g. Nrd1 and Pcf11) to the RNA pol II complex. Results shown here provide the first mechanistic evidence for this type of regulation. Taken together with genetics studies, our results demonstrate that Ess1 functions along with the known CTD-specific kinases and phosphatases in controlling progression through the various stages of the transcription cycle, and is thus a key player in writing and interpreting the “CTD code.”
Experimental Procedures
Yeast Strains and Plasmids
Yeast strains are listed in Table S4. Plasmids obtained from other laboratories are described in Supplementary Experimental Procedures. For this study, pRS424-PCF11 (2μ, TRP1) was made by insertion of a BamH1-Sac1 fragment from pRS415-PCF11 into the same sites of pRS424. pRS424-NAB3 was made by insertion of a 4.3 kb Nsi1 fragment from pNAB3.14 into the Pst1 site of pRS424.
RNA analysis
RNA was prepared and Northern analysis was performed using standard methods. RNA for RT-PCR and qRT-PCR was prepared using the Masterpure RNA purification method (Epicenter) followed by additional purification and concentration using RNeasy (Qiagen). cDNA was synthesized using random hexamers (Bioline) and used for real-time quantitation with SYBR green (USB) and an ABI Prism 7000 detection system.
Chromatin Immunoprecipitation
ChIP was performed by standard methods as described in the Supplement. Nrd1 antibody was a gift of D. Brow (U. Wisconsin) and Rpb3 antibody was from Neoclone.
Microarray analysis
Expression analysis was done using Affymetrix YG98S arrays, GeneSpring software (Agilent Technologies), and Microsoft Excel. Tiling analysis was done using Affymetrix Yeast 1.0 Tiling Microarrays, Tiling Array Software (TAS) and Integrated Genome Browser (IGB, version 5.12). Gene coordinates were based on the 10/2003 release of the S. cerevisiae genome. Details are available in the Supplement. Microarray data are available at the Gene Expression Omnibus (GSE17638).
Supplementary Material
Acknowledgments
We are grateful to David Brow, Steve Buratowski, and Jeff Corden for plasmids, strains and antibodies, to the Wadsworth Center Molecular Genetics and Microarray Cores and Media Facility, Stuart Lehrman in Illustrations, and to Steve Buratowski, Mike Hampsey and Joe Wade for helpful discussions and sharing of unpublished data. Pilot microarrays (not shown) were done with the help of Miguel Arévalo-Rodríguez and Joseph Heitman (Duke University). This work was funded by grants from the NIH (GM055108 to SDH), the NSF (MCB-0613001 to SDH) and the NSF-REU Program (DBI9987844) for support of C. Rabeler and O. Beane.
Footnotes
Supplemental Data: The supplemental data include Supplemental Experimental Procedures, five tables and seven figures.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aranda A, Proudfoot N. Transcriptional termination factors for RNA polymerase II in yeast. Mol Cell. 2001;7:1003–1011. doi: 10.1016/s1097-2765(01)00235-0. [DOI] [PubMed] [Google Scholar]
- Arigo JT, Eyler DE, Carroll KL, Corden JL. Termination of cryptic unstable transcripts is directed by yeast RNA-binding proteins Nrd1 and Nab3. Mol Cell. 2006;23:841–851. doi: 10.1016/j.molcel.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Barabino SM, Keller W. Last but not least: regulated poly(A) tail formation. Cell. 1999;99:9–11. doi: 10.1016/s0092-8674(00)80057-4. [DOI] [PubMed] [Google Scholar]
- Bird G, Zorio DA, Bentley DL. RNA polymerase II carboxy-terminal domain phosphorylation is required for cotranscriptional pre-mRNA splicing and 3′-end formation. Mol Cell Biol. 2004;24:8963–8969. doi: 10.1128/MCB.24.20.8963-8969.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birse CE, Minvielle-Sebastia L, Lee BA, Keller W, Proudfoot NJ. Coupling termination of transcription to messenger RNA maturation in yeast. Science. 1998;280:298–301. doi: 10.1126/science.280.5361.298. [DOI] [PubMed] [Google Scholar]
- Buratowski S. The CTD code. Nat Struct Biol. 2003;10:679–680. doi: 10.1038/nsb0903-679. [DOI] [PubMed] [Google Scholar]
- Buratowski S. Connections between mRNA 3′ end processing and transcription termination. Curr Op Cell Biol. 2005;17:257–261. doi: 10.1016/j.ceb.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Carroll KL, Ghirlando R, Ames JM, Corden JL. Interaction of yeast RNA-binding proteins Nrd1 and Nab3 with RNA polymerase II terminator elements. RNA. 2007;13:361–373. doi: 10.1261/rna.338407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll KL, Pradhan DA, Granek JA, Clarke ND, Corden JL. Identification of cis elements directing termination of yeast nonpolyadenylated snoRNA transcripts. Mol Cell Biol. 2004;24:6241–6252. doi: 10.1128/MCB.24.14.6241-6252.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman RD, Heidemann M, Albert TK, Mailhammer R, Flatley A, Meisterernst M, Kremmer E, Eick D. Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science. 2007;318:1780–1782. doi: 10.1126/science.1145977. [DOI] [PubMed] [Google Scholar]
- Cho EJ. RNA polymerase II carboxy-terminal domain with multiple connections. Exp & Mol Med. 2007;39:247–254. doi: 10.1038/emm.2007.28. [DOI] [PubMed] [Google Scholar]
- Corden JL. Yeast Pol II start-site selection: the long and the short of it. EMBO Reports. 2008;9:1084–1086. doi: 10.1038/embor.2008.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer P. Recent structural studies of RNA polymerases II and III. Bioch Soc Trans. 2006;34:1058–1061. doi: 10.1042/BST0341058. [DOI] [PubMed] [Google Scholar]
- David L, Huber W, Granovskaia M, Toedling J, Palm CJ, Bofkin L, Jones T, Davis RW, Steinmetz LM. A high-resolution map of transcription in the yeast genome. Proc Nat Acad Sci USA. 2006;103:5320–5325. doi: 10.1073/pnas.0601091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichtl B. Transcriptional ShortCUTs. Mol Cell. 2008;31:617–618. doi: 10.1016/j.molcel.2008.08.014. [DOI] [PubMed] [Google Scholar]
- Dichtl B, Blank D, Ohnacker M, Friedlein A, Roeder D, Langen H, Keller W. A role for SSU72 in balancing RNA polymerase II transcription elongation and termination. Mol Cell. 2002;10:1139–1150. doi: 10.1016/s1097-2765(02)00707-4. [DOI] [PubMed] [Google Scholar]
- Egloff S, Murphy S. Cracking the RNA polymerase II CTD code. Trends Genet. 2008;24:280–288. doi: 10.1016/j.tig.2008.03.008. [DOI] [PubMed] [Google Scholar]
- Egloff S, O'Reilly D, Chapman RD, Taylor A, Tanzhaus K, Pitts L, Eick D, Murphy S. Serine-7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science. 2007;318:1777–1779. doi: 10.1126/science.1145989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimori F, Takahashi K, Uchida C, Uchida T. Mice lacking Pin1 develop normally, but are defective in entering cell cycle from G(0) arrest. Biochem Biophys Res Commun. 1999;265:658–663. doi: 10.1006/bbrc.1999.1736. [DOI] [PubMed] [Google Scholar]
- Ganem C, Devaux F, Torchet C, Jacq C, Quevillon-Cheruel S, Labesse G, Facca C, Faye G. Ssu72 is a phosphatase essential for transcription termination of snoRNAs and specific mRNAs in yeast. EMBO J. 2003;22:1588–1598. doi: 10.1093/emboj/cdg141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemmill TR, Wu X, Hanes SD. Vanishingly low levels of Ess1 prolyl-isomerase activity are sufficient for growth in Saccharomyces cerevisiae. J Biol Chem. 2005;280:15510–15517. doi: 10.1074/jbc.M412172200. [DOI] [PubMed] [Google Scholar]
- Gudipati RK, Villa T, Boulay J, Libri D. Phosphorylation of the RNA polymerase II C-terminal domain dictates transcription termination choice. Nat Struct & Mol Biol. 2008;15:786–794. doi: 10.1038/nsmb.1460. [DOI] [PubMed] [Google Scholar]
- Hanes SD, Shank PR, Bostian KA. Sequence and mutational analysis of ESS1, a gene essential for growth in Saccharomyces cerevisiae. Yeast. 1989;5:55–72. doi: 10.1002/yea.320050108. [DOI] [PubMed] [Google Scholar]
- Hani J, Stumpf G, Domdey H. PTF1 encodes an essential protein in Saccharomyces cerevisiae, which shows strong homology with a new putative family of PPIases. FEBS Let. 1995;365:198–202. doi: 10.1016/0014-5793(95)00471-k. [DOI] [PubMed] [Google Scholar]
- Hirose Y, Ohkuma Y. Phosphorylation of the C-terminal domain of RNA polymerase II plays central roles in the integrated events of eucaryotic gene expression. J Biochem. 2007;141:601–608. doi: 10.1093/jb/mvm090. [DOI] [PubMed] [Google Scholar]
- Joseph JD, Yeh ES, Swenson KI, Means AR, Winkler The peptidyl-prolyl isomerase Pin1. Prog Cell Cycle Res. 2003;5:477–487. [PubMed] [Google Scholar]
- Keogh MC, Podolny V, Buratowski S. Bur1 kinase is required for efficient transcription elongation by RNA polymerase II. Mol Cell Biol. 2003;23:7005–7018. doi: 10.1128/MCB.23.19.7005-7018.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Krogan NJ, Vasiljeva L, Rando OJ, Nedea E, Greenblatt JF, Buratowski S. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature. 2004;432:517–522. doi: 10.1038/nature03041. [DOI] [PubMed] [Google Scholar]
- Kim M, Vasiljeva L, Rando OJ, Zhelkovsky A, Moore C, Buratowski S. Distinct pathways for snoRNA and mRNA termination. Mol Cell. 2006;24:723–734. doi: 10.1016/j.molcel.2006.11.011. [DOI] [PubMed] [Google Scholar]
- Kobor MS, Greenblatt J. Regulation of transcription elongation by phosphorylation. Biochim Biophys Acta. 2002;1577:261–275. doi: 10.1016/s0167-4781(02)00457-8. [DOI] [PubMed] [Google Scholar]
- Krishnamurthy S, He X, Reyes-Reyes M, Moore C, Hampsey M. Ssu72 Is an RNA polymerase II CTD phosphatase. Mol Cell. 2004;14:387–394. doi: 10.1016/s1097-2765(04)00235-7. [DOI] [PubMed] [Google Scholar]
- Krishnamurthy S, Ghazy MA, Moore C, Hampsey M. Functional Interaction of the Ess1 Prolyl Isomerase with Components of the RNA Polymerase II Initiation and Termination Machineries. Mol Cell Biol. 2009;29:2925–2934. doi: 10.1128/MCB.01655-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehner JN, Brow DA. Regulation of a eukaryotic gene by GTP-dependent start site selection and transcription attenuation. Mol Cell. 2008;31:201–211. doi: 10.1016/j.molcel.2008.05.018. [DOI] [PubMed] [Google Scholar]
- Lee W, Tillo D, Bray N, Morse RH, Davis RW, Hughes TR, Nislow C. A high-resolution atlas of nucleosome occupancy in yeast. Nat Genet. 2007;39:1235–1244. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- Licatalosi DD, Geiger G, Minet M, Schroeder S, Cilli K, McNeil JB, Bentley DL. Functional interaction of yeast pre-mRNA 3′ end processing factors with RNA polymerase II. Mol Cell. 2002;9:1101–1111. doi: 10.1016/s1097-2765(02)00518-x. [DOI] [PubMed] [Google Scholar]
- Liou YC, Ryo A, Huang HK, Lu PJ, Bronson R, Fujimori F, Uchida T, Hunter T, Lu KP. Loss of Pin1 function in the mouse causes phenotypes resembling cyclin D1-null phenotypes. Proc Nat Acad Sci USA. 2002;99:1335–1340. doi: 10.1073/pnas.032404099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu KP, Hanes SD, Hunter T. A human peptidyl-prolyl isomerase essential for regulation of mitosis. Nature. 1996;380:544–547. doi: 10.1038/380544a0. [DOI] [PubMed] [Google Scholar]
- Lu KP, Zhou XZ. The prolyl isomerase PIN1: a pivotal new twist in phosphorylation signalling and disease. Nat Rev Mol Cell Biol. 2007;8:904–916. doi: 10.1038/nrm2261. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen S, Jensen TH. Overlapping pathways dictate termination of RNA polymerase II transcription. Biochimie. 2007;89:1177–1182. doi: 10.1016/j.biochi.2007.05.007. [DOI] [PubMed] [Google Scholar]
- Martens JA, Laprade L, Winston F. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature. 2004;429:571–574. doi: 10.1038/nature02538. [DOI] [PubMed] [Google Scholar]
- McCracken S, Fong N, Rosonina E, Yankulov K, Brothers G, Siderovski D, Hessel A, Foster S, Shuman S, Bentley DL. 5′-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 1997;11:3306–3318. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinhart A, Kamenski T, Hoeppner S, Baumli S, Cramer P. A structural perspective of CTD function. Genes Dev. 2005;19:1401–1415. doi: 10.1101/gad.1318105. [DOI] [PubMed] [Google Scholar]
- Morris DP, Phatnani HP, Greenleaf AL. Phospho-carboxyl-terminal domain binding and the role of a prolyl isomerase in pre-mRNA 3′-End formation. J Biol Chem. 1999;274:31583–31587. doi: 10.1074/jbc.274.44.31583. [DOI] [PubMed] [Google Scholar]
- Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedea E, He X, Kim M, Pootoolal J, Zhong G, Canadien V, Hughes T, Buratowski S, Moore CL, Greenblatt J. Organization and function of APT, a subcomplex of the yeast cleavage and polyadenylation factor involved in the formation of mRNA and small nucleolar RNA 3′-ends. J Biol Chem. 2003;278:33000–33010. doi: 10.1074/jbc.M304454200. [DOI] [PubMed] [Google Scholar]
- Neil H, Malabat C, d'Aubenton-Carafa Y, Xu Z, Steinmetz LM, Jacquier A. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature. 2009;457:1038–1042. doi: 10.1038/nature07747. [DOI] [PubMed] [Google Scholar]
- Palancade B, Bensaude O. Investigating RNA polymerase II carboxyl-terminal domain (CTD) phosphorylation. E J Biochem FEBS. 2003;270:3859–3870. doi: 10.1046/j.1432-1033.2003.03794.x. [DOI] [PubMed] [Google Scholar]
- Pandit S, Wang D, Fu XD. Functional integration of transcriptional and RNA processing machineries. Curr Opin Cell Biol. 2008;20:260–265. doi: 10.1016/j.ceb.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patturajan M, Schulte RJ, Sefton BM, Berezney R, Vincent M, Bensaude O, Warren SL, Corden JL. Growth-related changes in phosphorylation of yeast RNA polymerase II. J Biol Chem. 1998;273:4689–4694. doi: 10.1074/jbc.273.8.4689. [DOI] [PubMed] [Google Scholar]
- Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- Rondon AG, Mischo HE, Proudfoot NJ. Terminating transcription in yeast: whether to be a ‘nerd’ or a ‘rat’. Nat Struct Mol Biol. 2008;15:775–776. doi: 10.1038/nsmb0808-775. [DOI] [PubMed] [Google Scholar]
- Schroeder SC, Schwer B, Shuman S, Bentley D. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev. 2000;14:2435–2440. doi: 10.1101/gad.836300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon KE, Mauger DM, Arndt KM. A Requirement for the Saccharomyces cerevisiae Paf1 complex in snoRNA 3′ end formation. Mol Cell. 2005;20:225–236. doi: 10.1016/j.molcel.2005.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz EJ, Brow DA. Repression of gene expression by an exogenous sequence element acting in concert with a heterogeneous nuclear ribonucleoprotein-like protein, Nrd1, and the putative helicase Sen1. Mol Cell Biol. 1996;16:6993–7003. doi: 10.1128/mcb.16.12.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz EJ, Brow DA. Ssu72 protein mediates both poly(A)-coupled and poly(A)-independent termination of RNA polymerase II transcription. Mol Cell Biol. 2003;23:6339–6349. doi: 10.1128/MCB.23.18.6339-6349.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz EJ, Conrad NK, Brow DA, Corden JL. RNA-binding protein Nrd1 directs poly(A)-independent 3′-end formation of RNA polymerase II transcripts. Nature. 2001;413:327–331. doi: 10.1038/35095090. [DOI] [PubMed] [Google Scholar]
- Sun ZW, Hampsey M. Synthetic enhancement of a TFIIB defect by a mutation in SSU72, an essential yeast gene encoding a novel protein that affects transcription start site selection in vivo. Mol Cell Biol. 1996;16:1557–1566. doi: 10.1128/mcb.16.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiebaut M, Colin J, Neil H, Jacquier A, Seraphin B, Lacroute F, Libri D. Futile cycle of transcription initiation and termination modulates the response to nucleotide shortage in S. cerevisiae. Mol Cell. 2008;31:671–682. doi: 10.1016/j.molcel.2008.08.010. [DOI] [PubMed] [Google Scholar]
- Thiebaut M, Kisseleva-Romanova E, Rougemaille M, Boulay J, Libri D. Transcription termination and nuclear degradation of cryptic unstable transcripts: a role for the nrd1-nab3 pathway in genome surveillance. Mol Cell. 2006;23:853–864. doi: 10.1016/j.molcel.2006.07.029. [DOI] [PubMed] [Google Scholar]
- Vasiljeva L, Buratowski S. Nrd1 interacts with the nuclear exosome for 3′ processing of RNA polymerase II transcripts. Mol Cell. 2006;21:239–248. doi: 10.1016/j.molcel.2005.11.028. [DOI] [PubMed] [Google Scholar]
- Vasiljeva L, Kim M, Mutschler H, Buratowski S, Meinhart A. The Nrd1-Nab3-Sen1 termination complex interacts with the Ser5-phosphorylated RNA polymerase II C-terminal domain. Nat Struct Mol Biol. 2008;15:795–804. doi: 10.1038/nsmb.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velculescu VE, Zhang L, Zhou W, Vogelstein J, Basrai MA, Bassett DE, Jr, Hieter P, Vogelstein B, Kinzler KW. Characterization of the yeast transcriptome. Cell. 1997;88:243–251. doi: 10.1016/s0092-8674(00)81845-0. [DOI] [PubMed] [Google Scholar]
- Verdecia MA, Bowman ME, Lu KP, Hunter T, Noel JP. Structural basis for phosphoserine-proline recognition by group IV WW domains. Nat Struct Biol. 2000;7:639–643. doi: 10.1038/77929. [DOI] [PubMed] [Google Scholar]
- Wilcox CB, Rossettini A, Hanes SD. Genetic interactions with C-terminal domain (CTD) kinases and the CTD of RNA Pol II suggest a role for ESS1 in transcription initiation and elongation in Saccharomyces cerevisiae. Genetics. 2004;167:93–105. doi: 10.1534/genetics.167.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Chang A, Sudol M, Hanes SD. Genetic interactions between the ESS1 prolyl-isomerase and the RSP5 ubiquitin ligase reveal opposing effects on RNA polymerase II function. Curr Genet. 2001;40:234–242. doi: 10.1007/s00294-001-0257-8. [DOI] [PubMed] [Google Scholar]
- Wu X, Rossettini A, Hanes SD. The ESS1 prolyl isomerase and its suppressor BYE1 interact with RNA pol II to inhibit transcription elongation in Saccharomyces cerevisiae. Genetics. 2003;165:1687–1702. doi: 10.1093/genetics/165.4.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Wilcox CB, Devasahayam G, Hackett RL, Arevalo-Rodriguez M, Cardenas ME, Heitman J, Hanes SD. The Ess1 prolyl isomerase is linked to chromatin remodeling complexes and the general transcription machinery. EMBO J. 2000;19:3727–3738. doi: 10.1093/emboj/19.14.3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Regnault B, Devaux F, Namane A, Seraphin B, et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- Xu YX, Hirose Y, Zhou XZ, Lu KP, Manley JL. Pin1 modulates the structure and function of human RNA polymerase II. Genes Dev. 2003;17:2765–2776. doi: 10.1101/gad.1135503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Munster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–1037. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe MB, Schutkowski M, Shen M, Zhou XZ, Stukenberg PT, Rahfeld JU, Xu J, Kuang J, Kirschner MW, Fischer G, et al. Sequence-specific and phosphorylation-dependent proline isomerization: a potential mitotic regulatory mechanism. Science. 1997;278:1957–1960. doi: 10.1126/science.278.5345.1957. [DOI] [PubMed] [Google Scholar]
- Yeh ES, Means AR. PIN1, the cell cycle and cancer. Nat Rev Cancer. 2007;7:381–388. doi: 10.1038/nrc2107. [DOI] [PubMed] [Google Scholar]
- Yuryev A, Corden JL. Suppression analysis reveals a functional difference between the serines in positions two and five in the consensus sequence of the C-terminal domain of yeast RNA polymerase II. Genetics. 1996;143:661–671. doi: 10.1093/genetics/143.2.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Fussel S, Reimer U, Schutkowski M, Fischer G. Substrate-based design of reversible Pin1 inhibitors. Biochem. 2002;41:11868–11877. doi: 10.1021/bi0262395. [DOI] [PubMed] [Google Scholar]
- Zhou XZ, Kops O, Werner A, Lu PJ, Shen M, Stoller G, Kullertz G, Stark M, Fischer G, Lu KP. Pin1-dependent prolyl isomerization regulates dephosphorylation of Cdc25C and tau proteins. Mol Cell. 2000;6:873–883. doi: 10.1016/s1097-2765(05)00083-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.