Summary
Development of the vertebrate pancreas is a complex stepwise process comprising regionalization, cell differentiation, and morphogenesis. Studies in zebrafish are contributing to an emerging picture of pancreas development in which extrinsic signaling molecules influence intrinsic transcriptional programs to allow ultimate differentiation of specific pancreatic cell types. Zebrafish experiments have revealed roles for several signaling molecules in aspects of this process; for example our own work has shown that Retinoic Acid signals specify the pre-pancreatic endoderm. Time-lapse imaging of live zebrafish embryos has started to provide detailed information about early pancreas morphogenesis. In addition to modeling embryonic development, the zebrafish has recently begun to be used as a model for pancreas regeneration studies. Here we review the significant progress in these areas and consider the future potential of zebrafish as a diabetes research model.
1. Introduction
Studies of the development, function and regeneration of the pancreas are not only relevant to our basic understanding of this critical organ, but also have the potential to facilitate dramatic improvements in the lives of diabetic patients. While diabetes can be effectively managed with insulin therapy, this is far from providing a cure. In an ideal situation the diseased pancreatic islets of diabetic patients would be replaced by healthy functional cells. The success of the Edmonton protocol(1,2), where cadaver-derived islets were transplanted into diabetic patients, shows that this is a feasible therapeutic avenue. However, such transplantation efforts are limited by the availability of harvested islets, encouraging major efforts to derive functional islets from stem cells or to induce regeneration of pancreatic cells. This is where developmental biology intersects with both diabetes and stem cell research. For example, we may be able to learn from the embryo, which has fine-tuned the process of building a pancreas over millions of years of evolution, how to coax undifferentiated embryonic stem cells into the differentiated cell-types that make a functional pancreas.
Much of the effort to understand pancreatic development has made use of the mouse as a model system. The mouse has several advantages for such studies: as a mammal it shares many characteristics with humans, and the availability of well-established molecular genetic tools allows gene function to be tested directly using homologous recombination to generate knock-out or conditional knock-out lines of mice. Such knock-out approaches have been particularly useful in establishing the hierarchy of transcription factors that specify and differentiate pancreatic cell types (reviewed in(3)). However, other model systems each offer their own particular advantages, and here we focus on the unique contributions that the zebrafish model is making to our understanding of pancreas development and function. The accessibility and optical clarity of zebrafish embryos allows cell manipulation and sophisticated in vivo imaging approaches that would be far more difficult in a mammal. We highlight how such experimental approaches have been coupled with forward and reverse genetics to provide new insights into various aspects of pancreas biology, including the role of cell signaling in pancreas development, and the processes of morphogenesis and regeneration.
2. Pancreas structure
The zebrafish pancreas shares a basic structure and cellular makeup with the mammalian pancreas, implying that the developmental lessons we learn in zebrafish will be broadly applicable. The pancreas, like the rest of the digestive tract and associated organs, develops from the endoderm germ layer(4), and the basic organization of the digestive tract is conserved from mammals to fish (Fig. 1). In both mammals and zebrafish the pancreas consists of an exocrine compartment (the acinar cells), which produces digestive enzymes that are secreted into a ductal system that transports these enzymes to the digestive tract, and an endocrine compartment (the islets), which is critical for blood sugar homeostasis. The pancreatic islet consists of a central core of insulin-producing β-cells, surrounded by, in fish as in mammals, glucagon-secreting β-cells, somatostatin producing β-cells, and ghrelin-producing ε-cells. Whether pancreatic polypeptide producing PP-cells also make up part of the zebrafish islet remains unclear, although PP cells have been described in a number of teleosts(5). During zebrafish embryonic development a single primary islet forms initially, with α-, β- and δ-cell types already present (as assessed by hormone gene expression) within the first day of development(6,7). By the adult stage several secondary islets and some scattered insulin-positive cells can also be recognized(8,9), again similar to the mammalian pancreas. The molecular markers available for differentiated zebrafish pancreas cell types and their progenitors are summarized in Table 1.
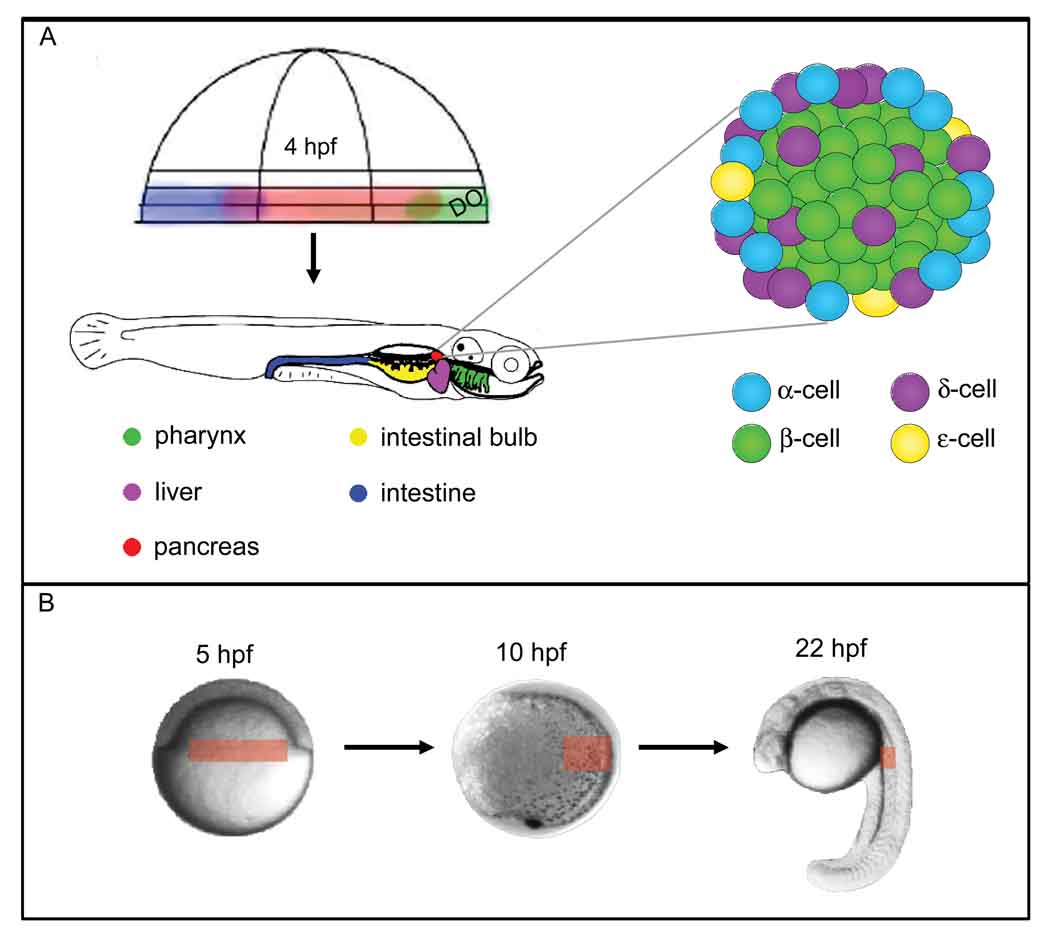
Figure 1. Organization of the zebrafish digestive tract and pancreatic islet.
A) Fate mapping from 4 hpf shows that anterior endodermal organs derive from precursors close to the dorsal organizer whereas posterior organs derive from more ventral locations(4). The islet is organized similarly to the mammalian islet. DO: Dorsal Organizer. B) Fate mapping shows that pancreatic precursors (red box, 5 hpf) derive from the blastoderm margin, in bilateral domains. By the end of gastrulation (10 hpf), the precursors have converged towards the trunk region. By the end of day 1 of development, differentiated endocrine cells cluster to form the islet in the anterior trunk region.
Table 1.
Gene expression and function in the developing zebrafish pancreas.
| gene | onset | function | expression domain | knock-down phenotype | mammalian disease association |
|---|---|---|---|---|---|
| sox4b | 11.5 hpf | transcription factor | pancreas primordium (78) | α-cell reduction (78) | α-, β-cells reduced*(91) |
| mnx1 (was hlxb9, hb9) | 14 hpf | transcription factor | endocrine pancreas (79) | β-cell reduction or loss (79) | pancreas dorsal lobe agenesis, abnormal islets*(92,93) |
| pdx1 | 14 hpf | transcription factor | pancreas primordium, later endocrine specific (7,59,80) | reduced or absent pancreas (90) | IDDM, MODY4, pancreatic agenesis, pancreatic hypoplasia (94) |
| nkx2.2a | 14 hpf | transcription factor | endocrine pancreas, pancreatic duct (81) | α-, β-cells reduced, ε-cells increased, duct absent (81) | hyperglycemia*(95) |
| isl1 (islet1) | 15 hpf | transcription factor | endocrine pancreas (7,82) | islet absent, exocrine pancreas reduced (39,82) | islet agenesis*(96) |
| pax6b (was pax6.2) | 15 hpf | transcription factor | pancreas primordium (7,83,84) | ND | hypomorphic islet, reduced hormone production*(97) |
| ins (preproinsulin) | 15 hpf | hormone | endocrine pancreas, β-cells (7,80) | ND | diabetes mellitus, etc.‡ |
| neurod (was neuroD) | 16 hpf | transcription factor | pancreas primordium (81,85) | ND | MODY6 (98) |
| sst2 (somatostatin 2) | 17 hpf | hormone | endocrine pancreas, δ-cells (6,7,86) | ND | |
| hhex | 18 hpf | transcription factor | pancreas primordium (18) | ND | diabetes mellitus, noninsulin dependent (99) |
| ghrl (ghrelin) | 18 hpf | hormone | endocrine pancreas, ε-cells (81,87) | ND | Prader-Willi syndrome (100) |
| gcga (glucagon a) | 21 hpf | hormone | endocrine pancreas, α-cells (6,7) | ND | |
| hlxb9la (was mnr2a) | 24 hpf | transcription factor | exocrine pancreas (79) | acinar cells reduced(79) | |
| sst1 (somatostatin 1) | 24 hpf | hormone | endocrine pancreas, δ-cells (86) | ND | |
| insb (preproinsulin b) | 24 hpf | hormone | endocrine pancreas (88) | ND | |
| hnf1b (was vhnf1,tcf2) | 26 hpf | transcription factor | general pancreas (89) | ND† | MODY5 (101) |
| ptf1a (p48) | 32 hpf | transcription factor | exocrine, later endocrine pancreas (58,65) | exocrine pancreas absent, duct reduced(58,59,81) | pancreatic agenesis, permanent neonatal diabetes mellitus (102) |
| try (trypsin) | 48 hpf | digestive enzyme | exocrine pancreas (7,65) | ND | |
Onset refers to earliest reported onset of pancreatic expression, to date.
Disease association known from mouse model.
Effects of a morpholino have not been determined, but mislet mutants show a severely reduced pancreas (see Table 2 for mutant phenotypes).
Defects in insulin are associated with multiple human diseases.
For more information, see the Online Mendelian Inheritance in Man (OMIM) database http://www.ncbi.nlm.nih.gov/sites/entrez?db=omim. IDDM: Insulin Dependent Diabetes Mellitus. ND: pancreas phenotype Not Determined.
3. Developmental origins of the pancreas: regionalization of the endoderm
In all vertebrates establishment of the three germ layers (endoderm, mesoderm and ectoderm) occurs immediately before, or at the onset of, the morphogenetic process of gastrulation. Gastrulation movements internalize the endoderm cells, eventually bringing them into the appropriate location to generate the digestive tract and associated organs. During this process endodermal precursors differentiate into a wide variety of cell types, to make up the various complex tissues and organs of the digestive tract. For example, the most anteriorly localized (cranial) endoderm cells will contribute to the pharynx, thyroid, thymus, and esophagus, while more posteriorly located (trunk) endoderm will contribute to the foregut (including the liver and pancreas), midgut and hindgut. Complete development of endoderm-derived structures can be considered a stepwise process: the endoderm first becomes regionalized into specific territories, then within these territories further differentiation and organogenesis occurs. For example, following initial regionalization to produce the foregut, the pancreas and liver develop as out-pocketings of the gut, and their specific cell types differentiate (reviewed in(10,11)). Thus the appropriate combination of regionalization, cell differentiation, and morphogenesis ultimately produces fully functional organs with critical roles in metabolism and digestion.
The initial process of establishing the endoderm germ layer is well understood at both the morphological and molecular levels in the zebrafish model. Early in zebrafish gastrulation both endoderm and mesoderm cells, which lie at the blastula margin, begin to internalize through a process of involution. The endodermal cells rapidly disperse through a “random walk” mechanism(12), develop a characteristic flattened morphology, and begin to express endodermal marker genes(4,13). As gastrulation proceeds, convergence-extension movements carry cells of all three germ layers to the dorsal side of the embryo where they intercalate to elongate the anteroposterior (AP) axis of the body. This process brings the endoderm cells towards the midline, where they will later form the gut tube on the ventral surface of the embryo, in between the yolk and the overlying mesoderm (Fig. 1b). Genetic approaches have shown that in zebrafish both formation of the endoderm and its unique cell behaviors rely on nodal-related TGFβ molecules, which ultimately regulate expression of the casanova/sox32 transcription factor gene (reviewed in(14,15)). Sox32 is both necessary for endoderm development and sufficient to switch mesoderm to an endodermal fate(16,17).
While we have a detailed understanding of endoderm formation in zebrafish, we know less about the regionalization of that endoderm along the AP axis in this system or any other. The zebrafish endoderm gives rise to a similar repertoire of structures to the mammalian endoderm (Fig. 1), with the exception of a swimbladder rather than lungs, and an enlarged proximal intestinal structure, termed the intestinal bulb, rather than a stomach and duodenum. Another difference is that the zebrafish gut tube forms via cavitation rather than by folding of a sheet as in other vertebrates(18). We also know from the Warga and Nusslein-Volhard(4) fate map that structures in the anterior of the animal (such as pharynx) derive from endoderm close to the dorsal organizer (or “shield”) whereas structures in the posterior of the animal derive from endoderm that lies far from the organizer (Fig. 1). What mechanisms function to subdivide the early endoderm into the territories that will later produce specific AP structures? Studies of axial patterning of the nervous system have implicated the Vitamin A derivative Retinoic Acid (RA) in the process of AP regionalization(19). This knowledge inspired us to ask whether RA might play a similar role in endoderm regionalization.
3.1 Retinoic Acid
RA is synthesized in animals from its Vitamin A precursor through a two-step process, with the second rate-limiting step catalyzed by aldehyde dehydrogenase (Aldh). During early development Aldh function is provided by the Raldh2 enzyme. Two mutations in zebrafish raldh2 have been described, neckless (nls) and no-fin (nof) mutants(20,21). We found that RA-deficient embryos, either mutant for nls or treated with the BMS493 inverse agonist of RA receptor (RAR) function, lack both endocrine and exocrine pancreatic cell types, as well as markers of pancreatic progenitors. However, endoderm is present and other derivatives such as pharynx and thyroid are able to differentiate as normal(22). Importantly, timed treatments with BMS493 revealed that RA functions to specify the pancreatic domain between 8 and 13 hours post fertilization (hpf), at the end of the gastrulation process. Further, we found that treatment of late gastrulae with exogenous RA has the remarkable capacity to expand the pancreatic field throughout the anterior endoderm, acting in a dose-responsive manner to expand foregut fates at the expense of pharyngeal structures(22). In a comparative study we confirmed that this critical role for RA is not confined to zebrafish; both quail and Xenopus embryos respond similarly to RA(23). RA was similarly shown to be essential for Xenopus pancreas development by Chen and colleagues(24), who additionally found that RA promotes endocrine cells in the dorsal pancreatic bud. Analyses of mouse Raldh2−/− mutants, which again lack early RA production, showed that RA is similarly required for dorsal pancreatic bud development in a mammal(25,26). These studies confirm that molecular mechanisms to regionalize endoderm are conserved across vertebrates, although they also highlight that there can be small but significant differences in these mechanisms. For example, while all pancreatic derivatives, as well as liver fates, require RA signaling in the zebrafish, in mouse this requirement is limited to the dorsal pancreatic bud. Nevertheless, RA is an important promoter of vertebrate pancreatic fate, and consistent with this finding RA treatment is an integral component of three recently reported protocols for the differentiation of human stem cells to endocrine pancreas cells(27–29). In summary, the RA signaling story represents a nice example of how lessons from zebrafish can be highly applicable to mammalian diabetes and stem cell research.
As RA is a diffusible signal produced in the mesoderm, where raldh2 is expressed, the question arises of how it exerts its effects on endoderm. To study this, we used a cell transplantation strategy that takes advantage of the Sox32 transcription factor. As Sox32 is both necessary and sufficient to specifiy endoderm from mesendodermal progenitors (17,30), we are able to microinject Sox32 mRNA into one-cell stage zebrafish embryos and thus direct cells to an endoderm fate(31). Co-injected reagents are also directed to the endoderm, and this allows us to determine whether a molecule of interest functions tissue-autonomously. As Sox32-overexpressing embryos lack mesoderm they do not undergo normal gastrulation. To study the endoderm, these embryos are therefore used as cell donors: endoderm cells are removed from a pre-gastrula stage donor and transplanted to a pre-gastrula stage host embryo. We can microinject the host embryo with an antisense morpholino to disrupt Sox32 function, thus blocking its capacity to make endoderm of its own. Transferring cells from the Sox32-positive donor to the Sox32-negative host produces a genetic chimera in which all endoderm is donor-derived.
Using this strategy, we expressed a dominant-negative RA receptor (RAR) specifically in the endoderm and found that RARs function in endoderm to receive the RA signals from adjacent mesoderm(31). However, as diffusible RA is synthesized throughout the anterior mesoderm, additional mechanisms must exist to precisely limit the endodermal domain specified as pancreatic. As the four zebrafish RARs are broadly expressed(32), it is unlikely that this process relies exclusively on limited RAR expression. Consistent with this hypothesis, in recent studies we have found that the Cdx4 transcription factor plays an important role in setting the normal posterior limit of the pancreatic field. Zebrafish mutant for cdx4/kgg (Table 2) not only have an abnormally posteriorly-located pancreas, but also an excess number of β-cells(33). As cdx4 is expressed in all three germ layers during early development, we asked whether Cdx4 functions directly within the endoderm to localize the pancreas. Again using cell transplantation, we found that mesodermal expression of Cdx4 is not critical for localizing the pancreas, but rather that endodermal Cdx4 is required(33). We predicted that in the absence of Cdx4 function exogenous RA would have the capacity to promote pancreatic fates not only in anterior endoderm, but also in the posterior. Consistent with this model, RA treatment of Cdx4-deficient specimens induces scattered insulin-expression throughout the posterior endoderm(33). Taken together, our data show that mesodermal RA and endodermal Cdx4 work positively and negatively, respectively, to establish the pre-pancreatic region within the endoderm (model summarized in Fig. 2).
Table 2.
Zebrafish pancreatic mutants
| mutant | gene | molecular function | Phenotype | reference |
|---|---|---|---|---|
| akreas (akr) | ptf1a | transcription factor activity | exocrine pancreas severely reduced, delayed | (60) |
| angelina | agl | ND | β-cells severely reduced | (70) |
| cheetah | chee | ND | α-, β-, δ-cells scattered | (70) |
| daedalus (dae) | fgf10a | growth factor activity | dysmorphic ductal system, ectopic endocrine cells | (39,40) |
| dalmatian | dal | ND | α-, β-, δ-cells scattered | (70) |
| ductjam | djm | ND | exocrine pancreas reduced, ductal defect | (71) |
| ducttrip | dtp | ND | exocrine pancreas reduced, ductal defect | (71) |
| earl grey (egy) | sart3 | calcium ion binding | exocrine pancreas reduced or absent | (103) |
| floating head | flh | DNA binding | β-cells reduced | (7) |
| flotte lotte | flo | ND | exocrine pancreas reduced, ductal defect | (71,104) |
| heart and soul (has) | prkci (was aPKCλ) | ATP binding | duplicated exocrine pancreas, endocrine bud not fused with exocrine | (50,51) |
| ikarus (ika) | fgf24 | growth factor activity | exocrine pancreas reduced | (39) |
| knypek | kny | protein binding | β-cells bilateral | (7) |
| lazarus (lzr) | pbx4 | DNA binding | anteriorly shifted endocrine pancreas | (105,106) |
| mind bomb | mib | protein binding | exocrine pancreas reduced, β-, δ-cells increased | (65,107) |
| minime | mnm | ND | α-, β-cells scattered, β-cells reduced, exocrine pancreas reduced | (70) |
| mislet (mis or msl) | hnf1b (was vhnf1, tcf2) | transcription factor activity | pancreatic precursors and derivatives severely reduced | (70) |
| mitomess | mms | ND | exocrine pancreas reduced, ductal defect | (71) |
| neckless (nls) | aldh1a2 (was raldh2) | oxidoreductase activity | pancreas absent | (20,22) |
| nil per os (npo) | rbm19 | RNA binding | exocrine pancreas development arrested | (108) |
| no relief | nor | ND | exocrine pancreas absent | (109) |
| peppershaker | pps or ppp | ND | α-, β-, δ-cells scattered, exocrine pancreas disorganized | (70) |
| piebald | pie | ND | exocrine pancreas reduced, ductal defect | (71,109) |
| scarlet | sle | ND | α-, β-, δ-cells scattered | (70) |
| schmalspur (sur) | foxh1 | sequence-specific DNA binding | β-cells slightly reduced | (7) |
| sea dragon | sdr | ND | α-, β-, δ-cells reduced; duplicated exocrine pancreas | (70) |
| slimjim (slj) | polr3b | tRNA transcription | exocrine pancreas reduced, ductal defect | (71,109) |
| slow-muscle-omitted | smo (was smu) | G-protein coupled receptor activity | endocrine pancreas severely reduced, bilateral ventral bud | (43,44,58) |
| spadetail (spt) | tbx16 | DNA binding | β-cells bilateral or absent | (7,110) |
| straight shot | sst | ND | exocrine pancreas absent | (109) |
| sweetbread | swd | ND | exocrine pancreas reduced, ductal defect | (71) |
For specific alleles and a list of additional mutants, see the Zebrafish Information Network (ZFIN). Molecular function terms are from the Gene Ontology Consortium; some genes have multiple functions, not listed. ND, Not Determined.
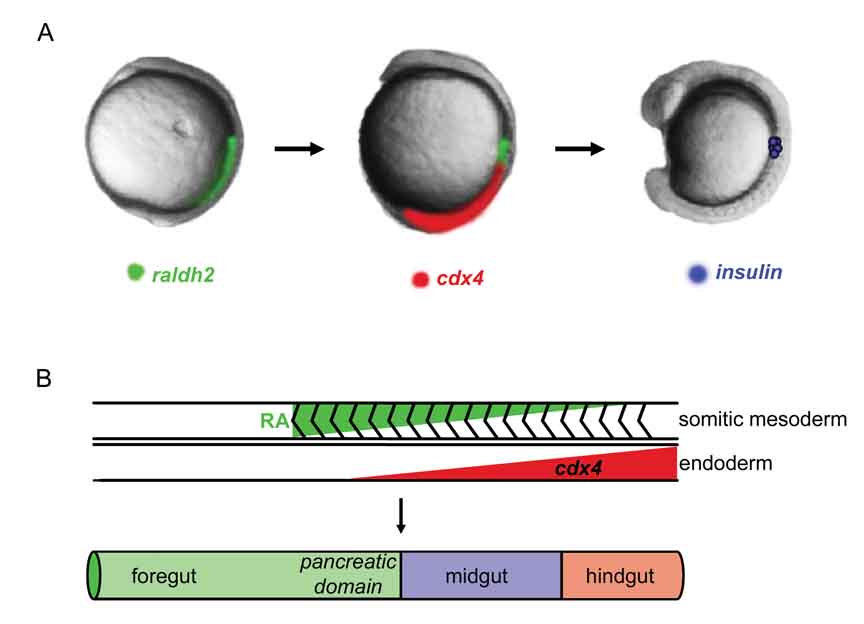
Figure 2. Model of endoderm regionalization.
A) Retinoic Acid (RA) is required between 8–13 hpf to specify the pancreas. RA is synthesized in the paraxial mesoderm by raldh2. Endodermal expression of the cdx4 transcription factor prevents posterior endoderm from expressing insulin. The β-cells are located anterior to the domain of high cdx4 expression. B) Opposing gradients of Retinoic Acid (RA) and cdx4 limit insulin expression to the foregut. The posterior limit of the pancreas is set by cdx4. Endodermal factors that limit the anterior boundary of the pancreas have not been identified.
3.2 FGFs
Beyond RA signaling, what other diffusible signals might function to regionalize the endoderm? Data from several systems implicate secreted FGF signals in this process. Wells and Melton(34) showed that FGF4 patterns cultured E7.5 mouse endoderm in a concentration dependent manner, with higher concentrations promoting intestinal fates and lower concentrations promoting pancreas/duodenal fates. Wells and colleagues(35) went on to show that local application of FGF to chick embryos, using beads at early somite stages, shifts the expression domain of CdxB (the homolog of zebrafish Cdx4) towards the anterior. Concomitantly, there is an anterior shift in the expression domain of the pancreas progenitor marker Pdx1. Interestingly, our own mis-expression experiments with zebrafish cdx4 led to a similar anterior shift of insulin expressing β-cells. Conversely, when Dessimoz and colleagues inhibited FGF signaling in chick endoderm at early somite stages, by applying a bead carrying the SU5402 FGF receptor inhibitor posterior to the pre-pancreatic domain, Pdx1 expression expanded posteriorly. Again, this result is reminiscent of Cdx4-deficiency in zebrafish, where we similarly find that the Pdx1 progenitor marker domain expands towards the posterior. Whether zebrafish Pdx1 also negatively regulates Cdx expression, as demonstrated in the chick(36), has not been tested. Taken together, available data suggest that regulation of Cdx4 may be a major mechanism through which FGF4 regionalizes endoderm, and further suggest that this regulatory pathway is conserved across vertebrates. The regulation of Cdx gene expression by FGF signaling is likely direct, as has been demonstrated for Xenopus Xcad3(37).
Interestingly, FGFs seem to play multiple sequential roles in pancreas development. In addition to the role of FGF4 in endoderm patterning, Hebrok et al.(38) have reported a slightly later role for chick FGF2 in repressing Shh expression in the posterior foregut that gives rise to the dorsal pancreas. Manfroid et al.(39) reported that endodermal expression of zebrafish fgf24 plays a later role in patterning the adjacent pancreatic lateral plate mesoderm, and subsequently, fgf24 and fgf10 from the lateral plate signal back to the endoderm to help specify the exocrine pancreas. Further, analysis of a zebrafish fgf10 mutant (Table 3) has revealed a critical role for this signal in patterning the hepatopancreatic duct(40).
Table 3.
Pancreas-related transgenic lines.
| line | transgene | expression | reference |
|---|---|---|---|
| ins:GFP | insulin driving Green Fluorescent Protein (GFP) | β-cells | (111) |
| pdx1:GFP | pancreatic duodenal homeobox-1 driving GFP | pancreas progenitors | (111) |
| XlEef1a1:GFP (gutGFP) | Xenopus laevis elongation factor-1a driving GFP* | throughout endoderm from 22 hpf | (11,112) |
| ptf1a:eGFP | pancreas specific transcription factor, 1a driving enhanced GFP | exocrine pancreas | (113) |
| sox17:GFP | SRY-box containing gene 17 driving GFP | endoderm percursors | (114) |
| sox17:dsRed | SRY-box containing gene 17 driving dsRed | endoderm percursors | (45) |
| elaA:EGFP | elastaseA driving GFP | exocrine pancreas | (67) |
| ins:eGFP | insulin driving enhanced GFP | β-cells | (9) |
| ins:mCherry | insulin driving mCherry | β-cells | (9) |
| ins:nfsB-mCherry | insulin driving nfsB-mCherry fusion protein† | β-cells | (9) |
| glu:GFP | glucagon driving GFP | α-cells | (9) |
| ins:gal4 | insulin driving Gal4 | β-cells | (9) |
| ins:kaede | insulin driving photoconvertable Kaede | β-cells | (9) |
| nkx2.2a:GFP | NK2 transcription factor related 2a driving GFP | endocrine pancreas | (55,81) |
| nkx2.2a:mEGFP | as above, with enhanced GFP localized to the membrane | endocrine pancreas | (112) |
| ins:CFP-NTR | insulin driving Cyan Fluorescent Protein (CFP) fused to Nitroreductase | β-cells | (68) |
| hsp70l:nog3 | noggin3 is inducible via heat shock cognate 70-kd protein, like | inducible, ubiquitous‡ | (115) |
For specific alleles and further information, see the Zebrafish Information Network (ZFIN).
The transgene integrated into the 3’UTR of Foxa3.
nsfB (nitrofurazone sensitivity) is an E. coli gene with nitroreductase (NTR) activity. Addition of the drug metronidazole induces NTR-mediated ablation.
Noggin3 inhibits BMP signaling, resulting in failure of gut looping and pancreas duplication.
3.3 Sonic Hedgehog (Shh)
As mentioned above, in chick explant experiments FGF2 inhibits Shh expression(38). This inhibition of early endodermal Shh function is a critical step in amniote pancreas development(38,41,42). By contrast, in zebrafish Hh signaling from the notochord is required for pancreas development: embryos treated with the Hh inhibitor cyclopamine, or mutant for the Hh receptor Smoothened, do not develop a pancreas(43,44). This apparent species-specific difference in the role of Shh reflects, at least in part, a difference in developmental timing; expression of zebrafish shh within the endoderm does not commence until 24 hpf, long after the onset of endocrine marker gene expression, and the related twhh(shhb) gene shows an even later onset of endodermal expression(43). However, as the mouse Shh mutant is early lethal just prior to pancreas specification, we do not know if mouse pancreas development also has an early requirement for notochord-derived Hh signaling. Interestingly, Chung and Stainier(45) have recently used a cell transplantation approach to reveal that the zebrafish Shh receptor Smo functions non-autonomously in endocrine pancreas development: its function is not required in β–cell progenitors themselves, but rather in more laterally located endoderm cells that are fated to form exocrine pancreas, intestine, and occasionally liver. This finding reveals that in addition to signals from mesoderm, intra-endodermal signals from lateral cells to more medial cells are also important in pancreas development. While the nature of these signals is not yet known, they seem to act at close range. Chung and Stainier(45), like diIorio et al.(44), suggest that notochord-derived Shh plays an early positive role, while endoderm-derived Shh plays a later negative role in controlling pancreas development. In summary, similar to FGFs, Hh signaling likely plays sequential roles in pancreas development, with cell culture experiments suggesting a requirement for Hh signaling in adult endocrine pancreas cell function (reviewed in(42)).
3.4 BMPs
TGFβ family growth factors, including BMPs, have also been implicated in the earliest stages of pancreas development. For example, mutants in zebrafish bmp2b/swirl show a reduced pancreatic progenitor domain, whereas mutants in the bmp antagonist chordin/dino show a reciprocal phenotype(46). These data may reflect a general early role for BMP signaling in setting up the basic body plan, or may suggest that BMP signals directly promote pancreas development. The latter hypothesis is supported by data from chick explant studies. Kumar et al.(47) showed that chick pre-pancreatic lateral plate mesoderm (LPM) has the capacity to instruct anterior endoderm to express the pancreas progenitor marker Pdx1; this capacity is blocked by the TGFβ inhibitors Follistatin and Noggin. More anterior LPM cannot play this role, unless RA, Activin A, or one of several BMP class molecules is also provided. Further, mouse mutations in the Activin receptors ActRIIA and ActRIIB(48), which can transduce signals from Activins and a subset of BMPs, lead to reduced pancreatic islets. Taken together, these data implicate one or more TGFβ family molecules in promoting pancreatic fates. Conversely, Spagnoli and Brivanlou(49) recently described a Xenopus BMP antagonist, TGIF2, which negatively regulates BMP function to promote pancreatic fate. It remains to be determined whether this apparently opposite role for BMP signaling in Xenopus versus other species represents a true species difference, or similar to the situation with HH, rather reflects different roles for TGFβ signals at different stages of pancreatic development.
4. Morphogenesis of the pancreas
Transgenic zebrafish carrying fluorescent reporter genes provide a powerful method to follow morphogenesis over real time. In recent years the number of transgenic lines relevant to studies of the pancreas has increased substantially as transgenic methods have improved; these lines are summarized in Table 3. Field et al.(50) used the convenient gutGFP transgenic line, in which the entire digestive tract expresses Green Fluorescent Protein due to a fortuitous transgene integration into the endoderm gene foxa3, to follow the details of zebrafish pancreas morphogenesis. The mammalian pancreas is initially formed as two separate buds that grow out from the developing gut tube; both dorsal and ventral pancreatic buds contribute endocrine and exocrine cells, with the separate buds merging to form the complete organ (reviewed in(10)). Field and colleagues showed that the zebrafish pancreas similarly derives from two pancreatic buds: a dorsal bud is apparent by 24 hpf, with a secondary ventral bud emerging slightly to the anterior at 40 hpf. By 52 hpf the two buds have merged to form a single pancreas located on the right hand side of the larval fish, as a consequence of gut rotation. Analysis of the zebrafish mutant heart and soul (has) in which gut looping fails to initiate revealed that the dorsal bud produces almost all of the endocrine cells, with the ventral bud being the sole source of exocrine cell types(50,51). This separation of endocrine and exocrine tissue may prove useful in allowing the different genetic programs underlying differentiation of endocrine versus exocrine cell types to be unraveled.
To investigate cell behavior at earlier stages of zebrafish pancreas development we recently generated a detailed fate map focused specifically on pancreatic cell types(52). Similar to other studies(4), we fate mapped by labeling single cells close to the blastoderm margin at late blastula stage, localizing them relative to the shield at early gastrula stage, then following the labeled clones to 40 or 72 hpf. This strategy enabled us to identify each labeled cell’s contributions to the dorsal versus ventral pancreatic bud, or to endocrine versus exocrine pancreas tissues. We found no intrinsic asymmetry in the disposition of pancreas progenitors, however we did uncover a tendency for dorsal bud/endocrine progenitors to derive from a location closer to the shield than the ventral bud/exocrine progenitors. While a single clone can contribute to to both the endocrine and exocrine buds as well as to endocrine and acinar cell types, our data suggest that even before cells have committed to a particular germ layer, biases are being established that will influence later pattern formation.
Additional fate map data recently reported by Chung and Stainier(45) show that the endocrine pancreas cells derive from the most medial aspect of the pdx1 expression domain, with more lateral cells contributing to exocrine pancreas and intestine. Expression of pdx1 is initially located in bilateral domains in the anterior trunk, as predicted by the fate maps(4,52). These domains later merge as convergence brings foregut cells to the midline. Interestingly, Matsui et al.(53) have described a role for non-canonical Wnt signaling in the process of foregut convergence. This group demonstrated that a dominant-negative Disheveled molecule, designed to specifically block non-canonical Wnt signaling, prevented Pdx1-positive progenitors or later differentiated pancreas and liver cell types from reaching the midline; rather, bilateral domains of expression were maintained for all foregut markers. They went on to demonstrate that three Wnt ligands (Wnt4a, Wnt11 and Wnt11r) function redundantly, through the RhoA kinase pathway, to mediate foregut convergence.
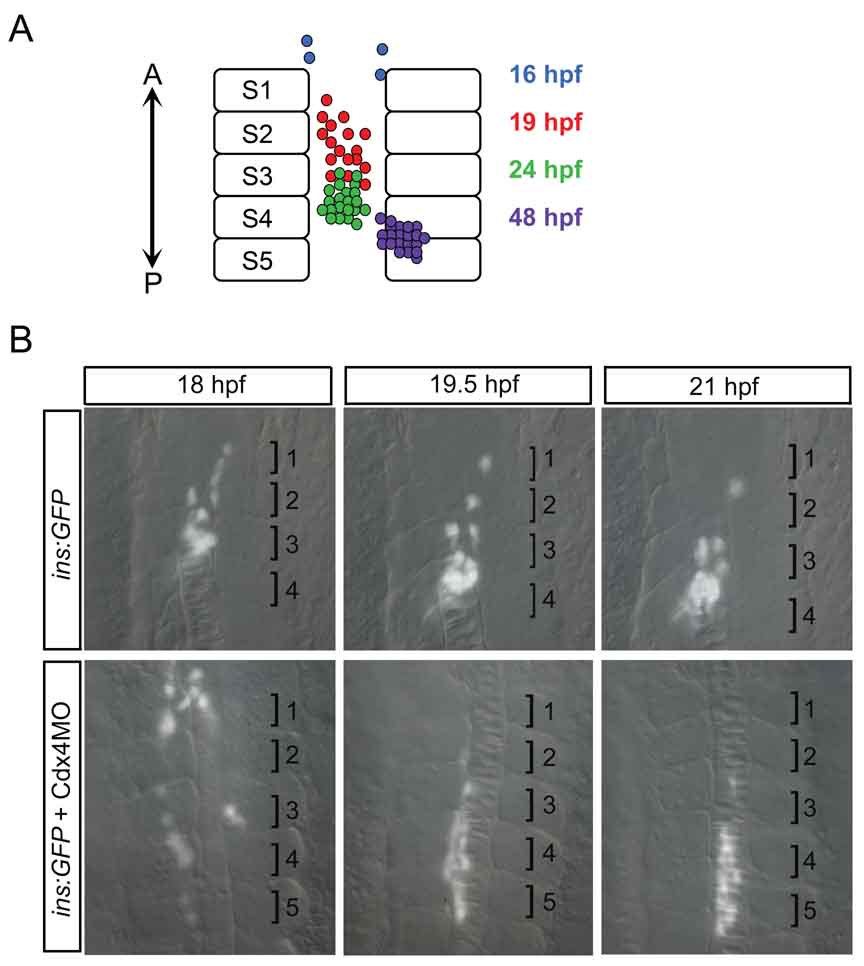
By following β-cell development during early zebrafish embryogenesis, we have obtained a fairly detailed picture of endocrine pancreas organogenesis. Insulin-positive β-cells are first detectable at 15 hpf, at which time they are located in scant numbers in bilateral domains in the anterior trunk(7). Over the next 4 hours, β-cell number increases and the cells converge to the midline, forming a nascent islet by 19 hpf(33). As the β-cells converge to the midline they also move posteriorly. Thus, insulin-positive cells that are initially detected adjacent to somite 1 are located adjacent to somites 3–4 by 24 hpf and somites 4–5 by 48 hpf (Fig. 3A). This change in location of the β-cells was first detected by in situ hybridization to locate insulin transcripts. Subsequently, transgenic zebrafish have been utilized to follow β-cell movements in live embryos via time-lapse imaging (e.g. Fig. 3). Notably, Kim et al.,(54) used insulin:GFP transgenics to track individual β-cells over the first 8 hours of their development following the onset of GFP expression. This study confirmed the active movement of β-cells suggested by in situ hybridization studies. They further studied β-cell movement by disrupting non-canonical Wnt signaling. Morpholino knockdown of Wnt5 or the Fz2 Wnt receptor results in a duplicated islet. Live time-lapse imaging of insulin:GFP transgenics in which Fz2 was knocked down revealed that this was caused by the failure of individual β-cells to move towards the posterior. Rather, cell movements were randomized along the AP axis, although midline convergence was normal. Thus non-canonical Wnt signaling plays separable roles in foregut convergence(53) and islet coalescence and migration(54). Finally, our own time-lapse studies of β-cell movement using the insulin:GFP transgenics have demonstrated the importance of the Cdx4 transcription factor in correctly localizing the pancreas along the AP axis(33). We observed that loss of Cdx4 results in movement of the β-cell population to an abnormally posterior location (Fig. 3B). Taken together, these studies demonstrate how the optically transparent zebrafish embryo, when coupled with transgenic technology, provides a powerful tool for studying pancreas morphogenesis.
Figure 3. β-cell movement during islet formation.
A) Schematic of β-cell location during islet formation. Over time, insulin-positive β-cells increase in number, converge to form the islet, and move posteriorly. A, Anterior; P, Posterior; S, Somite. B) Live timelapse imaging reveals that β-cells move posteriorly during islet formation. Cdx4 knockdown results in farther movement of β-cells. Based on in situ hybridization, glucagon-positive β-cells and somatostatin2-positive β-cells are also initially located anteriorly in the trunk and subsequently surround the β-cell cluster within the more posteriorly located islet. It is thus likely that α- and δ-cells move posteriorly, similar to the β-cell movement revealed by live timelapse imaging. Brightfield and fluorescent images of insulin:GFP transgenics taken in dorsal view at 200x magnification and merged. Numbered brackets indicate somites.
5. Cell fate decisions
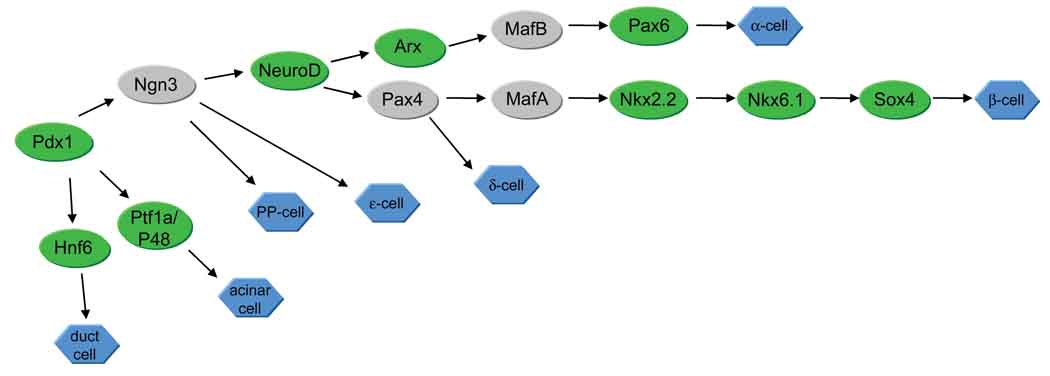
As the pre-pancreatic domain is established, precursors differentiate into the mature pancreatic cell types in a sequential fashion (Table 1). Mouse studies have demonstrated that each of the islet endocrine cell types differentiates independently from a common precursor pool. Lineage-specific transcription factors have been identified for some cell types, summarized in Figure 4. Much of this developmental program may be conserved between mice and zebrafish, as many of the transcription factors that characterize mouse pancreatic cell lineages have been identified in zebrafish lineages as well (Figure 4). A notable difference is that ngn3, a marker of endocrine pancreas precursors in mouse, is apparently not expressed in zebrafish pancreas until well after the endocrine cell types have differentiated(55). By contrast, mice lacking ngn3 expression fail to differentiate pancreatic endocrine cell types(56). Thus, although zebrafish and mouse have many similarities with respect to pancreas development, the developmental program is not identical.
Figure 4. Pancreatic cell lineages.
A simplified summary of transcription factors identified for mouse and zebrafish pancreatic cell differentiation. Lineages are based on mouse data. Green indicates that a zebrafish homolog has been identified in the pancreas (see Table 1 for more details). Function of a particular transcription factor is not necessarily conserved between mouse and zebrafish. Gray indicates mouse pancreatic transcription factors that have not been identified in the zebrafish pancreas. Pancreatic Polypeptide-expressing cells (PP-cells) have not been identified in the zebrafish pancreas, although immunostaining with a rabbit PP antibody has revealed immunopositive cells in the intestine(116).
These differences can be exploited to reveal general aspects of vertebrate pancreas development that might otherwise be difficult to detect in a mammalian model. For example, because the two zebrafish pancreatic buds consist of separate pools of endocrine cell types (dorsal bud) versus largely exocrine cells (ventral bud) the zebrafish has been useful for dissecting the differences between endocrine versus exocrine cell fate decisions. In mice, both pancreatic buds comprise mixed endocrine and exocrine progenitors, and both require the transcription factor gene Ptf1a for differentiation(57). However, in zebrafish ptf1a is required only for the later developing ventral bud(58,59). Loss of ptf1a results in loss of the exocrine population, and the ventral-derived endocrine population is greatly reduced, revealing a later requirement for ptf1a in endocrine differentiation(59). Recently, Dong and colleagues have added to this picture by showing that the level of ptf1a expression influences cell fate choices within the pancreatic progenitor pool. Using the akreas mutant, which is a ptf1a hypomorph, this group showed that in the ventral bud, cells with low levels of ptf1a activity adopt an endocrine fate, whereas cells with high levels adopt an exocrine fate(60).
In the pancreas, ptf1a appears to exert its influence in the context of the Notch activation state. When Notch signaling is active, ptf1a-positive cells are maintained in a proliferative state, and when Notch signaling is inactivated, ptf1a-positive cells become terminally differentiated (reviewed in(61)). Notch signaling is also important for the sequential differentiation of pancreatic cell types(55,62–65) and may function to limit the number of specific endocrine cell types via lateral inhibition, in a similar fashion to its role in neuronal development (reviewed in(66)). In mouse and zebrafish, exocrine pancreas precursors co-express ptf1a and hes1, a downstream target of the Notch pathway(65). Mouse studies have shown that inhibition of Notch signaling results in accelerated endocrine differentiation and inhibition of exocrine differentiation(62). These studies suggested that excessive differentiation of endocrine cells resulted in a depletion of the common pancreatic precursor pool, thus leaving few precursors available to differentiate into mature exocrine cells. However, Esni et al(65), using zebrafish, showed that there is a specific role for Notch signaling in acinar differentiation. Notch signaling must be inhibited in the precursors for exocrine cells to differentiate (reviewed in(61)). Use of the zebrafish avoided confounding factors presented by the mixed precursor population in the mouse pancreatic buds.
6. Regenerative capacity of β-cells
When zebrafish exocrine cells are ablated, by expressing diptheria toxin A (DTA) under control of the acinar-cell-specific elastaseA promoter, the endocrine population appears to be unaffected (67). At 6 days post-fertilization, α-, β-, and δ-cells are present in normal numbers in pElaA-DTA larvae. This indicates that endocrine cells are not dependent on the exocrine cells for maintenance or differentiation, and is consistent with the differentiation of the dorsal endocrine population in the absence of ptf1a. Two groups have used a similar strategy to ablate β-cells in the pancreas, but have gone a step farther by using transgenes that are responsive to drugs, enabling the temporal control of βcell ablation(9,68). Curado et al. and Pisharath et al. have generated transgenic lines encoding nitroreductase (NTR) in β-cells (ins:CFP-NTR and ins:nfsB-mCherry, see Table 3). NTR converts metronidazole to a cytotoxin, resulting in β-cell death. Both groups elegantly used this strategy to show that β-cells regenerate following drug removal. In addition, Pisharath et al. blocked exocrine pancreas development in their transgenic embryos by using a morpholino against ptf1a, and demonstrated that β-cell regeneration is independent of the exocrine pancreas. As ptf1a-morphant embryos have variable loss of the pancreatic duct, it will be important for future studies to determine whether the duct is required for β-cell regeneration. When coupled with lineage tracing, these new transgenic tools will allow the important question of which cells are progenitors of the regenerating β-cells to be addressed.
7. Harnessing the power of zebrafish genetics
One of the great advantages of the zebrafish model is its powerful genetics. Many of the studies mentioned above have relied either on mutant lines of zebrafish, or on reverse genetic approaches such as morpholino knock-down to disrupt candidate gene function (reviewed in (69)). The forward genetic approach has the advantage that it allows unbiased identification of genes based solely on the phenotypes that result from their disruption. In Table 2 we summarize the known mutants that have a major pancreas phenotype. As any mutant lacking endoderm will by definition also lack pancreas we have not included pan-endoderm mutants such as casanova/sox32 in this list.
The original large-scale genetic screens carried out in Tubingen and Boston and reported in 1996 produced several mutants that disrupt pancreas development, despite the difficulty of visualizing changes in the endoderm derivatives based on a pure morphology screen. Notably, the two raldh2 mutant alleles, nls and nof, were isolated during these screens, although the absence of pancreas in nls RA-deficient embryos was only recognized several years later(22).
A directed screen based on in situ hybridization for insulin identified a new mutant allele in tcf2/hnf1b (the mislet mutant), which has a severely reduced pancreas. This screen also identified another 8 mutants with reduced islets, disorganized endocrine cells, or both(70) (see Table 2). Other screens have focused on the exocrine pancreas(71), where reduction or occasional complete absence is a common phenotype. A recent screen using the gutGFP transgenic line yielded the prometheus/wnt2bb mutant, which shows defects in liver specification(72), as well as the akreas/ptf1a hypomorph described above(60). It seems likely that this screen and others based on available transgenic reporter lines will provide new mutants affecting pancreas development in the future. The paucity of mutants to date that completely lack endocrine pancreas, or even a specific endocrine cell type, is noteworthy. This is unlikely to reflect genetic redundancy as a consequence of the ancient gene duplication event that occurred in the lineage of the zebrafish, as, in general, duplicate genes have either been lost or have sub-functionalized. Rather, this lack of mutants suggests that after the pre-pancreatic domain has been appropriately regionalized there are complex and redundant mechanisms to produce the normal complement of endocrine pancreas cell types.
8. Conclusions/Perspectives
Our understanding of some of the earliest events in pancreas development, in particular the initial regionalization of the endoderm to produce the pre-pancreatic field, has been moved forward by studies in the zebrafish. While work in the mouse has been particularly helpful in establishing the intrinsic transcription factor networks critical to differentiation of pancreatic cell types, the zebrafish has contributed to our understanding of the extrinsic signaling pathways that influence these networks. For example, we now know that regulated RA signaling is critical to establishing the pre-pancreatic field, and that signaling molecules such as Shh and FGF can function more than once in pancreatic development, playing sequential roles at different developmental stages. In addition, the accessibility of zebrafish embryos is beginning to provide new insights into the morphogenesis of the pancreas. Nevertheless, despite significant progress in multiple models, much remains to be discovered regarding the details of the signaling pathways that allow differentiation of particular pancreatic cell types. Interactions between different signaling pathways have not been fully explored, and in many cases we lack a full understanding of the temporal sequence of events. Similarly, we do not yet fully understand the morphogenetic processes that allow the endocrine cells to arrange into functional islets, or indeed to build the pancreatic organ as a whole. Future studies using the zebrafish system will likely focus on these unanswered questions, as well as on understanding and overcoming limitations to pancreas regeneration.
Studies of pancreas developmental biology are also vital for stem cell biology; we should be able to learn from the embryo how to coax undifferentiated embryonic stem cells into the differentiated cell-types that make a functional pancreas. Consistent with this idea, several groups have applied knowledge gained from study of the embryonic pathways discussed above, to differentiate stem cells into β-cells(27–29,73,74). The protocols involve progressive administration of signaling molecules including Activin, FGFs and RA, to step the stem cells through a differentiation pathway from definitive endoderm, to primitive gut, to foregut, to pancreas progenitor, to differentiated β-cells. While many hurdles remain before stem cell-derived islets can be used to cure diabetic patients, these new findings do give significant hope that this is a plausible therapeutic avenue. In addition, they highlight the critical importance of understanding the normal progression of pancreatic development through the study of animal models including the zebrafish.
We predict that because of the tractability of the zebrafish model with respect to imaging and genetic manipulations, studies on the function of the zebrafish pancreas will become increasingly useful. Do zebrafish β-cells secrete insulin? Is regulation of blood glucose by the pancreas important in zebrafish? Although these questions are beginning to be addressed, insulin release has yet to be directly measured in zebrafish. Recent studies have measured blood glucose and shown that glucose levels drop in response to various anti-diabetic drugs that are routinely used in mammals(75). Changes in blood glucose levels have also been monitored by Gleeson et al.,(76), who demonstrated that hyperglycemia in zebrafish larvae induces retinal dysmorphology, reminiscent of retinopathy associated with both type I and type II diabetes in humans. In addition to use of the zebrafish for studies of pancreas function, we expect that the zebrafish will also become increasingly useful for other types of diabetes-related research beyond its role as a model for vertebrate pancreas development. For example, the zebrafish lends itself well to high throughput approaches, such as small molecule screens (reviewed in(77)). Small molecule screens can potentially be coupled with transgenic tools, such as reporters of gene function, or perhaps of regenerating β-cells. Such approaches will allow zebrafish to become a useful drug discovery and screening tool of the future.
Acknowledgements
We are extremely grateful for research funding from the Juvenile Diabetes Research Foundation (grants 1-2003-257 and 5-2007-970 to VEP), the National Institutes of Health (NIDDK DK064973 to VEP and NICHD HD050031 to MDK), the Digestive Diseases Research Core Center of The University of Chicago (DK42086), and through The University of Chicago Imaging Institute and The University of Chicago Clinical and Translational Science Award (National Center for Research Resources Grant UL1RR024999). We thank our colleagues Gokhan Dalgin, Stefani Eames, Elizabeth Sefton, Isaac Skromne and James Wells for helpful discussions and comments on the manuscript. We also thank two anonymous reviewers and editor Adam Wilkins for constructive criticism of the manuscript.
Abbreviations
- AP
anteroposterior
- DTA
diptheria toxin A
- GFP
Green Fluorescent Protein
- hpf
hours post-fertilization
- LPM
lateral plate mesoderm
- NTR
nitroreductase
- RA
Retinoic Acid
- RAR
Retinoic Acid Receptor
References
- 1.Shapiro AM, Lakey JR, Ryan EA, Korbutt GS, Toth E, et al. Islet transplantation in seven patients with type 1 diabetes mellitus using a glucocorticoid-free immunosuppressive regimen. N Engl J Med. 2000;343:230–238. doi: 10.1056/NEJM200007273430401. [DOI] [PubMed] [Google Scholar]
- 2.Shapiro AM, Ricordi C, Hering BJ, Auchincloss H, Lindblad R, et al. International trial of the Edmonton protocol for islet transplantation. N Engl J Med. 2006;355:1318–1330. doi: 10.1056/NEJMoa061267. [DOI] [PubMed] [Google Scholar]
- 3.Jorgensen MC, Ahnfelt-Ronne J, Hald J, Madsen OD, Serup P, et al. An illustrated review of early pancreas development in the mouse. Endocr Rev. 2007;28:685–705. doi: 10.1210/er.2007-0016. [DOI] [PubMed] [Google Scholar]
- 4.Warga RM, Nusslein-Volhard C. Origin and development of the zebrafish endoderm. Development. 1999;126:827–838. doi: 10.1242/dev.126.4.827. [DOI] [PubMed] [Google Scholar]
- 5.Falkmer S. Comparative morphology of pancreatic islets in animals. In: Volk BW, Arquilla ER, editors. The Diabetic Pancreas. 2nd ed. New York: Plenum Medical Book Company; 1985. pp. 17–52. [Google Scholar]
- 6.Argenton F, Zecchin E, Bortolussi M. Early appearance of pancreatic hormone-expressing cells in the zebrafish embryo. Mech Dev. 1999;87:217–221. doi: 10.1016/s0925-4773(99)00151-3. [DOI] [PubMed] [Google Scholar]
- 7.Biemar F, Argenton F, Schmidtke R, Epperlein S, Peers B, et al. Pancreas development in zebrafish: early dispersed appearance of endocrine hormone expressing cells and their convergence to form the definitive islet. Dev Biol. 2001;230:189–203. doi: 10.1006/dbio.2000.0103. [DOI] [PubMed] [Google Scholar]
- 8.Chen S, Li C, Yuan G, Xie F. Anatomical and histological observation on the pancreas in adult zebrafish. Pancreas. 2007;34:120–125. doi: 10.1097/01.mpa.0000246661.23128.8c. [DOI] [PubMed] [Google Scholar]
- 9.Pisharath H, Rhee JM, Swanson MA, Leach SD, Parsons MJ. Targeted ablation of beta cells in the embryonic zebrafish pancreas using E. coli nitroreductase. Mech Dev. 2007;124:218–229. doi: 10.1016/j.mod.2006.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edlund H. Pancreatic organogenesis--developmental mechanisms and implications for therapy. Nat Rev Genet. 2002;3:524–532. doi: 10.1038/nrg841. [DOI] [PubMed] [Google Scholar]
- 11.Field HA, Ober EA, Roeser T, Stainier DY. Formation of the digestive system in zebrafish. I. Liver morphogenesis. Dev Biol. 2003;253:279–290. doi: 10.1016/s0012-1606(02)00017-9. [DOI] [PubMed] [Google Scholar]
- 12.Pezeron G, Mourrain P, Courty S, Ghislain J, Becker TS, et al. Live analysis of endodermal layer formation identifies random walk as a novel gastrulation movement. Curr Biol. 2008;18:276–281. doi: 10.1016/j.cub.2008.01.028. [DOI] [PubMed] [Google Scholar]
- 13.Alexander J, Stainier DY. A molecular pathway leading to endoderm formation in zebrafish. Curr Biol. 1999;9:1147–1157. doi: 10.1016/S0960-9822(00)80016-0. [DOI] [PubMed] [Google Scholar]
- 14.Stainier DY. A glimpse into the molecular entrails of endoderm formation. Genes Dev. 2002;16:893–907. doi: 10.1101/gad.974902. [DOI] [PubMed] [Google Scholar]
- 15.Grapin-Botton A, Constam D. Evolution of the mechanisms and molecular control of endoderm formation. Mech Dev. 2007;124:253–278. doi: 10.1016/j.mod.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 16.Dickmeis T, Mourrain P, Saint-Etienne L, Fischer N, Aanstad P, et al. A crucial component of the endoderm formation pathway, CASANOVA, is encoded by a novel sox-related gene. Genes Dev. 2001;15:1487–1492. doi: 10.1101/gad.196901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kikuchi Y, Agathon A, Alexander J, Thisse C, Waldron S, et al. casanova encodes a novel Sox-related protein necessary and sufficient for early endoderm formation in zebrafish. Genes Dev. 2001;15:1493–1505. doi: 10.1101/gad.892301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wallace KN, Pack M. Unique and conserved aspects of gut development in zebrafish. Dev Biol. 2003;255:12–29. doi: 10.1016/s0012-1606(02)00034-9. [DOI] [PubMed] [Google Scholar]
- 19.Glover JC, Renaud JS, Rijli FM. Retinoic acid and hindbrain patterning. J Neurobiol. 2006;66:705–725. doi: 10.1002/neu.20272. [DOI] [PubMed] [Google Scholar]
- 20.Begemann G, Schilling TF, Rauch GJ, Geisler R, Ingham PW. The zebrafish neckless mutation reveals a requirement for raldh2 in mesodermal signals that pattern the hindbrain. Development. 2001;128:3081–3094. doi: 10.1242/dev.128.16.3081. [DOI] [PubMed] [Google Scholar]
- 21.Grandel H, Lun K, Rauch GJ, Rhinn M, Piotrowski T, et al. Retinoic acid signalling in the zebrafish embryo is necessary during pre-segmentation stages to pattern the anterior-posterior axis of the CNS and to induce a pectoral fin bud. Development. 2002;129:2851–2865. doi: 10.1242/dev.129.12.2851. [DOI] [PubMed] [Google Scholar]
- 22.Stafford D, Prince VE. Retinoic acid signaling is required for a critical early step in zebrafish pancreatic development. Curr Biol. 2002;12:1215–1220. doi: 10.1016/s0960-9822(02)00929-6. [DOI] [PubMed] [Google Scholar]
- 23.Stafford D, Hornbruch A, Mueller PR, Prince VE. A conserved role for retinoid signaling in vertebrate pancreas development. Dev Genes Evol. 2004;214:432–441. doi: 10.1007/s00427-004-0420-6. [DOI] [PubMed] [Google Scholar]
- 24.Chen Y, Pan FC, Brandes N, Afelik S, Solter M, et al. Retinoic acid signaling is essential for pancreas development and promotes endocrine at the expense of exocrine cell differentiation in Xenopus. Dev Biol. 2004;271:144–160. doi: 10.1016/j.ydbio.2004.03.030. [DOI] [PubMed] [Google Scholar]
- 25.Molotkov A, Molotkova N, Duester G. Retinoic acid generated by Raldh2 in mesoderm is required for mouse dorsal endodermal pancreas development. Dev Dyn. 2005;232:950–957. doi: 10.1002/dvdy.20256. [DOI] [PubMed] [Google Scholar]
- 26.Martin M, Gallego-Llamas J, Ribes V, Kedinger M, Niederreither K, et al. Dorsal pancreas agenesis in retinoic acid-deficient Raldh2 mutant mice. Dev Biol. 2005;284:399–411. doi: 10.1016/j.ydbio.2005.05.035. [DOI] [PubMed] [Google Scholar]
- 27.D'Amour KA, Bang AG, Eliazer S, Kelly OG, Agulnick AD, et al. Production of pancreatic hormone-expressing endocrine cells from human embryonic stem cells. Nat Biotechnol. 2006;24:1392–1401. doi: 10.1038/nbt1259. [DOI] [PubMed] [Google Scholar]
- 28.Jiang W, Shi Y, Zhao D, Chen S, Yong J, et al. In vitro derivation of functional insulin-producing cells from human embryonic stem cells. Cell Res. 2007;17:333–344. doi: 10.1038/cr.2007.28. [DOI] [PubMed] [Google Scholar]
- 29.Shim JH, Kim SE, Woo DH, Kim SK, Oh CH, et al. Directed differentiation of human embryonic stem cells towards a pancreatic cell fate. Diabetologia. 2007;50:1228–1238. doi: 10.1007/s00125-007-0634-z. [DOI] [PubMed] [Google Scholar]
- 30.Sakaguchi T, Kuroiwa A, Takeda H. A novel sox gene, 226D7, acts downstream of Nodal signaling to specify endoderm precursors in zebrafish. Mech Dev. 2001;107:25–38. doi: 10.1016/s0925-4773(01)00453-1. [DOI] [PubMed] [Google Scholar]
- 31.Stafford D, White RJ, Kinkel MD, Linville A, Schilling TF, et al. Retinoids signal directly to zebrafish endoderm to specify insulin-expressing β-cells. Development. 2006;133:949–956. doi: 10.1242/dev.02263. [DOI] [PubMed] [Google Scholar]
- 32.Waxman JS, Yelon D. Comparison of the expression patterns of newly identified zebrafish retinoic acid and retinoid X receptors. Dev Dyn. 2007;236:587–595. doi: 10.1002/dvdy.21049. [DOI] [PubMed] [Google Scholar]
- 33.Kinkel MD, Eames SC, Alonzo MR, Prince VE. Cdx4 is required in the endoderm to localize the pancreas and limit beta-cell number. Development. 2008;135:919–929. doi: 10.1242/dev.010660. [DOI] [PubMed] [Google Scholar]
- 34.Wells JM, Melton DA. Early mouse endoderm is patterned by soluble factors from adjacent germ layers. Development. 2000;127:1563–1572. doi: 10.1242/dev.127.8.1563. [DOI] [PubMed] [Google Scholar]
- 35.Dessimoz J, Opoka R, Kordich JJ, Grapin-Botton A, Wells JM. FGF signaling is necessary for establishing gut tube domains along the anterior-posterior axis in vivo. Mech Dev. 2006;123:42–55. doi: 10.1016/j.mod.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 36.Grapin-Botton A, Majithia AR, Melton DA. Key events of pancreas formation are triggered in gut endoderm by ectopic expression of pancreatic regulatory genes. Genes Dev. 2001;15:444–454. doi: 10.1101/gad.846001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haremaki T, Tanaka Y, Hongo I, Yuge M, Okamoto H. Integration of multiple signal transducing pathways on Fgf response elements of the Xenopus caudal homologue Xcad3. Development. 2003;130:4907–4917. doi: 10.1242/dev.00718. [DOI] [PubMed] [Google Scholar]
- 38.Hebrok M, Kim SK, Melton DA. Notochord repression of endodermal Sonic hedgehog permits pancreas development. Genes Dev. 1998;12:1705–1713. doi: 10.1101/gad.12.11.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Manfroid I, Delporte F, Baudhuin A, Motte P, Neumann CJ, Voz ML, Martial JA, Peers B. Reciprocal endoderm-mesoderm interactions mediated by fgf24 and fgf10 govern pancreas development. Development. 2007;134:4011–4021. doi: 10.1242/dev.007823. [DOI] [PubMed] [Google Scholar]
- 40.Dong PD, Munson CA, Norton W, Crosnier C, Pan X, et al. Fgf10 regulates hepatopancreatic ductal system patterning and differentiation. Nat Genet. 2007;39:397–402. doi: 10.1038/ng1961. [DOI] [PubMed] [Google Scholar]
- 41.Apelqvist A, Ahlgren U, Edlund H. Sonic hedgehog directs specialised mesoderm differentiation in the intestine and pancreas. Curr Biol. 1997;7:801–804. doi: 10.1016/s0960-9822(06)00340-x. [DOI] [PubMed] [Google Scholar]
- 42.Hebrok M. Hedgehog signaling in pancreas development. Mech Dev. 2003;120:45–57. doi: 10.1016/s0925-4773(02)00331-3. [DOI] [PubMed] [Google Scholar]
- 43.Roy S, Qiao T, Wolff C, Ingham PW. Hedgehog signaling pathway is essential for pancreas specification in the zebrafish embryo. Curr Biol. 2001;11:1358–1363. doi: 10.1016/s0960-9822(01)00402-x. [DOI] [PubMed] [Google Scholar]
- 44.diIorio PJ, Moss JB, Sbrogna JL, Karlstrom RO, Moss LG. Sonic hedgehog is required early in pancreatic islet development. Dev Biol. 2002;244:75–84. doi: 10.1006/dbio.2002.0573. [DOI] [PubMed] [Google Scholar]
- 45.Chung WS, Stainier DY. Intra-endodermal interactions are required for pancreatic beta cell induction. Dev Cell. 2008;14:582–593. doi: 10.1016/j.devcel.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tiso N, Filippi A, Pauls S, Bortolussi M, Argenton F. BMP signalling regulates anteroposterior endoderm patterning in zebrafish. Mech Dev. 2002;118:29–37. doi: 10.1016/s0925-4773(02)00252-6. [DOI] [PubMed] [Google Scholar]
- 47.Kumar M, Jordan N, Melton D, Grapin-Botton A. Signals from lateral plate mesoderm instruct endoderm toward a pancreatic fate. Dev Biol. 2003;259:109–122. doi: 10.1016/s0012-1606(03)00183-0. [DOI] [PubMed] [Google Scholar]
- 48.Kim SK, Hebrok M, Li E, Oh SP, Schrewe H, et al. Activin receptor patterning of foregut organogenesis. Genes Dev. 2000;14:1866–1871. [PMC free article] [PubMed] [Google Scholar]
- 49.Spagnoli FM, Brivanlou AH. The Gata5 target, TGIF2, defines the pancreatic region by modulating BMP signals within the endoderm. Development. 2008;135:451–461. doi: 10.1242/dev.008458. [DOI] [PubMed] [Google Scholar]
- 50.Field HA, Dong PD, Beis D, Stainier DY. Formation of the digestive system in zebrafish. II. Pancreas morphogenesis. Dev Biol. 2003;261:197–208. doi: 10.1016/s0012-1606(03)00308-7. [DOI] [PubMed] [Google Scholar]
- 51.Horne-Badovinac S, Lin D, Waldron S, Schwarz M, Mbamalu G, et al. Positional cloning of heart and soul reveals multiple roles for PKC lambda in zebrafish organogenesis. Curr Biol. 2001;11:1492–1502. doi: 10.1016/s0960-9822(01)00458-4. [DOI] [PubMed] [Google Scholar]
- 52.Ward AB, Warga RM, Prince VE. Origin of the zebrafish endocrine and exocrine pancreas. Dev Dyn. 2007;236:1558–1569. doi: 10.1002/dvdy.21168. [DOI] [PubMed] [Google Scholar]
- 53.Matsui T, Raya A, Kawakami Y, Callol-Massot C, Capdevila J, et al. Noncanonical Wnt signaling regulates midline convergence of organ primordia during zebrafish development. Genes Dev. 2005;19:164–175. doi: 10.1101/gad.1253605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim HJ, Schleiffarth JR, Jessurun J, Sumanas S, Petryk A, et al. Wnt5 signaling in vertebrate pancreas development. BMC Biol. 2005;3:23. doi: 10.1186/1741-7007-3-23. 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zecchin E, Filippi A, Biemar F, Tiso N, Pauls S, et al. Distinct delta and jagged genes control sequential segregation of pancreatic cell types from precursor pools in zebrafish. Dev Biol. 2007;301:192–204. doi: 10.1016/j.ydbio.2006.09.041. [DOI] [PubMed] [Google Scholar]
- 56.Gradwohl G, Dierich A, LeMeur M, Guillemot F. neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc Natl Acad Sci U S A. 2000;97:1607–1611. doi: 10.1073/pnas.97.4.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kawaguchi Y, Cooper B, Gannon M, Ray M, MacDonald RJ, et al. The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nat Genet. 2002;32:128–134. doi: 10.1038/ng959. [DOI] [PubMed] [Google Scholar]
- 58.Zecchin E, Mavropoulos A, Devos N, Filippi A, Tiso N, et al. Evolutionary conserved role of ptf1a in the specification of exocrine pancreatic fates. Dev Biol. 2004;268:174–184. doi: 10.1016/j.ydbio.2003.12.016. [DOI] [PubMed] [Google Scholar]
- 59.Lin JW, Biankin AV, Horb ME, Ghosh B, Prasad NB, et al. Differential requirement for ptf1a in endocrine and exocrine lineages of developing zebrafish pancreas. Dev Biol. 2004;270:474–486. doi: 10.1016/j.ydbio.2004.02.023. [DOI] [PubMed] [Google Scholar]
- 60.Dong PD, Provost E, Leach SD, Stainier DY. Graded levels of Ptf1a differentially regulate endocrine and exocrine fates in the developing pancreas. Genes Dev. 2008;22:1445–1450. doi: 10.1101/gad.1663208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Leach SD. Epithelial differentiation in pancreatic development and neoplasia: new niches for nestin and Notch. J Clin Gastroenterol. 2005;39:S78–S82. doi: 10.1097/01.mcg.0000155547.83901.a3. [DOI] [PubMed] [Google Scholar]
- 62.Apelqvist A, Li H, Sommer L, Beatus P, Anderson DJ, et al. Notch signaling controls pancreatic cell differentiation. Nature. 1999;400:877–881. doi: 10.1038/23716. [DOI] [PubMed] [Google Scholar]
- 63.Hald J, Hjorth JP, German MS, Madsen OD, Serup P, Jensen J. Activated Notch1 prevents differentiation of pancreatic acinar cells and attenuate endocrine development. Dev Biol. 2003;260:426–437. doi: 10.1016/s0012-1606(03)00326-9. [DOI] [PubMed] [Google Scholar]
- 64.Murtaugh LC, Stanger BZ, Kwan KM, Melton DA. Notch signaling controls multiple steps of pancreatic differentiation. Proc Natl Acad Sci U S A. 2003;100:14920–14925. doi: 10.1073/pnas.2436557100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Esni F, Ghosh B, Biankin AV, Lin JW, Albert MA, et al. Notch inhibits Ptf1 function and acinar cell differentiation in developing mouse and zebrafish pancreas. Development. 2004;131:4213–4224. doi: 10.1242/dev.01280. [DOI] [PubMed] [Google Scholar]
- 66.Lai EC. Notch signaling: control of cell communication and cell fate. Development. 2004;131:965–973. doi: 10.1242/dev.01074. [DOI] [PubMed] [Google Scholar]
- 67.Wan H, Korzh S, Li Z, Mudumana SP, Korzh V, et al. Analyses of pancreas development by generation of gfp transgenic zebrafish using an exocrine pancreas-specific elastaseA gene promoter. Exp Cell Res. 2006;312:1526–1539. doi: 10.1016/j.yexcr.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 68.Curado S, Anderson RM, Jungblut B, Mumm J, Schroeter E, et al. Conditional targeted cell ablation in zebrafish: a new tool for regeneration studies. Dev Dyn. 2007;236:1025–1035. doi: 10.1002/dvdy.21100. [DOI] [PubMed] [Google Scholar]
- 69.Skromne I, Prince VE. Current perspectives in zebrafish reverse genetics: moving forward. Dev Dyn. 2008;237:861–882. doi: 10.1002/dvdy.21484. [DOI] [PubMed] [Google Scholar]
- 70.Kim HJ, Sumanas S, Palencia-Desai S, Dong Y, Chen JN, et al. Genetic analysis of early endocrine pancreas formation in zebrafish. Mol Endocrinol. 2006;20:194–203. doi: 10.1210/me.2005-0189. [DOI] [PubMed] [Google Scholar]
- 71.Yee NS, Lorent K, Pack M. Exocrine pancreas development in zebrafish. Dev Biol. 2005;284:84–101. doi: 10.1016/j.ydbio.2005.04.035. [DOI] [PubMed] [Google Scholar]
- 72.Ober EA, Verkade H, Field HA, Stainier DY. Mesodermal Wnt2b signaling positively regulates liver specification. Nature. 2006;442:688–691. doi: 10.1038/nature04888. [DOI] [PubMed] [Google Scholar]
- 73.Serafimidis I, Rakatzi I, Episkopou V, Gouti M, Gavalas A. Novel effectors of directed and Ngn3-mediated differentiation of mouse embryonic stem cells into endocrine pancreas progenitors. Stem Cells. 2008;26:3–16. doi: 10.1634/stemcells.2007-0194. [DOI] [PubMed] [Google Scholar]
- 74.Kroon E, Martinson LA, Kadoya K, Bang AG, Kelly OG, et al. Pancreatic endoderm derived from human embryonic stem cells generates glucose-responsive insulin-secreting cells in vivo. Nat Biotechnol. 2008;26:443–452. doi: 10.1038/nbt1393. [DOI] [PubMed] [Google Scholar]
- 75.Elo B, Villano CM, Govorko D, White LA. Larval zebrafish as a model for glucose metabolism: expression of phosphoenolpyruvate carboxykinase as a marker for exposure to anti-diabetic compounds. J Mol Endocrinol. 2007;38:433–440. doi: 10.1677/JME-06-0037. [DOI] [PubMed] [Google Scholar]
- 76.Gleeson M, Connaughton V, Arneson LS. Induction of hyperglycaemia in zebrafish (Danio rerio) leads to morphological changes in the retina. Acta Diabetol. 2007;44:157–163. doi: 10.1007/s00592-007-0257-3. [DOI] [PubMed] [Google Scholar]
- 77.Murphey RD, Zon LI. Small molecule screening in the zebrafish. Methods. 2006;39:255–261. doi: 10.1016/j.ymeth.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 78.Mavropoulos A, Devos N, Biemar F, Zecchin E, Argenton F, et al. sox4b is a key player of pancreatic alpha cell differentiation in zebrafish. Dev Biol. 2005;285:211–223. doi: 10.1016/j.ydbio.2005.06.024. [DOI] [PubMed] [Google Scholar]
- 79.Wendik B, Maier E, Meyer D. Zebrafish mnx genes in endocrine and exocrine pancreas formation. Dev Biol. 2004;268:372–383. doi: 10.1016/j.ydbio.2003.12.026. [DOI] [PubMed] [Google Scholar]
- 80.Milewski WM, Duguay SJ, Chan SJ, Steiner DF. Conservation of PDX-1 structure, function, and expression in zebrafish. Endocrinology. 1998;139:1440–1449. doi: 10.1210/endo.139.3.5768. [DOI] [PubMed] [Google Scholar]
- 81.Pauls S, Zecchin E, Tiso N, Bortolussi M, Argenton F. Function and regulation of zebrafish nkx2.2a during development of pancreatic islet and ducts. Dev Biol. 2007;304:875–890. doi: 10.1016/j.ydbio.2007.01.024. [DOI] [PubMed] [Google Scholar]
- 82.Inoue A, Takahashi M, Hatta K, Hotta Y, Okamoto H. Developmental regulation of islet-1 mRNA expression during neuronal differentiation in embryonic zebrafish. Dev Dyn. 1994;199:1–11. doi: 10.1002/aja.1001990102. [DOI] [PubMed] [Google Scholar]
- 83.Krauss S, Maden M, Holder N, Wilson SW. Zebrafish pax[b] is involved in the formation of the midbrain-hindbrain boundary. Nature. 1992;360:87–89. doi: 10.1038/360087a0. [DOI] [PubMed] [Google Scholar]
- 84.Nornes S, Clarkson M, Mikkola I, Pedersen M, Bardsley A, et al. Zebrafish contains two pax6 genes involved in eye development. Mech Dev. 1998;77:185–196. doi: 10.1016/s0925-4773(98)00156-7. [DOI] [PubMed] [Google Scholar]
- 85.Korzh V, Sleptsova I, Liao J, He J, Gong Z. Expression of zebrafish bHLH genes ngn1 and nrd defines distinct stages of neural differentiation. Dev Dyn. 1998;213:92–104. doi: 10.1002/(SICI)1097-0177(199809)213:1<92::AID-AJA9>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 86.Devos N, Deflorian G, Biemar F, Bortolussi M, Martial JA, et al. Differential expression of two somatostatin genes during zebrafish embryonic development. Mech Dev. 2002;115:133–137. doi: 10.1016/s0925-4773(02)00082-5. [DOI] [PubMed] [Google Scholar]
- 87.Delporte F. 2008 pers comm. [Google Scholar]
- 88.Papasani MR, Robison BD, Hardy RW, Hill RA. Early developmental expression of two insulins in zebrafish (Danio rerio) Physiol Genomics. 2006;27:79–85. doi: 10.1152/physiolgenomics.00012.2006. [DOI] [PubMed] [Google Scholar]
- 89.Sun Z, Hopkins N. vhnf1, the MODY5 and familial GCKD-associated gene, regulates regional specification of the zebrafish gut, pronephros, and hindbrain. Genes Dev. 2001;15:3217–3229. doi: 10.1101/gad946701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yee NS, Yusuff S, Pack M. Zebrafish pdx1 morphant displays defects in pancreas development and digestive organ chirality, and potentially identifies a multipotent pancreas progenitor cell. Genesis. 2001;30:137–140. doi: 10.1002/gene.1049. [DOI] [PubMed] [Google Scholar]
- 91.Wilson ME, Yang KY, Kalousova A, Lau J, Kosaka Y, et al. The HMG Box Transcription Factor Sox4 Contributes to the Development of the Endocrine Pancreas. Diabetes. 2005;54:3402–3409. doi: 10.2337/diabetes.54.12.3402. [DOI] [PubMed] [Google Scholar]
- 92.Harrison KA, Thaler J, Pfaff SL, Gu H, Kehrl JH. Pancreas dorsal lobe agenesis and abnormal islets of Langerhans in Hlxb9-deficient mice. Nat Genet. 1999;23:71–75. doi: 10.1038/12674. [DOI] [PubMed] [Google Scholar]
- 93.Li H, Arber S, Jessell TM, Edlund H. Selective agenesis of the dorsal pancreas in mice lacking homeobox gene Hlxb9. Nat Genet. 1999;23:67–70. doi: 10.1038/12669. [DOI] [PubMed] [Google Scholar]
- 94.Stoffers DA, Ferrer J, Clarke WL, Habener JF. Early-onset type-II diabetes mellitus (MODY4) linked to IPF1. Nat Genet. 1997;17:138–139. doi: 10.1038/ng1097-138. [DOI] [PubMed] [Google Scholar]
- 95.Sussel L, Kalamaras J, Hartigan-O'Connor DJ, Meneses JJ, Pedersen RA, et al. Mice lacking the homeodomain transcription factor Nkx2.2 have diabetes due to arrested differentiation of pancreatic beta cells. Development. 1998;125:2213–2221. doi: 10.1242/dev.125.12.2213. [DOI] [PubMed] [Google Scholar]
- 96.Ahlgren U, Pfaff SL, Jessell TM, Edlund T, Edlund H. Independent requirement for ISL1 in formation of pancreatic mesenchyme and islet cells. Nature. 1997;385:257–260. doi: 10.1038/385257a0. [DOI] [PubMed] [Google Scholar]
- 97.Sander M, Neubuser A, Kalamaras J, Ee HC, Martin GR, et al. Genetic analysis reveals that PAX6 is required for normal transcription of pancreatic hormone genes and islet development. Genes Dev. 1997;11:1662–1673. doi: 10.1101/gad.11.13.1662. [DOI] [PubMed] [Google Scholar]
- 98.Fajans SS, Bell GI, Polonsky KS. Molecular mechanisms and clinical pathophysiology of maturity-onset diabetes of the young. N Engl J Med. 2001;345:971–980. doi: 10.1056/NEJMra002168. [DOI] [PubMed] [Google Scholar]
- 99.Sladek R, Rocheleau G, Rung J, Dina C, Shen L, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 100.Haqq AM, Farooqi IS, O'Rahilly S, Stadler DD, Rosenfeld RG, et al. Serum ghrelin levels are inversely correlated with body mass index, age, and insulin concentrations in normal children and are markedly increased in Prader-Willi syndrome. J Clin Endocrinol Metab. 2003;88:174–178. doi: 10.1210/jc.2002-021052. [DOI] [PubMed] [Google Scholar]
- 101.Lindner TH, Njolstad PR, Horikawa Y, Bostad L, Bell GI, et al. A novel syndrome of diabetes mellitus, renal dysfunction and genital malformation associated with a partial deletion of the pseudo-POU domain of hepatocyte nuclear factor-1beta. Hum Mol Genet. 1999;8:2001–2008. doi: 10.1093/hmg/8.11.2001. [DOI] [PubMed] [Google Scholar]
- 102.Sellick GS, Barker KT, Stolte-Dijkstra I, Fleischmann C, Coleman RJ, et al. Mutations in PTF1A cause pancreatic and cerebellar agenesis. Nat Genet. 2004;36:1301–1305. doi: 10.1038/ng1475. [DOI] [PubMed] [Google Scholar]
- 103.Trede NS, Medenbach J, Damianov A, Hung LH, Weber GJ, et al. Network of coregulated spliceosome components revealed by zebrafish mutant in recycling factor p110. Proc Natl Acad Sci U S A. 2007;104:6608–6613. doi: 10.1073/pnas.0701919104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chen JN, Haffter P, Odenthal J, Vogelsang E, Brand M, et al. Mutations affecting the cardiovascular system and other internal organs in zebrafish. Development. 1996;123:293–302. doi: 10.1242/dev.123.1.293. 293–302. [DOI] [PubMed] [Google Scholar]
- 105.Popperl H, Rikhof H, Chang H, Haffter P, Kimmel CB, et al. lazarus is a novel pbx gene that globally mediates hox gene function in zebrafish. Mol Cell. 2000;6:255–267. doi: 10.1016/s1097-2765(00)00027-7. [DOI] [PubMed] [Google Scholar]
- 106.diIorio P, Alexa K, Choe SK, Etheridge L, Sagerstrom CG. TALE-family homeodomain proteins regulate endodermal sonic hedgehog expression and pattern the anterior endoderm. Dev Biol. 2007;304:221–231. doi: 10.1016/j.ydbio.2006.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hegde A, Qiu NC, Qiu X, Ho SH, Tay KQ, et al. Genomewide expression analysis in zebrafish mind bomb alleles with pancreas defects of different severity identifies putative Notch responsive genes. PLoS ONE. 2008;3:e1479. doi: 10.1371/journal.pone.0001479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mayer AN, Fishman MC. Nil per os encodes a conserved RNA recognition motif protein required for morphogenesis and cytodifferentiation of digestive organs in zebrafish. Development. 2003;130:3917–3928. doi: 10.1242/dev.00600. [DOI] [PubMed] [Google Scholar]
- 109.Pack M, Solnica-Krezel L, Malicki J, Neuhauss SC, Schier AF, et al. Mutations affecting development of zebrafish digestive organs. Development. 1996;123:321–328. doi: 10.1242/dev.123.1.321. 321–328. [DOI] [PubMed] [Google Scholar]
- 110.Kinkel MD. 2008 unpublished observation. [Google Scholar]
- 111.Huang H, Vogel SS, Liu N, Melton DA, Lin S. Analysis of pancreatic development in living transgenic zebrafish embryos. Mol Cell Endocrinol. 2001;177:117–124. doi: 10.1016/s0303-7207(01)00408-7. [DOI] [PubMed] [Google Scholar]
- 112.Ng AN, de Jong-Curtain TA, Mawdsley DJ, White SJ, Shin J, et al. Formation of the digestive system in zebrafish: III. Intestinal epithelium morphogenesis. Dev Biol. 2005;286:114–135. doi: 10.1016/j.ydbio.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 113.Godinho L, Mumm JS, Williams PR, Schroeter EH, Koerber A, et al. Targeting of amacrine cell neurites to appropriate synaptic laminae in the developing zebrafish retina. Development. 2005;132:5069–5079. doi: 10.1242/dev.02075. [DOI] [PubMed] [Google Scholar]
- 114.Sakaguchi T, Kikuchi Y, Kuroiwa A, Takeda H, Stainier DY. The yolk syncytial layer regulates myocardial migration by influencing extracellular matrix assembly in zebrafish. Development. 2006;133:4063–4072. doi: 10.1242/dev.02581. [DOI] [PubMed] [Google Scholar]
- 115.Chocron S, Verhoeven MC, Rentzsch F, Hammerschmidt M, Bakkers J. Zebrafish Bmp4 regulates left-right asymmetry at two distinct developmental time points. Dev Biol. 2007;305:577–588. doi: 10.1016/j.ydbio.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 116.Wallace KN, Akhter S, Smith EM, Lorent K, Pack M. Intestinal growth and differentiation in zebrafish. Mech Dev. 2005;122:157–173. doi: 10.1016/j.mod.2004.10.009. [DOI] [PubMed] [Google Scholar]