Abstract
The oxidative phosphorylation (OXPHOS) system consists of five multimeric complexes embedded in the mitochondrial inner membrane. They work in concert to drive the aerobic synthesis of ATP. Mitochondrial and nuclear DNA mutations affecting the accumulation and function of these enzymes are the most common cause of mitochondrial diseases and have also been associated with neurodegeneration and aging. Several approaches for the assessment of the OXPHOS system enzymes have been developed. Based on the methods described elsewhere, we present here the creation and study of yeast models of mitochondrial OXPHOS deficiencies.
Keywords: Electron transport chain, Mitochondria, OXPHOS deficiencies, Yeast Model
INTRODUCTION
In this unit we present a series of methods (Basic Protocol and Alternate Protocols 1, 2, and 3) useful for creating yeast models of human mitochondrial disorders and determining the cause/effect relationship between a disease associated mutation and the pathological phenotype observed (validation of pathogenicity). The Basic Protocol describes the simpler scenario in which the respiratory deficiency of a yeast mutant in a conserved non essential gene can be complemented by its human homologue. Alternative approaches are required when heterologous complementation does not occur (Alternate Protocols 1 and 2) or when modelling mutations in yeast essential genes (Alternate protocol 3). We will try to guide the reader through the choice of the more appropriate approaches and the design of experimental plans adequate to each specific situation (see Strategic Planning).
We would like to emphasize that, in addition to the use of yeast models presented in this unit to validate allegedly pathogenic human mutations, further analysis of respiratory defects in yeast can give insight into the cellular pathological mechanisms responsible for the disease phenotype. This has been facilitated by the fact that many human genes associated with mitochondrial disorders have been first identified and characterized in yeast (Barrientos, 2003). The significance of yeast models is further supported by their extensive use in identifying genetic and pharmacological suppressors of respiratory deficiencies (Barrientos et al., 2009).
STRATEGIC PLANNING
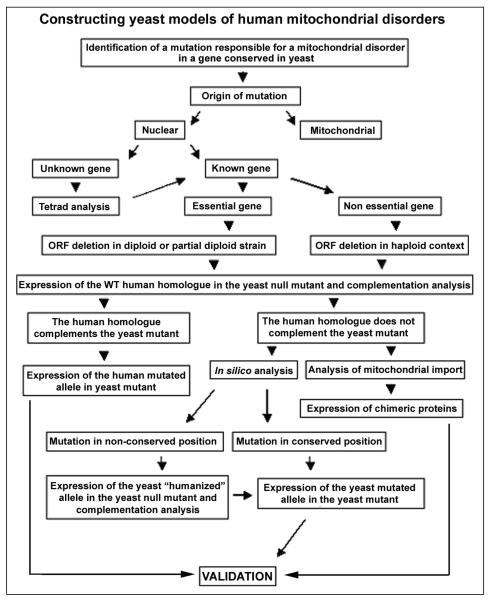
Constructing a yeast model of a human mitochondrial disorder does not present extreme technical difficulties but requires a carefully designed multistep plan (Figure 1).
Figure 1.
Strategic planning for the creation of yeast models of mitochondrial disorders
Mitochondrial disorders result from mutations in both the mitochondrial and the nuclear genomes. The study of pathogenic mutations in mitochondrial genes is not trivial because it involves the transformation of yeast mitochondria using biolistic approaches (Butow et al., 1996) which, due to space limitation, will not be described in this chapter. The existing yeast models of mitochondrial disorders associated with mitochondrial DNA mutations are however included in Table I.
Table I.
Existing yeast models of mitochondrial disorders. NP, unpublished
| Human gene |
Yeast homologue |
Disease | Literature |
|---|---|---|---|
| Nuclear origin | |||
| ABC7 | ATM1 | Sideroblastic anemia and ataxia | (Allikmets et al., 1999) |
| ANT1 | AAC2 | adPEO, hypertrophic cardiomyopathy | (Kaukonen et al., 2000, Fontanesi et al., 2004, Lodi et al., 2006) |
| BCS1L | BCS1 | Encephalopathy, GRACILE syndrome | (de Lonlay et al., 2001) |
| COX6B1 | COX12 | Leukodystrophic encephalopathy and myopathy |
(Massa et al., 2008) |
| DDP1 | TIM8 | Mohr-Tranebjaerg syndrome | (Hofmann et al., 2002) |
| EFG1 | MEF1 | Leigh syndrome | (Valente et al., 2007) |
| EFTu | TUF1 | Leukodystrophic encephalopathy | (Valente et al., 2007) |
| FXN | YFH1 | Friedreich ataxia | (Cavadini et al., 2000, Krasilnikova and Mirkin, 2004) |
| GFER | ERV1 | Progressive myopathy with cataract | (Di Fonzo et al., 2009) |
| MPV17 | SYM1 | Infantile hepatic mtDNA depletion syndrome | (Spinazzola et al., 2006) |
| ORNT1 | ARG11 | Hyperammonemia | (Morizono et al., 2005) |
| POLG | MIP1 | adPEO, arPEO, Alpers syndrome | (Baruffini et al., 2006, Stuart et al., 2006) |
| SDH Fp | SDHA | Leigh syndrome | (Bourgeron et al., 1995) |
| SDHAF1 | YDR379C-A | Infantile leukoencephalomyopathy | (Ghezzi et al., 2009) |
| SDHB | SDH2 | Glomus tumor | (Goffrini et al., 2009) |
| SLC25A3 | MIR1-PIC2 | Hypertrophic cardiomyopathy | (Mayr et al., 2007) |
| SOD1 | SOD1 | ALS | (Goto et al., 2000) |
| SUCLG1 | LSC1 | mtDNA depletion syndrome | Fontanesi, NP |
| TAZ | TAZ1 | Barth syndrome | (Claypool et al., 2006) |
|
| |||
| Mitochondrial origin | |||
| ATP6 | ATP6 | NARP syndrome and MILS | (Rak et al., 2007, Kucharczyk et al., 2009) |
| CYB | CYB | Exercise intolerance and myopathy, encephalopathy, cardiomyopathy, septo-optic dysplasia |
(Lemesle-Meunier et al., 1993, Fisher et al., 2004) |
| tRNALeu | tRNALeu | MELAS | (Feuermann et al., 2003) |
Modelling mitochondrial disorders involving mutations in a conserved nuclear gene is simpler and will be the focus of this unit. The first step involves the construction of the appropriate yeast strain in which the effects of the mutated human gene will be tested. To this purpose, it is crucial to know whether the gene is essential to support yeast life. With the exception of a few dubious open reading frames, the information is available at the S. cerevisiae web site.
Alternatively, the essential nature of a gene can be easily determined by tetrad analysis of the diploid heteroallelic strain. If the gene is non-essential, it can be deleted in a haploid context and the cells will survive in media containing fermentable carbon sources (Basic Protocol). If the gene is essential, the deletion of a single copy of the ORF needs to be performed in the presence of a second wild-type copy, either carried on a replicative vector (partial diploid context) or in a diploid context, to allow for the survival of the mutant cells (Alternate Protocol 3).
A collection of yeast null mutants in non-essential genes obtained thanks to the effort of the world wide yeast gene deletion project is also commercially available in a particular genetic background (see commentary). Once obtained, the appropriate yeast strain can be used as an in vivo cellular “laboratory” to test the effect of the single disease mutation on respiratory metabolism thus determining its specific role in the respiratory defect. If the wild type human gene is able to functionally complement the respiratory defect of the corresponding yeast mutant, the analysis of the residual function retained by the disease gene can be determined using this heterologous complementation test (Basic Protocol).
In some cases, if heterologous complementation cannot be properly assessed because the human protein fails to be imported into yeast mitochondria, the use of chimeric proteins targeted to mitochondria by specific yeast mitochondrial import sequences can allow to bypass this limitation (Alternate Protocol 1). If the protein is imported but complementation does not occur, the yeast gene can be used to reproduce and study the mutations reported in patients. In particular, when the disease mutation affects a residue conserved between the yeast and human proteins, the mutation can be introduced at the equivalent position of the yeast gene, by site-directed mutagenesis. Instead, when alignments show that the mutations localize in non-conserved residues, it is necessary to previously construct yeast alleles carrying the wild-type human amino-acids at the correspondent position and verify that these mutations do not affect protein function (Alternate Protocol 2).
ANALYSIS OF THE MUTATION EFFECTS
Yeast strains carrying null alleles of non-essential genes involved in mitochondrial function and biogenesis are respiratory deficient. In most cases, the respiratory growth defect is complete. However, point mutant alleles frequently retain partial function, conferring the cells with a residual respiratory growth. While semi-quantitative analysis of the respiratory growth capacity of wild type and mutant strains, as described in the Basic Protocol, provide very useful information at gross, it is always indicated the use of quantitative methods to evaluate more precisely the effects of the null and point mutations on respiration. Among the quantitative methods useful for this purpose are all the polarographic and spectrophotometric analyses described in UNIT 19.3. Yeast mitochondria can be purified as described elsewhere (Herrmann et al., 1994) and additionally used to spectrophotometrically analyze the profile of mitochondrial cytochromes (Tzagoloff et al., 1975).
BASIC PROTOCOLS
Basic Protocol 1
MODELLING PATHOGENIC MUTATIONS IN A CONSERVED NON-ESSENTIAL GENE
The first task when creating a yeast model of human mitochondrial disorder is to remove or knockout the wild type gene. This can be easily accomplished in the yeast S. cerevisiae by taking advantage of its ability to perform homologous recombination with very short regions of homology (~40 nucleotides). Following the one-step gene insertion method (Rothstein, 1983), the gene to be disrupted will be substituted by a disruptor prototrophic marker gene, commonly coding for a factor necessary for the synthesis of an amino acid (i.e. histidine, leucine or tryptophan) or a nucleobase (i.e. adenine or uracil) or that confers resistance to an antibiotic (i.e. geneticin). The yeast null mutant is then used to test in vivo the pathogenic effects of mitochondrial disease associated alleles on functionality and biogenesis of the mitochondrial respiratory chain and oxidative phosphorylation system using the biochemical analyses described above.
Materials
Agarose
Tris-Acetate-EDTA (TAE)
DNA extraction kit
YP-D solid and liquid media
Tris-EDTA-Lithium Acetate (TEL) solution
2 mg/ml salmon sperm carrier DNA
40% polyethylene glycol (PEG) solution
dH2O
WO-D solid and liquid media + amino-acids and nucleobases
Geneticin/G418 (Gibson)
Solubilization solution
10% Sodium dodecyl sulfate (SDS) (w/v)
8M Ammonium acetate
Isopropanol
80% Ethanol (v/v)
Restriction enzymes and digestion buffers
T4 DNA ligase and buffer
E. coli competent cells and selective media
DNA Miniprep kit
DNA Maxiprep kit
QuickChange II Site-Directed Mutagenesis kit (Stratagene)
YP and WO + E/G/L/P/A solid media + amino-acids and nucleobases
A 30°C incubator with shaker.
Preparation of yeast null mutants
- 1. Design primers to amplify by PCR the disruptor gene flanked by 40-60 nucleotides of DNA sequence homologous to the DNA sequence flanking the gene that is to be knocked out. Use commercially available plasmids containing the disruptor gene or wild type genomic DNA as the template for the PCR reaction. One of the most commonly used disruptor marker gene is the KANRMX4 cassette conferring resistance to geneticin.
- For information on setting and optimizing PCR amplifications see Current Protocols in Molecular Biology UNIT 15.1.
2. Purify the PCR fragment directly from the reaction suspension. If required to eliminate unspecific co-amplification products, separate the DNA electrophoretically in a 0.8% agarose gel (w/v) in TAE 1X, excise the band of interest and extract the DNA from agarose using a commercially available DNA extraction kit.
- 1. Use the purified PCR fragment to transform the wild-type yeast cells.
- Several methods can be used to transform yeast cells. Here we describe the transformation by lithium acetate/single stranded DNA carrier/Polyethylene glycol method (Schiestl and Gietz, 1989) because it is highly efficient and relatively quick and simple to perform. For more yeast transformation techniques see current Protocols in Molecular Biology UNIT 13.7.
Yeast Transformation
2. Grow 10 ml pre-culture of wild type cells overnight in complete YP-D media at 30°C with constant shaking at 200 × g.
- 3. Transfer an aliquot of the pre-culture to a fresh 10 ml flask of YP-D to obtain a final OD600 of ~ 0.1-0.2. Grow the cells until the culture reaches a confluency at OD600 of ~ 0.4-0.5 (approximately 2-3 hours).
- This is crucial because only cells in the mid logarithmic phase of growth will become significantly competent for transformation.
4. Transfer 1.5 ml of the culture to a microcentrifuge tube and pellet the cells at 1,500 × g for 5 minutes at room temperature.
5. Wash the cells in 1 ml of sterile TEL and resuspend them in 0.1 ml of sterile TEL.
6. Denature the salmon sperm carrier DNA (2 mg/ml) by boiling an aliquot at 95°C for 5 minutes and cool it in ice.
7. To the cell suspension, add 5 μl of salmon sperm carrier DNA premixed with 1-4 μg of transforming DNA in a volume of less than 15 μl. Include a negative control (sample without transforming DNA) to check for contaminations and the possible reversion of the marker locus and a positive control (sample transform with an empty circular plasmid of known concentration) to evaluate the transformation efficiency.
8. Incubate for 30 minutes at 30°C without shaking.
- 9. Add 0.7 ml of sterile 40% polyethylene glycol (PEG) prepared in TEL and mix it by pipetting.
- Because the PEG solution is very viscous, it is essential to mix the cells well after adding the solution.
10. Incubate for 30-60 minutes at room temperature without shaking.
11. Heat-shock the cells for 10 minutes at 42°C and place them on ice for 2 minutes. If the KANRMX4 cassette is used as deletion marker, resuspend the cells in 3 ml of YP-D liquid media and incubate at 30°C with constant shaking for 2 hours prior to proceed.
12. Pellet the cells by centrifugation at 1,500 × g for 1 minute at room temperature and wash them in 0.5 ml of sterile dH2O.
13. Resuspend the cells in 0.1 ml of sterile dH2O and plate them on selective WO-D solid media lacking the appropriate amino acid, nucleobase or containing the selective antibiotic. In the case the KANRMX4 cassette is used as disruptor marker gene, add 200 μg/ml geneticin.
14. Incubate the plates at 30°C. After 3-4 days single colonies start to appear. Transfer a few transformant clones on selective WO-D solid media to obtain small patches of cells. Allow them to grow at 30°C.
15. Confirm the selected clones by testing for integration of the selectable marker in the correct locus and consequent disruption of the desired gene. This can be done by amplifying the locus of interest by PCR using genomic DNA as the template.
Genomic DNA extraction
16. Yeast genomic DNA is extracted from 1 ml of a 10 ml overnight confluent culture in YP-D media.
17. Pellet the cells and wash them once with 0.5 ml of dH2O.
18. Resuspend the cells in 150 μl of cell solubilization solution and incubate them for 1 hour at 37°C without shaking to digest the cell wall.
21. Solubilize the cell membranes by adding 20 μl of 10% SDS (w/v) and vortex.
22. To facilitate the precipitation of the DNA, add 100 μl of 8M ammonium acetate, vortex and incubate the sample at −20°C for 15 minutes.
23. Centrifuge at 10,000 × g for 10 minutes at 4°C.
24. Transfer 180 μl of the supernatant fraction to a new tube and precipitate the DNA by adding 120 μl of isopropanol and incubate 5 minutes at room temperature.
25. Pellet the DNA by centrifugation at 10,000 × g for 15 minutes at 4°C and wash it once with 300 μl 80% ethanol.
26. Air-dry the pellet of DNA and resuspend it in 30 μl of sterile water.
27. Analyze the genetic locus of interest by PCR using primers that allow for the amplification of fragments of different sizes in the mutant and in the wild type strains.
Cloning of wild-type human cDNA
- 28. Amplify by PCR the cDNA of the human gene of interest, the homologue of the yeast gene that was previously disrupted, with primers that include the appropriate restriction sites.
- Human cDNA clones to use as template can be obtained from Open Biosystems.
- 29. Digest with the appropriate restriction enzymes the PCR product and a suitable yeast integrative vector carrying a selective marker appropriate for the yeast strain to transform.
- Yeast vectors with different selective markers and replication origin are listed in Current Protocols in Molecular Biology UNIT 13.4. Restriction enzymes characteristics can be found in Current Protocols in Molecular Biology UNIT 3.1.
30. Purify the digested DNA from a 0.8% agarose gel in TAE 1X using a commercially available DNA Gel Extraction Kit.
31. Ligate the purified DNA fragments at 16°C overnight with T4 DNA ligase, transform the ligation into E. coli competent cells, commercially available, and plate the bacteria on selective media.
32. Test the transformant clones for the presence of the correct construct by PCR and/or appropriate restriction digestion of isolated plasmid minipreps.
33. Grow overnight the positive clones and isolate the constructs by standard plasmid DNA maxipreps.
34. Linearize 1-4 μg of the construct by digestion with an enzyme that cuts only once and within the selectable prototrophic marker.
35. Transform the linearized construct into the yeast knockout strain by the Lithium Acetate method as described above.
36. Test the transformants for plasmid integration by PCR using as the template genomic DNA extracted as described above.
37. Test the ability of the human gene to complement the deletion mutant phenotype by serial dilutions growth test.
Serial dilution growth test
38. Grow overnight the yeast strain of interest, in this case the null mutant expressing the human homologue gene, and the control strains: the wild-type (positive control) and the null mutant (negative control), in YP-D liquid media at 30°C with constant shaking at 200 × g. The cultures should reach late logarithmic phase of growth.
- 39. Estimate cells concentration by chamber counting or by spectrophotometric measurement of absorbance at 600 nm.
- The conversion absorbance/cells concentration should be empirically determine in the laboratory and is expected to be approximately: 1 OD600 equal to 2 × 107 cells/ml.
40. Prepare for each strain 3-5 serial 10x dilutions in sterile dH2O. The samples concentration should be between 107 and 103 cells/ml.
- 41. Deposit a 5-10 μl drop of each sample on solid media in an ordered grid to obtain a series of spots of known cellular concentration.
- Sample concentration and drops volume are chosen to obtain a series of spot between 105 and 10 cells/spot. Drops of volume less then 5 μl increase the pipetting error, drops of volume higher than 10 μl cannot be easily absorbed on the solid media.
42. Repeat the step on different solid media: YP-D used as loading control, YP and/or WO media containing respiratory carbon sources such as ethanol, glycerol, lactate, pyruvate and acetate, alone or in combination (see commentary).
- 43. Incubate the plates at 30°C for 2-6 days depending on the strains doubling time in the different media.
- On YP-D media all the strains should grow equally, producing energy by fermentation. On respiratory media the respiratory deficient null mutant strain will have reduced or abolished growth compared to wild-type. Heterologous functional complementation is assessed by the ability of the human gene to partially or totally restore respiratory growth to the null mutant strain. When complementation is observed, the human gene construct can be used to reproduce the mutations reported in patients by site-directed mutagenesis.
Site-directed Mutagenesis
- 44. Use the QuickChange II Site-Directed Mutagenesis kit (Stratagene), the appropriate oligonucleotide primers and the double strand circular vector containing the wild-type human gene as the template, for the site-directed mutagenesis reaction following the manufacturer instruction.
- The QuickChange II Site-Directed Mutagenesis kit (Stratagene) is a three step procedure able to generate point mutations and short insertion/deletions. The procedure is based on the PCR amplification of a circular template using two complementary primers containing the desired mutation. The PCR product is then digested with the DpnI restriction enzyme that recognize methylated and hemimethylated DNA and allows to eliminate the non mutated template DNA. Finally the mutated molecules are transformed into E. coli cells for nick repair and selection.
45. Isolate the construct from the E. coli clones by standard plasmid DNA minipreps. Since the high efficiency of the site-directed mutagenesis kit (approximately 80%), in the majority of the cases the screening of 4-6 clones is sufficient to isolate the correct mutagenized allele.
- 46. Sequence the entire insert using universal primers to verify the presence of the desire mutation and the absence of unwanted second site errors.
- For extended information on sequence techniques see Current Protocols in Molecular Biology Chapter 7.
Validation of the mutation pathogenic effect
47. Linearize 1-4 μg of the mutated construct by digestion with an enzyme that cuts only once and within the selectable prototrophic marker.
48. Transform the linearized construct into the yeast knockout strain by the Lithium Acetate Method as described above.
49. Test the transformants for plasmid integration by PCR using as the template genomic DNA extracted as described above.
50. Test the ability of the mutated allele to complement the deletion mutant phenotype by serial dilutions growth test as described above.
Alternate Protocol 1
MODELLING PATHOGENIC MUTATIONS IN A NONESSENTIAL GENE BY CONSTRUCTION OF CHIMERIC PROTEINS
Mislocalization and lack of import of the human protein into yeast mitochondria can prevent heterologous functional complementation. For this reason, in the cases in which heterologous complementation does not occur, it is recommended to test the steady-state levels and topology of the human protein in yeast mitochondria. Defective mitochondrial import can be solved in some cases by the use of yeast-human chimeric proteins (see example in Ref (Lodi et al., 2006)). To this purpose, the sequence coding for the yeast mitochondrial N-terminus import sequence of the homologous yeast protein is fused in frame to the human gene. If the targeting sequence is known, it is also possible to replace the human by the yeast counterpart. Alternatively, yeast mitochondrial import sequences frequently used for this purpose are the ones of the COX4 gene for targeting to the matrix and the CYTB2 for targeting to the inter-membrane space (Hu et al., 2008, Sturtz et al., 2001).
Materials
See Basic Protocol
Chimera construction
The steady-state level of the human protein and its mitochondrial localization are tested by western blot analysis using high quality yeast mitochondria purified as previously reported (Barrientos et al., 2004, Herrmann et al., 1994). If the human homologue protein is not correctly imported into yeast mitochondria, it would be indicated to proceed with the creation of a yeast-human chimeric protein.
Obtain oligonucleotide primers to amplify the yeast mitochondrial import sequence of choice and the human cDNA designed to have at their 5′-end restriction enzyme sequences that allow a three fragment cloning in the appropriate yeast integrative vector and the in frame fusion of import sequence and gene ORF.
Amplify by PCR reaction the yeast import sequence and the human gene using as template yeast genomic DNA and the human cDNA clone of interest, respectively.
Clone the two PCR fragments in the appropriate yeast integrative vector as described above.
Linearize 1-4 μg of the construct by digestion with an enzyme that cuts only once and within the selectable prototrophic marker.
Transform the linearized chimeric construct into the yeast knockout strain by the Lithium acetate method as described above.
Test the transformants for plasmid integration by PCR using as the template genomic DNA extracted as described above.
Test the import into yeast mitochondria and the sub-mitochondrial localization of the yeast-human chimeric protein as described elsewhere (Hatanaka et al., 2001).
Test the ability of the yeast-human chimeric gene to complement the deletion mutant phenotype by serial dilution growth test as described above.
If heterologous complementation is obtained, use the chimeric construct to introduce and to test the effects of allegedly pathogenic mutations as described above.
Alternate Protocol 2
MODELLING PATHOGENIC MUTATIONS IN A NON-ESSENTIAL GENE LACKING HETEROLOGUOS COMPLEMENTATION
Protein function frequently involves transient or permanent interactions with other partners. These interactions can be disrupted when a protein is expressed in a heterologous background. Probably for this reason, it is frequently observed that even when human and yeast homologue proteins have highly similar sequences and share conserved mitochondrial localization and function, the human wild-type gene fails to complement the yeast mutant strain. The creation of yeast models to evaluate allegedly pathogenic mutations is still possible by constructing yeast alleles equivalent to those of human patients.
Materials
See Basic Protocol
Sequence alignment
1. Prepare all protein sequences in the same file under the FASTA format.
2. Copy the sequences (with their corresponding headline) and paste them into the ClustalW2 program available on line.
- 3. Run alignment.
- For the ClustalW2 program more extended information see the European Bioinformatics Institute at http://www.ebi.ac.uk/Tools/clustalw2/index.html.
Construction of yeast alleles
4. Amplify by PCR the wild-type yeast gene with primers that include the appropriate restriction sites and using as template yeast genomic DNA.
5. Clone the PCR fragment in the appropriate yeast integrative vector as described above.
- 6. Introduce the mutations reported in patients in the wild-type yeast gene, at the equivalent position of the human gene, by site-directed mutagenesis. In the case in which the wild-type amino-acid at the equivalent position in the human and in the yeast proteins is not conserved, prepare an additional construct introducing the wild-type human amino-acid in the wild-type yeast gene.
- In yeast, redundant codons are used with different frequency, in particular for highly expressed genes (Bennetzen and Hall, 1982). This codon bias varies among organisms and has to be taken into account when designing the site-directed mutagenesis primers to guarantee efficient translation, in particular when the yeast gene is used to mimic the mutations observed in patients.
7. Linearize 1-4 μg of the constructs by digestion with an enzyme that cuts only once and within the selectable prototrophic marker.
8. Transform the linearized constructs into the yeast knockout strain by the Lithium acetate method as described above.
9. Test the transformants for plasmid integration by PCR using as the template genomic DNA extracted as described above.
10. Test the ability of the wild-type yeast gene and, in case of not conserved residues, of the “humanized” yeast allele to complement the deletion mutant phenotype by serial dilution growth tests as described above.
11. Validate the pathogenic effect of the mutation reported in patients and reproduced in the yeast allele as described above.
Alternate Protocol 3
MODELLING PATHOGENIC MUTATIONS IN AN ESSENTIAL GENE
Most genes primarily involved in the biogenesis of the mitochondrial respiratory chain are not essential in yeast. Exceptions include those required for more general functions such as protein import and folding. Mutations in these genes have been associated with human disease (Di Fonzo et al., 2009).
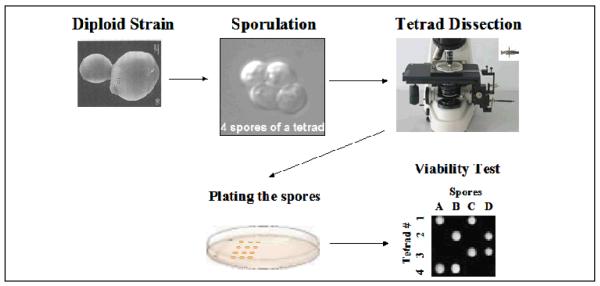
Yeast cells exist in both haploid and diploid state. While mutations in essential genes are lethal in haploid yeast cells, they can exist in heterozygosis in a diploid strain. Diploid cells can undergo induced meiosis producing 4 haploid spores (a tetrad) contained inside a sac called ascus. Upon tetrad dissection, characterization of the genotype and phenotype of the four spores allows for the analysis of the single meiotic event. During meiosis of an heteroallelic diploid strain two ascospores of each tetrad receive a copy of the wild-type allele and grow forming a colony while the other two ascospores receive a copy of the deleted allele and are able to grow only if the gene tested is not essential. In the case of essential genes, all the tetrads present a viability segregation pattern 2+:2−, as shown in Figure 2.
Figure 2.
Scheme of tetrad dissection analysis and vitality test of a heterozygous diploid strain deleted in an essential gene. Most micromanipulators used a fiberglass needle to separate the four haploid spores and allow them to germinate individually forming isolated colonies. The spores adhere to the needle due to the formation of a water meniscus between the agar and the needle.
Plasmid shuffling is the strategy more commonly used to study the effects of mutations in essential genes (Sikorski and Boeke, 1991). It takes advantage of the possibility to delete a copy of an essential gene in a diploid strain producing a heteroallelic strain. This strain is then transformed with a replicative plasmid carrying the wild-type gene and a marker which can be both selected for and against, such as the URA3 gene. Sporulation is induced by nitrogen starvation and a haploid spore is isolated by tetrad analysis containing the disrupted allele and remaining viable only by virtue of the wild-type gene on the replicative plasmid.
After introducing in this strain the mutated alleles on integrative vectors carrying a different selection marker, the lost of the wild-type gene is induced by plating the cells in the presence of the appropriate inhibitor, an inert compound that is transformed into a toxic product by the enzymatic activity of the marker on the plasmid carrying the wild-type gene thus allowing to select against the cells that retained the construct. The most commonly used inhibitor for the specificity and efficacy of its selection, is 5-fluoroorotic acid (5-FOA). 5-FOA prevents the growth of URA3 strains because the orotin-5′-phosphate decarboxylase encoded by the URA3 gene converts the inert compound 5-FOA to the toxic 5-fluorouracil (Boeke et al., 1984). Other useful but less frequently used compounds include ureidosuccinate, α-aminoadipate and methyl mercury which inhibit the growth of URA3, (Bach and Lacroute, 1972), LYS2 and LYS5 (Chattoo et al., 1979), and MET2 and MET5 strains (Singh and Sherman, 1974), respectively.
The major limitation of this strategy is the assumption that the mutant alleles partially retain a residual function and can support cellular life. In cases in which the mutations completely abolish protein function, a more appropriate but time consuming approach consists in following the segregation of the mutated gene by performing tetrad analysis of a heteroallelic strain transformed with an integrative vector carrying the mutated alleles.
Materials
YP-D liquid and solid media
WO-D solid media
Amino acids + nucleobases
K-Acetate solid media
β-glucuronidase (Sigma)
dH2O
5-FOA solid media
Micromanipulator
Crossing of haploid strains and heterozygous diploid strain preparation
- 1. Grow overnight at 30°C with constant shaking at 200 × g, 10 ml pre-cultures in complete YP-D media of two haploid wild type strains of opposite mating type (a and α), same genetic background and both ura3 mutant, but at least one different auxotrophy each.
- The presence of a different auxotrophic marker for each strain allows for a fast and simple selection of the diploid. It can be obtained, if necessary, by transforming the wild-type strains with empty plasmids carrying different markers.
2. Plate on YP-D solid media a 10 μl drop of each strain as control and a 10 μl drop of the two strains mix. Incubate the plate at 30°C until the cells grow forming a homogeneous patch (approximately 1-2 days).
- 3. Replicate the cells by replica plating on selective WO-D solid media and incubate the plate at 30°C for a couple of days.
- The selective medium is chosen to allow exclusively the growth of diploid cells. The construction of diploid strains can be beneficial not only to study mutations in essential genes, but also to determine mutations dominance/recessivity and to analyze the combined effect of mutations in two alleles.
4. Transform by lithium acetate method the diploid wild-type strain with the PCR product designed to delete the gene of interest as described above.
- 5. Test the integration of the selectable marker in the correct locus and consequent disruption of the gene of interest by PCR, using as template genomic DNA as described above.
- In this case, it is expected to obtain by PCR amplification both the fragment corresponding to the wild-type locus and the fragment corresponding to the deleted locus since a diploid heterozygous strain should be selected.
6. Clone the wild-type yeast gene in a URA3 replicative (centromeric or episomal) vector as described above.
7. Using the lithium acetate method described above, transform the diploid heterozygous strain with the construct expressing the yeast wild-type gene from a replicative URA3 plasmid.
Tetrad analysis
- 8. Plate the transformant strains on K-Acetate solid media and incubate the plate at 30°C for 5-6 days until a high percentage of diploid cells start to sporulate.
- After 4 days of incubation, check daily the formation of tetrads under a regular optic microscope using a 40X or 100X magnification objective lens (Figure 2).
- 9. Resuspend in a 15 ml falcon tube a pinhead size sample of the culture using a flamed loop in 1 ml sterile dH2O. Add 20 μl of β-glucuronidase and incubate 10-20 minute at room temperature to degrade the ascus wall.
- The digestion of the ascus wall can be followed under the optic microscope. Tetrads start to stretch and the four spores become clearly distinguishable. Avoid over-digestion to prevent damaging or loosing the haploid cells.
10. Add 5 ml sterile dH2O to dilute the enzyme. Pellet the tetrads by centrifugation at 1500 × g for 5 minute at room temperature. Carefully remove the supernatant and gently resuspend the cells in 1 ml sterile dH2O.
- 11. Micro-dissect at least 10 tetrads for each transformant strains on YP-D solid media using a micromanipulator. Incubate the plate at 30°C for 3-4 days, until the colonies are visible.
- Dissection of tetrads obtained from the heterozygous non-transformed diploid strain allows for the analysis/confirmation of the essentiality of a yeast gene (viability test shown in Figure 2).
- 12. Replicate the tetrads on selective WO-D solid media to test the presence of the wild-type construct and the segregation of the deleted allele.
- The deleted allele segregates during meiosis following Mendelian distribution, distribution 2+:2− that can be follow by testing the deletion marker phenotype.
Plasmid shuffling
13. Select a spore carrying the deleted allele in its genome and the wild-type construct.
14. Transform this strain by lithium acetate method with the appropriate integrative vectors expressing either the human mutated alleles or the yeast mutated alleles, previously described.
15. Test the transformants for plasmid integration by PCR using as the template genomic DNA extracted as described above.
16. Plate the transformant on selective 5-FOA solid media by spotting drops of cells suspension obtained by growing overnight at 30°C a 10 ml pre-culture in selective WO-D media.
17. Incubate the plates at 30°C for 4-7 days until the colonies that had lost the replicative plasmid carrying the wild-type gene start to appear.
18. Pick the colonies and confirm the Ura- phenotype by growing them in selective WO-D solid media ± uracil.
- 19. Once obtained the correct strain, analyze the effect of the pathological mutations on the respiratory phenotype by serial dilutions growth test as described above.
- In cases of point mutations in essential genes that cause only partial respiratory defects, it can be convenient to apply more stringent conditions and further characterize the mutations effects by testing the thermo-sensitivity of the mutations by incubating the cells at 24°C, 30°C and 37°C.
REAGENTS AND SOLUTIONS
Cell solubilization solution: 50 mM Tris-HCl, 10 mM ethylenediamine tetraacetic acid (EDTA) pH 8, 0.3% β-mercaptoethanol, and 0.5 mg/ml Zymolyase-20T. Prepare the solution freshly for each experiment.
DNA carrier solution: 2 mg/ml deoxyribonucleic acid sodium salt type III from Salmon testes in sterile TE solution (10 mM Tris-HCl, pH 8.0, 1 mM EDTA). Disperse the DNA into solution by passing it up and down repeatedly in a 10 ml pipette and mix vigorously in a magnetic stirrer until it dissolves completely. Aliquot and store at −20°C.
Lithium acetate solution (TEL): 10 mM Tris-HCl, pH 7.5, 1 mM EDTA, 100 mM Lithium Acetate. Sterilize and store at room temperature.
Polyethylene glycol solution (PEG): 40% Polyethylene glycol 4,000 (w/v) in TEL. Sterilize by filtering (do not autoclave). Because PEG easily degenerates, it is advised to prepare the solution freshly for each experiment or to store it at room temperature for a short period of time.
Tris-Acetate-EDTA (TAE): 40 mM Tris-acetate, 1mM EDTA pH 8. Store at room temperature
- Yeast media: the compositions of the yeast growth media routinely used were previously described (Myers et al., 1985).
- Complete media (YP): 1% yeast extract (w/v), 2% peptone (w/v).
- Minimum media (WO): 0.67% yeast nitrogen base (w/v).
- Different carbon sources to be added to YP and WO media:
- 2% glucose (D) (v/v)
- 2% ethanol (E) (v/v)
- 2% glycerol (G) (v/v)
- 2% galactose (GAL) (v/v)
- 2% DL-lactate (L) (v/v) pH 4.5
- 2% pyruvate (P) (v/v)
- 2% acetate (A) (v/v).
K-Acetate media: 10 g/l potassium acetate, 0.5 g/l glucose, 1 g/l yeast extract. 5-FOA media: 0.67% yeast nitrogen base (w/v), 2% glucose (v/v), 1 mg/ml 5-fluoroorotic acid (Sigma), 100 μg/ml uracil.
Agar (20 g/l) is added when required to obtain solid media. All culture media is prepared in distilled water. Complete media is autoclaved during 20 minutes at 20 lbs/sq. inch prior to use. 10X minimum media is sterilized by filtration and diluted with warm sterile dH2O ± agar. Carbon sources, amino acids and nucleobases are sterilized independently by autoclaving (D, G, GAL, L, P, A) or by filtration (amino acids and nucleobases). The commonly used yeast lab strains required addition to the minimum media of different combinations of nucleobases: 100 μg/ml adenine, 150 μg/ml uracil; and amino acids 50 μg/ml: leucine, tryptophan, histidine, methionine, lysine. Store at room temperature for short periods of time.
COMMENTARY
Background Information
To understand the mechanism and eventually treat genetic disorders, it is of primary importance the precise determination of the disease genetic cause. The establishment of genotype/phenotype relationships is frequently difficult in the absence of functional studies and can be challenging when new mutations in known disease associated genes present unexpected deficient phenotypes. Researchers have taken advantage of the similarity between yeast and human mitochondrial OXPHOS system to use the facultative aerobe/anaerobe yeast Saccharomyces cerevisiae as a model organism to assess the effect of mutations in conserved nuclear genes in OXPHOS enzyme assembly and function (Barrientos, 2003, Valnot et al., 2000). Because most basic metabolic pathways are largely conserved from yeast to human and yeast cells can be genetically manipulated with ease, they constitute suitable research models to study the mechanisms underlying human disorders. This is particularly true for the use of the yeast Saccharomyces cerevisiae to model diseases stemming from impaired mitochondrial metabolism, since this facultative aerobe/anaerobe organism is able to produce energy by substrates fermentation and to survive mutations abolishing its respiratory capacity (Barrientos, 2003).
Critical Parameters and Troubleshooting
The complete collection of yeast null mutants in non-essential genes is commercially available (EUROSCARF and Open Biosystems) in the BY4741/2 parental strain, a derivate of the S288c genetic background. To construct yeast models of human mitochondrial disorders, it is important to evaluate which yeast parental strain is the more appropriate to use. Several yeast strains, with different genetic backgrounds, commonly used in research laboratories, differ on their rate of endogenous cell respiration, level of cytochromes and several others parameters.
For example the S288c strain and its BY4741/42 derivates used in the yeast collection, but not the W303 strain, carry a mutation in the transcriptional activator Hap1p, that induces the expression of several respiratory genes depending on oxygen availability (Zitomer and Lowry, 1992). For this reason, S288c cells have a respiratory rate and cytochromes c + c1 accumulation significantly lower than cells with W303 and D273 genetic backgrounds (Gaisne et al., 1999). Mutations affecting other functions, such as cellular copper uptake, required for mitochondria biogenesis, are also known to be present in S288c wild-type strains. Even if these mutations are not completely compromising mitochondrial respiration and oxidative phosphorylation, they can reduce the respiratory growth of the wild-type strain thus making more difficult the assessment of mutant alleles retaining partial activity.
In the choice of the parental strain, it is also important to take into account its mitochondrial DNA stability. Yeast wild-type strains present a frequency of spontaneous mitochondrial mutations that ranges between approximately 0.5 and 2.5% in about 15 cells divisions at 30°C. In all S288 derivates including W303 the petite generation frequency increases up to approximately 15% at 37°C due to the presence in these strains of a mip1 (DNA polymerase gamma) termosensitive allele (Baruffini et al., 2007, Young and Court, 2008).
In S. cerevisiae, the heteroplasmic condition (presence of different populations of mtDNA molecules in the same cell) is transient and in a few generations mtDNA molecules segregate and cells become homoplasmic. In cases in which the gene of interest is involved in mtDNA maintenance and stability, the null mutant presents a complete or dramatic lost of mtDNA that does not allow the analysis of the direct effect of the mutated alleles on the mitochondrial respiratory chain. To bypass this limitation, it is possible, following the strategy design by Dr. TD Fox (Steele et al., 1996), to use a parental strain carrying in its mtDNA a selection marker that ensures mtDNA maintenance such as the mtARG8 gene in a nuclear arg8 mutated background.
Due to the different characteristics of the wild-type strains and to variations in the utilization of different respiratory carbon sources, it is appropriate, prior to testing the effects of mutated alleles, to carefully characterize the growth phenotype of the mutated strain in different respiratory media. In this way, it is possible to establish the best conditions to evaluate the respiratory defect. For example, even if both ethanol and lactate are respiratory carbon sources, ethanol can enter inside the cells by diffusion through the membrane while lactate import and utilization are tightly regulated at the transcriptional level (Lodi et al., 2002). In addition, the use of minimum synthetic media results in more stringent growth condition than complete media in which the large excess of amino-acids can increase an eventual background growth.
Heterologous complementation can be achieved by the use of replicative plasmids either centromeric or episomal that allow for a 2-5 fold to a 25-50 fold human gene overexpression, respectively. This is particularly indicated in cases in which only a weak complementation is observed by gene single copy expression. However, additional gene-dosage effects on mitochondrial function and biogenesis need to be carefully evaluated.
Construction of diploid strains and tetrad analysis are basic yeast genetic techniques useful not only in the study of mutations in essential genes but also to analyze the effects of combined mutations and mutations dominance/recessivity. However, to avoid these relative longer procedures, in the case of mutations in essential genes, it is possible to use haploid strains, commercially available, in which the essential gene of interest is under the control of a repressible promoter and its expression can be turn off just by addition of the appropriate inhibitor in the growth media. However, because residual gene expression is frequently observed, the use of these strains needs to be carefully evaluated.
Anticipated Results
In the cases in which human or yeast alleles carrying the allegedly pathogenic mutations reported in patients are unable to complement the respiratory deficient/lethal phenotype of yeast null mutants in the equivalent genes (e.g. the strains expressing mutated alleles grow less in respiratory media than the strains expressing the wild-type genes), it is possible to conclude that there is a cause-effect relationship between the analyzed mutations and the pathological phenotype observed. The measurement of the respiratory capacity conferred by the mutated alleles gives quantitative information concerning whether the function of the mutated protein is partially or totally abolished. In the case that the respiratory capacity is not compromised by the mutant allele, a direct involvement of the single mutation in alterations of the mitochondrial respiratory capacity cannot be claimed.
Time Considerations
To complete Basic Protocol 1, it is reasonable to consider an investment of approximately 2 months. When heterologous complementation is not observed, 6-8 additional weeks are required to analyze mitochondrial localization and to construct and test chimeric alleles. Alternatively, the use of yeast mutated alleles can be accomplished in approximately 4-6 weeks. More time consuming are all the experimental plans that include tetrad analysis. The Alternate Protocol 3 will take approximately 3 months for completion.
AKCNOWLEDGEMENTS
We thank Dr. Tiziana Lodi and Darryl Horn for critically reading the manuscript. The following foundations are acknowledged for their financial support: NIH (RO1 Grant GM071775A to A.B.), Muscular Disease Association (MDA Research Grant to A.B.). The James and Esther King Biomedical Research Program, Florida Department of Health 08KN-01 (to F.D.).
REFERENCES
- Allikmets R, Raskind WH, Hutchinson A, Schueck ND, Dean M, Koeller DM. Mutation of a putative mitochondrial iron transporter gene (ABC7) in X-linked sideroblastic anemia and ataxia (XLSA/A) Hum. Mol. Genet. 1999;8:743–9. doi: 10.1093/hmg/8.5.743. [DOI] [PubMed] [Google Scholar]
- Bach ML, Lacroute F. Direct selective techniques for the isolation of pyrimidine auxotrophs in yeast. Mol. Gen. Genet. 1972;115:126–30. doi: 10.1007/BF00277292. [DOI] [PubMed] [Google Scholar]
- Barrientos A. Yeast models of human mitochondrial diseases. IUBMB Life. 2003;55:83–95. doi: 10.1002/tbmb.718540876. [DOI] [PubMed] [Google Scholar]
- Barrientos A, Zambrano A, Tzagoloff A. Mss51p and Cox14p jointly regulate mitochondrial Cox1p expression in Saccharomyces cerevisiae. EMBO J. 2004;23:3472–82. doi: 10.1038/sj.emboj.7600358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrientos A, Gouget K, Horn D, Soto IC, Fontanesi F. Suppression mechanisms of COX assembly defects in yeast and human: Insights into the COX assembly process. Biochim. Biophys. Acta. 2009;1793:97–107. doi: 10.1016/j.bbamcr.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baruffini E, Lodi T, Dallabona C, Puglisi A, Zeviani M, Ferrero I. Genetic and chemical rescue of the Saccharomyces cerevisiae phenotype induced by mitochondrial DNA polymerase mutations associated with progressive external ophthalmoplegia in humans. Hum. Mol. Genet. 2006;15:2846–55. doi: 10.1093/hmg/ddl219. [DOI] [PubMed] [Google Scholar]
- Baruffini E, Lodi T, Dallabona C, Foury F. A single nucleotide polymorphism in the DNA polymerase gamma gene of Saccharomyces cerevisiae laboratory strains is responsible for increased mitochondrial DNA mutability. Genetics. 2007;177:1227–31. doi: 10.1534/genetics.107.079293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen JL, Hall BD. Codon selection in yeast. J. Biol. Chem. 1982;257:3026–31. [PubMed] [Google Scholar]
- Boeke JD, LaCroute F, Fink GR. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 1984;197:345–6. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- Bourgeron T, Rustin P, Chretien D, Birch-Machin M, Bourgeois M, Viegas-Pequignot E, Munnich A, Rotig A. Mutation of a nuclear succinate dehydrogenase gene results in mitochondrial respiratory chain deficiency. Nat. Genet. 1995;11:144–9. doi: 10.1038/ng1095-144. [DOI] [PubMed] [Google Scholar]
- Butow RA, Henke RM, Moran JV, Belcher SM, Perlman PS. Transformation of Saccharomyces cerevisiae mitochondria using the biolistic gun. Methods Enzymol. 1996;264:265–78. doi: 10.1016/s0076-6879(96)64026-9. [DOI] [PubMed] [Google Scholar]
- Cavadini P, Gellera C, Patel PI, Isaya G. Human frataxin maintains mitochondrial iron homeostasis in Saccharomyces cerevisiae. Hum. Mol. Genet. 2000;9:2523–30. doi: 10.1093/hmg/9.17.2523. [DOI] [PubMed] [Google Scholar]
- Chattoo BB, Sherman F, Azubalis DA, Fjellstedt TA, Mehnert D, Ogur M. Selection of lys2 Mutants of the Yeast SACCHAROMYCES CEREVISIAE by the Utilization of alpha-AMINOADIPATE. Genetics. 1979;93:51–65. doi: 10.1093/genetics/93.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claypool SM, McCaffery JM, Koehler CM. Mitochondrial mislocalization and altered assembly of a cluster of Barth syndrome mutant tafazzins. J. Cell Biol. 2006;174:379–90. doi: 10.1083/jcb.200605043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lonlay P, Valnot I, Barrientos A, Gorbatyuk M, Tzagoloff A, Taanman JW, Benayoun E, Chretien D, Kadhom N, Lombes A, de Baulny HO, Niaudet P, Munnich A, Rustin P, Rotig A. A mutant mitochondrial respiratory chain assembly protein causes complex III deficiency in patients with tubulopathy, encephalopathy and liver failure. Nat. Genet. 2001;29:57–60. doi: 10.1038/ng706. [DOI] [PubMed] [Google Scholar]
- Di Fonzo A, Ronchi D, Lodi T, Fassone E, Tigano M, Lamperti C, Corti S, Bordoni A, Fortunato F, Nizzardo M, Napoli L, Donadoni C, Salani S, Saladino F, Moggio M, Bresolin N, Ferrero I, Comi GP. The mitochondrial disulfide relay system protein GFER is mutated in autosomal-recessive myopathy with cataract and combined respiratory-chain deficiency. Am. J. Hum. Genet. 2009;84:594–604. doi: 10.1016/j.ajhg.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuermann M, Francisci S, Rinaldi T, De Luca C, Rohou H, Frontali L, Bolotin-Fukuhara M. The yeast counterparts of human ‘MELAS’ mutations cause mitochondrial dysfunction that can be rescued by overexpression of the mitochondrial translation factor EF-Tu. EMBO Rep. 2003;4:53–8. doi: 10.1038/sj.embor.embor713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher N, Castleden CK, Bourges I, Brasseur G, Dujardin G, Meunier B. Human disease-related mutations in cytochrome b studied in yeast. J. Biol. Chem. 2004;279:12951–8. doi: 10.1074/jbc.M313866200. [DOI] [PubMed] [Google Scholar]
- Fontanesi F, Palmieri L, Scarcia P, Lodi T, Donnini C, Limongelli A, Tiranti V, Zeviani M, Ferrero I, Viola AM. Mutations in AAC2, equivalent to human adPEO-associated ANT1 mutations, lead to defective oxidative phosphorylation in Saccharomyces cerevisiae and affect mitochondrial DNA stability. Hum. Mol. Genet. 2004;13:923–34. doi: 10.1093/hmg/ddh108. [DOI] [PubMed] [Google Scholar]
- Gaisne M, Becam AM, Verdiere J, Herbert CJ. A ‘natural’ mutation in Saccharomyces cerevisiae strains derived from S288c affects the complex regulatory gene HAP1 (CYP1) Curr. Genet. 1999;36:195–200. doi: 10.1007/s002940050490. [DOI] [PubMed] [Google Scholar]
- Ghezzi D, Goffrini P, Uziel G, Horvath R, Klopstock T, Lochmuller H, D'Adamo P, Gasparini P, Strom TM, Prokisch H, Invernizzi F, Ferrero I, Zeviani M. SDHAF1, encoding a LYR complex-II specific assembly factor, is mutated in SDH-defective infantile leukoencephalopathy. Nat. Genet. 2009 doi: 10.1038/ng.378. [DOI] [PubMed] [Google Scholar]
- Goffrini P, Ercolino T, Panizza E, Giache V, Cavone L, Chiarugi A, Dima V, Ferrero I, Mannelli M. Functional study in a yeast model of a novel succinate dehydrogenase subunit B gene germline missense mutation (C191Y) diagnosed in a patient affected by a glomus tumor. Hum. Mol. Genet. 2009;18:1860–8. doi: 10.1093/hmg/ddp102. [DOI] [PubMed] [Google Scholar]
- Goto JJ, Zhu H, Sanchez RJ, Nersissian A, Gralla EB, Valentine JS, Cabelli DE. Loss of in vitro metal ion binding specificity in mutant copper-zinc superoxide dismutases associated with familial amyotrophic lateral sclerosis. J. Biol. Chem. 2000;275:1007–14. doi: 10.1074/jbc.275.2.1007. [DOI] [PubMed] [Google Scholar]
- Hatanaka T, Takemoto Y, Hashimoto M, Majima E, Shinohara Y, Terada H. Significant expression of functional human type 1 mitochondrial ADP/ATP carrier in yeast mitochondria. Biol. Pharm. Bull. 2001;24:595–9. doi: 10.1248/bpb.24.595. [DOI] [PubMed] [Google Scholar]
- Herrmann JM, Stuart RA, Craig EA, Neupert W. Mitochondrial heat shock protein 70, a molecular chaperone for proteins encoded by mitochondrial DNA. J. Cell Biol. 1994;127:893–902. doi: 10.1083/jcb.127.4.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann S, Rothbauer U, Muhlenbein N, Neupert W, Gerbitz KD, Brunner M, Bauer MF. The C66W mutation in the deafness dystonia peptide 1 (DDP1) affects the formation of functional DDP1.TIM13 complexes in the mitochondrial intermembrane space. J. Biol. Chem. 2002;277:23287–93. doi: 10.1074/jbc.M201154200. [DOI] [PubMed] [Google Scholar]
- Hu J, Dong L, Outten CE. The redox environment in the mitochondrial intermembrane space is maintained separately from the cytosol and matrix. J. Biol. Chem. 2008;283:29126–34. doi: 10.1074/jbc.M803028200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaukonen J, Juselius JK, Tiranti V, Kyttala A, Zeviani M, Comi GP, Keranen S, Peltonen L, Suomalainen A. Role of adenine nucleotide translocator 1 in mtDNA maintenance. Science. 2000;289:782–5. doi: 10.1126/science.289.5480.782. [DOI] [PubMed] [Google Scholar]
- Krasilnikova MM, Mirkin SM. Replication stalling at Friedreich's ataxia (GAA)n repeats in vivo. Mol. Cell Biol. 2004;24:2286–95. doi: 10.1128/MCB.24.6.2286-2295.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucharczyk R, Rak M, di Rago JP. Biochemical consequences in yeast of the human mitochondrial DNA 8993T>C mutation in the ATPase6 gene found in NARP/MILS patients. Biochim Biophys Acta. 2009 Mar 6; doi: 10.1016/j.bbamcr.2009.02.011. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Lemesle-Meunier D, Brivet-Chevillotte P, di Rago JP, Slonimski PP, Bruel C, Tron T, Forget N. Cytochrome b-deficient mutants of the ubiquinol-cytochrome c oxidoreductase in Saccharomyces cerevisiae. Consequence for the functional and structural characteristics of the complex. J. Biol. Chem. 1993;268:15626–32. [PubMed] [Google Scholar]
- Lodi T, Fontanesi F, Guiard B. Co-ordinate regulation of lactate metabolism genes in yeast: the role of the lactate permease gene. JEN1. Mol. Genet. Genomics. 2002;266:838–47. doi: 10.1007/s00438-001-0604-y. [DOI] [PubMed] [Google Scholar]
- Lodi T, Bove C, Fontanesi F, Viola AM, Ferrero I. Mutation D104G in ANT1 gene: complementation study in Saccharomyces cerevisiae as a model system. Biochem. Biophys. Res. Commun. 2006;341:810–5. doi: 10.1016/j.bbrc.2006.01.034. [DOI] [PubMed] [Google Scholar]
- Massa V, Fernandez-Vizarra E, Alshahwan S, Bakhsh E, Goffrini P, Ferrero I, Mereghetti P, D'Adamo P, Gasparini P, Zeviani M. Severe infantile encephalomyopathy caused by a mutation in COX6B1, a nucleus-encoded subunit of cytochrome c oxidase. Am J Hum Genet. 2008;82:1281–9. doi: 10.1016/j.ajhg.2008.05.002. Epub 2008 May 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr JA, Merkel O, Kohlwein SD, Gebhardt BR, Bohles H, Fotschl U, Koch J, Jaksch M, Lochmuller H, Horvath R, Freisinger P, Sperl W. Mitochondrial phosphate-carrier deficiency: a novel disorder of oxidative phosphorylation. Am. J. Hum. Genet. 2007;80:478–84. doi: 10.1086/511788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morizono H, Woolston JE, Colombini M, Tuchman M. The use of yeast mitochondria to study the properties of wild-type and mutant human mitochondrial ornithine transporter. Mol. Genet. Metab. 2005;86:431–40. doi: 10.1016/j.ymgme.2005.08.013. [DOI] [PubMed] [Google Scholar]
- Myers AM, Pape LK, Tzagoloff A. Mitochondrial protein synthesis is required for maintenance of intact mitochondrial genomes in Saccharomyces cerevisiae. EMBO J. 1985;4:2087–92. doi: 10.1002/j.1460-2075.1985.tb03896.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rak M, Tetaud E, Godard F, Sagot I, Salin B, Duvezin-Caubet S, Slonimski PP, Rytka J, di Rago JP. Yeast cells lacking the mitochondrial gene encoding the ATP synthase subunit 6 exhibit a selective loss of complex IV and unusual mitochondrial morphology. J. Biol. Chem. 2007;282:10853–64. doi: 10.1074/jbc.M608692200. [DOI] [PubMed] [Google Scholar]
- Rothstein RJ. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–11. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Schiestl RH, Gietz RD. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr. Genet. 1989;16:339–46. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Boeke JD. In vitro mutagenesis and plasmid shuffling: from cloned gene to mutant yeast. Methods Enzymol. 1991;194:302–18. doi: 10.1016/0076-6879(91)94023-6. [DOI] [PubMed] [Google Scholar]
- Singh A, Sherman F. Characteristics and relationships of mercury-resistant mutants and methionine auxotrophs of yeast. J. Bacteriol. 1974;118:911–8. doi: 10.1128/jb.118.3.911-918.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinazzola A, Viscomi C, Fernandez-Vizarra E, Carrara F, D'Adamo P, Calvo S, Marsano RM, Donnini C, Weiher H, Strisciuglio P, Parini R, Sarzi E, Chan A, DiMauro S, Rotig A, Gasparini P, Ferrero I, Mootha VK, Tiranti V, Zeviani M. MPV17 encodes an inner mitochondrial membrane protein and is mutated in infantile hepatic mitochondrial DNA depletion. Nat. Genet. 2006;38:570–5. doi: 10.1038/ng1765. [DOI] [PubMed] [Google Scholar]
- Steele DF, Butler CA, Fox TD. Expression of a recoded nuclear gene inserted into yeast mitochondrial DNA is limited by mRNA-specific translational activation. Proc. Natl. Acad. Sci. U. S. A. 1996;93:5253–7. doi: 10.1073/pnas.93.11.5253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart GR, Santos JH, Strand MK, Van Houten B, Copeland WC. Mitochondrial and nuclear DNA defects in Saccharomyces cerevisiae with mutations in DNA polymerase gamma associated with progressive external ophthalmoplegia. Hum. Mol. Genet. 2006;15:363–74. doi: 10.1093/hmg/ddi454. [DOI] [PubMed] [Google Scholar]
- Sturtz LA, Diekert K, Jensen LT, Lill R, Culotta VC. A fraction of yeast Cu,Zn-superoxide dismutase and its metallochaperone, CCS, localize to the intermembrane space of mitochondria. A physiological role for SOD1 in guarding against mitochondrial oxidative damage. J. Biol. Chem. 2001;276:38084–9. doi: 10.1074/jbc.M105296200. [DOI] [PubMed] [Google Scholar]
- Tzagoloff A, Akai A, Needleman RB, Zulch G. Assembly of the mitochondrial membrane system. Cytoplasmic mutants of Saccharomyces cerevisiae with lesions in enzymes of the respiratory chain and in the mitochondrial ATPase. J. Biol. Chem. 1975;250:8236–42. [PubMed] [Google Scholar]
- Valente L, Tiranti V, Marsano RM, Malfatti E, Fernandez-Vizarra E, Donnini C, Mereghetti P, De Gioia L, Burlina A, Castellan C, Comi GP, Savasta S, Ferrero I, Zeviani M. Infantile encephalopathy and defective mitochondrial DNA translation in patients with mutations of mitochondrial elongation factors EFG1 and EFTu. Am. J. Hum. Genet. 2007;80:44–58. doi: 10.1086/510559. Epub 2006 Nov 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valnot I, von Kleist-Retzow JC, Barrientos A, Gorbatyuk M, Taanman JW, Mehaye B, Rustin P, Tzagoloff A, Munnich A, Rotig A. A mutation in the human heme A:farnesyltransferase gene (COX10) causes cytochrome c oxidase deficiency. Hum. Mol. Genet. 2000;9:1245–9. doi: 10.1093/hmg/9.8.1245. [DOI] [PubMed] [Google Scholar]
- Young MJ, Court DA. Effects of the S288c genetic background and common auxotrophic markers on mitochondrial DNA function in Saccharomyces cerevisiae. Yeast. 2008;25:903–12. doi: 10.1002/yea.1644. [DOI] [PubMed] [Google Scholar]
- Zitomer RS, Lowry CV. Regulation of gene expression by oxygen in Saccharomyces cerevisiae. Microbiol. Rev. 1992;56:1–11. doi: 10.1128/mr.56.1.1-11.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
Internet Resources
- The Saccharomyces genome database: http://www.yeastgenome.org/, is a scientific database of the molecular biology and genetics of the yeast Saccharomyces cerevisiae, containing the most current and accurate data relative to genomic sequences, gene annotation, protein function and expression and yeast literature.
- The EUROSCARF (European Saccharomyces cerevisiae archive for functional analysis): http://web.uni-frankfurt.de/fb15/mikro/euroscarf/index.html, collects yeast strains and plasmids that were generated during various yeast functional analysis projects, including the complete knockout collection of non-essential genes. EUROSCARF is run by the Institute of Microbiology, University of Frankfurt.
- The Open Biosystems company ( http://www.openbiosystems.com) sells the entire S. cerevisiae knockout collection of non-essential genes and knockout inducible essential genes, yeast vectors, proteins tagged constructs and human cDNA clones.