Abstract
Peptide nucleic acid (PNA) is a successful DNA/RNA mimic. A major challenge for research is to invent chemically modified PNAs that retain the favorable properties of the parent compound while improving biological recognition. Here we test modified PNAs containing [bis-o-(aminoethoxy)phenyl]pyrrolocytosine bases designed to engage guanine with an additional hydrogen bond. We observe elevated melting temperatures, localization to cellular compartments, and allele-selective inhibition of mutant huntingtin protein expression.
Peptide nucleic acids (PNAs) are among the most successful biomimetic compounds.1 PNA is a DNA/RNA mimic in which the phosphate backbone has been replaced by an amide backbone containing N-(2-aminoethyl)glycine linkages.2 In spite of this dramatic departure from the normal structure of DNA and RNA, PNA binds complementary sequences according to normal Watson-Crick base rules.2,3
The PNA backbone is uncharged and, unlike DNA:DNA or RNA:DNA duplexes, there is no phosphate repulsion. The lack of repulsion leads to high rates of hybridization, especially during strand invasion of duplex DNA4 and higher affinities for hybridization.3 These favorable properties have led to PNAs being used in several applications including inhibition of human telomerase,5 blocking miRNAs,6 hybridization to RNA structure,7 inhibition of gene expression by recognition of mRNA8 or chromosomal DNA,9 and fluorescence in situ hybridization (FISH).10
Ironically, the success of PNA poses a significant challenge for ongoing PNA research. How can chemical modifications be used to improve PNA without disrupting its many outstanding properties? The synthesis of PNAs is based on peptide chemistry and is robust and versatile. One strategy for improving PNAs is to use standard synthetic procedures to introduce one or more modified PNA bases into an oligomer that is mostly unmodified PNA. Such alterations might preserve the basic recognition properties of the parent PNA while allowing the affinity of hybridization to be tailored for specific applications. A similar judicious “sprinkling” of locked nucleic acid (LNA) bases among DNA or RNA has proven to be a highly successful strategy for the discovery of broadly useful LNA oligomers.11,12
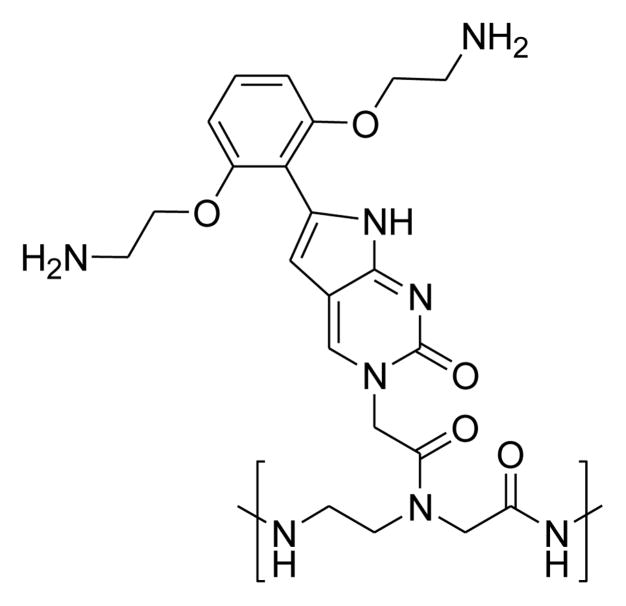
Recently, the Hudson laboratory has described modified PNAs containing [bis-o-(aminoethoxy)phenyl]pyrrolocytosine (PhpC) (Fig. 1).13 Introduction of PhpC into PNAs increases melting temperature (Tm) and affords good discrimination against binding to mismatched sequences. These favorable properties suggested that incorporation of PhpC bases into PNAs complementary to target sequences inside cells might lead to more potent inhibition of gene expression.
Figure 1.
Structure of [bis-o-(aminoethoxy)phenyl]pyrrolocytosine.
The Corey laboratory has recently shown that PNAs complementary to the mRNA encoding the protein huntingtin (HTT) can selectively inhibit expression of mutant HTT protein.14,15 Mutant HTT is responsible for Huntington’s Disease (HD), an incurable neurological disorder.16,17 HD patients are heterozygotes, expressing one mutant and one wild-type copy of the HTT gene.
Inhibition of HTT expression may be a useful strategy for treating HD. The wild-type protein, however, is involved in normal function and blocking its expression may be deleterious. Selective inhibition of the mutant allele, while retaining wild-type expression, might minimize side effects and facilitate translation into the clinic.
HD is caused by an expansion of a trinucleotide CAG repeat within the mutant HTT allele. CAG repeats are known to form hairpin structures when characterized in cell free systems.18 We reasoned that elongated hairpins might be preferentially susceptible to binding by PNAs because the mutant gene has more binding sites for complementary PNA or because of energetic differences between the mutant and wild-type repeat mRNA sequences. Our experiments showed that PNAs could achieve allele-selective inhibition of mutant HTT expression.14,15
Clinical development of anti-HTT PNAs would require that potency and allele-selectivity be optimized. We examined modified PNAs with PhpC bases because of their potential to tailor the affinity of PNA oligomers. Here we show the effects of PhpC substitution on allele selective inhibition and use the fluorescent properties of PhpC to follow intracellular localization.
We synthesized thirteen base PNAs containing one, two, three, or four PhpC substitutions (Table 1). Their sequences were complementary to the CAG repeat within HTT mRNA. All PNAs were synthesized to contain eight lysine residues in the D-configuration (D-K8) to facilitate cellular uptake.14,15 Many peptides can facilitate uptake of PNAs. We chose D-K8 because it was both effective and synthetically simple to add.
Table 1.
Tm data for PNA/RNA duplexes and IC50 values for inhibition of HTT expression in fibroblast cells.
| PNA | Sequence | # of PhpC bases | Tm (ΔTm) °C | IC50/mut (μM) | IC50/wt (μM) |
|---|---|---|---|---|---|
| I | GCTGCTGCTGCTG | 0 | 82.9 | 0.47±0.2 | > 2 |
| II | GXTGCTGCTGCTG | 1 | 84.5 (1.6) | 0.54±0.05 | 1.68±0.7 |
| III | GCTGCTGXTGCTG | 1 | 86.4 (3.5) | 0.71±0.07 | 1.86±0.1 |
| IV | GXTGCTGXTGCTG | 2 | 83.9 (1.0) | 0.58±0.05 | 1.3±0.1 |
| V | GXTGXTGXTGCTG | 3 | >87 (>4.0) | 0.97±0.2 | > 4 |
| VI | GXTGXTGXTGXTG | 4 | >87 (>4.0) | 2.6±0.7 | > 4 |
PNAs are listed N to C terminal. All PNAs have one D-lysine at the N terminus, and eight D-lysines at the C terminus. PhpC bases (X) are underlined. Tm measurements used complementary RNA oligomers. Mismatch control PNA GCCACTACTGATA was used for comparison.
Introduction of PhpC bases increased thermal stability (Table 1) 0.5 to 1°C per substitution and from 1 to ≥4°C overall. Tm values reflect both PNA-RNA basepairing and the interactions between the cationic peptide and the phosphodiester backbone of RNA.
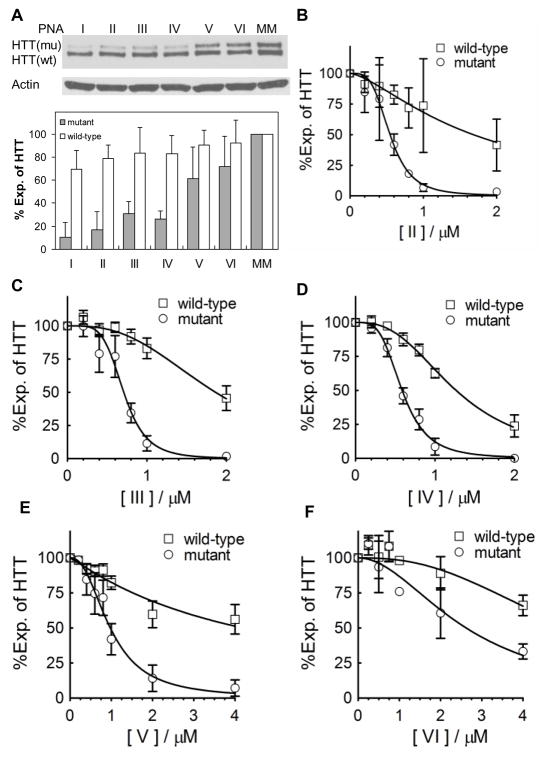
To examine inhibition of HTT we introduced the modified PNAs into GM04281 cells. GM04281 is a patient derived fibroblast cell line with 17 CAG repeats within the wild-type allele and 69 repeats within the mutant allele. When added at 1 μM PhpC-modified PNAs selectively inhibited HTT expression, but selectivity and/or potency appeared to decrease with the number of PhpC substitutions (Fig. 2A).
Figure 2.
Modified PNAs selectively inhibit mutant (mu) relative to wild-type (wt) HTT expression in fibroblast cell line GM04281. A. Top, western analysis the effects of PNAs I–VI on HTT expression. Bottom, quantitation of inhibition of mutant and wild-type HTT by PNAs I–VI. PNAs were added at 1 μM concentration. B–F. Effects of PNAs II–VI on HTT expression at varied concentrations. Experiments were performed in triplicate. Expression is relative to expression to untreated cells.
We then examined inhibition over a range of concentrations (0 – 4 μM) (Fig. 2B–F, Supp. Fig. 1B–F). PNA I with no PhpC substitutions selectively inhibited mutant HTT expression with an IC50 value of 0.47 μM and a >4 fold selectivity relative inhibition of the wild-type allele (Table 1).14 Introduction of one or two PhpC bases (PNAs II, III, and IV) did not greatly affect the potency of inhibition of mutant HTT or improve selectivity. Introduction of three or four PhpC bases (PNAs V and VI) significantly decreased the potency of both mutant and wild-type HTT expression. Triply-substituted PNA V had the best selectivity profile. Greater than 50 % inhibition of mutant HTT was achieved at concentrations above 1 μM, while greater than 50 % inhibition of wild-type HTT was not observed at any concentration tested.
An advantage of PhpC bases is that they are fluorescent, allowing oligomers to be tracked inside cells by fluorescence microscopy without the need to attach an additional fluorophore. Previous studies have used microscopy to track PNAs and have suggested localization to endosomes.19–24 These studies, however, have used PNAs tagged with fluorescent groups that might alter localization. An example of how fluorescent tags can alter localization is provided by one recent study revealing that fluoroscein can redirect a ruthenium-octaarginine conjugate from endosomal to nuclear localization.25 Another report noted that fluorescent dyes can alter intracellular localization of cell-penetrating peptides.26 PhpC allows the same oligomer to be used for both gene silencing and localization, permitting more definitive conclusions.
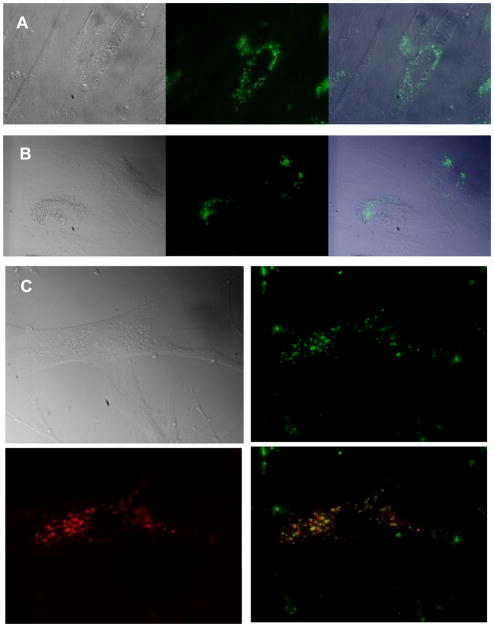
We added PNA II to GM04281 cells and used confocal fluorescent microscopy to visualize uptake. Live cells were used because chemical fixation can cause fluorescent compounds to spread and prevent accurate assessment of localization.27 One day after addition, PNA II was concentrated in compartments outside the periphery of the nucleus (Fig. 3A). A similar pattern of fluorescence could be observed at nine days (Fig. 3B), even though cells double 3–4 times during this period, substantially diluting the PNA. These data suggest that PNA-peptide conjugates are long-lived but that simply allowing them to remain inside cells over long periods of time and repeated cell divisions is not sufficient to release much of the conjugate into the cytosol.
Figure 3.
Fluorescent microscopy of PNA II in living fibroblasts. PNA was added at 1 μM concentration. A. One day or B. Nine days after PNA transfection. Left, Differential interference contrast microscopy (DIC) image; middle, PNA fluorescent; right, overlay of DIC and fluorescent images. C. PNA was co-localized with endosome marker Transferrin. 1 μM of Htt2 was co-incubated with 25 μg/mL of Transferrin-Alexa Fluor 633 for 15 h in fibroblast cells. Upper left, DIC image; upper right, PNA alone; lower left, transferrin fluorescence; lower right, overlay of PNA and transferrin images.
Our results have significant implications for interpreting prior studies of PNA localization. The finding that PNAs modified with PhpC show the same intracellular distribution as PNAs modified with terminal fluorescent groups suggests that previous observations of PNA localization reflect the localization of the PNAs themselves and were not substantially influenced by the attached fluorophores.
To estimate the location of cellular uptake we treated cells with both PhpC-modified PNA and transferrin, a marker for endosomal localization. We observed that uptake of PhpC-modified PNA and transferrin were co-localized, suggesting that both largely reside in the endosome and enter cells through similar uptake mechanisms (Fig. 3). Images were obtained fifteen hours after treatment with PNA/transferrin, and some of the overlap may arise from lysosomes. Endosomal/lysosomal localization for fluorescently-labeled PNA-peptide conjugates has been reported previously,19–24 and our results suggest that PNA conjugates containing PhpC follow a similar uptake route.
While these data indicate that most PNA is confined within endosomes, our observation of PNA-mediated inhibition demonstrates that some PNA escapes. Increasing the efficiency endosomal escape, either through addition of compounds that promote endosomal release19 or through chemical modification to the PNA,22,28 remains a significant goal for research. We also note that fluorescence is not a quantitative tool for judging the relative amount of PNA in the cytosol and endosomes because fluorescent material in the cytosol might be quenched by association with nucleic acids. The actual distribution of PNA to the cytosol may be higher than is apparent from the micrographs.
Our data show that PhpC bases can increase Tm values for antisense PNAs and modify their activities inside cells. The sensitivity of both Tm and IC50 values to the exact number and placement of PhpC bases show the usefulness of the modification as a tool for tailoring PNA properties.
Addition of PhpC bases to PNAs targeting the CAG repeat within HTT mRNA did not increase allele-selectivity, and in some cases reduced either potency or selectivity or both. It is not obvious why modifications that increase binding affinity should decrease the potency of recognition. One possibility is the modified PNAs form stronger self-complementary interactions (either aggregate or hairpin formation) that compete with intermolecular binding to mRNA. This explanation is especially relevant to the sequence used in this study because it contains a triplet repeat that tends to form a self-complementary hairpin structure. Tm curves for the single strands were broad, preventing conclusive evaluation of the hypothesis.
The triplet repeat within HTT mRNA is a special target. Other nucleic acid targets, such as nonrepetitive sequences chromosomal DNA or mRNA, might be more advantageous ones for recognition by PNAs modified with PhpC bases. Alternatively, the exact placement of PhpC bases may not be optimal. We have previously demonstrated that attaching the D-K8 peptide to the PNA N- rather than C-termini can dramatically enhance allele-selectivity.14,15 Similar simple changes in PhpC placement may also yield improved results.
Supplementary Material
Acknowledgments
We thank Dr. Filip Wojciechowski for providing the PhpC monomer. This work was supported by grants from the National Institutes of Health (NIGMS 73042 to DRC), The Robert A. Welch Foundation (I-1244 to DRC), an award from the McKnight Endowment Fund for Neuroscience (DRC), and the Natural Sciences and Engineering Research Council of Canada (RHEH). JH and DWD contributed equally to this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nielsen PE. Mol Biotech. 2004;26:233. doi: 10.1385/MB:26:3:233. [DOI] [PubMed] [Google Scholar]
- 2.Nielsen PE, Egholm M, Berg RH, Buchardt O. Science. 1991;254:1497. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 3.Egholm M, Buchardt O, Christensen L, Behrens C, Freier SM, Driver DA, Berg RH, Kim SK, Norden B, Nielsen PE. Nature. 1993;365:566. doi: 10.1038/365566a0. [DOI] [PubMed] [Google Scholar]
- 4.Smulevitch SV, Simmons CG, Norton JC, Wise TW, Corey DR. Nat Biotech. 1996;14:1700. doi: 10.1038/nbt1296-1700. [DOI] [PubMed] [Google Scholar]
- 5.Herbert BS, Pitts AE, Baker SI, Hamilton SE, Wright WE, Shay JW, Corey DR. Proc Natl Acad Sci USA. 1999;96:14726. doi: 10.1073/pnas.96.25.14276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fabani MM, Gait MJ. RNA. 2008;14:336. doi: 10.1261/rna.844108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marin VL, Armitage BA. Biochemistry. 2006;45:1745. doi: 10.1021/bi051831q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knudsen H, Nielsen PE. Nucl Acids Res. 1996;24:494. doi: 10.1093/nar/24.3.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Janowski BA, Huffman KE, Schwartz JC, Ram R, Hardy DB, Shames DS, Minna JD, Corey DR. Nat Chem Biol. 2005;1:216. doi: 10.1038/nchembio725. [DOI] [PubMed] [Google Scholar]
- 10.Baerlocher GM, Vulto I, de Jong G, Lansdorp PM. Nat Protoc. 2006;1:2365. doi: 10.1038/nprot.2006.263. [DOI] [PubMed] [Google Scholar]
- 11.Veedu RN, Wengel J. RNA Biol. 2009;6:321. doi: 10.4161/rna.6.3.8807. [DOI] [PubMed] [Google Scholar]
- 12.Braasch DB, Corey DR. Chem Biol. 2001;8:1. doi: 10.1016/s1074-5521(00)00058-2. [DOI] [PubMed] [Google Scholar]
- 13.Wojciechowski F, Hudson RHE. J Am Chem Soc. 2008;130:12574. doi: 10.1021/ja804233g. [DOI] [PubMed] [Google Scholar]
- 14.Hu J, Matsui M, Gagnon KT, Schwartz JC, Gabillet S, Arar K, Wu J, Bezprozvanny I, Corey DR. Nat Biotech. 2009;27:478. doi: 10.1038/nbt.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu J, Matsui M, Corey DR. Annals New York Acad Sci. 2009 in Press. [Google Scholar]
- 16.Walker FO. Lancet. 2007;369:218. doi: 10.1016/S0140-6736(07)60111-1. [DOI] [PubMed] [Google Scholar]
- 17.Gusella JF, MacDonald ME. Trends Biochem Sci. 2006;31:533. doi: 10.1016/j.tibs.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 18.Sobczak K, de Mezer M, Michlewski G, Krol J, Kryzosiak WJ. Nucl Acids Res. 2003;31:5469. doi: 10.1093/nar/gkg766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shiraishi T, Pankratova S, Nielsen PE. Chem Biol. 2005;12:923. doi: 10.1016/j.chembiol.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 20.Kaihatsu K, Huffman KE, Corey DR. Biochemistry. 2004;43:14340. doi: 10.1021/bi048519l. [DOI] [PubMed] [Google Scholar]
- 21.Lebleu B, Moulton HM, Abes R, Ivanova GD, Abes S, Stein DA, Iversen PL, Arzumanov AA, Gait MJ. Adv Drug Deliv Rev. 2007;60:517. doi: 10.1016/j.addr.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koppelhus U, Shiraishi T, Zachar V, Pankratova S, Nielsen PE. Bioconjugate Chem. 2008;19:1526. doi: 10.1021/bc800068h. [DOI] [PubMed] [Google Scholar]
- 23.Wolf Y, Pritz S, Abes S, Bienert M, Lebleu B, Oehlke J. Biochemistry. 2006;45:14944–14954. doi: 10.1021/bi0606896. [DOI] [PubMed] [Google Scholar]
- 24.Abes S, Williams D, Prevot P, Thierry A, Gait MJ, Lebleu BJ. Controlled Release. 2006;110:595. doi: 10.1016/j.jconrel.2005.10.026. [DOI] [PubMed] [Google Scholar]
- 25.Puckett CA, Barton JK. J Am Chem Soc. 2009;131:8738. doi: 10.1021/ja9025165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szeto HH, Schiller PW, Zhao K, Luo G. FASEB J. 2005;19:118. doi: 10.1096/fj.04-1982fje. [DOI] [PubMed] [Google Scholar]
- 27.Belitsky JM, Leslie SJ, Arora PS, Beerman TA, Dervan PB. Bioorg Med Chem. 2002;10:3313. doi: 10.1016/s0968-0896(02)00204-3. [DOI] [PubMed] [Google Scholar]
- 28.Hu J, Corey DR. Biochemistry. 2007;46:7581. doi: 10.1021/bi700230a. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.