Abstract
The contractile ring (CR) consists of bundled actin filaments and myosin II; however, the actin-bundling factor remains elusive. We show that the fission yeast Schizosaccharomyces pombe IQGAP Rng2 is involved in the generation of CR F-actin and required for its arrangement into a ring. An N-terminal fragment of Rng2 is necessary for the function of Rng2 and is localized to CR F-actin. In vitro the fragment promotes actin polymerization and forms linear arrays of F-actin, which are resistant to the depolymerization induced by the actin-depolymerizing factor Adf1. Our findings indicate that Rng2 is involved in the generation of CR F-actin and simultaneously bundles the filaments and regulates its dynamics by counteracting the effects of Adf1, thus enabling the reconstruction of CR F-actin bundles, which provides an insight into the physical properties of the building blocks that comprise the CR.
Keywords: actin cytoskeleton, contractile ring, cytokinesis, fission yeast, IQGAP
Introduction
The bundling and crosslinking of actin filaments are crucial for organizing three-dimensional actin cytoskeletons, which are in contrast with the thin filaments in muscle cells in which the filaments align one-dimensionally and are discrete from each other. Cells use a variety of actin-binding proteins (ABPs) to organize single actin filaments into bundled, crosslinked, or branched structures to perform cellular processes, such as motility, adhesion, vesicle trafficking, and division.
During the cytokinesis of many eukaryotes, a contractile ring (CR) is formed in the equatorial cortex and is believed to divide the cell in two by actomyosin-based contraction (Satterwhite and Pollard, 1992). The accumulation of F-actin and myosin II in the CR has been confirmed in many types of cells by fluorescence microscopy (FM). In some cells, the CR was also examined by electron microscopy (EM), which showed the bundling of F-actin (Sanger and Sanger, 1980; Yasuda et al, 1980; Mabuchi et al, 1988; Kamasaki et al, 2007). It should be underlined that the molecular entity responsible for the bundling of CR actin filaments remains to be determined. The actin-crosslinking protein α-actinin accumulates in the cleavage furrow of animal cells (Fujiwara et al, 1978; Mukhina et al, 2007) and is localized to the CR of fission yeast cells (Wu et al, 2001). Although mammalian and fission yeast α-actinins are involved in re-arrangement of the actin cytoskeleton during cytokinesis, their functions in CR formation seem subsidiary. Fimbrin and eukaryotic translation elongation factor 1A can bundle F-actin and are also present in the division furrow of Tetrahymena cells (Numata et al, 2000); however, their functions in CR formation are still unknown. Fimbrin also forms a medial ring during mitosis in fission yeast cells, but the ring does not contract and is not required for cytokinesis (Nakano et al, 2001; Wu et al, 2001).
To date, the fission yeast Schizosaccharomyces pombe is the best model system for studying CR formation and cytokinesis. Molecular genetics has identified >50 genes involved in cytokinesis (Balasubramanian et al, 2004). Many accumulate in the CR and their temporal order (Wu et al, 2003), and local concentrations (Wu and Pollard, 2005) were determined by FM using green fluorescent protein (GFP) fusion proteins. Specifically, the incorporation of actin filaments into the CR was shown to occur in two phases: (1) accumulation of fine F-actin bundles in a mesh-like fashion and (2) actomyosin-driven compaction of the bundles into a tight ring (Arai and Mabuchi, 2002; Vavylonis et al, 2008).
ABP, including tropomyosin Cdc8 (Balasubramanian et al, 1992), profilin Cdc3 (Balasubramanian et al, 1994), formin Cdc12 (Chang et al, 1997), IQGAP Rng2 (Eng et al, 1998), the actin-depolymerizing factor Adf1 (Nakano and Mabuchi, 2006), and myosin II Myo2 (Kitayama et al, 1997) localize to the CR and are required for CR formation in fission yeast. Taken together, the following picture of CR assembly emerges: in early mitosis, Cdc12 and Cdc3 nucleate and grow de novo actin filaments (Kovar et al, 2003), which are composed of a loose mesh, and then myosin II (Vavylonis et al, 2008) and Rng2 (Eng et al, 1998) organize this mesh into a ring. Adf1 is believed to (1) provide a source of G-actin, which is required for de novo actin ring formation, by breaking actin patches and (2) promote the turnover of actin in the CR (Nakano and Mabuchi, 2006). The functions of Cdc3 (Lu and Pollard, 2001; Takaine and Mabuchi, 2007), Cdc8 (Skoumpla et al, 2007; Skau et al, 2009), Cdc12 (Kovar et al, 2003), and Adf1 (Andrianantoandro and Pollard, 2006) in actin assembly were biochemically examined, whereas those of Rng2 and Myo2 remain to be elucidated.
To understand the mechanisms of actin filament bundling in the CR of fission yeast, we attempted to elucidate the function of Rng2 in this study. By close observation of temperature-sensitive rng2 mutant cells, Rng2 was found to be involved in both the generation (i.e. nucleation and/or elongation) of CR F-actin and its organization into a ring. Domain analysis showed that a short N-terminal fragment of Rng2 is required for these functions and is localized to CR F-actin. Moreover, we biochemically examined the fragment and found that it promotes actin polymerization and bundles F-actin. Finally, we shed some light on the physical properties of the bundles, which seemed to be flexible, consisting of subdividable arrays of F-actin under EM, and were able to counteract Adf1-induced disassembly. These results suggest that Rng2 generates bundles of actin filaments that are used to produce the CR.
Results
Rng2 is involved in both the generation and convergence of F-actin, which comprises the CR
Reportedly, temperature-sensitive rng2 mutant cells (rng2ts cells) show defects in the arrangement of F-actin into a ring after growth for 2–4 h at 36°C (restrictive temperature), displaying disorganized actin cables in the middle of the cell instead of a compact ring (Chang et al, 1996; Eng et al, 1998). We followed changes in the actin cytoskeleton of mutant cells for 2 h. In rng2ts cells at 25°C (permissive temperature), an F-actin ring was formed in the middle of cells (0 min in Figure 1A). Thirty minutes after the shift to 36°C, a trace of F-actin had accumulated in the middle of some binucleate cells instead of a ring, and grew to wrap randomly around the equator of the cell over 90 min (Figure 1A). Wild-type cells had normal medial F-actin rings at 25 and 36°C (Figure 1B). The increase in rng2ts cells without normal F-actin rings coincided with an increase in binucleate rng2ts cells (Figure 1C and D).
Figure 1.
Temperature-sensitive defects in actin ring formation of rng2 mutant cells. (A, B) After growing at 25°C, rng2ts (A) and wild-type (B) cells were shifted to 36°C and fixed after 0, 30, 60, or 90 min incubation before being stained for DNA and F-actin. Arrowheads indicate abnormal actin assembly in the medial region. (C) Percentage of binucleate cells. (D) Percentage of binucleate cells without normal CR. As these data include late telophase and septating cells, the CR was not found in about 20% of wild-type cells. Data are expressed as the means±s.d. (error bars) of three independent experiments. More than 120 cells were counted for each point. Note that rng2ts cells tended to aggregate, which prevented their correct observation, so we analysed cells outside of aggregates. (E, F) Phenotypes of rng2ts mutant cells at anaphase. Wild-type (E) or rng2ts (F) cells expressing GFP-atb2 (α-tubulin) were fixed and then stained for DNA and F-actin after 30 min incubation at 36°C. Cells with a long mitotic spindle were photographed. The percentages of cells with the depicted phenotypes are shown. About 40 cells from two independent experiments were observed for each strain. Asterisks and an arrowhead indicate the CR and the abnormal accumulation of F-actin, respectively. (G, H) Localization of Rlc1-GFP in the CR. After growing at 25°C, wild-type (G) or rng2ts (H) cells expressing Rlc1-GFP were fixed and then stained for DNA and F-actin. The percentages of cells with the depicted phenotypes are shown. More than 60 cells from two independent experiments were observed for each strain. Bars: 5 μm.
We examined further when the phenotype appeared during mitosis. Most wild type and rng2ts cells with long mitotic spindles, indicating that they were in anaphase, had F-actin rings in their centre at 25°C (data not shown). Even at 36°C, most wild-type cells in anaphase still had equatorial F-actin rings (asterisks in Figure 1E). In contrast, 62% of rng2ts cells showed no accumulation of F-actin (Figure 1F), and 36% showed abnormal accumulation of F-actin (arrowhead in Figure 1F) at 36°C, indicating that the generation of CR F-actin was disturbed in anaphase.
Even at 25°C, the fraction of binucleate rng2ts cells was significantly higher than that of wild-type cells (28±3% versus 19±1%, Figure 1C), suggesting delayed CR assembly or contraction in rng2ts cells because of the weakened function of Rng2. Thus, we examined the localization of Rlc1 (a light chain of myosin II) to the CR at 25°C. In wild-type cells at mitosis, Rlc1-GFP was observed in rings with uniform fluorescence intensity, and the rings were superimposable on F-actin rings in all cases (Figure 1G). In rng2ts cells, however, Rlc1-GFP was sometimes (45%) observed in rings with uneven fluorescence intensity (Figure 1H), even though F-actin formed uniform rings in such cells. The uneven distribution of Rlc1 in the CRs was observed also in living rng2ts cells (Supplementary Figure S1). We sometimes observed aggregate(s) of GFP fluorescence inside an Rlc1 ring in mutant cells.
We also observed the actin cytoskeleton of wild type and rng2-null cells after spore germination (Supplementary Figure S2A and B). In cells derived from wild-type spores, a medial F-actin ring was formed at mitosis. In contrast, as described in the earlier study (Eng et al, 1998), a spot of F-actin was formed instead of the CR in rng2-null cells. In these cells, cytokinesis and septation were blocked, whereas polar growth and nuclear division progressed. The F-actin spot formed once at the first mitosis in the medial region and remained there.
These results suggest that Rng2 is involved in the generation of medial F-actin in early mitosis, in the arrangement of the filaments into a ring before telophase, and in the uniform distribution of Rlc1 in CR.
Rng2 interacts with F-actin through its N-terminal region
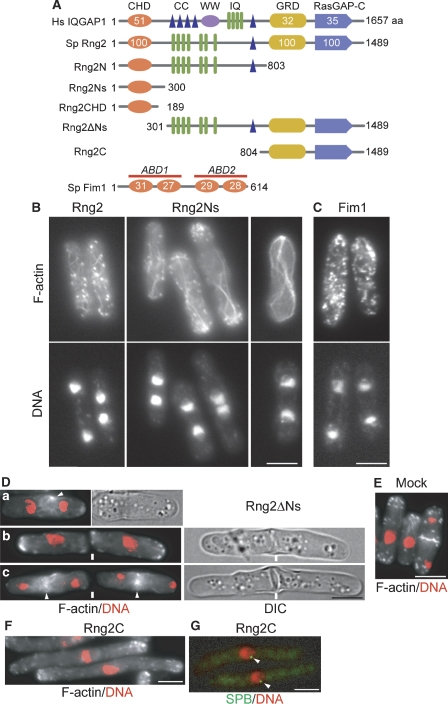
Human IQGAP1 (Figure 2A, Hs IQGAP1) contains six protein-interacting domains: a single calponin homology domain (CHD, one of the common F-actin-binding domains (ABDs)) at its N-terminal; four coiled coil regions (CCs), which mediate dimerization (Fukata et al, 1997), a WW domain in its middle; a single CC domain, a Ras GTPase-activating protein (GAP)-related domain (GRD); and a RasGAP-C homology domain (RasGAP-C) in its C-terminal region. Although Hs IQGAP1 is a mammalian orthologue of Rng2, Rng2 lacks the four CCs and the WW domain: seven IQ motifs directly follow the CHD, and we failed to detect any CC in the N-terminal half of Rng2 (residues 1–745) using the COILS program (Lupas et al, 1991). Thus, the N-terminal region of Rng2 seems to have specific functions that are different from those of Hs IQGAP1. In this study, we sometimes compared Rng2 with fission yeast fimbrin, Fim1, as a control actin-bundling protein. Fim1 can bundle F-actin using two ABDs, each of which is composed of two CHD in tandem (Nakano et al, 2001; Figure 2A).
Figure 2.
Overexpression of proteins containing the N-terminal region of Rng2 produced aberrant actin cables and blocked CR formation. (A) The domain organizations of human IQGAP1, S. pombe Rng2, truncated proteins of Rng2, and S. pombe fimbrin Fim1. White numbers show relative amino-acid sequence homology in percent. (B, D, F, G) Phenotype of cells overexpressing Rng2 or its truncated proteins or (C) Fim1. (E) Mock overexpression was performed using an empty vector. Constructs were expressed from plasmids under the control of the strongest nmt1 promoter in wild-type cells (B–F) or cells expressing Sid4-mRFP (G). Expression was induced for 15 h at 30°C, and then the cells were processed for FM as described in Figure 1. Overlaid images of DNA (red) and F-actin are shown in (D–F). In (G), the RFP signals and DNA staining of living cells are shown. Bars: 5 μm.
We observed the effects of the overexpression (OE) of Rng2 constructs (Figure 2A). OE of Rng2 (amino-acid residues 1–1489), Rng2N (residues 1–803), or Rng2Ns (residues 1–300) produced numerous thick actin cables, prevented CR formation, and dispersed actin patches over the cell cortex (Figure 2B; data for Rng2N is not shown). In these cells, cytokinesis and septation were blocked, whereas polar growth and mitosis were progressed. Although Rng2 seems to interact specifically with CR F-actin at mitosis (Eng et al, 1998), abnormally thick F-actin bundles were induced by OE of Rng2, Rng2N or Rng2Ns also in cdc25-22 cells arrested at the G2/M boundary (Supplementary Figure S3A). Thus, it was suggested that the bundling effects of these constructs are not necessarily specific to mitosis and to CR F-actin when they are overexpressed. In contrast, OE of Fim1 also prevented the formation of F-actin rings but produced abundant actin patches (Figure 2C). OE of Rng2ΔNs (residues 301–1489) did not disturb mitosis but specifically blocked CR formation: F-actin accumulated in the middle of cells during mitosis, but failed to form rings (Figure 2D). We noted that the accumulated F-actin was eventually cleared (i.e. turned over) and cells failed to complete septation (thick bars in Figure 2D). Interphase cells had normal actin patches and cables but not abnormal F-actin structures (b in Figure 2D; data not shown), which indicated that Rng2ΔNs essentially does not interfere with interphase F-actin structures. Mock OE through the pREP1 vector had no effect on the actin cytoskeleton (Figure 2E).
On the other hand, OE of Rng2C (residues 804–1489) blocked mitosis but did not disturb polar growth (Figure 2F). In these cells, actin patches were normally located at the cell ends, and actin cables and rings were absent. All mononucleate cells had only one spindle pole body (SPB) on the nucleus, which confirmed that they were held in interphase (Figure 2G). OE of Rng2GRD (residues 804–1205) or Rng2RasGAP-C (residues 1168–1489) caused no growth or morphological defect (data not shown).
These results suggest that the N- and C-terminal halves of Rng2 have different functions and that the former can produce thick F-actin bundles through the Rng2Ns moiety. Moreover, complementation assays using rng2ts and rng2-null cells showed that the Rng2Ns moiety is indispensable for the function of Rng2 during cytokinesis (see Supplementary data; Supplementary Figures S2 and S4).
Rng2 contains multiple localization domains
We examined the localization of various GFP-tagged Rng2 constructs in wild-type cells, as summarized in Figure 3J. The localization of various constructs is described also in the Supplementary data. GFP-Rng2 localized to the CR as reported earlier (Figure 3A; Eng et al, 1998; Wu et al, 2003). In addition to the rings, we detected GFP-Rng2 dots, as observed by Eng et al (1998), in some cells with prolonged induction, and they partially colocalized with SPB (Supplementary Figure S5A). GFP-Rng2Ns also localized to the medial rings that were able to contract (Figure 3B). GFP-Rng2ΔNs tended to aggregate in the cytoplasm and formed a dim medial ring during mitosis in some cells (Figure 3C). We noted that the localization of GFP-Rng2Ns was different from that of GFP-Rng2CHD (residues 1–189), which nonspecifically labelled all whole F-actin structures (Wachtler et al, 2003; Figure 3D), and that GFP-Rng2Ns did not mark actin patches or cables, even when it was abundantly expressed with prolonged induction (data not shown).
Figure 3.
Localization of Rng2 and its truncated proteins. All constructs were expressed in wild-type cells from plasmids under the control of the strongest nmt1 promoter. We observed cells that expressed the GFP fusions before the effects of OE became apparent. Images of (A) GFP-Rng2, (B) GFP-Rng2Ns, (C) GFP-Rng2ΔNs, and (D) GFP-Rng2CHD. (E–G) Localization of GFP-Rng2, GFP-Rng2ΔNs, and GFP- Rng2Ns in the absence of F-actin. Cells expressing each construct were treated with 0.2% DMSO alone (–Lat-A) or with 0.2% DMSO plus 4 μM Lat-A (+Lat-A) for 10 min at 25°C, before being stained for DNA. (H) Images of GFP-Rng2 or GFP-Rng2ΔNs expressed at a low level in the presence of 5 μM thiamine (repressed conditions). (I) Cells bearing pREP1-GFP-Rng2ts were grown under repressed conditions at 25°C, shifted to 25 or 36°C for 30 min, and stained for DNA. Arrowheads and small bars indicate the CR and bands of dots, respectively. Double arrowheads indicate the CR during contraction. (J) Summary of the localization of Rng2 constructs. Bars: 5 μm.
We verified whether the assembly of these rings depends on F-actin using Latrunculin A (Lat-A), which completely disassembled all F-actin structures within 10 min whereas DMSO, the solvent for Lat-A, did not affect F-actin organization (data not shown). In Lat-A, GFP-Rng2 or GFP-Rng2ΔNs formed a band of dots or a discontinuous ring at the division site (+Lat-A in Figure 3E and F). In contrast, the rings of GFP-Rng2Ns (+Lat-A in Figure 3G) and the localization of GFP-Rng2CHD (data not shown) were completely cleared by Lat-A. DMSO did not affect the localization of the proteins tested (−Lat-A in Figure 3E–G). GFP-Rng2 and GFP-Rng2ΔNs formed similar equatorial bands of dots even in the absence of Lat-A when they were expressed at lower levels (Figure 3H), whereas other constructs did not form such bands under any conditions (data not shown).
GFP-Rng2C accumulated around the nucleus and formed a single dot in both interphase and mitotic cells (Supplementary Figure S5C and D) independently of F-actin (data not shown). The dots were multiplied in some cells and colocalized with SPB (Supplementary Figure S5E). GFP-Rng2C was also weakly localized to the medial region as a discontinuous band in septum-forming cells.
These observations show that Rng2 (1) accumulates at the division site through Rng2ΔNs independently of F-actin and (2) probably associates with CR F-actin through Rng2Ns, and (3) the C-terminal half of Rng2 alone is able to accumulate in the perinucleus, including around SPB and the septum.
We also addressed the localization of temperature-sensitive mutant Rng2 protein in wild-type cells. We analysed the DNA sequence of the rng2-D5 allele and identified a single mutation that substitutes the relatively conserved glycine residue at position 1032 for glutamic acid (Supplementary Figure S4A). The GFP-tagged mutant Rng2 (GFP-Rng2ts) formed dim medial rings during cytokinesis at 25°C (Figure 3I). We failed to find GFP-Rng2ts forming an equatorial band of dots, and the rings of GFP-Rng2ts disappeared in Lat-A (Figure 3J; data not shown), indicating that mutant Rng2 forms an F-actin-dependent medial ring. At 36°C, the rings of GFP-Rng2ts completely disappeared within 30 min (Figure 3I), whereas those of GFP-Rng2 remained intact (data not shown). Thus, the mutant Rng2 is unable to interact with CR F-actin at 36°C for some unknown reason, which causes defects in CR formation in rng2ts cells (Figure 1).
Rng2Ns bundles F-actin
To elucidate how Rng2 interacts with F-actin during CR formation, we considered two constructs, Rng2Ns and Rng2CHD. N-terminally His6-tagged Rng2Ns or Rng2CHD was expressed in bacteria and successfully purified (Figure 4A). In gel filtration chromatography, Rng2Ns was eluted as a single peak (Figure 4B), and the apparent molecular weight was estimated to be 41k (Figure 4C), which was only 10% larger than the deduced molecular weight of the Rng2Ns monomer (37k). Thus, Rng2Ns appeared monomeric at an ionic strength of 0.05 M and a pH of 7.5.
Figure 4.
Cooperative association of Rng2Ns or Rng2CHD with F-actin. (A) Purified proteins resolved by SDS–PAGE and stained with CBB. (B) Gel filtration chromatography of Rng2Ns. Arrowheads indicate the peak elution volumes of the following molecular weight markers from left to right: Blue dextran 2000 (void volume), BSA (68k), ovalbumin (45k), chymotrypsinogen A (25k), and RNase A (14k). (C) The peak elution volume of Rng2Ns was plotted on a calibration line as a square, giving an apparent molecular weight of 41k. (D, E) High- (D) or low-speed (E) co-sedimentation of Rng2Ns, Rng2CHD, or Fim1 (1.5 μM) with F-actin (3 μM). Equal portions of the supernatants (S) and pellets (P) were subjected to SDS–PAGE and CBB-stained. (F) The high-speed co-sedimentation assay shown in (D) was performed for various concentrations of Rng2Ns, Rng2CHD, or Fim1 (0.3–3.5 μM). The concentration of the bound Rng2Ns (•), Rng2CHD (⧫), or Fim1 (▴) was plotted against the concentration of the free portion and fitted with a hyperbolic function, and representative plots are shown. (G) The low-speed co-sedimentation assay shown in (F) was performed for various concentrations of Rng2Ns, Rng2CHD, or Fim1. The amount of actin pelleted was plotted against the Rng2Ns/actin, Rng2CHD/actin, or Fim1/actin concentration ratio. Data are expressed as the means±s.d. (error bars) of three independent experiments. (H) Summary of the properties of the actin-bundling proteins tested. The dissociation constants (Kd) and stoichiometries were calculated from data obtained by three independent experiments.
By high-speed sedimentation, in which F-actin mostly sedimented (Figure 4D, lane A), a significant amount of Rng2Ns, Rng2CHD, or Fim1 co-sedimented with F-actin (Figure 4D, lanes A+N, A+C, and A+F), whereas they barely sedimented at all in the absence of F-actin (Figure 4D, lanes N, C, and F). Quantitative analysis showed that Rng2Ns, Rng2CHD, and Fim1 bind with F-actin with Kd values of 0.88, 0.21, and 0.42 μM, respectively (Figure 4F). At saturation, the amount of bound Rng2Ns, Rng2CHD, and Fim1 per 1 mol actin was estimated to be 1.4, 1.4, and 1.6 mol, respectively.
Next, we assessed whether Rng2Ns or Rng2CHD can crosslink F-actin using a low-speed sedimentation assay in which F-actin sediments only when it aggregates (Figure 4E, lane A). In the presence of Rng2Ns, Rng2CHD, or Fim1, a considerable amount of F-actin was recovered in the pellet (Figure 4E, lanes A+N, A+C, and A+F). The amount of F-actin sedimented showed a sigmoidal dependence on the Rng2Ns or Rng2CHD concentration, suggesting that they crosslinked the filaments in a cooperative manner (Figure 4G). Rng2Ns sedimented F-actin more effectively than Rng2CHD. Meanwhile, F-actin was sedimented with hyperbolic dependence on the Fim1 concentration, indicating its non-cooperative crosslinking of F-actin. The molecular ratios of Rng2Ns, Rng2CHD, or Fim1 to actin that gave half-maximal sedimentation were 0.2, 0.4, and 0.03, respectively. Figure 4H summarizes the properties of the actin-bundling proteins.
We examined the aggregates of F-actin formed by Rng2Ns, Rng2CHD, or Fim1 by FM. In their presence, thick bundles of rhodamine–phalloidin-labelled F-actin were detected (insets in Figure 5A–C), whereas only single filaments were observed for F-actin alone (Figure 5D). Bundles formed by Rng2Ns (Rng2Ns/actin bundles) were evenly distributed in the field, and some formed radial arrays (asterisks in Figure 5A). The bundles formed by Fim1 (Fim1/actin bundles) were often entangled and formed larger clots (Figure 5B), which may indicate that Fim1 can crosslink the filaments at various angles. Some single filaments were still present in the presence of Rng2Ns or Fim1 (dim filaments in the background of Figure 5A and B). The bundles formed by Rng2CHD were often loose (arrowheads in Figure 5C), and numerous single filaments always coexisted around them, suggesting that bundling by Rng2CHD was weaker than by Rng2Ns or Fim1. We further examined the ultrastructures of the bundles by EM. Rng2Ns/actin bundles appeared to be both thick bundles and radial arrays under EM (Figure 5E and F). Fim1/actin bundles seemed rather disentangled under EM: single bundles stood out, and large clots were rarely seen (Figure 5G). Only single filaments with a mean width of 6.9±1.2 nm were observed for F-actin alone (Figure 5H). Rng2Ns/actin bundles were more closely packed than Fim1/actin bundles: individual filaments in Fim1/actin bundles were distinguishable from each other, whereas those in Rng2Ns/actin bundles were obscure (Figure 5E′–H′). The mean width of Rng2Ns/actin bundles was 26±8 nm, whereas that of Fim1/actin bundles was distributed more widely with a mean of 36±17 nm (Figure 5I). Unlike Fim1/actin bundles, Rng2Ns/actin bundles were curvy, often smoothly split (Figure 5J), and sometimes twisted (Figure 5K), suggesting that they were flexible and could be divided into smaller units.
Figure 5.
Rng2Ns bundles F-actin. (A–D) Fluorescence micrographs of actin bundles. G-actin (2 μM) was polymerized alone (D), or with 0.6 μM Rng2Ns (A), Fim1 (B), or 1.8 μM Rng2CHD (C) for 10 min (A) or 50 min (B, C), and then stained with rhodamine–phalloidin. Asterisks indicate the centres of the radial arrays of actin bundles. Arrowheads indicate points where the bundles were loosened. (E–H) Electron micrographs of actin bundles. (I) Width distribution of Rng2Ns/actin (black bars) and Fim1/actin (white bars) bundles. The means±s.d. are depicted in the graph. The widths of the bundles were measured at more than 170 points for each condition. (J) Rng2Ns/actin bundles splitting. (K) Rng2Ns/actin bundles twisting (a) and simultaneously splitting (b). Bars: 10 μm (A–D) or 200 nm (E–H, J, and K).
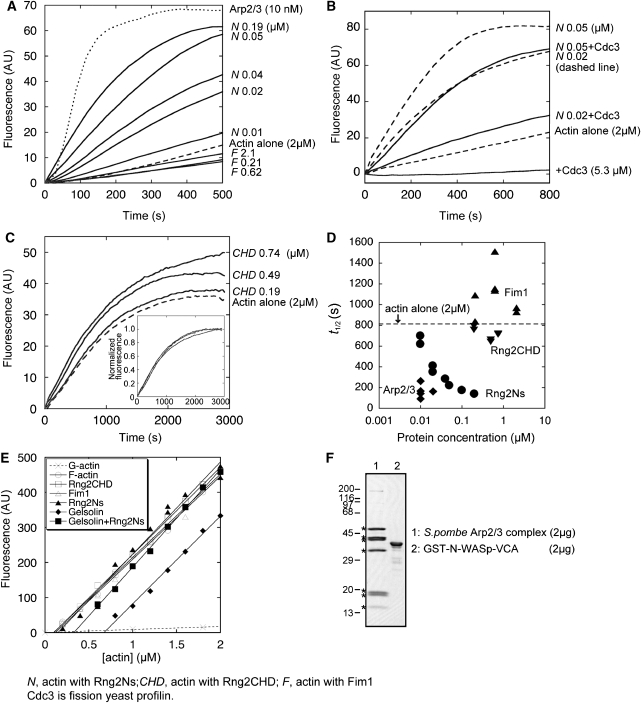
Rng2Ns accelerates spontaneous actin polymerization
From the result suggesting the involvement of Rng2 in the generation of CR F-actin (Figure 1), we inferred that Rng2 might have an effect on the kinetics of actin polymerization. Rng2Ns promotes actin polymerization by decreasing the initial lag because of nucleation (Figure 6A). The effect of Rng2Ns became apparent at a more than 1:100 molar ratio to actin, and was comparable to that of the activated fission yeast Arp2/3 complex as a positive control (Figure 6F) at higher concentrations. In contrast, Fim1 decelerated actin polymerization at a more than 1:10 molar ratio to actin. It is noteworthy that Rng2Ns/actin bundles emerged in 10 min (Figure 5A) when Rng2Ns-promoted polymerization was nearly complete, indicating that in the presence of Rng2Ns, actin polymerization and F-actin bundling occur simultaneously.
Figure 6.
Rng2Ns accelerates spontaneous actin polymerization. (A) Time course of the polymerization of 2 μM Mg-G-actin (1% pyrene labelled) in the presence of Rng2Ns, Fim1 or fission yeast Arp2/3 complex (10 nM, activated by 67 nM GST-N-WASp-VCA). (B) Time course of the polymerization of 2 μM Mg-G-actin in the presence of Rng2Ns and Cdc3. Dashed and solid lines indicate samples without and with Cdc3 (5.3 μM), respectively. The corresponding time courses until 3000 s are shown in Supplementary Figure S6. (C) The time course of the polymerization of 2 μM Mg-G-actin in the presence of Rng2CHD. Inset shows the curves normalized to fluorescence intensities at the plateau. (D) The time to half-maximal polymerization (t1/2) was plotted versus Rng2Ns, Fim1, Rng2CHD and Arp2/3 complex concentrations. Dashed line indicates the average t1/2 for 2 μM actin alone (815±104 s, n=8). The average t1/2 for 10 nM Arp2/3 complex activated by 67 nM GST-N-WASp-VCA was 168±72 s (n=4). (E) Effects of actin-bundling proteins on the critical concentration (Cc) for actin polymerization. Actin (5% pyrene labelled) was polymerized for 16 h at 25°C, and pyrene fluorescence was plotted versus total actin concentration. Cc was determined from the intersection of F-actin fluorescence with G-actin fluorescence. Cc values were 0.10 μM for F-actin alone (open circle), 0.70 μM with 10 nM gelsolin–actin seeds (closed diamond), 0.11 μM with 0.5 μM Rng2CHD (open square), 0.12 μM with 0.4 μM Fim1 (open triangle), 0.15 μM with 0.4 μM Rng2Ns (closed triangle), and 0.34 μM with 0.4 μM Rng2Ns and 10 nM gelsolin–actin seeds (closed square). Two or three independent experiments were performed for each condition, which gave essentially the same result. (F) Purified fission yeast Arp2/3 complex and GST-N-WASp-VCA were analysed by SDS–PAGE and CBB-staining. Asterisks indicate the seven subunits of Arp2/3 complex. Molecular weights × 10−3 are shown on the left.
Fission yeast profilin Cdc3 suppressed spontaneous actin polymerization similar to other profilins (Figure 6B; Lu and Pollard, 2001; Takaine and Mabuchi, 2007) whereas Cdc3 promoted formin (Cdc12)-stimulated actin assembly (Kovar et al, 2003). Unlike formins, Rng2Ns counteracted the effects of Cdc3; it accelerated actin polymerization in the presence of Cdc3 to a lesser extent than in the absence of Cdc3 (compare solid and dashed lines in Figure 6B). Rng2CHD did not affect actin-polymerization kinetics but slightly increased the fluorescence intensity from pyrene–actin (Figure 6C). We plotted the time to half-maximal polymerization (t1/2) as a function of Rng2Ns, Fim1, Rng2CHD, and Arp2/3 complex concentrations to compare nucleation activities of these proteins more precisely (Figure 6D). The activity of 100 nM Rng2Ns was approximately equal to that of 10 nM-activated Arp2/3 complex. As the nucleation activity of formin or Spir becomes equivalent to that of Arp2/3 complex only at 100-times higher concentration (Quinlan et al, 2005), Rng2Ns seems to be a stronger nucleator.
We also investigated the effect of actin-bundling proteins on the critical concentration (Cc) for F-actin assembly. Even an excess amount of Fim1, Rng2CHD, or Rng2Ns increased the Cc only slightly from 0.10 μM for F-actin with free ends to 0.12, 0.11, and 0.15 μM, respectively (Figure 6E, left lines), implying that they hardly interact with either end of the filament and do not interfere with F-actin dynamics. Gelsolin capped the barbed ends of F-actin and thus shifted the Cc up to 0.70 μM, a value close to that for the free pointed end (Figure 6E, rightmost line). Rng2Ns decreased the Cc of gelsolin-capped F-actin from 0.70 to 0.34 μM (Figure 6E, middle line), indicating that Rng2Ns decreases the Cc of the pointed ends.
These results suggest that Rng2Ns promotes spontaneous actin polymerization without capping the ends of F-actin. Promotion of actin assembly antagonized the effect of profilin and was specific to Rng2Ns rather than a common feature of actin-bundling proteins that possess CHD.
Rng2Ns/actin bundles are resistant to Adf1-induced actin depolymerization
In fission yeast cells, Adf1 localized to F-actin patches, and formed medial rings during mitosis in an F-actin-dependent manner (Figure 7A), suggesting that Adf1 and Fim1 or Rng2 simultaneously act on F-actin in patches or the CR, respectively. We also found that Adf1 genetically interacts with Rng2 (Supplementary Figure S7), which also supports their cooperative function in the CR. We therefore examined the effect of F-actin bundling by Rng2Ns or Fim1 against Adf1-induced depolymerization.
Figure 7.
Rng2Ns and Fim1 stabilize actin filaments against Adf1-induced depolymerization. (A) Localization of GFP-Adf1. Cells expressing GFP-Adf1 were stained for DNA. (B) Time course of the depolymerization of F-actin after dilution in the presence of Adf1. G-actin (3 μM, 20% pyrene labelled) plus 15 nM gelsolin was polymerized with Rng2Ns or Fim1. F-actin was diluted to 0.1 μM with 0.11 μM Adf1, and the decrease in pyrene fluorescence was monitored. (C) Data in B were fitted to single exponentials, and the disassembly rate was plotted as a function of Rng2Ns or Fim1 concentration. (D–G) Fluorescence micrographs of actin filaments during depolymerization. F-actin (3 μM in G-actin) plus 15 nM gelsolin was mixed with 1.8 μM Rng2Ns (F) or Fim1 (G) for 1 h, diluted 30-fold with 0.12 μM Adf1, removed from the reactions at the indicated time points after dilution, and stained with rhodamine–phalloidin. F-actin plus gelsolin alone was also diluted in the absence (D) or presence (E) of Adf1. (H, J) Co-sedimentation of Rng2Ns or Fim1 with F-actin prebound to Adf1. F-actin (2 μM) was mixed with 4 μM Adf1 (+Adf1) or buffer (–Adf1) for 30 min at pH6.0 or 8.0 (track 7 in H) and then mixed with 1 μM Rng2Ns or Fim1, before being centrifuged at (H) high speed or (J) low speed. The proteins in the supernatants (S) and pellets (P) were resolved by SDS–PAGE and CBB-stained. (I, K) The co-sedimentation assays shown in (H) and (J) were performed for various concentrations of Adf1. The molar ratios of Rng2Ns or Fim1 to actin in the pellet produced by high-speed sedimentation (I) or the amount of actin in the pellet produced by low-speed sedimentation (K) was normalized to that in the absence of Adf1 and plotted versus the Adf1/actin concentration ratio. Data are expressed as the means±s.d. (error bars) of three independent experiments. Bars: 5 μm.
Dilution of pyrene-labelled F-actin assembled from gelsolin–actin seeds to 0.1 μM caused a gradual decrease in fluorescence, which was mainly because of depolymerization from the pointed ends of the filaments, and Adf1 greatly promoted the decrease (black solid line in Figure 7B). Preincubation of F-actin with either Rng2Ns or Fim1 decelerated Adf1-induced depolymerization (Figure 7B). Fim1 suppressed depolymerization more efficiently than Rng2Ns (Figure 7C). We also confirmed these results by FM. After dilution to 0.1 μM, the number and length of actin filaments did not significantly decrease over 10 min (Figure 7D). Adf1 induced intensive fragmentation and depolymerization of filaments within 150 s, leaving only diminutive particles of residual filaments at 10 min (Figure 7E). Rng2/actin bundles were neither fragmented nor shortened even by Adf1 (Figure 7F), although the number of bundles seemed to incrementally decrease over time. Fim1 produced aggregations of F-actin, and both the number and diameter of the aggregates remained unchanged in the presence of Adf1 (Figure 7G). Thus, bundling of F-actin either by Rng2Ns or by Fim1 resisted the disassembly of the filaments caused by Adf1.
We further examined whether Adf1 prebound to F-actin disturbed F-actin binding or F-actin bundling of Rng2Ns or Fim1 using high- and low-speed sedimentation assays. Samples were prepared at pH 6.0, where Adf1 bound to F-actin, but its assembly was hardly affected, whereas Adf1 effectively disassembled F-actin at pH 8.0 (compare track 4 with 7 in Figure 7H). Adf1 inhibited F-actin binding of Fim1 but not of Rng2Ns (Figure 7H and I). Correspondingly, Adf1 reduced the F-actin-bundling efficiency of Rng2Ns and Fim1 by 28 and 77%, respectively (Figure 7J and K).
Discussion
Localization of Rng2 domains
During mitosis, GFP-Rng2 localized to the CR dependently and independently of F-actin as described earlier (Eng et al, 1998; Wu et al, 2003). F-actin-dependent localization is probably mediated by an Rng2Ns moiety that specifically sticks to CR F-actin rather than an Rng2CHD moiety that labels all F-actin structures. GFP-Rng2Ns-C (residues 190–300) or Rng2Ns-C itself is unlikely to associate with F-actin both in vivo and in vitro (Figure 3J; Supplementary data; Supplementary Figure S8). Moreover, the binding stoichiometries for actin were unchanged between Rng2CHD and Rng2Ns. On the Rng2Ns molecule, therefore, the Rng2Ns-C moiety must alter the binding mode of the Rng2CHD moiety for F-actin, yielding the definite targeting of GFP-Rng2Ns to CR F-actin. The probable difference in their binding modes is supported by their different biochemical functions (Figures 4, 5 and 6). The structural basis for the differential F-actin recognition of the two fragments remains to be determined.
GFP-Rng2 and GFP-Rng2ΔNs accumulated in the equatorial region as a band of dots during mitosis in the absence of F-actin or when expressed at a low level. The dots are probably essentially equivalent to ‘nodes' that act as progenitors of the CR (Wu et al, 2006) and emerge as a broad band in the medial cortex at metaphase and then condense into a continuous ring. The mitotic node contains some components of the CR (Mid1, Myo2, Cdc4, Rng2, Cdc12, and Cdc15), and Mid1, S. pombe anillin (Paoletti and Chang, 2000), is responsible for its organization. Cdr2 kinase anchors Mid1 at the medial cortex during interphase (Almonacid et al, 2009). Domain analysis of Rng2 showed that the medial node localization of GFP-Rng2 is attributed to residues 301–803, which contain IQ motifs. Rng2 associates with the node structure or directly with Mid1 through this region. Cdc4, an essential light chain of Myo2, is one of the possible candidates for tethering Rng2 to the node because Rng2 associates with Cdc4 through IQ motifs (D'souza et al, 2001) and requires Cdc4 for its accumulation at the division site (Wu et al, 2003). OE of Rng2ΔNs resulted in a failure to form the CR, which may be due to interference with the function of Mid1, Myo2, or Cdc4. As the CR is assembled, Rng2 will move from the nodes into CR F-actin because the ring of Rng2 or F-actin, but not that of Mid1, contracts (Paoletti and Chang, 2000). If interactions of Rng2 with CR F-actin and Mid1 (or the nodes) are mutually exclusive, Rng2 may simply transfer from the nodes to the F-actin ring gradually through the Rng2Ns moiety that specifically sticks to CR F-actin. In a case in which Rng2 can potentially interact with F-actin and Mid1 at the same time, active regulation seems to be necessary to release Rng2 from the nodes. Rng2–Mid1 interaction might be temporally regulated by phosphorylation of Rng2, as reported in the Myo2–Mid1 interaction (Motegi et al, 2004).
The effects of the Rng2C moiety on the localization and function of Rng2 are discussed further in the Supplementary data.
Functions of Rng2 in CR assembly
We showed that Rng2 is involved not only in the construction of CR but also partially in the generation of its filaments and that OE of Rng2 produces many aberrant thick actin cables. These results suggest that Rng2 functions as an F-actin bundler and nucleator. We also found that Rng2Ns bundles F-actin and promotes actin polymerization in vitro and attaches Rng2 to CR-F-actin (see above). Thus, we conclude that Rng2 assists the generation of CR F-actin by Cdc12 and simultaneously bundles them into a compact ring through the Rng2Ns moiety (Figure 8). The requirement of the Rng2Ns moiety to restore the cytokinesis defect in rng2ts or rng2-null cells (Supplementary Figures S2D and S4C–E) also suggests that CR F-actin bundling is at least one of the actual functions of Rng2 during cytokinesis.
Figure 8.
Model of Rng2 functions on CR formation and contraction. (A) Function and localization of each Rng2 domain. (B) Model depicting the roles of Rng2 in CR formation and contraction. During early mitosis, nodes appear as a broad band in the medial region of the cell, dependent on anillin Mid1. The nodes contain Rng2, formin Cdc12, and myosin II. Cdc12 forms actin nuclei in each node by cooperating with profilin Cdc3 in the cytoplasm and elongates filaments from the nuclei. Rng2 assists in the generation (nucleation and/or elongation) of filaments and simultaneously arranges them into a ring, while moving gradually from the nodes and onto the filaments. Rng2 may distribute myosin II evenly over the CR to ensure its efficient contraction. F-actin bundling by Rng2 regulates F-actin dynamics in the CR by counteracting the depolymerization/severing mediated by Adf1 for proper contraction/disassembly of the ring.
Indeed, the bundling of filaments is crucial for production of the F-actin ring from its intermediates (Vavylonis et al, 2008). The process is rapid; a mesh of thin filaments became a thick ring within 10 min. The cooperative interaction of Rng2 with F-actin might plausibly explain this remarkable change.
In rng2-null cells, CR formation seems to be impaired more severely than in rng2ts cells because F-actin does not even accumulate in the medial region whereas the F-actin spot is formed only once at the first mitosis (compare Supplementary Figure S2B and C). These results would support our claim that Rng2 is involved in the generation of CR F-actin.
Rng2Ns/actin bundles counteracted Adf1-driven disassembly. Rng2Ns and Adf1 did not compete to bind to F-actin, which allowed the simultaneous localization of Rng2 and Adf1 on CR F-actin. In addition, the genetic interaction of Rng2 with Adf1 implies their collaborative function in the CR. Therefore, the depolymerization and severing of CR F-actin mediated by Adf1 is dynamically and adequately tuned by Rng2-mediated bundling to achieve proper disassembly of the ring (Nakano and Mabuchi, 2006). Although Rng2Ns seems constitutively active in F-actin binding and bundling in vitro, the activity of the Rng2Ns moiety of endogenous Rng2 would be suppressed at interphase and be put into effect during mitosis.
The genetic interaction of Rng2 with formin Cdc12 (Eng et al, 1998) also supports the participation of Rng2 in the production of CR F-actin. Cdc3 profilin suppresses spontaneous actin nucleation (Takaine and Mabuchi, 2007) and antagonizes Rng2Ns-mediated actin polymerization, whereas it enhances Cdc12-mediated actin polymerization (Kovar et al, 2003). In addition, no accumulation of CR F-actin is observed in temperature-sensitive cdc12 mutant cells (Chang et al, 1996). Cdc12 would therefore has a critical role in nucleating and elongating CR F-actin in vivo (i.e. in the presence of Cdc3). On the other hand, elongation of CR F-actin occurs but is disturbed in rng2ts cells (Figure 1E–F), and hence Rng2 would have a subsidiary role in generating CR F-actin to assist Cdc12.
Reportedly, the polymorphic fungus Candida albicans IQGAP Iqg1 physically and genetically interacts with formins during cell division (Li et al, 2008). Mammalian IQGAP1 also interacts with formin Dia1 and is required for its localization in cell migration and phagocytosis (Brandt et al, 2007). Cooperation between formin and IQGAP might be an evolutionarily conserved method for remodelling the actin cytoskeleton.
Rng2Ns did not cap either end of F-actin and thus must bind to the sides of filaments (including oligomers) and stabilize them to promote nucleation or elongation. We suppose that the stabilization mediated by Rng2 decreases the Cc only when the barbed ends are capped because the Cc of the barbed end is low enough (Figure 6D). Rng2 is unlikely to compete with Cdc12 to bind to F-actin because Cdc12 interacts preferentially with the barbed ends (Kovar et al, 2003), which again assures cooperation between them. Although Cdc12 promotes the growth of filaments from the barbed ends, Rng2 may stabilize the filaments, especially near the pointed ends, and facilitates polymerization from these ends.
The domain organizations and effects of Rng2Ns, Rng2CHD and Rng2Ns-C to actin are summarized in Supplementary Figure S8. Although Rng2Ns or Rng2CHD contains only one CHD, they can bundle F-actin, suggesting that they possess additional ABDs outside the CHD, as observed for calponin or SM/transgelin (Nakano et al, 2005; Wu and Jin, 2008). We speculate that Rng2Ns binds to the side of an actin oligomer (or a filament) through at least a pair of ABDs. Rng2Ns may stabilize the oligomer, working as a clamp that binds its subunits, and hence facilitates the formation of actin nuclei. A similar mechanism was proposed for the function of Salmonella invasion protein A (Lilic et al, 2003). Rng2CHD, Rng2Ns lacking residues 190–300, can bundle F-actin but fails to promote actin polymerization. We therefore speculate that these residues are required for the stable side binding of Rng2Ns to actin oligomers but not for F-actin bundling. Future studies will identify the residues responsible for each ABD.
In rng2ts cells with delayed cytokinesis, Rlc1-GFP formed rings with uneven fluorescent intensity, showing that Rng2 regulates the correct distribution of myosin II in the CR to ensure its efficient contraction. As F-actin rings are normally formed even in these cells, Rng2 may tether myosin II to the medial ring independently of F-actin or mediate the association of myosin II with CR F-actin. The genetic interactions of Rng2 with Myo2 (Eng et al, 1998) also support this idea. In addition, we found that Rng2 genetically interacts with Mid1 and that Rlc1-GFP was not observed as a medial band of nodes in rng2ts cells (our unpublished data). We speculate that Rng2 may be involved in Myo2–Mid1 interaction or the localization of Myo2 to the nodes. The perturbed distribution of Rlc1 on the CR in rng2ts cells might be a consequence of the defective formation of Myo2 nodes. The molecular details of Rng2-mediated regulation of myosin II localization (rings or nodes) remain to be elucidated. The proposed functions of Rng2 are summarized in Figure 8.
Actin-bundling proteins feature F-actin structures
Cellular F-actin structures generally consist of fascicles, which are constructed by actin-bundling or -crosslinking proteins that are specifically responsible for their formation, for example, villin and fimbrin for the microvilli of the brush border (Glenney et al, 1981; Matsudaira and Burgess, 1982), Arp2/3 complex for lamellipodia (Higgs and Pollard, 2001), fascin for filopodia (Vignjevic et al, 2006), scruin for the acrosomal process of Limulus sperm (Schmid et al, 2004), and α-actinin and filamin for stress fibres (Naumanen et al, 2008). Each of the proteins with a unique bundling mode features this structure.
In this study, we showed that Rng2 bundles F-actin and is required to arrange CR actin filaments into a ring, which makes this the first report on the actin-bundling protein that is solely responsible for CR assembly. FM and EM showed that the bundling mode of Rng2Ns is clearly distinguishable from that of Fim1, which generates F-actin patches in vivo; Fim1 can crosslink F-actin at a variety of angles and produces F-actin bundles with a wide range of thicknesses, whereas Rng2Ns forms linear arrays of filaments of constant thickness. Rng2Ns/actin bundles appeared to be flexible and were convertible into smaller bundles, which would be suitable for the expected function of Rng2 (see above). In addition, the bundles are twistable; thus, we speculate that they are made up of bands of multi-layered F-actin rather than cylinders of hexagonally packed F-actin, which would verify their physical properties and be reasonable for the constituents of the CR as it appears as a closed band by EM (Sanger and Sanger, 1980; Yasuda et al, 1980; Kamasaki et al, 2007). Further structural investigation of the F-actin-bundling modes of Rng2 and Fim1will provide concrete insight into the structures of the CR and actin patches, respectively.
In vitro, Rng2Ns forms ‘asters' of F-actin in addition to bundles (Figure 5A). We speculate that these asters derive from clusters of short F-actin crosslinked by Rng2Ns. Similar structures were also observed in vitro in the presence of both Arp2/3 complex and fascin (an F-actin-bundling protein) (Ideses et al, 2008).
IQGAP has been reported to localize to the CR or be involved in cytokinesis in Dictyostelium, budding yeast, sea urchin eggs, and dividing mouse oocytes (Adachi et al, 1997; Epp and Chant, 1997; Nishimura and Mabuchi, 2003; Bielak-Zmijewska et al, 2008), although its mode of action on CR actin filaments has been unclear until this study. Thus, IQGAP may regulate re-arrangement of the actin cytoskeleton on cytokinesis in diverse cell types. It is also likely that functional homologues of Rng2 are universally responsible for CR assembly because numerous actin-bundling proteins remain unidentified in eukaryotic cells. As being bundled is a universal feature of CR actin filaments, their common characteristics can be deduced from the physical and biochemical properties of Rng2Ns/actin bundles, which can now be determined in vitro. Moreover, examination of the interactions among fission yeast actin filaments bundled by Rng2, myosin II, and Adf1 would provide insight into the formation and contraction mechanisms of the CR at the molecular level.
Materials and methods
Molecular biological and genetic methods
Standard media and genetic methods for S. pombe were used. Strain leu1-32 (h−leu1-32) or JY333 (h−ade6-M216 leu1-32) was used as the wild-type S. pombe strain. Strain rng2ts (h−leu1-32 rng2-D5) was used as a temperature-sensitive rng2 mutant strain in this study. Strain FY15527 (h−ade6-M210 leu1-32 lys1-131 ura4-D18 sid4+∷sid4-mRFP-kanR) was provided by the Yeast Genetic Resource Center (Osaka City University, Japan). Construction of the expression vectors is described in the Supplementary data.
Protein purification
Actin was prepared from rabbit skeletal muscle (Spudich and Watt, 1971). Pyrene–actin was produced by labelling muscle actin with N-(1-pyrene) iodoacetamide (Molecular Probes) (Cooper et al, 1983). Both unlabelled and labelled actins were further purified by gel filtration and stored in G-buffer (0.1 mM CaCl2, 0.5 mM DTT, 0.2 mM ATP, and 2 mM Tris–HCl, pH 8.0) on ice.
N-terminally His6-tagged Rng2Ns was expressed in BL21 Escherichia coli (Stratagene) transformed with pET28a-Rng2Ns. The cells were lysed by sonication in 1% NP-40, 1 mM DTT, 1 μg/ml pepstatin A, 0.5 mM PMSF, 0.3 × PBS, and 20 mM imidazole–HCl, pH 7.5 on ice. The homogenate was clarified by centrifugation and mixed with Ni-NTA agarose (Qiagen) for 1.5 h at 4°C. The beads were washed sequentially three times with a 20-fold volume of HS (0.2% NP-40, 0.35 M NaCl, 1 × PBS, and 20 mM imidazole–HCl, pH 8.0), AM (3 mM ATP, 15 mM MgSO4, and 1 × PBS), and LS (1 × PBS and 20 mM imidazole–HCl, pH 8.0). The proteins were eluted from the beads with 1 × PBS and 0.5 M imidazole–HCl, pH 8.0. The peak fractions were dialysed against 1 × PBS, 0.5 mM DTT, and 50 mM imidazole–HCl, pH 8.0 and were subsequently concentrated by dialysis against solid sucrose to make at least 60 μM Rng2Ns. The concentrate was centrifuged at 200 000 g for 20 min, and the supernatant (purified Rng2Ns) was stored at −80°C. N-terminally His6-tagged Rng2CHD was expressed and purified in the same way. Recombinant Fim1, Adf1, and Cdc3 were bacterially expressed with GST fusion and purified after the removal of GST by thrombin cleavage (Nakano et al, 2001; Nakano and Mabuchi, 2006; Takaine and Mabuchi, 2007). Rabbit plasma gelsolin was purified according to the method of Kurokawa et al (1990). Stock solutions of ABPs were precleared by centrifugation at 200 000 g for 10 min before use, especially after thawing or long storage (for more than 1 week) on ice.
Fluorescence spectroscopy of actin polymerization
In the polymerization assay, G-actin (with added 1–5% pyrene–actin) was dissolved in G-MEI (0.1 mM MgCl2, 0.5 mM EGTA, 0.5 mM DTT, 0.2 mM ATP, and 10 mM imidazole–HCl, pH 7.5) with additional proteins in a cuvette and converted to Mg2+-G-actin for 2 min. Polymerization was then initiated by the addition of a 1/29 volume of salts (1.5 M KCl and 30 mM MgCl2). The amount of polymerized actin was monitored through changes in pyrene fluorescence (excitation at 365 nm and emission at 407 nm) using an RF-5300PC spectrofluorophotometer (Shimadzu Corp) at 23°C. Numerical data were processed using Excel, and graphs were drawn using KaleidaGraph (Synergy Software).
Sedimentation assay
F-actin (3 μM) and various concentrations of Rng2Ns, Rng2CHD, or Fim1 were incubated together in KMEI (0.1 M KCl, 2 mM MgCl2, 0.5 mM EGTA, 0.5 mM DTT, 0.2 mM ATP, and 10 mM imidazole–HCl, pH 7.5) for 1 h at 25°C and then centrifuged at 20 000 g for 15 min (low-speed conditions) or at 200 000 g for 20 min (high-speed conditions). The supernatants and pellets were subjected to SDS–PAGE, and the gels were stained with Coomassie brilliant blue (CBB). The concentration of Rng2Ns, Rng2CHD, or Fim1 in each fraction was quantified by densitometry. Kd values for the actin-bundling proteins bound to F-actin were calculated by fitting plots of [protein bound to F-actin] versus [protein free] with a hyperbolic function using KaleidaGraph.
Microscopy
Fluorescence images were acquired with a CCD camera (ORCAII-ER-1394; Hamamatsu Photonics) on an Axioskop microscope (Carl Zeiss) equipped with a × 100/1.30 NA PlanNeofluar objective lens (Carl Zeiss) using Aquacosmos (Hamamatsu). The images were processed in Adobe Photoshop and analysed using Image J if necessary. For confocal microscopy, samples were imaged by a confocal microscope (LSM 510; Carl Zeiss, Inc) equipped with an alpha Plan-Fluar × 100/1.45 NA objective lens (Carl Zeiss, Inc), and the images were processed with Image J. Fission yeast cells were fixed with formaldehyde and processed for FM (Arai and Mabuchi, 2002). The actin cytoskeleton and nuclear DNA were stained with rhodamine–phalloidin or BODIPY–phallacidin (Molecular Probes), and DAPI (0.5 μg/ml for fixed cells) or Hoechst 33342 (0.5 μg/ml for living cells), respectively. F-actin in vitro was processed for FM as described earlier (Takaine and Mabuchi, 2007). For EM viewing of F-actin, the samples were stained with uranyl acetate and observed with an electron microscope (JEM1010; JEOL).
Supplementary Material
Supplementary Data
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Figures S7, S8, S9 and S10
Review Process File
Acknowledgments
We thank Mohan K Balasubramanian (Tamasek Life Sciences Laboratory, Singapore) and Yeast Genetic Resource Center (Osaka City University, Japan) for providing yeast strains, and Tadaomi Takenawa (Kobe University, Japan) for providing N-WASp plasmids. This work was supported by a Grant-in-Aid for Scientific Research to KN from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) (Nos. 19037004 and 20770150).
References
- Adachi H, Takahashi Y, Hasebe T, Shirouzu M, Yokoyama S, Sutoh K (1997) Dictyostelium IQGAP-related protein specifically involved in the completion of cytokinesis. J Cell Biol 137: 891–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almonacid M, Moseley JB, Janvore J, Mayeux A, Fraisier V, Nurse P, Paoletti A (2009) Spatial control of cytokinesis by Cdr2 kinase and Mid1/anillin nuclear export. Curr Biol 19: 961–966 [DOI] [PubMed] [Google Scholar]
- Andrianantoandro E, Pollard TD (2006) Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol Cell 24: 13–23 [DOI] [PubMed] [Google Scholar]
- Arai R, Mabuchi I (2002) F-actin ring formation and the role of F-actin cables in the fission yeast Schizosaccharomyces pombe. J Cell Sci 115: 887–898 [DOI] [PubMed] [Google Scholar]
- Balasubramanian MK, Bi E, Glotzer M (2004) Comparative analysis of cytokinesis in budding yeast, fission yeast and animal cells. Curr Biol 14: R806–R818 [DOI] [PubMed] [Google Scholar]
- Balasubramanian MK, Helfman DM, Hemmingsen SM (1992) A new tropomyosin essential for cytokinesis in the fission yeast S pombe. Nature 360: 84–87 [DOI] [PubMed] [Google Scholar]
- Balasubramanian MK, Hirani BR, Burke JD, Gould KL (1994) The Schizosaccharomyces pombe cdc3+ gene encodes a profilin essential for cytokinesis. J Cell Biol 125: 1289–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielak-Zmijewska A, Kolano A, Szczepanska K, Maleszewski M, Borsuk E (2008) Cdc42 protein acts upstream of IQGAP1 and regulates cytokinesis in mouse oocytes and embryos. Dev Biol 322: 21–32 [DOI] [PubMed] [Google Scholar]
- Brandt DT, Marion S, Griffiths G, Watanabe T, Kaibuchi K, Grosse R (2007) Dia1 and IQGAP1 interact in cell migration and phagocytic cup formation. J Cell Biol 178: 193–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F, Woollard A, Nurse P (1996) Isolation and characterization of fission yeast mutants defective in the assembly and placement of the contractile actin ring. J Cell Sci 109: 131–142 [DOI] [PubMed] [Google Scholar]
- Cooper JA, Walker SB, Pollard TD (1983) Pyrene actin: documentation of the validity of a sensitive assay for actin polymerization. J Muscle Res Cell Motil 4: 253–262 [DOI] [PubMed] [Google Scholar]
- D'souza VM, Naqvi NI, Wang H, Balasubramanian MK (2001) Interactions of Cdc4p, a myosin light chain, with IQ-domain containing proteins in Schizosaccharomyces pombe. Cell Struct Funct 26: 555–565 [DOI] [PubMed] [Google Scholar]
- Eng K, Naqvi NI, Wong KC, Balasubramanian MK (1998) Rng2p, a protein required for cytokinesis in fission yeast, is a component of the actomyosin ring and the spindle pole body. Curr Biol 8: 611–621 [DOI] [PubMed] [Google Scholar]
- Epp JA, Chant J (1997) An IQGAP-related protein controls actin-ring formation and cytokinesis in yeast. Curr Biol 7: 921–929 [DOI] [PubMed] [Google Scholar]
- Fujiwara K, Porter ME, Pollard TD (1978) Alpha-actinin localization in the cleavage furrow during cytokinesis. J Cell Biol 79: 268–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukata M, Kuroda S, Fujii K, Nakamura T, Shoji I, Matsuura Y, Okawa K, Iwamatsu A, Kikuchi A, Kaibuchi K (1997) Regulation of cross-linking of actin filament by IQGAP1, a target for Cdc42. J Biol Chem 272: 29579–29583 [DOI] [PubMed] [Google Scholar]
- Glenney JR, Kaulfus P, Matsudaira P, Weber K (1981) F-actin binding and bundling properties of fimbrin, a major cytoskeletal protein of microvillus core filaments. J Biol Chem 256: 9283–9288 [PubMed] [Google Scholar]
- Higgs HN, Pollard TD (2001) Regulation of actin filament network formation through ARP2/3 complex: activation by a diverse array of proteins. Annu Rev Biochem 70: 649–676 [DOI] [PubMed] [Google Scholar]
- Ideses Y, Brill-Karniely Y, Haviv L, Ben-Shaul A, Bernheim-Groswasser A (2008) Arp2/3 branched actin network mediates filopodia-like bundles formation in vitro. PLos ONE 3: e3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamasaki T, Osumi M, Mabuchi I (2007) Three-dimensional arrangement of F-actin in the contractile ring of fission yeast. J Cell Biol 178: 765–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitayama C, Sugimoto A, Yamamoto M (1997) Type II myosin heavy chain encoded by the myo2 gene composes the contractile ring during cytokinesis in Schizosaccharomyces pombe. J Cell Biol 137: 1309–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar DR, Kuhn JR, Tichy AL, Pollard TD (2003) The fission yeast cytokinesis formin Cdc12p is a barbed end actin filament capping protein gated by profilin. J Cell Biol 161: 875–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa H, Fujii W, Ohmi K, Sakurai T, Nonomura Y (1990) Simple and rapid purification of brevin. Biochem Biophys Res Commun 168: 451–457 [DOI] [PubMed] [Google Scholar]
- Li CR, Wang YM, Wang Y (2008) The IQGAP Iqg1 is a regulatory target of CDK for cytokinesis in Candida albicans. EMBO J 27: 2998–3010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilic M, Galkin VE, Orlova A, VanLoock MS, Egelman EH, Stebbins CE (2003) Salmonella Sip A polymerizes actin by stapling filaments with nonglobular protein arms. Science 301: 1918–1921 [DOI] [PubMed] [Google Scholar]
- Lu J, Pollard TD (2001) Profilin binding to poly-L-proline and actin monomers along with ability to catalyze actin nucleotide exchange is required for viability of fission yeast. Mol Biol Cell 12: 1161–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J (1991) Predicting coiled coils from protein sequences. Science 252: 1162–1164 [DOI] [PubMed] [Google Scholar]
- Mabuchi I, Tsukita S, Tsukita S, Sawai T (1988) Cleavage furrow isolated from newt eggs: contraction, organization of the actin filaments, and protein components of the furrow. Proc Natl Acad Sci USA 85: 5966–5970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsudaira PT, Burgess DR (1982) Partial reconstruction of the microvillus core bundle: characterization of villin as a Ca++-dependent, actin-bundling/depolymerizing protein. J Cell Biol 92: 648–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motegi F, Mishra M, Balasubramanian MK, Mabuchi I (2004) Myosin-II reorganization during mitosis is controlled temporally by its dephosphorylation and spatially by Mid1 in fission yeast. J Cell Biol 165: 685–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhina S, Wang YL, Murata-Hori M (2007) Alpha-actinin is required for tightly regulated remodeling of the actin cortical network during cytokinesis. Dev Cell 13: 554–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano K, Bunai F, Numata O (2005) Stg1 is a novel SM22/transgelin-like actin-modulating protein in ssion yeast. FEBS Lett 579: 6311–6316 [DOI] [PubMed] [Google Scholar]
- Nakano K, Mabuchi I (2006) Actin-depolymerizing protein adf1 is required for formation and maintenance of the contractile ring during cytokinesis in fission yeast. Mol Biol Cell 17: 1933–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano K, Satoh K, Morimatsu A, Ohnuma M, Mabuchi I (2001) Interactions among a fimbrin, a capping protein, and an actin-depolymerizing factor in organization of the fission yeast actin cytoskeleton. Mol Biol Cell 12: 3515–3526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumanen P, Lappalainen P, Hotulainen P (2008) Mechanisms of actin stress fibre assembly. J Microsc 231: 446–454 [DOI] [PubMed] [Google Scholar]
- Nishimura Y, Mabuchi I (2003) An IQGAP-like protein is involved in actin assembly together with Cdc42 in the sea urchin egg. Cell Motil Cytoskeleton 56: 207–218 [DOI] [PubMed] [Google Scholar]
- Numata O, Gonda K, Watanabe A, Kurasawa Y (2000) Cytokinesis in tetrahymena: determination of division plane and organization of contractile ring. Microsc Res Tech 49: 127–135 [DOI] [PubMed] [Google Scholar]
- Paoletti A, Chang F (2000) Analysis of mid1p, a protein required for placement of the cell division site, reveals a link between the nucleus and the cell surface in fission yeast. Mol Biol Cell 11: 2757–2773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan ME, Heuser JE, Kerkhoff E, Mullins RD (2005) Drosophila Spire is an actin nucleation factor. Nature 433: 382–388 [DOI] [PubMed] [Google Scholar]
- Sanger JM, Sanger JW (1980) Banding and polarity of actin filaments in interphase and cleaving cells. J Cell Biol 86: 568–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterwhite LL, Pollard TD (1992) Cytokinesis. Curr Opin Cell Biol 4: 43–52 [DOI] [PubMed] [Google Scholar]
- Schmid MF, Sherman MB, Matsudaira P, Chiu W (2004) Structure of the acrosomal bundle. Nature 431: 104–107 [DOI] [PubMed] [Google Scholar]
- Skoumpla K, Coulton AT, Lehman W, Geeves MA, Mulvihill DP (2007) Acetylation regulates tropomyosin function in the fission yeast Schizosaccharomyces pombe. J Cell Sci 120: 1635–1645 [DOI] [PubMed] [Google Scholar]
- Skau CT, Neidt EM, Kovar DR (2009) Role of tropomyosin in formin-mediated contractile ring assembly in fission yeast. Mol Biol Cell 20: 2160–2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich JA, Watt S (1971) The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J Biol Chem 246: 4866–4871 [PubMed] [Google Scholar]
- Takaine M, Mabuchi I (2007) Properties of actin from the fission yeast Schizosaccharomyces pombe and interaction with fission yeast profilin. J Biol Chem 282: 21683–21694 [DOI] [PubMed] [Google Scholar]
- Vavylonis D, Wu JQ, Hao S, O'Shaughnessy B, Pollard TD (2008) Assembly mechanism of the contractile ring for cytokinesis by fission yeast. Science 319: 97–100 [DOI] [PubMed] [Google Scholar]
- Vignjevic D, Kojima S, Aratyn Y, Danciu O, Svitkina T, Borisy GG (2006) Role of fascin in filopodial protrusion. J Cell Biol 174: 863–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachtler V, Rajagopalan S, Balasubramanian MK (2003) Sterol-rich plasma membrane domains in the fission yeast Schizosaccharomyces pombe. J Cell Sci 116: 867–874 [DOI] [PubMed] [Google Scholar]
- Wu JQ, Bähler J, Pringle JR (2001) Roles of a fimbrin and an alpha-actinin-like protein in fission yeast cell polarization and cytokinesis. Mol Biol Cell 12: 1061–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu KC, Jin JP (2008) Calponin in non-muscle cells. Cell Biochem Biophys 52: 139–148 [DOI] [PubMed] [Google Scholar]
- Wu JQ, Kuhn JR, Kovar DR, Pollard TD (2003) Spatial and temporal pathway for assembly and constriction of the contractile ring in fission yeast cytokinesis. Dev Cell 5: 723–734 [DOI] [PubMed] [Google Scholar]
- Wu JQ, Pollard TD (2005) Counting cytokinesis proteins globally and locally in fission yeast. Science 310: 310–314 [DOI] [PubMed] [Google Scholar]
- Wu JQ, Sirotkin V, Kovar DR, Lord M, Beltzner CC, Kuhn JR, Pollard TD (2006) Assembly of the cytokinetic contractile ring from a broad band of nodes in fission yeast. J Cell Biol 174: 391–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda T, Numata O, Ohnishi K, Watanabe Y (1980) A contractile ring and cortical changes found in the dividing Tetrahymena pyriformis. Exp Cell Res 128: 407–417 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Figures S7, S8, S9 and S10
Review Process File