Abstract
E2F1 is a key positive regulator of human cell proliferation and its activity is altered in essentially all human cancers. Deregulation of E2F1 leads to oncogenic DNA damage and anti-oncogenic apoptosis. The molecular mechanisms by which E2F1 mediates these two processes are poorly understood but are important for understanding cancer progression. During the G1-to-S phase transition, E2F1 associates through a short DHQY sequence with the cell-cycle regulator HCF-1 together with the mixed-lineage leukaemia (MLL) family of histone H3 lysine 4 (H3K4) methyltransferases. We show here that the DHQY HCF-1-binding sequence permits E2F1 to stimulate both DNA damage and apoptosis, and that HCF-1 and the MLL family of H3K4 methyltransferases have important functions in these processes. Thus, HCF-1 has a broader role in E2F1 function than appreciated earlier. Indeed, sequence changes in the E2F1 HCF-1-binding site can modulate both up and down the ability of E2F1 to induce apoptosis indicating that HCF-1 association with E2F1 is a regulator of E2F1-induced apoptosis.
Keywords: apoptosis, DNA damage, E2F1, HCF-1, MLL
Introduction
DNA damage and resulting mutations promote tumour progression (Halazonetis et al, 2008). A key source of DNA damage is inappropriate DNA replication, often induced by inactivation of tumour suppressors or activation of oncogenes. Key regulators of human DNA replication are the E2F proteins, which comprise both transcriptional activators (E2F1–3) and repressors (E2F4–5) of genes required for DNA replication (reviewed by Trimarchi and Lees, 2002; Blais and Dynlacht, 2007). Under healthy circumstances, the tumour suppressor pRb associates with and prevents the activator E2F proteins from stimulating DNA replication at inappropriate times. If, however, pRb is inactivated, as it is in nearly all human tumours, or activator E2F proteins, particularly E2F1, are overexpressed, inappropriate DNA replication and subsequent formation of DNA double-stranded breaks (DSBs) result in DNA damage (Bartkova et al, 2005; Pickering and Kowalik, 2006).
To suppress the deleterious effects of such oncogene-induced DNA damage, the cell can in turn induce a DNA-damage response (DDR) to either arrest the cell cycle and repair the DNA, or induce apoptosis or senescence and thus eliminate or inactivate, respectively, the damaged cell (Bartek et al, 2007; Halazonetis et al, 2008). The function of E2F1 in the oncogene-induced DDR leading to senescence is not well understood (Bartkova et al, 2006; Di Micco et al, 2006). In contrast, E2F1 is well known to support an apoptotic DDR, through p53-dependent and p53-independent pathways (Bates et al, 1998; Irwin et al, 2000; Stiewe and Pützer, 2000; Moroni et al, 2001; Powers et al, 2004; Radhakrishnan et al, 2004; Rogoff et al, 2004; Frame et al, 2006). Thus, E2F1 can both stimulate DNA damage by activating DNA replication at inappropriate times and suppress its ill effects by stimulating an apoptotic DDR. Understanding how E2F1 stimulates these oncogenic (DNA damage) and tumour-suppressive (e.g. apoptosis) activities is important for understanding cancer progression. We show here that E2F1 depends on association with the herpes–simplex virus (HSV) host-cell factor HCF-1 and the mixed-lineage leukaemia (MLL) family of trithorax-related histone H3 lysine 4 (H3K4) methyltransferases to stimulate both DNA damage and apoptosis.
HCF-1, originally discovered as a cellular co-activator of HSV transcription, has been linked to cell-cycle progression (Goto et al, 1997; Tyagi et al, 2007), embryonic stem-cell pluripotency (Dejosez et al, 2008), and poor cancer prognosis (Glinsky et al, 2005). It is a heterodimeric complex of N- and C-terminal subunits arising from proteolytic cleavage of a single precursor protein (see Wysocka and Herr, 2003). The N-terminal subunit recognizes and binds to a short tetrapeptide HCF-1-binding motif (HBM) with numerous transcription factors, including E2F1, E2F3, and E2F4 involved in cell-cycle regulation (Knez et al, 2006; Tyagi et al, 2007) and Ronin involved in the maintenance of embryonic stem-cell pluripotency (Dejosez et al, 2008). HCF-1 also associates with histone modifying complexes involved in both activation (e.g. MLL family of H3K4 methyltransferases) and repression [e.g. Sin3 histone deacetylase (HDAC)] of transcription and selectively recruits them to sequence-specific activators (e.g. VP16 and E2F1) or repressors (e.g. E2F4) of transcription, respectively (Wysocka et al, 2003; Yokoyama et al, 2004; Tyagi et al, 2007).
Here, we have studied the function of HCF-1 in promoting the ability of E2F1 to induce apoptosis. Numerous co-regulators of E2F1 have been implicated in its downstream function of regulating transcription. These include histone acetyltransferases (Hsu et al, 2001; Lang et al, 2001; Louie et al, 2004; El Messaoudi et al, 2006) and HCF-1 and the MLL family of H3K4 methyltransferases (Tyagi et al, 2007). A different set of co-regulators (e.g. Jab1, TIP49, MCPH1/BRIT1) has been implicated in supporting E2F1-induced apoptosis but their mechanisms of action are generally poorly understood (Dugan et al, 2002; Hallstrom and Nevins, 2006; Yang et al, 2008). Here, we have taken advantage of the ability of induced overexpression of E2F1 to produce DNA damage and apoptosis (Rogoff et al, 2004) to determine whether HCF-1 and the MLL family of H3K4 methytransferases have functions as co-regulators of E2F1 induction of DNA damage and apoptosis.
Results
Disruption of the HCF-1-binding site in E2F1 interferes with E2F1-induced apoptosis
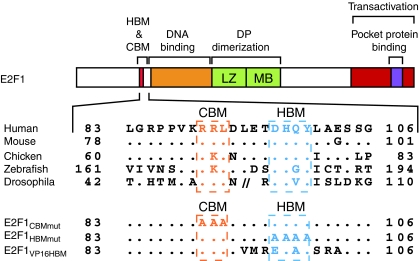
Figure 1 (adapted from Tyagi et al, 2007) illustrates the structure of human E2F1 and the conservation of sequences within and surrounding its HCF-1-binding site, thus highlighting the conserved HBM (consensus sequence D/EHxY, where x is any amino acid) (see Wysocka and Herr, 2003) and ‘cyclin A-binding motif' (CBM) (consensus sequence RxL) (Lowe et al, 2002). To study the function of the HBM in E2F1-induced apoptosis, we prepared three E2F1 mutants: E2F1CBMmut, in which the three amino-acid CBM has been changed to three alanines; E2F1HBMmut, in which the four amino-acid HBM has been changed to four alanines; and E2F1VP16HBM, in which the E2F1 HBM and flanking sequences have been replaced by the corresponding HSV VP16 HBM sequence (Figure 1, bottom).
Figure 1.
Schematic structure of E2F1 protein showing conserved HCF-1-binding motif (HBM) and Cyclin-binding motif (CBM) sequences and E2F1 mutants used in this study. (Top) Amino-acid sequence alignment of E2F1 HBM regions from the species listed are adapted from Tyagi et al (2007). Dots indicate identity with the human sequence. //denotes amino acids 53–99 of Drosophila E2F1. LZ, Leucine zipper; MB, marked box. (Bottom) Alanine substitutions made in the E2F1 CBM (E2F1CBMmut) or HBM (E2F1HBMmut) are shown. E2F1VP16HBM replacement of the E2F1 HBM and surrounding sequence by corresponding HSV-1 VP16 sequences.
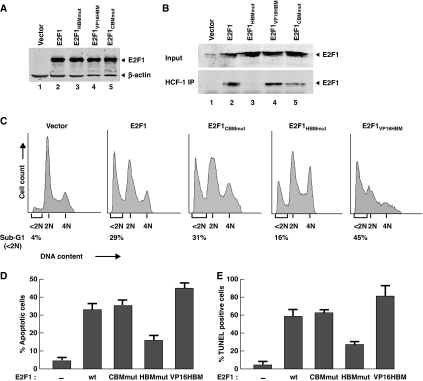
We first examined the function of the E2F1 CBM and HBM elements in E2F1-mediated apoptosis by transfecting expression constructs for the E2F1 wild-type protein and the E2F1CBMmut or E2F1HBMmut alanine substitution mutants into U2OS cells and measuring apoptosis by the appearance of a ‘sub-G1' population of cells 40 h after transfection. Figure 2A shows that the wild-type and mutant proteins were synthesized to equivalent levels. Furthermore, as hypothesized and shown in Figure 2B, HCF-1 immunoprecipitation revealed HCF-1 association with the wild-type and E2F1CBMmut proteins (compare lanes 2 and 5 with 1) but not effectively with the E2F1HBMmut protein (lane 3).
Figure 2.
Alterations in the E2F1 HCF-1-binding site can modulate induction of apoptosis. (A) Anti-E2F1 and β-actin immunoblot analysis from U2OS-cell extracts transfected with empty plasmid (lane 1) or plasmids encoding E2F1 (lane 2) or indicated E2F1 mutants (lanes 3–5) and harvested 40-h post-transfection. (B) Nuclear extracts of U2OS cells transfected as in (A) were prepared, immunoprecipitated with anti-HCF-1 antisera (bottom panel), and recovered E2F1 proteins were visualized by immunoblot with anti-E2F1antisera. Input (top panel) corresponds to 10% of the nuclear extract used for the immunoprecipitation. (C) U2OS cells were transfected as in (A), stained with propidium iodide, and analysed for DNA content by flow cytometry. Apoptosis was measured by the sub-G1 DNA content (<2N) population. 2N, G1-phase cells; 4N, G2/M-phase cells. (D) Transfected cells were analysed for the apoptotic sub-G1 DNA content cell population as in (C) and the averaged data are derived from three independent experiments. (E) Quantification of cells transfected as in (A), and scored positive for TdT-mediated dUTP nick end-labelling (TUNEL) assay. The percentage was determined by counting 200 cells per sample in each of two different experiments. Data are represented as mean±s.d.
The sub-G1 analysis showed that both wild-type E2F1 and E2F1CBMmut overexpression induced significant and equivalent levels of apoptosis (29 and 31%, respectively) compared with vector alone (4%) (Figure 2C). In contrast, the E2F1HBMmut mutant was significantly attenuated for apoptosis (16%). Consistent with the single samples shown in Figure 2C are the results of multiple samples shown in Figure 2D. To substantiate these results, the relative apoptotic activities of wild-type and mutant E2F1 proteins were reproduced by TUNEL analysis as shown in Figure 2E. These results show that the HBM sequence influences the ability of E2F1 to induce apoptosis and suggest that HCF-1 association with E2F1 is important for its full pro-apoptotic activity. As the HBM lies outside of the C-terminal transactivation domain (Figure 1), these results are also consistent with the earlier finding that C-terminal E2F1-truncation mutants can still stimulate apoptosis (Hsieh et al, 1997).
Replacement of the wild-type E2F1 HBM and flanking sequences by the equivalent viral VP16 sequences enhances E2F1 activation of apoptosis in U2OS cells
We have shown earlier that E2F1 displays autoinhibitory association with HCF-1. For example, full-length wild-type human E2F1 does not interact with HCF-1 in a yeast two-hybrid assay but either N- or C-terminal truncations of E2F1 activate the ability of E2F1 to interact with HCF-1 in an HBM-dependent manner (Tyagi et al, 2007). Furthermore, replacement of the E2F1 HBM and its three flanking amino acids on each side by the equivalent HSV VP16 sequences likewise activated E2F1 interaction with HCF-1 in the yeast two-hybrid assay. These results suggested that the VP16 HBM represents a more effective HCF-1-binding site in the context of human E2F1 (Tyagi et al, 2007).
We took advantage of this unusual E2F1 HBM mutant, in which 8 out of 10 residues within and surrounding the E2F1 HBM are changed, to ask whether the pro-apoptotic activity of E2F1 might be modulated by the quality of its HBM. Indeed, while synthesized at similar levels (Figure 2A, lane 4) and as expected maintaining association with HCF-1 (Figure 2B, lane 4), the E2F1VP16HBM mutant displayed enhanced induction of apoptosis compared with wild-type E2F1 (45 versus 30%; see also Figure 2D and E). Thus, the wild-type E2F1 HBM, perhaps owing to some element of autoinhibition for HCF-1 association, displays important but not a maximal potential for sustaining E2F1-induced apoptosis.
Two important conclusions can be made from this experiment. First, because the E2F1VP16HBM mutant maintains HCF-1 association with E2F1 and yet differs at 8 out of 10 positions within and surrounding the E2F1 HBM (every possible variable position), it is unlikely that the loss of apoptosis induction in the E2F1HBMmut is due to disruption of some unrelated (and unknown) E2F1 protein association. Second, the increase in apoptotic activity of the E2F1VP16HBM mutant, which mirrors the increased interaction of the E2F1VP16HBM mutant in the yeast two-hybrid assay, indicates that either the level or quality of HCF-1 association with E2F1 limits the pro-apoptotic properties of E2F1.
The E2F1 HBM contributes to induction of apoptosis in MCF7 and IMR-90tert cells
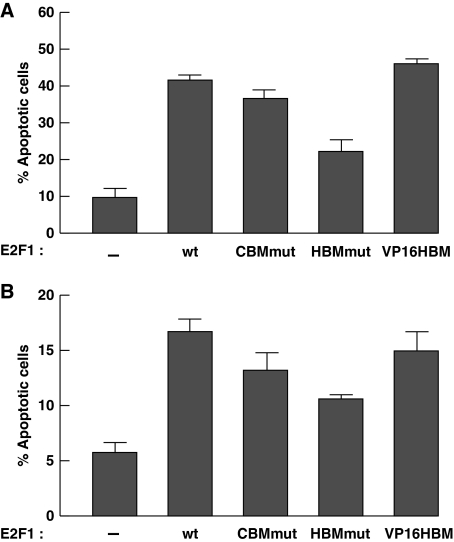
To determine whether a function for the E2F1 HCF-1-binding site in induction of apotosis is limited to U2OS cells, we assayed E2F1-induced apoptosis in p53- and pRb-positive MCF7 breast carcinoma cells and IMR-90 embryonic lung cells with telomerase to maintain viability (called IMR-90tert). As shown in Figure 3, the E2F1HBMmut is unable to support wild-type levels of apoptosis induction in both cell lines. As in U2OS cells, the E2F1VP16HBM displayed higher activity in the MCF7 cells (Figure 3A), suggesting that here too HCF-1 interaction with E2F1 can modulate apoptosis induction. In contrast, the E2F1VP16HBM mutant did not display enhanced activity in IMR-90tert cells (Figure 3B). This latter result suggests that the influence of the quality of the interaction of HCF-1 with E2F1 on apoptosis can be cell-type specific.
Figure 3.
E2F1 HCF-1-binding site supports induction of apoptosis in MCF7 and IMR-90tert cells. (A, B) Cells, MCF7 (A) or IMR-90tert (B), were transfected as in Figure 2A, stained with propidium iodide, and analysed for DNA content by flow cytometry. Apoptosis was measured by the sub-G1 DNA content (<2N) population and the averaged data are derived from two independent experiments.
The HCF-1-binding site in E2F1 contributes to DNA-damage induction
The ability of E2F1 to induce apoptosis has been linked to its ability to cause DNA damage by DSB accumulation (Rogoff et al, 2004). To test whether the apoptosis-defective E2F1HBMmut is also compromised for induction of DNA damage, we prepared cells stably expressing wild-type E2F1 (ER–E2F1) or the E2F1HBMmut mutant (ER–E2F1HBMmut) fused to the 4-hydroxytamoxifen (OHT)-sensitive estrogen-receptor ligand-binding domain in U2OS cells. Application of OHT to these cells permits activation of the recombinant ER–E2F1 protein (Vigo et al, 1999). Immunoblot analysis of our cell lines with an E2F1 antibody showed that the recombinant proteins are synthesized to levels equivalent to two-fold above the endogenous E2F1 protein level (Supplementary Figure S1A); thus application of innocuous OHT to these cells will provide a two- to three-fold combined endogenous and ectopic E2F1 overexpression in an otherwise stable environment.
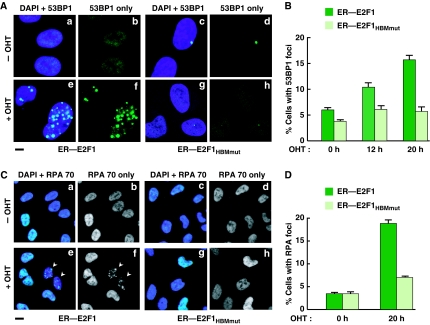
To monitor ER–E2F1- and ER–E2F1HBMmut-induced DNA damage on OHT application, we assayed over time the generation of DSBs and single-stranded DNA by staining for p53-binding protein 1 (53BP1; Figure 4A and B) (Schultz et al, 2000) and Replication Protein A (RPA; Figure 4C and D) (Zou and Elledge, 2003) foci, respectively. In contrast to wild-type E2F1, in both assays activation of E2F1HBMmut exhibited little effect, indicating that the E2F1 HBM and HCF-1 association have a function in the induction of DNA damage.
Figure 4.
The E2F1 HCF-1-binding site contributes to DNA-damage induction. (A, C) Immunoflourescence analysis of U2OS cells stably expressing ER–E2F1 or ER–E2F1HBMmut with or without OHT induction and stained with anti-53BP1 (A) or anti-RPA70 (C) shown with or without the result of DAPI co-staining. Scale bar represents 10 μm (A) or 40 μm (C). Arrowheads in (C) point to cells considered positive for RPA foci. (B, D) Quantification of cells positive for 53BP1-foci (B) or RPA-foci (D) after induction with OHT for the indicated times. The percentage was determined by counting 200 cells per sample in each of the two different experiments. Data are represented as mean±s.d.
E2F1HBMmut is defective for S-phase activation
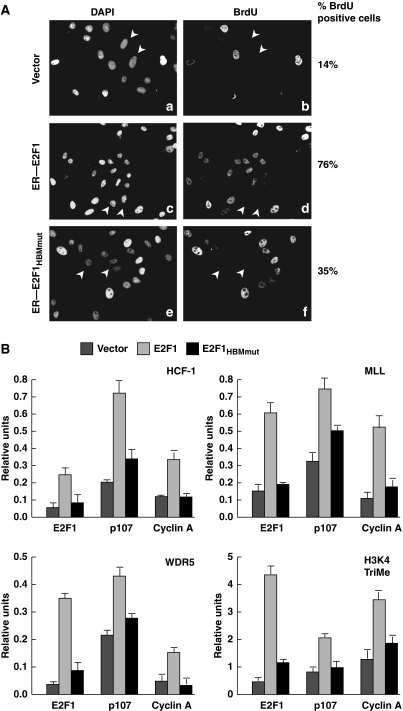
HCF-1 is important for cell-cycle progression from the G1-to-S phases (Goto et al, 1997; Julien and Herr, 2003) and has a function in the activation of E2F1-regulated S-phase promoters (Tyagi et al, 2007). The requirement if any, however, for the E2F1 HBM in cell-cycle progression is not known. As E2F1 induction of DNA damage is linked to its ability to promote DNA replication during the S phase (Bartkova et al, 2005) and the E2F1HBMmut is defective for DNA-damage induction, we asked whether the E2F1HBMmut is also defective for promoting G1-to-S phase progression. To answer this question, we (i) depleted the ER–E2F1 and ER–E2F1HBMmut cell lines of endogenous E2F1 (Supplementary Figure S1B), and then (ii) activated the siRNA-resistant ER–E2F1 and ER–E2F1HBMmut proteins with 24 h OHT treatment in the presence of BrdU, thus identifying cells that enter S phase during the 24-h period by BrdU incorporation. As the recombinant ER–E2F1 and ER–E2F1HBMmut proteins are present at similar levels to the endogenous protein (see Supplementary Figure S1B), the simultaneous depletion of the endogenous protein and activation of the recombinant proteins leads to similar levels of potentially active E2F protein. siRNA-induced depletion of E2F1 in vector control U2OS cells resulted in a reduction of BrdU incorporation from about 100% (data not shown) to 14% (Figure 5A) of cells, indicating an effective cell-cycle arrest owing to the absence of E2F1. Under these conditions, the wild-type ER–E2F1 cells maintained S-phase entry in 76% of cells, whereas the E2F1HBMmut cells only exhibited S-phase entry in 35% of cells, suggesting that the E2F1 HBM is necessary for full S-phase activation.
Figure 5.
E2F1HBMmut is defective for S-phase activation. (A) Long-term BrdU incorporation assay of E2F1 siRNA-treated cells stably expressing vector alone, ER–E2F1, or ER–E2F1HBMmut. The cells were harvested 72 h after the first siRNA transfection (with 24 h OHT induction) and stained with DAPI or anti-BrdU antibody. Arrowheads identify cells that lack BrdU incorporation. Percentage of BrdU positive cells was determined by counting 200 cells per sample. A second experiment yielded similar relative values. (B) Quantitation of HCF-1, MLL, WDR5, and H3K4 trimethylation (TriMe) ChIP analyses of U2OS cells transfected with plasmids encoding empty vector, E2F1, or E2F1HBMmut, by triplicate real-time PCR of the indicated promoters is shown. The cells were harvested 24 h post-transfection. The signals in cells immunoprecipitated using the anti-E2F1 antibody are set to 1 for each transfected sample with all other signals in that transfected sample indicated as relative-fold differences (relative units).
One explanation for why the E2F1HBMmut mutant is defective for activation of DNA replication is that it is unable to recruit HCF-1 and its associated H3K4 methyltransferases to activate transcription. To test this hypothesis, we overexpressed the E2F1 and E2F1HBMmut proteins in U2OS cells without altering endogenous HCF-1 and WDR5 (a universal component of MLL family H3K4 methyltransferases) levels (Supplementary Figure S2A) and measured HCF-1, MLL, and WDR5 occupancy, and H3K4 trimethylation at a collection of S-phase promoters (E2F1, p107, and cyclin A) described (Takahashi et al, 2000) and used (Tyagi et al, 2007) earlier by chromatin immunoprecipitation (ChIP; Figure 5B). Consistent with the aforementioned hypothesis, HCF-1, MLL, WDR5, and H3K4 trimethylation were all more enriched with the wild-type E2F1 as opposed to E2F1HBMmut mutant sample, suggesting that the E2F1 HBM is directly responsible for recruiting MLL family methytransferases through association with HCF-1. We also observed the same relative effects on the pro-apoptotic promoters described below (Supplementary Figure S2B). Taken together with the S-phase progression experiment (see Figure 5A), these HCF-1 promoter recruitment analyses suggest that E2F1–HCF-1 association is a critical element of cell-cycle progression.
HCF-1 and the H3K4 methytransferase component WDR5 are required for induction of E2F1-mediated apoptosis
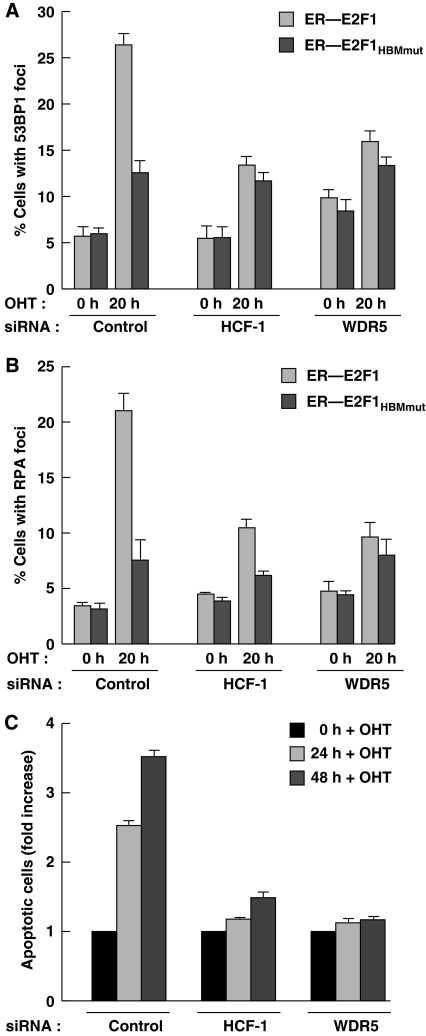
To elucidate the function of HCF-1 and the MLL family of H3K4 methyltransferases in E2F1-mediated DNA damage and apoptosis, we studied the effects of loss by siRNA treatment of HCF-1 and the global H3K4 methyltransferase complex component WDR5 on E2F1- and E2F1HBMmut-mediated DNA damage and apoptosis in the ER–E2F1 and ER–E2F1HBMmut U2OS cells (Figure 6). siRNA treatment per se (control sample) did not affect the relative induction of DNA damage (53BP1 and RPA foci) in the ER–E2F1 and ER–E2F1HBMmut U2OS cells (compare control 0 h in Figure 6A and B with Figure 4B and D). HCF-1 and WDR5 siRNA treatment, however, reduced DNA damage after 20 h OHT treatment of the ER–E2F1 cells, indicating a function for HCF-1 and the WDR5 component of MLL-related H3K4 methyltransferases in E2F1-mediated DNA damage. Consistent with this hypothesis, the activity of E2F1HBMmut for which HCF-1 association is abrogated is minimally affected by loss of either HCF-1 or WDR5, indicating that HCF-1 and WDR5 work through the HBM to stimulate DNA damage. The simplest interpretation of these results is that, to activate DNA damage, E2F1 recruits HCF-1 and one or more members of the MLL family of H3K4 methyltransferases to activate transcription.
Figure 6.
HCF-1 and the WDR5 component of the MLL family of H3K4 methytransferases are required for E2F1-mediated apoptosis induction. (A, B) ER–E2F1 or ER–E2F1HBMmut U2OS cells were treated with HCF-1 or WDR5 siRNAs for 72 h and quantified for 53BP1- (A) or RPA- (B) foci positive cells after induction with OHT for 0 or 20 h. The percentage was determined by counting 200 cells per sample in each of two different experiments. Data are represented as mean±s.d. (C) Apoptosis measurement of ER—E2F1 U2OS cells treated with HCF-1 or WDR5 siRNAs and induced with OHT for 24 or 48 h. Apoptosis, as measured by cells with a sub-G1 DNA content, was determined by flow cytometry. Apoptotic cell population at 0 h induction with OHT in each siRNA treatment was set to 1 with all other signals in that siRNA treatment indicated as relative-fold changes. The schematics and raw data for this experiment are shown in Supplementary Figure S3C.
To study the effects of loss of HCF-1 and WDR5 (Supplementary Figure S3A) on E2F1-induced apoptosis, 24 h after initiation of specific or control siRNA treatment, wild-type ER–E2F1 was induced in the U2OS cells by 24 or 48 h OHT treatment and apoptosis measured by sub-G1-cell accumulation by flow cytometry (see Supplementary Figure S3B). Owing to an induction of apoptosis by loss of HCF-1 and WDR5 in the absence of any E2F1 overexpression (see Figure 6; Supplementary Figure S3C), the samples were normalized to the corresponding 0 h OHT control. As expected, the control sample showed an induction of apoptosis at 24 and 48 h OHT treatment (Figure 6C; Supplementary Figure S3C). In contrast, ER–E2F1 U2OS cells that had been depleted of HCF-1 or WDR5 exhibited little response to E2F1 overexpression. These results suggest that both HCF-1 and the MLL family of H3K4 methyltransferases are necessary for full E2F1-mediated apoptosis.
HCF-1 activates transcription of pro-apoptotic E2F-target promoters but not a cell-cycle arrest promoter after DNA-damage induction
The aforementioned studies indicate that HCF-1 and the MLL family of H3K4 methyltransferases have a function in E2F1-mediated DNA damage and subsequent apoptosis but do not reveal whether the function they have in apoptosis is direct or simply a reflection of sustaining DNA damage by E2F1. To distinguish between these two possibilities, we studied the function of HCF-1 and the MLL family of H3K4 methyltransferases in E2F1-mediated apoptosis in response to DNA damage induced by the genotoxic drug adriamycin.
To stimulate apoptosis directly, E2F1 activates transcription of both p53-dependent (e.g. p14ARF) (Bates et al, 1998) and -independent (e.g. Apaf1 and p73) (Irwin et al, 2000; Stiewe and Pützer, 2000; Moroni et al, 2001) pro-apoptotic genes. We therefore examined the occupancy of HCF-1, MLL, WDR5, and trimethylated H3K4 on the E2F1, Apaf1, p73, and p14ARF promoters in response to adriamycin treatment. For comparison, we similarly analysed the E2F-responsive p21 promoter (Hiyama et al, 1998), which is involved in cell-cycle arrest instead of apoptosis in response to DNA damage (Harper et al, 1993).
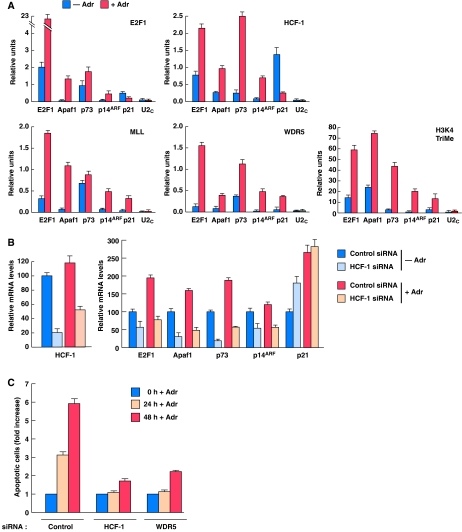
Consistent with a function of HCF-1 in E2F1-mediated apoptosis in response to DNA damage, HCF-1 can associate with E2F1 after adriamycin treatment (Supplementary Figure S4A) and like E2F1, on the E2F1, Apaf1, p73, and p14ARF promoters HCF-1, MLL, WDR5, and trimethylated H3K4 all increase in promoter occupancy on adriamycin treatment (Figure 7A). Interestingly, in the case of HCF-1, this increased recruitment is not due to increased levels of HCF-1 on adriamycin treatment (Supplementary Figure S4B) and is promoter selective because, whereas E2F1 can recruit HCF-1 to the p21 promoter (Supplementary Figure S2B), together with E2F1, HCF-1 occupancy of the cell-cycle arrest p21 promoter actually decreases on adriamycin treatment (Figure 7A). These results emphasize the parallel and intimate functions of E2F1 and HCF-1 in induction of apoptosis.
Figure 7.
HCF-1 regulates the transcriptional activation of pro-apoptotic E2F-target promoters and apoptosis after DNA-damage induction. (A) ChIP analyses of U2OS cells treated with or without adriamycin (Adr, 2 μg/ml) for 16 h using the indicated antisera (top right corner of each panel) and probed by Q-PCR for the indicated promoters are shown. PCR quantification was done using Delta Relative CT Quantification, where the values are calculated relative to input as follows: Delta CT=CT (input)−CT (sample); Relative Unit=2Delta CT. 0.3% of total ChIP input DNA was used in the input amplification. Triplicate real-time PCR data are represented as mean±s.d. A second experiment yielded similar results. (B) Quantitative real-time RT–PCR, in triplicate, was used to measure relative mRNA levels of indicated transcripts in cells treated with control or HCF-1 siRNA followed by treatment with or without adriamycin (16 h). Cells were harvested 48 h after the first siRNA transfection. (C) Apoptosis measurement of U2OS cells treated with HCF-1 or WDR5 siRNAs followed by treatment with adriamycin (5 μg/ml) for 24 or 48 h. Apoptosis, as measured by cells with a sub-G1 DNA content, was determined by flow cytometry. The apoptotic cell population at 0 h adriamycin treatment in each different siRNA sample was set to 1 with all other signals in that siRNA treatment indicated as relative-fold changes. The raw data for this experiment are shown in Supplementary Figure S4C.
Consistent with this apoptotic versus cell-cycle arrest pathway selectivity, depletion of HCF-1 by siRNA treatment in the absence or presence of adriamycin (Figure 7B, left) leads to a reduction in E2F1, Apaf1, p73, and p14ARF mRNA levels (Figure 7B, right). In contrast, in the same samples, HCF-1 depletion leads to an increased p21 mRNA level in the absence of adriamycin and no effect in the presence of adriamycin, suggesting a repressive function in p21-gene transcription during proliferation of normal cells and no p21 regulatory function during a DDR. Thus, together with E2F1, HCF-1 appears to be selectively involved in the transcriptional activation of pro-apoptotic-genes in a DDR.
HCF-1 and the H3K4 methytransferase component WDR5 are required for a DDR leading to apoptosis
The aforementioned studies suggest that HCF-1 and H3K4 methytransferases have a function in an E2F1-induced apoptotic response. To address whether HCF-1 and H3K4 methytransferases are important for mediating apoptosis in response to DNA damage generally, we asked whether they are required for the induction of apoptosis in response to the DNA damaging agent adriamycin as shown in Figure 7C and Supplementary Figure S4C. Indeed, depletion of either HCF-1 or WDR5 results in an attenuated induction of apoptosis in response to 24 or 48 h adriamycin treatment. Thus, HCF-1 and, as indicated by WDR5 depletion, one or more H3K4 methytransferases have an apparent function in the induction of apoptosis in response to DNA damage.
Discussion
Cancer is a complex set of diseases in which genetic and epigenetic alterations of cells have an important part in its progression. To combat cancer it is important to understand in molecular detail the effector mechanisms that lead to activation and inhibition of tumour progression because it is these interactions that can be potentially exploited for therapy. We have described here a precise region in E2F1, a tetrapeptide HBM, by which E2F1 activates DNA damage and apoptosis, two complementary oncogenic and tumour suppressor pathways, and how HCF-1 interaction with this sequence together with the MLL family H3K4 methyltransferases has an important function in these two activities. Indeed, alterations in the E2F1 HBM that can modulate E2F1 interaction with HCF-1 have the potential to correspondingly modulate E2F1-induced apoptosis, suggesting that HCF-1 in its association with E2F1 has a controlling function in these E2F1 responses. Thus, in contrast to other co-activators of E2F1-induced apoptosis [i.e. Jab1 (Hallstrom and Nevins, 2006), TIP49 (Dugan et al, 2002), and MCPH1/BRIT1 (Yang et al, 2008)], the target of E2F1 association (i.e. the HBM) and mechanism of E2F1 activation (i.e. downstream H3K4 methylation) by HCF-1 is known in much greater molecular detail.
We have shown earlier that in proliferating HeLa cells, E2F1 association with HCF-1 is cell-cycle regulated (Tyagi et al, 2007). Thus, even though HCF-1 is present throughout the cell cycle and E2F1 increases in abundance during the G1 phase, HCF-1–E2F1 association is relatively low in the G1 phase and yet increases as cells enter the S phase coincident with activation of S-phase promoters. Therefore, it was not a priori evident that HCF-1 association with E2F1 would have a function in induction of DNA damage by E2F1 overexpression outside of the G1–S phase transition. The finding here that HCF-1 indeed has a function in DNA damage mediated by inappropriate E2F1 expression indicates that HCF-1 can function with E2F1 outside of the G1-to-S phase transition and S phase itself. This finding suggests that, inside a cancer cell where E2F1 activities are deregulated, HCF-1 can have a broad role in its activities.
In addition to E2F1, HCF-1 interacts with the E2F3 and E2F4 members of the E2F family (Knez et al, 2006; Tyagi et al, 2007) and yet these other E2F members do not have defined functions in induction of apoptosis. E2F4 inhibits cell-cycle progression (Trimarchi and Lees, 2002) and E2F3-induced apoptosis is dependent on the presence of E2F1, suggesting that it is E2F1 that is the immediate activator of apoptosis (Lazzerini Denchi and Helin, 2005). What might be special about HCF-1's interaction with E2F1 that it would support induction of apoptosis with E2F1 and not with other E2F factors? In partial answer, we note interesting differences in how E2F3 and E2F4 associate with HCF-1. First, E2F3 possesses a noncanonical HBM with a glycine residue in place of an aspartic or glutamic acid residue in the first position of the D/EHxY consensus HBM. We have shown here that the sequences within and surrounding the E2F1 HBM can modulate its ability to induce apoptosis (the E2F1VP16HBM mutant). Thus, although HCF-1 associates with this noncanonical E2F3 HBM (Tyagi et al, 2007), the noncanonical sequence may determine different functions for HCF-1 association. Second, when HCF-1 associates with E2F4 in the early G1 phase where E2F4 represses cell-cycle progression, there is little E2F4–HCF-1 association of the MLL family of histone methyltransferases but rather a preferential association with the HCF-1-associated repressive Sin3 HDAC (Tyagi et al, 2007). These observations suggest that HCF-1 can have differing functions with individual members of the E2F factor family and thus be a determinant of the diversity of their functions.
A potential function for HCF-1 in oncogenesis and tumour suppression
There are two major tumour-suppressive responses to potentially oncogenic DNA damage—one leading to repair and cell survival and the other leading to cell removal (i.e. apoptosis) or inactivation (i.e. senescence). The studies described here implicate HCF-1 in sustaining oncogenic E2F1-induced DNA damage while at the same time aiding in E2F1 induction of tumour-suppressive apoptosis in response to DNA damage. Here, we have focused our studies on the apoptotic pathway because, unlike the case of oncogene-induced senescence, the function of E2F1 in induction of apoptosis has been extensively studied and many promoter targets well defined.
HCF-1 and E2F1 appear to discriminate between cell survival and cell loss by favouring an apoptotic response—they associate with and HCF-1 activates transcription of pro-apoptotic E2F1-target genes but not the cell-cycle arrest promoter p21. For E2F1 and HCF-1 to promote apoptosis rather than cell survival in response to DNA damage, the lack of activation of the p21 promoter is probably important for two reasons: First, p21 is important to inhibit DNA replication and thus stimulate cell-cycle arrest in response to DNA damage (Harper et al, 1993) and, second, p21 appears to have a critical function in inhibiting apoptosis (see Coqueret, 2003).
In addition to implicating HCF-1 in E2F1-mediated DNA damage and apoptosis, these results described here implicate the MLL family of H3K4 methyltransferases in tumour suppression. The MLL gene was originally discovered because it undergoes translocations in MLLs (Hess, 2004). Its relationship to the Drosophila Trithorax regulator of homeo-domain protein function led to studies of MLL in the regulation of human homeo-domain gene expression. More recently, it has been revealed that MLL associates with multiple E2F proteins and promotes cell-cycle progression (Takeda et al, 2006; Tyagi et al, 2007). Interestingly, although it interacts with many E2F proteins directly (Takeda et al, 2006), MLL does not associate with E2F1 directly but instead is brought to E2F1 in coordination with HCF-1 (Tyagi et al, 2007). The results provided here suggest that HCF-1 and the MLL family of H3K4 methytransferases come together to potentiate the preferential ability of E2F1 to activate DNA damage and apoptosis.
In conclusion, our studies have identified critical players—HCF-1 and the MLL family of H3K4 methyltransferases—in the activation of DNA damage and apoptosis by E2F1. Although negative regulators of E2F-1 (e.g. pRb) have been known for many years, an understanding of the proteins required for E2F1 activation of oncogenic processes (e.g. DNA damage) has been less evident in part because, unlike the negative regulators, these are not consistently inactivated in cancers. Indeed, the discovery of HCF-1 did not result from studies of cancer but rather of HSV infection. Even though more difficult to decipher, understanding the mechanism of activation of an oncoprotein and identifying its co-regulators are important for cancer therapy because the pharmacological intervention against these proteins that are still active in cancers may be key to cancer treatment.
Materials and methods
Cell culture, transfection, and generation of cell lines
U2OS, MCF7, and IMR-90Tert cell lines were used. Cells were maintained in Dulbecco's modified Eagle medium (DMEM; GIBCO) medium with 10% fetal bovine serum (FBS) and, for IMR-90Tert cells, 0.1 mM of non-essential amino acids. Cells in 10-cm plates were transfected with 8 μg of empty vector, E2F1 or E2F1 mutant plasmids using Lipofectamine (Invitrogen; or Lipofectamine LTX for MCF7 cells). After every transfection, protein synthesis was monitored by immunoblot analysis. Nuclear extracts were prepared from transfected U2OS cells and immunoprecipitations were performed using anti-HCF-1 sera as described (Tyagi et al, 2007).
Inducible cell lines were established in U2OS cells by transfection with pBabeHAER–E2F1, pBabeHAER–E2F1HBMmut, or pCMVHAER–E2F1. Cells were selected in DMEM medium with 10% FBS, supplemented with Puromycin (0.4 μg/ml, for pBabe) or G418 (500 μg/ml, for pCMV). The cell lines were induced by addition of 4-OHT (Sigma) to a final concentration of 1 μM. For induction of DNA damage using adriamycin, cells were treated with 2 μg/ml (or 5 μg/ml) adriamycin (Doxorubicin, Sigma) for the indicated times.
Apoptotic cell analysis by flow cytometry
At indicated time points, floating and adherent cells were harvested, washed in PBS and fixed in 70% methanol at 4°C overnight. The fixed cells were washed in PBS, and incubated with RNAse (50 mg/ml) and Propidium iodide (50 ng/ml) for 60 min at 37°C. The flow cytometry profiles were acquired on LSRII Becton Dickinson flow cytometer using BD FACSDiva Software.
Immunoblot analysis
Immunoblots with E2F1 and β-actin antisera were performed for analysis with an Odyssey Infrared Imager (LI-COR) as described (Tyagi et al, 2007).
TUNEL assay
Transfected cells were stained using in situ cell death detection kit (Roche cat no. 11684 795 910) according to the manufacturer's protocol.
siRNA transfections
Human HCF-1, control siRNAs (Julien and Herr, 2003), human WDR5 siRNA SMART pool consisting of four siRNA duplexes (Wysocka et al, 2005) and human E2F1 siRNA duplexes targeted against E2F1 mRNA sequence AAGCUUUAAUGGAGCGUUA (nucleotides 1997–2015 in 3′UTR) were purchased from Dharmacon. Double serial transfection was performed for all siRNAs as described before (Julien and Herr, 2003). Cells were harvested 48 or 72 h after the first transfection and either subjected to RNA preparation, or fixed for propidium iodide staining, immunostaining, or BrdU incorporation assay. BrdU incorporation assay was performed on E2F1 siRNA-treated cells as described (Julien and Herr, 2003).
Immunofluorescence
At indicated time points, cells were fixed in 1% paraformaldehyde for 15 min followed by extraction on ice for 20 min in 0.2% TritonX-100/PBS as described (Schultz et al, 2000). Incubations with primary antibody against 53BP1 (53BP1,1:50) and RPA 70 (RPA 70-9, 1:100) were conducted at room temperature for 60 min. Images were acquired on a Confocal Microscope Zeiss LSM 510 Meta (INVERTED) Scanhead using an EC Plan-NEOFLUAR 40 × /1,30 Oil or Plan-APOCHROMAT 63 × /1,40 Oil objective and the Zeiss LSM software.
ChIP and reverse-transcriptase PCR
ChIP, antibodies used and reverse-transcriptase PCR have been described earlier (Tyagi et al, 2007). Real-time PCR of ChIP DNAs was performed in triplicate using a SYBER Green quantitative PCR kit (Applied Biosystem) and a Rotor-geneTM RG300A Sequence Detector (Corbett Research) under conditions standardized for each primer set described in Supplementary data. PCR quantification was done using Delta Relative CT Quantification, where the values are calculated relative to input as follows: Delta CT=CT (input)−CT (sample); Relative Unit=2Delta CT. 0.3% of total ChIP input DNA was used in the input amplification. Representative results are shown in the figures.
Supplementary Material
Supplementary Data
Review Process File
Acknowledgments
We thank T Halazonetis for the 53BP1 antibody and MCF7 cell line; B Stillman for the RPA antibodies; K Helin for the pCMVER–E2F1 plasmid; J Lees for the pCMVHA-E2F1 plasmid; P L'Hôte for assistance with cell culture; F Lammers for technical support; T Halazonetis and N Hernandez for suggestions and critical reading of the manuscript. These studies were supported by the Swiss National Science Foundation, Oncosuisse, and the University of Lausanne. ST was supported by an EMBO long-term fellowship.
References
- Bartek J, Bartkova J, Lukas J (2007) DNA damage signalling guards against activated oncogenes and tumour progression. Oncogene 26: 7773–7779 [DOI] [PubMed] [Google Scholar]
- Bartkova J, Horejsí Z, Koed K, Krämer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, Ørntoft T, Lukas J, Bartek J (2005) DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 434: 864–870 [DOI] [PubMed] [Google Scholar]
- Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, Vassiliou LV, Kolettas E, Niforou K, Zoumpourlis VC, Takaoka M, Nakagawa H, Tort F, Fugger K, Johansson F, Sehested M, Andersen CL, Dyrskjot L, Ørntoft T, Lukas J et al. (2006) Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature 444: 633–637 [DOI] [PubMed] [Google Scholar]
- Bates S, Phillips AC, Clark PA, Stott F, Peters G, Ludwig RL, Vousden KH (1998) p14ARF links the tumour suppressors RB and p53. Nature 395: 124–125 [DOI] [PubMed] [Google Scholar]
- Blais A, Dynlacht BD (2007) E2F-associated chromatin modifiers and cell cycle control. Curr Opin Cell Biol 19: 658–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coqueret O (2003) New roles for p21 and p27 cell-cycle inhibitors: a function for each cell compartment? Trends Cell Biol 13: 65–70 [DOI] [PubMed] [Google Scholar]
- Dejosez M, Krumenacker JS, Zitur LJ, Passeri M, Chu LF, Songyang Z, Thomson JA, Zwaka TP (2008) Ronin is essential for embryogenesis and the pluripotency of mouse embryonic stem cells. Cell 133: 1162–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, Schurra C, Garre' M, Nuciforo PG, Bensimon A, Maestro R, Pelicci PG, d'Adda di Fagagna F (2006) Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature 444: 638–642 [DOI] [PubMed] [Google Scholar]
- Dugan KA, Wood MA, Cole MD (2002) TIP49 but not TRRAP, modulates c-Myc and E2F1 dependent apoptosis. Oncogene 29: 5835–5843 [DOI] [PubMed] [Google Scholar]
- El Messaoudi S, Fabbrizio E, Rodriguez C, Chuchana P, Fauquier L, Cheng D, Theillet C, Vandel L, Bedford MT, Sardet C (2006) Coactivator associated arginine methyltransferase 1 (CARM1) is a positive regulator of the Cyclin E1 gene. Proc Natl Acad Sci USA 103: 13351–13356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frame FM, Rogoff HA, Pickering MT, Cress WD, Kowalik TF (2006) E2F1 induces MRN foci formation and a cell cycle checkpoint response in human fibroblasts. Oncogene 25: 3258–3266 [DOI] [PubMed] [Google Scholar]
- Glinsky GV, Berezovska O, Glinskii AB (2005) Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. J Clin Invest 115: 1503–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto H, Motomura S, Wilson AC, Freiman RN, Nakabeppu Y, Fukushima K, Fujishima M, Herr W, Nishimoto T (1997) A single-point mutation in HCF causes temperature-sensitive cell-cycle arrest and disrupts VP16 function. Genes Dev 11: 726–737 [DOI] [PubMed] [Google Scholar]
- Halazonetis TD, Gorgoulis VG, Bartek J (2008) An oncogene-induced DNA damage model for cancer development. Science 319: 1352–1355 [DOI] [PubMed] [Google Scholar]
- Hallstrom TC, Nevins JR (2006) Jab1 is a specificity factor for E2F1-induced apoptosis. Genes Dev 20: 613–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ (1993) The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75: 805–816 [DOI] [PubMed] [Google Scholar]
- Hess JL (2004) MLL: a histone methyltransferase disrupted in leukemia. Trends Mol Med 10: 500–507 [DOI] [PubMed] [Google Scholar]
- Hiyama H, Iavarone A, Reeves SA (1998) Regulation of the cdk inhibitor p21 gene during cell cycle progression is under the control of the transcription factor E2F. Oncogene 16: 1513–1523 [DOI] [PubMed] [Google Scholar]
- Hsieh JK, Fredersdorf S, Kouzarides T, Martin K, Lu X (1997) E2F1-induced apoptosis requires DNA binding but not transactivation and is inhibited by the retinoblastoma protein through direct interaction. Genes Dev 11: 1840–1852 [DOI] [PubMed] [Google Scholar]
- Hsu SI, Yang CM, Sim KG, Hentschel DM, O'Leary E, Bonventre JV (2001) TRIP-Br: a novel family of PHD zinc finger- and bromodomain-interacting proteins that regulate the transcriptional activity of E2F-1/DP-1. EMBO J 20: 2273–2285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin M, Marin MC, Phillips AC, Seelan RS, Smith DI, Liu W, Flores ER, Tsai KY, Jacks T, Vousden KH, Kaelin WG Jr (2000) Role for the p53 homologue p73 in E2F-1-induced apoptosis. Nature 407: 645–648 [DOI] [PubMed] [Google Scholar]
- Julien E, Herr W (2003) Proteolytic processing is necessary to separate and ensure proper cell growth and cytokinesis functions of HCF-1. EMBO J 22: 2360–2369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knez J, Piluso D, Bilan P, Capone JP (2006) Host cell factor-1 and E2F4 interact via multiple determinants in each protein. Mol Cell Biochem 288: 79–90 [DOI] [PubMed] [Google Scholar]
- Lang SE, McMahon SB, Cole MD, Hearing P (2001) E2F transcriptional activation requires TRRAP and GCN5 cofactors. J Biol Chem 276: 32627–32634 [DOI] [PubMed] [Google Scholar]
- Lazzerini Denchi E, Helin K (2005) E2F1 is crucial for E2F-dependent apoptosis. EMBO Rep 6: 661–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louie MC, Zou JX, Rabinovich A, Chen HW (2004) ACTR/AIB1 functions as an E2F1 coactivator to promote breast cancer cell proliferation and antiestrogen resistance. Mol Cell Biol 24: 5157–5171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe ED, Tews I, Cheng KY, Brown NR, Gul S, Noble ME, Gamblin SJ, Johnson LN (2002) Specificity determinants of recruitment peptides bound to phospho-CDK2/cyclin A. Biochemistry 41: 15625–15634 [DOI] [PubMed] [Google Scholar]
- Moroni MC, Hickman ES, Lazzerini Denchi E, Caprara G, Colli E, Cecconi F, Müller H, Helin K (2001) Apaf-1 is a transcriptional target for E2F and p53. Nat Cell Biol 3: 552–558 [DOI] [PubMed] [Google Scholar]
- Pickering MT, Kowalik TF (2006) Rb inactivation leads to E2F1-mediated DNA double-strand break accumulation. Oncogene 25: 746–755 [DOI] [PubMed] [Google Scholar]
- Powers JT, Hong S, Mayhew CN, Rogers PM, Knudsen ES, Johnson DG (2004) E2F1 uses the ATM signaling pathway to induce p53 and Chk2 phosphorylation and apoptosis. Mol Cancer Res 2: 203–214 [PubMed] [Google Scholar]
- Radhakrishnan SK, Feliciano CS, Najmabadi F, Haegebarth A, Kandel ES, Tyner AL, Gartel AL (2004) Constitutive expression of E2F-1 leads to p21-dependent cell cycle arrest in S phase of the cell cycle. Oncogene 23: 4173–4176 [DOI] [PubMed] [Google Scholar]
- Rogoff HA, Pickering MT, Frame FM, Debatis ME, Sanchez Y, Jones S, Kowalik TF (2004) Apoptosis associated with deregulated E2F activity is dependent on E2F1 and Atm/Nbs1/Chk2. Mol Cell Biol 24: 2968–2977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz LB, Chehab NH, Malikzay A, Halazonetis TD (2000) p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J Cell Biol 151: 1381–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiewe T, Pützer BM (2000) Role of the p53-homologue p73 in E2F1-induced apoptosis. Nat Genet 26: 464–469 [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Rayman JB, Dynlacht BD (2000) Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev 14: 804–816 [PMC free article] [PubMed] [Google Scholar]
- Takeda S, Chen DY, Westergard TD, Fisher JK, Rubens JA, Sasagawa S, Kan JT, Korsmeyer SJ, Cheng EH, Hsieh JJ (2006) Proteolysis of MLL family proteins is essential for taspase1-orchestrated cell cycle progression. Genes Dev 20: 2397–2409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimarchi JM, Lees JA (2002) Sibling rivalry in the E2F family. Nat Rev Mol Cell Biol 3: 11–20 [DOI] [PubMed] [Google Scholar]
- Tyagi S, Chabes AL, Wysocka J, Herr W (2007) E2F activation of S phase promoters via association with HCF-1 and the MLL family of histone H3K4 methyltransferases. Mol Cell 27: 107–119 [DOI] [PubMed] [Google Scholar]
- Vigo E, Müller H, Prosperini E, Hateboer G, Cartwright P, Moroni MC, Helin K (1999) CDC25A phosphatase is a target of E2F and is required for efficient E2F-induced S phase. Mol Cell Biol 19: 6379–6395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J, Herr W (2003) The herpes simplex virus VP16-induced complex: the makings of a regulatory switch. Trends Biochem Sci 28: 294–304 [DOI] [PubMed] [Google Scholar]
- Wysocka J, Myers MP, Laherty CD, Eisenman RN, Herr W (2003) Human Sin3 deacetylase and trithorax-related Set1/Ash2 histone H3-K4 methyltransferase are tethered together selectively by the cell-proliferation factor HCF-1. Genes Dev 17: 896–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, Roeder RG, Brivanlou AH, Allis CD (2005) WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell 121: 859–872 [DOI] [PubMed] [Google Scholar]
- Yang SZ, Lin FT, Lin WC (2008) MCPH1/BRIT1 cooperates with E2F1 in the activation of checkpoint, DNA repair and apoptosis. EMBO Rep 9: 907–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama A, Wang Z, Wysocka J, Sanyal M, Aufiero DJ, Kitabayashi I, Herr W, Cleary ML (2004) Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol Cell Biol 24: 5639–5649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Elledge SJ (2003) Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300: 1542–1548 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data
Review Process File