Abstract
The CLC ‘Cl− channel' family consists of both Cl−/H+ antiporters and Cl− channels. Although CLC channels can undergo large, conformational changes involving cooperativity between the two protein subunits, it has been hypothesized that conformational changes in the antiporters may be limited to small movements localized near the Cl− permeation pathway. However, to date few studies have directly addressed this issue, and therefore little is known about the molecular movements that underlie CLC-mediated antiport. The crystal structure of the Escherichia coli antiporter ClC-ec1 provides an invaluable molecular framework, but this static picture alone cannot depict the protein movements that must occur during ion transport. In this study we use fluorine nuclear magnetic resonance (NMR) to monitor substrate-induced conformational changes in ClC-ec1. Using mutational analysis, we show that substrate-dependent 19F spectral changes reflect functionally relevant protein movement occurring at the ClC-ec1 dimer interface. Our results show that conformational change in CLC antiporters is not restricted to the Cl− permeation pathway and show the usefulness of 19F NMR for studying conformational changes in membrane proteins of known structure.
Keywords: channel, Cl−, NMR, transporter
Introduction
Members of the CLC family have indispensible ion transport functions in organisms ranging from bacteria to mammals (reviewed in Jentsch, 2008). Despite its name, the CLC ‘Cl− channel' family includes Cl−/H+ anitporters as well as Cl− channels. This scenario provides a unique opportunity to investigate the relationship between these two ostensibly distinct ion transport mechanisms (DeFelice, 2004; Miller, 2006; Lisal and Maduke, 2008). Channels need only change conformation to remove the physical constraints (the ‘gates') blocking ions from entering and exiting the protein's pore. Electrodiffusion then occurs rapidly, allowing 106–108 ions to move across the membrane every second (Hille, 2001). Transporters, however, work by coordinating a series of protein conformational changes with every round of ion transport. The necessity for coordinated conformational change places an upper limit on the speed of transport, with the fastest known transporter topping out at 12 000 ions/s (Brahm, 1977). A hypothetical model for how this process could occur in the CLC antiporters is presented in Figure 1.
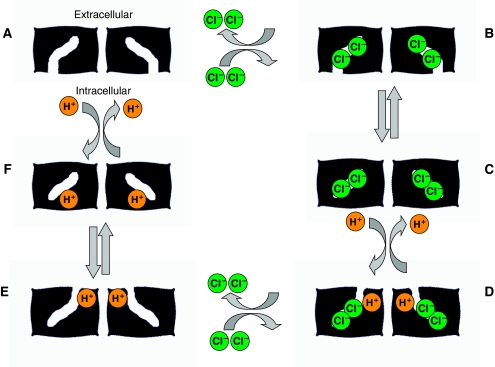
Figure 1.
Basic model for CLC-mediated Cl−/H+ antiport. This hypothetical model illustrates the essential features of antiport, which occurs through coupling of ion binding and unbinding to protein conformational changes. CLC proteins are homodimers, with each subunit containing its own Cl− and H+ permeation pathways. In this model (moving clockwise), Cl− ions enter the permeation pathway of an empty CLC homodimer (state A) from the intracellular side of the membrane to form a Cl−-filled state open to the intracellular side of the membrane (state B). A conformational change in the protein leads to closing of the Cl− pathway from the intracellular face and generates the Cl−-occluded state seen in the crystal structure (state C). Subsequent binding of H+ from the extracelluar side of the membrane (forming state D) then leads to another conformational change that opens the extracellular side of the Cl− permeation pathway and allows extracellular release of the Cl− ions (state E). Finally the protons transit through their permeation pathway to the intracellular face (state F) leading to their release and return to the empty state with the Cl− permeation pathway open to the intracellular face. For simplicity, this diagram depicts conformational changes occurring simultaneously in the two subunits; however, it is likely that both subunits may also operate asynchronously (Zdebik et al, 2008). In this particular model, coupling arises from the requirement that protons must bind to allow the Cl−-free transporter to reorient; however, the order of the ion-binding steps is not yet known.
The two CLC subtypes share a similar architecture, with the functional unit consisting of a homodimer in which each subunit possesses its own ion permeation pathway. At the functional level, the conformational changes involved in ion transport by the CLC channels have been more extensively studied than those of the CLC antiporters. The channels are regulated by two different gating mechanisms. The ‘fast gate' mechanism occurs because of the independent opening and closing of each of the two individual pores within the dimer (Miller, 1982; Accardi and Pusch, 2000) whereas the ‘slow-gate' process involves simultaneous opening and closing of the two pores, and entails large cooperative conformational changes between the subunits (Miller, 1982; Bykova et al, 2006). Although the relationship between gating in CLC channels and transport in CLC antiporters is not well understood, both processes share a common bond in that they are strongly modulated by changes in Cl− and H+ concentration (Pusch, 2004; Zifarelli et al, 2008). Recent work has shown that in the case of the ClC-0 channel, the slow-gating process involves H+ permeation through the protein (Lisal and Maduke, 2008). This suggests that the gating mechanisms for CLC channels are a vestige of the antiport mechanism used by their ancestors, which could occur if mutation caused one of the conformations involved in the antiport cycle to become leaky for Cl−.
Just as CLC channels can act as broken antiporters, CLC antiporters can also be ‘broken' and made to display channel-like properties through point mutations made at constriction points in the Cl− permeation pathway (Jayaram et al, 2008). These critical residues were identified using the X-ray crystallographic structure of the Escherichia coli Cl−/H+ antiporter ClC-ec1 (Dutzler et al, 2003), which is currently the only CLC for which an atomic resolution structure is known. On the exterior side of ClC-ec1, a conserved glutamate residue (E148) occludes Cl− from exiting the protein (Figure 2A). Mutation of this glutamate to alanine or glutamine abolishes H+ transport while retaining Cl− transport, resulting in a completely uncoupled protein in which Cl− movement occurs down its electrochemical gradient (Accardi and Miller, 2004). On the intracellular side of the protein, the Cl− is physically occluded from exiting by a tyrosine residue (Y445, shown in Figure 2A). Decreasing the side chain volume at this site to serine partially disrupts H+ transport, resulting in partial uncoupling (a decrease in the Cl−/H+ transport stoichiometry) (Walden et al, 2007). Alone, the Y445S or E148A mutations result in little to moderate effects on turnover rate. However, when combined, the E148A/Y445S ‘doubly ungated' mutant causes the Cl− turnover rate to skyrocket to 70 000 ions/s per dimer—almost 20 times faster than wild-type ClC-ec1 (Jayaram et al, 2008). Although this number falls shy of what is expected for a conventional Cl− channel, it is close to six times faster than the fastest known transporter (Brahm, 1977). Thus, it is highly likely that the doubly ungated ClC-ec1 mutant behaves similarly to an electrodiffusive channel, facilitating Cl− movement without relying on coordinated conformational changes.
Figure 2.
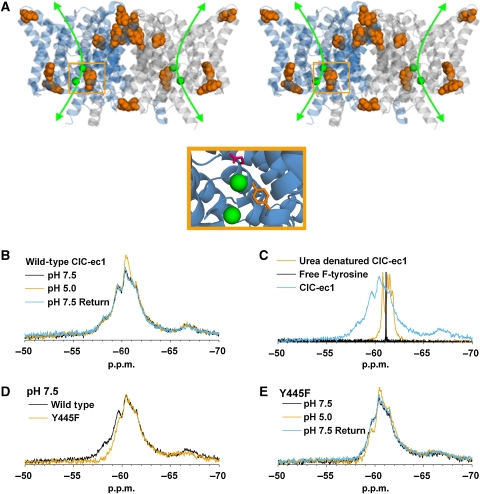
19F NMR detects H+-dependent conformational changes in ClC-ec1. (A) ClC-ec1 stereoview illustrating the two identical subunits (blue and grey). The bound Cl− ions together with presumed Cl− permeation pathway are shown in green, with the expanded view below illustrating coordination of one of these ions by Y445 (orange) and E148 (pink). The protein's tyrosine residues (nine per subunit) are shown space filled and highlighted in orange. (B) 19F spectra of wild-type ClC-ec1 labelled with F-tyrosine. The appearance of new intensities at −60.4 and −61.4 p.p.m. when the pH is shifted from 7.5 to 5.0 indicates that at least one of the nine F-tyr residues in ClC-ec1 experiences a change in its chemical environment. The change in pH is experienced by both the extracellular and intracellular faces of protein as these experiments are performed using detergent-solubilized ClC-ec1. (C) Denaturation of ClC-ec1 with 6 M urea results in a collapsed spectrum centred at the same chemical shift as free F-tyrosine. (D) The Y445F mutation removes fluorine labelling at the Cl−-coordinating Y445 position. Comparison of wild type and Y445F spectra at pH 7.5 highlights the decrease of intensity in the −57.0 to −60.0 p.p.m. range in the Y445F mutant. (E) The Y445F mutation does not abolish H+-dependent spectral changes; therefore, at least one other tyrosine residue is experiencing a change in chemical environment.
Although noteworthy, these functional observations do not directly address the nature of conformational change in the channel-like ClC-ec1 mutant or the more general, related question of what types of protein movements are involved in the normal CLC antiport cycle. A recent crosslinking study on ClC-ec1 has shown that unlike slow gating in the CLC channels, antiport does not require a large cooperative movement between the subunits as is observed in the slow-gating process of ClC-0 (Nguitragool and Miller, 2007). But how can we observe other, more subtle movements? Although a static crystal structure cannot on its own give dynamic information about conformational changes, it can provide a basis on which to model conformational changes observed by other means. The crystal structure of ClC-ec1 depicts an ‘occluded' state, in which a bound Cl− ion does not have access to either the extracellular or intracellular solutions. In order for transport to occur, additional protein conformations must exist, which allow the transporter to pick up and subsequently deliver Cl− from one side of the membrane to the other (Matulef and Maduke, 2007). However, despite the use of crystallization conditions spanning a wide range of Cl− and H+ concentrations, to date only the occluded conformation has been observed in the wild-type protein (Dutzler et al, 2002, 2003; Accardi et al, 2005, 2006; Lobet and Dutzler, 2006).
This problem is not unique to the CLC proteins. Although the growing database of high-resolution membrane protein structures provides molecular details fundamental to understanding membrane protein function (White, 2004), it also highlights the necessity for complementary studies to assess protein conformational changes (Elvington and Maduke, 2008). Solution-state nuclear magnetic resonance (NMR) techniques allow proteins to be observed at ambient temperature, without the physical constraints present in a crystal, and have the potential to provide tremendous insight into protein dynamics. Although there have been great advances in determining structures of membrane proteins by solution NMR (Hwang and Kay, 2005; Poget et al, 2007; Wang, 2008), the necessity of solubilizing the protein in micelles or bicelles greatly increases the effective size of the complex and currently limits the use of NMR for full structure determination of large integral membrane proteins. However, for cases such as ClC-ec1 in which the X-ray crystallographic structure of one conformation is already known, NMR has the potential to build on this knowledge by identifying regions of conformational change. Once these regions are identified, NMR can be used to study the dynamics of these changes, thus taking us well beyond the level of crystallography's frozen snapshots (Henzler-Wildman and Kern, 2007).
In this study, we use solution-state 19F NMR to identify regions of conformational change in ClC-ec1. The fluorine nucleus possesses several useful properties that make it ideally suited to such studies (reviewed in Gerig, 1994; Danielson and Falke, 1996; Gakh et al, 2000). First, the 19F chemical shift is highly sensitive to its chemical environment and thus is an excellent reporter of conformational change (Luck and Falke, 1991; Danielson et al, 1994; Rozovsky et al, 2001). Second, fluorine (1.35 Å radius) is nearly isosteric to hydrogen (1.2 Å), so the labelling itself does not tend to perturb protein structure (Danielson and Falke, 1996). Finally, the NMR sensitivity of 19F is high, and there is no signal contribution from any natural source (O'Hagan and Harper, 1999).
Here, we exploit these properties of the fluorine nucleus to probe conformational change in ClC-ec1. To ensure proper antiport function, these movements must be coupled to binding and unbinding of substrate ions (as illustrated in the model in Figure 1). On manipulating either [H+] or [Cl−] to drive these conformational changes, we observe concurrent changes in the 19F-NMR spectrum of fluorotyrosine-labelled ClC-ec1. Using site-directed mutagenesis, we have determined that some of this spectral change is due to structural changes occurring in regions outside the Cl− permeation pathway, near the subunit interface. These substrate-dependent spectral changes are absent in a high-turnover ClC-ec1 mutant acting in ‘channel-mode'. However, spectral changes persist with the addition of uncoupling mutations that degrade the Cl−/H+ transport stoichiometry but retain a normal Cl− turnover rate. These results provide our first glimpse of the conformational changes that govern the transport cycle in the CLC antiporters.
Results
19F NMR detects H+-dependent conformational changes in ClC-ec1
For an antiporter such as ClC-ec1, conformational change is driven by the coordinated binding and unbinding of substrate ions. Hence, we would expect that a change in substrate concentration would lead to a change in the ensemble of protein conformations in solution. We first examined the effect of [H+] on ClC-ec1 conformation under conditions where Cl− is present at saturating levels (Lobet and Dutzler, 2006)—a manipulation that should limit the number of significantly populated conformations. In terms of the model shown in Figure 1, increasing the H+ concentration at saturating Cl− would shift the population from states B and C to state D. As these different conformations are adopted, the changing chemical environment surrounding fluorinated residues will result in a change in the protein's 19F-NMR spectrum.
We identified tyrosine as an attractive candidate for fluorine labelling in ClC-ec1 because of the location of these residues in regions of interest. As illustrated in Figure 2A, ClC-ec1's nine tyrosine residues are distributed throughout the protein, thus providing probes at key regions near the subunit interface and lining the Cl− permeation pathway. Fluorotyrosine (Supplementary Figure 1) is readily incorporated into proteins biosynthetically by excluding the unlabelled amino acid from the growth media while supplementing with the fluorinated analog (Danielson and Falke, 1996). Following labelling in this manner, F-tyr-ClC-ec1 was found to be fully functional in assays for Cl− and H+ flux (Supplementary Figure 2).
As shown in Figure 2B, shifting the pH of the F-tyr-ClC-ec1 sample from 7.5 to 5.0 causes the appearance of new intensities at −60.4 and −61.4 p.p.m. in the 1D 19F-NMR spectrum. The pH shift used here represents a functionally significant change from an environment supporting negligible transporter activity (pH 7.5) to one promoting high activity (pH 5.0) (Accardi et al, 2004). This result indicates that at least one of the nine F-tyr residues in ClC-ec1 is subject to an H+-dependent change in chemical environment. The observed H+-dependent spectral changes are not the result of irreversible aggregation of the protein, as spectral changes seen at low pH are reversible on return to neutral pH. Such reversibility was confirmed in all experiments presented in this study (data not shown).
The spectra of F-tyr-ClC-ec1 presented in Figure 2 show significant chemical shift dispersion between −55 and −70 p.p.m. range, indicating the presence of a folded protein in which F-tyrosine residues are experiencing different chemical environments. This is in contrast to the signal observed for free F-tyrosine, which exhibits a sharp H+-insensitive peak at −61.1 p.p.m. (Figure 2C). Denaturation of ClC-ec1 with 6 M urea results in a collapsed spectrum centred at the same chemical shift as free F-tyrosine. This spectral collapse indicates unfolding of the protein and elimination of the signal dispersion observed for the intact protein. Although the pH shift used for these experiments represents a functionally significant change, we must consider the possibility that changes in [H+] could induce artifactual changes that are not necessarily relevant to transporter function. However, the absence of H+-dependent spectral changes in one of the mutants used in this study (see below) argues strongly against this possibility.
The spectral changes observed for F-tyr-ClC-ec1 in Figure 2B indicate that we are successfully monitoring changes in the chemical environment surrounding tyrosine residues in ClC-ec1. As NMR measures an ensemble of protein conformations over time, these spectral changes may arise from a combination of several events. A change in structure of one or more of the kinetically stable conformational states or a shift in equilibrium distribution between these states would be observed as a change in chemical shift. Structural changes may result from both local perturbations in regions surrounding the fluorine label as well as from larger alterations to the protein's tertiary structure. Local changes may arise from many sources, including protonation of nearby residues, binding of a nearby Cl− ion, or changes in mobility allowing tyrosine ring flipping. Both local and gross changes to protein structure have the potential to impact protein function. In addition to changes in chemical shift, some of the spectral changes we see may arise from changes in line width because of changes in residue mobility or the exchange rate between conformational species. Although our resolution is not sufficient to determine line widths and thus quantify the relative contribution of this effect, the conclusions drawn in our studies do not require such a distinction. Regardless of the underlying reason, a residue that contributes to the observed spectral changes participates in conformational change in some capacity. Thus, we define all of these events (changes in exchange rate, equilibrium distribution, local and gross structural changes) as reflecting ‘conformational change' in our studies.
The Cl−-coordinating residue Y445 is an obvious candidate for participation in conformational changes. As depicted in the crystal structure, Y445 occludes a bound Cl− ion from exiting the protein (Figure 2A). Therefore, Y445 must move to allow Cl− transport. To determine the contribution of F-Y445 to the ClC-ec1 spectrum, we mutated this residue to phenylalanine, thus removing the corresponding resonance from the F-tyr-ClC-ec1 19F-NMR spectrum. Importantly, this mutation does not seriously impair ClC-ec1 function (Supplementary Figure 2; Walden et al, 2007). The Y445F mutation results in a decrease of intensity in the −57.0 to −60.0 p.p.m. range as compared with wild-type ClC-ec1, but maintains a qualitatively similar H+ response (Figure 2D and E). The persistence of these H+-dependent spectral changes in the Y445F mutant indicates that the F-Y445 signal alone cannot account for the changes observed in wild-type F-tyr-ClC-ec1. As Y445 is the only tyrosine residue present in the Cl− permeation pathway, H+-dependent structural changes must also be occurring elsewhere in the protein.
H+-dependent conformational changes occur at the ClC-ec1 dimer interface
As there is no comprehensive method for theoretically predicting 19F chemical shift based on protein environment, peak assignments are usually performed using site-directed mutagenesis (Frieden et al, 2004). Owing to the large overlap in signal from the multiple F-tyrosine residues in our sample, we sought to simplify the spectrum by eliminating multiple tyrosine residues simultaneously. We reasoned that the F-tyrosine residues located on the surface of ClC-ec1 likely experience greater mobility than those buried in the protein's interior. Such mobile residues would yield strong peaks that could potentially mask the lower intensity, broader peaks produced by the less mobile interior tyrosine residues. To better observe the resonance of these interior residues, we engineered a mutant ClC-ec1 termed ‘Buried Only' in which those tyrosine residues with a solvent accessible surface area of >10% were mutated to phenylalanine. Accessibility was determined using ASAVIEW (Ahmad et al, 2004) from the crystal structure of ClC-ec1 (pdb 1OTS). As highlighted in Figure 3A, this leaves a total of five tyrosine residues per monomer intact and subject to fluorine labelling. The ‘Buried Only' mutant maintains wild-type function, as shown with Cl− and H+ flux assays (Supplementary Figure 2). A summary of all proteins and functional consequences of mutations used in this study is presented in Table I.
Figure 3.
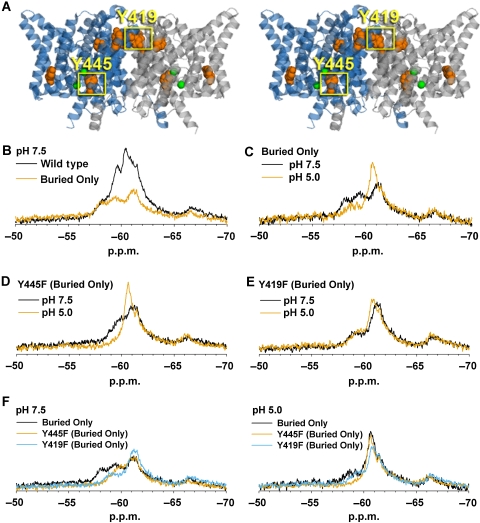
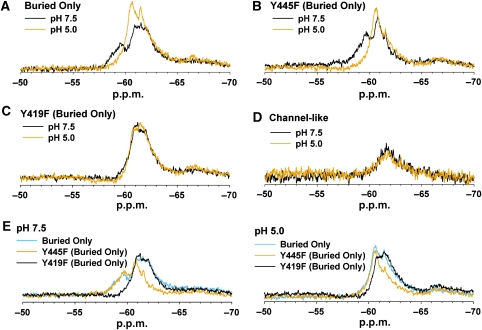
ClC-ec1 undergoes H+-dependent conformational change at the subunit interface. (A) Stereoview of ClC-ec1 illustrating the location of the tyrosine residues present in the ‘Buried Only' mutant. This mutant contains only those five tyrosine residues interior to the protein with a solvent accessible surface area <10%. All other tyrosine residues have been mutated to phenylalanine, and thus are not fluorine labelled in all subsequent experiments. The Cl−-coordinating residue Y445 and subunit interface residue Y419 are highlighted. (B) Compared with wild-type ClC-ec1 at pH 7.5, the Buried Only mutant produces more discrete, (albeit broad) peaks, with much of the signal intensity in the −59 to −62 range being eliminated. (C) H+-dependent spectral changes are retained in the Buried Only mutant. (D) Removal of fluorine labelling at the Y445 position in the Buried Only background does not eliminate H+-dependent spectral changes. (E) Removal of labelling at the Y419 position eliminates much of the H+-dependent response observed in the Buried Only mutant. (F) Comparison of Buried Only mutant spectra at pH 7.5 and 5.0. Mutation of either Y445 or Y419 to phenylalanine results in loss of signal in the 57–58 p.p.m. region at pH 7.5. Hence, mutation of one of these residues influences the chemical shift of the other.
Table 1.
ClC-ec1 proteins used in this study
| Protein name | Mutation(s) | Functional effect(s) |
|---|---|---|
| Wild-type ClC-ec1 | None | NA |
| Y445F | Y445F | Slightly reduced Cl− transport rate (Walden et al, 2007) |
| Buried Only | Y75F, Y100F, Y210F, Y382F | None |
| Y445F Buried Only | Y445F (Buried Only background) | Slightly reduced Cl− transport rate (Walden et al, 2007) |
| Y419F Buried Only | Y419F (Buried Only background) | None |
| Y419F/Y445F Buried Only | Y419F, Y445F (Buried Only background) | Slightly reduced Cl− transport rate (Walden et al, 2007) |
| Channel like | E148A, Y445S (Buried Only background) | High Cl− transport rate (Jayaram et al, 2008) |
| No H+ transport (Jayaram et al, 2008) | ||
| Uncoupled Transporter #1 | E148A, Y445F (Buried Only background) | No H+ transport |
| Uncoupled Transporter #2 | Y445S (Buried Only background) | Low Cl−/H+ coupling ratio (Walden et al, 2007) |
| Protein names (as given in figures), associated mutations, and functional effects of these mutations are presented. Function was assayed as reported in Supplementary Figure 2 and supported by earlier work as cited. |
As compared with wild-type ClC-ec1 at pH 7.5, the Buried Only mutant produces more discrete (albeit broad) peaks, with much of the signal intensity in the −59 to −62 p.p.m. range being eliminated (Figure 3B). When the pH of the Buried Only sample is shifted to 5.0, the peaks at −58.2 and −59.5 decrease in intensity, and a higher intensity emerges at −60.4 p.p.m. (Figure 3C). The emergence of this higher intensity at pH 5.0 is qualitatively similar to what is observed in the wild-type ClC-ec1 spectrum (Figure 2B). The retention of H+-dependent spectral change in the Buried Only mutant indicates that structural changes are occurring within the interior of the protein, and are not simply because of changes occurring on the protein surface.
Removal of labelling at Y445 in the Buried Only background results in a decrease in signal in the −57 to −60 p.p.m. range, but does not abolish [H+] responsiveness (Figure 3D). This result is qualitatively similar to that observed for the Y445F mutation in the wild-type background (Figure 2D and E). The persistence of H+-dependent spectral change in the Y445F Buried Only mutant allows us to further localize the structural changes to the four internal tyrosine residues that remain labelled in this protein. One of these residues is Y419, which lies at the ClC-ec1 subunit interface (Figure 3A). Mutating this residue to phenylalanine in the Buried Only background eliminates much of the observed H+-dependent spectral change (Figure 3E), thus indicating that the ClC-ec1 dimer interface is subject to H+-dependent structural changes. It is of note that because mutation of either Y445 or Y419 to phenylalanine results in loss of signal in the −57.0 to −60.0 p.p.m. region at pH 7.5, mutation of one of these residues must influence the chemical shift of another residue (Figure 3F). Although this interdependency complicates the assignment of specific resonance to a particular residue, it does not diminish the conclusions drawn here: protein movement is occurring outside the Cl− permeation pathway, and the subunit interface participates in this movement. Although the remaining buried tyrosine residues were not examined, the subtlety of changes persistent in the Y419F Buried Only mutant indicates that spectral contribution from these residues will be relatively small. Therefore, assessment of their contribution will require a higher resolution technique in which resonance from single residues can be identified unambiguously.
The ClC-ec1 ‘Channel-like' mutant is not subject to H+-dependent changes associated with ClC-ec1 transporters
The E148 residue of ClC-ec1 lies in the Cl− permeation pathway and acts as the external Cl− gatekeeper (Dutzler et al, 2003) (Figure 2A). Mutation of this residue to alanine uncouples Cl−/H+ antiport, resulting in a protein that can transport Cl− down its electrochemical gradient without transporting H+ in the opposite direction (Accardi and Miller, 2004). It was hypothesized that as this mutant allows uncoupled Cl− movement to occur, it may act as a Cl− channel. Contrary to this proposition, the Cl− turnover rate of the E148A mutant is no higher than wild type. However, when the E148 mutation is combined with mutation of Y445 to a smaller residue, the turnover rate of the double mutant jumps to a level nearing that of a bona fide channel (Jayaram et al, 2008). This led to the conclusion that the mechanism of Cl− flux through this double mutant involves channel-like electrodiffusion rather than the alternating-access cycle of conformational changes that must occur in the wild-type protein (Jayaram et al, 2008). To test the behaviour of this mutant using our 19F-NMR assay, we generated it in the Buried Only background and verified the high-turnover rate for the labelled protein (Supplementary Figure 2). In this uncoupled ‘Channel-like' mutant (E148A/Y445S Buried Only), the H+-dependence of the protein's 19F-NMR spectrum is eliminated (Figure 4A). The absence of H+-dependent spectral change indicates that this mutant is not subject to the same conformational changes as wild-type ClC-ec1 and thus argues that 19F spectral changes reflect functionally relevant protein movement.
Figure 4.
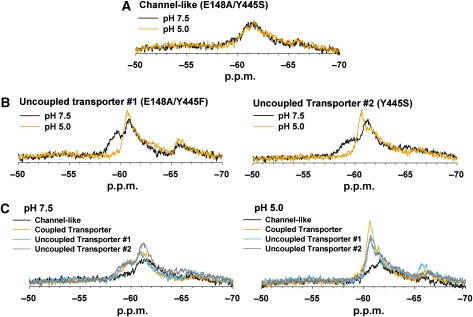
The ClC-ec1 Channel-like mutant is not subject to H+-dependent conformational changes associated with ClC-ec1 transporters. (A) The Channel-like mutant (E148A/Y445S, Buried Only) exhibits no H+-dependent spectral change. (B) In contrast, spectra from both Uncoupled Transporter mutants (E148A/Y445F Buried Only and Y445S Buried Only) change with [H+]. (C) Comparison of the Channel-like mutant to Uncoupled and Coupled Transporter proteins. The Coupled Transporter protein corresponds to Y445F Buried Only (data presented in Figure 3D). All mutants in this panel are F-tyr labelled at the same four positions (mutation at Y445 eliminates labelling of this residue in all four constructs). The spectra of the ClC-ec1 Transporters are similar to one another at each pH and markedly different from the spectra of the Channel-like mutant.
We next sought to determine whether the lack of H+-dependent structural change in the Channel-like mutant was due to the increase in Cl− turnover rate or rather to individual contributions from the E148A and Y445S mutations. To address whether uncoupling through E148A impacts H+-dependent conformational changes, we examined this mutation in the Y445F Buried Only background. In this ‘Uncoupled Transporter #1' protein, F-tyr labelling occurs at the same four positions as its Channel-like counterpart, thus facilitating comparison between structural changes because of E148A uncoupling alone versus changes because of increased Cl− transport rate. The E148A/Y445F Buried Only mutant retains the uncoupling phenotype but has a turnover rate similar to that of wild-type ClC-ec1 (Supplementary Figure 2). As shown in Figure 4B (left panel), the Uncoupled Transporter #1 spectrum remains H+-dependent; therefore, uncoupling through E148A alone is not sufficient to abolish H+-dependent conformational changes.
Similar to the E148A mutation, the Y445S mutation alone results in a severely uncoupled transporter (Walden et al, 2007). In addition, it has been crystallographically observed that mutations at the Y445 position result in decreased Cl− occupancy at the ClC-ec1 central Cl−-binding site (Accardi et al, 2006). To determine whether these effects account for the abolition of H+-dependent spectral changes in the Channel-like mutant, we examined the Y445S mutation alone in the Buried Only background. As expected, the Y445S mutation reduces the protein's H+ transport ability without having much effect on the Cl− transport rate (Supplementary Figure 2). The Y445S Buried Only protein (Uncoupled Transporter #2) exhibits H+-dependent spectral changes similar to those observed for the E148A mutant (Figure 4B, right panel). This indicates the functional effects conferred by the Y445S mutation alone cannot account for the abolition of H+-dependent spectral changes observed in the Channel-like protein.
A comparison of the H+-dependent spectra of the Uncoupled Transporter proteins, the Channel-like protein, and the fully Coupled Transporter is shown in Figure 4C. These proteins are labelled at the same tyrosine positions, and differ only in the mutations present at the Y445 and E148 positions. The observation that the spectra of the Uncoupled Transporters and the Coupled Transporter are very similar indicates that the E148A and Y445 mutations alone do not cause a substantial change to protein structure or the exchange rate between conformations at either [H+]. This strongly suggests that—although Cl−/H+ coupling is impaired in these proteins—the conformational changes induced by H+ binding at high [Cl−] remain largely intact. In contrast, in the Channel-like mutant, much of the resonance is broadened at both pH 7.5 and 5.0. This indicates that either the structure of individual conformations, the steady-state distribution of conformations, and/or the rate of interconversion between conformations is altered in this protein. The lack of spectral H+ dependence of this Channel-like protein indicates that it is not subject to the same H+-induced conformational changes that occur in the coupled or uncoupled transporters.
H+-dependent conformational changes occur at the subunit interface in the absence of Cl−
The experiments described above were performed at saturating concentrations of Cl−, thus allowing us to examine H+-dependent conformational changes occurring in ClC-ec1 in the Cl−-bound form. Performing similar experiments under zero-Cl− conditions allows us to access additional conformations achieved by the Cl−-unloaded protein. In terms of the model presented in Figure 1, this would include states A, F, and E.
As seen in Figure 5A, when the pH of the zero-Cl− Buried Only sample is shifted to 5.0, the peak at −59.5 decreases in intensity and a higher intensity emerges at −60.4 p.p.m. The persistence of these H+-dependent spectral changes indicates that protonation confers ClC-ec1 conformational change independent of Cl− binding. The changes include regions outside the chloride permeation pathway, as the spectral change persists even when fluorine labelling is removed at the Cl−-coordinating residue Y445 (Figure 5B). Removal of labelling at the Y419 position abolishes much of the spectral change observed in the Buried Only protein, which indicates that the subunit interface participates in H+-dependent conformational change even in the absence of Cl− (Figure 5C). The functional significance of the observed spectral changes is supported by their absence in the ClC-ec1 Channel-like mutant (Figure 5D).
Figure 5.
H+-dependent conformational changes occur in ClC-ec1 in the absence of Cl−. All spectra are from protein samples prepared in buffer containing zero Cl−. (A) H+-dependent spectral changes are observed in the Buried Only mutant. (B) H+-dependent spectral changes persist with the removal of fluorine labelling at the Y445 position. (C) Removal of labelling at the Y419 position eliminates much of the H+-dependent response observed in the Buried Only mutant. (D) The Channel-like mutant (E148A/Y445S Buried Only) does not experience H+-dependent spectral changes. (E) Comparison between Buried Only, Y419F Buried Only, and Y445F Buried Only in zero Cl− highlights the location of Y419 and Y445 resonance within the Buried Only spectrum.
Unlike experiments performed under saturating Cl− conditions (Figure 3), at zero Cl−, mutation of either Y419 or Y445 results in loss of signal in distinct regions of the spectrum. Comparison between spectra from the Buried Only and Y445F Buried Only proteins illustrates that the broad peak between −60.5 and −65.0 p.p.m. disappears in the Y445F mutant and thus likely corresponds to Y445 (Figure 5E). Likewise, comparing spectra from the Buried Only and Y419F Buried Only proteins indicates that at pH 7.5, Y419 is responsible for the peak centred at −59.5 p.p.m. and that this peak shifts to −60.4 p.p.m. at pH 5.0.
19F NMR detects Cl− binding within the Cl− permeation pathway and Cl−-dependent conformational change outside this region
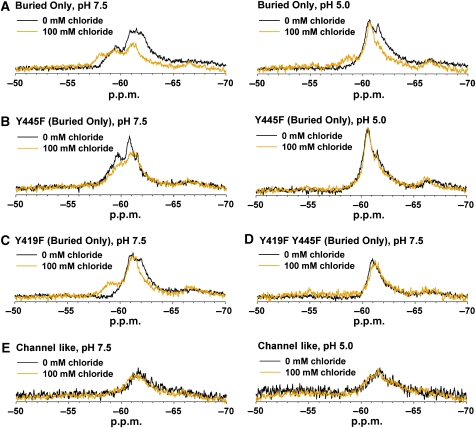
Comparison of spectra obtained in the presence and absence of Cl− also allows for examination of changes induced by Cl− binding. As seen in Figure 6A, the Buried Only 19F spectrum is highly sensitive to Cl− concentration. At pH 7.5, the peak corresponding to Y445 at zero Cl− (between −60.5 and −65.0 p.p.m.) shifts downfield to −58.2 p.p.m. in 100 mM Cl−. At pH 5.0, a similar Cl−-dependent downfield shift is observed. Among other contributions, this shift could result from the fluorine nucleus being in proximity to a negatively charged Cl− ion. As Y445 lines the Cl− permeation pathway (Figure 2A), it is not surprising to observe these Cl−-dependent spectral changes. Therefore, in addition to reflecting conformational changes that occur on Cl− binding, changes in the 19F spectrum also reflect Cl− binding itself.
Figure 6.
19F NMR detects Cl− binding within the Cl− permeation pathway and Cl−-dependent conformational change outside this region. (A) Cl− induces a change in chemical environment of one or more tyrosine residues in the Buried Only protein at both pH 7.5 and 5.0. (B) Removal of fluorine labelling at the Y445 position abolishes the Cl−-dependent spectral effect at pH 5.0 (right panel) but not at pH 7.5 (left). (C) In the absence of Y419 labelling, Cl−-dependent spectral changes are observed at pH 7.5. (D) Removal of labelling at both the Y419 and Y445 positions eliminates the Cl−-dependent spectral changes at pH 7.5. (E) The ClC-ec1 Channel-like mutant (E148A/Y445S Buried Only) does not experience a Cl−-dependent spectral change at either pH.
To distinguish Cl−-binding effects occurring outside the permeation pathway, we examined the impact of Cl− on the 19F spectrum of Y445F Buried Only. At pH 7.5, Cl− causes a decrease in intensity in the peaks occurring at −59.6 and −60.9 p.p.m. (Figure 6B, left). This indicates that under these conditions, Cl− has an effect on protein conformation outside the Cl− permeation pathway. At pH 5.0, however, the Y445F Buried Only spectra with and without Cl− remain nearly identical (Figure 6C, right). Comparing this to the Cl−-dependent change observed in the Buried Only spectrum at pH 5.0 (Figure 6A, right) illustrates that at this lower pH, the effect of Cl− binding is localized to the permeation pathway.
To determine whether the Cl−-dependent spectral change observed at pH 7.5 represents conformational change occurring at the subunit interface, we examined the effect of [Cl−] on proteins containing the Y419F mutation. As seen in Figure 6C, addition of the Y419F mutation yields a Cl−-dependent spectral shift that is qualitatively similar to that observed with the Buried Only protein. The persistence of spectral change in this single mutant is not surprising, as the Cl−-coordinating residue Y445F is still intact and fluorine labelled. We therefore examined the Y419F/Y445F double mutant to determine whether these two residues together could account for the Cl−-dependent spectral change observed at pH 7.5. As seen in Figure 6D, mutating both residues simultaneously removes the Cl− dependence. Thus, Cl− binding exerts an effect both inside the Cl− permeation pathway and at the subunit interface. Importantly, the Channel-like mutant does not display Cl−-dependent spectral change at either pH (Figure 6E), which indicates that Cl−-dependent conformational changes observed outside the permeation pathway in ClC-ec1 transporters are no longer present in this protein. Direct Cl− binding or changes within the permeation pathway cannot be observed in the Channel-like mutant because of lack of labelling at the Y445 position.
Discussion
We have described here the use of solution-state 19F NMR to monitor substrate-driven conformational changes occurring in ClC-ec1. This approach was inspired by the observation that despite being crystallized under a variety of conditions, only one ClC-ec1 conformation has ever been observed. The fluorine nucleus' small size, exquisite sensitivity to chemical environment, and ease of incorporation make it an excellent reporter of conformational change and ideally suited for overcoming this predicament. Although 19F NMR has been used primarily on soluble proteins (Hull and Sykes, 1975; Gerig, 1994; Danielson and Falke, 1996), its feasibility has also been shown with membrane proteins up to at least the size of the 65 kDa D-lactate dehydrogenase (Sun et al, 1996; Loewen et al, 2001; Oxenoid et al, 2002; Prosser et al, 2007). The focus of many of these earlier studies has been on structural questions related to single conformations, where the fluorine nucleus has been used as a probe to ascertain the topological disposition or membrane immersion depth of different test residues. Assessment of membrane protein conformational change has been performed using solid- state 19F NMR, but to date these methods have been limited to very small proteins such as the 25-residue peptide corresponding to the transmembrane domain of the M2 proton channel (Hong, 2007; Luo et al, 2007; Witter et al, 2008). Here, we show that solution-state 19F NMR can also be used to identify regions of conformational change in an integral membrane protein as large as the 102 kDa ClC-ec1 homodimer.
Functional significance of observed spectral changes
As our experiments have been performed on detergent-solubilized protein, it is necessary to question whether the spectral changes observed in our system report on conformational changes that are relevant to the ClC-ec1 transport cycle. Several pieces of evidence indicate this is indeed the case. First, we show that in the Channel-like mutant, Cl− and H+ binding do not cause the same conformational change induced in the transporters, as illustrated by the lack of H+ and Cl− dependency of the Channel-like 19F spectra (Figures 4A and 6E). Our data therefore provide strong support for the conclusion that this protein allows Cl− to move through its pore in an electrodiffusive manner that does not rely on coordinating a series of conformational changes with every round of ion transport (Jayaram et al, 2008).
Second, we have shown that the loss of substrate influence on conformation is specific to the high-turnover aspect of the Channel-like mutant (E148A/Y445S, Buried Only), as introduction of the E148A and Y445S uncoupling mutations individually does not eliminate H+-dependent spectral changes (Figure 4B). Additionally, the Uncoupled Transporter spectra are similar to coupled ClC-ec1 under both H+ conditions (Figure 4C). In the case of E148A, this indicates that the mutant protein remains conformationally sensitive to pH despite being functionally insensitive to pH. This result parallels observations from an earlier report, in which the E148A uncoupling mutation had no effect on the H+-induced spectral changes of fluorescent labels along ‘helix R,' which lines the Cl− permeation pathway (Bell et al, 2006). Likewise, the persistence of H+-dependent spectral changes in the Y445S mutant indicates that neither the uncoupling nor the changes in Cl− occupancy at this site abolish the protein's conformational sensitivity to [H+]. Thus (for both single mutations), despite the loss of H+/Cl− coupling, the observed antiporter conformational changes remain largely intact.
Finally, we have shown that changing the concentration of Cl− (a much less promiscuous protein binder than H+) also results in changes to ClC-ec1's 19F spectrum (Figure 6). Although the majority of this spectral change can be attributed to Y445 (which is expected to be highly sensitive to Cl− concentration because of its proximity to a bound chloride ion), we also observe Cl−-dependent spectral effects outside the Cl− permeation pathway (Figure 6B, left) including at the subunit interface (Figure 6D). Thus, binding of either substrate ion induces a conformational change that is observable in our system. Furthermore, the substrate-dependent spectral changes observed outside the Cl− permeation pathway are not likely to be due to electrostatic effects of ion binding, as the major contributor (Y419) is 20 Å away from the Cl−-binding sites.
Characteristics of ligand-driven conformational change in ClC-ec1
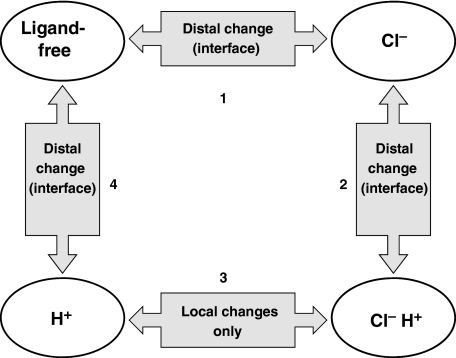
CLC-mediated antiport occurs through a series of conformational changes coupled to binding and unbinding of Cl− and H+. Manipulating the concentration of these substrates will therefore drive the protein to adopt different conformations corresponding to various states of ion occupancy. In this study, we have manipulated [Cl−] and [H+] to examine the conformational changes involved in transition of ClC-ec1 between four major states: ligand-free, Cl−-bound, Cl−/H+-bound, and H+-bound (Figure 7). The ligand-free state predominates at pH 7.5 and zero Cl−. Transition from the ligand-free state to the Cl−-bound state is observed on increasing the Cl− concentration to a saturating level (Step 1 in Figure 7, moving clockwise). This transition was found to involve regions distal to the Cl− permeation pathway (including the subunit interface), as indicated by the Cl− dependence of the Y445F Buried Only spectrum (Figure 6B) and the abolition of spectral changes in the Y419F/Y445F double mutation (Figure 6D). On increasing [H+], the Cl−-bound protein is converted to the doubly liganded form (Step 2 in Figure 7). This conformational change also involves movement outside the chloride permeation pathway, as shown by the persistence of H+-dependent spectral changes in Y445F Buried Only (Figure 3D). We have further localized part of this conformational change to the subunit interface (Y419F Buried Only, Figure 3E). Transition between the doubly liganded state and the protonated state is driven by removing Cl− under high [H+] (Step 3 in Figure 7). Unlike the other transitions observed in our study, Cl− binding to protonated ClC-ec1 seems to only induce local changes within the Cl− permeation pathway. This hypothesis is based on the fact that no Cl−-dependent spectral change is detected in the Y445F Buried Only protein at pH 5.0 (Figure 6B, right). However, we cannot rule out the possibility that movement occurs in regions of the protein that are not near any of the F-tyr reporters in our system. Lowering [H+] under zero Cl− drives ClC-ec1 to return to the ligand-free state (Step 4 in Figure 7). Similar to Step 2, this transition also involves movement distal to the Cl− permeation pathway, including movement at the subunit interface. This is evidenced by the spectral [H+] dependence of Y445F Buried Only under zero Cl− (Figure 5B) and the abolition of such dependence on mutation of Y419F (Figure 5C).
Figure 7.
Ligand-binding events and their associated conformational changes in ClC-ec1. In this schematic, ClC-ec1 is depicted in four major states corresponding to the four permutations of ion occupancy explored in our study. With the exception of Step 3, all transitions involve conformational change outside the chloride permeation pathway, near the subunit interface.
In summary, we have shown that conformational change outside the Cl− permeation pathway of ClC-ec1 is involved in three of the four transitional steps examined in this study. The functional consequences of such distant communications have previously been observed for a human Cl−/H+ antiporter (hClC-4) as well as two CLC channels (ClC-0 and ClC-2). In hClC-4, inhibition by zinc was drastically reduced by alterations in the permeation pathway (either by mutagenesis or by changes in the permeant ion), yet the proposed zinc-binding site was localized through mutagenesis to an extracellular loop region (Osteen and Mindell, 2008). This loop connects to a helix that extends into the core of the protein near the ion-binding sites, presumably enabling coupling of zinc binding and ion permeation. Although the influence of zinc binding on protein conformation was not directly examined, this study supports the idea that regions distal to the ion permeation pathway can have important functions in CLC transporter function. Similarly, in both ClC-0 (the Torpedo electric organ CLC channel) and the ClC-2 channel from guinea pig, mutations at the extracellular face of the protein and distant from the Cl− permeation pathway have been shown to affect properties of the channel pore (Ludewig et al, 1997; Niemeyer et al, 2009). Our own data suggest that such long-range communications also exist in ClC-ec1 and provide the first structural evidences that functionally relevant conformational changes occur outside the Cl− permeation pathway.
Inter-subunit crosstalk in ClC-ec1
Removal of labelling at the Y419 position (located at the subunit interface) decreases the spectral impact of both H+ and Cl− (Figures 3E, 5C, and 6D). This indicates that the subunit interface participates in both Cl− and H+-driven conformational changes. However, this does not mean that Y419 is the only residue involved in these changes. The Y419F mutation alone does not abolish the Cl− dependence of the ClC-ec1 spectrum (Figure 6C). It is only when labelling at the Cl−-coordinating residue Y445 is concurrently removed that the spectral Cl− dependence is abolished (Figure 6D). This indicates that both the Cl− permeation pathway and the subunit interface are influenced by Cl− binding.
H+-dependent spectral changes also persist in the Y419F mutant, although these changes are more subtle than those observed on changes in [Cl−] (Figures 3E and 5C). Although higher resolution studies will be needed to determine with certainty which residues account for these changes, we hypothesize that Y445 is a likely contributor. Although the contribution of Y445 to the spectral H+ dependence seems small in comparison with that of Y419, the magnitude of chemical shift does not necessarily reflect the magnitude of underlying protein movement. Therefore, although a change in chemical shift indicates a change in environment, it does not provide information concerning the scale of this change.
Removal of tyrosine labelling at either the Y445 or the Y419 position results in the disappearance of signal in the −57.0 to −60.0 p.p.m. range at pH 7.5 under saturating Cl− (Figure 3D and E). This indicates that mutation of one of these residues influences the signal of the other labelled residue, further supporting the idea of crosstalk between the subunit interface and the permeation pathway. The spectral interdependence of Y445 and Y419 at saturating Cl− may arise from either a change in structure of one or more of the kinetically stable conformational states or from a shift in the equilibrium distribution of these states. In the crystallographically accessible ‘occluded' state, the Y445F mutation alone does not change the observed ClC-ec1 structure (Accardi et al, 2006). However, this static picture cannot exclude the possibility that the Y445F mutation could influence the structure of ClC-ec1 in other conformations or the movements necessary to achieve these conformations. Indeed, the possibility that Y445F influences protein dynamics is supported by the fact that this mutant has a decreased Cl− turnover rate (Walden et al, 2007) (Supplementary Figure 2). Y419F, on the other hand, behaves more like the wild-type protein in assays for Cl− and H+ flux. As there is no crystal structure available for this mutant, we do not know whether there is any change to the occluded structure.
Interestingly, the spectral interdependence of Y445 and Y419 is not observed at zero Cl−, where mutation of each residue results in loss of signal in distinct regions of the spectrum (Figure 5E). This indicates that the effect of mutation of Y445 or Y419 may only be translated to the other residue in the presence of Cl−. Although the underlying reasons for the spectral interdependence of Y445 and Y419 at saturating Cl− remain unknown, this information is not necessary for the conclusions drawn here. In the absence of 19F labelling at the Y445 position, substrate-dependent spectral changes persist. This indicates protein movement must occur outside the Cl− permeation pathway of ClC-ec1. In contrast, the Y419F mutation eliminates much of the substrate-dependent spectral change observed in the Buried Only mutant without altering function, and thus this residue must be subject to changes in chemical environment.
We have shown that functionally relevant conformational change occurs at the ClC-ec1 dimer interface, but our NMR data alone do not allow us to draw conclusions concerning the nature or magnitude of these changes. However, information obtained from earlier studies can be used to limit the possibilities. One such study showed that constraining movement at the ClC-ec1 dimer interface through covalent crosslinking does not impair protein function (Nguitragool and Miller, 2007). Therefore, a large reorientation of the subunits relative to each other cannot be necessary for coordinated ion transport. This indicates that—unlike the large movement observed during gating of the CLC channels—relative inter-subunit movements in the CLC antiporters are likely to be small. Additional insight into the nature of inter-subunit communication comes from recent work with concatemers of a human CLC antiporter, hClC-5. Here, it was shown that antiport activity persists even when concatemers consist of one wild-type subunit and one non-functional subunit (Zdebik et al, 2008). This result indicates that any functionally relevant inter-subunit crosstalk cannot rely on ion permeation—a concept that is supported by our observation that even in mutant forms of ClC-ec1 in which H+ transport is lost or impaired, H+-dependent conformational changes remain largely intact (Figure 4B and C). Considered in light of these earlier studies, our results show that there are small conformational changes occurring at the ClC-ec1 dimer interface. These changes are absent in the Channel-like mutant protein, which suggests a link between the observed conformational changes and CLC antiport activity.
Conclusion
Although the details of the molecular movements involved in ClC-ec1 conformational cycling remain to be fully elucidated, here we have made several important observations concerning the nature of these movements. First, we have shown that substrate-driven conformational change is not constrained to the Cl− permeation pathway alone as was hypothesized earlier. Second, we have shown that the interface between the subunits of the ClC-ec1 homodimer participates in substrate-dependent protein movement. Third, we found that removal of the protein's H+ transport ability does not eliminate the H+-dependent protein movement observed in the intact antiporter. Finally, we observed that the ClC-ec1 Channel-like mutant is not subject to the same substrate-dependent conformational changes that occur in the ClC-ec1 transporters. Together, these results provide new insight into conformational change in ClC-ec1 and lay an essential foundation for future studies on ClC-ec1 protein dynamics.
Materials and methods
Protein expression and purification
ClC-ecl fused to a C-terminal polyhistidine (His) tag was produced in E. coli BL21(DE3) using the pASK expression vector. Mutations to the ClC-ec1 sequence were introduced using QuikChange mutagenesis (Stratagene) and confirmed by sequencing. Cells containing the expression construct were grown to OD550 1.0 in rich media, pelleted by centrifugation at 1700 g for 20 min at room temperature, and transferred to M9 minimal media supplemented with 0.4% glycerol, 1 mM MgSO4, 1 mM CaCl2, 0.5 mg/l thiamine, 100 mg/l ampicillin, and 40 mg/ml of each amino acid (excluding tyrosine). Fluorotyrosine labelling was achieved with the addition of 1 g/l glyphosate and 100 mg/l 3-fluoro-DL-tyrosine. Cells were grown for 30 min at room temperature at which time protein production was induced with 0.2 μg/ml anhydrotetracycline for 12 h. After this incubation, additional anhydrotetracycline and ampicillin were added, and induction continued for an additional 5 h.
After expression, ClC-ec1 was purified essentially as described (Accardi et al, 2004). Briefly, sonicated cells were extracted for 2 h at room temperature in the presence of 50 mM n-decyl-β-D-maltopyranoside. The extract was centrifuged at 20 000 g for 45 min and purified over a Co2+ column. The His-tag was then cleaved using Endoproteinase Lys-C, and the protein was further purified on a Superdex 200 size exclusion column. Analysis of purified protein through mass spectrometry indicates F-tyr labelling efficiency of >75% (data not shown).
NMR spectroscopy
For all 19F NMR experiments, purified ClC-ec1 was concentrated to 0.3–0.5 mM in a total volume of ∼350 μl including 10% (v:v) D2O. Samples were placed in the outer tube of Shigemi symmetrical microtubes matched to D2O to allow for reduction of necessary sample volume compared with standard 5 mm NMR tubes. To avoid sample frothing, the Shigemi insert was not used. Samples were prepared in 100 mM NaCl (or 100 mM Na+/K+ tartarate for Cl−-free experiments), 10 mM HEPES, pH 7.5, and n-decyl-β-D-maltopyranoside ranging from 50–100 mM (quantified using a sugar assay as described, Dubois et al, 1951). E. coli polar lipids (Avanti Polar Lipids) were added in a 1:80 lipid:detergent molar ratio to stabilize the protein and prevent aggregation over the course of the NMR experiments. The pH of the sample was lowered to 5.0 using citric acid and raised to 7.5 using tris-acetate. As our experiments are performed using detergent-solubilized ClC-ec1, the change in [H+] is experienced by both the extracellular and intracellular faces of the protein. Data were collected using a 5 mm H/F probe on a Bruker Avance 500 MHz NMR spectrometer running Topspin version 1.3 with variable temperature control. Data represent acquisition of 30–50k transients at 470 MHz; 12 kHz spectral width; 45° pulse; 0.17 s acquisition time; 1.8–2.8 s recycle delay; 25°C; 15 Hz line broadening; referenced to TFA.
Functional reconstitution and flux assays
ClC-ec1 functional reconstitutions and Cl−/H+ flux assays were performed essentially as described (Nguitragool and Miller, 2007; Walden et al, 2007). Briefly, proteoliposomes prepared for Cl− flux assays were reconstituted at density of 0.4 μg protein per mg lipid and loaded with 300 mM KCl, 25 mM citric acid–NaOH (pH 5.0). Valinomycin-initiated Cl− efflux from the vesicles was monitored using an Ag/AgCl electrode. Vesicles prepared for H+ flux assays were reconstituted at a density of 2.0 μg protein per mg lipid and loaded with 300 mM KCl and 75 mM glutamic acid–NaOH (pH 4.8). H+ pumping from a weakly buffered solution was initiated by addition of valinomycin and monitored over time.
Supplementary Material
Supplementary Material
Acknowledgments
We thank Rudi Nunlist, College of Chemistry NMR facility, University of California at Berkeley for use of the H/F NMR probe, Li Tan for her excellent technical assistance, and C Miller and A Accardi for sharing the high-turnover ClC-ec1 mutant before publication. We also thank William Kobertz, Jody Puglisi, Michael Reese, Michael Bokoch, Richard Reimer, and members of the Maduke laboratory for comments on the paper. This work was supported by National Institutes of Health Grant R01 GM070773, a Stanford OTL Research Incentive Award, and the Mathers Foundation. SE was supported by Award Number F32 GM080840 from the National Institute Of General Medical Sciences as well as a Dean's Fellowship and a Katherine McCormick Fellowship from Stanford University.
References
- Accardi A, Kolmakova-Partensky L, Williams C, Miller C (2004) Ionic currents mediated by a prokaryotic homologue of CLC Cl- channels. J Gen Physiol 123: 109–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Accardi A, Lobet S, Williams C, Miller C, Dutzler R (2006) Synergism between halide binding and proton transport in a CLC-type exchanger. J Mol Biol 362: 691–699 [DOI] [PubMed] [Google Scholar]
- Accardi A, Miller C (2004) Secondary active transport mediated by a prokaryotic homologue of ClC Cl-channels. Nature 427: 803–807 [DOI] [PubMed] [Google Scholar]
- Accardi A, Pusch M (2000) Fast and slow gating relaxations in the muscle chloride channel CLC-1. J Gen Physiol 116: 433–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Accardi A, Walden M, Nguitragool W, Jayaram H, Williams C, Miller C (2005) Separate ion pathways in a cl−/h+ exchanger. J Gen Physiol 126: 563–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad S, Gromiha M, Fawareh H, Sarai A (2004) ASAView: database and tool for solvent accessibility representation in proteins. BMC Bioinformatics 5: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SP, Curran PK, Choi S, Mindell JA (2006) Site-directed fluorescence studies of a prokaryotic ClC antiporter. Biochemistry 45: 6773–6782 [DOI] [PubMed] [Google Scholar]
- Brahm J (1977) Temperature-dependent changes of chloride transport kinetics in human red cells. J Gen Physiol 70: 283–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bykova EA, Zhang XD, Chen TY, Zheng J (2006) Large movement in the C terminus of CLC-0 chloride channel during slow gating. Nat Struct Mol Biol 13: 1115–1119 [DOI] [PubMed] [Google Scholar]
- Danielson MA, Biemann HP, Koshland DE Jr, Falke JJ (1994) Attractant- and disulfide-induced conformational changes in the ligand binding domain of the chemotaxis aspartate receptor: a 19F NMR study. Biochemistry 33: 6100–6109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielson MA, Falke JJ (1996) Use of 19F NMR to probe protein structure and conformational changes. Annu Rev Biophys Biomol Struct 25: 163–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFelice LJ (2004) Transporter structure and mechanism. Trends Neurosci 27: 352–359 [DOI] [PubMed] [Google Scholar]
- Dubois M, Gilles K, Hamilton JK, Rebers PA, Smith F (1951) A colorimetric method for the determination of sugars. Nature 168: 167. [DOI] [PubMed] [Google Scholar]
- Dutzler R, Campbell EB, Cadene M, Chait BT, MacKinnon R (2002) X-ray structure of a ClC chloride channel at 3.0 A reveals the molecular basis of anion selectivity. Nature 415: 287–294 [DOI] [PubMed] [Google Scholar]
- Dutzler R, Campbell EB, MacKinnon R (2003) Gating the selectivity filter in ClC chloride channels. Science 300: 108–112 [DOI] [PubMed] [Google Scholar]
- Elvington SM, Maduke M (2008) Thinking outside the crystal: complementary approaches for examining transporter conformational change. Channels (Austin) 2: 373–379 [DOI] [PubMed] [Google Scholar]
- Frieden C, Hoeltzli SD, Bann JG (2004) The preparation of 19F-labeled proteins for NMR studies. Methods Enzymol 380: 400–415 [DOI] [PubMed] [Google Scholar]
- Gakh YG, Gakh AA, Gronenborn AM (2000) Fluorine as an NMR probe for structural studies of chemical and biological systems. Magn Reson Chem 38: 551–558 [Google Scholar]
- Gerig J (1994) Fluorine NMR of proteins. Prog NMR Spectrosc 26: 293–370 [Google Scholar]
- Henzler-Wildman K, Kern D (2007) Dynamic personalities of proteins. Nature 450: 964–972 [DOI] [PubMed] [Google Scholar]
- Hille B (2001) Ion Channels of Excitable Membranes. Sinauer: Sunderland, MA [Google Scholar]
- Hong M (2007) Structure, topology, and dynamics of membrane peptides and proteins from solid-state NMR spectroscopy. J Phys Chem B 111: 10340–10351 [DOI] [PubMed] [Google Scholar]
- Hull WE, Sykes BD (1975) Fluorotyrosine alkaline phosphatase: internal mobility of individual tyrosines and the role of chemical shift anisotropy as a 19F nuclear spin relaxation mechanism in proteins. J Mol Biol 98: 121–153 [DOI] [PubMed] [Google Scholar]
- Hwang PM, Kay LE (2005) Solution structure and dynamics of integral membrane proteins by NMR: a case study involving the enzyme PagP. Methods Enzymol 394: 335–350 [DOI] [PubMed] [Google Scholar]
- Jayaram H, Accardi A, Wu F, Williams C, Miller C (2008) Ion permeation through a Cl—selective channel designed from a CLC Cl−/H+ exchanger. Proc Natl Acad Sci USA 105: 11194–11199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentsch TJ (2008) CLC chloride channels and transporters: from genes to protein structure, pathology and physiology. Crit Rev Biochem Mol Biol 43: 3–36 [DOI] [PubMed] [Google Scholar]
- Lisal J, Maduke M (2008) The ClC-0 chloride channel is a ‘broken' Cl(−)/H(+) antiporter. Nat Struct Mol Biol 15: 805–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobet S, Dutzler R (2006) Ion-binding properties of the ClC chloride selectivity filter. EMBO J 25: 24–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewen MC, Klein-Seetharaman J, Getmanova EV, Reeves PJ, Schwalbe H, Khorana HG (2001) Solution 19F nuclear Overhauser effects in structural studies of the cytoplasmic domain of mammalian rhodopsin. Proc Natl Acad Sci USA 98: 4888–4892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck LA, Falke JJ (1991) Open conformation of a substrate-binding cleft: 19F NMR studies of cleft angle in the D-galactose chemosensory receptor. Biochemistry 30: 6484–6490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludewig U, Jentsch TJ, Pusch M (1997) Inward rectification in ClC-0 chloride channels caused by mutations in several protein regions. J Gen Physiol 110: 165–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Mani R, Hong M (2007) Side-chain conformation of the M2 transmembrane peptide proton channel of influenza a virus from 19F solid-state NMR. J Phys Chem B 111: 10825–10832 [DOI] [PubMed] [Google Scholar]
- Matulef K, Maduke M (2007) The CLC ‘chloride channel' family: revelations from prokaryotes. Mol Membr Biol 24: 342–350 [DOI] [PubMed] [Google Scholar]
- Miller C (1982) Open-state substructure of single chloride channels from Torpedo electroplax. Philos Trans R Soc Lond B Biol Sci 299: 401–411 [DOI] [PubMed] [Google Scholar]
- Miller C (2006) ClC chloride channels viewed through a transporter lens. Nature 440: 484–489 [DOI] [PubMed] [Google Scholar]
- Nguitragool W, Miller C (2007) Inaugural article: CLC Cl/H+ transporters constrained by covalent cross-linking. Proc Natl Acad Sci USA 104: 20659–20665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemeyer MI, Cid LP, Yusef YR, Briones R, Sepulveda FV (2009) Voltage-dependent and -independent titration of specific residues accounts for complex gating of a ClC chloride channel by extracellular protons. J Physiol 587: 1387–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hagan D, Harper DB (1999) Fluorine-containing natural products. J Fluor Chem 100: 127–133 [Google Scholar]
- Osteen JD, Mindell JA (2008) Insights into the ClC-4 transport mechanism from studies of Zn2+ inhibition. Biophys J 95: 4668–4675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxenoid K, Sonnichsen FD, Sanders CR (2002) Topology and secondary structure of the N-terminal domain of diacylglycerol kinase. Biochemistry 41: 12876–12882 [DOI] [PubMed] [Google Scholar]
- Poget SF, Cahill SM, Girvin ME (2007) Isotropic bicelles stabilize the functional form of a small multidrug-resistance pump for NMR structural studies. J Am Chem Soc 129: 2432–2433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser RS, Evanics F, Kitevski JL, Patel S (2007) The measurement of immersion depth and topology of membrane proteins by solution state NMR. Biochim Biophys Acta 1768: 3044–3051 [DOI] [PubMed] [Google Scholar]
- Pusch M (2004) Structural insights into chloride and proton-mediated gating of CLC chloride channels. Biochemistry 43: 1135–1144 [DOI] [PubMed] [Google Scholar]
- Rozovsky S, Jogl G, Tong L, McDermott AE (2001) Solution-state NMR investigations of triosephosphate isomerase active site loop motion: ligand release in relation to active site loop dynamics. J Mol Biol 310: 271–280 [DOI] [PubMed] [Google Scholar]
- Sun ZY, Pratt EA, Simplaceanu V, Ho C (1996) A 19F-NMR study of the equilibrium unfolding of membrane-associated D-lactate dehydrogenase of Escherichia coli. Biochemistry 35: 16502–16509 [DOI] [PubMed] [Google Scholar]
- Walden M, Accardi A, Wu F, Xu C, Williams C, Miller C (2007) Uncoupling and turnover in a Cl−/H+ exchange transporter. J Gen Physiol 129: 317–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G (2008) NMR of membrane-associated peptides and proteins. Curr Protein Pept Sci 9: 50–69 [DOI] [PubMed] [Google Scholar]
- White SH (2004) The progress of membrane protein structure determination. Protein Sci 13: 1948–1949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witter R, Nozirov F, Sternberg U, Cross TA, Ulrich AS, Fu R (2008) Solid-state 19F NMR spectroscopy reveals that Trp41 participates in the gating mechanism of the M2 proton channel of influenza A virus. J Am Chem Soc 130: 918–924 [DOI] [PubMed] [Google Scholar]
- Zdebik AA, Zifarelli G, Bergsdorf EY, Soliani P, Scheel O, Jentsch TJ, Pusch M (2008) Determinants of anion-proton coupling in mammalian endosomal CLC proteins. J Biol Chem 283: 4219–4227 [DOI] [PubMed] [Google Scholar]
- Zifarelli G, Murgia AR, Soliani P, Pusch M (2008) Intracellular proton regulation of ClC-0. J Gen Physiol 132: 185–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material