Summary
Campylobacter jejuni, a prevalent cause of bacterial gastroenteritis, must adapt to different environments to be a successful pathogen. We previously identified a C. jejuni two-component regulatory system (Cj1226/7c) as upregulated during cell infections. Analyses described herein led us to designate the system CprRS (Campylobacter planktonic growth regulation). While the response regulator was essential, a cprS sensor kinase mutant was viable. The ΔcprS mutant displayed an apparent growth defect and formed dramatically enhanced and accelerated biofilms independent of upregulation of previously characterized surface polysaccharides. ΔcprS also displayed a striking dose-dependent defect for colonization of chicks and was modestly enhanced for intracellular survival in INT407 cells. Proteomics analyses identified changes consistent with modulation of essential metabolic genes, upregulation of stress tolerance proteins, and increased expression of MOMP and FlaA. Consistent with expression profiling, we observed enhanced motility and secretion in ΔcprS, and decreased osmotolerance and oxidative stress tolerance. We also found that C. jejuni biofilms contain a DNase I-sensitive component and that biofilm formation is influenced by deoxycholate and the metabolic substrate fumarate. These results suggest that CprRS influences expression of factors important for biofilm formation, colonization and stress tolerance, and also add to our understanding of C. jejuni biofilm physiology.
Introduction
Campylobacter jejuni is a Gram-negative food-borne pathogen now recognized as the leading cause of bacterial gastroenteritis in the developed world, infecting approximately 1% of the Canadian and US populations annually (Altekruse et al., 1999). C. jejuni is a commensal in many animal species, yet infection in humans commonly presents as acute gastroenteritis with symptoms of campylobacteriosis including nausea, vomiting, and intense, often bloody, diarrhoea. While usually self-limiting, this can proceed to bacteraemia in immunocom-promised individuals, and C. jejuni infection is a common antecedent to serious medical sequelae such as Guillain–Barré syndrome, a sometimes fatal ascending bilateral paralysis (Young and Mansfield, 2005; Helms et al., 2006). Strains resistant to antibiotics such as fluoroquinolones are becoming more common, and at present a vaccine is not available (Butzler, 2004); thus, new avenues for control are needed.
A key reservoir for C. jejuni infection is avian species, where it resides asymptomatically as a component of the intestinal microflora. This is likely responsible for its wide-spread presence in commercial poultry products (Jacobs-Rietsma, 2000). C. jejuni is also present in the intestinal mucosa of other animals; therefore, water contaminated with agricultural run-off and raw milk are also responsible for outbreaks. Consequently, as a zoonotic pathogen, C. jejuni encounters a variety of in vivo and transmission-related environmental challenges, and its presence as a serious public health concern suggests that it can overcome these challenges sufficiently (Chan et al., 2001). This is intriguingly inconsistent with its fastidious and fragile behaviour: culture of C. jejuni in the laboratory requires rich media, moderately increased (37–42°C) temperatures, and a low O2/high CO2 atmosphere. This suggests the existence of survival mechanisms that may not be immediately apparent from experiments performed solely in pure broth culture.
Compared with other enteric pathogens, relatively little is known about the molecular mechanisms underlying C. jejuni pathogenesis. This is due in large part to its recalcitrance to molecular genetics techniques developed in other bacteria and fastidious requirements for growth in the lab. Sequence analysis also suggests the absence of many classical enteric pathogen virulence factors, thus limiting the utility of comparative genomics (Parkhill et al., 2000; Hofreuter et al., 2006; Pearson et al., 2007). Work to date has demonstrated the importance of motility and surface carbohydrates, and has identified a cytolethal distending toxin involved in host cell cycle arrest (Young et al., 2007). Like Salmonella spp., the ability of C. jejuni to invade cells of the gastrointestinal epithelium is important for virulence; however, while the recently identified C. jejuni-containing vacuole (CCV) also avoids the canonical endocytic pathway and fusion with lysosomes, its maturation and trafficking appears to be unique in that it utilizes the microtubule network and resides near the Golgi apparatus (Watson and Galan, 2008). Furthermore, experiments have suggested that in the absence of a prototypical, dedicated type III secretion system such as those utilized by pathogenic Escherichia coli, C. jejuni uses the flagellar export apparatus to secrete effectors such as CiaB that aid host cell invasion and colonization (Konkel et al., 2004). Taken together, this suggests that C. jejuni uses distinct paradigms from prototypical enteric pathogens such as E. coli and Salmonella.
In addition to the environmental challenges of its zoonotic lifestyle, the absence of established enteric virulence factors in C. jejuni suggests that features contributing to its survival play a central role in its success as a pathogen. In support of this, several global regulatory and stress survival factors in C. jejuni (i.e. the stringent response and polyphosphate metabolism) also have been shown to control phenotypes directly linked to virulence (Gaynor et al., 2005; Candon et al., 2007). The importance of broad changes in physiology such as the viable but non-culturable state and, more definitively, biofilm formation to C. jejuni resilience have also been noted (Trachoo et al., 2002; Joshua et al., 2006). Upwards of 99% of bacterial species, including C. jejuni, exist outside of the laboratory primarily not as free-swimming, planktonic cells, but as part of communities called biofilms (Ehrlich et al., 2005), which exhibit marked differences in metabolism, cell physiology, and importantly, stress resistance from their broth-grown counterparts (O'Toole and Kolter, 1998). While analysis of biofilm formation in C. jejuni is in its infancy, work to date suggests that biofilms contribute to stress tolerance during pathogenesis (Joshua et al., 2006; Kalmokoff et al., 2006; McLennan et al., 2007; Fields and Thompson, 2008).
Adaptation of a bacterium to its environment requires regulation of gene expression. Two-component regulatory systems (TCRSs) are ubiquitous systems used by prokaryotes to respond to environmental changes (Eppinger et al., 2004) and achieve transduction of information from the environment via phosphorelay between two proteins, a membrane-bound sensor histidine kinase (SK) and its cognate cytoplasmic DNA-binding response regulator (RR). Activation of the RR results in expression changes that bring forth adaptation to the current challenge. The C. jejuni genome encodes a limited repertoire of regulatory elements (Parkhill et al., 2000), including a relatively small number of TCRSs (seven SKs and 12 RRs). TCRSs in many pathogens control phenotypes closely associated with virulence or survival; likewise, in C. jejuni, all TCRSs characterized to date (FlgRS, RacRS, DccRS and the orphan RR CbrR) (Bras et al., 1999; MacKichan et al., 2004; Wosten et al., 2004; Raphael et al., 2005) are required for optimal colonization of chicks except PhosSR (Wosten et al., 2006). Thus, TCRSs are attractive candidates for study of paradigms underlying C. jejuni survival and pathogenesis.
The availability of C. jejuni genome sequences has aided the development of new genetic tools to explore the molecular mechanisms mediating survival during pathogenesis. Microarray analysis of the transcriptional response of C. jejuni to INT407 epithelial cell monolayers (Gaynor et al., 2005) identified a TCRS encoded by Cj1226c and Cj1227c that may contribute to adaptation to pathogenesis-related environments. We have designated this system CprRS (Campylobacter planktonic growth regulation). In this study, we provide genetic, molecular, proteomic and phenotypic evidence demonstrating that this TCRS likely co-ordinates survival- and pathogenesis-related phenotypes in C. jejuni through control of essential biological processes, stress tolerance and biofilm formation, thereby contributing to the success of this apparently fragile zoonotic pathogen.
Results
The C. jejuni genes Cj1226c and Cj1227c encode a TCRS pair, CprRS; the CprR RR is essential for viability
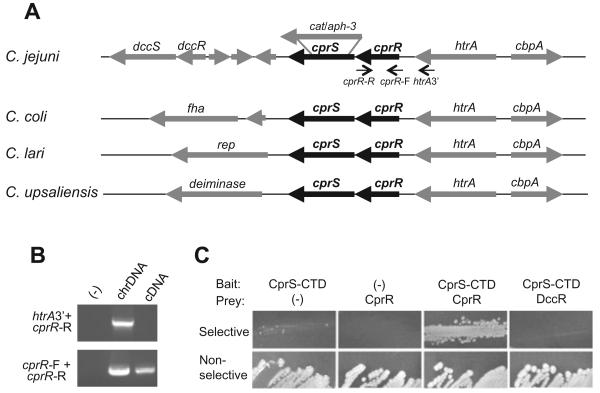
The annotated C. jejuni NCTC 11168 genome contains seven SKs and 12 RRs (Parkhill et al., 2000), and among these are five TCRS pairs in which the SK is encoded immediately downstream of the RR. One such pair, encoded by Cj1226c and Cj1227c, was previously identified as upregulated in the presence of live epithelial cells in vitro (Gaynor et al., 2005). CD-Search analyses of each component suggest that Cj1226c encodes an EnvZ-family SK, and the protein encoded by Cj1227c contains domains characteristic of the OmpR family of DNA-binding RRs (Marchler-Bauer and Bryant, 2004). The organization of the region surrounding Cj1226c and Cj1227c in various Campylobacter spp. is shown in Fig. 1A.
Fig. 1. Identification and genetic analysis of a Campylobacter-specific TCRS, cprRS.
A. Location of the cprR response regulator and cprS sensor kinase homologues (annotated as Cj1226c and Cj1227c in strain 11168) in the genomes of Campylobacter spp., identified using CampyDB (http://xbase.bham.ac.uk/campydb/). Approximate location of cat- and aph-3-marked deletions in cprS are shown, resulting in strains designated ΔcprS and ΔcprS::KmR respectively.
B. cprR and htrA are transcribed independently. Chromosomal DNA (chrDNA) or cDNA was used as a template for PCR using the primer combinations indicated in (A).
C. Bacterial two-hybrid analysis demonstrates that the CprS C-terminal domain (CTD) interacts with CprR, but not a control C. jejuni response regulator, DccR.
The TCRS encoded by Cj1226c and Cj1227c appears to be well conserved only among the campylobacters. BLAST analysis of ε-proteobacterial genomes sequenced to date identified putative orthologues of Cj1226c and Cj1227c in other Campylobacteraceae (C. coli, 100% and 97% amino acid similarity respectively; C. upsaliensis, 99% and 89% similarity; C. lari, 94% and 81% similarity; C. fetus, 82% and 69% similarity; Arcobacter butzleri, 71% and 52% similarity). In each case, the genes are encoded adjacent to the htrA gene, strongly suggesting orthology (Fig. 1A). In contrast, within the Helicobacteraceae, numerous TCRS genes within the same organism are all similarly unrelated to Cj1226c and Cj1227c (the closest Cj1227c and Cj1226c homologues are respectively: Sulfuromonas denitrificans, 73% and 56% similarity; Helicobacter pylori, 69% and 58% similarity; Wolinella succinogenes, 68% and 53% similarity). This suggests homology, but prevents distinction of clear orthologues from paralogues. In addition, both Cj1226c and Cj1227c showed higher similarity to other C. jejuni TCRS homologues, such as RacRS, than to potential orthologues in other taxa. Furthermore, none of the sequenced genera outside the Campylobacteraceae (Helicobacter, Wolinella, Sulfuromonas) have a TCRS encoded adjacent to htrA except for Nitratiruptor, a deep-sea vent ε-proteobacterium that appears to have diverged before the split of the Campylobacterales (Nakagawa et al., 2007), making solid evidence for Cj1226c and Cj1227c orthologues even more elusive. Based on conservation of this TCRS among members of the Campylobacter genus and potential divergence or absence of homologues in other ε-proteobacterial species, together with the phenotypes observed for Cj1226c mutants (see below), we propose renaming the system CprRS (Campylobacter planktonic growth regulation RR and SK).
To explore the physiological role of CprRS in C. jejuni, we set out to perform mutational analysis on each gene. Numerous efforts to delete the RR gene using different mutagenesis constructs were unsuccessful. Mutagenesis of Cj1227c (cprR) has been attempted by three other groups (Raphael et al., 2005; J. Ketley, pers. comm.; J.K. MacKichan, E.C. Gaynor and S. Falkow, unpublished), with Raphael et al. reporting that Cj1227c could only be inactivated when a second copy was present at a heterologous location. These results thus strongly suggest that activity of this RR is essential for viability of C. jejuni under laboratory conditions. In contrast, deletion mutagenesis of Cj1226c (cprS) was achieved using two different constructs, one with a CmR cassette replacing the entire cprS coding region and one with a non-polar KmR cassette, suggesting that the activity of the SK is dispensable. The resulting strains were designated ΔcprS and ΔcprS::KmR; as they behaved similarly in subsequent experiments, they were used interchangeably.
The non-essential nature of the SK, together with the fact that the RR is essential, raised the question of whether these proteins in fact form a cognate system. In all sequenced Campylobacter species, the genes encoding CprRS are encoded next to each other with a 4-base-pair overlap, and in all cases, are encoded adjacent to htrA. Operon prediction (Price et al., 2005) and microarray expression analysis (E. Gaynor, unpublished; Gaynor et al., 2004) suggest that they are encoded in a stand-alone two-gene operon. Nonetheless, because of the conservation of this genomic organization within the campylobacters, we explored the transcriptional (and thus the possible functional) relationship between cprRS and htrA. RT-PCR was performed using primers annealing to the RR gene only, or the 3′ end of htrA and the 3′ end of the RR (Fig. 1A). While amplicons were observed for both primer sets when chromosomal DNA was used as template, PCR products using cDNA as a template were only obtained for the cprR primer set (Fig. 1B), suggesting that cprR is transcribed independently from htrA.
Although the above transcriptional and operon analyses strongly suggest that these genes encode a cognate TCRS, we also wished to demonstrate a direct physical interaction between CprR and CprS. We used a bacterial two-hybrid system to measure physical association between the two proteins as has been previously employed (Kulasekara et al., 2005). Following co-transformation of bait and prey constructs (see Experimental procedures), colonies on selective media were obtained for CprS and CprR, but not CprS and a control RR (DccR) (Fig. 1C), indicating that only interaction of CprS with CprR was strong enough to activate expression of the reporter genes.
ΔcprS displays an apparent growth defect in broth culture
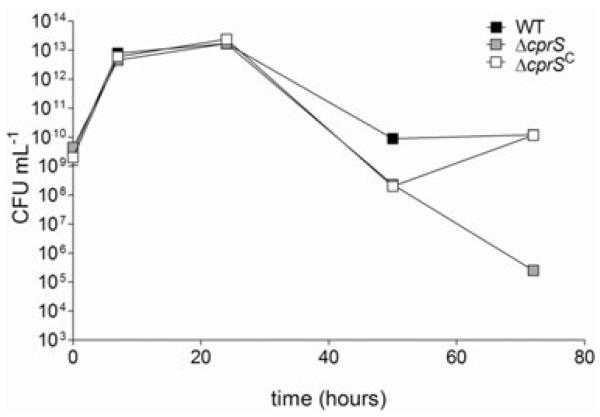
The role of CprRS in C. jejuni biology and pathogenesis was explored via extensive phenotypic characterization of the ΔcprS mutant. In shaking broth culture, ΔcprS appeared to have a late-stage culturability defect (Fig. 2), with a 4-log decrease in recovery of culturable bacteria [colony-forming units (cfu) ml−1] beyond 48 h of culture. These initial results were not surprising, given the essential nature of the cognate RR. The KmR cassette used to make one ΔcprS strain is non-polar, and our data suggest that cprRS is a stand-alone two-gene operon (Fig. 1B); furthermore, the flanking genes are likely essential (tRNA-Asn, cprR) or are involved in distinct phenotypes such as heat sensitivity not observed in ΔcprS (htrA) (Brondsted et al., 2005) (Table S1). Nonetheless, we addressed the possibility that the growth phenotype was not linked to the targeted mutation by inserting a wild-type (WT) copy of cprS into a heterologous location in the chromosome of ΔcprS to create the complemented strain ΔcprS C, which complemented the growth phenotype.
Fig. 2.
A ΔcprS sensor kinase mutant displays a growth defect compared with the WT strain in rich broth culture. WT (black squares), ΔcprS (grey squares) and ΔcprS C (white squares) were cultured in MH broth; at indicated time points, samples were taken to measure colony-forming units (cfu). Error bars are present but in most cases are too small to see.
ΔcprS exhibits enhanced and accelerated biofilm formation
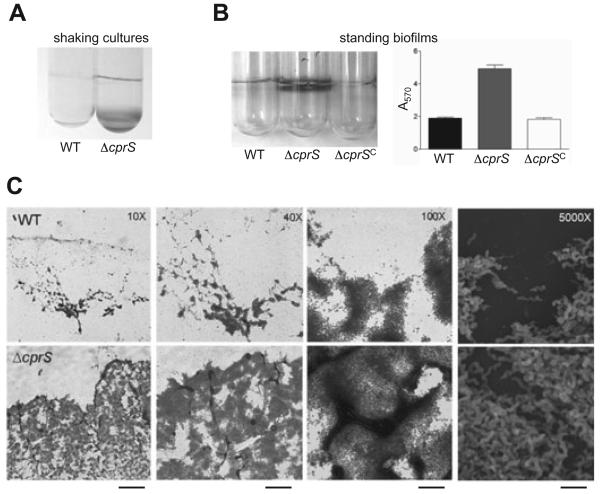
Upon closer observation of broth cultures prepared for further experiments, aggregates of bacteria were clearly visible in the flasks containing ΔcprS. We also noted a 30% increase in autoagglutination for ΔcprS compared with WT (data not shown), and crystal violet staining suggested that ΔcprS was adhering to shaking culture tubes (Fig. 3A). Taken together, this raised the possibility that the apparent growth defect shown in Fig. 2 may partially reflect aggregation of ΔcprS both with other bacteria and with abiotic surfaces, resulting in the loss of organisms from the media/planktonic fraction where samples were harvested for growth curve analysis.
Fig. 3. ΔcprS exhibits increased surface growth characteristics compared with WT.
A. ΔcprS adheres to shaking broth culture tubes. WT and ΔcprS were grown overnight with shaking at 200 r.p.m. in MH broth, and adherent bacteria were visualized by staining cultures directly with 0.2% crystal violet.
B. ΔcprS shows enhanced biofilm formation. Overnight liquid cultures of WT, ΔcprS, and ΔcprS C were diluted to an optical density of 0.0002 in MH broth, added to borosilicate glass tubes and incubated without shaking for 2 days. Biofilms were stained by the addition of crystal violet to a final concentration of 0.2% and tubes were photographed (left). Biofilms formed by WT (black bar), ΔcprS (grey bar) and ΔcprS C (white bar) were quantified (right) by dissolving adhered crystal violet with 30% methanol/10% acetic acid and measuring the A570 of the resulting solution. Quantifications were performed in triplicate.
C. ΔcprS biofilms are enhanced and accelerated compared with WT and are comprised of spiral-shaped bacteria. Biofilms were grown on glass coverslips in tubes prepared as in Fig. 4, and the region at the air-liquid interface was either stained with crystal violet for bright field microscopy (10×, 40×, 100×; first three panels) or prepared for scanning electron microscopy (SEM) (5000×; far right panels). Pictures are representative of a large region of each slide. From left to right, the bars represent ~400 μm, 100 μm, 40 μm, and 10 μm.
An ensuing hypothesis to these observations is that ΔcprS may favour surface-attached biofilm growth rather planktonic growth. Indeed, standing culture biofilm crystal violet assays (O'Toole and Kolter, 1998; McLennan et al., 2007) clearly demonstrated that ΔcprS exhibited a visible enhancement of biofilm formation compared with WT after only 1 day of incubation (Fig. 3B, left). Not surprisingly, when adhered crystal violet was dissolved and quantified, a statistically significant (P < 0.005) difference for ΔcprS compared with the WT strain was seen (Fig. 3B, right). Again, the complemented ΔcprS C strain exhibited biofilm formation at levels comparable to WT, supporting the hypothesis that CprS controls aspects of C jejuni physiology related to biofilm formation and/or planktonic growth.
We performed microscopy to confirm that the crystal violet-stained material represented healthy C. jejuni biofilms present at the air-liquid interface. Bright field microscopy and crystal violet staining (10×−100× magnifications) clearly identified ordered microcolony structures, suggesting active biofilm development rather than non-specific aggregation and adherence to the surface or acellular material (Fig. 3C). Biofilm formation appeared to be both accelerated and enhanced for ΔcprS, where much more biofilm material was observed compared with WT. Furthermore, a larger proportion of the ΔcprS biofilms exhibited mature biofilm characteristics such as mushroom-like architecture and water channels. There have been numerous reports that C. jejuni enters a viable but nonculturable state at later growth stages, which is often defined by a switch from spiral to coccoid physiology. As the ΔcprS mutant appeared to display a late-stage culturability defect (Fig. 2), we wanted to observe the morphology of the bacteria within the ΔcprS biofilms in order to solidify our conclusion that the growth ‘defect’ in liquid culture may at least in part be attributed to increased sessile growth rather than a loss in culturability per se. Scanning electron microscopy (SEM) (5000× magnification) confirmed spiral morphology (i.e. not coccoid) for both strains (Fig. 3C, far right panels).
Accelerated and enhanced ΔcprS biofilms form independent of changes in surface carbohydrates; C. jejuni biofilms are DNase I-sensitive and contain fibres that react with DNA stains
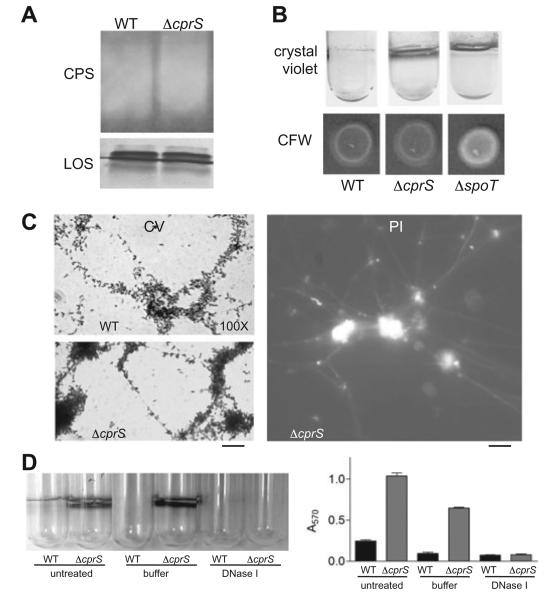
We performed further analyses to explore the molecular bases underlying the enhanced biofilm phenotype of ΔcprS. As the importance of surface carbohydrates in C. jejuni biofilms has been demonstrated (Kalmokoff et al., 2006; McLennan et al., 2007), we hypothesized that changes in such polysaccharides may be occurring in ΔcprS. We first extracted total carbohydrates from both WT and ΔcprS for separation by polyacrylamide gel electrophoresis, followed by silver staining to visualize lipooligosaccharide (LOS) and Western blotting with Penner antiserum to compare levels of capsular polysaccharide (CPS). The mutant strain had a similar LOS profile to the parental strain, both in amount and in species present; levels of CPS likewise appeared to be similar (Fig. 4A). Recent work has shown that a C. jejuni ΔspoT mutant forms enhanced biofilms commensurate with upregulation of a calcofluor white (CFW)-reactive polysaccharide (McLennan et al., 2007). However, CFW reactivity for ΔcprS was nearly identical to the WT strain (Fig. 4B). Similar results were obtained with Congo Red, another carbohydrate-binding dye (data not shown), further suggesting that the enhanced biofilm phenotype of ΔcprS was independent of previously characterized surface carbohydrates.
Fig. 4. Enhanced biofilm formation in ΔcprS is independent of changes in surface polysaccharides; the C. jejuni biofilm matrix exhibits a fibrous appearance and contains DNA.
A. Enhanced biofilm formation in ΔcprS is independent of changes in lipooligosaccharide (LOS) and capsular polysaccharide (CPS). Total carbohydrates were extracted from equal numbers of plate-grown bacteria and subjected to SDS-PAGE followed by silver staining to visualize LOS and Western blotting with Penner anti-O36 serotype antibody to visualize CPS.
B. ΔcprS does not overproduce a CFW-reactive surface polysaccharide. Overnight cultures of WT, ΔcprS and the positive control ΔspoT were spotted on BHI agar supplemented with 0.002% calcofluor white (CFW) for assessment of production of surface carbohydrates with β 1-3 and β 1-4 linkages. Crystal violet-stained biofilms for each strain are also shown for comparison.
C. Fibres can be seen in both ΔcprS (shown) and WT (not shown) biofilms. Biofilms were grown on glass coverslips as in Fig. 3C and stained with either crystal violet (CV, left) or propidium iodide (PI, right) and visualized by microscopy at 100× magnification. Bars represent ~25 μm (CV) and ~20 μm (PI).
D. C. jejuni biofilms can be disrupted by treatment with DNase I. WT and ΔcprS biofilms were grown for 2 days and either left untreated or rinsed with PBS, then incubated for 3 h in buffer alone or buffer with DNase I, followed by crystal violet staining.
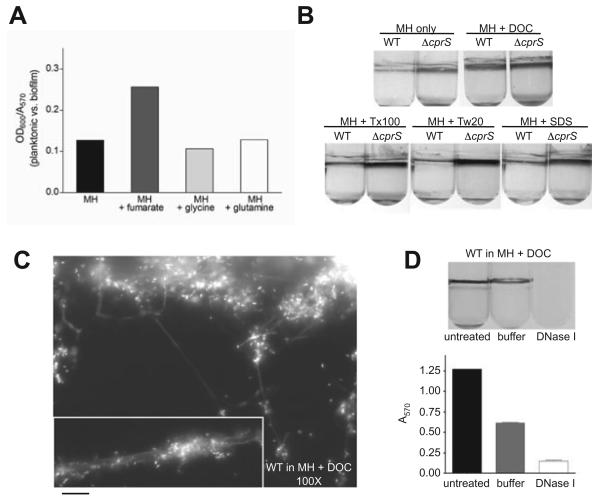
Microscopy of crystal violet-stained biofilms consistently suggested that the bacteria and microcolonies were connected by fibrous-like structures as exemplified in Fig. 3C, WT panel, 40× magnification. Closer examination clearly showed the presence of fibres connecting bacteria and microcolonies (Fig. 4C, left). We also observed these fibres under non-fixed conditions, and noted that the fibres also stained with propidium iodide (PI) (Fig. 4C, right). Shown are ΔcprS samples; similar fibres were also observed for WT biofilms. As both crystal violet and propidium iodide can stain DNA, we hypothesized that the fibres were, at least in part, composed of DNA. To provide support for this, 2-day-old biofilms were rinsed with PBS and incubated with DNase I. We found that DNase I treatment resulted in a striking decrease in ΔcprS biofilms compared with ΔcprS biofilms treated with buffer alone (P < 0.0001). WT biofilms were also disrupted by DNase I treatment despite not reaching statistical significance compared with buffer-treated biofilms. No difference in survival of each strain in the presence of similar concentrations of DNase I was observed (data not shown). These data provide the first evidence for the presence of DNA within the matrix surrounding C. jejuni biofilms.
ΔcprS shows modestly increased intracellular survival but is defective for colonization of 1-day-old chicks
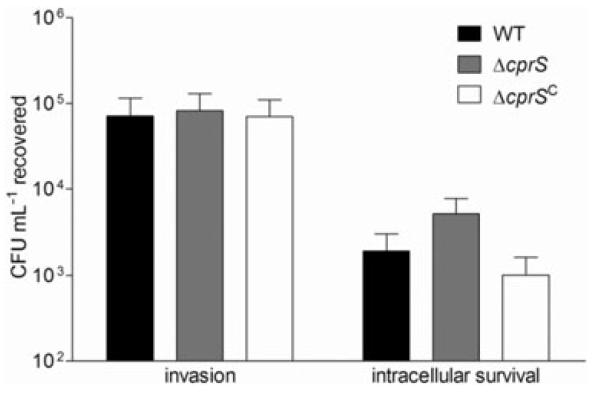
Because of its initial identification in a cell infection screen, we next explored the role of CprS in host-related phenotypes. We first assessed invasion and intracellular survival of ΔcprS in human epithelial cell monolayers. INT407 cell monolayers were infected with WT, ΔcprS or ΔcprS C bacteria, and there was no significant difference in the number of bacteria recovered at the invasion time point following 3 h of infection and 2 h of gentamicin treatment (Fig. 5). However, a surprising and reproducible twoto fivefold increase in the number of bacteria surviving an additional 5 h incubation within the INT407 cells was observed for ΔcprS compared with both WT and the complemented ΔcprS C strain. Both WT and ΔcprS exhibited the same gentamicin susceptibilities and tolerance of the INT407 cell water and syringe lysis procedures; likewise, no differences were observed for bacterial survival in media above the cells during the infection period, or for bacterial adherence to INT407 monolayers (data not shown).
Fig. 5.
ΔcprS invades INT407 cells at levels comparable to WT but exhibits a modest increase in intracellular survival. WT (black bars), ΔcprS (grey bars) and ΔcprS C were grown biphasically overnight and used to infect semi-confluent monolayers of INT407 intestinal epithelial cells at a multiplicity of infection of ~100. Following 3 h of infection and 2 h of gentamicin treatment, monolayers were lysed and serial dilutions were plated to determine intracellular bacteria (invasion). Intracellular survival was determined after an additional 5 h of incubation.
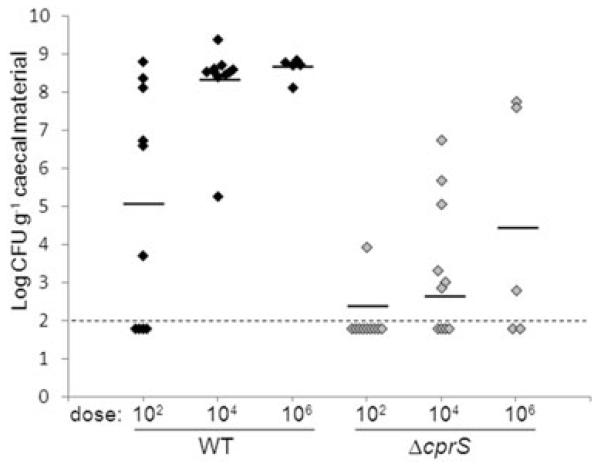
Next, to assess the contribution of CprS to in vivo fitness of C. jejuni, we tested the ability of ΔcprS to colonize 1-day-old chicks (Fig. 6). Groups of birds were infected with WT and ΔcprS at increasing inoculation levels; 6 days post infection, birds were sacrificed and caecal contents assayed for viable C. jejuni. Compared with WT, ΔcprS exhibited approximately 2.7-log, 5.7-log and 4.2-log lower average levels of colonization at doses of 102, 104 and 106 cfu, respectively, with corresponding statistically significant P-values of 0.008, 6.8 × 10−7 and 0.013. Furthermore, at doses of 104 and 106, several ΔcprS-inoculated chicks did not harbour detectable levels of C. jejuni whereas the WT strain colonized all chicks to very high levels. Both WT and ΔcprS colonized equally well at doses above 106 cfu (data not shown). These data suggest that ΔcprS is required for optimal colonization of chicks at lower doses, and that this defect can be overcome when chicks are administered higher doses of bacteria.
Fig. 6.
ΔcprS displays a dose-dependent chick colonization defect. One-day-old chicks were orally challenged with the indicated number of cfu of WT (black diamonds) or ΔcprS (grey diamonds) bacteria. Six days post infection, birds were sacrificed, and caecal colonization levels were determined by plating on C. jejuni-selective MH agar. Each data point represents the log cfu g−1 recovered from an individual chick, with the average recovery for each dosage denoted by a black bar. The limit of detection (100 cfu) is denoted by the thin dashed line.
Proteomics identifies specific protein expression differences between WT and ΔcprS strains
We next hypothesized that specific and/or global protein differences may underlie the striking biofilm and chick colonization observations; thus, we explored global protein expression profiles of WT, ΔcprS and ΔcprS C. Two-dimensional gel electrophoresis and mass spectrometry analyses revealed that numerous proteins varied significantly in expression between WT and ΔcprS, most of which are involved in stress tolerance, cell surface structures, regulation, and metabolic pathways (Fig. 7). Furthermore, the majority of expression changes were rescued in the ΔcprS C complemented strain.
Fig. 7. Proteomic analysis of WT, ΔcprS and ΔcprS C identifies expression differences that may underlie the phenotypes of ΔcprS. Cells were grown in MH broth to mid-log phase and harvested for proteomic analyses as described in Experimental procedures.
A. Two-dimensional SDS-PAGE analysis of the proteomes of WT, ΔcprS and ΔcprS C.
B. Proteins showing significant increases or decreases in expression in ΔcprS compared with the WT strain. Note that in most cases, complementation of ΔcprS (ΔcprS C) resulted in approximately WT levels of protein expression.
Several oxidative stress tolerance proteins were upregulated in ΔcprS compared with WT including catalase (KatA), thioredoxin reductase (TrxB) and alkyl hydroperoxide reductase (AhpC). In contrast, superoxide dismutase (SodB) and the non-haem iron protein Rrc were expressed at lower levels in ΔcprS, although the change in SodB expression was not complemented in ΔcprS C. Interestingly, both the major outer membrane protein (MOMP) (encoded by porA) and the flagellar filament protein FlaA were upregulated in ΔcprS compared with WT and ΔcprS C Cj0998 (YceI-like, isoprenoid transport and/or metabolism) was also upregulated. Many metabolic proteins were downregulated in ΔcprS, including fructose bisphosphate aldolase (Fba), fumarate hydratase (FumC), succinyl-CoA synthetase alpha chain (SucD) and aspartate-semialdehyde dehydrogenase (Asd). Also showing lower expression in ΔcprS versus WT were proteins involved in translation, such as ribosomal protein S1, trigger factor, EF-Tu, EF-G, EF-P and EF-Ts; however, a histidyl-tRNA was more highly expressed in ΔcprS. The nutrient acquisition protein ferric binding protein (FbpA) and a putative enoyl-[acyl-carrier-protein] reductase (FabI) were also lower in ΔcprS, although expression of FabI was not complemented in ΔcprS C. Also interesting was downregulation of Cj0355c, encoding an orphan RR, as well as the autoinducer-2 (AI-2) synthase, LuxS. Finally, two unknown proteins encoded by Cj0706 and Cj0092 were also disregulated in ΔcprS. The diversity of disregulated proteins in ΔcprS suggested that CprRS controls numerous aspects of C. jejuni biology, including essential metabolic functions, some of which may be involved in biofilm formation and/or the in vitro and host-related phenotypes of ΔcprS.
Consistent with several proteomics observations, ΔcprS exhibits enhanced motility, protein secretion, and osmotic and oxidative stress susceptibilities
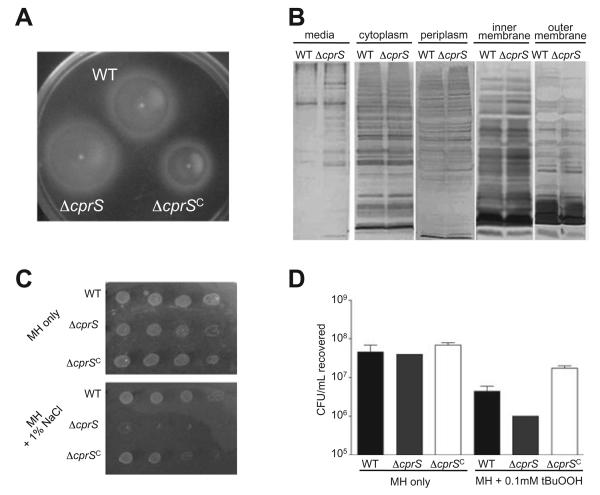
To explore and strengthen connections between the proteomics data and phenotypes observed for ΔcprS, a number of follow-up experiments were performed. Because of the higher FlaA expression seen in ΔcprS, we assessed this strain for motility. Microscopy suggested that ΔcprS was highly motile, and agar stab assays likewise showed an increase in motility in this strain (Fig. 8A). Because the flagellar apparatus has been proposed to function in protein secretion in C. jejuni (Konkel et al., 2004), and given the FlaA and MOMP protein expression differences, we also investigated whether subcellular protein localization or secreted protein profiles were altered in ΔcprS. Interestingly, while subcellular fractionation did not reveal notable differences in the cellular distribution of proteins, we did observe an increase in several specific proteins in the media fraction of ΔcprS compared with WT (Fig. 8B).
Fig. 8. Proteomics follow-up studies reveal enhanced motility, protein secretion, and osmotic and oxidative sensitivity differences between WT and ΔcprS.
A. ΔcprS shows an increase in motility compared with WT. Overnight cultures of WT, ΔcprS and ΔcprS C were stabbed into MH plates with 0.4% agar and incubated microaerobically for 24 h.
B. Protein secretion is increased in ΔcprS. The media fraction was clarified by centrifugation from overnight cultures of WT and ΔcprS at approximately equal optical densities. The collected cells were then subjected to subcellular fractionation to allow analysis of the secreted (media), cytosolic, periplasmic, inner membrane and outer membrane protein profiles. Equal volumes of sample were separated by SDS-PAGE followed by silver staining.
C. The ΔcprS mutant displays a decreased ability to grow under osmotic stress. Overnight cultures of WT and ΔcprS in MH broth were diluted to an optical density of 0.05 in MH broth, serially diluted 10-fold, spotted on MH plates or MH plates supplemented with 1% (w/v) NaCl and incubated for 24 h microaerobically. Dilutions were spotted left to right, with the highest starting CFUs on the left most side of the plate. D. The ΔcprS mutant shows decreased ability to survive in the presence of oxidative stress. Bacteria were suspended at an OD600 of 0.02 in MH broth or MH broth supplemented with 0.1 mM t-butylhydroperoxide (tBuOOH). Following 30 min of incubation at 37°C microaerobically, samples were serially diluted and plated for cfu counts.
Initial extensive assessments of the ΔcprS mutant strain for stress-related phenotypes surprisingly revealed no obvious differences between ΔcprS and the parental strain for many conditions tested (Table S1). However, in keeping with changes in the twofold increase in MOMP porin overexpression, ΔcprS exhibited a marked decrease in its ability to form colonies on media containing moderate (1%) levels of NaCl (Fig. 8C), as well as decreased survival in broth culture supplemented with NaCl (data not shown). Complementation restored the osmotic stress defect of ΔcprS to WT levels (Fig. 8C). These data indicate that ΔcprS is defective for osmotolerance, which was further supported by a two- to fourfold decrease in the minimum inhibitory concentration (MIC) for other salts such as MgCl2 and KCl (data not shown). After noting changes in expression of oxidative stress proteins, we also tested survival of ΔcprS in broth culture supplemented with the oxidative stress agent t-butylhydroperoxide, and noted a small but reproducible increase in sensitivity (P < 0.05) in the ΔcprS mutant compared with WT (Fig. 8D). We have also observed a slight increase in sensitivity to both hydrogen peroxide and paraquat (data not shown).
Biofilm formation in C. jejuni is suppressed by fumarate and promoted by deoxycholate
In Pseudomonas aeruginosa, the availability of carbon sources such as glucose, succinate and glutamate directs the maturation of biofilms (Shrout et al., 2006). Because we identified metabolic changes in the ΔcprS hyperbiofilm-forming strain by proteomics, we tested biofilm formation in C. jejuni of media supplemented with different metabolic substrates (Fig. 9A). Whereas glutamine and glycine did not influence biofilm formation, C. jejuni grown in the presence of 50 mM fumarate visibly favoured growth in the planktonic fraction, with a twofold increase in the ratio of planktonic (OD600) to biofilm (A570) bacteria. In contrast, the C2-dicarboxylate pyruvate had no effect on biofilm formation (data not shown).
Fig. 9. C. jejuni biofilm formation is inhibited by fumarate and enhanced by deoxycholate (DOC).
A. Fumarate promotes planktonic growth of C. jejuni. Biofilms were grown in MH broth or MH broth supplemented with 50 mM fumarate, glycine or glutamine. After 2 days of microaerobic incubation, samples were carefully removed from the media (planktonic) fraction to measure OD600, and biofilms were then quantified by crystal violet staining. Shown is an average of the planktonic (OD600) to biofilm (A570) ratio for each strain.
B. DOC enhances biofilm formation in C. jejuni. Biofilms were grown in MH broth supplemented with sub-MIC levels of various detergents. Concentrations used were as follows: DOC, 0.05%; Triton X-100 (Tx-100), 0.0005%; Tween-20 (Tw-20), 0.002%; and sodium dodecyl sulphate (SDS), 0.00025%.
C. DOC-induced biofilms exhibit normal architecture and fibre formation. WT (shown) and ΔcprS (not shown) biofilms were grown on glass coverslips in MH + 0.05% DOC, stained with propidium iodide, and visualized by microscopy at 100× magnification. The bar represents ~40 μm.
D. WT biofilms grown in the presence of DOC are visibly disrupted by DNase I treatment. WT (shown) and ΔcprS (not shown) biofilms were grown for 2 days in MH + 0.05% DOC and either left untreated or rinsed with PBS, then incubated for 3 h in buffer alone or buffer with DNase I, followed by crystal violet staining.
Finally, because of the expression changes in surface proteins in ΔcprS, we tested the effect of various detergents on biofilm formation (Fig. 9B). Biofilms were grown in the presence of sub-MIC levels of the bile salt deoxycho-late (DOC), Triton X-100, Tween-20 and sodium dodecyl sulphate (SDS). Interestingly, DOC significantly increased biofilm formation, causing WT bacteria to make biofilms at levels similar to ΔcprS. In contrast, the other detergents had no effect. No differences were observed in MICs between strains for each of these compounds (data not shown). Propidium iodide staining of WT (Fig. 9C) and ΔcprS (not shown) biofilms formed in the presence of DOC showed that they exhibit normal biofilm architecture and form an extensive fibrous network, consistent with observations shown in Fig. 4C. No fibres were observed by propidium iodide staining of coverslips incubated overnight in MH broth with DOC alone (data not shown). WT biofilms formed in the presence of DOC were also visibly and quantitatively disrupted with DNase I (Fig. 9D; P < 0.005 for DNase I-treated versus buffer-treated biofilms), with A570 values for all samples similar to those shown in Fig. 4D for ΔcprS biofilms. Together, these data indicate that DOC enhances biofilm formation in C. jejuni and provides further evidence for DNA as a component of the C. jejuni biofilm matrix.
Discussion
In C. jejuni, as in other zoonotic bacteria, comprehensive shifts in physiology may be required to adapt to either a transmission or host environment. The capacity for gene regulation in C. jejuni is limited; nonetheless, its prevalence suggests it must harbour mechanisms that allow such changes. Here we report the characterization of the C. jejuni CprS sensor kinase, which may influence such aspects of C. jejuni pathogenesis through control of biofilm or planktonic growth and modulation of essential biological functions. This work has also identified new paradigms for biofilm formation in C. jejuni.
Our preliminary genetic analyses indicated that the activity of the CprR RR, but not the CprS SK, was essential for viability of C. jejuni in the laboratory. It is possible that CprR mutants enter into a viable but non-culturable state and are not recoverable by colony isolation except under specific unknown conditions. Nonetheless, it is intriguing that only the RR appears to be required for viability. The predicted operonic structure of cprRS suggests a functional relationship, and while phosphotransfer could not be directly shown due to insolubility of CprS in vitro (S. Svensson and E. Gaynor, unpubl. obs.), the C-terminal domain of CprS interacted specifically with CprR by two-hybrid analysis. Based on work in other systems (Kulasekara et al., 2005) this interaction strongly supports a functional relationship between the two components.
The essential nature of CprR suggests that CprRS might control global aspects of physiology central to the biology of C. jejuni, similar to the essential Caulobacter RR CtrA (Quon et al., 1996) or the YycF RR in Bacillus subtilis (Szurmant et al., 2005). Consistent with control of essential processes, we observed apparent growth defects for ΔcprS in rich broth, and our data also suggest that CprS may influence global changes such as the biofilm-planktonic switch. The essential nature of only the RR is intriguing, but can be explained by a variety of scenarios. For instance, cross-talk between TCRSs has been demonstrated in the absence of the cognate SK in other bacteria (Verhamme et al., 2002). This may be especially significant for a TCRS (such as CprRS) that regulates genes required for viability, where non-cognate SK(s) may effect phosphotransfer to the RR when the native cognate SK is absent. Furthermore, acetyl phosphate can also phosphorylate RRs in E. coli (McCleary and Stock, 1994), and in C. jejuni, this may allow basal levels of RR phosphorylation that permit viability. Alternatively, phosphotransfer may not be required for CprR to modulate essential genes, similar to the H. pylori essential RRs HP1021 and HP1043 (Muller et al., 2007). H. pylori also encodes a TCRS, HP165-HP166, where like CprRS, only the RR is essential for growth (Schar et al., 2005). In this system, there are two species of RR-controlled promoters – one recognized by HP166 (which presumably regulates essential genes) and a second, lower-affinity class of promoters bound by phospho-HP166 (which activates dispensable genes) only when environmental conditions stimulate RR phosphorylation. In a bacterium with limited regulatory capacity such as C. jejuni, this scenario is especially attractive, as it would allow different forms of the same RR to be devoted to separate regulons.
There is debate as to whether the biofilm lifestyle is the default lifestyle for bacteria, or whether planktonic growth is simply an artefact of laboratory culture. The hyperbiofilm phenotype of ΔcprS suggests that this phenomenon is a regulated process, and this work suggests that CprS may play a role in its regulation. Numerous TCRSs have been implicated in regulation of biofilm formation in other pathogens through control of phenomena such as adhesin expression (Kulasekara et al., 2005), exopolysaccharide production (Hussa et al., 2008) and quorum sensing (Dong et al., 2008). Direct involvement of a TCRS in C. jejuni biofilms is supported by observations of defective biofilm formation upon inactivation of Cj0688, encoding a likely phosphate acetyltransferase (Joshua et al., 2006) which may affect RR phosphorylation. Furthermore, the C. jejuni orphan RR CbrR contains a putative GGDEF domain (Raphael et al., 2005) which may be involved in production of c-di-GMP, a mediator of biofilm formation in other pathogens (Romling and Amikam, 2006). A role for CbrR in control of C. jejuni biofilm phenotypes has not been demonstrated; however, the CprS SK is an appealing candidate cognate SK for this RR. Finally, downregulation of the Cj0355c RR, which is also essential (Raphael et al., 2005; J.A. Fields and S.A. Thompson, unpublished), in the 81-176 ΔcprS mutant, while in apparent contrast to previous work where Cj0355c was upregulated in C. jejuni NCTC 11168 biofilms (Kalmokoff et al., 2006), is consistent with CprRS affecting transcription of and thereby interacting with other regulators of both key biological processes and biofilm formation.
Mechanistic insight into C. jejuni biofilm formation is relatively limited, and we hypothesized that identifying specific physiological changes in the hyperbiofilm-forming ΔcprS mutant could be used to understand C. jejuni biofilms in more detail. Biofilm bacteria typically secrete an extracellular matrix, often composed of polysaccharides (Branda et al., 2005). While H. pylori secretes an exopolysaccharide during biofilm growth (Stark et al., 1999) and other enteric pathogens such as Salmonella and E. coli also utilize carbohydrates extensively (Romling, 2005), the matrix surrounding C. jejuni biofilms remains ill-defined. A ΔspoT stringent response mutant overproduces a CFW-reactive exopolysaccharide, production of which correlates with biofilm formation (McLennan et al., 2007). In contrast, analyses of LOS profile, capsule production and CFW reactivity of ΔcprS revealed no differences from WT. Furthermore, analysis of the C. jejuni genome did not identify homologues of biofilm-associated proteins such as Bap or Esp (Cucarella et al., 2001; Toledo-Arana et al., 2001).
A component of the biofilm matrix in other bacteria is extracellular DNA (Whitchurch et al., 2002). We observed fibres extending between microcolonies in C. jejuni biofilms stained with crystal violet or propidium iodide, both of which stain DNA. The presence of DNA in the C. jejuni biofilm matrix was further supported by our observations that the extensive biofilms formed by ΔcprS could be visibly disrupted by treatment with DNase I, and that WT (and ΔcprS) biofilms formed in the presence of DOC likewise exhibited striking DNase I sensitivity. Interestingly, strain 81-176 forms more robust biofilms than strain 11168 (S. Svensson and E. Gaynor, unpublished) and also harbours a plasmid (pVIR) containing components of a putative type IV secretion system (T4SS) not found in strain 11168 (Bacon et al., 2000). It will be interesting, in future work, to explore a possible involvement of this plasmid in secretion of DNA to the biofilm matrix.
Because our interest in CprRS was initially piqued by its identification in a cell infection screen, we hypothesized that CprRS may control phenotypes specifically related to pathogenesis. In spite of several growth- and stress-related defects in vitro, the ΔcprS mutant showed no obvious defect in epithelial cell invasion. Interestingly, however, we consistently recovered more ΔcprS than WT bacteria from INT407 cells following several hours of intra-cellular survival. It remains to be seen whether this is due to increased bacterial or epithelial cell survival, either of which could result in enhanced recovery of intracellular bacteria. Nonetheless, to the best of our knowledge, this is the first report of a C. jejuni mutant exhibiting enhanced longer-term intracellular recovery, particularly when coupled with no invasion differences from WT. It has been reported that exposure of certain clinical strains of C. jejuni to an aerobic atmosphere prior to infection of epithelial cells in vitro enhances intracellular survival (Mihaljevic et al., 2007). In light of our proteomics data demonstrating upregulation of oxidative stress tolerance proteins such as AhpC and KatA in ΔcprS, the stress resulting from disregulation of essential processes in this mutant may ‘prime’ the bacteria for survival within the stressful intraepithelial cell environment.
In contrast to the in vitro infection data, the ΔcprS mutant was significantly impaired for chick colonization, consistent with several other C. jejuni TCRS mutants (Bras et al., 1999; MacKichan et al., 2004; Wosten et al., 2004; 2006; Raphael et al., 2005). Inactivation of ppk1 in C. jejuni also results in a colonization defect that, like ΔcprS, is rescued by increasing the inoculating dose. A correlative dose-dependent increase in biofilm formation for the ppk1 mutant was proposed as potentially responsible for restoration of WT colonization levels at higher doses (Candon et al., 2007). In addition, a proteomics comparison of robust and poor chicken-colonizing strains of C. jejuni suggested that many expression trends in the robust colonizer mirrored those previously identified in biofilm- or agar-grown bacteria (Seal et al., 2007). Together, this suggests that at higher infective doses, the enhanced biofilm forming ability of ΔcprS may allow the mutant to overcome planktonic sensitivities (i.e. stress survival defects) that may occur with disregulation of essential genes.
We took a global proteomics-based approach to identify protein expression changes that might help explain the diverse pathogenesis-related phenotypes observed for ΔcprS. This identified approximately 20 differentially expressed proteins in the ΔcprS mutant compared with WT, with the vast majority of these proteins present at WT levels in ΔcprS C. We detected differences in proteins involved in several distinct aspects of physiology, including oxidative stress tolerance, metabolism and cell surface characteristics, as well as regulatory proteins. Many expression changes correlated well with previous biofilms studies. For instance, increased MOMP, FlaA, Cj0998 and succinyl-CoA synthetase expression was consistent with proteomics analyses of C. jejuni peb4 mutants, which are defective for biofilm formation and have decreased expression of these proteins (Asakura et al., 2007), and those of agar-grown (sessile) C. jejuni, which also identified succinyl-CoA synthetase and trigger factor as upregulated (Sampathkumar et al., 2006). Increased expression of TrxB (Asakura et al., 2007), AhpC and FlaA in ΔcprS is also consistent with proteomics analyses of WT C. jejuni biofilms (Kalmokoff et al., 2006). MOMP and FlaA also serve as adhesins in C. jejuni and therefore have relevance to biofilm formation (Moser et al., 1997), and previous work suggests that unlike other bacteria such as P. aeruginosa, biofilm-residing C. jejuni maintain their flagella (Kalmokoff et al., 2006). In contrast, downregulation of LuxS in ΔcprS was a surprising finding. While a role for AI-2 in quorum sensing in C. jejuni has not been demonstrated, it has been reported that C. jejuni luxS mutants exhibit decreased motility, autoagglutination (Jeon et al., 2003) and biofilm formation (Reeser et al., 2007), which we have also noted (S. Svensson and E. Gaynor, unpublished). Our expression profiling thus supports an AI-2-independent mechanism for the biofilm phenotype of ΔcprS.
Altered expression of metabolic proteins is consistent with several of the phenotypes associated with deletion of cprS. For instance, the essential nature of many metabolic genes makes them ideal candidates for regulation by an essential RR, and proteins representing of a number of metabolic pathways such as glycolysis (Fba), the trichloroacetic acid (TCA) cycle (FumC, SucD), fatty acid bio-synthesis (FabI), amino acid metabolism and biosynthesis (Asd), and protein synthesis (elongation factors, tRNA synthetase, ribosomal protein S1) were expressed differently in ΔcprS. Some of the metabolic differences observed might also reflect shifts that occur when C. jejuni enters or exits a biofilm. Likewise, metabolic alterations may help explain the modest increase in recovery of ΔcprS from within INT407 cells, as C. jejuni intracellular survival has recently been shown to involve adaptation to the likely very low-oxygen CCV (Watson and Galan, 2008) which is also likely to be a nutrient-poor environment (Gaynor et al., 2005; Candon et al., 2007). Consistent with this, microarray analyses suggest that C. jejuni undergoes global expression changes in numerous metabolic genes during cell infection (E. Gaynor, unpublished), and real-time quantitative PCR data suggest that expression of dcuA, encoding a likely C4-dicarboxylate transporter, is reduced 10-fold in ΔcprS (S. Svensson and E. Gaynor, unpublished). Finally, distinct metabolic pathways are initiated within the avian gastrointestinal tract (Woodall et al., 2005); thus the metabolic changes in ΔcprS may also account for its decreased chick colonization.
Importantly, the proteomics analysis also led us to perform additional experiments which provided further insight not only into the colonization and biofilm phenotypes of ΔcprS but also into factors affecting biofilm formation of WT C. jejuni. For instance, elevated MOMP porin expression in ΔcprS led us to hypothesize that outer membrane permeability may be altered, consistent with subsequent observations that ΔcprS exhibited a striking osmotolerance defect. Interestingly, we also observed a slight increase in sensitivity to oxidative stress agents such as t-butylhydroperoxide. This appears to contrast the observation that C. jejuni residing in biofilms are more resistant to oxidative stress than their planktonic counterparts (Trachoo and Frank, 2002) as well as our proteomics data, where numerous proteins involved in oxidative stress tolerance were upregulated in ΔcprS. However, it is possible that upregulation of oxidative stress proteins may be the result of a general stress response to disregulation of essential genes. Alternatively, increased levels of MOMP may increase outer membrane permeability to oxidative agents, resulting in increased oxidative stress sensitivity and upregulation of stress response proteins. It is nonetheless interesting to hypothesize that the increased osmotic and oxidative stress susceptibilities of ΔcprS may in part account for its chick colonization defect.
FlaA was also present at higher levels in the mutant strain, which was supported by our observation of a slight increase in motility in ΔcprS compared with WT. As noted, the flagellar filament is important for C. jejuni bioflm formation, and while a role for motility per se (i.e. via analysis of a filament-positive, motility-minus mutant) has not yet been proven for C. jejuni, it is not unreasonable to predict, based on work in other bacteria, that motility will prove important for C. jejuni biofilm formation as well. The increased expression of FlaA in ΔcprS is also consistent with our observation of an increased amount and number of protein species in the media fraction of ΔcprS cultures. As noted above, C. jejuni lacks dedicated type III secretion systems; however, secretion of C. jejuni virulence factors called Cia (Campylobacter invasion antigen) proteins is dependent on a functional flagellar apparatus (Konkel et al., 2004). At present, we cannot rule out the possibility that the appearance of proteins in the ΔcprS media fraction may represent loss of membrane integrity; however, there are several striking dissimilarities between the pattern of media versus periplasmic or cytoplasmic proteins, suggesting some specificity in the proteins present in the ΔcprS media. Future studies to identify these proteins will allow us to explore the connection between this phenotype and those involving biofilms and host-related properties.
Interestingly, Cia protein expression and secretion is stimulated by 0.05% DOC (Malik-Kale et al., 2008). We found that a similar concentration of DOC stimulated ΔcprS-level biofilm formation in WT C. jejuni. DOC also promotes biofilm formation in other enteric pathogens such as Vibrio cholerae (Hung et al., 2006). Thus in addition to the DNA component mentioned above, it is possible that the synthesis and/or secretion of proteins present in the media fraction of ΔcprS that contribute to biofilm formation are also induced by DOC, accounting at least in part for increased biofilm formation of WT C. jejuni in the presence of sub-MIC levels of DOC.
Our observation that numerous metabolic genes were altered in ΔcprS led us to hypothesize that nutrient availability may influence C. jejuni biofilm formation. Assays investigating the effect of a variety of carbon sources on WT biofilms demonstrated that the bacteria favoured planktonic growth when fumarate was added to the media. This correlates with observations in other bacteria where nutrient availability can trigger biofilm dispersal (Gjermansen et al., 2005). For example, control of swarming motility and biofilm architecture by quorum sensing in P. aeruginosa is nutritionally conditional (Shrout et al., 2006). Likewise, in Shewanella, different electron acceptors (i.e. nitrate versus fumarate) result in changes in carbohydrate exopolymer production and surface-associated behaviour (Neal et al., 2007). In C. jejuni, biofilm formation also appears to be promoted by differences in nutrient availability (Reeser et al., 2007). It is conceivable that CprRS allows C. jejuni to respond to nutrient availability cues by inducing and/or repressing genes related to biofilm formation.
In summary, our genetic analysis of a TCRS in C. jejuni, within the confines of an essential RR, has identified complex phenotypic and protein expression changes which point to a role for CprRS in controlling changes in physiology and metabolism involved in biofilm dynamics, stress tolerance and pathogenesis-related phenotypes. Strong conservation of CprRS in only the Campylobacteraceae suggests that CprRS controls campylobacterspecific phenomena, many of which may be shared by related, and even more understudied, pathogens that cause severe enteric disease (i.e. C. coli, C. upsaliensis and A. butzleri) and spontaneous abortion in livestock (C. fetus). Absence of CprRS from H. pylori, essentially host-restricted, is consistent with control of phenotypes relating to passage between hosts and/or phenotypes relating specifically to adaptation to an avian host. Our data also contribute to the understanding of other pathogens that use different paradigms from model bacteria. For instance, while genes involved in processes such as stress tolerance and biofilm formation may not fit the definition of virulence factors set out in molecular Koch's postulates (Falkow, 2004), they may allow survival of an apparently fastidious pathogen such as C. jejuni within a commensal reservoir, between hosts during transmission, or persistence within a susceptible host long enough to cause significant inflammation. The diverse phenotypes displayed by ΔcprS suggest that this TCRS is pleiotropic and likely controls many phenomena in C. jejuni, providing intriguing hypotheses for further testing.
Experimental procedures
Bacterial strains and growth conditions
Studies were performed using the C. jejuni WT strain 81-176, a highly invasive isolate from a raw milk outbreak (Korlath et al., 1985). All strains are listed in Table S2. C. jejuni was routinely cultured in Mueller–Hinton broth (MH; Oxoid, Hampshire, England) or agar plates. All incubations were performed at 37°C under microaerobic conditions (6% O2, 12% CO2) in a Heraeus tri-gas incubator (plates and biofilms) or generated using the Oxoid CampyGen system (broth cultures). Media used to culture C. jejuni was supplemented with 10 μg ml−1 vancomycin and 5 μg ml−1 trimethoprim. Where appropriate, the selective antibiotics (Sigma, Oakville, ON) kanamycin and chloramphenicol were added to a final concentration of 40 μg ml−1 and 25 μg ml−1 respectively. C. jejuni-selective MH agar plates were used for growth of C. jejuni for the chick colonization studies (see below). E. coli strain DH5α was used for recombinant DNA manipulations and was routinely cultured in LB broth (Sigma) with antibiotics added at the following concentrations: ampicillin, 100 μg ml−1, kanamycin 50 μg ml−1, chloramphenicol, 25 μg ml−1, and tetracycline, 20 μg ml−1.
General recombinant DNA techniques
Recombinant DNA techniques were performed according to Sambrook and Russell (2001). Restriction enzymes and DNA modifying enzymes were purchased from New England Biolabs (Mississauga, ON) or Invitrogen (Burlington, ON). Plasmids were isolated from bacteria using the Qiagen Qiaprep Spin miniprep kit (Qiagen, Mississauga, ON). Primers are listed in Table S3.
Targeted mutagenesis of C. jejuni
Targeted deletion mutagenesis of C. jejuni was achieved by double-cross-over homologous recombination with constructs prepared in a C. jejuni suicide vector containing approximately 500 base pairs of homologous DNA of the target gene region flanking either a cat (chloramphenicol acetyltransferase, CmR) or aph-3 (aminoglycoside 3′-phosphotransferase, KmR) cassette (Ounissi et al., 1990; Yao et al., 1993), with the majority of coding sequences removed by inverse PCR. Unless otherwise stated, experiments were performed using a cprS::cat strain (ΔcprS) constructed by replacing the majority of cprS coding sequence with a CmR marker, as follows. Briefly, the entire coding sequence of cprS, along with approximately 500 base pairs of flanking sequence on each side, was amplified by PCR using primers cprS-TOPO FWD/REV from genomic DNA isolated from C. jejuni 81-176 (DNeasy Kit, Qiagen), and this approximately 2300-base-pair product was ligated directly into the pCR2.1-Topo vector (Invitrogen). Inverse PCR using primers cprS-INV FWD/REV was then used to remove the majority of the cprS gene and engineer MfeI sites. A cat cassette was excised from plasmid pRY109 (Yao et al., 1993) using EcoRI and ligated into the MfeI-digested inverse PCR product. The resulting construct, containing approximately 86 and 137 base pairs of 5′ and 3′ coding sequence of cprS, was then used to naturally transform C. jejuni 81-176, and CmR recombinants were recovered and confirmed to be ΔcprS by PCR. A cprS::aph-3 strain (ΔcprS::KmR) was also constructed in a similar manner to the cat strain, using the primers cprS-pGEM FWD/REV and cprS-INV2 FWD/REV.
Complementation of ΔcprS
Insertion of copies of cprS for complementation was achieved using pRRK [J. Ketley, unpublished, based on pRRC integration vectors (Karlyshev and Wren, 2005)]. The coding region of cprS was amplified by PCR using primers cprS-pRRC FWD/REV, which introduced 5′-XbaI sites and 3′-MfeI sites, as well as the native ribosome binding site of cprR (Wosten et al., 1998). Following digestion with MfeI and XbaI, this product was ligated into the pRRK vector. The resulting construct was naturally transformed into C. jejuni ΔcprS::KmR, and putative ΔcprSC complemented clones were recovered on plates containing kanamycin. Single insertions of cprS in the rRNA spacer region adjacent to cj0029 were confirmed by PCR using primers ak233, ak234, ak235 and PKanF.
Bacterial two-hybrid analysis
Protein–protein interactions were determined using the Bacteriomatch II Two-hybrid System (Stratagene, La Jolla, CA). Coding regions for CprR and DccR were amplified by PCR using primers BT-cprR FWD/REV or BT-dccR FWD/REV, digested with BamHI and XhoI, and ligated into similarly digested pBT. The C-terminal domain (amino acids 187–415) of CprS was amplified by PCR using primers TRG-cprS-CTD FWD/REV, digested with BamHI and SpeI, and ligated into the similarly digested plasmid pTRG. Electrocompetent E. coli Bacteriomatch II cells were co-transformed with 50 ng of each bait and prey plasmid, followed by recovery for 90 min in LB broth at 37°C. Cells were then washed with M9+ His dropout broth and conditioned for 2 h in this medium at 37°C prior to plating dilutions on Non-selective and Selective (3-AT) plates.
RNA extraction, cDNA synthesis and quantification
RNA was extracted from mid-log-phase broth cultures (OD600 0.2–0.5) or overnight cultures on MH agar as previously described (Gaynor et al., 2004). Briefly, 1/10 volume of 10× Stop solution (5% buffer-saturated phenol in ethanol) was added to 0.5 OD600 of bacteria, and cells were collected by centrifugation at 10 000 g for 5 min at room temperature followed by immediate freezing at −80°C. Cells were thawed at room temperature, re-suspended in 50 μl of 0.4 mg ml−1 lysozyme (Sigma) in 10 mM Tris 1 mM EDTA, pH 8.0 and incubated at room temperature for 5 min. Cells were then lysed by addition of 950 μl of Tri-reagent (Sigma) and vortexing for 1 min, followed by the addition of 200 μl of chloroform. After centrifugation at 14 000 g for 15 min at 4°C, the top phase was transferred to a new tube. An equal volume of 70% ethanol was added dropwise, followed by application to an RNeasy column and washing according to the manufacturer's instructions. Samples were digested using the Qiagen on-column RNase-free DNase kit according to the manufacturer's instructions, and RNA was eluted in 30 μl of RNase-free water. cDNA was generated from the above preparation of total RNA using SSII enzyme (Invitrogen) and cleaned up using the Qiagen Qiaquick PCR purification kit. Concentrations of DNA and RNA were quantified using a NanoDrop ND-1000 spectrophotometer (Wilmington, DE), according to the manufacturer's instructions, and RNA quality was assessed by electrophoresis on 1% agarose TAE gels. Absence of genomic DNA within RNA samples was confirmed by PCR.
Transcript analysis
Independent transcription of cprR and the upstream htrA gene was achieved using cDNA extracted as described above. PCR reactions with Taq DNA polymerase using primer sets htrA-3′/cprR-BT REV and cprR BT FWD/REV were performed using cDNA or genomic DNA from WT C. jejuni 81-176, and the presence of amplicons was determined by agarose gel electrophoresis.
Growth curve assays
Campylobacter jejuni strains were grown in MH broth overnight to mid-log phase then diluted to an OD600 of 0.1 in MH broth. Cultures were incubated microaerobically at 37°C with shaking at 200 r.p.m. Growth and viability were assessed at various times post dilution by measuring the OD600 and plating serial 10-fold dilutions on MH agar.
Biofilm assays and broth culture tube staining
Biofilm formation was assessed as previously described (O'Toole and Kolter, 1998; McLennan et al., 2007) with modifications. Briefly, overnight broth cultures of C. jejuni strains were diluted to an OD600 of 0.002 in fresh media, and 1 ml was added to borosilicate glass tubes. Tubes were incubated without agitation in a Trigas incubator under microaerobic conditions for 1 or 2 days, followed by staining for 10 min by addition of 250 μl of 1% crystal violet (Sigma) dissolved in 100% ethanol. Tubes were then rinsed with distilled water and dried, followed by photographic documentation and/or quantification of adhered crystal violet by dissolving with 30% methanol/10% acetic acid and measuring the absorbance at 570 nm. Where indicated in figure legend 9, chemicals were included in the MH broth of suspensions introduced into borosilicate tubes prior to initiation of biofilm growth. Shaking broth culture tubes were stained with crystal violet as above.
Bright field and scanning electron microscopy of biofilms
Campylobacter jejuni biofilms were grown in borosilicate tubes as described above, with a glass coverslip standing upright in the culture. After 48 h of incubation, coverslips were either stained with crystal violet as described above, stained with PI using the LIVE/DEAD BacLight Bacterial Viability Kit according to the manufacturer's instructions, or removed and processed for SEM as follows. The coverslip was removed and gently rinsed once in 0.1 M cacodylate buffer, and the biofilms were fixed (2.5% glutaraldehyde in 0.1 M cacodylate) for 1 h. Cover glasses containing the biofilms were processed and visualized using SEM.
Carbohydrate analyses
Campylobacter jejuni strains were grown on MH agar for 48 h microaerobically and prepared based on the method of Hitchcock and Brown (1983). Briefly, bacteria were harvested with PBS and diluted to an OD600 of 10 in 100 μl. An equal volume of 2× lysis buffer [4% SDS, 8% β-mercaptoethanol, 20% glycerol, 0.125 M Tris (pH 6.8), 0.025% bromophenol blue] was added to the cell suspension, which was then heated at 95°C for10 min. After the samples cooled to room temperature, 10 μl of 10 mg ml−1 proteinase K was added and samples were incubated overnight at 37°C or 55°C. Samples were then incubated at 65°C for 1 h or 95°C for 5 min and separated by SDS-PAGE and silver-stained as previously described (Tsai and Frasch, 1982) or electroblotted to PVDF membrane for Western blotting with an α-O36 serotype antibody (a gift from Kris Rahn, Public Health Agency Canada Laboratory for Foodborne Zoonoses, Guelph, ON). Assessment of CFW reactivity has been described elsewhere (McLennan et al., 2007). Briefly, overnight cultures of each strain were diluted to similar OD600 and spotted on BHI agar containing 0.002% CFW (Sigma).
DNase I treatment of biofilms
Biofilms were grown as described above. Following 2 days of incubation, the medium was removed by pipetting, and tubes were washed with 1.5 ml of PBS, pH 7.4 (Invitrogen). DNase I buffer alone, or buffer containing DNase I (Fermentas) at 4 U ml−1 in PBS was added to each tube, followed by incubation for 3 h at 38°C under microaerophilic conditions and staining with crystal violet as described above. Bacterial viability under each condition was also assessed by quantifying cfu of each strain in an OD600 0.02 suspension in each condition after 3 h of incubation.
Adherence, invasion and intracellular survival in INT407 cells
In vitro gentamicin protection assays were performed essentially as described previously (Gaynor et al., 2005). Briefly, INT407 cells in Minimal Essential Medium (MEM) with 15% fetal bovine serum (FBS) (Invitrogen) were seeded into 24-well plates 16 h before infection (~105 cells per well) and incubated in a humidified 5% CO2 atmosphere. C. jejuni strains were harvested from healthy MH plates and inoculated into MH biphasic tubes to an OD600 of 0.002. After approximately 16 h of growth, bacteria were washed two times with MEM, and 1 ml of bacteria in MEM without FBS was used to infect INT407 cells, which had been washed once with MEM without FBS before infection, at a multiplicity of infection of ~100 (~107 bacteria per well). Infections were carried out in a 5% CO2 incubator at 37°C. Survival in media above cells and the number of adhered and/or invaded bacteria (‘adherence’ time point) were assayed after 3 h; gentamicin treatment (150 μg ml−1) was initiated at this point. Two hours following initiation of gentamicin treatment, invaded/intracellular bacteria were harvested by lysing the INT407 cells with sterile distilled water and a 27-gauge syringe and viability of the bacteria was assessed by plating serial dilutions on MH agar (‘invasion’ time point). Intracellular survival was tested by replacing the medium with MEM with 10 μg ml−1 gentamicin to prevent growth of bacteria released from lysing INT407 cells, followed by further incubation for 5 h and harvesting of intra-cellular bacteria as in above (‘intracellular survival’ time point). Experiments were performed in triplicate.
Chick colonization assays
The chick colonization assay was performed as previously described (Hendrixson and DiRita, 2004). Briefly, day-of-hatch chicks (white-leghorn, Charles River Laboratories) were orally inoculated with 102,104 or 106 cfu C. jejuni diluted in PBS. The C. jejuni strains were grown on MH agar containing 10 μg ml−1 trimethoprim for 18 h in a trigas incubator at 37°C. After 6 days, chicks were euthanized, and their caeca removed. Caecal contents were weighed, diluted, and plated onto MH agar containing 10 μg ml−1 trimethoprim and 30 μg ml−1 cefoperazone. C. jejuni colonies were counted, cfu g−1 caecal matter recorded and log cfu g−1 reported. To quantify the average recovered bacteria from each infected group, chicks harbouring fewer cfu than the detection limit were assigned a log cfu g−1 value of 2 (i.e. 100 cfu g−1, the limit of detection).
Proteomic comparison of WT, ΔcprS and ΔcprSC
Proteome analysis was performed using Differential In-Gel Electrophoresis (DIGE) technology (GE Biosystems, Piscataway, NJ). Briefly, C. jejuni WT, ΔcprS and ΔcprSC cells were grown to mid-log phase at 37°C in MH broth. Protein lysates were prepared by washing cells three times in wash buffer (10 mM Tris, pH 8.0, 5 mM magnesium acetate) at 4°C for 4 min at 12 000 g. One millilitre of lysis buffer [8 M urea, 30 mM Tris, pH 8.5, 5 mM magnesium acetate, 4% (w/v) CHAPS] was used to re-suspend the pellet, and the suspension incubated for 30 min on ice, then sonicated for six 10 s bursts (Model 100 Sonic Dismembrator, Fisher Scientific). Insoluble material was removed by centrifugation at 4°C for 10 min at 12 000 g and the soluble component used for further studies. Protein concentrations were determined using a BCA assay kit (Pierce, Rockford, IL). Protein samples from WT, ΔcprS and ΔcprSC were labelled individually with Cy2, Cy3 and Cy5 dyes according to the protocol supplied by the manufacturer (GE Biosystems). Briefly, 25 mg of each protein sample (WT, ΔcprS and ΔcprSC) were labelled at lysine residues with 1 ml Cy2, Cy3 and Cy5 dye conjugates respectively (for 10 min in the dark). The reactions were stopped by addition of 1 mM lysine. The Cy2-, Cy3- and Cy5-labelled proteins were then mixed with an equal amount of unlabelled protein, and finally all proteins mixed together (total of 150 mg of proteins in a single mixture). The protein mixture was subjected to isoelectric focusing (IEF) using IPGPhor IEF strips (range of 3–10, non-linear). After IEF, the strip was rinsed in equilibration buffer (6 M urea, 10 mM Tris, pH 6.8, 30% glycerol, 1% SDS), and then placed onto a 12% SDS-polyacrylamide gel for second dimension separation.
Following separation, the gel was scanned on a Typhoon fluorescent scanner (GE Biosystems), at the following wavelengths: Cy2, 488 nm excitation, 520 nm emission; Cy3, 532 nm excitation, 580 nm emission; Cy5, 633 nm excitation, 670 nm emission. Images were overlaid and analysed with Decyder Differential In-Gel Analysis (DIA) software (version 4.0, GE Biosystems) for pairwise identification of proteins with higher expression in pairs of the three strains. Proteins were designated as having statistically significant expression differences among strains if the abundance of an individual protein spot was more than two standard deviations greater or lesser than the mean variance of the abundance of all cellular proteins.
Differentially expressed proteins were excised, digested with trypsin (Invitrogen), and tryptic peptides were analysed using a matrix-associated laser desorption ionization – time-of-flight/time-of-flight (MALDI-ToF/ToF) spectrometer (Applied Biosystems, Foster City, CA). Protein identifications resulted from querying protein databases with both tryptic fingerprint data as well as primary amino acid sequence of peptides following collision-induced fragmentation and MS/MS.
In vitro stress tolerance and phenotyping assays
Growth under hyperosmotic stress was assessed by observing growth of serial 10-fold dilutions of overnight cultures on MH agar supplemented with 1% NaCl (Sigma). Oxidative stress tolerance was assessed by suspending each strain at an optical density of 0.02 in MH broth, followed by the addition of t-butylhydroperoxide (Sigma) to a final concentration of 0.1 mM. Following 30 min of incubation at 37°C microaerobically, samples of each strain were serially diluted and plated on MH plates for determination of cfu. Motility was determined by inspection of wet mounts by microscopy or measuring the halo of growth, after 24 h of microaerobic incubation, surrounding the point of inoculation of equal numbers (as determined by OD600) of bacteria into MH plates containing 0.4% agar.
Subcellular fractionation and analysis of secreted proteins
Culture supernatants (Poly et al., 2007) and subcellular fractions (Leon-Kempis Mdel et al., 2006) were prepared essentially as described previously. Each strain of C. jejuni was grown microaerobically overnight in 100 ml of MH broth and cells were collected by centrifugation at 4800 g for 30 min at 4°C. The supernatant was filtered through a 0.22 μM filter (Millipore, Bellerica, MA), proteins were precipitated by the addition of TCA (Sigma) to a final concentration of 10% and incubation at 4°C overnight, and precipitated material was collected by centrifugation at 9800 g for 1 h. Prior to loading, the sample was washed 2× with acetone and then re-suspended in 100 μl of SDS-PAGE sample buffer. The cell pellet was re-suspended in 10 μl of ST buffer (20% sucrose, 20 mM Tris-HCl, pH 8.0) and EDTA was added to a final concentration of 1 mM. Following gentle agitation for 10 min at room temperature, cells were collected by centrifugation at 8000 g for 10 min and re-suspended in 5 ml of ice-cold 10 mM Tris, pH 7.5 and incubated on ice for 10 min. The supernatant, containing periplasmic proteins, was collected by centrifugation at 15 000 g for 15 min at 4°C and then proteins were precipitated with TCA as described above. The cell pellet was then suspended in 2.5 ml of ice-cold 10 mM Tris, pH 8.0, sonicated for 3 × 30 s, and then subjected to centrifugation for 10 min at 13 000 g. The pellet was discarded, and the supernatant was subjected to centrifugation on a Beckman Optima L-90K Ultracentrifuge at 100 000 g to collect cell membranes. The supernatant was saved as the cytoplasmic fraction, and the pellet was washed and re-suspended in 100 μl of 10 mM HEPES, pH 7.4, followed by the addition of an equal volume of 2% sarcosine (Sigma) in HEPES buffer. After incubation at room temperature for 30 min to solubilize inner membranes, the insoluble outer membranes were collected by centrifugation at 15 600 g for 30 min at 4°C and re-suspended in 100 μl of HEPES buffer. Equal volumes of each fraction were separated by SDS-PAGE followed by silver staining.
Growth of biofilms in the presence of carbon sources and antimicrobial agents
To test carbon sources, biofilms were grown as in above in MH broth or MH broth supplemented with 50 mM fumarate, glycine or glutamine. After 2 days of microaerobic incubation, samples were carefully removed from the media (planktonic) fraction to measure OD600, and biofilms were then quantified by crystal violet staining. Shown is an average of the planktonic (OD600) to biofilm (A570) ratio for each strain. To test the effect of antimicrobial compounds, biofilms were again grown as in above in MH broth or MH broth supplemented with the following concentrations of each compound: DOC, 0.05%; Triton X-100, 0.0005%; Tween-20, 0.002%; and SDS, 0.00025%.
Supplementary Material
Acknowledgements
We gratefully acknowledge the technical assistance of C. Reamer and the VIDO animal care unit. We also thank Lindsay Wilson and Rosie Redfield for assistance with the bacterial two-hybrid studies, the UBC Bioimaging Facility for assistance with electron microscopy, Chris Chang for mouse infections and members of the Gaynor lab for helpful discussions. We are particularly grateful to Vic DiRita for supporting chick infection studies. M.P. and S.A.T. were supported by US National Institutes of Health Grants AI055715, AI058284 and AI061026 (to S.A.T.). S.L.S. is supported by a Natural Sciences and Engineering Research Council of Canada PGS-D traineeship. L.M.D. was supported by the NIH National Research Service Award 5-T32-GM07544 from the National Institute of General Medicine Services and funding from the U.S.D.A. (to Vic DiRita). E.C.G. is supported by a Canada Research Chair award, the Michael Smith Foundation for Health Research, and a Burroughs Wellcome Fund Career Development Award in the Biomedical Sciences. This work was funded by the Burroughs Wellcome Fund and Canadian Institutes of Health Research Grant MOP-68981 to E.C.G.
Footnotes
Publisher's Disclaimer: Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting information
Additional supporting information may be found in the online version of this article.
References
- Altekruse SF, Stern NJ, Fields PI, Swerdlow DL. Campylobacter jejuni – an emerging foodborne pathogen. Emerg Infect Dis. 1999;5:28–35. doi: 10.3201/eid0501.990104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura H, Yamasaki M, Yamamoto S, Igimi S. Deletion of peb4 gene impairs cell adhesion and biofilm formation in Campylobacter jejuni. FEMS Microbiol Lett. 2007;275:278–285. doi: 10.1111/j.1574-6968.2007.00893.x. [DOI] [PubMed] [Google Scholar]
- Bacon DJ, Alm RA, Burr DH, Hu L, Kopecko DJ, Ewing CP, et al. Involvement of a plasmid in virulence of Campylobacter jejuni 81-176. Infect Immun. 2000;68:4384–4390. doi: 10.1128/iai.68.8.4384-4390.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda SS, Vik S, Friedman L, Kolter R. Biofilms: the matrix revisited. Trends Microbiol. 2005;13:20–26. doi: 10.1016/j.tim.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Bras AM, Chatterjee S, Wren BW, Newell DG, Ketley JM. A novel Campylobacter jejuni two-component regulatory system important for temperature-dependent growth and colonization. J Bacteriol. 1999;181:3298–3302. doi: 10.1128/jb.181.10.3298-3302.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brondsted L, Andersen MT, Parker M, Jorgensen K, Ingmer H. The HtrA protease of Campylobacter jejuni is required for heat and oxygen tolerance and for optimal interaction with human epithelial cells. Appl Environ Microbiol. 2005;71:3205–3212. doi: 10.1128/AEM.71.6.3205-3212.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butzler JP. Campylobacter, from obscurity to celebrity. Clin Microbiol Infect. 2004;10:868–876. doi: 10.1111/j.1469-0691.2004.00983.x. [DOI] [PubMed] [Google Scholar]
- Candon HL, Allan BJ, Fraley CD, Gaynor EC. Polyphosphate kinase 1 is a pathogenesis determinant in Campylobacter jejuni. J Bacteriol. 2007;189:8099–8108. doi: 10.1128/JB.01037-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KF, Le Tran H, Kanenaka RY, Kathariou S. Survival of clinical and poultry-derived isolates of Campylobacter jejuni at a low temperature (4 degrees C) Appl Environ Microbiol. 2001;67:4186–4191. doi: 10.1128/AEM.67.9.4186-4191.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cucarella C, Solano C, Valle J, Amorena B, Lasa I, Penades JR. Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J Bacteriol. 2001;183:2888–2896. doi: 10.1128/JB.183.9.2888-2896.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong YH, Zhang XF, Soo HM, Greenberg EP, Zhang LH. The two-component response regulator PprB modulates quorum-sensing signal production and global gene expression in Pseudomonas aeruginosa. Mol Microbiol. 2008;69:780. doi: 10.1111/j.1365-2958.2005.04612.x. [DOI] [PubMed] [Google Scholar]
- Ehrlich GD, Hu FZ, Shen K, Stoodley P, Post JC. Bacterial plurality as a general mechanism driving persistence in chronic infections. Clin Orthop Relat Res. 2005;437:20–24. doi: 10.1097/00003086-200508000-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppinger M, Baar C, Raddatz G, Huson DH, Schuster SC. Comparative analysis of four Campylobacterales. Nat Rev Microbiol. 2004;2:872–885. doi: 10.1038/nrmicro1024. [DOI] [PubMed] [Google Scholar]
- Falkow S. Molecular Koch's postulates applied to bacterial pathogenicity – a personal recollection 15 years later. Nat Rev Microbiol. 2004;2:67–72. doi: 10.1038/nrmicro799. [DOI] [PubMed] [Google Scholar]
- Fields JA, Thompson SA. Campylobacter jejuni CsrA mediates oxidative stress responses, biofilm formation, and host cell invasion. J Bacteriol. 2008;190:3411–3416. doi: 10.1128/JB.01928-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynor EC, Cawthraw S, Manning G, MacKichan JK, Falkow S, Newell DG. The genome-sequenced variant of Campylobacter jejuni NCTC 11168 and the original clonal clinical isolate differ markedly in colonization, gene expression, and virulence-associated phenotypes. J Bacteriol. 2004;186:503–517. doi: 10.1128/JB.186.2.503-517.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynor EC, Wells DH, MacKichan JK, Falkow S. The Campylobacter jejuni stringent response controls specific stress survival and virulence-associated phenotypes. Mol Microbiol. 2005;56:8–27. doi: 10.1111/j.1365-2958.2005.04525.x. [DOI] [PubMed] [Google Scholar]
- Gjermansen M, Ragas P, Sternberg C, Molin S, Tolker-Nielsen T. Characterization of starvation-induced dispersion in Pseudomonas putida biofilms. Environ Microbiol. 2005;7:894–904. doi: 10.1111/j.1462-2920.2005.00775.x. [DOI] [PubMed] [Google Scholar]
- Helms M, Simonsen J, Molbak K. Foodborne bacterial infection and hospitalization: a registry-based study. Clin Infect Dis. 2006;42:498–506. doi: 10.1086/499813. [DOI] [PubMed] [Google Scholar]
- Hendrixson DR, DiRita VJ. Identification of Campylobacter jejuni genes involved in commensal colonization of the chick gastrointestinal tract. Mol Microbiol. 2004;52:471–484. doi: 10.1111/j.1365-2958.2004.03988.x. [DOI] [PubMed] [Google Scholar]
- Hitchcock PJ, Brown TM. Morphological heterogeneity among Salmonella lipopolysaccharide chemo-types in silver-stained polyacrylamide gels. J Bacteriol. 1983;154:269–277. doi: 10.1128/jb.154.1.269-277.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofreuter DJ, Tsai RO, Watson V, Novik B, Altman M, Benitez C, et al. Unique features of a highly pathogenic Campylobacter jejuni strain. Infect Immun. 2006;74:4694–4707. doi: 10.1128/IAI.00210-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung DT, Zhu J, Sturtevant D, Mekalanos JJ. Bile acids stimulate biofilm formation in Vibrio cholerae. Mol Microbiol. 2006;59:193–201. doi: 10.1111/j.1365-2958.2005.04846.x. [DOI] [PubMed] [Google Scholar]
- Hussa EA, Darnell CL, Visick KL. RscS functions upstream of SypG to control the syp locus and biofilm formation in Vibrio fischeri. J Bacteriol. 2008;190:4576–4583. doi: 10.1128/JB.00130-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs-Rietsma W. Campylobacter in the food supply. In: Nachamkin MJBI, editor. Campylobacter. American Society for Microbiology Press; Washington, DC: 2000. pp. 467–482. [Google Scholar]
- Jeon B, Itoh K, Misawa N, Ryu S. Effects of quorum sensing on flaA transcription and autoagglutination in Campylobacter jejuni. Microbiol Immunol. 2003;47:833–839. doi: 10.1111/j.1348-0421.2003.tb03449.x. [DOI] [PubMed] [Google Scholar]
- Joshua GW, Guthrie-Irons C, Karlyshev AV, Wren BW. Biofilm formation in Campylobacter jejuni. Microbiology. 2006;152:387–396. doi: 10.1099/mic.0.28358-0. [DOI] [PubMed] [Google Scholar]
- Kalmokoff M, Lanthier P, Tremblay TL, Foss M, Lau PC, Sanders G, et al. Proteomic analysis of Campylobacter jejuni 11168 biofilms reveals a role for the motility complex in biofilm formation. J Bacteriol. 2006;188:4312–4320. doi: 10.1128/JB.01975-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlyshev AV, Wren BW. Development and application of an insertional system for gene delivery and expression in Campylobacter jejuni. Appl Environ Microbiol. 2005;71:4004–4013. doi: 10.1128/AEM.71.7.4004-4013.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konkel ME, Klena JD, Rivera-Amill V, Monteville MR, Biswas D, Raphael B, Mickelson J. Secretion of virulence proteins from Campylobacter jejuni is dependent on a functional flagellar export apparatus. J Bacteriol. 2004;186:3296–3303. doi: 10.1128/JB.186.11.3296-3303.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korlath JA, Osterholm MT, Judy LA, Forfang JC, Robinson RA. A point-source outbreak of campylobacteriosis associated with consumption of raw milk. J Infect Dis. 1985;152:592–596. doi: 10.1093/infdis/152.3.592. [DOI] [PubMed] [Google Scholar]
- Kulasekara HD, Ventre I, Kulasekara BR, Lazdunski A, Filloux A, Lory S. A novel two-component system controls the expression of Pseudomonas aeruginosa fimbrial cup genes. Mol Microbiol. 2005;55:368–380. doi: 10.1111/j.1365-2958.2004.04402.x. [DOI] [PubMed] [Google Scholar]
- Leon-Kempis Mdel R, Guccione E, Mulholland F, Williamson MP, Kelly DJ. The Campylobacter jejuni PEB1a adhesin is an aspartate/glutamate-binding protein of an ABC transporter essential for microaerobic growth on dicarboxylic amino acids. Mol Microbiol. 2006;60:1262–1275. doi: 10.1111/j.1365-2958.2006.05168.x. [DOI] [PubMed] [Google Scholar]
- McCleary WR, Stock JB. Acetyl phosphate and the activation of two-component response regulators. J Biol Chem. 1994;269:31567–31572. [PubMed] [Google Scholar]
- MacKichan JK, Gaynor EC, Chang C, Cawthraw S, Newell DG, Miller JF, Falkow S. The Campylobacter jejuni dccRS two-component system is required for optimal in vivo colonization but is dispensable for in vitro growth. Mol Microbiol. 2004;54:1269–1286. doi: 10.1111/j.1365-2958.2004.04371.x. [DOI] [PubMed] [Google Scholar]
- McLennan MK, Ringoir DD, Frirdich E, Svensson SL, Wells DH, Jarrell H, et al. Campylobacter jejuni biofilms up-regulated in the absence of the stringent response utilize a calcofluor white-reactive surface polysaccharide. J Bacteriol. 2007;190:1097–1107. doi: 10.1128/JB.00516-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik-Kale P, Parker CT, Konkel ME. Culture of Campylobacter jejuni with sodium deoxycholate induces virulence gene expression. J Bacteriol. 2008;190:2286–2297. doi: 10.1128/JB.01736-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Bryant SH. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 2004;32:W327–W331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaljevic RR, Sikic M, Klancnik A, Brumini G, Mozina SS, Abram M. Environmental stress factors affecting survival and virulence of Campylobacter jejuni. Microb Pathog. 2007;43:120–125. doi: 10.1016/j.micpath.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Moser I, Schroeder W, Salnikow J. Campylobacter jejuni major outer membrane protein and a 59-kDa protein are involved in binding to fibronectin and INT 407 cell membranes. FEMS Microbiol Lett. 1997;157:233–238. doi: 10.1111/j.1574-6968.1997.tb12778.x. [DOI] [PubMed] [Google Scholar]
- Muller S, Pflock M, Schar J, Kennard S, Beier D. Regulation of expression of atypical orphan response regulators of Helicobacter pylori. Microbiol Res. 2007;162:1–14. doi: 10.1016/j.micres.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Nakagawa S, Takaki Y, Shimamura S, Reysenbach AL, Takai K, Horikoshi K. Deep-sea vent epsilon-proteobacterial genomes provide insights into emergence of pathogens. Proc Natl Acad Sci USA. 2007;104:12146–12150. doi: 10.1073/pnas.0700687104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neal AL, Dublin SN, Taylor J, Bates DJ, Burns JL, Apkarian R, DiChristina TJ. Terminal electron acceptors influence the quantity and chemical composition of capsular exopolymers produced by anaerobically growing Shewanella spp. Biomacromolecules. 2007;8:166–174. doi: 10.1021/bm060826e. [DOI] [PubMed] [Google Scholar]
- O'Toole GA, Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol. 1998;28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- Ounissi H, Derlot E, Carlier C, Courvalin P. Gene homogeneity for aminoglycoside-modifying enzymes in gram-positive cocci. Antimicrob Agents Chemother. 1990;34:2164–2168. doi: 10.1128/aac.34.11.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhill J, Wren BW, Mungall K, Ketley JM, Churcher C, Basham D, et al. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hyper-variable sequences. Nature. 2000;403:665–668. doi: 10.1038/35001088. [DOI] [PubMed] [Google Scholar]
- Pearson BM, Gaskin DJH, Segers RPAM, Wells JM, Nuijten PJM, van Vliet AHM. The complete genome sequence of Campylobacter jejuni strain 81116 (NCTC11828) J Bacteriol. 2007;189:8402–8403. doi: 10.1128/JB.01404-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poly F, Ewing C, Goon S, Hickey TE, Rockabrand D, Majam G, et al. Heterogeneity of a Campylobacter jejuni protein that is secreted through the flagellar filament. Infect Immun. 2007;75:3859–3867. doi: 10.1128/IAI.00159-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Huang KH, Alm EJ, Arkin AP. A novel method for accurate operon predictions in all sequenced prokaryotes. Nucleic Acids Res. 2005;33:880–892. doi: 10.1093/nar/gki232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quon KC, Marczynski GT, Shapiro L. Cell cycle control by an essential bacterial two-component signal transduction protein. Cell. 1996;84:83–93. doi: 10.1016/s0092-8674(00)80995-2. [DOI] [PubMed] [Google Scholar]
- Raphael BH, Pereira S, Flom GA, Zhang Q, Ketley JM, Konkel ME. The Campylobacter jejuni response regulator, CbrR, modulates sodium deoxycholate resistance and chicken colonization. J Bacteriol. 2005;187:3662–3670. doi: 10.1128/JB.187.11.3662-3670.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeser RJ, Medler RT, Billington SJ, Jost BH, Joens LA. Characterization of Campylobacter jejuni biofilms under defined growth conditions. Appl Environ Microbiol. 2007;73:1908–1913. doi: 10.1128/AEM.00740-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romling U. Characterization of the rdar morphotype, a multicellular behaviour in Enterobacteriaceae. Cell Mol Life Sci. 2005;62:1234–1246. doi: 10.1007/s00018-005-4557-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romling U, Amikam D. Cyclic di-GMP as a second messenger. Curr Opin Microbiol. 2006;9:218–228. doi: 10.1016/j.mib.2006.02.010. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. [Google Scholar]
- Sampathkumar B, Napper S, Carrillo CD, Willson P, Taboada E, Nash JH, et al. Transcriptional and translational expression patterns associated with immobilized growth of Campylobacter jejuni. Microbiology. 2006;152:567–577. doi: 10.1099/mic.0.28405-0. [DOI] [PubMed] [Google Scholar]
- Schar J, Sickmann A, Beier D. Phosphorylation-independent activity of atypical response regulators of Helicobacter pylori. J Bacteriol. 2005;187:3100–3109. doi: 10.1128/JB.187.9.3100-3109.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seal BS, Hiett KL, Kuntz RL, Woolsey R, Schegg KM, Ard M, Stintzi A. Proteomic analyses of a robust versus a poor chicken gastrointestinal colonizing isolate of Campylobacter jejuni. J Proteome Res. 2007;6:4582–4591. doi: 10.1021/pr070356a. [DOI] [PubMed] [Google Scholar]
- Shrout JD, Chopp DL, Just CL, Hentzer M, Givskov M, Parsek MR. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol Microbiol. 2006;62:1264–1277. doi: 10.1111/j.1365-2958.2006.05421.x. [DOI] [PubMed] [Google Scholar]
- Stark RM, Gerwig GJ, Pitman RS, Potts LF, Williams NA, Greenman J, et al. Biofilm formation by Helicobacter pylori. Lett Appl Microbiol. 1999;28:121–126. doi: 10.1046/j.1365-2672.1999.00481.x. [DOI] [PubMed] [Google Scholar]
- Szurmant H, Nelson K, Kim E-J, Perego M, Hoch JA. YycH regulates the activity of the essential YycFG two-component system in Bacillus subtilis. J Bacteriol. 2005;187:5419–5426. doi: 10.1128/JB.187.15.5419-5426.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Arana A, Valle J, Solano C, Arrizubieta MJ, Cucarella C, Lamata M, et al. The enterococcal surface protein, Esp, is involved in Enterococcus faecalis biofilm formation. Appl Environ Microbiol. 2001;67:4538–4545. doi: 10.1128/AEM.67.10.4538-4545.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachoo N, Frank JF. Effectiveness of chemical sanitizers against Campylobacter jejuni-containing biofilms. J Food Prot. 2002;65:1117–1121. doi: 10.4315/0362-028x-65.7.1117. [DOI] [PubMed] [Google Scholar]
- Trachoo N, Frank JF, Stern NJ. Survival of Campylobacter jejuni in biofilms isolated from chicken houses. J Food Prot. 2002;65:1110–1116. doi: 10.4315/0362-028x-65.7.1110. [DOI] [PubMed] [Google Scholar]
- Tsai CM, Frasch CE. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal Biochem. 1982;119:115–119. doi: 10.1016/0003-2697(82)90673-x. [DOI] [PubMed] [Google Scholar]
- Verhamme DT, Arents JC, Postma PW, Crielaard W, Hellingwerf KJ. Investigation of in vivo crosstalk between key two-component systems of Escherichia coli. Microbiology. 2002;148:69–78. doi: 10.1099/00221287-148-1-69. [DOI] [PubMed] [Google Scholar]
- Watson RO, Galan JE. Campylobacter jejuni survives within epithelial cells by avoiding delivery to lysosomes. PLoS Pathog. 2008;4:e14. doi: 10.1371/journal.ppat.0040014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295:1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- Woodall CA, Jones MA, Barrow PA, Hinds J, Marsden GL, Kelly DJ, et al. Campylobacter jejuni gene expression in the chick cecum: evidence for adaptation to a low-oxygen environment. Infect Immun. 2005;73:5278–5285. doi: 10.1128/IAI.73.8.5278-5285.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wosten MM, Boeve M, Koot MG, van Nuene AC, van der Zeijst BA. Identification of Campylobacter jejuni promoter sequences. J Bacteriol. 1998;180:594–599. doi: 10.1128/jb.180.3.594-599.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wosten MM, Wagenaar JA, van Putten JP. The FlgS/FlgR two-component signal transduction system regulates the fla regulon in Campylobacter jejuni. J Biol Chem. 2004;279:16214–16222. doi: 10.1074/jbc.M400357200. [DOI] [PubMed] [Google Scholar]
- Wosten MM, Parker CT, van Mourik A, Guilhabert MR, van Dijk L, van Putten JP. The Campylobacter jejuni PhosS/PhosR operon represents a non-classical phosphate-sensitive two-component system. Mol Microbiol. 2006;62:278–291. doi: 10.1111/j.1365-2958.2006.05372.x. [DOI] [PubMed] [Google Scholar]
- Yao R, Alm RA, Trust TJ, Guerry P. Construction of new Campylobacter cloning vectors and a new mutational cat cassette. Gene. 1993;130:127–130. doi: 10.1016/0378-1119(93)90355-7. [DOI] [PubMed] [Google Scholar]
- Young KT, Davis LM, Dirita VJ. Campylobacter jejuni: molecular biology and pathogenesis. Nat Rev Microbiol. 2007;5:665–679. doi: 10.1038/nrmicro1718. [DOI] [PubMed] [Google Scholar]
- Young VB, Mansfield . Campylobacter infection – clinical context. In: Ketley JM, Konkel ME, editors. Campylobacter, Molecular and Cellular Biology. Horizon Bioscience; Norfolk: 2005. pp. 1–12. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.