Abstract
Inappropriate kinase expression and subsequent promiscuous activity defines the transformation of many solid tumors including renal cell carcinoma (RCC). Thus, the expression of novel tumor-associated kinases has the potential to dramatically shape tumor cell behavior. Further, identifying tumor-associated kinases can lend insight into patterns of tumor growth and characteristics. Here, we report the identification of Ror2, a new tumor-associated kinase in RCC cell lines and primary tumors. Ror2 is an orphan receptor tyrosine kinase with physiological expression normally seen in the embryonic kidney. However, in RCC, Ror2 expression correlated with expression of genes involved at the extracellular matrix, including Twist and matrix metalloprotease-2 (MMP2). Expression of MMP2 in RCC cells was suppressed by Ror2 knockdown, placing Ror2 as a mediator of MMP2 regulation in RCC and a potential regulator of extracellular matrix remodeling. The suppression of Ror2 not only inhibited cell migration, but also inhibited anchorage independent growth in soft agar and growth in an orthotopic xenograft model. These findings suggest a novel pathway of tumor-promoting activity by Ror2 within a subset of renal carcinomas, with significant implications for unraveling the tumorigenesis of RCC.
Keywords: Renal Cell Carcinoma, Ror2, tyrosine kinase, cancer, cell signaling
INTRODUCTION
Although tumor biology is tightly linked in many ways to the processes of embryonic development, a largely untapped area of solid tumor oncology is the specific examination of growth promoting pathways that are unique and critical to early developmental patterning. One such signaling pathway, Wnt, is a process common to both developmental and malignant states (Coombs et al., 2008). The identification of these signaling molecules that play a physiologic role in early developmental processes, with potential to promote tumor growth or other pathologic characteristics in the adult, provides an opportunity to define biological cause for various tumor attributes (Nakagawara, 2001). Further, identifying such molecules with restricted tissue expression in the adult organism provides a valuable chance for diagnostic or therapeutic implementation.
Epithelial cells require modulations in diverse signaling pathways to support the numerous cellular changes necessary to attain metastatic potential. Specifically, receptor tyrosine kinase (RTK) activation causes deregulated signaling and can defines tumor subsets across a broad spectrum of tumors (Stommel et al., 2007). Such variations in kinase activation have the potential to facilitate the cellular adaptations that determine patterns of tumor growth and can provide tremendous insight into the cell biology of that transformative process (de Castro-Carpeno et al., 2008; De Luca et al., 2008).
Among the cellular changes associated with tumor progression is tissue remodeling compatible with the extensive migration necessary for the progression of metastatic disease. This cellular transition is shared by the developmentally regulated processes which direct limb formation or other tissue patterning events. Tissue remodeling is driven by matrix metalloproteases (MMPs), a family of enzymes that degrade the extracellular matrix (ECM), and is integral to both normal tissue development and cancer progression (Liotta & Kohn, 2001). These processes signal cellular alterations that allow single cells to detach from the epithelial tumor collective, interrupting cell-cell junctions and increasing cell motility. Taken together, this highly coordinated process is critical for the lethality of most epithelial malignancies.
One such lethal epithelial malignancy, renal cell carcinoma (RCC), affects over 40,000 individuals in the US annually (Cancer Facts and Figures, (2008). Nearly a third of these patients present with unresectable or metastatic disease, for which antiangiogenic therapies are the mainstay of therapy (Cowey & Rathmell, 2008; Rathmell et al., 2005). RCC describes a group of histologically and genetically distinct neoplasms, the most common being clear cell RCC, with the remainder being papillary and chromophobe RCCs (Linehan et al., 2003). The mechanisms that promote tumor invasiveness or the acquisition of metastatic potential for the subtypes of RCC remain undefined.
ECM remodeling, a common step towards invasiveness, is promoted by tumor cells through changes in intracellular signaling and commonly acquiring characteristics of less differentiated cellular precursors (Liotta & Kohn, 2001). One developmentally-regulated pathway inappropriately activated in many epithelial tumors is Wnt protein signaling via the frizzled and LRP co-receptors (Atkins et al., 2004; van Amerongen et al., 2008). Here we report the identification of a Wnt receptor of embryonic mesenchymal origin expressed in RCC cell lines and tumors that defines a subgroup of human tumors with an invasive growth profile of gene expression. The novel receptor tyrosine kinase-like orphan receptor 2 (Ror2) is part of a family of orphan RTKs (Deloukas et al., 1998; Oldridge et al., 2000). Ror2 is characterized by an intracellular tyrosine kinase domain (Masiakowski & Carroll, 1992) and an extracellular Frizzled-like cysteine-rich domain (CRD), shown to act as a receptor for Wnt ligands (Oishi et al., 1997). Mouse Ror2 (mRor2) expression is normally observed in the developing heart, brain and lungs (Matsuda et al., 2001; Schwabe et al., 2004; Takeuchi et al., 2000; Yoda et al., 2003), with the greatest expression seen in migrating neural crest and mesenchymal tissues (Yoda et al., 2003). In the developing kidney, we have observed Ror2 expression in the migrating nests of mesenchymal cells. Further, Ror2 expression was identified in RCC tumors and is coordinated with invasive growth in culture. Coupled with the observations that Ror2 impacts expression of MMP2 and tumor cell growth in vitro and in vivo, this study identifies a novel developmentally regulated receptor that enhances RCC tumor cell growth capabilities.
RESULTS
Identification of Ror2 in renal carcinoma cells and human RCC tumors
To identify activated kinases in RCC cell lines, a phosphorylated-tyrosine kinase array was used to screen for kinases with a basal level of tyrosine phosphorylation in unstimulated RCC lysates. Tyrosine kinases expected to be activated in RCC such as the epithelial growth factor receptor (EGFR) were detected on the array. Among the phosphorylated-tyrosine kinases interrogated on the array, Ror2 was positively identified upon multiple examinations of the RCC cell line, 786-0 RC3, a control vector-transfected derivative of the VHL- mutant RCC cell line 786-0 (Figure 1A). Abundant expression of the endogenous protein in 786-0 RC3 cells was confirmed by immunoblot analysis. Immunoprecipitation of Ror2 with independent antibodies confirms high protein levels and basal autophosphorylation of the tyrosine kinase (Supplementary Figure 1A). Upon examination of other cell lines, we observed Ror2 expression in the parental RCC cell line, 786-0, using two independent antibodies targeting Ror2, but not in the SV-40-immortalized human proximal tubule cell line, HKC (Supplementary Figure 1B). Additionally, we demonstrated that Ror2 expression was present in an unrelated RCC cell line, RCC4 2-1 (Supplementary Figure 1B) further signifying Ror2′s role in RCC tumorigenesis deserved further exploration.
Figure 1. Human RCC tumors express Ror2.
A. Human Phospho-RTK array screen for activated kinases in RCC. Utilizing a phospho-RTK array, 42 RTKs were screened for phosphorylation in RCC using the 786-0 RC3 cell line. Activated kinases were identified using an HRP-conjugated pan-phosphotyrosine antibody. From this screen, one novel RTK identified was Receptor Tyrosine Kinase-like Orphan Receptor 2 (Ror2). Additional spots demonstrating positive identification include a kinase known to be activated in RCC cells (EGFR) and negative control spots (Goat IgG).
B. RT-PCR of archival human tumor samples shows mRNA expression of Ror2. Nineteen (19) archival tumor samples from the UNC Tumor Bank (1994-2004) were amplified with primers against the CRD domain of Ror2 (HPRT gene used as a control to assess quality of RNA). Direct sequencing of the PCR product confirmed the product as Ror2. Representative examples of Ror2 expression seen in RCC tumors are shown. A tabular distribution of tumors expressing a Ror2 transcript is shown for the individual RCC histological subtypes.
C. Ror2 is expressed in fetal kidney tissues. Left panel, H&E stain of fetal kidney, Middle panel, Ror2 expression detected by immunohistochemistry on primitive migrating mesenchymal cells, Right panel, CD117 is used as a negative control for staining specificity in the fetal kidney and (expected to be negative as shown) is a control for staining specificity for the secondary antibody.
Ror2 is a developmentally regulated kinase, with expression primarily relegated to embryonic mesenchymal tissues (Yoda et al., 2003). To examine a possible role in malignant tissue, Ror2 mRNA expression was analyzed by RT-PCR in 19 primary human RCC tumors. Four representative tumor specimens are demonstrated in Figure 1B with the HPRT gene used as a control for RNA abundance and integrity. The band detected as Ror2 in Figure 1B was purified and sequenced, confirming identity to the Ror2 CRD domain as predicted (data not shown). Overall, Ror2 transcript was detected in more than 55% of an unselected set of archival renal tumors (Figure 1B). Ror2 mRNA was detected even in some tumors of non-clear cell histology, suggesting that various genetic mechanisms may have the potential to promote the induction of Ror2 in renal cancers.
Detecting Ror2 expression in a malignant context prompted an evaluation of Ror2 expression in the normal human kidney. Ror2 is well known to be expressed in developing tissues with expression often restricted to mesenchymal cells (Yoda et al., 2003). Consistent with this profile, Ror2 protein was detected in human fetal kidneys in the migrating nests of mesenchymal cells that give rise to the bulk of the cell types of the developing kidney (Figure 1C).
Ror2 protein expression is detected on tumor cells of primary human tumors
As Ror2 mRNA expression was found in RCC tumors, we asked if Ror2 can also be identified at the protein level. Protein expression of Ror2 was analyzed by immunostaining two tumors demonstrating high levels of Ror2 transcript and two Ror2 transcript-negative tumors (one representative Ror2 negative tumor shown). Hematoxylin and eosin (H&E) serial sections demonstrate the clear cell histology of each of these tumors and lack of significant non-tumor cell infiltration. Immunofluorescent detection of Ror2 demonstrates tumor cell specific expression of Ror2 protein (Figure 2). The immunofluorescence signal was undetectable in tumors identified as lacking Ror2 mRNA expression based on the RT-PCR evaluation, or in the absence of Ror2 primary antibody. Nuclear stain shown from the same field demonstrates the presence of cellular constituents. Ror2 protein is, therefore, present in a subset of primary RCC tumors and can be detected specifically on the tumor cells of the specimen.
Figure 2. Immunofluorescence detects Ror2 in human RCC tumors.
Three individual RCC tumors were stained to determine the cellular expression of Ror2. H&E of the 3 tumors verifies clear cell histology. Two tumors were selected having high expression of Ror2 transcript (+), and a third tumor identified as having undetectable levels of Ror2 transcript (-) was used to verify antibody specificity. An Ror2 antibody was used to assay for Ror2 expression in the RCC tumors. Top and middle panels show that Ror2, a transmembrane cell surface receptor, is mainly expressed in the cell surface or cytoplasm, with no detectable protein in the control tumor in the bottom panel. Control panels show negative staining in the no primary control. DAPI is included for both primary and no primary antibody stained sections to demonstrate nuclear staining. All pictures were taken at 20X and the black bar represents 34 uM.
Ror2 expression in RCC tumors defines a tumor genetic profile of ECM genes
To analyze the role of Ror2 signaling in human RCC, an initial set of 13 human tumors were examined for gene expression using the 44K Agilent array platform. Tumors were classified as having undetectable, low, or high expression of Ror2 based on absolute Ror2 probe values on the array. Ror2 expression on the array was congruent with expression patterns observed by conventional RT-PCR. Based on these designations, a three-way analysis for differentially expressed targets was performed. When the 250 genes that associate most significantly with Ror2 expression (False Discovery Rate (FDR) < 0.07) were analyzed by hierarchical clustering, a subset of genes including extracellular matrix (ECM) candidates Twist1 and Twist2 were identified (Supplementary Figure 2). In a larger, but still highly significant gene cluster for association with Ror2 (FDR < 0.15), the ECM factor MMP2 was also identified (data not shown). Other genes identified in this significant association included those linked with Wnt signaling such as catenin δ2, frizzled related protein and cathepsin K.
In support of these data, additional RCC tumors were analyzed and clustered based on Ror2 expression. Independent microarray analysis was performed using the same 44K Agilent array platform revealing a similar set of highly coordinately regulated genes which were identified in all RCC histologies with a FDR < 0.045 (Figure 3A) and in clear cell with a FDR < 0.039 (Figure 3B). Again, Twist1 and MMP2 were found to be tightly associated with Ror2 expression. Altered expression for these transcripts (Twist1 and MMP2) was confirmed by quantitative RT-PCR of four representative tumors (Ror2 + and Ror2 -), confirming co-regulation (Figure 3C). Gene set analysis further supports an association between Ror2 expression and MMP2, identifying gene associations with the extended list of MMP family members and members of the Reck pathway. Additionally, in its physiologic role, Ror2 is tightly linked to skeletal development, thus it is intriguing to see a gene set reminiscent of that process associated with high statistical significance with a subset of human tumors of epithelial origin. An expanded listing of the members of the associated gene sets is shown in Figure 3D. These gene sets validate the initial gene expression study and support a thematic program of gene expression in these tumors associated with extracellular matrix remodeling. Together, these data extend a potential role for Ror2 as a mediator of tumor characteristics normally reserved for early and highly regulated developmental processes.
Figure 3. Microarray analysis of RCC tumors defines a tumor genetic phenotype.
Tumor mRNA from 40 RCC tumors were analyzed by Agilent 4×44K gene expression microarray and separated into groups based on their Ror2 expression. SAM analysis was performed to identify differentially expressed genes, and the most significant in (A) all histologies (FDR <0.045) and (B) clear cell only histology (FDR<0.0388) were analyzed by two-way clustering.
C. Ror2 (+) RCC tumors have increased mRNA expression of genes involved at the ECM. Quantitative RT-PCR analysis of 2 Ror2 (-) and 2 Ror2 (+) human RCC tumors were analyzed to verify microarray expression. Transcript values are normalized to β-actin RNA internal standard and are shown relative to an Ror2 (-) tumor (Tumor A). Error bars represent SEM. Significant differences were observed in Twist1 (*p<0.001) and MMP2 (**p<0.0001) in the Ror2 (+) tumors (*p<0.001) compared to Ror2 (-) tumor (Tumor A), p-values are based on cT values.
D. Table showing the significant gene set list with corresponding genes involved at the ECM that clustered with Ror2 expression. Tumor mRNA were analyzed by multiclass comparison using SAM, and the resulting Ror2 cluster was further analyzed for over-represented gene categories using the Expression Analysis Systematic Explorer (EASE) (Hosack et al., 2003). Genes from the three most tightly associated ontogenies are listed in the table.
Ror2 expression directs MMP2 gene expression, cell migration and anchorage independent growth in RCC
Based on the expression array findings, we sought to explore a direct relationship between Ror2 expression and the expression of genes involved in ECM remodeling. Suppression of Ror2 protein in the 786-0 RCC cell line with two independent shRNAs targeting distinct regions of Ror2, in comparison to the scramble shRNA and control empty vector pRS, is demonstrated by immunoblot in Figure 4A. The levels of Twist1 protein were also examined and no change in expression was observed by immunoblot analysis (Figure 4A). This result suggests that Ror2 may be part of a larger program of gene expression but does not play a direct role in regulating Twist1 expression. Ror2-deficient cells were examined by quantitative RT-PCR for effects on other genes expressed as a part of the ECM phenotype. Transcript levels of MMP2 were highly suppressed by Ror2 shRNA (Figure 4B). Thus, Ror2 may participate in the coordinated events involved in renal epithelial ECM remodeling by regulating the expression of MMP2 in tumor cells derived from this tissue.
Figure 4. Ror2 expression directs MMP2 expression, anchorage independent growth and invasive potential in vitro.
A. Ror2 expression is suppressed in 786-0 cells by shRNA. Whole cell protein extracts from RCC cells (786-0) were infected with a scramble short hairpin retrovirus, a pRS control virus or Ror2 short hairpin retroviruses and immunoblotted with polyclonal Ror2 antibody, Twist1 antibody or Ku80 antibody as a loading control (LC).
B. MMP2 expression is suppressed when Ror2 levels are knocked down. Quantitative RT-PCR analysis of two 786-0 Ror2 knockdown cell lines demonstrates MMP2 suppression coordinate with the degree of suppression of Ror2. Significant differences were observed in MMP2 (**p<0.001 for both comparisons) in the 786-0 Ror2 suppressed cell lines compared to the Ror2 expressing cell lines (**p<0.001 for both comparisons). Transcript values are normalized to β-actin RNA internal standard and are shown relative to 786-0 scramble RNA. Error bars represent SEM.
C. Ror2 knockdown decreases cell migration. RCC cells (786-0) infected with a control virus pRS or Ror2 short hairpin RNA retroviruses were plated and allowed to grow overnight. Cells were scratched and the length of the scratch was observed for both 0 hr and 16hr timepoints (left). Significant differences were observed when comparing the % invasion of the Ror2 knockdown cell lines compared to the empty vector pRS control (right) - (**p<0.0001, *p=0.009). Error bars represent SEM.
D. Ror2 knockdown inhibits anchorage independent growth. RCC cells (786-0) infected with a scramble short hairpin RNA retrovirus, a control virus pRS or Ror2 short hairpin RNA retroviruses were allowed to grow in soft agar over a period of 3-4 weeks. Ror2 knockdown cells have inhibited growth in comparison to the scramble short hairpin retrovirus and the empty vector pRS control. Multicellular colonies >150 uM were counted from triplicate plates. Data shown is from the combination of two representative experiments (**p<0.0001). Error bars represent SEM. Inset - Cells were stained with MTT dye, and pictures taken at 10x magnification. Black bar represents 67.3 um.
The functional consequence of Ror2 suppression was examined in assays of tumor cell invasive properties, beginning with cell migration. Wound healing was measured as a means to quantify cellular migration because suppression of Ror2 by shRNA did not directly impact doubling time or viability of cultured cells (Supplementary Figure 3). We examined wound healing in two independent stable shRNA Ror2 knockdowns generated in 786-0 subclones. Healing was assayed in triplicate at 0, 8, 16, and 24 hour timepoints. Cells lacking Ror2 expression failed to close the wound as efficiently as their isogenic counterparts, scramble shRNA (data not shown) or pRS (Figure 4C), suggesting that Ror2 may play an important role in promoting cell migration.
Another tumor cell characteristic of metastatic RCC and tumor invasiveness associated with the ECM is anchorage independent growth. To determine whether Ror2 suppression also impacts this mode of cell growth, we assessed the growth of stable shRNA Ror2 knockdowns in soft agar (Figure 4D). Colonies greater than 150 uM in diameter were counted from triplicate plates in duplicate assays. Cells lacking Ror2 expression retained the ability to survive as single cells in soft agar, but failed to proliferate. These cells, therefore, failed to exhibit the anchorage independent growth of their Ror2-expressing isogenic counterparts, suggesting that Ror2 may play an important role in supporting cellular invasion in RCC tumorigenesis.
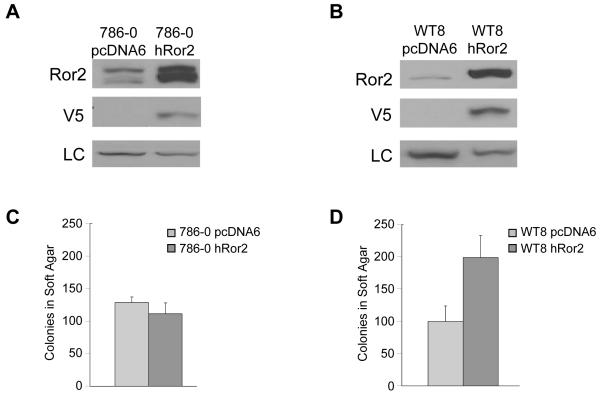
As Ror2 suppression is shown to be necessary for anchorage independent growth, we also asked if Ror2 expression was sufficient to induce growth. Human Ror2 (hRor2) expression plasmids were generated for expression in two RCC cell lines - 786-0 where Ror2 is highly expressed (Figure 5A) and a derivative line, WT8, where Ror2 expression is negligible (Figure 5B). Adding more hRor2 to the 786-0 cell line did not change anchorage independent growth (Figure 5C) suggesting that increasing the level of Ror2 beyond a threshold fails to impact this feature of tumorigenicity. However, when hRor2 was added to an RCC cell line that lacks robust Ror2 expression, an increase in anchorage independent growth was observed (Figure 5D), suggesting that Ror2 is sufficient to induce anchorage independent growth. Taken together, these data suggest that Ror2 expression is both necessary and sufficient for the anchorage independent growth characteristic of RCC.
Figure 5. Ror2 overexpression enhances anchorage independent growth in vitro.
A. Ror2 expression is increased in 786-0 cells with an overexpression plasmid. RCC cells (786-0) were transfected with control plasmid pcDNA6 or hRor2 cDNA and immunoblotted with polyclonal Ror2 antibody, a V5 antibody or Ku80 antibody as a loading control (LC).
B. Ror2 expression is amplified in the 786-0 derivative cell line WT8, a clone expressing a VHL cDNA which expresses reduced levels of Ror2. The RCC cell line WT8 was transfected with a control plasmid pcDNA6 or hRor2 cDNA and immunoblotted with polyclonal Ror2 antibody, a V5 antibody or Ku80 antibody as a loading control (LC). Panel A and B represent a contiguous blot with intervening lanes removed.
C. An increase in Ror2 expression in Ror2 expressing cells does not affect anchorage independent growth. RCC cells with high levels of Ror2 already expressed (786-0) transfected with a control plasmid pcDNA6 or hRor2 cDNA were allowed to grow in soft agar over a period of 3-4 weeks. Multicellular colonies >150 uM were counted from triplicate plates. An increase in Ror2 expression did not change colony growth. Data shown is from two representative experiments.
D. Ror2 overexpression increases anchorage independent growth in cells lacking Ror2 expression. RCC cells lacking high level of expression of Ror2 (WT8) transfected with a control plasmid pcDNA6 or hRor2 cDNA were allowed to grow in soft agar over a period of 3-4 weeks. Multicellular colonies >150 uM were counted from triplicate plates. Ror2 overexpression cells displayed enhanced growth in comparison to the control empty vector pcDNA6. Data shown is from one representative experiment (p=0.079).
Ror2 suppression reduces tumor growth in vivo
As our data showed that 786-0 Ror2 shRNA suppressed anchorage independent growth in vitro in RCC cells, we also sought to assess tumor growth in vivo. Two independently derived Ror2 deficient cell lines and an isogenic control shRNA cell line were injected orthotopically in a cohort of athymic nude (nu/nu) female mice. Macroscopically visible tumors were only detected in mice injected with 786-0 isogenic control cells. Additionally, the control cell line had significantly more tumors than the two shRNA cell lines combined. The histology of a control engrafted tumor with adjacent normal kidney tissue is shown in Figure 6A. By comparison, tumor cells derived from the Ror2 suppressed cells were present on microscopic evaluation of two kidneys in the vicinity of the kidney subcapsule (Figure 6B, arrows), but failed to form clearly discrete masses. The extent of tumor growth, including tumors which implanted outside the area of injection, is quantified in the panel of Figure 6C and the average of all ten implants (in cm) was larger in the isogenic control cell line than in the two shRNA cell lines (Figure 6D). This experiment was replicated independently with xenografts grown longer than 3 months, and the results mirrored these findings (data not shown). MMP2 protein levels were also examined in these engrafted tumors. As previously shown at the transcription level, when Ror2 is expressed there is an abundance of MMP2 protein expression (Figure 6E, F). In tumors where Ror2 expression is suppressed, MMP2 protein levels are decreased (Figure 6G, H). This data confirms in vivo the findings of the experiments examining anchorage independent growth, validating a role for Ror2 in promoting tumor cell growth independent of tumor cell survival in RCC. Further, this data provides concrete evidence that links Ror2 to RCC tumor development and places this signaling pathway as an immediate facilitator of tumor growth.
Figure 6. Ror2 suppression reduces tumor growth in vivo.
Two independent shRNA cell lines were injected orthotopically under the kidney capsule of athymic nude mice, compared to 786-0 pRS control cells.
A. Histology of control engrafted tumor with adjacent normal kidney tissue is shown.
B. H&E of the kidney surface demonstrated that Ror2-suppressed engrafted tumor cells were present in the vicinity of the subcapsule microscopically but failed to form distinct tumors.
C. Macroscopically visible tumors were only detected in mice injected with 786-0 pRS control cells. Macroscopic and microscopic evidence of tumor cells was tabulated, demonstrating significantly more tumors identified in the control cell implanted mice than those implanted with either of the two Ror2 shRNA cell lines.
D. Average tumor diameter of each implant from the 786-0 pRS control cell line and the two Ror2 shRNA cell lines (in cm).
E. The control engrafted tumor, 786-0 pRS, displays increased levels of MMP2 as determined by immunohistochemistry.
F. Control panel for 786-0 pRS shows negative staining in the no primary control.
G. The Ror2-suppressed engrafted tumor, 786-0 shRor2.1, has diminished levels of MMP2 expression compared to the empty vector control, 786-0 pRS.
H. Control panel for 786-0 shRor2.1 shows insignificant background staining in the no primary control.
DISCUSSION
In a strategy to uncover potential signaling kinases for RCC, we have identified the developmentally regulated orphan kinase Ror2 as an activated tyrosine kinase in RCC cancer cell lines and renal tumors. Ror2 is physiologically expressed primarily on embryonic mesenchymal cells and developmentally expressed on migrating mesenchymal cells of the fetal kidney. The aberrant expression of Ror2 in human RCC tumors has not been specifically described to date, although evidence of active Ror2 has been observed on kinase arrays from glioblastoma cell lines (Stommel et al., 2007). In this study, however, for the first time we show that Ror2 expression was detected in the majority of human RCC tumors, and protein expression was confirmed at the tumor cell level. These data support the identification of a tumor cell intrinsic receptor tyrosine kinase, usually limited to developing mesenchymal cell subsets, that is detectable on the majority of renal cell carcinomas.
Ror2 expression in RCC may herald the activation of many signalling events, particularly in the aberrant context of a tumor cell environment. For epithelial cells to metastasize, they must acquire mesenchymal characteristics that permit proliferation outside of the usual niche. Ror2 is primarily expressed in mesenchymal cells, immediately suggesting a possible role for this kinase as part of the ECM remodeling process in tumor development. Expression of the Ror2 kinase in human tumors was found to be tightly correlated with genes involved at the ECM, in particular MMP2, linking this epithelially derived tumor with mesenchymal phenotypic markers and a potentially important transitioning event in the development of invasive renal cell carcinoma. Matrix metalloprotease-2 (MMP2) has previously been shown to be overexpressed in many metastatic cancers including RCC (Kurban et al., 2006; Zhang et al., 2002) and Ror2 may define an important and unique pathway for this process in kidney cancers (Kurban et al., 2006). MMP2, in particular among the MMP family, is essential for the renal tubular cell epithelial to mesenchymal transformation (EMT) specific to the recovery from renal tubular injury (Irwin et al., 2003). This process requires adaptations of the extracellular matrix as well as mechanisms of promoting cellular migration. Using wound healing as a surrogate for cell migration, we found that Ror2-suppressed cells migrated at a slower rate than their isogenic partners. As a critical test of tumor cell growth in a three-dimensional environment, Ror2 expression supported anchorage independent cell growth, an indicator of tumor cell growth and a critical quality of metastatic tumors. We further demonstrated that suppression of Ror2 was sufficient to inhibit tumor growth in xenografts, suggesting a key role in permitting the 3-dimensional growth essential for tumor formation.
It is additionally intriguing to consider the unique embryonic origins of the kidney, in which the epithelial tissues are entirely derived from a mesenchymal source. The process of developing the excretory tubules has in fact been termed a “mesenchymal to epithelial transition” (Davies, 1996). The kidney is derived from the ureteric bud and the metanephric mesoderm (Saxen & Sariola, 1987), thus the emergence of Ror2 as a part of a dedifferentiation profile may provide the embryonic signal necessary to reinact a mesenchymal program in malignant epithelial cells derived from the kidney. Taken together, these data suggest that Ror2 can play an important role in renal cell carcinoma growth patterns by providing tumor cells with an immature phenotype primed for invasive growth.
Further, the regulation of Wnt signaling via Wnt receptors such as Ror2 can be greatly influenced by the availability of Wnt ligands, Frizzled family members, and factors such as the secreted frizzled related protein 1, which may influence the activity of Ror2 in tumorigenicity (Billiard et al., 2005). It remains uncertain which specific Wnt signaling pathway Ror2 engages as a cancer promoting factor; thus, the identification of Ror2 as a feature of RCC presents a novel avenue for future integrated studies of developmental and cancer biology.
Ror2 activity thus defines a novel component of the signaling machinery in place to promote the growth of many renal tumors and may define a unique tumor subset with distinct pathologic or physiologic characteristics. Finally, this newly identified kinase in RCC provides an ideal candidate to explore as a marker of tumors with aggressive growth potential or as a putative target to disable tumor growth.
MATERIALS AND METHODS
Human Phospho-Receptor Tyrosine Kinase (RTK) array
A human phospho-RTK array kit (RND Systems, Minneapolis, MN) was used according to the manufacturer’s instruction. Phosphorylated RTKs were detected using ECL-Plus chemiluminescence reagents (GE, Pittsburg, PA).
Cell culture and treatments
786-0 and RCC4 cell lines, and derivatives 786-0 RC3 and WT8 (vector control transfected and VHL transfected clones, respectively, kindly provided by Dr. W. Kaelin, Boston, MA), and RCC4 2-1 (vector transfected clone, kindly provided by Dr. M.C. Simon and Dr. B. Keith, Philadelphia, PA), were maintained in Dulbecco’s Modified Eagle Medium (DMEM) with 10% fetal bovine serum (FBS), L-glutamine and non essential amino acids. The shRNA knockdown cells were generated by introducing commercial shRNAs in pRetroSuper (pRS) for Ror2 (Origene Technologies Inc, Rockville, MD), a control virus delivering pRS (empty vector) or a scramble short hairpin retrovirus. Puromycin selection was used to maintain the shRNAs. Overexpression cell lines were generated by transferring hRor2 cDNA (Origene Technologies Inc, Rockville, MD) into the pcDNA6/V5-HisA plasmid (Invitrogen, Carlsbad, CA) via SacI/XhoI restriction enzyme sites (New England BioLabs Inc, Ipswich, MA). The HKC line (kindly provided by Dr. L. Racusen, Baltimore, MD (Racusen et al., 1997)) was maintained in DMEM/F12 supplemented with 10% FBS.
RT-PCR and qRT-PCR
Total RNA was prepared using Gentra Systems RNA Extraction Cell Kit (Minneapolis, MN). cDNA was made from 0.5μg total RNA by reverse transcriptase (RT) using oligo dT primers and Superscript II RT-PCR reagents (Invitrogen, Carlsbad, CA). For automated TaqMan qRT-PCR, proprietary commercial FAM labeled primers were used targeting Ror2, MMP2, Twist1 and β-actin (Applied Biosystems, Foster City, CA) and analyzed on the 7900H Fast Real-Time PCR System (Applied Biosystems, Foster City, CA). For RT-PCR, primers targeting the Ror2 CRD domain (Left: 5′ tttcaggatgattaccacgag 3′, Right: 5′ ctcacacttgggcagctgaa 3′) were generated. The Ror2 CRD domain PCR fragment was confirmed by bidirectional DNA sequencing and BLAST sequence homology analysis. Standard HPRT primers (Left: 5′ cctgctggattacatataagcactg 3′, Right: 5′ gtcaagggcatatccaacaacaaac 3′) were used to analyze the RNA integrity.
Immunoblotting
Cells were lysed in whole cell extraction buffer containing 20mM Tris, 100mM NaCl, 1 mM EDTA, 1% NP-40 and protease inhibitor cocktail (Roche Diagnostics, Indianapolis, IN). Bradford reagent was used to assess the concentration. Protein samples were separated on 8% SDS-polyacrylamide gels and transferred to nitrocellulose (GE, Pittsburg, PA). The following antibodies were used to assess for expression: Monoclonal Ror2 at 1:250 (RND Systems, Minneapolis, MN), Polyclonal Ror2 at 1:3000 (RND Systems, Minneapolis, MN), Twist1 at 1:1000 (Cell Signaling Technology Inc, Danvers, MA), V5-HRP at 1:2500 (Invitrogen, Carlsbad, CA), Ku80 at 1:2000 (Genetex Inc, San Antonio, TX) as a loading control (LC). Appropriate horseradish peroxidase conjugated secondary antibodies were used and proteins detected using ECL-Plus chemiluminescence reagents (GE, Pittsburg, PA).
Immunoprecipitation
500μg of cell lysates were immunoprecipitated using a polyclonal biotinylated Ror2 antibody (RND Systems, Minneapolis, MN), Epo (N-19) nonspecific IgG goat antibody (Santa Cruz, Santa Cruz, CA) and a bead only control using streptavidin conjugated Dynabeads-280 (Invitrogen, Carlsbad, CA). Immunoblotting was performed as described above using monoclonal anti-Ror2 at 1:250 (RND Systems, Minneapolis, MN) and pY99 anti-phosphotyrosine at 1:1000 (Santa Cruz, Santa Cruz, CA).
Wound Healing Assay
35,000 cells were plated in triplicate on 6-well plates and allowed to adhere overnight at 37°C in 10% FBS media. The monolayer was then “wounded” in perpendicular directions with a 200ul pipet tip (forming a cross) and 2% FBS media was added. Pictures were taken at each of the cross junctions at 4x magnification at 0, 8, 16, and 24 hours.
Soft Agar Colony Formation Assay
0.6% bottom agar was plated onto 6-well plates. 10,000 cells were plated in triplicate, placed at 37°C overnight and covered with 0.2mL RCC media. Colonies were allowed to form over 3-4 weeks and then stained with 2mg/mL MTT (Sigma-Aldrich, St. Louis, MO) for 1-4hrs. Images were taken at 10x magnification. Black bar represents 67.3μm.
Cell Viability Assay
A modified MTT assay (Promega, Madison, WI) was used to assess cell viability. 5,000 cells were plated with 50uL selection media for 5 days. The MTT dye containing the tetrazolium salt was added according to manufacturer instructions and absorbance measured at 570nm.
Doubling Time Assay
300,000 cells were plated and after 96hrs, the total number of cells was counted. This process was repeated 10 times and the doubling time calculated.
Immunohistochemistry and Immunofluorescence
RCC tumor blocks were obtained from the UNC Tissue Procurement Facility with approval by the institutional review board (IRB) and serially sectioned onto unprepared glass slides. The first section was stained with hematoxylin and eosin (H&E) for tumor verification, and a subsequent section probed with a primary mouse anti-human Ror2 monoclonal antibody (generated by Drs. Mikels and Nusse). Detection was performed according to the instructions of the TSA™-plus Fluorescien System (NEL 741001 KT from PerkinElmer Labs, Inc, Waltham, MA). Slides were mounted with Vectashield (H-1200, Vector Laboratories, Burlingame, CA) and imaged using fluorescence microscopy at 20X magnification. Black bar represents 34uM. Immunohistochemical stain on human fetal kidney sections acquired through an IRB-approved protocol was performed using the same Ror2 monoclonal antibody. Standard antigen unmasking procedures were used and protein was detected using di-amino benzidine.
Xenograft Analysis
500,000 cells were injected orthotopically under the kidney capsule in a cohort of athymic nude (nu/nu) female mice and aged 2.5 or 3 months. The xenografted tumors were formalin fixed, paraffin embedded and tissue blocks were serially sectioned onto unprepared glass slides. Xenografted tumors were stained with hematoxylin and eosin (H&E) for tumor verification. The slides were immunostained with MMP2 antibody 3158 (Abcam Inc, Cambridge, MA) and detected as above.
Microarray Analysis
Fresh frozen tumors from the UNC Tissue Procurement Facility tumor bank (1994-2007) were used for these studies. Total mRNA was prepared as described above, incorporating a Rotor-Stator for tissue homogenization. Quantitation was performed by NanoDrop (NanoDrop Technologies, Wilmington, DE). RNA quality check, amplification, labeling, and hybridization was performed by the UNC Genomics Core Facility. Two-color hybridization was performed, with a modified reference human mRNA derived from the Stratagene Universal Human Tumor Reference RNA (kindly provided by Dr. C. Perou, Chapel Hill, NC).
Raw data was derived using Agilent feature extraction software, and results were expressed as a Log (2)-transformed ratio between tumor and control mRNA. Three-way comparison was performed using Significance Analysis of Microarrays (SAM) version 3.01 (Tusher et al., 2001) for Supplementary Figure 2. Duplicate genes were removed, targets were median centered and complete linkage analysis was performed with Pearson’s Correlation with Cluster 3.0. For Figure 3, quantitative analysis of data according to Ror2 gene expression was performed using SAM. Genes with a false discovery rate <5% were then used for average linkage analysis with Cluster 3.0. Gene set analysis was also performed in SAM according to Ror2 gene expression values, using a cutoff of FDR=0. Gene sets analyzed were from Erin Segal or the MSigDB collection at the Broad Institute.
Statistical Analysis
The standardized cycle threshold (cT) values were fit with a one-way ANOVA model with the experimental variable of interest (Ror2, EGLN3, MMP2, etc) as the fixed effect. To verify the one-way ANOVA model, a nonparametric statistical procedure, Kruskal-Wallis test, was performed to compare the standardized cT among the cell lines. Error bars are shown as standard error of the mean (SEM).
Ror2 expression was evaluated across RCC tumor histologies using a Student’s T test to assign p values as shown. Because clear cell tumors are more common than other histologies, the difference in sample size is reflected in the standard deviations using an adjustment to account for unequal variances. A non-parametric Wilcoxon Rank Sum procedure was performed to confirm the significant result found using the parametric two sample t-test.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank the Rathmell Lab members and the Lineberger Cancer Center community for thoughtful insights/advice. Additional thanks to the Microscopy Services Laboratory for use of their facilities and W. Wilson (Baldwin Lab) for use of reagents. We would also like to thank Dr. H. Shelton Earp, Dr. Ian Davis and Dr. Jon Serody for paper critiques. Funding for this project was provided in part by the Doris Duke Charitable Fund and the V foundation (WKR), Initiative for Maximizing Student Diversity Training Grant NIH-R25GM055336 (TMW and CM) and NCI-F31CA132543 (TMW). Funding for JDG was provided by a MSTP grant and a T32 grant in hemostasis and thrombosis. Human tumor samples were collected by the UNC Tumor Procurement Facility, supported by the UNC Lineberger Comprehensive Cancer Center.
Support for the UNC Tissue procurement facility, the Genomic Core facility, and the Animal studies core facility was provided by the Lineberger Cancer Center Core grant from the NCI.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- Atkins MB, Hidalgo M, Stadler WM, Logan TF, Dutcher JP, Hudes GR, et al. Randomized phase II study of multiple dose levels of CCI-779, a novel mammalian target of rapamycin kinase inhibitor, in patients with advanced refractory renal cell carcinoma. J Clin Oncol. 2004;22:909–18. doi: 10.1200/JCO.2004.08.185. [DOI] [PubMed] [Google Scholar]
- Billiard J, Way DS, Seestaller-Wehr LM, Moran RA, Mangine A, Bodine PV. The orphan receptor tyrosine kinase Ror2 modulates canonical Wnt signaling in osteoblastic cells. Mol Endocrinol. 2005;19:90–101. doi: 10.1210/me.2004-0153. [DOI] [PubMed] [Google Scholar]
- Cancer Facts and Figures. American Cancer Society, Inc; Atlanta, GA: 2008. [Google Scholar]
- Coombs GS, Covey TM, Virshup DM. Wnt signaling in development, disease and translational medicine. Curr Drug Targets. 2008;9:513–31. doi: 10.2174/138945008784911796. [DOI] [PubMed] [Google Scholar]
- Cowey CL, Rathmell WK. Using molecular biology to develop drugs for renal cell carcinoma. Expert Opinion on Drug Discovery. 2008;3:311–327. doi: 10.1517/17460441.3.3.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies JA. Mesenchyme to epithelium transition during development of the mammalian kidney tubule. Acta Anat (Basel) 1996;156:187–201. doi: 10.1159/000147846. [DOI] [PubMed] [Google Scholar]
- de Castro-Carpeno J, Belda-Iniesta C, Casado Saenz E, Hernandez Agudo E, Feliu Batlle J, Gonzalez Baron M. EGFR and colon cancer: a clinical view. Clin Transl Oncol. 2008;10:6–13. doi: 10.1007/s12094-008-0147-3. [DOI] [PubMed] [Google Scholar]
- De Luca A, Carotenuto A, Rachiglio A, Gallo M, Maiello MR, Aldinucci D, et al. The role of the EGFR signaling in tumor microenvironment. J Cell Physiol. 2008;214:559–67. doi: 10.1002/jcp.21260. [DOI] [PubMed] [Google Scholar]
- Deloukas P, Schuler GD, Gyapay G, Beasley EM, Soderlund C, Rodriguez-Tome P, et al. A physical map of 30,000 human genes. Science. 1998;282:744–6. doi: 10.1126/science.282.5389.744. [DOI] [PubMed] [Google Scholar]
- Hosack DA, Dennis G, Jr., Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin MS, Kondo K, Marin MC, Cheng LS, Hahn WC, Kaelin WG., Jr. Chemosensitivity linked to p73 function. Cancer Cell. 2003;3:403–10. doi: 10.1016/s1535-6108(03)00078-3. [DOI] [PubMed] [Google Scholar]
- Kurban G, Hudon V, Duplan E, Ohh M, Pause A. Characterization of a von Hippel Lindau pathway involved in extracellular matrix remodeling, cell invasion, and angiogenesis. Cancer Res. 2006;66:1313–9. doi: 10.1158/0008-5472.CAN-05-2560. [DOI] [PubMed] [Google Scholar]
- Linehan WM, Walther MM, Zbar B. The genetic basis of cancer of the kidney. J Urol. 2003;170:2163–72. doi: 10.1097/01.ju.0000096060.92397.ed. [DOI] [PubMed] [Google Scholar]
- Liotta LA, Kohn EC. The microenvironment of the tumour-host interface. Nature. 2001;411:375–9. doi: 10.1038/35077241. [DOI] [PubMed] [Google Scholar]
- Masiakowski P, Carroll RD. A novel family of cell surface receptors with tyrosine kinase-like domain. J Biol Chem. 1992;267:26181–90. [PubMed] [Google Scholar]
- Matsuda T, Nomi M, Ikeya M, Kani S, Oishi I, Terashima T, et al. Expression of the receptor tyrosine kinase genes, Ror1 and Ror2, during mouse development. Mech Dev. 2001;105:153–6. doi: 10.1016/s0925-4773(01)00383-5. [DOI] [PubMed] [Google Scholar]
- Nakagawara A. Trk receptor tyrosine kinases: a bridge between cancer and neural development. Cancer Lett. 2001;169:107–14. doi: 10.1016/s0304-3835(01)00530-4. [DOI] [PubMed] [Google Scholar]
- Oishi I, Sugiyama S, Liu ZJ, Yamamura H, Nishida Y, Minami Y. A novel Drosophila receptor tyrosine kinase expressed specifically in the nervous system. Unique structural features and implication in developmental signaling. J Biol Chem. 1997;272:11916–23. doi: 10.1074/jbc.272.18.11916. [DOI] [PubMed] [Google Scholar]
- Oldridge M, Fortuna AM, Maringa M, Propping P, Mansour S, Pollitt C, et al. Dominant mutations in ROR2, encoding an orphan receptor tyrosine kinase, cause brachydactyly type B. Nat Genet. 2000;24:275–8. doi: 10.1038/73495. [DOI] [PubMed] [Google Scholar]
- Racusen LC, Monteil C, Sgrignoli A, Lucskay M, Marouillat S, Rhim JG, et al. Cell lines with extended in vitro growth potential from human renal proximal tubule: characterization, response to inducers, and comparison with established cell lines. J Lab Clin Med. 1997;129:318–29. doi: 10.1016/s0022-2143(97)90180-3. [DOI] [PubMed] [Google Scholar]
- Rathmell WK, Wright TM, Rini BI. Molecularly targeted therapy in renal cell carcinoma. Expert Rev Anticancer Ther. 2005;5:1031–40. doi: 10.1586/14737140.5.6.1031. [DOI] [PubMed] [Google Scholar]
- Saxen L, Sariola H. Early organogenesis of the kidney. Pediatr Nephrol. 1987;1:385–92. doi: 10.1007/BF00849241. [DOI] [PubMed] [Google Scholar]
- Schwabe GC, Trepczik B, Suring K, Brieske N, Tucker AS, Sharpe PT, et al. Ror2 knockout mouse as a model for the developmental pathology of autosomal recessive Robinow syndrome. Dev Dyn. 2004;229:400–10. doi: 10.1002/dvdy.10466. [DOI] [PubMed] [Google Scholar]
- Stommel JM, Kimmelman AC, Ying H, Nabioullin R, Ponugoti AH, Wiedemeyer R, et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science. 2007;318:287–90. doi: 10.1126/science.1142946. [DOI] [PubMed] [Google Scholar]
- Takeuchi S, Takeda K, Oishi I, Nomi M, Ikeya M, Itoh K, et al. Mouse Ror2 receptor tyrosine kinase is required for the heart development and limb formation. Genes Cells. 2000;5:71–8. doi: 10.1046/j.1365-2443.2000.00300.x. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Amerongen R, Mikels A, Nusse R. Alternative Wnt Signaling Is Initiated by Distinct Receptors. Sci. Signal. 2008;1:re9. doi: 10.1126/scisignal.135re9. [DOI] [PubMed] [Google Scholar]
- Yoda A, Oishi I, Minami Y. Expression and function of the Ror-family receptor tyrosine kinases during development: lessons from genetic analyses of nematodes, mice, and humans. J Recept Signal Transduct Res. 2003;23:1–15. doi: 10.1081/rrs-120018757. [DOI] [PubMed] [Google Scholar]
- Zhang X, Yamashita M, Uetsuki H, Kakehi Y. Angiogenesis in renal cell carcinoma: Evaluation of microvessel density, vascular endothelial growth factor and matrix metalloproteinases. Int J Urol. 2002;9:509–14. doi: 10.1046/j.1442-2042.2002.00511.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.