Abstract
Mucins form a group of heavily O-glycosylated biologically important glycoproteins that are involved in a variety of biological functions, including modulating immune response, inflammation, and adhesion. Mucins are also involved in cancer and metastasis, and often express diagnostic cancer antigens. Recently, a modified porcine submaxillary mucin (Tn-PSM) was shown to bind to soybean agglutinin (SBA) with ~106-fold enhanced affinity relative to GalNAcα1-O-Ser, the pancarcinoma carbohydrate antigen. In the present study, dynamic force spectroscopy (DFS) is used to investigate molecular pairs of SBA and Tn-PSM. A number of force jumps that demonstrate unbinding/rebinding events were observed up to a distance equal to 2.0 micrometers, consistent with the length of the mucin chain. The unbinding force increased from 103 pN to 402 pN with increasing force loading rate. The position of the activation barrier in the energy landscape of the interaction was 0.1 nm. The lifetime of the SBA – TnPSM complex in the absence of applied force was determined to be is in the range 1.3 – 1.9 s. Kinetic parameters describing the rate of dissociation of other sugar lectin interactions are in the range 3.3 × 10−3 − 2.5 × 10−3 s. The long lifetime of the SBA – TnPSM complex is compatible with a binding model in which lectin molecules “bind and jump” from α-GalNAc residue to α-GalNAc residue along the polypeptide chain of Tn-PSM before dissociating. These findings have important implications for the molecular recognition properties of mucins.
Keywords: AFM, DFS, SBA, lectin, ConA, mucin
Introduction
Mucins are heavily O-glycosylated glycoproteins that are secreted in higher organisms to protect and lubricate epithelia cell surfaces. Mucin and mucin-like domains are also involved in modulating immune response, inflammation, adhesion and tumorigenesis.1–3 The tandem repeat domains of mucins and mucin-like glycoproteins contain high contents of clustered Ser and Thr residues with many of these residues O-linked to a variety of “core” oligosaccharide structures including linear and branched chain blood group determinants.2 The oligosaccharide chains on mucins have been shown to be important for a variety of their biological properties including their interactions with animal lectins including selectins, siglecs and galectins, 3 as well as their linear structures.4
In addition to the variety of O-linked carbohydrates found on mucins, there are at least seventeen mucin gene products (MUC1-MUC17).1 These gene products represent two structurally and functionally distinct classes of mucins: secreted gel-forming mucins and transmembrane mucins. The transmembrane mucins such as MUC1 possess a cytoplasmic domain in addition to the extracellular O-glycosylated polypeptide tandem repeat domains.5 Evidence indicates that the C-terminal cytoplasmic domain of MUC1 is involved in signal transduction mechanisms including T-cell activation and inhibition, and adhesion signaling responses.6, 7
In addition to their normal biological properties, mucins appear to play important roles in cancer and metastasis.3, 4 In particular, the levels of expression of mucin peptide antigens and type of carbohydrate chains of mucins are diagnostic markers for a variety of cancers.1, 8 For example, MUC1 expression as detected immunologically is increased in colon cancers and is associated with a poor diagnosis.1 Colon cancer associated mucins also have differences in their core carbohydrate structures, often presenting shorter chain versions of normal mucins. Colon cancer mucins often have increased expression of the GalNAcαThr/Ser (Tn-antigen, GalNAc = N-acetyl-D-galactosamine ), Galβ3GalNAc (T or TF antigen, Gal = D-galactose) and NeuAcα6GalNAc (sialyl Tn-antigen).1 Importantly, recent studies have shown that binding of galectin-3, an endogenous Gal specific animal lectin, to cancer associated MUC1 causes increased endothelial cancer cell adhesion.9 Thus, the molecular recognition properties of cancer related mucins including their truncated carbohydrates are important in terms of gaining insight into the structure-activity properties of these aberrantly expressed glycoproteins.
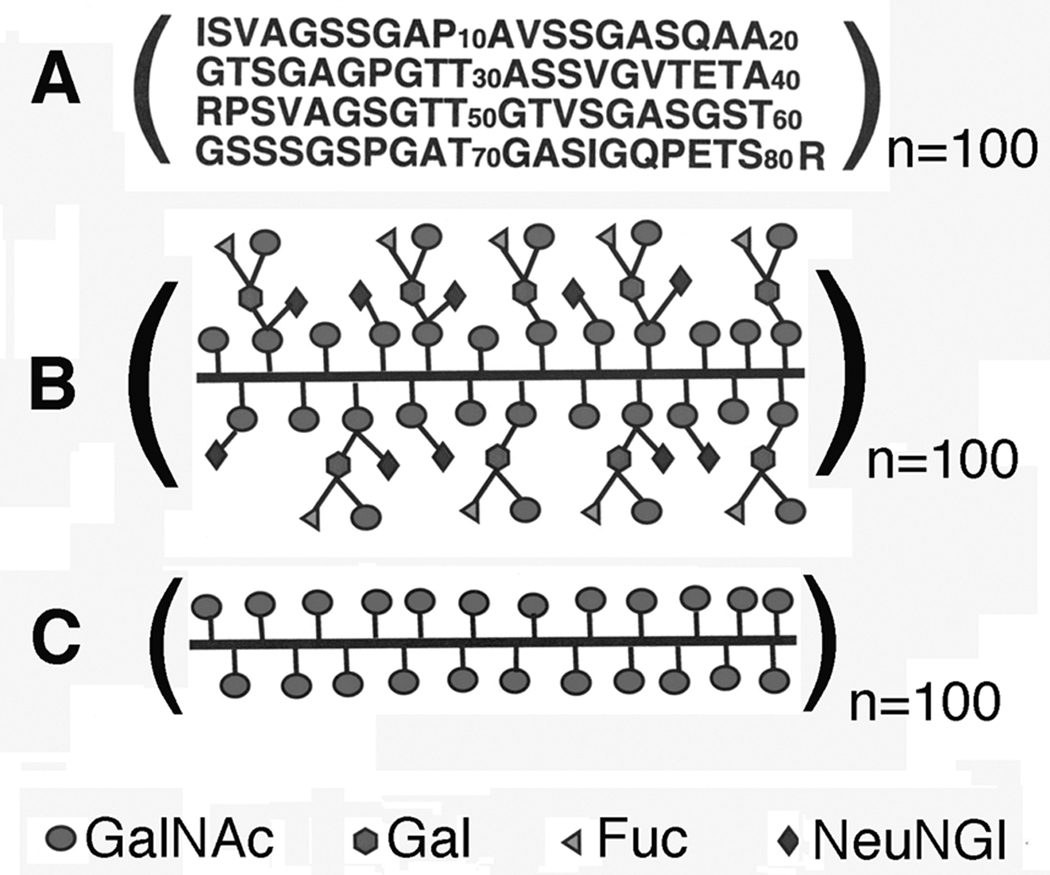
Porcine submaxillary mucin (PSM) is a physically well-characterized mucin, and the subject of studies of the regulation of O-glycosylation with glycosyl transferases.10 The cDNA sequence of porcine submaxillary mucin (PSM) has been determined,11 and the structures of the carbohydrate chains determined by chemical12 and NMR techniques.13 Figure 1a shows the amino acid sequence of the 100-repeat 81-residue polypeptide O-glycosylation domain of intact PSM. Gerken and Jentoft13 have isolated the O-glycosylated domain of PSM that possesses a molecular mass of ~106 daltons and is fully decorated with naturally occurring carbohydrates (Fd-PSM) (Figure 1b), which is a mixture of peptide linked α-GalNAc residues and the core 1 blood group A tetrasaccharide. Each of the mono- and tetrasaccharides is potentially glycosylated by a N-glycolylneuraminic acid (NeuNGl) residue. The O-glycosylation domain of PSM possessing only α-GalNAc residues (Tn-PSM) (Figure 1c) was also obtained using chemical and enzymatic treatments.14 The GalNAcα1-O-Ser/Thr residue(s) in Tn-PSM is the pancarcinoma carbohydrate antigen Tn that is aberrantly expressed in mucins such as MUC1 in adenocarcinomas.15
Figure 1.
Structural representations of A) the amino acid sequence of the 100-repeat 81-residue polypeptide O-glycosylation domain of intact PSM; B) the fully carbohydrate decorated form of the 100-repeat 81-residue polypeptide O-glycosylation domain of PSM (Fd-PSM); C) the 100-repeat 81-residue polypeptide O-glycosylation domain of PSM containing only peptide linked α-GalNAc residues (Tn-PSM). The value n is the number of tandem repeat domains.
Until recently, little has been known about the binding properties of mucins with other macromolecules and the effects of altered carbohydrate expression on these properties. However, studies of the binding of two GalNAc specific lectins, the soybean agglutinin (SBA) and Vatairea macrocarpa lectin (VML), to Fd-PSM, Tn-PSM, and two smaller fragments of Tn-PSM by isothermal titration microcalorimetry (ITC) measurements have been reported.16 Differences in the affinities and thermodynamic binding parameters were reported for SBA and VML binding to Tn-PSM as compared to Fd-PSM, indicating the importance of carbohydrate composition and epitope density of the affinities of these mucins for the lectins.16 Higher affinities of the two lectins for Tn-PSM relative to the two smaller fragments of Tn-PSM were also observed. A mechanism involving the lectins binding and jumping from GalNAc to GalNAc residue along the backbone of the mucins was proposed, similar to that observed for the binding of protein ligands to DNA.17
The traditional techniques used to investigate interactions between biological macromolecules such as kinetic binding studies, isothermal titration calorimetry, x-ray crystallography, nuclear magnetic resonance and other spectroscopic analyses provide indirect measurements of intermolecular binding forces. During the last decades ultrasensitive probes allowing direct measurements of intermolecular bonds at the single molecule level by means of force-distance curves have been developed 18–20. In order to study intermolecular interactions using these force probes the interacting molecules must be immobilised onto surfaces.21 Numerous studies have concentrated on both the theoretical and the experimental aspects of such measurements, and the use of this approach in studies of the structure and function of biological macromolecules, referred to as dynamic force spectroscopy (DFS), has evolved as an important and fascinating field. The present study reports DFS investigations of the binding of SBA to Tn-PSM.
Materials and Methods
Lectins and Tn-PSM
ConA was obtained from Sigma Chemical Company, and SBA was obtained from EY Biochemicals, Inc. The mucin sample used in this study was the O-glycosylation domain of PSM possessing only α-GalNAc residues (Tn-PSM) (Figure 1c). The sample was obtained using chemical and enzymatic treatments as previously described.14 The preparation protocoll and basic sample characterisation applying NMR, gel-filtration, and sequence analysis of both the polypeptide backbone and sugars were according to previous reports. 22,23 It has a length dispersity of 100 – 120 tandem repeat domains. All other reagents were of analytical grade.
Immobilization of lectins on mica substrates
Functionalization of the mica substrate and AFM tip were performed based on a previously reported method suitable for immobilization of a C5-epimerase and an alginate polysaccharide for determination of their interactions.24 In that study, the enzyme was conjugated by a primary amine to the aldehyde group of the topmost layer of the substrate, and the alginate by its carboxylate groups to the amin groups on funcionalised tips using EDC (N-Ethyl-N′-(3-dimethylaminopropyl)carbodiimide) as an activating agent. This method is well suited to immobilise molecules carrying carboxylate or amin groups. Freshly cleaved mica (mineral muscovite) was cleaned in 1:1 v/v concentrated HCl:MeOH (30 min), rinsed in MQ-water and silanized (freshly prepared 1% (v/v) solution of EDC in 1 mM acetic acid, for 20 min. at room temperature and rinsed in MQ-water). The amine-silanized layer was transformed to an aldehyde layer by incubating with glutaraldehyde (12.5% (v/v) in MQ-water, 14 hours, rinsed in MQ-water and dried under N2). ConA and SBA lectin were immobilized to the surface by conjugating their primary amine groups to the aldehyde groups of the immobilized cross linking agent (0.5 mg/ml lectin in 100 mM Hepes buffer pH 7.2 containing 1mM CaCl2 and 1mM MnCl2, 14 hours, room temperature).
Immobilization of Tn-PSM on AFM Tips
Tn-PSM was covalently anchored to the AFM tips (Si3N4 contact mode tips with nominal spring constants k = 0.06 nN/nm, silanized as the mica substrate) using EDAC (1-(3-dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride) (20 – 100 µg/ml Tn-PSM in 50 mM boric acid, pH 5.8, and 0.5–1.0 mg/ml EDAC, 1 hour). The density of Tn-PSM on the AFM tip was controlled by selecting the Tn-PSM concentration and incubation duration.
Dynamic Force Spectroscopy
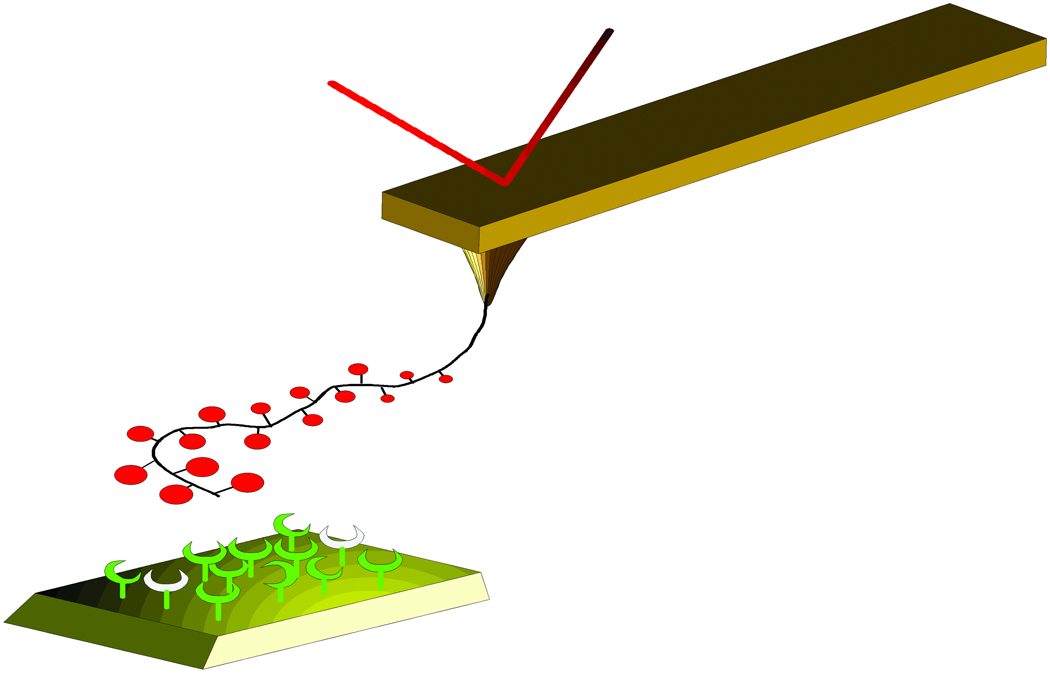
Force spectroscopy was carried out using a Digitial Instruments Multimode IIIa equipped with a PicoForce controller (Santa Barbara, CA). The force-distance curves were obtained at room temperature using the AFM with a picoForce scanner and a liquid cell filled with aqueous 100 mM Hepes buffer pH 7.2 containing 1mM CaCl2 and 1mM MnCl2. The spring constant of the AFM tips functionalised with Tn-PSM was determined using the Nanoscope software prior to each series of measurements. The determination is based on the observation of the thermal fluctuations of the cantilever. 25 The Tn-PSM functionalised AFM tips were then brought in contact with the lectins on mica and immediately retracted (Figure 2). The z-piezo retraction speed was varied from vret= 0.2 µm/s to 7.0 µm/s.
Figure 2.
Schematic illustration of the dynamic force spectroscopy experiments. The lectins are immobilized onto a mica surface and the Tn-PSM molecules are immobilized to the tip of an AFM cantilever which serves as a force transducer. The distance between the mica surface and tip is controlled by the z-piezo element of the AFM scanner. The surface is pulled down at a constant speed v. When the tip reaches the mica surface the Tn-PSM molecules attached to the tip comes in contact with, and may bind to, the lectins on the surface. As the tip is retracted from the surface, monotonically increasing forces act on the Tn-PSM-SBA complex.
Analyses
Unbinding between molecular pairs under external force is viewed as a scenario where the external force aids the thermal activation in the unbinding process and decreases the activation barrier(s) separating the bound from the free states. Consequently, the rate of dissociation under a constant loading force f, koff(f), increases as
| (1) |
, where xβ is the thermally averaged distance from the bound complex to the transition state projected along the direction of the applied force and kB T is the thermal energy. When the applied force along the unbinding pathway exceeds the force fβ governed by the distance xβ, i.e.,fβ = kB T / xβ, an exponential increase in the most likely unbinding force is predicted26–28:
| (2) |
Parameter rf is the actual force loading rate, and rf0 a thermal scale for loading rate, rf 0 = fβ / t0 where t0 is the inverse of the transition rate. The probability density P(f) for observing a bond rupture between a molecular pair at the force f subjected to constant force loading rate rf is:
| (3) |
The experiments carried out using retractive piezo velocity between 0.2 and 7 µm/s yielded observations of unbinding forces for various force loading rates. The loading rate acting at a molecular bond was determined for each force jump from the slope ΔF/Δt prior to each observed bond dissociation event. Force jumps that were located closer to the surface than 30 nm or that were not well separated from previous or subsequent unbinding events were not included in the analysis. The theoretical probability density function P(f) (Eq. 3) was fitted to the experimentally determined distributions of unbinding forces within an interval of force loading rates, using the parameters koff and xβ as fitting parameters. The most probable unbinding force f*, the rupture force, was determined from the fit of P(f) to the observed unbinding forces.
Results and Discussion
Design of Lectin and Tn-PSM Terminated Surfaces
A key prerequisite for successful investigation of molecular recognition forces by AFM is that receptor and ligand molecules are firmly anchored to solid surfaces while keeping sufficient mobility.29 In this study, Con A and SBA were immobilized on mica surfaces by allowing amino groups on the proteins to react covalently with aldehyde groups on functionalised mica surfaces. This approach has previously been shown to lead to a dense layer of active proteins on the mica surface.24 The Tn-PSM covalently anchored to the AFM tips using experimental conditions as described in the materials and methods section yielded functionalised samples suited for the interaction studies.
Observation of Single Molecular Pair Unbinding Events
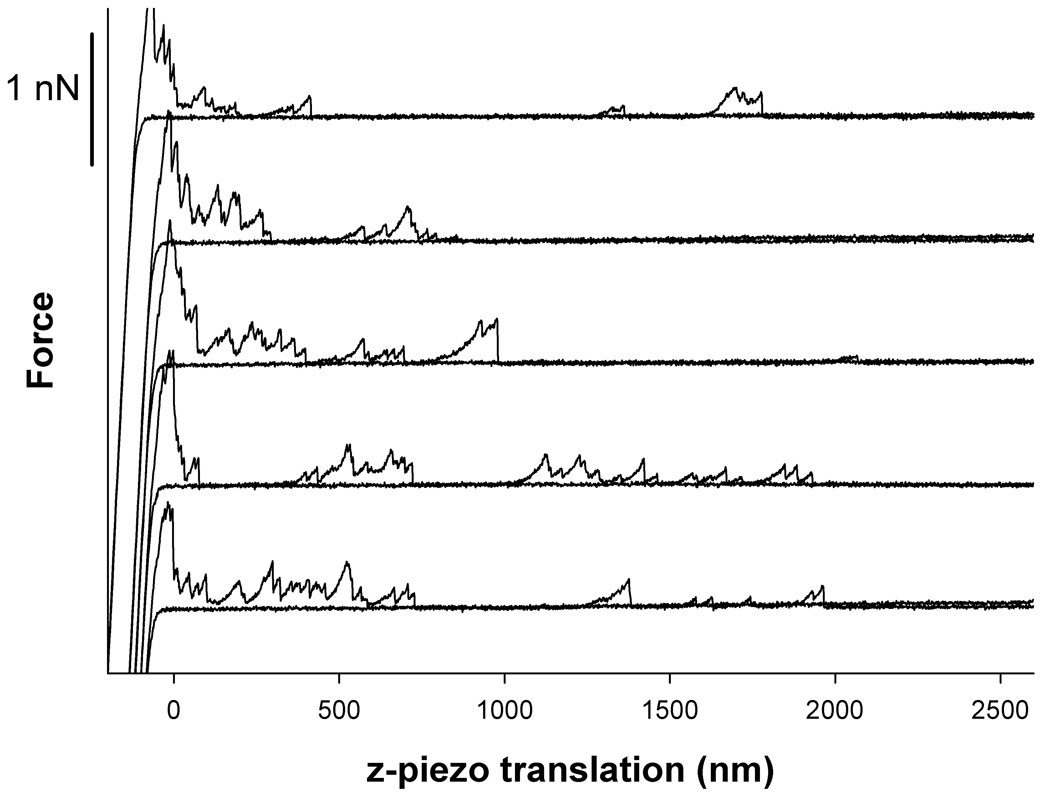
Figure 3 displays representative force – z-piezo retraction curves obtained for Tn-PSM covalently attached to the AFM tips interacting with SBA lectins immobilized onto mica. The curves display signatures of stretching of Tn-PSM molecules prior to force induced dissociation of the Tn-PSM – SBA lectin bound. The unbinding events were observed up to tip-surface separations up to about 2000 nm. This is about 40% of the chain length of Tn-PSM , which was found to be ~5 µm calculated from information related to the mass per unit length of Tn-PSM and the molecular length of the sample used. Existence of unbinding events up to such tip-surface separations are judged qualitatively to be consistent with the total chain length of the mucin sample when using the anchoring procedure yielding covalent binding to a point randomly along the chain
Figure 3.
Examples of force – z-piezo retraction curves for Tn-PSM functionalised AFM tips against SBA lectin immobilized on mica. The force-distance curves were obtained at room temperature in aqueous 100 mM Hepes buffer pH 7.2 containing 1mM CaCl2 and 1mM MnCl2. The AFM tips were functionalised by immersing them in solutions containing EDAC (0.5 mg/mL) and mucin (40 µg/mL) for 1 hour. Both approach and retrace curves are shown. The retract curves includes signatures of polymer stretching and forced unbinding of Tn-PSM chains that have bound to lectins on the mica surface.
The probability of observing an unbinding event will in AFM measurements depend on several parameters, including the density of active interacting molecules, both on tip and surface, as well as properties of the environment (pH, temperature, etc) and the delay time of the tip at the surface before retraction. To provide an experimental basis dominated by single-molecular pair unbinding events corresponding to the situation valid for the theoretical expressions used in the analysis, the fraction of force curves containing anchoring events should be kept low.28
In the present study well-isolated unbinding events were sometimes observed but the unbinding events showed a clear tendency of appearing in groups of ~3 – 4 events occurring within a small distance-interval from the surface. Reducing the density of Tn-PSM molecules on the tip was found to lead to a decreased probability to observe interaction events but did not influence on the probability of the unbinding events to arise in series. This behavior is not observed in previous studies focusing on the binding between a polysaccharide and an enzyme.24 SBA is a tetrameric lectin with one carbohydrate binding site per subunit.30 The unbinding events observed in the force curves may thus be due to different subunits of SBA binding to neighboring GalNAc residues along the Tn-PSM chain. Alternative explanations include the scenario where the same polymer chain is attached to several lectin molecules or because different Tn-PSM chains attached to the tip are anchored to different SBA molecules. The lack of correlation between the frequency of multiple interactions and the density of Tn-PSM molecules immobilized on the AFM tip is not compatible with the latter scenario. The observed behavior may be explained by the fact that once a Tn-PSM molecule has bound to one lectin molecule on the surface, it is kept in close proximity to the surface, and this fact leads to a large probability for neighboring segments along the Tn-PSM chain to bind to neighboring lectins on the surface. This mechanism is consistent with the bind and slide mechanism for SBA binding to Tn-PSM postulated by Dam et al.16
Specificity of Unbinding Events
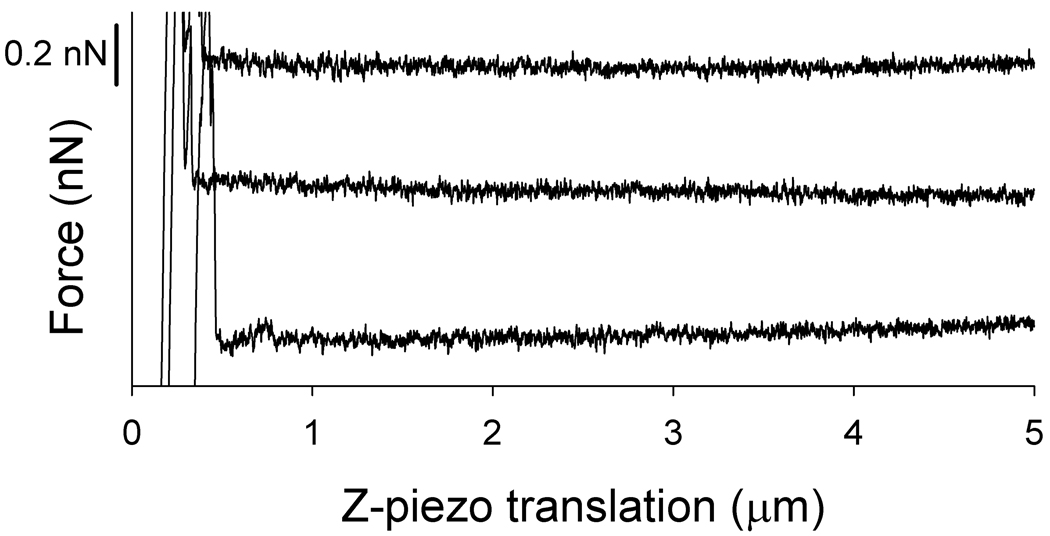
Tn-PSM – functionalised tips were retracted from both SBA lectin functionalised surfaces and from ConA functionalised surfaces. Whereas the force-curves obtained when retracting the tips from the SBA-functionalised surfaces contained numerous events of forced unbinding of Tn-PSM molecules, the curves obtained when retracting the Tn-PSM functionalised tips from ConA functionalised surfaces did not contain such events (Figure 4). The activity of the immobilised ConA proteins was affirmed by obtaining force curves upon retraction of ConA functionalised AFM tips from SBA functionalised surfaces. These curves cointained adhesive interactions reflecting the expected interaction between the ConA proteins and the mannose nonamer structures found on the surface of SBA (data not shown) The observed absence of interaction between ConA and Tn-PSM is in accordance with the previously determined binding specificities of ConA, which binds mannose and glucose residues, whereas SBA binds to galactose and N-acetylgalactosamine residues.30 The absence of forced unbinding events when retracting the functionalised AFM tips from the ConA surfaces is thus an indication of the specificity of the binding events observed when retracting tips from SBA functionalised surfaces and interpreted as specific SBA lectin – Tn-PSM interactions.
Figure 4.
Examples of force – z-piezo retraction curves for Tn-PSM functionalised AFM tips against ConA lectin immobilized on mica. The force-distance curves were obtained at room temperature in aqueous 100 mM Hepes buffer pH 7.2 containing 1mM CaCl2 and 1mM MnCl2. Only the retract curves are shown in the figure.
Dynamic Force Spectroscopy Measurements of SBA – Tn-PSM Interactions
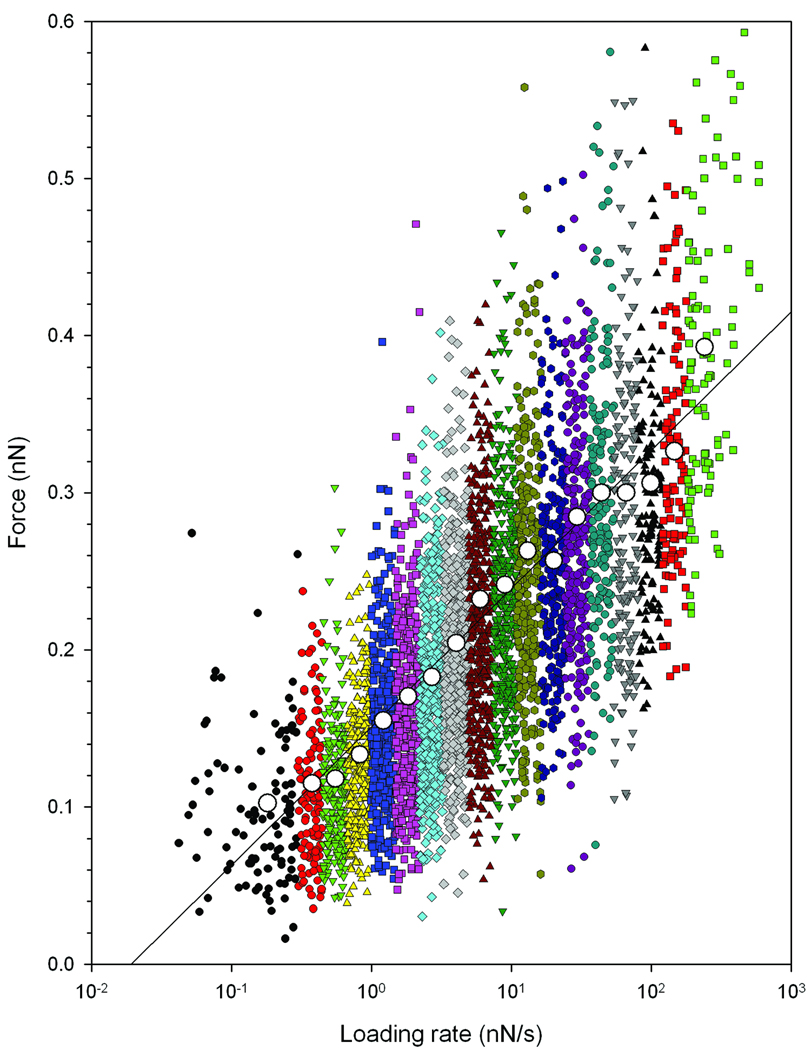
Force curves were obtained using retractive piezo velocities in the interval 0.2 to 7 µm/s. In total 4826 force jumps were identified and for each of these force jumps the unbinding force as well as the loading rate was determined. This analysis indicated that the data included observations of loading rates from 0.05 nN/s to 500 nN/s. A linear increase of the unbinding forces with the logarithm of the loading rates was found (Figure 5). This behaviour is characteristic for a thermally activated dissociation under an applied load.27, 28, 31
Figure 5.
Distribution of experimentally determined Tn-PSM – SBA unbinding forces at increasing force loading rates. The data are collected from 1260 force curves obtained using six different tip retraction speeds in the interval 0.2 – 7 µm/s. The loading rate acting at a molecular bond was determined for each force jump from the slope Δf/Δt prior to each observed bond dissociation event. Based on the determined loading rate, the continuous distribution of observations was divided into 18 subgroups within a range of loading rates. The most probable unbinding force f*, determined from the peak in the histograms as explained in figure 6 is visualized in the plot (large points).
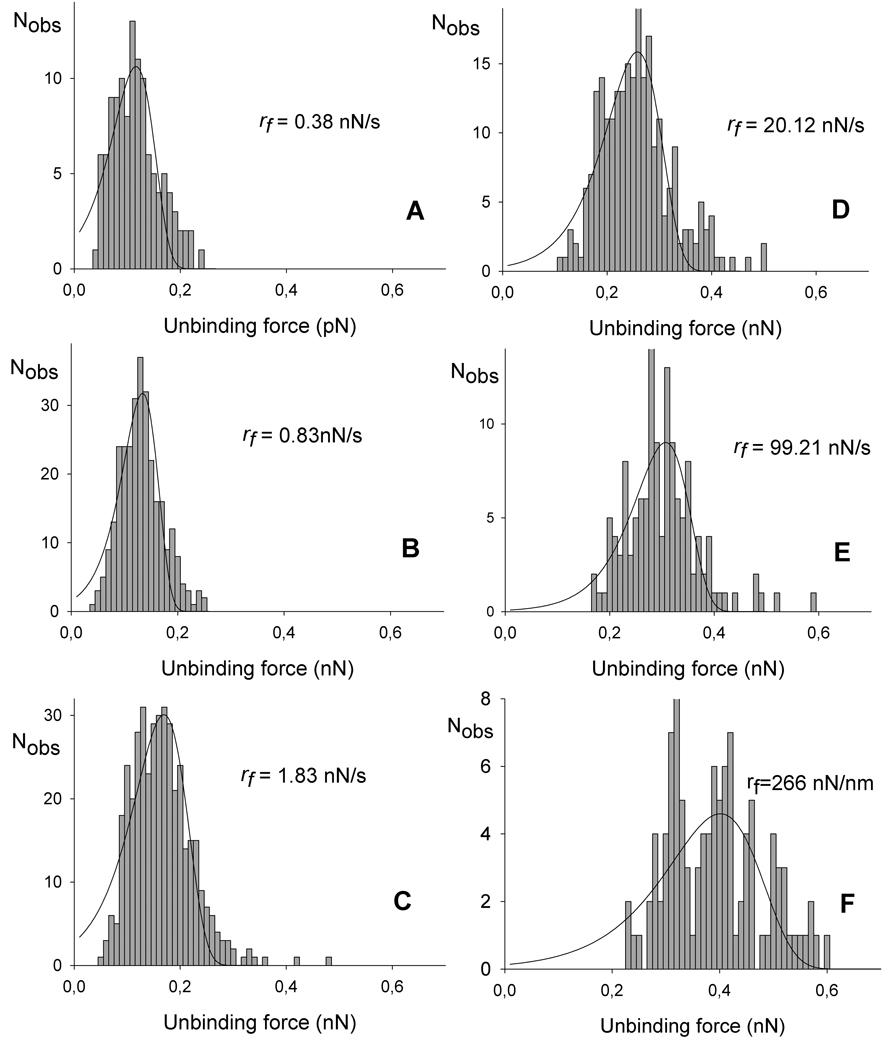
The observations were grouped into 18 subgroups based on the determined loading rate as illustrated in Figure 5. For each subinterval a mean loading rate was calculated, and the values obtained are presented in Table 1. As shown the mean loading rates calculated for the intervals increased from 0.18 nN/s to 266 nN/s. The magnitude of the observed unbinding forces increased with increasing loading rate. For a mean force loading rate equal to 0.38 nN/s the observed unbinding forces were in the interval 35 to 237 pN with a most probable unbinding force equal to 115 pN, while at the higest loading rate investigated, i.e. 266 nN/s the most probable unbinding force was equal to 402 pN (see Figure 6 and Table 1). These values are within the range observed for other protein – carbohydrate interactions 32. The observed unbinding forces are also in all cases less than 20% of the unbinding forces of covalent bonds reported from 2.0± 0.3 nN for Si-C to 4.1 – 4.3 nN for C-C and C-O bonds, respectively.33 Potential interference from covalent interactions in the distributions of the unbinding forces are therefore not likely.
Table 1.
Estimated parameters characterising the energy-landscape of the Tn-PSM – SBA interaction.
| Intervall | Number of observations |
rf | koff | xβ | f* | r |
|---|---|---|---|---|---|---|
| (nN/s) | s-1 | nm | pN | |||
| 1 | 98 | 0.18 | 0.68 ± 0.10 | 0.09 ± 0.008 | 103 | 0.92 |
| 2 | 117 | 0.38 | 0.53 ± 0.07 | 0.10 ± 0.006 | 115 | 0.94 |
| 3 | 214 | 0.55 | 0.59 ± 0.10 | 0.12 ± 0.007 | 118 | 0.93 |
| 4 | 299 | 0.83 | 0.46 ± 0.07 | 0.12 ± 0.006 | 133 | 0.96 |
| 5 | 392 | 1.2 | 0.43 ± 0.09 | 0.12 ±0.006 | 155 | 0.95 |
| 6 | 408 | 1.8 | 1.28 ± 0.17 | 0.08 ± 0.004 | 171 | 0.96 |
| 7 | 489 | 2.7 | 1.31 ± 0.24 | 0.09 ± 0.005 | 183 | 0.93 |
| 8 | 511 | 4.0 | 1.44 ± 0.28 | 0.08 ± 0.005 | 204 | 0.94 |
| 9 | 415 | 6.0 | 2.78 ± 0.41 | 0.06 ± 0.003 | 232 | 0.94 |
| 10 | 384 | 9.0 | 2.50 ± 0.48 | 0.07 ± 0.004 | 241 | 0.94 |
| 11 | 319 | 13 | 3.43 ± 0.80 | 0.06 ± 0.004 | 263 | 0.90 |
| 12 | 242 | 20 | 3.01 ± 0.69 | 0.08 ± 0.004 | 257 | 0.94 |
| 13 | 243 | 30 | 5.39 ± 1.34 | 0.06 ± 0.004 | 285 | 0.91 |
| 14 | 173 | 44 | 6.23 ± 2.12 | 0.06 ± 0.005 | 300 | 0.85 |
| 15 | 165 | 99 | 5.67 ± 2.20 | 0.08 ± 0.006 | 306 | 0.89 |
| 16 | 134 | 147 | 13.1 ± 5.43 | 0.07 ± 0.006 | 326 | 0.83 |
| 17 | 115 | 216 | 12.8 ± 7.33 | 0.06 ± 0.007 | 383 | 0.76 |
| 8 | 108 | 266 | 31.1 ± 13.2 | 0.05 ± 0.005 | 402 | 0.80 |
Figure 6.
Histograms based on the observed unbinding forces within each subgroup displayed in figure 5. Fitting Eq. 3 to the experimental data allows determination of the parameters xβ, koff and f*. The fitted curves are overlaid on the distributions presented.
The probability density function P(f) (Eq. 3) was fitted to the experimentally determined distributions of unbinding forces within an interval of force loading rates, using the parameters koff and xβ as fitting parameters (Figure 6). The most probable unbinding force f*, the rupture force, determined from the fit of P(f) to the observed unbinding forces was found to increase from 103 pN at a mean loading rate of 0.18 nN/s to 402 pN at a loading rate of 266 nN/s for Tn-PSM - SBA interactions (Table 1).
The results of the determination of separation distances xβ from the fit of Eq. 3 to the experimental data are presented in Table 1. The results indicate that parameter xβ is constant independent of the loading rates in the experimental range investigated in this study, with an average value of xβ =0.09± 0.02nm. This number is determined based on the fits giving a coefficient of determination R equal to or greater than 0.9. An alternative approach to determine xβ is based on the slope of the linear regimes seen in the dynamic force spectra (Figure 5). This slope is defined as kBT/xβ, and thus reflects the position of the activation barriers situated along the direction of applied force. The slope observed in Figure 5 positions the rate limiting barrier in the energy landscape of the Tn-PSM – SBA complex at xβ = 0.1 nm. This way of estimating the value of xβ yields consistent results with the approach based on analysis of the distributions of the unbinding events within various intervals of force loading rates. The distance from the bound complex to the activation barrier indicated by xβ = 0.1 nm governing the strength of the Tn-PSM - SBA interaction is within the range reported for other biomolecular interactions. One example in this respect is the xβ ≈ 0.12 nm, xβ ≈ 0.3 nm and xβ ≈ 3 nm for the high, intermediate and low strength regime reported for the avidin-biotin interaction34 as well as xβ ≈ 0.26 nm determined for the mannuronan AlgE4 interaction. 24
The values of koff obtained by fitting the parameters in Eq. 3 to the distributions of experimentally determined unbinding forces are presented in Table 1. The values obtained when looking at observations collected when applying a loading rate up to 1.2 nN/s give a mean value koff equal to 0.54 ± 0.04 s−1. The values determined when fitting Eq. 3 to distributions of unbinding forces for rf larger than 1.2nN/s increases with increasing loading rate. This behaviour is expected in dynamic force spectroscopy since an applied force f decreases the activation energy for dissociation from ΔE‡ to ΔE‡(f) with the relation ΔE‡ - ΔE‡(f) = −xβf, leading to an exponential increase of the off-rate with increasing force, as described in Eq. 1. The value of koff was also obtained by extrapolating the rupture force to zero loading rate following the procedure of Schwesinger and coworkers. 35 It was found to be 0.76 ± 0.09 s−1 assuming xβ ≈ 0.09 nm.
The value obtained for koff, reflects an average duration of the Tn-PSM-lectin interaction in the range 1.3 –1.9 seconds. The available literature related to the kinetics of saccharide – lectin binding is limited. However, data have been obtained for affinity constants (Ka) as well as the kinetic parameters kon and koff describing the forward and reverse rate constants, respectively, in solution. The Ka values for the binding of monosaccharides to ConA and mono- and disaccharides to lysozyme have been published, as well as the forward and reverse rate constants. 36 For the binding of methyl α- and β-D-glucopyranoside to ConA, koff equal to 30 s−1 and 400 s−1, respectively.These values are significantly larger than the value koff = 0.76 ± 0.09 s−1 determined in the present study. The long lifetime indicated by koff = 0.76 ± 0.09 s−1 is compatible with a binding model in which lectin molecules “bind and jump” from α-GalNAc residue to α-GalNAc residue along the polypeptide chain of Tn-PSM before dissociating, as previously proposed for this system.16
It should be noted that some challenges are faced when attempting to compare information about lifetimes of intermolecular bonds obtained in dynamic force spectroscopy experiments with Ka, kon and koff determined in solution studies. The kinetics of binding and dissociation of carbohydrates from lectins have been observed to be different in 2-D systems (i.e., surface plasmon resonance and other solid phase binding assays) compared to solution kinetics, with much slower kinetics for both rate constants in the 2-D systems.37, 38 AFM based DFS studies usually necessities immobilisation of the interacting molecules onto surfaces, and effects of constraining the interacting molecular pair to the surfaces should be taken into account when comparing data with data obtained from experiments in solutions. Off-rates determined in AFM studies for simple sugars binding to lectins and antibodies reveal that dissociation rate constants obtained when using these techniques 39 are several orders of magnitude smaller than those determined in solution studies. 36 Other factors that may explain the deviating observations are related to the fact that association between bound molecules may be heavily dependent on properties such as binding length or flexibility that are not important in solution.40
Conclusions
Dynamic force microscopy experiments in the present study have provided insight into the physical interactions of SBA with Tn-PSM, a high molecular weight mucin that possesses the Tn blood group pancarcinoma antigen. Force profiles of the interactions show multiple unbinding/rebinding events of SBA with Tn-PSM that occur over a length of 2.0 µm, which is consistent with the length of the mucin polypeptide chain. The kinetics of complete dissociation of the lectin from the mucin is also consistent with repeated unbinding/rebinding events between the two molecules. These findings are consistent with a binding model that involves “binding and jumping” of SBA from GalNAc to GalNAc residue along the mucin chain. This mechanism has previously been suggested for SBA binding to Tn-PSM based on thermodynamic binding data obtained by ITC.16 Furthermore, the bind and jump mechanism is similar to that observed for the binding of protein ligands to DNA, and suggest a common conserved binding mechanism of ligands to the two biopolymers and possibly between ligands and all biopolymers, as recently suggested.41
Acknowledgements
This work was supported by Grant CA-16054 from the National Cancer Institute, Department of Health, Education and Welfare, Core Grant P30 CA-13330 from the same agency (C. F. B.), Grant CA-78834 from the National Institues of Health, National Cancer Institute (T. A. G.) and Grant 121894/420 from the Norwegian Research Council (M.S. and B.T.S.).
Abbreviations
- SBA
soybean agglutinin
- VML
Vatairea macrocarpa lectin
- ConA
concanavalin A
- PSM
porcine submaxillary mucin
- Fd-PSM
fully carbohydrate decorated porcine submaxillary mucin
- Tn-PSM
porcine submaxillary mucin containing GalNAcα1-O-Ser/Thr residues
- AFM
atomic force microscopy
- DFS
dynamic force spectroscopy
References
- 1.Byrd JC, Bresalier RS. Cancer Metastasis Rev. 2004;23:77–99. doi: 10.1023/a:1025815113599. [DOI] [PubMed] [Google Scholar]
- 2.Fukuda M. Biochim. Biophys. Acta. 2002;1573:394–405. doi: 10.1016/s0304-4165(02)00409-9. [DOI] [PubMed] [Google Scholar]
- 3.Hollingsworth MA, Swanson BJ. Nat. Rev. Cancer. 2004;4:45–60. doi: 10.1038/nrc1251. [DOI] [PubMed] [Google Scholar]
- 4.Hang HC, Bertozzi CR. Bioorg. Med. Chem. 2005;13:5021–5034. doi: 10.1016/j.bmc.2005.04.085. [DOI] [PubMed] [Google Scholar]
- 5.Hanisch F-G, Muller S. Glycobiology. 2000;10:439–449. doi: 10.1093/glycob/10.5.439. [DOI] [PubMed] [Google Scholar]
- 6.Hakomori S. Proc. Nat. Acad. Sci. USA. 2002;99:225–232. doi: 10.1073/pnas.012540899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh PK, Hollingsworth MA. TRENDS Cell Biol. 2006;16:467–476. doi: 10.1016/j.tcb.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 8.Dube DH, Bertozzi CR. Nature Rev. Drug Disc. 2005;4:477–488. doi: 10.1038/nrd1751. [DOI] [PubMed] [Google Scholar]
- 9.Yu L-G, Andrews N, Zhao Z, McKean D, Williams JF, Connor LJ, Gerasimenko OV, Hilkens J, Hirabayashi J, Kasai K, Rhodes JM. J. Biol. Chem. 2007;282:773–781. doi: 10.1074/jbc.M606862200. [DOI] [PubMed] [Google Scholar]
- 10.Gerken TA. Biochemistry. 2004;43:4137–4142. doi: 10.1021/bi036306a. [DOI] [PubMed] [Google Scholar]
- 11.Eckhardt AE, Timpte CS, DeLuca AW, Hill RL. J Biol. Chem. 1997;272:33204–33210. doi: 10.1074/jbc.272.52.33204. [DOI] [PubMed] [Google Scholar]
- 12.Carlson D. J. Biol. Chem. 1968;243:616–626. [PubMed] [Google Scholar]
- 13.Gerken TA, Jentoft N. Biochemistry. 1987;26:4689–4699. doi: 10.1021/bi00389a015. [DOI] [PubMed] [Google Scholar]
- 14.Gerken TA, Owens CL, Pasumarthy M. J. Biol. Chem. 1997;272:9709–9719. doi: 10.1074/jbc.272.15.9709. [DOI] [PubMed] [Google Scholar]
- 15.Sorensen AL, Reis CA, Tarp MA, Sankaranarayanan V, Schwientek T, Graham R, Taylor-Papadimitriou J, Hollingsworth MA, Burchell J, Clausen H. Glycobiology. 2006;16:96–107. doi: 10.1093/glycob/cwj044. [DOI] [PubMed] [Google Scholar]
- 16.Dam TK, Gerken TA, Cavada BS, Nascimento KS, Moura TR, Brewer CF. J. Biol. Chem. 2007;282:28256–28263. doi: 10.1074/jbc.M704677200. [DOI] [PubMed] [Google Scholar]
- 17.von Hippel PH. Annu. Rev. Biophys. Biomolec. Struct. 2007;36:79–105. doi: 10.1146/annurev.biophys.34.040204.144521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Evans EA, Calderwood DA. Science. 2007:1148–1153. doi: 10.1126/science.1137592. [DOI] [PubMed] [Google Scholar]
- 19.Deniz AA, Mukhopadhyay S, Lemke EA. J. Royal Soc. Interface. 2008;5:15–45. doi: 10.1098/rsif.2007.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bustamante C, Chemla YR, Forde NR, Izhaky D. Annu. Rev. Biochem. 2004;73:705–748. doi: 10.1146/annurev.biochem.72.121801.161542. [DOI] [PubMed] [Google Scholar]
- 21.Florin E-L, Moy VT, Gaub HE. Science. 1994;264:415–417. doi: 10.1126/science.8153628. [DOI] [PubMed] [Google Scholar]
- 22.Gerken TA, Owens CL, Pasumarthy M. J. Biol. Chem. 1998;273:26580–26588. doi: 10.1074/jbc.273.41.26580. [DOI] [PubMed] [Google Scholar]
- 23.Gerken TA, Gilmore M, Zhang J. J. Biol. Chem. 2002;277:7736–7751. doi: 10.1074/jbc.M111690200. [DOI] [PubMed] [Google Scholar]
- 24.Sletmoen M, Skjåk-Braek G, Stokke BT. Biomacromolecules. 2004;5:1288–1295. doi: 10.1021/bm0345211. [DOI] [PubMed] [Google Scholar]
- 25.Florin E-L, Rief M, Lehmann H, Ludwig M, Dornmair C, Moy VT, Gaub HE. Biosens. Bioelectron. 1995;10:895–901. [Google Scholar]
- 26.Bell GI. Science. 1978;200:618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- 27.Evans E, Ritchie K. Biophys. J. 1997;72:1541–1555. doi: 10.1016/S0006-3495(97)78802-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Evans E. Faraday Discuss. 1998;111:1–16. doi: 10.1039/a809884k. [DOI] [PubMed] [Google Scholar]
- 29.Hinterdorfer P, Baumgartner W, Gruber HJ, Schilcher K, Schindler H. Proc. Nat. Acad. Sciences USA. 1996;93:3477–3481. doi: 10.1073/pnas.93.8.3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goldstein IJ, Poretz RD. In: The Lectins. Liener IE, Sharon N, Goldstein IJ, editors. New York: Academic Press; 1986. pp. 35–244. [Google Scholar]
- 31.Merkel R, Nassoy P, Leung A, Ritchie K, Evans E. Nature. 1999;397:50–53. doi: 10.1038/16219. [DOI] [PubMed] [Google Scholar]
- 32.Lee C-K, Wang Y-M, Huang L-S, Lin S. Micron. 2007;38:446–461. doi: 10.1016/j.micron.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 33.Grandbois M, Beyer M, Rief M, Clausen-Schumann H, Gaub HE. Science. 1999;283(5408):1727–1730. doi: 10.1126/science.283.5408.1727. [DOI] [PubMed] [Google Scholar]
- 34.Moy VT, Florin EL, Gaub HE. Science. 1994;264:415–417. doi: 10.1126/science.8153628. [DOI] [PubMed] [Google Scholar]
- 35.Schwesinger F, Ros R, Strunz T, Anselmetti D, Guntherodt H-J, Honegger A, Jermutus L, Tiefenauer L, Pluckthun A. Proc. Nat. Acad. Sci. USA. 2000;97:9972–9977. doi: 10.1073/pnas.97.18.9972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brewer CF, Marcus D, Grollman AP, Sternlicht H. In: Lysozyme. Osserman EF, Canfield RE, Beychok S, editors. New York: Academic Press; 1974. pp. 239–250. [Google Scholar]
- 37.Dusten ML, Furguson LM, Chan P-Y, Springer TA, Golan DE. J. Cell Biol. 1996;132:465–474. doi: 10.1083/jcb.132.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pierres A, Benoliel AM, Zhu C, Bongrand P. Diffusion of microspheres in shear flow near a wall: use to measure binding rates between attached molecules. Biophys. J. 2001;81:25–42. doi: 10.1016/S0006-3495(01)75677-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dettmann W, Grandbois M, Andre S, Benoit M, Wehle AK, Kaltner H, Gabius H-J, Gaub HE. Arch. Biochem. Biophys. 2000;383:157–170. doi: 10.1006/abbi.2000.1993. [DOI] [PubMed] [Google Scholar]
- 40.Robert P, Benoliel A-M, Pierres A, Bongrand P. J. Molec. Recogn. 2007;20:432–447. doi: 10.1002/jmr.827. [DOI] [PubMed] [Google Scholar]
- 41.Dam TK, Brewer CF. Biochemistry. 2008;47:8470–8476. doi: 10.1021/bi801208b. [DOI] [PubMed] [Google Scholar]