Abstract
Chloroquine (CQ) resistance (CQR) in Plasmodium falciparum originated from at least six foci in South America, Asia, and Oceania. Malaria parasites from these locations exhibit contrasting resistance phenotypes that are distinguished by point mutations and microsatellite polymorphisms in and near the CQR transporter gene, pfcrt, and the multidrug resistance transporter gene, pfmdr1. Amodiaquine (AQ), a 4-aminoquinoline related to CQ, is recommended and often used successfully against CQ-resistant P. falciparum in Africa, but it is largely ineffective across large regions of South America. The relationship of different pfcrt and pfmdr1 combinations to these drug-resistant phenotypes has been unclear. In two P. falciparum genetic crosses, particular pfcrt and pfmdr1 alleles from South America interact to yield greater levels of resistance to monodesethylamodiaquine (MDAQ; the active metabolite of AQ) than to CQ, whereas a pfcrt allele from Southeast Asia and Africa is linked to greater CQ than MDAQ resistance with all partner pfmdr1 alleles. These results, together with (i) available haplotype data from other parasites; (ii) evidence for an emerging focus of AQ resistance in Tanzania; and (iii) the persistence of 4-aminoquinoline-resistant parasites in South America, where CQ and AQ use is largely discontinued, suggest that different histories of drug use on the two continents have driven the selection of distinct suites of pfcrt and pfmdr1 mutations. Increasing use of AQ in Africa poses the threat of a selective sweep of highly AQ-resistant, CQ-resistant parasites with pfcrt and pfmdr1 mutations that are as advantaged and persistent as in South America.
Keywords: malaria, pfcrt, pfmdr1
Chloroquine (CQ) and amodiaquine (AQ) are structurally related 4-aminoquinoline drugs (1) that have had important applications against malaria for >70 years (2). Although both of these compounds are thought to have similar mechanisms of action against Plasmodium falciparum (3), differential clinical responses have been observed with their use (4–6). In Africa, where CQ-resistant strains of P. falciparum are now prevalent, AQ has remained sufficiently efficacious for the World Health Organization to recommend it as a partner in drug combinations including artemisinin combination therapy (ACT) (7). However, poor in vivo responses and outright failures of AQ against P. falciparum were described decades ago in India, Southeast Asia, the Philippines, South America, and Papua New Guinea (PNG) (8–12) and have been seen more recently in Africa (13, 14).
Reports have pointed to a number of puzzling observations on CQ resistance (CQR) and AQ resistance (AQR) and the relationship between them. In South America, where in vitro IC50 levels indicate only moderate levels of resistance to CQ but high levels of resistance to monodesethylamodiaquine (MDAQ) (15), the principal active metabolite of AQ (16), drug-resistant P. falciparum strains have stably saturated large regions of the continent for half a century even where use of these drugs was discontinued. Such dominance in South America contrasts dramatically with changes that have been observed after discontinued drug pressure in certain countries of Africa and Southeast Asia. For example, a region of Malawi known for highly prevalent CQR was repopulated with drug-sensitive parasites within 10 years after CQ use was stopped (17). Similar recovery of CQ-sensitive P. falciparum populations was recently reported in Kenya and has also been observed in China (18, 19). These changes in the absence of drug pressure have been explained by fitness costs that are carried by CQ-resistant mutants (20). However, such a selective disadvantage has been less apparent in South America where CQ-sensitive parasites have not replaced their CQ-resistant counterparts. A satisfactory explanation for this difference between Southeast Asian/African and South American forms of CQR has not been proposed.
4-Aminoquinoline-resistant parasites from different continents can be distinguished in some cases by chemosensitization with verapamil (VP), a “reversal” phenomenon characteristic of CQ-resistant but not CQ-sensitive P. falciparum (21). For example, African CQ-resistant parasites have been found to be more readily reversed than CQ-resistant parasites from South America (22).
Molecular-epidemiological studies have identified at least six independent origins of CQR in different regions of the world (23). These origins are distinguished by codon mutations and flanking polymorphisms of pfcrt, the transporter-encoding gene responsible for CQR (24, 25). Additional P. falciparum genes may modulate resistance once it is established; among these is the P-glycoprotein homolog (Pgh-1)-encoding gene, pfmdr1, which has been linked to higher IC50 levels of drug response in some resistant strains, but not others (26, 27). These different results have led to alternative conclusions about the relationships of pfcrt and pfmdr1 mutations in the response phenotypes of malaria parasites from different geographical origins.
Here, we explore the genetics of the different features of CQ- and AQ-resistant P. falciparum parasites from different continents. Using a genetic cross between parasite clones from Brazil and Ghana (7G8×GB4) (28), we provide information on the linkage of pfcrt and pfmdr1 haplotypes from these geographical regions to the levels of response to CQ, AQ, and their respective metabolites monodesethlychloroquine (MDCQ) and MDAQ. From this information, we suggest why AQ treatment is often useful against CQ-resistant P. falciparum in Africa but is generally ineffective against parasites that show lower levels of CQR in South America and other malarious regions such as Papua New Guinea. Finally, we hypothesize why South America remains saturated with CQ-resistant P. falciparum in the absence of CQ pressure, whereas removal of CQ pressure from certain African and Southeast Asian regions has led to their relatively rapid repopulation with CQ-sensitive P. falciparum.
Results
Inherited Features of 4-Aminoquinoline Resistance Associated with pfcrt and pfmdr1 Mutations.
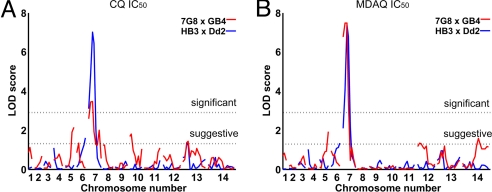
To identify mutations associated with the different features of 4-aminoquinoline drug resistance in parasites from South America and Africa, we performed quantitative trait loci analysis (QTL) on the in vitro IC50 and IC90 drug responses of 32 previously described progeny from the 7G8×GB4 cross (7G8, Brazil; GB4, Ghana) (28). Responses to CQ, AQ, and their respective active metabolites, MDCQ and MDAQ (Fig. 1, Fig. S1, and Table S1) were scanned against full ≈5-cM linkage maps of the 7G8×GB4 progeny (www.cell.com/cell-host-microbe/supplemental/S1931–3128(08)00185-6). Results from these scans showed consistent significant or suggestive associations of pfcrt and pfmdr1 peaks with 4-aminoquinoline responses in the 7G8×GB4 cross and association of only the pfcrt peak with VP reversal (Fig. 2 and Fig. S2; encoded amino acid polymorphisms are listed in Table 1). Copy number variations of the pfcrt or pfmdr1 alleles could not be associated with the different 7G8 and GB4 drug response phenotypes, because the 7G8 and GB4 parents carry single copies of these genes (H. Jiang and X. Su, personal communication). We also performed linkage scans of drug responses from an earlier cross between a CQ-sensitive clone from Central America [HB3; which carries a single copy of pfmdr1 (29)] and a CQ-resistant clone from Southeast Asia [Dd2; which carries three or four copies of pfmdr1 (29)]. Consistent with previous results from this HB3×Dd2 cross (30), CQR was associated only with a peak of pfcrt inheritance, with no significant linkage to pfmdr1 inheritance (Fig. 2 and Fig. S2).
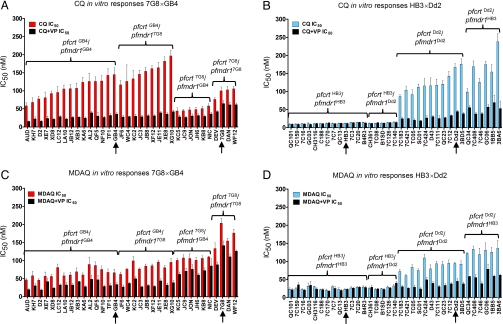
Fig. 1.
In vitro CQ and MDAQ responses of individual P. falciparum clones. Mean CQ IC50 values of parents (arrows) and progeny from the 7G8×GB4 (A) and HB3×Dd2 (B) crosses and mean MDAQ IC50 values of parents (arrows) and progeny from the 7G8×GB4 (C) and HB3×Dd2 (D) crosses are shown. The mean values and standard errors are from at least six independent drug assays in the absence (red, for 7G8×GB4 and blue, for HB3×Dd2) or presence (black) of 0.8 μM VP. Inherited pfcrt and pfmdr1 alleles are indicated above each group.
Fig. 2.
QTL analysis of in vitro drug responses from the 7G8×GB4 and HB3×Dd2 genetic crosses. Results of QTL scans using the IC50 values of CQ (A) and MDAQ (B) are shown. Each graph represents the logarithm of odds scores of the individual markers ordered along the 14 P. falciparum chromosomes. Dashed lines correspond to the 95% C.I. threshold (significant) and 63% C.I. threshold (suggestive) calculated from 1,000 permutations. Small differences on the exact placement of peaks reflect differences in the genetic distances of linkage markers observed in the 7G8×GB4 and HB3×Dd2 crosses.
Table 1.
Predicted PfCRT and Pgh-1 polymorphisms, drug responses, and VP response modification indices of P. falciparum clones 7G8, GB4, HB3, and Dd2
| Parasite clone | PfCRT |
Pgh-1 |
IC50,nM |
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 72 | 74 | 75 | 76 | 97 | 220 | 271 | 326 | 356 | 371 | 86 | 184 | 1034 | 1042 | 1246 | CQ | CQ + VP | RMI* | MDAQ | MDAQ + VP | RMI* | |
| 7G8 | S | M | N | T | H | S | Q | D | L | R | N | F | C | D | Y | 99.8 ± 10.8 | 64.4 ± 8.3 | 0.64 | 203.8 ± 13.2 | 141.4 ± 13.0 | 0.69 |
| GB4 | C | I | E | T | H | S | E | S | T | I | Y | F | S | N | D | 144.8 ± 18.8 | 35.3 ± 4.2 | 0.30 | 66.6 ± 9.6 | 33.8 ± 4.5 | 0.50 |
| HB3 | C | M | N | K | H | A | Q | N | I | R | N | F | S | D | D | 12.3 ± 0.5 | 11.7 ± 0.8 | 0.95 | 22.8 ± 1.1 | 23.0 ± 0.6 | 1.00 |
| Dd2 | C | I | E | T | H | S | E | S | T | I | Y | Y | S | N | D | 167.1 ± 10.4 | 44.2 ± 3.6 | 0.26 | 94.1 ± 3.7 | 45.2 ± 6.7 | 0.48 |
*Response modification index defined as the ratio of the IC50 in the presence of VP over that without VP (22); values closer to 1 are associated with lower susceptibility to VP chemosensitization. Amino acid polymorphisms are indicated under the codon position using standard one-letter notation.
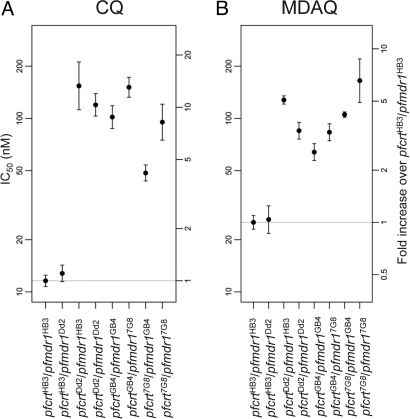
Having confirmed pfcrt and pfmdr1 as determinants of CQR and AQR in the 7G8×GB4 cross, we sought additional insights on the contributions of these loci to the drug responses of the 7G8×GB4 and HB3×Dd2 progeny phenotypes. Considering the haploid state of mammalian stage Plasmodium, we assigned the progeny into eight groups according to their pfcrt and pfmdr1 alleles and calculated the geometric mean drug responses of each group: (i) pfcrtHB3 + pfmdr1HB3; (ii) pfcrtHB3 + pfmdr1Dd2; (iii) pfcrtDd2 + pfmdr1HB3; (iv) pfcrtDd2 + pfmdr1Dd2; (v) pfcrtGB4 + pfmdr1GB4; (vi) pfcrtGB4 + pfmdr17G8; (vii) pfcrt7G8 + pfmdr1GB4; and (viii) pfcrt7G8 + pfmdr17G8 (Table S2). Evaluation of these groups in a linear model based on the log-transformed IC50 values showed that 7G8×GB4 progeny carrying the pfcrt7G8 allele possessed on average approximately half of the CQ IC50 of pfmdr1-matched progeny carrying pfcrtGB4 [fold-change = 0.55, 95% C.I. (0.47, 0.64); Fig. 3A and Table S3]. Mean CQ IC50 values were 76–105 nM for parasites with the pfcrt7G8 + pfmdr17G8 combination of alleles, whereas lower mean CQ IC50s of 44–58 nM were obtained for clones with the pfcrt7G8 + pfmdr1GB4 combination of alleles (Table S1), a range above the IC50s of CQ-sensitive isolates (e.g., HB3; Fig. 1B) but well below the IC50s of standard strains of resistant clones (31). On average, progeny with pfmdr17G8 showed a 1.71 [95% C.I. (1.46, 2.00)] fold higher CQ IC50 than pfcrt-matched progeny with pfmdr1GB4 (Fig. 3A and Table S3). In the HB3×Dd2 cross, the CQ IC50 values of CQ-resistant parasites carrying the pfcrtDd2 and pfmdr1Dd2 combination were 86–176 nM, whereas the IC50s of progeny with the pfcrtDd2 and pfmdr1HB3 combination showed a greater range of 99–234 nM (Fig. 1B and Table S1). In overall averages, the mean CQ IC50 of the pfcrtDd2/pfmdr1HB3 parasites was calculated to be 1.29-fold higher than the mean CQ IC50 of pfcrtDd2/pfmdr1Dd2-carrying parasites [95% C.I. (1.05, 1.58); Fig. 3A and Table S3].
Fig. 3.
Average in vitro CQ and MDAQ responses of P. falciparum clones grouped according to their pfcrt and pfmdr1 haplotypes. Average IC50 responses to CQ (A) and MDAQ (B) are shown. IC50 values and 95% C.I.s are indicated along with fold increases over the group of parasites carrying the pfcrt HB3and pfmdr1HB3 alleles. Note the log scale of IC50 values along the vertical axes.
MDCQ, which along with CQ contributes significantly to in vivo action against CQ-sensitive parasites but has effectively no action on CQ-resistant parasites because of their very high MDCQ IC50s (32), yielded patterns of IC50 response in 7G8×GB4 progeny that were relatively similar to the patterns of CQ (compare Fig. 1 A and B with Fig. S1 A and B). In the progeny of the HB3×Dd2 cross, significantly different MDCQ IC50s were detected with the pfmdr1Dd2 or pfmdr1HB3 allele in association with the pfcrtDd2 allele, although not in 4-aminoquinoline-sensitive progeny carrying the pfcrtHB3 allele (Fig. S1B and Fig. S3A). Because the IC50 levels of CQ-resistant parasites are ≈10-fold greater with MDCQ than with CQ and have little relevance to our practical conclusions about clinical drug resistance, the MDCQ findings are recorded here for informational purposes only and will not be discussed further.
In contrast to the relatively low CQR levels of 7G8×GB4 parasites carrying the pfcrt7G8 allele (Fig. 1A), MDAQ resistance levels of these clones included the highest of any parasites in our study, with an average IC50 1.81-fold [95% C.I. (1.60, 2.04)] higher than parasites carrying pfcrtGB4 (Figs. 1C and 3B). The MDAQ IC50 of the 7G8 parent, 204 nM (Table S1) is >3-fold higher than that of the Ghana GB4 parent [67 nM, just above the arbitrary in vitro resistance threshold of 60 nM suggested for this metabolite (33)]. Higher MDAQ IC50s were also obtained from progeny carrying pfmdr17G8 than from pfcrt-matched progeny carrying pfmdr1GB4 [1.43-fold higher, 95% C.I. (1.27, 1.61)].
HB3×Dd2 parasites carrying the pfcrtDd2 allele exhibited MDAQ IC50s indicative of resistance, although not at the same high average levels of 7G8×GB4 progeny carrying the pfcrt7G8 + pfmdr17G8 combination (Fig. 1D). These IC50s were detectably affected by the parental type of pfmdr1 allele (Fig. 3B and Table S2): HB3×Dd2 parasites carrying pfcrtDd2 and pfmdr1HB3 averaged IC50 values 1.51-fold higher [95% C.I. (1.29, 1.76); Table S3] than parasites carrying pfcrtDd2 and pfmdr1Dd2. No significant effect of the pfmdr1 allele was observed on the responses of drug-sensitive progeny carrying the pfcrtHB3 allele (Figs. 1D and 3B).
In agreement with previous reports describing in vitro responses of P. falciparum to AQ (34, 35), our IC50 values for this prodrug fell within narrow ranges of 6 to 16 nM and 5 to 15 nM for the 7G8×GB4 and HB3×Dd2 crosses, respectively (Fig. S1 and Table S1). Consistent with the results of QTL scans, 7G8×GB4 parasites carrying the pfmdr17G8 allele and HB3×Dd2 parasites carrying the pfcrtDd2 allele exhibited slightly increased average AQ IC50 values over other progeny from the 7G8×GB4 and HB3×Dd2 crosses (Fig. S3B). However, because AQ is a rapidly metabolized prodrug, these AQ findings are presented here for informational purposes and will not be discussed further.
Distinct Features of VP Reversal Linked to pfcrt Inheritance.
We also evaluated the effect of 0.8 μM VP in combination with the 4-aminoquinoline drugs. In assays with CQ, VP reduced the IC50 levels of clones carrying the pfcrtGB4 or pfcrtDd2 alleles by 70–80% (Fig. 1 A and B and Table S4). Clones carrying the pfcrt7G8 allele show reductions of only ≈40%, consistent with the reported lower susceptibility of CQ-resistant parasites from South America to VP chemosensitization (22). As expected, VP showed no detectable chemosensitization of CQ-sensitive clones carrying the pfcrtHB3 allele from the HB3×Dd2 cross.
In assays with MDAQ, VP showed significant effects on the IC50 values of all CQ-resistant clones, including 50–60% reductions of clones inheriting pfcrtGB4 or pfcrtDd2 allele and ≈30% reductions of clones carrying the pfcrt7G8 allele (P < 0.0001 in all cases; Fig. 1 and Table S4). There was no significant effect of VP on the IC50 values of the CQ-sensitive clones carrying the pfcrtHB3 allele. QTL scans of our test results from VP in combination with MDAQ or CQ identified only a single, highly significant peak on chromosome 7 associated with pfcrt in the 4-aminoquinoline-resistant progeny of both genetic crosses (Fig. S2).
To further investigate the relationship of particular pfcrt-encoded polymorphisms to VP chemosensitization, we examined the drug responses of an additional collection of parasite lines distinguished by their different pfcrt alleles (24). Among the CQ-resistant parasites, a line from Ecuador, Ecu1110, was found to be similar to 7G8 in its VP chemosensitization profile but differed by the lack of mutation C72S (7G8 pfcrt codons 72–76 SVMNT; Ecu1110 pfcrt codons 72–76 CVMNT; Table S5).
Discussion
In drug-resistant P. falciparum malaria, different phenotypes of CQ and AQ resistance are now prevalent in regions of Asia, Africa, and South America. Here, we have described the contributions of two key determinants of drug resistance, pfcrt and pfmdr1, to these different drug-resistant phenotypes. In two genetic crosses containing resistance to the 4-aminoquinolines as segregating phenotypes, mutations in the pfcrt and pfmdr1 alleles were linked to different levels of resistance to CQ relative to MDAQ, the active metabolite of AQ. Particularly striking results were obtained from the cross of CQ-resistant lines from Brazil (7G8) and Ghana (GB4). In this 7G8×GB4 cross, the South American parental combination of pfcrt7G8and pfmdr17G8 alleles exhibited high levels of MDAQ resistance but only moderate levels of CQR. In contrast, the combination of South American pfcrt7G8 and African pfmdr1GB4 alleles was linked to moderate levels of MDAQ resistance and the lowest range of CQ IC50 levels that our laboratory has observed for P. falciparum parasites carrying a resistant pfcrt allele. Our findings therefore indicate that the high MDAQ resistance and moderate CQR of the 7G8 South American parasite are highly dependent upon joint contributions of the pfcrt7G8 and pfmdr17G8 alleles.
Our results from the cross between the CQ-sensitive HB3 clone from Central America and the CQ-resistant Dd2 clone from Southeast Asia (HB3×Dd2) exhibited less robust, although detectable, effects of pfmdr1 on CQ and MDAQ response levels. In our statistical model of CQ IC50 responses, HB3×Dd2 parasites carrying the pfcrtDd2/pfmdr1HB3 combination showed on average a 1.29-fold greater IC50 than parasites carrying the pfcrtDd2/pfmdr1Dd2 combination [95% C.I. (1.05, 1.58), P = 0.02]. Whole-genome QTL primary scans did not detect this effect; in this study and previous work (30) these scans did not identify a significant association of the pfmdr1 locus with CQ responses of the HB3×Dd2 progeny. Similarly, in analysis of MDAQ IC50 levels, our model detected a 1.51-fold greater IC50 for HB3×Dd2 parasites carrying the pfcrtDd2/pfmdr1HB3 combination relative to parasites carrying pfcrtDd2/pfmdr1Dd2 [95% C.I. (1.29, 1.76); P < 0.0001], despite a lack of association with the pfmdr1 locus in whole-genome QTL scans of the HB3×Dd2 progeny. These observations point to the value of alternative statistical models to detect the effects of candidate genes when numbers of cross progeny are limited, particularly when these effects are not large and the involvement of the candidate genes is suggested by indicators from independent results. We note that a number of additional transporter genes are reported to be associated with drug responses in P. falciparum populations (36), although these genes were not detected by QTL analyses of progeny available from the HB3×Dd2 and 7G8×GB4 crosses. The use of alternative statistical models that focus on these and other genes implicated by independent evidence may be helpful for the evaluation of additional loci that can contribute or interact in drug resistance and other P. falciparum phenotypes.
Previous reports examined pfmdr1 allelic modifications for effects on responses to CQ and certain other antimalarial drugs including quinine, mefloquine, halofantrine, and artemisinin (26, 27). In one of these studies, the introduction of three codon replacements 1034C → S, 1042D → N, and 1246Y → D in the Brazilian 7G8 clone reduced the original CQ IC50 by ≈40% (26), consistent with the 2-fold difference we have observed between progeny of the 7G8×GB4 cross that inherited the pfcrt7G8/pfmdr17G8 vs. the pfcrt7G8/pfmdr1GB4 alleles. In a second allelic modification study, Sidhu et al. (27) found little shift of CQ IC50 with changes of pfmdr1-encoded amino acids (1042D → N or introduction of the three residues 1034C, 1042D, and 1246Y) in a CQ-resistant progeny clone (3BA6) from the HB3×Dd2 cross, also consistent with our findings of small average difference between HB3×Dd2 progeny carrying the pfcrtDd2/pfmdr1HB3 or pfcrtDd2/pfmdr1Dd2 alleles. Taken together, these observations suggest that pfmdr1 mutations differentially affect the CQ responses of CQ-resistant parasites and that their activities depend on the particular pfcrt haplotype with which they are associated.
In addition to the above findings on the genetics of CQR, our results provide information on high-level resistance to MDAQ modulated by the South American pfmdr17G8 and Central American pfmdr1HB3 alleles. The N1042D polymorphism is encoded by both of these pfmdr1 alleles (Table 1), suggesting that this amino acid change can have an important role in boosting the drug resistance levels. The effect of N1042D is particularly prominent in parasites carrying the 7G8 pfcrt allele linked to low-level CQR and relatively poor VP chemosensitization. This observation agrees with previous findings that pfmdr1 genes encoding N1042D can have a strong effect on parasite responses to various antimalarial drugs such as quinine, mefloquine, and artemisinin (26, 27), and certain other chemical response phenotypes that have been identified by high-throughput chemical screens (37). Results from the 7G8×GB4 and HB3×Dd2 crosses do not support an effect from N86Y that is as influential as N1042D on the CQ- or MDAQ-resistance phenotypes (N86Y is encoded by pfmdr1GB4 and pfmdr1Dd2 and not by the pfmdr17G8 or pfmdr1HB3 alleles that encode N1042D; Table 1). Remarkably, after a full literature search (Table S6) we were unable to find any report of P. falciparum isolates with a Pgh-1 sequence containing both N86Y and N1042D. It will be interesting to test whether these two polymorphisms might continue to show mutually exclusivity in additional surveys or in attempts to engineer both polymorphisms into a single allele.
A large study of CQ-resistant P. falciparum in PNG identified parasites carrying the same pfcrt polymorphisms and VP chemosensitization phenotype as the 7G8 CQ-resistant parasite from South America (22). However, none of the 902 clinical isolates from three PNG field sites contained the N86, 1034C, 1042D, and 1246Y polymorphisms encoded by the South American pfmdr17G8 allele (Table S6). Only one reported culture-adapted line from PNG, identified as 1905 and characterized by Foote et al. (38), carried the same pfcrt and pfmdr1 alleles as 7G8, and this exhibited the highest CQ IC50 of the PNG series; the extent to which parasites of this 1905 or 7G8 type might occur in PNG remains to be determined. IC50s indicative of low-level CQR were found for all other PNG isolates carrying a SVMNT-type pfcrt allele (i.e., 7G8 type, encoding amino acids SVMNT at codons 72–76) and a pfmdr1 allele encoding 86Y, S1034, N1042, and D1246; the lowest CQ IC50 of all (50 ng/mL) was obtained from an isolate carrying neither 86Y nor 1042D (PNG 1917; Table S6) (22, 38). Those findings taken together with our results from the 7G8×GB4 and HB3×Dd2 crosses are consistent for parasites with greater levels of AQR than CQR. Observations from the field support this conclusion, as clinical results in PNG have shown AQR at higher rates than CQR and high level (RIII) clinical failures only with AQ treatment (39).
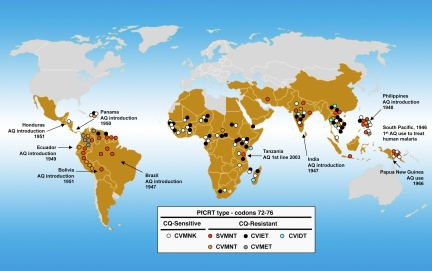
What selective factors account for the distributions of distinct pfcrt and pfmdr1 haplotypes associated with different phenotypes of CQ and MDAQ resistance in the parasite populations of South America, Africa, Asia, PNG, and other malaria endemic regions? One likely influence is the history of AQ and CQ use in these regions and the consequent selective pressure for pfcrt and pfmdr1 polymorphisms of one type or another. Haplotypes of pfcrt-encoding polymorphisms similar to those of the pfcrt alleles in South America and PNG have been reported from East Timor, Indonesia, the Philippines, Laos, India, and Iran (40–49). In many of these regions AQ was widely used in the 1940s (50) and early 1950s before the advent of CQR (Fig. 4). Although at least six foci (origins) of CQR have previously been inferred, including a major prevalent CVIET form in Africa that evidently entered from Southeast Asia, growing databases from surveys suggest that additional foci may also have occurred in the Americas, Africa, and Asia (Fig. 4). Two such additional foci may be suggested by the presence of CVIET and CVMET CQ-resistant parasites in Haiti and Northern areas of South America (Fig. 4). Worrisomely, an increasing prevalence of SVMNT parasites in Tanzania (7G8-type PfCRT; possibly another new focus) has been associated with a switch from CQ to AQ use (2001) and more frequent observations of AQR from that country in recent years (51, 52).*
Fig. 4.
AQ introduction and PfCRT types in malaria-endemic regions. Parasites with the CQ-sensitive PfCRT type (wild-type CVMNK parasites) are prevalent in most malaria endemic areas except in large areas of South America. P. falciparum parasites with the CVIET (i.e., Dd2 or GB4) PfCRT type are found frequently in Africa, areas of Southeast Asia, and Northern areas of South America, where resistance to CQ is high. Parasites with the SVMNT (i.e., 7G8) PfCRT types are found frequently in South America and other regions where AQ was frequently used. One of the earliest recorded uses of AQ was in 1946 in the South Pacific against P. vivax malaria [personal communication from L. T. Coggeshall cited by WHO (50) and Payne et al. (62)]. Between 1946 and 1951 AQ was introduced in India (63), Brazil (50, 64, 65), the Philippines (50, 66), Ecuador (67), Panama (68), Bolivia (69), and Honduras (70). Other regions of more recent use of AQ and prevalence of PfCRT- type SVMNT include PNG (71) and Tanzania (51). Data points have been included only if confirmed by more than a single observation; details and references for the individual points are provided in Table S6.
Patterns of pfcrt and pfmdr1 haplotypes in malaria endemic regions also depend on the fitness costs of these haplotypes relative to the advantages they offer to parasite populations. In Malawi and Hainan Island, China, where CQ-resistant CVIET parasites (Dd2- or GB4-type pfcrt) from the large Southeast Asian–African sweep had once been selected almost completely, populations of wild-type CQ-sensitive parasites characterized by the haplotype CVMNK (HB3-type pfcrt) returned after withdrawal of CQ (17, 18). These findings suggest that CVIET mutants are at a fitness disadvantage to wild-type forms in the absence of drug pressure (20, 54). However, a lower fitness cost appears to be associated with the SVMNT mutants. Except for a recent report of a single sample from Brazil containing the wild-type CQ-sensitive haplotype CVMNK (55), a significant return of CQ-sensitive parasites has not yet occurred among the MDAQ-resistant populations of SVMNT parasites in South America or in other regions where SVMNT is present and CQ has been either withdrawn or is no longer recommended (56). Consistent with these observations, Mehlotra et al. (57) have found that variation in the pfcrt and pfmdr1 loci of Asian and African parasites populations is maintained by substantially different mechanisms than in South American populations where the haplotypes of pfcrt and pfmdr1 both exhibit relatively low levels of diversity. We also note the recent observation of increasingly prevalent SVMNT parasites in Tanzania where AQ pressure has presumably facilitated their spread (51, 52). In areas of Africa where AQ is used as monotherapy or in combination with partner antimalarial drugs, CVIET parasites may be under displacement by highly AQ-resistant, CQ-resistant SVMNT parasites that are as advantaged and persistent as in South America. SVMNT parasites in Africa may therefore present a particular threat where AQ-containing combinations are being used in coordinated malaria control efforts; these parasites warrant careful surveillance and appropriate adjustment of drug use policy where their presence gives rise to prevalent AQ treatment failures.
Our observations also suggest that the various amino acid substitutions encoded by pfcrt and pfmdr1 alleles are tuned to the chemical and structural differences of CQ and MDAQ and contribute distinct levels of resistance to these compounds. Further, certain amino acid polymorphisms in PfCRT are linked to different levels of chemosensitization by VP. Available data on P. falciparum isolates and in vitro-adapted lines support an association of PfCRT N75 with reduced VP chemosensitization of CQ-resistant P. falciparum (a possible requirement of PfCRT 72S for reduced VP chemosensitization is not supported because the Ecuador CVMNT-type isolate Ecu1110 has a phenotype similar to 7G8). Polymorphisms in the Pgh-1 protein encoded by pfmdr1 have also been shown to affect P. falciparum responses to a variety of compounds including quinine, mefloquine, halofantrine, and artemisinin (26, 27, 37). However, CQ stands out in this picture, because parasite responses to CQ are altered more by the Pgh-1 mutations in CQ-resistant parasites that carry 7G8 (South American) than Dd2 or GB4 (Southeast Asian, African) forms of PfCRT, and no effect of Pgh-1 mutations is seen in CQ-sensitive parasites that harbor wild-type forms of PfCRT. These findings raise fascinating questions about the molecular nature of the interactions between PfCRT and Pgh-1, the different functional and structural effects of mutations on transport processes involved in CQ and MDAQ resistance, and the evolution and spread of different pfcrt and pfmdr1 alleles in P. falciparum populations under changing conditions of drug pressure.
Materials and Methods
P. falciparum In Vitro Culture.
P. falciparum clones were as reported: HB3, Dd2, and 33 progeny clones (29, 30); 7G8, GB4, and 32 progeny (28); D10, M5, Haiti, 106-1, Ecu1110, FCB, JAV, TM284, and RB8 (24). Parasites were cultivated using standard methods (ref. 58 and www.mr4.org/Portals/3/Methods_In_Malaria_Research-5theditionv5–2.pdf). AL2 is a clone from a cross in Aotus of the 7G8 and GB4 parents.
Antimalarial Drugs and In Vitro Drug Response Assays.
CQ, AQ, and VP were purchased from Sigma. MDAQ and MDCQ were synthesized by S. Ward (Liverpool School of Tropical Medicine) and AsisChem, respectively. Drug assays were performed as described (59). Initial drug concentrations in the serial dilutions were 5 μM CQ, 20 μM MDCQ, 0.5 μM AQ, and 0.7 μM MDAQ. When used, VP was incorporated at a concentration 0.8 μM in combination with CQ, MDCQ, AQ, or MDAQ.
Statistical Analysis.
To evaluate the relative contribution of pfcrt and pfmdr1 to the in vitro drug responses, we placed the progeny clones of the genetic crosses into eight groups (four groups for each cross) according to the pfcrt and pfmdr1 alleles they presented: (i) pfcrtHB3 + pfmdr1HB3; (ii) pfcrtHB3 + pfmdr1Dd2; (iii) pfcrtDd2 + pfmdr1HB3; (iv) pfcrtDd2 + pfmdr1Dd2; (v) pfcrtGB4 + pfmdr1GB4; (vi) pfcrtGB4 + pfmdr17G8; (vii) pfcrt7G8 + pfmdr1GB4; and (viii) pfcrt7G8 + pfmdr17G8. Geometric means of log transformed IC50 values from these groups were calculated, and 95% C.I.s were based on the associated t test. To evaluate the effect of VP on each drug response, we plotted the log-transformed IC50 values of each group of clones according to their pfcrt and pfmdr1 alleles and estimated the mean ratio of drug+VP/drug IC50 values by using a linear model with the response equal to the difference in log-transformed values. Similar linear models were used to test for average fold changes in one allele when holding the other constant. The 95% C.I. and P values were calculated from the linear models by a two-sided test to check for ratios significantly different from 1. Calculations were done with R version 2.8.0 (R Development Core Team 2008).
QTL Analysis.
QTL analysis was performed by using the R/qtl and J/qtl packages (60)) and Pseudomarker using Matlab software (61) as described (30).
Supplementary Material
Acknowledgments.
We thank Jianbing Mu for assistance in genotyping the P. falciparum clones; Hongying Jiang, Xinzhuan Su, Jeff Skinner, Vivek Gopalan, Jason Barnett, Yentram Huyen, and William Knight for assistance with in vitro drug assays and automated analysis of IC50s and IC90s; and Erika Phelps, Sarnia Laurent, Michael Krause, and Rick Fairhurst for assistance with QTL mapping, DNA sequencing, and discussions. The Intramural Research Program of the National Institutes of Health, National Institute of Allergy and Infectious Diseases, supported this research.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0911317106/DCSupplemental.
Consistent with the pfcrt–pfmdr1 interactions described in this study, pfmdr1 codon 1246Y with possible synergistic or compensatory influence of 86Y or 184Y has also been associated with treatment failures after AQ monotherapy or after AQ plus ACT in Tanzania (53).
References
- 1.Burckhalter JH, et al. Aminoalkylphenols as antimalarials. II. (Heterocyclic-amino)-a-amino-o-cresols: The synthesis of camoquin. J Am Chem Soc. 1948;70:1363–1373. doi: 10.1021/ja01184a023. [DOI] [PubMed] [Google Scholar]
- 2.Ridley RG, Hudson AT. Quinoline antimalarials. Exp Opin Ther Patents. 1998;8:121–136. [Google Scholar]
- 3.Fitch CD, Chevli R, Gonzalez Y. Chloroquine-resistant Plasmodium falciparum: Effect of substrate on chloroquine and amodiaquin accumulation. Antimicrob Agents Chemother. 1974;6:757–762. doi: 10.1128/aac.6.6.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spencer HC, et al. Amodiaquine more effective than chloroquine against Plasmodium falciparum malaria on Kenya coast. Lancet. 1984;1:956–957. doi: 10.1016/s0140-6736(84)92406-1. [DOI] [PubMed] [Google Scholar]
- 5.Olliaro P, et al. Systematic review of amodiaquine treatment in uncomplicated malaria. Lancet. 1996;348(9036):1196–1201. doi: 10.1016/S0140-6736(96)06217-4. [DOI] [PubMed] [Google Scholar]
- 6.Adjetey TA, et al. Evaluation of the therapeutic efficacy of amodiaquine versus chloroquine in the treatment of uncomplicated malaria in Abie, Cote-d'Ivoire. Bull Soc Pathol Exot. 2005;98:193–196. [PubMed] [Google Scholar]
- 7.WHO. Guidelines for the Treatment of Malaria. Geneva: WHO; 2006. [Google Scholar]
- 8.Patel JC, Dalal SD. Treatment of malaria with a single dose of amodiaquin (Camoquin) Ind J Malariol. 1954;8:71–76. [PubMed] [Google Scholar]
- 9.Shute GT, Ray AP, Sangalang R. Preliminary studies on a Philippine strain of Plasmodium falciparum resistant to amodiaquine. J Trop Med Hyg. 1972;75:125–132. [PubMed] [Google Scholar]
- 10.Hall AP, Segal HE, Pearlman EJ, Phintuyothin P, Kosakal S. Amodiaquine-resistant falciparum malaria in Thailand. Am J Trop Med Hyg. 1975;24:575–580. doi: 10.4269/ajtmh.1975.24.575. [DOI] [PubMed] [Google Scholar]
- 11.Kremsner PG, et al. A comparative trial of three regimens for treating uncomplicated falciparum malaria in Acre, Brazil. J Infect Dis. 1988;158:1368–1371. doi: 10.1093/infdis/158.6.1368. [DOI] [PubMed] [Google Scholar]
- 12.Sapak P, et al. Ineffectiveness of amodiaquine against Plasmodium falciparum malaria in symptomatic young children living in an endemic malarious area of Papua New Guinea. J Trop Pediatr. 1991;37:185–190. doi: 10.1093/tropej/37.4.185. [DOI] [PubMed] [Google Scholar]
- 13.Lemnge M, et al. High reinfection rate and treatment failures in children treated with amodiaquine for falciparum malaria in Muheza villages, Northeastern Tanzania. Am J Trop Med Hyg. 2006;75:188–193. [PubMed] [Google Scholar]
- 14.Sasi P, et al. In vivo and in vitro efficacy of amodiaquine against Plasmodium falciparum in an area of continued use of 4-aminoquinolines in East Africa. J Infect Dis. 2009;199:1575–1582. doi: 10.1086/598862. [DOI] [PubMed] [Google Scholar]
- 15.Echeverry DF, et al. Short report: Polymorphisms in the pfcrt and pfmdr1 genes of Plasmodium falciparum and in vitro susceptibility to amodiaquine and desethylamodiaquine. Am J Trop Med Hyg. 2007;77:1034–1038. [PubMed] [Google Scholar]
- 16.Churchill FC, et al. Amodiaquine as a prodrug: Importance of metabolite(s) in the antimalarial effect of amodiaquine in humans. Life Sci. 1985;36:53–62. doi: 10.1016/0024-3205(85)90285-1. [DOI] [PubMed] [Google Scholar]
- 17.Kublin JG, et al. Reemergence of chloroquine-sensitive Plasmodium falciparum malaria after cessation of chloroquine use in Malawi. J Infect Dis. 2003;187:1870–1875. doi: 10.1086/375419. [DOI] [PubMed] [Google Scholar]
- 18.Wang X, et al. Decreased prevalence of the Plasmodium falciparum chloroquine resistance transporter 76T marker associated with cessation of chloroquine use against P. falciparum malaria in Hainan, People's Republic of China. Am J Trop Med Hyg. 2005;72:410–414. [PubMed] [Google Scholar]
- 19.Mwai L, et al. Chloroquine resistance before and after its withdrawal in Kenya. Malar J. 2009;8:106. doi: 10.1186/1475-2875-8-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laufer MK, et al. Return of chloroquine antimalarial efficacy in Malawi. N Engl J Med. 2006;355:1959–1966. doi: 10.1056/NEJMoa062032. [DOI] [PubMed] [Google Scholar]
- 21.Martin SK, Oduola AM, Milhous WK. Reversal of chloroquine resistance in Plasmodium falciparum by verapamil. Science. 1987;235:899–901. doi: 10.1126/science.3544220. [DOI] [PubMed] [Google Scholar]
- 22.Mehlotra RK, et al. Evolution of a unique Plasmodium falciparum chloroquine-resistance phenotype in association with pfcrt polymorphism in Papua New Guinea and South America. Proc Natl Acad Sci USA. 2001;98:12689–12694. doi: 10.1073/pnas.221440898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wellems TE, Hayton K, Fairhurst RM. The impact of malaria parasitism: From corpuscles to communities. J Clin Invest. 2009;119:2496–2505. doi: 10.1172/JCI38307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fidock DA, et al. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol Cell. 2000;6:861–871. doi: 10.1016/s1097-2765(05)00077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Naude B, Brzostowski JA, Kimmel AR, Wellems TE. Dictyostelium discoideum expresses a malaria chloroquine resistance mechanism upon transfection with mutant, but not wild type, Plasmodium falciparum transporter PfCRT. J Biol Chem. 2005;280:25596–25603. doi: 10.1074/jbc.M503227200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reed MB, Saliba KJ, Caruana SR, Kirk K, Cowman AF. Pgh1 modulates sensitivity and resistance to multiple antimalarials in Plasmodium falciparum. Nature. 2000;403:906–909. doi: 10.1038/35002615. [DOI] [PubMed] [Google Scholar]
- 27.Sidhu AB, Valderramos SG, Fidock DA. pfmdr1 mutations contribute to quinine resistance and enhance mefloquine and artemisinin sensitivity in Plasmodium falciparum. Mol Microbiol. 2005;57:913–926. doi: 10.1111/j.1365-2958.2005.04729.x. [DOI] [PubMed] [Google Scholar]
- 28.Hayton K, et al. Erythrocyte binding protein PfRH5 polymorphisms determine species-specific pathways of Plasmodium falciparum invasion. Cell Host Microbe. 2008;4:40–51. doi: 10.1016/j.chom.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wellems TE, et al. Chloroquine resistance not linked to mdr-like genes in a Plasmodium falciparum cross. Nature. 1990;345:253–255. doi: 10.1038/345253a0. [DOI] [PubMed] [Google Scholar]
- 30.Ferdig MT, et al. Dissecting the loci of low-level quinine resistance in malaria parasites. Mol Microbiol. 2004;52:985–997. doi: 10.1111/j.1365-2958.2004.04035.x. [DOI] [PubMed] [Google Scholar]
- 31.Wellems TE, Plowe CV. Chloroquine-resistant malaria. J Infect Dis. 2001;184:770–776. doi: 10.1086/322858. [DOI] [PubMed] [Google Scholar]
- 32.Aderounmu AF. In vitro assessment of the antimalarial activity of chloroquine and its major metabolites. Ann Trop Med Parasitol. 1984;78:581–585. doi: 10.1080/00034983.1984.11811868. [DOI] [PubMed] [Google Scholar]
- 33.Ringwald P, Bickii J, Basco LK. In vitro activity of antimalarials against clinical isolates of Plasmodium falciparum in Yaounde, Cameroon. Am J Trop Med Hyg. 1996;55:254–258. doi: 10.4269/ajtmh.1996.55.254. [DOI] [PubMed] [Google Scholar]
- 34.Rieckmann KH. Determination of the drug sensitivity of Plasmodium falciparum. J Am Med Assoc. 1971;217:573–578. [PubMed] [Google Scholar]
- 35.Ramanamirija JA, Deloron P, Biaud JM, Le Bras J, Coulanges P. In vivo and in vitro sensitivity to 4-aminoquinolines of Plasmodium falciparum in Madagascar: Results of 2 years' study. Bull Soc Pathol Exot Filiales. 1985;78:606–614. [PubMed] [Google Scholar]
- 36.Mu J, et al. Multiple transporters associated with malaria parasite responses to chloroquine and quinine. Mol Microbiol. 2003;49:977–989. doi: 10.1046/j.1365-2958.2003.03627.x. [DOI] [PubMed] [Google Scholar]
- 37.Yuan J, et al. Genetic mapping of targets mediating differential chemical phenotypes in Plasmodium falciparum. Nat Chem Biol. 2009;5:765–771. doi: 10.1038/nchembio.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Foote SJ, et al. Several alleles of the multidrug-resistance gene are closely linked to chloroquine resistance in Plasmodium falciparum. Nature. 1990;345:255–258. doi: 10.1038/345255a0. [DOI] [PubMed] [Google Scholar]
- 39.Schuurkamp GJ, Kereu RK. Resistance of Plasmodium falciparum to chemotherapy with 4-aminoquinolines in the Ok Tedi area of Papua New Guinea. Papua New Guinea Med J. 1989;32:33–44. [PubMed] [Google Scholar]
- 40.Mehlotra RK, et al. Insight into the early spread of chloroquine-resistant Plasmodium falciparum infections in Papua New Guinea. J Infect Dis. 2005;192:2174–2179. doi: 10.1086/497694. [DOI] [PubMed] [Google Scholar]
- 41.DaRe JT, et al. Microsatellite polymorphism within pfcrt provides evidence of continuing evolution of chloroquine-resistant alleles in Papua New Guinea. Malar J. 2007;6:34. doi: 10.1186/1475-2875-6-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen N, et al. Short report: Molecular evaluation of the efficacy of chloroquine treatment of uncomplicated Plasmodium falciparum malaria in East Timor. Am J Trop Med Hyg. 2002;67:64–66. doi: 10.4269/ajtmh.2002.67.64. [DOI] [PubMed] [Google Scholar]
- 43.Huaman MC, Yoshinaga K, Suryanatha A, Suarsana N, Kanbara H. Short report: Polymorphisms in the chloroquine resistance transporter gene in Plasmodium falciparum isolates from Lombok, Indonesia. Am J Trop Med Hyg. 2004;71:40–42. [PubMed] [Google Scholar]
- 44.Chen N, et al. pfcrt allelic types with two novel amino acid mutations in chloroquine-resistant Plasmodium falciparum isolates from the Philippines. Antimicrob Agents Chemother. 2003;47:3500–3505. doi: 10.1128/AAC.47.11.3500-3505.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen N, et al. Origin and dissemination of chloroquine-resistant Plasmodium falciparum with mutant pfcrt alleles in the Philippines. Antimicrob Agents Chemother. 2005;49:2102–2105. doi: 10.1128/AAC.49.5.2102-2105.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dittrich S, et al. Falciparum malaria in the north of Laos: The occurrence and implications of the Plasmodium falciparum chloroquine resistance transporter (pfcrt) gene haplotype SVMNT. Trop Med Int Health. 2005;10:1267–1270. doi: 10.1111/j.1365-3156.2005.01514.x. [DOI] [PubMed] [Google Scholar]
- 47.Vathsala PG, et al. Widespread occurrence of the Plasmodium falciparum chloroquine resistance transporter (pfcrt) gene haplotype SVMNT in P. falciparum malaria in India. Am J Trop Med Hyg. 2004;70:256–259. [PubMed] [Google Scholar]
- 48.Ursing J, Zakeri S, Gil JP, Bjorkman A. Quinoline resistance-associated polymorphisms in the pfcrt, pfmdr1, and pfmrp genes of Plasmodium falciparum in Iran. Acta Trop. 2006;97:352–356. doi: 10.1016/j.actatropica.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 49.Zakeri S, et al. Association of pfcrt but not pfmdr1 alleles with chloroquine resistance in Iranian isolates of Plasmodium falciparum. Am J Trop Med Hyg. 2008;78:633–640. [PubMed] [Google Scholar]
- 50.WHO Expert Committee on Malaria. Summary Review of the Literature on Camoquin. Geneva: WHO; 1950. [Google Scholar]
- 51.Alifrangis M, et al. Occurrence of the Southeast Asian/South American SVMNT haplotype of the chloroquine-resistance transporter gene in Plasmodium falciparum in Tanzania. J Infect Dis. 2006;193:1738–1741. doi: 10.1086/504269. [DOI] [PubMed] [Google Scholar]
- 52.Mutabingwa TK, et al. Amodiaquine alone, amodiaquine+sulfadoxine-pyrimethamine, amodiaquine+artesunate, and artemether-lumefantrine for outpatient treatment of malaria in Tanzanian children: A four-arm randomised effectiveness trial. Lancet. 2005;365:1474–1480. doi: 10.1016/S0140-6736(05)66417-3. [DOI] [PubMed] [Google Scholar]
- 53.Holmgren G, et al. Selection of pfmdr1 mutations after amodiaquine monotherapy and amodiaquine plus artemisinin combination therapy in East Africa. Infect Genet Evol. 2007;7:562–569. doi: 10.1016/j.meegid.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 54.Mita T, et al. Expansion of wild-type allele rather than back mutation in pfcrt explains the recent recovery of chloroquine sensitivity of Plasmodium falciparum in Malawi. Mol Biochem Parasitol. 2004;135:159–163. doi: 10.1016/j.molbiopara.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 55.Gama BE, et al. Chloroquine and sulphadoxine-pyrimethamine sensitivity of Plasmodium falciparum parasites in a Brazilian endemic area. Malar J. 2009;8:156. doi: 10.1186/1475-2875-8-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.de Almeida A, Arez AP, Cravo PV, do Rosario VE. Analysis of genetic mutations associated with antimalarial drug resistance in Plasmodium falciparum from the Democratic Republic of East Timor. Malar J. 2009;8:59. doi: 10.1186/1475-2875-8-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mehlotra RK, et al. Discordant patterns of genetic variation at two chloroquine resistance loci in worldwide populations of the malaria parasite Plasmodium falciparum. Antimicrob Agents Chemother. 2008;52:2212–2222. doi: 10.1128/AAC.00089-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moll K, et al. Methods in Malaria Research. Manassas, VA: American Type Culture Collection; 2008. [Google Scholar]
- 59.Smilkstein M, Sriwilaijaroen N, Kelly JX, Wilairat P, Riscoe M. Simple and inexpensive fluorescence-based technique for high-throughput antimalarial drug screening. Antimicrob Agents Chemother. 2004;48:1803–1806. doi: 10.1128/AAC.48.5.1803-1806.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- 61.Sen S, Churchill GA. A statistical framework for quantitative trait mapping. Genetics. 2001;159:371–387. doi: 10.1093/genetics/159.1.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Payne EH, Sharp EA, Nickel MD. Parenteral use of camoquin hydrochloride as an antimalarial. Am J Trop Med Hyg. 1949;29:353–368. doi: 10.4269/ajtmh.1949.s1-29.353. [DOI] [PubMed] [Google Scholar]
- 63.Simeons ATW, Chhatre KD. Preliminary report on a new synthetic antimalarial. Ind Med Gaz. 1947;82:255–257. [PMC free article] [PubMed] [Google Scholar]
- 64.Mein RM, Rosado PNS. Experiences with new medicines against malaria in the Amazon. Rev Serv Espec Saude Publ May. 1948:1059–1069. (in Portuguese) [Google Scholar]
- 65.Mein RM. Camoquin in the treatment of human malaria. Am J Trop Med Hyg. 1951;31:212–217. doi: 10.4269/ajtmh.1951.s1-31.212. [DOI] [PubMed] [Google Scholar]
- 66.Ejercito A, Duque M. Preliminary report on CAM-AQI dihydrochloride (Miaquin, Camoquin) in treatment of human malaria. Phil Med Ass J Manila. 1948;24:633. [Google Scholar]
- 67.Hoekenga MT. The treatment of malaria with a single dose of camoquin. Am J Trop Med Hyg. 1951;S1-31:139–143. doi: 10.4269/ajtmh.1951.s1-31.139. [DOI] [PubMed] [Google Scholar]
- 68.Hoekenga MF. Camoquin treatment of malaria, a preliminary report. Am J Trop Med Hyg. 1950;30:63–69. doi: 10.4269/ajtmh.1950.s1-30.63. [DOI] [PubMed] [Google Scholar]
- 69.Victor M, Villarejos M. Experiences with amodiaquin (camoquin) a new synthetic antimalarial. Am J Trop Med Hyg. 1951;31:703–706. [Google Scholar]
- 70.Hoekenga MT. Treatment of malaria with a single dose of amodiaquine or chloroquine. J Am Med Assoc. 1952;149:1369–1371. doi: 10.1001/jama.1952.02930320009003. [DOI] [PubMed] [Google Scholar]
- 71.Rieckmann KH. A field study on the effects of a combination of cycloguanil pamoate and amodiquine against malaria in the Rabaul area of New Guinea. Am J Trop Med Hyg. 1966;15:833–837. doi: 10.4269/ajtmh.1966.15.833. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.