Abstract
The objective of this study was to determine the effects of binge-like alcohol drinking on gene expression changes in the nucleus accumbens (ACB) of alcohol-preferring (P) rats. Adult male P rats were given ethanol under multiple scheduled access (MSA; three 1-hr dark-cycle sessions/day) conditions for 8 weeks. For comparison purposes, a second ethanol drinking group was given continuous/daily alcohol access (CA; 24 hr/day). A third group was ethanol-naïve (W group). Average ethanol intakes for the CA and MSA groups were approximately 9.5 and 6.5 g/kg/day, respectively. Fifteen hr after the last drinking episode, rats were euthanized, the brains extracted, and the ACB dissected. RNA was extracted and purified for microarray analysis. The only significant differences were between the CA and W groups (p < 0.01; Storey false discovery rate = 0.15); there were 374 differences in named genes between these 2 groups. There were 20 significant Gene Ontology (GO) categories, which included negative regulation of protein kinase activity, anti-apoptosis, and regulation of G-protein-coupled receptor signaling. Ingenuity® analysis indicated a network of transcription factors, involving oncogenes (Fos, Jun, Junb had higher expression in the ACB of the CA group), suggesting increased neuronal activity. There were 43 genes located within rat QTLs for alcohol consumption and preference; 4 of these genes (Tgfa, Hspa5, Mtus1 and Creb3l2) are involved in anti-apoptosis and increased transcription, suggesting that they may be contributing to cellular protection and maintaining high alcohol intakes. Overall, these findings suggest that chronic CA drinking results in genomic changes that can be observed during the early acute phase of ethanol withdrawal. Conversely, chronic MSA drinking, with its associated protracted withdrawal periods, results in genomic changes that may be masked by tight regulation of these genes following repeated experiences of ethanol withdrawal.
Keywords: Alcohol-Preferring rats, Nucleus accumbens, Gene Expression, Microarrays, Alcohol drinking, Self-administration, Ethanol responsive genes, Ethanol withdrawal
1. Introduction
Microarray analysis has emerged as a tool to study the multiple, complex effects of pharmacological treatments on changes in gene expression. Examining innate differences and changes in gene expression in response to ethanol in lines or strains of mice and rats with divergent responses to ethanol has provided important clues toward identifying genes and gene networks involved in vulnerability to high ethanol-drinking behavior. Given this, examining changes in gene expression following chronic ethanol drinking will, presumably, provide information to identify genes and gene networks involved in maintaining this behavior, as well as the consequences of chronic ethanol exposure.
Many innate genetic expression differences between high and low ethanol-consuming rodent lines have been indentified. For example, Edenberg et al. (2005) examined differences in gene expression in the hippocampus of inbred alcohol-preferring (iP) and inbred alcohol-non-preferring (iNP) rats, and reported differences for genes involved in cell growth and adhesion, cellular stress reduction and anti-oxidation, protein trafficking, cellular signaling pathways, and synaptic function. In a subsequent study, Kimpel et al. (2007) reported on innate differences in gene expression between iP and iNP rats in 5 CNS regions, including the nucleus accumbens (ACB). These authors indicated that genes associated with anti-apoptosis, axon guidance, nerve transmission as well as synaptic structure and function displayed expression differences between the rat strains. Worst et al. (2005) reported on the transcriptome analysis of the frontal cortex from ethanol-naïve AA (Alko, alcohol) and ANA (Alko, non-alcohol) rats, with mRNA level differences found that could reflect altered neurotransmitter release. Using a whole brain analysis of inbred long-sleep and inbred short-sleep mice, Xu et al. (2001) detailed expression differences for metabolic-associated genes with higher levels seen in the long-sleep mice. In a comprehensive transcriptome meta-analysis of gene expression differences across a number of different mouse strains, Mulligan et al. (2006) identified several cis-regulated candidate genes for an ethanol preference QTL on chromosome 9.
Alterations in gene expression produced by exposure to ethanol have been reported in a few studies. Acute ethanol injections (6 g/kg; i.p.) produced gene expression changes associated with cell signal regulation, gene regulation, and homeostasis/stress responses in the whole brain from C57BL/6J and DBA/2J, high- and low-ethanol drinking mice, respectively (Treadwell and Singh, 2004). Kerns et al. (2005) reported that acute i.p. ethanol injections altered the expression of genes involved in glucocorticoid signaling, neurogenesis, myelination, neuropeptide signaling, and retinoic acid signaling in the ACB, prefrontal cortex and VTA of C57BL/6J and DBA/2J mice. Differences in the expression levels of genes coding for oxido-reductases and ADP-ribosylation factors have also been found in the dorsal hippocampus of Lewis rats given 12% ethanol or water for 15 months (Saito et al., 2002). In a recent study, Bowers et al. (2006) reported that chronic ethanol consumption, in a liquid diet, altered the expression of over 100 genes in the cerebellum of PKCgamma wild-type and mutant mice. In contrast, Saito et al. (2004) in a previous study found no statistically significant effects of chronic free-choice ethanol drinking on gene expression in the striatum of C57BL/6By mice. The above studies were conducted using ethanol injections or 24-hr free- or forced-choice drinking. A recent study from our laboratory (Rodd et al., 2008) reported that operant ethanol self-administration produced approximately 500 significant changes in gene expression in the ACB when measured 24 hr after the last 1-hr operant session, whereas saccharin self-administration produced less than 60 significant changes, suggesting that chronic ethanol consumption was producing persisting effects on gene expression in the ACB of P rats. However, it is important to determine if the effects of ethanol drinking alone (absence of operant responding) produce similar changes in gene expression in limbic regions that are involved in regulating ethanol drinking.
In an initial study from our laboratory, Bell et al. (2006a) examined protein expression changes in the ACB and amygdala of iP rats given 24-hr continuous access (CA) or multiple scheduled access (MSA; four 1-hr sessions during the dark cycle) to ethanol for 6 weeks. The results of this study indicated that ethanol drinking conditions differentially changed protein expression in the ACB and amygdala. However, a relatively insensitive 2-dimensional gel electrophoresis procedure was used in this study and only the most abundant proteins found in tissue from the whole ACB or amygdala could be detected. The microarray procedure offers a potentially more sensitive method to measure changes resulting from ethanol drinking under different conditions of availability, which produce different patterns of ethanol intake and associated blood alcohol levels (c.f., Bell et al., 2006a, 2006b). Therefore, the objective of this study was to examine changes in gene expression associated with chronic ethanol drinking under binge-like ethanol drinking conditions. For comparison purposes, the effects of 24-hr free-choice drinking on gene expression were also determined. Gene expression changes were determined the next day after the binge-like group's last scheduled access period of the previous day. Ethanol was removed from both groups at the same time to control for the length of ethanol deprivation before brain tissue was harvested. The hypothesis to be tested was that chronic binge-like ethanol drinking would produce significant persisting effects on gene expression in the ACB of P rats that would not be observed with 24-hr continuous ethanol access drinking.
2. Method
2.1. Animals and ethanol drinking procedures
Subjects were adult (> 90 days old), ethanol-naïve, male P rats from the S52 generation. The rats were single-housed in hanging stainless steel wire-mesh (bottom and front) cages on a reverse 12 hr/12 hr dark-light cycle (light offset at 1000 hr). Animals had ad libitum access to food. Rats were randomly divided into three groups (n = 9/group): the 1st group had access to water as their sole fluid, the 2nd group had continuous/daily, concurrent, free-choice access to 15% and 30% (v/v) ethanol and water, and the 3rd group had bout-like, concurrent access to 15% and 30% ethanol, with water available ad libitum. The bout-like group experienced a multiple scheduled access (MSA) protocol, such that they received three 1-hr access periods each separated by 2 hr starting at the beginning of the dark cycle (i.e., 1000-1100, 1300-1400, and 1600-1700 hr). The MSA animals were given ethanol access in 5-day blocks (Monday—Friday), with each block separated by 2 days without ethanol. Measurements of water and ethanol intake, and body weights were taken Monday through Friday at 0900 hr; ethanol intakes, for MSA animals, were also taken at the end of each 1-hr access period. After the MSA group's 1st day of re-exposure to ethanol access, of the 9th week, both groups of ethanol-drinking rats had ethanol removed at 1700 hr. To ensure that ethanol blood levels were absent and the deprivation period was equivalent for both groups, all rats were killed the next day (15 hr after removing ethanol). Rats were killed by decapitation and their brains processed for microarray analyses, as described below.
2.2. Brain dissections
Rats were killed by decapitation within the same 2-hr time frame over 2 days with equal number of animals from each group being killed on each day to minimize differences in time of sacrifice and dissection, and maintain the experimental balance across groups. During the 7th and 8th weeks, the rats in the MSA group dissected on the 2nd day had ethanol access moved to Tuesday through Saturday to preserve the 5-day a week schedule. The head was immediately placed in a cold box maintained at -15°C, where the brain was rapidly removed and placed on a glass plate for dissection. All equipment used to obtain tissue were treated with RNAse Zap (Ambion, Inc. Austin, TX) to prevent RNA degradation. The ACB was dissected according to the coordinates of Paxinos and Watson (1998). Briefly, the ACB was dissected from a 2-mm section generated by a coronal cut at 2 mm anterior to the optic chiasm (Bregma 1.70 mm) and a coronal cut at the optic chiasm (Bregma −0.26 mm). Dissected tissue was immediately homogenized in Trizol reagent (Invitrogen, Carlsbad, CA) and processed according to the manufacturer's protocol, but with twice the suggested ratio of Trizol to tissue, as discussed previously (Edenberg et al., 2005). Ethanol precipitated RNA was further purified through RNeasy® columns (Qiagen, Valencia, CA), according to the manufacturer's protocol. The yield, concentration and purity of the RNA were determined by running a spectrum from 210 to 350 nm, and analyzing the ratio of large and small ribosomal RNA bands using an Agilent Bioanalyzer. Yields and purity of the RNA were deemed excellent.
2.3. Microarray procedures
Separate preparations of total RNA were made from the ACB of individual animals. Samples were not pooled. Standard Affymetrix protocols (GeneChip® Expression Analysis Technical Manual, Rev. 5 and updates) were used to synthesize biotinylated cRNA, starting with 5 μg total RNA from each region, using the Affymetrix kits for cDNA synthesis, in vitro transcription and sample cleanup. Fifteen μg of fragmented, biotinylated cRNA from each independent sample were mixed into 300 μl of hybridization cocktail, of which 200 μl were used for each hybridization. Hybridization was for 17 hr at 42°C. Samples were hybridized to the Affymetrix GeneChip® (Rat Genome 230 2.0 array GeneChips). Washing and scanning of the GeneChips were carried out according to standard protocols, as previously described (Edenberg et al., 2005; McClintick et al., 2003). To minimize potential systematic errors, all stages of the experiment were balanced across experimental groups. That is, equal numbers of animals in each group were sacrificed within the same 2-hr time frame each day, and equal numbers of RNA preparations from the representative groups were processed through the labeling, hybridization, washing and scanning protocols on a given day, in a counterbalanced order, using premixes of reagents.
2.4. Statistical and neuroinformatic analyses of microarray data
Each GeneChip® was scanned using an Affymetrix Model 3000 scanner and underwent image analysis using Affymetrix GCOS software. Microarray data are available at the National Center for Biotechnology Information's Gene Expression Omnibus, http://www.ncbi.nlm.nih.gov/geo/, under series accession no. GSE13524 [GSM341183…GSM341211] (Barrett et al., 2005; Edgar et al., 2002). Raw .cel files were then imported into the statistical programming environment R (R: A language and environment for statistical computing Ver 2.2.0; R Foundation for Statistical Computing, 2005) for further analysis with tools available from the Bioconductor Project (Gentleman et al., 2004), with these further expanded by the authors using the R language. Expression data from the 27 arrays of the ACB region were normalized and converted to log(2) using the Robust Multi-chip Average (RMA) method (Irizarry et al., 2003) implemented in the Bioconductor package RMA. As a standardization step to facilitate later comparisons with other experiments, expression levels were scaled such that the mean expression of all arrays was log2(1000). Because the primary objective was identifying genes that could be subjected to further bioinformatic analysis, all probesets currently annotated by Affymetrix as “expressed sequence tags” or whose gene names contained the words “riken”, “predicted”, or “similar to” were filtered out. Next, probe sets with a very low likelihood of actual expression in our samples were removed, with this accomplished by the Bioconductor package “genefilter.” Probe sets that did not have at least 25% of samples with normalized scaled expression greater than 64 were filtered out as well. Linear modeling to calculate gene-wise p-values for the contrasts of the CA versus W group and the MSA versus W group was performed using the package Limma (Smyth, 2004); probe sets were considered to be statistically significant at p < 0.01, with a false discovery rate (FDR) less than 0.15. An FDR of 0.15 was used as a cutoff because this allowed a significant number of genes to be included in the Gene Ontology (GO) and Ingenuity® Pathways Analysis to help identify networks of genes that changed. This FDR value is a reasonably stringent cutoff value that provides a good balance between allowing more thorough gene network analyses (limiting beta-error) without including too many false positives (limiting alpha-error) in the analyses. Thus, to facilitate discussion of the present results in the context of our laboratory's previously published work with microarray data (c.f., Edenberg et al., 2005; Kimpel et al., 2007; Rodd et al., 2008), we have used the same standard statistical procedures used previously, and by the field (Gentleman et al., 2004; Irizarry et al., 2003; Smyth, 2004), to determine the effects of the two different ethanol drinking conditions on changes in gene expression.
Testing for over-representation of Gene Ontology (GO) biologic process categories (Harris et al., 2004; Ashburner et al., 2000) was performed using the Bioconductor package GOstats (Gentleman, 2004). Briefly, for each gene set tested, a list of unique Entrez-Gene identifiers was constructed. This list was then compared to the list of all known Entrez-Gene identifiers that are represented on the Affymetrix chipset Rat Genome 230 2.0. Identification of over-represented GO categories was then accomplished within GOstats using the hypergeometric distribution. To filter out uninteresting categories, only those categories with greater than 9 and less than 300 genes represented on the chipset were included in the analysis. GO categories were called significant at p < 0.05. In addition, network analyses were conducted with Ingenuity® Pathway Analysis (Ingenuity® Systems, www.Ingenuity.com). Briefly, a data set containing gene identifiers and corresponding fold-changes was uploaded into the application. Each gene identifier was mapped to its corresponding gene object in the Ingenuity® Pathways Knowledge Base. An FDR cutoff of 0.15 was set to identify genes with expression levels that were significantly altered. These genes, called focus genes, were overlaid onto a global molecular network developed from information contained in the Ingenuity® Pathways Knowledge Base. Networks of these focus genes were then algorithmically generated based on their connectivity.
2.5. Quantitative Real-Time PCR
Real-Time PCR was carried out using SybrGreen chemistry and the ABI Prism 7300 Sequence Detection System (Applied Biosystems Inc. Foster City, CA). The amplification primers were designed using Vector NTI (Invitrogen, Carlsbad, CA). Total RNA, isolated for the microarray analyses, was treated with DNase I for these analyses. Following reverse transcription of the RNA (SuperScript™ III First-Strand Synthesis System for RT-PCR, Invitrogen, Carlsbad, CA), an aliquot of each reverse transcription reaction was amplified in triplicate. This reaction was repeated to generate 6 values for each test group. Two control reactions were run for each RNA preparation: 1) a reverse transcription and PCR reaction with no added RNA to control for contamination of the reagents; and 2) a PCR reaction without the reverse transcription reaction in the presence of RNA to detect DNA contamination of the RNA preparation. To correct for sample-to-sample variation, an endogenous control (GAPDH) was amplified with the target and served as an internal reference to normalize the data. Relative quantification of data from the ABI Prism 7300 Sequence Detection System was performed using the standard curve method (Applied Biosystems, User Bulletin #2; htpp://www.appliedbiosystems.com). Quantitative RT-PCR (qRT-PCR) measurements were conducted on genes to verify differences observed with microarray hybridization. These genes were selected on the basis of significant differential expression, relatively large fold changes, and the availability of primers.
3. Results
3.1. Ethanol drinking
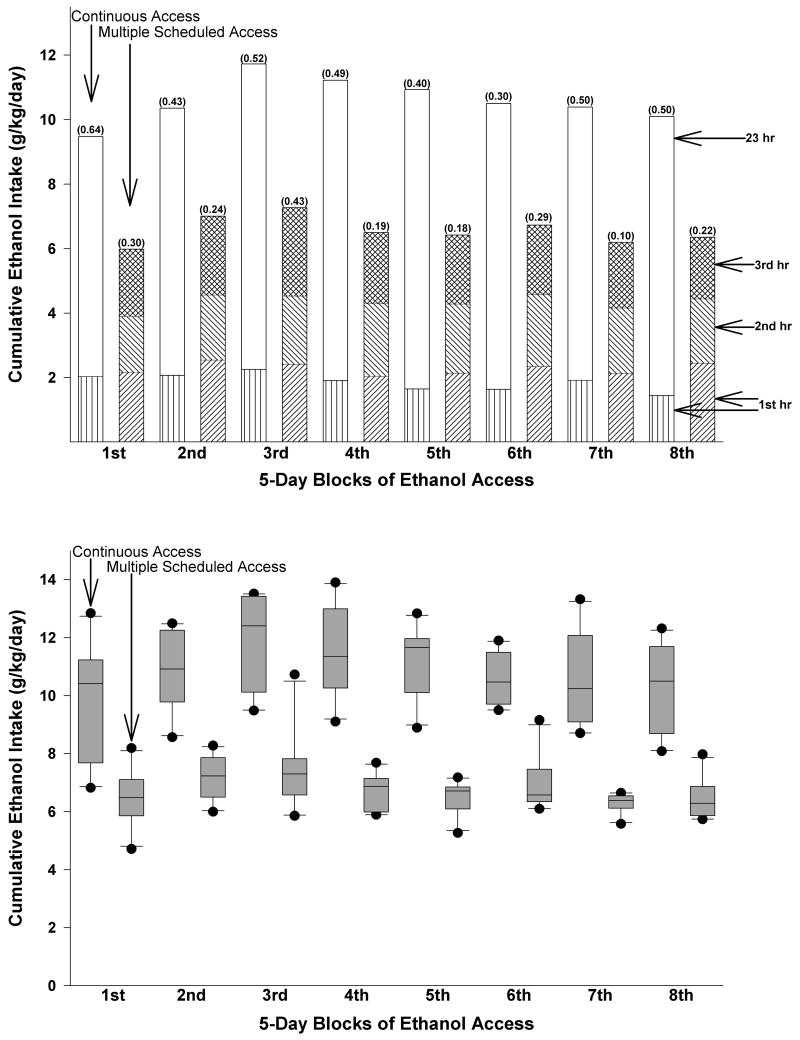
Daily ethanol intakes (mean ± S.E.M.) of the CA group averaged 9.6 ± 0.9 g/kg/day across the 8 weeks, whereas daily ethanol intakes of the MSA group averaged 6.4 ± 0.3 g/kg/day, with hourly intakes ranging between 1.7 and 2.7 g/kg (Fig. 1, upper panel). Overall, the distributions of drinking scores for the two groups did not overlap (Fig. 1, lower panel: the boxplot depicts the means, interquartile ranges, and the most extreme scores). In the present study, the average daily intakes of the CA group and average hourly intakes of the MSA are higher than previously reported (Bell et al., 2006a), despite the fact that the MSA group had access to only three 1-hr sessions compared with four 1-hr access sessions in the Bell et al. (2006a) study. These differences may have been due to the use of selectively bred P rats in the present study versus inbred P rats used in the previous study and/or the use of male P rats in the present study versus the use of female inbred P rats in the previous study.
Fig. 1.
The upper panel depicts the 5-day average ethanol intake values (± SEM of the total daily intake indicated in the parentheses) for the first and subsequent 23 hours of the continuous access (CA) P rats and the 5-day average ethanol intake values for the three 1-hour access periods of the multiple scheduled access (MSA) P rats. The box plot in the lower panel indicates that the distributions (CA versus MSA) of drinking scores, for the most part, did not overlap, which includes the most extreme scores. Note: the 5-day averages represent the days that ethanol (15% and 30% concurrent with water) was available to both CA and MSA P rats.
3.2. Effects of ethanol on gene expression in the ACB
Comparison of the CA versus W group indicated 406 probe sets of named genes were significantly (p < 0.01; FDR ≤ 0.15) different between the two groups, with 233 probe sets higher and 173 probe sets lower in the CA group (Table 1). These 406 significantly different probe sets represented 374 uniquely named genes. Most of the gene expression differences (Table 1) were in the range of 1.1- to 1.3- fold.
Table 1.
Significant Effects of Alcohol Drinking under 24-hr Continuous Access on Changes in Gene Expression in the Nucleus Accumbens of P rats
| Symbol | Gene Description | CA/W | FDR q | p-value | QTL | Chr | LOD |
|---|---|---|---|---|---|---|---|
| Hmgcr | 3-hydroxy-3-methylglutaryl-Coenzyme A reductase | 1.08 | 0.15 | 0.010 | |||
| Pfkfb2 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 | 1.11 | 0.11 | 0.002 | |||
| Acta1 | actin, alpha 1, skeletal muscle | 1.12 | 0.12 | 0.004 | |||
| Actl6a | Actin-like 6A | 1.10 | 0.12 | 0.003 | |||
| Abt1 | activator of basal transcription 1 | 1.07 | 0.12 | 0.005 | |||
| --- | Activity and neurotransmitter-induced early gene 1 (ania-1) mRNA, 3′UTR | 1.23 | 0.12 | 0.004 | |||
| Arc | activity regulated cytoskeletal-associated protein | 1.53 | 0.01 | 0.000 | |||
| Adamts1 | a disintegrin-like and metallopeptidse (reprolysin type) with thrombospondin type 1 motif, 1 | 1.37 | 0.09 | 0.001 | |||
| Arf6 | ADP-ribosylation factor 6 | 1.17 | 0.14 | 0.008 | |||
| Arl1 | ADP-ribosylation factor-like 1 | 1.08 | 0.11 | 0.002 | |||
| Afg3l2 | AFG3(ATPase family gene 3)-like 2 (yeast) | 1.08 | 0.12 | 0.004 | |||
| Appbp2 | amyloid beta precursor protein (cytoplasmic tail) binding protein 2 | 1.17 | 0.13 | 0.006 | |||
| Apex1 | apurinic/apyrimidinic endonuclease 1 | 1.07 | 0.15 | 0.011 | |||
| Actr10 | ARP10 actin-related protein 10 homolog (S. cerevisiae) | 1.09 | 0.15 | 0.011 | |||
| Btg1 | B-cell translocation gene 1, anti-proliferative | 1.10 | 0.14 | 0.008 | |||
| Btg2 | B-cell translocation gene 2, anti-proliferative | 1.35 | 0.02 | 0.000 | |||
| Btg2 | B-cell translocation gene 2, anti-proliferative | 1.66 | 0.00 | 0.000 | |||
| St6gal2 | Beta galactoside alpha 2,6 sialyltransferase 2 | 1.26 | 0.09 | 0.001 | |||
| Bex1 | brain expressed X-linked 1 | 1.09 | 0.13 | 0.006 | |||
| Bckdhb | branched chain keto acid dehydrogenase E1, beta polypeptide | 1.11 | 0.11 | 0.003 | |||
| C1galt1c1 | C1GALT1-specific chaperone 1 | 1.08 | 0.15 | 0.011 | |||
| Cdh20 | cadherin 20 | 1.11 | 0.12 | 0.004 | |||
| Crcp | calcitonin gene-related peptide-receptor component protein | 1.07 | 0.14 | 0.008 | Alc10 | 12 | 2.2 |
| Cant1 | calcium activated nucleotidase 1 | 1.07 | 0.12 | 0.005 | |||
| Cacybp | calcyclin binding protein | 1.18 | 0.11 | 0.003 | |||
| Chst1 | carbohydrate (keratan sulfate Gal-6) sulfotransferase 1 | 1.13 | 0.15 | 0.011 | |||
| Cdv1 | carnitine deficiency-associated gene expressed in ventricle 1 | 1.18 | 0.15 | 0.009 | Alc6 | 12 | 4.7 |
| Cebpd | CCAAT/enhancer binding protein (C/EBP), delta | 1.13 | 0.14 | 0.008 | |||
| Cdc42 | Cell division cycle 42 homolog (S. cerevisiae) | 1.09 | 0.15 | 0.010 | |||
| Cep57 | centrosomal protein 57 | 1.11 | 0.15 | 0.010 | |||
| Chrna7 | Cholinergic receptor, nicotinic, alpha polypeptide 7 | 1.11 | 0.15 | 0.010 | |||
| Cbx3 | chromobox homolog 3 (HP1 gamma homolog, Drosophila) | 1.09 | 0.11 | 0.003 | Alc18 | 4 | 9.2 |
| Coq3 | coenzyme Q3 homolog, methyltransferase (yeast) | 1.10 | 0.10 | 0.002 | |||
| Cirbp | cold inducible RNA binding protein | 1.16 | 0.14 | 0.008 | |||
| Commd10 | COMM domain containing 10 | 1.16 | 0.13 | 0.006 | |||
| Commd3 | COMM domain containing 3 | 1.06 | 0.12 | 0.004 | |||
| Ctgf | connective tissue growth factor | 1.24 | 0.09 | 0.001 | |||
| Crh | corticotropin releasing hormone | 1.24 | 0.12 | 0.004 | |||
| Ddx21a | DEAD (Asp-Glu-Ala-Asp) box polypeptide 21a | 1.16 | 0.09 | 0.001 | |||
| Dnd1 | dead end homolog 1 (zebrafish) | 1.13 | 0.10 | 0.001 | |||
| Dhrs7b | dehydrogenase/reductase (SDR family) member 7B | 1.06 | 0.14 | 0.007 | |||
| Dlc1 | deleted in liver cancer 1 | 1.14 | 0.11 | 0.003 | Alc11 | 16 | 3.2 |
| Dnttip1 | deoxynucleotidyltransferase, terminal, interacting protein 1 | 1.09 | 0.13 | 0.006 | |||
| Dld | dihydrolipoamide dehydrogenase | 1.13 | 0.15 | 0.010 | |||
| Dcx | doublecortin | 1.20 | 0.14 | 0.007 | |||
| Dr1 | down-regulator of transcription 1 | 1.13 | 0.09 | 0.001 | |||
| Dph5 | DPH5 homolog (S. cerevisiae) | 1.11 | 0.14 | 0.007 | Alc15 | 2 | 4.1 |
| Dsm-1 | D-serine modulator-1 | 1.06 | 0.13 | 0.006 | |||
| Dusp1 | dual specificity phosphatase 1 | 1.42 | 0.00 | 0.000 | Alc5 | 10 | 2.4 |
| Dusp1 | dual specificity phosphatase 1 | 1.42 | 0.02 | 0.000 | |||
| Dusp6 | dual specificity phosphatase 6 | 1.18 | 0.07 | 0.000 | |||
| Dusp6 | dual specificity phosphatase 6 | 1.13 | 0.10 | 0.001 | |||
| Dusp6 | Dual specificity phosphatase 6 | 1.13 | 0.14 | 0.008 | |||
| Ep300 | E1A binding protein p300 | 1.17 | 0.11 | 0.002 | |||
| Egln3 | EGL nine homolog 3 (C. elegans) | 1.10 | 0.15 | 0.011 | |||
| Efna3 | ephrin A3 | 1.13 | 0.10 | 0.002 | |||
| Eif2a | eukaryotic translation initiation factor 2A | 1.18 | 0.14 | 0.009 | |||
| Eif2s1 | eukaryotic translation initiation factor 2, subunit 1 alpha | 1.12 | 0.10 | 0.002 | |||
| Nme1 | expressed in non-metastatic cells 1 | 1.05 | 0.15 | 0.011 | |||
| Fos | FBJ murine osteosarcoma viral oncogene homolog | 1.72 | 0.00 | 0.000 | |||
| Frag1 | FGF receptor activating protein 1 | 1.13 | 0.14 | 0.007 | |||
| Fkbp1b | FK506 binding protein 1b | 1.14 | 0.09 | 0.001 | |||
| Galk2 | galactokinase 2 | 1.11 | 0.12 | 0.003 | |||
| Gtf2a2 | general transcription factor IIa 2 | 1.08 | 0.15 | 0.009 | |||
| Gpm6a | glycoprotein m6a | 1.11 | 0.15 | 0.009 | Alc11 | 16 | 3.2 |
| Gpr85 | G protein-coupled receptor 85 | 1.11 | 0.12 | 0.005 | |||
| Gdf11 | growth differentiation factor 11 | 1.09 | 0.12 | 0.004 | |||
| Grb2 | growth factor receptor bound protein 2 | 1.07 | 0.11 | 0.003 | |||
| Gng10 | guanine nucleotide binding protein (G protein), gamma 10 | 1.08 | 0.12 | 0.003 | |||
| Gucy1b3 | guanylate cyclase 1, soluble, beta 3 | 1.09 | 0.15 | 0.010 | |||
| Hes1 | hairy and enhancer of split 1 (Drosophila) | 1.17 | 0.15 | 0.009 | |||
| Hsf2 | heat shock factor 2 | 1.13 | 0.10 | 0.001 | |||
| Havcr2 | hepatitis A virus cellular receptor 2 | 1.18 | 0.15 | 0.010 | |||
| Hnrpab | heterogeneous nuclear ribonucleoprotein A/B | 1.10 | 0.11 | 0.003 | |||
| Hnrpk | heterogeneous nuclear ribonucleoprotein K | 1.08 | 0.15 | 0.009 | |||
| Hdac2 | histone deacetylase 2 | 1.13 | 0.13 | 0.005 | |||
| Hdac5 | Histone deacetylase 5 | 1.14 | 0.11 | 0.003 | |||
| Homer1 | homer homolog 1 (Drosophila) | 1.71 | 0.01 | 0.000 | |||
| Hrpap20 | hormone-regulated proliferation associated protein 20 | 1.11 | 0.13 | 0.005 | |||
| Hagh | hydroxyacyl glutathione hydrolase | 1.09 | 0.14 | 0.007 | Alc5 | 10 | 2.4 |
| Hcn1 | Hyperpolarization-activated cyclic nucleotide-gated potassium channel 1 | 1.33 | 0.11 | 0.003 | |||
| Ier2 | immediate early response 2 | 1.33 | 0.01 | 0.000 | |||
| Itga6 | integrin, alpha 6 | 1.11 | 0.11 | 0.003 | |||
| Itgb3bp | integrin beta 3 binding protein (beta3-endonexin) | 1.14 | 0.12 | 0.004 | |||
| Ift74 | intraflagellar transport 74 homolog (Chlamydomonas) | 1.14 | 0.13 | 0.006 | |||
| Nyw1 | ischemia related factor NYW-1 | 1.15 | 0.07 | 0.000 | |||
| Idi1 | isopentenyl-diphosphate delta isomerase | 1.14 | 0.02 | 0.000 | |||
| Icmt | isoprenylcysteine carboxyl methyltransferase | 1.07 | 0.13 | 0.006 | |||
| Junb | Jun-B oncogene | 1.27 | 0.00 | 0.000 | |||
| Jun | Jun oncogene | 1.15 | 0.14 | 0.009 | |||
| Jun | Jun oncogene | 1.23 | 0.01 | 0.000 | |||
| Kab | KARP-1 binding protein 1 | 1.10 | 0.11 | 0.002 | |||
| Khdrbs2 | KH domain containing, RNA binding, signal transduction associated 2 | 1.35 | 0.11 | 0.002 | |||
| Kif2 | kinesin heavy chain family, member 2 | 1.13 | 0.12 | 0.003 | |||
| Lactb2 | lactamase, beta 2 | 1.14 | 0.14 | 0.007 | |||
| Lactb2 | Lactamase, beta 2 | 1.10 | 0.15 | 0.011 | |||
| Lap3 | leucine aminopeptidase 3 | 1.20 | 0.15 | 0.010 | |||
| Lrrc23 | leucine rich repeat containing 23 | 1.15 | 0.13 | 0.006 | |||
| LOC246187 | liver regeneration-related protein | 1.12 | 0.11 | 0.002 | |||
| Mgat2 | mannoside acetylglucosaminyltransferase 2 | 1.16 | 0.11 | 0.002 | |||
| Mkks | McKusick-Kaufman syndrome protein | 1.07 | 0.12 | 0.004 | |||
| Med4 | mediator of RNA polymerase II transcription, subunit 4 homolog (yeast) | 1.11 | 0.12 | 0.004 | |||
| Mtf2 | metal response element binding transcription factor 2 | 1.39 | 0.13 | 0.005 | |||
| Mtx2 | metaxin 2 | 1.16 | 0.10 | 0.001 | |||
| Mbd1 | Methyl-CpG binding domain protein 1 | 1.12 | 0.14 | 0.008 | |||
| Mtfmt | mitochondrial methionyl-tRNA formyltransferase | 1.10 | 0.14 | 0.008 | |||
| Mcl1 | myeloid cell leukemia sequence 1 | 1.14 | 0.14 | 0.007 | |||
| Mx2 | myxovirus (influenza virus) resistance 2 | 1.21 | 0.04 | 0.000 | |||
| Ndufa9 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9 | 1.17 | 0.11 | 0.003 | |||
| Ndufv3l | NADH dehydrogenase (ubiquinone) flavoprotein 3-like | 1.13 | 0.12 | 0.004 | |||
| LOC493574 | notch1-induced protein | 1.24 | 0.11 | 0.003 | |||
| Nip7 | nuclear import 7 homolog (S. cerevisiae) | 1.15 | 0.09 | 0.001 | |||
| Nrbf2 | nuclear receptor binding factor 2 | 1.15 | 0.09 | 0.001 | |||
| Nr4a1 | nuclear receptor subfamily 4, group A, member 1 | 1.62 | 0.00 | 0.000 | |||
| Nr4a3 | nuclear receptor subfamily 4, group A, member 3 | 1.46 | 0.04 | 0.000 | |||
| Nup155 | Nucleoporin 155 | 1.13 | 0.14 | 0.008 | |||
| Nupl1 | nucleoporin like 1 | 1.15 | 0.11 | 0.003 | |||
| Nap1l2 | nucleosome assembly protein 1-like 2 | 1.25 | 0.14 | 0.007 | |||
| Nap1l3 | nucleosome assembly protein 1-like 3 | 1.17 | 0.10 | 0.001 | |||
| Numbl | Numb-like | 1.16 | 0.15 | 0.010 | |||
| Orc6l | origin recognition complex, subunit 6-like (S. cerevisiae) | 1.20 | 0.04 | 0.000 | |||
| Orc6l | origin recognition complex, subunit 6-like (S. cerevisiae) | 1.22 | 0.12 | 0.004 | |||
| Pspc1 | paraspeckle protein 1 | 1.13 | 0.15 | 0.010 | |||
| Psip1 | PC4 and SFRS1 interacting protein 1 | 1.39 | 0.08 | 0.000 | |||
| Ppig | peptidylprolyl isomerase G | 1.22 | 0.10 | 0.001 | |||
| Phf12 | PHD finger protein 12 | 1.10 | 0.12 | 0.004 | |||
| Phf12 | PHD finger protein 12 | 1.12 | 0.15 | 0.010 | |||
| Pigk | phosphatidylinositol glycan, class K | 1.10 | 0.14 | 0.008 | |||
| Pfkl | phosphofructokinase, liver, B-type | 1.11 | 0.15 | 0.010 | |||
| Pgam5 | phosphoglycerate mutase family member 5 | 1.10 | 0.11 | 0.003 | |||
| Pik3c3 | phosphoinositide-3-kinase, class 3 | 1.07 | 0.13 | 0.005 | |||
| Plaa | phospholipase A2, activating protein | 1.12 | 0.11 | 0.002 | |||
| Ppcs | phosphopantothenoylcysteine synthetase | 1.08 | 0.15 | 0.009 | |||
| Prps2 | phosphoribosyl pyrophosphate synthetase 2 | 1.22 | 0.09 | 0.001 | |||
| Prpsap2 | phosphoribosyl pyrophosphate synthetase-associated protein 2 | 1.11 | 0.10 | 0.001 | |||
| Psph | phosphoserine phosphatase | 1.13 | 0.13 | 0.005 | Alc10 | 12 | 2.2 |
| Plagl1 | pleiomorphic adenoma gene-like 1 | 1.14 | 0.14 | 0.008 | |||
| Pcyox1 | prenylcysteine oxidase 1 | 1.20 | 0.11 | 0.002 | Alc18 | 4 | 9.2 |
| Pnrc1 | proline rich 2 | 1.16 | 0.11 | 0.003 | |||
| Psmc6 | proteasome (prosome, macropain) 26S subunit, ATPase, 6 | 1.28 | 0.09 | 0.001 | |||
| Psmc6 | proteasome (prosome, macropain) 26S subunit, ATPase, 6 | 1.21 | 0.03 | 0.000 | |||
| Psmc6 | proteasome (prosome, macropain) 26S subunit, ATPase, 6 | 1.20 | 0.09 | 0.001 | |||
| Psmd12 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 12 | 1.12 | 0.10 | 0.001 | |||
| Psmd6 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 | 1.07 | 0.12 | 0.003 | |||
| Psma4 | proteasome (prosome, macropain) subunit, alpha type 4 | 1.09 | 0.09 | 0.001 | |||
| Pkib | protein kinase inhibitor beta, cAMP dependent, catalytic | 1.23 | 0.11 | 0.003 | |||
| Pkib | protein kinase inhibitor beta, cAMP dependent, catalytic | 1.36 | 0.15 | 0.010 | |||
| Prkra | protein kinase, interferon inducible double stranded RNA dependent activator | 1.11 | 0.12 | 0.004 | |||
| Pofut1 | Protein O-fucosyltransferase 1 | 1.13 | 0.15 | 0.010 | |||
| Ppp2ca | Protein phosphatase 2 (formerly 2A), catalytic subunit, alpha isoform | 1.20 | 0.08 | 0.000 | |||
| Ppp2ca | Protein phosphatase 2 (formerly 2A), catalytic subunit, alpha isoform | 1.10 | 0.04 | 0.000 | |||
| Pcdha1 /// Pcdha10 /// Pcdha11 /// Pcdha12 /// Pcdha13 /// Pcdha2 /// Pcdha3 protocadherin alpha 4 /// protocadherin alpha 13 /// protocadher i/n// aPlpcdhhaa 140 / // /P pcrdohtoac5a /d/h/ ePrcind haalp6h | 1.15 | 0.15 | 0.010 | ||||
| P2ry2 | purinergic receptor P2Y, G-protein coupled 2 | 1.13 | 0.10 | 0.002 | |||
| LOC289809 | putatative 28 kDa protein | 1.24 | 0.10 | 0.001 | |||
| Rab1 | RAB1, member RAS oncogene family | 1.10 | 0.14 | 0.007 | |||
| Rab21 | RAB21, member RAS oncogene family | 1.14 | 0.14 | 0.007 | |||
| Rab35 | RAB35, member RAS oncogene family | 1.14 | 0.13 | 0.006 | |||
| Rab3c | RAB3C, member RAS oncogene family | 1.11 | 0.14 | 0.007 | |||
| Rad17 | RAD17 homolog (S. pombe) | 1.13 | 0.11 | 0.003 | |||
| Rdx | Radixin | 1.12 | 0.12 | 0.003 | |||
| Rae1 | RAE1 RNA export 1 homolog (S. pombe) | 1.07 | 0.14 | 0.008 | |||
| Ripx | rap2 interacting protein x | 1.12 | 0.10 | 0.001 | |||
| Rdbp | RD RNA-binding protein | 1.09 | 0.10 | 0.001 | |||
| Rcn2 | reticulocalbin 2 | 1.14 | 0.10 | 0.002 | |||
| Rbbp6 | Retinoblastoma binding protein 6 | 1.17 | 0.12 | 0.003 | |||
| Rnf149 | ring finger protein 149 | 1.15 | 0.14 | 0.007 | |||
| Rnf2 | ring finger protein 2 | 1.14 | 0.13 | 0.005 | |||
| --- | RM2 mRNA, partial sequence | 1.39 | 0.04 | 0.000 | |||
| Rbm13 | RNA binding motif protein 13 | 1.13 | 0.14 | 0.007 | |||
| Rbm14 | RNA binding motif protein 14 | 1.11 | 0.15 | 0.011 | |||
| Rbm17 | RNA binding motif protein 17 | 1.08 | 0.14 | 0.007 | |||
| Rbm3 | RNA binding motif protein 3 | 1.14 | 0.11 | 0.002 | |||
| Rg9mtd1 | RNA (guanine-9-) methyltransferase domain containing 1 | 1.10 | 0.14 | 0.008 | |||
| Rpo1-1 | RNA polymerase 1-1 | 1.08 | 0.15 | 0.011 | |||
| Rwdd3 | RWD domain containing 3 | 1.18 | 0.14 | 0.008 | Alc15 | 2 | 4.1 |
| Rwdd3 | RWD domain containing 3 | 1.17 | 0.14 | 0.009 | |||
| Sec23ip | SEC23 interacting protein | 1.06 | 0.15 | 0.009 | |||
| Scg2 | secretogranin 2 | 1.12 | 0.13 | 0.006 | |||
| Siah1a | seven in absentia 1A | 1.13 | 0.14 | 0.007 | |||
| Siah2 | seven in absentia 2 | 1.13 | 0.12 | 0.004 | |||
| Srp54 | signal recognition particle 54 | 1.16 | 0.12 | 0.004 | |||
| Six3 | Sine oculis homeobox homolog 3 (Drosophila) | 1.19 | 0.13 | 0.006 | Alc17 | 6 | 3.1 |
| Ssbp1 | single-stranded DNA binding protein 1 | 1.16 | 0.09 | 0.001 | Alc18 | 4 | 9.2 |
| Ssbp1 | single-stranded DNA binding protein 1 | 1.15 | 0.09 | 0.001 | |||
| Scye1 | small inducible cytokine subfamily E, member 1 | 1.08 | 0.15 | 0.011 | Alc15 | 2 | 4.1 |
| Slc4a10 | Solute carrier family 4, sodium bicarbonate transporter-like, member 10 | 1.15 | 0.14 | 0.007 | |||
| Slc6a15 | solute carrier family 6 (neurotransmitter transporter), member 15 | 1.10 | 0.14 | 0.009 | |||
| Snx16 | sorting nexin 16 | 1.13 | 0.13 | 0.006 | |||
| Snx16 | sorting nexin 16 | 1.13 | 0.11 | 0.003 | |||
| Spg3a | spastic paraplegia 3A homolog (human) | 1.12 | 0.11 | 0.003 | |||
| Spdy1 | speedy homolog 1 (Drosophila) | 1.17 | 0.14 | 0.007 | Alc17 | 6 | 3.1 |
| Sf4 | splicing factor 4 | 1.09 | 0.14 | 0.009 | Alc11 | 16 | 3.2 |
| Spon1 | spondin 1 | 1.12 | 0.10 | 0.001 | |||
| Spry2 | sprouty homolog 2 (Drosophila) | 1.10 | 0.15 | 0.011 | |||
| Sod2 | superoxide dismutase 2, mitochondrial | 1.14 | 0.10 | 0.001 | |||
| Syncrip | Synaptotagmin binding, cytoplasmic RNA interacting protein | 1.19 | 0.15 | 0.010 | |||
| Taf9l | TAF9-like RNA polymerase II, TATA box binding protein (TBP)-associated factor, 31kDa | 1.21 | 0.11 | 0.002 | |||
| Tfpt | TCF3 (E2A) fusion partner | 1.11 | 0.11 | 0.003 | |||
| Tcp11l2 | t-complex 11 (mouse) like 2 | 1.11 | 0.14 | 0.007 | |||
| Tgif | TG interacting factor | 1.21 | 0.02 | 0.000 | |||
| Txnl1 | thioredoxin-like 1 | 1.13 | 0.14 | 0.008 | |||
| Txnl2 | thioredoxin-like 2 | 1.08 | 0.15 | 0.010 | |||
| Tmsbl1 | thymosin beta-like protein 1 | 1.22 | 0.11 | 0.003 | |||
| Tceal8 | transcription elongation factor A (SII)-like 8 | 1.09 | 0.13 | 0.006 | |||
| Tceb3 | Transcription elongation factor B (SIII), polypeptide 3 | 1.10 | 0.10 | 0.002 | |||
| Tgfa | transforming growth factor alpha | 1.13 | 0.14 | 0.008 | Alc18 | 4 | 9.2 |
| Tgfa | transforming growth factor alpha | 1.14 | 0.15 | 0.010 | |||
| Tomm20 | translocase of outer mitochondrial membrane 20 homolog (yeast) | 1.07 | 0.12 | 0.005 | |||
| Tmem17 | transmembrane protein 17 | 1.10 | 0.12 | 0.003 | |||
| Tmeff1 | transmembrane protein with EGF-like and two follistatin-like domains 1 | 1.13 | 0.11 | 0.002 | |||
| Tpm3 | tropomyosin 3, gamma | 1.11 | 0.07 | 0.000 | |||
| Tpm3 | tropomyosin 3, gamma | 1.15 | 0.15 | 0.010 | |||
| Tubb2b | tubulin, beta 2b | 1.10 | 0.10 | 0.001 | |||
| Uchl5 | ubiquitin carboxyl-terminal hydrolase L5 | 1.11 | 0.04 | 0.000 | |||
| Ube2d2 | ubiquitin-conjugating enzyme E2D 2 | 1.17 | 0.14 | 0.007 | |||
| Ube2d3 | Ubiquitin-conjugating enzyme E2D 3 (UBC4/5 homolog, yeast) | 1.07 | 0.14 | 0.007 | Alc15 | 2 | 4.1 |
| LOC500954 | Ubiquitin-Like 5 Protein | 1.12 | 0.09 | 0.001 | |||
| Ufc1 | Ufm1-conjugating enzyme 1 | 1.07 | 0.13 | 0.005 | |||
| Vps33a | vacuolar protein sorting 33A (yeast) | 1.21 | 0.13 | 0.006 | Alc6 | 12 | 4.7 |
| Vkorc1l1 | vitamin K epoxide reductase complex, subunit 1-like 1 | 1.14 | 0.10 | 0.001 | |||
| Wdfy1 | WD repeat and FYVE domain containing 1 | 1.11 | 0.11 | 0.003 | |||
| Wdr39 | WD repeat domain 39 | 1.08 | 0.14 | 0.009 | |||
| Wdr77 | WD repeat domain 77 | 1.18 | 0.11 | 0.002 | Alc15 | 2 | 4.1 |
| Wdr77 | WD repeat domain 77 | 1.12 | 0.10 | 0.001 | |||
| Wrnip1 | Werner helicase interacting protein 1 | 1.12 | 0.11 | 0.003 | |||
| Zcchc10 | zinc finger, CCHC domain containing 10 | 1.11 | 0.10 | 0.001 | |||
| Zcrb1 | zinc finger CCHC-type and RNA binding motif 1 | 1.20 | 0.10 | 0.001 | |||
| Zipro1 | zinc finger proliferation 1 | 1.14 | 0.12 | 0.004 | |||
| Zfp297 | zinc finger protein 297 | 1.09 | 0.15 | 0.010 | |||
| Zfp367 | zinc finger protein 367 | 1.18 | 0.12 | 0.004 | |||
| Zfp403 | zinc finger protein 403 | 1.15 | 0.14 | 0.009 | |||
| Zfp451 | zinc finger protein 451 | 1.10 | 0.15 | 0.010 | |||
| Zfp91 | zinc finger protein 91 | 1.13 | 0.09 | 0.001 | |||
| Htr4 | 5-hydroxytryptamine (serotonin) receptor 4 | -1.14 | 0.11 | 0.002 | |||
| Nt5e | 5′ nucleotidase, ecto | -1.15 | 0.10 | 0.002 | |||
| Actn1 | actinin, alpha 1 | -1.12 | 0.09 | 0.001 | |||
| Actn1 | Actinin, alpha 1 | -1.16 | 0.14 | 0.007 | |||
| Apeh | acylpeptide hydrolase | -1.07 | 0.15 | 0.011 | |||
| Adam17 | a disintegrin and metalloproteinase domain 17 (tumor necrosis factor, alpha, converting enzyme) | -1.08 | 0.15 | 0.010 | |||
| Akap8l | A kinase (PRKA) anchor protein 8-like | -1.12 | 0.15 | 0.010 | |||
| Aldh6a1 | aldehyde dehydrogenase family 6, subfamily A1 | -1.13 | 0.10 | 0.001 | |||
| Amt | aminomethyltransferase (glycine cleavage system protein T) | -1.10 | 0.12 | 0.004 | |||
| Angptl4 | angiopoietin-like 4 | -1.24 | 0.01 | 0.000 | |||
| Armc5 | armadillo repeat containing 5 | -1.11 | 0.10 | 0.002 | |||
| Aspa | aspartoacylase | -1.17 | 0.13 | 0.005 | |||
| Atp6v0e1 | ATPase, H+ transporting, V0 subunit E isoform 1 | -1.13 | 0.14 | 0.009 | Alc5 | 10 | 2.4 |
| Abcg2 | ATP-binding cassette, sub-family G (WHITE), member 2 | -1.14 | 0.13 | 0.006 | Alc18 | 4 | 9.2 |
| St6gal1 | beta galactoside alpha 2,6 sialyltransferase 1 | -1.15 | 0.14 | 0.007 | |||
| Cast | calpastatin | -1.10 | 0.15 | 0.010 | |||
| Cml3 | camello-like 3 | -1.16 | 0.02 | 0.000 | Alc18 | 4 | 9.2 |
| Creb3l2 | cAMP responsive element binding protein 3-like 2 | -1.17 | 0.02 | 0.000 | Alc18 | 4 | 9.2 |
| Car11 | carbonic anhydrase 11 | -1.10 | 0.11 | 0.002 | |||
| Cpt1c | carnitine palmitoyltransferase 1c | -1.09 | 0.14 | 0.009 | |||
| Csnk1g1 | casein kinase 1, gamma 1 | -1.13 | 0.12 | 0.005 | |||
| Catna1 | catenin (cadherin-associated protein), alpha 1 | -1.08 | 0.14 | 0.009 | |||
| Cav | caveolin | -1.18 | 0.13 | 0.005 | |||
| Cav2 | caveolin 2 | -1.19 | 0.11 | 0.002 | |||
| Cd97 | CD97 antigen | -1.22 | 0.08 | 0.000 | |||
| Cdc37l1 | cell division cycle 37 homolog (S. cerevisiae)-like 1 | -1.07 | 0.14 | 0.007 | |||
| Ccr5 | Chemokine (C-C motif) receptor 5 | -1.11 | 0.12 | 0.004 | |||
| Cldn1 | claudin 1 | -1.15 | 0.15 | 0.011 | |||
| Cldn5 | claudin 5 | -1.16 | 0.11 | 0.002 | |||
| Creld2 | cysteine-rich with EGF-like domains 2 | -1.10 | 0.12 | 0.004 | |||
| Cyp4f2 | cytochrome P450, family 4, subfamily F, polypeptide 2 | -1.11 | 0.11 | 0.002 | |||
| Dock9 | dedicator of cytokinesis 9 | -1.21 | 0.10 | 0.001 | |||
| Degs2 | degenerative spermatocyte homolog 2 (Drosophila), lipid desaturase | -1.16 | 0.07 | 0.000 | |||
| Dhrs3 | dehydrogenase/reductase (SDR family) member 3 | -1.18 | 0.11 | 0.003 | |||
| Dsp | desmoplakin | -1.15 | 0.10 | 0.002 | |||
| Dgka | diacylglycerol kinase, alpha | -1.12 | 0.15 | 0.009 | |||
| Dlgap2 | Discs, large (Drosophila) homolog-associated protein 2 | -1.26 | 0.11 | 0.002 | |||
| Dlgh1 | Discs, large homolog 1 (Drosophila) | -1.09 | 0.09 | 0.001 | |||
| Dscr1l1 | Down syndrome critical region gene 1-like 1 | -1.18 | 0.05 | 0.000 | |||
| Dscr1l1 | Down syndrome critical region gene 1-like 1 | -1.19 | 0.11 | 0.003 | |||
| Eltd1 | EGF, latrophilin and seven transmembrane domain containing 1 | -1.29 | 0.01 | 0.000 | |||
| Ehd2 | EH-domain containing 2 | -1.27 | 0.09 | 0.001 | |||
| Ecam | endothelial cell adhesion molecule | -1.18 | 0.07 | 0.000 | Alc20 | 8 | 2 |
| Tek | endothelial-specific receptor tyrosine kinase | -1.13 | 0.15 | 0.009 | |||
| Ece1 | endothelin converting enzyme 1 | -1.10 | 0.13 | 0.005 | |||
| Epim | epimorphin | -1.14 | 0.14 | 0.008 | Alc10 | 12 | 2.2 |
| Epn2 | epsin 2 | -1.09 | 0.14 | 0.009 | |||
| Eef2k | Eukaryotic elongation factor-2 kinase | -1.11 | 0.14 | 0.007 | |||
| Ecm1 | extracellular matrix protein 1 | -1.19 | 0.13 | 0.005 | |||
| Fbxl20 | F-box and leucine-rich repeat protein 20 | -1.09 | 0.15 | 0.010 | |||
| Fkbp5 | FK506 binding protein 5 | -1.14 | 0.10 | 0.001 | |||
| Fkbp5 | FK506 binding protein 5 | -1.18 | 0.07 | 0.000 | |||
| Fmo2 | flavin containing monooxygenase 2 | -1.25 | 0.12 | 0.003 | |||
| Foxo1a | Forkhead box O1A | -1.15 | 0.13 | 0.006 | |||
| Fut2 | fucosyltransferase 2 (secretor status included) | -1.15 | 0.13 | 0.005 | |||
| Galt | galactose-1-phosphate uridyl transferase | -1.08 | 0.11 | 0.003 | |||
| Gja1 | gap junction membrane channel protein alpha 1 | -1.11 | 0.15 | 0.010 | |||
| Gpt1 | glutamic pyruvic transaminase 1, soluble | -1.10 | 0.11 | 0.002 | |||
| Gpd1 | glycerol-3-phosphate dehydrogenase 1 (soluble) | -1.22 | 0.10 | 0.001 | |||
| Gpd1 | glycerol-3-phosphate dehydrogenase 1 (soluble) | -1.18 | 0.10 | 0.001 | |||
| Gramd3 | GRAM domain containing 3 | -1.09 | 0.15 | 0.011 | |||
| Gramd3 | GRAM domain containing 3 | -1.13 | 0.11 | 0.003 | |||
| Garnl1 | GTPase activating RANGAP domain-like 1 | -1.25 | 0.12 | 0.004 | |||
| Gimap6 | GTPase, IMAP family member 6 | -1.11 | 0.11 | 0.003 | Alc18 | 4 | 9.2 |
| Gng7 | guanine nucleotide binding protein, gamma 7 | -1.12 | 0.15 | 0.010 | |||
| Hspa5 | heat shock 70kDa protein 5 (glucose-regulated protein) | -1.10 | 0.11 | 0.002 | Alc8 | 3 | 2.7 |
| Hspa1b | heat shock 70kD protein 1B (mapped) | -1.28 | 0.15 | 0.010 | |||
| Hbp1 | high mobility group box transcription factor 1 | -1.07 | 0.15 | 0.010 | |||
| Homer1 | homer homolog 1 (Drosophila) | -1.72 | 0.00 | 0.000 | |||
| Hip1 | huntingtin interacting protein 1 | -1.14 | 0.10 | 0.002 | Alc10 | 12 | 2.2 |
| Hdh | Huntington disease gene homolog | -1.15 | 0.11 | 0.002 | |||
| Hyal2 | hyaluronoglucosaminidase 2 | -1.17 | 0.09 | 0.001 | |||
| Igj | immunoglobulin joining chain | -1.11 | 0.15 | 0.009 | |||
| Irs3 | insulin receptor substrate 3 | -1.14 | 0.09 | 0.001 | |||
| Ifngr1 | interferon gamma receptor 1 | -1.07 | 0.15 | 0.010 | |||
| Irf3 | interferon regulatory factor 3 | -1.11 | 0.12 | 0.004 | |||
| Il16 | interleukin 16 | -1.09 | 0.13 | 0.005 | |||
| Nyw1 | ischemia related factor NYW-1 | -1.14 | 0.11 | 0.002 | |||
| Jam2 | junction adhesion molecule 2 | -1.13 | 0.14 | 0.008 | |||
| Kif11 | kinesin family member 11 | -1.12 | 0.12 | 0.005 | |||
| Kif1b | Kinesin family member 1B | -1.10 | 0.15 | 0.010 | |||
| Kif6 | kinesin family member 6 | -1.09 | 0.14 | 0.009 | |||
| Klc1 | kinesin light chain 1 | -1.08 | 0.14 | 0.008 | |||
| Lphn2 | latrophilin 2 | -1.08 | 0.14 | 0.007 | |||
| Leng8 | leukocyte receptor cluster (LRC) member 8 | -1.12 | 0.14 | 0.006 | |||
| Lims2 | LIM and senescent cell antigen like domains 2 | -1.23 | 0.01 | 0.000 | |||
| LMO7 | LIM domain only protein 7 | -1.19 | 0.12 | 0.004 | |||
| Lcn7 | lipocalin 7 | -1.24 | 0.09 | 0.001 | |||
| Lsr | lipolysis stimulated lipoprotein receptor | -1.21 | 0.14 | 0.006 | |||
| Lef1 | Lymphoid enhancer binding factor 1 | -1.12 | 0.09 | 0.001 | Alc15 | 2 | 4.1 |
| Man2a1 | mannosidase 2, alpha 1 | -1.16 | 0.09 | 0.001 | |||
| Mcpt1 | mast cell protease 1 | -1.14 | 0.10 | 0.001 | |||
| Mxi1 | Max interacting protein 1 | -1.07 | 0.14 | 0.008 | |||
| Mbc2 | membrane bound C2 domain containing protein | -1.15 | 0.14 | 0.008 | |||
| Mpp4 | membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4) | -1.08 | 0.12 | 0.005 | |||
| Mgea5 | Meningioma expressed antigen 5 (hyaluronidase) | -1.14 | 0.11 | 0.003 | |||
| Mat2a | methionine adenosyltransferase II, alpha | -1.11 | 0.14 | 0.007 | |||
| Mettl7b | methyltransferase like 7B | -1.12 | 0.12 | 0.004 | |||
| Mtus1 | mitochondrial tumor suppressor 1 | -1.09 | 0.13 | 0.005 | Alc11 | 16 | 3.2 |
| Mobp | myelin-associated oligodendrocytic basic protein | -1.22 | 0.10 | 0.001 | |||
| Ntrk2 | Neurotrophic tyrosine kinase, receptor, type 2 | -1.67 | 0.00 | 0.000 | |||
| Nexn | nexilin | -1.15 | 0.14 | 0.008 | |||
| Ng3 | Ng3 protein | -1.11 | 0.11 | 0.003 | |||
| Nmnat1 | nicotinamide nucleotide adenylyltransferase 1 | -1.11 | 0.15 | 0.011 | |||
| Nos3 | nitric oxide synthase 3, endothelial cell | -1.21 | 0.11 | 0.003 | |||
| Nxf1 | nuclear RNA export factor 1 | -1.08 | 0.14 | 0.007 | |||
| Numb | numb gene homolog (Drosophila) | -1.12 | 0.12 | 0.004 | |||
| Ocln | occludin | -1.14 | 0.10 | 0.002 | |||
| Pthr1 | Parathyroid hormone receptor 1 | -1.20 | 0.07 | 0.000 | |||
| Per2 | period homolog 2 (Drosophila) | -1.17 | 0.04 | 0.000 | |||
| Ppap2b | phosphatidic acid phosphatase type 2B | -1.12 | 0.11 | 0.002 | |||
| Plce1 | phospholipase C, epsilon 1 | -1.12 | 0.14 | 0.009 | |||
| Plcg1 | phospholipase C, gamma 1 | -1.12 | 0.12 | 0.004 | |||
| RGD1303232 | Phytn_dehydro and Pyr_redox domain containing protein RGD1303232 | -1.10 | 0.13 | 0.006 | |||
| Pkp2 | plakophilin 2 | -1.14 | 0.10 | 0.001 | |||
| Podxl | podocalyxin-like | -1.11 | 0.12 | 0.004 | Alc18 | 4 | 9.2 |
| Pola2 | Polymerase (DNA directed), alpha 2 | -1.16 | 0.13 | 0.005 | |||
| Polm | polymerase (DNA directed), mu | -1.11 | 0.13 | 0.006 | |||
| Kcnt1 | potassium channel, subfamily T, member 1 | -1.12 | 0.14 | 0.008 | Alc8 | 3 | 2.7 |
| Kcnq3 | Potassium voltage-gated channel, subfamily Q, member 3 | -1.22 | 0.10 | 0.001 | |||
| Plod1 | procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1 | -1.13 | 0.15 | 0.009 | |||
| Col4a1 | procollagen, type IV, alpha 1 | -1.14 | 0.12 | 0.004 | |||
| Psrc2 | proline/serine-rich coiled-coil 2 | -1.07 | 0.14 | 0.008 | |||
| Pcsk7 | proprotein convertase subtilisin/kexin type 7 | -1.11 | 0.11 | 0.003 | |||
| Ptpn5 | protein tyrosine phosphatase, non-receptor type 5 | -1.08 | 0.15 | 0.009 | |||
| Ptprg | Protein tyrosine phosphatase, receptor type, G | -1.15 | 0.10 | 0.001 | |||
| Ptprv | protein tyrosine phosphatase, receptor type, V | -1.10 | 0.14 | 0.007 | |||
| Pxk | PX domain containing serine/threonine kinase | -1.08 | 0.13 | 0.006 | |||
| Rad9b | RAD9 homolog B (S. cerevisiae) | -1.19 | 0.11 | 0.002 | Alc6 | 12 | 4.7 |
| Ralgds | ral guanine nucleotide dissociation stimulator | -1.16 | 0.09 | 0.001 | Alc8 | 3 | 2.7 |
| Rap1ga1 | RAP1, GTPase activating protein 1 | -1.11 | 0.14 | 0.007 | |||
| Rasd2 | RASD family, member 2 | -1.17 | 0.10 | 0.001 | |||
| Rasd2 | RASD family, member 2 | -1.20 | 0.08 | 0.000 | |||
| Rasgrp4 | RAS guanyl releasing protein 4 | -1.16 | 0.07 | 0.000 | |||
| Ramp2 | receptor (calcitonin) activity modifying protein 2 | -1.11 | 0.13 | 0.006 | |||
| Rgs8 | regulator of G-protein signaling 8 | -1.12 | 0.14 | 0.008 | |||
| Rgs9 | regulator of G-protein signaling 9 | -1.15 | 0.11 | 0.002 | |||
| Rgs3 | Regulator of G-protein signalling 3 | -1.12 | 0.14 | 0.007 | |||
| Rcn2 | Reticulocalbin 2 | -1.10 | 0.12 | 0.004 | |||
| Arhgap17 | Rho GTPase activating protein 17 | -1.09 | 0.13 | 0.006 | |||
| Serpina1 | serine (or cysteine) proteinase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 | -1.12 | 0.14 | 0.009 | |||
| Shank1 | SH3 and multiple ankyrin repeat domains 1 | -1.14 | 0.15 | 0.010 | |||
| Cr16 | SH3 domain binding protein CR16 | -1.13 | 0.13 | 0.005 | Alc18 | 4 | 9.2 |
| Sipa1l1 | Signal-induced proliferation-associated 1 like 1 | -1.21 | 0.04 | 0.000 | |||
| Srpr | signal recognition particle receptor (‘docking protein’) | -1.10 | 0.15 | 0.010 | Alc20 | 8 | 2 |
| Stat4 | signal transducer and activator of transcription 4 | -1.18 | 0.12 | 0.004 | |||
| Smr2 | SMR2 | -1.12 | 0.15 | 0.010 | |||
| Slc14a2 | solute carrier family 14 (urea transporter), member 2 | -1.21 | 0.11 | 0.003 | |||
| Slc29a1 | solute carrier family 29 (nucleoside transporters), member 1 | -1.10 | 0.10 | 0.002 | |||
| Slc2a1 | solute carrier family 2 (facilitated glucose transporter), member 1 | -1.16 | 0.04 | 0.000 | |||
| Slc9a3r2 | Solute carrier family 9 (sodium/hydrogen exchanger), isoform 3 regulator 2 | -1.14 | 0.07 | 0.000 | Alc5 | 10 | 2.4 |
| Slco2b1 | solute carrier organic anion transporter family, member 2b1 | -1.16 | 0.11 | 0.002 | |||
| Sox18 | SRY-box containing gene 18 | -1.16 | 0.12 | 0.005 | |||
| Sts | steroid sulfatase | -1.11 | 0.12 | 0.003 | |||
| Stom | stomatin | -1.13 | 0.14 | 0.007 | Alc8 | 3 | 2.7 |
| Sulf2 | Sulfatase 2 | -1.13 | 0.14 | 0.007 | |||
| Sdc1 | syndecan 1 | -1.13 | 0.10 | 0.002 | Alc17 | 6 | 3.1 |
| Tec | tec protein tyrosine kinase | -1.09 | 0.15 | 0.011 | |||
| Ttc7 | tetratricopeptide repeat domain 7 | -1.12 | 0.14 | 0.006 | Alc17 | 6 | 3.1 |
| Tef | thyrotroph embryonic factor | -1.14 | 0.14 | 0.008 | |||
| pur-beta | Transcription factor Pur-beta | -1.13 | 0.15 | 0.009 | |||
| Tgfbr3 | transforming growth factor, beta receptor III | -1.14 | 0.13 | 0.005 | |||
| Tmed7 | Transmembrane emp24 protein transport domain containing 7 | -1.11 | 0.15 | 0.010 | |||
| Tsc22d3 | TSC22 domain family 3 | -1.10 | 0.09 | 0.001 | |||
| Tyrp1 | tyrosinase-related protein 1 | -1.13 | 0.12 | 0.004 | |||
| Tie1 | tyrosine kinase with immunoglobulin-like and EGF-like domains 1 | -1.21 | 0.10 | 0.002 | |||
| Ubadc1 | Ubiquitin associated domain containing 1 | -1.11 | 0.15 | 0.010 | Alc8 | 3 | 2 |
| Utrn | utrophin | -1.10 | 0.09 | 0.001 | |||
| Vamp1 | vesicle-associated membrane protein 1 | -1.12 | 0.13 | 0.005 | |||
| Vwa1 | von Willebrand factor A domain containing 1 | -1.13 | 0.12 | 0.004 | |||
| Wnk4 | WNK lysine deficient protein kinase 4 | -1.21 | 0.02 | 0.000 | |||
| Wbp1 | WW domain binding protein 1 | -1.08 | 0.12 | 0.004 | Alc18 | 4 | 9.2 |
| Wbp1 | WW domain binding protein 1 | -1.08 | 0.14 | 0.008 | |||
CA/W = fold change of continuous access group to water group; Chr = chromosome number; QTL = rat alcohol preference QTL; LOD = LOD score for QTL; Bold type indicates genes in common with MSA vs. W differences.
There were 20 significant Gene Ontology (GO) Biological Processes categories with over-representation of gene differences between the CA and W groups (Table 2, which lists GO category name and GO identification number, as well as member gene names and symbols). Some of these GO categories included (a) ‘anti-apoptosis’ with a total of 10 gene expression differences, 5 higher (Frag1, Hrpap20, Sod2, Tgfa, Zfp91) and 5 lower (Adam17, Foxo1a, Hspa5, Hdh, Tsc22d3) in the CA group; (b) ‘negative regulation of programmed cell death’ with a total of 13 gene expression differences, 7 higher (Btg2, Frag1, Hrpap20, Scg2, Sod2, Tgfa, zfp91) and 6 lower (Adam17, Angptl4, fox01a, Hspa5, Hdh, Tsc22d3) in the CA group; (c) ‘microtubule-based movement’ with a total of 8 gene expression differences, 3 higher (Actr10, Kif2, Tubb2b) and 5 lower (Hdh, Kif11, Kif1b, Kif6, Klc1) in the CA group; (d) ‘regulation of kinase activity’ with a total of 11 gene expression differences, with 7 higher (Cdc42, Dusp6, Pkib, Spdy1, Spry2, Tgfa) and 4 lower (Cav, Dgka, Plce1, Rgs3) in the CA group; (e) ‘negative regulation of protein kinase activity’ with a total of 5 gene expression differences, 3 higher (Dusp6, Pkib, Spry2) and 2 lower (Cav, Rgs3) in the CA group; (f) ‘regulation of MAPK activity’ with a total of 7 gene expression differences, 4 higher (Chrna7, Dusp6, Spry2, Tgfa) and 3 lower (cav, Plce1, Rgs3) in the CA group; (g) ‘regulation of G-protein coupled receptor protein signaling pathway’ with a total of 6 genes displaying decreased expression levels (Ece1, Plce1, Rasgrp4, Ramp2, Rgs3, Rgs9) in the CA group; and (h) ‘anatomical structure formation’ with a total of 11 gene expression differences, 7 higher (Btg1, Ctgf, Nr4a3, Pofut1, Scg2, Scye1, Tgfa) and 4 lower (Angptl4, Tek, Hdh, Tie1) in the CA group.
Table 2.
Gene Ontology (GO) categories, with names and identification numbers, displaying significant over-representation of genes, and the respective gene names and their symbols
| GO Category/GOBPID | Genes | Symbol |
|---|---|---|
| blood vessel development/GO:0001568 | angiopoietin-like 4 | Angptl4 |
| B-cell translocation gene 1, anti-proliferative | Btg1 | |
| connective tissue growth factor | Ctgf | |
| endothelial-specific receptor tyrosine kinase | Tek | |
| forkhead box O1A | Foxo1a | |
| gap junction membrane channel protein alpha 1 | Gja1 | |
| neurotrophic tyrosine kinase, receptor, type 2 | Ntrk2 | |
| phosphatidic acid phosphatase type 2B | Ppap2b | |
| protein O-fucosyltransferase 1 | Pofut1 | |
| secretogranin 2 | Scg2 | |
| small inducible cytokine subfamily E, member 1 | Scye1 | |
| transforming growth factor alpha | Tgfa | |
| tyrosine kinase with immunoglobulin-like and EGF-like domains 1 | Tie1 | |
| regionalization/GO:0003002 | activity regulated cytoskeleton-associated protein | Arc |
| B-cell translocation gene 2, anti-proliferative | Btg2 | |
| E1A binding protein p300 | Ep300 | |
| Huntington disease gene homolog | Hdh | |
| lymphoid enhancer binding factor 1 | Lef1 | |
| protein O-fucosyltransferase 1 | Pofut1 | |
| ring finger protein 2 | Rnf2 | |
| negative regulation of protein kinase activity/GO:0006469 | caveolin | Cav |
| dual specificity phosphatase 6 | Dusp6 | |
| protein kinase inhibitor beta, cAMP dependent, catalytic | Pkib | |
| regulator of G-protein signalling 3 | Rgs3 | |
| sprouty homolog 2 | Spry2 | |
| isoprenoid metabolic process/GO:0006720 | 3-hydroxy-3-methylglutaryl-coenzyme A reductase | Hmgcr |
| dehydrogenase/reductase (SDR family) member 3 | Dhrs3 | |
| isopentenyl-diphosphate delta isomerase notch1-induced protein phytn-dehydro and Pyr-redox domain containing protein | Idi1 | |
| one-carbon compound metabolic process/GO:0006730 | B-cell translocation gene 1, anti-proliferative | Btg1 |
| carbonic anhydrase 11 | Car11 | |
| FBJ murine osteosarcoma viral oncogene homolog | Fos | |
| isoprenylcysteine carboxyl methyltransferase | Icmt | |
| methionine adenosyltransferase II, alpha | Mat2a | |
| methyl-CpG binding domain protein 1 | Mbd1 | |
| protein tyrosine phosphatase, receptor type, G | Ptprg | |
| anti-apoptosis/GO:0006916 | a disintegrin and metalloproteinase domain 17 | Adam17 |
| FGF receptor activating protein 1 | Frag1 | |
| Forkhead box O1A | Foxo1a | |
| heat shock 70kDa protein 5 (glucose-regulated protein) | Hspa5 | |
| hormone-regulated proliferation associated protein 20 | Hrpap20 | |
| Huntington disease gene homolog | Hdh | |
| superoxide dismutase 2, mitochondrial | Sod2 | |
| transforming growth factor alpha | Tgfa | |
| TSC22 domain family 3 | Tsc22d3 | |
| zinc finger protein 91 | Zfp91 | |
| microtubule-based movement/GO:0007018 | actin-related protein 10 homolog | Actr10 |
| Huntington disease gene homolog | Hdh | |
| kinesin family member 1B | Kif1b | |
| kinesin family member 6 | Kif6 | |
| kinesin family member 11 | Kif11 | |
| kinesin heavy chain family, member 2 | Kif2 | |
| kinesin light chain 1 | Klc1 | |
| tubulin, beta 2b | Tubb2b | |
| gamete generation/GO:0007276 | 3-hydroxy-3-methylglutaryl-coenzyme A reductase | Hmgcr |
| a disintegrin-like and metallopeptidse with thrombospondin type 1 motif, 1 | Adamts1 | |
| B-cell translocation gene 1, anti-proliferative | Btg1 | |
| carnitine deficiency-associated gene expressed in ventricle 1 | Cdv1 | |
| dead end homolog 1 | Dnd1 | |
| heat shock factor 2 | Hsf2 | |
| hydroxyacyl glutathione hydrolase | Hagh | |
| kinesin family member 6 | Kif6 | |
| McKusick-Kaufman syndrome protein | Mkks | |
| nitric oxide synthase 3, endothelial cell | Nos3 | |
| seven in absentia 1A | Siah1a | |
| zinc finger protein 403 | Zfp403 | |
| gastrulation/GO:0007369 | camello-like 3 | Cml3 |
| Huntington disease gene homolog | Hdh | |
| nuclear receptor subfamily 4, group A, member 3 | Nr4a3 | |
| phosphatidic acid phosphatase type 2B | Ppap2b | |
| ring finger protein 2 | Rnf2 | |
| heart development/GO:0007507 | E1A binding protein p300 | Ep300 |
| endothelial-specific receptor tyrosine kinase | Tek | |
| gap junction membrane channel protein alpha 1 | Gja1 | |
| histone deacetylase 5 | Hdac5 | |
| McKusick-Kaufman syndrome protein | Mkks | |
| plakophilin 2 | Pkp2 | |
| protein O-fucosyltransferase 1 | Pofut1 | |
| transforming growth factor, beta receptor 3 | Tgfbr3 | |
| regulation of G-protein coupled receptor protein signaling/GO:0008277 | endothelin converting enzyme 1 | Ece1 |
| phospholipase C, epsilon 1 | Plce1 | |
| RAS guanyl releasing protein 4 | Rasgrp4 | |
| receptor (calcitonin) activity modifying protein 2 | Ramp2 | |
| regulator of G-protein signaling 3 | Rgs3 | |
| regulator of G-protein signaling 9 | Rgs9 | |
| protein catabolic process/GO:0030163 | calpastatin | Cast |
| gap junction membrane channel protein alpha 1 | Gja1 | |
| prenylcysteine oxidase 1 | Pcyox1 | |
| proteasome (prosome, macropain) 26S subunit, ATPase, 6 | Psmc6 | |
| proteasome (prosome, macropain) 26S subunit, non-ATPase, 12 | Psmd12 | |
| proteasome (prosome, macropain) subunit, alpha type 4 | Psma4 | |
| seven in absentia 1A | Siah1a | |
| seven in absentia 2 | Siah2 | |
| ubiquitin associated domain containing 1 | Ubadc1 | |
| ubiquitin carboxyl-terminal hydrolase L5 | Uchl5 | |
| ubiquitin-conjugating enzyme E2D 3 | Ube2d3 | |
| protein modification by small protein conjugation/GO:0032446 | calcyclin binding protein | Cacybp |
| LIM domain only protein 7 | Lmo7 | |
| retinoblastoma binding protein 6 | Rbbp6 | |
| ring finger protein 2 | Rnf2 | |
| seven in absentia 1A | Siah1a | |
| seven in absentia 2 | Siah2 | |
| ubiquitin associated domain containing 1 | Ubadc1 | |
| negative regulation of programmed cell death/GO:0043069 | a disintegrin and metalloproteinase domain 17 | Adam17 |
| angiopoietin-like 4 | Angptl4 | |
| B-cell translocation gene 2, anti-proliferative | Btg2 | |
| FGF receptor activating protein 1 | Frag1 | |
| Forkhead box O1A | Foxo1a | |
| heat shock 70kDa protein 5 (glucose-regulated protein) | Hspa5 | |
| hormone-regulated proliferation associated protein 20 | Hrpap20 | |
| Huntington disease gene homolog | Hdh | |
| secretogranin 2 | Scg2 | |
| superoxide dismutase 2, mitochondrial | Sod2 | |
| transforming growth factor alpha | Tgfa | |
| TSC22 domain family 3 | Tsc22d3 | |
| zinc finger protein 91 | Zfp91 | |
| regulation of MAPK activity/GO:0043405 | caveolin | Cav |
| cholinergic receptor, nicotinic, alpha polypeptide 7 | Chrna7 | |
| dual specificity phosphatase 6 | Dusp6 | |
| phospholipase C, epsilon 1 | Plce1 | |
| regulator of G-protein signaling 3 | Rgs3 | |
| sprouty homolog 2 | Spry2 | |
| transforming growth factor alpha | Tgfa | |
| biopolymer methylation/GO:0043414 | B-cell translocation gene 1, anti-proliferative | Btg1 |
| B-cell translocation gene 2, anti-proliferative | Btg2 | |
| FBJ murine osteosarcoma viral oncogene homolog | Fos | |
| isoprenylcysteine carboxyl methyltransferase | Icmt | |
| methyl-CpG binding domain protein 1 | Mbd1 | |
| regulation of kinase activity/GO:0043549 | caveolin | Cav |
| cell division cycle 42 homolog | Cdc42 | |
| cholinergic receptor, nicotinic, alpha polypeptide 7 | Chrna7 | |
| diacylglycerol kinase, alpha | Dgka | |
| dual specificity phosphatase 6 | Dusp6 | |
| phospholipase C, epsilon 1 | Plce1 | |
| protein kinase inhibitor beta, cAMP dependent, catalytic | Pkib | |
| regulator of G-protein signaling 3 | Rgs3 | |
| speedy homolog 1 | Spdy1 | |
| sprouty homolog 2 | Spry2 | |
| transforming growth factor alpha | Tgfa | |
| cell maturation/GO:0048469 | actin, alpha 1, skeletal muscle | Acta1 |
| calpastatin | Cast | |
| gap junction membrane channel protein alpha 1 | Gja1 | |
| growth differentiation factor 11 | Gdf11 | |
| hairy and enhancer of split 1 | Hes1 | |
| parathyroid hormone receptor 1 | Pthr1 | |
| anatomical structure formation/GO:0051603 | angiopoietin-like 4 | Angptl4 |
| B-cell translocation gene 1, anti-proliferative | Btg1 | |
| connective tissue growth factor | Ctgf | |
| endothelial-specific receptor tyrosine kinase | Tek | |
| Huntington disease gene homolog | Hdh | |
| nuclear receptor subfamily 4, group A, member 3 | Nr4a3 | |
| protein O-fucosyltransferase 1 | Pofut1 | |
| secretogranin 2 | Scg2 | |
| small inducible cytokine subfamily E, member 1 | Scye1 | |
| transforming growth factor alpha | Tgfa | |
| tyrosine kinase with immunoglobulin-like and EGF-like domains 1 | Tie1 |
Among the named genes that were significantly different between the CA and W groups (Table 1), there were 43 genes located within rat QTLs for alcohol consumption and preference (Bice et al., 1998; Carr et al., 1998, 2003; Foroud et al., 2003). There were 11 genes located within Alc18 on chromosome 4, 6 genes located within Alc15 on chromosome 2, and 5 genes located within Alc8 on chromosome 3. Four or fewer genes were located within each of the following: Alc6 on chromosome 12, Alc11 on chromosome 16, Alc17 on chromosome 6, Alc5 on chromosome 10, Alc10 on chromosome 12, and Alc20 on chromosome 8 (Table 1).
Comparison of the MSA and W groups revealed 87 probe sets (representing 84 individual named genes) were significantly different at p < 0.01. However, because this number is less than 1% of the identifiable genes on the Affymetrix GeneChip® (Rat Genome 230 2.0 array GeneChips) and the overall estimated FDR rate was greater than 0.90, these differences could have occurred by chance alone (data not shown). Of the 84 named genes, there were 35 differences in common with differences observed between the CA vs. W groups, some of which included Arc, Cav2, Fos, Jun, Junb, Nos3, Pik3c3, and Plce1 (indicated in bold type in Table 1).
Comparison of the CA versus MSA group revealed 51 significant differences (p < 0.01). However, the FDR-values for these findings were approximately 0.4, suggesting a high number of false positives. Therefore, this analysis indicated there were few genes with significant expression differences between the two alcohol drinking groups, even though there were a significant number of differences between the CA versus W group but not between the MSA versus W group.
3.3. RT-PCR confirmation
Eight genes from the list of significant genes in Table 1 were selected for RT-PCR. The criteria for selection were fold change (at least 20% difference), that the gene was neurobiologically interesting (e.g., Fos and Jun changes may be related to neuronal activity), and the availability of primers. The microarray results indicated that 6 of the 8 genes were up regulated in the alcohol drinking group and 2 of the genes were down regulated. RT-PCR confirmed the direction and magnitude of the changes observed with the microarray analysis between the CA and W control groups (Table 3).
Table 3.
Quantitative RT-PCR confirmation of microarray results for respective genes
| Gene Symbol | CA Expression | W Expression | qRT-PCR Fold Δ | Microarray Fold Δ |
|---|---|---|---|---|
| Fos | 2.02 | 1.46 | 1.38* | 1.72 |
| Homer1 | 3.60 | 2.13 | 1.69* | 1.71 |
| Nr4a1 | 2.62 | 1.27 | 2.07* | 1.62 |
| Dusp1 | 1.16 | 0.78 | 1.49* | 1.42 |
| Junb | 3.31 | 1.80 | 1.84* | 1.27 |
| Mobp | 1.23 | 1.50 | -1.22* | -1.22 |
| Jun | 1.01 | 0.93 | 1.09* | 1.23 |
| Ntrk2 | 3.18 | 6.09 | -1.92* | -1.67 |
CA = Continuous access group; W = Water control group;
indicates qRT-PCR probability value < 0.001.
3.4. Supplemental tables
See supplemental tables A and B for more complete information on data for gene expression differences in the ACB between the CA, MSA and W groups.
4. Discussion
The major finding of this study was that chronic, continuous/daily ethanol drinking under 24-hr free-choice conditions (CA) altered the expression of over 370 uniquely named genes in the ACB of P rats, whereas intermittent ethanol drinking, using a multiple scheduled access protocol (MSA), with three 1-hr sessions each day for 5 days per week did not produce a significant number of gene expression differences. These results do not support our hypothesis that binge-like alcohol-drinking would produce significant changes in gene expression in the ACB of P rats. The disparity in findings could be due to the higher daily intakes of the CA group (∼9.5 g/kg/day) versus the MSA group (∼6.5 g/kg/day). However, the MSA group consumed their ethanol in distinct bouts of 1.7—2.7 g/kg/hr with most of the intake expected to occur within the first 15 min of each access session (Bell et al., 2006b), mimicking binge-like drinking, with BACs approximating 80 mg% or greater (Bell et al., 2006b, 2008). The relative lack of effect of ethanol in the MSA group versus the W group suggests that gene expression in the ACB may be tightly regulated, such that, with intermittent ethanol exposure under a regimented protocol (i.e., with an inherently strong time-of-day conditioning component), the genetic machinery may adjust to ethanol-induced alterations (e.g., neuroadaptations) and be able to maintain new steady-state protein levels with basal levels of gene expression. This conclusion is supported by the finding that there were very few differences (approximately 50) between the 2 alcohol drinking groups. This suggests that the MSA procedure may be producing similar changes as the CA procedure but the effects of binge drinking may be much smaller compared with the effects of 24-hr continuous access drinking. It is noteworthy that ∼ 40% of the genes with significant differences in expression between the MSA and W groups were also similarly different between the CA and W groups. Therefore, increasing power, by increasing the number of animals in the MSA and W groups, in future studies may result in detecting a significant number of differences in gene expression between these two groups.
The lack of a significant number of differences in gene expression between the MSA and W groups of the present study, such that the number of gene expression differences (with p-values less than 0.01) was less than that expected by chance (i.e., less than 1% of the total number of genes), appears to disagree with a recent genomic study examining inbred P rats in a 1-hr operant ethanol self-administration procedure (Rodd et al., 2008). These authors reported that inbred P rats responding for ethanol displayed a significant number (> 200) of gene expression differences, in the ACB, compared to a water control group, when animals were killed the day after the last operant session. This lack of agreement suggests that multiple factors, other than temporal (i.e., time-of-day) conditioning and ethanol alone, are influencing the number of gene expression differences between the effects of MSA sessions of oral self-administration of ethanol per day and daily sessions of operant ethanol self-administration, compared with their respective controls. Some of these factors may include the role of Pavlovian and instrumental conditioning or lever pressing in the operant study (Rodd et al., 2008), as well as total ethanol consumed or expected peak BACs achieved and reduced conditioning to environmental cues found in the home-cage setting of the present study (i.e., animals were habituated to the wire mesh home cages before ethanol was made available).
Although 24-hr free-choice ethanol access is not regimented in the same manner as the MSA procedure, the same routine of body and fluid measurements are used each day and, based upon the results between the MSA and W groups, the expectation is that the genetic machinery of the CA group would also adjust to chronic ethanol-drinking conditions, with a corresponding modest number of gene expression changes. The chronic ethanol-drinking conditions experienced by the CA group should produce tolerance (Gatto et al., 1987; Lumeng and Li, 1986; Stewart et al., 1991) and possibly dependence (Kampov-Polevoy et al., 2000; Waller et al., 1982). Even though similar studies have yet to be conducted with P rats consuming ethanol under MSA conditions, it is anticipated that tolerance, and possibly dependence, would also develop in the MSA group, because BACs of 80 mg% or greater are expected during each ethanol access session, when using the present MSA protocol (Bell et al., 2006b, 2008; Murphy et al., 2002). Gene expression was measured 15 hr after the MSA group's last drinking episode when ethanol was also removed from the CA group. It is noteworthy that 10 of these hours occurred during the daily light-cycle, when P rats normally drink limited amounts of ethanol (Bell et al., 2006b, 2006c; Murphy et al., 1986). Nevertheless, it is likely that the CA group experienced symptoms of its first protracted withdrawal at this time point (Kampov-Polevoy et al., 2000; Waller et al., 1982). Therefore, the gene expression differences observed between the CA and W groups may be due in part to ethanol withdrawal. It would be difficult to resolve the issue of withdrawal effects from continuous chronic alcohol drinking without undertaking a more detailed time-course study with this alcohol drinking protocol. However, since the MSA group did not show a significant number of differences in gene expression compared to the W control group (suggesting little effect of repeated BACs, that exceeded 50 mg% per access period, five days per week), the differences between the CA and W groups may reflect the effects of removal of the ethanol. A recent study (Bell et al., 2009) indicated significant behavioral changes (alterations in motor activity and rearing behavior) occur between 9 and 13 hr after removal of ethanol in P rats that had 24-hr continuous/daily free-choice access to ethanol for approximately 6 months. These studies suggest some behavioral alterations are occurring after removal of ethanol from P rats given continuous access, and there may be a relationship between the changes in gene expression within the ACB observed in the present study and these changes in general motor activity.
A proteomics study of the ACB using similar drinking procedures with inbred P rats indicated that the levels of 12 proteins were altered by MSA drinking compared to the W group and 8 proteins were altered by CA drinking compared to the W group (Bell et al., 2006a). None of the proteins that were different in the ACB between the CA and W groups of the proteomics study (Bell et al., 2006a) were found to be different between the CA and W groups in the gene expression data of the present study (Table 1). These results suggest that a direct relationship between changes in mRNA and protein levels may not be necessary (for an example of dissociations between DNA, RNA and protein levels in the brain after ethanol exposure see Babu et al., 1994) within this brain region. This could be due to a number of factors, not the least of which is procedural differences between the studies, but, in addition, proteins may be synthesized in other regions and transported to the ACB. Another possibility is that there is temporal discontinuity between changes in the expression levels of mRNA and protein, such that protein levels may increase (or decrease) leading to their accumulation (or reduction) because of post-translational modifications and/or changes in chaperoning or trafficking.
The Gene Ontology (GO) analysis indicated several significant biological processes categories. The categories of ‘anti-apoptosis’ and ‘negative regulation of programmed cell death’ suggest that cellular changes may have occurred to counter any neurotoxic effects of chronic ethanol exposure. A number of the genes identified in the CA group of the present study as having significantly changed expression levels (Table 1) and were members of over-represented GO categories (Table 2) have also been reported in the literature as genes altered by or associated with high ethanol-consumption. For example, (a) Btg2 gene expression, elevated in the CA group, is greater in inbred P versus inbred NP rats as well (Edenberg et al., 2005); (b) Scg2 gene expression, elevated in the CA group, is also increased in the frontal cortex, but decreased in the motor cortex of alcoholics (Mayfield et al., 2002); (c) Tgfa, with gene expression increased in the CA group, over-expressing mice display greater ethanol preference than their wild-type counterparts (Hilakivi-Clarke and Goldberg, 1995); and (d) a gene moderately similar to Zfp91 is altered in the prefrontal cortex of alcoholics (Flatscher-Bader et al., 2005), with Zfp91 gene expression increased in the CA group of the present study as well. The results of the GO analysis (Table 2) also suggest that significant changes are occurring in intracellular signaling systems, involving protein kinase activity, G-protein coupled receptor protein signaling, and MAPK activity. These changes in intracellular signaling systems may indicate that major neuronal alterations occurred in the ACB of the CA group.
Several of the kinase activity-related genes (Table 2) identified as having altered expression levels in the CA group (Table 1) have been implicated in alcohol abuse. For instance, (a) Cav2 gene expression, which was reduced in the present study as was gene expression of the family member Cav, is increased in the ACB of iP rats after operant self-administration of ethanol (Rodd et al., 2008); (b) Dusp6 gene expression, which was increased in the CA group of the present study, is greater in iNP than iP rats (Kimpel et al., 2007), with gene expression differences also found between high and low alcohol-consuming mice (Kerns et al., 2005); and (c) Pkib gene expression is increased in the frontal cortex of alcoholics vs. nonalcoholics (Liu et al., 2006), which was elevated in the CA group of the present study as well. Interestingly, inhibition of PKA in the ACB shell increases ethanol intake (Misra and Pandey, 2006), and family member Pkia (cAMP-dependent, regulatory) gene expression is decreased in the frontal and motor cortices of alcoholics (Mayfield et al., 2002).
Chrna7 gene expression was increased in the CA group (Table 1) and was identified in the over-represented GO category “regulation of MAPK activity” (Table 2). It is noteworthy that several reports support a role for Chrna7 in substance abuse, for instance, (a) Chrna7 knock out mice display greater sensitivity to lower dose ethanol-induced motor activation and higher dose ethanol-induced hypothermia and loss of righting reflex compared with their wild-type counterparts (Bowers et al., 2005); (b) Chrna7 has been proposed to reduce ethanol-induced neurotoxicity (de Fiebre and de Fiebre, 2003); and (c) a significant association between the Chrna7 gene, altered cognitive (response inhibition and sustained attention) function (Rigbi et al., 2008) or psychological characteristics (Greenbaum et al., 2006), and smoking behavior have been reported in humans.
Although the Arc gene was identified under the GO category ‘regionalization’ (Table 2), this gene was one of the most cited genes both detected as significantly changed in the present study (its gene expression was increased 1.5-fold in the CA group, Table 1) and implicated in substance abuse for morphine (Ammon et al., 2003), amphetamine (Gonzalez-Nicolini and McGinty, 2002), cocaine (Freeman et al., 2002; Samaha et al., 2004) and nicotine (Schochet et al., 2005; Samaha et al., 2005). Arc is an immediate early gene found in soma and dendrites and is involved in, or associated with, synaptic modification and learning/memory (e.g., Guzowski et al., 2006). In a recent study (Pandey et al., 2008), the BDNF-Arc signaling pathway has been implicated in both alcohol dependence and the comorbid expression of anxiety with alcohol abuse.
Among the 43 genes that were located within rat QTLs for alcohol consumption and preference, some were evident in certain GO categories and gene networks. Tgfa (located within Alc18 on chromosome 4) appears to be associated with ethanol preference in mice (Hilakivi-Clark & Goldberg, 1995), anti-apoptosis (Table 2) and up-regulation of Fos-related transcription factors (Fig. 2). Hspa5 (located within Alc8 on chromosome 3) is also involved in anti-apoptosis (Table 2). Mtus1 (located within Alc11 on chromosome 16) and Creb3l2 (located within Alc18 on chromosome 4) are involved in transcription (Fig. 3). The anti-apoptosis involvement and increased transcription functions of these genes suggest that increased cellular protection may be occurring in the ACB of the CA group, which could be factors contributing to high alcohol intakes.
Fig. 2.
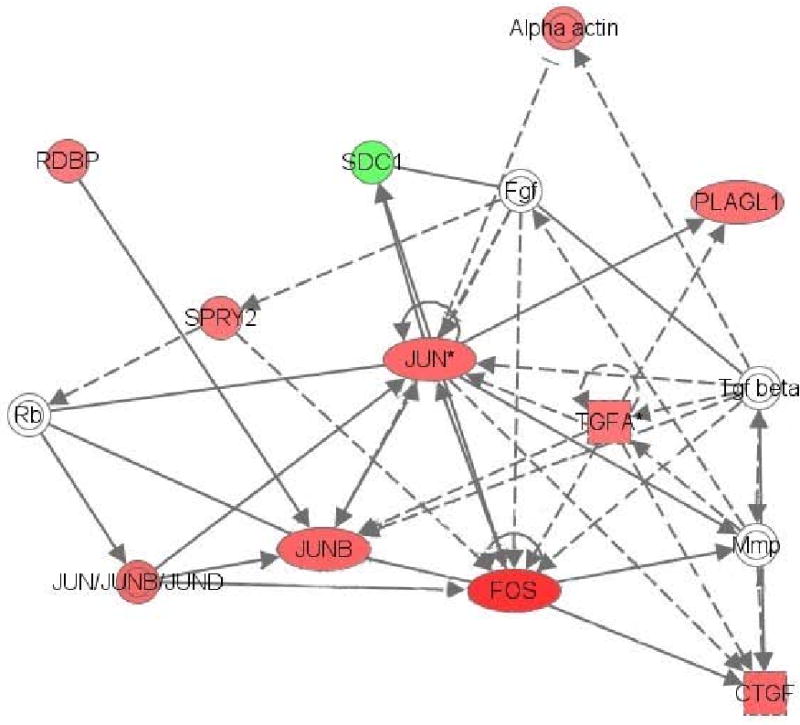
Abridged Ingenuity® network analysis revealed up-regulation of Fos-related transcription factors. Red indicates up-regulation and green down-regulation of the associated genes. Genes not colored indicate no change in expression but they are highly linked to genes that did change. Solid lines indicate a direct interaction between genes, whereas dashed lines indicate an indirect interaction between the genes. [For interpretation of the references to color in this figure, the reader is referred to the web version of the article] Abbreviations used: Ctgf – connective tissue growth factor; Fos – FBJ osteosarcoma oncogene; Fgf – fibroblast growth factor; Jun – Jun oncogene; Junb -Jun B oncogene; Mmp – matrix metallopeptidase; Plagl1 – pleiomorphic adenoma gene-like 1; Rdbp – RD RNA-binding protein; Rb - retinoblastoma; Spry2 – sprouty homolog 2; Sdc1 – syndecan 1; Tgfa – transforming growth factor alpha.
Fig. 3.
Ingenuity® network analysis revealed primarily up-regulation of interacting pathways involved in transcription, calcium signaling, oxidative stress response and glucocorticoid receptor signaling. Red indicates up-regulation and green down-regulation of the associated genes. Genes not colored indicate no change in expression but they are highly linked to genes that did change. Solid lines indicate a direct interaction between genes, whereas dashed lines indicate an indirect interaction between the genes. [For interpretation of the references to color in this figure, the reader is referred to the web version of the article] Abbreviations used: Acta1 – actin alpha 1 skeletal muscle; Actl6a – actin-like 6A; Adam17 – a disintegrin and metallopeptidase domain 17; Adamts1 – a disintegrin-like and metallopeptidase; Apex1 – a purinic/pyrimidinic endonuclease 1; Cdc37l1 – cell division cycle 37 homolog-like 1; Cbx3 – chromobox homolog 3; Cbp/p300 – CREB binding protein; Dr1 – down-regulator of transcription 1; Ep300 – E1A binding protein p300; Gtf2a2 – general transcription factor IIA 2; Hsp70 – heat shock protein 70; Hnrpk – heterogeneous nuclear ribonucleoprotein K; Hdac2 – histone deacetylase 2; Hdac5 – histone deacetylase 5; Irf3 – interferon regulatory factor 3; Kcnq3 – potassium voltage-gated channel subfamily Q member 3; Mxl1 – Max interacting protein 1; Med4 – mediator of RNA polymerase II transcription subunit 4 homolog; Mbd1 – methyl-CpG binding domain protein 1; Mtus1 – mitochondrial tumor suppressor 1; Pi3k – phosphatidylinositol 3-kinase; Pik3c3 – phosphoinositide-3-kinase class 3; Ppp2ca – protein phosphatase 2 catalytic subunit alpha isoform; Rbbp6 – retinoblastoma binding protein 6; Rnf2 – ring finger protein 2; Rbm3 – RNA binding motif protein 3; Tceb3 – transcription elongation factor B polypeptide 3; Tpm3 – tropomyosin 3 gamma.
Ingenuity® pathway analyses uncovered networks overlapping and extending those detected with the GO analysis. In agreement with the GO biological processes category of ‘anti-apoptosis’ genes, the Ingenuity® pathway analysis revealed a network of 11 genes involved in apoptosis, 8 of which were reduced in the CA group (Cast, Ccr5, Ece1, Nos3, Ntrk2, Plce1, Slc2a1, Tgfbr3). Several of these genes have been implicated in alcoholism and drug abuse, including reports that (a) Cast gene expression is reduced in the frontal cortex of alcoholics vs. nonalcoholics (Liu et al., 2006); (b) female, but not male, Ccr5 knock-out mice display greater ethanol intake, but not preference, as well as ethanol-induced conditioned taste aversion than their wild type counterparts (Blednov et al., 2005); and (c) Slc2a1 (facilitated glucose transporter) gene expression is increased in the ACB of iP rats operantly self-administering ethanol, although, at the same time, family member Slc2a3 (facilitated glucose transporter) gene expression is decreased (Rodd et al., 2008). Regarding Ntrk2, its gene expression is decreased in the frontal and motor cortices of alcoholics (Mayfield et al., 2002). Moreover, single nucleotide polymorphism (SNP)-based analyses implicate the Ntrk2 gene in alcohol dependence (Xu et al., 2007). The Ntrk2 gene has been implicated in nicotine abuse as well (Beuten et al., 2007; Sun et al., 2007). Several genes responding to glucocorticoid receptor signaling were also altered in the CA versus W groups with 3 genes having reduced expression levels (Fkbp5, Hspa12b, Tsc22d3), whereas only one was increased (Dusp1).
In addition to the oncogenes, Fos, Jun and Junb, there were several other genes in the oncogene network that were up regulated in the ACB of the CA group compared with the control animals (See Fig. 2). These genes included Ctgf, Tgfa, Plagl1, Spry2 and Rdbp. The up-regulation of Fos and other transcription factors in the CNS are often associated with increased neuronal activity (Greenberg et al., 1986; Herrera and Robertson, 1996; Morgan and Curran, 1989). Fos (along with Jun and JunB) is regulated by, or mediates the effects of, ethanol, for example, Fos is induced in the ACB shell by acute ethanol and 80% of the Fos-positive cells labeled for GAD as well (Leriche et al., 2008). Fos expression has also been associated with morphine (Taracha et al., 2008), cocaine (Zhang et al., 2006) and nicotine (Schochet et al., 2005) abuse. Ctgf and Tgfa are growth factors and their increased expression may indicate neuronal alterations are occurring as well. These results are in agreement with the GO analysis (Table 2), suggesting that anatomical structural alterations may be occurring in the CA group. It is noteworthy that CNS Ctgf gene expression levels differ between high alcohol-consuming AA vs low alcohol-consuming ANA rats (Sommer et al., 2006) and this gene has been linked with cocaine abuse (Mash et al., 2007), and, as indicated above, Tgfa over-expressing mice display greater ethanol preference than their wild-type counterparts (Hilakivi-Clarke and Goldberg, 1995).
The higher expression of Plagl1 [a zinc finger protein (Yang et al., 2005)] and Rdbp [a nuclear RNA-binding protein (Surowy et al., 1990)] are also consistent with increased transcription associated with neuronal activity, with Plag1 gene expression increased in the accumbens of iP rats operantly self-administering ethanol (Rodd et al., 2008). Moreover, the higher expression (Table 1) of several members of the oncogene family (Rab1, Rab3c, Rab21, Rab35) and RNA binding motif proteins (Rbm3, Rbm13, Rbm17) are also consistent with an interpretation of increased transcription activity. Figure 3 shows a network of genes involved in calcium signaling, oxidative stress response, and transcription. In the calcium-signaling network, there were 4 genes (Acta1, Ep300, Hdac5, Tpm3), and, in the oxidative stress network, there were 4 genes (Acta1, Bex1, Ep300, Pik3c3) that were up regulated in the CA group. In the transcription network, there were 12 genes that were different between the CA and W groups with 10 genes higher (Cbx3, Dr1, Ep300, Hdac2, Hdac5, Mbd1, Med4, Rbbp6, Rnf2, Tceb3) and only 2 genes lower (Irf3, Mxi1) in the CA group; these results are consistent with the findings for the oncogenes (Fig. 2) and also support an interpretation of increased transcription.
A number of the genes in Figure 3 have been implicated in alcohol abuse, such as (a) Acta1 gene expression differences are found between iP and iNP rats (Kimpel et al., 2007); (b) Irf3 gene expression differences are found between iP and iNP rats (Kimpel et al., 2007); (c) Tceb3 gene expression is increased in the frontal cortex of cirrhotic alcoholics vs. controls (Liu et al., 2007); and (d) Tpm3 gene expression is decreased in the ACB of iP rats operantly self-administering ethanol (Rodd et al., 2008), with Tpm3 protein expression levels decreased in the amygdala of chronic ethanol-drinking iP rats, but increased in the ACB of iP rats given multiple scheduled-access sessions of ethanol per day as well (Bell et al., 2006a).
Although not identified by the GO or Ingenuity® analyses, Crh gene expression was increased in the chronic ethanol drinking P rats of the present study and has been implicated in substance abuse (c.f., Heilig and Koob, 2007; Koob and Le Moal, 2008). For example, (a) Crh knockout mice display greater ethanol preference and limited access ethanol intake than their wild-type counterparts (Olive et al., 2003); (b) Crh over-expressing mice display lower ethanol preference and reduced 24-hr ethanol intake than their wild-type counterparts (Palmer et al., 2004); (c) chronic ethanol drinking increases preproCrh mRNA in the CNS of Sprague-Dawley rats (Lack et al., 2005); and (d) Crh levels predict intensity of craving and probability of relapse to drug use after acute detoxification (Kiefer and Wiedemann, 2004; see also Goeders, 2002a, 2002b).
In summary, the results of the present study suggest that, under intermittent ethanol drinking conditions, gene expression levels may reach a near normal steady state level, which may be sufficient to maintain altered protein levels in the ACB. Because gene expression was determined 15 hr after removal of ethanol in the CA group, these changes may also reflect withdrawal-responsive genes rather than purely ethanol-responsive genes. Nevertheless, because a number of the genes identified as significant in the present study have also been described in the literature on drug and/or alcohol abuse, these genes may serve as candidates for continued research into the neurobiology of drug and/or alcohol abuse.
Supplementary Material
Acknowledgments
The present study was supported in part by AA07611, AA13521 [INIA Project], AA13522 [INIA Project], AA016652 [INIA Project], AA16660 [INIA Project] and INGEN® (which is partially funded by Lilly Endowment Inc.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ammon S, Mayer P, Riechert U, Tischmeyer H, Hollt V. Microarray analysis of genes expressed in the frontal cortex of rats chronically treated with morphine and after naloxone precipitated withdrawal. Brain Res Mol Brain Res. 2003;112:113–125. doi: 10.1016/s0169-328x(03)00057-3. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu PP, Kumari LR, Vemuri MC. Differential changes in cell morphology, macromolecular composition and membrane protein profiles of neurons and astrocytes in chronic ethanol treated rats. Mol Cell Biochem. 1994;130:29–40. doi: 10.1007/BF01084265. [DOI] [PubMed] [Google Scholar]
- Barrett T, Suzek TO, Troup DB, Wilhite SE, Ngau WC, Ledoux P, et al. NCBI GEO: mining millions of expression profiles--database and tools. Nucleic Acids Res. 2005;33:D562–6. doi: 10.1093/nar/gki022. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell RL, Hauser SR, Smith RJ, McConnell KK, Rodd ZA, McBride WJ. Evaluating withdrawal-like behavior 8 to 15 hours after cessation of chronic 24-hr free-choice ethanol drinking in alcohol-preferring (P) rats. Alcohol Clin Exp Res. 2009;33(S1):139A. [Google Scholar]
- Bell RL, Kimpel MW, Rodd ZA, Strother WN, Bai F, Peper CL, et al. Protein expression changes in the nucleus accumbens and amygdala of inbred alcohol-preferring rats given either continuous or scheduled access to ethanol. Alcohol. 2006a;40:3–17. doi: 10.1016/j.alcohol.2006.10.001. [DOI] [PubMed] [Google Scholar]
- Bell RL, Rodd ZA, Lumeng L, Murphy JM, McBride WJ. The alcohol-preferring P rat and animal models of excessive alcohol drinking. Addict Biol. 2006b;11:270–288. doi: 10.1111/j.1369-1600.2005.00029.x. [DOI] [PubMed] [Google Scholar]
- Bell RL, Rodd ZA, Sable HJK, Schultz JA, Hsu CC, Lumeng L, et al. Daily patterns of ethanol drinking in peri-adolescent and adult alcohol-preferring (P) rats. Pharmacol Biochem Behav. 2006c;83:35–46. doi: 10.1016/j.pbb.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Bell RL, Rodd ZA, Schultz JA, Peper CL, Lumeng L, Murphy JM, et al. Effects of short deprivation and re-exposure intervals on the ethanol drinking behavior of selectively bred high alcohol-consuming rats. Alcohol. 2008;42:407–416. doi: 10.1016/j.alcohol.2008.03.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuten J, Ma JZ, Payne TJ, Dupont RT, Lou XY, Crews KM, et al. Association of specific haplotypes of neurotrophic tyrosine kinase receptor 2 gene (NTRK2) with vulnerability to nicotine dependence in African-Americans and European-Americans. Biol Psychiatry. 2007;61:48–55. doi: 10.1016/j.biopsych.2006.02.023. [DOI] [PubMed] [Google Scholar]
- Bice P, Foroud T, Bo R, Castelluccio P, Lumeng L, Li TK, et al. Genomic screen for QTLs underlying alcohol consumption in the P and NP rat lines. Mamm Genome. 1998;9:949–955. doi: 10.1007/s003359900905. [DOI] [PubMed] [Google Scholar]
- Blednov YA, Bergeson SE, Walker D, Ferreira VM, Kuziel WA, Harris RA. Perturbation of chemokine networks by gene deletion alters the reinforcing actions of ethanol. Behav Brain Res. 2005;165:110–125. doi: 10.1016/j.bbr.2005.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers BJ, McClure-Begley TD, Keller JJ, Paylor R, Collins AC, Wehner JM. Deletion of the alpha7 nicotinic receptor subunit gene results in increased sensitivity to several behavioral effects produced by alcohol. Alcohol Clin Exp Res. 2005;29:295–302. doi: 10.1097/01.alc.0000156116.40817.a2. [DOI] [PubMed] [Google Scholar]
- Bowers BJ, Radcliffe RA, Smith AM, Miyamoto-Ditmon J, Wehner JM. Microarray analysis identifies cerebellar genes sensitive to chronic ethanol treatment in PKCgamma mice. Alcohol. 2006;40:19–33. doi: 10.1016/j.alcohol.2006.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr LG, Foroud T, Bice P, Gobbett T, Ivashina J, Edenberg H, et al. A quantitative trait locus for alcohol consumption in selectively bred rat lines. Alcohol Clin Exp Res. 1998;22:884–887. [PubMed] [Google Scholar]
- Carr LG, Habegger K, Spence J, Ritchotte A, Liu L, Lumeng L, et al. Analyses of quantitative trait loci contributing to alcohol preference in HAD1/LAD1 and HAD2/LAD2 rats. Alcohol Clin Exp Res. 2003;27:1710–1717. doi: 10.1097/01.ALC.0000097161.51093.71. [DOI] [PubMed] [Google Scholar]
- de Fiebre NEC, de Fiebre CM. Alpha-7 nicotinic acetylcholine receptor-mediated protection against ethanol-induced neurotoxicity. Alcohol. 2003;31:149–153. doi: 10.1016/j.alcohol.2003.08.006. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ, Strother WN, McClintick JN, Tian H, Stephans M, Jerome RE, et al. Gene expression in the hippocampus of inbred alcohol-preferring and -nonpreferring rats. Genes Brain Behav. 2005;4:20–30. doi: 10.1111/j.1601-183X.2004.00091.x. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatscher-Bader T, van der Brug M, Hwang JW, Gochee PA, Matsumoto I, Niwa S, et al. Alcohol-responsive genes in the frontal cortex and nucleus accumbens of human alcoholics. J Neurochem. 2005;93:359–370. doi: 10.1111/j.1471-4159.2004.03021.x. [DOI] [PubMed] [Google Scholar]
- Foroud T, Ritchotte A, Spence J, Liu L, Lumeng L, Li TK, et al. Confirmation of alcohol preference quantitative trait loci in the replicate high alcohol drinking and low alcohol drinking rat lines. Psychiatr Genet. 2003;13:155–161. doi: 10.1097/00041444-200309000-00004. [DOI] [PubMed] [Google Scholar]
- Freeman WM, Brebner K, Lynch WJ, Patel KM, Robertson DJ, Roberts DC, et al. Changes in rat frontal cortex gene expression following chronic cocaine. Brain Res Mol Brain Res. 2002;104:11–20. doi: 10.1016/s0169-328x(02)00197-3. [DOI] [PubMed] [Google Scholar]
- Gatto GJ, Murphy JM, Waller MB, McBride WJ, Lumeng L, Li TK. Chronic ethanol tolerance through free-choice drinking in the P line of alcohol-preferring rats. Pharmacol Biochem Behav. 1987;28:111–5. doi: 10.1016/0091-3057(87)90021-9. [DOI] [PubMed] [Google Scholar]
- Gentleman RC. Using GO for statistical analysis. Proc COMPSTAT. 2004:171–180. [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeders NE. Stress and cocaine addiction. J Pharmacol Exp Ther. 2002a;301:785–9. doi: 10.1124/jpet.301.3.785. [DOI] [PubMed] [Google Scholar]
- Goeders NE. The HPA axis and cocaine reinforcement. Psychoneuroendocrin. 2002b;27:13–33. doi: 10.1016/s0306-4530(01)00034-8. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Nicolini V, McGinty JF. Gene expression profile from the striatum of amphetamine-treated rats: a cDNA array and in situ hybridization histochemical study. Brain Res Gene Expr Patterns. 2002;1:193–8. doi: 10.1016/s1567-133x(02)00017-0. [DOI] [PubMed] [Google Scholar]
- Greenbaum L, Kanyas K, Karni O, Merbl Y, Olender T, Horowitz A, et al. Why do young women smoke? I. Direct and interactive effects of environment, psychological characteristics and nicotine cholinergic receptor genes. Mol Psychiatry. 2006;11:312–322. doi: 10.1038/sj.mp.4001774. [DOI] [PubMed] [Google Scholar]
- Greenberg ME, Ziff EG, Greene LA. Stimulation of neuronal acetylcholine receptors induces rapid gene transcription. Science. 1986;234:80–3. doi: 10.1126/science.3749894. [DOI] [PubMed] [Google Scholar]
- Guzowski JF, Miyashita T, Chawla MK, Sanderson J, Maes LI, Houston FP, et al. Recent behavioral history modifies coupling between cell activity and Arc gene transcription in hippocampal CA1 neurons. Proc Natl Acad Sci USA. 2006;103:1077–1082. doi: 10.1073/pnas.0505519103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–261. doi: 10.1093/nar/gkh036. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilig M, Koob GF. A key role for corticotrophin-releasing factor in alcohol dependence. Trends Neurosci. 2007;30:399–406. doi: 10.1016/j.tins.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera DG, Robertson HA. Activation of c-fos in the brain. Prog Neurobiol. 1996;50:83–107. doi: 10.1016/s0301-0082(96)00021-4. [DOI] [PubMed] [Google Scholar]
- Hilakivi-Clarke L, Goldberg R. Gonadal hormones and aggression-maintaining effect of alcohol in male transgenic transforming growth factor-alpha mice. Alcohol Clin Exp Res. 1995;19:708–713. doi: 10.1111/j.1530-0277.1995.tb01571.x. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Kampov-Polevoy AB, Matthews DB, Gause L, Morrow AL, Overstreet DH. P rats develop physical dependence on alcohol via voluntary drinking: Changes in seizure thresholds, anxiety, and patterns of alcohol drinking. Alcohol Clin Exp Res. 2000;24:278–284. [PubMed] [Google Scholar]
- Kerns RT, Ravindranathan A, Hassan S, Cage MP, York T, Sikela JM, et al. Ethanol-responsive brain region expression networks: Implications for behavioral responses to acute ethanol in DBA/2J versus C57BL/6J mice. J Neurosci. 2005;25:2255–2266. doi: 10.1523/JNEUROSCI.4372-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer F, Wiedemann K. Neuroendocrine pathways of addictive behavior. Addict Biol. 2004;9:205–212. doi: 10.1080/13556210412331292532. [DOI] [PubMed] [Google Scholar]
- Kimpel MW, Strother WN, McClintick JN, Carr LG, Edenberg HJ, McBride WJ. Functional gene expression differences between inbred alcohol-preferring (iP) and –non-preferring (iNP) rats in five brain regions. Alcohol. 2007b;41:95–132. doi: 10.1016/j.alcohol.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob GF, Le Moal M. Review. Neurobiological mechanisms for opponent motivational processes in addiction. Philos Trans R Soc Lond B Biol Sci. 2008;363:3113–3123. doi: 10.1098/rstb.2008.0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lack AK, Floyd DW, McCool BA. Chronic ethanol ingestion modulates proanxiety factors expressed in rat central amygdala. Alcohol. 2005;36:83–90. doi: 10.1016/j.alcohol.2005.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leriche M, Mendez M, Zimmer L, Berod A. Acute ethanol induces Fos in GABAergic and non-GABAergic forebrain neurons: a double-labeling study in the medial prefrontal cortex and extended amygdala. Neuroscience. 2008;153:259–267. doi: 10.1016/j.neuroscience.2008.01.069. [DOI] [PubMed] [Google Scholar]
- Liu J, Lewohl JM, Harris RA, Iyer VR, Dodd PR, Randall PK, et al. Patterns of gene expression in the frontal cortex discriminate alcoholic from nonalcoholic individuals. Neuropsychopharmacology. 2006;30:1574–1582. doi: 10.1038/sj.npp.1300947. [DOI] [PubMed] [Google Scholar]
- Liu J, Lewohl JM, Harris RA, Dodd PR, Mayfield RD. Altered gene expression profiles in the frontal cortex of cirrhotic alcoholics. Alcohol Clin Exp Res. 2007;31:1460–1466. doi: 10.1111/j.1530-0277.2007.00444.x. [DOI] [PubMed] [Google Scholar]
- Lumeng L, Li TK. The development of metabolic tolerance in the alcohol-preferring P rats: Comparison of forced and free-choice drinking of ethanol. Pharmacol Biochem Behav. 1986;25:1013–1020. doi: 10.1016/0091-3057(86)90079-1. [DOI] [PubMed] [Google Scholar]
- Mash DC, Ffrench-Mullen J, Adi N, Qin Y, Buck A, Pablo J. Gene expression in human hippocampus from cocaine abusers identifies genes which regulate extracellular matrix remodeling. PLoS ONE. 2007;2:e1187. doi: 10.1371/journal.pone.0001187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield RD, Lewohl JM, Dodd PR, Herlihy A, Liu J, Harris RA. Patterns of gene expression are altered in the frontal and motor cortices of human alcoholics. J Neurochem. 2002;81:802–813. doi: 10.1046/j.1471-4159.2002.00860.x. [DOI] [PubMed] [Google Scholar]
- McClintick JN, Jerome RE, Nicholson CR, Crabb DW, Edenberg HJ. Reproducibility of oligonucleotide arrays using small samples. BMC Genomics. 2003;4:1–15. doi: 10.1186/1471-2164-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra K, Pandey SC. The decreased cyclic-AMP dependent-protein kinase A function in the nucleus accumbens: a role in alcohol drinking but not in anxiety-like behaviors in rats. Neuropsychopharmacology. 2006;31:1406–1419. doi: 10.1038/sj.npp.1300900. [DOI] [PubMed] [Google Scholar]
- Morgan JI, Curran T. Stimulus-transcription coupling in neurons: role of cellular immediate-early genes. Topics Neurosci. 1989;12:459–462. doi: 10.1016/0166-2236(89)90096-9. [DOI] [PubMed] [Google Scholar]
- Mulligan MK, Ponomerav I, Hitzemann RJ, Belknap JK, Tabakoff B, Harris RA, et al. Toward understanding the genetics of alcohol drinking through transcriptome meta-analysis. Proc Natl Acad Sci USA. 2006;103:6368–6373. doi: 10.1073/pnas.0510188103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy JM, Gatto GJ, Waller MB, McBride WJ, Lumeng L, Li TK. Effects of scheduled access on ethanol intake by the alcohol-preferring (P) line of rats. Alcohol. 1986;3:331–6. doi: 10.1016/0741-8329(86)90010-8. [DOI] [PubMed] [Google Scholar]
- Murphy JM, Stewart RB, Bell RL, Badia-Elder NE, Carr LG, McBride WJ, et al. Phenotypic and genotypic characterization of the Indiana University rat lines selectively bred for high and low alcohol preference. Behav Genet. 2002;32:363–388. doi: 10.1023/a:1020266306135. [DOI] [PubMed] [Google Scholar]
- Olive MF, Mehmert KK, Koenig HN, Camarini R, Kim JA, Nannini MA, et al. A role for corticotropin releasing factor (CRF) in ethanol consumption, sensitivity, and reward as revealed by CRF-deficient mice. Psychopharmacology. 2003;165:181–7. doi: 10.1007/s00213-002-1248-2. [DOI] [PubMed] [Google Scholar]
- Palmer AA, Sharpe AL, Burkhart-Kasch S, McKinnon CS, Coste SC, Stenzel-Poore MP, et al. Corticotropin-releasing factor overexpression decreases ethanol drinking and increases sensitivity to the sedative effects of ethanol. Psychopharmacology. 2004;176:386–397. doi: 10.1007/s00213-004-1896-5. [DOI] [PubMed] [Google Scholar]
- Pandey SC, Zhang H, Ugale R, Prakash A, Xu T, Misra K. Effector immediate-early gene arc in the amygdala plays a critical role in alcoholism. J Neurosci. 2008;28:2589–2600. doi: 10.1523/JNEUROSCI.4752-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G, Watson C. The Rat Brain in Stereotaxic Coordinates. Academic Press; New York: 1998. [Google Scholar]
- Rigbi A, Kanyas K, Yakir A, Greenbaum L, Pollak Y, Ben-Asher E, et al. Why do young women smoke? V. Role of direct and interactive effects of nicotinic cholinergic receptor gene variation on neurocognitive function. Genes Brain Behav. 2008;7:164–172. doi: 10.1111/j.1601-183X.2007.00329.x. [DOI] [PubMed] [Google Scholar]
- Rodd ZA, Kimpel MW, Edenberg HJ, Bell RL, Strother WN, McClintick JN, et al. Differential gene expression in the nucleus accumbens with ethanol self-administration in inbred alcohol-preferring rats. Pharmacol Biochem Behav. 2008;89:491–8. doi: 10.1016/j.pbb.2008.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito M, Smiley J, Toth R, Vadasz C. Microarray analysis of gene expression in rat hippocampus after chronic ethanol treatment. Neurochem Res. 2002;27:1221–9. doi: 10.1023/a:1020937728506. [DOI] [PubMed] [Google Scholar]
- Saito M, Szakall I, Toth R, Kovacs KM, Oros M, Prasad VV, et al. Mouse striatal transcriptome analysis: effects of oral self-administration of alcohol. Alcohol. 2004;32:223–241. doi: 10.1016/j.alcohol.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Samaha AN, Mallet N, Ferguson SM, Gonon F, Robinson TE. The rate of cocaine administration alters gene regulation and behavioral plasticity: implications for addiction. J Neurosci. 2004;24:6362–6370. doi: 10.1523/JNEUROSCI.1205-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaha AN, Yau WY, Yang P, Robinson TE. Rapid delivery of nicotine promotes behavioral sensitization and alters its neurobiological impact. Biol Psychiatry. 2005;57:351–360. doi: 10.1016/j.biopsych.2004.11.040. [DOI] [PubMed] [Google Scholar]
- Schochet TL, Kelley AE, Landry CF. Differential expression of arc mRNA and other plasticity-related genes induced by nicotine in adolescent rat forebrain. Neurosci. 2005;135:285–297. doi: 10.1016/j.neuroscience.2005.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical Bayes methods for assessing differntial expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3(no 1) doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Sommer W, Hyytia P, Kiianmaa K. The alcohol-preferring AA and alcohol-avoiding ANA rats: neurobiology of the regulation of alcohol drinking. Addict Biol. 2006;11:289–309. doi: 10.1111/j.1369-1600.2006.00037.x. [DOI] [PubMed] [Google Scholar]
- Stewart RB, McBride WJ, Lumeng L, Li TK, Murphy JM. Chronic alcohol consumption in alcohol-preferring P rats attenuates subsequent conditioned taste aversion produced by ethanol injections. Psychopharmacology. 1991;105:530–534. doi: 10.1007/BF02244375. [DOI] [PubMed] [Google Scholar]
- Sun D, Huang W, Hwang YY, Zhang Y, Zhang Q, Li MD. Regulation by nicotine of Gpr51 and Ntrk2 expression in various rat brain regions. Neuropsychopharmacology. 2007;32:110–6. doi: 10.1038/sj.npp.1301134. [DOI] [PubMed] [Google Scholar]
- Surowy CS, Hoganson G, Gosink J, Strunk K, Spritz RA. The human RD protein is closely related to nuclear RNA-binding proteins and has been highly conserved. Gene. 1990;90:299–302. doi: 10.1016/0378-1119(90)90194-v. [DOI] [PubMed] [Google Scholar]
- Taracha E, Chrapusta SJ, Lehner M, Skorzewska A, Maciejak P, Szyndler J, et al. Morphine and methadone pre-exposure differently modify brain regional Fos protein expression and locomotor activity responses to morphine challenge in the rat. Drug Alcohol Depend. 2008;97:21–32. doi: 10.1016/j.drugalcdep.2008.03.013. [DOI] [PubMed] [Google Scholar]
- Treadwell JA, Singh SM. Microarray analysis of mouse brain gene expression following acute ethanol treatment. Neurochem Res. 2004;29:357–369. doi: 10.1023/b:nere.0000013738.06437.a6. [DOI] [PubMed] [Google Scholar]
- Waller MB, McBride WJ, Lumeng L, Li TK. Induction of dependence on ethanol by free-choice drinking in alcohol-preferring rats. Pharmacol Biochem Behav. 1982;16:501–7. doi: 10.1016/0091-3057(82)90459-2. [DOI] [PubMed] [Google Scholar]
- Worst TJ, Tan JC, Robertson DJ, Freeman WM, Hyytia P, Kiianmaa K, et al. Transcriptome analysis of frontal cortex in alcohol-preferring and -non-preferring rats. J Neurosci Res. 2005;80:529–538. doi: 10.1002/jnr.20496. [DOI] [PubMed] [Google Scholar]
- Xu K, Anderson TR, Neyer KM, Lamparella N, Jenkins G, Zhou Z, et al. Nucleotide sequence variation within the human tyrosine kinase B neurotrophin receptor gene: association with antisocial alcohol dependence. Pharmacogenomics. 2007;7:368–379. doi: 10.1038/sj.tpj.6500430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Ehringer M, Yang F, Sikela JM. Comparison of global brain gene expression profiles between inbred long-sleep and inbred short-sleep mice by high-density gene array hybridization. Alcohol Clin Exp Res. 2001;25:810–8. [PubMed] [Google Scholar]
- Yang MC, Weissler JC, Terada LS, Deng F, Yang YS. Pleiomorphic adenoma gene-like-2, a zinc finger protein, transactivates the surfactant protein-C promoter. Am J Respir Cell Mol Biol. 2005;32:35–43. doi: 10.1165/rcmb.2003-0422OC. [DOI] [PubMed] [Google Scholar]
- Zhang J, Zhang L, Jiao H, Zhang Q, Zhang D, Lou D, et al. c-Fos facilitates the acquisition and extinction of cocaine-induced persistent changes. J Neurosci. 2006;26:13287–13296. doi: 10.1523/JNEUROSCI.3795-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.