Abstract
Androgen deprivation is the mainstay of therapy for progressive prostate cancer. Despite initial and dramatic tumor inhibition, most men eventually fail therapy and die of metastatic castration-resistant (CR) disease. Here, we characterize the profound degree of genomic alteration found in CR tumors using array CGH, gene expression arrays, and FISH. By cluster analysis, we show that the similarity of the genomic profiles from primary and metastatic tumors is driven by the patient. Using data adjusted for this similarity, we identify numerous high-frequency alterations in the CR tumors, such as 8p loss and chromosome 7 and 8q gain. By integrating array CGH and expression array data, we reveal genes whose correlated values suggest they are relevant to prostate cancer biology. We find alterations that are significantly associated with the metastases of specific organ sites, and others with CR tumors versus the tumors of patients with localized prostate cancer not treated with androgen deprivation. Within the high-frequency sites of loss in CR metastases, we find an over-representation of genes involved in cellular lipid metabolism, including PTEN. Finally, using FISH we verify the presence of a gene fusion between TMPRSS2 and ERG suggested by chromosome-21 deletions detected by array CGH. We find the fusion in 54% of our CR tumors, and 81% of the fusion-positive tumors contain cells with multiple copies of the fusion. Our investigation lays the foundation for a better understanding of and possible therapeutic targets for CR disease, the poorly responsive and final stage of prostate cancer.
Keywords: Prostate cancer, castration-resistant metastases, metastatic prostate cancer, array CGH, expression, genomic alterations, TMPRSS2-ERG fusion
INTRODUCTION
Genomic analyses of malignant disease are intended to distinguish the molecular features that underlie carcinogenesis and identify clinical targets. Moreover, comparing primary and metastatic tumors is an exceptionally useful way to assess the molecular alterations associated with stage or progression.
The plethora of molecular events that occur in prostate cancer and no single ubiquitous alteration illustrate the complex biology of this disease. This complexity is likely to result, in part, from different genetic backgrounds and environmental exposures of the patients. One way to reduce this heterogeneity when comparing primaries to metastases is to assay both tumor types from the same patient, but these matching sets are difficult to obtain. A decade or more can elapse between the resection of the primary tumor by prostatectomy, detection of overt metastases, and death from castration-resistant (CR) disease. To address this deficiency, we set out to obtain sets of primary and metastatic tumors from the same patient. Although technical limitations prevented us from including bone metastases in this study, we present here analyses of an extraordinary set of multiple soft-tissue metastases.
To block the effects of androgens on tumor growth, patients with advanced disease are often deprived of androgen by surgical or chemical castration. However, aggressive and ultimately lethal CR disease inevitably develops. Few treatment options with clinical benefits exist (1–3), and most represent palliative interventions once this CR state is achieved.
One potential diagnostic marker and treatment target, the fusion of the TMPRSS2 and ERG genes, has generated considerable interest (4–10). Deletion of the 3-Mbp region of 21q22.2 between the promoter of the androgen-regulated serine-protease TMPRSS2 and the 3′ exons of ERG, a member of the oncogenic ETS family of transcription factors, is the principal mechanism for this gene fusion (7, 10). Androgens are presumed to drive the expression of this oncogenic fusion, which is found in ~30% of prostate cancers.
Here we characterize the genomic changes in CR prostate cancer using matching sets of primary prostate tumors and metastases. This work reveals alterations that might have causative properties, clinical value, or site-specific consequences for this end-stage prostate cancer.
METHODS
Sample acquisition
Use of human samples was approved by the Institutional Review Boards of participating institutions. Tumor samples were collected by radical prostatectomy or from autopsies performed at the University of Washington Medical Center under the rapid autopsy program as described previously (11). Available clinical data (stage, Gleason grade, treatment, etc.) are provided in Supplemental Table 1. Autopsies were performed within 2–4 h of death on fourteen patients (median (range) age at death 67 y (47–83)) with clinically diagnosed CR disease. Fifty-four tumors were obtained from various organ sites (Supplemental Table 2). Radical prostatectomy specimens from 19 individuals with organ-confined (i.e., localized) prostate cancer not treated by androgen deprivation were also collected.
Specimens (n=73) were embedded in freezing media (Tissue-Tek OCT Compound, Sakura Finetek, Torrance, CA) and stored in liquid nitrogen. Cells from the 54 CR tumors, nine localized prostate cancers (LocPCs), or normal stromal of 10 LocPC patients were isolated by laser-capture microdissection (LCM) as described previously (12). A pathologist (L.D.T.) reviewed all LCM images.
DNA isolation and amplification
DNA from LCM-collected samples was isolated using the QIAamp DNA Micro Kit (Qiagen Inc, Valencia, CA). DNA from 42 tumors (Supplemental Table 2) was amplified by ligation-mediated PCR (LMP) as described previously (13). DNA samples from 12 CR tumors and 9 LocPC tumors could not successfully be amplified by LMP and instead were amplified using a whole genome amplification (WGA) kit, WGA2 (Sigma-Aldrich, St. Louis, MO). DNAs amplified from these two methods are comparable (14). Of the 10 normal samples, 5 were amplified by LMP and 5 by WGA.
Reference DNA was obtained from the peripheral blood of a single female individual isolated using the QIAamp DNA Blood Mini Kit (Qiagen Inc). The amplification method for all reference samples was matched with the test sample.
Array comparative genomic hybridization (array CGH) analysis
The bacterial artificial chromosome (BAC) clones that make up the array, array CGH methods and analyses have been described previously (14, 15). Clone coordinates given refer to the May 2004 sequence assembly (Build 35).
The log2-ratio array data were normalized with a block-level Loess algorithm (16) and processed by Circular Binary Segmentation (CBS, (17)) to organize the output into segments of approximately equal copy number. Thresholds for calling loss and gain in arrays of LMP- or WGA-amplified material were determined using the array results obtained from the normal-cell samples amplified by LMP or WGA, respectively, as described previously (14).
CBS data were subjected to hierarchical/complete linkage clustering (similarity metric was correlation centered) using Gene Cluster software (18). The tree was produced using TreeView (http://rana.lbl.gov/EisenSoftware.htm).
Significant associations between alterations and tumor state (i.e., CR primaries or CR metastases) or CR organ site (i.e., prostate, lymph-node metastasis, or liver metastasis) were identified using the Significance Analysis of Microarrays (SAM) method (19) employing response formats two-class (unpaired) and multiclass, respectively. SAM is based on a modified t-statistic and uses random permutations of class labels to estimate a false discovery rate (FDR). For each analysis, 1000 permutations were performed.
The methods for GO analyses and tests of statistical significance are as described previously (14, 20).
RNA isolation and amplification
Total RNA from LCM samples was isolated using the Arcturus PicoPure RNA Isolation kit (Molecular Devices, Sunnyvale, CA) and DNAse-treated using the RNase-Free DNase Set (Qiagen Inc.) The reference RNA was a pool of equal amounts of total RNA isolated from the LNCaP, DU145, PC3, and CWR22 cell lines (American Type Culture Collection, Manassas, VA). Experimental and reference total RNA samples were subjected to two rounds of amplification using the MessageAmp aRNA Kit (Applied Biosystems/Ambion Inc, Austin, TX).
Expression array analysis
cDNA probe pairs were prepared by amino-allyl reverse transcription using 2 μg of amplified RNA and labeling with Cy3-dCTP (experimental) or Cy5-dCTP (reference) (Amersham Bioscience, Piscataway, NJ). Custom microarrays composed of 6,760 cDNA clones selected from the Prostate Expression Database (PEDB, www.pedb.org) repository of human prostate expressed sequence tag data were constructed and hybridized as previously described (21).
Combined DNA/RNA analysis
Measurements for genes represented by multiple clones on the PEDB expression array were averaged. Each gene-expression measurement was paired with a copy-number measurement from the BAC probe nearest to that gene. Correlated data were identified by performing a Pearson’s correlation for the CBS-determined BAC clone measurement and gene expression level for each gene. Statistical significance was determined using a Bonferroni correction to adjust for multiple hypothesis testing.
To identify genes with significant differences in both copy number and gene expression between primary and metastatic prostate cancer specimens, the DNA/RNA-SAM (DR-SAM) method of DR-Integrator was used (Salari et al., in preparation)9. Briefly, for each gene, a modified Student’s t-test was applied to generate a copy-number score and a gene-expression score for the primary or metastatic samples. The two scores were summed and weighted to favor genes with substantial differences between the two tumor types. Significance was established by recalculating the scores on 1000 random permutations of the sample labels (FDR of 5%).
TMPRSS2:ERG FISH
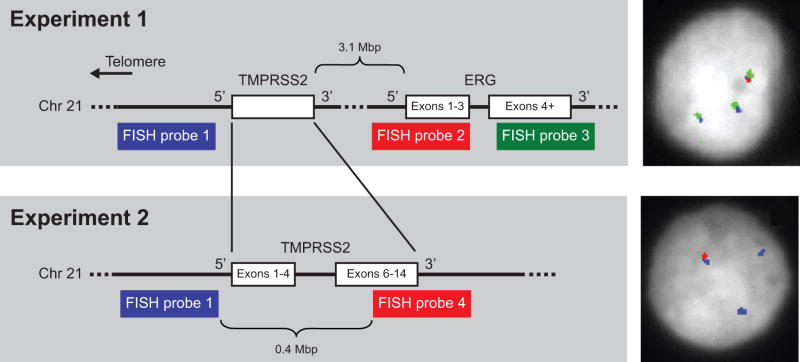
Five-μm tumor sections from frozen tissue blocks were fixed in 3:1 methanol:acetic acid. Four BAC DNAs were used as probes, RP11–35C4 (probe 1), RP11–95I21 (probe 2), RP11–476D17 (probe 3), and RP11–120C17 (probe 4) (Figure 4). BAC DNA was labeled using a nick-translation kit and either Spectrum Red-dUTP, Spectrum Green-dUTP, or Spectrum Aqua-dUTP (Abbot Molecular, Abbott Park, IL). Hybridization was performed as described previously (22).
Figure 4.
Experimental designs to detect the presence of the TMPRSS2:ERG fusion by FISH. In Experiment 1, three probes were used to detect the fusion: a probe 5′ of TMPRSS2 (blue, probe 1), one encompassing the 5′ exons of ERG (red, probe 2), and one encompassing the 3′ exons of ERG (green, probe 3). In the normal configuration, the signals of all three probes overlap. The fusion is indicated by overlapping signals of probes 1 and 3 with loss or dissociation of probe 2. A CR tumor nucleus with two fusions and one normal probe configuration is shown to the right. In Experiment 2, we confirmed the presence of fusion with probe 1 and a second probe encompassing the 3′ exons of TMPRSS2 (red, probe 4) in adjacent tissue sections. Lone probe 1 signals indicate a deletion consistent with a TMPRSS2:ERG fusion. The panel to the right shows a positive nucleus from a section adjacent to the section used to capture the nucleus shown for Experiment 1. DAPI staining is indicated in gray; the hybridization signals are pseudocolored to correspond to the experimental schematics.
FISH signals were scored manually (100x oil immersion). Fusion-positive tumors had positive nuclei (i.e., juxtaposition of probe 1 and probe 3 with concurrent loss or dissociation of probe 2) in at least 20 of 50 cells analyzed. We designated the tumor as possessing multiple copies of the fusion if at least five of 20 positive nuclei showed at least two gene-fusion probe conformations. The results were confirmed in a second FISH experiment (using the same scoring criteria) testing for retention of probe 1 concurrent with dissociation or loss of probe 4.
RESULTS
Hierarchical clustering groups tumors from the same patient together
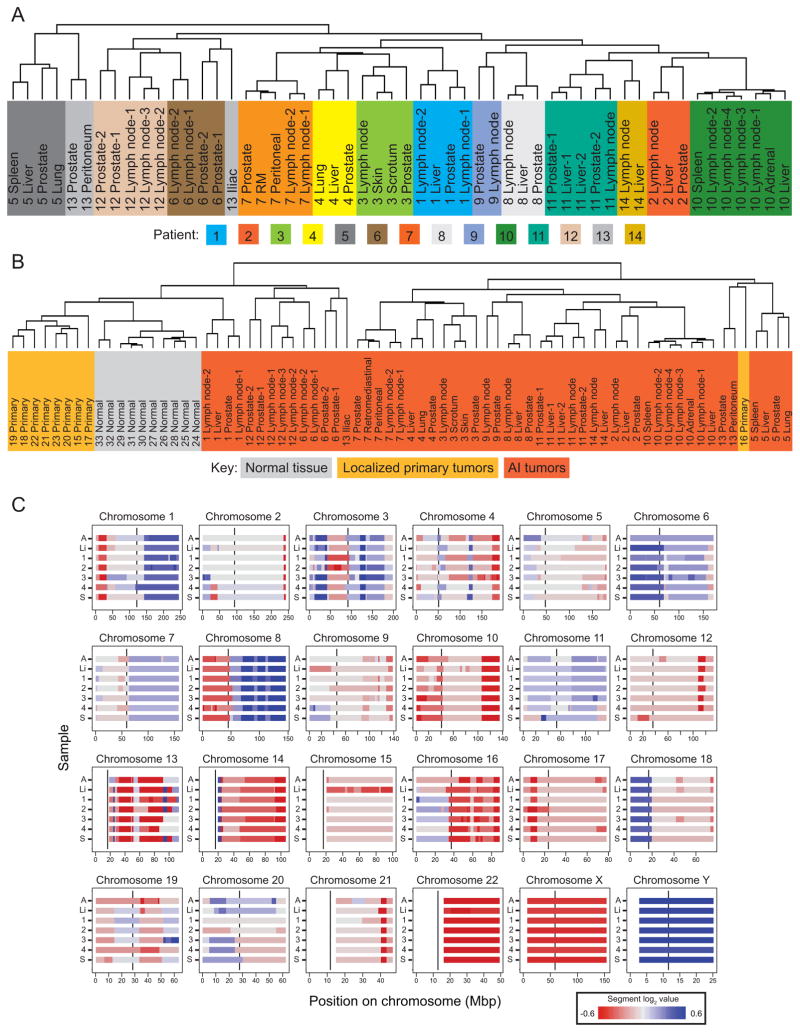
We performed hierarchical cluster analysis to evaluate relationships among the 54 CR tumors from 14 patients based on their genomic alterations. These CR tumors include those resected from the prostate (and called CR primaries here) and multiple soft-tissue metastatic sites. Tumors most often group with other tumors from the same patient, rather than cluster by organ of origin (Figure 1A). The tumors for 11 of the 14 (~79%) patients define patient-specific clusters. The closest neighbor in the hierarchy for 53 out of 54 tumors is a tumor from the same patient. Including LocPC and normal arrays does not alter the relationships observed for the CR tumors (Figure 1B). The consistency of alterations for the tumors of a given patient is illustrated in Figure 1C and emphasized by the separation of the distributions of pairwise correlation coefficients calculated for all intra-tumor pairs versus for all inter-tumor pairs (Supplemental Figure 1).
Figure 1.
Similarity of the tumors of a given patient. Trees from hierarchical clustering of segmented data from (A) the CR tumors and (B) the CR tumors, LocPCs, and normal prostate stromal tissue. In both trees, the patient number precedes the organ description, and numerical suffixes indicate multiple tumors from the same organ. Each color in A indicates tumors from a different patient; in B, the three sample types are each shaded with a different color. The third panel (C) shows heat maps by chromosome of the CBS segment data for each of the 7 tumors from patient 10. Red denotes negative segment values (regions of copy-number loss), and blue denotes positive segment values (regions of copy-number gain). Note the expected relative loss of X-chromosome sequences and gain of Y chromosome in this male:female comparison. For each chromosome, the vertical black line within each box indicates centromere position, and the large gaps without data indicate unsurveyed repetitive regions. The y-axis indicates the sample: A, adrenal metastasis; Li, liver metastasis; 1–4, the four lymph-node metastases; and S, spleen metastasis. The color bar below the figure indicates the range of colors representing the segment values shown.
Tumor-related loci and candidate genes in the frequent alterations of CR tumors
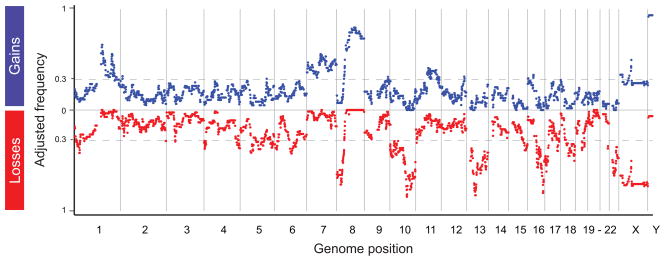
Every position in the genome is represented in the cumulative spectrum of changes observed in one or more of the 54 CR tumors (Figure 2). To summarize these results, we first adjusted for the variation in number of tumors evaluated for each patient before calculating the frequency of genomic alterations across the CR patients. BAC clones encompassed by a lost or gained segment were assigned a value of −1 or 1, respectively; no change was assigned a value of 0. For each loss or gain, we first calculated the average value for each patient and then averaged the resultant fractions across all 14 patients to represent the frequency of a given deviation in this patient set. All frequencies noted in this work are these adjusted frequencies.
Figure 2.
Deviations in CR disease. The y-axis indicates the adjusted frequency with which each BAC clone on the array was included in a deviant segment across the 54 CR tumors from 14 individuals. Gains are plotted in blue, and losses are plotted in red. The hatched grey lines are drawn at a frequency of 0.3 (i.e., the minimum frequency for significance p < 0.01). The x-axis is the genomic position of each BAC clone. Note that loss in X- and gain in Y-chromosome sequences are expected in these male:female comparisons.
To define deviations of interest, we used the one-sample binomial test to calculate a threshold frequency such that for any deviation with frequency exceeding this threshold, the 99% confidence interval of its frequency did not include 0. This frequency is z2/(n + z2), where z is the critical value (Standard normal table) and n is the sample size. For n=14 and p < 0.01, i.e., z=2.56, the threshold frequency is approximately 0.3. The dashed grey lines in Figure 2 mark this threshold. High-frequency sites of loss or gain exceeding this threshold are listed in Table 1.
Table 1.
High-frequency deviations and minimally overlapping regions (MOR) in CR tumors. Losses (A) and gains (B) are listed separately. High-frequency is defined as observed with an adjusted frequency of ≥30%. The start position, size, and any constituent MORs observed at higher frequency than the larger deviation are given for each deviation. Mbp positions are rounded to nearest 0.1 Mbp. MORs observed at ≥50% are in bold. The start site and size of CR-associated alterations are given.
| A. Losses | B. Gains | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Deviation |
MOR |
CR-associated |
Deviation |
MOR |
CR-associated |
||||||||||
| Band | Start (Mbp) |
Size (Mbp) |
(%) | Start (Mbp) |
Size (Mbp) |
Start (Mbp) |
Size (Mbp) |
Band | Start (Mbp) |
Size (Mbp) |
(%) | Start (Mbp) |
Size (Mbp) |
Start (Mbp) |
Size (Mbp) |
| 1p36.23-p35.1 | 7.2 | 25.2 | 36 | 16.3 | 3.5 | 7.2 | 25.2 | 1p12-q43 | 120.0 | 117.7 | 65 | 147.9 | 2.8 | 153.6 | 84.3 |
| 43 | 26.7 | 2.5 | 52 | 155.2 | 0.7 | ||||||||||
| 1p33-p32.3 | 48.4 | 6.8 | 49 | 157.6 | 6.9 | ||||||||||
| 2q37.3 | 238.6 | 3.7 | 238.6 | 3.7 | 55 | 200.3 | 1.9 | ||||||||
| 3p21.31 | 46.3 | 1.8 | 31 | 46.3 | 0.6 | 46.3 | 1.8 | 45 | 209.4 | 3.0 | |||||
| 4p16.3-p16.1 | 0.1 | 10.6 | 38 | 2.7 | 0.3 | 0.1 | 4.4 | 1q41–43 | 215.6 | 22.1 | 33 | 220.4 | 7.6 | ||
| 5.1 | 5.6 | 2p25.1 | 9.6 | 1.8 | 33 | 9.5 | 1.2 | ||||||||
| 4q33-q34.3 | 171.7 | 9.9 | 35 | 175.5 | 3.5 | 171.7 | 9.9 | 5q31.3 | 140.1 | 0.8 | |||||
| 4q34.3-q35.2 | 182.2 | 8.2 | 37 | 184.3 | 0.9 | 182.2 | 5.2 | 7p22.3-q36.3 | 0.1 | 158.5 | 47 | 5.5 | 0.5 | 6.4 | 16.4 |
| 38 | 188.0 | 2.4 | 45 | 26.8 | 2.5 | 27.7 | 44.2 | ||||||||
| 5q11.2-q13.2 | 55.1 | 15.9 | 42 | 55.3 | 7.5 | 47 | 62.0 | 4.0 | 75.3 | 23.6 | |||||
| 40 | 67.8 | 2.4 | 55 | 72.6 | 2.6 | 105.6 | 42.1 | ||||||||
| 5q14.2-q22.2 | 81.7 | 31.0 | 39 | 96.0 | 13.7 | 55 | 78.9 | 1.3 | |||||||
| 5q23.1-q23.2 | 115.4 | 11.8 | 35 | 117.6 | 7.3 | 53 | 99.1 | 3.1 | |||||||
| 5q34-q35.2 | 160.7 | 15.6 | 37 | 165.0 | 6.1 | 171.3 | 5.0 | 44 | 121.3 | 5.4 | |||||
| 6p25.3-p22.3 | 0.2 | 21.5 | 39 | 12.3 | 1.2 | 5.8 | 9.1 | 46 | 128.7 | 0.8 | |||||
| 6q13-q22.31 | 75.2 | 43.7 | 43 | 88.7 | 5.2 | 48 | 136.2 | 0.4 | |||||||
| 36 | 103.9 | 9.8 | 50 | 148.2 | 5.2 | ||||||||||
| 8p23.3-p11.21 | 0.3 | 40.3 | 67 | 1.5 | 1.7 | 8p12-q24.3 | 36.5 | 109.6 | 64 | 43.3 | 6.0 | 42.9 | 103.2 | ||
| 66 | 4.6 | 1.6 | 69 | 57.2 | 8.2 | ||||||||||
| 74 | 19.8 | 3.5 | 81 | 79.3 | 0.5 | ||||||||||
| 71 | 25.5 | 1.8 | 82 | 93.3 | 6.0 | ||||||||||
| 10p15.3-q26.3 | 0.2 | 135.1 | 39 | 0.2 | 0.1 | 0.2 | 42.6 | 79 | 102.3 | 2.5 | |||||
| 47 | 9.0 | 3.3 | 50.4 | 25.7 | 78 | 115.1 | 6.5 | ||||||||
| 44 | 32.4 | 2.8 | 79.1 | 15.7 | 73 | 131.1 | 2.4 | ||||||||
| 42 | 43.0 | 1.0 | 95.9 | 39.4 | 9q33.2 | 122.4 | 1.1 | ||||||||
| 52 | 72.0 | 2.4 | 9q33.2-q33.3 | 124.3 | 3.8 | 33 | 124.6 | 1.1 | |||||||
| 86 | 89.4 | 1.2 | 9q34.11 | 129.5 | 0.8 | ||||||||||
| 68 | 98.2 | 3.7 | 11q12.1-q24.1 | 59.4 | 63.0 | 42 | 65.8 | 1.2 | 77.5 | 39.5 | |||||
| 68 | 103.4 | 0.4 | 41 | 73.5 | 1.3 | ||||||||||
| 79 | 105.9 | 5.0 | 39 | 83.8 | 18.4 | ||||||||||
| 68 | 121.7 | 6.9 | 40 | 105.3 | 4.2 | ||||||||||
| 11p15.4 | 4.2 | 0.3 | 33 | 117.8 | 0.4 | ||||||||||
| 13q11-q34 | 18.4 | 95.0 | 50 | 18.4 | 4.4 | 18.4 | 9.7 | 11q23.3-q24.1 | 119.0 | 3.4 | |||||
| 45 | 27.0 | 1.1 | 97.2 | 16.2 | 11q24.1-q24.2 | 123.1 | 2.7 | 32 | 123.1 | 2.3 | 123.1 | 2.3 | |||
| 80 | 40.6 | 0.8 | 12q24.31-q24.32 | 119.8 | 4.8 | 33 | 119.8 | 2.8 | |||||||
| 85 | 44.6 | 3.1 | 16p13.3 | 0.9 | 2.4 | ||||||||||
| 64 | 52.1 | 0.1 | 16p12.1-p11.1 | 27.6 | 6.9 | 35 | 29.7 | 4.8 | |||||||
| 63 | 72.4 | 2.6 | 18p11.32 | 0.2 | 2.0 | ||||||||||
| 59 | 101.3 | 2.8 | Xp22.31-p22.12 | 7.5 | 11.8 | ||||||||||
| 15q24.3-q25.1 | 75.1 | 1.9 | 71.4 | 13.9 | Xp11.22-q13.1 | 51.5 | 16.5 | 50 | 65.1 | 2.8 | 65.1 | 2.8 | |||
| 15q25.1-q26.3 | 78.7 | 21.3 | 40 | 85.4 | 7.4 | 86.5 | 4.1 | ||||||||
| 96.6 | 3.4 | ||||||||||||||
| 16q11.2-q24.3 | 45.3 | 43.3 | 57 | 49.4 | 5.3 | 55.0 | 4.3 | ||||||||
| 47 | 63.6 | 0.2 | 65.0 | 2.2 | |||||||||||
| 82 | 80.7 | 1.6 | 82.4 | 6.2 | |||||||||||
| 17p13.3-p11.2 | 0.1 | 19.5 | 51 | 0.1 | 0.5 | 0.1 | 15.2 | ||||||||
| 61 | 7.6 | 0.3 | 15.4 | 4.2 | |||||||||||
| 17q24.2-q25.3 | 64.6 | 8.9 | 41 | 66.1 | 2.6 | 64.6 | 8.9 | ||||||||
| 17q25.3 | 74.4 | 1.6 | 73.8 | 2.1 | |||||||||||
| 18q12.1-q23 | 28.2 | 47.9 | 75.4 | 0.7 | |||||||||||
| 19p12-q13.11 | 19.9 | 20.3 | 40 | 34.3 | 2.2 | ||||||||||
| 19q13.2-q13.31 | 46.1 | 2.5 | 36 | 47.4 | 0.6 | 46.1 | 2.5 | ||||||||
| 21q22.2-q22.3 | 38.7 | 4.5 | 38 | 38.7 | 3.0 | ||||||||||
| 22q11.1-q13.33 | 16.2 | 33.2 | 65 | 24.6 | 1.6 | 16.2 | 8.2 | ||||||||
| 46 | 43.4 | 6.0 | 26.7 | 23.2 | |||||||||||
Abbreviations: “Chr.” = chromosome, “Min”. = the minimum observed frequency for the larger deviation, and “MOR (%)”= the peak frequency that defines the MOR.
Table 1 also provides the locations of the minimally overlapping regions (MORs), which are the most frequently deviant subregions within each set of overlapping deviations. We have identified novel MORs on the two chromosome arms that are thought to play an important role in the biology of prostate cancer, 8p and 8q. Of the four MORs identified for 8p, two have been frequently identified in prostate cancer (23–27), but the more distal MORs in 8p23 have not. We identified four previously reported MORs on 8q (between 8q21.3-q24.22) (23) and three MORs (between 8q11.1–8q21.12) that are heretofore unreported for prostate cancer.
We also examined single BAC clones with extreme log2-ratio values (i.e., those in the upper and lower 95th percentile) prior to smoothing by the CBS method. These singleton deviations, when high, might represent focal sites of gene amplification or, when low, homozygous deletions or loss of multiple copies from polyploid tumor cells. (BAC clones whose normalized log2 ratio value was outside the 95%ile in two or more normal control arrays were excluded from this analysis.) Supplemental Table 3 lists the singletons identified as deviant in ≥30% of the CR tumors (adjusted frequency) and the genes encompassed by them. The BAC clones that overlap sites of normal copy-number variation (http://hgsv.washington.edu/) are noted. We observed singleton deviations that encompass known cancer-related genes (AR, C13, CDH13, MMP16, MYC, PTEN, and PTK2) and androgen-regulated genes (AR, MMP16, MYC, NDRG1, and TSC22D1).
Genomic profiles of CR tumors and untreated localized primaries are significantly different
To distinguish sites that relate to metastasis or CR disease, we identified those alterations that occur in significantly greater numbers in the CR tumors (n=54) versus the LocPCs (n=9) and vice versa. We found 26 losses spanning a total of 306 Mbp and eight gains spanning a total of 255 Mbp that were significantly (Fisher’s exact p<0.05) associated with the CR tumors (Table 1). One of the gains encompasses the androgen receptor, an amplification specifically associated with CR disease (28). In the converse analysis, we found only three losses spanning 20 Mbp (5q22.2-q23.1, 11q14.1, and 20q11.23-q13.12) and six gains spanning 69 Mbp (1p36.33-p34.3, 3p21.31-p21.1, 10q21.3-q22.1, 12q13.11-q14.1, 15q15.1-q15.3, and 16q22.1-q22.2) in significantly greater numbers of LocPC than CR tumors.
To verify our findings for CR-associated loci, we compared our results to a recent survey summarizing 41 CGH studies examining 872 prostate tumors (23). All of the MORs encompassed by the seven alterations that this survey called common (>10%) to primary prostate cancer (loss at 5q15, 6q15, 8p21.3, 13q21.33, 16q22.1, and 18q21.33–22.1 and gain at 8q22.2) were observed in our LocPCs and were in high frequency in our CR tumors (≥30%). Of these, our analysis found that the MORs on 16q and 8q were significantly more associated with CR tumors than with LocPCs (Table 1).
Stratification by CR tumor location identifies organ-specific alterations
We stratified the CR tumors to look for differences that might relate to particular states (i.e., CR primaries or CR metastases) or sites (i.e., CR primaries, lymph node, or liver metastases). We found that the CR primaries possessed significantly fewer gains and losses (average of 14 gains and 18 losses; Student’s p-values = 0.008 and 0.004, respectively) than the CR metastases (average of 23 gains and 24 losses).
We used SAM (19) to identify alterations with significantly different frequencies between the CR primaries and metastases. Only two alterations were found, both in significantly more metastases, deletions of 10p15.2–10p15.1 (FDR = 0%, 20% of primaries and 56% of metastases) and 22q12.1–22q12.3 (FDR = 0%, 27% of primaries and 80% of metastases). These deletions were also found in significantly fewer LocPCs (11% and 33%, respectively) as compared to the entire set of CR tumors. Among the genes encompassed by these loci are the tumor suppressors PRKCQ (10p15.1), MYO18B (20q12.1) and SEZ6L (20q12.1).
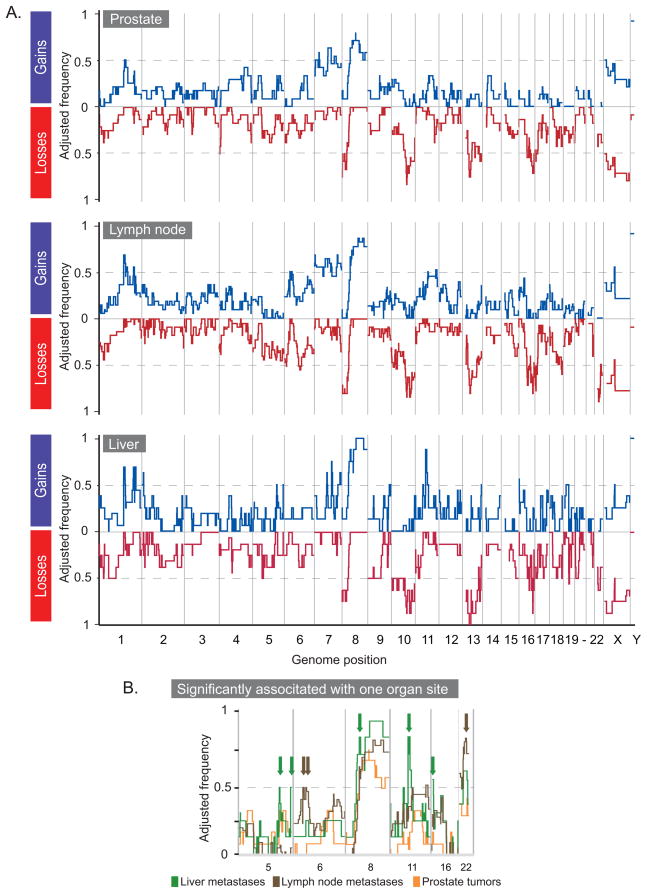
We next asked what differences might exist between the genomic profiles of the CR tumors from the three organ sites that make up 80% of our CR tumors, i.e., prostate (n=15 from 12 patients), lymph node (n=19 from 11 patients), and liver (n=9 from 8 patients). Figure 3A shows the frequencies of alterations for each set. We found that the prostate tumors had significantly fewer gains (average 14; Student’s p-value = 0.03 and 0.005, respectively) and losses (average 18; Student’s p-value = 0.02 and 0.04, respectively) than lymph node (averages 21 and 22, respectively) or liver (averages 30 and 24, respectively) metastases. Lymph node metastases had significantly fewer gains (Student’s p-value = 0.04) but not losses than liver metastases.
Figure 3.
Deviations in CR tumors stratified by organ of origin. (A) The adjusted frequencies of deviation (y-axis) for gain (blue) or loss (red) for prostate tumors (n=15), lymph-node (n=19), and liver (n=9) metastasis, respectively. (B) Zoom in for gains on chromosomes 5, 6, 8, 11, and 16 and losses on chromosome 22. Alterations significantly associated with lymph node (brown) and liver (green) metastases are indicated by arrows.
Using SAM, we identified five alterations in significantly more liver metastases, three in lymph node metastases, and none with the primary tumors (Figure 3B). The sites for liver metastases were gains at 5q31.2–5q31.3, 5q35.2–5q35.3, 8p11.21, 11q13.2, and 16p13.3. The gain at 8p encompasses a single gene ANK1. The sites for lymph node metastases were gains at 6p21.32–6p21.2 and 6p21.1 and loss at 22q12.1. Among the genes encompassed by the gains were the oncogene ETV7 (6p21.31) and the tumor-related genes PTK7 (29) and VEGF (30).
Merged copy-number and expression results reveal candidate genes
Genomic alterations in cancer can encompass many genes. To identify those that might relate to tumor phenotypes, we integrated our array CGH data with microarray analyses of expression levels. We were able to obtain copy-number and expression data for 51 of our CR tumors.
We identified 131 genes that showed correlated array CGH segment and micro-array expression values at a significance level of 0.05 (Supplemental Table 4; tumor-related and androgen-regulated genes are highlighted). Supplemental Figure 2 illustrates the correlation between copy-number and expression for the following eight genes. The gene with the third highest correlation value was TMPRSS2. A significant subset of tumors exhibited negative CBS segment values and lower expression (n=15) or positive CBS segment values and higher expression (n=19) of TMPRSS2. Other genes identified were the tumor suppressor retinoblastoma 1 (RB1, deleted in 79% of the CR tumors), MYC binding protein 2 (MYCBP2, deleted in 60% of the CR tumors), mucin 1 (MUC1, gained in 57% of CR tumors), PBX1 (gained in 49% of the CR tumors), PARP1 (gained in 33%), LSM1 (gained in 31%), and TP53 binding protein 2 (TP53BP2, gained in 30%).
To identify genes with significant differences in both copy number and gene expression between primary and metastatic prostate cancer specimens, we used the DNA/RNA-Significance Analysis of Microarrays (DR-SAM) method of DR-Integrator (Salari et al., manuscript in preparation)9. At an FDR of 5%, we found 19 genes with higher copy-number and expression in CR metastases versus CR primaries and six with higher copy-number and expression in CR primaries versus CR metastases (Supplemental Table 5). Supplemental Figure 3 illustrates the correlation between selected genes.
Over-representation of ontologically related genes in regions frequently altered in CR tumors
To further prioritize possible genes of interest, we looked for over-representation of genes of particular gene-ontology (GO) categories in regions frequently (≥30%, adjusted frequencies) altered in the CR tumors. Lost and gained segments were evaluated separately. GO categories and genes are given in Supplemental Table 6. The p-value “SimPValue” is a statistic that corrects for GO categories that might be over-represented due to gene clustering. Categories with a SimPValue > 0.05 should be considered with caution.
Cellular lipid metabolism and catabolic process were two of the four categories with genes over-represented in CR tumor losses. The former category included the tumor suppressors PTEN and WWOX, and the latter category included RBX1, which functions in a complex with the von Hippel-Lindau tumor suppressor gene. Seven of the genes belonging to GO categories enriched in regions of loss (BNIP3, ECHS1, HDLBP, RNF6, UBB, UBE2L3, and USP10) and two (AGT and PCDHGC3) belonging to categories enriched in gains were also identified in our analysis of correlated copy-number alterations and expression levels (Supplemental Table 4).
The TMPRSS2:ERG gene fusion is prevalent in CR tumors
One MOR, a deletion in 21q22.2-q22.3, was detected in ~40% (22/54) of the CR tumors and 50% (7/14) of the patients with CR disease and suggested the presence of a fusion of the 5′ portion of the TMPRSS2 gene and the 3′ exons of the ERG gene (7, 10).
We used FISH to confirm the presence and assess the copy-number of the fusion in the cells of our CR tumors. Figure 4 illustrates our FISH strategies. Of the 52 CR tumors for which we could obtain reliable FISH results, 27 (54%) were positive for the TMPRSS2:ERG fusion. All tumors from five patients were positive, and 8 patients had at least one fusion-positive tumor. All fusions observed were a result of loss of the 5′ ERG probe (i.e., deletion). Of the 27 tumors FISH-positive for the fusion, 21 had concordant array CGH results. All tumors with a deletion in 21q22.2-q22.3 were confirmed by FISH.
We had FISH, array CGH, and microarray expression analysis for 49 CR tumors, of which 25 were fusion-positive. Tumors with correlated negative array CGH and expression values (n=15) showed a significant association with possession of the fusion (14 of 15) relative to the tumors with any other trend in array CGH and expression values (Fisher’s exact p=0.00006). Of the other 11 fusion-positive tumors, nine showed negative array CGH and positive expression values and two showed positive array CGH and expression values.
Two or more copies of the TMPRSS2:ERG fusions were detected by FISH for 22 (42%) of the CR tumors tested. Thus, multiple copies of the fusion were seen in 22 of the 27 fusion-positive tumors (81%), at least one CR tumor from eight of the 14 patients (57%), the majority of tumors from three of the patients (21%), and all of the tumors from three of the patients (21%).
DISCUSSION
Genomic alterations within CR tumors might reveal important biological insights into this ultimately lethal stage of prostate cancer. Our cluster analysis shows that CR tumors from a given patient are more similar to each other than they are to tumors from matching organ sites of other patients. These results argue for a monoclonal origin of metastases, consistent with a recent publication by Liu et al. who conclude that most, if not all, metastatic prostate cancers have monoclonal origins (31). As in our study, Liu et al. found that metastatic tumors possess genomic profiles that reflect that of the originating tumor cell.
Androgen deprivation also undoubtedly played a part in generating the similarity of tumors of a given patient. Abrogation of androgen, a hormone with profound effects on tumor biology, places a strong selective pressure on the malignant cell population likely increasing the homogeneity of the tumor population. However, this homogenizing force appears insufficient to generate a single CR genomic signature, as the tumor sets are sufficiently distinct to cluster in patient-specific groups. These differences must reflect the uniqueness of each of our CR patients in terms of his genetic risk factors, environmental exposures, history of therapeutic modalities, and chance events.
Across patients, we find a multitude of high-frequency alterations, some encompassing candidate genes that might relate to prostate cancer, metastases, and CR disease. Within these alterations were several cancer-related genes with correlated DNA and mRNA values, including RB1, the first recognized tumor suppressor, and MUC1, a marker of prostate cancer progression and a novel therapeutic target (32). PARP1 was also identified; it is involved in DNA repair and inhibitors of it have received considerable attention as novel therapy of breast cancer (33).
We found that majority of our CR tumors possessed a fusion between TMPRSS2 and ERG as a result of the deletion. The high frequency of CR tumors with multiple copies of the TMPRSS2:ERG fusion is further evidence of the association of amplification of the fusion gene and poor clinical outcome (5). These findings support the idea that it is a promising target for therapeutic interventions (34, 35). Given that the deletion that generates the TMPRSS2:ERG fusion was found to encompass the majority of the TMPRSS2 gene, it was not surprising that we found a significant association between the fusion and negative array CGH and expression values TMPRSS2. However, a subset of fusion-positive tumors showed higher expression of TMPRSS2. This finding may indicate that expression from an intact copy of this androgen-regulated gene might be biologically relevant for some prostate tumors.
Our analysis of high-frequency alterations in CR tumors helps refine prostate cancer- related loci and narrow in on additional candidate genes. No consensus has yet been reached about the critical locus (loci) affected by the two most common alterations seen in prostate cancer, loss at 8p and gain at 8q (28). Our study identifies four distinct MORs within 8p and seven MORs within 8q. Of the less well-characterized MORs on 8p, one of them encompasses a single gene, CSMD1, a candidate suppressor of multiple cancer types (36). AEG1 (a.k.a. Lyric and MTDH) is among the genes encompassed by an MOR at 8q21.3-q22.2. AEG1 is over-expressed in breast, brain and prostate cancers (37–39) and is thought to promote tumor progression (38–40). The novel MOR at 8q21.12 contains a single gene, PKIA, an extremely potent competitive inhibitor of cAMP-dependent protein kinase activity (41). The role of PKIA in prostate tumorigenesis merits exploration.
The most frequent MORs in our CR dataset might help identify other genes relevant to tumorigenesis. The most frequent MOR in our CR tumors was loss at 10q23.31 (86%), which encompasses only PTEN, the well-characterized tumor suppressor. Twelve known genes are encompassed by the MOR at 13q14.13-q14.2 (85%), including the ITM2B gene, a tumor suppressor (42). Loss at 16q23.3 (82%) encompasses only CDH13 whose reduced expression in primary prostate tumors is associated with an increased risk of biochemical failure (43). Methylation of CDH13, assessed in primary prostate cancer, is generally considered the primary mechanism of gene silencing (44, 45). Our results suggest that methylation in premetastatic states might precede deletion in later stages or that deletion is an alternate mechanism of silencing.
Our GO analysis of the high-frequency alterations in CR tumors also provides insight into candidate genes. The genes that overlapped between our GO analysis and the integration of copy-number and expression warrant particular attention. One of these genes encompassed by CR deletion encodes BNIP3, a Bcl-2 family member that can promote apoptosis (46). RNF6 and USP10, which modulate androgen receptor function (47, 48), showed correlated genomic loss and lower expression in a subset of tumors and gain and higher expression in others. This finding underscores the complexity of the relationship between CR disease and tumor processes controlled by the androgen receptor.
Our analysis of CR tumors provides evidence of accumulation of genomic change with disease progression and outgrowth towards CR disease. We found significantly fewer alterations in CR primaries than in CR metastases and no alterations significantly more associated with CR primaries relative to the matched set of lymph node and liver metastases. However, we did find eight alterations significantly more associated with those metastatic sites (three and five, respectively). Moreover, we identified several alterations significantly more associated with CR tumors versus LocPCs.
To expand on the significance of our analysis, future work will need to include bone metastasis, a clinically important entity in prostate cancer, and validation of candidate genes. For example, it would be interesting to see the effect of PARP1 inhibitors on models of prostate cancer. The androgen-dependent LNCaP cells and the androgen-independent derivative line (49) would be particularly useful in assessing the role of genes encompassed by CR-associated alterations identified in our study. Direct injection methods for studying prostate cancer cell-bone interactions and the effects that drugs have on these interactions could be used to validate candidate genes and their therapeutic potential (50).
We have shown that CR tumors possess a profound degree of genomic change encompassing many regions that could contain therapeutic targets for metastatic prostate cancer, or at least illuminate the biology of this lethal disease. By combining the results of array-CGH and expression microarrays, we have identified numerous candidate regions and genes. Moreover, we have verified that the TMPRSS2:ERG fusion, a promising target of cancer therapeutics, is highly prevalent in CR tumors. This extensive and in-depth investigation of the alterations found in CR disease lays the foundation for a better understanding of this final stage of prostate cancer.
Supplementary Material
Acknowledgments
Grant Support: Pacific Northwest Prostate Cancer SPORE CA97186; NIH PO1 CA085859; NIH T32 HG00035; NIH RO1 DC004209; Fred Hutchinson Cancer Research Center Institutional Funds; NIH R01 DK069690; DOD PC060595; Paul & Daisy Soros Foundation; NIH GM07365; NIH RO1 AG14358; NIH RO1 CA098415; NIH RO1 CA95717; VA Merit Award; NIH U24 CA80295
The authors gratefully acknowledge the support and technical expertise of other members of the Trask, Vessella, and Nelson laboratories, especially David Gifford and Tom Kim, and the members of the Tissue Acquisition Necropsy team in the Department of Urology, University of Washington.
Footnotes
Salari K and Pollack J. DNA/RNA-Significance Analysis of Microarrays (DR-SAM). In preparation 2009.
References
- 1.Garmey EG, Sartor O, Halabi S, Vogelzang NJ. Second-line chemotherapy for advanced hormone-refractory prostate cancer. Clin Adv Hematol Oncol. 2008;6:118–22. 27–32. [PubMed] [Google Scholar]
- 2.Drake CG. Immunotherapy for metastatic prostate cancer. Urol Oncol. 2008;26:438–44. doi: 10.1016/j.urolonc.2007.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dreicer R. Current status of cytotoxic chemotherapy in patients with metastatic prostate cancer. Urol Oncol. 2008;26:426–9. doi: 10.1016/j.urolonc.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 4.Ahlers CM, Figg WD. ETS-TMPRSS2 fusion gene products in prostate cancer. Cancer Biol Ther. 2006;5:254–5. doi: 10.4161/cbt.5.3.2603. [DOI] [PubMed] [Google Scholar]
- 5.Attard G, Clark J, Ambroisine L, et al. Duplication of the fusion of TMPRSS2 to ERG sequences identifies fatal human prostate cancer. Oncogene. 2008;27:253–63. doi: 10.1038/sj.onc.1210640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mehra R, Tomlins SA, Yu J, et al. Characterization of TMPRSS2-ETS gene aberrations in androgen-independent metastatic prostate cancer. Cancer Res. 2008;68:3584–90. doi: 10.1158/0008-5472.CAN-07-6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perner S, Demichelis F, Beroukhim R, et al. TMPRSS2:ERG fusion-associated deletions provide insight into the heterogeneity of prostate cancer. Cancer Res. 2006;66:8337–41. doi: 10.1158/0008-5472.CAN-06-1482. [DOI] [PubMed] [Google Scholar]
- 8.Tomlins SA, Rhodes DR, Perner S, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–8. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 9.Tomlins SA, Laxman B, Varambally S, et al. Role of the TMPRSS2-ERG gene fusion in prostate cancer. Neoplasia. 2008;10:177–88. doi: 10.1593/neo.07822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoshimoto M, Joshua AM, Chilton-Macneill S, et al. Three-color FISH analysis of TMPRSS2/ERG fusions in prostate cancer indicates that genomic microdeletion of chromosome 21 is associated with rearrangement. Neoplasia. 2006;8:465–9. doi: 10.1593/neo.06283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morrissey C, True LD, Roudier MP, et al. Differential expression of angiogenesis associated genes in prostate cancer bone, liver and lymph node metastases. Clin Exp Metastasis. 2008;25:377–88. doi: 10.1007/s10585-007-9116-4. [DOI] [PubMed] [Google Scholar]
- 12.Lin DW, Coleman IM, Hawley S, et al. Influence of surgical manipulation on prostate gene expression: implications for molecular correlates of treatment effects and disease prognosis. J Clin Oncol. 2006;24:3763–70. doi: 10.1200/JCO.2005.05.1458. [DOI] [PubMed] [Google Scholar]
- 13.Klein CA, Schmidt-Kittler O, Schardt JA, Pantel K, Speicher MR, Riethmuller G. Comparative genomic hybridization, loss of heterozygosity, and DNA sequence analysis of single cells. Proc Natl Acad Sci U S A. 1999;96:4494–9. doi: 10.1073/pnas.96.8.4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holcomb IN, Grove DI, Kinnunen M, et al. Genomic alterations indicate tumor origin and varied metastatic potential of disseminated cells from prostate cancer patients. Cancer Res. 2008;68:5599–608. doi: 10.1158/0008-5472.CAN-08-0812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loo LW, Grove DI, Williams EM, et al. Array comparative genomic hybridization analysis of genomic alterations in breast cancer subtypes. Cancer Res. 2004;64:8541–9. doi: 10.1158/0008-5472.CAN-04-1992. [DOI] [PubMed] [Google Scholar]
- 16.Yang YH, Dudoit S, Luu P, et al. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–72. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 18.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Young JM, Endicott RM, Parghi SS, Walker M, Kidd JM, Trask BJ. Extensive copy-number variation of the human olfactory receptor gene family. Am J Hum Genet. 2008;83:228–42. doi: 10.1016/j.ajhg.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin B, Ferguson C, White JT, et al. Prostate-localized and androgen-regulated expression of the membrane-bound serine protease TMPRSS2. Cancer Res. 1999;59:4180–4. [PubMed] [Google Scholar]
- 22.Trask BJ. Genome Analysis: A Laboratory Manual. Vol. 4. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1999. pp. 303–413. [Google Scholar]
- 23.Sun J, Liu W, Adams TS, et al. DNA copy number alterations in prostate cancers: a combined analysis of published CGH studies. Prostate. 2007;67:692–700. doi: 10.1002/pros.20543. [DOI] [PubMed] [Google Scholar]
- 24.He WW, Sciavolino PJ, Wing J, et al. A novel human prostate-specific, androgen-regulated homeobox gene (NKX3.1) that maps to 8p21, a region frequently deleted in prostate cancer. Genomics. 1997;43:69–77. doi: 10.1006/geno.1997.4715. [DOI] [PubMed] [Google Scholar]
- 25.Dong JT. Chromosomal deletions and tumor suppressor genes in prostate cancer. Cancer Metastasis Rev. 2001;20:173–93. doi: 10.1023/a:1015575125780. [DOI] [PubMed] [Google Scholar]
- 26.Paris PL, Andaya A, Fridlyand J, et al. Whole genome scanning identifies genotypes associated with recurrence and metastasis in prostate tumors. Hum Mol Genet. 2004;13:1303–13. doi: 10.1093/hmg/ddh155. [DOI] [PubMed] [Google Scholar]
- 27.Perinchery G, Bukurov N, Nakajima K, et al. Loss of two new loci on chromosome 8 (8p23 and 8q12–13) in human prostate cancer. Int J Oncol. 1999;14:495–500. doi: 10.3892/ijo.14.3.495. [DOI] [PubMed] [Google Scholar]
- 28.Visakorpi T, Kallioniemi AH, Syvanen AC, et al. Genetic changes in primary and recurrent prostate cancer by comparative genomic hybridization. Cancer Res. 1995;55:342–7. [PubMed] [Google Scholar]
- 29.Boudeau J, Miranda-Saavedra D, Barton GJ, Alessi DR. Emerging roles of pseudokinases. Trends Cell Biol. 2006;16:443–52. doi: 10.1016/j.tcb.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 30.Ferrara N. VEGF and the quest for tumour angiogenesis factors. Nat Rev Cancer. 2002;2:795–803. doi: 10.1038/nrc909. [DOI] [PubMed] [Google Scholar]
- 31.Liu W, Laitinen S, Khan S, et al. Copy number analysis indicates monoclonal origin of lethal metastatic prostate cancer. Nat Med. 2009;15:559–65. doi: 10.1038/nm.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y, Cozzi PJ. MUC1 is a promising therapeutic target for prostate cancer therapy. Curr Cancer Drug Targets. 2007;7:259–71. doi: 10.2174/156800907780618338. [DOI] [PubMed] [Google Scholar]
- 33.Lord CJ, Ashworth A. Targeted therapy for cancer using PARP inhibitors. Curr Opin Pharmacol. 2008;8:363–9. doi: 10.1016/j.coph.2008.06.016. [DOI] [PubMed] [Google Scholar]
- 34.Rubin MA. Targeted therapy of cancer: new roles for pathologists--prostate cancer. Mod Pathol. 2008;21 (Suppl 2):S44–55. doi: 10.1038/modpathol.2008.11. [DOI] [PubMed] [Google Scholar]
- 35.Bjorkman M, Iljin K, Halonen P, et al. Defining the molecular action of HDAC inhibitors and synergism with androgen deprivation in ERG-positive prostate cancer. Int J Cancer. 2008 doi: 10.1002/ijc.23885. [DOI] [PubMed] [Google Scholar]
- 36.Sun PC, Uppaluri R, Schmidt AP, et al. Transcript map of the 8p23 putative tumor suppressor region. Genomics. 2001;75:17–25. doi: 10.1006/geno.2001.6587. [DOI] [PubMed] [Google Scholar]
- 37.Ash SC, Yang DQ, Britt DE. LYRIC/AEG-1 overexpression modulates BCCIPalpha protein levels in prostate tumor cells. Biochem Biophys Res Commun. 2008;371:333–8. doi: 10.1016/j.bbrc.2008.04.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Emdad L, Sarkar D, Su ZZ, et al. Astrocyte elevated gene-1: recent insights into a novel gene involved in tumor progression, metastasis and neurodegeneration. Pharmacol Ther. 2007;114:155–70. doi: 10.1016/j.pharmthera.2007.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li J, Zhang N, Song LB, et al. Astrocyte elevated gene-1 is a novel prognostic marker for breast cancer progression and overall patient survival. Clin Cancer Res. 2008;14:3319–26. doi: 10.1158/1078-0432.CCR-07-4054. [DOI] [PubMed] [Google Scholar]
- 40.Emdad L, Sarkar D, Su ZZ, et al. Activation of the nuclear factor kappaB pathway by astrocyte elevated gene-1: implications for tumor progression and metastasis. Cancer Res. 2006;66:1509–16. doi: 10.1158/0008-5472.CAN-05-3029. [DOI] [PubMed] [Google Scholar]
- 41.Olsen SR, Uhler MD. Inhibition of protein kinase-A by overexpression of the cloned human protein kinase inhibitor. Mol Endocrinol. 1991;5:1246–56. doi: 10.1210/mend-5-9-1246. [DOI] [PubMed] [Google Scholar]
- 42.Latil A, Chene L, Mangin P, Fournier G, Berthon P, Cussenot O. Extensive analysis of the 13q14 region in human prostate tumors: DNA analysis and quantitative expression of genes lying in the interval of deletion. Prostate. 2003;57:39–50. doi: 10.1002/pros.10272. [DOI] [PubMed] [Google Scholar]
- 43.Alumkal JJ, Zhang Z, Humphreys EB, et al. Effect of DNA Methylation on Identification of Aggressive Prostate Cancer. Urology. 2008 doi: 10.1016/j.urology.2007.12.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Esteller M. CpG island hypermethylation and tumor suppressor genes: a booming present, a brighter future. Oncogene. 2002;21:5427–40. doi: 10.1038/sj.onc.1205600. [DOI] [PubMed] [Google Scholar]
- 45.Maruyama R, Toyooka S, Toyooka KO, et al. Aberrant promoter methylation profile of prostate cancers and its relationship to clinicopathological features. Clin Cancer Res. 2002;8:514–9. [PubMed] [Google Scholar]
- 46.Burton TR, Gibson SB. The role of Bcl-2 family member BNIP3 in cell death and disease: NIPping at the heels of cell death. Cell Death Differ. 2009;16:515–23. doi: 10.1038/cdd.2008.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu K, Shimelis H, Linn DE, et al. Regulation of androgen receptor transcriptional activity and specificity by RNF6-induced ubiquitination. Cancer Cell. 2009;15:270–82. doi: 10.1016/j.ccr.2009.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Faus H, Meyer HA, Huber M, Bahr I, Haendler B. The ubiquitin-specific protease USP10 modulates androgen receptor function. Mol Cell Endocrinol. 2005;245:138–46. doi: 10.1016/j.mce.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 49.van Steenbrugge GJ, van Uffelen CJ, Bolt J, Schroder FH. The human prostatic cancer cell line LNCaP and its derived sublines: an in vitro model for the study of androgen sensitivity. J Steroid Biochem Mol Biol. 1991;40:207–14. doi: 10.1016/0960-0760(91)90184-7. [DOI] [PubMed] [Google Scholar]
- 50.Corey E, Brown LG, Quinn JE, et al. Zoledronic acid exhibits inhibitory effects on osteoblastic and osteolytic metastases of prostate cancer. Clin Cancer Res. 2003;9:295–306. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.