Summary
The eukaryotic circadian oscillators consist of autoregulatory negative feedback loops. However, little is known about the role of post-transcriptional regulation of RNA in circadian oscillators. In the Neurospora circadian negative feedback loop, FRQ and FRH form the FFC complex that represses frq transcription. Here we show that FFC also binds frq RNA and interacts with the exosome to regulate frq RNA decay. Consequently, frq RNA is robustly rhythmic as it is more stable when FRQ levels are low. Silencing of RRP44, the catalytic subunit of the exosome, elevates frq RNA levels and impairs clock function. In addition, rrp44 is a clock-controlled gene and a direct target of the WHITE COLLAR complex, and RRP44 controls the circadian expression of at least one ccg. Taken together, these results suggest that FFC and the exosome are part of a post-transcriptional negative feedback loop that regulates frq transcript levels and the circadian output pathway.
Introduction
The eukaryotic circadian oscillators are composed of autoregulatory negative feedback loops that function to regulate circadian gene expression (Allada et al., 2001; Bell-Pedersen et al., 2005; Dunlap, 1999; Sehgal, 2004; Young and Kay, 2001). In Neurospora, Drosophila, and mammals, heterodimeric complexes, consisting of two PAS domain-containing transcription factors, act as positive elements in the negative feedback loop by binding cis-elements in the promoter and activating the expression of negative elements. These negative elements repress their own transcription by inhibiting the activity of the positive elements to close the negative feedback loop. In addition, the positive elements also activate the transcription of clock-controlled genes (ccgs) to drive rhythmic gene expression. Our current models of the eukaryotic clock mechanisms are largely transcription-based and very little is known about the role of post-transcriptional regulation of RNA in clocks. Post-transcriptional regulation has been shown to regulate circadian clocks and rhythm of mRNA stability in animals and plants (Garbarino-Pico and Green, 2007; Lidder et al., 2005; So and Rosbash, 1997). The mRNA half-life of the Drosophila clock gene period (per) is rhythmic and the 3′UTR-mediated mRNA decay of mouse Per3 is required for its mRNA oscillation (Kwak et al., 2006; So and Rosbash, 1997). However, the post-transcriptional mechanism involved is unknown.
Despite the evolutionary distances between the filamentous fungus Neurospora crassa and higher eukaryotes, mechanisms of their circadian oscillators are remarkably similar (Dunlap, 2006; Heintzen and Liu, 2007; Liu and Bell-Pedersen, 2006). In the Neurospora circadian negative feedback loop, the heterodimeric WCC complex formed by two PAS-domain transcription factors WHITE COLLAR-1 (WC-1) and WC-2 acts as the positive element and the complex of FREQUENCY (FRQ) and FRH (FRQ-interacting RNA helicase) functions as the negative element. In darkness, WCC activates the transcription of the frq gene by binding to the Clock box (C-box) in the frq promoter (Belden et al., 2007; Cheng et al., 2002; Cheng et al., 2001b; Crosthwaite et al., 1997; Froehlich et al., 2003; He et al., 2006; He and Liu, 2005; Loros and Dunlap, 2001). After the synthesis of FRQ protein, FRQ self associates and forms a complex (called FFC) with FRH. FFC represses WCC activity on frq transcription by promoting WC phosphorylation (Aronson et al., 1994a; Cha et al., 2008; Cheng et al., 2005; Cheng et al., 2001a; He et al., 2006; He and Liu, 2005; He et al., 2005; Huang et al., 2007; Schafmeier et al., 2005).
Microarray and other methods identified more than 150 clock- controlled genes (ccgs) which are involved in a wide variety of processes (Correa et al., 2003; de Paula et al., 2007; Liu and Bell-Pedersen, 2006; Nowrousian et al., 2003). In all cases examined, the circadian oscillator functioned normally in strains with disrupted copies of the ccgs, indicating that they are part of the output pathway and are not part of the oscillator. Some of these ccgs contain promoter elements similar to the frq C-box or ACE (activating clock element found in ccg-2), suggesting that they are direct targets of WCC or unknown rhythmically-activated transcription factors, respectively (Bell-Pedersen et al., 1996a; Correa et al., 2003; He and Liu, 2005; Lewis et al., 2002). How the circadian oscillator regulates other ccgs is unknown.
FRH is a homolog of Mtr4p in Saccharomyces cerevisiae and has sequence homologs in from fungi to mammals (Cheng et al., 2005). Mtr4p is an essential nuclear co-factor for the yeast exosome complex, which is a large complex consisting of several 3′ -> 5′ exonucleases and an important regulator of RNA (rRNA, mRNA, and noncoding RNA) metabolism by mediating 3′ to 5′ processing and degradation (Houseley et al., 2006; LaCava et al., 2005; Vanacova et al., 2005). The diversity of functions, including cell cycle and development, assumed by the exosome in RNA degradation is analogous in eukaryotic organisms to the proteasome in protein degradation (Lorentzen and Conti, 2006 ).
In Neurospora, the entire pool of FRQ is in complex with FRH (Cheng et al., 2005). Down-regulation of frh abolished circadian rhythmicity and resulted in high frq RNA levels. These results suggest that FRH functions as the circadian negative element. Despite its importance in the clock, the circadian role of FRH as an RNA helicase is not known. Approximately 40% of FRH are in complex with FRQ (Cheng et al., 2005). Unlike Mtr4p in yeast, a significant portion of FRH was also found in the cytosol, consistent with the fact that more than 90% of FRQ is cytoplasmically localized (Cheng et al., 2005; Schafmeier et al., 2005). Furthermore, due to the rhythmic FRQ levels, the amounts of FFC complex oscillate daily. These results raise the possibility that FFC may have a role in rhythmic RNA processing. In this study, we show that in addition to its role as a negative element in the transcription-based circadian negative feedback loop, FFC regulates frq RNA post-transcriptionally by mediating its rhythmic decay through the exosome. Down-regulation of the core catalytic subunit of the Neurospora exosome RRP44 results in elevated frq RNA and impairs clock functions. rrp44 is also a ccg that is directly regulated by the WCC. In addition to its role in regulating frq RNA, RRP44/exosme is also important for the rhythmic expression of some ccgs. Therefore, FFC and the exosome are part of a novel post-transcriptional negative feedback loop, which is interlocked with the transcriptional negative feedback loop to regulate frq levels and circadian gene expression.
Results
FRH negatively regulates frq mRNA levels post-transcriptionally
To understand the role of FRH in regulating frq mRNA levels, frh expression was down-regulated by inducibly expressing an inverted-repeat specific for frh in a wt,dsfrh strain with quinic acid (QA). As shown in Figure 1A, down-regulation of FRH resulted in a dramatic increase of frq mRNA levels. Similar to our previous results (Cheng et al., 2005), FRQ protein levels were low under this condition despite elevated frq mRNA levels. The low FRQ levels in the induced knockdown strain are most likely due to FRQ instability when it cannot form a complex with FRH. FRQ levels are similarly low in frq mutants in which FRQ cannot interact with FRH (Cheng et al., 2005). In addition, WC-1 and WC-2 levels exhibited modest reduction after the down-regulation of FRH, suggesting that like FRQ, FRH also participates in the positive feedback loops to up-regulate WC expression. Consistent with FRH as being a negative component of the circadian negative feedback loop, hypophosphorylation of WC-1 was also observed when FRH was down-regulated.
Figure 1.
FRH regulates frq RNA post-transcriptionally. (A) Northern and Western blot analyses showing the levels frq RNA, FRQ and WC levels in dsfrh cultures with/without the addition of QA. The cultures were harvested at the indicated time points. (B) Northern blot analyses showing the levels of frq RNA in the dsfrh strain. The densitometric analyses of the results are shown in the bottom. The asterisk indicates the p value (a student t- test) < 0.01. (C &D) ChIP assays using the WC-2 antibody showing the relative binding of the WCC to the frq C-box in the indicated strains. The densitometric analysis of the results from three experiments are shown in the bottom.
To further confirm the role of FRH in the circadian negative feedback loop, chromatin immunoprecipiation (ChIP) assay with the WC-2 antiserum was performed to examine the levels of WCC binding to the C-box of the frq promoter. As shown in Figure 1B and S1, WCC binding in the absence of the QA was rhythmic and peaked at DD14, consistent with previously results (He et al., 2006; Huang et al., 2007). But the down-regulation of FRH in the wt,dsfrh strain resulted in constantly high levels of WCC C-box binding. Together with our previous results, these data indicate that FRH is an essential component in the Neurospora circadian negative feedback loop.
To determine whether these high levels of frq mRNA after down-regulation of FRH are solely due to an impaired circadian negative feedback loop, we compared the frq mRNA levels in the wild-type, wt,dsfrh, frq9, and frq9,dsfrh strains. In frq9 strains, a premature stop-codon mutation results in a truncated FRQ protein and subsequent disruption of the circadian negative feedback loop (Aronson et al., 1994b). The addition of QA to strains without the dsfrh construct did not affect the levels of frq (Figure 1C), indicating that the effects triggered by dsfrh were not due to a non-specific QA response. As predicted, levels of frq mRNA in the frq9 strains were higher than those in the wild-type strains in the absence of QA. However, the knock-down of FRH in the wild-type strain resulted in frq transcript levels which were significantly higher than those in the frq9 strain. In addition, the knock-down of FRH in the frq9 strain also resulted in a significant increase of frq mRNA levels. ChIP assays in the frq9, dsfrh strain showed that the down-regulation of FRH did not result in a further increase of WCC binding to the C-box of frq promoter (Figure 1D & S1), indicating that the high frq levels after FRH down-regulation is due to both the loss of the circadian negative feedback loop and a post-transcriptional mechanism involving FRH.
FRH promotes frq mRNA decay and interacts with the exosome components
Because Mtr4p functions as a cofactor of the yeast exosome in RNA processing and degradation in yeast and other eukaryotes, we wondered whether FRH promotes frq mRNA decay. After screening several transcription inhibitors, we found that thiolutin, a transcription inhibitor used successfully in yeast and other organisms (Das et al., 2003), efficiently blocks transcription in Neurospora. As shown in Figure 2A, the addition of thiolutin to the wild-type strain resulted in rapid decrease of frq levels in the absence of QA. However, when frh expression was knocked-down, frq degradation rate was significantly reduced. QA had no affect on frq levels in the wild-type strain without the dsfrh construct (Figure 1B). These data indicates that FRH promotes the degradation of frq mRNA. Thus, the high levels of frq mRNA after the down-regulation of frh is in part due to increased frq mRNA stability.
Figure 2.
FRH regulates the frq RNA decay by the exosome. (A) Northern blot analysis showing the decay of frq RNA after the addition of thiolutin. Due to high levels of frq RNA after down-regulation of FRH, a shorter exposure of the Northern blots for the QA treated samples was used. The densitometric analysis of the results from three independent experiments are shown in the bottom. (B) Northern blot analysis showing the results of the RNAse H assay for frq RNA. RNA samples from the wild-type and dsfrh strains were used. Due to high levels of frq RNA in the dsfrh samples, less RNA for the dsfrh samples was applied to bring frq signals to comparable levels for both sets of the samples. (C) Western blot analysis showing the results of immunoprecipitation assay using the FRH antibody. Immunoprecipitation using the FRH pre-immune (PI) serum was used as the control. Myc-RRP44, Myc-RRP6 and Myc-QIP (the negative control) strains were used.
Recent studies in yeast have shown that Mtr4p is part of the TRAMP complex, which polyadenylates rRNAs and tRNAs and mediates the processing of RNA by the exosome (LaCava et al., 2005; Vanacova et al., 2005). Polyadenylation of pre-rRNA is significantly increased in yeast mtr4 mutants, suggesting that Mtr4p couples polyadenylated RNAs to degradation (Houseley and Tollervey, 2006). These studies prompted us to investigate whether FRH regulates frq mRNA stability using a similar mechanism. Thus we performed a RNase H assay to compare the poly(A) length of frq RNA in the dsfrh strain in the presence or absence of QA. The RNA samples were first incubated with a DNA oligonucleotide probe complementary to a region upstream of the frq 3′UTR and were then digested by RNase H (which cleaves DNA/RNA hybrids) so that the poly(A) tail and the 3′UTR of frq RNA were removed from the full-length mRNA. The RNA samples were then analyzed by Northern blot analysis using a probe specific for the frq 3′UTR. Oligo d(T) was also added to some of samples before the RNase H treatment, so that the poly(A) tail was removed. As shown in Figure 2B, the down-regulation of frh led to significant increase of poly(A) tail length of frq RNA, suggesting that FRH regulate the degradation of frq mRNA by coupling the degradation of polyadenylated frq mRNA to the exosome complex.
To further confirm the role of FRH in the exosome-mediated RNA processing, we examined the interaction between FRH and two key exosome components, RRP44 and RRP6 (Buttner et al., 2006). A construct that expresses either Myc-tagged RRP44 or RRP6 was transformed into a wild-type strain and the resulting strains were used in immunoprecipitation (IP) assays with an FRH antibody. As shown in Figure 2C, the immunoprecipitation of FRH specifically pulled down both Myc-RRP44 and RRP6. On the other hand, FRH did not pull-down Myc-QIP, a component of the Neurospora RNAi pathway (Maiti et al., 2007), despite its high expression level. The substoichiometric interaction between FRH and RRP44 and RRP6 indicates that FRH is not a core component of the exosome complex, consistent with its predicted role as a co-factor.
FFC interacts with the exosome and associates with frq mRNA to promote frq mRNA decay as part of the post-transcriptional negative feedback loop
All available FRQ protein is complexed with a significant portion of FRH to form FFC (Cheng et al., 2005), this raises the possibility that FFC also interacts with the exosome and regulates the decay of frq mRNA. To test this we examined the interaction between FRQ and exosome components. As shown in Figure 3A & S2, immunoprecipiation of FRQ specifically pulled-down Myc-tagged RRP44 and RRP6, indicating that FFC interacts with the exosome. This result suggests that FFC may have functions outside of the transcription-based circadian negative feedback loop.
Figure 3.
FFC interacts with the exosome components, associates with frq RNA, and promotes the rhythmic decay of frq RNA. (A) Western blot analysis showing the results of immunoprecipitation assay using the FRQ antibody. Immunoprecipitation using the FRQ pre-immune (PI) serum was used as the control. Myc-RRP44 and Myc-RRP6 strains were used. (B) RT-PCR result showing that FFC is associated with frq RNA. The cell extracts were immunoprecipitated with our FRQ antibody and RNA was extracted from the immunoprecipitates. RT-PCR was performed with/without RT (reverse transcriptase). (C &D) Northern blot analyses showing the frq RNA decay in the wild-type and frq9 strains (C) or at the indicated time points in the wild-type strain (D). Thiolutin was added to LL cultures at time 0. (E) Western blot analysis showing the levels of FRQ at DD12 and DD22. (F) The frq 3′ UTR confers the FRQ-dependent RNA decay for a reporter gene. Top, a diagram showing the firefly luciferase gene reporter construct. Bottom: Q-PCR results showing the levels of luc RNA after the addition of thiolutin. Three independent experiments were performed. The asterisk indicates the p value (a student t- test) < 0.01.
To further test this hypothesis, we investigated whether FFC can associate with RNA in vivo by immunoprecipitation (Peritz et al., 2006). After the preparation of cell extracts of the wild-type strain, FRQ protein was immunoprecipitated using the FRQ antibody. RNA and then cDNA were then prepared from the immunoprecipitates. We analyzed the cDNA samples for the presence of frq cDNA by PCR. As shown in Figure 3B, frq cDNA was only amplified from the FRQ IP sample and only when the reverse transcriptase (RT) was used to produce cDNAs. In contrast, no product was detected in a mock (without antibody) and a control (with tubulin antibody) IP samples, indicating that frq RNA associates specifically with FFC. In addition, only background levels of wc-2 and ccg-1 RNA were found to be non-specifically associated with FFC (Supplemental Figure 3), indicating that not all mRNAs bind to FFC.
The association of FFC and frq mRNA suggests that FFC can regulate frq mRNA decay. To test this, we compared the frq mRNA decay between the wild-type strain and frq9 strain. As shown in Figure 3C, frq mRNA in the frq9 strain was significantly more stable than in the wild-type strain, indicating that FRQ promotes frq mRNA decay. It is worth noting that in the frq9 strain the premature stop codon in the FRQ open reading frame should also trigger the nonsense-mediated decay pathway, resulting in increased rate of frq mRNA degradation. Thus, the post-transcriptional regulation of frq by FFC contributes to the high levels of frq mRNA in the frq9 strain.
The comparison of frq RNA and FRQ protein oscillations previously revealed that frq RNA levels rise and fall rapidly during a circadian cycle, suggesting that frq is regulated post-transcriptionally (Garceau et al., 1997). The rapid rising phase of frq RNA rhythm corresponds to time points when FRQ levels are low while the FRQ levels are relatively high when levels of frq RNA rapidly decrease. Since the amount of FFC oscillates daily due to the daily rhythm of FRQ, we then asked whether the stability of frq RNA exhibits a circadian rhythm. We compared frq RNA stability of the wild-type strain at two different time points (DD12 and DD20), which have similar amounts of frq RNA but have significantly different amounts of FRQ protein (Figure 3D & 3E). At DD12, FRQ protein levels are low while the amount of frq mRNA increases towards its peak level. At DD20, FRQ protein is near its peak level whereas frq mRNA amount is rapidly decreasing (Garceau et al., 1997). Despite similar frq levels, after the addition of thiolutin, frq RNA levels decreased significantly faster at DD20 than DD12, suggesting that frq RNA is less stable when FRQ levels are high. These results provide a molecular explanation for the rapid rise and fall of frq RNA during a circadian cycle. When FRQ levels are low, frq RNA is more stable which leads to the accumulation of frq mRNA. In contrast, during peak levels of FRQ frq RNA is less stable which results in decline in frq mRNA.
To further demonstrate the role of FRQ in the posttranscriptional control of its own transcript, we created a construct in which the frq 3′ UTR is placed downstream of the luciferase ORF (LUC) (Gooch et al., 2008) under the control of the qa-2 promoter (Figure 3F). As a control, we made a construct in which the 3′ UTR of actin gene is placed downstream LUC. These constructs were transformed into the wild-type and frq10 strains. In the presence of QA, the qa-2 promoter is constitutively activated and luc RNA stability was compared in these two strains by the addition of thiolutin. As expected, the decay rate of luc RNA was significantly slower in the frq10, LUC-frq strain than that in the wt, LUC-frq strain, while there was no significant difference in luc RNA decay rate in the frq10, LUC-actin and wt, LUC-actin strains. This result indicates that the cis-element regulating frq RNA stability is located in its 3′ UTR. Together, these results uncovered a novel post-transcriptional negative feedback loop that is interlocked with the transcription-based feedback loop. Within these negative feedback loops, FFC performs dual roles in the frq transcription inhibition process by inhibiting WCC activity while promoting the post-transcriptionally decay of frq RNA.
Down-regulation of RRP44, the core exosome catalytic subunit, impairs frq mRNA decay and circadian clock function
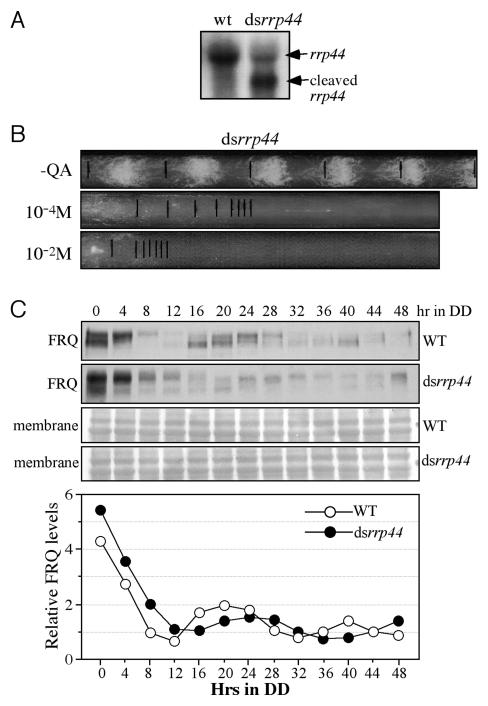
The results presented above suggest that FFC promotes the frq RNA decay by the exosome. To directly examine the role of exosome in the clock, a construct (dsrrp44) that expresses an inverted repeat specifically for rrp44 was introduced into a wild-type strain. RRP44 (also called Dis3), a member of the RNase II family that is essential for cell survival, is the core catalytic subunit of yeast exosome (Dziembowski et al., 2007; Schneider et al., 2007). As shown in Figure 4A, the expression of the dsrrp44 construct resulted in the significant decrease of full-length rrp44 transcript and the appearance of a truncated rrp44 RNA. We also examined the circadian conidiation rhythm of the dsrrp44 strain by race tube assay. Although a robust rhythm of conidiation was observed in the absence of QA, the rhythm was abolished when rrp44 was down-regulated (Figure 4B). In addition, the presence of QA also resulted in significant reduction of growth and reduction of aerial hyphae and conidia (Figure 4B).
Figure 4.
Down-regulation of rrp44 impairs the circadian clock function. (A) Northern blot analysis showing the levels of rrp44 in the wild-type and dsrrp44 strains. (B) Race tube assay showing the conidiation rhythm of the dsrrp44 strain in the presence of different concentrations of QA. (C) Western blot analyses showing the circadian oscillation of FRQ in the wild-type and dsrrp44 strains.
To investigate whether the lack of overt circadian rhythm in the dsrrp44 strain is due to impaired clock function, we examined the rhythmic FRQ expression in liquid cultures grown in constant darkness. Although a low amplitude rhythm of FRQ expression could still be observed in the dsrrp44 strain, the period of rhythm was 4-6 hrs longer than that in the wild-type strain (Figure 4C). Note that the gene silencing efficiency in liquid culture is not as effective as on race tube, as reflected by only partial reduction of rrp44 RNA (Figure 4A) and a modest growth inhibition in liquid cultures.
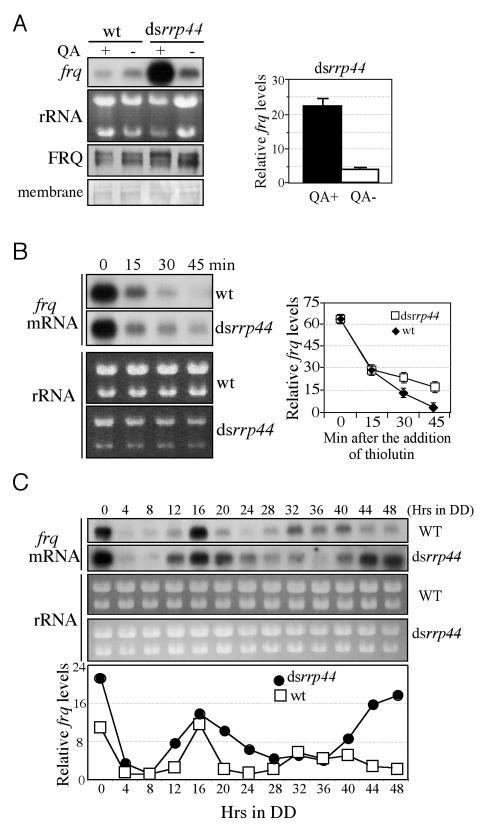
Regulation of RNA turnover is an important process in determining levels of gene expression. To examine the role of RRP44 and the exosome in regulating the clock, frq RNA levels were measured by Northern blot analysis (Figure 5A). As expected, the down-regulation of rrp44 resulted in a significant increase of frq RNA level and frq RNA was also more stable (Figure 5B). Furthermore, the frq RNA levels in the dsrrp44 strain were elevated in constant darkness and oscillated with a period 4-6 hrs longer than observed in the wild-type strain (Figure 5C). The longer period of the frq mRNA oscillation in the silenced rrp44 mutant is consistent with its longer FRQ protein oscillation. ChIP assay using WC-2 antibody showed that the down-regulation of RRP44 did not increase WCC binding to the frq promoter, suggesting that the effects of RRP44 on frq RNA is mostly post-transcriptional (Supplemental Figure 4). Taken together, these results demonstrate that RRP44 play an important role in the post-transcriptional negative feedback loop by regulating frq RNA decay.
Figure 5.
Silencing of rrp44 expression results in the elevation of frq RNA by blocking its decay. (A) Northern and Western blot analysis showing the levels of frq RNA and FRQ in the wild-type and dsrrp44 strains. The densitometric analysis of the frq RNA levels from three independent experiments were shown on the right. (B) Northern blot analyses showing the circadian oscillation of frq RNA in the wild-type and dsrrp44 strains. (C) Northern blot analyses showing the frq RNA decay in the wild-type and dsrrp44 strains after the addition of thiolutin. The densitometric analysis of the frq RNA levels from three independent experiments were shown on the right.
Circadian regulation of rrp44 expression by the WCC
Since RRP44 is the catalytic subunit of exosome, we then investigated whether the expression of rrp44 is also under circadian control. Levels of rrp44 and frq RNA were monitored by Northern blot analyses in the wild-type, frq7 (a long period mutant) and frq9 strains. As shown in Figure 6A, levels of rrp44 RNA oscillated in both the wild-type and frq7 strains with a period that was consistent with the genotype of the strains. In addition, the phases of rrp44 rhythms were very similar to those of frq. In the frq9 strain, the rhythmic expression of rrp44 and frq was abolished. These results demonstrated that the expression of rrp44 is clock-controlled.
Figure 6.
rrp44 is a ccg directly controlled by the WCC. (A) Densitometric analysis of the Northern blot results showing the transcript levels of frq and rrp44 over two circadian cycles in the wild-type, frq7 and frq9 strains. (B) Northern blot analysis showing the levels of frq and rrp44 RNA in the wild-type, frq10, wc-1 and wc-2 strains. Cultures were harvested in LL. (C) Sequence alignment of the C-box-like elements in the promoters of frq, rrp44 and other genes. (D) ChIP assays using WC-2 antibody showing that WCC binds to the rrp44 and frq promoters specifically and rhythmically in the wild-type strain. For the control, a wc-2ko strain was used for the ChIP assay.
The near identical phase relationship between rrp44 and frq RNA suggests that their circadian expression is regulated by the same mechanism. To test this hypothesis, we compared the levels of rrp44 in the frq10, wc-1 and wc-2 mutants. As shown in Figure 6B, the rrp44 RNA level was high in the frq10 strain but low in the wc mutants, indicating that rrp44 transcription is regulated by the WCC. However, unlike the extremely low frq RNA levels seen in the wc mutants, the rrp44 levels were found to be maintained at a modest level, suggesting that rrp44 transcription was also independently regulated by other mechanisms. This result is also consistent with rrp44 being an essential gene in Neurospora.
To further determine whether rrp44 is a direct target of WCC, we searched the promoter of rrp44 for C-box like elements and one such element was identified (Figure 6C). To show that WCC binds to this element in vivo, ChIP assays were performed using the WC-2 antibody and the immunoprecipitated DNA was amplified by primers flanking the putative C-box. As shown in Figure 6D and S5, WCC bound specifically and rhythmically to the C-box-like element in the rrp44 promoter in a manner that was very similar to the frq C-box by peaking near DD14. Together, these results demonstrate that rrp44 is a ccg directly activated by WCC.
The exosome regulates the rhythmic expression of at least one ccg
The rhythmic expression of rrp44 suggests that the exosome activity is rhythmic. Since exosome plays a major in role in RNA degradation, we hypothesized that the rhythmic activity of exosome will contribute to the rhythmic gene expression in Neurospora. Although some of the ccgs have either C-box elements or activating clock element (ACE, discovered in ccg-2) in their promoters, a majority of ccgs lack both of these elements (Bell-Pedersen et al., 1996a; Correa et al., 2003; He and Liu, 2005; Lewis et al., 2002; Liu and Bell-Pedersen, 2006). To investigate whether the exosome contributes to the rhythmic levels of some ccgs at the post-transcriptional level, we surveyed the transcript levels of a few known ccgs after down-regulation of rrp44 (Bell-Pedersen et al., 1996b; Shinohara et al., 1998). We found that the RNA level of ccg-8, which encodes a transcriptional repressor-like protein, respectively, was significantly elevated in LL in the presence of QA in the dsrrp44 strain (Figure 7A and supplemental Figure 6). On the other hand, the RNA levels in LL of other ccgs (such as ccg-7, ccg-9), wc-1 and wc-2 remained unchanged after the silencing of rrp44, indicating that the exosome only regulates some mRNAs. In addition, the knock-down of rrp44 also resulted in the increase of stability of ccg-8 RNA (Figure 7B). These results suggest that the exosome post-transcriptionally regulates the levels of at least one ccg.
Figure 7.
RRP44 is required for the rhythmic expression of some ccgs. (A) Northern blot analysis showing the levels of a few known ccgs, wc-1 and wc-2 in the wild-type and dsrrp44 strains. The levels of RNA in the -QA wild-type samples was used as the bench mark to measure relative RNA levels in other samples. The error bars are standard deviations of RNA levels from 4-6 independent experiments. (B) Northern blot analysis showing the ccg-8 RNA decay in the wild-type and dsrrp44 strains after the addition of thiolutin. The densitometric analysis of the results from three independent experiments were shown in the bottom. (C) Northern blot results showing the RNA levels of ccg-7, ccg-8 and ccg-9 over two circadian cycles in the wild-type and dsrrp44 strains. QA was added to all cultures. (D) A model depicting the interlocked transcriptional and post-transcriptional negative feedback loops in the Neurospora clock. The question mark indicates ccgs that are not the direct targets of WCC.
To further determine the role of the exosome in the rhythmic expression of ccgs, we compared their rhythmic expression in DD in the wild-type and dsrrp44 strains. While the levels of ccg-7, ccg-8 and ccg-9 transcripts were rhythmic in the wild-type strain, their rhythmic expression was strongly perturbed in the dsrrp44 strain (Figure 7C and supplemental Figure 7). Interestingly, although the down-regulation of rrp44 did not significantly affect the levels of ccg-9 in LL, its level was significantly elevated in DD in the dsrrp44 strain. On the other hand, although the levels of ccg-7 were comparable in both strains, its rhythmic expression was severely impaired in the dsrrp44 strain. These results suggest that exosome can directly (for ccg-8) or indirectly (for ccg-7) regulate the rhythmic expression of some ccgs. Thus, exosome is also required for the proper function of the circadian output pathway.
Discussion
Interlocked transcriptional and post-transcriptional circadian negative feedback loops
The transcription-based negative feedback loops have been shown to be essential for the eukaryotic circadian oscillators. In this study, we showed that a post-transcriptional negative feedback loop that is interlocked with the transcriptional loop also plays an important role in the Neurospora circadian clock (Figure 7D). In both of these negative feedback loops, FFC acts as the negative element. In the transcriptional loop, FFC inhibits WCC activity and consequently represses frq transcription, by promoting the phosphorylation of the WC proteins (He et al., 2006; He and Liu, 2005; He et al., 2005; Schafmeier et al., 2005). In the post-transcriptional loop, FFC promotes frq RNA decay through the exosome. Thus, the decrease in frq RNA is a combination of inhibition of WCC activity by FFC and the FFC-mediated degradation of frq RNA via the exosome. This conclusion is supported by several lines of evidence.
First, FRQ associates with the core exosome components and FFC binds to frq RNA. Because FRQ lacks a putative nucleic acid binding domain, the interaction between FRQ and frq RNA is most likely mediated by FRH, a RNA helicase. Second, FRQ promotes frq RNA decay and the level of FRQ is a determinant of frq RNA stability, resulting in a circadian rhythm of frq RNA stability. frq RNA is more stable in the frq9 strain at a time point when FRQ level is low (Figure 3). Third, the down-regulation of FRH also results in stabilization of frq RNA and an increase of its poly(A) length, suggesting the involvement of the exosome in frq RNA decay (Figure 2). Fourth, FRQ and WCC regulate the expression of rrp44, which encodes the catalytic subunit of exosome. Finally, the down-regulation of RRP44 led to an increase of frq RNA and severe impairment of the conidiation and molecular rhythms. Taken together, these results strongly suggest that FFC and the exosome are part of a post-transcriptional negative feedback loop that regulates frq RNA levels. Thus, both the transcriptional and the post-transcriptional processes mediated by FFC contribute to the circadian negative feedback loop process. When FRQ levels are high, FFC inhibits WCC activity and promotes rapid degradation of frq RNA. When FRQ levels are low, WCC activates frq transcription and frq RNA is more stable. The combination of these two actions on frq RNA results in the observed rapid increase and decrease in frq levels and a robust frq rhythm during a circadian cycle (Garceau et al., 1997).
Based on our results, we propose that the FFC may recruit frq RNA to the exosome for degradation. Although the biochemical function of FRQ in the post-transcriptional regulation is presently unclear, it is possible that FRQ can promote the RNA helicase activity of FRH for certain RNA substrates. On the other hand, FRQ may regulate the intracellular localization of FRH due to their physical interaction. Since the levels of FRH do not exhibit a circadian rhythm (Cheng et al., 2005), the rhythmic expression of FRQ may drive the rhythmic modulation of FRH activity by forming FFC. It should be noted that the down-regulation of FRH in the frq9 strain also resulted in the increase of frq RNA levels (Figure 1B), suggesting that FRH and the exosome can also function independently of FRQ and that the role of FRQ on FRH activity is only regulatory. In addition, there is still a substantial frq RNA degradation after the knock-down of FRH (Figure 2A). While this is part due to partial silencing of frh expression, it is likely that other pathways can also regulate frq RNA degradation.
Like FRQ, a putative RNA-binding protein NONO was found to associate with mouse PER1 and is important for clock functions in mammals and fly (Brown et al., 2005). Thus, PER, via its interaction with NONO, may also participate in the post-transcriptional regulation of RNA in animals. Importantly, it has been previously demonstrated that the Drosophila per is regulated at both transcriptional and post-transcriptional levels and the latter makes a major contribution to per mRNA oscillation (So and Rosbash, 1997). Like frq, the half-life of per mRNA also exhibits a rhythm over the course of the circadian cycle and per is more stable during the rising phase and less stable during the declining phase. This indicates that per RNA decay is under circadian control. In mammals, the 3′UTR-mediated mRNA decay of mPer3 is important for its mRNA cycling (Kwak et al., 2006). In Arabidopsis, two ccgs, CCL and SEN1, also exhibit circadian control of their mRNA stability (Lidder et al., 2005). Therefore, similar to the Neurospora system, post-transcriptional negative feedback loops may also exist in animal and plant clocks.
rrp44, the exosome catalytic subunit, is a ccg that feeds back to the circadian oscillator and controls the expression of some ccgs
Our data here demonstrated that knock-down of RRP44 led to reduced mRNA degradation for frq and at least one ccg in Neurospora. We showed here that rrp44 is rhythmically expressed and its circadian regulation is directly controlled by the WCC. Unlike other known ccgs in Neurospora, RRP44 feeds back into circadian oscillator by regulating the levels of frq RNA levels. Down-regulation of rrp44 impairs clock function and results in the elevation of frq RNA. Together with FFC, RRP44 acts as a negative element in the post-transcriptional negative feedback loop. The rhythmic expression of rrp44 also suggests that the activity of the Neurospora exosome is rhythmic. Thus, the rhythmic activity of the exosome may contribute to rhythmic frq RNA stability.
In addition to its role in the post-transcriptional negative feedback loop, the exosome also determines the circadian gene expression for at least one ccg. By examining the effect of rrp44 knock-down on the circadian expression of some known ccgs, we found that RRP44 plays a major role in controlling the rhythmic expression of ccg-8 and ccg-9 by regulating their RNA decay. The elevated RNA levels of these two genes after the silencing of rrp44 suggest that they are directly regulated by the exosome. Notably, the circadian oscillations of these genes were abolished when rrp44 was silenced. On the other hand, although the silencing of rrp44 did not affect the levels of ccg-7 RNA, its circadian expression was also impaired, suggesting that ccg-7 is indirectly regulated by the exosome. These results also suggest that the post-transcriptional control of some ccgs by the exosome is an important part of the Neurospora circadian output pathway (Figure 7D).
Several lines of evidence also suggest the involvement of post-transcriptional regulation in other eukaryotic circadian systems. A few genes involved in RNA metabolism from plant to animals were previously shown to be under the circadian control (Garbarino-Pico and Green, 2007). In Arabidopsis, AtGRP7 is a circadian-regulated RNA-binding protein that regulates the alternative splicing of its own transcript (Heintzen et al., 1997; Staiger et al., 2003). In animals, Nocturnin, a circadian-regulated gene originally identified in Xenopus, is an mRNA deadenylase that confers resistance to diet-induced obesity in mice (Garbarino-Pico and Green, 2007; Green et al., 2007). The RNA-binding protein LARK, which exhibits circadian rhythms despite its constant RNA levels in animals, regulates the circadian output in Drosphila and post-transcriptionally regulates the expression of mPER1 in mice (Kojima et al., 2007; Newby and Jackson, 1996). Together with our results presented here, these studies indicate that the post-transcriptional mechanisms play important roles in the eukaryotic circadian clocks. The conservation of the circadian mechanisms from fungi to animals suggests that the highly conserved exosome may be an important post-transcriptional regulator in other eukaryotic clocks.
Experimental Procedures
Strains and growth conditions
The 87-3 (ras-1bd, a) strain was used as the wild-type strain. frq7 is a strain showing extra long circadian period (~28hr). The frq9 strain bears a frameshift mutation in the frq ORF (Aronson et al., 1994b). 301-6 strain (ras-1bd, his-3, A) was used as host strain for his-3 targeting constructs. Liquid cultures were grown in minimal medium (1X Vogel’s, 2% glucose). When QA was used, liquid cultures were grown in 0.01 M QA (pH 5.8), 1X Vogel’s, 0.1% glucose, and 0.17% arginine. Race tube medium contained 1X Vogel’s, 0.1% glucose (0% when QA was used), 0.17% arginine, 50 ng/mL biotin, and 1.5% agar.
The dsrrp44 strain was generated by introducing a plasmid expressing a rrp44-specific RNA hairpin into a wild-type strain (Cheng et al., 2005). The plasmid pdsrrp44 containing the hairpin sequence was constructed by inserting an inverted repeat (~500 bp) corresponding to rrp44 gene downstream of the qa-2 promoter. The resulting plasmid was targeted to the his-3 locus by transformation into 301-6 (bd, his-3, A). For the luciferase reporter construct, a PCR fragment containing the fusion between the luciferase ORF (from pfrq-luc-I) (Gooch et al., 2008) and 3′ UTR of frq (1.1 kb) or actin (0.1kb) was inserted downstream of the qa-2 promoter into the vector pDE3qa-2. The resulting construct was transformed into a bd, his-3 and a bd, frq10, his-3 strains at the his-3 locus.
For mRNA decay assay, Neurospora mats were cut into discs and transferred to flasks with liquid medium with or without QA (10-2 M QA). After growth overnight, thiolutin (a gift from Pfizer co., final concentration 12μg/ml) was added. Afterwards, the tissues were harvested at different time points, and total RNA was extracted and subjected for Northern blot analyses or reverse transcription and quantitative real-time PCR (Q-PCR).
Protein and RNA analyses
Protein extraction, western blot analysis, and immunoprecipitation assays were performed as previously described (Cheng et al., 2001a; Garceau et al., 1997). Equal amounts of total protein (40 μg) were loaded in each protein lane of SDS-PAGE (7.5% SDS-PAGE gels containing a ratio of 37.5:1 acrylamide/bisacrylamide). RNA extraction and northern blot analyses were performed as described previously (Aronson et al., 1994a). Equal amounts of total RNA (20 μg) were loaded onto agarose gels for electrophoresis, and the gels were blotted and probed with RNA probes specific for the gene of interest. To compare protein or RNA levels from different strains, experiments were performed side-by-side and the protein or RNA samples were transferred to the same membrane for western or northern blot analysis.
RNase H assay
20 μg total RNA was treated with RNase H (Unit/μl, Ambion) at 37oC for 1hr in the presence of 200 ng of oligonucleotide complementary to the cleavage site, and separated by agarose gel electrophoresis. To determine if the mobility variation is due to the difference of poly(A) tail length, the same samples were treated with both oligonucleotide complementary to the cleavage site and oligo d(T)50. The 1.0% agarose gel contains 6% formaldehyde, 20 mM MOPS, 8 mM sodium acetate, 1 mM EDTA. After transfer, the membrane was hybridized with a frq RNA probe corresponding to the sequence downstream the RNase H cleavage site. The sequence of the complementary oligonucleotide: 5′-TCAGAAGCTAGATCATCGCCATCGTCCCAGCTGGAATCGGTACTGAACGG-3′.
Chromatin immunoprecipitation assay
The ChIP assay was performed as previously described (He et al., 2006; He and Liu, 2005). The immunoprecipitation was performed using a WC-2 antibody. Each experiment was independently performed three times and immunoprecipitation without the WC-2 antibody or using the wc-2ko extract was used as the negative control.
mRNA-protein immunoprecipitation
RNA IP was performed as previously described (Niranjanakumari et al., 2002; Peritz et al., 2006) with following modifications. Freshly harvested tissues were grounded into powders and homogenized by the IP buffer (25mM Tris pH7.5, 150mM NaCl, 1.5mM MgCl2, 1% NP40, 1mM DTT plus protease inhibitors: 1μg /ml Pepstatin A, 1μg /ml Leupeptin, 1mM PMSF and RNase inhibitor RNaseOUT, 100U/ml final concentration, Ambion USA). After centrifugation at 15 000g for 10 min, 1 ml of the resulting supernatant was incubated with FRQ antibody. A mock (without the antibody) and a control (with tubulin antibody) IP were used as the negative controls. Total cDNA synthesized from the total RNA of the wild-type strain was amplified as the positive control. The beads were then washed three times with the IP buffer before total RNA was extracted and subjected to reverse transcription using random primers. PCR was performed with frq primers (forward: 5′-ACGACAGCCCATTGGCAGCCG-3′; reverse: 5′-ATGAGACGTCCTCCATCGAAC-3′).
Supplementary Material
Acknowledgements
We thank Dr. Guocun Huang for technical assistance and Dr. Chi-Tai Tang for critical comments. We thank Dr. Jay Dunlap for providing the luciferase-containing plasmid. This research was supported by grants from National Institutes of Health and Welch Foundation to Yi Liu.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allada R, Emery P, Takahashi JS, Rosbash M. Stopping time: the genetics of fly and mouse circadian clocks. Annu Rev Neurosci. 2001;24:1091–1119. doi: 10.1146/annurev.neuro.24.1.1091. [DOI] [PubMed] [Google Scholar]
- Aronson B, Johnson K, Loros JJ, Dunlap JC. Negative feedback defining a circadian clock: autoregulation in the clock gene frequency. Science. 1994a;263:1578–1584. doi: 10.1126/science.8128244. [DOI] [PubMed] [Google Scholar]
- Aronson BD, Johnson KA, Dunlap JC. The circadian clock locus frequency: A single ORF defines period length and temperature compensation. Proc Natl Acad Sci USA. 1994b;91:7683–7687. doi: 10.1073/pnas.91.16.7683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belden WJ, Loros JJ, Dunlap JC. Execution of the circadian negative feedback loop in Neurospora requires the ATP-dependent chromatin-remodeling enzyme CLOCKSWITCH. Mol Cell. 2007;25:587–600. doi: 10.1016/j.molcel.2007.01.010. [DOI] [PubMed] [Google Scholar]
- Bell-Pedersen D, Cassone VM, Earnest DJ, Golden SS, Hardin PE, Thomas TL, Zoran MJ. Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat Rev Genet. 2005;6:544–556. doi: 10.1038/nrg1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell-Pedersen D, Dunlap JC, Loros JJ. Distinct cis-acting elements mediate clock, light, and developmental regulation of the Neurospora crassa eas (ccg-2) gene. Mol Cell Biol. 1996a;16:513–521. doi: 10.1128/mcb.16.2.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell-Pedersen D, Shinohara M, Loros J, Dunlap JC. Circadian clock-controlled genes isolated from Neurospora crassa are late night to early morning specific. Proc Natl Acad Sci USA. 1996b;93:13096–13101. doi: 10.1073/pnas.93.23.13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SA, Ripperger J, Kadener S, Fleury-Olela F, Vilbois F, Rosbash M, Schibler U. PERIOD1-associated proteins modulate the negative limb of the mammalian circadian oscillator. Science. 2005;308:693–696. doi: 10.1126/science.1107373. [DOI] [PubMed] [Google Scholar]
- Buttner K, Wenig K, Hopfner KP. The exosome: a macromolecular cage for controlled RNA degradation. Mol Microbiol. 2006;61:1372–1379. doi: 10.1111/j.1365-2958.2006.05331.x. [DOI] [PubMed] [Google Scholar]
- Cha J, Chang SS, Huang G, Cheng P, Liu Y. Control of WHITE COLLAR localization by phosphorylation is a critical step in the circadian negative feedback process. Embo J. 2008;27:3246–3255. doi: 10.1038/emboj.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P, He Q, He Q, Wang L, Liu Y. Regulation of the Neurospora circadian clock by an RNA helicase. Genes Dev. 2005;19:234–241. doi: 10.1101/gad.1266805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P, Yang Y, Gardner KH, Liu Y. PAS domain-mediated WC-1/WC-2 interaction is essential for maintaining the steady state level of WC-1 and the function of both proteins in circadian clock and light responses of Neurospora. Mol Cell Biol. 2002;22:517–524. doi: 10.1128/MCB.22.2.517-524.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P, Yang Y, Heintzen C, Liu Y. Coiled-coil domain mediated FRQ-FRQ interaction is essential for its circadian clock function in Neurospora. EMBO J. 2001a;20:101–108. doi: 10.1093/emboj/20.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P, Yang Y, Liu Y. Interlocked feedback loops contribute to the robustness of the Neurospora circadian clock. Proc Natl Acad Sci USA. 2001b;98:7408–7413. doi: 10.1073/pnas.121170298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa A, Lewis ZA, Greene AV, March IJ, Gomer RH, Bell-Pedersen D. Multiple oscillators regulate circadian gene expression in Neurospora. Proc Natl Acad Sci USA. 2003;100:13597–13602. doi: 10.1073/pnas.2233734100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosthwaite SK, Dunlap JC, Loros JJ. Neurospora wc-1 and wc-2: Transcription, photoresponses, and the origins of circadian rhythmicity. Science. 1997;276:763–769. doi: 10.1126/science.276.5313.763. [DOI] [PubMed] [Google Scholar]
- Das B, Butler JS, Sherman F. Degradation of normal mRNA in the nucleus of Saccharomyces cerevisiae. Mol Cell Biol. 2003;23:5502–5515. doi: 10.1128/MCB.23.16.5502-5515.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Paula RM, Vitalini MW, Gomer RH, Bell-Pedersen D. Complexity of the Neurospora crassa circadian clock system: multiple loops and oscillators. Cold Spring Harb Symp Quant Biol. 2007;72:345–351. doi: 10.1101/sqb.2007.72.002. [DOI] [PubMed] [Google Scholar]
- Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- Dunlap JC. Proteins in the Neurospora circadian clockworks. J Biol Chem. 2006;281:28489–28493. doi: 10.1074/jbc.R600018200. [DOI] [PubMed] [Google Scholar]
- Dziembowski A, Lorentzen E, Conti E, Seraphin B. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat Struct Mol Biol. 2007;14:15–22. doi: 10.1038/nsmb1184. [DOI] [PubMed] [Google Scholar]
- Froehlich AC, Loros JJ, Dunlap JC. Rhythmic binding of a WHITE COLLAR-containing complex to the frequency promoter is inhibited by FREQUENCY. Proc Natl Acad Sci U S A. 2003;100:5914–5919. doi: 10.1073/pnas.1030057100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbarino-Pico E, Green CB. Posttranscriptional regulation of mammalian circadian clock output. Cold Spring Harb Symp Quant Biol. 2007;72:145–156. doi: 10.1101/sqb.2007.72.022. [DOI] [PubMed] [Google Scholar]
- Garceau N, Liu Y, Loros JJ, Dunlap JC. Alternative initiation of translation and time-specific phosphorylation yield multiple forms of the essential clock protein FREQUENCY. Cell. 1997;89:469–476. doi: 10.1016/s0092-8674(00)80227-5. [DOI] [PubMed] [Google Scholar]
- Gooch VD, Mehra A, Larrondo LF, Fox J, Touroutoutoudis M, Loros JJ, Dunlap JC. Fully codon-optimized luciferase uncovers novel temperature characteristics of the Neurospora clock. Eukaryot Cell. 2008;7:28–37. doi: 10.1128/EC.00257-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green CB, Douris N, Kojima S, Strayer CA, Fogerty J, Lourim D, Keller SR, Besharse JC. Loss of Nocturnin, a circadian deadenylase, confers resistance to hepatic steatosis and diet-induced obesity. Proc Natl Acad Sci U S A. 2007;104:9888–9893. doi: 10.1073/pnas.0702448104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Cha J, He Q, Lee H, Yang Y, Liu Y. CKI and CKII mediate the FREQUENCY-dependent phosphorylation of the WHITE COLLAR complex to close the Neurospora circadian negative feedback loop. Genes & Dev. 2006;20:2552–2565. doi: 10.1101/gad.1463506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Liu Y. Molecular mechanism of light responses in Neurospora: from light-induced transcription to photoadaptation. Genes & Dev. 2005;19:2888–2899. doi: 10.1101/gad.1369605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Shu H, Cheng P, Chen S, Wang L, Liu Y. Light-independent phosphorylation of WHITE COLLAR-1 regulates its function in the Neurospora circadian negative feedback loop. J Biol Chem. 2005;280:17526–17532. doi: 10.1074/jbc.M414010200. [DOI] [PubMed] [Google Scholar]
- Heintzen C, Liu Y. The Neurospora crassa circadian clock. Adv Genet. 2007;58:25–66. doi: 10.1016/S0065-2660(06)58002-2. [DOI] [PubMed] [Google Scholar]
- Heintzen C, Nater M, Apel K, Staiger D. AtGRP7, a nuclear RNA-binding protein as a component of a circadian-regulated negative feedback loop in Arabidopsis thaliana. Proc Natl Acad Sci USA. 1997;94:8515–8520. doi: 10.1073/pnas.94.16.8515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, LaCava J, Tollervey D. RNA-quality control by the exosome. Nat Rev Mol Cell Biol. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- Houseley J, Tollervey D. Yeast Trf5p is a nuclear poly(A) polymerase. EMBO Rep. 2006;7:205–211. doi: 10.1038/sj.embor.7400612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Chen S, Li S, Cha J, Long C, Li L, He Q, Liu Y. Protein kinase A and casein kinases mediate sequential phosphorylation events in the circadian negative feedback loop. Genes Dev. 2007;21:3283–3295. doi: 10.1101/gad.1610207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S, Matsumoto K, Hirose M, Shimada M, Nagano M, Shigeyoshi Y, Hoshino S, Ui-Tei K, Saigo K, Green CB, et al. LARK activates posttranscriptional expression of an essential mammalian clock protein, PERIOD1. Proc Natl Acad Sci U S A. 2007;104:1859–1864. doi: 10.1073/pnas.0607567104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak E, Kim TD, Kim KT. Essential role of 3′-untranslated region-mediated mRNA decay in circadian oscillations of mouse Period3 mRNA. J Biol Chem. 2006;281:19100–19106. doi: 10.1074/jbc.M511927200. [DOI] [PubMed] [Google Scholar]
- LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- Lewis ZA, Correa A, Schwerdtfeger C, Link KL, Xie X, Gomer RH, Thomas T, Ebbole DJ, Bell-Pedersen D. Overexpression of White Collar-1 (WC-1) activates circadian clock-associated genes, but is not sufficient to induce most light-regulated gene expression in Neurospora crassa. Mol Microbiol. 2002;45:917–931. doi: 10.1046/j.1365-2958.2002.03074.x. [DOI] [PubMed] [Google Scholar]
- Lidder P, Gutierrez RA, Salome PA, McClung CR, Green PJ. Circadian control of messenger RNA stability. Association with a sequence-specific messenger RNA decay pathway. Plant Physiol. 2005;138:2374–2385. doi: 10.1104/pp.105.060368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Bell-Pedersen D. Circadian rhythms in Neurospora crassa and other filamentous fungi. Eukaryot Cell. 2006;5:1184–1193. doi: 10.1128/EC.00133-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorentzen E, Conti E. The exosome and the proteasome: nano-compartments for degradation. Cell. 2006;125:651–654. doi: 10.1016/j.cell.2006.05.002. [DOI] [PubMed] [Google Scholar]
- Loros JJ, Dunlap JC. Genetic and molecular analysis of circadian rhythms in NEUROSPORA. Annu Rev Physiol. 2001;63:757–794. doi: 10.1146/annurev.physiol.63.1.757. [DOI] [PubMed] [Google Scholar]
- Maiti M, Lee HC, Liu Y. QIP, a putative exonuclease, interacts with the Neurospora Argonaute protein and facilitates conversion of duplex siRNA into single strands. Genes Dev. 2007;21:590–600. doi: 10.1101/gad.1497607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newby LM, Jackson FR. Regulation of a specific circadian clock output pathway by lark, a putative RNA-binding protein with repressor activity. J Neurobiol. 1996;31:117–128. doi: 10.1002/(SICI)1097-4695(199609)31:1<117::AID-NEU10>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Niranjanakumari S, Lasda E, Brazas R, Garcia-Blanco MA. Reversible cross-linking combined with immunoprecipitation to study RNA-protein interactions in vivo. Methods. 2002;26:182–190. doi: 10.1016/S1046-2023(02)00021-X. [DOI] [PubMed] [Google Scholar]
- Nowrousian M, Duffield GE, Loros JJ, Dunlap JC. The frequency gene is required for temperature-dependent regulation of many clock-controlled genes in Neurospora crassa. Genetics. 2003;164:923–933. doi: 10.1093/genetics/164.3.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peritz T, Zeng F, Kannanayakal TJ, Kilk K, Eiriksdottir E, Langel U, Eberwine J. Immunoprecipitation of mRNA-protein complexes. Nat Protoc. 2006;1:577–580. doi: 10.1038/nprot.2006.82. [DOI] [PubMed] [Google Scholar]
- Schafmeier T, Haase A, Kaldi K, Scholz J, Fuchs M, Brunner M. Transcriptional feedback of neurospora circadian clock gene by phosphorylation-dependent inactivation of its transcription factor. Cell. 2005;122:235–246. doi: 10.1016/j.cell.2005.05.032. [DOI] [PubMed] [Google Scholar]
- Schneider C, Anderson JT, Tollervey D. The exosome subunit Rrp44 plays a direct role in RNA substrate recognition. Mol Cell. 2007;27:324–331. doi: 10.1016/j.molcel.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehgal A. Molecular biology of circadian rhythms. John Wiley & Sons; Hoboken, NJ: 2004. [Google Scholar]
- Shinohara M, Loros JJ, Dunlap JC. Glyceraldehyde-3-phosphate dehydrogenase is regulated on a daily basis by the circadian clock. J Biol Chem. 1998;273:446–452. doi: 10.1074/jbc.273.1.446. [DOI] [PubMed] [Google Scholar]
- So WV, Rosbash M. Post-transcriptional regulation contributes to Drosophila clock gene mRNA cycling. Embo J. 1997;16:7146–7155. doi: 10.1093/emboj/16.23.7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger D, Zecca L, Kirk D.A. Wieczorek, Apel K, Eckstein L. The circadian clock regulated RNA-binding protein AtGRP7 autoregulates its expression by influencing alternative splicing of its own pre-mRNA. Plant J. 2003;33:361–371. doi: 10.1046/j.1365-313x.2003.01629.x. [DOI] [PubMed] [Google Scholar]
- Vanacova S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MW, Kay SA. Time zones: a comparative genetics of circadian clocks. Nat Rev Genet. 2001;2:702–715. doi: 10.1038/35088576. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.