Abstract
Summary: The introduction of in vitro nucleic acid amplification techniques, led by real-time PCR, into the clinical microbiology laboratory has transformed the laboratory detection of viruses and select bacterial pathogens. However, the progression of the molecular diagnostic revolution currently relies on the ability to efficiently and accurately offer multiplex detection and characterization for a variety of infectious disease pathogens. Microarray analysis has the capability to offer robust multiplex detection but has just started to enter the diagnostic microbiology laboratory. Multiple microarray platforms exist, including printed double-stranded DNA and oligonucleotide arrays, in situ-synthesized arrays, high-density bead arrays, electronic microarrays, and suspension bead arrays. One aim of this paper is to review microarray technology, highlighting technical differences between them and each platform's advantages and disadvantages. Although the use of microarrays to generate gene expression data has become routine, applications pertinent to clinical microbiology continue to rapidly expand. This review highlights uses of microarray technology that impact diagnostic microbiology, including the detection and identification of pathogens, determination of antimicrobial resistance, epidemiological strain typing, and analysis of microbial infections using host genomic expression and polymorphism profiles.
INTRODUCTION
Molecular detection techniques continue to increase in utility in clinical microbiology laboratories. The implementation of in vitro nucleic acid amplification techniques, led by real-time PCR, in diagnostic laboratories has transformed viral detection and select bacterial detection. Although not likely to completely replace culture techniques in the near future, molecular applications in the diagnosis of infectious diseases have become commonplace in academic medical centers and tertiary-care facilities and are becoming more tangible in community-based settings as more FDA-cleared products are available. The further advancement of molecular infectious disease diagnostics is dependent on the ability of multiplexing technologies, or the ability to detect and identify more than one pathogen simultaneously from the same specimen, to be implemented in clinical microbiology laboratories with ease and accuracy. One approach to multiplex detection and characterization is microarray analysis.
Simply defined, a microarray is a collection of microscopic features (most commonly DNA) which can be probed with target molecules to produce either quantitative (gene expression) or qualitative (diagnostic) data. Although other types of microarrays exist, such as protein microarrays (122, 125), this review will focus on DNA microarrays. The initial production of arrays in the research arena included radiolabeled macroarrays such as Southern blots and dot blots (91, 177). Scientific ingenuity in research laboratories in the 1990s led to the development of modern two-dimensional hybridization microarrays (167, 172). Largely due to advances in fabrication, robotics, and bioinformatics, microarray technology has continued to improve in terms of efficiency, discriminatory power, reproducibility, sensitivity, and specificity (135). In addition, microarray platforms have expanded to include three-dimensional arrays or suspension bead arrays. These improvements have allowed the transition of microarrays from strictly research settings to clinical diagnostic applications. The number of articles on microarrays and articles describing their use in microbiology and infectious diseases has rapidly increased over the past 9 years (Fig. 1). Although many of these articles can still be attributed to clinical microbiology or infectious disease “research,” research in the diagnostic realm has led to the optimization of the diagnostic potential of microarrays and has led to the development of commercially available qualitative detection platforms. Thus, we have now entered a new era in molecular diagnostics where the use of microarray technology in clinical microbiology is a reality.
FIG. 1.
Microarray publications. The number of primary manuscripts published using microarray technology (bars) and the number of microarray publications that have infectious disease and/or microbiology applications (line) are depicted.
BASIC CONCEPTS OF MICROARRAYS
Microarrays can be distinguished based upon characteristics such as the nature of the probe, the solid-surface support used, and the specific method used for probe addressing and/or target detection (135). Below, we review the methodologies of printed and in situ-synthesized microarrays, high-density bead arrays, and electronic and suspension bead microarrays. In all of these approaches, the probe refers to the DNA sequence bound to the solid-surface support in the microarray, whereas the target is the “unknown” sequence of interest. In general terms, probes are synthesized and immobilized as discrete features, or spots. Each feature contains millions of identical probes. The target is fluorescently labeled and then hybridized to the probe microarray. A successful hybridization event between the labeled target and the immobilized probe will result in an increase of fluorescence intensity over a background level, which can be measured using a fluorescent scanner (135). The fluorescence data can then be analyzed by a variety of methods. Experimental details including probe length and synthesis, number of possible features (i.e., density of the microarray), and the solid surface used vary depending on the type of microarray employed and are discussed below and summarized in Table 1. The goals of the manuscript are to review the concepts behind each of these microarray technologies, highlighting their benefits and disadvantages, as well as provide a detailed review of the applications of these techniques in clinical microbiology.
TABLE 1.
Comparison of microarray platformsa
| Microarray | Principle(s) | Format(s) | Density | Relative cost | Diagnostic application(s) | References |
|---|---|---|---|---|---|---|
| Printed | Glass slides are used as the solid support for printing DNA probes | For dsDNA, PCR amplicons (200-800 bp) from known genomic sequence, shotgun library clones, or cDNA are used; for oligonucleotides, 25-80-bp probes are synthesized | Moderate (∼10,000-30,000) | $$$ | No commercially available applications; pathogen detection and identification, antimicrobial resistance detection, viral discovery, molecular surveillance | 21, 32, 153, 167, 172, 195, 206-208 |
| In situ synthesized | Oligonucleotides are synthesized directly on the surface of a quartz wafer using photochemistry; multiple probe sets (one perfect-match probe and one mismatch probe) are included per target | Affymetrix GeneChips, 20-25-bp probes; Roche NimbleGen, 60-100-bp probes; Agilent, 60-bp probes | High (Affymetrix, >106; NimbleGen and Agilent, 15,000->106) | $$$$ | No commercially available applications; pathogen detection and identification, antimicrobial resistance detection, viral discovery, molecular surveillance, strain typing | 59, 117, 150, 151, 157, 200, 215 |
| High-density bead arrays | Sequence-tagged beads are randomly assorted onto fiber-optic bundles or silicon slides | SAM, 96 samples; Sentrix BeadChip; 1-16 samples | High (∼50,000-106) | $$$ | No commercially available applications; potential use in microbiology but no studies published to date | 52, 71, 146 |
| Electronic | Electric fields are used to promote active hybridization of nucleic acids on a microelectronic device; streptavidin-biotin bonds immobilize the probes on the array surface | NanoChip 400; capture probe down; amplicon down; sandwich assays | Low (400 max) | $$ | Commercially available products discontinued; pathogen detection and identification | 10, 108, 185, 226 |
| Liquid-bead suspension | Spectrally unique microspheres provide solid support for application of probes or universal sequence tags; bead hybridization with fluorescently labeled target DNA is measured using flow cytometry | Direct DNA hybridization; competitive DNA hybridization; solution-based chemistries (ASPE/TSPE, OLA, SBCE) | Low (100 max) | $$ | FDA-cleared xTAG RVP assay; pathogen detection and identification, antimicrobial resistance detection, strain typing | 18, 73, 79, 109, 112, 148, 191 |
Data from reference 135. $$, low-moderate cost; $$$, moderate cost; $$$$, high cost.
Printed Microarrays
Printed arrays were the first microarrays utilized in research laboratories and are so called because of the “printing” or spotting of the probes onto the microarray surface, which is most commonly a glass microscope slide. Glass slides are an attractive medium for microarrays because they are economical; are stable throughout high temperatures and stringent washes; are nonporous, allowing for efficient kinetics during hybridization; and have minimal background fluorescence (25). The probe spots, or features, can be applied by either noncontact or contact printing. A noncontact printer uses the same technology as computer printers (i.e., bubble jet or inkjet) to expel small droplets of probe solution onto the glass slide. In contact printing, each print pin directly applies the probe solution onto the microarray surface. The result in both cases is the application of a few nanoliters of probe solution per spot to create an array of 100- to 150-μm features. During the printing process it is imperative to control for cross-contamination and printing consistency to preserve the integrity of the microarray and subsequent hybridization data. Due to the relatively large size of the features, printed microarrays are of lower density (∼10,000 to 30,000 features) than in situ-synthesized microarrays and high-density bead arrays (discussed below) but offer considerably more features than either electronic microarrays or suspension bead arrays. The general workflow for the processing of printed microarrays is depicted in Fig. 2.
FIG. 2.
Workflow summary of printed microarrays. Probes are PCR amplified (or oligonucleotides are synthesized) and subsequently spotted onto a glass slide. In this example, two samples to be compared undergo RNA extraction, cDNA production, and differential fluorescent labeling. Hybridization of labeled target nucleic acids to the probe array allows fluorescent scanning to provide data for analysis. (Adapted from reference 51 [Fig. 1A, © Springer-Verlag 2006] with kind permission from Springer Science and Business Media.)
Printed arrays can be further classified based upon the nature of the probes: double-stranded DNA (dsDNA) or oligonucleotide microarrays. For dsDNA microarrays, the probes consist of amplification products (amplicons) obtained by PCR using primers designed from a known genomic sequence, shotgun library clones, or cDNA (74, 167, 194). The double-stranded amplicons are denatured, either in print buffer or after immobilization, which allows the probes to be available for hybridization. Amplicons can be attached to the glass slide surface by the electrostatic interaction of the negative charge of the phosphate backbone of the DNA with a positively charged coating of the slide surface (51) or by UV-cross-linked covalent bonds between the thymidine bases in the DNA and amine groups on treated slides (25). Typically, each 200- to 800-bp dsDNA probe represents a different gene. Ideally, PCR amplicons for microarrays should have high specificity and yield but no contamination, including nonspecific amplification and contaminants that affect attachment to the microarray surface or that autofluoresce (19). Unfortunately, dsDNA probes generally have a high sensitivity but suffer in specificity. For example, in a report by Hager, 21 to 34% of probes did not match the intended target and/or were contaminated (72). The ultimate assessment of probe specificity is sequencing of the products. However, due to financial constraints, most laboratories test the purity and quantity of the amplified product by agarose gel electrophoresis. The specificity of hybridization data can be improved by incorporating redundancy by the inclusion of multiple gene segments among the probes. Although decreased specificity can be beneficial when analyzing a genomic sequence rich in natural polymorphisms, it is disadvantageous when trying to discriminate among highly similar target sequences and unacceptable for clinical diagnostic applications (105).
The spotted probes of oligonucleotide microarrays consist of short, chemically synthesized sequences. In contrast to the relatively large size of dsDNA probes discussed above, the length of probes typically used in printed oligonucleotide microarrays ranges from 25 to 80 bp but may be as long as 150 bp for gene expression microarrays (30). The use of shorter probe lengths allows fewer errors to be introduced during probe synthesis and facilitates the interrogation of small genomic regions, including polymorphisms. While the decreased probe length may adversely affect sensitivity compared to dsDNA probes, specificity is often greater when short, specific genomic regions are interrogated. Longer probes have higher melting temperatures and greater mismatch tolerance, leading to decreased specificity. Generally, the strength of the hybridization signal and the sensitivity increase with an increasing length of the probe. The addition of spacers coupled to very short oligonucleotides or the application of a higher concentration of probe during printing can improve the hybridization signal strength (30). Ramdas et al. reported an eightfold increase in sensitivity with 70-mer oligonucleotides relative to the sensitivity of 30-mer probes, especially for low-expression genes (159). Since, as with dsDNA probes, very long oligonucleotide probes may decrease specificity due to random hybridization to nontarget sequences, it is critical to cautiously determine the optimal probe length for each microarray design. Although easier to manufacture than dsDNA probes, oligonucleotide probes require careful design so that all probes have comparable melting temperatures (within 5°C) and lack palindromic sequences. Preferably, each probe should be tested experimentally to guarantee nonbiased hybridization data, although this may not always be financially possible (30). Oligonucleotide probes are attached to glass slides by covalent linkage because, due to their small size, a significant quantity of probes would be lost during wash steps after noncovalent electrostatic immobilization and cross-linking. The probes are coupled to the microarray surface via modified 5′ or 3′ ends (most commonly a 5′ amino group) on coated slides that provide aldehyde or epoxy functional groups.
Compared to the in situ-synthesized microarrays discussed below, printed microarrays are relatively simple and inexpensive. However, the initial setup of microarray facilities is costly and requires dedicated space in which environmental variables such as dust, humidity, and temperature are well controlled. Dedicated microarray core facilities are available at many universities, making these challenges minimal for individual investigators. A major advantage of printed microarrays is flexibility. The ability to quickly adjust spotted probes based upon updated annotations or the discovery of new, emerging pathogens or resistance mechanisms makes printed microarrays attractive for use in clinical microbiology. However, the implementation of a printed microarray in a clinical diagnostic laboratory is complicated by the arduous and expensive tasks of monitoring production reproducibility, performing clinical validation studies, and continuously assessing the quality of downstream data. A major drawback in the manufacturing of printed dsDNA microarrays is the enormous scale of amplicon production and the associated difficulties of quality control, information management, efficiency, and accuracy. Likewise, the design of oligonucleotide probes is labor-intensive, and errors introduced from probe synthesis are a problem. Although printed microarrays are conducive to “homebrew” or user-defined testing, their use in diagnostic microbiology remains limited to specific research applications. Printed dsDNA microarrays are also crucial to the study of organisms that have not been fully sequenced. The lack of commercially available printed microarrays for use in clinical infectious disease diagnostics makes it unlikely that printed microarrays will soon transition to clinical microbiology laboratories. There are, however, commercially available whole-genome microarrays for select organisms that are useful for research endeavors.
In Situ-Synthesized Oligonucleotide Microarrays
In situ-synthesized arrays are extremely-high-density microarrays that use oligonucleotide probes, of which GeneChips (Affymetrix, Santa Clara, CA) are the most widely known. Unlike the printed oligonucleotide arrays described above, the oligonucleotide probes are synthesized directly on the surface of the microarray, which is typically a 1.2-cm2 quartz wafer. Because in situ-synthesized probes are typically short (20 to 25 bp), multiple probes per target are included to improve sensitivity, specificity, and statistical accuracy. Classically, 11 probes are used per 600 bases being examined (38). The use of probe sets further increases the specificity. A probe set includes one perfect-match probe and one mismatch probe that contains a 1-bp difference in the middle position of the probe (i.e., position 13 of a 25-bp probe). Each member of the probe set is located in a separate feature, which allows the mismatch probe to act as a negative control to identify possible nonspecific cross-hybridization events. Recent advances in GeneChips include the use of longer probes, the design of arrays that interrogate across entire genes or exons, and the implementation of multiple independent and nonoverlapping perfect-match probes in lieu of classic probe sets.
Affymetrix GeneChips typically have >106 features per microarray depending on the interfeature distance (38, 59). GeneChip oligonucleotide probes are synthesized using semiconductor-based photochemical synthesis. On the quartz surface are synthesis linkers modified with light-sensitive protecting groups (59). Thus, the microarray surface is chemically protected from a nucleotide addition until deprotected by light. When the array surface is exposed to UV light, reactive nucleotides modified with a photolabile protecting group can be added to growing oligonucleotide chains. To target specific nucleotides to exact probe sites, photolithographic masks are used. Each photolithographic mask has a defined pattern of windows, which acts as a filter to either transmit or block UV light from specific features on the chemically protected microarray surface. Areas of the microarray surface in which UV light has been blocked will remain protected from the addition of nucleotides, whereas areas exposed to light will be deprotected, and specific nucleotides can be added. The pattern of windows in each mask directs the order of nucleotide addition. In situ probe synthesis is therefore accomplished through the cycling of masking, light exposure, and the addition of either A, C, T, or G bases to the growing oligonucleotide (Fig. 3) (38, 51).
FIG. 3.
Affymetrix GeneChip oligonucleotide microarray. (Top) Photolithography. UV light is passed through a lithographic mask that acts as a filter to either transmit or block the light from the chemically protected microarray surface (wafer). The sequential application of specific lithographic masks determines the order of sequence synthesis on the wafer surface. (Bottom) Chemical synthesis cycle. UV light removes the protecting groups (squares) from the array surface, allowing the addition of a single protected nucleotide as it is washed over the microarray. Sequential rounds of light deprotection, changes in the filtering patterns of the masks, and single nucleotide additions form microarray features with specific 25-bp probes. (Adapted from reference 38 with permission of the publisher [copyright Elsevier Inc. 2006].)
Other high-density oligonucleotide microarrays include those manufactured by Roche NimbleGen (Madison, WI) and Agilent Technologies (Palo Alto, CA). Both platforms use longer oligonucleotide probes (60 to 100 bp), but NimbleGen uses maskless photo-mediated synthesis, and Agilent employs inkjet technology for the in situ manufacturing of the probes. While experiments performed with GeneChips are limited to one label, the NimbleGen and Agilent platforms allow multicolor hybridizations. As mentioned above for printed microarrays, the use of longer oligonucleotides increases sensitivity. The Roche NimbleGen approach to in situ synthesis is similar to that of the GeneChips described above, but photolithographic masks are replaced by “virtual” or digital masks in Roche NimbleGen's maskless array synthesizer technology. Maskless array synthesizer technology uses an array of programmable micromirrors to create digital masks that reflect the desired pattern of UV light to deprotect the features where the next nucleotide will be coupled (Fig. 4). Each NimbleGen microarray can contain >106 features and can be purchased in the following formats per slide: 1 × 2.1 million features, 3 × 720,000 features, 1 × 385,000 features, 4 × 72,000 features, and 12 × 135,000 features. In contrast to the quartz wafers used for the above-described technologies, Agilent microarrays use glass slides and inkjet printing, which eliminates the need for either lithographic or digital masks (Fig. 5). The in situ synthesis of 60-mer oligonucleotides is achieved using five-“ink” (4 bases plus catalyst) printing of nucleotide precursors combined with coupling and deprotection steps (83). Agilent microarrays are available in the following formats: 1 × 244,000 features, 2 × 105,000 features, 4 × 44,000 features, and 8 × 15,000 features.
FIG. 4.
Roche NimbleGen oligonucleotide microarray. Maskless array synthesizer technology is depicted, which utilizes a digital micromirror device (DMD) to create virtual masks. The DMD forms the pattern of UV light needed to direct the specific nucleic acid addition during photo-mediated synthesis. UV light removes the photolabile protecting group (circles) from the microarray surface, allowing the addition of a single protected nucleotide to the growing oligonucleotide chain. The cycling of DMD filtering, light deprotection, and nucleotide addition creates oligonucleotide features 60 to 100 bp in length on the NimbleGen microarray. (Courtesy of Roche NimbleGen [copyright Roche NimbleGen, Inc.].)
FIG. 5.
Agilent oligonucleotide microarray. (A) Noncontact inkjet printing technology delivers a small and accurate volume (picoliters) of nucleotides to the first layer on the microarray surface. (B) Repeated rounds of base-by-base printing extend the length of specific oligonucleotide probes. (C) Close-up of growing oligonucleotide chain with a base being added. (D) The final product is a 60-mer in situ-synthesized probe as a feature on a microarray containing thousands of specifically synthesized probes. (Images courtesy of Agilent Technologies.)
Due to the complex nature of chemical synthesis and the expense involved in production, synthesized microarrays are dependent on commercial manufacturing and are therefore not conducive to user-defined development. There is a growing number of microbial genome microarrays available commercially for gene expression studies. Also, several resequencing microarrays have been developed by TessArae (Potomac Falls, VA) on the Affymetrix GeneChip platform to simultaneously detect and differentiate large numbers of microbial pathogens (115, 116). Currently, a synthesized oligonucleotide array appropriate for use in a diagnostic microbiology laboratory would need to be ordered as a custom microarray. The expense of a custom Affymetrix microarray and the inherent inflexibility of its custom mask make the use of an Affymetrix-synthesized array impractical for the clinical laboratory. In contrast, other in situ-synthesized platforms (Nimblegen and Agilent) can be easily customized with unique oligonucleotide sequence content. Furthermore, a Web-based tool provided by Agilent (eArray) allows users to custom design microarrays with no minimum manufacturing batch size, making Agilent microarrays a primary choice for homebrew and pilot applications. Although it is relatively expensive to obtain a custom oligonucleotide array, many universities already have one or more of these platforms on campus for hybridization and analysis, which may offset the upfront costs. The major advantages to these systems are the reproducibility of the manufacturing process and the standardization of reagents, instrumentation, and data analysis, all of which are critical for methodologies to transition to the clinical laboratory (38). Further advantages that make this approach attractive for clinical diagnostics include controls, such as reference probes for intensity normalization; internal standards of known concentrations; and probes arranged in a checkerboard pattern that are homologous to an internal control included in the hybridization mix.
Whether printed or synthesized, oligonucleotide microarrays generally allow much cleaner downstream hybridization data than do amplicon-based microarrays. With oligonucleotide arrays, the ability to standardize probe concentrations and hybridization temperatures while avoiding or controlling for significant nonspecific hybridization has resulted in considerable improvements in the accuracy and reproducibility of microarray data (105). Although in situ-synthesized oligonucleotide microarrays are very robust systems and have significant control measures included, there are currently none with direct diagnostic infectious disease applications that are commercially available.
High-Density Bead Arrays
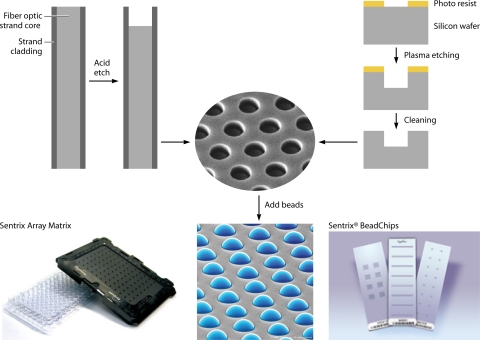
Similar to the printed and in situ-hybridized microarrays discussed above, BeadArrays (Illumina, San Diego, CA) provide a patterned substrate for the high-density detection of target nucleic acids. However, instead of glass slides or silicon wafers as direct substrates, BeadArrays rely on 3-μm silica beads that randomly self-assemble onto one of two available substrates: the Sentrix Array Matrix (SAM) or the Sentrix BeadChip (Fig. 6) (52, 146). The SAM contains 96 1.4-mm fiber-optic bundles. Each bundle is an individual array consisting of 50,000 5-μm light-conducting fibers, each of which is chemically etched to create a microwell for a single bead (52). In the universal BeadArray, up to 1,536 bead types (each with a unique capture sequence) assemble onto each fiber bundle, resulting in ∼30 beads of each type in the array (146). Each SAM allows the analysis of 96 independent samples. The BeadChip can be used to assay 1 to 16 samples at a time on a silicon slide that has been processed by microelectromechanical systems technology to provide microwells for individual beads (53). BeadChips are more appropriate for very-high-density applications such as whole-genome genotyping, which requires 105 to 106 features for determining genome-wide single nucleotide polymorphisms (SNP) (Infinium assay; Illumina) (70).
FIG. 6.
Illumina BeadArray. The SAM contains 96 1.4-mm fiber-optic bundles (bottom left). Each bundle is an individual array consisting of 50,000 5-μm fiber-optic strands, each of which is chemically etched to create a microwell for a single bead (top left). The Sentrix BeadChips can assay 1 to 16 samples at a time on a silicon slide (bottom right) that has been processed to provide microwells for individual beads (top right). Both BeadArray platforms rely on 3-μm silica beads that randomly self-assemble (center). (Adapted from reference 53 with permission of the publisher. © 2009 BioTechniques.)
Unlike the known locations of printed and in situ-hybridized microarray features, the beads in BeadArrays randomly assort to their final location on the array. Thus, the bead location must be mapped, which is accomplished by a decoding process (71). This “decoding” process is in contrast to the use of internal dyes for “encoding” the Luminex microspheres discussed in “Suspension Bead Arrays” below. Each bead has ∼700,000 copies of a unique capture oligonucleotide covalently attached to it, which serves as the bead's identifier (107). In the universal arrays, the capture sequences specifically avoid homology with human and mouse nucleic acid sequences and are referred to as IllumiCodes. The mapping of the Illumina beads is accomplished by a series of hybridization and rinse steps, allowing fluorescently labeled complementary oligonucleotides to bind to their specific bead sequence (IllumiCode) and thus track the location of the bead type (52). An additional advantage to the decoding process is the quality control provided for each feature of the microarray (53).
The SAM can be processed using a standard microtiter plate, which makes it amenable to standard automation and high-throughput processing. The distance between individual arrays on the 16-sample BeadChip is identical to that of a standard multichannel pipettor, thereby facilitating ease of use. BeadArrays can support up to 105 to 106 features and have built-in redundancy. This redundancy is a crucial experimental control for intermicroarray comparative data since each manufactured microarray will not be identical. An additional advantage to the uniqueness of each microarray is that altering the bead pattern provides a means to identify spatial bias. Although the analysis tools available for BeadArray-specific data analysis, background correction, and spatial artifact recognition have been lagging behind those provided by other microarray manufacturers, independent researchers have begun to fill the gaps (20, 47, 49, 50, 182, 217). BeadArrays have been successfully applied to DNA methylation studies (12, 13), gene expression profiling (14, 54, 107), and SNP genotyping, including the International HapMap Project (www.hapmap.org) (23, 53, 70).
Electronic Microarrays
The printed and in situ-synthesized microarrays and BeadArrays described above rely on passive transport for the hybridization of nucleic acids. In contrast, electronic microarrays utilize active hybridization via electric fields to control nucleic acid transport. Microelectronic cartridges (NanoChip 400; Nanogen, San Diego, CA) use complementary metal oxide semiconductor technology for the electronic addressing of nucleic acids (175). Each NanoChip cartridge has 12 connectors that control 400 individual test sites. Negatively charged nucleic acids are transported to specific sites, or features, when a positive current is applied to one or more test sites on the microarray. The surface of the microarray contains streptavidin, which allows for the formation of streptavidin-biotin bonds once electronically addressed biotinylated probes reach their targeted location. The positive current is then removed from the active features, and new test sites can be activated by the targeted application of a positive current. Once the probes have been hybridized at discrete features, the microarray is ready for the application of fluorescently labeled target DNA. Typically, target DNA passively hybridizes with the immobilized probes on the microarray but can also be concentrated electronically (Fig. 7). Although addressing the capture probe down first is the most commonly used format, amplicon-down and sandwich assays have also been utilized. Regardless of the addressing format used, if hybridization occurs between the probe and the target DNA, fluorescent reporters will be present at the positive test, which will be detected when the electronic microarray is scanned and analyzed.
FIG. 7.
Electronic microarray. (A) A positive electric current is applied to test sites, facilitating the active movement and concentration of negatively charged DNA probes to the activated locations. (B) Once the first probe is bound to its targeted location(s) by streptavidin-biotin bonds, the test site(s) can be deactivated, and current can be applied to a different test site. This process is repeated until all the probes are arrayed. (C) Nanogen's RVA ASR. Upon application of the probes to targeted test sites, extracted and amplified nucleic acids from a respiratory sample passively hybridize to the microarray surface. If hybridization occurs, secondary probes that are specific for the target and that contain a nonspecific detector sequence will bind. Secondary fluorescent detector oligonucleotides are used to measure positive hybridization reactions. Multiple probes can be used per site when multiple fluorophores are incorporated. P1, parainfluenza virus type 1; P2, parainfluenza virus type 2; P3, parainfluenza virus type 3; FB, influenza B virus; FA, influenza A virus; RSV, respiratory syncytial virus; BKGD, background. (Images courtesy of Nanogen.)
Electronic microarrays offer several advantages. For example, multiplex detection can be accomplished at an individual test site since multiple probes, each with a distinct fluorophore, can be sequentially addressed to the same feature. The flexibility of this platform allows nucleic acids from a single sample to be hybridized to multiple (but not necessarily all) test sites for the detection of multiple targets, or nucleic acids from multiple samples can be analyzed on the same microarray cartridge, minimizing waste. Furthermore, the NanoChip is a universal blank chip, and the content of the microarray is specified directly by the user, which allows more flexibility in assay design and decreases costs associated with microarray manufacturing. Although the density of electronic microarrays is currently limited to 400 spots, this is sufficient for the majority of diagnostic microbiology applications. In 2007, Nanogen announced the termination of its microarray business. Nonetheless, this technology demonstrates the evolution of microarray technology to a platform that is more practical for diagnostic applications.
Suspension Bead Arrays
In contrast to the two-dimensional, or planar, arrays discussed above, suspension bead arrays are essentially three-dimensional arrays based on the use of microscopic polystyrene spheres (microspheres or beads) as the solid support and flow cytometry for bead and target detection. Furthermore, they are distinct from the high-density Illumina BeadArrays discussed above, in which the beads are immobilized on fiber-optic strands or silicon slides. Suspension-bead-based assays were initially described in 1977 and focused on the detection of antigens and antibodies (78). Multiplexing was first achieved by using different-sized microsphere sets for the simultaneous detection of multiple antibodies (130, 169). Currently, more-robust multiplexing is accomplished using different microsphere sets based on color. Red (658-nm emission) and infrared (712-nm emission) fluorochromes are used at various concentrations to fill 5.6-μm microspheres. Each bead of the 100-microsphere set has a distinct red-to-infrared ratio, and therefore, each bead has a unique spectral address (Fig. 8A). Microspheres with a specific spectral address coupled to a specific probe are equivalent to a feature in a planar microarray. Once multiple individual microspheres have been coupled to separate specific probes, a mixture of microspheres (in theory, up to 100) can be used to interrogate extracted and amplified nucleic acids (Fig. 8B). The subsequent detection of a fluorescent reporter that indicates probe-target DNA hybridization is accomplished using a bench-top flow cytometer. A single-file microsphere suspension passes by two lasers. A 635-nm laser excites the red and infrared fluorochromes impregnated in the microspheres, which allows the classification of the bead and therefore the identity of the probe-target being analyzed. A 532-nm laser excites reporter fluorochromes such as R-phycoerythrin and Alexa 532 to quantify any hybridization that occurs on the microsphere (Fig. 8C).
FIG. 8.
Suspension bead array. (A) Microspheres 5.6 μm in diameter are filled with different relative concentrations of an infrared dye and a red dye to create 100 beads, each with a unique spectral identity. (B) Potential targets are amplified using a biotinylated primer and then denatured and hybridized to microspheres tagged with target-specific sequence probes. Probe-target hybridization is measured using a streptavidin-bound green fluorophore. (C) Flow cytometry is used to analyze the microsphere suspension. A red laser is used to determine the spectral identity of the bead and, therefore, the probe being analyzed. The reporter fluorochrome is excited by a green laser, which quantifies the probe-target reaction on the microsphere surface. (Panels A and C courtesy of Luminex Corporation; panel B adapted from reference 48 with permission from Elsevier [copyright Elsevier Inc. 2006].)
Several chemistries have been developed for nucleic acid detection by suspension bead arrays, including direct DNA hybridization, competitive DNA hybridization, and solution-based chemistries with microsphere capture (48). In direct DNA hybridization, PCR amplicons hybridize directly to probe capture sequences immobilized on the microspheres (Fig. 8B) (8, 179). Generally, a biotinylated primer is used during amplification, which allows streptavidin-R-phycoerythrin to bind and label hybridized microspheres. Competitive DNA hybridization utilizes unlabeled PCR amplicons and biotinylated competitor oligonucleotides. In contrast to the direct hybridization method, competitive DNA hybridization yields high fluorescence in the absence of target DNA. When target DNA is present, it binds the labeled competitor DNA, which, in turn, is not available to hybridize to the microsphere, yielding low fluorescence. Allele-specific primer extension (ASPE) or target-specific primer extension (TSPE), oligonucleotide ligation assay (OLA), and single-base-chain extension (SBCE) are solution-based chemistries coupled with subsequent microsphere capture. By exploiting the natural properties of DNA polymerases and ligases, these chemistries incorporate a capture sequence during the solution-based reaction (48). Both ASPE or TSPE and OLA use a capture primer, which contains a unique 5′ sequence followed by a target-specific sequence. In ASPE and/or TSPE, the primer can be extended by DNA polymerase only if target DNA is present to supply the complementary base for the 3′ nucleotide. The label in ASPE and/or TSPE is provided by a biotinylated deoxynucleotide triphosphate. The OLA reaction is ligase dependent. In addition to the capture primer, a biotinylated probe homologous to target DNA is present during an OLA. The capture primer and reporter probes can be ligated only if target DNA is present in the sample. Used for multiplex SNP detection, SBCE requires independent reactions for each nucleotide query. For every SNP being interrogated, one probe with a unique capture sequence is used to assay the possible alleles in separate wells containing a different dideoxynucleoside triphosphate per well (24). When the capture and target sequences are homologous, a biotinylated dideoxynucleoside triphosphate is incorporated, thereby terminating further extension.
The solution-based chemistries described above all take advantage of universal microspheres with nonspecific capture sequences. The first universal sequences used to tag microspheres were ZipCode/cZipCode capture sequences originally used with SBCE in SNP genotyping assays (24, 84, 192, 219). The 25-bp ZipCode sequences are based on random genomic sequences from Mycobacterium tuberculosis (24). A unique ZipCode sequence is included in the 5′ end of the capture probe used in the chemistries described above, while microspheres are tagged with the complementary sequence (cZipCode). Additional sets of universal capture sequences have been developed, including those by Tm Biosciences (xTAG; Luminex Molecular Diagnostics, Inc., Toronto, Canada) and EraGen (Madison, WI) (EraCode) (90). The sequences of the xTAG (also Tag-It and FlexMAP) system consist of 3 of the 4 nucleotides, thereby decreasing the likelihood of nonspecific hybridization to naturally occurring sequences. Since all of the xTAG sequences are thermodynamically matched, variability in hybridization efficiency is not an issue. The xTAG universal bead technology is used for all commercial assays available through Luminex. Based on the expanded genetic alphabet of MultiCode technology, EraCode sequences incorporate synthesized isoguanosine and 5-Me-isocytosine bases. EraCode sequences are highly specific since the isoguanosine and 5-Me-isocytosine bases will pair with each other but not naturally occurring bases.
Although the feature density of suspension bead arrays is the lowest of all the platforms reviewed, advantages abound that make this platform the most practical for clinical microbiology applications. The availability of universal bead sets and their inherent flexibility make the development of user-defined applications feasible and relatively inexpensive. Although users must carefully validate the positive fluorescent threshold for each analyte in the multiplex, user-defined bead-based assays provide experienced users a multitude of clinically relevant applications (see below). Importantly, in 2008, Luminex obtained FDA clearance for the first infectious-disease suspension bead array (xTAG RVP), which detects 12 respiratory viruses and subtypes (106, 132). Although analyte-specific reagents (ASRs) also exist, the availability of FDA-cleared products is a critical step in getting this technology into less-experienced diagnostic microbiology laboratories. However, many established clinical molecular microbiology laboratories rely heavily on real-time PCR, which has minimal contamination risks. In contrast, the opening of postamplification tubes and the subsequent pipetting steps in the workflow of suspension arrays increase the risk for intra- and interrun contamination. Careful consideration should be paid to contamination control measures and the reestablishment of postamplification laboratory space in the era of real-time PCR. Nonetheless, the relative simplicity, powerful multiplexing capabilities, and affordability of suspension bead arrays make this platform the most attractive for high-throughput nucleic acid detection in clinical infectious disease diagnostics.
POTENTIAL APPLICATIONS IN CLINICAL MICROBIOLOGY
Microarray technology has been used for over a decade to investigate the differential gene expression of pathogens. Although gene expression analyses have contributed significantly to our understanding of pathogenic mechanisms, pathogen responses to environmental stimuli, and host-pathogen interactions, one could argue that the data from these investigations have little direct impact on diagnostic microbiology. However, as outlined below, microarray technology has been applied to the detection and identification of various pathogens, pathogen discovery, antimicrobial resistance monitoring, and strain typing. In addition, the monitoring of host responses to infection and therapy represents a burgeoning field that, when coupled with pathogen-specific detection and monitoring, will be the ultimate diagnostic platform for infectious diseases.
Microbial Detection and Identification
Perhaps the most promising area in applying microarray technology in clinical microbiology is the use of low- or middle-density microarrays for the simultaneous assessment of large numbers of microbial genetic targets (64, 183). Specific microbial gene amplification by either a broad-range or a multiplex PCR prior to microarray analysis enhances test sensitivity. The amplification of universal microorganism targets by broad-range PCR followed by sequencing analysis has been considered a standard procedure (190); however, microarrays have emerged as potential tools for bacterial detection and identification given their high parallelism in screening for the presence of a wide diversity of genes. The most commonly used gene targets have been the 16S bacterial and 28S fungal and intergenic transcribed spacers (ITSs) in rRNA genes, and microarray technology has been incorporated to compensate for the time-consuming sequencing identification procedure (190). An oligonucleotide microarray targeting the 16S rRNA gene was developed for the detection of a panel of 40 predominant human intestinal bacterial pathogens in human fecal samples (208). Assays using broad-range PCR incorporated with microarrays have been shown to allow rapid bacterial detection and identification with positive blood cultures (5, 128). A similar procedure was developed and used for the rapid diagnosis of bloodstream infections caused by common bacterial pathogens in the pediatric and general populations (32, 173). PCR amplification, in combination with an oligonucleotide microarray, was used to identify Bacillus anthracis based on the rRNA ITS region (144). Several studies reported the use of microarrays to identify pathogenic yeasts and molds by targeting the ITS regions in fungal rRNA genes (80, 81, 110). Recently, a DNA microarray was established to detect and identify 14 commonly encountered fungal pathogens in clinical specimens collected from neutropenic patients (178).
The key for broad-range PCR amplification followed by microarray identification to work is to target the right gene. It is critical to use a gene “broad” enough so that most related microorganisms can be covered in one amplification reaction. On the other hand, the targeted gene should possess enough polymorphic information to supply sufficient discriminatory power to differentiate and characterize related microorganisms. Degenerate primer sets can be designed to increase the coverage of relatively variable genes. Other universal bacterial genes have been used to detect and identify organisms using microarrays. For mycobacterial detection and identification, the gyrB, rpoB, and katG genes have been targeted by using microarrays (61, 197). Microarrays targeting the 23S rRNA and gyrB genes for bacterial detection and identification using clinical specimens have been described (92, 102, 136). In addition to bacterial and mycobacterial organisms, microarrays following broad-range PCR amplification have been used to detect and identify fungal, parasitic, and viral pathogens (43, 101, 210).
Microarrays have also been incorporated with multiplex PCR amplification for the simultaneous detection and identification of a panel of microbial pathogens in a single reaction. Khodakov et al. described a novel microarray-based approach for the simultaneous identification and quantification of human immunodeficiency virus type 1 (HIV-1) and hepatitis B and C viruses in donor plasma specimens (96). A microarray technique for the detection and identification of enteropathogenic bacteria at the species and subspecies levels was developed, covering pathogenic Escherichia coli, Vibrio cholerae, Vibrio parahaemolyticus, Salmonella enterica, Campylobacter jejuni, Shigella spp., Yersinia enterocolitica, and Listeria monocytogenes (220). A microarray-based multiplexed assay was developed to detect foot-and-mouth disease virus with rule-out assays for two other foreign animal diseases and four domestic animal diseases that cause vesicular or ulcerative lesions that are indistinguishable from those of foot-and-mouth disease virus infection of cattle, sheep, and swine (111). Bøving et al. reported the development of a novel multiplex PCR with product detection by the Luminex suspension array system covering a panel of bacterial and viral pathogens causing meningitis. This system detected and identified nine microorganisms including Neisseria meningitidis, Streptococcus pneumoniae, E. coli, Staphylococcus aureus, L. monocytogenes, Streptococcus agalactiae, herpes simplex virus types 1 and 2, and varicella zoster virus directly from cerebrospinal fluid (15). The ResPlex I system, manufactured by Qiagen (Valencia, CA), was used to detect a panel of bacterial pathogens related to community-acquired pneumonia from tracheal aspirates collected from hospitalized antibiotic-treated children. The data indicated that the ResPlex I system significantly enhanced the pathogen-specific diagnosis of community-acquired pneumonia in children (39). This gene-specific, multiplex amplification followed by a microarray identification system provides a great example for additional clinical diagnostic applications such as the detection and differentiation of respiratory viral pathogens, which is described in detail below.
Respiratory Viral Pathogen Detection in Connection with Multiplex PCR Amplification
Respiratory infections caused by a panel of bacterial, viral, and fungal pathogens usually present with similar signs and symptoms that are nearly indistinguishable by clinical diagnosis. Simultaneous testing for all possible pathogens is an efficient means to obtain a conclusive result. In addition, assaying for all potential pathogens may yield information regarding possible coinfections or induced secondary infections. The first promising respiratory microarray system was described in 2002, which incorporated 1,600 unique 70-mer-long oligonucleotide probes covering approximately 140 viral genome sequences (206, 207). This ViroChip system was used to identify the severe acute respiratory syndrome virus as a coronavirus (163), for the discovery of a human parainfluenza virus type 4 infection associated with respiratory failure (27) and human coronavirus and rhinovirus in nonasthmatic patients (98), and for the diagnosis of a human metapneumovirus causing critical respiratory illness (26). A resequencing microarray based on the Affymetrix GeneChip platform that used short oligonucleotides to simultaneously provide both species-level and strain-level identification of respiratory pathogens was developed (127, 209). The system was able to detect and identify 26 respiratory pathogens including the novel influenza virus subtypes H5N1 and H1N1 (115, 116). Another comprehensive and panmicrobial microarray, the GreeneChipResp system, was developed and later used for the detection of respiratory viruses and the subtype identification of influenza A viruses (150, 157). In addition to the detection and identification of respiratory pathogens, several formats of microarrays that detect the whole coronavirus genus (42) and detect and type influenza viruses (113, 131, 170, 227) have been described.
Several commercial products are available for the detection of a panel of respiratory viruses, which incorporate microarrays as the identification method (18, 106, 109, 112, 126, 132, 143, 148, 160, 185). These products include the Infiniti RVP from AutoGenomics, Inc. (Carlsbad, CA); the MultiCode-PLx RVP from EraGen Biosciences (Madison, WI); the ResPlex II assay from Qiagen (Valencia, CA); the Ngen respiratory virus ASR assay from Nanogen (San Diego, CA); and the xTAG RVP from Luminex Molecular Diagnostics (Toronto, Canada). Table 2 contrasts these commercially available kits. Among them, the EraGen, Qiagen, and Luminex molecular diagnostics systems incorporate multianalyte profiling by a liquid-bead microarray system developed by Luminex (discussed above) (48). Specific applications of this technology for nucleic acid detection include SNP genotyping, genetic disease screening, gene expression profiling, and microbial detection and typing. Although suspension bead arrays are amenable to high-throughput nucleic acid detection, the efficiency of the front-end multiplex PCR amplification limits the number of pathogens that can be included in one reaction. With the implementation of novel multiplex amplification procedures, numbers of targets included in one reaction can be significantly increased without a significant loss of sensitivity (112, 166).
TABLE 2.
Comparison of commercially available, microarray-based kits for detection and identification of respiratory virusesa
| Product | Company | Viruses and/or genotypes detected | Amplification platform(s) | Microarray platform | Characteristic(s) | Reference(s) |
|---|---|---|---|---|---|---|
| Infiniti RVP | AutoGenomics, Inc. (Carlsbad, CA) | Flu-A, Flu-B, PIV-1, PIV-2, PIV-3, PIV-4, RSV-A, RSV-B, hMPV-A, hMPV-B, RhV-A, RhV-B, EnV, CoV, and Adv | Multiplex PCR and RT-PCR | Infiniti analyzer (solid chip) | The detection step by the Infiniti analyzer is completely automatic | 160 |
| MultiCode-PLx RVP | EraGen Biosciences (Madison, WI) | Flu-A, Flu-B, PIV-1, PIV-2, PIV-3, PIV-4, RSV, hMPV, RhV, AdV, and CoV | Multiplex PCR and RT-PCR | Luminex (liquid chip) | Universal beads used in detection employ EraCode sequences | 109, 143 |
| ResPlex II assay | Qiagen (Valencia, CA) | Flu-A, Flu-B, PIV-1, PIV-2, PIV-3, PIV-4, RSV-A, RSV-B, hMPV, RhV, EnV, and severe acute respiratory CoV | Multiplex RT-PCR (Tem-PCR) | Luminex (liquid chip) | A unique Tem-PCR allows large numbers of targets included in one reaction without significant loss of sensitivity | 18, 112 |
| NGEN respiratory virus ASR | Nanogen (San Diego, CA) | Flu-A, Flu-B, PIV-1, PIV-2, PIV-3, and RSV | Multiplex RT-PCR | NanoChip (solid chip) | Probe labeling, target capture, and detection are accomplished using electronic microarray technology | 112, 185 |
| xTAG RVP | Luminex Molecular Diagnostics (Toronto, Ontario, Canada) | Flu-A, Flu-B, PIV-1, PIV-2, PIV-3, PIV-4, RSV-A, RSV-B, hMPV, AdV, EnV, CoV, and RhV | Multiplex PCR and RT-PCR | Luminex (liquid chip) | TSPE is used in combination with universal detection beads | 126, 132, 148 |
Abbreviations: Tem, target-enriched multiplex; Flu, influenza virus; PIV, parainfluenza virus; RSV, respiratory syncytial virus; hMPV, human metapneumovirus; RhV, rhinoviruses; EnV, enteroviruses; CoV, coronavirus; RT, reverse transcription.
Simultaneous Detection and Typing of Human Papillomaviruses
Persistent infection with known high-risk human papillomavirus (HPV) types is a significant risk factor for cervical cancer and is increasingly being recognized as playing a role in other cancers. Recently, HPV vaccines have demonstrated effectiveness in preventing type-specific persistent infection and disease. To monitor the impact of vaccine implementation strategies, determine type-specific persistence, and evaluate the clinical significance of coinfection with multiple genotypes, HPV testing will require type-specific results. A high-throughput, sensitive, specific, and reproducible HPV detection and typing assay is therefore highly desirable. Most established HPV typing assays are based on consensus PCR to amplify the relatively conserved L1 gene region with hybridization, restriction enzyme digestion, or sequencing of the amplicon to determine type(s). Recently, several studies were aimed at evaluating the usefulness of microarray technology for the simultaneous detection and typing of HPV in routine clinical specimens. A user-developed HPV DNA microarray for high-risk HPV genotyping was evaluated by using a panel of malignant and nonmalignant cervical smears. This approach provides the potential to improve the clinical management of patients with cervical cytological abnormalities (3). Several systems that combine multiplex PCR amplification and microarray identification have been reported to provide rapid and reliable diagnostic tools for HPV detection and typing that are amenable to automation (73, 119, 140, 145). Additional studies that incorporated microarrays to detect and characterize high-risk mucosal HPV types (66), betapapillomavirus types (65), and the frequencies of 23 HPV types in women with and without cytological anomalies (193) have been reported. A novel DNA detection assay incorporating the Luminex suspension array was reported and was applied to the genotyping of HPV in cervical samples (141). The molecular inversion probe microarray assay, originally applied to large-scale human SNP detection, has been used for HPV detection and typing to demonstrate the potential of the method for the detection and characterization of any microbe (1).
Rapid Detection and Characterization of Methicillin-Resistant Staphylococcus aureus
Staphylococcus aureus, including methicillin-resistant S. aureus (MRSA), is an important pathogen in hospitals and, increasingly, in communities around the world. Advanced laboratory techniques, including diagnostic microarray analysis, have been sought to rapidly identify staphylococcal isolates and determine antimicrobial susceptibility patterns. DNA microarray analyses of large samples of clinically characterized community-acquired MRSA strains have been reported, which provide broad insights into evolution, pathogenesis, and disease emergence (57, 99, 168). DNA microarrays based on the Array-Tube platform (ClonDiag Chip Technologies, Jena, Germany) have been used for characterizing and genotyping staphylococcal DNA, including their relevant resistance determinants and virulence factors (137-139). Microarrays provide a valuable epidemiological tool for the detailed characterization of MRSA isolates and comparison of strains at a global level (137). In addition, several techniques incorporating peptide and/or nucleic acid probes and conventional and real-time PCR have been used to take advantage of the rapid enrichment of automated blood culture instruments to rapidly identify MRSA from flagged blood cultures when gram-positive cocci in clusters are observed. The combination of novel multiplex PCR amplification and suspension bead array detection (StaphPlex) for the rapid detection and characterization of staphylococci directly from positive blood culture bottles was described (191). The StaphPlex system provides simultaneous staphylococcal identification, antibiotic resistance determinant detection, detection of Panton-Valentine leukocidin, and determination of staphylococcal cassette chromosome mec types I to IV within 5 h. This approach potentially impacts antibiotic usage when gram-positive cocci in clusters are detected by reducing the unnecessary use of vancomycin, which is often used empirically to treat patients until susceptibility results are available (191). A similar system (MVPlex) was developed and was used to screen for MRSA in nasal swabs (154). Notably, the MVPlex system detects 13 different molecular targets including vancomycin-resistant Enterococcus.
Determination of Antimicrobial Drug Resistance
Another successful application of microarray techniques in clinical microbiology is the determination of antimicrobial resistance by simultaneously detecting a panel of drug resistance-related mutations in microbial genomes (21, 36, 72, 153, 196, 224, 225). The emergence of multidrug-resistant tuberculosis and extensively drug-resistant tuberculosis and time-consuming phenotypic antimycobacterial susceptibility procedures have stimulated the pursuit of microarray platforms in antituberculosis drug resistance determinations. High-density DNA oligonucleotide arrays have been used for parallel species identification and rifampin resistance-related mutations in mycobacteria (197) and, more specifically, for the detection of M. tuberculosis strains that are resistant to rifampin (40, 176, 222) or isoniazid, kanamycin, streptomycin, pyrazinamide, and ethambutol (41, 189, 204). Oligonucleotide microarrays were developed to analyze and identify drug-resistant M. tuberculosis strains, and it was found that the results were comparable with those of standard antimicrobial susceptibility testing (69, 134, 184). A low-cost and -density DNA microarray was designed to detect mutations that confer isoniazid and rifampin resistance in M. tuberculosis isolates. The low-cost and -density array protocol takes 45 min after PCR amplification, with only minimal laboratory equipment required (7). Antonova and colleagues developed a method for the detection and identification of mutations in the M. tuberculosis genome determining resistance to fluoroquinolones by hybridization on biological microchips (6). A recently developed QIAplex system combines a novel multiplex PCR amplification and suspension bead array identification for the simultaneous detection of 24 M. tuberculosis gene mutations responsible for resistance to isoniazid, rifampin, streptomycin, and ethambutol (63). Several studies that detected antibiotic resistance-related mutations in bacterial genomes have been reported (2, 68, 153, 202, 221, 225).
Microarray-based techniques face several application challenges to determine antimicrobial resistance in the clinical setting. First, genomes of some pathogens continue to mutate under natural and therapeutic selective pressures, which is well demonstrated by HIV-1. An Affymetrix microarray was developed to provide HIV-1 antiretroviral-drug-resistant profiles (104, 198, 213). The product was discontinued due to rapidly emerging HIV-1 genome mutations. The company now has a comprehensive, high-density microarray available to identify every mutation in resistance-related HIV-1 genomes. Second, molecular mechanisms for many antimicrobial drug resistances remain to be discovered while novel resistance genes and mutations continue to emerge. It takes considerable time and effort to decipher all of the resistance-related mutations and transfer the basic science findings to clinical applications. For M. tuberculosis, until such knowledge is available, the currently used phenotypic methods for identifying resistance will continue to play an invaluable role in optimizing the therapy of persons with tuberculosis.
Microbial Typing
Numerous studies that use microarrays for microorganism typing by taking advantage of its simultaneous detection of a variety of genomes have been reported. The accurate identification and prompt typing of pathogens causing diarrheal diseases are critical for directing clinical intervention, including appropriate antibiotic administration, and facilitating epidemiological investigations. Microarray-based approaches along with other genetic approaches that can be used to support or replace the classical serotyping method for several conventional diarrhea bacterial pathogens have already been offered. The use of microarrays has included Salmonella, Helicobacter, and Campylobacter species (46, 56, 155, 164, 201, 212). PCR followed by a microarray hybridization step has been used for the detection and typing of E. coli virulence genes (28, 199). A serotype-specific DNA microarray for the identification of clinically encountered Shigella and pathogenic E. coli strains was recently described (114). Diagnostic microarrays based on the ArrayTube format were devised for virulence determinant detection as well as for protein-based serotyping of E. coli (4, 100). A novel ArrayTube assay, which incorporates oligonucleotide DNA probes representing 24 of the most epidemiologically relevant O antigens and 47 H antigens, has been described for fast DNA serotyping of E. coli (9). Microarrays have also been used to characterize and type other gastroenteritis-causing viral pathogens including rotavirus, norovirus, and astrovirus (29, 77, 86, 103, 123). Beyond diarrheal illnesses, Pas et al. reported the comparison of reverse hybridization, microarray, and sequence analysis for hepatitis B virus (HBV) genotyping, suggesting that the InnoLipa HBV genotyping strip assay, a microarray-based system, detected dual infections and was an easy and quick tool for HBV genotyping (152).
Microbial Gene Expression Profiling
The quantification of multiple microorganisms simultaneously using microarray techniques has rarely been reported, probably due to technical difficulties. Instead, the detection and monitoring of the gene expressions of individual microorganism genomes during infection have begun to generate meaningful data (16, 62). Whole-genome microarrays for M. tuberculosis were first described using the amplicon arrays developed at Stanford to define gene expression responses to isoniazid and ethambutol (214). Subsequently, microarrays have been used to monitor M. tuberculosis gene expression responses to a variety of environmental conditions and exposure to antibiotics (11, 93, 161, 180, 203). M. tuberculosis gene expression patterns associated with resistance and susceptibility and mycobacterial survival during infection have been investigated by use of oligonucleotide microarrays (60, 94, 165). The transcriptional profile of M. tuberculosis from human lung samples has been studied; during pulmonary tuberculosis, M. tuberculosis actively transcribes a number of genes involved in active fortification and evasion from host defense systems (158). Microbial candidate genes have been studied by differential-expression microarrays for discrimination between infection and disease caused by M. tuberculosis (87).
Gene expression profiles of other bacterial and fungal infections have also been studied by microarrays (17, 37, 44, 45, 67, 85, 121, 124, 162, 181, 218). A concordance of the gene expression data between intracellular Shigella and Salmonella has been noted, although they colonize different niches inside the cell (124). So far, most studies of microbial gene expression profiling have been limited in research. Techniques used in these studies need to be validated to ensure that sufficient amounts of mRNA are extracted such that gene expression data are not compromised. The host-pathogen interactions that define a disease are clearly complex, and other genotypic and phenotypic data need to be integrated to clarify the intricate cross talk from host to pathogen and the environmental cues that lead to the expression of bacterial virulence factors in vivo. Nevertheless, microbial gene expression profiles reveal a complete picture of the metabolic state of bacteria under a particular condition, thereby providing a potential tool for the diagnosis and monitoring of microbial infection and disease.
Host Gene Expression Profiling during Microbial Infections
Pathogen-induced phenotypic changes in a host are often accompanied by marked changes in host gene expression. Genome-wide expression profiling of the hosts, in addition to the pathogens, has become increasingly important for studying host-pathogen interactions (88). The advent of microarray technology has greatly expanded our ability to monitor changes in host gene expression. The cellular transcriptional response to human cytomegalovirus was globally monitored with an oligonucleotide array in 1998 (223). Subsequently, oligonucleotide microarrays have been generated to measure host gene expression profiles in response to E. coli, Candida albicans, L. monocytogenes, influenza virus, and respiratory syncytial virus infections (34, 58, 82). A microarray was generated to incorporate a series of host response genes including those involved in inflammation and chemotaxis as well as those involved in the synthesis of prostaglandins, Toll-like receptors, and T-cell regulation (97). Such a microarray system has been used to determine immune responses in normal human monocytes after fungal pathogen infections and antifungal drug inoculation (35, 97, 174). Microarrays have been implemented to generate gene expression profiles for viral hepatitis infections, which provide enormous diagnostic and therapeutic potential (75, 76, 147, 171, 211).
Several studies that used microarray-based techniques to detect and characterize host gene expression profiles for sepsis have been reported. Microarray technology was first used to analyze tissue-specific changes in gene expression induced by sepsis in animal models (33). Subsequently, numerous studies that used host gene expression profiling toward sepsis diagnostics, pathogen type differentiation, and clinical outcome prediction have been described (Table 3) (55). The rapid determination of a host sepsis transcriptome provides an early differential diagnosis and clinical outcome prediction. Current microarray-based techniques using host gene expression profiles are limited due to the background variation among and within the individuals studied and poor quality control built into the microarrays. Therefore, the gene expression differences notified by the microarrays have to be verified by gene-specific quantitative real-time PCR assays (95).
TABLE 3.
Selected sepsis studies using microarray-based host gene expression
| Chip used | Subject(s) | Main findings and conclusions | Reference |
|---|---|---|---|
| Atlas array, Clontech Laboratories (Mountain View, CA) | Mouse | Microarray technology provides a powerful new tool for rapidly analyzing tissue-specific changes in gene expression induced by sepsis | 33 |
| Hu95aVer2 GeneChip, Affymetrix (Santa Clara, CA) | Adult patients | The host inflammatory responses to gram-negative and gram-positive stimuli share some common response elements but also exhibit distinct patterns of cytokine appearance and leukocyte gene expression | 55 |
| Image consortium libraries, Livermore National Laboratory (Livermore, CA)a | Mouse | Both gram-positive sepsis and gram-negative sepsis share a final common pathway involved in the pathogenesis of sepsis, but certain genes are differentially expressed under distinct regulation | 221 |
| Arraytor human 500-1 cDNA, SIRS-Laboratory (Jena, Germany) | Adult patients | Microarrays can identify typical gene expression profiles for blood samples from patients with severe sepsis | 156 |
| Hu 133A and 133B GeneChip, Affymetrix | Healthy adult blood leukocytes receiving bacterial endotoxin stimulus | Human blood leukocyte response to acute systemic inflammation includes the transient dysregulation of leukocyte bioenergetics and modulation of translational machinery; these findings provide insight into the regulation of global leukocyte activities as they relate to innate immune system tolerance and increased susceptibility to infection in humans | 22 |
| MGU74Av2 GeneChip, Affymetrix | Mouse | A(2A)R blockade may be useful for treatment of infection and sepsis | 142 |
| HG-U133A GeneChip, Affymetrix | Adult patients | Blood transcriptional profiling is a valuable approach not only for patient stratification but also to identity new genes possibly involved in sepsis pathophysiology | 149 |
| Mouse 430 2.0 GeneChip, Affymetrix | Mouse | T-cell receptor signaling and mitogen-activated protein kinase signaling were significantly altered by sepsis | 129 |
| U74Av2 GeneChip, Affymetrix | Mouse | Sepsis induces common inflammatory response gene changes in mouse leukocyte gene expression that can be used to diagnose sepsis | 31 |
| Adelaide Microarray, Compugen, San Jose, CA) | Adult patients | The signature genes reflect suppression of neutrophils’ immune and inflammatory function by sepsis; gene expression profiling therefore provides a novel approach to advance our understanding of the host response to sepsis | 188 |
| 430A GeneChip, Affymetrix | Mouse | Sepsis induces alterations in balance of pro- and antiapoptotic transcriptional networks, and bcl-2 overexpression improves survival in sepsis | 205 |
| U133 Plus 2.0 GeneChip, Affymetrix | Pediatric patients | Genome-level alterations of zinc homeostasis may be prevalent in clinical pediatric septic shock | 216 |
| U133 Plus 2.0 GeneChip, Affymetrix | Adult patients | Sepsis has a unique gene expression profile that is different from that for uninfected inflammation and becomes apparent prior to expression of the clinical sepsis phenotype | 89 |
| U133 Plus 2.0 GeneChip, Affymetrix | Adult patients | Toll-like receptors and downstream signaling genes are differentially expressed in critically ill patients developing sepsis compared with those with sterile inflammation; these expression differences occur before phenotype-based diagnosis of clinical sepsis | 120 |
| U133 Plus 2.0 GeneChip, Affymetrix | Adult patients | There was evidence of sepsis-related immunosuppression and reduced inflammatory response in mononuclear cells on a transcriptome level; these characteristic transcriptional changes can be used to aid the diagnosis of sepsis | 187 |
Distributed by ResGen Invitrogen (Huntsville, AL).
Host Genomic Polymorphism Determination
When infections, especially chronic infections, are viewed as horizontally acquired genetic diseases, it makes sense to view the pathogen and host as an integrated system. Host genetic polymorphisms that influence the host immune response to infectious agents, thereby determining susceptibility to certain diseases and pathological conditions, which has been well explored in sepsis, have been described (118). SNP analysis is a powerful tool for the mapping and diagnosing disease-related alleles. While sequencing remains the “gold standard” to determine host genetic variabilities, microarray-based techniques may become a simple, rapid, automatic, and user-friendly format for screening and detecting a large panel of related SNPs simultaneously. An Affymetrix HuSNP assay was used to study the role of human genomic SNPs in the pathogenesis of human parvovirus B19 infection, and relevant SNPs revealed by the microarray study were further confirmed by allele-specific real-time PCR assays (95). A microarray was developed for the simultaneous genotyping of four host SNPs associated with the therapeutic effect of interferon in hepatitis C virus patients (186). These preliminary data suggest that a genetic predisposition is associated with the pathogenesis and development of microbial infections.
CONCLUDING REMARKS
Microarrays have the unprecedented potential to simultaneously detect and identify thousands of microbial genes, which provides another evolutionary technical advance in the field of clinical microbiology. Although, historically, microarrays have been used largely for gene expression studies, microarrays have gradually been applied in the detection and characterization of microbial pathogens, determination of antimicrobial resistance, typing of microbial pathogens, and monitoring of microbial infections by investigating host genomic expression and polymorphism profiles. Even with these major advances, the potential power behind microarray applications in clinical microbiology has yet to be fully realized. The ability to detect multiple pathogens and/or monitor the variability of normal microbial populations in a disease process could transform our current understanding of infectious diseases. In addition, massively parallel sequencing performed by microarray analysis offers the opportunity of sequencing directly from complex clinical specimens. This metagenomics approach will allow a comprehensive analysis of every nucleic acid in the specimen. For these robust applications, high-density microarray platforms must be able to transition from translational research laboratories to the clinical laboratory. It is unlikely that traditional, planar microarrays will soon appear in clinical microbiology laboratories due to their high cost, relative lack of flexibility, and limited throughput. The ideal microarray platform for the diagnostic laboratory is a low- to medium-density array that offers limited, reliable, and straightforward results without the need for sophisticated equipment and data management (133). Indeed, platforms that have begun to meet these criteria have been developed, such as electronic microarrays and suspension bead arrays.
With the potential power of microarray analysis comes abundant challenges, particularly in relation to the diagnostic laboratory. Several critical issues need to be resolved before microarray-based techniques can be widely implemented in clinical microbiology services. Due to the potential variability in multiple steps included in the microarray analysis, it is difficult to compare quantitative data between, and even within, microarray experiments. Substantial obstacles still exist along the entire spectrum of preanalytical-to-postanalytical analysis. Heterogeneous clinical specimens present unique challenges with respect to sensitivity, specificity, quantification, and data analysis of microarrays that are not encountered during the analysis of pure cultures. In addition, optimization of extraction, labeling, and hybridization; incorporation of appropriate quality controls, design, and implementation of clinical validation studies; and management and interpretation of data remain challenges in a clinical setting. Moreover, laboratories must account for microarray reproducibility in production and analysis, cost of implementation, acquisition of appropriately skilled laboratorians, as well as intellectual property and reimbursement issues. Compared to real-time PCR, microarray analysis requires additional manipulations including hybridization and washing, which increase the contamination risk and the amount of hands-on time needed, both steps backwards in diagnostic molecular microbiology.
Although improvements are still needed to make the majority of microarray applications amenable to clinical microbiology laboratories, the future role of these robust technologies in diagnostic microbiology is indisputable. Microarray-based analyses will revolutionize infectious disease diagnostics through the detection and identification of previously unknown or unsuspected pathogens, by transforming our current view of multiplexed laboratory testing, and by expanding pathogen detection to include bacterial population-based analyses and host-specific responses (135). As more pathogen genomes and targeted genes are sequenced, costs associated with microarray production decrease, and FDA-cleared products become available, diagnostic applications of microarray-based analyses will continue to expand. As PCR has done in the last 25 years, and more recently real-time PCR, microarray technology will undoubtedly transform the diagnostic capabilities of clinical laboratories, ushering us into a new molecular revolution.
Biography
 Melissa B. Miller, Ph.D., D(ABMM), is an Assistant Professor of Pathology and Laboratory Medicine at the University of North Carolina at Chapel Hill School of Medicine. She is also the Director of the Molecular Microbiology Laboratory and Associate Director of the Microbiology-Immunology Laboratory at the UNC Hospitals. Dr. Miller received her Ph.D. in Molecular Biology from Princeton University and completed the Medical and Public Health Microbiology Fellowship at the UNC Hospitals. She is a member of the Board of American College of Microbiology and the editorial board for the Journal of Clinical Microbiology. Dr. Miller was honored as the 2009 recipient of ASM's Siemens Healthcare Diagnostics Young Investigator Award. Dr. Miller's research focus is the development and assessment of molecular diagnostic assays for the detection of pathogens and the study of microbial epidemiology and antimicrobial resistance.
Melissa B. Miller, Ph.D., D(ABMM), is an Assistant Professor of Pathology and Laboratory Medicine at the University of North Carolina at Chapel Hill School of Medicine. She is also the Director of the Molecular Microbiology Laboratory and Associate Director of the Microbiology-Immunology Laboratory at the UNC Hospitals. Dr. Miller received her Ph.D. in Molecular Biology from Princeton University and completed the Medical and Public Health Microbiology Fellowship at the UNC Hospitals. She is a member of the Board of American College of Microbiology and the editorial board for the Journal of Clinical Microbiology. Dr. Miller was honored as the 2009 recipient of ASM's Siemens Healthcare Diagnostics Young Investigator Award. Dr. Miller's research focus is the development and assessment of molecular diagnostic assays for the detection of pathogens and the study of microbial epidemiology and antimicrobial resistance.
 Yi-Wei Tang, M.D., Ph.D., is an Associate Professor of Pathology and Medicine and Director of the Molecular Infectious Disease Laboratory at Vanderbilt University Medical Center. Dr. Tang received his medical degree from Fudan University School of Medicine in Shanghai and his Ph.D. in microbiology and immunology from Vanderbilt University. He has received postdoctoral training at the U.S. Centers for Disease Control and Prevention and a clinical microbiology fellowship at the Mayo Clinic. Dr. Tang is an editor for the Journal of Clinical Microbiology and a fellow of the American Academy for Microbiology and of the Infectious Disease Society of America. Dr. Tang has research interests in the development and validation of molecular techniques and has published over 100 peer-reviewed original articles, reviews, and book chapters in the field of diagnostic molecular microbiology.
Yi-Wei Tang, M.D., Ph.D., is an Associate Professor of Pathology and Medicine and Director of the Molecular Infectious Disease Laboratory at Vanderbilt University Medical Center. Dr. Tang received his medical degree from Fudan University School of Medicine in Shanghai and his Ph.D. in microbiology and immunology from Vanderbilt University. He has received postdoctoral training at the U.S. Centers for Disease Control and Prevention and a clinical microbiology fellowship at the Mayo Clinic. Dr. Tang is an editor for the Journal of Clinical Microbiology and a fellow of the American Academy for Microbiology and of the Infectious Disease Society of America. Dr. Tang has research interests in the development and validation of molecular techniques and has published over 100 peer-reviewed original articles, reviews, and book chapters in the field of diagnostic molecular microbiology.
REFERENCES
- 1.Akhras, M. S., S. Thiyagarajan, A. C. Villablanca, R. W. Davis, P. Nyren, and N. Pourmand. 2007. PathogenMip assay: a multiplex pathogen detection assay. PLoS ONE 2:e223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albert, T. J., D. Dailidiene, G. Dailide, J. E. Norton, A. Kalia, T. A. Richmond, M. Molla, J. Singh, R. D. Green, and D. E. Berg. 2005. Mutation discovery in bacterial genomes: metronidazole resistance in Helicobacter pylori. Nat. Methods 2:951-953. [DOI] [PubMed] [Google Scholar]
- 3.Albrecht, V., A. Chevallier, V. Magnone, P. Barbry, F. Vandenbos, A. Bongain, J. C. Lefebvre, and V. Giordanengo. 2006. Easy and fast detection and genotyping of high-risk human papillomavirus by dedicated DNA microarrays. J. Virol. Methods 137:236-244. [DOI] [PubMed] [Google Scholar]
- 4.Anjum, M. F., M. Mafura, P. Slickers, K. Ballmer, P. Kuhnert, M. J. Woodward, and R. Ehricht. 2007. Pathotyping Escherichia coli by using miniaturized DNA microarrays. Appl. Environ. Microbiol. 73:5692-5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anthony, R. M., T. J. Brown, and G. L. French. 2000. Rapid diagnosis of bacteremia by universal amplification of 23S ribosomal DNA followed by hybridization to an oligonucleotide array. J. Clin. Microbiol. 38:781-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Antonova, O. V., D. A. Gryadunov, S. A. Lapa, A. V. Kuz'min, E. E. Larionova, T. G. Smirnova, E. Y. Nosova, O. I. Skotnikova, L. N. Chernousova, A. M. Moroz, A. S. Zasedatelev, and V. M. Mikhailovich. 2008. Detection of mutations in Mycobacterium tuberculosis genome determining resistance to fluoroquinolones by hybridization on biological microchips. Bull. Exp. Biol. Med. 145:108-113. [DOI] [PubMed] [Google Scholar]
- 7.Aragon, L. M., F. Navarro, V. Heiser, M. Garrigo, M. Espanol, and P. Coll. 2006. Rapid detection of specific gene mutations associated with isoniazid or rifampicin resistance in Mycobacterium tuberculosis clinical isolates using non-fluorescent low-density DNA microarrays. J. Antimicrob. Chemother. 57:825-831. [DOI] [PubMed] [Google Scholar]
- 8.Armstrong, B., M. Stewart, and A. Mazumder. 2000. Suspension arrays for high throughput, multiplexed single nucleotide polymorphism genotyping. Cytometry 40:102-108. [PubMed] [Google Scholar]
- 9.Ballmer, K., B. M. Korczak, P. Kuhnert, P. Slickers, R. Ehricht, and H. Hachler. 2007. Fast DNA serotyping of Escherichia coli by use of an oligonucleotide microarray. J. Clin. Microbiol. 45:370-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barlaan, E. A., M. Sugimori, S. Furukawa, and K. Takeuchi. 2005. Electronic microarray analysis of 16S rDNA amplicons for bacterial detection. J. Biotechnol. 115:11-21. [DOI] [PubMed] [Google Scholar]
- 11.Betts, J. C., A. McLaren, M. G. Lennon, F. M. Kelly, P. T. Lukey, S. J. Blakemore, and K. Duncan. 2003. Signature gene expression profiles discriminate between isoniazid-, thiolactomycin-, and triclosan-treated Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 47:2903-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bibikova, M., and J. B. Fan. 2009. GoldenGate assay for DNA methylation profiling. Methods Mol. Biol. 507:149-163. [DOI] [PubMed] [Google Scholar]
- 13.Bibikova, M., Z. Lin, L. Zhou, E. Chudin, E. W. Garcia, B. Wu, D. Doucet, N. J. Thomas, Y. Wang, E. Vollmer, T. Goldmann, C. Seifart, W. Jiang, D. L. Barker, M. S. Chee, J. Floros, and J. B. Fan. 2006. High-throughput DNA methylation profiling using universal bead arrays. Genome Res. 16:383-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bibikova, M., D. Talantov, E. Chudin, J. M. Yeakley, J. Chen, D. Doucet, E. Wickham, D. Atkins, D. Barker, M. Chee, Y. Wang, and J. B. Fan. 2004. Quantitative gene expression profiling in formalin-fixed, paraffin-embedded tissues using universal bead arrays. Am. J. Pathol. 165:1799-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bøving, M. K., L. N. Pedersen, and J. K. Moller. 2009. Eight-plex PCR and liquid-array detection of bacterial and viral pathogens in cerebrospinal fluid from patients with suspected meningitis. J. Clin. Microbiol. 47:908-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boyce, J. D., P. A. Cullen, and B. Adler. 2004. Genomic-scale analysis of bacterial gene and protein expression in the host. Emerg. Infect. Dis. 10:1357-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boyce, J. D., I. Wilkie, M. Harper, M. L. Paustian, V. Kapur, and B. Adler. 2002. Genomic scale analysis of Pasteurella multocida gene expression during growth within the natural chicken host. Infect. Immun. 70:6871-6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brunstein, J., and E. Thomas. 2006. Direct screening of clinical specimens for multiple respiratory pathogens using the Genaco Respiratory Panels 1 and 2. Diagn. Mol. Pathol. 15:169-173. [DOI] [PubMed] [Google Scholar]
- 19.Burr, A., K. Bogart, J. Conaty, and J. Andrews. 2006. Automated liquid handling and high-throughput preparation of polymerase chain reaction-amplified DNA for microarray fabrication. Methods Enzymol. 410:99-120. [DOI] [PubMed] [Google Scholar]
- 20.Cairns, J. M., M. J. Dunning, M. E. Ritchie, R. Russell, and A. G. Lynch. 2008. BASH: a tool for managing BeadArray spatial artefacts. Bioinformatics 24:2921-2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Call, D. R., M. K. Bakko, M. J. Krug, and M. C. Roberts. 2003. Identifying antimicrobial resistance genes with DNA microarrays. Antimicrob. Agents Chemother. 47:3290-3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Calvano, S. E., W. Xiao, D. R. Richards, R. M. Felciano, H. V. Baker, R. J. Cho, R. O. Chen, B. H. Brownstein, J. P. Cobb, S. K. Tschoeke, C. Miller-Graziano, L. L. Moldawer, M. N. Mindrinos, R. W. Davis, R. G. Tompkins, and S. F. Lowry. 2005. A network-based analysis of systemic inflammation in humans. Nature 437:1032-1037. [DOI] [PubMed] [Google Scholar]
- 23.Chen, J., L. Guo, D. A. Peiffer, L. Zhou, O. T. Chan, M. Bibikova, E. Wickham-Garcia, S. H. Lu, Q. Zhan, J. Wang-Rodriguez, W. Jiang, and J. B. Fan. 2008. Genomic profiling of 766 cancer-related genes in archived esophageal normal and carcinoma tissues. Int. J. Cancer 122:2249-2254. [DOI] [PubMed] [Google Scholar]
- 24.Chen, J., M. A. Iannone, M. S. Li, J. D. Taylor, P. Rivers, A. J. Nelsen, K. A. Slentz-Kesler, A. Roses, and M. P. Weiner. 2000. A microsphere-based assay for multiplexed single nucleotide polymorphism analysis using single base chain extension. Genome Res. 10:549-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheung, V. G., M. Morley, F. Aguilar, A. Massimi, R. Kucherlapati, and G. Childs. 1999. Making and reading microarrays. Nat. Genet. 21:15-19. [DOI] [PubMed] [Google Scholar]
- 26.Chiu, C. Y., A. A. Alizadeh, S. Rouskin, J. D. Merker, E. Yeh, S. Yagi, D. Schnurr, B. K. Patterson, D. Ganem, and J. L. DeRisi. 2007. Diagnosis of a critical respiratory illness caused by human metapneumovirus by use of a pan-virus microarray. J. Clin. Microbiol. 45:2340-2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chiu, C. Y., S. Rouskin, A. Koshy, A. Urisman, K. Fischer, S. Yagi, D. Schnurr, P. B. Eckburg, L. S. Tompkins, B. G. Blackburn, J. D. Merker, B. K. Patterson, D. Ganem, and J. L. DeRisi. 2006. Microarray detection of human parainfluenzavirus 4 infection associated with respiratory failure in an immunocompetent adult. Clin. Infect. Dis. 43:e71-e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chizhikov, V., A. Rasooly, K. Chumakov, and D. D. Levy. 2001. Microarray analysis of microbial virulence factors. Appl. Environ. Microbiol. 67:3258-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chizhikov, V., M. Wagner, A. Ivshina, Y. Hoshino, A. Z. Kapikian, and K. Chumakov. 2002. Detection and genotyping of human group A rotaviruses by oligonucleotide microarray hybridization. J. Clin. Microbiol. 40:2398-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chou, C. C., C. H. Chen, T. T. Lee, and K. Peck. 2004. Optimization of probe length and the number of probes per gene for optimal microarray analysis of gene expression. Nucleic Acids Res. 32:e99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chung, T. P., J. M. Laramie, D. J. Meyer, T. Downey, L. H. Tam, H. Ding, T. G. Buchman, I. Karl, G. D. Stormo, R. S. Hotchkiss, and J. P. Cobb. 2006. Molecular diagnostics in sepsis: from bedside to bench. J. Am. Coll. Surg. 203:585-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cleven, B. E., M. Palka-Santini, J. Gielen, S. Meembor, M. Kronke, and O. Krut. 2006. Identification and characterization of bacterial pathogens causing bloodstream infections by DNA microarray. J. Clin. Microbiol. 44:2389-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cobb, J. P., J. M. Laramie, G. D. Stormo, J. J. Morrissey, W. D. Shannon, Y. Qiu, I. E. Karl, T. G. Buchman, and R. S. Hotchkiss. 2002. Sepsis gene expression profiling: murine splenic compared with hepatic responses determined by using complementary DNA microarrays. Crit. Care Med. 30:2711-2721. [DOI] [PubMed] [Google Scholar]
- 34.Cohen, P., M. Bouaboula, M. Bellis, V. Baron, O. Jbilo, C. Poinot-Chazel, S. Galiegue, E. H. Hadibi, and P. Casellas. 2000. Monitoring cellular responses to Listeria monocytogenes with oligonucleotide arrays. J. Biol. Chem. 275:11181-11190. [DOI] [PubMed] [Google Scholar]
- 35.Cortez, K. J., C. A. Lyman, S. Kottilil, H. S. Kim, E. Roilides, J. Yang, B. Fullmer, R. Lempicki, and T. J. Walsh. 2006. Functional genomics of innate host defense molecules in normal human monocytes in response to Aspergillus fumigatus. Infect. Immun. 74:2353-2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crameri, A., J. Marfurt, K. Mugittu, N. Maire, A. Regos, J. Y. Coppee, O. Sismeiro, R. Burki, E. Huber, D. Laubscher, O. Puijalon, B. Genton, I. Felger, and H. P. Beck. 2007. Rapid microarray-based method for monitoring of all currently known single-nucleotide polymorphisms associated with parasite resistance to antimalaria drugs. J. Clin. Microbiol. 45:3685-3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cui, L., J. Q. Lian, H. M. Neoh, E. Reyes, and K. Hiramatsu. 2005. DNA microarray-based identification of genes associated with glycopeptide resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 49:3404-3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dalma-Weiszhausz, D. D., J. Warrington, E. Y. Tanimoto, and C. G. Miyada. 2006. The Affymetrix GeneChip platform: an overview. Methods Enzymol. 410:3-28. [DOI] [PubMed] [Google Scholar]
- 39.Deng, J., Y. Zheng, R. Zhao, P. F. Wright, C. W. Stratton, and Y. W. Tang. 2009. Culture versus polymerase chain reaction for the etiologic diagnosis of community-acquired pneumonia in antibiotic-pretreated pediatric patients. Pediatr. Infect. Dis. J. 28:53-55. [DOI] [PubMed] [Google Scholar]
- 40.Deng, J.-Y., X.-E. Zhang, H.-B. Lu, Q. Liu, Z.-P. Zhang, Y.-F. Zhou, W.-H. Xie, and Z.-J. Fu. 2004. Multiplex detection of mutations in clinical isolates of rifampin-resistant Mycobacterium tuberculosis by short oligonucleotide ligation assay on DNA chips. J. Clin. Microbiol. 42:4850-4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Denkin, S., D. Volokhov, V. Chizhikov, and Y. Zhang. 2005. Microarray-based pncA genotyping of pyrazinamide-resistant strains of Mycobacterium tuberculosis. J. Med. Microbiol. 54:1127-1131. [DOI] [PubMed] [Google Scholar]
- 42.de Souza Luna, L. K., V. Heiser, N. Regamey, M. Panning, J. F. Drexler, S. Mulangu, L. Poon, S. Baumgarte, B. J. Haijema, L. Kaiser, and C. Drosten. 2007. Generic detection of coronaviruses and differentiation at the prototype strain level by reverse transcription-PCR and nonfluorescent low-density microarray. J. Clin. Microbiol. 45:1049-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Diaz, M. R., and J. W. Fell. 2005. Use of a suspension array for rapid identification of the varieties and genotypes of the Cryptococcus neoformans species complex. J. Clin. Microbiol. 43:3662-3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dietrich, G., S. Kurz, C. Hubner, C. Aepinus, S. Theiss, M. Guckenberger, U. Panzner, J. Weber, and M. Frosch. 2003. Transcriptome analysis of Neisseria meningitidis during infection. J. Bacteriol. 185:155-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Domenech-Sanchez, A., V. J. Benedi, L. Martinez-Martinez, and S. Alberti. 2006. Evaluation of differential gene expression in susceptible and resistant clinical isolates of Klebsiella pneumoniae by DNA microarray analysis. Clin. Microbiol. Infect. 12:936-940. [DOI] [PubMed] [Google Scholar]
- 46.Dorrell, N., J. A. Mangan, K. G. Laing, J. Hinds, D. Linton, H. Al-Ghusein, B. G. Barrell, J. Parkhill, N. G. Stoker, A. V. Karlyshev, P. D. Butcher, and B. W. Wren. 2001. Whole genome comparison of Campylobacter jejuni human isolates using a low-cost microarray reveals extensive genetic diversity. Genome Res. 11:1706-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Du, P., W. A. Kibbe, and S. M. Lin. 2008. lumi: a pipeline for processing Illumina microarray. Bioinformatics 24:1547-1548. [DOI] [PubMed] [Google Scholar]
- 48.Dunbar, S. A. 2006. Applications of Luminex xMAP technology for rapid, high-throughput multiplexed nucleic acid detection. Clin. Chim. Acta 363:71-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dunning, M. J., N. L. Barbosa-Morais, A. G. Lynch, S. Tavare, and M. E. Ritchie. 2008. Statistical issues in the analysis of Illumina data. BMC Bioinformatics 9:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dunning, M. J., M. L. Smith, M. E. Ritchie, and S. Tavare. 2007. beadarray: R classes and methods for Illumina bead-based data. Bioinformatics 23:2183-2184. [DOI] [PubMed] [Google Scholar]
- 51.Ehrenreich, A. 2006. DNA microarray technology for the microbiologist: an overview. Appl. Microbiol. Biotechnol. 73:255-273. [DOI] [PubMed] [Google Scholar]
- 52.Fan, J. B., K. L. Gunderson, M. Bibikova, J. M. Yeakley, J. Chen, E. Wickham Garcia, L. L. Lebruska, M. Laurent, R. Shen, and D. Barker. 2006. Illumina universal bead arrays. Methods Enzymol. 410:57-73. [DOI] [PubMed] [Google Scholar]
- 53.Fan, J. B., S. X. Hu, W. C. Craumer, and D. L. Barker. 2005. BeadArray-based solutions for enabling the promise of pharmacogenomics. BioTechniques 39:583-588. [PubMed] [Google Scholar]
- 54.Fan, J. B., J. M. Yeakley, M. Bibikova, E. Chudin, E. Wickham, J. Chen, D. Doucet, P. Rigault, B. Zhang, R. Shen, C. McBride, H. R. Li, X. D. Fu, A. Oliphant, D. L. Barker, and M. S. Chee. 2004. A versatile assay for high-throughput gene expression profiling on universal array matrices. Genome Res. 14:878-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Feezor, R. J., C. Oberholzer, H. V. Baker, D. Novick, M. Rubinstein, L. L. Moldawer, J. Pribble, S. Souza, C. A. Dinarello, W. Ertel, and A. Oberholzer. 2003. Molecular characterization of the acute inflammatory response to infections with gram-negative versus gram-positive bacteria. Infect. Immun. 71:5803-5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fitzgerald, C., M. Collins, S. van Duyne, M. Mikoleit, T. Brown, and P. Fields. 2007. Multiplex, bead-based suspension array for molecular determination of common Salmonella serogroups. J. Clin. Microbiol. 45:3323-3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fitzgerald, J. R., D. E. Sturdevant, S. M. Mackie, S. R. Gill, and J. M. Musser. 2001. Evolutionary genomics of Staphylococcus aureus: insights into the origin of methicillin-resistant strains and the toxic shock syndrome epidemic. Proc. Natl. Acad. Sci. USA 98:8821-8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fjaerli, H. O., G. Bukholm, A. Krog, C. Skjaeret, M. Holden, and B. Nakstad. 2006. Whole blood gene expression in infants with respiratory syncytial virus bronchiolitis. BMC Infect. Dis. 6:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fodor, S. P., J. L. Read, M. C. Pirrung, L. Stryer, A. T. Lu, and D. Solas. 1991. Light-directed, spatially addressable parallel chemical synthesis. Science 251:767-773. [DOI] [PubMed] [Google Scholar]
- 60.Fu, L. M., and T. M. Shinnick. 2007. Understanding the action of INH on a highly INH-resistant Mycobacterium tuberculosis strain using Genechips. Tuberculosis (Edinburgh) 87:63-70. [DOI] [PubMed] [Google Scholar]
- 61.Fukushima, M., K. Kakinuma, H. Hayashi, H. Nagai, K. Ito, and R. Kawaguchi. 2003. Detection and identification of Mycobacterium species isolates by DNA microarray. J. Clin. Microbiol. 41:2605-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gao, H., Y. Wang, X. Liu, T. Yan, L. Wu, E. Alm, A. Arkin, D. K. Thompson, and J. Zhou. 2004. Global transcriptome analysis of the heat shock response of Shewanella oneidensis. J. Bacteriol. 186:7796-7803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gegia, M., N. Mdivani, R. E. Mendes, H. Li, M. Akhalaia, J. Han, G. Khechinashvili, and Y. W. Tang. 2008. Prevalence of and molecular basis for tuberculosis drug resistance in the Republic of Georgia: validation of a QIAplex system for detection of drug resistance-related mutations. Antimicrob. Agents Chemother. 52:725-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gentry, T. J., and J. Zhou. 2006. Microarray-based microbial identification and characterization, p. 276-290. In Y. W. Tang and C. W. Stratton (ed.), Advanced techniques in diagnostic microbiology. Springer Science and Business Media, New York, NY.
- 65.Gheit, T., G. Billoud, M. N. de Koning, F. Gemignani, O. Forslund, B. S. Sylla, S. Vaccarella, S. Franceschi, S. Landi, W. G. Quint, F. Canzian, and M. Tommasino. 2007. Development of a sensitive and specific multiplex PCR method combined with DNA microarray primer extension to detect betapapillomavirus types. J. Clin. Microbiol. 45:2537-2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gheit, T., S. Landi, F. Gemignani, P. J. Snijders, S. Vaccarella, S. Franceschi, F. Canzian, and M. Tommasino. 2006. Development of a sensitive and specific assay combining multiplex PCR and DNA microarray primer extension to detect high-risk mucosal human papillomavirus types. J. Clin. Microbiol. 44:2025-2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grifantini, R., E. Bartolini, A. Muzzi, M. Draghi, E. Frigimelica, J. Berger, F. Randazzo, and G. Grandi. 2002. Gene expression profile in Neisseria meningitidis and Neisseria lactamica upon host-cell contact: from basic research to vaccine development. Ann. N. Y. Acad. Sci. 975:202-216. [DOI] [PubMed] [Google Scholar]
- 68.Grimm, V., S. Ezaki, M. Susa, C. Knabbe, R. D. Schmid, and T. T. Bachmann. 2004. Use of DNA microarrays for rapid genotyping of TEM beta-lactamases that confer resistance. J. Clin. Microbiol. 42:3766-3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gryadunov, D., V. Mikhailovich, S. Lapa, N. Roudinskii, M. Donnikov, S. Pan'kov, O. Markova, A. Kuz'min, L. Chernousova, O. Skotnikova, A. Moroz, A. Zasedatelev, and A. Mirzabekov. 2005. Evaluation of hybridisation on oligonucleotide microarrays for analysis of drug-resistant Mycobacterium tuberculosis. Clin. Microbiol. Infect. 11:531-539. [DOI] [PubMed] [Google Scholar]
- 70.Gunderson, K. L. 2009. Whole-genome genotyping on bead arrays. Methods Mol. Biol. 529:197-213. [DOI] [PubMed] [Google Scholar]
- 71.Gunderson, K. L., S. Kruglyak, M. S. Graige, F. Garcia, B. G. Kermani, C. Zhao, D. Che, T. Dickinson, E. Wickham, J. Bierle, D. Doucet, M. Milewski, R. Yang, C. Siegmund, J. Haas, L. Zhou, A. Oliphant, J. B. Fan, S. Barnard, and M. S. Chee. 2004. Decoding randomly ordered DNA arrays. Genome Res. 14:870-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hager, J. 2006. Making and using spotted DNA microarrays in an academic core laboratory. Methods Enzymol. 410:135-168. [DOI] [PubMed] [Google Scholar]
- 73.Han, J., D. C. Swan, S. J. Smith, S. H. Lum, S. E. Sefers, E. R. Unger, and Y. W. Tang. 2006. Simultaneous amplification and identification of 25 human papillomavirus types with Templex technology. J. Clin. Microbiol. 44:4157-4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hayward, R. E., J. L. Derisi, S. Alfadhli, D. C. Kaslow, P. O. Brown, and P. K. Rathod. 2000. Shotgun DNA microarrays and stage-specific gene expression in Plasmodium falciparum malaria. Mol. Microbiol. 35:6-14. [DOI] [PubMed] [Google Scholar]
- 75.Helbig, K. J., A. Ruszkiewicz, R. E. Lanford, M. D. Berzsenyi, H. A. Harley, S. R. McColl, and M. R. Beard. 2009. Differential expression of the CXCR3 ligands in chronic hepatitis C virus (HCV) infection and their modulation by HCV in vitro. J. Virol. 83:836-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Helbig, K. J., A. Ruszkiewicz, L. Semendric, H. A. Harley, S. R. McColl, and M. R. Beard. 2004. Expression of the CXCR3 ligand I-TAC by hepatocytes in chronic hepatitis C and its correlation with hepatic inflammation. Hepatology 39:1220-1229. [DOI] [PubMed] [Google Scholar]
- 77.Honma, S., V. Chizhikov, N. Santos, M. Tatsumi, M. D. C. S. T. Timenetsky, A. C. Linhares, J. D. Mascarenhas, H. Ushijima, G. E. Armah, J. R. Gentsch, and Y. Hoshino. 2007. Development and validation of DNA microarray for genotyping group A rotavirus VP4 (P[4], P[6], P[8], P[9], and P[14]) and VP7 (G1 to G6, G8 to G10, and G12) genes. J. Clin. Microbiol. 45:2641-2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Horan, P. K., and L. L. Wheeless, Jr. 1977. Quantitative single cell analysis and sorting. Science 198:149-157. [DOI] [PubMed] [Google Scholar]
- 79.Hou, X. L., H. L. Jiang, Q. Y. Cao, L. Y. Zhao, B. J. Chang, and Z. Chen. 2008. Using oligonucleotide suspension arrays for laboratory identification of bacteria responsible for bacteremia. J. Zhejiang Univ. Sci. B 9:291-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hsiao, C. R., L. Huang, J.-P. Bouchara, R. Barton, H. C. Li, and T. C. Chang. 2005. Identification of medically important molds by an oligonucleotide array. J. Clin. Microbiol. 43:3760-3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Huang, A., J.-W. Li, Z.-Q. Shen, X.-W. Wang, and M. Jin. 2006. High-throughput identification of clinical pathogenic fungi by hybridization to an oligonucleotide microarray. J. Clin. Microbiol. 44:3299-3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Huang, Q., D. Liu, P. Majewski, L. C. Schulte, J. M. Korn, R. A. Young, E. S. Lander, and N. Hacohen. 2001. The plasticity of dendritic cell responses to pathogens and their components. Science 294:870-875. [DOI] [PubMed] [Google Scholar]
- 83.Hughes, T. R., M. Mao, A. R. Jones, J. Burchard, M. J. Marton, K. W. Shannon, S. M. Lefkowitz, M. Ziman, J. M. Schelter, M. R. Meyer, S. Kobayashi, C. Davis, H. Dai, Y. D. He, S. B. Stephaniants, G. Cavet, W. L. Walker, A. West, E. Coffey, D. D. Shoemaker, R. Stoughton, A. P. Blanchard, S. H. Friend, and P. S. Linsley. 2001. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat. Biotechnol. 19:342-347. [DOI] [PubMed] [Google Scholar]
- 84.Iannone, M. A., J. D. Taylor, J. Chen, M. S. Li, P. Rivers, K. A. Slentz-Kesler, and M. P. Weiner. 2000. Multiplexed single nucleotide polymorphism genotyping by oligonucleotide ligation and flow cytometry. Cytometry 39:131-140. [PubMed] [Google Scholar]
- 85.Israel, D. A., N. Salama, C. N. Arnold, S. F. Moss, T. Ando, H. P. Wirth, K. T. Tham, M. Camorlinga, M. J. Blaser, S. Falkow, and R. M. Peek, Jr. 2001. Helicobacter pylori strain-specific differences in genetic content, identified by microarray, influence host inflammatory responses. J. Clin. Investig. 107:611-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jaaskelainen, A. J., and L. Maunula. 2006. Applicability of microarray technique for the detection of noro- and astroviruses. J. Virol. Methods 136:210-216. [DOI] [PubMed] [Google Scholar]
- 87.Jacobsen, M., D. Repsilber, A. Gutschmidt, A. Neher, K. Feldmann, H. J. Mollenkopf, A. Ziegler, and S. H. Kaufmann. 2007. Candidate biomarkers for discrimination between infection and disease caused by Mycobacterium tuberculosis. J. Mol. Med. 85:613-621. [DOI] [PubMed] [Google Scholar]
- 88.Jenner, R. G., and R. A. Young. 2005. Insights into host responses against pathogens from transcriptional profiling. Nat. Rev. Microbiol. 3:281-294. [DOI] [PubMed] [Google Scholar]
- 89.Johnson, S. B., M. Lissauer, G. V. Bochicchio, R. Moore, A. S. Cross, and T. M. Scalea. 2007. Gene expression profiles differentiate between sterile SIRS and early sepsis. Ann. Surg. 245:611-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Johnson, S. C., D. J. Marshall, G. Harms, C. M. Miller, C. B. Sherrill, E. L. Beaty, S. A. Lederer, E. B. Roesch, G. Madsen, G. L. Hoffman, R. H. Laessig, G. J. Kopish, M. W. Baker, S. A. Benner, P. M. Farrell, and J. R. Prudent. 2004. Multiplexed genetic analysis using an expanded genetic alphabet. Clin. Chem. 50:2019-2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kafatos, F. C., C. W. Jones, and A. Efstratiadis. 1979. Determination of nucleic acid sequence homologies and relative concentrations by a dot hybridization procedure. Nucleic Acids Res. 7:1541-1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kakinuma, K., M. Fukushima, and R. Kawaguchi. 2003. Detection and identification of Escherichia coli, Shigella, and Salmonella by microarrays using the gyrB gene. Biotechnol. Bioeng. 83:721-728. [DOI] [PubMed] [Google Scholar]
- 93.Kaushal, D., B. G. Schroeder, S. Tyagi, T. Yoshimatsu, C. Scott, C. Ko, L. Carpenter, J. Mehrotra, Y. C. Manabe, R. D. Fleischmann, and W. R. Bishai. 2002. Reduced immunopathology and mortality despite tissue persistence in a Mycobacterium tuberculosis mutant lacking alternative sigma factor, SigH. Proc. Natl. Acad. Sci. USA 99:8330-8335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Keller, C., J. Lauber, A. Blumenthal, J. Buer, and S. Ehlers. 2004. Resistance and susceptibility to tuberculosis analysed at the transcriptome level: lessons from mouse macrophages. Tuberculosis (Edinburgh) 84:144-158. [DOI] [PubMed] [Google Scholar]
- 95.Kerr, J. R., N. Kaushik, D. Fear, D. A. Baldwin, E. F. Nuwaysir, and I. M. Adcock. 2005. Single-nucleotide polymorphisms associated with symptomatic infection and differential human gene expression in healthy seropositive persons each implicate the cytoskeleton, integrin signaling, and oncosuppression in the pathogenesis of human parvovirus B19 infection. J. Infect. Dis. 192:276-286. [DOI] [PubMed] [Google Scholar]
- 96.Khodakov, D. A., N. V. Zakharova, D. A. Gryadunov, F. P. Filatov, A. S. Zasedatelev, and V. M. Mikhailovich. 2008. An oligonucleotide microarray for multiplex real-time PCR identification of HIV-1, HBV, and HCV. BioTechniques 44:241-246, 248. [DOI] [PubMed] [Google Scholar]
- 97.Kim, H. S., E. H. Choi, J. Khan, E. Roilides, A. Francesconi, M. Kasai, T. Sein, R. L. Schaufele, K. Sakurai, C. G. Son, B. T. Greer, S. Chanock, C. A. Lyman, and T. J. Walsh. 2005. Expression of genes encoding innate host defense molecules in normal human monocytes in response to Candida albicans. Infect. Immun. 73:3714-3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kistler, A., P. C. Avila, S. Rouskin, D. Wang, T. Ward, S. Yagi, D. Schnurr, D. Ganem, J. L. Derisi, and H. A. Boushey. 2007. Pan-viral screening of respiratory tract infections in adults with and without asthma reveals unexpected human coronavirus and human rhinovirus diversity. J. Infect. Dis. 196:817-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Koessler, T., P. Francois, Y. Charbonnier, A. Huyghe, M. Bento, S. Dharan, G. Renzi, D. Lew, S. Harbarth, D. Pittet, and J. Schrenzel. 2006. Use of oligoarrays for characterization of community-onset methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 44:1040-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Korczak, B., J. Frey, J. Schrenzel, G. Pluschke, R. Pfister, R. Ehricht, and P. Kuhnert. 2005. Use of diagnostic microarrays for determination of virulence gene patterns of Escherichia coli K1, a major cause of neonatal meningitis. J. Clin. Microbiol. 43:1024-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Korimbocus, J., N. Scaramozzino, B. Lacroix, J. M. Crance, D. Garin, and G. Vernet. 2005. DNA probe array for the simultaneous identification of herpesviruses, enteroviruses, and flaviviruses. J. Clin. Microbiol. 43:3779-3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kostic, T., A. Weilharter, S. Rubino, G. Delogu, S. Uzzau, K. Rudi, A. Sessitsch, and L. Bodrossy. 2007. A microbial diagnostic microarray technique for the sensitive detection and identification of pathogenic bacteria in a background of nonpathogens. Anal. Biochem. 360:244-254. [DOI] [PubMed] [Google Scholar]
- 103.Kostrzynska, M., and A. Bachand. 2006. Application of DNA microarray technology for detection, identification, and characterization of food-borne pathogens. Can. J. Microbiol. 52:1-8. [DOI] [PubMed] [Google Scholar]
- 104.Kozal, M. J., N. Shah, N. Shen, R. Yang, R. Fucini, T. C. Merigan, D. D. Richman, D. Morris, E. Hubbell, M. Chee, and T. R. Gingeras. 1996. Extensive polymorphisms observed in HIV-1 clade B protease gene using high-density oligonucleotide arrays. Nat. Med. 2:753-759. [DOI] [PubMed] [Google Scholar]
- 105.Kreil, D. P., R. R. Russell, and S. Russell. 2006. Microarray oligonucleotide probes. Methods Enzymol. 410:73-98. [DOI] [PubMed] [Google Scholar]
- 106.Krunic, N., T. D. Yager, D. Himsworth, F. Merante, S. Yaghoubian, and R. Janeczko. 2007. xTAG RVP assay: analytical and clinical performance. J. Clin. Virol. 40(Suppl. 1):S39-S46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kuhn, K., S. C. Baker, E. Chudin, M. H. Lieu, S. Oeser, H. Bennett, P. Rigault, D. Barker, T. K. McDaniel, and M. S. Chee. 2004. A novel, high-performance random array platform for quantitative gene expression profiling. Genome Res. 14:2347-2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kumar, S., L. Wang, J. Fan, A. Kraft, M. E. Bose, S. Tiwari, M. Van Dyke, R. Haigis, T. Luo, M. Ghosh, H. Tang, M. Haghnia, E. L. Mather, W. G. Weisburg, and K. J. Henrickson. 2008. Detection of 11 common viral and bacterial pathogens causing community-acquired pneumonia or sepsis in asymptomatic patients by using a multiplex reverse transcription-PCR assay with manual (enzyme hybridization) or automated (electronic microarray) detection. J. Clin. Microbiol. 46:3063-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lee, W. M., K. Grindle, T. Pappas, D. J. Marshall, M. J. Moser, E. L. Beaty, P. A. Shult, J. R. Prudent, and J. E. Gern. 2007. High-throughput, sensitive, and accurate multiplex PCR-microsphere flow cytometry system for large-scale comprehensive detection of respiratory viruses. J. Clin. Microbiol. 45:2626-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Leinberger, D. M., U. Schumacher, I. B. Autenrieth, and T. T. Bachmann. 2005. Development of a DNA microarray for detection and identification of fungal pathogens involved in invasive mycoses. J. Clin. Microbiol. 43:4943-4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lenhoff, R. J., P. Naraghi-Arani, J. B. Thissen, J. Olivas, A. C. Carillo, C. Chinn, M. Rasmussen, S. M. Messenger, L. D. Suer, S. M. Smith, L. F. Tammero, E. A. Vitalis, T. R. Slezak, P. J. Hullinger, B. J. Hindson, S. K. Hietala, B. M. Crossley, and M. T. McBride. 2008. Multiplexed molecular assay for rapid exclusion of foot-and-mouth disease. J. Virol. Methods 153:61-69. [DOI] [PubMed] [Google Scholar]
- 112.Li, H., M. A. McCormac, R. W. Estes, S. E. Sefers, R. K. Dare, J. D. Chappell, D. D. Erdman, P. F. Wright, and Y. W. Tang. 2007. Simultaneous detection and high-throughput identification of a panel of RNA viruses causing respiratory tract infections. J. Clin. Microbiol. 45:2105-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Li, J., S. Chen, and D. H. Evans. 2001. Typing and subtyping influenza virus using DNA microarrays and multiplex reverse transcriptase PCR. J. Clin. Microbiol. 39:696-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Li, Y., D. Liu, B. Cao, W. Han, Y. Liu, F. Liu, X. Guo, D. A. Bastin, L. Feng, and L. Wang. 2006. Development of a serotype-specific DNA microarray for identification of some Shigella and pathogenic Escherichia coli strains. J. Clin. Microbiol. 44:4376-4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lin, B., K. M. Blaney, A. P. Malanoski, A. G. Ligler, J. M. Schnur, D. Metzgar, K. L. Russell, and D. A. Stenger. 2007. Using a resequencing microarray as a multiple respiratory pathogen detection assay. J. Clin. Microbiol. 45:443-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lin, B., A. P. Malanoski, Z. Wang, K. M. Blaney, N. C. Long, C. E. Meador, D. Metzgar, C. A. Myers, S. L. Yingst, M. R. Monteville, M. D. Saad, J. M. Schnur, C. Tibbetts, and D. A. Stenger. 2009. Universal detection and identification of avian influenza virus by use of resequencing microarrays. J. Clin. Microbiol. 47:988-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lin, B., G. J. Vora, D. Thach, E. Walter, D. Metzgar, C. Tibbetts, and D. A. Stenger. 2004. Use of oligonucleotide microarrays for rapid detection and serotyping of acute respiratory disease-associated adenoviruses. J. Clin. Microbiol. 42:3232-3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lin, M. T., and T. E. Albertson. 2004. Genomic polymorphisms in sepsis. Crit. Care Med. 32:569-579. [DOI] [PubMed] [Google Scholar]
- 119.Lin, Z. H., X. H. Shen, Z. Jin, Y. Kim, E. Lee, H. Kim, and I. Kim. 2005. Human papillomavirus genotyping by oligonucleotide microarray and p16 expression in uterine cervical intraepithelial neoplasm and in invasive carcinoma in Korean women. Pathol. Int. 55:491-496. [DOI] [PubMed] [Google Scholar]
- 120.Lissauer, M. E., S. B. Johnson, G. V. Bochicchio, C. J. Feild, A. S. Cross, J. D. Hasday, C. C. Whiteford, W. A. Nussbaumer, M. Towns, and T. M. Scalea. 2009. Differential expression of Toll-like receptor genes: sepsis compared with sterile inflammation 1 day before sepsis diagnosis. Shock 31:238-244. [DOI] [PubMed] [Google Scholar]
- 121.Liu, T. T., R. E. Lee, K. S. Barker, R. E. Lee, L. Wei, R. Homayouni, and P. D. Rogers. 2005. Genome-wide expression profiling of the response to azole, polyene, echinocandin, and pyrimidine antifungal agents in Candida albicans. Antimicrob. Agents Chemother. 49:2226-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lopez, M. F., and M. G. Pluskal. 2003. Protein micro- and macroarrays: digitizing the proteome. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 787:19-27. [DOI] [PubMed] [Google Scholar]
- 123.Lovmar, L., C. Fock, F. Espinoza, F. Bucardo, A. C. Syvanen, and K. Bondeson. 2003. Microarrays for genotyping human group A rotavirus by multiplex capture and type-specific primer extension. J. Clin. Microbiol. 41:5153-5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lucchini, S., H. Liu, Q. Jin, J. C. Hinton, and J. Yu. 2005. Transcriptional adaptation of Shigella flexneri during infection of macrophages and epithelial cells: insights into the strategies of a cytosolic bacterial pathogen. Infect. Immun. 73:88-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.MacBeath, G. 2002. Protein microarrays and proteomics. Nat. Genet. 32(Suppl.):526-532. [DOI] [PubMed] [Google Scholar]
- 126.Mahony, J., S. Chong, F. Merante, S. Yaghoubian, T. Sinha, C. Lisle, and R. Janeczko. 2007. Development of a respiratory virus panel test for detection of twenty human respiratory viruses by use of multiplex PCR and a fluid microbead-based assay. J. Clin. Microbiol. 45:2965-2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Malanoski, A. P., B. Lin, Z. Wang, J. M. Schnur, and D. A. Stenger. 2006. Automated identification of multiple micro-organisms from resequencing DNA microarrays. Nucleic Acids Res. 34:5300-5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Marlowe, E. M., J. J. Hogan, J. F. Hindler, I. Andruszkiewicz, P. Gordon, and D. A. Bruckner. 2003. Application of an rRNA probe matrix for rapid identification of bacteria and fungi from routine blood cultures. J. Clin. Microbiol. 41:5127-5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.McDunn, J. E., I. R. Turnbull, A. D. Polpitiya, A. Tong, S. K. MacMillan, D. F. Osborne, R. S. Hotchkiss, M. Colonna, and J. P. Cobb. 2006. Splenic CD4+ T cells have a distinct transcriptional response six hours after the onset of sepsis. J. Am. Coll. Surg. 203:365-375. [DOI] [PubMed] [Google Scholar]
- 130.McHugh, R. S., W. D. Ratnoff, R. Gilmartin, K. W. Sell, and P. Selvaraj. 1998. Detection of a soluble form of B7-1 (CD80) in synovial fluid from patients with arthritis using monoclonal antibodies against distinct epitopes of human B7-1. Clin. Immunol. Immunopathol. 87:50-59. [DOI] [PubMed] [Google Scholar]
- 131.Mehlmann, M., A. B. Bonner, J. V. Williams, D. M. Dankbar, C. L. Moore, R. D. Kuchta, A. B. Podsiad, J. D. Tamerius, E. D. Dawson, and K. L. Rowlen. 2007. Comparison of the MChip to viral culture, reverse transcription-PCR, and the QuickVue influenza A+B test for rapid diagnosis of influenza. J. Clin. Microbiol. 45:1234-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Merante, F., S. Yaghoubian, and R. Janeczko. 2007. Principles of the xTAG respiratory viral panel assay (RVP assay). J. Clin. Virol. 40(Suppl. 1):S31-S35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Mikhailovich, V., D. Gryadunov, A. Kolchinsky, A. A. Makarov, and A. Zasedatelev. 2008. DNA microarrays in the clinic: infectious diseases. Bioessays 30:673-682. [DOI] [PubMed] [Google Scholar]
- 134.Mikhailovich, V., S. Lapa, D. Gryadunov, A. Sobolev, B. Strizhkov, N. Chernyh, O. Skotnikova, O. Irtuganova, A. Moroz, V. Litvinov, M. Vladimirskii, M. Perelman, L. Chernousova, V. Erokhin, A. Zasedatelev, and A. Mirzabekov. 2001. Identification of rifampin-resistant Mycobacterium tuberculosis strains by hybridization, PCR, and ligase detection reaction on oligonucleotide microchips. J. Clin. Microbiol. 39:2531-2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Miller, M. B. 2009. Solid and liquid phase array technologies. In D. Persing, F. Tenover, R. Hayden, F. Nolte, Y. W. Tang, and A. Van Belkum (ed.), Molecular microbiology: diagnostic principles and practice, 2nd ed., in press. ASM Press, Washington, DC.
- 136.Mitterer, G., M. Huber, E. Leidinger, C. Kirisits, W. Lubitz, M. W. Mueller, and W. M. Schmidt. 2004. Microarray-based identification of bacteria in clinical samples by solid-phase PCR amplification of 23S ribosomal DNA sequences. J. Clin. Microbiol. 42:1048-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Monecke, S., B. Berger-Bachi, G. Coombs, A. Holmes, I. Kay, A. Kearns, H. J. Linde, F. O'Brien, P. Slickers, and R. Ehricht. 2007. Comparative genomics and DNA array-based genotyping of pandemic Staphylococcus aureus strains encoding Panton-Valentine leukocidin. Clin. Microbiol. Infect. 13:236-249. [DOI] [PubMed] [Google Scholar]
- 138.Monecke, S., and R. Ehricht. 2005. Rapid genotyping of methicillin-resistant Staphylococcus aureus (MRSA) isolates using miniaturised oligonucleotide arrays. Clin. Microbiol. Infect. 11:825-833. [DOI] [PubMed] [Google Scholar]
- 139.Monecke, S., P. Slickers, H. Hotzel, G. Richter-Huhn, M. Pohle, S. Weber, W. Witte, and R. Ehricht. 2006. Microarray-based characterisation of a Panton-Valentine leukocidin-positive community-acquired strain of methicillin-resistant Staphylococcus aureus. Clin. Microbiol. Infect. 12:718-728. [DOI] [PubMed] [Google Scholar]
- 140.Muldrew, K. L., S. H. Beqaj, J. Han, S. H. Lum, V. Clinard, S. J. Schultenover, and Y. W. Tang. 2007. Evaluation of a Digene-recommended algorithm for human papillomavirus low-positive results present in a “retest zone.” Am. J. Clin. Pathol. 127:97-102. [DOI] [PubMed] [Google Scholar]
- 141.Nazarenko, I., L. Kobayashi, J. Giles, C. Fishman, G. Chen, and A. Lorincz. 2008. A novel method of HPV genotyping using Hybrid Capture sample preparation method combined with GP5+/6+ PCR and multiplex detection on Luminex XMAP. J. Virol. Methods 154:76-81. [DOI] [PubMed] [Google Scholar]
- 142.Nemeth, Z. H., B. Csoka, J. Wilmanski, D. Xu, Q. Lu, C. Ledent, E. A. Deitch, P. Pacher, Z. Spolarics, and G. Hasko. 2006. Adenosine A2A receptor inactivation increases survival in polymicrobial sepsis. J. Immunol. 176:5616-5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Nolte, F. S., D. J. Marshall, C. Rasberry, S. Schievelbein, G. G. Banks, G. A. Storch, M. Q. Arens, R. S. Buller, and J. R. Prudent. 2007. MultiCode-PLx system for multiplexed detection of seventeen respiratory viruses. J. Clin. Microbiol. 45:2779-2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Nubel, U., P. M. Schmidt, E. Reiss, F. Bier, W. Beyer, and D. Naumann. 2004. Oligonucleotide microarray for identification of Bacillus anthracis based on intergenic transcribed spacers in ribosomal DNA. FEMS Microbiol. Lett. 240:215-223. [DOI] [PubMed] [Google Scholar]
- 145.Oh, Y., S. M. Bae, Y. W. Kim, H. S. Choi, G. H. Nam, S. J. Han, C. H. Park, Y. Cho, B. D. Han, and W. S. Ahn. 2007. Polymerase chain reaction-based fluorescent Luminex assay to detect the presence of human papillomavirus types. Cancer Sci. 98:549-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Oliphant, A., D. L. Barker, J. R. Stuelpnagel, and M. S. Chee. 2002. BeadArray technology: enabling an accurate, cost-effective approach to high-throughput genotyping. BioTechniques 2002(Suppl.):56-58, 60-61. [PubMed] [Google Scholar]
- 147.Otsuka, M., H. Aizaki, N. Kato, T. Suzuki, T. Miyamura, M. Omata, and N. Seki. 2003. Differential cellular gene expression induced by hepatitis B and C viruses. Biochem. Biophys. Res. Commun. 300:443-447. [DOI] [PubMed] [Google Scholar]
- 148.Pabbaraju, K., K. L. Tokaryk, S. Wong, and J. D. Fox. 2008. Comparison of the Luminex xTAG respiratory viral panel with in-house nucleic acid amplification tests for diagnosis of respiratory virus infections. J. Clin. Microbiol. 46:3056-3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Pachot, A., A. Lepape, S. Vey, J. Bienvenu, B. Mougin, and G. Monneret. 2006. Systemic transcriptional analysis in survivor and non-survivor septic shock patients: a preliminary study. Immunol. Lett. 106:63-71. [DOI] [PubMed] [Google Scholar]
- 150.Palacios, G., P. L. Quan, O. J. Jabado, S. Conlan, D. L. Hirschberg, Y. Liu, J. Zhai, N. Renwick, J. Hui, H. Hegyi, A. Grolla, J. E. Strong, J. S. Towner, T. W. Geisbert, P. B. Jahrling, C. Buchen-Osmond, H. Ellerbrok, M. P. Sanchez-Seco, Y. Lussier, P. Formenty, M. S. Nichol, H. Feldmann, T. Briese, and W. I. Lipkin. 2007. Panmicrobial oligonucleotide array for diagnosis of infectious diseases. Emerg. Infect. Dis. 13:73-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Palmer, C., E. M. Bik, M. B. Eisen, P. B. Eckburg, T. R. Sana, P. K. Wolber, D. A. Relman, and P. O. Brown. 2006. Rapid quantitative profiling of complex microbial populations. Nucleic Acids Res. 34:e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Pas, S. D., N. Tran, R. A. de Man, C. Burghoorn-Maas, G. Vernet, and H. G. Niesters. 2008. Comparison of reverse hybridization, microarray, and sequence analysis for genotyping hepatitis B virus. J. Clin. Microbiol. 46:1268-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Perreten, V., L. Vorlet-Fawer, P. Slickers, R. Ehricht, P. Kuhnert, and J. Frey. 2005. Microarray-based detection of 90 antibiotic resistance genes of gram-positive bacteria. J. Clin. Microbiol. 43:2291-2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Podzorski, R. P., H. Li, J. Han, and Y.-W. Tang. 2008. MVPlex assay for direct detection of methicillin-resistant Staphylococcus aureus in naris and other swab specimens. J. Clin. Microbiol. 46:3107-3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Porwollik, S., R. M. Wong, and M. McClelland. 2002. Evolutionary genomics of Salmonella: gene acquisitions revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 99:8956-8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Prucha, M., A. Ruryk, H. Boriss, E. Moller, R. Zazula, I. Herold, R. A. Claus, K. A. Reinhart, P. Deigner, and S. Russwurm. 2004. Expression profiling: toward an application in sepsis diagnostics. Shock 22:29-33. [DOI] [PubMed] [Google Scholar]
- 157.Quan, P.-L., G. Palacios, O. J. Jabado, S. Conlan, D. L. Hirschberg, F. Pozo, P. M. J. Jack, D. Cisterna, N. Renwick, J. Hui, A. Drysdale, R. Amos-Ritchie, E. Baumeister, V. Savy, K. M. Lager, J. A. Richt, D. B. Boyle, A. García-Sastre, I. Casas, P. Perez-Breña, T. Briese, and W. I. Lipkin. 2007. Detection of respiratory viruses and subtype identification of influenza A viruses by GreeneChipResp oligonucleotide microarray. J. Clin. Microbiol. 45:2359-2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Rachman, H., M. Strong, T. Ulrichs, L. Grode, J. Schuchhardt, H. Mollenkopf, G. A. Kosmiadi, D. Eisenberg, and S. H. Kaufmann. 2006. Unique transcriptome signature of Mycobacterium tuberculosis in pulmonary tuberculosis. Infect. Immun. 74:1233-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ramdas, L., D. E. Cogdell, J. Y. Jia, E. E. Taylor, V. R. Dunmire, L. Hu, S. R. Hamilton, and W. Zhang. 2004. Improving signal intensities for genes with low-expression on oligonucleotide microarrays. BMC Genomics 5:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Raymond, F., J. Carbonneau, N. Boucher, L. Robitaille, S. Boivert, W. K. Wu, G. De Serres, G. Boivin, and J. Corbeil. 2009. Comparison of automated microarray detection with real-time PCR assays for detection of respiratory viruses in specimens obtained from children. J. Clin. Microbiol. 47:743-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Rengarajan, J., B. R. Bloom, and E. J. Rubin. 2005. Genome-wide requirements for Mycobacterium tuberculosis adaptation and survival in macrophages. Proc. Natl. Acad. Sci. USA 102:8327-8332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Revel, A. T., A. M. Talaat, and M. V. Norgard. 2002. DNA microarray analysis of differential gene expression in Borrelia burgdorferi, the Lyme disease spirochete. Proc. Natl. Acad. Sci. USA 99:1562-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Rota, P. A., M. S. Oberste, S. S. Monroe, W. A. Nix, R. Campagnoli, J. P. Icenogle, S. Penaranda, B. Bankamp, K. Maher, M. H. Chen, S. Tong, A. Tamin, L. Lowe, M. Frace, J. L. DeRisi, Q. Chen, D. Wang, D. D. Erdman, T. C. Peret, C. Burns, T. G. Ksiazek, P. E. Rollin, A. Sanchez, S. Liffick, B. Holloway, J. Limor, K. McCaustland, M. Olsen-Rasmussen, R. Fouchier, S. Gunther, A. D. Osterhaus, C. Drosten, M. A. Pallansch, L. J. Anderson, and W. J. Bellini. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394-1399. [DOI] [PubMed] [Google Scholar]
- 164.Salama, N., K. Guillemin, T. K. McDaniel, G. Sherlock, L. Tompkins, and S. Falkow. 2000. A whole-genome microarray reveals genetic diversity among Helicobacter pylori strains. Proc. Natl. Acad. Sci. USA 97:14668-14673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Sassetti, C. M., and E. J. Rubin. 2003. Genetic requirements for mycobacterial survival during infection. Proc. Natl. Acad. Sci. USA 100:12989-12994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Sato, M., H. Li, M. R. Ikizler, J. A. Werkhaven, J. V. Williams, J. D. Chappell, Y. W. Tang, and P. F. Wright. 2009. Detection of viruses in human adenoid tissues by use of multiplex PCR. J. Clin. Microbiol. 47:771-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Schena, M., D. Shalon, R. W. Davis, and P. O. Brown. 1995. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270:467-470. [DOI] [PubMed] [Google Scholar]
- 168.Scherl, A., P. Francois, Y. Charbonnier, J. M. Deshusses, T. Koessler, A. Huyghe, M. Bento, J. Stahl-Zeng, A. Fischer, A. Masselot, A. Vaezzadeh, F. Galle, A. Renzoni, P. Vaudaux, D. Lew, C. G. Zimmermann-Ivol, P. A. Binz, J. C. Sanchez, D. F. Hochstrasser, and J. Schrenzel. 2006. Exploring glycopeptide-resistance in Staphylococcus aureus: a combined proteomics and transcriptomics approach for the identification of resistance-related markers. BMC Genomics 7:296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Scillian, J. J., T. M. McHugh, M. P. Busch, M. Tam, M. J. Fulwyler, D. Y. Chien, and G. N. Vyas. 1989. Early detection of antibodies against rDNA-produced HIV proteins with a flow cytometric assay. Blood 73:2041-2048. [PubMed] [Google Scholar]
- 170.Sengupta, S., K. Onodera, A. Lai, and U. Melcher. 2003. Molecular detection and identification of influenza viruses by oligonucleotide microarray hybridization. J. Clin. Microbiol. 41:4542-4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Shackel, N. A., P. H. McGuinness, C. A. Abbott, M. D. Gorrell, and G. W. McCaughan. 2002. Insights into the pathobiology of hepatitis C virus-associated cirrhosis: analysis of intrahepatic differential gene expression. Am. J. Pathol. 160:641-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Shalon, D., S. J. Smith, and P. O. Brown. 1996. A DNA microarray system for analyzing complex DNA samples using two-color fluorescent probe hybridization. Genome Res. 6:639-645. [DOI] [PubMed] [Google Scholar]
- 173.Shang, S., G. Chen, Y. Wu, L. Du, and Z. Zhao. 2005. Rapid diagnosis of bacterial sepsis with PCR amplification and microarray hybridization in 16S rRNA gene. Pediatr. Res. 58:143-148. [DOI] [PubMed] [Google Scholar]
- 174.Simitsopoulou, M., E. Roilides, C. Likartsis, J. Ioannidis, A. Orfanou, F. Paliogianni, and T. J. Walsh. 2007. Expression of immunomodulatory genes in human monocytes induced by voriconazole in the presence of Aspergillus fumigatus. Antimicrob. Agents Chemother. 51:1048-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Sosnowski, R. G., E. Tu, W. F. Butler, J. P. O'Connell, and M. J. Heller. 1997. Rapid determination of single base mismatch mutations in DNA hybrids by direct electric field control. Proc. Natl. Acad. Sci. USA 94:1119-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Sougakoff, W., M. Rodrigue, C. Truffot-Pernot, M. Renard, N. Durin, M. Szpytma, R. Vachon, A. Troesch, and V. Jarlier. 2004. Use of a high-density DNA probe array for detecting mutations involved in rifampicin resistance in Mycobacterium tuberculosis. Clin. Microbiol. Infect. 10:289-294. [DOI] [PubMed] [Google Scholar]
- 177.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 178.Spiess, B., W. Seifarth, M. Hummel, O. Frank, A. Fabarius, C. Zheng, H. Morz, R. Hehlmann, and D. Buchheidt. 2007. DNA microarray-based detection and identification of fungal pathogens in clinical samples from neutropenic patients. J. Clin. Microbiol. 45:3743-3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Spiro, A., M. Lowe, and D. Brown. 2000. A bead-based method for multiplexed identification and quantitation of DNA sequences using flow cytometry. Appl. Environ. Microbiol. 66:4258-4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Stewart, G. R., B. D. Robertson, and D. B. Young. 2004. Analysis of the function of mycobacterial DnaJ proteins by overexpression and microarray profiling. Tuberculosis (Edinburgh) 84:180-187. [DOI] [PubMed] [Google Scholar]
- 181.Stintzi, A., D. Marlow, K. Palyada, H. Naikare, R. Panciera, L. Whitworth, and C. Clarke. 2005. Use of genome-wide expression profiling and mutagenesis to study the intestinal lifestyle of Campylobacter jejuni. Infect. Immun. 73:1797-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Stokes, T. H., X. Han, R. A. Moffitt, and M. D. Wang. 2007. Extending microarray quality control and analysis algorithms to Illumina chip platform. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2007:4637-4640. [DOI] [PubMed] [Google Scholar]
- 183.Stover, A. G., E. Jeffery, J. C. Xu, and D. H. Persing. 2003. Hybridization array, p. 619-639. In D. H. Persing, F. C. Tenevor, J. Versalovic, Y. W. Tang, E. R. Unger, D. A. Relman, and T. J. White (ed.), Molecular microbiology: diagnostic principles and practice. ASM Press, Washington, DC.
- 184.Strizhkov, B. N., A. L. Drobyshev, V. M. Mikhailovich, and A. D. Mirzabekov. 2000. PCR amplification on a microarray of gel-immobilized oligonucleotides: detection of bacterial toxin- and drug-resistant genes and their mutations. BioTechniques 29:844-848, 850-852, 854. [DOI] [PubMed] [Google Scholar]
- 185.Takahashi, H., S. A. Norman, E. L. Mather, and B. K. Patterson. 2008. Evaluation of the NanoChip 400 system for detection of influenza A and B, respiratory syncytial, and parainfluenza viruses. J. Clin. Microbiol. 46:1724-1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Takahashi, M., J. Okada, K. Ito, M. Hashimoto, K. Hashimoto, Y. Yoshida, Y. Furuichi, Y. Ohta, S. Mishiro, and N. Gemma. 2004. Electrochemical DNA array for simultaneous genotyping of single-nucleotide polymorphisms associated with the therapeutic effect of interferon. Clin. Chem. 50:658-661. [DOI] [PubMed] [Google Scholar]
- 187.Tang, B. M., A. S. McLean, I. W. Dawes, S. J. Huang, and R. C. Lin. 2009. Gene-expression profiling of peripheral blood mononuclear cells in sepsis. Crit. Care Med. 37:882-888. [DOI] [PubMed] [Google Scholar]
- 188.Tang, B. M., A. S. McLean, I. W. Dawes, S. J. Huang, and R. C. Lin. 2007. The use of gene-expression profiling to identify candidate genes in human sepsis. Am. J. Respir. Crit. Care Med. 176:676-684. [DOI] [PubMed] [Google Scholar]
- 189.Tang, X., S. L. Morris, J. J. Langone, and L. E. Bockstahler. 2005. Microarray and allele specific PCR detection of point mutations in Mycobacterium tuberculosis genes associated with drug resistance. J. Microbiol. Methods 63:318-330. [DOI] [PubMed] [Google Scholar]
- 190.Tang, Y.-W., N. M. Ellis, M. K. Hopkins, D. H. Smith, D. E. Dodge, and D. H. Persing. 1998. Comparison of phenotypic and genotypic techniques for identification of unusual aerobic pathogenic gram-negative bacilli. J. Clin. Microbiol. 36:3674-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Tang, Y.-W., A. Kilic, Q. Yang, S. K. McAllister, H. Li, R. S. Miller, M. McCormac, K. D. Tracy, C. W. Stratton, J. Han, and B. Limbago. 2007. StaphPlex system for rapid and simultaneous identification of antibiotic resistance determinants and Panton-Valentine leukocidin detection of staphylococci from positive blood cultures. J. Clin. Microbiol. 45:1867-1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Taylor, J. D., D. Briley, Q. Nguyen, K. Long, M. A. Iannone, M. S. Li, F. Ye, A. Afshari, E. Lai, M. Wagner, J. Chen, and M. P. Weiner. 2001. Flow cytometric platform for high-throughput single nucleotide polymorphism analysis. BioTechniques 30:661-666, 668-669. [DOI] [PubMed] [Google Scholar]
- 193.Tempfer, C., C. Grimm, C. Harwanegg, M. Huber, M. W. Mueller, B. Buerkle, A. Reinthaller, and L. A. Hefler. 2007. Frequency of 23 human papillomavirus types using DNA microarray in women with and without cytological anomalies. Anticancer Res. 27:1721-1726. [PubMed] [Google Scholar]
- 194.Tomiuk, S., and K. Hofmann. 2001. Microarray probe selection strategies. Brief. Bioinform. 2:329-340. [DOI] [PubMed] [Google Scholar]
- 195.Townsend, M. B., E. D. Dawson, M. Mehlmann, J. A. Smagala, D. M. Dankbar, C. L. Moore, C. B. Smith, N. J. Cox, R. D. Kuchta, and K. L. Rowlen. 2006. Experimental evaluation of the FluChip diagnostic microarray for influenza virus surveillance. J. Clin. Microbiol. 44:2863-2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Tran, N., R. Berne, R. Chann, M. Gauthier, D. Martin, M. A. Armand, A. Ollivet, C. G. Teo, S. Ijaz, D. Flichman, M. Brunetto, K. P. Bielawski, C. Pichoud, F. Zoulim, and G. Vernet. 2006. European multicenter evaluation of high-density DNA probe arrays for detection of hepatitis B virus resistance mutations and identification of genotypes. J. Clin. Microbiol. 44:2792-2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Troesch, A., H. Nguyen, C. G. Miyada, S. Desvarenne, T. R. Gingeras, P. M. Kaplan, P. Cros, and C. Mabilat. 1999. Mycobacterium species identification and rifampin resistance testing with high-density DNA probe arrays. J. Clin. Microbiol. 37:49-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Vahey, M., M. E. Nau, S. Barrick, J. D. Cooley, R. Sawyer, A. A. Sleeker, P. Vickerman, S. Bloor, B. Larder, N. L. Michael, and S. A. Wegner. 1999. Performance of the Affymetrix GeneChip HIV PRT 440 platform for antiretroviral drug resistance genotyping of human immunodeficiency virus type 1 clades and viral isolates with length polymorphisms. J. Clin. Microbiol. 37:2533-2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.van Ijperen, C., P. Kuhnert, J. Frey, and J. P. Clewley. 2002. Virulence typing of Escherichia coli using microarrays. Mol. Cell. Probes 16:371-378. [DOI] [PubMed] [Google Scholar]
- 200.van Leeuwen, W. B., C. Jay, S. Snijders, N. Durin, B. Lacroix, H. A. Verbrugh, M. C. Enright, A. Troesch, and A. van Belkum. 2003. Multilocus sequence typing of Staphylococcus aureus with DNA array technology. J. Clin. Microbiol. 41:3323-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Volokhov, D., V. Chizhikov, K. Chumakov, and A. Rasooly. 2003. Microarray-based identification of thermophilic Campylobacter jejuni, C. coli, C. lari, and C. upsaliensis. J. Clin. Microbiol. 41:4071-4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Vora, G. J., C. E. Meador, M. M. Bird, C. A. Bopp, J. D. Andreadis, and D. A. Stenger. 2005. Microarray-based detection of genetic heterogeneity, antimicrobial resistance, and the viable but nonculturable state in human pathogenic Vibrio spp. Proc. Natl. Acad. Sci. USA 102:19109-19114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Voskuil, M. I., D. Schnappinger, K. C. Visconti, M. I. Harrell, G. M. Dolganov, D. R. Sherman, and G. K. Schoolnik. 2003. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J. Exp. Med. 198:705-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Wade, M. M., D. Volokhov, M. Peredelchuk, V. Chizhikov, and Y. Zhang. 2004. Accurate mapping of mutations of pyrazinamide-resistant Mycobacterium tuberculosis strains with a scanning-frame oligonucleotide microarray. Diagn. Microbiol. Infect. Dis. 49:89-97. [DOI] [PubMed] [Google Scholar]
- 205.Wagner, T. H., A. M. Drewry, S. Macmillan, W. M. Dunne, K. C. Chang, I. E. Karl, R. S. Hotchkiss, and J. P. Cobb. 2007. Surviving sepsis: bcl-2 overexpression modulates splenocyte transcriptional responses in vivo. Am. J. Physiol. Regul. Integr. Comp. Physiol. 292:R1751-R1759. [DOI] [PubMed] [Google Scholar]
- 206.Wang, D., L. Coscoy, M. Zylberberg, P. C. Avila, H. A. Boushey, D. Ganem, and J. L. DeRisi. 2002. Microarray-based detection and genotyping of viral pathogens. Proc. Natl. Acad. Sci. USA 99:15687-15692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Wang, D., A. Urisman, Y. T. Liu, M. Springer, T. G. Ksiazek, D. D. Erdman, E. R. Mardis, M. Hickenbotham, V. Magrini, J. Eldred, J. P. Latreille, R. K. Wilson, D. Ganem, and J. L. DeRisi. 2003. Viral discovery and sequence recovery using DNA microarrays. PLoS Biol. 1:E2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Wang, R. F., M. L. Beggs, B. D. Erickson, and C. E. Cerniglia. 2004. DNA microarray analysis of predominant human intestinal bacteria in fecal samples. Mol. Cell. Probes 18:223-234. [DOI] [PubMed] [Google Scholar]
- 209.Wang, Z., L. T. Daum, G. J. Vora, D. Metzgar, E. A. Walter, L. C. Canas, A. P. Malanoski, B. Lin, and D. A. Stenger. 2006. Identifying influenza viruses with resequencing microarrays. Emerg. Infect. Dis. 12:638-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Wang, Z., P. A. Orlandi, and D. A. Stenger. 2005. Simultaneous detection of four human pathogenic microsporidian species from clinical samples by oligonucleotide microarray. J. Clin. Microbiol. 43:4121-4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Wieland, S., R. Thimme, R. H. Purcell, and F. V. Chisari. 2004. Genomic analysis of the host response to hepatitis B virus infection. Proc. Natl. Acad. Sci. USA 101:6669-6674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Willse, A., T. M. Straub, S. C. Wunschel, J. A. Small, D. R. Call, D. S. Daly, and D. P. Chandler. 2004. Quantitative oligonucleotide microarray fingerprinting of Salmonella enterica isolates. Nucleic Acids Res. 32:1848-1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Wilson, J. W., P. Bean, T. Robins, F. Graziano, and D. H. Persing. 2000. Comparative evaluation of three human immunodeficiency virus genotyping systems: the HIV-GenotypR method, the HIV PRT GeneChip assay, and the HIV-1 RT line probe assay. J. Clin. Microbiol. 38:3022-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Wilson, M., J. DeRisi, H. H. Kristensen, P. Imboden, S. Rane, P. O. Brown, and G. K. Schoolnik. 1999. Exploring drug-induced alterations in gene expression in Mycobacterium tuberculosis by microarray hybridization. Proc. Natl. Acad. Sci. USA 96:12833-12838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Wong, C. W., C. L. Heng, L. W. Yee, S. W. Soh, C. B. Kartasasmita, E. A. Simoes, M. L. Hibberd, W. K. Sung, and L. D. Miller. 2007. Optimization and clinical validation of a pathogen detection microarray. Genome Biol. 8:R93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Wong, H. R., T. P. Shanley, B. Sakthivel, N. Cvijanovich, R. Lin, G. L. Allen, N. J. Thomas, A. Doctor, M. Kalyanaraman, N. M. Tofil, S. Penfil, M. Monaco, M. A. Tagavilla, K. Odoms, K. Dunsmore, M. Barnes, and B. J. Aronow. 2007. Genome-level expression profiles in pediatric septic shock indicate a role for altered zinc homeostasis in poor outcome. Physiol. Genomics 30:146-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Xie, Y., X. Wang, and M. Story. 2009. Statistical methods of background correction for Illumina BeadArray data. Bioinformatics 25:751-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Xu, Q., M. Dziejman, and J. J. Mekalanos. 2003. Determination of the transcriptome of Vibrio cholerae during intraintestinal growth and midexponential phase in vitro. Proc. Natl. Acad. Sci. USA 100:1286-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Ye, F., M. S. Li, J. D. Taylor, Q. Nguyen, H. M. Colton, W. M. Casey, M. Wagner, M. P. Weiner, and J. Chen. 2001. Fluorescent microsphere-based readout technology for multiplexed human single nucleotide polymorphism analysis and bacterial identification. Hum. Mutat. 17:305-316. [DOI] [PubMed] [Google Scholar]
- 220.You, Y., C. Fu, X. Zeng, D. Fang, X. Yan, B. Sun, D. Xiao, and J. Zhang. 2008. A novel DNA microarray for rapid diagnosis of enteropathogenic bacteria in stool specimens of patients with diarrhea. J. Microbiol. Methods 75:566-571. [DOI] [PubMed] [Google Scholar]
- 221.Yu, X., M. Susa, C. Knabbe, R. D. Schmid, and T. T. Bachmann. 2004. Development and validation of a diagnostic DNA microarray to detect quinolone-resistant Escherichia coli among clinical isolates. J. Clin. Microbiol. 42:4083-4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Yue, J., W. Shi, J. Xie, Y. Li, E. Zeng, L. Liang, and H. Wang. 2004. Detection of rifampin-resistant Mycobacterium tuberculosis strains by using a specialized oligonucleotide microarray. Diagn. Microbiol. Infect. Dis. 48:47-54. [DOI] [PubMed] [Google Scholar]
- 223.Zhu, H., J. P. Cong, G. Mamtora, T. Gingeras, and T. Shenk. 1998. Cellular gene expression altered by human cytomegalovirus: global monitoring with oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95:14470-14475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Zhu, L.-X., Z.-W. Zhang, D. Liang, D. Jiang, C. Wang, N. Du, Q. Zhang, K. Mitchelson, and J. Cheng. 2007. Multiplex asymmetric PCR-based oligonucleotide microarray for detection of drug resistance genes containing single mutations in Enterobacteriaceae. Antimicrob. Agents Chemother. 51:3707-3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Zhu, L.-X., Z.-W. Zhang, C. Wang, H.-W. Yang, D. Jiang, Q. Zhang, K. Mitchelson, and J. Cheng. 2007. Use of a DNA microarray for simultaneous detection of antibiotic resistance genes among staphylococcal clinical isolates. J. Clin. Microbiol. 45:3514-3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Zimmermann, K., T. Eiter, and F. Scheiflinger. 2003. Consecutive analysis of bacterial PCR samples on a single electronic microarray. J. Microbiol. Methods 55:471-474. [DOI] [PubMed] [Google Scholar]
- 227.Zou, S., J. Han, L. Wen, Y. Liu, K. Cronin, S. H. Lum, L. Gao, J. Dong, Y. Zhang, Y. Guo, and Y. Shu. 2007. Human influenza A virus (H5N1) detection by a novel multiplex PCR typing method. J. Clin. Microbiol. 45:1889-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]