Abstract
The mammalian sterol regulatory element-binding protein (SREBP) homolog, Sre1, is important for adaptation and growth of Cryptococcus neoformans in the mouse brain, where oxygen concentration and nutritional conditions are suboptimal for fungal growth. The extent of conservation of the SREBP pathway in C. neoformans or in any other fungi, however, has not been investigated. We generated mutants susceptible to low oxygen and identified six genes that play a role in the SREBP pathway. Three of these genes (SFB2, KAP123, and GSK3) are not known to be involved in the SREBP pathway in other fungi. Furthermore, we show that C. neoformans contains an additional gene, DAM1, which functions in the SREBP pathway but is yet to be described. Mutants associated with the steps prior to formation of the nuclear Sre1 form dramatically reduced accumulation of the nuclear form under low-oxygen conditions. Concurrently, two mutant strains, scp1Δ and stp1Δ, and the previously isolated sre1Δ strain showed reduction in ergosterol levels, hypersensitivity to several chemical agents, including azole antifungals, CoCl2, and compounds producing reactive oxygen or nitrogen species, and most importantly, reduced virulence in mice. Mutants affecting genes involved in later steps of the Sre1 pathway, such as those required for import and phosphorylation of proteins in the nucleus, showed less compelling phenotypes. These findings suggest that the SREBP pathway is highly conserved in C. neoformans and it serves as an important link between sterol biosynthesis, oxygen sensing, CoCl2 sensitivity, and virulence in C. neoformans.
Cryptococcus neoformans is an opportunistic fungal pathogen that causes life-threatening meningoencephalitis, primarily in immunocompromised patients (22). Cryptococcosis is initiated by inhalation of airborne C. neoformans cells, which spread to the central nervous system by hematogenous dissemination. Upon entry into the brain, the sterol regulatory element-binding protein (SREBP) homolog, Sre1, is required for cells to adapt to the host environment and cause fulminating meningoencephalitis (3, 5). Mutations in SRE1 and the gene encoding a homolog of the SREBP cleavage-activating protein (SCAP), SCP1, resulted in reduced growth under low-oxygen conditions in vitro. The sre1 mutant was either significantly reduced in virulence or unable to cause fatal CNS infection in mice, depending on the strain's genetic background (3, 5).
SREBPs, a family of membrane-bound transcription factors, are involved in the control of cholesterol and lipid metabolism in mammalian systems (7, 34), and they are known to directly enhance the transcription of more than 30 genes required in these processes (15). These unique transcription factors are themselves subject to positive and negative feedback regulation at the transcriptional, translational, and posttranslational levels (34). SREBPs are synthesized as inactive precursors with two transmembrane helices and reside in the endoplasmic reticulum (ER) membrane. The N-terminal domain of SREBP is a basic helix-loop-helix leucine zipper transcription factor. The C terminus forms a tight complex with the tryptophan-aspartate repeat (WD) domain of SCAP, which functions as a sensor for membrane cholesterol levels. In sterol-replete cells, SCAP binds to cholesterol in the ER membrane and assumes a conformation that promotes its binding to the ER-resident protein Insig (for insulin-induced gene) (33). In sterol-depleted cells, this binding to Insig is disrupted and SREBP-SCAP is sorted from the ER to Golgi complex via coat protein complex II-coated transport vesicles, which contain Sar1, Sec23, and Sec24 (8, 12, 35, 38). In the Golgi complex, the N-terminal transcription factor domain of SREBP is released from the membrane by two sequential proteolytic cleavage events mediated by the site 1 and site 2 proteases (S1P and S2P) (12). The released SREBP is transported into the nucleus as a dimer by importin β through interactions with the helix-loop-helix domain (24, 30). In the nucleus, SREBP executes its transcriptional regulatory function. Turnover of SREBP involves phosphorylation by Gsk3, whose activity is inhibited by insulin signaling (10, 21), followed by its binding to the E3 ubiquitin ligase SCFFbw7, which leads to rapid degradation of SREBP (39).
In fungi, the SREBP pathway appears to be diverse among different species. For example, SREBP and SCAP homologs exist in C. neoformans and Schizosaccharomyces pombe but are absent in both Saccharomyces cerevisiae and Candida albicans, while Aspergillus fumigatus contains the SREBP but not SCAP homolog (3, 16, 43). Moreover, no gene other than homologs of SREBP, SCAP, and S2P has been identified to play a role in the fungal SREBP pathway (3, 5).
We used a genetic approach to identify C. neoformans genes involved in adaptation to low-oxygen conditions by screening insertional mutants under low oxygen at 37°C. We found a class of mutants showing sensitivity to low oxygen due to mutations in the genes that are homologous to mammalian genes in the SREBP pathway. Most of these mutants showed sensitivity to CoCl2, azole antifungals, and various reactive oxygen species (ROS)-generating chemicals. Interestingly, all but one of these mutants showed reduced virulence compared to the wild type in a murine model. We also identified the gene Dam1, which functions in the Sre1 pathway but had not been described previously. Our findings show that the SREBP pathway is well conserved in C. neoformans, and they underscore its importance in the pathobiology of C. neoformans.
MATERIALS AND METHODS
Strains, media, and growth conditions.
Strains used in this study are listed in Table 1. All strains were derived from B-3501A, a serotype D strain whose genome was sequenced and annotated (25). Yeast extract-peptone-dextrose (YEPD) contained 1% yeast extract, 2% Bacto peptone, and 2% dextrose. YES (yeast extract with supplements) medium consisted of 0.5% (wt/vol) yeast extract and 2% dextrose and was supplemented with uracil, adenine, leucine, histidine, and lysine (225 μg/ml). Low-oxygen conditions (5% CO2 and 1% O2) were maintained at 37°C using an Invivo2 400 workstation (Ruskinn, United Kingdom). YES media containing 0.5 mM H2O2 or 1 mM NaNO2 were buffered to pH 4 with 25 mM sodium dihydrogen citrate. Plates for assays to test the sensitivity of strains to various chemicals were made using YES with CoCl2 (0.7 mM), diethyl maleate (DEM; 2 mM), menadione (2-methyl-1,4-naphthoquinone; 5 μg/ml), or paraquat (1,1′-dimethyl-4,4′-bipyridinium dichloride; 0.25 mM). Fluconazole (50 mg/ml) and voriconazole (2 mg/ml) were each dissolved in dimethyl sulfoxide (DMSO) and incorporated into the media at 250 ng/ml and 7.5 ng/ml, respectively. For growth phenotype analysis, serial dilutions of cultures were spotted on YES plates containing the indicated chemicals and incubated at 30°C for 3 to 6 days.
TABLE 1.
List of strains used in this study
| Clone no. | Genotype of strain | Reference |
|---|---|---|
| C1037 | sre1Δ::NEO | Chang. et al. (3) |
| C1051 | sre1Δ::NEO SRE1 | Chang. et al. (3) |
| C1052 | sre1Δ::NEO pYCC744a | Chang. et al. (3) |
| C813 | scp1Δ::NEO | This study |
| C1046 | scp1Δ::NEO SCP1 | This study |
| C1044 | scp1Δ::NEO pYCC744 | This study |
| C730 | sfb2*b | This study |
| C906 | sfb2Δ::NEO | This study |
| C1095 | sfb2Δ::NEO SFB2 | This study |
| C1097 | sfb2Δ::NEO pYCC744 | This study |
| C1110 | stp1Δ::NEO | This study |
| C1295 | stp1Δ::NEO STP1 | This study |
| C1302 | stp1Δ::NEO pYCC744 | This study |
| C743 | kap123* | Ingavale et al. (18) |
| C1313 | kap123Δ::NEO | This study |
| C1328 | kap123Δ::NEO KAP123 | This study |
| C1330 | kap123Δ::NEO pYCC744 | This study |
| C768 | gsk3* | This study |
| C1233 | gsk3Δ::NEO | This study |
| C1276 | gsk3Δ::NEO GSK3 | This study |
| C1274 | gsk3Δ::NEO pYCC744 | This study |
| C697 | rsp5* | Ingavale et al. (18) |
| C1127 | rsp5 RSP5 | This study |
| C1129 | rsp5 pYCC744 | This study |
| C778 | dam1* | This study |
| C1108 | dam1Δ::NEO | This study |
| C1120 | dam1Δ::NEO DAM1 | This study |
| C1134 | dam1Δ::NEO pYCC744 | This study |
pYCC744 denotes the strains that were transformed with empty vector.
*, strain isolated from the T-DNA insertion library.
Identification of C. neoformans genes important for growth under low-oxygen conditions.
A T-DNA insertion library of B-3501A was made using Agrobacterium tumefaciens-mediated transformation (ATMT) (26). Primary screening of the mutants sensitive to low oxygen was carried out by replica spotting of the library consisting of 22,000 clones on YEPD plus 50 μg/ml Geneticin and 200 μM cefotaxime plates and incubated at 37°C in 1% O2 and 5% CO2 for 3 to 5 days. Mutants that failed to grow were selected for further analysis. A total of 173 mutants were obtained, of which 58 were temperature sensitive at 37°C. Genomic DNA was isolated from all the mutants, and the T-DNA insertion site was mapped by using the PCR-based Vectorette system (Sigma, Woodlands, TX). Of the 173 mutants, 12 failed to produce clear PCR products and 33 produced more than one PCR product. Genomic sequences flanking the insertion sites were subsequently obtained by sequencing the PCR products. BLAST analysis of these sequences was carried out to identify loci containing the T-DNA inserts using the B-3501A database (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi?organism=fungi).
Screening of Sre1 pathway mutants.
To identify strains with a defect in the Sre1 pathway, selected low-oxygen-sensitive mutants were further analyzed by immunoblot assays. We chose only those strains in which the T-DNA-inserted loci were clearly annotated in the database. For the initial screen, strains were grown in YES medium under normoxic conditions at 37°C. Cells in log phase were transferred to 1% oxygen at 37°C for 1.5 h and harvested. For chase experiments, the cells grown with 1% oxygen for 1.5 h were transferred to normoxic conditions and grown further for 15, 30, or 60 min prior to harvesting. Protein extracts were obtained, and immunoblot assays were performed as described previously (3). Strains showing altered levels of the Sre1 precursor or the nuclear form at 1% oxygen or during chase conditions compared to wild type were chosen for further studies.
Gene deletion and complementation.
Deletants were generated for all the genes used in this study, except for RSP5. Biolistic transformation was used to generate deletion strains via homologous recombination (42). Wild-type genes from the strain B-3501A were PCR amplified, cloned, and sequenced. All the original mutants and the deletion strains were complemented with their respective wild-type genes using biolistic transformation or the ATMT method (26). The gene used for complementation was inserted in the multiple cloning site of pYCC744, which contained the NAT gene as a selectable marker. Stable transformants were selected after repeated transfers on medium without antibiotics.
Sterol analysis.
Sterols were extracted from 1 × 108 cells in log phase that had been grown in YES medium for 3 h under ambient or low-oxygen conditions (1% O2) at 30°C. Cells were washed with sterile H2O and collected by centrifugation. Cell pellets were resuspended in 9 ml methanol, 4.5 ml 60% (wt/vol) KOH, and 5 μg cholesterol (used as an internal recovery standard). Samples were then saponified for 2 h at 75°C with agitation, cooled to room temperature, and extracted with 4 ml of petroleum ether. Sterols were recovered by evaporation of the petroleum ether under a stream of nitrogen gas. Sterols were resuspended in 200 μl heptane, and 2 μl was injected into an Agilent 6850 gas chromatograph with an HP-1 column and flame ionization detection. Sterol species and squalene were identified by comparisons with known standards, and peak areas for each species were normalized to the internal cholesterol standard.
Confocal microscopy.
Cells were stained with MitoTracker Red CMXRos stain (Molecular Probes, Eugene, OR) at a final concentration of 100 nM as described previously (18). Before proceeding with microscopy, cells were washed and suspended in HEPES buffer. Images were collected on a Leica SP5 confocal microscope (Leica Microsystems, Exton, PA) using a 100× oil immersion objective, numerical aperture 1.4, zoom 4. Fluorochromes were excited using a 561-nm diode laser for MitoTracker Red CMXRos. Differential interference contrast images were collected simultaneously with fluorescence images by using a transmitted light detector. Images were processed using Leica LAS-AF software (version 1.8.2, build 1465).
Oxygen consumption assays.
Cells were grown and harvested for oxygen consumption assays as described previously, with some modifications (18). Changes in oxygen tension were measured at 37°C with a Clark-type oxygen electrode, and the respiratory rate was calculated as the change in oxygen tension over time and reported as a relative value compared to the wild-type levels. In order to quantify mitochondrial respiratory oxygen consumption, the respiration inhibitor antimycin A (1 μg/ml) was used to block mitochondrial respiration.
Virulence studies.
Female BALB/c mice (6 to 8 weeks old) were injected via the lateral tail vein with 0.2 ml of suspension of the indicated yeast strain (2.5 × 106 cells/ml in 0.9% NaCl), and mortality was monitored as previously described (4). Kaplan-Meier analysis of survival was performed using JMP software for Macintosh (SAS Institute, Cary, NC).
RESULTS
Sre1 pathway mutants manifest growth defects under low-oxygen conditions.
To identify the genes important for C. neoformans to grow under low-oxygen conditions, we used the ATMT system and generated an insertional mutant library composed of 22,000 clones (18). The library was screened for strains susceptible to 1% oxygen and 5% CO2 at 37°C. Although 1% oxygen conditions rarely occur in a healthy human body, we chose such a stringent oxygen condition as our screening criterion in order to exclude mutants which might display a marginal phenotype under slightly relaxed oxygen conditions. Mutants showing a growth defect phenotype under such parameters were selected to identify the genes containing the T-DNA insertion by using PCR and DNA sequencing (see Materials and Methods). Sequence analysis indicated that two of the mutated genes encode products with similarity to importin β and Gsk3, which are important for the SREBP pathway in mammals. Since the SREBP homolog Sre1 is important for the response to low-oxygen conditions in C. neoformans (3, 5), we focused on characterizing mutants associated with the Sre1 pathway as the initial step toward understanding the oxygen response process in this fungus.
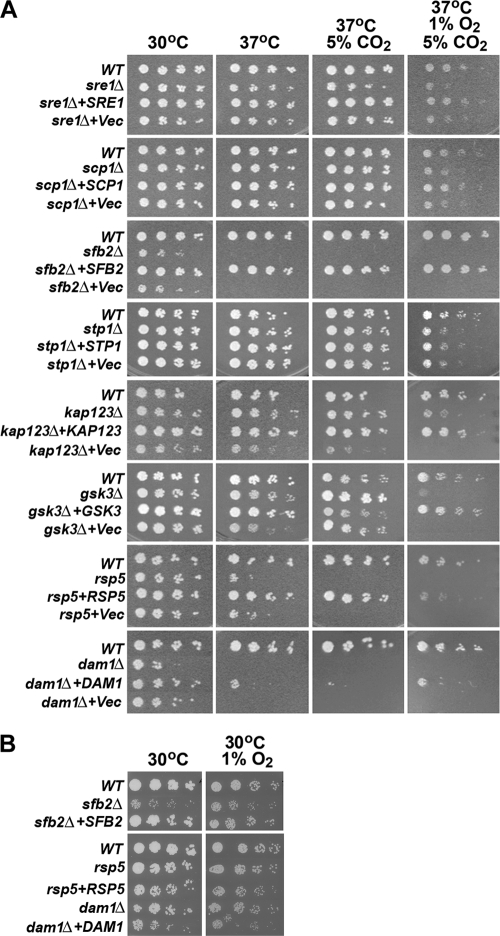
Figure 1A shows the growth phenotype of the putative Sre1 pathway mutants under low-oxygen conditions. The strains are arranged and described according to their presumed function in the Sre1 pathway. Sre1 is synthesized in the ER and forms a complex with Scp1. The sre1 and scp1 deletants showed slightly reduced growth in 1% oxygen and 5% CO2 at 37°C, which was similar to the phenotype described previously under 1% oxygen at 30°C (3). The exit of SREBP/SCAP from the ER in mammalian cells requires Insig, but a homolog of Insig was not found either in our mutant library or in the C. neoformans database.
FIG. 1.
Sre1 pathway mutants exhibit a low-oxygen-sensitive phenotype. (A) Threefold serial dilutions of the indicated strains (starting with approximately 500 cells) were spotted on YES medium and incubated at 30°C, 37°C, and 37°C plus 5% CO2 for 3 days and 37°C plus 5% CO2 plus 1% O2 for 5 days. The names of the genes studied are indicated on the left. (B) Strains showing temperature sensitivity were further tested on YES medium at 30°C for 3 days and 30°C plus 1% O2 for 4 days.
After leaving the ER, SREBP in mammalian systems is transported to the Golgi complex by the conserved coat protein complex II-coated vesicles, which contain Sar1, Sec23, and Sec24. The C. neoformans database contains genes related to SAR1 (CNB03430), SEC23 (CNJ01150), and SEC24 (CNB00260). We were unable to recover mutants for any one of these three genes in our screen. However, we identified an sfb2 (CNM01000) mutant in our screen which is related to the transport protein Sec24 in Saccharomyces cerevisiae (32). Growth of the sfb2 deletant was reduced dramatically at 30°C, while it was inviable at 37°C as well as under low-oxygen conditions (Fig. 1A). However, the sfb2 deletant was not sensitive to 1% oxygen conditions at 30°C (Fig. 1B).
After transport into the Golgi complex, two sequential proteolytic cleavages by the S1P and the S2P proteases are required to release the N-terminal transcription factor domain of the membrane-bound SREBP (12). The C. neoformans genome contains putative homologs of S1P and S2P, CNH01120 (26.7% similarity to human S1P) and CNF02350 (25.4% similarity to human S2P), respectively. Mutants of these two genes, however, were not found among our mutants susceptible to low oxygen. Despite repeated attempts using the biolistic transformation method, we failed to delete the putative S1P homolog, CNH01120. Deletion of the S2P homolog, Stp1, in a serotype A strain of C. neoformans resulted in a growth defect under low-oxygen conditions (5). Similarly, deletion of STP1 in the wild-type serotype D strain, B-3501A, resulted in a growth defect that was slightly more severe than sre1 and scp1 under the same environmental conditions (Fig. 1).
After proteolytic cleavage in the Golgi complex, the N-terminal transcription factor domain of SREBP migrates into the nucleus via nucleocytoplasmic transport factors that mostly belong to the family of karyopherin β proteins, also known as importin β-like proteins (reviewed in reference 28). We found a T-DNA insertional mutant with a defect in the gene (CNF00890) that shares similarity with a human importin β-4 subunit (45.8% similarity) and the S. cerevisiae karyopherin β, Kap123 (48.9% similarity). The deletion strain of KAP123 showed reduced growth under low-oxygen conditions, similar to the original T-DNA insertional mutant (Fig. 1 and data not shown).
The N-terminal transcription factor domain of SREBP migrates into the nucleus and is phosphorylated by glycogen synthase kinase, Gsk3 (21). Gsk3 is a serine/threonine kinase that has been implicated in multiple cellular processes, including the Wnt and insulin signaling pathways in mammals (10). We found a strain containing a T-DNA insertion in GSK3 (CNB00720), and deletion of GSK3 resulted in normal growth under all other conditions tested except under low-oxygen conditions, where only a trace amount of growth was seen at 1% oxygen (Fig. 1A).
The nuclear form of SREBP is rapidly degraded by the ubiquitin-proteasome pathway involving the SCF (Skp1-Cul1-F-box protein) ubiquitin ligase complex (21). Fbw7 is an F-box protein within the SCF modular complex which interacts with the nuclear SREBP and enhances its ubiquitination and degradation in a Gsk3-dependent manner. The C. neoformans CNA06710 sequence shares similarity with Fbw7/Cdc4 (34.6% similarity) and Met30 (28.5% similarity) of S. cerevisiae. Since both CDC4 and MET30 are essential in S. cerevisiae, we did not attempt to delete CNA06710. However, we identified a mutant, C697, in our screen which harbors a T-DNA insertion in the ubiquitin-protein ligase gene, CNH00220, which shares similarity to Rsp5 of S. cerevisiae (71.8% similarity). Rsp5 functions in multivesicular body sorting, heat shock response, and ubiquitylation of arrested RNA polymerase II (RNAP II) (37). The rsp5 mutant showed reduced growth at 37°C and failed to grow under both 5% CO2 and low-oxygen conditions (Fig. 1A). However, the rsp5 mutant did not show a clear reduction of growth at 30°C with 1% oxygen (Fig. 1B). When the mutants mentioned above were transformed with their corresponding wild-type gene, the growth of these complemented strains was restored close to that of the wild-type levels under low-oxygen conditions (Fig. 1). These results demonstrate the importance of the genes related to Sre1 processing with respect to growth of C. neoformans under low-oxygen conditions.
Processing of Sre1 is affected in mutants sensitive to a low oxygen level.
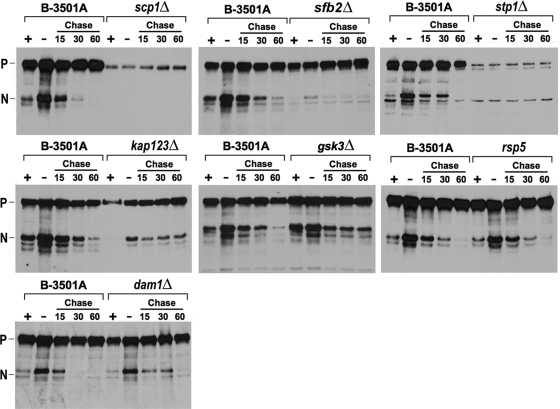
To establish a direct relationship between Sre1 production and the putative cryptococcal SREBP pathway genes, the Sre1 protein levels were monitored in these mutants. Since some of the mutants had a growth defect at 37°C, crude protein extracts were prepared from wild-type and mutant strains grown at 30°C. Sre1 protein levels were determined using an antibody against the N-terminal portion of Sre1 which detected both its full-length precursor and the mature nuclear form, Sre1N (5). Under normoxic conditions, the wild-type cells of B-3501A had considerably higher amounts of the precursor form compared to the nuclear form (Fig. 2). When the wild-type cells were transferred to 1% oxygen, the amount of nuclear form increased significantly. Returning the cells back to normoxic conditions resulted in the disappearance of the nuclear form within an hour. Mutations in SCP1 and STP1 significantly reduced the accumulation of both precursor and nuclear forms under either 21% or 1% oxygen conditions. In the sfb2 mutant, levels of the precursor form were not affected but accumulation of the nuclear form was markedly reduced under all conditions compared to the wild type. In the kap123 mutant, the amounts of both precursor and nuclear forms were reduced under both 21% and 1% oxygen conditions, and the turnover rate of the nuclear form was slightly slower compared to the wild type. In contrast, both the precursor and nuclear forms were unaffected in the gsk3 mutant, but disappearance of the Sre1 nuclear form was slightly delayed in the gsk3 mutant. Levels of the Sre1 precursor and nuclear forms in the rsp5 mutant were similar to the wild type under all conditions, suggesting that RSP5 is not involved in the Sre1 pathway.
FIG. 2.
Sre1 pathway mutants have defects in Sre1 production. Log-phase cells of B-3501A and the indicated mutant strains were incubated with 21% (+) or 1% (−) oxygen for 4 h at 30°C in YES medium. Then, the cultures were shifted to 21% oxygen for 15, 30, or 60 min in YES medium. Whole-cell extracts were prepared and subjected to immunoblot analysis using anti-Sre1 antiserum. P and N denote the Sre1 precursor and its cleaved nuclear forms, respectively.
We next sought to determine if Sre1 production was compromised in any of the low-oxygen-sensitive mutants in which the affected genes bore no similarity to the genes known for SREBP processing. We screened 50 low-oxygen-sensitive mutants in which the affected genes had clearly been annotated in the database. Only one mutant which had a T-DNA insertion in the DAM1 (CNF02850) gene, which comprises a domain called breast carcinoma amplified sequence 2 (BCAS2), was identified to have a defect in Sre1 production. The dam1 deletant had a slightly slower turnover rate of Sre1N when cells were transferred from 1% oxygen to 21% oxygen levels (Fig. 2). This deletant also displayed a growth defect at 37°C, but not at 30°C, with 1% oxygen (Fig. 1). Thus, including SRE1, we have identified a total of seven genes that function in the Sre1 pathway of C. neoforman, of which six are homologous to the mammalian SREBP pathway genes.
Sre1 pathway mutants affect ergosterol production.
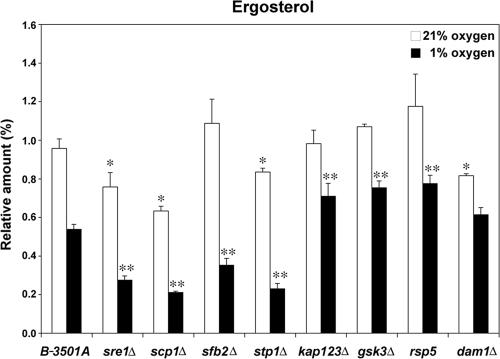
Sterols are essential for fungal cells, and ergosterol is the final product of sterol biosynthesis. Given that Sre1 regulates gene expression of multiple ergosterol biosynthetic enzymes and the sre1 mutant has reduced ergosterol content (3), we examined whether mutations in the Sre1 pathway genes all affected ergosterol biosynthesis. Figure 3 shows ergosterol content in wild-type and mutant strains. Like in the sre1 mutant (3), the amount of ergosterol was lower in the scp1, stp1, and dam1 mutants compared to the wild-type strain under normoxic conditions. After 3 h at 1% oxygen, ergosterol content decreased in wild-type cells due to the requirement of oxygen for sterol synthesis. Interestingly, like in the sre1 mutant, ergosterol content decreased significantly in scp1, sfb2, and stp1 mutants, corroborating the dramatically reduced amounts of the nuclear Sre1 form in these mutants (Fig. 2). These data confirm the importance of the Sre1 pathway in ergosterol biosynthesis in C. neoformans.
FIG. 3.
Sterol synthesis is altered in Sre1 pathway mutants. Cells of the indicated strains were grown in YES medium at 21% or at 1% oxygen for 3 h at 30°C. Total sterols were extracted from each sample and analyzed by gas chromatography. Relative amounts of ergosterol were determined by normalizing data to an internal cholesterol standard. The experiment was repeated three times, and error bars indicate standard deviations. * and ** indicate that the difference is statistically significant compared to the wild type under normoxic and low-oxygen conditions, respectively (P < 0.05).
Sre1 pathway mutants are hypersensitive to different chemicals.
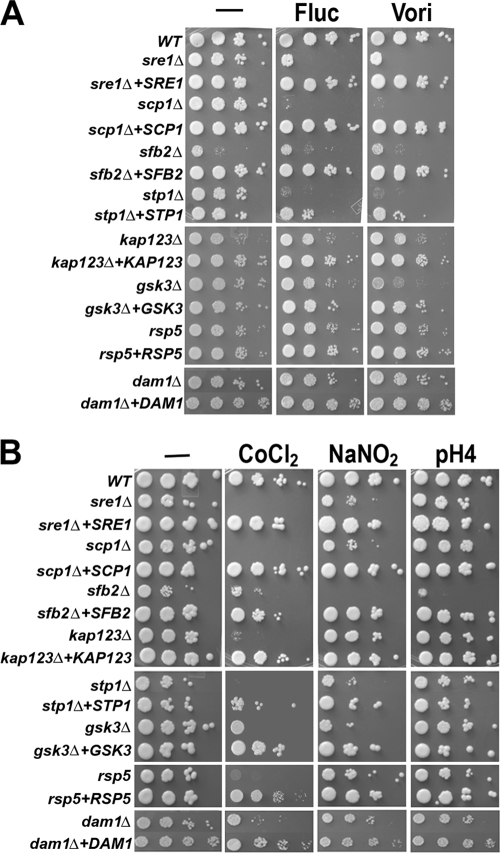
Since several aforementioned mutants showed altered sterol content, we speculated that as in the sre1 mutants (3, 5), susceptibility to inhibitors of ergosterol synthesis in these mutants could have been altered. Figure 4A shows the sensitivity of these mutants to the triazole class of antifungal drugs. It is clear that the scp1 and stp1 mutants are hypersensitive to fluconazole and voriconazole at 250 ng/ml and 7.5 ng/ml, respectively. Complementation of each mutant with the wild-type gene restored sensitivity to near-wild-type levels. In contrast, no clear sensitivity to azoles was observed in the other mutants except that growth of the gsk3 mutant was slightly reduced in the presence of the drugs. These data show that hypersensitivity to drugs that target ergosterol biosynthesis is manifested only in strains that lack or have reduced Sre1 and not in the other mutants of the Sre1 pathway.
FIG. 4.
Sre1 pathway mutants are sensitive to different chemical agents. Tenfold serial dilutions of the indicated strains were spotted on YES medium without supplement (−) or YES medium supplemented with different chemicals and incubated at 30°C for 3 days. (A) Fluc, fluconazole (250 ng/ml); Vori, voriconazole (7.5 ng/ml). (B) The CoCl2 concentration was 0.7 mM. The pH 4 YES medium was included as control, since the YES medium was adjusted to pH 4 before NaNO2 (1 mM final concentration) was added. The names of the genes studied are indicated on the left.
We showed previously that sre1 and scp1 mutants are sensitive to medium containing the hypoxia-mimicking agent CoCl2 (23). To assess the response to CoCl2, mutants were inoculated on rich medium containing 0.7 mM CoCl2. Each mutant was sensitive to CoCl2 and could be generally categorized into one of two groups (Fig. 4B). One group showed a severe growth reduction in the presence of CoCl2 and included the sre1, scp1, stp1, and kap123 mutants. The other group, which included sfb2, gsk3, and dam1 mutants, showed moderate sensitivity to CoCl2. Although the rsp5 mutant did not influence Sre1 levels (Fig. 2), it showed sensitivity to CoCl2. These results demonstrate a close relationship between the Sre1 pathway and CoCl2 sensitivity in C. neoformans.
CoCl2-sensitive mutants are compromised in handling the stress exerted by reagents that generate reactive nitrogen species (RNS) and ROS (18). Since all mutants in the present study were sensitive to CoCl2, we hypothesized that our mutants would also exhibit sensitivity to RNS and ROS reagents. We examined the mutants in the presence of 1 mM NaNO2 at pH 4.0, which is known to generate nitric oxide (14, 20). Nitric oxide forms RNS by reacting with different chemicals present inside cells. While most of the mutants showed sensitivity to the presence of 1 mM NaNO2, no effect was observed in the dam1 and kap123 mutants (Fig. 4B). To test the sensitivity of the Sre1 pathway mutants to ROS-producing chemicals, each strain was grown in the presence of different reagents, including DEM (2 mM), hydrogen peroxide (0.5 mM), menadione (5 μg/ml), and paraquat (0.25 mM). Diethyl maleate is a glutathione-depleting agent, and exposure of cells to diethyl maleate leads to protein denaturation and increased intracellular ROS levels (11). Hydrogen peroxide has the ability to directly damage nucleic acids, proteins, and lipids (1, 19). Menadione is a quinone, and through the redox cycle of quinone, superoxide anion radical hydrogen peroxide and other reactive oxygen species are generated (27). Paraquat is widely used to induce superoxide generation in cells and causes extensive damage of mitochondria (6). Table 2 summarizes the effects of these ROS-producing agents on cell growth. Collectively, all mutants were susceptible to at least one of the ROS-producing reagents, but their tolerance to these compounds returned to wild-type levels when the mutations were complemented with their respective wild-type genes. These results indicate that all mutants were defective in handling the stress generated by ROS-producing reagents.
TABLE 2.
Growth phenotypes of Sre1-related hypoxia-sensitive mutants in the presence of ROS- or RNS-generating agents
| Strain genotype | Growth ona: |
|||||
|---|---|---|---|---|---|---|
| DEM | H2O2 | Menadione | Paraquat | NaNO2 | Control, pH 4.0 | |
| sre1Δ | 1 | 2 | 2 | 2 | 1 | 3 |
| sre1Δ SRE1 | 3 | 3 | 3 | 3 | 3 | 3 |
| scp1Δ | 1 | 2 | 2 | 2 | 1 | 3 |
| scp1Δ SCP1 | 3 | 3 | 3 | 3 | 3 | 3 |
| sfb2Δ | 1 | 1 | 1 | 3 | 0 | 0 |
| sfb2Δ SFB2 | 3 | 3 | 3 | 3 | 3 | 3 |
| stp1Δ | 1 | 1 | 3 | 2 | 1 | 2 |
| stp1Δ STP1 | 2 | 3 | 3 | 3 | 3 | 2 |
| kap123Δ | 2 | 3 | 1 | 2 | 2 | 3 |
| kap123Δ KAP123 | 3 | 3 | 2 | 3 | 3 | 3 |
| gsk3Δ | 0 | 3 | 3 | 3 | 1 | 3 |
| gsk3Δ GSK3 | 3 | 3 | 3 | 3 | 3 | 3 |
| rsp5 | 1 | 2 | 1 | 2 | 2 | 3 |
| rsp5 RSP5 | 3 | 3 | 3 | 3 | 3 | 3 |
| dam1Δ | 0 | 3 | 3 | 3 | 3 | 3 |
| dam1Δ DAM1 | 3 | 3 | 3 | 3 | 3 | 3 |
Scoring: 3, growth similar to wild type; 2, slight reduction in growth compared to wild type; 1, significant reduction in growth compared to wild type; 0, no growth.
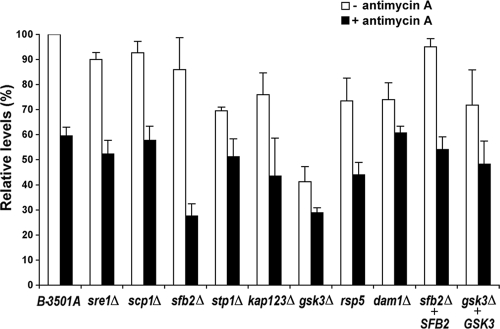
Many of the previously studied CoCl2-sensitive C. neoformans mutants showed mitochondrial dysfunction, reflected by perturbed mitochondrial membrane potential and respiration efficiency (18). Since all the mutants in the present study were sensitive to CoCl2, it is possible that our mutants also have mitochondrial dysfunction. First, we examined the mutants for any perturbation in mitochondrial membrane potential using the MitoTracker dye CMXRos. Confocal microscopy did not show a clear reduction of CMXRos staining in all the mutants compared to wild-type cells, suggesting the presence of an intact mitochondrial membrane potential (data not shown). When the rate of oxygen consumption was measured in the mutants, only the gsk3 mutant showed a greater-than-50% reduction in the presence or absence of antimycin A compared to the wild type. Complementation of the gsk3 mutation restored oxygen consumption close to the wild-type level (Fig. 5, gsk3Δ GSK3). Since antimycin A blocks mitochondrial respiration by inhibiting complex III of the electron transport chain, it was used to reveal the rate of nonrespiratory oxygen consumption. Addition of antimycin A resulted in a 40% reduction of oxygen consumption in wild-type cells, whereas the sfb2 mutant showed a greater-than-50% reduction. Complementation of the sfb2 mutation (sfb2Δ SFB2) restored oxygen consumption close to wild-type levels. These data suggest that except for sfb2 and gsk3 mutants, the other Sre1 pathway mutants have no clear defect in mitochondrial function.
FIG. 5.
Oxygen consumption is not significantly altered in Sre1 pathway mutants. Yeast cells of each strain were grown as described in Materials and Methods. A total of 5 × 107 washed cells were suspended in 2 ml of medium, and changes in oxygen tension were measured using a Clark-type electrode. Samples were measured with (closed bars) or without (open bars) antimycin A. Data are expressed as levels relative to the wild-type strain.
Virulence of Sre1 pathway mutants.
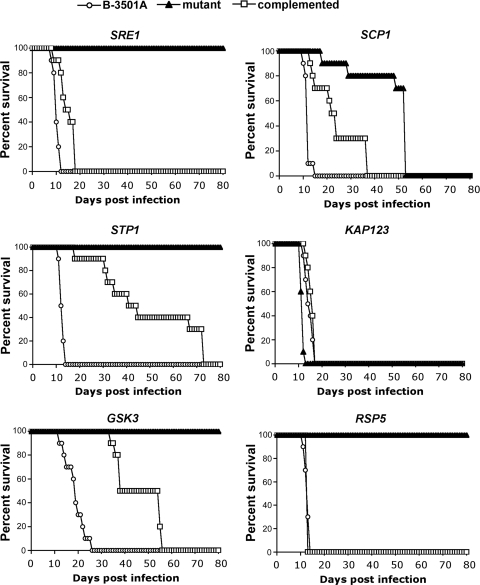
The importance of SRE1 in causing fulminating infection in a murine intravenous model has been demonstrated previously (3, 5). We examined the effects of mutations in the other Sre1 pathway genes by using an intravenous murine model. Since sfb2 and dam1 mutants grew poorly at 37°C, they were excluded from the virulence test. While the rsp5 mutant showed some reduction of growth at 37°C, this mutant was included in the virulence test. The published data of an SRE1 mutant from a serotype D strain were included as a comparison (3). As in the case of sre1, mice challenged with stp1, gsk3, and rsp5 mutants failed to succumb to infection during 80 days of observation (Fig. 6). Complementation of the individual mutations restored virulence close to that of the wild-type strain, although the restoration was not as complete as in the cases of stp1 and gsk3 mutants. The incomplete restoration of virulence in these strains could be due to positional effects generated by random genomic integration during transformations to create the complemented strains. Although not completely avirulent, the scp1 mutant showed a significant reduction in virulence compared to the wild-type strain (P < 0.0001). Surprisingly, deletion of kap123 did not affect virulence, since mice infected with the kap123 strain showed mortality similar to that with the wild type. These data indicate that not all low-oxygen-sensitive mutants fail to cause fulminating infection in mice.
FIG. 6.
Sre1 pathway mutants affect virulence in mice. Female BALB/c mice (10 per group) were injected with C. neoformans strains via the tail vein, and mortality was monitored. The symbols in each panel represent survival of mice challenged with the wild type (circles), mutant strain (triangles), and complemented strain (squares).
DISCUSSION
In this study, we describe the function of six C. neoformans genes that share homology with those involved in the mammalian SREBP pathway. These include SRE1, SCP1, SFB2, STP1, KAP123, and GSK3. Involvement of SFB2, KAP123, and GSK3 in the SREBP pathway has not been reported in other fungi. In addition, we found a new gene, DAM1, whose mutation slightly affected transient accumulation of the nuclear form of Sre1 in C. neoformans. More importantly, mutations in these genes resulted in growth defects under low-oxygen conditions at 37°C. Mutations in SCP1 and STP1 caused a major decrease in the accumulation of both precursor and nuclear forms of Sre1. Concurrently, these mutants with a reduction in ergosterol content were hypersensitive to several chemical agents, including triazoles, CoCl2, and the agents generating ROS and RNS, and were less virulent. Table 3 summarizes all the phenotypes of the mutants in this study. These findings confirm the importance of the Sre1 pathway as a link between sterol biosynthesis, oxygen sensing, CoCl2 sensitivity, and virulence in C. neoformans.
TABLE 3.
Summary of mutant phenotypes compared to wild type
| Mutated gene | Putative role in SREBP pathway | Defective processing | Defective turnover | Sensitivity to low O2 | Ergosterol abnormality | Azole sensitivity | CoCl2 sensitivity | NaNO2 sensitivity | ROS sensitivityf | Virulence |
|---|---|---|---|---|---|---|---|---|---|---|
| sreI | Transcriptional factor | NDa | NDa | Yes | Yes | Yes | Yes | Yes | Yes | No |
| scpI | Cleavage activating protein | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Reduced |
| sfb2 | ER-to-Golgi complex transport | Yes | Yes | Nob | Yes | Yes | No | ?e | Yes | Not tested |
| stpI | Proteolytic cleavage | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | No |
| kap123 | Nuclear transport | Yes | Yes | Yes | No | No | Yes | No | Yes | Yes |
| gsk3 | Phosphorylation, turnover | No | Yes | Yes | No | Yesd | Yes | Yes | Yes | No |
| dam1 | Turnover? | No | Yes | Noc | No | No | Yes | No | Yes | Not tested |
ND, not detectable due to deletion.
Temperature sensitive at 37°C.
Mutation can only be partially complemented.
Slightly increased susceptibility.
Could not be determined due to sensitivity at pH 4.
Sensitive to at least one of the ROS reagents tested.
The existence of a conserved SREBP pathway in C. neoformans was demonstrated by the identification of genes whose mutations affected the maturation steps of Sre1 before and after production of the nuclear form of the protein. Since it is known that Sre1 regulates its own expression in mammals and in S. pombe (15, 16, 41), it is possible that a reduced amount of the nuclear Sre1 form in C. neoformans would also cause a reduction in the accumulated precursor as seen in scp1 and stp1 mutants. However, it is not clear why the Sre1 precursor levels were not affected in sfb2 mutants despite low levels of the nuclear Sre1 form in sfb2 mutants.
The other phenotypic defects, such as low ergosterol production or hypersensitivity to triazoles and CoCl2, that resulted from mutations in genes that function in steps after rather than before generation of the nuclear Sre1 were less compelling. It is possible that once the mature Sre1 nuclear form is produced, its transport into the nucleus or quick turnover has less of an impact on the various functions affected by Sre1. Alternatively, other proteins functionally related to the mutated protein might compensate for the defects in the mutants. For example, C. neoformans Kap123 shares similarity with karyopherin β, Kap123, and two other proteins of S. cerevisiae (Kap104 and Pse1). In S. cerevisiae, it has been proposed that in the absence of Kap123 these karyopherins are able to supplant the role of Kap123 in importing molecules (40). Although we did not detect any gene closely related to KAP123 in C. neoformans, other unidentified proteins may functionally substitute for Kap123 and reduce the severity of the phenotype under stressful conditions.
The present study has identified most of the key players of the SREBP pathway in C. neoformans except for Insig and S1P. In fission yeast, deletion of the INSIG homolog, ins1+, had no effect on Sre1 activation, and ins1+ is dedicated to regulation of the HMG-coenzyme A reductase, Hmg1 (2, 16). In budding yeast, a homolog of SREBP was not found, but the Insig homolog called Nsg1 acts as a chaperone and is involved in the regulation of sterol biosynthesis. It specifically stabilizes Hmg2, one of the two HMG-coenzyme A isoenzymes that catalyze the rate-limiting step in sterol biosynthesis (9). It is possible that exit of Sre1/Scp1 from the ER is regulated by a different protein or mechanism in different fungal species. We were unable to delete the putative S1P homolog, CNH01120, in a serotype D strain. In mice, homozygous germ line disruptions of S1P were embryonically lethal (44), suggesting that CNH01120 may also be an essential gene in serotype D strains. Thus, the function of CNH01120 in serotype D strains remains unclear. We successfully deleted the putative S1P homolog, CNAG_05446.2, in the serotype A strain H99 (data not shown). This deletant was resistant to CoCl2, which is inconsistent with the sensitive phenotype of all other Sre1 pathway mutants. Since Sre1 processing in the Golgi complex requires two sequential cleavage events by S1P and S2P in mammalian systems, it is possible that C. neoformans uses different proteins or mechanisms to release the N-terminal transcriptional factor domain of Sre1 in the Golgi complex.
DAM1 (CNF02850), the newly identified gene which slightly affected turnover of the Sre1 nuclear form, is annotated as a hypothetical gene in the C. neoformans database and is absent in S. cerevisiae. The BCAS2 family consists of several eukaryotic sequences of unknown function, and the human Dam1 is a putative spliceosome-associated protein (29). It is unclear how Dam1 operates in the Sre1 pathway of C. neoformans. Western blot analysis showed that mutations in both DAM1 and GSK3 affected the turnover rate of the nuclear Sre1 form, which suggests that Dam1 may function in the later steps of Sre1 processing.
In mammals, the central regulator of hypoxic gene expression is the heterodimeric transcription factor HIF (hypoxia-inducible factor) (for reviews, see references 13 and 36). The degradation and activity of the HIF-1α subunit are both regulated by oxygen-dependent posttranslational hydroxyl modifications catalyzed by enzymes belonging to the 2-oxoglutarate-Fe(II) dioxygenase family (31). In fission yeast, the Sre1 transcription factor is a principal regulator of low-oxygen gene expression (41). Recently, a prolyl 4-hydroxylase-like 2-oxoglutarate-Fe(II) dioxygenase, Ofd1, that accelerates Sre1 degradation in the presence of oxygen was identified in S. pombe (17). C. neoformans contains a gene, CNA04020, that shares 42.8% similarity to Ofd1, and its expression is regulated by oxygen levels (H. Lee and Y. Chang, unpublished data). However, unlike in S. pombe, deletion of CNA04020 did not affect Sre1 levels under 21% or 1% oxygen conditions. In the C. neoformans genome, there are two more prolyl 4-hydroxylase α-subunit domain (P4Hc)-containing genes, CNF02010 and CNBK2780, but the similarity to Ofd1 is limited only to the P4Hc domain in these two genes. It remains to be elucidated if C. neoformans utilizes oxygen-dependent posttranslational hydroxyl modifications to regulate Sre1 levels.
This study was undertaken to determine whether the ability to grow under low-oxygen conditions is an essential property in the pathobiology of C. neoformans. Five of the six low-oxygen-sensitive mutants for the SREBP pathway showed a significant reduction in virulence. However, the kap123 mutant, which is as sensitive to low oxygen as the stp1 mutant, showed virulence comparable to wild type. We also examined the virulence of six other low-oxygen-sensitive mutants from our collection that are not involved in the Sre1 pathway. Again, one of these mutants was as virulent as the wild-type strain, while the other five mutants showed a significant reduction in virulence (data not shown). Oxygen is a key requirement for many pathways, ranging from sterol to heme biosynthesis. Since sterol content of the kap123 mutant, one of the six that showed no reduction in virulence, was not significantly altered, it is possible that the kap123 mutation affects other cellular processes important for growth under low-oxygen conditions in vitro but can be circumvented in vivo. Therefore, the relationship between the ability to grow under low-oxygen conditions and virulence merits further study.
In other organisms, several of the genes we identified in the Sre1 pathway, such as SFB2, KAP123, and GSK3, are known to be involved in many other physiological processes besides Sre1 processing. Moreover, the sfb2 and dam1 mutants of C. neoformans were not sensitive to low-oxygen conditions at 30°C (Fig. 1B). In fact, most of the mutants that we obtained by screening for a growth defect under low-oxygen conditions did not influence Sre1 production. This observation suggests that only a limited overlap exists between the Sre1 pathway and oxygen-sensing pathways in C. neoformans. Interestingly, all the Sre1 pathway mutants were sensitive to CoCl2 and conversely, all the previously identified CoCl2-sensitive mutants showed sensitivity to low-oxygen conditions (18). Mutants sensitive to CoCl2 apparently are defective in multiple physiological processes that affect growth. Further studies of these mutants showed that mitochondria play a prominent role in CoCl2 sensitivity. Although most of the Sre1 pathway mutants displayed the CoCl2-sensitive phenotype, their mitochondrial function was not significantly impacted as assessed by their mitochondrial membrane potential and respiration efficiency. In addition, most of the low-oxygen-sensitive mutants obtained from our screening were not sensitive to CoCl2 (data not shown). Collectively, these observations indicate that the Sre1 pathway plays an important role in the pathobiology of C. neoformans, and cryptococcal cells employ complex processes to overcome unfavorable environmental conditions, such as the low oxygen levels they encounter in the animal host.
Acknowledgments
This study was supported by funds from the intramural program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD. This work was supported by grant AI072186 from the National Institutes of Health (to P.E.). P.E. is a Burroughs Wellcome Fund Investigator in Pathogenesis of Infectious Disease.
We are grateful to Ashok Varma for critical reading of the manuscript. We thank J. Schroth and T. Jones for technical assistance.
Footnotes
Published ahead of print on 11 September 2009.
REFERENCES
- 1.Berlett, B. S., and E. R. Stadtman. 1997. Protein oxidation in aging, disease, and oxidative stress. J. Biol. Chem. 272:20313-20316. [DOI] [PubMed] [Google Scholar]
- 2.Burg, J. S., D. W. Powell, R. Chai, A. L. Hughes, A. J. Link, and P. J. Espenshade. 2008. Insig regulates HMG-CoA reductase by controlling enzyme phosphorylation in fission yeast. Cell Metab. 8:522-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang, Y. C., C. M. Bien, H. Lee, P. J. Espenshade, and K. J. Kwon-Chung. 2007. Sre1p, a regulator of oxygen sensing and sterol homeostasis, is required for virulence in Cryptococcus neoformans. Mol. Microbiol. 64:614-629. [DOI] [PubMed] [Google Scholar]
- 4.Chang, Y. C., L. A. Penoyer, and K. J. Kwon-Chung. 2001. The second STE12 homologue of Cryptococcus neoformans is MATa-specific and plays an important role in virulence. Proc. Natl. Acad. Sci. USA 98:3258-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chun, C. D., O. W. Liu, and H. D. Madhani. 2007. A link between virulence and homeostatic responses to hypoxia during infection by the human fungal pathogen Cryptococcus neoformans. PLoS Pathog. 3:e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cocheme, H. M., and M. P. Murphy. 2008. Complex I is the major site of mitochondrial superoxide production by paraquat. J. Biol. Chem. 283:1786-1798. [DOI] [PubMed] [Google Scholar]
- 7.Espenshade, P. J., and A. L. Hughes. 2007. Regulation of sterol synthesis in eukaryotes. Annu. Rev. Genet. 41:401-427. [DOI] [PubMed] [Google Scholar]
- 8.Espenshade, P. J., W. P. Li, and D. Yabe. 2002. Sterols block binding of COPII proteins to SCAP, thereby controlling SCAP sorting in ER. Proc. Natl. Acad. Sci. USA 99:11694-11699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flury, I., R. Garza, A. Shearer, J. Rosen, S. Cronin, and R. Y. Hampton. 2005. INSIG: a broadly conserved transmembrane chaperone for sterol-sensing domain proteins. EMBO J. 24:3917-3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frame, S., and P. Cohen. 2001. GSK3 takes centre stage more than 20 years after its discovery. Biochem. J. 359:1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freeman, M. L., S. A. Huntley, M. J. Meredith, G. A. Senisterra, and J. Lepock. 1997. Destabilization and denaturation of cellular protein by glutathione depletion. Cell Stress Chaperones 2:191-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldstein, J. L., R. A. DeBose-Boyd, and M. S. Brown. 2006. Protein sensors for membrane sterols. Cell 124:35-46. [DOI] [PubMed] [Google Scholar]
- 13.Gordan, J. D., and M. C. Simon. 2007. Hypoxia-inducible factors: central regulators of the tumor phenotype. Curr. Opin. Genet. Dev. 17:71-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harvey, B. H. 2000. Acid-dependent dismutation of nitrogen oxides may be a critical source of nitric oxide in human macrophages. Med. Hypotheses 54:829-831. [DOI] [PubMed] [Google Scholar]
- 15.Horton, J. D., N. A. Shah, J. A. Warrington, N. N. Anderson, S. W. Park, M. S. Brown, and J. L. Goldstein. 2003. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc. Natl. Acad. Sci. USA 100:12027-12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hughes, A. L., B. L. Todd, and P. J. Espenshade. 2005. SREBP pathway responds to sterols and functions as an oxygen sensor in fission yeast. Cell 120:831-842. [DOI] [PubMed] [Google Scholar]
- 17.Hughes, B. T., and P. J. Espenshade. 2008. Oxygen-regulated degradation of fission yeast SREBP by Ofd1, a prolyl hydroxylase family member. EMBO J. 27:1491-1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ingavale, S., Y. C. Chang, H. Lee, C. M. McClelland, M. L. Leong, and K. J. Kwon-Chung. 2008. Importance of mitochondria in survival of Cryptococcus neoformans under low oxygen conditions and tolerance to cobalt chloride. PLoS Pathog. 4:e1000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jamieson, D. J. 1998. Oxidative stress responses of the yeast Saccharomyces cerevisiae. Yeast 14:1511-1527. [DOI] [PubMed] [Google Scholar]
- 20.Jolly, W. L. 1964. The inorganic chemistry of nitrogen, p.68-79. W.A. Benjamin, Inc., New York, NY.
- 21.Kim, K. H., M. J. Song, E. J. Yoo, S. S. Choe, S. D. Park, and J. B. Kim. 2004. Regulatory role of glycogen synthase kinase 3 for transcriptional activity of ADD1/SREBP1c. J. Biol. Chem. 279:51999-52006. [DOI] [PubMed] [Google Scholar]
- 22.Kwon-Chung, K. J., and J. E. Bennett. 1992. Medical mycology. Lea & Febiger, Philadelphia, PA.
- 23.Lee, H., C. M. Bien, A. L. Hughes, P. J. Espenshade, K. J. Kwon-Chung, and Y. C. Chang. 2007. Cobalt chloride, a hypoxia-mimicking agent, targets sterol synthesis in the pathogenic fungus Cryptococcus neoformans. Mol. Microbiol. 65:1018-1033. [DOI] [PubMed] [Google Scholar]
- 24.Lee, S. J., T. Sekimoto, E. Yamashita, E. Nagoshi, A. Nakagawa, N. Imamoto, M. Yoshimura, H. Sakai, K. T. Chong, T. Tsukihara, and Y. Yoneda. 2003. The structure of importin-beta bound to SREBP-2: nuclear import of a transcription factor. Science 302:1571-1575. [DOI] [PubMed] [Google Scholar]
- 25.Loftus, B. J., E. Fung, P. Roncaglia, D. Rowley, P. Amedeo, D. Bruno, J. Vamathevan, M. Miranda, I. J. Anderson, J. A. Fraser, J. E. Allen, I. E. Bosdet, M. R. Brent, R. Chiu, T. L. Doering, M. J. Donlin, C. A. D'Souza, D. S. Fox, V. Grinberg, J. Fu, M. Fukushima, B. J. Haas, J. C. Huang, G. Janbon, S. J. Jones, H. L. Koo, M. I. Krzywinski, J. K. Kwon-Chung, K. B. Lengeler, R. Maiti, M. A. Marra, R. E. Marra, C. A. Mathewson, T. G. Mitchell, M. Pertea, F. R. Riggs, S. L. Salzberg, J. E. Schein, A. Shvartsbeyn, H. Shin, M. Shumway, C. A. Specht, B. B. Suh, A. Tenney, T. R. Utterback, B. L. Wickes, J. R. Wortman, N. H. Wye, J. W. Kronstad, J. K. Lodge, J. Heitman, R. W. Davis, C. M. Fraser, and R. W. Hyman. 2005. The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science 307:1321-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McClelland, C. M., Y. C. Chang, and K. J. Kwon-Chung. 2005. High frequency transformation of Cryptococcus neoformans and Cryptococcus gattii by Agrobacterium tumefaciens. Fungal Genet. Biol. 42:904-913. [DOI] [PubMed] [Google Scholar]
- 27.Monks, T. J., R. P. Hanzlik, G. M. Cohen, D. Ross, and D. G. Graham. 1992. Quinone chemistry and toxicity. Toxicol. Appl. Pharmacol. 112:2-16. [DOI] [PubMed] [Google Scholar]
- 28.Mosammaparast, N., and L. F. Pemberton. 2004. Karyopherins: from nuclear-transport mediators to nuclear-function regulators. Trends Cell Biol. 14:547-556. [DOI] [PubMed] [Google Scholar]
- 29.Nagasaki, K., N. Maass, T. Manabe, H. Hanzawa, T. Tsukada, K. Kikuchi, and K. Yamaguchi. 1999. Identification of a novel gene, DAM1, amplified at chromosome 1p13.3-21 region in human breast cancer cell lines. Cancer Lett. 140:219-226. [DOI] [PubMed] [Google Scholar]
- 30.Nagoshi, E., N. Imamoto, R. Sato, and Y. Yoneda. 1999. Nuclear import of sterol regulatory element-binding protein-2, a basic helix-loop-helix-leucine zipper (bHLH-Zip)-containing transcription factor, occurs through the direct interaction of importin beta with HLH-Zip. Mol. Biol. Cell 10:2221-2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ozer, A., and R. K. Bruick. 2007. Non-heme dioxygenases: cellular sensors and regulators jelly rolled into one? Nat. Chem. Biol. 3:144-153. [DOI] [PubMed] [Google Scholar]
- 32.Peng, R., A. De Antoni, and D. Gallwitz. 2000. Evidence for overlapping and distinct functions in protein transport of coat protein Sec24p family members. J. Biol. Chem. 275:11521-11528. [DOI] [PubMed] [Google Scholar]
- 33.Peng, Y., E. J. Schwarz, M. A. Lazar, A. Genin, N. B. Spinner, and R. Taub. 1997. Cloning, human chromosomal assignment, and adipose and hepatic expression of the CL-6/INSIG1 gene. Genomics 43:278-284. [DOI] [PubMed] [Google Scholar]
- 34.Raghow, R., C. Yellaturu, X. Deng, E. A. Park, and M. B. Elam. 2008. SREBPs: the crossroads of physiological and pathological lipid homeostasis. Trends Endocrinol. Metab. 19:65-73. [DOI] [PubMed] [Google Scholar]
- 35.Rawson, R. B. 2003. The SREBP pathway: insights from Insigs and insects. Nat. Rev. Mol. Cell Biol. 4:631-640. [DOI] [PubMed] [Google Scholar]
- 36.Schofield, C. J., and P. J. Ratcliffe. 2005. Signalling hypoxia by HIF hydroxylases. Biochem. Biophys. Res. Commun. 338:617-626. [DOI] [PubMed] [Google Scholar]
- 37.Somesh, B. P., J. Reid, W. F. Liu, T. M. Sogaard, H. Erdjument-Bromage, P. Tempst, and J. Q. Svejstrup. 2005. Multiple mechanisms confining RNA polymerase II ubiquitylation to polymerases undergoing transcriptional arrest. Cell 121:913-923. [DOI] [PubMed] [Google Scholar]
- 38.Sun, L. P., L. Li, J. L. Goldstein, and M. S. Brown. 2005. Insig required for sterol-mediated inhibition of Scap/SREBP binding to COPII proteins in vitro. J. Biol. Chem. 280:26483-26490. [DOI] [PubMed] [Google Scholar]
- 39.Sundqvist, A., M. T. Bengoechea-Alonso, X. Ye, V. Lukiyanchuk, J. Jin, J. W. Harper, and J. Ericsson. 2005. Control of lipid metabolism by phosphorylation-dependent degradation of the SREBP family of transcription factors by SCF(Fbw7). Cell Metab. 1:379-391. [DOI] [PubMed] [Google Scholar]
- 40.Sydorskyy, Y., D. J. Dilworth, E. C. Yi, D. R. Goodlett, R. W. Wozniak, and J. D. Aitchison. 2003. Intersection of the Kap123p-mediated nuclear import and ribosome export pathways. Mol. Cell. Biol. 23:2042-2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Todd, B. L., E. V. Stewart, J. S. Burg, A. L. Hughes, and P. J. Espenshade. 2006. Sterol regulatory element binding protein is a principal regulator of anaerobic gene expression in fission yeast. Mol. Cell. Biol. 26:2817-2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Toffaletti, D. L., T. H. Rude, S. A. Johnston, D. T. Durack, and J. R. Perfect. 1993. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J. Bacteriol. 175:1405-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Willger, S. D., N. Grahl, and R. A. Cramer, Jr. 2009. Aspergillus fumigatus metabolism: clues to mechanisms of in vivo fungal growth and virulence. Med. Mycol. 47(Suppl. 1):S72-S79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang, J., J. L. Goldstein, R. E. Hammer, Y. A. Moon, M. S. Brown, and J. D. Horton. 2001. Decreased lipid synthesis in livers of mice with disrupted site-1 protease gene. Proc. Natl. Acad. Sci. USA 98:13607-13612. [DOI] [PMC free article] [PubMed] [Google Scholar]