Abstract
Under conditions of nitrogen stress, leguminous plants form symbioses with soil bacteria called rhizobia. This partnership results in the development of structures called root nodules, in which differentiated endosymbiotic bacteria reduce molecular dinitrogen for the host. The establishment of rhizobium-legume symbioses requires the bacterial synthesis of oligosaccharides, exopolysaccharides, and capsular polysaccharides. Previous studies suggested that the 3-deoxy-d-manno-oct-2-ulopyranosonic acid (Kdo) homopolymeric capsular polysaccharide produced by strain Sinorhizobium meliloti Rm1021 contributes to symbiosis with Medicago sativa under some conditions. However, a conclusive symbiotic role for this polysaccharide could not be determined due to a lack of mutants affecting its synthesis. In this study, we have further characterized the synthesis, secretion, and symbiotic function of the Kdo homopolymeric capsule. We showed that mutants lacking the enigmatic rkp-1 gene cluster fail to display the Kdo capsule on the cell surface but accumulate an intracellular polysaccharide of unusually high Mr. In addition, we have demonstrated that mutations in kdsB2, smb20804, and smb20805 affect the polymerization of the Kdo homopolymeric capsule. Our studies also suggest a role for the capsular polysaccharide in symbiosis. Previous reports have shown that the overexpression of rkpZ from strain Rm41 allows for the symbiosis of exoY mutants of Rm1021 that are unable to produce the exopolysaccharide succinoglycan. Our results demonstrate that mutations in the rkp-1 cluster prevent this phenotypic suppression of exoY mutants, although mutations in kdsB2, smb20804, and smb20805 have no effect.
When nitrogen is limited, many leguminous plants enter into symbiotic relationships with members of the rhizobia, a group of gram-negative soil bacteria. Rhizobia are able to elicit the formation of specialized symbiotic structures located on the plant root, referred to as nodules. The bacteria colonize the developing nodules via a complex series of developmental steps. Initially, the rhizobia adhere to root hairs, specialized epidermal cells located on the surface of the plant root. The adhered bacteria then alter the morphology of the root hairs, such that a curled structure called a shepherd's crook is formed, which entraps a microcolony of the bacteria. Bacteria entrapped within the shepherd's crook subsequently elicit the formation of a tubular invagination of the root hair, referred to as an infection thread. The rhizobia occupy the infection thread, which extends to the base of the root hair and penetrates into the developing nodule. Finally, the bacteria are released from the infection thread into the cytoplasm of the nodule cells, where they differentiate into bacteroids and reduce molecular dinitrogen for the plant (6, 7, 32, 60, 75).
The establishment of a successful rhizobium-legume symbiosis requires an exchange of molecular signals between the symbiotic partners. Plants secrete small molecules into the surrounding soil, such as flavonoids, isoflavonoids, and betaines. These compounds induce the transcription of nod genes within the bacteria (50, 51, 53, 56, 64, 66), which are responsible for the biosynthesis of Nod factor (16, 17, 43, 46, 63). Nod factor, in turn, elicits symbiotic responses within the plant that lead to cortical cell dedifferentiation and division, resulting in the formation of a nodule (15, 16, 19, 21, 46, 49, 69, 72).
Besides Nod factor, three additional classes of polysaccharides have been shown to be required for symbiosis: lipopolysaccharides (LPS), exopolysaccharides, and capsular polysaccharides (K antigens) (57). Mutations that affect the structure of LPS, or its modification by small molecules, result in defects in the ability of the bacteria to colonize root nodules or prevent the differentiation of intracytoplasmic bacteria into bacteroids (4, 14, 39, 68). Mutations in exo genes of Sinorhizobium meliloti strain Rm1021 prevent the biosynthesis of the acidic exopolysaccharide succinoglycan and result in a lack of infection thread extension into the developing nodule (10, 18, 40, 48, 70, 80). The requirement for succinoglycan can be bypassed by mutations that activate the production of a normally cryptic galactoglucan (also known as exopolysaccharide II), which can functionally replace succinoglycan during symbiosis (28-30, 48, 81).
Although exo mutants unable to produce succinoglycan display strong symbiotic phenotypes in strain Rm1021, the production of succinoglycan is not required for symbiosis in other S. meliloti strains. In S. meliloti strain Rm41, the lack of a symbiotic requirement for succinoglycan results from the synthesis of a capsular polysaccharide, which can functionally replace succinoglycan during symbiosis (59, 79). This capsular polysaccharide is a copolymer of glucuronic acid and pseudaminic acid (57-59), and its biosynthesis is dependent on the rkp-1, rkp-2, and rkp-3 gene clusters (36, 54). The rkp-1 and rkp-2 clusters appear to be present among other Sinorhizobium species, whereas the third gene cluster seems to be specific to S. meliloti strain Rm41 (37). The rkp-1 cluster includes the genes rkpA-F, which display significant sequence similarity to type I fatty acid synthases (27, 52) or polyketide synthases (4). Consistently with the proposed function of the rkpA-F cluster in the biosynthesis of fatty acids, the rkpF gene has been shown to undergo a posttranslational modification by 4′-phosphopantetheine, which is required for the biological activity of all fatty acid synthases studied to date (23). Additionally, the rkp-1 cluster includes rkpG, predicted to encode an acyl-transferase; rkpH, predicted to encode an oxidoreductase; and the rkpIJ genes, predicted to encode part of an export apparatus (38). Collectively, the genes of the rkp-1 cluster have been proposed to function in generating a lipid or polyketide necessary for the production of capsule (27, 38, 52). The rkp-2 gene cluster is necessary for the synthesis of the glucuronic acid component of the capsule and is composed of two genes: lpsL, which encodes a UDP-glucuronic acid epimerase, and rkpK, a gene encoding UDP-glucose dehydrogenase (36). Finally, the genes in the rkp-3 cluster encode proteins involved in the production of precursors of the capsule polymer (rkpL-Q) as well as proteins responsible for the export of the capsular polysaccharide to the cell surface (rkpR-T) (37).
Strain Rm41 normally produces a capsule of 20 to 25 kDa (59). Mutants of Rm41 lacking the rkpZ gene produce a capsule of approximately 25 to 35 kDa, which cannot replace succinoglycan during symbiosis (59). Consequently, the rkpZ gene has been suggested to encode a chain length determinator (59). The expression of rkpZ also can affect polysaccharide formation in other S. meliloti strains. The introduction of rkpZ into strain Rm1021 partially corrects the symbiotic defect of exo mutants (48, 59, 67, 79). These data suggested that strain Rm1021 produces the same capsular polysaccharide as Rm41, but the degree of polymerization of the capsule is inappropriate to promote symbiosis.
A recent study reported the characterization of a capsular polysaccharide derived from S. meliloti strain Rm1021 (25). Unlike the capsular polysaccharide detected in strain Rm41, this polysaccharide was reported to be a homopolymer of 3-deoxy-d-manno-oct-2-ulopyranosonic acid (Kdo) (25). The Kdo homopolymer produced by S. meliloti Rm1021 was reported to be of low Mr (3 to 6 kDa). However, the introduction of the rkpZ gene from strain Rm41 resulted in an increase in the Mr of this polysaccharide (3 to 30 kDa) as well as an increase in its abundance (67).
Because the introduction of rkpZ from Rm41 altered the Mr of the Kdo homopolymer and suppressed the symbiotic phenotype of Rm1021 exo mutants, the previous study concluded that this Kdo homopolymeric capsular polysaccharide could functionally replace succinoglycan during symbiosis (67). However, because of a lack of mutants that disrupt the synthesis of the Kdo homopolymer, the authors were unable to eliminate the possibility that the expression of rkpZ influences the biosynthesis of an alternative polysaccharide, which resulted in the observed symbiotic phenotypes.
In this communication, we describe a genetic characterization of capsular polysaccharide biosynthesis in S. meliloti strain Rm1021. We identified a gene cluster involved in the optimum polymerization of the Kdo homopolymeric capsule. Furthermore, we show that the rkp-1 cluster is required for the detection of the Kdo homopolymer on the cell surface. Finally, we demonstrate that the rkp-1 cluster is required for the suppression of the exo mutant symbiotic phenotype by rkpZ.
MATERIALS AND METHODS
Bacterial strains and media.
All Sinorhizobium meliloti strains used in this study are derivatives of either Rm1021 or Rm41 and are listed in Table 1. S. meliloti strains were cultured in Luria-Bertani (LB) (13) or tryptone yeast extract (TY) medium (5) with antibiotics as previously described (45).
TABLE 1.
Strains and plasmids
| Strain and plasmid | Genotype | Reference or source |
|---|---|---|
| S. meliloti | ||
| Rm1021 | SU47 Smr | 42 |
| MGM044 | Rm1021/pTE3 | This study |
| MGM312 | Rm1021/pMW23 | This study |
| MGM522 | Rm1021/pTE3, pMS04 | This study |
| MGM523 | Rm1021/pMW23, pMS04 | This study |
| MGM490 | Rm1021/pTE3, pMB393 | This study |
| MGM491 | Rm1021/pMW23, pMB393 | This study |
| MGM024 | rkpA::Nm | This study |
| MGM050 | rkpA::Nm/pTE3 | This study |
| MGM314 | rkpA::Nm/pMW23 | This study |
| MGM003 | rkpG::pDW33 | This study |
| MGM048 | rkpG::pDW33/pTE3 | This study |
| MGM388 | rkpG::pDW33/pMW23 | This study |
| MGM445 | rkpH::pDW33 | This study |
| MGM454 | rkpH::pDW33/pTE3 | This study |
| MGM455 | rkpH::pDW33/pMW23 | This study |
| MGM418 | rkpI::pDW33 | This study |
| MGM423 | rkpI::pDW33/pTE3 | This study |
| MGM424 | rkpI::pDW33/pMW23 | This study |
| MGM419 | rkpJ::pDW33 | This study |
| MGM425 | rkpJ::pDW33/pTE3 | This study |
| MGM426 | rkpJ::pDW33/pMW23 | This study |
| MGM532 | rkpA::Nm/pTE3, pMS04 | This study |
| MGM508 | rkpA::Nm/pTE3, pMS04::rkpGH | This study |
| MGM533 | rkpA::Nm/pMW23, pMS04 | This study |
| MGM509 | rkpA::Nm/pMW23, pMS04::rkpGH | This study |
| MGM528 | rkpG::pDW33/pTE3, pMS04 | This study |
| MGM510 | rkpG::pDW33/pTE3, pMS04::rkpGH | This study |
| MGM529 | rkpG::pDW33/pMW23, pMS04 | This study |
| MGM511 | rkpG::pDW33/pMW23, pMS04::rkpGH | This study |
| MGM530 | rkpH::pDW33/pTE3, pMS04 | This study |
| MGM512 | rkpH::pDW33/pTE3, pMS04::rkpGH | This study |
| MGM531 | rkpH::pDW33/pMW23, pMS04 | This study |
| MGM513 | rkpH::pDW33/pMW23, pMS04::rkpGH | This study |
| MGM395 | rkpG::pDW33 rkpA::Nm | This study |
| MGM400 | rkpG::pDW33 rkpA::Nm/pTE3 | This study |
| MGM402 | rkpG::pDW33 rkpA::Nm/pMW23 | This study |
| MGM464 | rkpH::pDW33 rkpA::Nm | This study |
| MGM465 | rkpH::pDW33 rkpA::Nm/pTE3 | This study |
| MGM466 | rkpH::pDW33 rkpA::Nm/pMW23 | This study |
| MGM427 | rkpI::pDW33 rkpA::Nm | This study |
| MGM429 | rkpI::pDW33 rkpA::Nm/pTE3 | This study |
| MGM430 | rkpI::pDW33 rkpA::Nm/pMW23 | This study |
| MGM428 | rkpJ::pDW33 rkpA::Nm | This study |
| MGM431 | rkpJ::pDW33 rkpA::Nm/pTE3 | This study |
| MGM432 | rkpJ::pDW33 rkpA::Nm/pMW23 | This study |
| MGM403 | kdsB2::pDW33 | This study |
| MGM404 | kdsB2::pDW33/pTE3 | This study |
| MGM405 | kdsB2::pDW33/pMW23 | This study |
| MGM420 | smb20804::pDW33 | This study |
| MGM433 | smb20804::pDW33/pTE3 | This study |
| MGM434 | smb20804::pDW33/pMW23 | This study |
| MGM365 | smb20805::pDW33 | This study |
| MGM383 | smb20805::pDW33/pTE3 | This study |
| MGM385 | smb20805::pDW33/pMW23 | This study |
| MGM494 | kdsB2::pDW33/pMW23, pMB393 | This study |
| MGM486 | kdsB2::pDW33/pMW23, pMB393::smb20805 | This study |
| MGM547 | smb20804::pDW33/pMW23, pMB393 | This study |
| MGM526 | smb20804::pDW33/pMW23, pMB393::smb20805 | This study |
| MGM492 | smb20805::pDW33/pMW23, pMB393 | This study |
| MGM488 | smb20805::pDW33/pMW23, pMB393::smb20805 | This study |
| DKR145 | exoY::pVO155 | 78 |
| MGM081 | exoY::pVO155/pTE3 | This study |
| MGM358 | exoY::pVO155/pMW23 | This study |
| MGM340 | exoY::pVO155 kdsB2::pDW33 | This study |
| MGM352 | exoY::pVO155 kdsB2::pDW33/pTE3 | This study |
| MGM354 | exoY::pVO155 kdsB2::pDW33/pMW23 | This study |
| DKR530 | exoY::pDW33 | This study |
| MGM359 | exoY::pDW33/pTE3 | This study |
| MGM361 | exoY::pDW33/pMW23 | This study |
| MGM349 | exoY::pDW33 rkpA::Nm | This study |
| MGM355 | exoY::pDW33 rkpA::Nm/pTE3 | This study |
| MGM357 | exoY::pDW33 rkpA::Nm/pMW23 | This study |
| Escherichia coli | ||
| DH5α | F− φ80lacZΔM15 Δ(lacZYA-argF)U169 deoR recA endA1 hsdR17(rK− mK+) phoA supE44 thi-1 gyrA96 relA1 | 65 |
| MT616 | E. coli pRK600 (conjugation helper strain) | 40 |
| Plasmids | ||
| pCR2.1 | TOPO cloning vector | Invitrogen |
| pVO155 | Insertional inactivation plasmid Nmr | 45 |
| pDW33 | Insertional inactivation plasmid Hygr | 12 |
| pJQ200 | Suicide plasmid GmrsacB | 55 |
| pTE3 | pLAFR containing the trp promoter from Salmonella typhi serovar Typhimurium | 3 |
| pMW23 | pRK404, harbors 4.0-kb PstI fragment containing rkpZ from Rm41 | 79 |
| pMB393 | Broad-host-range cloning vector; Spr | 2 |
| pMS04 | Broad-host-range vector, high-copy-number derivative of pMB393 containing the trp promoter from Salmonella typhi serovar Typhimurium | 35 |
| pMTO001 | pCR2.1::rkpA (2,156-bp internal fragment) | This study |
| pMTO002 | pCR2.1::rkpA (2,156-bp internal fragment, carrying a 1,258-bp deletion) | This study |
| pMG003 | pJQ200::rkpA (2,156-bp internal fragment, carrying a 1,258-bp deletion) | This study |
| pMG006 | pJQ200::rkpA::Nm (2,156-bp internal fragment, a 1,258-bp deletion with insertion of Nmr cassette) | This study |
| pMG043 | pVO155::rkpA (internal fragment) | This study |
| pMG042 | pDW33::rkpG (internal fragment) | This study |
| pMG510 | pDW33::rkpH (internal fragment) | This study |
| pMG511 | pDW33::rkpI (internal fragment) | This study |
| pMG512 | pDW33::rkpJ (internal fragment) | This study |
| pDKR520 | pDW33::kdsB2 (internal fragment) | This study |
| pMG502 | pDW33::smb20804 (internal fragment) | This study |
| pMG503 | pDW33::smb20805 (internal fragment) | This study |
| pDKR530 | pDW33::exoY (internal fragment) | This study |
| pMG060 | pMS04::rkpGH (full length) | This study |
| pMG052 | pMB393::smb20805 (full length) | This study |
Strain construction.
Plasmid DNA was introduced into all S. meliloti strains via triparental mating utilizing the Escherichia coli helper strain MT616, as previously described (20).
Strain MGM024 was constructed by introducing plasmid pMG006, a derivative of plasmid pJQ200 (55), containing a neomycin-resistant (Nmr) marked deletion fragment of rkpA, into strain Rm1021, followed by selection for gentamicin resistance. Plasmid pMG006 cannot replicate in S. meliloti and can be stably maintained only through a recombination event between the rkpA gene fragment on the plasmid and the rkpA gene on the chromosome. The gentamicin-resistant colonies then were streaked onto LB medium containing 5% sucrose. Plasmid pMG006 contains the sacB gene, which confers sensitivity to sucrose. Sucrose-resistant colonies that were sensitive to gentamicin but resistant to neomycin were presumed to have undergone an allelic exchange event that resulted in the replacement of the wild-type rkpA fragment on the chromosome with the Nmr-marked rkpA deletion. The mutant construct then was verified by PCR amplification and sequencing. Strain MGM164 was constructed in strain Rm41 by the same method.
Strain MGM260 was constructed by the introduction of plasmid pMG043 (a derivative of plasmid pVO155 [45], harboring an internal fragment of rkpA) into Rm1021 and selection for neomycin-resistant colonies. Plasmid pMG043 cannot replicate within Rm1021, therefore colonies resistant to neomycin can arise only following recombination between the rkpA gene fragment on plasmid pMG043 and the genomic copy of rkpA. Colonies that were neomycin resistant were presumed to have undergone recombination events that integrated the plasmid into the genome, thereby disrupting the rkpA gene. The insertion events then were confirmed via PCR. Strain DKR530 was constructed by the introduction of plasmid pDKR530 (a derivative of plasmid pDW33 [45], which contains an internal fragment of the exoY gene) into Rm1021 and selection for hygromycin-resistant colonies. Colonies that were hygromycin resistant were presumed to have undergone recombination events that integrated plasmid pDKR530 into the genome, disrupting the exoY gene, which was confirmed by PCR analysis and the examination of succinoglycan production. Strains MGM003, MGM445, MGM418, MGM419, DKR520, MGM420, and MGM365 were constructed in an equivalent manner via the introduction of the plasmids pMG042, pMG510, pMG511, pMG512, pDK520, pMG502, and pMG503, respectively, selection for hygromycin resistance, and confirmation by PCR.
Strain MGM349 was constructed via the introduction of rkpA::Nm (from strain MGM024) into strain DKR530 using bacteriophage N3. Similarly, strains MGM395, MGM464, MGM427, MGM428, MGM435, and MGM436 were constructed by the bacteriophage N3-mediated introduction of rkpA::Nm into strains MGM003, MGM445, MGM418, MGM419, MGM420, and MGM365, respectively.
The mutant strains MGM340 and MGM417 were constructed via the introduction of kdsB2::pDW33 with bacteriophage N3 into strains DKR145 and MGM024, respectively.
Plasmid construction.
All plasmids used in this study are listed in Table 1. Plasmid pMTO001 was constructed by amplifying a 2,156-bp internal fragment of rkpA from S. meliloti Rm1021 genomic DNA via PCR using the primers 5′-GGATCCGGGCACCCCCACGTCACCT-3′ and 5′-GGGCCCGGCTCCGGTACGCTCGCCT, followed by being cloned into plasmid pCR2.1 (Invitrogen). Using two SmaI restriction sites located within the cloned rkpA fragment, 1,258 bp of DNA was excised to yield plasmid pMTO002. The rkpA gene fragment containing the 1,258-bp deletion then was liberated from plasmid pMTO002 using restriction enzymes ApaI and BamHI and ligated into vector pJQ200 (digested with the same enzymes) to generate plasmid pMG003. A 1,497-bp fragment containing the neomycin resistance gene (nptII) was liberated from plasmid pVO155 through digestion with NruI. This fragment subsequently was ligated into plasmid pMG003 digested with SmaI to yield plasmid pMG006. The presence of the fragment was verified via PCR, restriction digestion, and the demonstration of neomycin resistance.
Plasmid pMG043 was constructed by excising a 2,156-bp internal rkpA fragment from plasmid pMTO001 using the restriction enzymes BamHI and XbaI and ligating it into vector pVO155, which had been digested with the same restriction enzymes. The presence of the fragment was verified by PCR and restriction digestion.
The plasmid pMG042 was constructed by amplifying an internal fragment of rkpG from Rm1021 genomic DNA using the primers 5′-GGATCCCCGCGTCGGCGAGCCGCCT-3′ and 5′-CTCGAGATCCGCGGTGTCGTTGTGC-3′. The resulting PCR product was cloned into plasmid pCR2.1, yielding plasmid pMTO509. The rkpG-containing fragment then was excised from plasmid pMTO509 via restriction digestion with the enzymes BamHI and XhoI and ligated into plasmid pDW33, which had been digested with the same enzymes. The presence of the fragment was verified via PCR and restriction digestion.
Plasmid pMG510 was constructed using the PCR primers 5′-CCGGATCCATTCGAGGCGATGCAGG-3′ and 5′-ACCTCGAGGCGCAGTAGGCGGTATA-3′ to amplify an internal fragment of the rkpH gene from Rm1021 genomic DNA, which subsequently was cloned into plasmid pCR2.1 to yield plasmid pMTO510. The resulting plasmid then was digested with the restriction enzymes BamHI and XhoI and ligated into plasmid pDW33, which had been digested with the same enzymes.
Plasmid pMG511 was constructed by the PCR amplification of an internal fragment of rkpI from Rm1021 genomic DNA using primers 5′-TCGGATCCGTCGTCTTCCATTTCAT-3′ and 5′-CCCTCGAGGACATTGCTCAAGCGCC-3′ and was cloned into plasmid pCR2.1 to yield plasmid pMTO511. The rkpI-containing fragment then was liberated from pMTO511 by digestion with restriction enzymes BamHI and XhoI and ligated into plasmid pDW33, which had been treated with the same enzymes.
The plasmid pMG512 was constructed by amplifying an internal fragment of rkpJ from Rm1021 genomic DNA using the PCR primers 5′-CCGGTCGACGAATATGCCGGGTGGA-3′ and 5′-TTCTCGAGCGGGTGCACCTTGACGG-3′. The resulting PCR product was cloned into plasmid pCR2.1 to yield plasmid pMTO512. The rkpJ fragment then was excised with the restriction enzymes SalI and XhoI and ligated into plasmid pDW33, which had been digested with the same enzymes.
Plasmid pDKR520 was constructed by PCR amplification of a kdsB2 internal fragment from Rm1021 genomic DNA utilizing the primers 5′-ACGTCGACAAGCTGTTCGCGCCGCA-3′ and 5′-TCTGGGATCCTCGAAGAGATTTCCGCAGCG-3′. The kdsB2-containing PCR fragment was subsequently digested with the enzymes SalI and BamHI and ligated into plasmid pDW33, which had been treated with the same enzymes.
The plasmid pMG502 was constructed by amplifying an internal fragment of ORF smb20804 from Rm1021 genomic DNA using the primers 5′-AGGTCGACGGAACCACTGCGAACGC-3′ and 5′-TTGGATCCGGCGTCCATCTCGAGGG-3′. The PCR product then was cloned into plasmid pCR2.1 to yield plasmid pMTO502. The smb20804-containing fragment then was excised from plasmid pMTO502 with the restriction enzymes SalI and BamHI and ligated into plasmid pDW33, which had been digested with the same enzymes.
The plasmid pMG503 was constructed by amplifying an internal fragment of open reading frame (ORF) smb20805 from Rm1021 genomic DNA using the primers 5′-TCCCAGCGTAATCGCAAAGCCTCTG-3′ and 5′-GGTGTCTAGACGTGCCACCCCGTAC-3′. The PCR product then was cloned into plasmid pCR2.1 to yield plasmid pMTO503. The smb20805-containing fragment then was excised with the restriction enzymes BamHI and XhoI and ligated into plasmid pDW33, which had been digested with the same enzymes.
Plasmid pDKR530 was constructed by the PCR amplification of an exoY internal fragment from Rm1021 genomic DNA utilizing the primers 5′-AGGTCGACGAAGAGTTTTTCAGGATCAACC-3′ and 5′-TGCCGGATCCCCGTCAGGCCGGGACGCGAT-3′. The exoY-containing PCR fragment subsequently was digested with the enzymes SalI and BamHI and ligated into plasmid pDW33, which had been treated with the same enzymes.
The plasmid pMG060 was constructed by amplifying ORFs rkpG and rkpH from Rm1021 genomic DNA using the primers 5′-ATTGTCTAGAACAAGCATGGCCGCGGGCGC-3′ and 5′-TGTCGGATCCGACAAGGAAAAGCA-3′. The PCR product then was cloned into plasmid pCR2.1 to yield plasmid pMTO060. The rkpGH-containing fragment then was excised with the restriction enzymes BamHI and XbaI and ligated into plasmid pMS04, which had been digested with the same enzymes.
The plasmid pMG052 was constructed by amplifying ORF smb20805 from Rm1021 genomic DNA using the primers 5′-TTGCTCGCTCAACGCCTGCGTTAGA-3′ and 5′-CTTTAACGTCAGCTAACGCAATCCC-3′. The PCR product then was cloned into plasmid pCR2.1 to yield plasmid pMTO052. The smb20805-containing fragment then was excised with the restriction enzymes XbaI and BamHI and ligated into plasmid pMB393, which had been digested with the same enzymes.
Extraction and analysis of cellular polysaccharides.
Cellular polysaccharides were obtained by phenol-water extraction as described previously (71) with the following modifications: cells were grown to stationary phase in 1 ml LB liquid medium (in the presence of antibiotics), pelleted, and washed in 1 ml of LB medium. The resulting cell pellet was resuspended in 180 μl of solution A (0.05 M Na2HPO4, 0.005 M EDTA, pH 7), and 180 μl of 90% phenol subsequently was added.
Alternatively, cellular polysaccharides were extracted from S. meliloti cells harvested either from 1-ml LB liquid cultures or directly from LB medium plates using EDTA-triethylamine (EDTA-TEA) buffer as described previously (74), with the following modifications. Briefly, cells were washed once in LB liquid medium and resuspended in 50 μl of 100 mM EDTA (titrated to pH 7.0 with TEA) per approximately 20 mg of wet cell mass. Samples then were incubated at room temperature for 15 min before centrifugation at 10,000 × g for 2 min. The supernatant was removed, 25 μl sodium dodecyl sulfate (SDS) sample buffer was added, and samples were incubated at 100°C for 5 min.
Detection of polysaccharides.
After the addition of SDS sample buffer, the polysaccharide extracts were fractionated via Tris-Tricine SDS-polyacrylamide gel electrophoresis (PAGE) as described previously (44), and polysaccharides were visualized by staining with Alcian blue (11).
Fractionation of polysaccharides and glycosyl composition analysis.
Polysaccharides from 1-liter cell cultures were extracted by the phenol-water procedure, and the water layers were treated sequentially with a mixture of DNase and RNase (20 μl/ml each) and Pronase (20 μl/ml). Following dialysis and lyophilization, the total yield of polysaccharide was approximately 80 mg from each extract. Portions (10 mg) of the polysaccharide extracts were fractionated by size-exclusion chromatography under dissociative conditions on a column of Sephacryl S-400 HR (1.1 by 100 cm) equilibrated in 10 mM Tris buffer (pH 9.2) containing 0.25% sodium deoxycholate, 0.2 M NaCl, and 1.0 mM EDTA. Chromatography was performed on an AKTA system (G.E. Healthcare/Amersham), and the eluant was monitored for the refractive index (RID-10A detector; Shimadzu Corp., Kyoto, Japan) and by UV absorbance at 215, 254, and 280 nm. The fractions obtained from size-exclusion chromatography were dialyzed to remove detergent (57) and then lyophilized. Aliquots (100 to 200 μg) were subjected to glycosyl composition analysis by preparing the trimethylsilyl methyl glycoside derivatives (70), and gas chromatography-mass spectrometry was performed as described previously (71).
In vivo labeling of cellular polysaccharides.
Wild-type and mutant S. meliloti cells were inoculated into 1 ml LB medium in the presence of 2 μCi of [U-14C]glucose and cultured to saturation (optical density at 600 nm of ∼3). The cells were washed once in LB medium, and the radiolabeled polysaccharides then were extracted via phenol-water or EDTA-TEA as described above. The radiolabeled polysaccharides were subsequently fractionated on Tris-Tricine SDS-PAGE, and the incorporation of 14C was detected by phosphorimaging.
Periplasmic fractionation.
The preparation of periplasmic fractions was carried out by osmotic shock treatment as described previously (41), with the following modifications. S. meliloti strains were cultured to stationary phase in 1 ml LB liquid medium (in the presence of antibiotics), and the cells were harvested via centrifugation at 16,000 × g. After being washed once in LB medium, the cells were resuspended in 0.4 ml of 50 mM Tris-HCl (pH 8.0), 20% (wt/vol) sucrose, 2 mM EDTA containing 1 mg/ml lysozyme and were incubated for 15 min on ice. NaCl then was added to a final concentration of 100 mM, and samples were incubated for an additional 10 min on ice. Samples were centrifuged for 10 min at 6,800 × g. Material from both the soluble and insoluble fractions then were extracted via the phenol-water method and fractionated by Tris-Tricine PAGE, and polysaccharides detected by Alcian blue staining or by phosphorimaging.
Subcellular fractionation of very-high-molecular-weight polysaccharide.
S. meliloti strains were cultured to stationary phase in 200 ml LB (in the presence of antibiotics). Cells were harvested at 5,500 × g and exposed to osmotic shock treatment (as described above, except that cells were washed in 25 ml LB medium and resuspended in 10 ml osmotic shock buffer). The insoluble pellet was frozen at −20°C, thawed, and resuspended in 15 ml buffer A. The suspensions were treated with DNase and RNase (20 μl/ml each) at 37°C for several hours, until they showed clearing. Pronase subsequently was added at 20 μl/ml, and suspensions were incubated again at 37°C for several hours. Suspensions were centrifuged at 8,000 × g for 15 min, and the supernatant was removed and centrifuged again to be cleared completely of debris. The supernatants then were exposed to ultracentrifugation for 30 min at 237,000 × g (Beckman TL-100 Ultracentrifuge, Rotor TLA-100.3). Polysaccharides were extracted from the resulting pellet fractions using phenol-water (as described above), while the supernatants were left untreated.
Nodulation assay.
Nodulation assays were performed as described previously (22) on Jensen's medium agar (76) in glass tubes. Plants were grown for 80 days.
Recovery of bacteria from nodules.
Nodules were removed from the root of the plant and pooled (15 plants for each treatment listed). The nodules then were treated with 20% sodium hypochlorite for 2 min, followed by three washes with sterile water. The surface-sterilized nodules were resuspended in LB liquid medium containing 0.3 M sucrose and homogenized. The resulting suspension then was serially diluted and plated on LB medium plates containing streptomycin. After 3 to 4 days the colonies were counted.
Dry weight measurements.
Eighty days after inoculation, the plants were dried for 5 h at 95°C and their dry weights determined.
β-Glucuronidase assay.
The β-glucuronidase activity was determined by a spectrophotometric assay, as described by Jefferson et al. (34).
RESULTS
Inactivation of rkpA-J in S. meliloti strain Rm1021.
To better understand the physiological function of polysaccharides in strain S. meliloti Rm1021, we sought to construct mutants deficient in the synthesis of the Kdo homopolymeric capsule. In strain Rm41, the production of capsular polysaccharide has been reported to require the rkp-1 cluster (38, 52, 54). Although the rkp-1 cluster also is present in strain Rm1021, to our knowledge a characterization of its physiological function has not been reported. We inactivated each of the five genes in the rkp-1 cluster and investigated the effects of the mutations on the synthesis of cell surface polysaccharides. We constructed a 1,258-bp deletion in the rkpA gene, which was replaced with a neomycin resistance cassette. Mutations in the remaining four genes (rkpG, rkpH, rkpI, and rkpJ) were generated via vector integration with plasmid pDW33, a derivative of plasmid pVO155 (45). Plasmid pDW33 contains a ColE1 origin (which is incapable of replication within S. meliloti) and harbors a promoterless copy of uidA, allowing for the construction of transcriptional fusions, and it encodes hygromycin resistance (12, 45).
Mutations in the rkp-1 gene cluster prevent detection of surface-associated Kdo homopolymeric capsule.
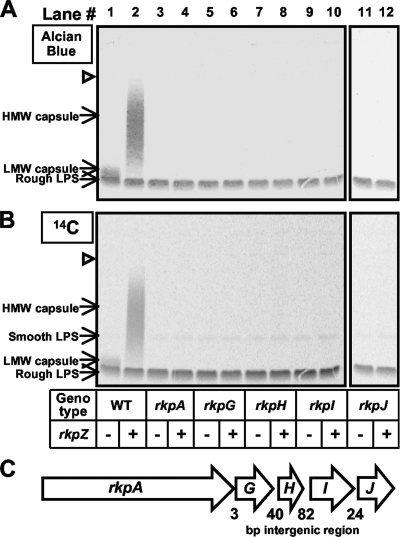
We cultured wild-type S. meliloti and each of the rkp-1 cluster mutants in LB medium in the presence of [U-14C]glucose, recovered cell surface polysaccharides via extraction with EDTA-TEA buffer, fractionated the extracts on Tris-Tricine SDS-PAGE, and visualized the polysaccharides by staining with Alcian blue (Fig. 1A) or by phosphorimaging (Fig. 1B). When fractionated on Tris-Tricine SDS-PAGE, LPS migrates as two species: rough LPS, which migrates at a lower Mr, and smooth LPS, which migrates at a higher Mr. Consistently with previous reports (25, 67), we observed a polysaccharide that migrated between the rough and smooth LPS, forming a light crown above the intensely stained rough LPS band (Fig. 1A and B).
FIG. 1.
Analysis of capsular polysaccharides from wild-type S. meliloti Rm1021 and mutants affecting the rkp-1 gene cluster. Wild-type and mutant strains were cultured in the presence of [U-14C]glucose, and cell surface polysaccharides were isolated via EDTA-TEA extraction, fractionated using Tris-Tricine SDS-PAGE, and detected by staining with Alcian blue (A) or by phosphorimaging (B) as described in Materials and Methods. Lane 1, strain MGM044 (wild-type Rm1021/pTE3 [WT/pTE3]); lane 2, MGM312 (WT/pMW23); lane 3, MGM050 (rkpA::Nm/pTE3); lane 4, MGM314 (rkpA::Nm/pMW23); lane 5, MGM048 (rkpG::pDW33/pTE3); lane 6, MGM388 (rkpG::pDW33/pMW23); lane 7, MGM454 (rkpH::pDW33/pTE3); lane 8, MGM455 (rkpH::pDW33/pMW23); lane 9, MGM423 (rkpI::pDW33/pTE3); lane 10, MGM424 (rkpI::pDW33 pMW23); lane 11, MGM425 (rkpJ::pDW33/pTE3); and lane 12, MGM426 (rkpJ::pDW33/pMW23). The presence of the plasmid containing rkpZ (pMW23) is indicated beneath each lane with a plus, while the presence of the control plasmid (pTE3) is indicated with a minus. The open triangle refers to the interface between the stacking and separating gel. (C) Organization of the rkp-1 gene cluster in S. meliloti Rm1021. Genes are represented as arrows, with the length of the intergenic regions depicted in base pairs. HMW, high molecular weight; LMW, low molecular weight.
Whereas wild-type strains produced detectable quantities of this polysaccharide species, it was undetectable in extracts derived from mutants affecting the rkp-1 gene cluster (rkpA-J) (Fig. 1A and B). Despite the lack of detectable capsular polysaccharide, the LPS of each of the rkp-1 mutant strains was present at levels similar to those of wild-type cells and exhibited a similar banding pattern. These results suggest that mutations in the rkp-1 gene cluster prevent the detection of the capsule but do not result in a gross perturbation of polysaccharide synthesis.
Previous studies showed that the introduction of the rkpZ gene (from S. meliloti strain Rm41) into strain Rm1021 results in a higher degree of the polymerization of the Kdo homopolymeric capsule as well as an increase in overall capsule production (67). To better understand the role of the rkp-1 cluster in the production of capsular polysaccharide, we introduced plasmid pMW23 (which contains the rkpZ gene from Rm41 [79]) into wild-type Rm1021 and the rkp-1 mutants. Similarly to what had been observed previously (67), when examined by Tris-Tricine SDS-PAGE and Alcian blue staining, extracts prepared from wild-type Rm1021 harboring plasmid pMW23 displayed a ladder of bands, which extended from the region corresponding to the rough LPS band to the top of the separating portion of the gel (Fig. 1A and B). In contrast, we did not observe the production of this ladder of bands in the rkp-1 mutants. We thus conclude that the rkp-1 gene cluster is required for the detection of the Kdo homopolymeric polysaccharide in strain Rm1021.
Function of individual rkp-1 cluster genes in expression of the Kdo homopolymer.
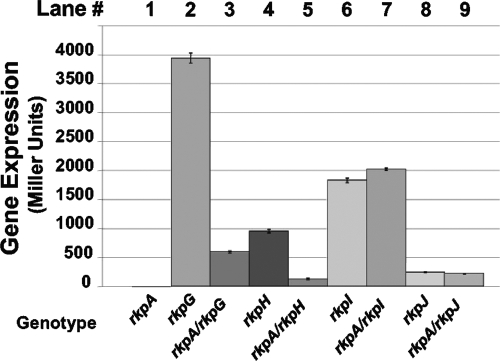
Mutations in each of the genes comprising the rkp-1 cluster resulted in a lack of detectable Kdo homopolymer. Although little is known about the structure of rhizobial promoters, the intergenic regions between the rkpA-J genes are short in length (Fig. 1C). Therefore, it was possible that the genes within the rkp-1 cluster are cotranscribed, and that mutations in the upstream genes of this cluster exert a polar effect on the transcription of the downstream genes. The rkpG::pDW33, rkpH::pDW33, rkpI::pDW33, and rkpJ::pDW33 insertions result in a transcriptional fusion of the respective gene to uidA. We measured the β-glucuronidase activity of the rkpG::uidA, rkpH::uidA, rkpI::uidA, and rkpJ::uidA transcriptional fusions in a wild-type background or in the presence of the rkpA::Nm mutation. In the presence of the rkpA::Nm mutation, the expression of rkpG::uidA and rkpH::uidA fusions was reduced ca. 83% with respect to the expression in a wild-type background (Fig. 2). These data suggest that the rkpA::Nm mutation is polar on the expression of the rkpG and rkpH genes. In contrast, the expression of the rkpI::uidA and rkpJ::uidA fusions was unaffected by the presence of the rkpA::Nm mutation (Fig. 2), suggesting that the rkpA and rkpIJ genes are transcribed independently. Complementation experiments with plasmid-borne rkpG and rkpGH were consistent with these findings (see Fig. S1 in the supplemental material).
FIG. 2.
Expression of rkp-1 cluster genes. Transcriptional fusions between the rkpG, rkpH, rkpI, and rkpJ genes and uidA (encoding β-glucuronidase) were constructed using plasmid pDW33. The strains were grown to stationary phase (optical density at 600 nm of ∼2.5) and assayed for activity as described in Materials and Methods. Enzyme activity is indicated in Miller units. Error bars represent standard deviations from experiments carried out in triplicate. Lane 1, MGM050 (rkpA::Nm/pTE3); lane 2, MGM048 (rkpG::pDW33/pTE3); lane 3, MGM400 (rkpG::pDW33 rkpA::Nm/pTE3); lane 4, MGM454 (rkpH::pDW33/pTE3); lane 5, MGM465 (rkpH::pDW33 rkpA::Nm/pTE3); lane 6, MGM423 (rkpI::pDW33/pTE3); lane 7, MGM429 (rkpI::pDW33 rkpA::Nm/pTE3); lane 8, MGM425 (rkpJ::pDW33/pTE3); and lane 9, MGM431 (rkpJ::pDW33 rkpA::Nm/pTE3). Plasmid pTE3 had no effect on the expression of any of the rkp::uidA fusions (M. G. Müller et al., unpublished).
Identification of a very-high-molecular-weight polysaccharide species in rkp-1 mutant strains harboring rkpZ.
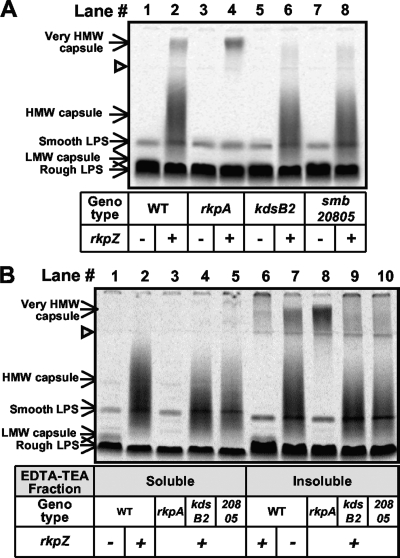
While mutations in the rkp-1 cluster resulted in a lack of detectable capsular polysaccharide under our assay conditions, the reason behind this lack of polysaccharide production was unclear. The method we employed for polysaccharide preparation involved the treatment of the cells with EDTA-TEA, which is expected primarily to extract macromolecules present in the outer membrane (39, 61, 73). We therefore utilized an alternative method, phenol-water extraction (71), to isolate total radiolabeled cellular polysaccharides, which then were fractionated on Tris-Tricine SDS-PAGE and visualized using phosphorimaging (Fig. 3A). Consistently with the results observed with the EDTA-TEA protocol, we did not detect the capsular polysaccharide in the rkpA::Nm mutant harboring vector alone when extracting with phenol-water. However, the phenol-water extraction of the rkpA::Nm mutant harboring pMW23 resulted in the detection of a very-high-molecular-weight species that migrated in the stacking portion of the Tris-Tricine gel. A similar phenotype was observed with the rkpA::pVO155, rkpG::pDW33, rkpH::pDW33, rkpI::pDW33, and rkpJ::pDW33 mutants harboring plasmid pMW23 (M. G. Müller, L. S. Forsberg, and D. H. Keating, unpublished data). Thus, rkp-1 mutant strains harboring plasmid pMW23 produce a very-high-molecular-weight polysaccharide that cannot be liberated via extraction with EDTA-TEA, suggesting that it is not present on the cell surface. Interestingly, we also observed the presence of the very-high-molecular-weight species in phenol-water extracts derived from wild-type Rm1021 containing pMW23 (Fig. 3A), although the species was present in reduced quantity compared to those of the rkp-1 mutants.
FIG. 3.
Mutants deficient in rkpA but harboring pMW23 produce a very-high-molecular-weight polysaccharide. (A) Wild-type and mutant strains, harboring the vector control or pMW23, were cultured in the presence of [U-14C]glucose. Polysaccharides then were extracted via phenol-water, fractionated by Tris-Tricine PAGE, and visualized by phosphorimaging. Lane 1, MGM044 (wild-type Rm1021/pTE3 [WT/pTE3]); lane 2, MGM312 (WT/pMW23); lane 3, MGM050 (rkpA::Nm/pTE3); lane 4, MGM314 (rkpA::Nm/pMW23); lane 5, MGM404 (kdsB2::pDW33/pTE3); lane 6, MGM405 (kdsB2::pDW33/pMW23); lane 7, MGM383 (smb20805::pDW33/pTE3); and lane 8, MGM385 (smb20805::pDW33 ::pDW33/pMW23). In panels A and B the presence of the plasmid containing rkpZ (pMW23) is indicated beneath each lane with a plus, and the presence of the control plasmid (pTE3) is indicated with minus. The open triangle refers to the interface between the stacking and separating gel. (B) Cells were treated with EDTA-TEA and polysaccharides from both the EDTA-TEA-soluble fractions (lanes 1 to 5) and EDTA-TEA-insoluble fractions (lanes 6 to 10) and then were extracted via phenol-water, fractionated by Tris-Tricine PAGE, and visualized by phosphorimaging. Lanes 1 and 6, MGM044; lanes 2 and 7, MGM312; lanes 3 and 8, MGM314; lanes 4 and 9, MGM405; and lanes 5 and 10, MGM385. HMW, high molecular weight; LMW, low molecular weight.
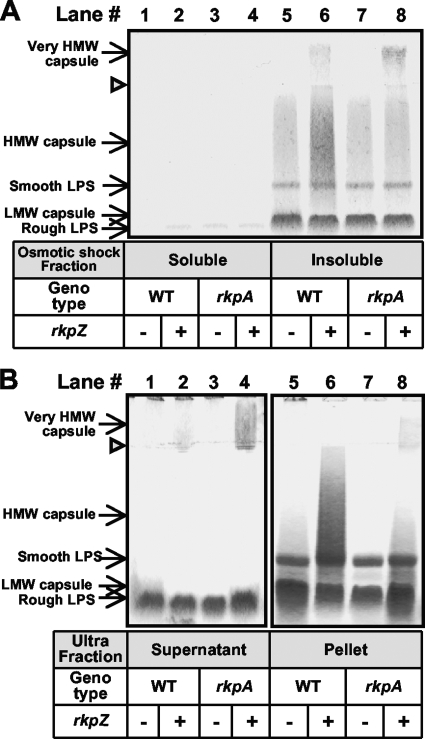
To determine if the very-high-molecular-weight species observed in rkp-1 mutants containing plasmid pMW23 represents an intracellular polysaccharide, we examined the subcellular localization of the polysaccharides produced by both wild-type and rkpA mutant strains harboring pMW23. Treatment with EDTA-TEA divides cellular material into two fractions: a soluble fraction, which is expected to consist of outer membrane components (and likely components of the periplasm), and an EDTA-TEA-insoluble fraction, expected to contain intact cells, components of the periplasm, and the cytoplasm and cytoplasmic membrane (39, 61, 73). We examined the polysaccharide components of the EDTA-TEA-insoluble fraction by extraction via phenol-water and detection by phosphorimaging (Fig. 3B). Using this approach, we were able to demonstrate that the EDTA-TEA-insoluble fraction of both wild-type and rkpA::Nm mutant strains harboring pMW23 contained the very-high-molecular-weight polysaccharide species. Our ability to extract the very-high-molecular-weight polysaccharide exclusively from this fraction suggested that it was located in the periplasm, the cytoplasmic membrane, or within the cytoplasm. To determine if the very-high-molecular-weight polysaccharide was present in the periplasm, we subjected wild-type cells and the rkpA::Nm mutant harboring pMW23 to osmotic shock. This method has been used previously with S. meliloti strain Rm1021 for the recovery of periplasmic contents (41). We then employed phenol-water extraction to prepare polysaccharides from the periplasmic fraction and detected the polysaccharides by staining with Alcian blue. We did not detect the very-high-molecular-weight polysaccharide in the osmotic shock fraction of either the wild type or the rkpA mutant (Fig. 4A). Instead, we detected the very-high-molecular-weight polysaccharide in the insoluble material remaining after osmotic shock extraction (Fig. 4B).
FIG. 4.
Cellular location of very-high-molecular-weight polysaccharides. (A) Analysis of periplasmic contents of wild-type and rkpA::Nm cells. Periplasmic polysaccharides were liberated from wild-type and mutant strains using osmotic shock as described in Materials and Methods. Polysaccharides recovered from both the soluble (lanes 1 to 4) and insoluble osmotic shock fractions (lanes 5 to 8) then were subjected to Tris-Tricine SDS-PAGE, and the polysaccharides were visualized by staining with Alcian blue. Lanes 1 and 5, strain MGM044 (wild-type Rm1021/pTE3 [WT/pTE3]); lanes 2 and 6, MGM312 (WT/pMW23); lanes 3 and 7, MGM050 (rkpA::Nm/pTE3); and lanes 4 and 8, MGM314 (rkpA::Nm/pMW23). The presence of the plasmid containing rkpZ (pMW23) is indicated beneath each lane with a plus, and the presence of the control plasmid (pTE3) is indicated with a minus. The open triangle refers to the interface between the stacking and separating gel. (B) Subcellular fractionation of very-high-molecular-weight capsular polysaccharide extracted from insoluble osmotic shock fractions. Wild-type and mutant cells were treated with osmotic shock. The insoluble pellets then were exposed to freeze-thaw followed by ultracentrifugation. Polysaccharides were extracted from the resulting pellet fraction via phenol-water extraction (lanes 5 to 8) or supernatants were left untreated (lanes 1 to 4). Finally, polysaccharides from both ultracentrifugation fractions were fractionated using Tris-Tricine SDS-PAGE and stained with Alcian blue. Lanes 1 and 5, strain MGM044; lanes 2 and 6, MGM312; lanes 3 and 7, MGM050; and lanes 4 and 8, MGM314. HMW, high molecular weight; LMW, low molecular weight.
Our inability to observe the very-high-molecular-weight polysaccharide in either the EDTA-TEA-soluble fraction or the osmotic shock fraction suggested that the polysaccharide was located in the cytoplasm or associated with the cytoplasmic membrane. Thus, we subjected the insoluble fraction after osmotic shock treatment to freeze-thaw, low-speed centrifugation to remove cell debris and unbroken cells, and then ultracentrifugation. The resulting pelleted material then was extracted using phenol-water. Utilizing this approach, we found that the very-high-molecular-weight polysaccharide was present in the supernatants resulting from the ultracentrifugation of both the wild type and the rkpA::Nm mutant harboring pMW23 (although more was observed in the supernatants derived from rkpA::Nm), and only a small portion was recovered from the ultracentrifuge-pelleted fractions (Fig. 4B). The ultracentrifuge pellets contained significant amounts of what appeared to be LPS, indicating the presence of outer membrane material in this fraction. However, these results suggest that the majority of the very-high-molecular-weight polysaccharide is found in the soluble fraction, consistently with a cytoplasmic location.
Compositional analysis of very-high-molecular-weight polysaccharide.
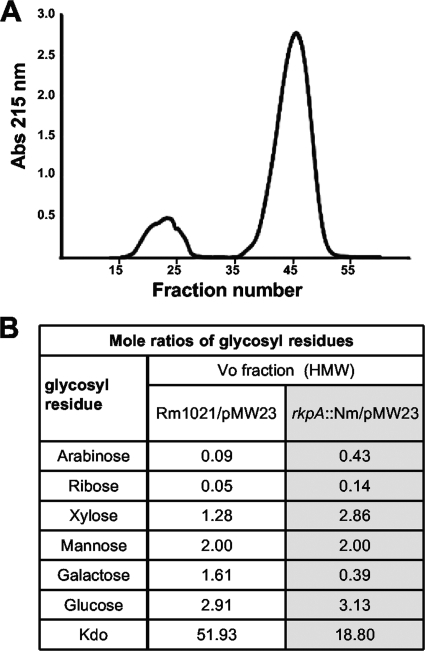
To determine if the very-high-molecular-weight polysaccharide observed in rkp-1 mutants harboring pMW23 was an intracellular form of the Kdo homopolymeric capsular polysaccharide normally on the cell surface, we prepared total cellular polysaccharides from the wild type and rkp-1 mutants harboring pMW23 and subjected them to additional analysis. The chromatography of the polysaccharide extracts derived from the wild-type Rm1021 and the rkpA mutant harboring plasmid pMW23 indicated that both strains produced very-high-molecular-weight material that migrated as a distinct peak at the exclusion volume of an S-400 column when chromatographed in the presence of detergent (dissociative conditions) (Fig. 5A). This indicates an apparent molecular mass of approximately 2 × 106 Da or greater, in agreement with the reported exclusion limit of this matrix as measured with polysaccharides. This fraction was isolated and subjected to glycosyl composition analysis (Fig. 5B). The predominant carbohydrate in both strains was Kdo, suggesting that the primary high-molecular-weight polysaccharide component was a Kdo-rich polysaccharide (K antigen). Small amounts of other glycosyl components also were present in this fraction; however, the structural relationships of these sugars with Kdo were not investigated further.
FIG. 5.
Compositional analysis of very high-molecular-weight (HMW) polysaccharide. Polysaccharide was prepared from the wild type and rkpA mutants harboring plasmid pMW23. (A) Fractionation of polysaccharide extract derived from Rm1021 containing pMW23 by size-exclusion chromatography. Chromatography was performed in the presence of detergent (dissociative conditions) on a column of Sephacryl S-400 HR. The high-molecular-weight polysaccharide (fractions 15 to 26) migrates at the column void volume, indicating an approximate molecular weight of ≥1 × 106 to 2 × 106. LPS migrated in fractions 37 to 53 as revealed by glycosyl composition analysis (M. G. Müller et al., unpublished). Abs, absorbance. (B) Compositional analysis of polysaccharide isolated from void volume (Vo). The molar ratios of glycosyl residues were normalized such that the amount of mannose was equivalent in both the Rm1021 and rkpA::Nm samples.
Identification of genes required for optimum polymerization of the very-high-molecular-weight polysaccharide.
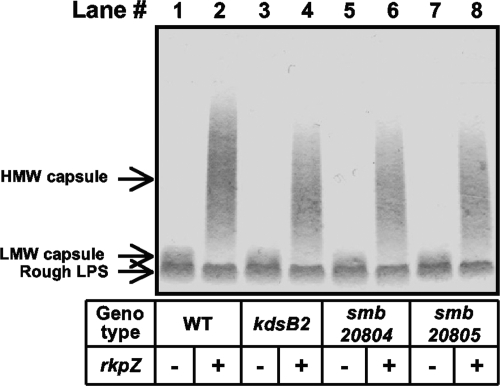
Our data suggested that rkp-1 mutants fail to express the capsular polysaccharide on their cell surface. Instead, the mutants produce a very-high-molecular-weight intracellular polysaccharide species. Compositional analysis demonstrated that the very-high-molecular-weight polysaccharide is enriched in Kdo, suggesting that it is an altered form of the Kdo homopolymer normally found on the cell surface. To determine if the two polysaccharides are related, we sought to construct mutants that affect the polymerization of the Kdo homopolymer. The identification of mutations that affect the Kdo homopolymeric capsule and the very-high-molecular-weight polysaccharide would provide evidence of a precursor-product relationship between the two. The S. meliloti gene kdsB2 was annotated as a CMP-Kdo cytidylyltransferase (1, 24, 26) based on sequence identity to known enzymes. We hypothesized that kdsB2 is necessary for the synthesis of the homopolymeric capsule. We generated a disruption of kdsB2 via vector integration with pDW33 and assayed the strain for the production of Kdo homopolymer in the presence of either plasmid pMW23 or vector control. The polysaccharide profile for kdsB2::pDW33 harboring the vector control was indistinguishable from that seen for the wild type (Fig. 6). However, in the presence of plasmid pMW23 the capsular polysaccharide isolated from the kdsB2::pDW33 mutant exhibited a small but reproducible decrease in Mr compared to that of wild-type strains harboring pMW23 (Fig. 6, compare lanes 2 and 4). Thus, we concluded that kdsB2 is required for the optimum polymerization of the Kdo homopolymer.
FIG. 6.
Analysis of capsular polysaccharides from wild-type S. meliloti and kdsB2, smb20804, and smb20805 mutant strains. Wild-type and mutant strains were cultured in liquid medium, cell surface polysaccharides then were isolated via EDTA-TEA extraction and fractionated using Tris-Tricine SDS-PAGE, and polysaccharides were visualized by staining with Alcian blue, as described in Materials and Methods. Lane 1, strain MGM044 (wild-type Rm1021/pTE3 [WT/pTE3]); lane 2, MGM312 (WT/pMW23); lane 3, MGM404 (kdsB2::pDW33/pTE3); lane 4, MGM405 (kdsB2::pDW33/pMW23); lane 5, MGM433 (smb20804::pDW33/pTE3); lane 6, MGM434 (smb20804::pDW33/pMW23); lane 7, MGM383 (smb20805::pDW33/pTE3); and lane 8, MGM385 (smb20805::pDW33 ::pDW33/pMW23). The presence of the plasmid containing rkpZ (pMW23) is indicated beneath each lane with a plus, and the presence of the control plasmid (pTE3) is indicated with a minus. HMW, high molecular weight; LMW, low molecular weight.
Interestingly, the kdsB2 gene is located directly upstream of two genes: smb20804 (predicted to encode a membrane protein of unknown function) and smb20805 (predicted to encode a glycosyltransferase). We constructed mutations in both smb20804 and smb20805 and visualized the cell surface polysaccharides by Tris-Tricine SDS-PAGE and staining with Alcian blue. Both the smb20804::pDW33 and smb20805::pDW33 mutations resulted in a phenotype similar to that observed in the kdsB2::pDW33 mutant in the presence of pMW23 (Fig. 6). Complementation experiments clearly demonstrated that smb20805 is required for the optimum synthesis of the Kdo homopolymer; results with kdsB2 and smb20804 were inconclusive (see Fig. S2 in the supplemental material).
KdsB2 and Smb20805 affect the synthesis of the very-high-molecular-weight polysaccharide. To determine if the kdsB2 gene cluster mutants affected synthesis of the very-high-molecular-weight polysaccharide, we used the phenol-water extraction method to recover cellular polysaccharides from wild-type Rm1021, the rkpA::Nm mutant, and the kdsB2::pDW33 and smb20805::pDW33 mutant strains, each harboring the pMW23 plasmid. We did not observe the presence of the very-high-molecular-weight polysaccharide species in the kdsB2::pDW33 and smb20805::pDW33 mutants harboring the pMW23 plasmid (Fig. 3A, compare lanes 2, 4, 6, and 8). The ability of the kdsB2::pDW33 and smb20805::pDW33 mutants to affect the polymerization of both polysaccharides suggests that they are variants of the same capsular species.
Symbiotic effects of mutations affecting Kdo homopolymer synthesis.
Our studies identified mutations that affect the polymerization (kdsB2, smb20804, and smb20805) or surface display (rkp-1) of the Kdo homopolymer. In addition, none of these mutants displayed obvious differences in growth in laboratory medium (M. G. Müller et al., unpublished). Thus, these mutants provided molecular tools to alter the production of the Kdo homopolymer and examine its symbiotic function. We inoculated Medicago sativa (alfalfa) seedlings with wild-type S. meliloti Rm1021, mutants affecting the production of the Kdo homopolymer (the rkpA::Nm and kdsB2::pDW33 mutants), and mutants affecting the production of succinoglycan (the exoY::pDW33 and exoY::pVO155 mutants), each harboring either the vector control or plasmid pMW23. Additionally, we inoculated plants with double mutants that affect both the Kdo homopolymer and succinoglycan production (the rkpA::Nm exoY::pDW33 double mutant and the exoY::pVO155 kdsB2::pDW33 double mutant), each carrying pMW23 or the vector control. At various time points postinoculation, we measured the rate of nodule formation. Consistently with what had been reported previously (67), all of the strains assayed were able to elicit the formation of root nodules.
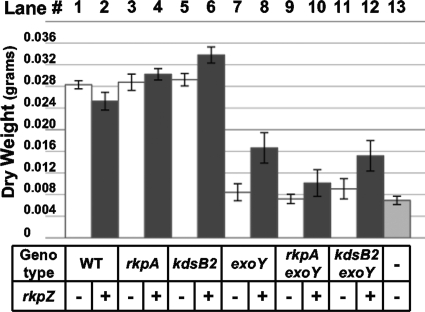
Although all strains could elicit nodules on M. sativa, they differed in their ability to fix nitrogen for the host. Wild-type Rm1021, as well as the rkpA::Nm and kdsB2::pDW33 mutant strains, elicited the formation of pink nodules on at least 14/15 plants, a result that was unaffected by the presence or absence of pMW23 (Table 2). Furthermore, all of the plants displayed robust growth on the nitrogen-free Jensen medium, generating healthy green shoots. Similar results were observed in plant dry weight measurements (Fig. 7), which have been used in several prior studies as an estimate of nitrogen fixation (31, 33, 76, 77). Collectively, these data suggest that the Kdo homopolymeric capsule is not essential for either nodule formation or nitrogen fixation in otherwise wild-type Rm1021.
TABLE 2.
Symbiosis of mutants affected in synthesis or display of the Kdo homopolymer
| S. meliloti strain | rkpZa | No. of green plants | No. of plants tested |
|---|---|---|---|
| Rm1021 | − | 15 | 15 |
| + | 14 | 15 | |
| Rm1021 rkpA::Nm | − | 15 | 15 |
| + | 15 | 15 | |
| Rm1021 kdsB2::pDW33 | − | 15 | 15 |
| + | 15 | 15 | |
| Rm1021 exoY::pDW33 | − | 1 | 15 |
| + | 7 | 15 | |
| RM1021 exoY::pDW33 rkpA::Nm | − | 0 | 15 |
| + | 1 | 15 | |
| Rm1021 exoY::VO155 kdsB2::pDW33 | − | 1 | 15 |
| + | 6 | 15 | |
| Mock inoculation | NA | 0 | 15 |
The presence of the plasmid containing rkpZ (pMW23) is indicated with a plus, while the presence of the control plasmid (pTE3) is indicated with a minus. NA, not applicable. Similar results were observed in two independent experiments.
FIG. 7.
Nitrogen-fixing ability of wild-type and mutant strains affected in capsule biosynthesis. Sterile M. sativa seedlings were placed onto Jensen agar slants. Plants were inoculated 48 h after planting with wild-type S. meliloti or mutant strains harboring either vector control (pTE3) or pMW23. Plants were harvested 80 days postinoculation, and their dry weight was measured. Lane 1, strain MGM044 (wild-type Rm1021/pTE3 [WT/pTE3]); lane 2, MGM312 (WT/pMW23); lane 3, MGM050 (rkpA::Nm/pTE3); lane 4, MGM314 (rkpA::Nm/pMW23); lane 5, MGM404 (kdsB2::pDW33/pTE3); lane 6, MGM405 (kdsB2::pDW33/pMW23); lane 7, MGM359 (exoY::pDW33/pTE3); lane 8, MGM361 (exoY::pDW33/pMW23); lane 9, MGM355 (exoY::pDW33 rkpA::Nm/pTE3); lane 10, MGM357 (exoY::pDW33 rkpA::Nm/pMW23); lane 11, MGM352 (exoY::pVO155 kdsB2::pDW33/pTE3), lane 12, MGM354 (exoY::pVO155 kdsB2::pDW33/pMW23); and lane 13, sample mock inoculated with sterile 10 mM MgSO4. The presence of the plasmid containing rkpZ (pMW23) is indicated beneath each lane with a plus, and the presence of the control plasmid (pTE3) is indicated with a minus. The exoY::pDW33 mutant and exoY::pVO155 mutant behave identically under all conditions tested (M. G. Müller et al., unpublished). Similar results were observed in an additional, independent experiment (M. G. Müller et al., unpublished).
In contrast, mutants unable to produce succinoglycan, both in a wild-type background (exoY::pDW33) or when combined with a mutation that alters Kdo capsule polymerization (exoY::pVO155 kdsB2::pDW33) or with a mutation preventing capsule surface display (exoY::pDW33 rkpA::Nm), triggered the formation of white nodules. The corresponding plants displayed stunted growth and were yellow in color. We observed only a single plant under these conditions that was green and carried a few pink nodules (Table 2). Consistently with these results, we observed a decrease in measurements of the dry weights of plants inoculated with these strains (Fig. 7). In the presence of pMW23, however, the number of green plants carrying pink nodules increased to 7/15 plants when inoculated with the exoY::pDW33 mutant and 6/15 plants when inoculated with the exoY::pVO155 kdsB2::pDW33 double mutant. We also observed an increase in mean plant dry weight, although it was reduced with respect to the wild type (Fig. 7). This result is similar to those of previous studies, which also reported only a partial restoration of nitrogen-fixing symbiosis in exo mutants carrying pMW23 (48, 59, 67, 79).
In contrast, we observed only a single green plant with pink nodules when inoculated with the exoY::pDW33 rkpA::Nm double mutant harboring pMW23 (Table 2). Plants inoculated with this strain displayed a mean dry weight similar to that of the same mutant harboring the vector control, which was reduced compared to the dry weight observed for the wild type or for the kdsB2::pDW33 and rkpA::Nm single-mutant strains harboring pMW23 (Fig. 7). We concluded that the rescue of the symbiotic phenotype of Rm1021 exoY mutants by rkpZ is dependent on the rkp-1 cluster. Interestingly, the kdsB2::pDW33 mutation did not affect the rescue of exoY::pVO155 mutants by rkpZ, suggesting that changes in the degree of polymerization do not affect symbiotic efficiency. In addition, we observed that plants displaying a reduced fixation of nitrogen contained nodules with fewer viable bacteria than those of plants inoculated with wild-type Rm1021 (see Table S1 in the supplemental material).
DISCUSSION
Cell surface polysaccharides, such as LPS and capsule polysaccharide, contribute to diverse microbe-host interactions, including the nitrogen-fixing symbioses between rhizobia and leguminous plants. Studies of S. meliloti strain Rm41 have demonstrated a clear role for capsule polysaccharidein symbiosis. In other S. meliloti strains, such as the well-studied model Rm1021, the symbiotic function of capsule polysaccharide is less clear. To address this gap in knowledge, we carried out a molecular analysis of the Kdo homopolymeric capsule produced by strain Rm1021. We identified three new genes that, when disrupted, affect the degree of the polymerization of the Kdo homopolymeric capsule. We also demonstrated that the rkp-1 cluster is required for the display of the Kdo homopolymeric capsule on the cell surface. The results of this study, combined with data from other laboratories, provide strong evidence that the Kdo homopolymeric capsule produced by strain Rm1021 can contribute to symbiosis with alfalfa.
Based on the reported capsule composition, we focused our attention on kdsB2, which is annotated as a CMP-Kdo cytidylyltransferase. In the presence of plasmid-borne rkpZ from strain Rm41, mutants disrupted for kdsB2 or the downstream genes smb20804 and smb20805, produced capsule with a reduction in degree of polymerization compared to that of the wild type. In addition to its role as a substituent of the S. meliloti capsule, the sugar Kdo is a component of S. meliloti LPS (9). We speculate that enzymes involved in LPS polymerization are able to functionally replace KdsB2, Smb20804, and Smb20805, which would explain the modest effects of the mutations on capsule polymerization. For example, the S. meliloti genome contains a predicted CMP-Kdo synthetase encoded by the kdsB gene. We were not able to generate a mutant deficient for kdsB, suggesting that it is an essential gene, which prevented any insight into its potential role in either LPS or capsule biosynthesis. Future studies will examine the effect of LPS mutants on the polymerization of the Kdo homopolymeric capsule.
A much stronger effect on the Kdo homopolymeric capsule was observed in mutants of the rkp-1 gene cluster. In the presence of the plasmid-borne rkpZ gene, rkp-1 mutants of strain Rm1021 accumulated a very-high-molecular-weight polysaccharide. Interestingly, this polysaccharide species also could be observed, albeit to a lesser extent, in wild-type Rm1021 cells harboring plasmid-borne rkpZ. Several lines of evidence suggest that the very-high-molecular-weight polysaccharide represents an intracellular form of the Kdo homopolymeric capsule normally present on the cell surface. First, a combination of EDTA-TEA extraction, osmotic shock treatment, and ultracentrifugation suggested that this very-high-molecular-weight polysaccharide was located in the cytoplasm, with a very small fraction being present in the cytoplasmic or outer membrane. Second, mutations in kdsB2 and smb20805 (which affect the polymerization of the Kdo homopolymer) prevented the accumulation of the very-high-molecular-weight polysaccharide. Third, an analysis of the composition of the very-high-molecular-weight polysaccharide showed that it was enriched in Kdo, which is consistent with it being a high-molecular-weight form of the Kdo homopolymer (Fig. 5). Collectively, these data strongly suggest that rkp-1 mutants accumulate an intracellular high-molecular-weight form of the Kdo homopolymeric capsule. Interestingly, the rkpA gene of strain Rm1021 consists of one large ORF, whereas the equivalent region of strain Rm41 has been reported to encode six individual ORFs (rkpA-F) (52). To further examine this surprising difference between Rm1021 and Rm41, we partially resequenced the rkpA region in Rm41. Although incomplete, our initial DNA analysis suggests that the rkpA region of Rm41 also consists of one large ORF that is similar to that of Rm1021 (M. G. Müller et al., unpublished).
The sequence similarity between rkpA and genes involved in fatty acid synthesis led to the proposal that the rkp-1 gene cluster was responsible for the synthesis, modification, or transfer of a lipid anchor or lipid carrier (38, 52). In this view, nascent cytoplasmic chains of polysaccharide (4 to 7 kDa) are transferred to an rkp-1-dependent lipid anchor/carrier prior to further polymerization, followed by export to the cell surface (59). A key prediction of this model is that only the 4- to 7-kDa nascent chains of polysaccharide accumulate in mutants of the rkp-1 cluster. Consistently with this model, in the absence of plasmid-borne rkpZ we failed to detect capsular polysaccharide in rkp-1 mutants (the 4- to 7-kDa polysaccharides likely are undetectable via our PAGE assay). However, in the presence of plasmid-borne rkpZ, we observed an accumulation of polysaccharides of very high molecular weight in rkp-1 mutants, which appeared to be cytoplasmically located. This result stands in contrast to the prediction of the model. Mutations affecting rkp-1 in S. meliloti strain Rm41 have been reported to result in a lack of detectable capsular polysaccharide (38, 52). However, our studies using [U-14C]glucose labeling and phenol-water extraction also resulted in the observation of a similar very-high-molecular-weight form of capsule in rkpA mutants of strain Rm41 (M. G. Müller et al., unpublished). Although preliminary in nature, these data suggest that rkp-1 mutants display similar phenotypes in both Rm1021 and Rm41.
In the presence of plasmid-borne rkpZ, S. meliloti strain Rm1021 produces Kdo homopolymeric capsule with an increased degree of polymerization (67). Within strain Rm41, however, mutants lacking rkpZ display an increased degree of the polymerization of capsular polysaccharide (59), suggesting that the protein functions to reduce the degree of polymerization. In either case, the results have led to the suggestion that RkpZ functions as a chain length regulator (37, 59). Interestingly, the expression of rkpZ in strain Rm1021 also resulted in an overall increase in the abundance of the Kdo homopolymer (67). These unexpected effects of rkpZ on capsule abundance led the authors of a previous study to suggest that RkpZ instead acts as a regulator of polysaccharide synthesis (67), with the altered degree of polymerization being a consequence of capsule overproduction. The results of our study are consistent with this idea. We propose that the heterologous expression of rkpZ leads to greatly increased capsule biosynthesis in wild-type strain Rm1021, which exceeds the ability of the cell to transport the polysaccharide from the cytoplasm. In the absence of efficient transport from the cytoplasm, this cytoplasmic Kdo homopolymeric polysaccharide continues to undergo polymerization, resulting in the production of the very-high-molecular-weight species observed in wild-type strains harboring rkpZ. Mutations in kdsB2, smb20804, and smb20805 decrease the rate of polymerization, which allows transport to remain balanced with synthesis, thereby preventing the accumulation of the very-high-molecular-weight polysaccharide. In rkp-1 mutants transport is blocked, leading to the production of exclusively very-high-molecular-weight polysaccharide.
While evidence from our group, as well as that from other groups (67), suggests that RkpZ acts as a regulator of Kdo synthesis, the mechanism of this regulation is unclear. The introduction of plasmid-borne rkpZ had no effect on the expression of kdsB2::uidA, smb20804::uidA, and smb20805::uidA transcriptional fusions, nor did it affect the expression of rkpA::uidA, rkpG::uidA, rkpH::uidA, rkpI::uidA, or rkpJ::uidA transcriptional fusions (M. G. Müller et al., unpublished data). Therefore, RkpZ does not appear to act as a regulator of transcription. Interestingly, RkpZ exhibits 42% amino acid identity to KpsC from E. coli (8, 47). When kpsC was mutated, it rendered the E. coli cell surface devoid of K5 antigen. Instead, the polysaccharide was present solely in the cell cytoplasm and lacked a phospholipid anchor, although its size was similar to that of wild-type E. coli K5 antigen (8). In addition, recent localization studies have suggested that KpsC functions as part of a complex connecting polysaccharide synthesis to transport (62), which could mean that KpsC and RkpZ carry out similar functions. Future studies will examine the role of RkpZ in polysaccharide synthesis and export.
Supplementary Material
Acknowledgments
We thank Guy Townsend for helpful discussions regarding the manuscript and Larissa Sharapova for providing plasmid pMW23.
This work was funded by Award 2005-35319-15304 from the U.S. Department of Agriculture. The Complex Carbohydrate Research Center was supported in part by Department of Energy grant DEFG02-93ER20097.
Footnotes
Published ahead of print on 4 September 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Barnett, M. J., R. F. Fisher, T. Jones, C. Komp, A. P. Abola, F. Barloy-Hubler, L. Bowser, D. Capela, F. Galibert, J. Gouzy, M. Gurjal, A. Hong, L. Huizar, R. W. Hyman, D. Kahn, M. L. Kahn, S. Kalman, D. H. Keating, C. Palm, M. C. Peck, R. Surzycki, D. H. Wells, K. C. Yeh, R. W. Davis, N. A. Federspiel, and S. R. Long. 2001. Nucleotide sequence and predicted functions of the entire Sinorhizobium meliloti pSymA megaplasmid. Proc. Natl. Acad. Sci. USA 98:9883-9888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnett, M. J., V. Oke, and S. R. Long. 2000. New genetic tools for use in the Rhizobiaceae and other bacteria. BioTechniques 29:240-245. [DOI] [PubMed] [Google Scholar]
- 3.Barnett, M. J., J. A. Swanson, and S. R. Long. 1998. Multiple genetic controls on Rhizobium meliloti syrA, a regulator of exopolysaccharide abundance. Genetics 148:19-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becker, A., N. Fraysse, and L. Sharypova. 2005. Recent advances in studies on structure and symbiosis-related function of rhizobial K-antigens and lipopolysaccharides. Mol. Plant-Microbe Interact. 18:899-905. [DOI] [PubMed] [Google Scholar]
- 5.Beringer, J. E. 1974. R factor transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 84:188-198. [DOI] [PubMed] [Google Scholar]
- 6.Brewin, N. J. 1991. Development of the legume root nodule. Annu. Rev. Cell Biol. 7:191-226. [DOI] [PubMed] [Google Scholar]
- 7.Brewin, N. J. 1992. Nodule formation in legumes, p. 229-248. In J. Lederberg (ed.), Encyclopedia of microbiology, vol. 3. Academic Press, San Diego, CA. [Google Scholar]
- 8.Bronner, D., V. Sieberth, C. Pazzani, I. S. Roberts, G. J. Boulnois, B. Jann, and K. Jann. 1993. Expression of the capsular K5 polysaccharide of Escherichia coli: biochemical and electron microscopic analyses of mutants with defects in region 1 of the K5 gene cluster. J. Bacteriol. 175:5984-5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carlson, R. W., B. Reuhs, T. B. Chen, U. R. Bhat, and K. D. Noel. 1995. Lipopolysaccharide core structures in Rhizobium etli and mutants deficient in O-antigen. J. Biol. Chem. 270:11783-11788. [DOI] [PubMed] [Google Scholar]
- 10.Cheng, H. P., and G. C. Walker. 1998. Succinoglycan is required for initiation and elongation of infection threads during nodulation of alfalfa by Rhizobium meliloti. J. Bacteriol. 180:5183-5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corzo, J., R. Perez-Galdona, M. Leon-Barrios, and A. M. Gutierrez- Navarro. 1991. Alcian blue fixation allows silver staining of the isolated polysaccharide component of bacterial lipopolysaccharides in polyacrylamide gels. Electrophoresis 12:439-441. [DOI] [PubMed] [Google Scholar]
- 12.Cronan, G. E., and D. H. Keating. 2004. Sinorhizobium meliloti sulfotransferase that modifies lipopolysaccharide. J. Bacteriol. 186:4168-4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis, R. W., D. Botstein, and J. R. Roth. 1980. Advanced bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 14.Dazzo, F. B., G. L. Truchet, R. I. Hollingsworth, E. M. Hrabak, H. S. Pankratz, S. Philip-Hollingsworth, J. L. Salzwedel, K. Chapman, L. Appenzeller, A. Squartini, et al. 1991. Rhizobium lipopolysaccharide modulates infection thread development in white clover root hairs. J. Bacteriol. 173:5371-5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Bruijn, F. 1991. Biochemical and molecular studies: symbiotic nitrogen fixation. Curr. Opin. Biotechnol. 2:184-192. [DOI] [PubMed] [Google Scholar]
- 16.Dénarié, J., and J. Cullimore. 1993. Lipo-oligosaccharide nodulation factors: a new class of signaling molecules mediating recognition and morphogenesis. Cell 74:951-954. [DOI] [PubMed] [Google Scholar]
- 17.Dénarié, J., F. Debelle, and C. Rosenberg. 1992. Signaling and host range variation in nodulation. Annu. Rev. Microbiol. 46:497-531. [DOI] [PubMed] [Google Scholar]
- 18.D'Haeze, W., J. Glushka, R. De Rycke, M. Holsters, and R. W. Carlson. 2004. Structural characterization of extracellular polysaccharides of Azorhizobium caulinodans and importance for nodule initiation on Sesbania rostrata. Mol. Microbiol. 52:485-500. [DOI] [PubMed] [Google Scholar]
- 19.D'Haeze, W., and M. Holsters. 2002. Nod factor structures, responses, and perception during initiation of nodule development. Glycobiology 12:79R-105R. [DOI] [PubMed] [Google Scholar]
- 20.Ditta, G., S. Stanfield, D. Corbin, and D. R. Helinski. 1980. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc. Natl. Acad. Sci. USA 77:7347-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Downie, J. A. 1994. Signalling strategies for nodulation of legumes by rhizobia. Trends Microbiol. 2:318-324. [DOI] [PubMed] [Google Scholar]
- 22.Ehrhardt, D. W., E. M. Atkinson, and S. R. Long. 1992. Depolarization of alfalfa root hair membrane potential by Rhizobium meliloti Nod factors. Science 256:998-1000. [DOI] [PubMed] [Google Scholar]
- 23.Epple, G., K. M. van der Drift, J. E. Thomas-Oates, and O. Geiger. 1998. Characterization of a novel acyl carrier protein, RkpF, encoded by an operon involved in capsular polysaccharide biosynthesis in Sinorhizobium meliloti. J. Bacteriol. 180:4950-4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Finan, T. M., S. Weidner, K. Wong, J. Buhrmester, P. Chain, F. J. Vorholter, I. Hernandez-Lucas, A. Becker, A. Cowie, J. Gouzy, B. Golding, and A. Puhler. 2001. The complete sequence of the 1,683-kb pSymB megaplasmid from the N2-fixing endosymbiont Sinorhizobium meliloti. Proc. Natl. Acad. Sci. USA 98:9889-9894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fraysse, N., B. Lindner, Z. Kaczynski, L. Sharypova, O. Holst, K. Niehaus, and V. Poinsot. 2005. Sinorhizobium meliloti strain 1021 produces a low-molecular-mass capsular polysaccharide that is a homopolymer of 3-deoxy-D-manno-oct-2-ulosonic acid harboring a phospholipid anchor. Glycobiology 15:101-108. [DOI] [PubMed] [Google Scholar]
- 26.Galibert, F., T. M. Finan, S. R. Long, A. Puhler, P. Abola, F. Ampe, F. Barloy-Hubler, M. J. Barnett, A. Becker, P. Boistard, G. Bothe, M. Boutry, L. Bowser, J. Buhrmester, E. Cadieu, D. Capela, P. Chain, A. Cowie, R. W. Davis, S. Dreano, N. A. Federspiel, R. F. Fisher, S. Gloux, T. Godrie, A. Goffeau, B. Golding, J. Gouzy, M. Gurjal, I. Hernandez-Lucas, A. Hong, L. Huizar, R. W. Hyman, T. Jones, D. Kahn, M. L. Kahn, S. Kalman, D. H. Keating, E. Kiss, C. Komp, V. Lelaure, D. Masuy, C. Palm, M. C. Peck, T. M. Pohl, D. Portetelle, B. Purnelle, U. Ramsperger, R. Surzycki, P. Thebault, M. Vandenbol, F. J. Vorholter, S. Weidner, D. H. Wells, K. Wong, K. C. Yeh, and J. Batut. 2001. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293:668-672. [DOI] [PubMed] [Google Scholar]
- 27.Geiger, O., and I. M. Lopez-Lara. 2002. Rhizobial acyl carrier proteins and their roles in the formation of bacterial cell-surface components that are required for the development of nitrogen-fixing root nodules on legume hosts. FEMS Microbiol. Lett. 208:153-162. [DOI] [PubMed] [Google Scholar]
- 28.Glazebrook, J., and G. C. Walker. 1989. A novel exopolysaccharide can function in place of the calcofluor-binding exopolysaccharide in nodulation of alfalfa by Rhizobium meliloti. Cell 56:661-672. [DOI] [PubMed] [Google Scholar]
- 29.González, J. E., B. L. Reuhs, and G. C. Walker. 1996. Low molecular weight EPS II of Rhizobium meliloti allows nodule invasion in Medicago sativa. Proc. Natl. Acad. Sci. USA 93:8636-8641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.González, J. E., G. M. York, and G. C. Walker. 1996. Rhizobium meliloti exopolysaccharides: synthesis and symbiotic function. Gene 179:141-146. [DOI] [PubMed] [Google Scholar]
- 31.Haydock, K. P., D. O. Norris, and L. T. Mannetje. 1980. The relation between nitrogen percent and dry weight of inoculated legumes. Plant Soil 57:353-362. [Google Scholar]
- 32.Hirsch, A. M. 1992. Developmental biology of legume nodulation. New Phytol. 122:211-237. [DOI] [PubMed] [Google Scholar]
- 33.Hozbor, D. F., A. J. Pich Otero, A. R. Lodeiro, M. F. Del Papa, M. Pistorio, and A. Lagares. 2004. The symbiotic defect in a Sinorhizobium meliloti lipopolysaccharide mutant can be overcome by expression of other surface polysaccharides. Res. Microbiol. 155:855-860. [DOI] [PubMed] [Google Scholar]
- 34.Jefferson, R. A., S. M. Burgress, and D. Hirsch. 1986. β-Glucuronidase from Escherichia coli as a gene-fusion marker. Proc. Natl. Acad. Sci. USA 83:8447-8451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keating, D. H., M. G. Willits, and S. R. Long. 2002. A Sinorhizobium meliloti lipopolysaccharide mutant altered in cell surface sulfation. J. Bacteriol. 184:6681-6689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kereszt, A., E. Kiss, B. L. Reuhs, R. W. Carlson, A. Kondorosi, and P. Putnoky. 1998. Novel rkp gene clusters of Sinorhizobium meliloti involved in capsular polysaccharide production and invasion of the symbiotic nodule: the rkpK gene encodes a UDP-glucose dehydrogenase. J. Bacteriol. 180:5426-5431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kiss, E., A. Kereszt, F. Barta, S. Stephens, B. L. Reuhs, A. Kondorosi, and P. Putnoky. 2001. The rkp-3 gene region of Sinorhizobium meliloti Rm41 contains strain-specific genes that determine K antigen structure. Mol. Plant-Microbe Interact. 14:1395-1403. [DOI] [PubMed] [Google Scholar]
- 38.Kiss, E., B. L. Reuhs, J. S. Kim, A. Kereszt, G. Petrovics, P. Putnoky, I. Dusha, R. W. Carlson, and A. Kondorosi. 1997. The rkpGHI and -J genes are involved in capsular polysaccharide production by Rhizobium meliloti. J. Bacteriol. 179:2132-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lagares, A., G. Caetano-Anolles, K. Niehaus, J. Lorenzen, H. D. Ljunggren, A. Puhler, and G. Favelukes. 1992. A Rhizobium meliloti lipopolysaccharide mutant altered in competitiveness for nodulation of alfalfa. J. Bacteriol. 174:5941-5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leigh, J. A., E. R. Signer, and G. C. Walker. 1985. Exopolysaccharide-deficient mutants of Rhizobium meliloti that form ineffective nodules. Proc. Natl. Acad. Sci. USA 82:6231-6235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Long, S., S. McCune, and G. C. Walker. 1988. Symbiotic loci of Rhizobium meliloti identified by random TnphoA mutagenesis. J. Bacteriol. 170:4257-4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meade, H. M., S. R. Long, G. B. Ruvkun, S. E. Brown, and F. M. Ausubel. 1982. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J. Bacteriol. 149:114-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mergaert, P., W. D'Haeze, D. Geelen, D. Prome, M. Van Montagu, R. Geremia, J. C. Prome, and M. Holsters. 1995. Biosynthesis of Azorhizobium caulinodans Nod factors. Study of the activity of the NodABC proteins by expression of the genes in Escherichia coli. J. Biol. Chem. 270:29217-29223. [DOI] [PubMed] [Google Scholar]
- 44.Niehaus, K., A. Largares, and A. Puhler. 1998. A Sinorhizobium meliloti Lipopolysaccharide mutant induces effective nodules on the host plant Medicago sativa (Alfalfa) but fails to establish a symbiosis with Medicago truncatula. Mol. Plant-Microbe Interact. 11:906-914. [Google Scholar]
- 45.Oke, V., and S. R. Long. 1999. Bacterial genes induced within the nodule during the Rhizobium-legume symbiosis. Mol. Microbiol. 32:837-849. [DOI] [PubMed] [Google Scholar]
- 46.Oldroyd, G. E., and J. A. Downie. 2004. Calcium, kinases and nodulation signalling in legumes. Nat. Rev. Mol. Cell Biol. 5:566-576. [DOI] [PubMed] [Google Scholar]
- 47.Pazzani, C., C. Rosenow, G. J. Boulnois, D. Bronner, K. Jann, and I. S. Roberts. 1993. Molecular analysis of region 1 of the Escherichia coli K5 antigen gene cluster: a region encoding proteins involved in cell surface expression of capsular polysaccharide. J. Bacteriol. 175:5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pellock, B. J., H. P. Cheng, and G. C. Walker. 2000. Alfalfa root nodule invasion efficiency is dependent on Sinorhizobium meliloti polysaccharides. J. Bacteriol. 182:4310-4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perret, X., C. Staehelin, and W. J. Broughton. 2000. Molecular basis of symbiotic promiscuity. Microbiol. Mol. Biol. Rev. 64:180-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peters, N. K., J. W. Frost, and S. R. Long. 1986. A plant flavone, luteolin, induces expression of Rhizobium meliloti nodulation genes. Science 233:977-980. [DOI] [PubMed] [Google Scholar]
- 51.Peters, N. K., and D. P. Verma. 1990. Phenolic compounds as regulators of gene expression in plant-microbe relations. Mol. Plant-Microbe Interact. 3:4-8. [DOI] [PubMed] [Google Scholar]
- 52.Petrovics, G., P. Putnoky, B. Reuhs, J. Kim, T. A. Thorp, K. D. Noel, R. W. Carlson, and A. Kondorosi. 1993. The presence of a novel type of surface polysaccharide in Rhizobium meliloti requires a new fatty acid synthase-like gene cluster involved in symbiotic nodule development. Mol. Microbiol. 8:1083-1094. [DOI] [PubMed] [Google Scholar]
- 53.Phillips, D. A., C. M. Joseph, and C. A. Maxwell. 1992. Trigonelline and stachydrine released from alfalfa seeds activate NodD2 protein in Rhizobium meliloti. Plant Physiol. 99:1526-1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Putnoky, P., G. Petrovics, A. Kereszt, E. Grosskopf, D. T. Ha, Z. Banfalvi, and A. Kondorosi. 1990. Rhizobium meliloti lipopolysaccharide and exopolysaccharide can have the same function in the plant-bacterium interaction. J. Bacteriol. 172:5450-5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Quandt, J., and M. F. Hynes. 1993. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127:15-21. [DOI] [PubMed] [Google Scholar]
- 56.Redmond, J. W., M. Batley, M. A. Djordjevic, R. W. Innes, P. L. Kuempel, and B. G. Rolfe. 1986. Flavones induce expression of nodulation genes in Rhizobium. Nature 323:632-635. [Google Scholar]
- 57.Reuhs, B. L., R. W. Carlson, and J. S. Kim. 1993. Rhizobium fredii and Rhizobium meliloti produce 3-deoxy-D-manno-2-octulosonic acid-containing polysaccharides that are structurally analogous to group II K antigens (capsular polysaccharides) found in Escherichia coli. J. Bacteriol. 175:3570-3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reuhs, B. L., D. P. Geller, J. S. Kim, J. E. Fox, V. S. Kolli, and S. G. Pueppke. 1998. Sinorhizobium fredii and Sinorhizobium meliloti produce structurally conserved lipopolysaccharides and strain-specific K antigens. Appl. Environ. Microbiol. 64:4930-4938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reuhs, B. L., M. N. Williams, J. S. Kim, R. W. Carlson, and F. Cote. 1995. Suppression of the Fix− phenotype of Rhizobium meliloti exoB mutants by lpsZ is correlated to a modified expression of the K polysaccharide. J. Bacteriol. 177:4289-4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ridge, R. 1992. A model for legume root hair growth and Rhizobium infection. Symbiosis 14:359-373. [Google Scholar]
- 61.Ridley, B. L., B. S. Jeyaretnam, and R. W. Carlson. 2000. The type and yield of lipopolysaccharide from symbiotically deficient rhizobium lipopolysaccharide mutants vary depending on the extraction method. Glycobiology 10:1013-1023. [DOI] [PubMed] [Google Scholar]
- 62.Rigg, G. P., B. Barrett, and I. S. Roberts. 1998. The localization of KpsC, S and T, and KfiA, C and D proteins involved in the biosynthesis of the Escherichia coli K5 capsular polysaccharide: evidence for a membrane-bound complex. Microbiology 144:2905-2914. [DOI] [PubMed] [Google Scholar]
- 63.Roche, P., F. Maillet, C. Plazanet, F. Debelle, M. Ferro, G. Truchet, J. C. Prome, and J. Denarie. 1996. The common nodABC genes of Rhizobium meliloti are host-range determinants. Proc. Natl. Acad. Sci. USA 93:15305-15310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rolfe, B. G. 1988. Flavones and isoflavones as inducing substances of legume nodulation. Biofactors 1:3-10. [PubMed] [Google Scholar]
- 65.Sambrook, J., E. F. Fritsch, and T. E. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed., vol. 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 66.Schlaman, H. R., R. J. Okker, and B. J. Lugtenberg. 1992. Regulation of nodulation gene expression by NodD in rhizobia. J. Bacteriol. 174:5177-5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sharypova, L. A., G. Chataigne, N. Fraysse, A. Becker, and V. Poinsot. 2006. Overproduction and increased molecular weight account for the symbiotic activity of the rkpZ-modified K polysaccharide from Sinorhizobium meliloti Rm1021. Glycobiology 16:1181-1193. [DOI] [PubMed] [Google Scholar]
- 68.Spaink, H. P., and B. J. Lugtenberg. 1994. Role of rhizobial lipo-chitin oligosaccharide signal molecules in root nodule organogenesis. Plant Mol. Biol. 26:1413-1422. [DOI] [PubMed] [Google Scholar]
- 69.Spaink, H. P., D. M. Sheeley, A. A. van Brussel, J. Glushka, W. S. York, T. Tak, O. Geiger, E. P. Kennedy, V. N. Reinhold, and B. J. Lugtenberg. 1991. A novel highly unsaturated fatty acid moiety of lipo-oligosaccharide signals determines host specificity of Rhizobium. Nature 354:125-130. [DOI] [PubMed] [Google Scholar]
- 70.Staehelin, C., L. S. Forsberg, W. D'Haeze, M. Y. Gao, R. W. Carlson, Z. P. Xie, B. J. Pellock, K. M. Jones, G. C. Walker, W. R. Streit, and W. J. Broughton. 2006. Exo-oligosaccharides of Rhizobium sp. strain NGR234 are required for symbiosis with various legumes. J. Bacteriol. 188:6168-6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Townsend, G. E., Jr., L. S. Forsberg, and D. H. Keating. 2006. Mesorhizobium loti produces nodPQ-dependent sulfated cell surface polysaccharides. J. Bacteriol. 188:8560-8572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Truchet, G., P. Roche, P. Lerouge, J. Vasse, S. Camut, F. de Billy, J.-C. Prome, and J. Denarie. 1991. Sulphated lipo-oligosaccharide signals of Rhizobium meliloti elicit root nodule organogenesis in alfalfa. Nature 351:670-673. [Google Scholar]
- 73.Vaara, M. 1992. Agents that increase the permeability of the outer membrane. Microbiol. Rev. 56:395-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Valverde, C., D. F. Hozbor, and A. Lagares. 1997. Rapid preparation of affinity-purified lipopolysaccharide samples for electrophoretic analysis. BioTechniques 22:230-236. [DOI] [PubMed] [Google Scholar]
- 75.Vijn, I., L. das Nevas, A. van Kammen, H. Franssen, and T. Bisseling. 1993. Nod factors and nodulation in plants. Science 260:1764-1765. [DOI] [PubMed] [Google Scholar]
- 76.Vincent, J. M. 1970. A manual for the practical study of the root-nodule bacteria. Blackwell Scientific, Oxford, United Kingdom.
- 77.Wang, C., M. Saldanha, X. Sheng, K. J. Shelswell, K. T. Walsh, B. W. Sobral, and T. C. Charles. 2007. Roles of poly-3-hydroxybutyrate (PHB) and glycogen in symbiosis of Sinorhizobium meliloti with Medicago sp. Microbiology 153:388-398. [DOI] [PubMed] [Google Scholar]
- 78.Wells, D. H., and S. R. Long. 2002. The Sinorhizobium meliloti stringent response affects multiple aspects of symbiosis. Mol. Microbiol. 43:1115-1127. [DOI] [PubMed] [Google Scholar]
- 79.Williams, M. N., R. I. Hollingsworth, S. Klein, and E. R. Signer. 1990. The symbiotic defect of Rhizobium meliloti exopolysaccharide mutants is suppressed by lpsZ+, a gene involved in lipopolysaccharide biosynthesis. J. Bacteriol. 172:2622-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yang, C., E. R. Signer, and A. M. Hirsch. 1992. Nodules initiated by Rhizobium meliloti exopolysaccharide mutants lack a discrete, persistent nodule meristem. Plant Physiol. 98:143-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhan, H. J., S. B. Levery, C. C. Lee, and J. A. Leigh. 1989. A second exopolysaccharide of Rhizobium meliloti strain SU47 that can function in root nodule invasion. Proc. Natl. Acad. Sci. USA 86:3055-3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.