Abstract
A common landmark of activated genes is the presence of trimethylation on lysine 4 of histone H3 (H3K4) at promoter regions. Set1/COMPASS was the founding member and is the only H3K4 methylase in Saccharomyces cerevisiae; however, in mammals, at least six H3K4 methylases, Set1A and Set1B and MLL1 to MLL4, are found in COMPASS-like complexes capable of methylating H3K4. To gain further insight into the different roles and functional targets for the H3K4 methylases, we have undertaken a genome-wide analysis of H3K4 methylation patterns in wild-type Mll1+/+ and Mll1−/− mouse embryonic fibroblasts (MEFs). We found that Mll1 is required for the H3K4 trimethylation of less than 5% of promoters carrying this modification. Many of these genes, which include developmental regulators such as Hox genes, show decreased levels of RNA polymerase II recruitment and expression concomitant with the loss of H3K4 methylation. Although Mll1 is only required for the methylation of a subset of Hox genes, menin, a component of the Mll1 and Mll2 complexes, is required for the overwhelming majority of H3K4 methylation at Hox loci. However, the loss of MLL3/MLL4 and/or the Set1 complexes has little to no effect on the H3K4 methylation of Hox loci or their expression levels in these MEFs. Together these data provide insight into the redundancy and specialization of COMPASS-like complexes in mammals and provide evidence for a possible role for Mll1-mediated H3K4 methylation in the regulation of transcriptional initiation.
The mixed-lineage leukemia gene (MLL1) is involved in numerous translocations found in several human acute leukemias (2, 43, 53). Chromosomal translocations in the MLL1 gene result in hematological malignancies, including acute myeloid and lymphoid leukemia. Although these cytogenetic abnormalities were discovered over 25 years ago, little is known about the biochemical functions of MLL1, its protein complexes, its translocation partners, and particularly, why these translocations result in leukemia. MLL1 is one of at least six genes encoding histone methyltransferases in mammals that methylate histone H3 on lysine 4 (H3K4), a posttranslational mark primarily associated with promoters of active genes (49). Much of our knowledge of the implementation of this mark comes from studies of Set1/COMPASS in the yeast Saccharomyces cerevisiae. The first H3K4 methylase complex to be identified, COMPASS, was purified from yeast and contains Set1 and seven other polypeptides, named Cps60 to Cps15 according to their size in yeast (30). COMPASS is capable of mono-, di-, and trimethylating H3K4 (20, 42, 48, 56). Subsequently, it was determined that six mammalian Set1 homologs, MLL1 to MLL4 (KMT2A to KMT2D) and human Set1A and human Set1B (KMT2F and KMT2G), are found in COMPASS-like complexes, all capable of methylating H3K4 (5, 15, 16, 23, 38, 49, 59).
We have recently shown that human Set1A/Set1B function most similarly to yeast COMPASS and mediate the bulk of the H3K4 trimethylation in mammalian cell extracts (57). In contrast, the members of the MLL family of proteins do not appear to individually mediate bulk changes in H3K4 methylation (9, 31, 57). A number of approaches have been used to investigate MLL1 and its target genes (3, 11, 31, 32, 44, 45). Collectively, however, there is no clear picture of whether members of the MLL family work globally on a common set of active genes (10-12) and/or more specifically on distinct subsets of targets (9, 32) to exert their regulatory activities. We have addressed this important issue for Mll1, the murine ortholog of human MLL1, by comparing gene expression profiles and genome-wide patterns of H3K4 trimethylation and dimethylation in Mll1+/+ and Mll1−/− mouse embryonic fibroblasts (MEFs) to identify the set of genes that require Mll1-dependent methyltransferase activity. Our analyses have revealed that only a few hundred genes in MEF cells require Mll1 to maintain the appropriate levels of H3K4 methylation and gene expression. Although the Hox genes represent a significant proportion of the target genes identified, only a subset of Hox genes is regulated by Mll1, while the Mll1/Mll2-interacting partner menin is required for essentially all H3K4 trimethylation across the Hox loci. In contrast, MLL3, the MLL3/MLL4 complex-interacting partner Pax transactivation domain-interacting protein (PTIP), and the Set1 complex have modest effects on H3K4 methylation or gene expression at the Hox loci. This suggests that each member of the Mll1-to-Mll4 family has distinct roles, with Mll1 and Mll2 being major regulators of Hox genes. Our work also reveals that many other regulators of transcription in development are Mll1-dependent target genes, which can further our understanding of the mechanistic role of MLL1 in development and in MLL1-mediated leukemogenesis. Furthermore, unlike H3K4 methylation by COMPASS, which correlates with the initiation of transcription and is subsequent to Pol II initiation, mammalian MLL1 functional targets require H3K4 methylation for recruitment of the basal transcription machinery and transcriptional initiation. This could be more important for specific classes of genes, including those lacking TATA or other core promoter elements that facilitate the recruitment of the basal transcription machinery (17).
(This work was done to fulfill, in part, requirements for C.L.s Ph.D. thesis research as a student registed with the Open University.)
MATERIALS AND METHODS
Antibodies, cell lines, and siRNAs.
The antibodies to dimethylated histone H3K4 (H3K4me2) (ab7766), trimethylated histone H3K4 (H3K4me3) (ab8580), histone H3 (ab1791), TATA-binding protein (TBP) (ab51841), and RNA Pol II C-terminal domain (ab5408; used for ChIP-chip experiment whose results are shown in Fig. 4) were purchased from Abcam. The rabbit anti-human RNA polymerase Rpb1 antibody (used in the experiment whose results are shown in Fig. 2) was generated by immunization with the synthetic peptide ERALRRTLQEDLVKDVLSNGC conjugated to keyhole lympet hemocyanin. All small interfering RNAs (siRNAs) were SMARTpool siRNAs from Dharmacon, Inc., and RNA interference analysis was done as previously described (57).
FIG. 4.
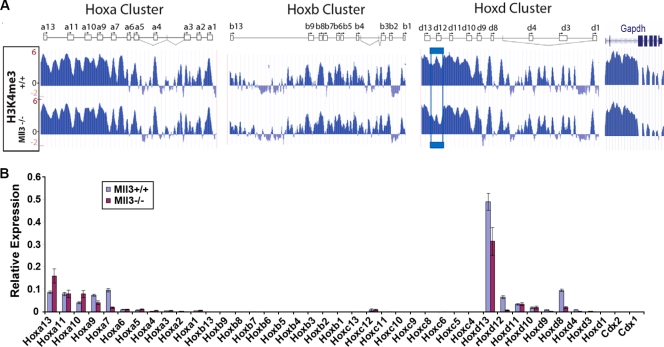
H3K4 methylation is broadly lost at some coding and intergenic regions of Hox genes in the absence of Mll1. H3K4me3, H3K4me2, RNA Pol II, and total H3 profiles were determined across the Hoxa, Hoxb, and Hoxd loci by using a custom Agilent tiling array in the presence and absence of Mll1. Blue boxes indicate regions showing large reductions in both H3K4me3 and Pol II occupancy in the absence of Mll1. Green boxes indicate regions with unchanged H3K4 methylation and unchanged RNA Pol II levels. The orange box indicates a region in the Hoxd locus which shows little change in the H3K4me3 level but notable increases in H3K4me2 and Pol II occupancy in the Mll1−/− cells. Red boxes indicate three regions of interest that correspond to the location of putative alternative promoters and enhancers regulated by Mll1. Also shown is the Gapdh gene, which is a control region on the Hox tiling array. Schematics for the Hox clusters are described in the Fig. 3A legend.
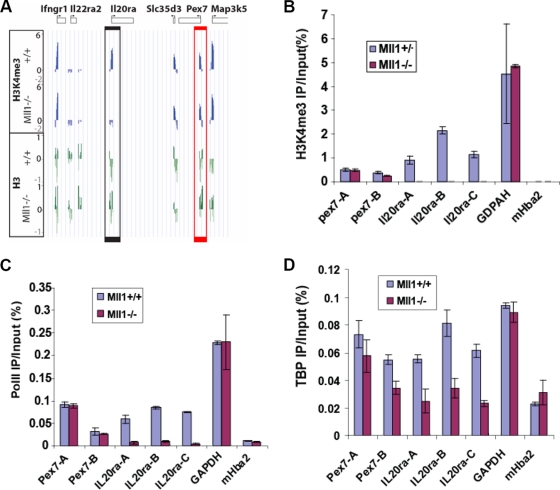
FIG. 2.
Mll1 is required for the recruitment of the basal transcription machinery to chromatin. (A) The H3K4me3 level in the absence of Mll1 as determined by ChIP-chip analysis is dramatically decreased at the Il20ra promoter region (black box) but not at the neighboring Pex7 gene (red box). Enrichment profiles for H3K4me3 and total H3 expressed as log2 ChIP/input. (B) ChIP-qPCR with H3K4me3 antibodies was used to validate the results shown in panel A, using three primer sets spanning the transcription start site. (C and D) The results of ChIP-qPCR with Pol II antibodies (C) or TBP antibodies (D) show that RNA polymerase and TBP recruitment to Il20ra are significantly decreased in the absence of Mll1. Pol II and TBP recruitment on the Pex7 and Gapdh genes, however, did not change in the absence of Mll1. The murine Hba2 gene (mHba2), which is not transcribed in these cells, is used as a negative control. Error bars show standard deviations.
The Mll1 and Men1 wild-type and knockout MEF cell lines are gifts from Jay L. Hess (University of Michigan Medical School) and were described previously (13, 15). For the gene expression microarray analyses, RNA was extracted from Mll1+/+ and Mll1−/− MEFs. Ten micrograms of RNA was labeled and hybridized to an Agilent microarray as described below. For the chromatin immunoprecipitation and hybridization microarray (ChIP-chip) analysis, approximately 3 × 108 to 5 × 108 cells were harvested and cross-linked with 1% formaldehyde. Preparation of ChIP-chip DNA was performed according to a published protocol (25).
The PTIP+/+ and PTIP−/− cell lines were generated by infecting the simian virus 40 T antigen-immortalized PTIP conditional knockout MEF cell line PTIPflox/flox with retroviruses expressing vector or Cre as described previously (5). Primary Mll3+/+ and Mll3−/− MEFs were isolated from wild-type or Mll3−/− littermate embryos at embryonic day 13.5 and immortalized by following the 3T3 protocol. The Mll3 knockout mice were derived from embryonic stem cell line XM083 obtained from BayGenomics. The generation and characterization of Mll3 knockout mice will be described in another manuscript.
ChIP-qPCR assay.
Cells were fixed in 1% formaldehyde and sonicated in lysis buffer using a Bioruptor (Diagenode, Inc.). Sonicated lysates equivalent to 4 × 106 cells were subjected to ChIP analysis. ChIP products were analyzed by quantitative real-time PCR (qPCR) using SYBR green and a Bio-Rad MyIQ. The comparative cycle threshold method was used to determine expression levels relative to the level of input or total histone H3, which was then averaged over three independent experiments.
qRT-PCR assay.
For quantitative reverse transcriptase PCR (qRT-PCR), RNA was isolated with RNeasy, treated with DNase I, and repurified with RNeasy. Hox expression assays were purchased from Applied Biosystems and included in a custom TaqMan array card arranged in an 8-by-48 format. Five reference controls were experimentally validated using geNorm. Actb and Tbp were found to be stably expressed in all experiments and were chosen for use in normalization. Data analysis was performed using the comparative cycle threshold method.
Data analysis.
All microarray data were analyzed by using R and the limma package. ChIP-chip experiments were performed on two different platforms, the commercially available Agilent mouse promoter arrays and the custom-designed mouse Hox tiling arrays. Two replicates were done on each platform. ChIP-chip data were median normalized within each array and aquantile normalized between arrays. Expression experiments were performed with two replicates on the Agilent mouse whole-genome platform. Expression data were loess normalized within each array and aquantile normalized between arrays (51). Dye swaps were performed for the expression data. After initial normalization and analysis, peaks were found in ChIP-chip data by using Splitter (http://zlab.bu.edu/splitter) with the following settings: Manual Cutoff, 1; Minrun, 5; and Maxgap, 200. Track files were created and viewed in the UCSC genome browser (18). Venn diagrams were created using Vennmaster (19) and VENNY (http://bioinfogp.cnb.csic.es/tools/venny/index.html).
Nomenclature for the MLL family.
Two numbering systems are currently in use for the four MLLs. In this paper, MLL2 (HRX2) refers to the closest paralog of MLL1, and both are putative orthologs of Drosophila melanogaster Trx. MLL4 (ALR) is most similar to MLL3, and these two are putative orthologs of the Drosophila trithorax-related protein Trr. According to the recently adopted nomenclature for histone-modifying enzymes, MLL1 (MLL, HRX) is KMT2A, MLL2 (HRX2) is KMT2B, MLL3 is KMT2C, and MLL4 (ALR) is KMT2D (1).
Microarray data accession numbers.
All ChIP-chip data have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE18258.
RESULTS
Genome-wide analysis of Mll1-regulated genes.
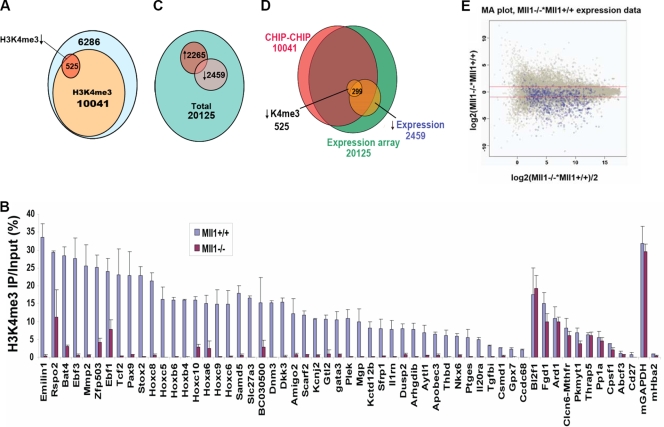
MLL1 has been shown to associate with most transcriptionally active genes, although Mll1-deficient cells show no bulk changes in H3K4 methylation (10-12). It is possible that loss of this specific methyltransferase activity can be compensated by other family members and that MLL1 activity is only required at a few specific sites. To define functional targets of the MLL family, we assessed H3K4me3 levels in Mll1+/+ and Mll1−/− MEFs (13, 60) by performing ChIP-chip analysis on microarrays containing 16,327 mouse gene promoters. Peaks of H3K4me3 were detected at 10,041 genes in the Mll1+/+ MEF cells. Reduction of H3K4me3 levels in the Mll1−/− MEFs was only observed at 525 promoters (Fig. 1A; also see Table S1 in the supplemental material), comprising roughly 5% of promoters harboring H3K4me3 peaks. This reflects a surprisingly small number of promoters affected by Mll1, considering its widespread association with active genes. We validated these results by repeating the ChIP experiment and by measuring H3K4me3 enrichments using qPCR. We chose 45 genes showing the largest decreases in H3K4me3 levels and 10 unchanged genes from the microarray analysis for validation by qPCR (Fig. 1B). The results of these “manual ChIP” analyses showed strong correspondence to changes observed in the ChIP-chip data, verifying the robustness of the microarray results.
FIG. 1.
Identification of genes requiring Mll1 for H3K4 methylation and gene expression. (A) Numbers of promoters with changes in H3K4me3 levels as assessed by ChIP-chip with H3K4me3 antibodies in Mll1+/+ and Mll1−/− MEFs. Ten thousand forty-one out of 16,852 gene promoters had detectable peaks of H3K4me3 in wild-type MEFs. Only 525 (5%) of these genes showed significant loss of H3K4me3 in the absence of Mll1. (B) ChIP-qPCR analysis confirmed the H3K4me3 changes on 45 of the genes found by ChIP-chip analysis to be most changed for H3K4me3, as well as the ChIP-chip results for 10 unchanged genes. The Hemoglobin gene, inactive in fibroblasts (Hba2), was used as a negative control for H3K4me3, and the housekeeping gene Gapdh was used as a positive control for the presence of H3K4me3. Error bars show standard deviations. (C) Gene expression analysis of 20,125 genes was performed with RNA isolated from Mll1+/+ and Mll1−/− fibroblasts. The expression of 2,265 genes is upregulated and that of 2,459 genes is downregulated in the absence of Mll1. (D) Overlap of H3K4me3 and gene expression. About 299 of the 525 genes with loss of H3K4me3 also have decreased expression in the absence of Mll1. (E) Microarray (MA) plot of the gene expression data, with the 525 genes with significantly reduced H3K4 methylation highlighted in blue. The y axis represents the intensity ratio of mutant over wild type, and the x axis represents the average intensity of each spot. The red lines indicate a twofold up- or downregulation of expression. Most of the genes dependent on Mll1 for H3K4me3 show reduced expression.
To determine the effect of loss of Mll1-dependent H3K4me3, we performed gene transcriptional profiling in Mll1+/+ and Mll1−/− MEFs. Out of 20,125 genes on the array, 2,265 genes were upregulated and 2,459 genes were downregulated upon loss of Mll1 (Fig. 1C; also see Table S2 in the supplemental material). Comparing these results with the ChIP-chip data, of the 525 genes with reduced levels of H3K4me3 in the absence of Mll1, 299 (60%) also displayed a reduction in the levels of gene expression (Fig. 1D and E; also see Tables S3 and S4 in the supplemental material). Therefore, it appears that in these MEFs, approximately 3% of genes require Mll1's methyltransferase activity for gene expression.
Role of H3K4me3 in gene regulation.
In the yeast S. cerevisiae, loss of the sole H3K4 methyltransferase (COMPASS) has little effect on the recruitment of RNA polymerase II (Pol II) and the expression of the majority of genes (21, 30, 37). COMPASS is recruited to active genes by polymerase-associated factor (the Paf1 complex), and H3K4me3 only marks actively transcribed genes (21, 55). This relationship was also observed in a reconstituted transcription system containing the mammalian homologs of these factors (39). However, given the reduced expression of genes in the absence of Mll1-dependent methylation, we investigated the recruitment of Pol II to Mll1-regulated genes by performing Pol II ChIP on two neighboring genes, Il20ra (whose H3K4 trimethylation is dependent on Mll1) and Pex7 (whose H3K4 trimethylation remains unchanged after loss of Mll1) (Fig. 2A and B). Primers spanning different regions of each gene were used to measure H3K4me3 levels and Pol II occupancy. While Pex7 had unchanged H3K4me3 and Pol II levels, both H3K4me3 and Pol II were markedly reduced at the Il20ra gene in the Mll1−/− cells (Fig. 2B and C). Based on the model by Vermeulen and colleagues that H3K4me3 recruits TFIID through the TAF3 plant homeodomain (PHD) finger (54), we performed TBP ChIP on the Pex7 and Il20ra genes and found that TBP recruitment to Il20ra is significantly reduced in the Mll1−/− cells (Fig. 2D). The requirement of Mll1 for recruitment of the basal transcriptional machinery and Pol II reveals a difference between the role of the mammalian Mll1 methyltransferase and that of the related Set1/COMPASS in yeast, where the primary function of H3K4me3 appears to occur after Pol II recruitment.
Mll1 targets transcriptional regulators.
Analysis of the 299 genes that require Mll1 for both H3K4 trimethylation and transcription reveals an abundance of transcriptional regulators (∼55), cellular signaling pathways (∼30), and signal transduction components (∼30) that play key roles in regulating developmental processes (see Table S4 in the supplemental material). For example, downregulated expression of genes encoding multiple extracellular (Dkk3, Sfrp1, Sfrp2, and Frzb) modulators and one intracellular (Frat2) modulator of the Wnt pathway are revealed. While the gene expression of representatives of many of the major classes of transcription factors is affected upon loss of Mll1 (e.g., Pax9, Pitx2, MafB, Egr3, Foxc1, Stat5a, Sox5, and Zic), members of the largest class (24 genes), including 11 members of the clustered Hox genes, encode homeobox-containing proteins. These observations are interesting in light of the established role of Mll1 and Hox genes in regulating hematopoiesis and the leukemogenic process (6, 22, 35, 36). Furthermore, the Wnt pathway plays an important role in specifying mesoderm differentiation toward a hematopoietic fate, in part through regulation of Hox genes (26, 28), suggesting that Mll1 might also modulate signaling pathways, such as Wnt, that have an impact upon Hox gene regulation.
Mll1 specialization at the Hox loci.
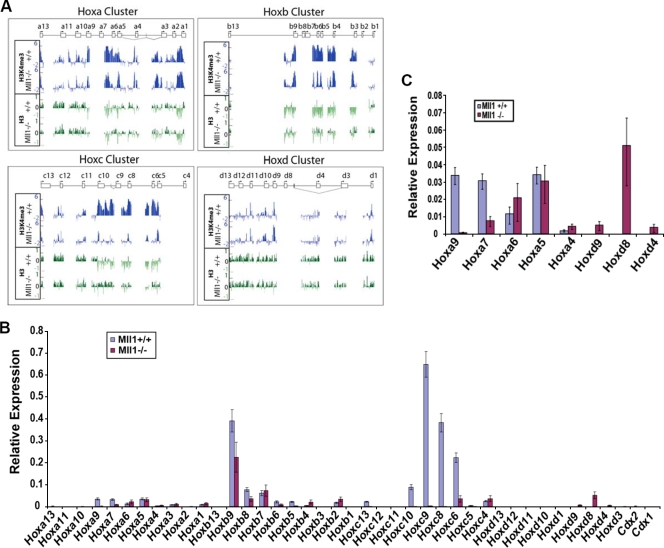
Mll1, like its Drosophila homolog Trithorax, is well-known as an important regulator of Hox gene expression. We therefore focused our attention on the changes in H3K4me3 levels at the promoters of the Hox genes by utilizing ChIP-chip on a promoter array. Comparison of the H3K4 trimethylation patterns of Mll1+/+ and Mll1−/− MEF cells revealed that Mll1 does not exert a common function in all four Hox clusters or for all genes within a cluster. Instead it appears to exert a more selective role in targeting the Hox clusters and genes. For example, the Hoxc complex is strongly affected by loss of Mll1 in these cells, with nearly all of the promoters showing markedly reduced H3K4me3 levels in the Mll1−/− cells (Fig. 3A). In sharp contrast, promoters in the Hoxd complex are largely insensitive to the loss of Mll1. The Hoxa and Hoxb clusters display a mixed pattern where a few of the promoters (e.g., Hoxa6 and Hoxb4) display Mll1-dependent H3K4 trimethylation (Fig. 3A). We also performed qRT-PCR analysis of the 39 murine Hox genes in Mll1+/+ and Mll1−/− MEFs to assess the role of Mll1 in Hox gene expression (Fig. 3B and C). Consistent with the analyses from the promoter array and transcriptional profiling, Mll1 has a major role at Hoxc in these cells. Several genes from the Hoxa and Hoxb clusters show reduced levels of expression in the absence of Mll1, although the expression levels of the Hoxa, Hoxb, and Hoxd genes are relatively low in these fibroblasts.
FIG. 3.
Mll1 is required for the H3K4 methylation and transcription of a subset of Hox genes. (A) Distribution of H3K4me3 and total H3 across 8-kb regions of each Hox gene promoter (5.5 kb before the transcription start site and 2.5 kb after the transcription start site) on the Agilent promoter array as determined by ChIP. Each individual Hox gene is represented as a box with an arrow indicating the major transcription start site and the direction of transcription. Vertical lines represent small alternative exons, and diagonal lines represent alternative splicing events. Reductions in H3K4me3 can be seen at several locations across the Hoxa, Hoxb, and Hoxc clusters. In contrast, Hoxd genes show little loss of H3K4me3 in Mll1−/− cells. (B) qRT-PCR analysis of the expression of Hox genes in Mll1+/+ and Mll1−/− fibroblasts normalized to the average value of reference genes Actb and Tbp. (C) Rescaling of the RT-PCR data presented in panel B. Shown are selected Hoxa and Hoxd genes, both of which have relatively low levels of expression in these fibroblasts. Some Hoxd genes that are not expressed in wild-type MEFs are expressed in the Mll1−/− MEFs. Error bars show standard deviations.
The presence of Mll1 activity at Hox promoters led us to more systematically investigate whether Mll1 affected other regions throughout the Hox clusters. To this end, we utilized a high-density tiling microarray, designed in-house, that covers the entire region of the murine Hoxa, Hoxb, and Hoxd complexes. These arrays were probed with DNA obtained with H3K4me3, H3K4me2, RNA Pol II, and total H3 antibodies from Mll1+/+ and Mll1−/− cells (Fig. 4 and 5). The results of our analyses reveal that the regions of H3K4 methylation dependent on Mll1 are quite extensive, including not just promoters but also coding and some intergenic regions (Fig. 4). At several Hox genes, particularly the Hoxa6-Hoxa9 and Hoxb5-Hoxb8 genes, the levels of both H3K4me3 and H3K4me2 decrease (Fig. 4), consistent with the ability of MLL1 to mediate di- and trimethylation of H3K4 in vitro (57). However, some regions showing loss of H3K4me3 in the absence of Mll1 still have residual peaks of this modification (Fig. 5). Sometimes, the region in between the H3K4me3 peaks and the H3K4me2 pattern “inverts” in the mutant, with valleys of H3K4me2 in the wild-type showing peaks of this modification, while peaks of H3K4me2 in the wild type decrease in the mutant. Together these results suggest some redundancy or interplay among different H3K4 methyltransferases at the Hox loci.
FIG. 5.
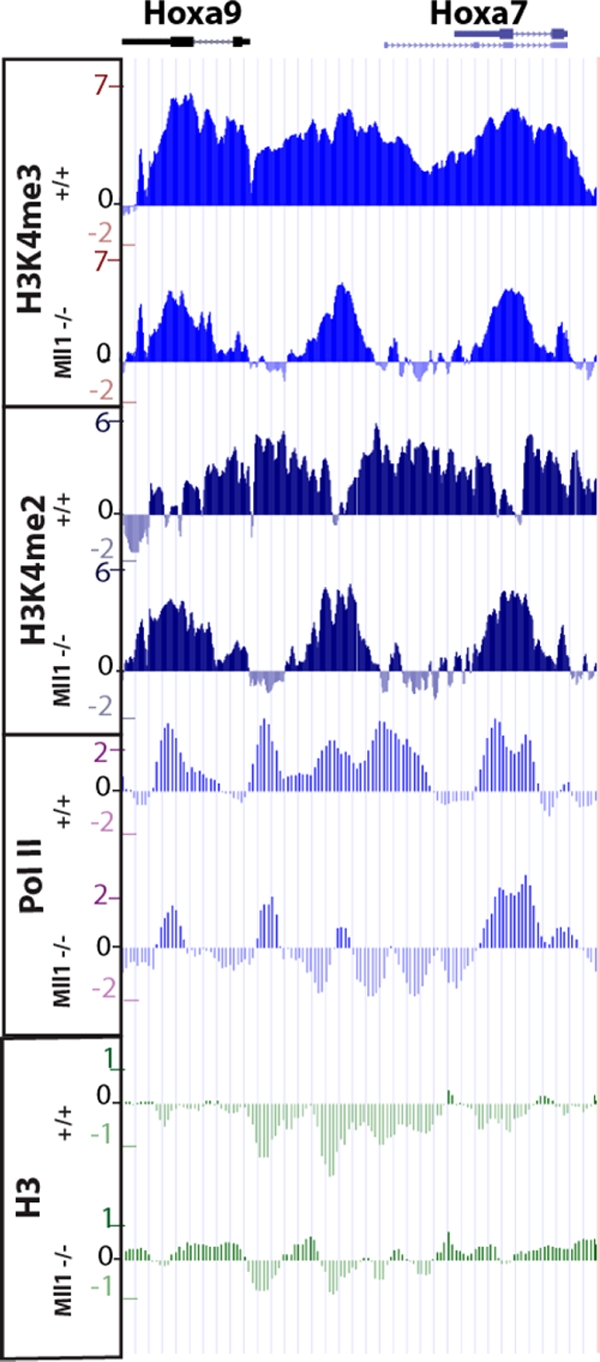
Enlargement of the results for the Hoxa9-Hoxa7 region shown in Fig. 4. Some regions showing loss of H3K4me3 in the absence of Mll1 still have residual peaks of this modification. In the region in between the H3K4me3 peaks, the H3K4me2 pattern “inverts” in the mutant, with valleys of H3K4me2 in the wild-type now showing peaks of this modification. Accordingly, peaks of H3K4me2 in the wild type decrease in the mutant. Together, these results suggest some redundancy or interplay among different H3K4 methyltransferases at the Hox loci.
Intriguingly, some genes that exhibit no significant or detectable decrease of H3K4me3 levels in the Mll1−/− cells show increased levels of H3K4me2. This is clearly observed for nearly all members of the Hoxd complex (for example, see the results for the Hoxd9 cluster in Fig. 4). These changes in dimethylation appear significant, as Hox genes and other loci showing selectively increased H3K4me2 frequently displayed increased expression (Fig. 4; also see Table S2 in the supplemental material). For example, expression of Hoxd9, Hoxd8, and Hoxd4 is not detectable in Mll1+/+ MEFs, but in the absence of Mll1, expression is observed for these genes (Fig. 3C).
Several regions of the Hox complexes with large reductions in H3K4 methylation in Mll1−/− MEFs also have significant drops in RNA Pol II levels (Fig. 4). This is in agreement with the hypothesis that in mammalian cells, MLL1-mediated H3K4 methylation helps to recruit RNA Pol II, in contrast to the established roles for COMPASS in yeast. Accordingly, those regions not dependent on Mll1 for H3K4 trimethylation have normal levels of Pol II (Fig. 4). Furthermore, some regions with increased H3K4me2 (e.g., Hoxd9) also have increased levels of Pol II (Fig. 4), thus showing a correlation between changes in H3K4 methylation and Pol II recruitment in mammals.
ChIP with an antibody that recognizes all forms of histone H3 shows no large-scale differences between Mll1+/+ and Mll1−/− MEFs, although regions showing loss of both H3K4me3 and RNA Pol II generally have low levels of H3 in wild-type cells and a modest increase in H3 levels in the Mll1−/− cells (Fig. 4). This is consistent with lower nucleosome density at actively transcribed genes that is due to nucleosome eviction (34). For example, in the Hoxd locus, where there are generally lower levels of transcription, we find higher levels of H3 and lower levels of RNA Pol II than at Hoxa and Hoxb.
Identifying new features of Hox complexes.
The organization and regulation of clustered Hox genes in vertebrates is a diverse and complex process, with alternative promoters and splicing, tissue-specific enhancers, global/long-range regulatory inputs, and the presence of noncoding RNA transcripts. While H3K4me3 is a common landmark for promoter regions of active genes and might serve to identify novel promoters within the Hox complexes, it might reveal additional features which participate in gene regulation. Therefore, we carefully examined results from the high-density tiling arrays for differences in H3K4me3 levels across the entirety of the complexes. In the intergenic region between Hoxb3 and Hoxb4, there is a region that is highly methylated at H3K4 in Mll1+/+ cells that has greatly reduced H3K4 methylation in Mll1−/− cells (Fig. 4). This region precisely corresponds to a previously identified alternative promoter for Hoxb3 in the nervous system (46), indicating that this promoter is also recognized in MEF cells. A similar change in pattern is seen at the corresponding location in the Hoxa locus where a peak of H3K4me3 in the intergenic region between Hoxa4 and Hoxa3 is reduced in Mll1−/− cells (Fig. 4). This suggests that this region corresponds to an alternative promoter for Hoxa3, similar to the alternative promoter at Hoxb3, which has been conserved following the duplication and divergence of the vertebrate clusters.
Another region of interest on the Hoxb tiling array is located between Hoxb9 and Hoxb8 (Fig. 4). While there is no evidence for alternative promoters in this location, this region is known to contain an enhancer for Hoxb8 that directs expression in mesodermal tissues (4). Genome-wide data suggest that monomethylation of H3K4, not trimethylation, typically marks enhancers (14), but this may vary for specific genes. The selective changes in H3K4 methylation that we observed upon loss of Mll1 not only point out the potentially unique and complex organization of the Hox loci but illustrate that novel noncoding functional elements within the Hox clusters might be discovered by analyzing histone modification changes in different genetic and developmental states.
The role of other MLL family members at Hox loci.
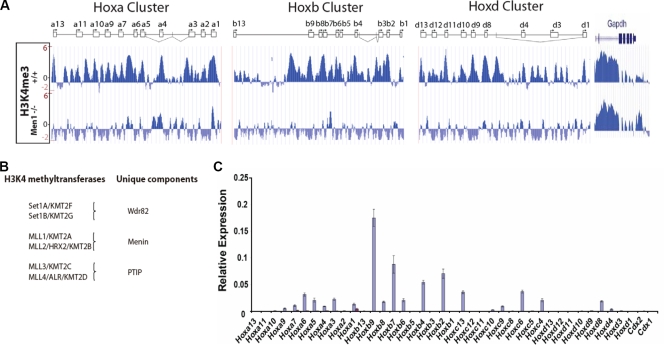
Our studies have uncovered a specialized or selective requirement for Mll1 in Hox regulation. However, Mll1 might play a larger role in regulating these genes which is masked by functional compensation from the other five H3K4 methylases. Based on common and distinct components of the protein complexes they participate in, the MLL family can be divided into two groups, Mll1/Mll2 and Mll3/Mll4 (Fig. 6B). Mll1 and Mll2 share the highest similarity to the Drosophila Hox gene regulator Trithorax and can form complexes with the tumor suppressor menin. Menin has previously been shown to be important for the recruitment of MLL1 to target genes (33). We performed H3K4me3 ChIP-chip analyses on a Hox tiling array of genes from wild-type and Men1−/− MEFs. In contrast to the loss of Mll1, the absence of menin results in a profound reduction in H3K4 methylation across the entirety of the Hoxa, Hoxb, and Hoxd clusters (Fig. 6A). The results of RT-PCR analyses also show that menin is required for the expression of all the detectably expressed Hox genes in these cells (Fig. 6C). Together, these results reveal that in MEFs, Mll1 and Mll2 function as the major H3K4 trimethylases at Hox loci and are key regulators of Hox gene expression.
FIG. 6.
Menin, a common component of Mll1 and Mll2 complexes, is required for the expression of nearly all Hox genes and for H3K4 trimethylation at Hox loci. (A) H3K4me3 ChIP profiles from Men1+/+ and Men1−/− fibroblasts show that the vast majority of H3K4 methylation at Hox loci depends on menin. Schematics for the Hox clusters are described in the Fig. 3A legend. (B) Schematic of the association of subunits unique to Mll1/Mll2, Mll3/Mll4, and Set1A/Set1B complexes. Menin is found in Mll1/Mll2 but not other H3K4 methyltransferase complexes, PTIP is unique to Mll3/Mll4 but not other H3K4 methyltransferase complexes, and WDR82 is unique to the Set1A/Set1B but not the Mll1-to-Mll4 complexes. Alternate names for MLL2 and MLL4 are included to avoid confusion over alternative naming systems. (C) qRT-PCR analysis of Hox gene expression shows near total loss of Hox gene expression in Men1−/− fibroblasts. Error bars show standard deviations.
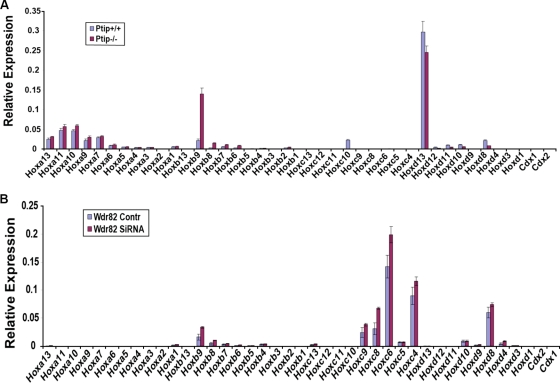
The other two MLL family members, MLL3 and MLL4, were recently found to form complexes with PTIP, the nuclear receptor coactivator Ncoa6, and the H3K27 demethylase UTX (5, 16, 38). To explore the roles of this subfamily in Hox regulation, we performed H3K4me3 ChIP-chip on the Hox tiling array using Mll3−/− and PTIP−/− fibroblasts. The loss of Mll3 exerted few or no changes in H3K4me3 levels across the Hox clusters (Fig. 7A). RT-PCR analysis of Hox genes from Mll3+/+ and Mll3−/− fibroblasts showed modest and variable changes in Hox gene expression in these cells (Fig. 7B). We also performed RT-PCR analysis of Hox gene expression in PTIP+/+ and PTIP−/− fibroblasts and saw few changes in the expression of Hox genes with the loss of PTIP, a unique component of the MLL3/MLL4 complexes shown to be required for H3K4 methylation at a Pax-dependent reporter gene (Fig. 8A) (38). Furthermore, RNA interference-mediated knockdown of a unique component of the Set1A/Set1B complex, the Wdr82 protein, resulted in few or no changes in Hox expression (Fig. 8B). We did not analyze Ash1, the remaining enzyme reported to methylate H3K4 in mammals; Ash1 has been shown previously to be present at Hox genes but did not appear to affect H3K4 methylation levels (10). Together, the results of these analyses suggest that the Set1A/Set1B and Mll3/Mll4 complexes have relatively minor roles in controlling Hox expression in MEFs, whereas they provide support for the menin-containing Mll1/Mll2 complexes in mediating the majority of H3K4 trimethylation at Hox loci and serving as key regulators of Hox transcription.
FIG. 7.
Mll3 is not a major H3K4 histone methyltransferase at Hox loci in fibroblasts. (A) H3K4me3 ChIP profiles from Mll3+/+ and Mll3−/− fibroblasts show relatively minor effects on the Hox gene H3K4me3 pattern by Mll3. A blue box outlines the results for the Hoxd12 gene, where a significant reduction in H3K4me3 can be observed in Mll3−/− fibroblasts. Schematics for the Hox clusters are described in the Fig. 3A legend. (B) qRT-PCR analysis of Hox gene expression reveals minor effects of Mll3. Hoxd12 shows reductions in both H3K4me3 and gene expression in Mll3−/− fibroblasts. Error bars show standard deviations.
FIG. 8.
qRT-PCR analysis of Hox gene expression in the presence and absence of H3K4 methylation regulators. (A) Results for RNA from PTIP+/+ and PTIP−/− fibroblasts. (B) Results for RNA from wild-type fibroblasts treated with an siRNA to Wdr82 or a nontargeting control siRNA. Error bars show standard deviations.
DISCUSSION
We have shown that a small but important subset of genes requires Mll1 for H3K4 trimethylation and gene expression. Although Mll1 is known as a major regulator of Hox gene transcription, we find a highly specialized role for Mll1 within the Hox clusters, affecting a subset of Hox genes. In addition to Hox genes, Mll1 is also a key regulator of other master regulators of transcription. Indeed, while the manuscript was under review, one of the 299 genes that we classify as functional targets of Mll1 was reported as an Mll1-regulated gene required for neurogenesis (27). Investigation of this set of Mll1-regulated genes could also potentially reveal additional effectors of leukemogenesis. The correlation between Mll1-dependent H3K4 methylation and Pol II recruitment reveals a key distinction between Mll1 and the yeast Set1/COMPASS.
Differences between MLL and COMPASS methyltransferases.
Most of the work on the implementation of H3K4 methylation comes from the yeast S. cerevisiae, where a single methyltransferase complex, Set1/COMPASS, is recruited to genes by the ubiquitination of histone H2B, requiring prior recruitment of Pol II and the PAF1 complex (21, 48, 55). Work in mammalian systems has primarily focused on MLL1, which is targeted by reciprocal translocations with heterogeneous partner genes fused in-frame to MLL1, encoding fusion proteins and associated with both acute lymphoblastic and myeloblastic leukemia (43, 53). Recent work by our lab indicates that although COMPASS and MLL1 form very similar complexes, they could have different functions (24, 57). COMPASS contains a unique subunit, Cps35, which mediates the recruitment of COMPASS to chromatin by ubiquitinated histone H2B (24). A human Set1/COMPASS complex containing a Cps35 homolog (WDR82) is also regulated by H2B ubiquitination and mediates the vast majority of H3K4 trimethylation in mammals (57). In contrast, Mll1−/− fibroblasts show no observable loss of H3K4 methylation in assays of bulk histones (31, 57). This may be explained by our detailed biochemical studies, which indicate that the MLL1 complex lacks a Cps35-like subunit (57), suggesting that it is recruited to chromatin by a different mechanism than Set1/COMPASS. In further support of these differences, the Set1/COMPASS in yeast is not important for Pol II occupancy on promoters, while the loss of Mll1 in MEFs results in reductions in Pol II occupancy on Mll1's functional targets.
The relationship between H3K4 methylation and gene activation could be explained by Vermeulen and colleagues' observation that the PHD domain of TAF3 is required for TFIID and, thereby, Pol II recruitment and transcriptional initiation (54). Similarly, several nucleosome remodeling complexes, such as NURF and chromodomain helicases, harbor H3K4me3 binding domains which could indirectly aid in the recruitment of the basal transcriptional machinery (41, 50, 58). ING proteins, bearing PHD fingers that have been shown to bind methylated H3K4, are found in many MYST family histone acetyltransferase complexes, so there appear to be multiple pathways for activating gene expression through H3K4 methylation (7, 40, 47). A requirement for MLL1-dependent H3K4 methylation could be restricted to a subset of genes lacking canonical core promoter elements (17). Interestingly, Hox genes are mostly TATA-less, suggesting that TFIID recruitment to these genes could be enhanced by prior methylation of H3K4 by MLL family proteins.
The MLL family and Hox gene regulation.
Mll1 is well known as an important regulator of Hox gene expression during development, and misexpression of Hox genes can lead to leukemia (6, 60). Previous studies have individually found that some Hox genes are affected and some other Hox genes are not affected by the loss of Mll1 or Mll2 (8, 9); however, a Hox-wide analysis of Mll1 and Mll2 regulation has not previously been fully addressed. Here, we have shown that Mll1 is responsible for diverse and selective changes in the patterns of H3K4 methylation in Hox complexes and that this affects the levels of gene expression of a fraction of the Hox genes. Our results open the possibility that specific subsets of Hox genes are differentially regulated by distinct members of the MLL family. Our analyses of Men1+/+ and Men1−/− MEFs implicate Mll2 as the other major agent of H3K4 trimethylation at these genes. Furthermore, our results with Mll3−/−, PTIP−/−, and Wdr82 knockdown cells further indicate that other MLL family members contribute to different subsets of targets than Mll1/2.
In these studies, we have taken advantage of the availability of fibroblast cell lines (MEFs) derived from knockout mouse embryos, some of which were derived by other laboratories. Isolation of MEFs from embryos of different ages and using different methods of cellular transformation will result in different Hox expression patterns in the resulting MEF lines. Although we cannot directly compare the regulation of a particular Hox gene in Mll1 MEFs with the same Hox gene in Men1 or Mll3 MEFs, we can compare, for example, Mll1+/+ to Mll1−/− MEFs because paired MEF cells are always derived from the same litter under identical conditions. Furthermore, by performing Hox-wide analysis with MEFs, we are able to obtain an overall picture of Hox gene regulation by the MLL family of proteins.
The four Hox clusters in vertebrates arose from an ancestral Hox cluster by successive duplication and divergence of the ancestral cluster (29). A large body of work has indicated that there is both functional redundancy and specialization among Hox paralogs in different clusters. Hoxa and Hoxb are generally more similar to each other and differ from Hoxd and Hoxc with respect to the numbers of genes, patterns of expression, and positions of conserved regulatory elements within the complexes. Although there are also four MLL family proteins in mammals, we do not see a major role of Mll3/Mll4 complexes in Hox gene regulation in fibroblasts. Mll3 has been shown to activate Hoxc8 in an Ap2-dependent manner (52). We also see a requirement for Mll3 in Hoxd12 H3K4 trimethylation and gene expression in our cells (Fig. 7). Therefore, the potential for combinations of functional compensation and specialization between MLL-related proteins and the different Hox loci is enormous.
Additional functions of MLL1-mediated methylation.
H3K4me3 has been strongly correlated with promoters of genes. In our analysis of the Hox loci, we observe a broad distribution of H3K4me3 across the entire complex, a result that has been previously observed (3). In the Mll1−/− cells, we observed loss of H3K4me3 from a subset of Hox promoters, coding regions, and intergenic regions, suggesting that Mll1 has multiple inputs into the regulation of these genes. Some of the intergenic regions methylated by Mll1 are notable because they correspond to known or suspected alternative promoters and tissue-specific enhancers. Another intriguing aspect revealed by our study is whether Mll1-mediated methylation has distinct functions at enhancers versus promoters versus intergenic regions. One potential role for intergenic methylation is in the transcription of noncoding RNAs. In humans, a noncoding RNA transcribed in Hoxc helps to repress genes in Hoxd. The study of functional noncoding RNAs is a rapidly growing field, in large part due to the growth of resources for genome-wide analysis.
In light of the importance of Hox genes in development and disease processes, our current level of knowledge of regulatory components and their organization in the clusters is limited to a small subset of elements identified by focused functional assays. The correlation between H3K4 methylation patterns and specific gene activity dependent upon Mll1 illustrates the potential of systematically repeating our analyses with all of the MLL family members to uncover novel features of the Hox clusters in different cellular and developmental states. This could yield a wealth of novel regulatory elements for Hox genes that, in combination with the results of functional studies, have the potential to rapidly expand our mechanistic understanding of Hox gene regulation in development and disease.
MLL1 target genes and leukemia.
MLL1's role in regulating Hox gene expression during development and the role of the translocation-associated MLL1 fusion proteins in leukemia may be tightly linked. The MLL1 portion of the fusion protein may lead to altered recruitment of complexes to Hox genes, resulting in altered gene expression as a key part of the multistep leukemogenic pathway (6, 12). Some of the Mll1-target genes identified by our analyses may be candidates for involvement in this process. For example, we found decreases in expression and H3K4 trimethylation at the loci of genes encoding extracellular (Dkk3, Sfrp1, Sfrp2, and Frzb) and intracellular (Frat2) proteins known to control the Wnt signaling pathway (Table 1; also see Table S4 in the supplemental material). Wnt signaling itself plays an important role in regulating Hox genes and other targets during hematopoietic differentiation (26, 28). Hence, in addition to the direct interactions we have described, Mll1 may have indirect inputs into modulating Hox expression in leukemia via the Wnt pathway. Since a large portion of the Mll1 target genes are known to be important transcriptional regulators of developmental processes, it would be valuable to analyze these regulators and their downstream targets in other contexts to determine whether they are also part of the gene regulatory network linked to leukemogenesis.
TABLE 1.
Top 20 genes with the K4me3 decreasing and transcriptional downregulation in the absence of MII1
| Gene | Product or other description | Expression level |
ChIP-chip peak score (log2) | ChIP-qPCR fold change (log2) | ||
|---|---|---|---|---|---|---|
| MII1−/−/MII1+/+ (log2) | P value | Adjusted P value | ||||
| Thbd | Thrombomodulin | −11.10 | 5.70E−05 | 1.39E−03 | −4.75 | −6.18 |
| Kctd12b | Potassium channel tetramerization domain containing 12b | −8.08 | 5.86E−03 | 2.26E−02 | −3.86 | −5.46 |
| Hoxc9 | Homeobox C9 | −6.53 | 7.14E−08 | 3.67E−04 | −4.25 | −6.04 |
| Rspo2 | R-Spondin 2 homolog (Xenopus laevis) | −6.52 | 4.64E−07 | 3.67E−04 | −4.10 | −1.37 |
| Mgp | Matrix Gla protein | −6.50 | 5.69E−06 | 5.00E−04 | −4.09 | −6.25 |
| Bnc1 | Basonuclin 1 | −8.28 | 1.91E−04 | 2.61E−03 | −2.10 | |
| Plek | Pleckstrin | −5.74 | 1.21E−06 | 3.67E−04 | −4.41 | −6.09 |
| Tcf2 | HNF1 homeobox B | −4.59 | 7.09E−06 | 5.43E−04 | −5.28 | −6.27 |
| C4bp | Complement component 4 binding protein | −6.88 | 3.60E−07 | 3.67E−04 | −2.99 | |
| Pax9 | Paired box gene 9 | −6.42 | 1.36E−07 | 3.67E−04 | −3.43 | −5.03 |
| 9030425E11Rik | RIKEN cDNA 9030425E11 gene | −7.63 | 1.18E−07 | 3.67E−04 | −2.16 | |
| II20ra | Interleukin 20 receptor alpha | −6.57 | 1.01E−06 | 3.67E−04 | −3.20 | −4.78 |
| Abi3bp | ABI gene family, member 3 (NESH) binding protein | −7.06 | 6.97E−03 | 2.56E−02 | −2.56 | |
| Dkk3 | Dickkopf homolog 3 (Xenopus laevis) | −5.95 | 4.85E−06 | 4.81E−04 | −3.49 | −5.80 |
| Mafb | v-maf musculoaponeurotic fibrosarcoma oncogene family, protein B (avian) | −6.37 | 5.14E−06 | 4.85E−04 | −3.01 | |
| Ebf3 | Early B-cell factor 3 | −5.10 | 5.32E−02 | 1.11E−01 | −4.26 | −5.52 |
| Tgfbi | Transforming growth factor, beta induced | −5.85 | 8.74E−07 | 3.67E−04 | −3.51 | −4.27 |
| Pitx2 | Paired-like homeodomain transcription factor 2 | −6.83 | 6.69E−03 | 2.49E−02 | −2.23 | |
| Sfrp1 | Secreted frizzled-related protein 1 | −5.18 | 1.18E−03 | 7.62E−03 | −3.79 | |
| Enpp3 | Ectonucleotide pyrophosphatase/phosphodiesterase 3 | −6.62 | 8.58E−03 | 2.96E−02 | −2.30 | |
Supplementary Material
Acknowledgments
We thank Jay Hess (University of Michigan, Ann Arbor) for the Mll1−/− and Mll1+/+ cell lines, Allison Peak for technical assistance and Laura Shilatifard for editorial assistance.
This work was supported by grants from the NCI to A.S. (5R01CA089455) and by support of A.S. and R.K. by the Stowers Institute for Medical Research.
Footnotes
Published ahead of print on 24 August 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Allis, C. D., S. L. Berger, J. Cote, S. Dent, T. Jenuwien, T. Kouzarides, L. Pillus, D. Reinberg, Y. Shi, R. Shiekhattar, A. Shilatifard, J. Workman, and Y. Zhang. 2007. New nomenclature for chromatin-modifying enzymes. Cell 131:633-636. [DOI] [PubMed] [Google Scholar]
- 2.Ayton, P. M., and M. L. Cleary. 2001. Molecular mechanisms of leukemogenesis mediated by MLL fusion proteins. Oncogene 20:5695-5707. [DOI] [PubMed] [Google Scholar]
- 3.Bernstein, B. E., M. Kamal, K. Lindblad-Toh, S. Bekiranov, D. K. Bailey, D. J. Huebert, S. McMahon, E. K. Karlsson, E. J. Kulbokas III, T. R. Gingeras, S. L. Schreiber, and E. S. Lander. 2005. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120:169-181. [DOI] [PubMed] [Google Scholar]
- 4.Charité, J., W. de Graaff, D. Consten, M. J. Reijnen, J. Korving, and J. Deschamps. 1998. Transducing positional information to the Hox genes: critical interaction of cdx gene products with position-sensitive regulatory elements. Development 125:4349-4358. [DOI] [PubMed] [Google Scholar]
- 5.Cho, Y. W., T. Hong, S. Hong, H. Guo, H. Yu, D. Kim, T. Guszczynski, G. R. Dressler, T. D. Copeland, M. Kalkum, and K. Ge. 2007. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J. Biol. Chem. 282:20395-20406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daser, A., and T. H. Rabbitts. 2005. The versatile mixed lineage leukaemia gene MLL and its many associations in leukaemogenesis. Semin. Cancer Biol. 15:175-188. [DOI] [PubMed] [Google Scholar]
- 7.Doyon, Y., C. Cayrou, M. Ullah, A. J. Landry, V. Cote, W. Selleck, W. S. Lane, S. Tan, X. J. Yang, and J. Cote. 2006. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol. Cell 21:51-64. [DOI] [PubMed] [Google Scholar]
- 8.Ernst, P., M. Mabon, A. J. Davidson, L. I. Zon, and S. J. Korsmeyer. 2004. An Mll-dependent Hox program drives hematopoietic progenitor expansion. Curr. Biol. 14:2063-2069. [DOI] [PubMed] [Google Scholar]
- 9.Glaser, S., J. Schaft, S. Lubitz, K. Vintersten, F. van der Hoeven, K. R. Tufteland, R. Aasland, K. Anastassiadis, S. L. Ang, and A. F. Stewart. 2006. Multiple epigenetic maintenance factors implicated by the loss of Mll2 in mouse development. Development 133:1423-1432. [DOI] [PubMed] [Google Scholar]
- 10.Gregory, G. D., C. R. Vakoc, T. Rozovskaia, X. Zheng, S. Patel, T. Nakamura, E. Canaani, and G. A. Blobel. 2007. Mammalian ASH1L is a histone methyltransferase that occupies the transcribed region of active genes. Mol. Cell. Biol. 27:8466-8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guenther, M. G., R. G. Jenner, B. Chevalier, T. Nakamura, C. M. Croce, E. Canaani, and R. A. Young. 2005. Global and Hox-specific roles for the MLL1 methyltransferase. Proc. Natl. Acad. Sci. USA 102:8603-8608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guenther, M. G., L. N. Lawton, T. Rozovskaia, G. M. Frampton, S. S. Levine, T. L. Volkert, C. M. Croce, T. Nakamura, E. Canaani, and R. A. Young. 2008. Aberrant chromatin at genes encoding stem cell regulators in human mixed-lineage leukemia. Genes Dev. 22:3403-3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanson, R. D., J. L. Hess, B. D. Yu, P. Ernst, M. van Lohuizen, A. Berns, N. M. van der Lugt, C. S. Shashikant, F. H. Ruddle, M. Seto, and S. J. Korsmeyer. 1999. Mammalian Trithorax and polycomb-group homologues are antagonistic regulators of homeotic development. Proc. Natl. Acad. Sci. USA 96:14372-14377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heintzman, N. D., R. K. Stuart, G. Hon, Y. Fu, C. W. Ching, R. D. Hawkins, L. O. Barrera, S. Van Calcar, C. Qu, K. A. Ching, W. Wang, Z. Weng, R. D. Green, G. E. Crawford, and B. Ren. 2007. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 39:311-318. [DOI] [PubMed] [Google Scholar]
- 15.Hughes, C. M., O. Rozenblatt-Rosen, T. A. Milne, T. D. Copeland, S. S. Levine, J. C. Lee, D. N. Hayes, K. S. Shanmugam, A. Bhattacharjee, C. A. Biondi, G. F. Kay, N. K. Hayward, J. L. Hess, and M. Meyerson. 2004. Menin associates with a trithorax family histone methyltransferase complex and with the hoxc8 locus. Mol. Cell 13:587-597. [DOI] [PubMed] [Google Scholar]
- 16.Issaeva, I., Y. Zonis, T. Rozovskaia, K. Orlovsky, C. M. Croce, T. Nakamura, A. Mazo, L. Eisenbach, and E. Canaani. 2007. Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Mol. Cell. Biol. 27:1889-1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang, C., and B. F. Pugh. 2009. Nucleosome positioning and gene regulation: advances through genomics. Nat. Rev. Genet. 10:161-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kent, W. J., C. W. Sugnet, T. S. Furey, K. M. Roskin, T. H. Pringle, A. M. Zahler, and D. Haussler. 2002. The human genome browser at UCSC. Genome Res. 12:996-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kestler, H. A., A. Muller, J. M. Kraus, M. Buchholz, T. M. Gress, H. Liu, D. W. Kane, B. R. Zeeberg, and J. N. Weinstein. 2008. VennMaster: area-proportional Euler diagrams for functional GO analysis of microarrays. BMC Bioinform. 9:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krogan, N. J., J. Dover, S. Khorrami, J. F. Greenblatt, J. Schneider, M. Johnston, and A. Shilatifard. 2002. COMPASS, a histone H3 (lysine 4) methyltransferase required for telomeric silencing of gene expression. J. Biol. Chem. 277:10753-10755. [DOI] [PubMed] [Google Scholar]
- 21.Krogan, N. J., J. Dover, A. Wood, J. Schneider, J. Heidt, M. A. Boateng, K. Dean, O. W. Ryan, A. Golshani, M. Johnston, J. F. Greenblatt, and A. Shilatifard. 2003. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol. Cell 11:721-729. [DOI] [PubMed] [Google Scholar]
- 22.Lawrence, H. J., C. D. Helgason, G. Sauvageau, S. Fong, D. J. Izon, R. K. Humphries, and C. Largman. 1997. Mice bearing a targeted interruption of the homeobox gene HOXA9 have defects in myeloid, erythroid and lymphoid hematopoiesis. Blood 89:1922-1930. [PubMed] [Google Scholar]
- 23.Lee, J. H., and D. G. Skalnik. 2005. CpG-binding protein (CXXC finger protein 1) is a component of the mammalian Set1 histone H3-Lys4 methyltransferase complex, the analogue of the yeast Set1/COMPASS complex. J. Biol. Chem. 280:41725-41731. [DOI] [PubMed] [Google Scholar]
- 24.Lee, J. S., A. Shukla, J. Schneider, S. K. Swanson, M. P. Washburn, L. Florens, S. R. Bhaumik, and A. Shilatifard. 2007. Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell 131:1084-1096. [DOI] [PubMed] [Google Scholar]
- 25.Lee, T. I., S. E. Johnstone, and R. A. Young. 2006. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat. Protoc. 1:729-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lengerke, C., S. Schmitt, T. V. Bowman, I. H. Jang, L. Maouche-Chretien, S. McKinney-Freeman, A. J. Davidson, M. Hammerschmidt, F. Rentzsch, J. B. Green, L. I. Zon, and G. Q. Daley. 2008. BMP and Wnt specify hematopoietic fate by activation of the Cdx-Hox pathway. Cell Stem Cell 2:72-82. [DOI] [PubMed] [Google Scholar]
- 27.Lim, D. A., Y. C. Huang, T. Swigut, A. L. Mirick, J. M. Garcia-Verdugo, J. Wysocka, P. Ernst, and A. Alvarez-Buylla. 2009. Chromatin remodelling factor Mll1 is essential for neurogenesis from postnatal neural stem cells. Nature 458:529-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindsley, R. C., J. G. Gill, M. Kyba, T. L. Murphy, and K. M. Murphy. 2006. Canonical Wnt signaling is required for development of embryonic stem cell-derived mesoderm. Development 133:3787-3796. [DOI] [PubMed] [Google Scholar]
- 29.McGinnis, W., and R. Krumlauf. 1992. Homeobox genes and axial patterning. Cell 68:283-302. [DOI] [PubMed] [Google Scholar]
- 30.Miller, T., N. J. Krogan, J. Dover, H. Erdjument-Bromage, P. Tempst, M. Johnston, J. F. Greenblatt, and A. Shilatifard. 2001. COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc. Natl. Acad. Sci. USA 98:12902-12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Milne, T. A., S. D. Briggs, H. W. Brock, M. E. Martin, D. Gibbs, C. D. Allis, and J. L. Hess. 2002. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol. Cell 10:1107-1117. [DOI] [PubMed] [Google Scholar]
- 32.Milne, T. A., Y. Dou, M. E. Martin, H. W. Brock, R. G. Roeder, and J. L. Hess. 2005. MLL associates specifically with a subset of transcriptionally active target genes. Proc. Natl. Acad. Sci. USA 102:14765-14770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Milne, T. A., C. M. Hughes, R. Lloyd, Z. Yang, O. Rozenblatt-Rosen, Y. Dou, R. W. Schnepp, C. Krankel, V. A. Livolsi, D. Gibbs, X. Hua, R. G. Roeder, M. Meyerson, and J. L. Hess. 2005. Menin and MLL cooperatively regulate expression of cyclin-dependent kinase inhibitors. Proc. Natl. Acad. Sci. USA 102:749-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mito, Y., J. G. Henikoff, and S. Henikoff. 2005. Genome-scale profiling of histone H3.3 replacement patterns. Nat. Genet. 37:1090-1097. [DOI] [PubMed] [Google Scholar]
- 35.Nakamura, T., D. A. Largaespada, M. P. Lee, L. A. Johnson, K. Ohyahiki, K. Toyama, S. J. Chen, C. L. Willman, I.-M. Chen, A. P. Feinberg, N. A. Jenkins, N. G. Copeland, and J. D. Shaughnessy, Jr. 1996. Fusion of the nucleoporin gene NUP98 to HOXA9 by the chromosome translocation t(7;11)(p15;p15) in human myeloid leukaemia. Nat. Genet. 12:154-158. [DOI] [PubMed] [Google Scholar]
- 36.Nakamura, T., D. A. Largaespada, J. D. Shaughnessy, N. A. Jenkins, and N. G. Copeland. 1996. Cooperative activation of Hoxa and Pbx1-related genes in murine myeloid leukaemias. Nat. Genet. 12:149-153. [DOI] [PubMed] [Google Scholar]
- 37.Ng, H. H., F. Robert, R. A. Young, and K. Struhl. 2003. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell 11:709-719. [DOI] [PubMed] [Google Scholar]
- 38.Patel, S. R., D. Kim, I. Levitan, and G. R. Dressler. 2007. The BRCT-domain containing protein PTIP links PAX2 to a histone H3, lysine 4 methyltransferase complex. Dev. Cell 13:580-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pavri, R., B. Zhu, G. Li, P. Trojer, S. Mandal, A. Shilatifard, and D. Reinberg. 2006. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell 125:703-717. [DOI] [PubMed] [Google Scholar]
- 40.Pena, P. V., F. Davrazou, X. Shi, K. L. Walter, V. V. Verkhusha, O. Gozani, R. Zhao, and T. G. Kutateladze. 2006. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature 442:100-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pray-Grant, M. G., J. A. Daniel, D. Schieltz, J. R. Yates III, and P. A. Grant. 2005. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature 433:434-438. [DOI] [PubMed] [Google Scholar]
- 42.Roguev, A., D. Schaft, A. Shevchenko, W. W. Pijnappel, M. Wilm, R. Aasland, and A. F. Stewart. 2001. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J. 20:7137-7148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rowley, J. D. 1998. The critical role of chromosome translocations in human leukemias. Annu. Rev. Genet. 32:495-519. [DOI] [PubMed] [Google Scholar]
- 44.Scacheri, P. C., S. Davis, D. T. Odom, G. E. Crawford, S. Perkins, M. J. Halawi, S. K. Agarwal, S. J. Marx, A. M. Spiegel, P. S. Meltzer, and F. S. Collins. 2006. Genome-wide analysis of menin binding provides insights into MEN1 tumorigenesis. PLoS Genet. 2:e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schraets, D., T. Lehmann, T. Dingermann, and R. Marschalek. 2003. MLL-mediated transcriptional gene regulation investigated by gene expression profiling. Oncogene 22:3655-3668. [DOI] [PubMed] [Google Scholar]
- 46.Sham, M.-H., P. Hunt, S. Nonchev, N. Papalopulu, A. Graham, E. Boncinelli, and R. Krumlauf. 1992. Analysis of the murine Hox-2.7 gene: conserved alternative transcripts with differential distributions in the nervous system and the potential for shared regulatory regions. EMBO J. 11:1825-1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi, X., T. Hong, K. L. Walter, M. Ewalt, E. Michishita, T. Hung, D. Carney, P. Pena, F. Lan, M. R. Kaadige, N. Lacoste, C. Cayrou, F. Davrazou, A. Saha, B. R. Cairns, D. E. Ayer, T. G. Kutateladze, Y. Shi, J. Cote, K. F. Chua, and O. Gozani. 2006. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature 442:96-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shilatifard, A. 2006. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu. Rev. Biochem. 75:243-269. [DOI] [PubMed] [Google Scholar]
- 49.Shilatifard, A. 2008. Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr. Opin. Cell Biol. 20:341-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sims, R. J., III, S. Millhouse, C. F. Chen, B. A. Lewis, H. Erdjument-Bromage, P. Tempst, J. L. Manley, and D. Reinberg. 2007. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol. Cell 28:665-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smyth, G. K., and T. Speed. 2003. Normalization of cDNA microarray data. Methods 31:265-273. [DOI] [PubMed] [Google Scholar]
- 52.Tan, C. C., K. V. Sindhu, S. Li, H. Nishio, J. Z. Stoller, K. Oishi, S. Puttreddy, T. J. Lee, J. A. Epstein, M. J. Walsh, and B. D. Gelb. 2008. Transcription factor Ap2delta associates with Ash2l and ALR, a trithorax family histone methyltransferase, to activate Hoxc8 transcription. Proc. Natl. Acad. Sci. USA 105:7472-7477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tenney, K., and A. Shilatifard. 2005. A COMPASS in the voyage of defining the role of trithorax/MLL-containing complexes: linking leukemogenesis to covalent modifications of chromatin. J. Cell. Biochem. 95:429-436. [DOI] [PubMed] [Google Scholar]
- 54.Vermeulen, M., K. W. Mulder, S. Denissov, W. W. Pijnappel, F. M. van Schaik, R. A. Varier, M. P. Baltissen, H. G. Stunnenberg, M. Mann, and H. T. Timmers. 2007. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 131:58-69. [DOI] [PubMed] [Google Scholar]
- 55.Wood, A., N. J. Krogan, J. Dover, J. Schneider, J. Heidt, M. A. Boateng, K. Dean, A. Golshani, Y. Zhang, J. F. Greenblatt, M. Johnston, and A. Shilatifard. 2003. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol. Cell 11:267-274. [DOI] [PubMed] [Google Scholar]
- 56.Wood, A., A. Shukla, J. Schneider, J. S. Lee, J. D. Stanton, T. Dzuiba, S. K. Swanson, L. Florens, M. P. Washburn, J. Wyrick, S. R. Bhaumik, and A. Shilatifard. 2007. Ctk complex-mediated regulation of histone methylation by COMPASS. Mol. Cell. Biol. 27:709-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu, M., P. F. Wang, J. S. Lee, S. Martin-Brown, L. Florens, M. Washburn, and A. Shilatifard. 2008. Molecular regulation of H3K4 trimethylation by Wdr82, a component of human Set1/COMPASS. Mol. Cell. Biol. 28:7337-7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wysocka, J., T. Swigut, H. Xiao, T. A. Milne, S. Y. Kwon, J. Landry, M. Kauer, A. J. Tackett, B. T. Chait, P. Badenhorst, C. Wu, and C. D. Allis. 2006. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 442:86-90. [DOI] [PubMed] [Google Scholar]
- 59.Yokoyama, A., Z. Wang, J. Wysocka, M. Sanyal, D. J. Aufiero, I. Kitabayashi, W. Herr, and M. L. Cleary. 2004. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol. Cell. Biol. 24:5639-5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu, B. D., J. L. Hess, S. E. Horning, G. A. Brown, and S. J. Korsmeyer. 1995. Altered Hox expression and segmental identity in Mll-mutant mice. Nature 378:505-508. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.